Significance
Mechanisms for why influenza A virus (IAV) infections sensitize for pneumococcal infections are not clear. Here, we show that IAV-induced capillary leakage results in influx of nutrients and antioxidants to the lungs, thereby promoting pneumococcal growth in the lower respiratory tract. The evoked inflammation leads to redox imbalances that require bacterial adaptation to the oxidized environment, including induction of the pneumococcal chaperone/protease HtrA that protects the bacteria from clearance by the immune system. The results give us insight into the delicate interplay between the bacteria and the host environment during coinfections that needs to be explored in order to find novel therapeutic approaches.
Keywords: Streptococcus pneumoniae, influenza A virus, pneumococci, redox imbalance, antioxidants
Abstract
Influenza A virus (IAV)-related mortality is often due to secondary bacterial infections, primarily by pneumococci. Here, we study how IAV-modulated changes in the lungs affect bacterial replication in the lower respiratory tract (LRT). Bronchoalveolar lavages (BALs) from coinfected mice showed rapid bacterial proliferation 4 to 6 h after pneumococcal challenge. Metabolomic and quantitative proteomic analyses demonstrated capillary leakage with efflux of nutrients and antioxidants into the alveolar space. Pneumococcal adaptation to IAV-induced inflammation and redox imbalance increased the expression of the pneumococcal chaperone/protease HtrA. Presence of HtrA resulted in bacterial growth advantage in the IAV-infected LRT and protection from complement-mediated opsonophagocytosis due to capsular production. Absence of HtrA led to growth arrest in vitro that was partially restored by antioxidants. Pneumococcal ability to grow in the IAV-infected LRT depends on the nutrient-rich milieu with increased levels of antioxidants such as ascorbic acid and its ability to adapt to and cope with oxidative damage and immune clearance.
Streptococcus pneumoniae (the pneumococcus) is a leading cause of lower respiratory tract (LRT) infections and a major contributor to morbidity and mortality globally (1). A major risk factor for development of invasive pneumococcal diseases (IPDs) is preceding infections with influenza A virus (IAV) (2). IAV is a common pathogen of the human respiratory tract, generating a large burden of morbidity and mortality during seasonal and pandemic outbreaks (3). A major part of the disease burden attributed to IAV is the result of superinfections with bacteria. Thus, the high mortality rates found during the Spanish flu pandemic 1918 to 1919, especially among younger individuals, was suggested to be due to severe secondary bacterial pneumonia caused by respiratory pathogens, primarily Streptococcus pneumoniae (4). Previous studies have shown that dysregulated lung physiology, including impaired mucociliary clearance and modulation of host immune responses caused by IAV, contributes to the increased susceptibility to secondary bacterial infections (5). IAV modifies IFN responses, leading to attenuated macrophage and neutrophil function (6–8), and viral sialidases remove sialic acids from epithelial surfaces, thereby exposing host receptors and promoting bacterial adhesion (9, 10). IAV-induced increase of sialic acid in the murine nasopharynx has been linked to enhanced pneumococcal colonization (11). Furthermore, virus-induced epithelial damage, mainly caused by significant activation of reactive oxygen species (ROS) and cell death responses (12, 13), was also shown to contribute to the severity of secondary bacterial pneumonia (10). However, the effects of IAV-induced modifications and stresses on bacterial adaption and growth in the LRT are poorly investigated.
S. pneumoniae is a fastidious organism, depending on the human host for a number of different amino acids and other nutrients (14). In its normal localization in the healthy nasopharynx, carbohydrates for fermentation are also limited and primarily attained by enzymatic retrieval of glycan sugars, such as galactose, from mucin after desialylation by neuraminidases (15, 16). The LRT is kept virtually free from inhaled microbes by the mucociliary clearance system. The constantly beating cilia generate transportation of mucus together with associated particles upwards, thereby reducing bacterial numbers (17). A lower mucin content in the noninflamed LRT might also prevent bacterial proliferation due to carbohydrate and likely other nutrient limitations.
For pneumococci to grow in the inflamed LRT they must adapt to the highly oxidizing environment prevailing under these conditions. Pneumococci are unusual in lacking endogenous production of catalase, and they produce high levels of hydrogen peroxide during all fermentative growth conditions, where pyruvate is oxidized into acetylphosphate and hydrogen peroxide by the pyruvate oxidase SpxB (18). A mutant in spxB is less resistant to oxidative stress mediated by exogenous hydrogen peroxide from the environment as compared to wild-type (WT) pneumococci. To adapt to oxidative stress, pneumococci have evolved different mechanisms, including the induction of the pneumococcal high-temperature requirement A (HtrA), that promotes pneumococcal virulence in vivo (19, 20). The htrA gene is transcriptionally induced by phosphorylated CiaR, the response regulator of the ciaRH two-component histidine kinase system (21, 22). It has been suggested that CiaR phosphorylation, and hence htrA induction, is accomplished by acetyl phosphate generated by SpxB oxidation of pyruvate (23, 24).
Here, we demonstrate how a profound shift in nutrients and redox balance in the LRT of IAV-infected lungs due to viral-induced inflammation and capillary leakage leads to rapid pneumococcal proliferation in this environment, which is promoted by the surface chaperone/protease HtrA. This mediates increased pneumococcal resistance to oxidative damage and protection from complement-mediated opsonophagocytosis.
Results
IAV Infection Increases Glucose Levels in the LRT, Thereby Influencing Pneumococcal Growth.
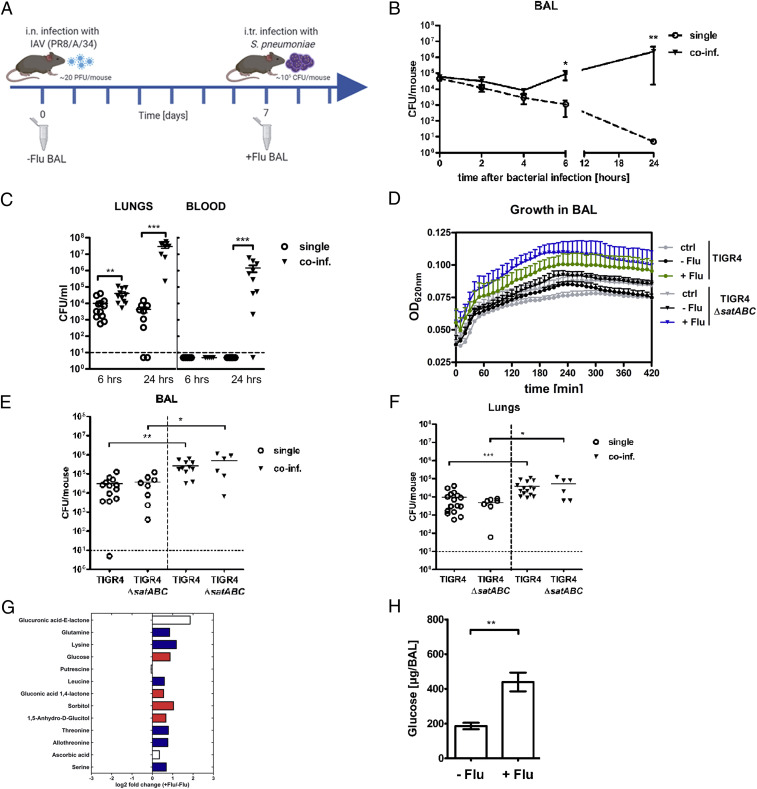
Intranasal IAV infection (10 to 30 plaque-forming units [PFU]) of mice led to efficient viral replication in the lungs with a peak at day 3 postinfection and complete clearance of the virus from the lungs at day 10 (SI Appendix, Fig. S1A). The IAV infection caused moderate weight loss between days 6 and 10 postinfection, after which the mice recovered (SI Appendix, Fig. S1B). To study bacterial growth in a coinfection model, noninfected mice, or mice first infected with IAV for 7 d, were infected with a low dose of TIGR4 pneumococci (105) intratracheally (i.tr.) to ensure reaching the LRT (Fig. 1A). Viable counts revealed similar bacterial numbers in the bronchoalveolar lavage (BAL) of single-infected and coinfected mice until 4 h postbacterial infection (Fig. 1B). However, at 6 h postinfection only about 2% of the bacteria remained in single-infected mice, whereas bacterial numbers in coinfected mice increased about 10-fold compared to the 4-h time point, implying a bacterial doubling time as short as 35 min between these two time points. The difference in bacterial numbers in the LRT was even more pronounced at 24 h after bacterial infection, with almost complete bacterial clearance in single-infected mice, and a 25-fold further increase of bacterial numbers in coinfected animals compared to the 6-h time point (Fig. 1B), suggesting a considerably slower proliferation between 6 and 24 h (doubling time about 230 min) compared to the 4- to 6-h time period. Bacterial numbers in the lungs were also significantly higher in coinfected compared to single-infected mice at both 6 and 24 h after infection (Fig. 1 C, Left). No bacteria were detected in the blood of IAV-infected mice at 6 h, suggesting that at this time point most bacteria in the IAV-infected lungs were confined to the airspace. At 24 h, however, coinfected, but not single-infected animals, had high CFU counts (∼106/mL) (Fig. 1 C, Right). The 40-fold increase of bacterial numbers in coinfected mice at 24 h compared to time 0, clearly demonstrates extensive bacterial proliferation under IAV-infected conditions, whereas no or only limited bacterial proliferation occurs in the LRT of single-infected mice. To get an estimate on whether IAV-infected BAL contains a more available carbon source than uninfected BAL for pneumococcal growth, we used uninfected (−Flu) and IAV-infected (+Flu) BAL supplemented with C + Y medium containing all growth requirements for pneumococci except for a carbon source. Even though the BALs were diluted when sampled, there was a difference in the maximum optical density (OD) when bacteria were grown in +Flu BAL compared to −Flu BAL, and no difference between −Flu BAL and the control (Fig. 1D), suggesting more available carbohydrates present in +Flu BAL compared to −Flu BAL.
Fig. 1.
IAV-induced increase of glucose in the LRT contributes to rapid bacterial growth. (A) Overview of the in vivo infection model. C57bl/6 mice were infected i.n. with a nonlethal dose of IAV (mouse-adapted PR8/A/34), followed by intratracheal infection with strain S. pneumoniae serotype 4 (TIGR4) at day 7 after IAV infection. BALs were collected from uninfected (day 0) and day 7 IAV-infected mice for ex vivo analysis and in vitro experiments. (B) Bacterial viable counts in BALs of mice infected with S. pneumoniae TIGR4 alone (single, open circle, n = 6 to 7; except t0 and 24h n = 3) or coinfected (coinf.) with IAV and S. pneumoniae (coinf., triangle, n = 6 to 7; except t0 n = 3) at the indicated time points after bacterial infection. *P < 0.05, **P < 0.01. (C) Bacterial viable counts in lungs and blood of mice infected with S. pneumoniae alone (single, open circle, n = 5 to 12) or coinfected with IAV and S. pneumoniae (coinf., triangle, n = 5 to 11) at 6 and 24 h after bacterial infection. Dotted lines indicate the detection limit. **P < 0.01. ***P < 0.001. (D) WT strain TIGR4 and mutant strain TIGR4ΔsatABC were grown in BALs from uninfected mice (−Flu) and day 7 IAV-infected mice (+Flu). Optical density at 620 nm was monitored over time. Data are presented as mean of three independent experiments (mean ± SEM). (E) Bacterial counts in BALs of mice infected with S. pneumoniae TIGR4 or TIGR4ΔsatABC alone (single, open symbols, n = 8 to 13) or coinfected with the respective strains 7 d after IAV infection (coinf., triangle, n = 6 to 11) at 6 h after bacterial infection. *P < 0.05, **P < 0.01. (F) Bacterial viable counts in the lungs of mice infected with S. pneumoniae WT TIGR4 (n = 15 to 16) or TIGR4ΔsatABC (n = 6 to 7) alone or coinfected with IAV and the respective strain (coinf., triangle). BAL was collected at 6 h after bacterial infection. *P < 0.05, ***P < 0.001. (G) Overview of differentially expressed metabolites presented as log2 fold change of +Flu/−Flu. Values <0 on the x axis indicate lower abundances in +Flu BAL; values >0 indicate a higher abundance in +Flu BAL. Amino acids are marked in blue and carbohydrates are marked in red. (H) Glucose levels in BAL from uninfected mice (−Flu, n = 8), and IAV-infected mice at day 7 (+Flu, n = 8) as determined by colorimetric assay. **P < 0.01. Data in graphs are presented as mean ± SEM. See also SI Appendix, Fig. S1.
A recent study showed enhanced pneumococcal growth in the upper respiratory tract (URT) of mice due to IAV-induced increase in sialic acids in the nasopharynx (11). To investigate whether increased sialic acid levels contribute to enhanced bacterial numbers in our model, we also analyzed sialic acid concentrations in the LRT and found higher levels of total and free sialic acid in IAV-infected compared to uninfected BAL (SI Appendix, Fig. S1C). However, in vitro growth analysis of pneumococci lacking the main sialic acid transporter SatABC (TIGR4ΔsatABC) also showed better growth in IAV-infected conditions as compared to without IAV, as was found for WT TIGR4 (Fig. 1D). This indicates that the enhanced bacterial growth observed under IAV-infected conditions was not due to higher levels of sialic acids. Moreover, coinfection with TIGR4ΔsatABC also exhibited increased bacterial numbers at 6 h compared to single bacterial infection (Fig. 1E), and there was no difference in the bacterial load in the LRT of single-infected mice using TIGR4 or TIGR4ΔsatABC (Fig. 1E). Bacterial numbers in the lungs were also significantly higher in coinfected compared to single-infected mice for both TIGR4 and the mutant TIGR4ΔsatABC (Fig. 1F). This argues that the rapid proliferation in the IAV-infected LRT is not mediated by bacterial utilization of sialic acid as a primary carbon source, as shown for the URT (11).
To determine other possible nutrient sources in the LRT, we used a metabolomic approach and identified and quantified 41 metabolites in mouse BAL with a clear separation between uninfected and IAV-infected samples, as shown using principal component analysis (PCA) (SI Appendix, Fig. S1D). Due to a dilution of the BAL samples during the collection of the samples, only compounds present in higher amounts were detected. The 41 metabolites identified represent 14 biochemical classes, with most compounds belonging to the class of carboxylic acids and organooxygen compounds, which include amino acids and carbohydrates, respectively (SI Appendix, Table S1). Among the 12 metabolites that were increased in IAV-infected BAL compared to uninfected BAL were six amino acids (marked in blue), and four carbohydrates (marked in red) including glucose, the generally preferred carbon source for the pneumococcus, and sorbitol, a reduced form of glucose (Fig. 1G). Quantification of absolute glucose levels in the LRT revealed a 2.4-fold increase in IAV-infected compared to uninfected BAL (Fig. 1H). The increased glucose and sorbitol levels likely contribute to the initially very high growth rate in the LRT of IAV-infected compared to single-infected mice. Furthermore, glucose-mediated catabolite repression might explain why sialic acid-dependent growth is not of major importance in the IAV-infected LRT environment at the 6-h time point, whereas it is in the URT (11).
IAV Infection Induces Capillary Leakage Leading to Changes in the Protein Composition in the LRT.
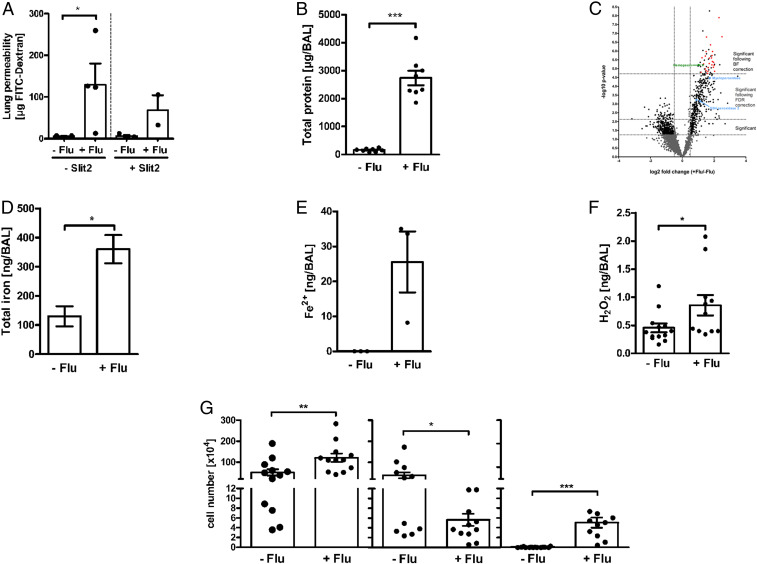
To test whether the increased levels of glucose, and other metabolites in the alveolar space, are a result of increased capillary permeability, we measured lung permeability using FITC-dextran. The lung permeability in IAV-infected mice was 22-fold higher compared to uninfected mice (Fig. 2 A, Left). It has been demonstrated that the soluble ligand Slit2, by interacting with Robo4, can modulate endothelial permeability and integrity in the lungs (25). Therefore, we administered Slit2 daily intravenously (i.v.) for 4 d (1.56 µg/mouse) (26) into IAV-infected mice. We found that Slit2 partly reduced the FITC-dextran leakage (Fig. 2 A, Right).
Fig. 2.
IAV-induced inflammation and capillary leakage leads to changes in protein composition and altered redox balance in the LRT. (A) Fluorescence intensity (FI) value of FITC-dextran (20 mg/mL via i.v. injection) in the BAL of uninfected (−Flu, n = 5) and IAV-infected mice at day 7 (+Flu, n = 4) with and without Slit2 treatment (n = 2 to 3). *P < 0.05. (B) Total protein levels in BAL from uninfected mice (−Flu, n = 10) and IAV-infected mice at day 7 (+Flu, n = 7) were determined using bicinchoninic acid. ***P < 0.001. (C) Volcano plot overview of the proteomic data. Values <0 on the x axis indicate lower protein abundances in +Flu BAL, values >0 indicate a higher protein abundance in +Flu BAL. Data were statistically analyzed by t test. P value cutoff following FDR and BF correction and two-fold change cutoff (at ± 0.5) are indicated with dotted lines. Plasma proteins are highlighted in red, hemopexin is highlighted in green, and redox-active proteins are highlighted in light blue. (D and E) Total iron levels and ferrous (Fe2+) in BAL from uninfected mice (−Flu, n = 3) and IAV-infected mice at day 7 (+Flu, n = 3) were determined by ICP-MS analysis. *P < 0.05. Data in graphs are presented as mean ± SEM. (F) Hydrogen peroxide (H2O2) levels in BAL from uninfected mice (−Flu, n = 13) and IAV-infected mice at day 7 (+Flu, n = 11) were determined according to a standard curve using known concentrations upon incubation with 2,2-azino-bis (3-ethylbenzothiazoline-6 sulfonic acid) and horseradish peroxidase by measuring optical densities at 560 nm. *P < 0.05. (G) Number of total cells (Left), pulmonary macrophages (CD11c+/F4/80+; Middle, and polymorphonuclear leukocytes (PMNs, CD11b+, Ly6G+; Right) in BAL of uninfected (−Flu, n = 12 to 14) and day 7 IAV infected mice (+Flu, n = 12–13). *P < 0.05, **P < 0.01, ***P < 0.001. See also SI Appendix, Fig. S2.
The increased lung permeability was linked to a 15-fold increase in total protein content in IAV-infected BAL compared to uninfected BAL (Fig. 2B). To investigate whether the increased protein content was due to plasma protein leakage into the alveolar space, we performed quantitative proteomics of BAL proteins where we found a clear separation of the proteins identified in uninfected compared to IAV-infected BAL at day 7, as shown by PCA (SI Appendix, Fig. S2A). Among the 2,723 proteins identified and quantified in total, 49 proteins were significantly different in the LRT of IAV-infected mice compared to uninfected mice following Bonferroni (BF) correction (Fig. 2 C, Top Right corner). STRING pathway analysis of these 49 proteins revealed that 26 are plasma proteins (marked red), confirming the leakage of plasma through capillary walls into the alveolar space. Together with plasma proteins, there was also an influx of iron as shown by increased levels of total iron and ferrous iron (Fe2+, below the detection limit in −Flu BAL) (Fig. 2 D and E). Ferrous iron likely enters the alveolar space together with hemopexin, an abundant plasma protein that efficiently scavenges heme-containing iron (27, 28), and that was higher in IAV-infected BAL (SI Appendix, Table S2 and Fig. 2C, green triangle). Out of the 26 plasma-derived proteins from the proteomic analyses, 20 are immune-reactive proteins involved in host responses, out of which 9 are proteins of the complement pathway (SI Appendix, Table S2 and Fig. S2B, blue circles).
IAV Infection Alters the Redox Balance in the LRT and Increases the Levels of Antioxidants.
Increased oxidative stress plays an important role in the pathogenesis of influenza infections (12, 13). We found higher levels of hydrogen peroxide (H2O2) in IAV-infected BAL at day 7 compared to uninfected BAL (Fig. 2F). The increased levels of H2O2 together with increased levels of ferrous iron led to enhanced ROS production and oxidative stress in the LRT environment via the Fenton reaction (Fig. 2 E and F). Additionally, the neutrophil-derived myeloperoxidase (MPO) that catalyzes the reduction of H2O2 to hypochlorite, which is extremely damaging to proteins and lipids (29), was increased in IAV-infected BAL (Fig. 2C, light blue).
The imbalanced redox milieu was a result of IAV-induced inflammation with increased numbers of neutrophils (Ly6G+, CD11b+), whereas the levels of macrophages (CD11c+, F4/80+) were lower (Fig. 2G). The altered cellular content in the LRT of IAV-infected mice was also in accordance with the quantitative proteomics data showing higher levels of neutrophil-specific elastase (green asterisk) and lower levels of the macrophages mannose receptor-1 (MRC-1, yellow asterisk) in IAV-infected BAL compared to noninfected BAL (SI Appendix, Fig. S2B). The combined data show that IAV-induced inflammation results in increased capillary permeability causing plasma leakage into the alveolar space. This allows nutrient influx and increases the total protein content in the alveolar space, but it also increases the level of microbial stressors and contributes to a redox imbalance with presence of both oxidants and antioxidants in the LRT microenvironment.
Enhanced Pneumococcal Growth in the IAV-Infected LRT Requires the Surface Chaperone/Protease HtrA.
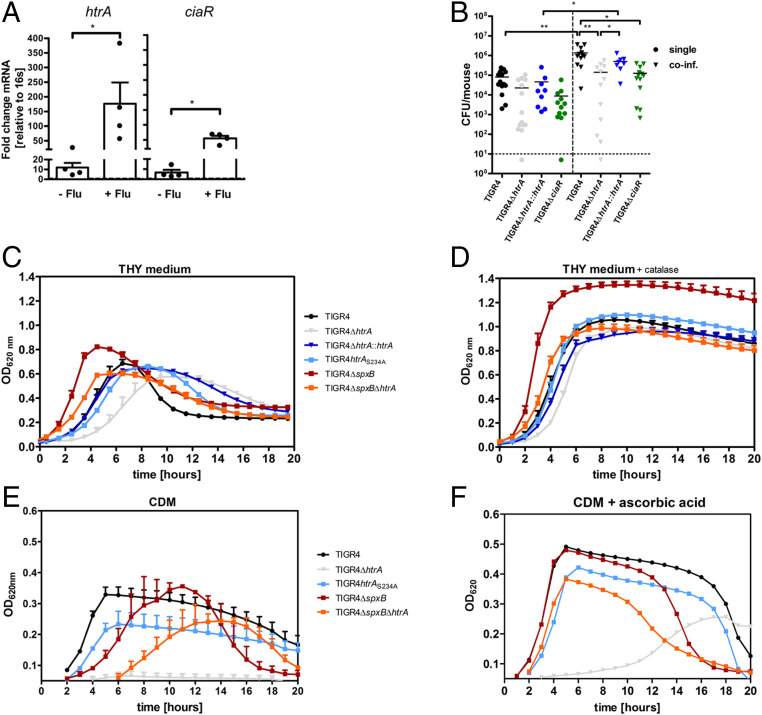
Pneumococci have developed strategies to alleviate the consequences of redox imbalances, including systems to repair or degrade damaged proteins. One such protein that helps pneumococci to resist oxidative damage and promotes pneumococcal virulence is the surface-exposed chaperone and serine protease HtrA (19). We found that htrA transcription was increased when pneumococci were incubated, both in BAL from mice not infected and in those infected with IAV at day 7, compared to incubation in phosphate-buffered saline (PBS), i.e., baseline level (dotted line = 1). HtrA transcription was highest under IAV-infected conditions (Fig. 3 A, Left). The transcriptional inducer of htrA, CiaR, was also increased in bacteria incubated in BAL from noninfected and IAV-infected mice, compared to incubation in PBS, with the highest transcription levels under IAV-infected conditions. This suggests that HtrA is induced via its regulator CiaR (Fig. 3 A, Right). We next tested whether HtrA is required for bacterial growth during coinfection. Coinfected mice challenged with WT TIGR4 showed considerably more bacteria in their BAL at 6 h compared to single-infected mice (17-fold increase). In contrast, mice coinfected with the mutant TIGR4ΔhtrA showed only a 6-fold increase in bacterial numbers compared to a single infection with TIGR4ΔhtrA. Interestingly, mice coinfected with TIGR4ΔhtrA had significantly fewer bacteria in their LRT compared to mice coinfected with TIGR4 (10-fold difference), whereas complementation of TIGR4ΔhtrA with the WT allele (TIGR4ΔhtrA::htrA) restored the WT TIGR4 phenotype (Fig. 3B). These results suggest that HtrA promotes the rapid early bacterial proliferation in the nutrient-rich, but highly oxidizing environment prevailing in the IAV-infected LRT. Similar to TIGR4ΔhtrA, mice coinfected with the mutant TIGR4ΔciaR also had significantly fewer bacteria in the LRT compared to mice coinfected with WT TIGR4 (11-fold difference) (Fig. 3B), which is in agreement with the data above, suggesting that induction of htrA during influenza-infected conditions occurs via ciaR.
Fig. 3.
HtrA-mediated oxidative stress adaptation increases the pneumococcal growth rate in the IAV-infected LRT and in vitro. (A) RNA levels of htrA and ciaR in S. pneumoniae WT TIGR4 after 90-min incubation in BAL from uninfected mice (−Flu; n = 4) or IAV-infected mice at day 7 (+Flu; n = 4). 16sRNA was used as an endogenous control for PCR. The fold change of transcription was normalized to bacteria incubated in PBS (dotted line = 1). *P < 0.05. (B) Bacterial viable counts in BAL of mice infected with S. pneumoniae WT TIGR4 (n = 12 to 14), TIGR4ΔhtrA (n = 12 to 15), the complemented strain TIGR4ΔhtrA::htrA (n = 8 to 9), or TIGR4ΔciaR (n = 12 to 13) alone or coinfected with IAV and the respective bacterial strain (coinf., triangle) at 6 h after bacterial infection. *P < 0.05, **P < 0.01. (C–F) WT TIGR4, mutant TIGR4ΔhtrA, the complemented strain of TIGR4ΔhtrA, TIGR4ΔhtrA::htrA, the proteolytic inactive mutant TIGR4htrAS234A, mutant strain TIGR4ΔspxB, and the double mutant TIGR4ΔspxBΔhtrA were grown on complete THY medium in absence (C) or presence (D) of catalase (2,500 U/mL) or on CDM in the presence of glucose as carbon source ± ascorbic acid (10 mM) (E and F). Data are presented as mean of three to four independent experiments (mean ± SEM). Optical density at 620 nm was monitored over time.
Both the Chaperone and Protease Role of HtrA Contribute to the Rapid Bacterial Proliferation in an Oxidizing Environment.
Pneumococci produce significant levels of hydrogen peroxide during aerobic growth in glucose containing complete media, mainly through its pyruvate oxidase SpxB (18). Since aerobic pneumococcal growth in vitro results in self-afflicted oxidative stress, a role for pneumococcal HtrA in oxidative stress adaptation can be studied under different in vitro growth conditions. We first analyzed growth of TIGR4 and its isogenic mutant derivatives in Todd Hewitt broth (THY), a complete broth plus yeast extract medium with added glucose that we reasoned reflect the nutritional composition of the fluid in the IAV-infected LRT. In THY medium during the exponential growth phase, WT TIGR4 showed a doubling time considerably longer (91 min) than for the same time period in the IAV-infected LRT (35 min) (Fig. 3C). TIGR4ΔhtrA showed a much longer doubling time (137 min) compared to WT TIGR4, and also a longer lag period before initiation of exponential growth (Fig. 3C). TIGR4 carrying a serine-alanine substitution in the proteolytic active site (TIGR4htrAS234A) showed a longer doubling time (117 min) than WT TIGR4, but shorter than TIGR4ΔhtrA, suggesting that both the chaperone and protease functions of HtrA contribute to pneumococcal proliferation in the THY medium. The TIGR4spxB mutant, producing low levels of H2O2, showed no lag period, a shorter doubling time (73 min), and also reached a higher maximum OD620 compared to WT TIGR4 (Fig. 3C). In the mutant lacking both spxB and htrA, TIGR4ΔspxBΔhtrA, the lag period was shorter and the exponential doubling time higher (89 min) than for TIGR4ΔhtrA and close to WT levels, demonstrating that HtrA is required for growth in an oxidizing environment (Fig. 3C).
We next used chemically defined medium (CDM) where glucose is used as a fermentable carbon source and analyzed growth over time and bacterial viability. While WT TIGR4 grew in this medium, there was a complete growth arrest for the TIGR4ΔhtrA mutant (Fig. 3E). This growth arrest was due to hydrogen peroxide production, as the double mutant TIGR4ΔspxBΔhtrA, after a ∼5-h lag period, resumed similar growth as WT TIGR4. A long lag period was also seen for the TIGR4ΔspxB mutant followed by a robust growth reaching a maximum density higher than for WT TIGR4. These results demonstrate that pneumococcal growth in glucose medium requires bacterial ability to cope with oxidative stress, and this is partly dependent on HtrA that mediates protection and/or degradation of oxidized proteins presented on the bacterial surface.
Pneumococcal Growth Is Promoted by Antioxidants.
To counteract the highly oxidative environment, pneumococci make use of available antioxidants in their host environment such as catalase that degrades hydrogen peroxide and ascorbic acid. Ascorbic acid is synthesized from glucose in the mouse liver, and our metabolomic data showed that ascorbic acid was increased in IAV-infected BAL (Fig. 1G). The increased level of this antioxidant is thus suggested to come from the circulation. To study the effect of antioxidants on pneumococcal growth in vitro, we first used THY medium, that unlike the IAV-infected LRT, lacks catalase. Addition of catalase shortened the doubling time and increased the maximal growth density for WT TIGR4 and its mutant derivatives (Fig. 3D). We then studied the effects of ascorbic acid, which protects extracellular and intracellular components from free radical damage, on pneumococcal growth in CDM + glucose medium. Addition of ascorbic acid had a profound effect on growth of all strains with faster growth and higher maximum ODs (Fig. 3F). Moreover, addition of ascorbic acid abolished the delayed growth onset for the spxB mutant and allowed a similar growth pattern as for WT TIGR4. Interestingly, the TIGR4∆htrA mutant in the presence of ascorbic acid resumed rapid growth, but only after a long lag period of 6 to 8 h.
To address the strain-specific characteristics of pneumococci, we next studied in vitro growth and bacterial viability of pneumococci of serotype 2 (D39), in addition to serotype 4 (TIGR4). D39 grew more slowly in CDM + glucose as compared to TIGR4 with an initial growth arrest of ∼6 to 7 h (SI Appendix, Fig. S3A). Bacterial viable counts supported the data of an initial growth arrest in absence of antioxidants for both D39 and TIGR4ΔhtrA with only 2-fold and 1.3-fold more bacteria, respectively, during the first 6 h. After the growth arrest period, the mutant D39ΔhtrA grew more slowly than WT D39, but did not show the complete growth arrest observed for TIGR4∆htrA. Addition of the antioxidant ascorbic acid enhanced growth considerably in all strains with a significant increase in bacterial numbers of TIGR4 between 0 and 6 h. Interestingly, the initial growth arrest of D39 was overcome in presence of ascorbic acid. In WT D39 and D39ΔhtrA mutant bacteria, which both showed growth arrest in absence of antioxidants, addition of ascorbic acid allowed growth resumption, with 203-fold and 41-fold more bacteria at 6 h compared to time point 0, respectively (SI Appendix, Fig. S3B and Table S3). These results suggest that different strains of pneumococci may differ in their ability to adapt and cope with oxidative stress beyond HtrA, but that presence of antioxidants in the IAV-infected LRT may be a common requirement for rapid early onset proliferation.
Pneumococcal HtrA Expression Promotes Antiphagocytic Capsule Production under Oxidative Stress Conditions.
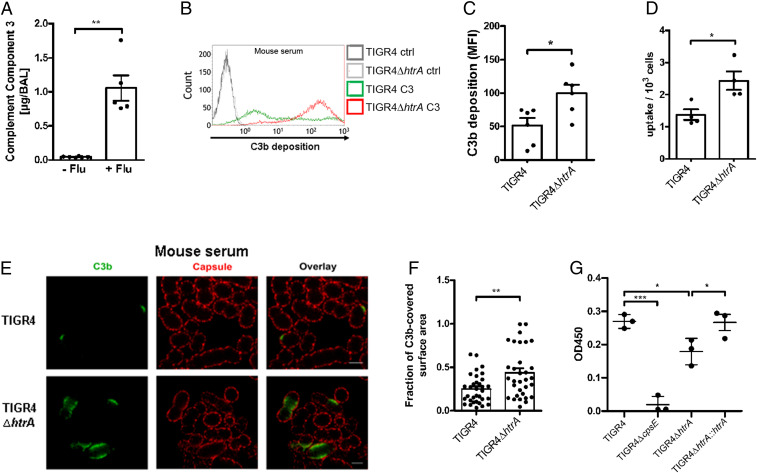
The pulmonary host defense, promoting bacterial clearance, was intact in the IAV-infected LRT as shown in the proteomic data by the increased presence of complement pathway proteins (SI Appendix, Fig. S2B and Table S2). To confirm and extend the proteomic analysis, we also measured levels of C3 in uninfected and IAV-infected BAL at day 7 by ELISA, showing a 15-fold higher C3b content in BAL from IAV-infected compared to uninfected mice (Fig. 4A). The presence of active complement C3b was also confirmed by Western blot (SI Appendix, Fig. S4A). Additionally, the increase in neutrophils (Ly6G+/CD11b+) (Fig. 2 G, Right) was linked to higher protein levels of the processed form of the most abundant antimicrobial peptide in the lungs, CRAMP (the mouse cathelicidin equivalent of human LL37), suggesting that the recruited neutrophils are functional (SI Appendix, Fig. S4B).
Fig. 4.
Stress-induced HtrA protects bacteria from opsonophagocytosis. (A) Expression levels of the complement C3 in BAL from uninfected (−Flu, n = 6) and IAV-infected mice at day 7 (+Flu, n = 5) as determined by sandwich ELISA. **P < 0.01. See also SI Appendix, Fig. S5. (B and C) Representative histogram of C3b deposition on WT TIGR4 and TIGR4ΔhtrA after incubation in 20% normal mouse serum using (B) flow cytometry and (C) quantification of mean fluorescence intensity (MFI) (n = 6), *P < 0.05. See also SI Appendix, Fig. S5. (D) Opsonophagocytosis of serum opsonized WT TIGR4 (n = 4) and TIGR4ΔhtrA (ΔhtrA, n = 4) by murine RAW267.4 macrophages. *P < 0.05. Data are presented as mean ± SEM. See also SI Appendix, Fig. S5. (E) Superresolution STED microscopy images of C3b deposition on WT TIGR4 and TIGR4ΔhtrA after incubation with 30% mouse serum (representative of two independent experiments). C3b was stained using goat anti-C3 antibody followed by Atto647N-labeled secondary antibody (green). The capsule was detected using TIGR4 rabbit anti-capsule serum and anti-rabbit Alexa Fluor 594 antibody (red). (Scale bar, 1 µm.) (F) Calculations of the fraction of C3b-covered surface area. The fraction of the area covered by C3b was calculated (see Materials and Methods for a more detailed description) for the images shown in E (TIGR4 n = 33 and TIGR4ΔhtrA, n = 32). **P < 0.01. (G) Quantification of capsular amount in the cell wall (CW) and pellet fraction of TIGR4, TIGR4ΔhtrA, and TIGR4ΔhtrA::htrA (negative ctrl = TIGR4ΔcpsE). Data are presented as mean ± SEM of three independent experiments. *P < 0.05; ***P < 0.001. See also SI Appendix, Fig. S6.
S. pneumoniae expresses polysaccharide capsules that prevent complement deposition and thereby complement mediated opsonophagocytosis. Considering that the IAV-infected LRT contains functional components of the host response which promote bacterial clearance, we next asked whether pneumococcal TIGR4 tolerance to the clearing environment in coinfected mice requires HtrA expression. Analyzes of the deposition of the active complement C3b on WT TIGR4 and TIGR4ΔhtrA bacteria by flow cytometry showed that the mutant deposited more C3b on its surface compared to WT TIGR4 when incubated in mouse or human serum (Fig. 4 B and C and SI Appendix, Fig. S5 A and B). Interestingly, increased C3b deposition correlated with higher bacterial uptake using murine RAW267.4 macrophages (Fig. 4D), and the human-derived macrophage cell line THP-1, in the presence of mice or human serum, respectively (SI Appendix, Fig. S5C). Fluorescence microscopy confirmed an increased C3b deposition on TIGR4ΔhtrA (∼20%) compared to WT bacteria (∼6%) in the presence of mouse serum (SI Appendix, Fig. S5D). Subsequent superresolution stimulated emission depletion (STED) microscopy extended these findings, showing that not only more bacteria showed C3b deposition in the absence of HtrA, but also a much larger area underneath the capsular layer contained deposited C3b when compared to WT TIGR4 (Fig. 4 E and F). The increased complement deposition on htrA-deficient bacteria was associated with a less efficient capsular barrier function as mutant bacteria showed lower levels of capsule on their surface (Fig. 4G and SI Appendix, Fig. S6). In the complemented strain of TIGR4ΔhtrA (TIGR4ΔhtrA::htrA) the levels of capsule were restored to that of WT TIGR4 (Fig. 4G). In conclusion, these findings suggest that the surface protease HtrA may play a significant role in oxidative damage control of enzymes required for capsular biosynthesis.
Discussion
IAV usually infects the upper airways, causing a self-limited disease. However, this virus also causes severe LRT infections with acute lung inflammation and lung damage (30, 31). Moreover, it has been shown that prior IAV infections sensitize for subsequent pneumococcal infections, but we still lack knowledge on mechanisms in the LRT that promote pneumococcal growth. Inflammation during LRT infections is a major reason for the observed symptomatology. A decrease in the endothelial barrier function is a hallmark in any acute inflammatory process mediating endothelial contraction and allowing movement of plasma constituents into the inflamed tissue. A controlled increase in vascular permeability is designed to counteract the inflammatory process, promoting clearance of the infecting agent (32).
We show here, by quantitative mass spectrometry-based proteomics, that mice recovering from a self-limiting IAV infection have a significant increase in plasma proteins in the BAL obtained on day 7 after IAV infection, which represents the time point of highest sensitivity to pneumococcal coinfection. We verified this suggested capillary leakage by showing increased leakage of FITC-dextran into the alveolar space at the same time point, and that Slit2, known to decrease leakage, partly decreased FITC-dextran leakage in our mouse IAV-infection model. Furthermore, we find that the plasma leakage provides enough nutrients and antioxidants for a burst of pneumococcal proliferation when the bacteria are instilled into the LRT at this time point. We record a pneumococcal doubling time of less than 40 min between 4 and 6 h after bacterial infection, a proliferation rate only achieved in vitro by a readily fermentable carbon source such as glucose and presence of antioxidants. This together with the increased glucose levels found in BAL from day 7 of IAV-infected mice support our conclusion that the rapid bacterial proliferation is fueled by glucose present in the accumulated plasma fluid in the LRT. We used a very low infectious dose to mimic the situation in human where bacteria present in the upper airways may reach the lower airways through aspiration and breathing. In mice, such low numbers of pneumococci are unable to replicate in the noninflamed LRT, likely due to a lack of a fermentable carbon source and possibly also antioxidants and are rapidly cleared from this environment. Instead, inoculation with a high dose of pneumococci into the noninflamed LRT will result in pneumococcal growth, pneumonia, and eventually invasive disease.
The IAV-infected LRT provides a more oxidative environment as compared to the noninfected LRT, as evidenced by increased levels of hydrogen peroxide, ferrous iron, infiltrating neutrophils, and MPO. Mechanisms by which bacteria diminish the damaging effects of redox stress include induction of bacterial chaperones, which control protein folding and prevent accumulation of protein aggregates as well as production of antioxidants. Pneumococci, however, lack endogenous production of catalase and cannot synthesize glutathione (GSH), making them dependent on antioxidants produced by the host. We find that catalase is present in the LRT both in the absence and presence of IAV infection. GSH peroxidase 3, that neutralizes H2O2 by oxidizing glutathione, is increased in IAV-infected BAL as compared to uninfected BAL, suggesting that pneumococci may employ this pathway, in addition to catalase, to reduce the oxidative stress in the IAV-inflamed LRT. Plasma leakage also caused a fourfold increase in the content of the potent antioxidant ascorbic acid (vitamin C) in the IAV-infected BAL, and our in vitro growth experiments reveal that WT pneumococci require not only glucose and other nutrients, but also host-derived antioxidants to achieve the early growth burst in the IAV-infected LRT. A surprising finding was the profound growth-promoting effect of ascorbic acid on pneumococci in a glucose-containing environment. Unlike mice, humans cannot synthesize ascorbic acid in the liver, but retrieve this antioxidant from the diet (33). A fourfold increase in ascorbic acid levels in bronchoalveolar lavage from patients with acute lung inflammation (acute respiratory distress syndrome) has been reported while the level of glutathione was not increased in these patients (34).
Pneumococci adapt to the redox-imbalanced inflamed environment in the IAV-infected LRT by inducing its surface localized chaperone/protease HtrA via the CiaRH two-component system that protects the organism against oxidative protein damage. No growth burst was observed for pneumococci lacking HtrA in the IAV-infected LRT, with partial growth arrest likely due to extensive protein oxidation. Our in vitro data support this hypothesis. In the defined CDM + glucose medium, the hydrogen peroxide-producing htrA mutant in serotype TIGR4 suffered complete growth arrest, that was restored by addition of ascorbic acid, but only after a lag phase. Another pneumococcal strain of serotype 2, strain D39, unlike TIGR4, suffered from a long growth arrest even in presence of HtrA. We hypothesize that the chaperone function of HtrA promotes protection of surface proteins from oxidation, and its protease activity removes oxidized and misfolded proteins that if not degraded would result in ribosome stalling and growth arrest. However, extensive intracellular protein oxidation may also result in growth arrest, if not in situ repaired by antioxidants, like ascorbic acid. A growth-promoting effect of HtrA has been suggested in previous studies where HtrA has been shown to be important for pneumococcal growth in vivo (19). In that study, deletion of HtrA had a more dramatic effect in a D39 strain as compared to TIGR4, and the D39ΔhtrA mutant was unable to survive in the blood. The inability to survive in an environment where opsonophagocytic activity is high, supports our data of a protective effect on complement-mediated opsonophagocytosis by HtrA. The protection found from complement deposition in presence of HtrA was associated with significantly higher levels of capsule. The level of capsule is affected by membrane-bound enzymes that might be susceptible to oxidative damage during infection, and therefore requiring HtrA protection. HtrA has also been shown to play a role in the virulence of Klebsiella pneumoniae (35). Furthermore, HtrA has been suggested as a promising antibacterial target as it is expressed by all prokaryotic cells (36). However, this important stress-induced class of serine proteases and chaperones are also expressed by eukaryotic species including humans, making it difficult to generate selective specificity (37).
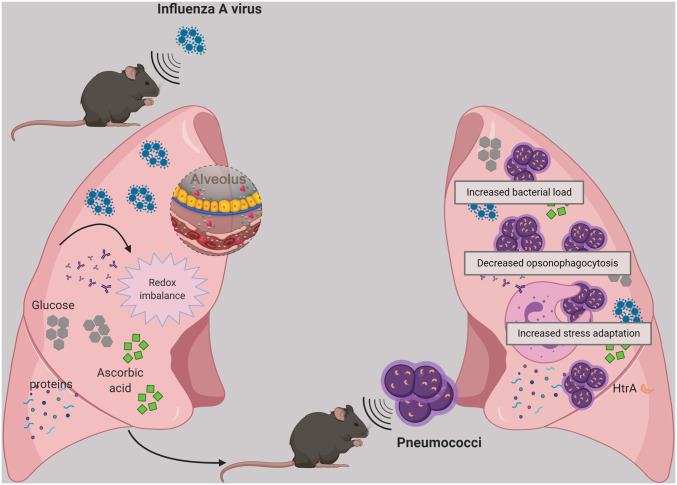
In conclusion, our results suggest that IAV infection provides a nutritional advantage for pneumococci to grow in the LRT due to virus-induced inflammation and capillary leakage. Furthermore, the enhanced pneumococcal proliferation observed requires presence of antioxidants, such as ascorbic acid, also emanating from the circulation, as well as stress-induced expression of the surface chaperone/protease HtrA. Together these components help to promote oxidative stress resistance and protect pneumococci from complement-mediated opsonophagocytosis in the lungs (Fig. 5).
Fig. 5.
Schematic summary of pneumococcal proliferation in influenza-infected lungs. Influenza-induced inflammation and capillary leakage leads to increased nutrient and antioxidant availability, thereby allowing pneumococcal outgrowth in the LRT. Pneumococcal adaptation to IAV-induced inflammation and redox imbalance is linked to higher expression of the pneumococcal chaperone/protease HtrA, which promotes pneumococcal growth under oxidative stress conditions and protects pneumococci from complement-mediated opsonophagocytosis by maintaining capsular production. Created with https://biorender.com/.
Moreover, we suggest that our findings in mice have direct bearings on the increased susceptibility of IAV-infected humans to develop pneumococcal pneumonia. In the absence of antibiotics, as the situation was during the Spanish flu pandemic, severe secondary pneumonia primarily caused by pneumococci, developed rapidly within a week following the onset of IAV symptoms, with high mortality, also in young individuals. At autopsy, the infected lungs were filled by bacteria (4), suggesting very high initial bacterial replication rates. It has been reported that severe influenza infections of the human lungs cause capillary leakage (26). We propose that the bacterial growth and clearing microenvironment in the LRT of IAV-infected individuals may differ between individuals, depending on the degree of plasma leakage into the LRT, its content of nutrients, such as glucose, and levels and types of antioxidants. The bacterial clearing environment in the virally infected LRT will also differ between individuals, depending on influx of phagocytic cells, complement factors, and bacterial-specific antibodies from the circulation. In our view any bacterial organism able to reach and grow to high numbers in the IAV-modulated LRT could contribute to lung damage by production of proinflammatory mediators such as cytotoxins for gram-positive organisms like pneumococci and endotoxin for gram-negatives. A bacterial growth-promoting environment, due to capillary leakage, will occur during all acute lung inflammatory conditions, including also severely ill Covid-19 patients, with acute respiratory distress syndrome.
Materials and Methods
Microbial Strains and Growth Conditions.
IAV PR8/A/34 (H1N1), kindly provided by Adolfo Garcia-Sastre, Icahn School of Medicine at Mount Sinai, New York, NY, was grown on Madin-Darby canine kidney (MDCK) cells. Briefly, cells were infected with the virus and incubated at 35 °C, 5% CO2, for 36 to 48 h. Supernatant was harvested, spun down to remove cellular debris, and the virus was then purified and concentrated on sucrose gradient (Virapur). Virus titers were determined by performing Avicel (FMC Biopolymer) plaque assays on MDCK cells as described previously (38). A dilution of 1:200 (corresponds to ∼10 to 30 PFU) of the virus stock was determined as a nonlethal dose for mouse infection experiments. S. pneumoniae serotype 4 (TIGR4, ATCC BAA-334) and its isogenic mutants (SI Appendix, Table S4) were grown overnight on blood agar plates (BD Diagnostic Systems, Columbia agar with 5% sheep blood) or in C medium with yeast extract (C + Y) until OD620 = 0.45. Stock cultures in logarithmic growth were frozen in 10% glycerol and stored at −80 °C. CFU were determined by plating on blood agar plates and optical density readings and bacteria were washed twice and diluted in PBS to obtain the appropriate concentrations for the mouse infections. Bacterial medium and blood agar plates were purchased from the Karolinska Clinical Microbiology Laboratory (Solna, Sweden).
Mutant Construction.
The bacterial mutants used in this study were constructed by fusion PCR mutagenesis as described previously (23). Briefly, upstream and downstream fragments of the target gene and the gene encoding for the appropriate antibiotic resistance were amplified with overlapping regions using the primers listed in SI Appendix, Table S5. The antibiotic marker was fused to the upstream region of the target gene, and, afterward, the downstream region was added by PCR. The correct fragment was gel purified and transformed into S. pneumoniae. To genetically complement the htrA-deficient strain, we inserted a fragment carrying the WT htrA gene as well as an adjacent erythromycin gene into the htrA locus of TIGR4ΔhtrA. Transformation was carried out in C + Y medium in presence of 100 ng µL−1 of the competence-stimulating peptide CSP-2. Bacteria were plated onto blood agar plates containing respective antibiotics.
Animals.
For all experiments 6- to 8-wk-old male C57BL/6J mice were used which were purchased from Charles River Laboratories and housed under specific pathogen-free conditions at Karolinska Institutet, Division of Comparative Medicine. All experiments were performed in accordance with the local ethical committee (Stockholms Norra djurfösöksetiska nämnd).
Cell Culture Models.
MDCK cells were cultivated in Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen) containing 10% heat-inactivated fetal bovine serum (FBS) and l-glutamine in a humidified incubator at 37 °C, 5% CO2. Human monocytic leukemia THP-1 cells (American Type Culture Collection [ATCC]) and RAW264.7 cells were cultivated in RPMI 1640 (Invitrogen) supplemented with 10% heat-inactivated FBS and 2 mM l-glutamine in a humidified incubator at 37 °C, 5% CO2. All cell lines had previously been used in the laboratory and no additional verification was performed.
In Vivo Infection Model.
C57BL/6 mice were infected intranasally (i.n.) with 10 to 30 PFU of IAV PR8/A/34 (H1N1) in 50 µL or treated with PBS (mock) under sedation with 80 mg/kg ketamine and 5 mg/kg Rompun (Bayer). At day 7 after IAV infection, mice were challenged i.tr. with ∼105 CFU S. pneumoniae TIGR4 in 50 µL. The technique for administration of the inoculum was described previously (39). BALs were obtained by rinsing the lungs twice with PBS. BAL samples were centrifuged at 4 °C, 2,900 rpm for 10 min to separate cells and supernatants. To determine the viral load in the lungs, supernatants of homogenized lungs were used for Avicel (FMC Biopolymer) plaque assays on MDCK cells. To determine bacterial numbers in BAL, blood, and lungs, serial dilutions of the samples were plated onto blood agar plates.
FITC-Dextran Permeability Assay.
Capillary permeability in the lung was assessed using FITC-dextran. For that, mice were given an i.v. injection of FITC-dextran (100 µL of 20 mg/mL) at day 7 after influenza infection. About 60 min after injection, the mice were killed, and blood and BAL were collected. Plasma was collected from ethylenediaminetetraacetic acid (EDTA) blood and each sample was diluted in an equivalent amount of PBS. BAL samples were centrifuged and FITC-dextran concentration in both BAL and serum was measured against a standard curve. The concentration of FITC-dextran in BAL was corrected by the concentration in the blood (FITC-dextran fluorescence in BAL/FITC-dextran fluorescence in blood). In some experiments, lung permeability in influenza-infected mice was inhibited by daily i.v. injection of Slit2 for 4 d (1.56 μg from day 3 to day 6 after IAV infection).
Flow Cytometry Analysis of Cells in BAL.
Cell suspension of BAL was centrifuged at 4 °C, 2,900 rpm for 10 min, and cell viability was determined by incubation with fixable viability dye (FVD) eFluor780 (eBiosciences) for 20 min. For subsequent flow cytometry (Gallios; Beckman Coulter), cells were fixed with 4% paraformaldehyde (PFA) and then incubated with Fc block (anti-mouse CD16/CD32; BD Biosciences) on ice for 20 min, followed by surface marker staining with anti-mouse Abs: F4/80 (PE; BD), CD11b (PE-CF594; BD), CD11c (PE-Cy7; eBiosciences), Ly6G (APC; BD), and Ly6C (V450; BD). The data were analyzed using Kaluza 1.2 (Beckman Coulter). The absolute numbers of different cell types were calculated based on the proportion of viable events analyzed by flow cytometry as related to the total number of viable cells per sample.
RNA Isolation, cDNA Synthesis, and Quantitative RT-PCR.
Bacterial RNA was isolated from samples lysed in Triton X-100 upon incubation in BAL ex vivo or from mouse lungs resuspended in RNeasy lysis buffer (RLT) (Qiagen RNeasy lysis buffer) using the RNeasy MiniKit (Qiagen). The concentration and purity of isolated RNA were determined spectrophotometrically with the NanoDrop ND 1000 apparatus. cDNA was synthesized from the isolated RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). RT-PCR was performed using the iTaq Universal SYBR green supermix (Bio-Rad) and the CFX Connect Real-Time PCR Detection System (Bio-Rad).
Primers used to amplify control and target genes are listed in SI Appendix, Table S4. For qPCR from mouse lungs, predesigned primer mixes containing forward and reverse primers for the specific RT-PCR target were purchased from Qiagen (QuantiTect primer assay) or Sigma (SI Appendix, Table S4). For each sample, the mRNA expression level was normalized to the level of beta-actin (host) or 16s (bacteria), and relative expression was determined via the ∆∆CT method. Each PCR run included no-template controls.
Western Blot Analysis from Tissue Lysates.
Whole cell lysates of mouse lungs were prepared using radioimmunoprecipitation assay buffer (RIPA) buffer supplemented with 1× protease inhibitors. Cell debris and nuclear material were pelleted by centrifugation (13,000 rpm, 10 min) and protein content was measured by the bicinchoninic acid reagent. Defined amounts of the protein lysates were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) on a NuPAGE 4 to 12% Bis-Tris gel (Thermo Fisher Scientific) and transferred onto polyvinylidene difluoride (PVDF) membrane (Bio-Rad). Membranes were incubated with a CRAMP antibody which is a custom-designed protein G purified polyclonal rabbit antiserum against CRAMP peptide, and synthetic CRAMP (ISRLAG LLRKGGEKIG EKLKKIGQKI KNFFQKLVPQ PE), (1/3000; Innovagen AB) or anti-beta-actin (1/1,000, Cell Signaling). Anti-rabbit IgG-horseradish peroxidase (HRP) (1/10000; GE Healthcare) served as secondary Ab. Immunoreactive proteins were visualized using the ECL Western blotting detection system.
Measurement of Carbohydrates in BAL Samples.
Sialic acid and glucose concentrations in BAL of mice were determined using a sialic acid or glucose assay kit (Sigma-Aldrich) following the manufacturer’s instruction. Briefly, to quantify total sialic acids, a hydrolysis was performed for 60 min at 80 °C in order to release bound sialic acids. Free sialic acid concentrations were determined by oxidizing the samples for 60 min at room temperature. Finally, the dye was added and the absorbance at 549 nm was measured. Glucose concentrations were measured by oxidizing the samples for 30 min at 37 °C after which the dye was added and the absorbance at 570 nm was measured. Absolute concentrations were calculated using a standard curve containing known concentrations.
Metabolomic Analyses of BAL Samples.
Samples for gas chromatography-mass spectrometry (GC-MS) analysis were prepared following a standard procedure (40). A total of 450 μL of extraction mix with labeled internal standards was added to 50 µL of each lavage sample in a microcentrifuge tube. Internal standards were: [2H4] butanediamine ·2HCl (500 ng/μL), [13C4] disodium-α-ketoglutarate (500 ng/μL), [13C6] glucose (500 ng/μL), [13C5,15N] glutamic acid (500 ng/μL), [13C5] proline (500 ng/μL), [2H4] succinic acid (500 ng/μL), [13C12] sucrose (500 ng/μL), [2H7] cholesterol (500 ng/μL), [1,2,3-13C3] myristic acid (500 ng/μL), [13C4] palmitic acid (500 ng/μL), and [2H6] salicylic acid (500 ng/μL). Metabolites were extracted in a bead mill for 2 min and frequency 30 Hz and allowed to stand on an ice bath for 1.5 h. Samples were then centrifuged for 10 min, 4 °C, 14,000 rpm; 100 μL of each supernatant was transferred to a separate GC vial and evaporated to dryness in a SpeedVac. Derivatization was done by adding 15 μL of O-methylhydroxylamine hydrochloride 98% (30 μL, 15 μg/μL) in pyridine followed by 15 min shaking, heating to 70 °C for 1 h and incubation at room temperature for approximately 16 h. N-methyl-N-trimethylsilyltrifluoroacetamide (MSTFA) +1% trimethylchlorosilane (TMCS) (15 μL) was added to the samples together with 15 ng methylstearate/μL in heptane (15 μL) and incubated at room temperature for 1 h prior to GC-MS analysis. For GC-MS, a 1-μL aliquot was injected (autosampler) into a gas chromatograph, Agilent 7890A, equipped with a 30-m fused-silica capillary column (J&W Scientific) and coupled to a Pegasus HT High-Throughput Time of Flight Mass Spectrometer (Leco). Masses were acquired in the mass-to-charge (m/z) range of 50 to 800. To be able to accurately analyze the metabolites present in high concentrations (e.g., glucose), the lavage samples where diluted five times in Milli-Q water. Diluted samples were prepared in duplicates and metabolites were extracted following the same protocol as for the undiluted samples.
In Vitro Growth Analysis and Bacterial Viable Counts of Bacteria.
To follow the growth over time, bacteria were grown on complete THY ± catalase (2,500 units/mL) or ±Flu BAL supplemented with C + Y without carbohydrates. Measurements were performed at OD620 every 10 min for 16 h and the experiment was repeated at least three times in triplicates. Growth curves were generated, and doubling times were calculated from the slope of the growth curves at midlog phase.
For experiments involving CDM as described previously (41) (SI Appendix, Table S6) bacteria were grown in 24-well polystyrene plates in presence of glucose as carbon source ± catalase (2,500 units/mL) or ascorbic acid (10 mM). Optical density was measured every 30 min at 620 nm over 20 h using the Infinite 200 Pro plate reader (Tecan) and the experiment was repeated three times in triplicates. For bacterial viability, samples were taken at different time points (0, 2, 4, 6, and 18.5 h) and serial dilutions of the samples were plated onto blood agar plates.
Sample Preparation for Proteomic Analysis.
Protein precipitation of BAL samples was performed by adding cold acetone with a volume corresponding to seven times the sample volume followed by incubation at −20 °C overnight. The precipitated pellet was first collected and washed with cold acetone using two centrifugation steps (10 min, 14,000 rpm). The precipitate was then dissolved in 50 μL 2% SDS and solubilization was aided by water bath sonication for 30 min. After measuring total protein concentration via micro BCA assay kit (Thermo Fisher Scientific), 50 µg of each sample was taken out, reduced in 8 mM dithiothreitol (DTT) (56 °C, 45 min) and alkylated in 25 mM iodoacetic acid (22 °C, 30 min in darkness). This step was followed by a second protein precipitation step using sample-cold acetone in a ratio of 1:9, −20 °C overnight. The precipitates were collected and washed with acetone via centrifugation (10 min, 14,000 rpm). Each dry pellet was first resuspended in 15 µL of 8 M urea in 20 mM 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid (EPPS) buffer, and then 16 µL of 20 mM EPPS buffer containing 0.67 µg LysC enzyme (Wako) was added (pH 8.2). The digestion with LysC was performed at 30 °C for 8 h. A total of 92 µL EPPS buffer containing 1 µg of sequencing grade modified trypsin (Promega) was then added and the sample was digested overnight at 37 °C. A total of 25 µg of each sample was subjected to Tandem Mass Tag 10 (TMT10) labeling (Thermo Fisher Scientific) and pooled to one TMT10 sample (250 µg) according to TMT10 standard procedures. Sample desalting and cleaning was performed using C18 (Sep-Pak, C18, Vac 1 cc, 50 mg, Waters). A total of 125 µg of the TMT-labeled sample pool was fractionated (Pierce High pH Reversed-Phase Peptide Fractionation Kit, Thermo Fisher Scientific) and dried. Sample fractions were dissolved into 10 µL of 0.1% formic acid and 2% acetonitrile prior to LC-MS proteomics analysis.
LC-MS/MS Proteomic Analysis.
Nano-HPLCy-electrospray ionization (HPLC-ESI)–MS/MS analyses were performed using a Q Exactive HF mass spectrometer (Thermo Scientific) equipped with an EASY Spray Source and connected to an UltiMate 3000 RSLCnano UPLC system (Thermo Scientific). Injected samples were preconcentrated and further desalted online using a PepMap C18 nano trap column (length, 2 cm; inner diameter, 75 µm; particle size, 3 µm; pore size, 100 Å; Thermo Scientific) with a flow rate of 3 µL min−1 for 5 min. Peptide separation was performed using an EASY-Spray C18 reversed-phase nano LC column (Acclaim PepMap RSLC; length, 50 cm; inner diameter, 2 µm; particle size, 2 µm; pore size, 100 Å; Thermo Scientific) at 55 °C and a flow rate of 300 nL min−1. Peptides were separated using a binary solvent system consisting of 0.1% (vol/vol) formic acid (FA), 2% (vol/vol) acetonitrile (ACN) (solvent A) and 98% ACN (vol/vol), 0.1% (vol/vol) FA (solvent B) and eluted with a gradient of 4 to 26% B in 120 min, 26 to 95% B in 10 min. Subsequently, the analytical column was washed with 95% B for 5 min before reequilibration with 4% B. For fractions 3 to 8, the first step of gradient was split into two parts, 4 to 10% B in 100 min plus 10 to 26% B in 20 min from fraction 3 to 5, and 4 to 14% in 100 min plus 14 to 26% B in 20 min for fractions 6 to 8. A total of 2 μg of unfractionated sample was also run using a longer gradient, of 4 to 26% B in 210 min and 26 to 95% B in 10 min, respectively. Mass spectra were acquired in a m/z range of 375 to 1,500 with a resolution of 120,000 at m/z 200. Automatic gain control target was set to 3 × 106 with a maximum injection time of 100 ms. The 17 most intense peptide peaks were selected for peptide fragmentation via higher-energy collision dissociation (HCD) with normalized collision energy value set at 33. The ion selection abundance threshold was set at 0.1% with charge exclusion of z = 1 ion. The MS/MS spectra were acquired at a resolution of 60,000, with a target value of 2 × 105 ions, and a maximum injection time of 120 ms. The fixed first m/z was 100, and the isolation window was 1.6 m/z. The instrument was operated in the positive ion mode for data-dependent acquisition of MS/MS spectra with a dynamic exclusion time of previously selected precursor ions of 45s.
Proteomic Analysis.
Acquired raw files (from fractionated and unfractionated samples) were analyzed as one single study with Proteome Discoverer 2.2 software (Thermo Fisher Scientific). Protein identification and quantification was carried out using the Mascot database search engine against SwissProt mouse proteome database (May 23, 2018, 16,983 entries) as well as a database for common protein contaminants. Applied value correction for TMT10 quantification was used based on values relative to the TMT10 lot provided by the manufacturer. Precursor mass tolerance was set to 10 ppm and fragment mass tolerance to 0.05 Da; a minimum peptide length of six amino acids and a maximum of two trypsin missed cleavages was allowed. Static modifications included carbamidomethylation of cysteine and residual modifications of TMT 10plex reagents on peptide N terminus and lysine. Dynamic modifications included methionine oxidation and deamidation of arginine and glutamine. For high confidence identifications, peptide groups were filtered for maximum 1% false discovery rate (FDR) based on the number of hits obtained for searches against the forward and decoy database. Only proteins with at least two unique peptides were used for final quantitative data analysis. In total 2,723 proteins were quantified. Each protein was normalized to the average ion intensities of the TMT tags from the five control mice (−Flu).
Complement C3 in BAL Samples.
Equivalent volumes (20 µL) of BAL were separated by SDS/PAGE, transferred to a PVDF membrane, and C3b was detected using an anti-C3 antibody (1/300; Calbiochem), recognizing C3 followed by HRP-conjugated goat anti-rabbit antibody (1/10,000; GE Healthcare) and Amersham Biosciences ECL Prime Western blotting detection reagent (GE Healthcare) to visualize the protein. Quantitative analysis of C3 levels in BAL was performed by sandwich ELISA (Nordic BioSite AB) following manufacturer’s instructions.
Measurement of Hydrogen Peroxide Levels.
Levels of H2O2 were measured in BAL supernatants isolated from uninfected and IAV-infected mice and in bacterial culture medium after 30 min of incubation using different bacterial strains. Briefly, 2,2′-azino-bis (3-ethylbenzthiazoline-6-sulfonic acid) and horseradish peroxidase (both Sigma-Aldrich) were added to the samples and upon incubation at room temperature for 20 min, optical densities at 560 nm were measured and hydrogen peroxide concentrations were calculated using a standard curve with known concentrations.
Determination of Iron Levels in BAL Samples by Inductive Coupled Plasma-MS.
Iron levels in BAL supernatants were determined by sector field (SF)-inductive coupled plasma (ICP)-MS analysis at the Wisconsin State Laboratory of Hygiene at University of Wisconsin-Madison. Briefly, samples were digested in a nitric acid/peroxide mixture followed by another digestion in hydrochloric acid/hydrofluoric acid mixture to determine total iron levels. For separation of dissolved from colloidal and particulate iron ultrafiltration with 10 kDa Amicon regenerated cellulose centrifugal ultrafiltration units was performed. Fe(II) levels were determined by ferrozine-spectrophotometry.
Ex Vivo Bacterial Gene Analysis.
Bacteria were incubated at 37 °C in either uninfected BAL or IAV-infected BAL supplemented with C + Y medium which allows us to mimic the in vivo conditions when using the diluted BAL samples. Bacteria incubated in PBS supplemented with C + Y medium were used as control. After 90 min the bacteria were pelleted down and RNA was isolated as described under RNA isolation, cDNA synthesis, and qRT-PCR.
Deposition of Complement C3b on Bacteria.
Deposition of complement C3b on the bacteria was performed as described previously (42). Briefly, bacteria were grown to midlog phase and then incubated with 20% normal human serum (Sigma) or mouse serum (Innovative Research) in PBS for 30 min at 37 °C, washed three times with PBS, and then sequentially stained with goat anti-C3 antibody (Calbiochem) followed by Alexa Fluor 488-labeled rabbit anti-goat antibody (Life Technologies). Fixed bacteria were analyzed by flow cytometry (Gallios; Beckman Coulter) and data were analyzed using Kaluza 1.2 (Beckman Coulter).
Phagocytosis Assay.
THP-1 cells were seeded in 24-well plates (5 × 105 cells per well) and differentiated for 48 h with 20 ng/mL of phorbol myristate acetate (PMA; Sigma). RAW264. cells were seeded in 24-well plates (5 × 105 cells per well) and grown to confluent monolayers. Cells were washed with PBS and infected at a multiplicity of infection (MOI) of 20 with serum-opsonized bacteria resuspended in Roswell Park Memorial Institute (RPMI) medium without FBS. After 1.5 h of incubation, cells were washed three times with PBS and incubated for 1 h in the presence of 300 μg/mL gentamicin to kill extracellular bacteria. The cells were then washed three times with PBS and lysed for 12 min at 37 °C with 1% saponin in PBS. Serial dilutions were plated on blood agar plates and incubated overnight at 37 °C and 5% CO2 to determine the number of phagocytosed bacteria.
STED Imaging and MATLAB Analysis.
STED imaging and image analysis was performed as described previously using an instrument from Abberior Instruments, built on a stand from Olympus (IX83), with a four-mirror beam scanner (Quad scanner, Abberior Instruments), and modified for two-color STED imaging (42). Capsule staining was performed using rabbit anti-capsule serum for serotype 4 (Statens Serum Institute) and anti-rabbit Alexa Fluor 594 antibody, and complement factor 3 was stained using goat anti-C3 antibody followed by anti-goat Atto647N-labeled secondary antibody (Sigma). Image acquisition, including laser timing/triggering and detector gating is controlled via a field-programmable gate array (FPGA) card and by the Imspector software (Abberior Instruments).
Analysis of STED images was carried out using custom-written code in MATLAB R2013b and an algorithm that estimated the relative area of the bacteria covered by complement. The algorithm worked as follows: A longitudinal line was manually drawn in the STED images, between the end-poles of each of the bacteria, along their symmetry axes. Along the lines the total fluorescence signal is integrated orthogonally to the line and over the bacteria’s width. In this way the total fluorescence distribution along the length of the bacteria can be stored and analyzed as a fluorescence trace for each selected bacterium. For area estimation, the patches along the symmetry line where the fluorescence distribution was nonzero was summed together, and the result was taken in relation to the total length of the bacteria as an estimation of the relative area of the bacteria covered by complement.
Analyses of Capsular Polysaccharide by ELISA and Immunoblot.
Capsular polysaccharide production was detected by immunoblotting as described before with minor modifications (43). Briefly, bacteria were grown in THY medium to OD620 = 0.2 and harvested by centrifugation at 16,100 × g for 1 min at room temperature and washed once with PBS. For ELISA analyses, 5 μL of proteinase K was added to the cell suspension and incubated at 56 °C for 1 h. The mixture was centrifuged at 16,100 × g for 1 min. The supernatant was discarded, and the pellet was resuspended in 50 mM carbonate-bicarbonate buffer (Sigma). Maxisorp 96-well immuno plates (Thermo Scientific) were coated with the samples and incubated overnight at 4 °C. Anti-CPS4 antisera (SSI Diagnostica) was cross-absorbed with heat-killed Δcps4E TIGR4 cell lysate in 1× PBS with 1% bovine serum albumin (BSA) at a 1:1,000 dilution at 4 °C overnight. The plates were blocked with 1× PBS with 1% BSA for 1 h at room temperature and then washed four times with phosphate buffered saline with Tween (PBST) (1× PBS, 0.05% [vol/vol] Tween 20). Cross-absorbed CPS4 antisera was added to the wells and incubated at room temperature for 1 h and then washed four times with PBST. Goat anti-rabbit HRP (Thermo Fisher A16110) in 1× PBS with 1% BSA at a 1:1,000 dilution was added to the wells and incubated at room temperature for 1 h and then washed four times with PBST. SIGMAFAST OPD (Sigma) was added to the wells and incubated for 30 min at room temperature as instructed by the manufacturer’s protocol. Optical density at 450 nm was recorded by an automated reader (Tecan).
For immunoblot analyses pellets were washed once with PBS and resuspended in protoplast buffer (50 mM of Tris⋅HCl pH 7, 50 mM MgCl2, and 20% sucrose). Forty units of mutanolysin (Sigma) and 25 μg of lysozyme (Sigma) were added and incubated at room temperature overnight. Formation of protoplasts were confirmed using light microscopy. The reaction mixture was centrifuged at 3,000 × g for 10 min at room temperature. Supernatant containing the cell wall fraction was collected and the pellet containing the protoplast fraction was resuspended in PBS. To each of the sample, equal volume of 2× Laemmli buffer and 2 μL of proteinase K were added and incubated at 56 °C for 1 h. The samples were resolved on a 10% SDS/PAGE, transferred onto a nitrocellulose membrane, and blocked for 30 min at room temperature in 5% skim milk in PBST (PBS with 0.05% Tween 20). The blots were probed with anti-CPS4 antisera (SSI Diagnostica) at a 1:5,000 dilution at 4 °C overnight. The blots were washed twice with PBST followed by incubation with goat anti-rabbit HRP (Thermo Fisher A16110) at a 1:10,000 dilution for 1 h at room temperature. The membranes were washed three times with PBST and developed by SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher).
Statistical Analysis.
All graphs show mean ± SEM if not stated otherwise and statistical analysis was performed using GraphPad Prism. For comparison between two groups an unpaired, two-tailed t test or Mann–Whitney U test was used as appropriate. For comparison between multiple groups, a one-way ANOVA followed by Dunn’s multiple comparison test was used. In all tests P < 0.05 was considered significant. The exact values for the number of experiments or mice are listed in each figure legend.
For statistical analysis of proteomics data −Flu and +Flu mice were compared via two-tailed Student’s t test (with equal or unequal variance depending on F test). P values were corrected by FDR analysis and BF. Functional classification of proteins was performed with the help of UniProtKB (https://www.uniprot.org/uniprot/). PCA was performed using SIMCA 15.0 following mean centering, log transformation, and unit variance scaling.
Supplementary Material
Acknowledgments
We thank Adolfo Garcia-Sastre (Icahn School of Medicine at Mount Sinai) for providing the viral strain. Proteomics was performed by the Chemical Proteomics facility at Biomedicum, Karolinska Institutet and part of SciLifeLab and the Swedish National Infrastructure for Biological Mass Spectrometry (BioMS). SF-ICP-MS analysis of metal ions in BAL samples was performed at the Wisconsin State Laboratory of Hygiene at University of Wisconsin-Madison. We thank Henrik Antti and Olena Rzhepishevska for their support with the MS analysis of the metabolites in the BALs; Priyanka Nannapaneni, Katrin Pütsep, and Ilias Galanis for helpful discussions; and we thank http://grafikalabs.com/ for their help with the graphical summary. This work was supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Research Council, and the Swedish Foundation for Strategic Research; a grant from the Stockholm County Council; a project grant from the National Technological University, Singapore; a National Research Foundation Fellowship, Singapore; a National University of Singapore start-up grant; a European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Research Grant (2015); a project support grant from BioMS; and by the National Medical Research Council under its Clinical Basic Research Grant (MOH-OFIRG18may-0005).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012265117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Troeger C., Jr, et al. , Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990-2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 18, P1191–P1210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCullers J. A., The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 12, 252–262 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Hayward A. C. et al.; Flu Watch Group , Comparative community burden and severity of seasonal and pandemic influenza: Results of the flu watch cohort study. Lancet Respir. Med. 2, 445–454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morens D. M., Taubenberger J. K., Fauci A. S., Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: Implications for pandemic influenza preparedness. J. Infect. Dis. 198, 962–970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris D. E., Cleary D. W., Clarke S. C., Secondary bacterial infections associated with influenza pandemics. Front. Microbiol. 8, 1041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura S., Davis K. M., Weiser J. N., Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J. Clin. Invest. 121, 3657–3665 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shahangian A., et al. , Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J. Clin. Invest. 119, 1910–1920 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun K., Metzger D. W., Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat. Med. 14, 558–564 (2008). [DOI] [PubMed] [Google Scholar]
- 9.McCullers J. A., Bartmess K. C., Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J. Infect. Dis. 187, 1000–1009 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Plotkowski M. C., Puchelle E., Beck G., Jacquot J., Hannoun C., Adherence of type I Streptococcus pneumoniae to tracheal epithelium of mice infected with influenza A/PR8 virus. Am. Rev. Respir. Dis. 134, 1040–1044 (1986). [DOI] [PubMed] [Google Scholar]
- 11.Siegel S. J., Roche A. M., Weiser J. N., Influenza promotes pneumococcal growth during coinfection by providing host sialylated substrates as a nutrient source. Cell Host Microbe 16, 55–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kash J. C., et al. , Treatment with the reactive oxygen species scavenger EUK-207 reduces lung damage and increases survival during 1918 influenza virus infection in mice. Free Radic. Biol. Med. 67, 235–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterhans E., Grob M., Bürge T., Zanoni R., Virus-induced formation of reactive oxygen intermediates in phagocytic cells. Free Radic. Res. Commun. 3, 39–46 (1987). [DOI] [PubMed] [Google Scholar]
- 14.Leonard A., Lalk M., Infection and metabolism–Streptococcus pneumoniae metabolism facing the host environment. Cytokine 112, 75–86 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Yesilkaya H., Manco S., Kadioglu A., Terra V. S., Andrew P. W., The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol. Lett. 278, 231–235 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Paixão L., et al. , Host glycan sugar-specific pathways in Streptococcus pneumoniae: Galactose as a key sugar in colonisation and infection [corrected]. PLoS One 10, e0121042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ermund A., et al. , The mucus bundles responsible for airway cleaning are retained in cystic fibrosis and by cholinergic stimulation. Eur. Respir. J. 52, 1800457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spellerberg B., et al. , Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19, 803–813 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Ibrahim Y. M., Kerr A. R., McCluskey J., Mitchell T. J., Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect. Immun. 72, 3584–3591 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cassone M., Gagne A. L., Spruce L. A., Seeholzer S. H., Sebert M. E., The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J. Biol. Chem. 287, 38449–38459 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ibrahim Y. M., Kerr A. R., McCluskey J., Mitchell T. J., Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae. J. Bacteriol. 186, 5258–5266 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sebert M. E., Palmer L. M., Rosenberg M., Weiser J. N., Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70, 4059–4067 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hentrich K., et al. , Streptococcus pneumoniae senses a human-like sialic acid profile via the response regulator CiaR. Cell Host Microbe 20, 307–317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pericone C. D., Park S., Imlay J. A., Weiser J. N., Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the fenton reaction. J. Bacteriol. 185, 6815–6825 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai C., Gong Q., Cheng Y., Su G., Regulatory mechanisms of Robo4 and their effects on angiogenesis. Biosci. Rep. 39, BSR20190513 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.London N. R., et al. , Targeting Robo4-dependent slit signaling to survive the cytokine storm in sepsis and influenza. Sci. Transl. Med. 2, 23ra19 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heide K., Haupt H., Stoeriko K., Schultze H. E., On the heme-binding capacity of hemopexin. Clin. Chim. Acta 10, 460–469 (1964). [DOI] [PubMed] [Google Scholar]
- 28.Morgan W. T., Smith A., Domain structure of rabbit hemopexin. Isolation and characterization of a heme-binding glycopeptide. J. Biol. Chem. 259, 12001–12006 (1984). [PubMed] [Google Scholar]
- 29.Hawkins C. L., Davies M. J., Hypochlorite-induced damage to proteins: Formation of nitrogen-centred radicals from lysine residues and their role in protein fragmentation. Biochem. J. 332, 617–625 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armstrong S. M., et al. , Influenza infects lung microvascular endothelium leading to microvascular leak: Role of apoptosis and claudin-5. PLoS One 7, e47323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh Y. C., et al. , Influenza pandemics: Past, present and future. J. Formos. Med. Assoc. 105, 1–6 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Kenne E., Lindbom L., Imaging inflammatory plasma leakage in vivo. Thromb. Haemost. 105, 783–789 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Grollman A. P., Lehninger A. L., Enzymic synthesis of L-ascorbic acid in different animal species. Arch. Biochem. Biophys. 69, 458–467 (1957). [DOI] [PubMed] [Google Scholar]
- 34.Schmidt R., et al. , Alveolar antioxidant status in patients with acute respiratory distress syndrome. Eur. Respir. J. 24, 994–999 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Cortés G., de Astorza B., Benedí V. J., Albertí S., Role of the htrA gene in Klebsiella pneumoniae virulence. Infect. Immun. 70, 4772–4776 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wessler S., Schneider G., Backert S., Bacterial serine protease HtrA as a promising new target for antimicrobial therapy? Cell Commun. Signal. 15, 4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D. Y., Kim K. K., Structure and function of HtrA family proteins, the key players in protein quality control. J. Biochem. Mol. Biol. 38, 266–274 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Matrosovich M., Matrosovich T., Garten W., Klenk H. D., New low-viscosity overlay medium for viral plaque assays. Virol. J. 3, 63 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sender V., Lang L., Stamme C., Surfactant protein-A modulates LPS-induced TLR4 localization and signaling via β-arrestin 2. PLoS One 8, e59896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wuolikainen A., et al. , Multi-platform mass spectrometry analysis of the CSF and plasma metabolomes of rigorously matched amyotrophic lateral sclerosis, Parkinson’s disease and control subjects. Mol. Biosyst. 12, 1287–1298 (2016). [DOI] [PubMed] [Google Scholar]
- 41.van de Rijn I., Kessler R. E., Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27, 444–448 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pathak A., et al. , Factor H binding proteins protect division septa on encapsulated Streptococcus pneumoniae against complement C3b deposition and amplification. Nat. Commun. 9, 3398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xayarath B., Yother J., Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. J. Bacteriol. 189, 3369–3381 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.