Significance
Helicobacter pylori, a gastric pathogen causing 800,000 deaths in the world annually, is encountered both in vitro and in patients as spiral-shaped bacteria and as round cells named coccoids. We discovered that the toxin from a chromosomal type I toxin−antitoxin system is targeting H. pylori membrane and acting as an effector of H. pylori morphological conversion to coccoids. We show that these round cells maintain their membrane integrity and metabolism, strongly suggesting that they are viable dormant bacteria. Oxidative stress is identified as a signal inducing toxin expression and coccoid formation. Our findings reveal insights into a form of dormancy of this bacterium that might be associated with H. pylori infections refractory to treatment.
Keywords: toxin–antitoxin, bacterial morphology, coccoids, dormant bacteria, Helicobacter pylori
Abstract
Toxin−antitoxin systems are found in many bacterial chromosomes and plasmids with roles ranging from plasmid stabilization to biofilm formation and persistence. In these systems, the expression/activity of the toxin is counteracted by an antitoxin, which, in type I systems, is an antisense RNA. While the regulatory mechanisms of these systems are mostly well defined, the toxins’ biological activity and expression conditions are less understood. Here, these questions were investigated for a type I toxin−antitoxin system (AapA1−IsoA1) expressed from the chromosome of the human pathogen Helicobacter pylori. We show that expression of the AapA1 toxin in H. pylori causes growth arrest associated with rapid morphological transformation from spiral-shaped bacteria to round coccoid cells. Coccoids are observed in patients and during in vitro growth as a response to different stress conditions. The AapA1 toxin, first molecular effector of coccoids to be identified, targets H. pylori inner membrane without disrupting it, as visualized by cryoelectron microscopy. The peptidoglycan composition of coccoids is modified with respect to spiral bacteria. No major changes in membrane potential or adenosine 5′-triphosphate (ATP) concentration result from AapA1 expression, suggesting coccoid viability. Single-cell live microscopy tracking the shape conversion suggests a possible association of this process with cell elongation/division interference. Oxidative stress induces coccoid formation and is associated with repression of the antitoxin promoter and enhanced processing of its transcript, leading to an imbalance in favor of AapA1 toxin expression. Our data support the hypothesis of viable coccoids with characteristics of dormant bacteria that might be important in H. pylori infections refractory to treatment.
Toxin−antitoxin (TA) systems are small genetic elements that are widely distributed on bacterial mobile genetic elements and among archaeal and bacterial genomes, often in multiple copies (1–4). They code for a small stable toxic protein, designated toxin, whose action or expression is counteracted by an unstable antitoxin molecule that can either be an RNA or a protein (3, 5). Under conditions in which the toxins can act, they target essential cellular processes or components (transcription, DNA replication, translation, cell wall, or membrane), resulting in growth arrest or cell death (3, 5). The molecular mechanism of the toxins’ activities and their regulation are often known in detail, while the biological function remains elusive. TA systems take part in plasmid maintenance and protection from phage infection. Less is known on the biological functions and targets of the chromosomally encoded TA systems. Some are important for bacterial survival in their mammalian host (6) or biofilm formation (7). There is accumulating evidence that, upon stress conditions, some TA systems play a role in the switch of actively growing bacteria to persisters or dormant cells (8–11). Persister bacteria constitute a subpopulation that is metabolically active but slow-growing and highly tolerant to antibiotics and stress conditions (11). Persistence is considered to be an important cause of recalcitrance of chronic bacterial infections to therapy. However, stress-induced activation of TA-encoded toxins does not necessarily cause persister formation (12), and it was more generally found that decreased intracellular adenosine 5′-triphosphate (ATP) concentration and/or low proton motive force (PMF) are landmarks of persister formation (13, 14).
In the present work, we explored the role of TA systems in Helicobacter pylori, a bacterium that colonizes the stomach of half of the human population worldwide and causes the development of gastritis. In some cases, gastritis evolves into peptic ulcer disease or gastric carcinoma that causes about 800,000 deaths in the world every year (15, 16). This microaerophilic bacterium is unique in its capacity to persistently colonize the stomach despite that organ’s extreme acidity and intense immune response (16). The molecular mechanisms at the origin of this exceptional adaptation capacity of H. pylori remain only partially understood. Their elucidation is crucial to understand H. pylori virulence and to improve its eradication in patients with recurrent peptic ulcer disease. H. pylori is a Gram-negative bacterium with a helical shape. Upon stress conditions (antibiotics, aerobic growth) or prolonged culture, the shape of H. pylori progressively evolves into a U shape followed by a spherical form designated coccoid (17, 18). H. pylori coccoids are nonculturable bacteria proposed to be dormant forms (18). Recently, dormant nonculturable cells were associated with a deeper state of dormancy as compared to persister cells (19).
H. pylori coccoids were observed in human gastric biopsies and, like spirals, adhere to gastric epithelial cells (20). Despite numerous reports on coccoid forms of H. pylori, no cellular effector of this conversion has been reported so far.
In type I TAs, the antitoxin is a small regulatory RNA inhibiting the synthesis of the toxin by base pairing the toxin-encoding messenger RNA (mRNA) (3–5, 21, 22). Four families (A−B−C−D) of conserved type I TA systems are expressed from the chromosome of H. pylori; only the A family was studied, as it is highly expressed and conserved among H. pylori strains (23–25). For the A1 TA system, the detailed mechanism by which transcription of the IsoA1 antitoxin RNA impairs AapA1 toxin synthesis by base pairing with its primary transcript, ensuring both translation inhibition of the AapA1 active message and leading to rapid degradation of the duplex by RNase III, has been recently published (24, 25). The H. pylori type I toxins are typically small hydrophobic peptides of 30 to 40 amino acids predicted to form alpha-helices (26). No clues on the mode of action or the physiological role of these systems have been reported.
Here we show that the AapA1 toxin induces a rapid morphological transformation of H. pylori from spirals to coccoids by targeting the inner membrane and probably interfering with cell elongation and division. Furthermore, oxidative stress induces coccoid formation and triggers imbalanced expression of the TA components in favor of toxin production.
Results
The AapA1 Toxic Peptide, Expressed by the AapA1/IsoA1 TA System, Triggers Rapid Transformation of H. pylori into Coccoids.
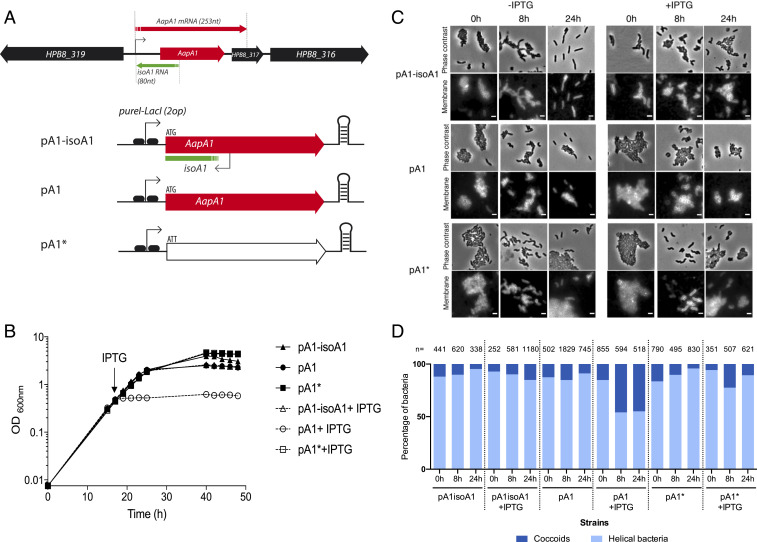
Recently, the study of the AapA1/IsoA1 TA system of H. pylori (Fig. 1A) showed that expression of the AapA1 toxin (a 30-amino acid-long hydrophobic peptide) leads to bacterial growth arrest (24). Given the genetic organization of the type I AapA1/IsoA1 locus in B128 strain, we decided to investigate the mechanism underlying the activity of AapA1 by using a strain in which AapA1 is under the control of an inducible promoter. We thus transformed H. pylori strain B128 deleted of the AapA1/IsoA1 chromosomal locus with each of the three plasmids constructed by Arnion et al. (24) (Fig. 1A). The first, pA1−IsoA1, derived of the pILL2157 vector (27), contains a functional TA locus in which the AapA1 gene is under the control of an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoter (24). In plasmid pA1, derived from this first construct, the antitoxin IsoA1 promoter has been inactivated by point mutations that do not interfere with transcription or translation of the AapA1 open reading frame (ORF) (24). Derived from this second construct, plasmid pA1* contains an additional mutation that inactivates the start codon of the AapA1 peptide (24). Under our experimental conditions, we observed, in agreement with data of ref. 24, that addition of IPTG did not significantly influence the growth rate of strains harboring pA1−IsoA1 or pA1*. In contrast, addition of the inducer causes a rapid growth arrest of H. pylori with pA1, indicating a toxic effect of AapA1 expression (Fig. 1B). The growth arrest was accompanied by loss of culturability of more than 103-fold 8 h after induction.
Fig. 1.
Expression of the AapA1 toxin from the AapA1/IsoA1 TA system induces growth arrest and transformation of H. pylori strain B128 into coccoids. (A) Genetic organization of AapA1/IsoA1 locus in H. pylori B128 strain with the AapA1 transcript encoding the toxin and the IsoA1 antitoxin RNA. Below is the representation of the inserts of the three plasmids used in this study (24), each derived from the pILL2157 E. coli/H. pylori vector (27). These inserts are expressed under the control of an IPTG inducible promoter. Plasmid pA1−IsoA1 expresses both the toxin and the first 30 nucleotides of the IsoA1 RNA antitoxin. In pA1, the promoter region of IsoA1 was mutated without affecting the amino acid sequence of AapA1. Plasmid pA1* was derived from pA1 and contained an additional mutation inactivating the start codon of AapA1. (B) Expression of the AapA1 toxin causes rapid growth arrest of H. pylori. Growth kinetics of H. pylori B128 strains carrying each of the three plasmids illustrated in A, grown in the absence (black symbols) or in the presence of (empty symbols) 1 mM IPTG that was added at the time indicated by an arrow (16 h). (C) Expression of the AapA1 toxin induces transformation of H. pylori into coccoids. Microscopy analysis of the H. pylori B128 strains carrying each of the three plasmids at 8 and 24 h following IPTG addition or grown without IPTG. (Upper) Phase contrast and (Lower) fluorescence images. H. pylori membranes were stained with the lipophilic dye FM4-64. (Scale bar, 2 μm.) (D) Quantification of the transformation of H. pylori into coccoids. The proportion of helical and coccoid bacteria under each condition was quantified with the phase contrast images using the MicrobeJ software. The number of analyzed cells is indicated for each condition. Cells were considered helical when they had an area of more than 0.70 μm2, a length between 1 and 5 μm, a width between 0.4 and 1 μm, and a circularity of less than 0.8, and they were considered coccoids when they had an area between 1 and 2 μm2, a width between 0.4 and 1.7 μm, and a circularity greater than 0.7.
In parallel to growth, we investigated the consequences of AapA1 expression on H. pylori morphology. Samples from bacterial cultures with these plasmids were grown in the presence or absence of IPTG, stained with a membrane-specific dye, FM4-64, and analyzed by fluorescence microscopy at 0, 8, and 24 h (Fig. 1C). H. pylori cells expressing the wild-type (WT) AapA1/IsoA1 locus (pA1−IsoA1) or the pA1* mutated locus present a classical helical rod-shaped phenotype upon IPTG exposure (Fig. 1C). Under the same conditions, we observed that cells carrying the pA1 plasmid, and thus expressing the AapA1 toxin, exhibit a rapid morphological conversion to spherical cells also known as coccoid forms (Fig. 1C). Using MicrobeJ (28) analysis of the phase contrast images, we quantified the relative proportions of coccoids vs. spiral bacteria under each condition (Fig. 1D). When the toxin is expressed (pA1+IPTG), about half of the cells have converted into coccoids as soon as 8 h postinduction, without further changes at 24 h postinduction. No bacterial lysis was detected even at 24 h postinduction. With strains carrying the pA1* plasmid, significant amounts of coccoids were only observed after 64 h of culture, similarly to the control WT strain containing an empty plasmid. These data show that the AapA1 toxin induces a fast conversion of H. pylori cells from spiral-shaped to coccoid forms.
Inner Membrane Targeting by the AapA1 Toxin.
To investigate the mode of action of the AapA1 toxin, we first analyzed its subcellular localization in H. pylori using a GFP reporter. A C-terminal fusion of the toxin with GFP (AapA1−GFP) was introduced into B128 strain deleted of the chromosomal AapA1/IsoA1 locus and expressed either from the chromosome under control of its native promoter or from the IPTG-inducible promoter of vector pILL2157 (27) (SI Appendix, Fig. S1). After IPTG induction, the plasmidic fusion did not affect H. pylori growth, indicating that a C-terminal GFP fusion prevents the toxicity of the toxin. Attempts to construct N-terminal−tagged AapA1 were unsuccessful.
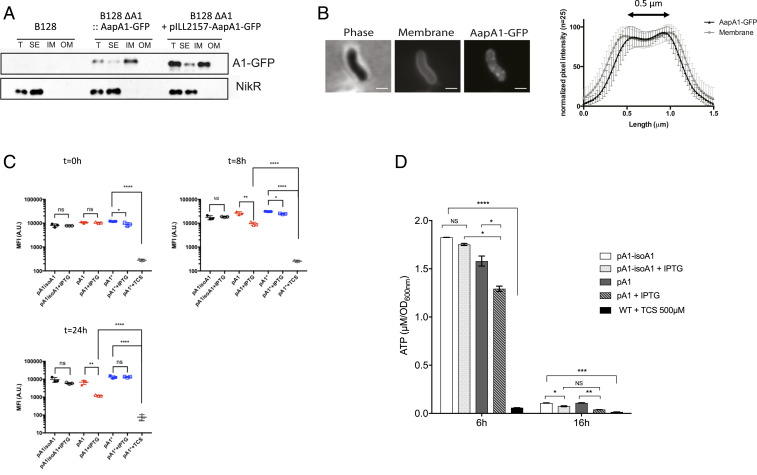
Cellular fractionation was then performed on the strains, and the subcellular localization of the fused toxin was analyzed by immunoblotting (fractionation controls are shown in SI Appendix, Fig. S2A). Fig. 2A shows that the AapA1−GFP fusions expressed from the chromosome or from the plasmid are almost exclusively present at the inner membrane of H. pylori. No AapA1−GFP peptide was detected in the culture medium. Fluorescence microscopy of live bacteria after induction of the AapA1−GFP fusion revealed a weak patchy pattern in the periphery of the cell (Fig. 2B), different from a GFP-alone control cell (SI Appendix, Fig. S2B), and compatible with membrane localization. This observation was confirmed by measuring the overlap of the fluorescence intensity profiles of the toxin and 1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene p-toluenesulfonate (TMA-DPH) membrane dye measured perpendicular to the length axis of H. pylori (Fig. 2B).
Fig. 2.
Analysis of the consequences of the AapA1 toxin targeting of the H. pylori inner membrane. (A) AapA1 toxin localizes to the H. pylori inner membrane. Western blot analysis of total extracts (T), soluble extracts (SE), inner membrane (IM), and outer membrane (OM) fractions prepared from H. pylori B128 strain expressing a GFP-tagged AapA1 toxin (AapA1−GFP) expressed either from the chromosome or from a plasmid (pILL2157) under the control of an IPTG-inducible promoter. Anti-NikR antibodies were used as a control of the cytoplasmic fraction. The fractionation procedure was validated as shown in SI Appendix, Fig. S2. (B) In live cells, AapA1−GFP localizes as discrete foci at the H. pylori membrane. Strain B128 expressing the AapA1−GPF fusion protein was analyzed on agarose pads by fluorescence microscopy 6 h after IPTG induction. Membranes were stained with TMA-DPH lipophilic dye. The membrane association of AapA1−GPF was quantified by measuring the fluorescence intensity profile perpendicular to the length axis of H. pylori. The graph shows the average fluorescence intensity profiles with SD (n = 25). The fluorescence maxima separated by 0.5 μm correlate with the H. pylori cell width. (Scale bar, 1 μm.) (C) Analysis of the effects of the AapA1 toxin on H. pylori membrane potential. MitoTracker Red CMXRos, a membrane potential reactive dye, was used to analyze live H. pylori B128 strains expressing each of the three plasmids illustrated in Fig. 1. Samples were taken at 0, 8, and 24 h postinduction and stained with MitoTracker Red CMXRos, and the PMF was measured as the mean fluorescence intensity (MFI) by flow cytometry. The cell population distribution histograms are presented in SI Appendix, Fig. S3. Cells expressing the toxin (pA1) present a moderate fluorescence reduction at 8 h that was more pronounced at 24 h of culture. This is indicative of the absence of major disturbance of their membrane potential. No changes were observed in cells containing either pA1−isoA1 or pA1* plasmid. As a control, pA1*-bearing cells were treated with 500 μM TCS, a protonophore active on H. pylori, resulting in a massive loss of membrane potential. The experiment was performed three times. The Student’s t test was used to determine significant differences of the means of the data. Error bars represent the SD, with * corresponding to P < 0.05, ** to P < 0.01, and **** to P < 0.0001, indicating that the mean values are significantly different, and with NS corresponding to nonsignificant (P > 0.05). (D) Measurement of intracellular ATP content. Intracellular ATP was extracted from B128 strains harboring pA1−IsoA1 or pA1 at 6 or 16 h postinduction growth in the presence or absence of IPTG. B128 WT strain treated with 500 μM TCS protonophore was used as a control of PMF dissipation. ATP concentrations were determined using a luciferase-based assay (BacTiter-Glo, Promega). Results from three independent experiments performed in triplicate are shown. After 6 h of culture, IPTG induction had a moderate effect on cellular ATP content of the pA1 strain as compared to the drastic consequences observed with the TCS control. After 16 h postinduction, both strains presented a strong drop in overall ATP content, and IPTG induction caused an additional minor decrease. The Student’s t test was used to determine significant differences of the means of the data. Error bars represent the SD, with * corresponding to P < 0.05, ** to P < 0.01, *** to P < 0.001, and **** to P < 0.0001, indicating that the mean values are significantly different, and with NS corresponding to nonsignificant (P > 0.05).
From these results, we conclude that the AapA1 toxin is specifically targeted to the inner membrane of H. pylori.
AapA1 Toxin Has a Moderate Impact on H. pylori Membrane Potential and Intracellular ATP Content.
Considering the membrane localization of the toxin, we hypothesized that AapA1 expression could affect the membrane potential and, by consequence, the ATP content of the cells. To explore this possibility, H. pylori cells harboring pA1−IsoA1, pA1*, or pA1 plasmids were taken at different culture time points (0, 8, and 24 h), exposed to MitoTracker Red CMXRos, a red-fluorescent dye whose accumulation inside live bacteria depends upon membrane potential, and fluorescence of individual cells was measured by flow cytometry. As a control, fluorescence was measured for the pA1* strain treated with 3,3′,4′,5-tetrachlorosalicylanilide (TCS), an effective H. pylori protonophore (29). The TCS treatment causes a massive drop in fluorescence due to dissipation of membrane potential (Fig. 2C and SI Appendix, Fig. S3). We observed that, when the AapA1 toxin is expressed in H. pylori, the membrane potential presented a weak drop at 8 h after induction that was more pronounced at 24 h (Fig. 2C).
Next, the ATP content of H. pylori strains carrying either pA1−IsoA1 or pA1 was measured from total metabolites extracted at different culture time points and quantified by a luciferase-based assay (Fig. 2D). After 6 h of culture, IPTG induction had a moderate effect on cellular ATP content of the pA1 strain as compared to the drastic consequences observed with the TCS control. At 6 h postinduction, many pA1-containing cells have already transformed into coccoids (see live imaging data below). At 16 h postinduction, in stationary phase, both strains presented a strong drop in overall ATP content, and IPTG induction caused an additional minor decrease (Fig. 2D).
Taken together, these results show that the AapA1 toxin moderately perturbs the H. pylori membrane potential with no major consequences on the cellular ATP content. We conclude that dissipation of membrane potential is not the major cause of toxin-induced bacterial growth arrest.
Ultrastructural Analysis of Toxin-Induced Coccoids.
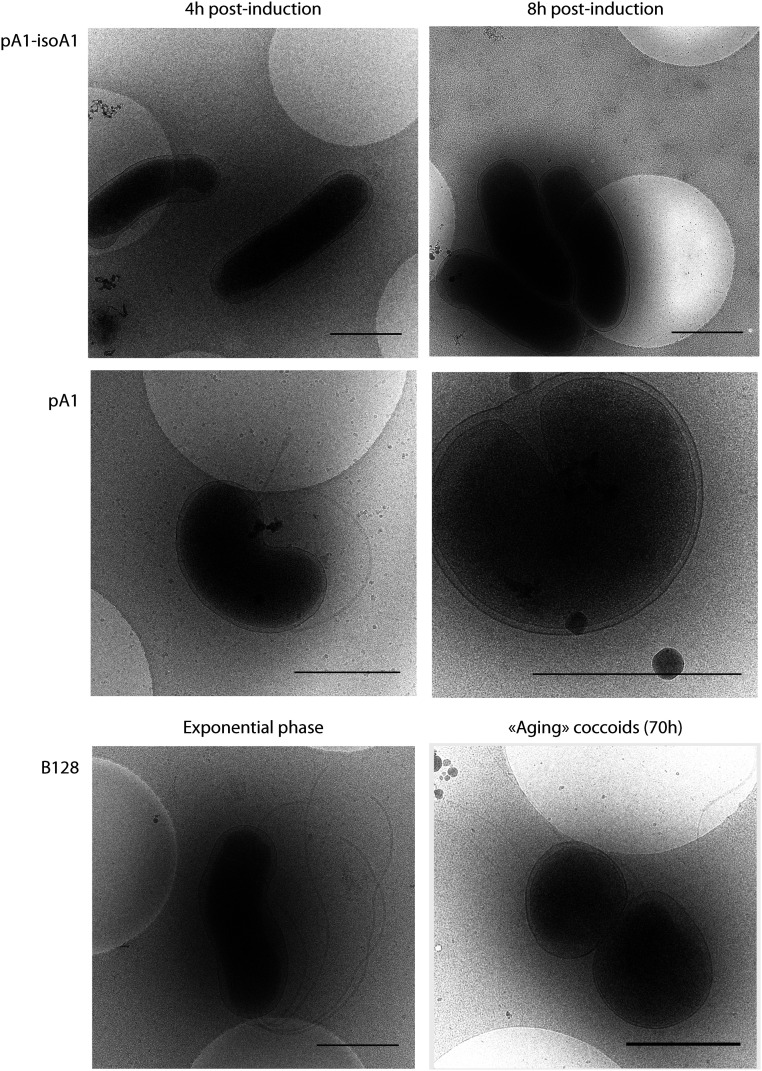
Cryoelectron microscopy (cryo-EM) was used to visualize H. pylori coccoids in near-native states. Using this method, we compared exponentially growing bacteria, toxin-induced coccoids and “aging coccoids” (70 h growth) (Fig. 3). In exponential phase, the characteristic helicoidal shape of H. pylori (strains B128 WT, 4 h, and pA1−IsoA1, 4 h to 8 h) was perfectly visible with a uniformly contrasted cytoplasm surrounded by two dense layers of membranes. The periplasm of variable thickness surrounding the cell is distinguishable as the low-density space between the inner and outer membrane. Strains expressing the toxin (pA1 at 4 h to 8 h) have intact flagella and are visible in two major morphotypes. In the first “U”-shaped morphology, a round intact outer membrane surrounds a bent intact inner membrane. The second morphotype corresponds to round coccoids of a diameter of ∼1 µm, with visible integrity of both membranes and a central uniformly stained dense cytoplasm. This second morphotype is similar to that of aging coccoids from the WT strain (70 h).
Fig. 3.
Ultrastructural analysis of toxin-induced coccoids and “aging” coccoids by cryo-EM. Cryo-EM was used to compare “aging” coccoids (70-h-old cultures) and exponentially growing B128 strains, either WT or carrying plasmid pA1−IsoA1 or pA1 grown for 4 or 8 h in the presence of IPTG. Expression of the toxin results in the formation of U-shaped bacteria at 4 h and round coccoid cells at 8 h with visible intact bacterial cell envelope similar to the aging coccoids. (Scale bar, 1 μm.)
In conclusion, cryo-EM images revealed that the toxin-induced H. pylori coccoids have no visible defect in membrane integrity and are ultrastructurally similar to “aging coccoid” forms.
Modification of Peptidoglycan Composition upon Transformation of H. pylori into Coccoids.
Our data showed that the AapA1 toxin targets H. pylori inner membrane without causing significant membrane potential collapse and without visible loss of integrity. Therefore, we tested whether the toxin-induced cell shape transformation could be associated with modifications in the cell wall and thus in peptidoglycan (PG) composition. PG extracted from B128 WT strain in exponential (24 h of culture) or stationary (36 h of culture) phase or after 72 h of growth (“aging coccoids”) was compared to PG extracted from the pA1-bearing strain either 8 h or 24 h after induction (toxin-induced coccoids, 24 and 36 h total culture time) and from the pA1* strain as a negative control. Samples were digested with mutanolysin and subjected to high-performance liquid chromatography/mass spectrometry (HPLC/MS) analyses. The relative abundance of muropeptides in each sample was calculated according to Glauner (30) (SI Appendix, Table S1). We found that the PG of B128 WT stationary phase and “aging coccoids” as well as the pA1-induced coccoids present enhanced amounts of GlcNAc−MurNAc dipeptide (GM2) as compared to helical exponential phase H. pylori and to the negative control with pA1*. This GM2 increase seems to be at the expense of the GM3 muropeptides, whose amount is reduced in “aging coccoids” and in pA1-induced bacteria where it reaches a minimal value 24 h postinduction. This shows that, during the transition from exponentially growing spiral bacteria to coccoids, the muropeptide composition undergoes important modifications that are comparable for the toxin-induced coccoids and “aging coccoids.” These changes, compatible with a looser PG macromolecule, suggest that the coccoid formation under these two conditions might be generated by similar mechanisms.
Kinetics of the Toxin-Induced H. pylori Morphological Transformation Assessed by Live-Cell Imaging.
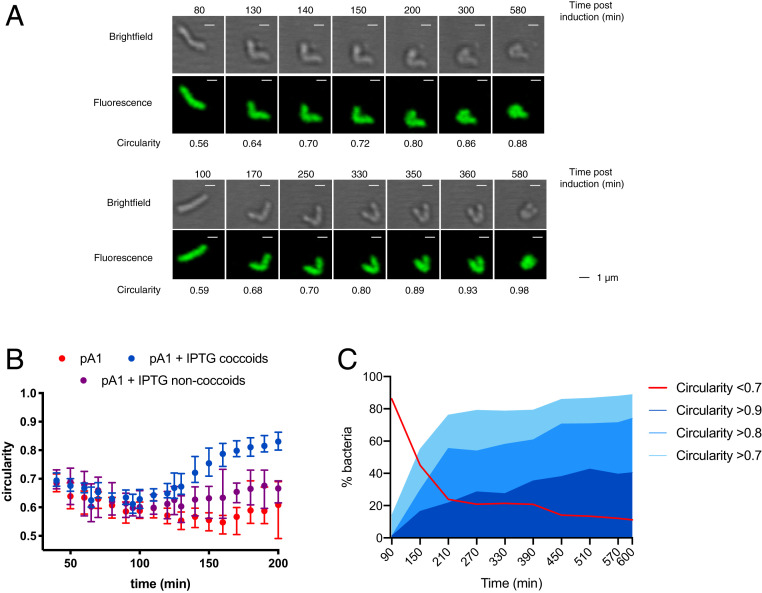
To progress in our understanding of the morphological conversion of H. pylori, we monitored the entire process of H. pylori toxin-induced conversion by live-cell time-lapse microscopy under physiological microaerobic conditions in a temperature-controlled chamber. The H. pylori B128 strain tested constitutively expressed cytoplasmic GFP, was impaired in motility (by deletion of flaA encoding the major flagellar protein), and carried the inducible toxin-expressing plasmid pA1. Individual live H. pylori bacteria were monitored in agarose pads by both bright-field and fluorescence microscopy.
First, the general growth features of the strain between cell birth and its next division were established on 61 bacteria in the absence of the IPTG inducer. Under these conditions, statistics of the data collected showed that the strain has a mean doubling time of 165 min (indicative of optimal growth conditions), with an initial bacterial length of 1.9 µm after division and a maximum length prior to division of 3.2 µm (SI Appendix, Fig. S4). In a second round of experiments, snapshots of the cells were taken at intervals of 10 min during 10 h under two conditions, one in which the AapA1 toxin was induced by IPTG addition to the pad and the other without inducer, as a control (Fig. 4). Three representative movies of the induced condition can be viewed in Movies S1–S3. To quantify the changes in shape of individual H. pylori cells over time, we used the MicrobeJ software (28) and applied circularity attributes to discriminate the populations. First, we compared the evolution over time of the median circularity of noninduced bacteria (n = 109) versus bacteria with IPTG induction of the A1 toxin (pA1 + IPTG) that either transformed into coccoids during the 10-h time frame (n = 93) or did not reach a completely round shape during this period (n = 49) (Fig. 4B). Since the noninduced bacteria were still dividing and rapidly formed dense colonies, individual cells could only be analyzed during 200 min (Fig. 4B). We observed that the median circularity of noninduced bacteria remained lower than 0.7 and close to 0.6, which corresponds to rod-shaped cells. For the induced bacteria that did not become coccoid over the course of the experiment, their circularity remained under 0.7 but tended to increase over time, suggesting that this population would probably become coccoid at later time points (Fig. 4B). In contrast, the median circularity of the IPTG-induced bacteria that became coccoid progressively increased to a value of >0.8, attesting of their rounding (Fig. 4B).
Fig. 4.
Time-lapse microscopy of H. pylori conversion to coccoid upon toxin expression. Live-cell time-lapse microscopy was used to record the morphological modifications of individual H. pylori B128 strain cells. Representative movies of the transformation into coccoids can be seen in Movies S1–S3. (A) Bright-field and fluorescence images of two examples of the kinetics of morphological modifications of individual H. pylori B128 strain cells after IPTG induction of the AapA1 toxin monitored every 10 min during 10 h. Circularity of the bacteria was measured with the MicrobeJ software. (Scale bar, 1 µm.) (B) Median circularity was measured over time for individual H. pylori B128 strain cells carrying the pA1 plasmid, either noninduced (n = 109, red dots) or IPTG induced that either become coccoids (n = 93, blue dots) or did not become coccoid (n = 49, purple dots) during the 10-h time frame. The time points correspond to those postinduction for the IPTG-treated bacteria, and only 200 min are presented, because of the multiplication of noninduced bacteria. The error bars correspond to the 95% CI. (C) Quantification of the evolution of the shape of individual H. pylori cells over time after IPTG induction. The percentage of each type of bacterium was classified as a function of its circularity (>0.7; >0.8; >0.9) during the 10-h time frame of the experiment. The red curve corresponds to those that did not become coccoid during the time of the experiment (circularity <0.7). For clarity, the graph only presents the values every 60 min postinduction.
To have a more precise view of the progression of the toxin-induced cell shape transformation into coccoids, the circularity of a total number of 142 cells was analyzed over a period of 10 h from two independent experiments that gave consistent results. Fig. 4C presents the circularity changes over time of the bacterial population under conditions of IPTG induction. During the 10 h of the experiment, we observed that about 89% of these bacteria reached a circularity of more than 0.7, which corresponds to cell shape rounding, among which about 66% of the total population became round or nearly round coccoids (circularity of >0.8) (Fig. 4C).
Fig. 4 provides an overall view of the progressive rounding of the analyzed bacteria. The kinetics of circularity changes reveal that the first step (>0.7) starts as early as at a median of 150 min postinduction, which is very close to the doubling time of H. pylori cells. Most interestingly, the length of bacteria just before they enter the first morphological transformation (circularity of >0.7) was 3.46 µm (±0.6), which is very close to the maximum length we measured before H. pylori division. In addition, we observed many examples of two bacteria that were dividing but not yet separated and from which one out of the two underwent morphological transformation (Movies S1–S3). This analysis gave us a dynamic picture of the morphological transformation of H. pylori. Importantly, it suggests that the observed morphological transformation is likely to be consecutive to a toxin-induced perturbation of cell elongation and division, through a mechanism that is still to be defined.
TA Imbalanced Expression upon Oxidative Stress.
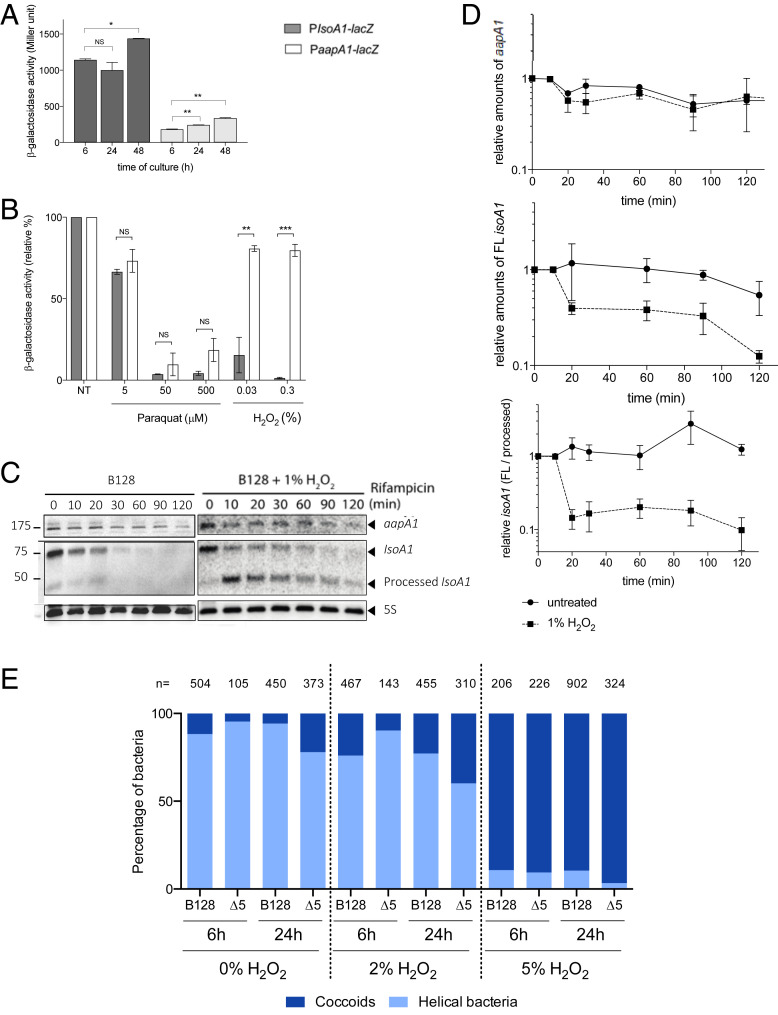
We then searched for the physiological signal that could trigger the induction of the AapA1 toxin expression in H. pylori. First, the respective activities of individual aapA1 and IsoA1 promoters were measured with chromosomally expressed transcriptional lacZ fusions (SI Appendix, Fig. S1). Under normal growth conditions, β-galactosidase activities indicated that the antitoxin promoter has a 10-fold stronger activity than the toxin promoter (Fig. 5A). This result is consistent with type I TA features where the antitoxin RNA is strongly expressed (but highly labile) compared to the more stable toxin transcript that is expressed at a lower level (8, 9, 22). The lacZ fusions were then used to follow the activity of the two promoters during H. pylori growth and to search for conditions relevant to H. pylori lifestyle that could lead to imbalanced expression of the two promoters. Only a slight increase in activity of both promoters was observed over time of growth (Fig. 5A). No significant differential expression of the promoters was observed upon acid, antibiotic stress, or high nickel concentration exposure (SI Appendix, Fig. S5). In contrast, oxidative stress induced by hydrogen peroxide (H2O2) resulted in a strong specific decrease in the IsoA1 promoter activity, while, at the same concentration, the toxin promoter activity was marginally reduced (Fig. 5B). Exposure to paraquat, another oxidative stress agent, resulted in a comparable strong decrease of both promoters. The expression patterns and stability of the aapA1 and isoA1 transcripts were then analyzed by Northern blot, with total RNA extracted at different time points after rifampicin addition. As shown in Fig. 5C and SI Appendix, Fig. S6, the aapA1 transcript was detected as a 175-nucleotide-long band with an estimated half-life of approximately 120 min. In contrast, the 75-nt isoA1 full-length transcript declines much faster, with an estimated 25-min half-life, and the production of an approximately 50-nt short processed form. Upon hydrogen peroxide exposure, the half-life of the AapA1 transcript was not significantly modified, while we observed a more rapid depletion of the full-length IsoA1 transcript with an almost immediate accumulation of the processed form (Fig. 5C). From three independent experiments, we quantified the relative amounts of the transcripts under normal growth conditions versus H2O2 exposure during 120 min after rifampicin addition (Fig. 5D). No significant change was observed for the aapA1 transcript, while H2O2 exposure resulted in a diminished half-life of the IsoA1 full-length transcript and in an imbalanced ratio of IsoA1 in favor of its processed form (Fig. 5D). Taken together, these results show that H2O2 causes both diminished IsoA1 transcription and increased degradation of IsoA1 full-length transcript. These observations suggest that, by decreasing the amounts of antitoxin transcript, exposure to oxidative stress favors translation of the AapA1 mRNA and thus toxin production that ultimately leads to coccoid transition.
Fig. 5.
Oxidative stress generated by hydrogen peroxide results in decreased antitoxin promoter activity, promotes IsoA1 transcript processing, and induces coccoid formation. (A) Activity of the AapA1 and IsoA1 promoters as a function of growth. β-galactosidase activities (expressed in Miller units) measured with H. pylori B128 strain carrying chromosomal PaapA1-lacZ and PIsoA1-lacZ fusions after 6, 24, and 48 h of growth. Results from three independent experiments performed in duplicate are shown. The Student’s t test was used to determine significant differences of the means of the data. Error bars represent the SD, with * corresponding to P < 0.05, and ** to P < 0.01, indicating that the mean values are significantly different, and with NS indicating that they are not significantly different (P > 0.05). The activity of the toxin fusion (aapA1-lacZ) is about 10 times that of the antitoxin fusion (IsoA1-lacZ), and the activity of both promoters slightly increases as a function of growth. (B) Oxidative stress generated by H2O2 decreases antitoxin promoter activity. β-galactosidase activities expressed by PaapA1-lacZ and PIsoA1-lacZ fusions were measured after 6-h treatment with oxidative stress generators, paraquat (5, 50, and 500 µM) and H2O2 (0.03% and 0.3%). β-galactosidase activities are presented as a ratio (expressed in percent) of activities measured with stress versus activities of untreated samples. Hydrogen peroxide strongly decreases the expression of the PIsoA1-lacZ fusion. Results from three independent experiments performed in duplicate are shown. The Student’s t test was used to determine significant differences of the means of the data. Error bars represent the SD, with ** corresponding to P < 0.01 and *** to P < 0.001, indicating that the mean values are significantly different, and with NS indicating that they are not significantly different (P > 0.05). (C) Determination of AapA1 mRNA and IsoA1 RNA half-lives in H. pylori strain B128 under normal conditions and upon hydrogen peroxide exposure. Northern blots of total RNA from B128 WT strain grown under normal conditions or exposed to 1% hydrogen peroxide, extracted at the indicated times after addition of 80 μg/mL rifampicin. Five micrograms of RNA were loaded in each lane, and the membranes were probed with [γ-32P] ATP-labeled oligonucleotides specific to the following RNAs: aapA1, IsoA1, and 5S rRNA as a loading control. In the presence of H2O2, the half-life of IsoA1 transcript is diminished in favor of the processed IsoA1 form. (D) Hydrogen peroxide promotes IsoA1 transcript processing. Graphic representation of the effects of hydrogen peroxide exposure on the amounts of aapA1 and IsoA1 transcripts as well as on IsoA1 processing during 120 min after rifampicin addition. Quantification was performed after scanning with an FLA-9000 Phospho Imager (Fujifilm), and normalization was performed with the 5S rRNA. The relative amounts of aapA1 (Upper) and full-length (FL) IsoA1 (Middle) RNAs versus 5S rRNA are shown. Lower presents the relative amounts of the two forms of IsoA1, FL versus the processed form. The results of three independent experiments are shown; error bars represent the SD analyzed by two-way ANOVA multiple comparisons. (E) Quantification of the proportion of coccoids and spiral bacteria of strain B128 and the ∆5 mutant at 6 and 24 h without treatment or posttreatment with 2% or 5% hydrogen peroxide. The images were analyzed with the MicrobeJ plugin from ImageJ. The number of analyzed cells is indicated for each condition. Cells were considered helical when they had an area of more than 0.70 μm2, a length between 1 and 5 μm, a width between 0.4 and 1 μm, and a circularity of less than 0.8, and they were considered coccoids when they had an area between 1 and 2 μm2, a width between 0.4 and 1.7 μm, and a circularity greater than 0.7.
Oxidative Stress Induction of Coccoids: Are Class A Type I TA the Only Effectors?
Since we established that oxidative stress results in imbalanced expression in favor of the expression of the AapA1 toxin, we examined the consequences of exposure to H2O2 on H. pylori morphology and viability (Fig. 5E and SI Appendix, Fig. S7). Upon treatment with 2% H2O2 of WT B128 strain, we measured a partial conversion to coccoids (around 25%) either after 6 h or 24 h. Treatment with 5% H2O2 resulted in a rapid and massive transformation (about 90% of cells) of this strain into coccoids after 6 h or 24 h, without detectable cell lysis (Fig. 5E). Strain B128 contains six class A type I TA systems (SI Appendix, Fig. S8). To assess their role, we constructed mutants carrying deletions of each of the active AapA/IsoA TA loci of strain B128, namely ∆A1, ∆A3, ∆A4 (including deletion of both A4-1 and A4-2 tandem systems), ∆A5, or ∆A6, or of the five loci collectively (∆5) using a nonmarked deletion strategy (31). A locus corresponding to the position of the A2 locus in other H. pylori strains was identified, but its corresponding A2 ORF was inactive in strain B128. Under normal conditions, the growth rate, culturability, and kinetics of occurrence of coccoids of the different mutants were comparable to that of the parental strain (SI Appendix, Fig. S7). Most interestingly, treatment of the ∆5 mutant with 2% H2O2 revealed minimal conversion after 6 h of treatment, with only 10% of the cells identified as coccoids as compared to 25% coccoid conversion for the WT B128 strain. After 24 h of treatment, about 40% of the ∆5 cells were coccoids. Upon treatment with 5% H2O2, both WT and ∆5 mutant strains displayed similar kinetics of conversion to coccoids (Fig. 5E). This suggests that, in the ∆5 strain, there is a significant delay in oxidative stress-induced coccoid formation.
We conclude that oxidative stress is triggering transformation into coccoids through type I class A TA systems but that other effectors are able to eventually drive this conversion. In some cases, TA systems have been shown to play a role in persister cell formation under stress conditions (8, 10). Therefore, the number of persisters after exposure to hydrogen peroxide was measured for the ∆A1 and ∆5 mutants and compared to the parental strain (SI Appendix, Fig. S7). No significant difference was observed, suggesting that, upon hydrogen peroxide stress, the class A TA systems are not promoting viable persisters in H. pylori.
Discussion
Here we established that the expression of a small toxin from a TA system triggers rapid morphological transformation of the spiral-shaped H. pylori bacterium into round coccoid cells. Transformation of spirals into coccoids has been observed after extended H. pylori growth (>70 h, “aging coccoids”) or upon stress conditions (17), but the process itself was poorly characterized. Here we found that the morphological transformation starts as soon as 2.5 h after toxin induction and that around 50% and 66% of the bacterial population convert into round coccoids, depending on the conditions (liquid culture vs. agar pads for live imaging). In liquid culture, the amount of coccoids is probably underestimated, since the vast majority of the cells had lost culturability, suggesting that they are indeed entering the process of dormancy. We consider that the amount of 66% coccoid conversion that we observe by live microscopy is a more accurate value, since, in these experiments, the shape evolution of single cells was precisely followed. These experiments also revealed that, upon toxin induction, as much as 89% of the total bacterial population showed significant shape rounding.
The toxin (AapA1) is a small hydrophobic peptide of a type I TA system that we showed here to target H. pylori inner membrane without being secreted. Toxins of type I TA systems from other organisms such as Escherichia coli, Bacillus subtilis, or Staphylococcus aureus have been shown to localize to the inner membrane, but there are no reports on induction of major cell shape modifications (4, 21, 32, 33). The E. coli TisB (34) and the S. aureus PepA1 and PepA2 (35, 36) toxins act by creating membrane pores (similar to phage holins), thereby disrupting the membrane potential, drastically impairing ATP synthesis and depending on the toxin concentration, either leading to the formation of persisters or to cell death (9, 33). One notable exception is the BsrG/SR4 Type I TA system of B. subtilis (32). The BsrG toxin was found to target the membrane without causing destruction or affecting the PMF, but it rather directly interferes with the cell envelope biosynthesis, indirectly delocalizes the cell wall synthesis machinery, and, ultimately, triggers bacterial autolysis (32). There are now two examples of type I TA hydrophobic toxins (BsrG and AapA1 reported here) that do not act by permeabilizing the membrane and probably reflect a novel mode of action for small peptidic toxins.
Using cryo-EM on H. pylori, we observed that toxin-induced H. pylori coccoids present neither visible membrane disruption nor pores even 8 h post toxin induction and are ultrastructurally comparable to 70-h-old “aging coccoids.” In addition, the membrane potential of coccoids was not dissipated but only partially affected. This is in contrast with a previous study that reported a total loss of membrane potential in H. pylori “aging coccoids,” associated with a complete loss of membrane integrity (37). Unlike the effects of the TisB toxin in E. coli (34), we found that the ATP content in our H. pylori toxin-induced coccoids is moderately affected and drops only after prolonged culture, as is the case for spiral-shaped cells (38).
Toxins expressed by TA systems cause growth arrest by interfering with conserved vital cellular processes such as translation, division, or PG synthesis (9, 33). In bacteria, PG cell wall dictates cell shape. We found that coccoid transformation was accompanied by changes in PG composition. Indeed, when compared to spirals, coccoids have a threefold increase in dipeptide monomers concomitant with a tripeptide monomers reduction. These changes are comparable to those previously reported for “aging” H. pylori coccoids (39) and were confirmed here. Our results point to a reduction in the potential for generating new cross-bridges in the PG; this is compatible with a looser PG macromolecule and could explain the morphological transition to coccoids.
Live imaging analysis of the toxin-induced conversion of H. pylori spirals into coccoids suggested that the AapA1 toxin could interfere with cell elongation and division. In E. coli, the CbtA toxin of a type IV TA system was found to inhibit cell division and cell elongation via direct and independent interactions with FtsZ and MreB (40). More work is needed to identify the molecular target(s) of the AapA1 toxin.
Under normal conditions, as for all TA systems, the IsoA1 antitoxin potently inhibits the AapA1 toxin, by preventing its expression through a multilayered mechanism (24, 25). We searched for physiological conditions that could result in toxin expression. We found that oxidative stress, generated by H2O2, causes a rapid and specific decline in the levels of IsoA1 antitoxin full-length transcript by both reducing its promoter activity and enhancing its degradation through a mechanism that remains to be precisely defined. Imbalanced expression of the AapA1/IsoA1 system in favor of the toxin mRNA should lead to toxin expression. Accordingly, we observed that H2O2 causes a rapid conversion of H. pylori into coccoids resembling those induced by the AapA1 toxin. Prolonged exposure to aerobic conditions was previously reported to lead to coccoid formation; however, in that case, these cells lost their membrane integrity (41). Regulation of type I TA systems in response to stress has been observed in other species. For the bsrE/SR5 type I system of B. subtilis, the SR5 antitoxin is affected by pH, anoxia, and iron limitation, while the BsrE toxin is sensitive to temperature shock and alkaline stress (42). In S. aureus, the SprA1/SprA1AS system is induced in response to acidic or oxidative stresses (35), and the antitoxin RNAs of the SprG/SprF systems are reduced upon oxidative stress exposure (43). In E. coli, several type I toxins are induced by the SOS response (8), a system that does not exist in H. pylori.
The biological function of H. pylori coccoids is still a matter of debate. Coccoids are observed in prolonged in vitro cultures and induced by stress conditions, mostly aerobic, anaerobic culture, or exposure to antibiotic or oxidative stress. However, these reports are difficult to compare, since no standard procedure was used and analysis was, most of the time, performed after more than a week of exposure to stress (17, 37, 44). H. pylori coccoids are nonculturable cells under standard laboratory growth conditions. Our analysis of both toxin-induced and 70-h “aging coccoids” is in favor of their viability despite their “unculturability.” We argue that contradictory conclusions on viability refer to “damaged” or so-called “fragmented” coccoid forms corresponding to membrane-less bacteria in prolonged 5-d-old H. pylori cultures (37). In favor of coccoid viability are previous reports showing that coccoids express virulence factors (17) and are capable, like spirals, of binding to host cells and inducing cellular changes (45). H. pylori coccoids were also visualized in human gastric biopsies (46), probably as part of biofilms (44); however, it cannot be excluded that they correspond to a transverse view of a spiral bacterium. In mice, coccoids induce gastritis (47) and can revert to colonizing spiral bacteria (48). Taken together, these observations suggest that coccoids are “dormant” viable forms of H. pylori that recover during host infection and might play a role in transmission or in treatment failure. In agreement with this view, Chaput et al. (49) showed that coccoids present decreased activation of NF-κB and might allow the bacterium to escape the host immune response.
We showed that H. pylori coccoids are induced by a class A type I TA toxin and by oxidative stress, probably through the imbalanced expression of TA systems. During infection, H. pylori is indeed exposed to harsh oxidative stress as a consequence of the chronic inflammation it generates (50). Deletion of the five “class A” type I TA clusters of H. pylori was found to delay the oxidative stress induction of coccoids but did not preclude it, nor did it change the number of persister cells. We concluded that these toxins are probably not the only triggers of coccoid transformation and suggest that the two other classes of chromosomally encoded type I TA systems (B and C representing nine systems) might also be important.
Dormant/persister bacteria present enhanced tolerance to antibiotics and, in some cases, have been shown to be induced by TA systems (8, 10), although other mechanisms resulting in lowered ATP content and/or lowered PMF are also associated with their occurrence (13, 14). We hypothesize that TA-induced H. pylori coccoids are dormant bacteria as defined in a recent consensus review (51). As previously proposed, nonculturable cells and “classical” dormant bacteria are part of a shared “dormancy continuum” (52). In our model, this continuum would depend on the intracellular toxin concentration as already suggested (53).
Stress-induced morphological transformation of bacteria into nonculturable coccoid-like cells has been reported in at least 85 bacterial species, among which are important bacterial pathogens such as Campylobacter jejuni, Vibrio cholerae, or Salmonella typhimurium (19). For these organisms, the inducing trigger and function remain to be characterized. We postulate that these forms are dormant bacteria and that our findings on H. pylori might be more generally relevant and play important roles in recurrent and persistent bacterial infections.
Materials and Methods
Bacterial Strains and Growth Conditions.
The H. pylori strains used in this study (SI Appendix, Table S2) were all derived of strain B128 (54, 55). Plasmids (SI Appendix, Table S3) used to create or complement H. pylori mutants were constructed and amplified using E. coli One-Shot TOP10 or DH5α strains (Thermo Fisher Scientific). H. pylori strains were grown on Blood Agar Base 2 (Oxoid) plates supplemented with 10% defibrinated horse blood and with the following antibiotics−antifungal mixture: amphotericin B 2.5 μg⋅mL−1, polymyxin B 0.31 μg⋅mL−1, trimethoprim 6.25 μg⋅mL−1, and vancomycin 12.5 μg⋅mL−1. Selection of H. pylori mutants was performed using kanamycin 20 μg⋅mL−1, streptomycin 10 μg⋅mL−1, apramycin 10 μg⋅mL−1, or chloramphenicol 8 μg⋅mL−1. For liquid cultures, we used Brain Heart Infusion (BHI) broth (Oxoid) supplemented with 10% Fetal Calf Serum (Eurobio), the antibiotics−antifungal mixture, and the selective antibiotic when necessary. H. pylori cells were grown at 37 °C under microaerophilic atmosphere conditions (6% O2, 10% CO2, 84% N2) using an Anoxomat (MART Microbiology) atmosphere generator. When indicated, 1 mM IPTG (EuroMedex) was added to agarose pads or culture media.
Molecular Techniques.
Molecular biology experiments were performed according to standard procedures and the supplier (Fermentas) recommendations. NucleoBond Xtra Midi Kit (Macherey-Nagel) and QIAamp DNA Mini Kit (Qiagen) were used for plasmid preparations and H. pylori genomic DNA extractions, respectively. PCR was performed with Taq Core DNA polymerase (MP Biomedicals), or with Phusion Hot Start DNA polymerase (Finnzymes) when the product required high-fidelity polymerase. The pGEM-T easy vector system (Promega) was used to construct, in E. coli, the suicide plasmids that served for markerless deletions of TA systems in H. pylori.
Construction of H. pylori Strains Carrying Mutations or Plasmids.
Mutations of H. pylori were introduced into strain B128, either WT or a streptomycin-resistant variant B128 rpsL1, for markerless mutagenesis (31, 56). Chromosomal deletions of the AapA1/IsoA1 locus, of the flaA gene, or of the different class A TA loci were performed either by insertion of a selectable antibiotic resistance marker to disrupt or replace the gene of interest or, when necessary, by the markerless counter-selected mutagenesis strategy (31, 56). Plasmids were derived from the pILL2157 E. coli/H. pylori shuttle vector that contains an IPTG-inducible promoter (27). Deletions were introduced by allelic exchange using suicide plasmids (SI Appendix, Table S3) or PCR fragments. Introduction of plasmids and construction of H. pylori mutants were obtained by natural transformation and selection with the corresponding antibiotic as described previously (31). PCR and sequencing of the regions of interest were used to validate the introduction of plasmids, deletion of genes of interest, and correct insertion of fusions. Primers used for these constructs or their validation are listed in SI Appendix, Table S4.
Fluorescence Microscopy.
Fluorescence microscopy was performed with an Axio Observer microscope (Zeiss) equipped with an Axiocam camera under a 100× magnification. Images acquisition was performed using the axiovision software. Images were cropped and adjusted using ImageJ v2.0.0 software. Cells were quantified and assigned to helical or coccoid categories based on phase contrast images by using the MicrobeJ plugin of ImageJ (28) with the following parameters: 1) Helical cells had an area of more than 0.7 μm2, a length between 1 and 5 μm, a width between 0.4 and 1 μm, and a circularity of less than 0.8; and 2) coccoid cells had an area between 1 and 2 μm2, a width between 0.4 and 1.7 μm, and a circularity greater than 0.7.
H. pylori B128 strains were grown in BHI medium with chloramphenicol (8 µg/mL) to maintain the plasmids. One millimolar IPTG was added to the culture, and samples were taken at different time points, concentrated by 2 min centrifugation at 3,000 × g and washed twice with PBS buffer. Cells were immobilized using 2% (wt/vol) agarose pads containing PBS before being imaged. In Fig. 1C, cell membranes were stained with 4% FM4-64 (Thermo Fisher Scientific). In Fig. 2B, cell membranes were stained with 0.01 mM TMA-DPH (Euromedex).
Flow Cytometry for PMF Measurement.
H. pylori strains were grown in triplicate for 16 h and then treated or not with 1 mM IPTG. Samples were taken at 0, 8, and 24 h postinduction, and 107 live cells per experimental condition were washed and stained with 25 nM MitoTracker Red CMXRos (Invitrogen), a PMF-sensitive dye. As a control for PMF depletion, pA1* cells were treated with 500 µM protonophore TCS (Fisher Scientific). The fluorescent signal from at least 100,000 individual bacteria per condition was measured by flow cytometry with an MACSQuant VYB analyzer (Miltenyi Biotec) (Y2 channel, λex = 561 nm and λem = 605 nm to 625 nm) after calibration. The experiment was performed three times. Data were analyzed with FlowJo V10.
ATP Extraction and Assay.
Exponentially growing H. pylori cells (6 h of culture) and stationary growing cells (16 h) were harvested by centrifugation at room temperature for 4 min at 5,000 × g. Metabolites from the resulting cell pellets were extracted immediately using 300 μL of a solvent mixture of Acetonitrile/Methanol/H2O (40/40/20) for 15 min at 4 °C and 10 min at 95 °C. Mixtures were subsequently spun in a microfuge for 5 min at maximum speed and 4 °C to separate insoluble materials from the extracted metabolites. The resulting pellets were then reextracted twice with 200 μL of solvent at 4 °C. The supernatants were pooled to yield 700 μL of final extract. Metabolites were lyophilized and subsequently diluted in water for ATP assays. ATP content was determined by a luciferase-based ATP bioluminescence assay kit (BacTiter-Glo Microbial cell viability assay, Promega). Luminescence values were determined using a 10-s RLU (relative light units) signal integration time and measured using a Centro XS3 LB960 Luminometer (Berthold Technologies). ATP concentrations were calculated based on values determined using serial dilutions of known amounts of ATP and expressed as a function of the optical density at 600 nm, OD600nm, of the corresponding culture.
Fractionation and Immunoblotting.
H. pylori fractionation was performed as in Tejada-Arranz et al. (57). H. pylori strains were grown in the absence or presence of 1 mM IPTG. When cultures reached OD600nm of 0.8, cells were harvested by centrifugation, and washed twice in PBS medium prior to their disruption by sonication in Buffer 1, 10 mM Tris⋅HCl, pH 7.5 containing 5 mM β-mercaptoethanol and proteases inhibitors (cOmplete, EDTA-free Protease Inhibitor Mixture, Roche). Cell debris were removed by centrifugation (10 min, 16,000 × g, 4 °C), and supernatants containing the soluble extract and membrane fractions were collected as total extracts. Samples of total extracts were frozen, and the remaining supernatants were subjected to ultracentrifugation (125,000 × g, 45 min, 4 °C). Samples from the supernatant containing the cytoplasm and periplasm were collected and frozen, while pellets containing total membranes were resuspended in buffer 1 supplemented with 0.1% N-lauroylsarcosine (buffer 2) prior to being ultracentrifuged (125,000 × g, 45 min, 4 °C). Samples of inner membrane fractions from supernatants were frozen. Outer membrane pellets were washed twice in buffer 2 to avoid inner membrane fractions contamination prior to freezing. For each cellular compartment, equal cellular amounts were loaded and separated on a 4 to 20% Mini-Protean TGX precast protein gel (BioRad) and subsequently electrotransferred on a polyvinylidene difluoride membrane (Biorad). The control H. pylori proteins PPB2, AlpA, AmiE, and NikR as well as the fusion protein AapA1−GFP were detected with rabbit polyclonal antibodies α-PBP2 (58), α-AlpA (57), α-AmiE (59), α-NikR (60) or with goat anti-GFP-HRP antibody (Abcam) at the respective dilutions of 1:2,000, 1:1,000, 1:100, 1:5,000, and 1:5,000. Goat anti-rabbit IgG-HRP (Santa Cruz) was used as secondary antibody (1:10,000 dilution), and the detection was achieved with the ECL (Enhanced ChemoLuminescent) reagent (Thermo Fisher Scientific).
Cryo-EM.
Four microliters of H. pylori bacteria were spotted on glow-discharged lacey grids (S166-3, EMS) or Quantifoil R2/2 (Quantifoil). The samples were cryofixed by plunge freezing at −180 °C in liquid ethane using a Leica EMGP (Leica). Grids were observed at 200 kV with a Tecnai F20 (Thermo Fisher Scientific). Images were acquired under low-dose conditions using the software EPU (Thermo Fisher Scientific) and a direct detector Falcon II (Thermo Fisher Scientific).
Time-Lapse Microscopy.
Confocal analysis of live cells was performed as previously described (61). Twenty microliters of exponentially grown bacteria (105 cells) suspended in BHI with or without 1 mM IPTG were deposited on 35-mm glass-bottomed Petri dishes. The suspension was covered with BHI medium in 1.5% low melting agarose supplemented with Chloramphenicol (8 µg/mL) with or without IPTG (1 mM). Live-cell imaging was performed with a Nikon A1R confocal laser scanning microscope system attached to an inverted ECLIPSE Ti (Nikon Corp.) and equipped with an environmental chamber allowing the control of temperature (37 °C), humidity (90%), and gas mixture (10% CO2, 3% O2). GFP fluorescence images were captured through a Plan APO 60× objective (NA: 1.40) by using optimal spatial resolution settings. The cytoplasm compartment volume was defined by using GFP staining (excitation with 488-nm laser, emission collected with a 500/50-nm filter set). Image captures were performed every 10 min during 10 h. Image treatment and analysis were performed using NIS (Nikon Imaging Software) elements (Nikon Corp.), and ImageJ and MicrobeJ software (28).
β-Galactosidase Activity Assays.
B128∆AapA1-IsoA1::PaapA1-lacZ-Kan and B128∆AapA1-IsoA1::PIsoA1-lacZ-Kan strains were grown to OD600nm 0.3 in BHI liquid medium, then divided into two samples, one of them being submitted to stress conditions during 6 h with 5, 50, and 500 µM paraquat (Sigma), 0.03% and 0.3% hydrogen peroxide (Sigma), 20 and 200 mM NiCl2 (Sigma), acidic pH, antibiotics with 0.1 and 1 mg/mL tetracycline, or 0.05 and 0.5 mg/mL rifampicin. Then 0.5-mL samples were taken, washed twice with 1× PBS, and further permeabilized with 100 µL of chloroform and 50 µL of sodium dodecyl sulfate (SDS) 0.1% in Z buffer containing 70 mM Na2HPO4, 30 mM NaH2PO4, 1 mM MgSO4, and 0.2 mM MnSO4 (62). Samples were briefly vortexed and incubated at 28 °C for 2 min. The β-galactosidase assay was started by adding 0.1 mL of ortho-nitrophenyl-β-galactoside (Thermo Fisher Scientific) at 4 mg/mL and stopped by the addition of 0.5 mL of 1M Na2CO3 when sufficient yellow color was reached. The β-galactosidase activity is expressed in Miller units (62) and represented in Fig. 5 and SI Appendix, Fig. S5 as percentage of activities relative to the control activity with the corresponding strains not exposed to stress and was calculated from three independent experiments performed in duplicate.
Total RNA Extraction and Northern Blotting.
Total RNAs were extracted from 10 mL of H. pylori cultures (OD600nm 0.5 to 0.9) exposed or not to 1% H2O2 using the NucleoSpin miRNA kit (Macherey-Nagel) at different time points after 80 µg/mL rifampicin addition (to stop transcription). For Northern blotting, 5 µg of total RNA were separated on 10% Mini protean TBE Urea gel (Biorad) and transferred to Hybond N+ (Amersham Biosciences) membrane using a Trans-Blot Turbo system (Biorad). A denaturated DNA marker was used for size estimation.
Transferred RNA was fixed to the membrane by ultraviolet irradiation for 2 min. The membrane was blocked for 45 min at 42 °C with ULTRAhyb Hybridization Buffer (Ambion), then 20 µL of 5′-labeled (γ32P) oligodeoxynucleotides (SI Appendix, Table S4) were added, and the membrane was further incubated overnight at the same temperature. After three washes for 10 min at 65 °C with 2× saline-sodium citrate (SSC) buffer and 0.2% SDS, the membrane was exposed to a phosphorimager screen (Kodak) and scanned with FLA-9000 Phospho Imager (Fujifilm). Normalization was performed with the 5S ribosomal RNA (rRNA).
PG Extraction and Muropeptides Analysis by HPLC/High-Resolution MS.
Samples corresponding to 2 mL of culture at OD600nm 10 of H. pylori were taken and chilled in an ice−ethanol bath, and each condition was prepared in triplicate. The samples were B128 WT strain in exponential phase (24-h culture), stationary phase (36-h culture), and after 72 h culture (“aging coccoids”). To examine the effect of the AapA1 toxin, we performed a culture of 16 h, added IPTG, and then took samples of the following strains: B128 ∆aapA1−IsoA1 + pA1 strain after 8 h of induction (equivalent to 24 h of total culture time) or 24 h of induction (equivalent to 36 h), and, as a negative control, of strain B128 ∆aapA1−IsoA1 + pA1* after 8 and 24 h of induction (total 24 and 36 h, respectively). Crude murein sacculi were immediately extracted in boiling SDS (4% final) (63). The resulting purified PG was digested overnight at 37 °C in 12.5 mM sodium phosphate buffer (pH 5.6) supplemented with 100 UI of mutanolysin from Streptomyces globisporus ATCC 21553 (Sigma). The reaction was stopped by heat inactivation of the enzyme, and insoluble material was removed by centrifugation. Soluble muropeptides were then reduced with sodium borohydride in borate buffer (pH 9). After centrifugation, reduced muropeptides were diluted 20-fold in water-formic acid 0.1% (vol/vol; solvent A). HPLC/high-resolution MS was performed on an Ultimate 3000 ultra-HPLC system coupled to a quadrupole-Orbitrap mass spectrometer (Q-Exactive Focus, Thermo Fisher Scientific). Muropeptides were separated on a Hypersil Gold aQ analytical column (1.9 µm, 2.1 × 150 mm) at 200 µL/min, column temperature at 50 °C. Muropeptides were separated using a total run of 45 minutes with the following linear gradient: 1) from 0 to 25 min, the gradient started with 0.1% (v/v) formic acid to finish with 12.5% acetonitrile - 0.1% formic acid; 2) from 20 to 30 min, the percentage of acetonitrile increased from 12.5% to 20% with 0.1% formic acid; 3) from 30 to 35 min, the column was kept on constant 20% acetonitrile with 0.1% formic acid; 4) from 35 to 45 min, the solvent was shifted back to 0.1% formic acid and kept constant to re-equilibrate the column under the initial buffer conditions. Eluted muropeptides were introduced into Q-Exactive instrument, operating in positive ion mode, and then were analyzed in the data-dependent acquisition mode (FullMSddMS2). The identification of muropeptides was performed after precursor ion fragmentation (MS2 analysis). Data were then processed with the software TraceFinder 3.3 (Thermo Fisher Scientific) for peak areas determination; the relative abundance of muropeptides in each sample was calculated according to Glauner (30).
Statistical Analysis.
The Student’s t test was used to determine significant differences of the means of the data for all of the experiments except for the Northern blots. Error bars represent the SD, with * (P < 0.05), ** (P < 0.01), *** (P < 0.001), **** (P < 0.0001) indicating that the mean values are significantly different, and NS indicating that they are not significantly different (P > 0.05). The two-way ANOVA multiple comparisons test was used to compare Northern blots band intensities under different conditions.
Supplementary Material
Acknowledgments
This work was funded by the Agence National de la Recherche (grants ANR 09 BLAN 0287 01 and ANR 12 BSV5-0025-02), the Laboratoire d’Excellence IBEID (Integrative Biology of Emerging Infectious Diseases) Grant ANR-10-LABX-62-IBEID from the French government’s Investissement d’Avenir program, and the Pasteur-Weizmann Consortium of “The Roles of Noncoding RNAs in Regulation of Microbial Life Styles and Virulence.” L.E.M. was funded by a Roux fellowship of the Institut Pasteur and by a Carnot MI (Maladies infectieuses) fellowship. A.T.-A. is part of the Pasteur Paris University International PhD Program. This program has received funding from the European Union Horizon 2020 research and innovation program under Marie Sklodowska-Curie Grant Agreement 665807, and from the Institut Carnot Pasteur Microbes and Santé. A.R. was supported by a postdoctoral fellowship from the Labex IBEID (Grant 10-LABX-62-IBEID). We also thank Janssen for financial support. We are grateful to F. Darfeuille, I. Iost, and H. Arnion for the gift of plasmids and to W. Fischer for the gift of anti-AlpA antibodies. We also appreciate the expertise and help of A. Chery-Faleme, M. Fromont-Racine, and A. Jacquier with Northern blotting, of R. Wheeler for PG extraction, of Z. Baharoglu and I. Santecchia for flow cytometry, and of A. Ducret for image analysis. We thank M. Denic and S. Kumar for comments on the manuscript, and J. Berry for discussions. Finally, we thank M. Nilges and the Equipex CACSICE (Centre d'analyse de systèmes complexes dans les environnements complexes) for providing the Falcon II direct detector.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016195117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Yamaguchi Y., Park J.-H., Inouye M., Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45, 61–79 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Goeders N., Van Melderen L., Toxin-antitoxin systems as multilevel interaction systems. Toxins (Basel) 6, 304–324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Page R., Peti W., Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12, 208–214 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Brantl S., Müller P., Toxin-antitoxin systems in Bacillus subtilis. Toxins (Basel) 11, 262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Q. E., Walsh T. R., Toxin-antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol. Rev. 41, 343–353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helaine S., et al. , Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X., et al. , Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7, 359–366 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berghoff B. A., Wagner E. G. H., RNA-based regulation in type I toxin-antitoxin systems and its implication for bacterial persistence. Curr. Genet. 63, 1011–1016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harms A., Brodersen D. E., Mitarai N., Gerdes K., Toxins, targets, and triggers: An overview of toxin-antitoxin biology. Mol. Cell 70, 768–784 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Ronneau S., Helaine S., Clarifying the link between toxin–antitoxin modules and bacterial persistence. J. Mol. Biol. 431, 3462–3471 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Gollan B., Grabe G., Michaux C., Helaine S., Bacterial persisters and infection: Past, present, and progressing. Annu. Rev. Microbiol. 73, 359–385 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Goormaghtigh F., et al. , Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. MBio 9, e00640-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan Y., et al. , ATP-dependent persister formation in Escherichia coli. MBio 8, e02267-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y., et al. , Inactivation of TCA cycle enhances Staphylococcus aureus persister cell formation in stationary phase. Sci. Rep. 8, 10849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaoka Y., Mechanisms of disease: Helicobacter pylori virulence factors. Nat. Rev. Gastroenterol. Hepatol. 7, 629–641 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amieva M., Peek R. M. Jr, Pathobiology of Helicobacter pylori–induced gastric cancer. Gastroenterology 150, 64–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loke M. F., Ng C. G., Vilashni Y., Lim J., Ho B., Understanding the dimorphic lifestyles of human gastric pathogen Helicobacter pylori using the SWATH-based proteomics approach. Sci. Rep. 6, 26784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reshetnyak V. I., Reshetnyak T. M., Significance of dormant forms of Helicobacter pylori in ulcerogenesis. World J. Gastroenterol. 23, 4867–4878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayrapetyan M., Williams T., Oliver J. D., The relationship between the viable but nonculturable state and antibiotic persister cells. J. Bacteriol. 200, e00249-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun H. J., et al. , Electron microscopic evaluation of adhesion of Helicobacter pylori to the gastric epithelial cells in chronic gastritis. Korean J. Intern. Med. (Korean. Assoc. Intern. Med.) 17, 45–50 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brantl S., Jahn N., sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 39, 413–427 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Masachis S., Darfeuille F., Type I toxin-antitoxin systems: Regulating toxin expression via Shine-Dalgarno sequence sequestration and small RNA binding. Microbiol. Spectr. 6, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma C. M., et al. , The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Arnion H., et al. , Mechanistic insights into type I toxin antitoxin systems in Helicobacter pylori: The importance of mRNA folding in controlling toxin expression. Nucleic Acids Res. 45, 4782–4795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masachis S., et al. , A genetic selection reveals functional metastable structures embedded in a toxin-encoding mRNA. eLife 8, e47549 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korkut D. N., et al. , Structural insights into the AapA1 toxin of Helicobacter pylori. Biochim. Biophys. Acta, Gen. Subj. 1864, 129423 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Boneca I. G., et al. , Development of inducible systems to engineer conditional mutants of essential genes of Helicobacter pylori. Appl. Environ. Microbiol. 74, 2095–2102 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ducret A., Quardokus E. M., Brun Y. V., MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stingl K., Altendorf K., Bakker E. P., Acid survival of Helicobacter pylori: How does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 10, 70–74 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Glauner B., Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172, 451–464 (1988). [DOI] [PubMed] [Google Scholar]
- 31.Fischer F., et al. , Characterization in Helicobacter pylori of a nickel transporter essential for colonization that was acquired during evolution by gastric Helicobacter species. PLoS Pathog. 12, e1006018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jahn N., Brantl S., Strahl H., Against the mainstream: The membrane-associated type I toxin BsrG from Bacillus subtilis interferes with cell envelope biosynthesis without increasing membrane permeability. Mol. Microbiol. 98, 651–666 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Brielle R., Pinel-Marie M.-L., Felden B., Linking bacterial type I toxins with their actions. Curr. Opin. Microbiol. 30, 114–121 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Unoson C., Wagner E. G. H., A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 70, 258–270 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Sayed N., Nonin-Lecomte S., Réty S., Felden B., Functional and structural insights of a Staphylococcus aureus apoptotic-like membrane peptide from a toxin-antitoxin module. J. Biol. Chem. 287, 43454–43463 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Germain-Amiot N., et al. , A novel Staphylococcus aureus cis−trans type I toxin-antitoxin module with dual effects on bacteria and host cells. Nucleic Acids Res. 47, 1759–1773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kusters J. G., Gerrits M. M., Van Strijp J. A., Vandenbroucke-Grauls C. M., Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect. Immun. 65, 3672–3679 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enroth H., et al. , In vitro aging of Helicobacter pylori: Changes in morphology, intracellular composition and surface properties. Helicobacter 4, 7–16 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Costa K., et al. , The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J. Bacteriol. 181, 3710–3715 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heller D. M., Tavag M., Hochschild A., CbtA toxin of Escherichia coli inhibits cell division and cell elongation via direct and independent interactions with FtsZ and MreB. PLoS Genet. 13, e1007007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng H., Guo G., Mao X. H., Tong W. D., Zou Q. M., Proteomic insights into Helicobacter pylori coccoid forms under oxidative stress. Curr. Microbiol. 57, 281–286 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Müller P., et al. , A multistress responsive type I toxin-antitoxin system: bsrE/SR5 from the B. subtilis chromosome. RNA Biol. 13, 511–523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riffaud C., Pinel-Marie M.-L., Pascreau G., Felden B., Functionality and cross-regulation of the four SprG/SprF type I toxin-antitoxin systems in Staphylococcus aureus. Nucleic Acids Res. 47, 1740–1758 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cellini L., Helicobacter pylori: A chameleon-like approach to life. World J. Gastroenterol. 20, 5575–5582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segal E. D., Falkow S., Tompkins L. S., Helicobacter pylori attachment to gastric cells induces cytoskeletal rearrangements and tyrosine phosphorylation of host cell proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 1259–1264 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldstein N. S., Chronic inactive gastritis and coccoid Helicobacter pylori in patients treated for gastroesophageal reflux disease or with H pylori eradication therapy. Am. J. Clin. Pathol. 118, 719–726 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Nagai S., et al. , Role of Peyer’s patches in the induction of Helicobacter pylori-induced gastritis. Proc. Natl. Acad. Sci. U.S.A. 104, 8971–8976 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cellini L., et al. , Coccoid Helicobacter pylori not culturable in vitro reverts in mice. Microbiol. Immunol. 38, 843–850 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Chaput C., et al. , Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog. 2, e97 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butcher L. D., den Hartog G., Ernst P. B., Crowe S. E., Oxidative stress resulting from Helicobacter pylori infection contributes to gastric carcinogenesis. Cell. Mol. Gastroenterol. Hepatol. 3, 316–322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balaban N. Q., et al. , Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ayrapetyan M., Williams T. C., Oliver J. D., Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 23, 7–13 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Rotem E., et al. , Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. U.S.A. 107, 12541–12546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farnbacher M., et al. , Sequencing, annotation, and comparative genome analysis of the gerbil-adapted Helicobacter pylori strain B8. BMC Genom. 11, 335 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McClain M. S., Shaffer C. L., Israel D. A., Peek R. M. J. Jr, Cover T. L., Genome sequence analysis of Helicobacter pylori strains associated with gastric ulceration and gastric cancer. BMC Genom. 10, 3 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Debowski A. W., et al. , Xer-cise in Helicobacter pylori: One-step transformation for the construction of markerless gene deletions. Helicobacter 17, 435–443 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Tejada-Arranz A., et al. , The RNase J-based RNA degradosome is compartmentalized in the gastric pathogen Helicobacter pylori. MBio 11, e01173-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.El Ghachi M., et al. , Characterization of the elongasome core PBP2:MreC complex of Helicobacter pylori. Mol. Microbiol. 82, 68–86 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Skouloubris S., Labigne A., De Reuse H., The AmiE aliphatic amidase and AmiF formamidase of Helicobacter pylori: Natural evolution of two enzyme paralogues. Mol. Microbiol. 40, 596–609 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Muller C., et al. , Hierarchical regulation of the NikR-mediated nickel response in Helicobacter pylori. Nucleic Acids Res. 39, 7564–7575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corbinais C., Mathieu A., Kortulewski T., Radicella J. P., Marsin S., Following transforming DNA in Helicobacter pylori from uptake to expression. Mol. Microbiol. 101, 1039–1053 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Miller J. H., A Short Course in Bacterial Genetics : A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 1992). [Google Scholar]
- 63.Kühner D., Stahl M., Demircioglu D. D., Bertsche U., From cells to muropeptide structures in 24 h: Peptidoglycan mapping by UPLC-MS. Sci. Rep. 4, 7494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.