Significance
Adult neural stem cells (NSC) are closely related to multiple neurological disorders and brain tumors. Comprehensive investigation of their composition, lineage, and aging will provide insights that may lead to enhanced patient treatment. This study applies a transgene to label and manipulate neural stem/progenitor cells and monitor their evolution during aging. Together with high-throughput single-cell RNA sequencing, we are able to analyze the subventricular zone cells from infancy to advanced age with unprecedented granularity. Diverse cell states are identified in the stem-cell niche, and an aging-related NSC deficiency in transition from quiescence to proliferation is identified. The related biological features provide rich resources to inspect adult NSC maintenance and neurogenesis.
Keywords: neural stem cell, single-cell RNA sequencing, subventricular zone, aging, transcriptome
Abstract
Adult neural stem cells (NSC) serve as a reservoir for brain plasticity and origin for certain gliomas. Lineage tracing and genomic approaches have portrayed complex underlying heterogeneity within the major anatomical location for NSC, the subventricular zone (SVZ). To gain a comprehensive profile of NSC heterogeneity, we utilized a well-validated stem/progenitor-specific reporter transgene in concert with single-cell RNA sequencing to achieve unbiased analysis of SVZ cells from infancy to advanced age. The magnitude and high specificity of the resulting transcriptional datasets allow precise identification of the varied cell types embedded in the SVZ including specialized parenchymal cells (neurons, glia, microglia) and noncentral nervous system cells (endothelial, immune). Initial mining of the data delineates four quiescent NSC and three progenitor-cell subpopulations formed in a linear progression. Further evidence indicates that distinct stem and progenitor populations reside in different regions of the SVZ. As stem/progenitor populations progress from neonatal to advanced age, they acquire a deficiency in transition from quiescence to proliferation. Further data mining identifies stage-specific biological processes, transcription factor networks, and cell-surface markers for investigation of cellular identities, lineage relationships, and key regulatory pathways in adult NSC maintenance and neurogenesis.
Most mammalian organs and tissues harbor stem cells that serve as a source of continual cell replenishment (1, 2). Adult rat neurogenesis was first documented in the 1960s, and since then the rodent brain has served as a useful model for study of neural stem cells (NSC) (3). In the mouse brain, two distinct pools of NSC, found in the subventricular zone (SVZ) of the lateral ventricle walls and in the dentate gyrus of the hippocampus, produce new neurons that functionally integrate into preexisting circuits during lifetime (4–6). In the SVZ, both type B1 cells and ependymal cells have been proposed as NSC (7–10). B1 cells are elongated stem cells that belong to the radial glia-astrocytic lineage and express markers characteristically found in astrocytes, such as glial fibrillary acidic protein (Gfap) or glutamate-aspartate transporter (GLAST-Slc1a3). They exhibit a thin cytoplasmic process that intercalates between ependymal cells to adorn the ventricle cavities and terminates in a sensing primary cilium and another long process that interacts with the basal lamina of irrigating vasculature. B1 cells give rise to migratory and dividing transient amplifying progenitor (TAP) cells that differentiate into interneurons destined to the olfactory bulb (OB) or, less frequently, into oligodendrocytes in the corpus callosum (11, 12).
Apart from their role in OB interneuron replacement and, to a lesser degree, in other neurogenesis and gliogenesis, it has been reported that SVZ NSC/progenitor cells can mount responses to brain-cell death caused by trauma (13, 14). In addition, increasing evidence points to NSC as a relevant source for malignant transformation into glioma. Direct oncogenic transformation of mouse NSC or progenitors has been demonstrated to mediate glioblastoma multiforme (GBM) and medulloblastoma (15, 16). Mouse-modeling studies have portrayed a direct lineage relationship between NSC/oligodendrocyte lineage cell stem/progenitor cells and GBM (17, 18). Thus, a complete understanding of the properties of SVZ NSC, their transition into TAPs, and finally into differentiated cells holds relevance to brain physiology and repair and to the etiology of malignant brain tumors.
Advances in genomics, deep sequencing, and microfluidics have allowed increased resolution of the SVZ stem-cell niche cell dynamics (6, 10, 19–24). NSC surface markers, such as Prom1-CD133 and Slc1a3-Glast, and transgenes encoding Cre-recombinase or fluorescent reporters regulated by Gfap promoters, have been used to partially enrich for prospective NSC from the SVZ for bulk RNA sequencing (RNAseq). The data yielded information consistent with the existence of a quiescent NSC population that transitions to an activated state (19–22). Single-cell transcriptomics of SVZ-derived cells have further enhanced resolution. Recent studies relying on cell sorting with NSC or TAP markers have yielded limited numbers of cells for single-cell sequencing ranging from tens to hundreds of cells (10, 23, 24).
To more efficiently label and study NSC, we constructed a transgene with a GFP reporter that highlights the vast majority of stem and progenitor cells within the adult SVZ. The sequencing of 12,200 GFP+ sorted or unselected SVZ cells from this transgenic mouse model has yielded a uniquely comprehensive and granular view of the SVZ stem-cell niche uncovering a previously under-appreciated heterogeneity. The resultant transcriptional landscape viewed horizontally across time uncovers features responsible for reduced neurogenesis with aging.
Results
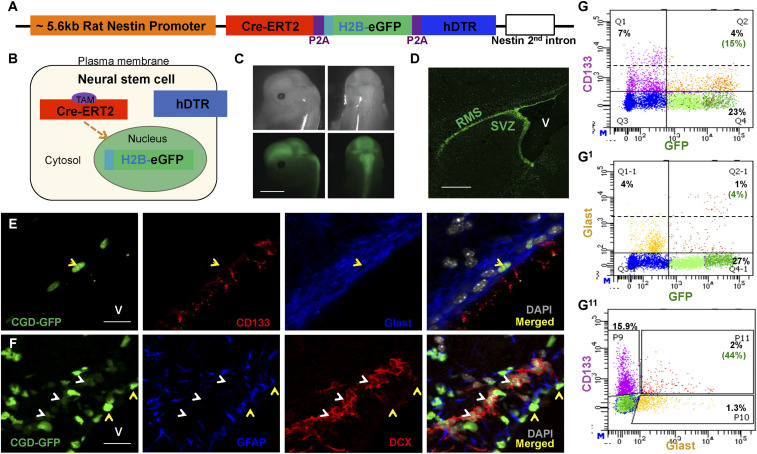
Defined components of the endogenous Nestin gene promoter/enhancer fragment (Nes) that specify expression in NSC have been identified and widely used to express transgenic proteins in the SVZ region (15, 25–29). To increase the power and versatility for identification, targeting, and isolation of adult NSC, we constructed a transgene, termed CGD (CreERT2, histone 2B tagged enhanced GFP, and human diphtheria toxin receptor), designed to express three proteins from one transcript (Fig. 1 A and B). The properties of the CGD transgene cassette, including appropriate protein expression, Cre recombinase activity, and diphtheria toxin receptor (DTR) function were verified in cell culture prior to development of transgenic mouse lines (SI Appendix, Fig. S1 A–E). Twenty-nine CGD transgenic founder lines were screened to identify a single founder that displayed the desired expression patterns in the embryonic and adult central nervous system stem-cell/progenitor compartments without detectable ectopic expression (Fig. 1 C and D).
Fig. 1.
CGD transgene labels adult subventricular zone neural stem cells and progenitors. (A) Diagram of the transgene construct. Purple blocks represent the P2A ribosomal skipping element insulating the three gene cassettes. (B) Cartoon illustrates localization of the three transgene products in NSC: Cre-ERT2 in the cytosol, H2B-eGFP in the nucleus, and hDTR on the plasma membrane. (C) Transgene GFP imaging of a 14-d embryo undergoing neurogenesis illustrates CGD transgene expression in developing central nervous system (Scale bar, 3 mm.) (D) Enhanced transgene GFP expression in adult SVZ and rostral migratory stream (RMS). V: lateral ventricle. (Scale bar, 500 μm.) (E) Coronal section staining of 2-mo-old mouse SVZ shows colocalization of CGD-GFP with stem-cell markers CD133 and Glast. V: lateral ventricle. (Scale bar, 20 μm.) (F) Relationship between CGD-GFPhi and stem-cell marker GFAP versus GFPlo and progenitor marker DCX in a sagittal section of adult SVZ (Scale bar, 20 μm.) (G, G1, and G11) A small fraction of CGD-GFP+ cells in the SVZ express CD133 (G, 15%) or Glast (G1, 4%), and 44% of all CD133+Glast+ double-positive cells express CGD-GFP (G11). V: lateral ventricle. See also SI Appendix, Fig. S1.
Functional Correlation of GFP+ Cells with NSC/Progenitor Markers.
Commonly used markers to identify and label the NSC lineage include the proteins GFAP, Glast, and CD133 (10, 19–21, 23, 24, 30–32). We investigated the expression of these markers in relationship to the CGD transgene nuclear GFP reporter. Immunohistochemical (IHC) staining of sagittal and coronal adult brain sections from CGD mice confirmed that Glast, CD133, and GFAP antibodies decorated the most intensive GFP+ nuclei (Fig. 1 E and F), and whole-mount staining of SVZ tissue identified the “pin wheel” structure formed by the GFP+/CD133+/Glast+ NSC surrounded by ependymal GFP– cells (SI Appendix, Fig. S1F) (33). Fluorescence-activated cell sorting (FACS) analysis of dissociated SVZ tissue indicated that only a small fraction of CGD-GFP+ cells coexpress CD133+ (15%), which coincides with the CD133lo category (Fig. 1G, quadrant 2). Similar analysis of GFP/Glast coexpression indicated that only 4% of GFP+ cells were double positive for Glast (Fig. 1G1, quadrant 2-1). Taking into account all CD133+/Glast+ SVZ cells (about 2% of the entire cell preparation), less than half were CGD-GFP+. Thus, in the SVZ, CGD-GFP reporter expression marks a comprehensive cohort of cells including a significant proportion of CD133- and Glast-expressing cells (Fig. 1G11, P11). We next examined the relationship of CGD-GFP expression to that of doublecortin (DCX) protein, a progenitor marker, and found that it was excluded from the intensely fluorescing GFPhi cells but instead decorated a cohort of less intense GFPlo fluorescent cells (Fig. 1F).
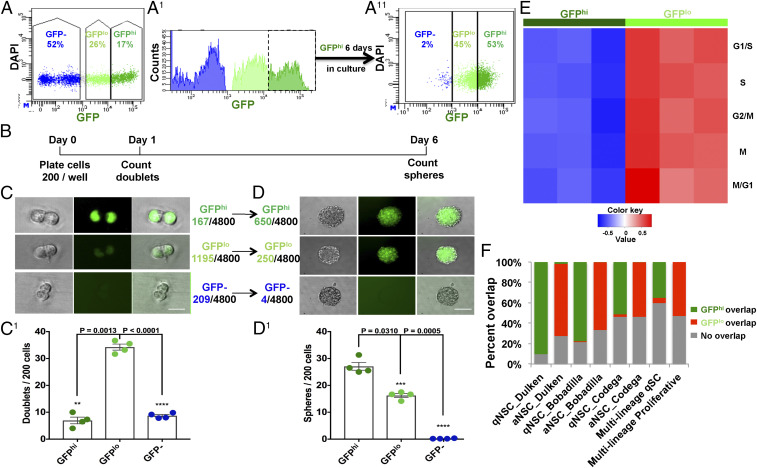
While IHC analysis provides a qualitative snapshot of CGD transgene expression in NSC/progenitor cells, FACS analysis allows quantification for the whole SVZ tissue. Consistent with the IHC studies, FACS sorting for GFP revealed three distinct populations: GFPhi, GFPlo, and GFP– cells (Fig. 2A). GFPhi-sorted SVZ neurospheres were cultured in defined serum-free medium with supplemental growth factors for 6 d and resorted for GFP. A large fraction (∼50%) of GFPhi cells transitioned to GFPlo and GFP– states (Fig. 2 A1 and A11). These data are consistent with a lineage relationship among the varying GFP levels. We next more rigorously tested the self-renewing properties of CGD-GFP–expressing stem/progenitor cells by evaluating low-density doublet formation and progression to neurospheres. Two hundred dissociated GFPhi, GFPlo, or GFP– cells were plated per well in 96-well plates. All wells were examined after 16 to 24 h for the presence of doublets and again at 6 d when neurosphere numbers were determined (Fig. 2 B–D). Within the first 24 h, the GFPlo cultures had the greatest proficiency for generating doublets. In contrast, the GFPhi and GFP– cells exhibited reduced capacity to form doublets. However, the situation shifted over the subsequent 5-d period. The greatest number of neurospheres arose among the GFPhi population (650) whereas the GFPlo doublets yielded fewer neurospheres (250). Thus, despite inefficient initial cell-cycle entry and doublet formation, the GFPhi SVZ population shows the greatest capacity to generate neurospheres while the GFPlo population has limited potential to endure in primary culture and form spheres. In this assay, CGD transgene expression (GFPhi plus GFPlo cells) accounts for essentially all (99.6%) of the primary neurospheres formed in these assays (Fig. 2D). In these studies, GFP– SVZ cells failed to form spheres and to thrive in serum-free medium (Fig. 2 D and D1). Time-lapse movies of freshly sorted CGD-GFPhi cells further revealed an inherent functional heterogeneity among the GFPhi cells. One group of cells formed doublets within 24 h while a second group remained as singlets for 2 d or more before entering cell division (Movie S1). These data indicate additional functional heterogeneity within the GFPhi cohort of SVZ cells.
Fig. 2.
Functional assays validate CGD expression in adult neural stem and progenitor cells. (A and A11) FACS analysis of whole adult SVZ delineates two GFP populations: GFPhi and GFPlo and GFP– cells. A1 presents a histogram view of GFP expression of the same data shown in A. (A11) When placed in serum-free culture, GFPhi cells sequentially progress to GFPlo and GFP– state. (B) Diagram of timeline for doublet and sphere formation assays below. (C and D1) Representative images and statistical analysis of C and C1 24-h doublet and subsequent (D and D1) 6-d sphere formation assays for FACS-sorted adult SVZ GFPhi, GFPlo, and GFP– cells. Note that, despite inefficient doublet formation, the more initially quiescent GFPhi cells are most efficient in forming neurospheres. Mean ± SEM, n = 4 biological replicate mice for each group (each representing six technical replicate wells). (Scale bars in C and D, 10 μm and 100 μm, respectively.) (E) Following RNAseq of GFPhi- and GFPlo-sorted cells, GSVA using cell-cycle signatures indicates a low proliferative status for GFPhi cells compared to GFPlo cells. (F) DEGs of GFPhi and GFPlo cells have high coincidence with published quiescent-stem and activated-progenitor cell signatures. qSC: quiescent stem cell (multilineage). See also SI Appendix, Fig. S2 and Dataset S1.
CGD-GFP FACS analysis of dissociated SVZ tissue identifies about 39.9 ± 2.4% (n = 4) of sorted cells as GFP+ with a GFPhi composition of ∼19.4 ± 1.5% (n = 4) and a GFPlo composition of about 20.5 ± 3.5% (n = 4). These data coincide with anatomical estimates of cell types present in the adult murine SVZ reporting that type B cells (stem cells) represent over 20% of the regional cell population (34). In contrast, other SVZ niche studies made use of combinations of markers and/or reporter transgenes to enrich for NSC or progenitors. These different combinations of markers, including Glast, CD133, and EGFR and reporter transgenes like GFAP-GFP, were reported to yield putative NSC populations that range from 2 to 4.4% of the SVZ preparation, indicating a significant underrepresentation of the entire SVZ population (21–23).
To further investigate the in vivo relatively quiescent versus proliferative state, freshly dissected and sorted SVZ GFPhi and GFPlo cells underwent bulk RNAseq transcriptome analysis. Gene set variation analysis (GSVA) using five sets of cell-cycle phase gene signatures indicated that, while the GFPhi cells uniformly exhibit low cell-cycle gene expression, GFPlo cells have robust expression (Fig. 2E) (35–37). In aggregate, these observations suggest a greater functional complexity among CGD-GFP transgene-expressing cells in the SVZ that goes beyond the discriminatory powers of established stem versus progenitor protein marker-based approaches.
We also performed GSVA analysis of the CGD-GFPhi and GFPlo bulk sequencing data using published SVZ stem/progenitor cell signatures (21, 23). The differentially expressed genes (DEGs) demonstrated substantial overlap between GFPhi and “quiescent” NSC populations, as did the GFPlo and “activated” NSC populations (Fig. 2F and SI Appendix, Figs. S2 A and B1). Gene ontology (GO; https://david-d.ncifcrf.gov/home.jsp) (38) analysis of the GFPhi/GFPlo DEGs identified several stem-cell–signaling pathways enriched in GFPhi cells, including glucose metabolism, lipid metabolic pathway, and oxidation reduction. The GFPlo cells presented cell-cycle–related signatures including transcription, translation, ribosome biogenesis. (SI Appendix, Fig. S2 C–E and Dataset S1 C and D). In sum, these results indicate that the CGD transgene provides an effective tool to identify and efficiently enrich for SVZ stem and progenitor cells in a comprehensive manner.
Discrimination of Cell Subgroups among SVZ NSC/Progenitors.
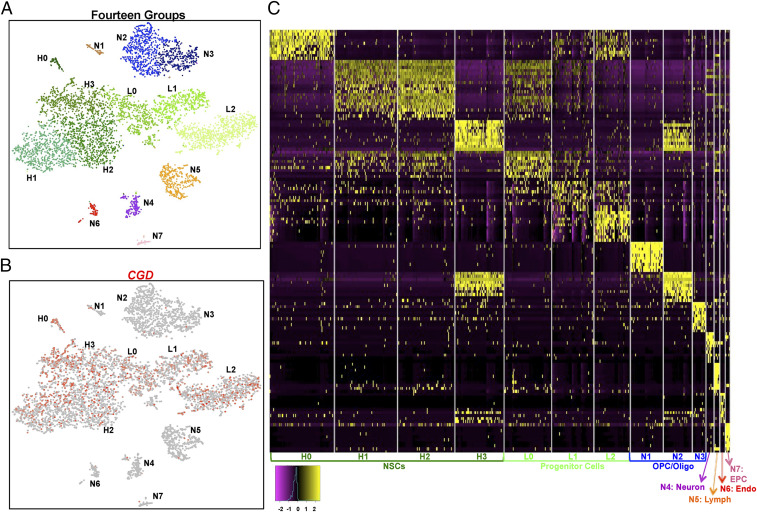
To examine whether CGD-GFP sorting introduces undue bias in representation of the scope of NSC/progenitor cells in SVZ tissue, we turned to single-cell RNA sequencing (SI Appendix, Supplemental Materials and Methods). Whole CGD transgenic SVZ from three adult mice was dissected, submitted to FACS for cell viability (DAPI–), and ∼2,000 DAPI– cells underwent single-cell RNA sequencing. This preparation contained a presumably unbiased mixture of GFPhi, GFPlo, and GFP– cells (termed UNB). In a parallel experiment, SVZs from six CGD transgenic mice were dissected, intermixed, and sorted for relative GFP levels (high, low, or negative). The Seurat package (https://satijalab.org/seurat/vignettes.html) was utilized to process the resulting transcriptomes, and cell groupings for both experiments were identified through clustering and visualized using t-Distributed Stochastic Neighbor Embedding (tSNE) projection. We then combined the entire single-cell sequencing data collected from the four samples (1,600 GFPhi cells; 1,900 GFPlo cells; 1,100 GFP– cells; and 2,000 UNB cells), resulting in 14 coherent groupings (Fig. 3 A and B). The GFPhi cells dominated four groups (H0 to H3); GFPlo cells were enriched in three separate groups (L0–L2); and GFP– cells were ascribed to the remaining seven groups (GFP–:N1–N7; SI Appendix, Fig. S3A). The CGD-GFP+ (GFP+:H0–L2) cells dominated the stem/progenitor cell lineage (Fig. 3 A and B), which accounted for 99% of GFPhi and 92% of GFPlo cells in the SVZ (SI Appendix, Fig. S3A). Conversely, GFPhi and GFPlo cells together represent 96% of the cells in subgroups GFP:H0–L2 (SI Appendix, Fig. S3A). We independently probed the UNB single-cell dataset to examine CGD transgene transcript expression. CGD transcripts could be appreciably detected in only seven subgroups that corresponded to the GFP+ subgroups (GFP:H0–H3 and GFP:L0–L2) ascribed to the sorted GFPhi and GFPlo single-cell sequencing datasets (SI Appendix, Fig. S3B). Thus, single-cell sequencing reveals underlying molecular heterogeneity within both the quiescent (GFPhi) and progenitor (GFPlo) cell groups that were previously masked in the bulk RNA sequencing of the samples depicted in Fig. 2E. We conclude that the CGD transgene exclusively and efficiently labels the majority of the SVZ-derived stem cell/progenitor cell pool.
Fig. 3.
Adult SVZ single-cell RNA sequencing analysis reveals seven distinguishable CGD-GFP+ groups. (A) tSNE projection of Seurat analysis from the sum of the sorted SVZ samples (UNB, GFPhi, GFPlo, and GFP– cells) reveals 14 populations. (B) Normalized CGD expression visualized on tSNE coordinates of SVZ cells demonstrates its preferential expression in stem/progenitor populations. (C) DEGs distinguish 14 SVZ cell types. Heatmap is generated using the top 10 unique genes for each group. EPC: endothelial precursor cells; Endo: endothelial cells; Lymph: lymphocytes. See also SI Appendix, Fig. S3 and Dataset S2.
We further analyzed the transcriptomes of the 14 SVZ subgroups to identify both DEGs and uniquely expressed genes (UEGs) in each subpopulation. The data revealed varying numbers of DEGs, ranging from 114 to 1454 genes, and the UEGs per subgroup ranged from 1 to 866 (Fig. 3C and SI Appendix, Dataset S2 B and C). Not surprisingly, the GFP– cells are the most heterogeneous (SI Appendix, Dataset S2B) with each of the seven subgroups exhibiting diverse specialized genes and transcriptional pathways representative of the spectrum of cells present in the normal brain parenchyma including myelination (oligodendrocyte-oligodendrocyte precursor cells; N1–N3), synaptic transmission (neurons: N4), immune regulatory pathways (lymphocytes: N5), and vasculature (endothelial cells and endothelial progenitors; N6–N7), respectively.
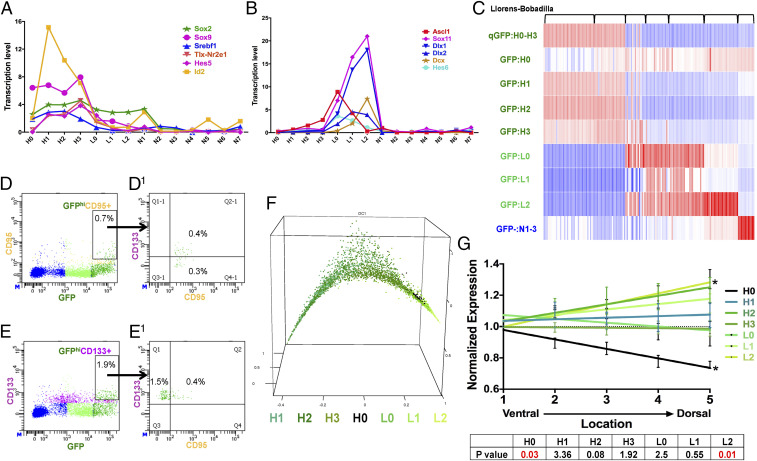
Among the GFP+ subgroups of the classic NSC marker genes, only GFAP messenger RNA (mRNA) is truly confined to the GFPhi:H0–H3 groups although it does not seem to discriminate between them (SI Appendix, Fig. S3C). CD133 mRNA showed persistent but low representation in all seven GFP+ subgroups and also in the GFP–: N7 endothelial precursor subgroup (SI Appendix, Fig. S3C). Glast transcripts are highest in the GFPhi:H0–H3 groups, but expression persists throughout the SVZ (SI Appendix, Fig. S3C). Similarly, all four GFPhi subpopulations express NSC-associated transcription factors including Sox2, Sox9, and Id4 (Fig. 4A and SI Appendix, Fig. S3C1). In contrast, transcription factors associated with progenitor cells such as Ascl1, Dlx1, and Dcx were absent in the four H0 to H3 groups, but were enriched in the GFPlo populations (Fig. 4B). The cell-cycle gene signatures used to identify the proliferative activity of GFPlo versus GFPhi SVZ cells (Fig. 2E) were applied to the single-cell sequencing data and uncovered greater resolution (SI Appendix, Fig. S4A). Among the three GFPlo groups, L1 is clearly the most mitotically active, and this activity is not uniform with the L0 and L2 groups as implied by the bulk sequencing data. In addition, the GFPhi group data reveal heterogeneity despite the overall limited cell-cycle gene expression. The GFPhi:H2 group also shows activity in the G1/S phase, which could represent self-renewal (SI Appendix, Fig. S4A). GO analysis (SI Appendix, Dataset S2D) associated the four GFPhi subgroups with stem-cell–related functions including cilium morphogenesis, oxidation reduction, homeostatic processes, generation of precursor metabolites. GFPlo cells are associated with cell cycle and proliferation genes as well as with signatures of progenitor cells. Thus, single-cell sequencing is consistent with the bulk sequencing data for GFPhi and GFPlo cell populations but additionally uncovers details of both the quiescent and the progenitor NSC states within the SVZ.
Fig. 4.
GFPhi cells resolve into four subgroups of quiescent neural stem cells. (A) NSC-specific transcription factors are enriched in the GFPhi subgroups (all SEs < 0.6; units are normalized values scaled to 10,000 counts/cell). (B) Neural progenitor-specific transcription factors are enriched in GFPlo subgroups (all SEs < 0.2; units are normalized values scaled to 10,000 counts/cell). (C) GFPhi and GFPlo subgroup-specific gene signatures improve resolution of published SVZ cell-sequencing profiles. Transcriptomes of 155 cells published in Llorens-Bobadilla et al. (23) were analyzed with our signatures. Each column represents one cell. (D, D1, E, and E1) FACS analysis showed (D and D1) approximately one-half of GFPhi;CD95+ cells are also CD133+, and (E and E1) about 20% of the GFPhi;CD133+ cells are CD95+. (F) Pseudotime analysis provides a nearest neighbor random walk of single-cell transcriptional profiles revealing a linear progression from GFPhi:H1 through GFPlo:L2. (G) H0 cells preferentially reside on the ventral aspect of adult SVZ as defined by Allen Brain Atlas expression profiles. * indicates P values <0.05. See also SI Appendix, Fig. S4.
Four Subgroups of Quiescent NSC.
To identify a core set of GFPhi NSC genes, we compared expression of each of the four GFPhi populations with the remaining subgroups. This yielded 121 commonly expressed genes among the four CGD-GFPhi groups (SI Appendix, Dataset S2G). A previous and related study (23) reported RNA sequencing of 130 adult murine SVZ single cells that were presorted for Glast+/CD133+ and further evaluated by EGFR levels (termed qNSC or aNSC, respectively). The study also presorted for neuroblasts using a PSA-NCAM antibody (23). We analyzed this published dataset by probing for the presence of the gene signatures derived from the 14 GFP+ and GFP– subgroups including the CGD-GFPhi 121 shared gene signature (Fig. 4C and SI Appendix, Dataset S2G). Significant correlations were observed between the signatures of the differentiated parenchymal cell populations (e.g., GFP:L2 to NB neuroblasts; GFP–:N1-3 to oligodendrocytes, Fig. 4C), but not the more quiescent subgroups. To examine CD133 protein expression in GFPhi populations, we performed FACS analysis using CD133 antibodies together with a GFPhi:H0-specific marker, Fas-CD95 (SI Appendix, Fig. S4B). The results indicate that only half of GFPhi;CD95+ (GFPhi:H0) cells coexpress CD133 protein (Fig. 4 D and D1). Conversely, among GFPhi;CD133+ cells, only 20% of the cells express CD95 (Fig. 4 E and E1). Thus CD133-based sorting of SVZ would preselect only a subset of GFP:H0 cells plus additional GFPhi cells.
We next collectively examined the four GFPhi groups (H0–H3) for expression of biological processes associated with quiescent NSC maintenance and proliferation. This resulted in a comprehensive list of 1,914 genes with 18 related biological processes (SI Appendix, Fig. S4D), some of which overlap with the bulk GFPhi vs. GFPlo analysis. Further analysis of the stringent 121 GFPhi subgroup gene list identified five transcription factors and networks including Sox9 and Srebf1 that potentially regulate qNSC maintenance (SI Appendix, Fig. S4 E and E11 and Dataset S2H).
A Linear Progression of the CGD-GFP+ Stem/Progenitor Populations.
The preceding data are consistent with a developmental relationship from quiescent (GFPhi) to proliferative (GFPlo) cells. To further examine this model, we applied the “Destiny” package designed by dimensional reduction to generate a transition probability from one cell to another according to a random diffusion model (39). Pseudotime ranking of SVZ CGD-GFP+ cells begins with the CGD-GFP:H1 subgroup and progresses to the L2 population (Fig. 4F). Notably, the H0 group did not separate well along the continuum. This is likely due to the relatively small size of this group which may not provide sufficient transcriptional depth and detail to permit this algorithm to distinguish it.
Recent anatomical and lineage SVZ studies have reported a previously unappreciated level of regional heterogeneity (4, 40). To evaluate whether the transcriptional heterogeneity observed in this study might be reflected spatially in the SVZ, we examined regional distribution of quiescent and progenitor GFPhi-lo (H0-L2)-specific transcripts. Ten UEGs from the CGD-GFP:H0-L2 groups were examined in the Allen Brain Atlas in situ hybridization brain panel collection (http://mouse.brain-map.org/) to assess their expression profile within SVZ. Using brain sagittal views, we partitioned the frontal wall of the SVZ into five equivalent regions with the most ventral assigned a value of 1 and the most dorsal assigned a value of 5. We found that H0 cells primarily reside in the ventral-most SVZ, while the L2 subgroup was enriched in the dorsal region (Fig. 4G).
Anatomical studies have pointed to the “type B cell” in the SVZ as the adult NSC (7, 21–23, 41). Reports have also pointed to ependymal cells as the source of NSC (8, 10). However, others argued against it (42). We examined the “ependymal NSC”-specific “hub” genes described by Luo et al. (10) and found them to be specifically enriched in the GFP–:N7 population, which our analysis identifies as endothelial progenitor cells unrelated to the GFP+ NSC/progenitor cells (SI Appendix, Fig. S5 A and B and Dataset S2D). GFP–:N7 cells do not have neural stem/progenitor cell properties in culture (Fig. 2D), and the Luo et al. (10) gene set is not present in GFPhi (H0–H3) or GFPlo (L0–L2) cells (SI Appendix, Fig. S5A). Thus, our analysis excludes the Luo et al. (10) ependymal cell signatures from the SVZ stem-cell GFPhi cell cohort.
GFP:H0 Cells Have Unique NSC Properties.
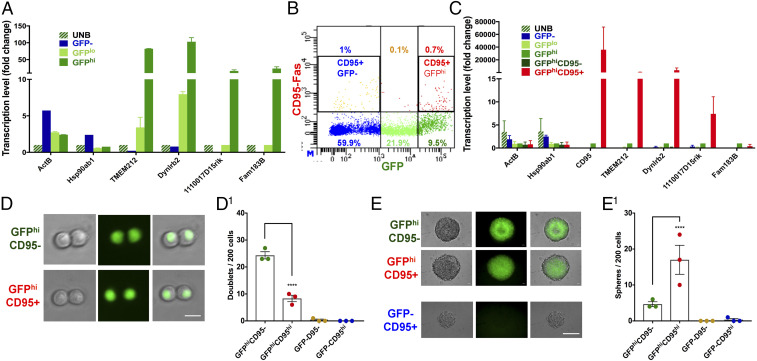
The presence of cilia in both type B NSC and ependymal cells is well established (19, 21, 23, 33). Among the GFP+ cells, our transcriptome analysis appoints a cilia signature specifically to the CGD-GFP:H0 subpopulation and, with the exception of CD133, is absent from the GFP–:N7 subgroup (SI Appendix, Fig. S5C and Dataset S2D). To further characterize the GFP:H0 subgroup, we confirmed its unique gene list with qRT-PCR using SVZ cells that were sorted for single-cell sequencing. UNB, GFPhi, GFPlo, and GFP– cells were examined for GFP:H0 unique gene expression. With some variability, all housekeeping genes (Actb and Hsp90ab1) tested were expressed in all samples (Fig. 5A). The four GFP:H0-specific genes, Tmem212, Dynlrb2, Fam183b, and 1110017D15Rik, were enriched in the GFPhi sample (Fig. 5A). The GFP– sample consistently showed the lowest transcription levels for all four genes, with Fam183b and 1110017D15Rik below detection. To specifically isolate GFP:H0 cells for functional analysis, the cell-surface marker CD95 proved effective for FACS analysis and CD95+/GFP+ sorting yielded a 0.7% population enriched within the GFPhi cells (Fig. 5B). This low percentage was concordant with the low representation of GFP:H0 cells in the entire preparation (SI Appendix, Fig. S3D). GFP– cells also contained around 1% of CD95+ cells, which were likely to be GFP–:N7 cells (SI Appendix, Fig. S5D). Further analysis of concordant CD95 expression suggested that the CD95+ cells trend with higher GFP protein levels, which coincides with the highest transcription level of CGD transgene in GFP:H0 cells (SI Appendix, Figs. S4B and S5 E and F).
Fig. 5.
H0 cells have unique stem-like properties. (A) Expression of four H0-specific genes (Tmem212, Dynlrb2, Fam183b, and 1110017D15rik) was verified by qRT-PCR in single-cell sequencing samples. Gene expression levels were normalized to UNB cells. Mean ± SEM, n = 2. (B) CD95-Fas receptor antibody labels one subgroup of GFPhi and one subgroup of GFP– cells. All of the numbers represent the percentage of each population among the whole SVZ. (C) GFPhi;CD95+ cells have enriched expression of the H0-specific genes. qRT-PCR was performed with GFP and CD95 sorted cells for the H0-specific genes. Gene expression levels were normalized to GFPhi cells. Mean ± SEM, n = 2. (D and D1) Representative images and quantification of doublet formation by GFP and CD95 sorted cells at 24 h. Note that GFPhi;CD95– cells are more efficient at forming doublets (Scale bar, 10 μm.) (E and E1) Representative images and quantification of sphere formation of GFP and CD95 sorted cells. Note that, reminiscent of Fig. 2 C and D, GFPhi;CD95+ cells, which were inefficient in doublet formation at 24 h, are most efficient at forming neurospheres 6 d later. Note also that GFP–;CD95+ cells do not form neurospheres. (Scale bar, 100 μm.) Mean ± SEM, n = 3 biological replicate mice for each group in D and E1. **** in D1 and E1 indicates P values <0.0001. See also SI Appendix, Fig. S5.
To further assess the identity of CGD-GFPhi;CD95+ cells as the GFP:H0 subgroup, the four candidate genes used for qRT-PCR validation were reemployed to analyze sorted GFPhi;CD95+ and GFPhi;CD95– cells. While housekeeping gene expression was present in all samples, the four GFP:H0-specific genes were significantly enriched in GFPhi cells (Fig. 5C). Three were further enriched in GFPhi;CD95+ cells (Fig. 5C).
We next compared GFPhi;CD95+, GFPhi;CD95–, GFP–;CD95+, and GFP–;CD95– cells in doublet and sphere formation assays. Consistent with the preceding observations, the two GFP– samples did not form doublets or spheres (Fig. 2 C and D and Fig. 5 D and E1). GFPhi;CD95+ cells formed fewer doublets, but more spheres than GFPhi;CD95– cells (Fig. 5 D and E1). These results are compatible with the hypothesis that GFP:H0 (CD95+) cells represent a discrete quiescent NSC population that encounters greater difficulty in entering the cell cycle when initially cultured but outcompetes type B-like GFP:H1–H3 cells in eventual neurosphere formation. Furthermore, immunostaining of the CD95 antibody with GFP fluorescence confirmed the existence of a double-positive cell population residing preferentially in the ventral SVZ (SI Appendix, Fig. S5G). Taken together, our data indicate that GFP:H0 cells represent a specialized, regionally discrete, quiescent subpopulation of SVZ stem cells.
CGD-GFP+ Cell Populations Decrease with Age.
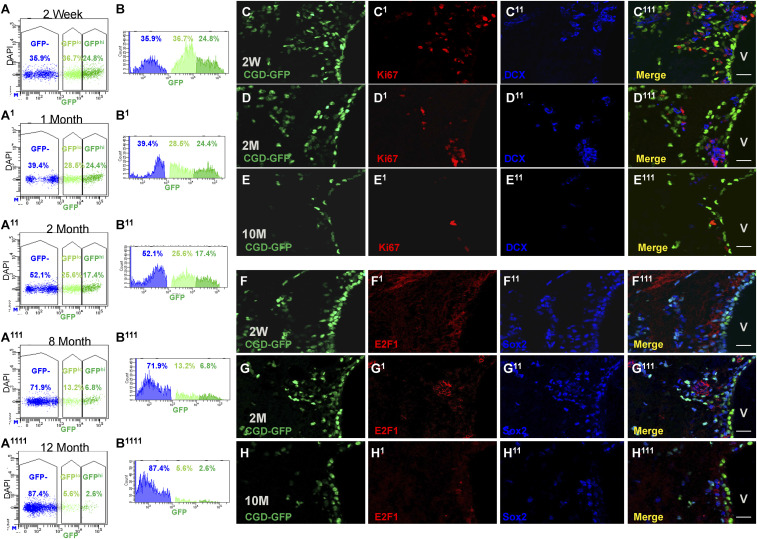
We extended the SVZ NSC/progenitor lineage single-cell analysis over the process of aging. For this study 2-wk and 1-, 2-, 8-, and 12-mo-old CGD SVZ samples were FACS-analyzed for GFP expression. The results indicated that both GFPhi and GFPlo populations decrease over age. By 12 mo, the GFPhi population (2.3 ± 0.1%, n = 2) was reduced more than 10-fold as compared to the 2-wk-old sample (24.9 ± 2.7%, n = 3), and the GFPlo population (5.4 ± 0.4%, n = 2 vs. 38 ± 2.2%, n = 2) dropped by more than 7-fold (Fig. 6 A and B). For further analysis, we selected genes with group-specific expression profiles in the 2-mo-old SVZ samples (SI Appendix, Fig. S6A). Among them, Sox2 was high in all GFP+ cells; E2f1 and Mki67 were specifically induced in GFP:L0–L1; and DCX was expressed in GFP:L1–L2 subpopulations (SI Appendix, Fig. S6A). We found that both GFP+ cells and the L1–L2 markers decreased significantly over age (Fig. 6 C–H). Cells that were double positive for CGD-GFP and these aforementioned genes were almost entirely depleted in 10-mo samples (Fig. 6 C–H and SI Appendix, Fig. S6 B and C11). These results indicate that quiescent SVZ NSC gradually lose capacity to enter the cell cycle over the process of aging.
Fig. 6.
CGD-GFP+ cells decrease over age. (A and B) Whole SVZ FACS analysis delineates the decrease of CGD-GFP+ cells as they age from 2 wk (A and B), 1 mo (A1 and B1), 2 mo (A11 and B11), 8 mo (A111 and B111), until 12 mo (A1111 and B1111). (C–E) IHC staining reveals Commensurate loss of CGD-GFP (C, D, and E) with Mki67 (C1–E1) and DCX (C11–E11) in 2-wk (C–C111), 2-mo (D–D111), and 10-mo mouse brain sagittal sections (E and E111). (Scale bar, 20 μm.) (F–H) IHC staining for CGD-GFP (F–H) and stem/progenitor markers E2F1 (F1–H1) and Sox2 (F11–H11) in 2-wk (F–F111), 2-mo (G–G111), and 10-mo mouse brains (H–H111) (Scale bar, 20 μm.) V: lateral ventricle in C111–H111. See also SI Appendix, Fig. S6.
To examine age-related changes for each of the seven CGD-GFP+ populations, we performed single-cell RNAseq analysis using SVZ tissues acquired at 2 wk and 2, 6, and 12 mo. Combined analysis of 5,600 cells identified 13 major subpopulations (SI Appendix, Fig. S7A). The list included 13 of the 14 subgroups identified in the original sampling at 2 mo of age, including all 7 GFP– subgroups (N1-N7), and 6 GFP+ subgroups corresponding to GFP:H1-H3 and GFP:L0–L2 (SI Appendix, Fig. S7 A–D). In this study, the GFP:H0 subgroup was underrepresented and did not form a separate cluster but instead merged with GFP:H3 subgroup (SI Appendix, Fig. S7 A–D).
Failure of Quiescent NSC to Transition to Progenitors in Aged SVZ.
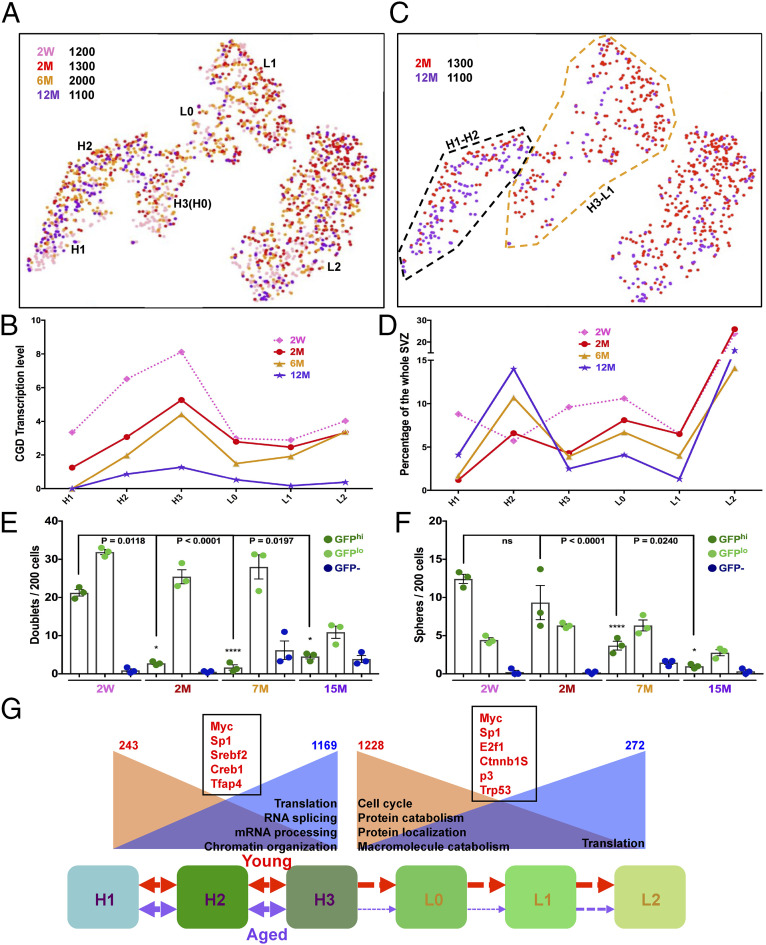
We then focused on the GFP+ subgroups and found that 2-wk SVZ contained the highest proportion of stem/progenitor cells (GFP:H3–L1) concordant with the highest CGD transgene expression (Fig. 7 A and B). Over a period of 12 mo, the GFP:H3–L1 subgroups were increasingly underrepresented (Fig. 7 C and D). Doublet/sphere formation assays with GFPhi/GFPlo/GFP– cells isolated from 2-wk and 2-, 7-, and 15-mo SVZs also indicated a continuous decline in sphere-formation capacity of GFPhi cells during aging (Fig. 7 E and F). Thus, the GFP:H3–L1 bridge, from quiescent to proliferative states, is disrupted, indicating that over age there is a deficit in quiescent cell activation.
Fig. 7.
Aging quiescent neural stem cells exhibit deficiency in transition to the progenitor state. (A) tSNE projection reveals the neural stem/progenitor lineage distribution of four aged samples: 2 wk and 2, 6, and 12 mo. (B) CGD transcription levels consistently decrease over age in each GFP+ subgroup. (C) Direct comparison of the neural stem/progenitor-cell distribution between 2-mo (red) and 12-mo (purple) samples. Note the stark underrepresentation of the GFP:H3–L1 lineage in the 12-mo sample. The black dashed line marks the H1–H2 lineage, and the brown dashed line characterizes the H3–L1 cluster. (D) Quantitative illustration of distribution of each GFP+ subgroup (H1–L2) at different age points. Note the continuous decrease of H3–L1 lineage from 2- to 12-mo adult SVZs. (E and F) Quantification of doublet and sphere formation assays for SVZ GFPhi, GFPlo, and GFP– cells over age. Note that, by 7 mo, the GFPhi population begins to lose its capacity to transition from doublets to neurospheres, consistent with the reduced capacity to transition from H3 to L1 in vivo. Mean ± SEM; n = 3 biological replicate mice for each group (each representing three technical replicate wells). (G) Cartoon describes the activation blockage of adult NSC in old mice compared to young mice. H1 cells in the SVZ progress into H2 and H3 and then become activated to form L0, L1, and L2 cells. This progress is robust in young mice (red arrows), but reduced significantly in aged mice (purple arrows). H1–H2 and H3–L1 cells from 2 and 12 mo are combined to perform differential gene analysis. The numbers on the top reflect the down-regulated (red) and up-regulated (blue) genes in aged groups. GO analysis from the DEGs unravels potential signaling pathways responsible for the changes. The down-regulated (brown background) or up-regulated (blue background) pathways during aging are summarized. The red genes in black boxes reflect the putative transcription factors associated with the age-related gene expression change in either H1–H2 or H3–L1 lineages. See also Fig. 4F and SI Appendix, Fig. S7 and Dataset S3.
To further examine the transition deficit, we compared pooled SVZ GFPhi subgroups (H1-H2, Fig. 7C, black dashed line) and the GFPhi-lo transition subgroup (H3-L1, Fig. 7C, brown dashed line) between 2 and 12 mo. The 12-mo H1–H2 lineage samples yielded 1,169 up- and 243 down-regulated genes while the H3–L1 lineage yielded 272 up-regulated genes and 1,228 down-regulated genes. GO analysis indicated key features of both aged SVZ cohorts including increased translation. The H1–H2 cluster presented additional activity in RNA splicing and mRNA processing and chromatin organization processes (Fig. 7G and SI Appendix, Dataset S3 D–J). The aged mice H3–L1 lineage showed down-regulation of the cell cycle, protein catabolism and localization, and macromolecule catabolism processes. The transcription factors responsible for the up- and down-regulation of the gene sets in each cluster were also determined. While Myc and Sp1 are related to the aging profile switch in both lineages, Srebf2-Creb1-Tfap4 and E2f1-Ctnnb1-Sp3-Trp53 are associated with either H1–H2 or H3–L1 clusters (Fig. 7G). These biological processes and transcription factors provide potential targets to overcome aging deficits of NSC.
Discussion
Single-cell tSNE analysis of the dissected SVZ region identified 14 distinguishable transcriptional subgroups of cells including 7 stem/progenitor subgroups. Among them, four were superimposed on the CGD-GFPhi subgroups, three on the CGD-GFPlo subgroups, and the remaining seven groups on the GFP– subgroups. This provides powerful validation that the CGD-GFP sorting process had indeed portrayed an unbiased representation of the stem/progenitor cell niche.
Functional assays including primary culture low-density doublet and neurosphere formation indicated that the sorted GFPhi cells represent a quiescent stem-like cell population. When placed in culture, this population is most efficient in establishing neurosphere cultures when compared to the GFPlo cells that efficiently form doublets but have reduced potential to evolve into neurospheres. Neurosphere formation is widely considered as a valid assay for stem-cell self-renewal (43–47). However, as we show, transient amplifying cells can also form and sustain neurospheres. Therefore, assays such as the low-density doublet assay and subsequent quantification of progression to neurospheres may provide a more stringent method for assessing self-renewal capability.
A key determinant that permitted analysis of single-cell sequence data from different experiments for each of the 14 SVZ subgroups was the consistent expression of cell-specific transcription factors and markers commonly used to define stem cells, progenitor cells, and specialized parenchymal cells (SI Appendix, Fig. S3). Moreover, molecular analysis further validated the identity of GFPhi and GFPlo cells as stem and progenitor cells, respectively. GO and cell-cycle analysis confirmed the differential status of these cells both in the metabolic and in the cell-cycle states (SI Appendix, Fig. S4D and Dataset S2). In contrast, the seven GFP negative (GFP–:N1–N7) cell subgroups aligned with parenchymal cell types included in the SVZ preparation. The increased granularity achieved with sequencing of many thousand cells enriched with a transgene revealed unexpected features. For example, only two of three proliferative GFPlo subgroups (GFP:L0–L1) are highly active in the cell cycle. Moreover, few of the standard markers for quiescent stem cells segregated at the transcriptional level to a specific subgroup.
The GFP:H0 subpopulation is particularly intriguing and has unique features. Anatomically, GFP:H0 genes preferentially localize in the ventral SVZ. This subgroup is the only one among GFP+ cells that has high transcription of cilia genes. Furthermore, isolation of GFP:H0 cells reveals that they are likely the subgroup within the GFP+ cells responsible for reduced low-density doublet formation but enhanced neurosphere development in primary culture. We suggest that H0 cells may represent a specialized quiescent NSC cell population, and whether this population participates actively in the homeostatic process of self-renewal and production of transient amplifying cells merits further investigation.
The uncovering of additional subgroups of GFPhi cells and of GFPlo subgroups underscores the heterogeneity underlying a complex biology. The identification of subgroup-specific transcriptional signatures and, importantly, of transmembrane-receptor–encoding genes, should provide the means to prospectively isolate these populations and better study their functions. Very recent studies have used related approaches to investigate the SVZ, and our present data serve to validate but importantly to greatly enrich and resolve the emerging picture of the heterogeneous SVZ. We cannot ignore a recent study that, through sequencing of 28 SVZ cells, reported a NSC-specific signature and proposed an ependymal origin for NSC (10). Our sequencing studies, which included 6,600 cells, ascribes the Luo et al. (10) NSC signature, not to NSC, but instead to endothelial progenitor cells (GFP–:N7 subgroup). Finally, unlike the relative facility in culturing doublets or neurospheres from GFP+ cultures (albeit with differing efficiencies), we were unable to obtain cultures from GFP– cells. This substantial discrepancy raises a cautionary note about the importance of validating single-cell data not only at the molecular level but also with meaningful functional assays.
It has long been appreciated that, over time, SVZ neurogenesis declines (48–50). However, the effect of aging on different NSC/progenitor populations is unresolved, in part due to methodological constraints of cell-identification criteria or limited cell number (49, 51). Our approach allows detailed transcriptional examination of the process and demonstrates the persistence of two qNSC populations (H1 and H2) from 2 to 12 mo. Instead, a particular bottleneck within the stem/progenitor lineage at the H3–L1 juncture becomes pronounced with age. The data provide a rich list of genes including transcription factors and signaling pathways that may be valuable in identifying potential molecular determinants of this deficiency and potential strategies to overcome it. Here, we focused analysis between adult SVZ of 2 and 12 mo. Additional comparisons focusing on the adolescent (2 wk) and aged (12 mo) data may reveal additional insights into NSC maturing and aging. Further analysis using the seven GFP– populations could identify aging-related changes in oligodendrocytes, neurons, immune cells, and endothelial cells.
NSC have important functions in normal physiology that warrant their thorough study and understanding. Furthermore, NSC may be among the few or unique cells in the several hundred million cells within the adult brain that have the capacity to engender malignant brain tumors such as glioblastoma (18). This disease stands out as one of the most intractable. In this study, we present seven stem/progenitor populations to lay the foundation for further investigation into the mechanism underlying the transition from a stem cell to a tumor cell of origin. Finally, the CGD transgene provides a practical GFP-reporter–based means to isolate adult neural stem cells and related cell types in tumors. This includes the capability to examine candidate gene function via CreERT2 conditional knockout and to investigate the potential roles of the transgene-expressing cells in brain pathology with DTR through targeted cell ablation.
Materials and Methods
Animal Studies.
All mouse experiments were approved and performed in accordance with the guidelines of the Institutional Animal Care and Research Advisory Committee at the University of Texas Southwestern Medical Center and the Institutional Animal Care and Use Committee of Memorial Sloan Kettering Cancer Center (MSKCC).
Generation of the CGD Transgenic Mice.
A CGD fragment was amplified and assembled from a CreERT2-containing construct, an H2B-eGFP vector (from Yan Jiang, Institutes of Brain Science, Fudan University, Shanghai, China), and genomic DNA from 293T cells (diphtheria toxin receptor). To ensure the efficient expression of all of the three proteins in the CGD transgene, a P2A fragment was used to bridge the three different genes (52–54). To verify the adequate expression of monoproteins from the multicistronic cassette, the coding region of the CGD fragment was inserted into a FETPTR vector to transfect 293T cells. Western blot was used to detect the comparative expression levels of monomers vs. polymers from the same open-reading frame with a GFP antibody (1:1,000, SI Appendix, Fig. S1C). Further analysis demonstrated the activity of the three proteins (SI Appendix, Fig. S1 B–E). The CGD fragment was then inserted into the pNERV vector (gift from Steve Kernie, Columbia University, New York, NY) (28) through NotI (New England Labs) sites. The pNERV-CGD plasmid was cut with SalI to release the Nestin-CGD fragment to generate transgenic mice by pronuclear injection into fertilized murine eggs in a C57BL/6 genetic background. A total of 82 founders were screened with PCR, and 29 lines were identified as positive with the CGD transgene. The primers used to verify the transgene were the following: forward primer, 5′-GTCTATATCATGGCCGACAAGC-3′; and the reverse primer, 5′-GAAAGAGCTTCAGCACCACC-3′.
Histology and Immunohistochemistry.
Mice were perfused and fixed in 4% paraformaldehyde, soaked in 30% sucrose, and imbedded in OCT compound (Tissue-Tek). Frozen brain tissues were sectioned (12 μm) and stained with the following antibodies: goat anti-GFAP (Santa Cruz, 1:500), mouse anti-GFAP (1:200, Millipore), goat anti-Doublecortin (Santa Cruz, 1:500), goat anti-Sox2 (Santa Cruz, 1:200), rabbit anti-Mki67 (Novus, 1:500), goat anti-GFP (Rockland Immunochemicals, 1:200), mouse anti-Glast (Miltenyi Biotec, 1:100), rat anti-CD133 (eBioscience, 1:100), and rabbit anti-E2f1 (Abnova, 1:100). DAPI was used to stain the nucleus (ThermoScientific, 1 μg/mL). The sections were then imaged with a Zeiss LSM 510 confocal microscope using Argon 488, He543, and He 633. Whole-mount SVZs were prepared and stained as described (55).
Tissue Culture and FACS Analysis.
Fresh mouse SVZ tissues were dissected and cultured for doublet and sphere formation analysis with a modified protocol as described (47, 56). Briefly, 200 cells were plated into a 96-well plate and counted for doublets within 16 to 24 h and for spheres 6 d later. When cell numbers were limited, 100 to 300 cells were sorted and plated. The data were then normalized to 200 cells. Antibody staining for FACS was performed in Flow Cytometry Staining Buffer (ThermoFischer Scientific), and the following antibodies were used: PE-GLAST (ACSA-1, Miltenyi Biotec, 1/40), APC-CD133 (13A4, Miltenyi Biotec, 1/75), APC-Cy7-CD45 (BD, 1/200), and APC-CD95-Fas (Miltenyi Biotec, 1:100). DAPI was used to stain the dead cells (ThermoScientific, 1 μg/mL). Viable cells were sorted in a FACSAriaII (BD Biosciences) and collected in complete serum-free medium for culture. For Movie S1, the cells were incubated in Cytation 5 (BioTek) and imaged every hour for GFP and cell morphology.
Total RNA Extraction and Gene Expression Profile Analysis.
Freshly prepared samples were sorted directly into concentrated TRIzol LS reagent (Invitrogen 10296028) and submitted to the Integrated Genomics Operation facility (MSKCC) for RNA extraction and sequencing with the paired-end 50 method. The results were processed by Bioinformatics Core of the MSKCC to identify differentially expressed genes.
Supplementary Material
Acknowledgments
We thank Omar Aly (Weill Cornell Medicine), Yanjiao Li, and Alicia Pedraza for technical support in single-cell sample preparation. The H2B-eGFP plasmid was a gift from Yan Jiang, and Nestin enhancer/promoter was a gift from Steven Kernie. We thank Elvin Feng (MSKCC Molecular Cytology Core Facility) for designing the ImageJ script used for quantification of gene expression in the Allen Mouse Brain Atlas. We thank Hui Julia Zhao and Nicholas Socci (MSKCC Bioinformatics Core Facility) for bioinformatics support. This work was supported by NIH Grants R01 CA131313-01A1 and NOA 1R35CA210100-01 and in part through NIH/National Cancer Institute Cancer Center Support Grant P30 CA008748. X.P.X. was supported by American Brain Tumor Association Basic Research Fellowship in honor of Joel A. Gingras, Jr. L.F.P. holds the Albert C. Foster Chair in Cancer Biology.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014389117/-/DCSupplemental.
Data Availability.
Single-cell sequence data have been deposited in Gene Expression Omnibus (accession no. GSE107220). All study data are included in the article and supporting information. Data analyzed in Fig. 4C were published in Llorens-Bobadilla et al. (23), deposited in Gene Expression Omnibus (accession no. GSE67833).
References
- 1.Yousefi M., Li L., Lengner C. J., Hierarchy and plasticity in the intestinal stem cell compartment. Trends Cell Biol. 27, 753–764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L. D., Wagers A. J., Dynamic niches in the origination and differentiation of haematopoietic stem cells. Nat. Rev. Mol. Cell Biol. 12, 643–655 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altman J., Das G. D., Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 124, 319–335 (1965). [DOI] [PubMed] [Google Scholar]
- 4.Merkle F. T., et al. , Adult neural stem cells in distinct microdomains generate previously unknown interneuron types. Nat. Neurosci. 17, 207–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ming G. L., Song H., Adult neurogenesis in the mammalian brain: Significant answers and significant questions. Neuron 70, 687–702 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeisel A., et al. , Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doetsch F., Caillé I., Lim D. A., García-Verdugo J. M., Alvarez-Buylla A., Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97, 703–716 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Johansson C. B., et al. , Identification of a neural stem cell in the adult mammalian central nervous system. Cell 96, 25–34 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Sirko S., et al. , Reactive glia in the injured brain acquire stem cell properties in response to sonic hedgehog. [corrected]. Cell Stem Cell 12, 426–439 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Luo Y., et al. , Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell 161, 1175–1186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim D. A., Alvarez-Buylla A., Adult neural stem cells stake their ground. Trends Neurosci. 37, 563–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjornsson C. S., Apostolopoulou M., Tian Y., Temple S., It takes a village: Constructing the neurogenic niche. Dev. Cell 32, 435–446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jessberger S., Stem cell-mediated regeneration of the adult brain. Transfus. Med. Hemother. 43, 321–326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon K. J., et al. , Endogenous neural stem/progenitor cells stabilize the cortical microenvironment after traumatic brain injury. J. Neurotrauma 32, 753–764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alcantara Llaguno S., et al. , Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15, 45–56 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li P., et al. , A population of Nestin-expressing progenitors in the cerebellum exhibits increased tumorigenicity. Nat. Neurosci. 16, 1737–1744 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alcantara Llaguno S. R., et al. , Adult lineage-restricted CNS progenitors specify distinct glioblastoma subtypes. Cancer Cell 28, 429–440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alcantara Llaguno S., et al. , Cell-of-origin susceptibility to glioblastoma formation declines with neural lineage restriction. Nat. Neurosci. 22, 545–555 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coskun V., et al. , CD133+ neural stem cells in the ependyma of mammalian postnatal forebrain. Proc. Natl. Acad. Sci. U.S.A. 105, 1026–1031 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckervordersandforth R., et al. , In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell 7, 744–758 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Codega P., et al. , Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron 82, 545–559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mich J. K., et al. , Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. eLife 3, e02669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llorens-Bobadilla E., et al. , Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17, 329–340 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Dulken B. W., Leeman D. S., Boutet S. C., Hebestreit K., Brunet A., Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 18, 777–790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmerman L., et al. , Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron 12, 11–24 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Mignone J. L., Kukekov V., Chiang A. S., Steindler D., Enikolopov G., Neural stem and progenitor cells in nestin-GFP transgenic mice. J. Comp. Neurol. 469, 311–324 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Imayoshi I., Ohtsuka T., Metzger D., Chambon P., Kageyama R., Temporal regulation of Cre recombinase activity in neural stem cells. Genesis 44, 233–238 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Yu T. S., Zhang G., Liebl D. J., Kernie S. G., Traumatic brain injury-induced hippocampal neurogenesis requires activation of early nestin-expressing progenitors. J. Neurosci. 28, 12901–12912 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J., Kwon C. H., Lin L., Li Y., Parada L. F., Inducible site-specific recombination in neural stem/progenitor cells. Genesis 47, 122–131 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zywitza V., Misios A., Bunatyan L., Willnow T. E., Rajewsky N., Single-cell transcriptomics characterizes cell types in the subventricular zone and uncovers molecular defects impairing adult neurogenesis. Cell Rep. 25, 2457–2469.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Mizrak D., et al. , Single-cell analysis of regional differences in adult V-SVZ neural stem cell lineages. Cell Rep. 26, 394–406.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ninkovic J., Mori T., Götz M., Distinct modes of neuron addition in adult mouse neurogenesis. J. Neurosci. 27, 10906–10911 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirzadeh Z., Merkle F. T., Soriano-Navarro M., Garcia-Verdugo J. M., Alvarez-Buylla A., Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell 3, 265–278 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doetsch F., García-Verdugo J. M., Alvarez-Buylla A., Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J. Neurosci. 17, 5046–5061 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitfield M. L., et al. , Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13, 1977–2000 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J. P., GSEA-P: A desktop application for gene set enrichment analysis. Bioinformatics 23, 3251–3253 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Macosko E. Z., et al. , Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Angerer P., et al. , destiny: Diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Fiorelli R., Azim K., Fischer B., Raineteau O., Adding a spatial dimension to postnatal ventricular-subventricular zone neurogenesis. Development 142, 2109–2120 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Lao C. L., Lu C. S., Chen J. C., Dopamine D3 receptor activation promotes neural stem/progenitor cell proliferation through AKT and ERK1/2 pathways and expands type-B and -C cells in adult subventricular zone. Glia 61, 475–489 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Shah P. T., et al. , Single-cell transcriptomics and fate mapping of ependymal cells reveals an absence of neural stem cell function. Cell 173, 1045–1057.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Román-Trufero M., et al. , Maintenance of undifferentiated state and self-renewal of embryonic neural stem cells by Polycomb protein Ring1B. Stem Cells 27, 1559–1570 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Delgado A. C., et al. , Endothelial NT-3 delivered by vasculature and CSF promotes quiescence of subependymal neural stem cells through nitric oxide induction. Neuron 83, 572–585 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Reynolds B. A., Rietze R. L., Neural stem cells and neurospheres: Re-evaluating the relationship. Nat. Methods 2, 333–336 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Pastrana E., Silva-Vargas V., Doetsch F., Eyes wide open: A critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 8, 486–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belenguer G., Domingo-Muelas A., Ferrón S. R., Morante-Redolat J. M., Fariñas I., Isolation, culture and analysis of adult subependymal neural stem cells. Differentiation 91, 28–41 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Enwere E., et al. , Aging results in reduced epidermal growth factor receptor signaling, diminished olfactory neurogenesis, and deficits in fine olfactory discrimination. J. Neurosci. 24, 8354–8365 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi Z., et al. , Single-cell transcriptomics reveals gene signatures and alterations associated with aging in distinct neural stem/progenitor cell subpopulations. Protein Cell 9, 351–364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leeman D. S., et al. , Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daynac M., Morizur L., Chicheportiche A., Mouthon M. A., Boussin F. D., Age-related neurogenesis decline in the subventricular zone is associated with specific cell cycle regulation changes in activated neural stem cells. Sci. Rep. 6, 21505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizuguchi H., Xu Z., Ishii-Watabe A., Uchida E., Hayakawa T., IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol. Ther. 1, 376–382 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Szymczak A. L., et al. , Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 22, 589–594 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Kim J. H., et al. , High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6, e18556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mirzadeh Z., Doetsch F., Sawamoto K., Wichterle H., Alvarez-Buylla A., The subventricular zone en-face: Wholemount staining and ependymal flow. J. Vis. Exp., 1938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferron S. R., et al. , A combined ex/in vivo assay to detect effects of exogenously added factors in neural stem cells. Nat. Protoc. 2, 849–859 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell sequence data have been deposited in Gene Expression Omnibus (accession no. GSE107220). All study data are included in the article and supporting information. Data analyzed in Fig. 4C were published in Llorens-Bobadilla et al. (23), deposited in Gene Expression Omnibus (accession no. GSE67833).