Significance
Dendritic cells (DC) play a crucial role in the immune system by bridging innate and adaptive immunity. In the spleen, a specific subset of DCs accumulates around the blood-filtering marginal zone to capture particulate antigens such as red blood cells. We show here that these specialized DCs develop in response to NOTCH2 instruction that is regulated by a poorly studied kinase, called Thousand and One Kinase 3 (TAOK3). This kinase controls DC development in a cell-intrinsic manner, rendering cells receptive to NOTCH2 signaling. Interfering with this kinase opens up ways to manipulate a specific subset of DCs.
Keywords: thousand and one kinase, dendritic cell, Notch signaling
Abstract
Antigen-presenting conventional dendritic cells (cDCs) are broadly divided into type 1 and type 2 subsets that further adapt their phenotype and function to perform specialized tasks in the immune system. The precise signals controlling tissue-specific adaptation and differentiation of cDCs are currently poorly understood. We found that mice deficient in the Ste20 kinase Thousand and One Kinase 3 (TAOK3) lacked terminally differentiated ESAM+ CD4+ cDC2s in the spleen and failed to prime CD4+ T cells in response to allogeneic red-blood-cell transfusion. These NOTCH2- and ADAM10-dependent cDC2s were absent selectively in the spleen, but not in the intestine of Taok3−/− and CD11c-cre Taok3fl/fl mice. The loss of splenic ESAM+ cDC2s was cell-intrinsic and could be rescued by conditional overexpression of the constitutively active NOTCH intracellular domain in CD11c-expressing cells. Therefore, TAOK3 controls the terminal differentiation of NOTCH2-dependent splenic cDC2s.
Conventional dendritic cells (cDCs) orchestrate innate and adaptive immunity by taking up and presenting soluble and particulate antigens at sites of high exposure. Two subtypes of cDCs differ in molecular signature, function, and lineage-defining transcription factors (1). Type 1 cDCs (cDC1s) are identified by high surface levels of XCR1 and CD24 and additionally express CD8α or the integrin αE (CD103) when they reside in lymphoid or nonlymphoid tissues, respectively. Conventional DC1s are indispensable for the generation of cytotoxic T cell responses against viruses and tumors and specialize in cross-presentation and loading of peptides on MHCI. Type 2 cDCs (cDC2s) are characterized by expression of signal regulatory protein α (SIRPα; CD172a) and the integrin αM (CD11b). They preferentially activate CD4+ T cells in response to extracellular antigens which are presented on MHCII (2–4) and can be further boosted to present antigens to CD8+ T cells in the presence of type 1 IFN and after TLR ligation (5).
Conventional DCs in peripheral tissues are constantly replenished from circulating precursor cells (6–8). Commitment of cDC precursors (precDCs) to the cDC1 or cDC2 fate begins in the bone marrow, but the terminal differentiation of cDCs requires tissue-specific cues in the periphery (9). While the transcriptional network controlling cDC1 development has been well delineated, involving ID2, NFIL3, BATF3, and IRF8 (10–15), the factors that govern cDC2 fate specification remain poorly characterized. The transcription factors IRF4, ZEB2, and KLF4 were shown to control the development of specific cDC2 subsets in different organs but cannot fully account for the observed heterogeneity among terminally differentiated cDC2s (16–19).
The development of specific cDC populations is also controlled by Notch signaling, an evolutionarily conserved pathway that controls dichotomous cell-fate decisions in immune cells via microenvironmental cues. In the spleen, membrane contacts between delta-like ligand 1 (DLL1) on fibroblasts and the NOTCH2 receptor on immune cells instruct the terminal differentiation of marginal zone B cells (MZB) and cDCs (20–24). Dendritic cell-specific loss of key Notch signaling intermediates has revealed that CD8α+ cDC1s and a major subset of cDC2s that coexpress CD4 and the endothelial cell-selective adhesion molecule (ESAM) depend on NOTCH2 instruction in the spleen (20, 21). The absence of ESAM+ CD4+ cDC2s in mice that lack the NOTCH2 receptor or the downstream transcriptional regulator RBPjκ in DCs impaired CD4+ T cell priming in response to blood-borne antigens that are captured in the splenic marginal zone (20, 25). NOTCH2 signaling also controls the development of highly unique CD103+ CD11b+ cDC2s in the intestinal lamina propria. These specialized cDC2s cells produce high amounts of IL-23 and are required for defense against attaching and effacing pathogens such as Citrobacter rodentium (26). Despite the clear role for NOTCH2 in terminal differentiation of cDCs, it is unclear how Notch pathway activation is regulated. Only a subset of cDCs becomes Notch-instructed, despite ubiquitous expression of Notch2 by all cDCs (27). Furthermore, although NOTCH2 ligation markedly improved the generation of cDC1s in vitro, the prototypical ESAM marker is not induced in cDC2s (28, 29). These findings suggest that additional mechanisms control Notch activation in vivo.
Thousand-and-one (TAO) kinases belong to a subfamily of Ste20-like proteins, serine/threonine kinases that activate MAP kinases in response to extracellular signals (30, 31). Three TAO kinase paralogs exist in mammals: TAOK1 (also known as MAP3K16, PSK2, or MARKK), TAOK2 (MAP3K17 or PSK1), and TAOK3 (MAP3K18, JIK, or DPK) (32). While the regulation and function of these kinases remain largely unknown, their role in the immune system is emerging. We have reported that Taok3 controls commitment to MZB cell fate by regulating the surface expression of the metalloprotease A Disintegrin and Metalloprotease 10 (ADAM10) on transitional B cells, thereby rendering these cells receptive to NOTCH instruction (33, 34). While ADAM10 controls NOTCH2 activation in MZB cells, its role in splenic cDC development is not known (35).
Here, using genetic approaches, we demonstrate that TAOK3 is needed for cell-intrinsic NOTCH2 signaling that leads to terminal differentiation of ESAM+ CD4+ cDC2s and programs these cells to present particulate antigens in the marginal zone of the spleen.
Results
Taok3−/− Mice Lack a Subset of Splenic cDC2s.
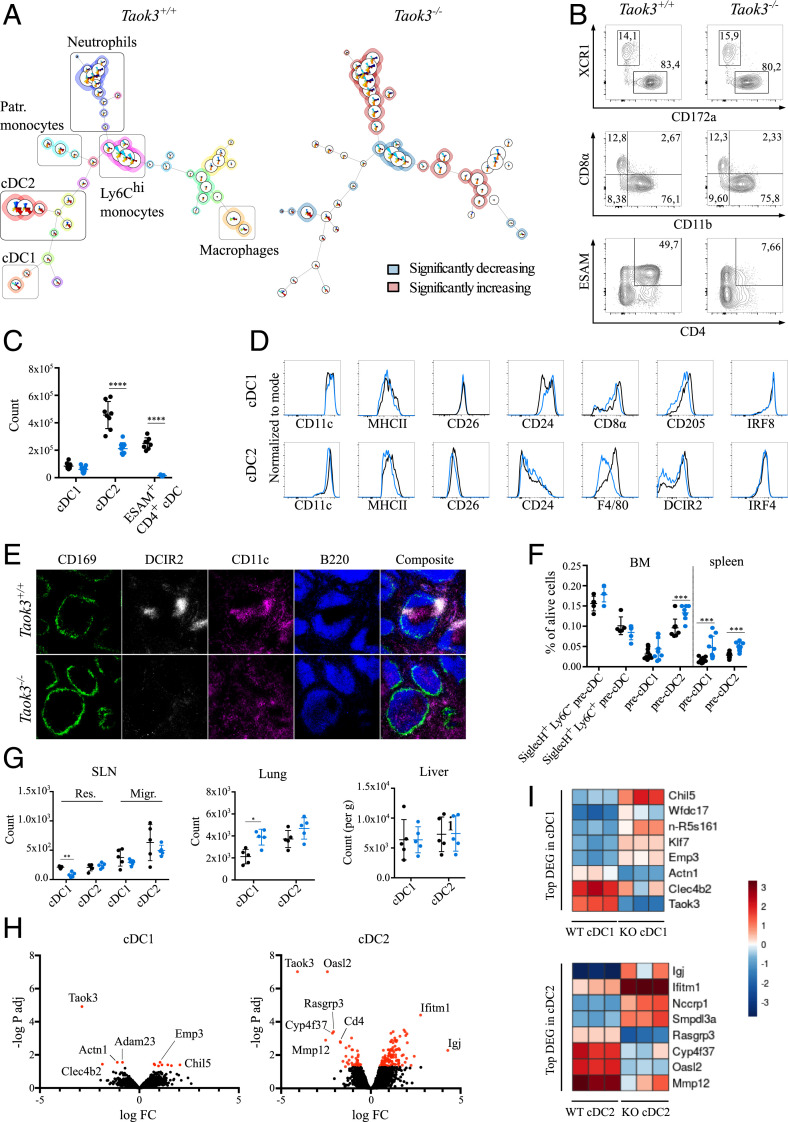
In order to identify how loss of TAOK3 affects cells of the mononuclear phagocyte system, we applied the unsupervised clustering algorithm FlowSOM to flow cytometry datasets from Taok3−/− and Taok3+/+ splenocytes. The algorithm clusters cells into nodes based on the expression level of distinct markers and organizes these nodes into a minimal spanning tree (36). After exclusion of B, T, and NK cells from analysis, remaining splenocytes were separated into 49 clusters (nodes) and 20 metaclusters in an unbiased manner (Fig. 1A and SI Appendix, Fig. S1A). Comparison with manual gating (SI Appendix, Fig. S1B) revealed robust separation of cDC1s, cDC2s, and Ly6Chi monocytes, patrolling monocytes, macrophages, and neutrophils into different metaclusters. Among cDCs, the manually gated XCR1+ CD172a− cDC1 population corresponded to one metacluster containing two nodes distinguished by their level of CD8α expression. In contrast, XCR1− CD172a+ cDC2s were automatically clustered into seven distinct nodes and three metaclusters, based on differential expression of the surface markers ESAM, CD4, CD11b, CD43, and F4/80. These findings illustrate the phenotypic heterogeneity of splenic cDC2s. Strikingly, there was a drastic and selective reduction in the frequency of ESAM+ CD4+ cDC2s (3.8-fold at the metacluster level) in Taok3−/− mice, whereas other cDC2 and cDC1 metaclusters did not change significantly (Fig. 1A and SI Appendix, Fig. S1 C and D). Among other metaclusters, less-pronounced yet significant reductions were observed in the percentage of CD64+ F4/80+ CD172a+ macrophages and Ly6Chi monocytes. In line with our previous findings, there was also a small increase in the percentage of splenic neutrophils in Taok3−/− mice (33).
Fig. 1.
Taok3−/− mice lack ESAM+ CD4+ splenic cDCs. (A) The FlowSOM algorithm was run on single, live, lineage (CD3e, CD19, NK1.1, Ter119) splenocytes from Taok3−/− mice and littermate controls. A minimal spanning tree with 49 nodes and 20 metaclusters was generated by automated, unsupervised clustering. The average FlowSOM tree is displayed for all Taok3+/+ (Left) and all Taok3−/− (Right) mice. Node size corresponds to the frequency of that cell cluster among all analyzed cells. Pie charts indicate the mean expression of the given surface markers for each node (SI Appendix, Fig. S1A). In the Taok3+/+ tree, background colors denote the different metaclusters while, in the Taok3−/− tree, the background color highlights metaclusters that are significantly decreased (blue) or enriched (red) in Taok3−/− mice. Two-tailed Mann–Whitney U test with Bonferroni correction for multiple testing. Data are representative of two biologically independent experiments (n = 8 to 9 mice per group). The FlowSOM algorithm was run five times to ensure reproducibility of the results. (B) Representative contour plots of splenic cDCs from Taok3+/+ and Taok3−/− mice (gating strategy, SI Appendix, Fig. S2A). Numbers adjacent to the gates denote percentage of total cDCs. (C) Number of cDC1s, cDC2s, and ESAM+ CD4+ cDCs in Taok3+/+ (black) and Taok3−/− (blue) spleen. Conventional DC1s are defined as XCR1+ CD172a− cDCs, and cDC2s as XCR1− CD172a+ cDCs. Data pooled from two independent experiments (n = 8 to 9 mice per group), two-tailed Mann–Whitney U test. (D) Histograms indicating expression levels on the surface of splenic cDC1s (Top) and cDC2s (Bottom) of Taok3+/+ (black) and Taok3−/− (blue) mice. Data are representative of at least two independent experiments (n = at least 8 mice per group). (E) Confocal image of spleens from Taok3+/+ and Taok3−/− mice. (Magnification: 10×.) Green: CD169; white: DCIR2; magenta: CD11c; blue: B220. Data are representative of three independent experiments (n = 13 mice per group). (F) Frequency of precDCs (gating strategy, SI Appendix, Fig. S2B) among live cells in bone marrow and spleen of Taok3+/+ (black) and Taok3−/− (blue) mice. Data pooled from two independent experiments (n = 10 per group). Two-tailed Mann–Whitney U test. (G) Absolute number of cDC1s and cDC2s in SLN (Left), lung (Middle), and liver (Right) of Taok3+/+ (black) and Taok3−/− (blue) mice. “Res.” and “Migr.” indicate resident and migratory cDC populations, respectively. Data are representative of at least two independent experiments (n = 8 to 10 for lung and liver) or are representative of 4 to 5 mice per group (SLN). Two-tailed Mann–Whitney U test. (H and I) cDNA microarray on sorted splenic cDC1s and cDC2s from Taok3−/− and Taok3+/+ mice. (H) Volcano plot based on fold change and P value of all transcripts in cDC1s (Left) and cDC2s (Right). (I) Heatmap of differentially expressed genes between Taok3+/+ and Taok3−/− cDC1s (Top) or cDC2s (Bottom) for the individual samples. Dendritic cells were sorted from three mice per group. Bars indicate mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To validate these findings, we analyzed the splenic cDC compartment using a manual gating strategy (SI Appendix, Fig. S2A). We confirmed that ESAM+ CD4+ cDCs, which consist entirely of cDC2s, were almost completely absent in Taok3−/− mice (Fig. 1 B and C). Moreover, the total number of CD172a+ cDC2s significantly decreased (Fig. 1C), implying that the effects of Taok3 are not limited to impaired expression of these surface markers. Remaining cDC2s also displayed lower median fluorescence intensities (MFIs) of F4/80, slightly decreased surface levels of MHCII, CD172a, and DCIR2 (33D1), and an increased MFI of CD11b (Fig. 1D and SI Appendix, Fig. S3A). While the number of XCR1+ cDC1s was not altered (Fig. 1C), Taok3−/− cDC1s expressed significantly lower levels of CD8α and CD205 and had a slightly higher MFI of CD24 compared to littermate controls (Fig. 1D). Loss of Taok3 did not impact the expression of the transcription factors IRF4 and IRF8 in cDC1s and cDC2s (Fig. 1D).
In secondary lymphoid organs, cDCs occupy specific anatomical locations crucial for their function (9). In the spleen, cDC2s have been shown to reside in the marginal zone and in bridging channels, which connect the T cell zone with the red pulp through interruptions in the ring of CD169+ macrophages surrounding the white pulp (37, 38). Immunostaining for CD11c and DCIR2 (33D1) allowed us to address the location of cDC2s in Taok3−/− mice (Fig. 1E). While DCIR2+ clusters were readily identified in bridging channels of wild-type (WT) littermates, they were absent in the spleen of Taok3−/− mice. Although the median DCIR2 expression was slightly lower on Taok3−/− cDC2s (Fig. 1D), this could not sufficiently explain the drastic loss of DCIR2 signal on histology. Accordingly, CD11c+ cells were scattered throughout the red pulp and did not aggregate in well-defined clusters at the bridging channels in Taok3−/− mice. In contrast, the staining of CD11c+ cells residing in the T cell zone was preserved in knockout mice, in line with the preserved cDC1 counts found by flow cytometry. Of note, no B220 staining was identified outside of the ring of the CD169+ marginal metallophilic macrophages, consistent with the absence of MZB cells in Taok3−/− mice (33).
cDC Development in Taok3−/− Mice.
We wondered whether the loss of ESAM+ CD4+ cDC2s in Taok3−/− mice reflected defects in the development of cDCs from progenitors in the bone marrow. Committed precDCs have been identified in bone marrow and spleen (12, 39). In the bone marrow, the absence of Taok3 had no effect on the frequency of bipotent SiglecH+ Ly6C− and SiglecH+ Ly6C+ precDCs (gating strategy, SI Appendix, Fig. S2B). SiglecH− Ly6C− CD24hi precDC1s were equally represented in the bone marrow but increased in the spleen of Taok3−/− mice compared to littermate controls. The percentage of SiglecH− Ly6C+ precDC2s, the direct precursors for mature cDC2s, was significantly elevated in bone marrow and spleen (Fig. 1F). These data suggest that the decreased cDC2 numbers in Taok3−/− mice are not due to defective development of DC progenitors. Rather, the accumulation of Taok3−/− precDC2s implies a failure of these cells to undergo terminal differentiation. The unexpected increase in splenic precDC1s supports subtle derangements in the differentiation of mature cDC1s as well. Alternatively, these effects could be the consequence of compensatory mechanisms serving to restore a mature cDC population with increased cell turnover. In the latter case, however, one would also expect to see an increase in the numbers of early bone marrow precDCs.
To study whether the loss of cDC2s was unique to spleen, we examined the frequency of cDCs in other lymphoid and nonlymphoid organs. We found intact numbers of cDC2s in the lung, liver, and skin-draining lymph node (SLN) of Taok3−/− mice (Fig. 1G). However, subtle changes were noted in the phenotype of cDC2s across different tissues: the expression levels of F4/80 and CD172a were consistently lower in spleen, lung, liver, and lymph nodes. In addition, the MFI of CD11b significantly increased in the liver but not in the lung of Taok3−/− mice (SI Appendix, Fig. S3 A–E). We found a small but significant increase in the number of cDC1s in the lung, while resident cDC1s in the SLN decreased in knockout mice (Fig. 1G). Thus, loss of Taok3 is associated with an altered cDC2 phenotype across different tissues.
Altered Gene Expression Profile in Taok3−/− cDCs.
To identify Taok3-regulated genes, we compared genome-wide expression profiles of splenic cDC1s and cDC2s from Taok3−/− mice and littermate controls. Principal component analysis demonstrated separation of cDC1 and cDC2 cell types along one direction, and separation of Taok3+/+ and Taok3−/− cells along another (SI Appendix, Fig. S3F). Gene expression analysis revealed 8 differentially expressed (DE) genes in cDC1s and 101 DE genes in cDC2s between wild-type and knockout cells, while Taok3 was effectively knocked out in both cDC subsets (Fig. 1H). These results are consistent with the minor impact of Taok3 loss on cDC1 homeostasis compared to the effects observed in cDC2s. Top DE genes for cDC1s and cDC2s are listed in Fig. 1I. Based on gene ontology (GO) enrichment analysis, the processes associated with down-regulated genes concern mainly regulation of interleukin-2 (IL-2) biosynthesis and leukocyte differentiation. In contrast, the main GO term associated with up-regulated genes in Taok3−/− cDC2s is regulation of cell adhesion. Indeed, many DE genes in cDC2s related to cell adhesion and migration such as L1cam, Epcam, Vim, Itgb5, Lgals1, and Lgals3 were up-regulated, while Mmp9 and Mmp12 were down-regulated in knockout cDC2s. Differentially expressed genes in cDC2s also comprised type 1 IFN-induced genes such as Ifitm1 and Oasl2. In addition, the expression of several genes associated with small GTPases increased (Rhob) or decreased (Arhgap6 and Rasgrp3) in Taok3−/− cDC2s.
Functional Responses of Taok3−/− cDCs.
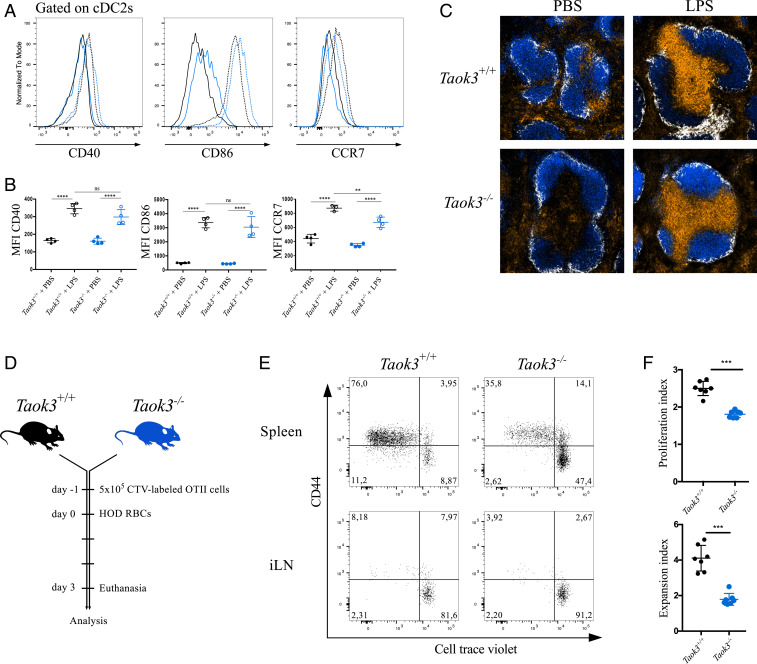
We reasoned that the defects in terminal differentiation might affect the maturation and function of Taok3-deficient cDCs. To assess their ability to mature and migrate in response to pathogen-associated molecular patterns, we injected mice with the TLR4 ligand lipopolysaccharide (LPS) intravenously (i.v.) (40). Upon exposure to LPS, cDCs from Taok3−/− mice up-regulated the costimulatory markers CD40, CD80 and CD86 to the same extent as littermate controls (Fig. 2 A and B). Mature Taok3−/− cDC2s also up-regulated the chemokine receptor CCR7, albeit not to the same level as wild-type cDC2s. To verify whether the difference in surface expression levels of CCR7 impacted the ability of DCs to migrate to the T cell zone, we assessed the position of CD11c+ cells in the spleen 8 h after LPS exposure in vivo (40). In wild-type mice, the CD11c+ clusters in the bridging channels had disappeared and the DCs had relocated to the T cell zone upon LPS injection. In Taok3−/− mice, similar accumulation of CD11c+ cells in the T cell zone was observed (Fig. 2C). Thus, Taok3 deficiency did not impair the normal maturation and migration of DCs to the T cell zone.
Fig. 2.
Loss of Taok3 hampers CD4+ T cell priming to allogeneic red blood cells. (A) Histograms of surface CD40, CD86, and CCR7 expression on splenic cDC2s from Taok3+/+ (black) and Taok3−/− mice (blue). Mice were injected intraperitoneally with 20 μg LPS in PBS (dotted line) or PBS only (solid line) and killed 8 h later. (B) Median fluorescence intensities of CD40, CD86, and CCR7 on cDC2s from A. One-way ANOVA with post hoc Tukey’s test. (C) Confocal images of spleens from Taok3−/− mice and littermate controls treated with LPS or PBS as in A. (Magnification: 10×.) B220 in blue, CD169 in white, CD11c in orange. Data in A–C are representative of two independent experiments (n = 7 to 8 per group in total). (D–F) Taok3−/− and littermate control mice were injected with 5 × 105 CellTrace Violet-labeled OTII-specific CD45.1+ CD4+ T cells and transfused with 100 μL of HOD-RBCs (Materials and Methods) 1 d later. (E) Representative dot plots of OTII cells in the spleen and inguinal lymph node (iLN) of Taok3+/+ (Left) and Taok3−/− (Right) acceptor mice 72 h after HOD-RBC transfusion. Plots are pregated on live CD19− NK1.1− Ter119− CD3e+ CD4+ CD45.1+ cells. Numbers adjacent to the gates indicate the percentage of OTII cells. (F) Proliferation and expansion index of OTII cells in the spleen of the acceptors, according to the Flowjo proliferation algorithm. Two-tailed Mann–Whitney U test. Data in E and F are representative of two independent experiments (n = 14 to 15 acceptor mice per group in total). Bars indicate mean ± SD; ns, nonsignificant; *P < 0.05, ***P < 0.001, and ****P < 0.0001.
Next, we wanted to probe the capacity of Taok3−/− cDCs to prime CD4+ T cell immune responses in vivo. The role of splenic cDCs in the generation of CD4+ T cell responses against blood-borne antigens and apoptotic and damaged cells is well established (25, 37, 41, 42). More specifically, efficient CD4+ T cell priming crucially depended on CD4+ cDC2s in a model of allogeneic red-blood-cell transfusion (43). Taok3−/− mice and littermate controls received a transfusion of stored red blood cells (RBCs) from congenic B6 mice that express a membrane-bound fusion protein containing antigenic epitopes of hen egg lysozyme (HEL), ovalbumin (OVA), and Duffyb blood group antigen (HOD-RBCs). Storage of these HOD-RBCs renders them immunogenic through an unknown mechanism. Transfusion of HOD-RBCs induced significantly less proliferation in transferred OVA-specific CD4+ T cells in Taok3−/− recipient mice compared to littermate control recipient mice (Fig. 2 D–F). These data indicate a crucial role for TAOK3- and TAOK3-dependent cDC2s in triggering CD4+ T cell responses to allogeneic RBCs.
Taok3 Is Required Intrinsically in cDC2s.
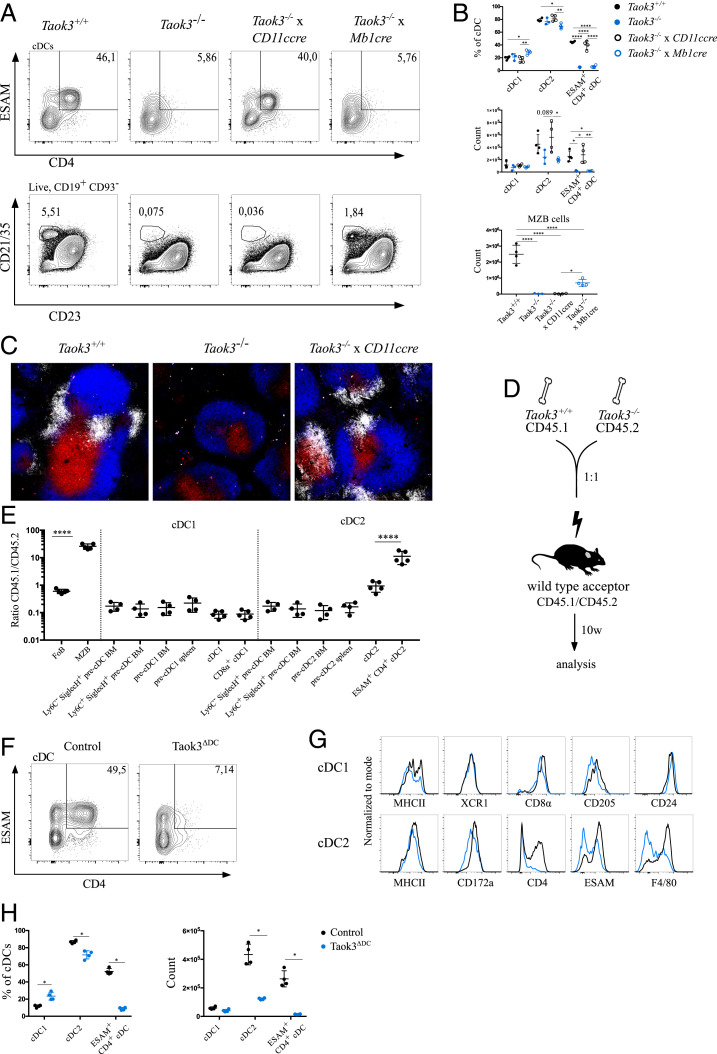
Normal cDC2 development relies on multiple environmental factors such as RELB-expressing stromal cells and DLL1 expression on fibroblastic cells (21, 44). To address whether cDC homeostasis requires Taok3 expression in hematopoietic or stromal cells, we crossed Taok3−/− mice to Vav-cre mice that express the CRE recombinase in all hematopoietic cells. Because the gene trap construct of Taok3−/− mice contains loxP sites, TAOK3 expression can be restored selectively by CRE-mediated recombination and gene trap reversal. We found that the percentage of ESAM+ CD4+ cDC2s was almost completely restored in the spleen of Taok3−/− x Vav-cre+ mice, ruling out a role for stromal Taok3 in cDC2 development (SI Appendix, Fig. S4 A and B). Conceivably, the observed cDC phenotype of Taok3−/− mice could be secondary to their MZB cell deficit. Marginal zone B cells do occupy a similar niche as bridging channel cDCs, and regulatory roles of B cells in cDC homeostasis have been described (6). To test this hypothesis, we crossed Taok3−/− mice to Mb1-cre mice allowing selective gene trap reversal in B cells (45). While the number of CD21/35+ CD23lo MZB cells was partially restored in Taok3−/− x Mb1-cre+ mice, the number and phenotype of splenic cDCs did not differ from littermate Taok3−/− x Mb1-cre− controls (Fig. 3 A and B). In order to address more directly whether the role of Taok3 in cDCs was cell-intrinsic, we generated Taok3−/− x CD11c-cre mice. Strikingly, the number of ESAM+ CD4+ cDC2s in Taok3−/− x Cd11c-cre+ mice was restored to wild-type levels (Fig. 3 A and B). As expected, MZB cells were still completely absent in these mice. Thus, selective restoration of Taok3 expression in CD11c+ cells was sufficient for ESAM+ CD4+ cDC2 development. We validated these findings in Taok3−/− x Cd11c-cre-GFP+ mice, where the CRE is expressed only after the precDC stage. While this still restored cDC2 numbers, it rescued ESAM and CD4 expression less efficiently than the earlier acting CD11c-cre, suggesting a role for Taok3 early in the commitment of precDC2s to cDC2s (SI Appendix, Fig. S4 C and D). Rescue of Taok3 expression in CD11c-positive cells was also sufficient to restore clustering of DCIR2+ dendritic cells at marginal zone bridging channels (Fig. 3C). Together, these data demonstrate independent and cell-intrinsic roles of Taok3 in DCs and MZB cells. Furthermore, they prove that the presence of splenic cDC2s and their correct positioning at bridging channels does not depend on the presence of marginal zone B cells.
Fig. 3.
The role of Taok3 in splenic cDC2s is cell-intrinsic. (A) Representative contour plots of cDCs (Top row) and marginal zone B cells (Bottom row) in the spleen of Taok3−/−, Taok3+/+, Taok3−/− x CD11ccre (where Taok3 expression is selectively restored in CD11c-expressing cells) and Taok3−/− x Mb1cre mice (where Taok3 expression is selectively restored in Mb1-expressing cells). The ESAM+ CD4+ cDC gate was pregated on cDCs, and the MZB cell gate was pregated on live CD19+, CD93− cells. (B) Frequency of the indicated cDC subsets among all cDCs (Top Left) and their absolute number (Bottom Left). Number of marginal zone B cells (Bottom Right). One-way ANOVA with Tukey’s post hoc test. Data in A and B are representative of two independent experiments (n = 7 to 8 per group). (C) Representative confocal images of spleens from Taok3+/+ (Left), Taok3−/− (Middle), and Taok3−/− x CD11ccre mice (Right). (Magnification: 10×.) White: DCIR2; blue: B220; red: CD3e. Data are representative of two independent experiments (n = 4 to 8 per group). (D) Schematic representation of CD45.1 WT:CD45.2 Taok3−/− mixed BM chimeras. (E) Contribution of donor cells to immune cell populations 10 wk after transplantation in BM and spleen. Results are expressed as the ratio between CD45.1 (WT) and CD45.2 (Taok3−/−) cells. Ratios of CD45.1/CD45.2 origin along cDC1 and cDC2 development are compared to follicular (FoB) and marginal zone (MZB) B cells. Unless otherwise indicated, cell populations originate from the spleen. The same Ly6C− SiglecH+ and Ly6C+ SiglecH+ BM precDC populations are depicted twice. One-way ANOVA with Bonferroni correction for multiple testing. Data representative of two independent experiments (n = 13 mice). (F) Representative contour plot of ESAM and CD4 expression on splenic cDCs from Taok3ΔDC mice. (G) Histograms illustrate surface expression of the indicated markers on splenic cDC1s (Top) and cDC2s (Bottom) from Taok3ΔDC mice (blue) and Cre-negative littermate controls (black). (H) Quantification of splenic cDC subsets as a percentage of all cDCs (Left) and in absolute numbers (Right). Two-tailed Mann–Whitney U test. Data in F and H are representative of four mice per group. Numbers adjacent to the gates (A and F) indicate the percentage of the parent population. Bars indicate mean ± SD; *P < 0.05, **P < 0.01, ****P < 0.0001.
To reveal even more subtle intrinsic differences in DC homeostasis and development, we also set up competitive bone marrow chimeric experiments, where cells of wild-type or Taok3−/− origin need to compete in the same host. Lethally irradiated wild-type mice were reconstituted with CD45.2 Taok3−/− and CD45.1 wild-type bone marrow in equal parts (Fig. 3D). After reconstitution, MZB cells were almost uniquely derived from the WT compartment, while bone marrow of both genotypes contributed equally to the follicular B-cell population in the spleen (Fig. 3E), confirming earlier results (33). Interestingly, Taok3 expression was not intrinsically required in cDC1s or in DC progenitors in spleen and bone marrow. In contrast, wild-type ESAM+ CD4+ cDC2s outcompeted Taok3−/− counterparts by a ratio of more than 10:1. This effect was less pronounced on the total cDC2 population, as Taok3-deficient bone marrow still gave rise to ESAMlo cDC2s. We finally also created conditional Taok3fl/fl mice and crossed them to CD11c-cre mice to generate Taok3ΔDC mice. We confirmed a significant decrease in the number and percentage of cDC2s in the spleen of Taok3ΔDC mice (Fig. 3 F–H). Again, this reduction was specific for a subpopulation of cDC2s that expressed high levels of CD4, ESAM, and F4/80. Despite having decreased numbers of ESAM+ CD4+ cDC2s, Taok3ΔDC mice were still able to induce CD4+ T cell responses to allogeneic HOD-RBCs (SI Appendix, Fig. S4 E and F), implying that additional defects contribute to the impaired T cell priming in Taok3−/− mice. While the frequency of cDC1s was comparable to littermate controls, the MFI of CD8α and CD205 was slightly lower in Taok3ΔDC mice (Fig. 3G), supporting our earlier findings. Based on these results, we conclude that Taok3 plays a nonredundant, cell-intrinsic role in the terminal differentiation of ESAM+ CD4+ cDCs.
Impaired Notch Signaling in Taok3−/− cDCs.
The homeostasis of ESAM+ CD4+ cDC2s requires trophic signals through the lymphotoxin β receptor (LTβR) (6, 20, 26). We hypothesized that TAOK3 might regulate signaling through the LTβR in DCs. However, surface expression of LTβR was comparable between cDCs from Taok3−/− mice and littermate controls (SI Appendix, Fig. S5A). In addition, treatment of Taok3−/− mice with an agonistic antibody against the LTβR could not restore cDC2 numbers or rescue the expression of ESAM and CD4 (SI Appendix, Fig. S5B). Therefore, it is unlikely that LTβR signaling functions downstream of TAOK3.
At the same time, the loss of ESAM+ cDC2s in Taok3-deficient mice is highly reminiscent of the phenotype of mice lacking Notch signaling in DCs (20, 26). Moreover, we previously demonstrated that Notch instruction is hampered in Taok3−/− B cells due to the lack of surface ADAM10 in transitional B cells, the immediate precursors of MZB cells (33). To assess whether loss of Taok3 also affected the NOTCH2 pathway in cDCs, we performed gene set enrichment analysis (GSEA) on the gene expression data from Taok3−/− mice using published gene expression profiles of Notch2−/− cDCs (26). We found that the down-regulated genes in Taok3−/− cDC1s and cDC2s were significantly enriched for the set of genes down-regulated in Notch2−/− cDC1s and cDC2s, respectively. Equally, the set of genes up-regulated in Notch2−/− cDC1s and cDC2s was strongly overrepresented among the up-regulated genes in Taok3+/+ cDC1s and cDC2s, respectively (SI Appendix, Fig. S6 A–D). These findings reveal considerable overlap between the gene expression signatures of Notch2−/− and Taok3−/− dendritic cells.
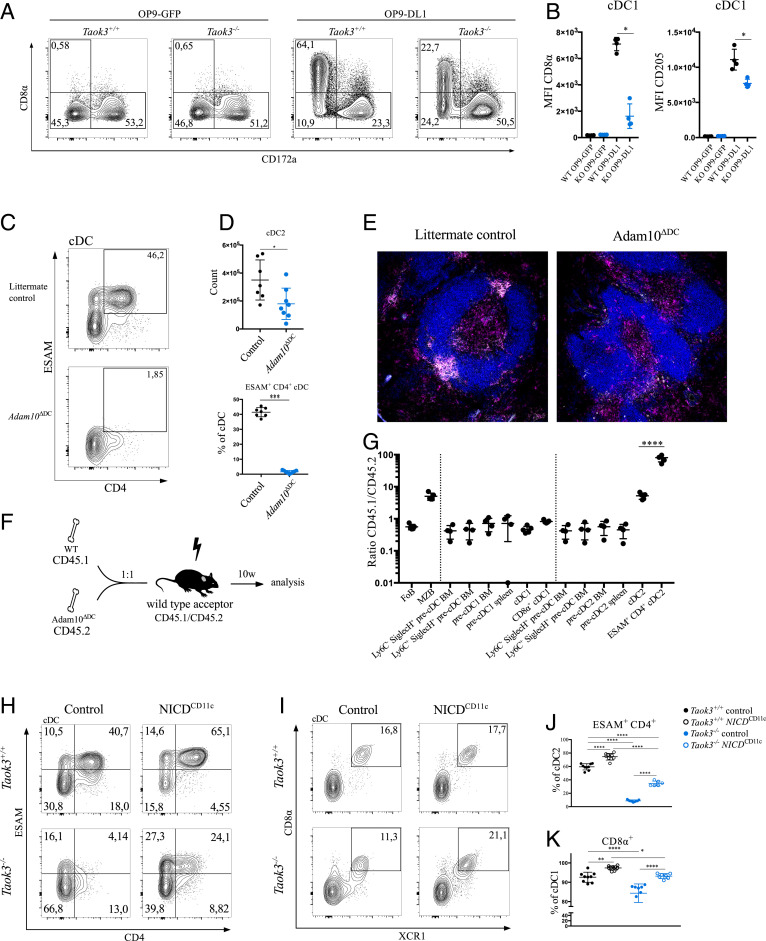
Recently, a method was described to generate CD8α+ cDC1s in vitro on DLL1-expressing fibroblasts in a NOTCH2-dependent manner (28, 29). To further assess the ability of Taok3−/− precDCs to undergo Notch instruction, we cultured Taok3+/+ and Taok3−/− bone marrow-derived DCs (BMDCs) on OP9 cells transduced with Dll1 or with a GFP construct as a control. We confirmed that WT BMDCs cultured on OP9-DLL1 fibroblasts generated higher numbers of DC1s and DC2s and lower numbers of plasmacytoid DCs (pDCs) than did OP9-GFP cocultures (SI Appendix, Fig. S6E). In line with previous findings (29), OP9-DLL1 strongly induced the expression of CD8α and CD205 on BM-derived cDC1s but failed to induce relevant levels of ESAM expression on DCs. Compared to littermate controls, cocultures of Taok3−/− BMDCs with OP9-DLL1 fibroblasts generated fewer XCR1+ DC1s and CD172a+ DC2s (SI Appendix, Fig. S6E). Additionally, loss of Taok3 markedly reduced the expression of levels of CD8α and CD205 on DC1s (Fig. 4 A and B).
Fig. 4.
Defective NOTCH signaling in Taok3−/− cDCs. (A) Representative contour plots of bone marrow-derived dendritic cells from Taok3−/− mice or littermate controls cocultured with OP9-GFP or OP9-DL1 cells. Plots are pregated on live GFP− lineage− B220− CD64− CD11c+ MHCII+ cells. (B) Median fluorescence intensity of CD8α and CD205 on the surface of cDC1s from Taok3−/− or Taok3+/+ BM-derived DCs cocultured with OP-DL1 fibroblasts. Two-tailed Mann–Whitney U test. Data in A and B are representative of two independent experiments (nine biological replicates per group). (C) Representative contour plot of ESAM and CD4 expression on splenic cDCs from Adam10ΔDC mice and Cre-negative littermate controls. (D) Absolute number of splenic cDC2s (Top) and percentage of ESAM+ CD4+ cells (Bottom) among cDCs in Adam10ΔDC mice. Two-tailed Mann–Whitney U test. Data in C and D are representative of four independent experiments (n = 15 to 16 per group). (E) Confocal image of spleens from Adam10ΔDC (Right) and littermate control (Left) mice. (Magnification: 10×.) White: DCIR2; blue: B220; magenta: CD11c. (F) Schematic representation of WT: Adam10ΔDC mixed bone marrow chimeras. (G) Contribution of donor cells to several cell populations during cDC1 and cDC2 development, 10 wk after mixed BM transplantation. Results are expressed as the ratio between CD45.1 (WT) and CD45.2 (Adam10ΔDC) cells. Unless otherwise indicated, cell populations originate from spleen. Ly6C− SiglecH+ and Ly6C+ SiglecH+ BM precDCs are depicted twice. Data are representative of five acceptor mice. (H and I) Representative contour plots of splenic cDCs from Taok3+/+ NICDTg/+ CD11ccre+/+ (Taok3+/+ control), Taok3+/+ NICDTg/+ CD11ccreTg/+ (Taok3+/+ NICDCD11c), Taok3−/− NICDTg/+ CD11ccre+/+ (Taok3−/− control), and Taok3−/− NICDTg/+ CD11ccreTg/+ (Taok3−/− NICDCD11ccre) mice. (J) Frequency of ESAM+ CD4+ cells among cDC2s in spleen. (K) Frequency of CD8α+ cells among cDC1s in spleen. One-way ANOVA with Tukey’s post hoc test (J and K). Data in H–K are representative of two independent experiments (n = 6 to 9 per group). Numbers adjacent to the gates in A, C, H, and I represent the percentage of total cDCs. Bars indicate mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, and ***P < 0.0001.
We next interrogated how Notch signaling might be affected by loss of TAOK3. Expression of the NOTCH2 receptor was intact in Taok3−/− cDCs at protein and messenger RNA levels (SI Appendix, Fig. S6 F and G). Activation of the canonical Notch pathway is regulated by specific proteolytic cleavage events that occur upon ligand binding (46). ADAM10 is a crucial NOTCH2 protease in vivo and has been shown to control the development of marginal zone B cells (34, 35). By analogy with transitional B cells, we wondered whether surface expression of ADAM10 was altered in Taok3−/− DCs or their progenitors. However, despite extensive efforts, we failed to demonstrate detectable levels of specific surface staining of ADAM10 in wild-type DCs or precDCs by flow cytometry (SI Appendix, Fig. S6H). To nevertheless assess a potential role of ADAM10 in the development of Taok3- and Notch2-dependent DCs, we crossed conditional Adam10fl/fl mice to CD11c-cre mice. In the spleen of these Adam10ΔDC mice, ESAM+ CD4+ cDC2s were completely absent (Fig. 4 C and D), and clustering of DCIR2+ cDC2s at the bridging channels was impaired (Fig. 4E). To address whether the role of ADAM10 in cDCs was cell-intrinsic, we generated mixed bone marrow chimeras. We lethally irradiated wild-type mice and reconstituted them with CD45.1 WT and CD45.2 Adam10 ΔDC bone marrow in a 1:1 ratio (Fig. 4F). After reconstitution, we found equal contributions of both donors to the follicular B-cell pool, while MZB cells were preferentially of WT origin (Fig. 4G). This suggests off-target effects of CD11c-cre in MZB cells, a finding supported by the reduced MZB rim in Adam10 ΔDC found on histological examination (Fig. 4E). Along the developmental continuum of cDC2s, a step-up in the WT/Adam10ΔDC ratio was seen from the precDC2 stage to the mature cDC2 stage. An additional step-up was seen in the ratio of ESAM+ CD4+ cDC2s, which were almost exclusively derived from Adam10-sufficient bone marrow. In contrast, mature cDC1s and precDCs were equally derived from wild-type and Adam10ΔDC bone marrow. The defect in ESAM+ CD4+ cDC2s in Adam10ΔDC mice was of functional importance, as the proliferation of OVA-specific CD4+ T cells upon transfusion of HOD-RBCs was significantly reduced (SI Appendix, Fig. S7 A and B). In conclusion, these observations underscore a cell-intrinsic role of ADAM10 in ESAM+ CD4+ cDC2s. Furthermore, the highly similar phenotypes of Taok3-, Adam10-, and Notch2-conditional knockout mice suggest regulation by a common molecular pathway, involved in MZB and cDC2 development in the splenic marginal zone, although the nature of the interaction remains elusive.
Taok3 Is Redundant for the Development of CD103+ CD11b+ cDC2s in the Small Intestine.
It has been reported that NOTCH2 also controls the expression of ESAM in a subset of cDC1s in the spleen and in resident cDC2s in SLN and mesenteric lymph nodes (MLN) (26). Upon careful examination, we confirmed ESAM expression on ±20% of cDC1s in wild-type spleen. Remarkably, this fraction was significantly reduced in Taok3ΔDC mice (SI Appendix, Fig. S8 A and B). Similarly, the percentage of ESAM-expressing cells decreased in resident cDC2s in SLN and MLN (SI Appendix, Fig. S8 C and D). Notch2 conditional knockout mice also lack a subset of cDC2s in the small intestine that express CD103. Given the importance of Taok3 for the development of Notch2-dependent cDC2s in spleen, we further investigated whether Taok3 is equally required for CD103+ CD11b+ cDCs in the small intestine. The percentage of CD103+ cDC2s in mesenteric lymph nodes and in the lamina propria of the small intestine was unaltered in Taok3−/− mice (SI Appendix, Fig. S9 A–C) and Taok3ΔDC mice (SI Appendix, Fig. S9D) compared to littermate controls. Next, we analyzed the contribution of Taok3-sufficient and Taok3−/− bone marrow to intestinal cDC subsets in mixed bone marrow chimeras. The WT:Taok3ΔDC origin ratio for CD103+ CD11b+ cDC2s was not different from the ratio of precDC2s in bone marrow (SI Appendix, Fig. S9E). Together, these data suggest that Taok3 is redundant for the development of CD103+ cDC2s in the small intestine.
Rescue of Taok3−/− Phenotype by NICD Overexpression.
We finally reasoned that if Taok3 controlled NOTCH2 signaling in cDC2s, forced overexpression of the downstream constitutively active notch intracellular domain (NICD) might override the dependency on Taok3. To test this hypothesis, we crossed Rosa26-LSL-NICD mice (47) onto a Taok3+/+ and Taok3−/− background. In these mice, NICD is overexpressed from the Rosa26 locus only after Cre-mediated recombination and removal of a transcriptional stop site. Because the original Taok3−/− mice contained a floxed gene trap, which would also be excised by Cre-mediated recombination, we first had to generate another Taok3 knockout mouse by removing exon 6 with CRISPR-Cas9 technology. We confirmed that the cDC phenotype of these Taok3ex6−/− mice was identical to that of the original Taok3−/− mice (SI Appendix, Fig. S10A). In wild-type littermates, CD11c-cre-driven NICD overexpression increased the fraction of ESAM+ CD4+ and ESAM+ CD4− cDC2s, confirming the role of NOTCH2 signaling in these cDC subsets. Overexpression of NICD in Taok3ex6−/− cDCs significantly rescued the differentiation of cDC2s. In particular, the percentage of cDCs expressing ESAM and CD4 increased more than five-fold (Fig. 4 H and J). In cDC1s, NICD overexpression restored the expression of CD8α to levels comparable to wild-type mice (Fig. 4 I and K). Accordingly, a normal proportion of cDC1s expressed ESAM again in Taok3ex6−/− NICDCD11c mice (SI Appendix, Fig. S10 B and C).
Based on the results from in vivo and in vitro experiments, we conclude that Taok3 controls terminal differentiation of NOTCH2- and ADAM10-dependent cDC2s in the spleen.
Discussion
In contrast to cDC1s, cDC2s harbor striking heterogeneity, determined at least partially by tissue-specific imprints. This allows cDC2s to acquire specific functions adapted to their environment (3, 4, 9, 48). One such factor that dictates tissue-specific differentiation in cDCs is the NOTCH pathway. In the spleen, NOTCH signaling instructs a specific transcriptional program in cDC2s that is essential for their function in capturing and processing blood-borne antigens and presenting them to CD4+ T cells (20, 25–27). However, the NOTCH2 receptor is widely expressed by cDCs, and it is unclear how the NOTCH pathway is selectively activated only in specific subsets of DCs (27). Here, we report a crucial and cell-intrinsic role for the Sterile-20–related kinase TAOK3 in the terminal differentiation of NOTCH2-dependent cDCs.
The function of Taok3 upstream of the Notch2 pathway is supported by several lines of evidence. First, we found that both genes control the development of an identical population of splenic cDC2s that is characterized by high levels of surface ESAM, CD4, and F4/80. Interestingly, loss of Taok3 affected the expression of F4/80 and CD172a across different tissues, implying that some of its effects might extend to all cDC2s. Whether the expression of F4/80 and CD172a is also affected by Notch2 deficiency is not known. Second, Taok3 controlled ESAM expression in a fraction of cDC1s and resident lymph node cDC2s, subpopulations that were selectively ablated in Notch2-deficient mice (26). Using gene set enrichment analysis, we found a significant overlap between the gene expression signatures from Taok3−/− and Notch2−/− cDCs and identified a set of core genes regulated by both Taok3 and Notch2 in cDC1s and cDC2s. Next, Taok3−/− bone marrow generated fewer DC1s and DC2s in an in vitro model of NOTCH-driven dendritic cell differentiation (29). Moreover, surface expression of CD8α and CD205, target genes of the NOTCH pathway, was lower in Taok3−/− cDC1s in vitro and in vivo. Finally, we demonstrated how conditional overexpression of the NICD rescued development of ESAM-expressing cDC1s and cDC2s in Taok3−/− mice, indicating a function of Taok3 upstream of NICD.
Surprisingly, loss of Taok3 did not affect the differentiation of NOTCH2-dependent CD103+ cDC2s in the small intestine. Similarly, Taok3−/− cDC1s did not exactly phenocopy the effect of Notch2 deficiency (20). In contrast to the pronounced and cell-intrinsic deficit of cDC1s in Notch2fl/fl x CD11ccre mice, the number of CD8α+ cDC1s did not significantly decrease in Taok3−/− or Taok3ΔDC mice. In addition, no intrinsic effects were observed in competitive bone marrow chimeras. These findings imply that Taok3 might differentially regulate NOTCH signaling in different subtypes of cDCs. Interestingly, no effect on splenic cDC1s and intestinal CD103+ cDC2s was observed in RBPjκfl/fl x CD11ccre mice, suggesting that in these cells NOTCH2 might signal through a noncanonical pathway that is not controlled by Taok3. Nonredundant roles for noncanonical NOTCH signaling have previously been demonstrated in immune cells, although the mechanisms remain unclear (49–51). To validate the coexistence of both pathways in cDCs, it would be interesting to assess whether RBPjκ controls ESAM expression in cDC1s. If Taok3 regulates only certain aspects of the NOTCH2 transcriptional program, that could explain why key NOTCH target genes such as Hes1 and Deltex1 were not differentially expressed in Taok3−/− cDC2s. Alternatively, these target genes might be expressed only briefly during a specific developmental window before the mature cDC stage. Nevertheless, the fact that Notch2−/− mice are embryonically lethal (52) while Taok3−/− mice are not further supports the idea that Taok3 only controls Notch signaling under highly specific conditions.
The role of Taok3 and NOTCH signaling seems to concentrate on the terminal differentiation of cDCs. The precDC1s and precDC2s numbers increased in the spleen of Taok3−/− mice, implying that the transition to mature cDCs requires Taok3. However, the effects of CD11c-cre–mediated gene trap reversal were more pronounced than those of the later-acting CD11c-cre-GFP, indicating that Taok3 activity in precDCs might already determine their late differentiation. In line with this, NOTCH also controlled target gene expression in ESAM− cDC2s (26). Analogous to our findings in MZB cell development (33), we hypothesized that Taok3 controlled the action of ADAM10 by regulating its translocation to the cell surface. However, we failed to identify a convincing ADAM10 signal on cDCs or precDCs by flow cytometry. The reasons behind this observation remain unclear. It is possible that the surface levels on precDCs are lower than those on transitional B cells or that ADAM10 is more rapidly down-regulated ex vivo. Alternatively, ADAM10 might be expressed only transiently during a brief developmental window. Yet, the drastic reduction in NOTCH2-dependent cDC1s and cDC2s in conditional ADAM10 knockout mice strongly suggests that ADAM10 is the essential NOTCH2 sheddase in DCs. In support of our findings, a role of dendritic cell ADAM10 in the differentiation of cDC2s and the generation of TH2 responses was previously reported by us and others (53, 54). Although off-target effects of the CD11c-driven Cre have been described (55, 56), our mixed bone marrow experiments clearly confirmed the effects of ADAM10 to be cell-intrinsic. Still, it remains unclear whether ADAM10 bioactivity in cDCs is controlled by TAOK3 and how this process is regulated at the molecular level. To advance our understanding of the role of TAOK3 in controlling ADAM10 localization and/or activity in cDCs, the development of additional tools such as reporter mice and in vivo assays for proteolytic activity is warranted.
The capacity of Taok3−/− mice to mount CD4+ T cell responses to transfused allogeneic red blood cells was impaired. This was not caused by an inability of cDC2s to undergo normal maturation and migrate to the T cell zone upon TLR activation. Conceivably, the remaining Taok3−/− cDCs might be unable to process or present antigens efficiently, a conclusion not supported by in vitro findings (25). Alternatively, the defective clustering of cDC2s in marginal-zone bridging channels could limit the exposure of Taok3-deficient cDC2s to antigens. Indeed, the precise location of cDC2 in bridging channels, which is controlled by sphingosine-1-phosphate (S1P) and by gradients of dihydroxycholesterols acting on the chemokine receptor EBI2 (GPR183), (37, 41, 57, 58) is required for efficient T cell priming to circulating particulate antigens (37, 41, 59). It is unclear whether EBI2 signals and Notch instruction are interrelated or simply control different aspects of cDC2 function. It has been reported that Gpr183−/− cDC2s still express ESAM and can be rescued by treatment with an LTβR agonist (37). These findings sharply contrast with the phenotype of Taok3−/− cDCs, implying that the effects of Taok3 cannot be completely explained by interference with the EBI2 pathway. Why CD4+ T cell priming was similarly impaired in Adam10ΔDC mice but not in Taok3ΔDC mice remains unclear. We speculate that some functional collaboration exists between marginal zone B cells, cDC2s, and macrophages in priming the T cell response to allogeneic RBCs. Indeed, MZB cells shuttle circulating antigens to the B- cell follicle (60, 61) and are required for antigen capture and for mounting immune responses to allogeneic RBCs (62, 63). In this regard, the simultaneous reduction in marginal zone B cells and cDC2s observed in Adam10ΔDC mice, which is also observed in full Taok3−/− mice, but not in Taok3ΔDC mice, might explain why T cell responses to RBC are reduced in the former two, but not in the latter, mouse strain. Alternatively, CD4+ T cell priming in Adam10ΔDC mice might be impaired due to defects in ADAM10 function that are not controlled by TAOK3 or that are more pronounced than in Taok3−/− mice.
Taken together, Taok3 controls the terminal differentiation of specialized cDC and B-cell subsets in the marginal zone that depend on ADAM10 and NOTCH2 signaling (33). Whether Taok3 controls ADAM10 activity directly or through indirect effects in DCs remains elusive and requires further investigation.
Materials and Methods
Mice.
The WT C57BL/6J mice were obtained from the Janvier laboratory. Taok3−/− mice were described previously (33). In brief, a loxP-flanked splicing acceptor was inserted as a gene trap in the intronic region between exon 1 and exon 2 of the Taok3 gene, leading to premature transcriptional termination. Conditional Taok3fl/fl mice were generated in-house with the Easi-CRISPR method (64). The guide RNAs (gRNAs) were designed using the CRISPOR webtool to target a region in intron 5 (5′ CCGTCGTTGACTCTGCACAT 3′) and a region in intron 6 (5′ GGAGGCTGAGGCGGAACCAA 3′) in order to introduce loxP sites flanking exon 6 (ENSMUSE00000373662.1) of the Taok3 gene. A long single-stranded DNA repair template containing exon 6, upstream and downstream intronic region with loxP sites inserted, was coinjected with the ribonucleoprotein (RNP) complexes in zygotes of C57BL/6J mice. F1 offspring of germline-transmitting founders were interbred until mice were homozygous for the floxed allele. Taok3−/− mice were generated by injecting C57BL/6J zygotes with RNP complexes with gRNAs targeting a region in intron 5 (5′ GGGTAACTGTGGTGACTTTG 3′) and a region in intron 6 (5′ GGAGGCTGAGGCGGAACCAA 3′), resulting in the deletion of exon 6. Adam10fl/fl mice (35) and HOD mice (65) have been previously described. In brief, HOD mice express a triple-fusion protein under the control of the human β-globin promotor. The fusion protein consists of HEL, a fragment of ovalbumin that contains both OT-I and OT-II–specific epitopes, and the Duffyb blood group antigen. Mb1-cre mice (45) were provided by Michael Reth, Max Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany. CD11c-cre (B6.Cg-Tg [Itgax-cre]1–1 Reiz/J) mice, CD11c-cre-GFP (B6.Cg-Tg[Itgax-cre,-EGFP]4097Ach/J) mice, OTII mice (B6.Cg-Tg[TcraTcrb]425Cbn/J), CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ) mice, and Rosa26-lox-stop-lox-NICD-EGFP mice (Gt[ROSA]26Sortm1(Notch1)Dam/J) were acquired from the Jackson Laboratory. All experimental mice were on a C57BL/6J background and were housed in the VIB-UGent animal facility under specific pathogen-free conditions. Housing conditions entailed individually ventilated cages in a controlled day–night cycle, and food and water intake was ad libitum. Both male and female mice, between 6 to 10 wk of age, were used. Mice were age- and sex-matched and randomly assigned to experimental groups. All animal experiments and procedures were approved by the animal ethics committee of the VIB Center for Inflammation Research and were in accordance with the Belgian animal protection law.
Isolation of Tissue Leukocytes.
Cell suspensions from spleen and bone marrow were obtained through mechanical disruption and filtering over a 70-μm mesh. Lung, liver, and lymph nodes were cut with scissors, digested for 30 min in RPMI-1640 (Gibco) containing 20 μg/mL Liberase TM (Roche) and 10 U/mL DNase I (Roche) at 37 °C, and filtered through a 70-μm nylon sieve. Intestines were soaked in RPMI-1640 with 2% fetal calf serum (Bodinco), opened longitudinally, and cut into 5-mm sections. These segments were incubated twice in phosphate-buffered saline (PBS) containing 2 mM EDTA at 37 °C for 20 min after which the supernatant epithelial cells were discarded. Remaining tissue was digested in RPMI-1640 containing 0.6 mg/mL Collagenase VIII (Sigma) and 10 U/mL DNase I (Roche) at 37 °C with shaking for 15 min and passed through a 40-μm filter afterward. Samples were incubated in ammonium chloride buffer (10 mM KHCO3, 155 mM NH4Cl, 0.1 mM EDTA in milliQ water) for erythrocyte lysis. Total cell counts were obtained by adding counting beads (123count eBeads, Thermo Fisher Scientific).
Flow Cytometry.
Single-cell suspensions were incubated with a fixable viability dye (eFluor506 or eFluor780 from eBioscience) to identify dead cells and with an FcγRII/III antibody (2.4G2) for 30 min to limit aspecific binding. After washing, the cells were incubated for 30 min at 4 °C with a mixture of fluorochrome- or biotin-labeled antibodies. The following antibodies were used: B220 (RA3-6B2), BST2 (120G8), CCR7 (4B12), CD3 (145-2C11), CD4 (GK1.5), CD4 (RM4-5), CD8α (53-6.7), CD11b (M1/70), CD11c (N418), CD19 (1D3), CD21/35 (4E3), CD23 (B3B4), CD24 (M1/69), CD26 (H194-112), CD40 (3/23), CD43 (S6), CD45 (30-F11), CD64 (X54-5/7.1), CD80 (16-10A1), CD86 (PO3), CD93 (AA4.1), CD103 (2-E7), CD135 (A2F10), CD172a (P84), CD205 (205yekta), DCIR2 (33D1), ESAM (1G8), F4/80 (BM8), IgM (II/41), LTβR (3C8), Ly6C (HK1.4 or AL-21), Ly6G (1A8), MHCII (M5/114.15.2), NK1.1 (PK136), Siglec H (ebio440c), Ter119 (TER-119), and XCR1 (ZET). When using biotin-labeled primary antibodies, a second staining step with BV786- or BV605-coupled streptavidin (BD Biosciences) was performed. For intracellular staining of the transcription factors IRF4 and IRF8, cells were fixed using the FoxP3 fixation/permeabilization kit (eBioscience) according to the manufacturer’s protocol. Cells were incubated with PerCP-efluor710–labeled anti-IRF8 (V3GYWCH) or unlabeled goat anti-mouse IRF4 (M-17). Unlabeled IRF4 was detected with an AF647-labeled donkey anti-goat antibody (Invitrogen) in an additional intracellular staining step. Samples were acquired on an LSRFortessa or BD Symphony cytometer with FACSDiva software (BD Biosciences) and analyzed using FlowJo software. Cell sorting was performed using a FACSAria II (BD Biosciences).
FlowSOM.
Splenocytes from Taok3−/− and Taok3+/+ mice were stained and acquired as described above. Automated analysis of flow cytometry samples was done with the FlowSOM algorithm (36). First, the compensated data were manually gated on single, live, Ter119− NK1.1− CD3e− CD19− splenocytes. The remaining cells from all samples were concatenated and assigned to a Self-Organizing Map with a 7 × 7 grid. Similar clusters of cells were thus grouped into 49 nodes. After mapping the data onto the grid, a Minimal Spanning Tree was built that connected all nodes based on similarities in the expression of surface markers. A metaclustering of the nodes into 20 metaclusters was calculated. Subsequently, Taok3−/− and Taok3+/+ samples were mapped individually to the same Self-Organizing Map and Minimal Spanning Tree. Because of the random initialization of the FlowSOM grid, each run of the algorithm will have slightly different results. Therefore, we confirmed that the results of the automated analysis varied only minimally in five runs of the algorithm.
Bone Marrow Chimeras.
Bone marrow cells were obtained from CD45.1 wild-type and CD45.2 Taok3−/− or CD45.2 Adam10fl/fl CD11ccreTg/+ donor mice. CD45.1 × CD45.2 F1 wild-type acceptor mice were irradiated using 9.5 Gy, followed by i.v. injection of 2 × 106 WT cells and 2 × 106 Taok3−/− or Adam10fl/fl CD11ccreTg/+ cells in endotoxin-free PBS at least 4 h after the irradiation. Acceptor mice were euthanized 8 to 10 wk after reconstitution. Bone marrow, spleen, and mesenteric lymph nodes were analyzed for the presence of dendritic cell progenitors and mature cDCs. Marrow reconstitution was validated using the ratio of CD45.1 wild-type to CD45.2 knockout cells in follicular B cells.
Treatment Protocols.
For in vivo maturation and migration assays, mice were i.v. injected with 20 μg LPS in PBS or with PBS only. After 8 h, the mice were killed, and their spleen was analyzed by flow cytometry and confocal imaging. For LTβR agonism in Taok3−/− mice, 100 μg of agonistic rat anti-mouse LTβR antibody (anti-LTβR, clone 4H8, provided by Carl Ware) was injected intraperitoneally on days 0, 3, 7, and 10 followed by euthanasia on day 12. Control mice were injected with PBS only.
Allotransfusion Assay.
OVA-specific naive CD4+ T cells were sorted from the spleen and lymph nodes of OTII mice. The OTII cells were labeled with Cell Proliferation Dye eFluor450 (eBioscience) at 100 μM, according to the manufacturer’s instructions. Approximately 5 × 105 OTII cells were injected i.v. into recipient mice. Erythrocytes were collected from HOD transgenic mice in 12% citrate phosphate dextrose adenine 1 anticoagulant and leukoreduced with a Pall neonatal filter, followed by 4°C storage for 12 d. Before transfusion, the RBCs were washed by centrifugation, and the packed RBCs were diluted 1:15 with sterile PBS. One day after OTII transfer, 100 µL of diluted HOD RBCs was transfused i.v. into recipient mice. Three days after erythrocyte transfer, the spleen and lymph nodes of acceptor mice were harvested and analyzed by flow cytometry.
Confocal Microscopy.
Spleens were harvested, embedded in OCT Tissue Tek medium (Takura), and immediately frozen by liquid nitrogen. Ten-micrometer-thick cryostat sections were fixed for 10 min by paraformaldehyde 2% in PBS, washed with PBS, and permeabilized/blocked with PBS containing 0,1% Triton X-100 (Roche) and 2% bovine serum albumin (Sigma) (PBT buffer) for 30 min. The following antibodies were used: fluorescein isothiocyanate-conjugated CD169 (MOMA-1) was obtained from Serotec Bio-Rad. PE-conjugated DCIR2 (33D1), eFluor660-conjugated CD11c (N418), and eFluor450-conjugated B220 (RA3-6B2) antibodies were obtained from eBioscience. APC-conjugated CD3e (145-2C11) was obtained from BD Biosciences. Sections were stained with the primary antibodies diluted in PBT buffer for 60 min at room temperature. Sections were counterstained with DAPI, washed, and coverslipped with antifading polyvinyl alcohol (Sigma-Aldrich). Images were acquired on a Zeiss LSM710 confocal microscope equipped with 488-, 561-, and 633-nm lasers and with a tunable two-photon laser. Emissions were recorded in three separate channels. Digital pictures at 1,024- × 1,024-pixel density and 8-bit depth were acquired with ZEN software (Zeiss). Images were analyzed with Imaris software.
RNA Extraction and qRT-PCR.
Whole-spleen tissue was homogenized with the Tissuelyser II (Qiagen) prior to RNA extraction using TriPure isolation reagent (Roche). Splenic cDC1s and cDC2s were sorted directly into RNeasy lysis buffer, and RNA was extracted with the RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. The complementary DNA (cDNA) was synthesized with the Sensifast cDNA synthesis kit (Bioline). qRT-PCR reactions were conducted in a LightCycler 480 (Roche) using the SensiFAST sybr no-ROX mix (Bioline). The following primer pairs were used: Gadph (5′ TGGTGC-TTGTCTCACTGACC, 3′ TTCAGTATGTTCGGCTTCCC), Ubc (5′ AAAGCCCCTCAA-TCTCTGGAC, 3′ TGCATCGTCTCTCTCACGGA), Hmbs (5′ GAAACTCTGCTTCGC-TGCATT, 3′ TGCCCATCTTTCATCACTGTATG), Actb (5′ GCTTCTAGGCGGACTGTT-ACTGA, 3′ GCCATGCCAATGTTGTCTCTTAT), Sdha (5′ TTTCAGAGACGGCCATGA-TCT, 3′ TGGGAATCCCACCCATGTT), Taok3 (5′ TTGCATGAAATTGGACATGGGA, 3′:CGATGGTGTTAGGATGCTTCAG), Notch2 (5′ GACTGCCAATACTCCACCTCT, 3′ CCATTTTCGCAGGGATGAGAT). Data were analyzed with qBase+ software (Biogazelle).
Culture of Bone Marrow-Derived Dendritic Cells on OP9-DL1 Cells.
OP9 cell lines transduced with GFP- or DL1-encoding retroviruses were cultured in MEMα medium containing 20% fetal calf serum (FCS, Bodinco), 56 μg/mL gentamycin (Gibco), and 1% GlutaMAX (Gibco) at 37 °C with 5% CO2. Single-cell suspensions of bone marrow were generated by crushing tibias and femurs and filtering them over a 70-μm nylon sieve. After erythrocyte lysis, cells were resuspended in complete RPMI-1640 medium supplemented with 10% FCS (Bodinco), 1,1 mg/mL β-mercaptoethanol, 56 μg/mL gentamycin, 1% GlutaMAX, and 250 ng/mL hFLT3L. The BM cells were plated at 2 × 106 per well in 24-well plates. On day 3 of the primary BM culture, half of the cells from every well were transferred to a single well that contained a monolayer of OP9-GFP or OP9-DL1 cells. OP9 cells were pretreated with 10 μg/mL mitomycin C for 2 h at 37 °C and plated on 24-well plates 1 d before the start of the coculture. Cell cultures were analyzed on day 7.
Microarray and GSEA.
Dendritic cells were sorted from the spleen of Taok3−/− and Taok3+/+ animals by fluorescence-activated cell sorting. Within the conventional dendritic cell gate, cDC1s were defined as XCR1+ CD172a− and cDC2s as XCR1− CD172a+. RNA was extracted using the QIAshredder spin column (Qiagen) and RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol and sent to the nucleomics facility, Flemish Institute for Biotechnology (VIB), Leuven, Belgium, where the microarrays were performed using the GeneChip Mouse Gene 1.0 ST arrays (Affymetrix). Samples were subsequently analyzed using R/Bioconductor. All samples passed quality control, and the Robust Multiarray Average (RMA) procedure was used to normalize data within arrays (probeset summarization, background correction, and log2 transformation) and between arrays (quantile normalization). Only probesets that mapped uniquely to one gene were kept, and for each gene, the probeset with the highest expression level was kept. Differentially expressed genes were identified with the Limma package from Bioconductor, and the resulting P values were corrected for multiple testing with the Benjamini–Hochberg procedure for control of the false-discovery rate. Probe sets with a corrected P value <0.05 and an increase or decrease in expression of at least two-fold were considered differentially expressed. A principal component analysis plot was created using the 15% of genes with the most variable expression. The heatmaps show the relative expression per gene, calculated by subtracting the mean normalized value of each normalized log2 value for every gene. GSEA was performed with software provided by the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp) using the default settings and gene-based permutations with n = 1,000. All transcripts from the above microarray were preranked for cDC1s and cDC2s separately, according to their fold change in Taok3−/− versus Taok3+/+ dendritic cells. Gene sets of differentially expressed genes in Notch2−/− cDC1s and cDC2s were retrieved from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE45681, as published by Satpathy et al. (26).
Statistical Analysis.
Statistical analyses were performed with Graphpad Prism 7.0 (Graphpad Software). For comparisons between two groups, a two-tailed Mann–Whitney U test was used. For comparisons between three or more groups, a one-way ANOVA with Tukey’s correction for multiple comparisons was used. Bonferroni correction was used when testing multiple selected comparisons. The significance level α was 0.05. Error bars represent mean ± SD of the mean. Significant differences are indicated as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Supplementary Material
Acknowledgments
We thank Julie Van Duyse and Gert Van Isterdael from the Flow Core of the Inflammation Research Center for their technical assistance. M. Vanderkerken was supported by a fellowship grant from Research Foundation-Flanders (FWO, Grant 3F023515W). B.M. was supported by a fellowship grant from FWO (Grant 11U0116N). L.V. was supported by a fellowship from the Agency for Innovation through Science and Technology (IWT, Grant 111581). H.H. is supported by a Ghent University Grant (GOA 01G02817). B.N.L. was supported by a European Research Council consolidator Grant 261231, the University of Ghent Multidisciplinary Research Platform (MRP-GROUP-ID), and Ghent University Grant GOA 01G02817.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009847117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Guilliams M., et al. , Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity 45, 669–684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merad M., Sathe P., Helft J., Miller J., Mortha A., The dendritic cell lineage: Ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31, 563–604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durai V., Murphy K. M., Functions of murine dendritic cells. Immunity 45, 719–736 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mildner A., Jung S., Development and function of dendritic cell subsets. Immunity 40, 642–656 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Bosteels C., et al. , Inflammatory type 2 cDCs acquire features of cDC1s and macrophages to orchestrate immunity to respiratory virus infection. Immunity 52, 1039–1056.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabashima K., et al. , Intrinsic lymphotoxin-beta receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity 22, 439–450 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Kamath A. T., et al. , The development, maturation, and turnover rate of mouse spleen dendritic cell populations. J. Immunol. 165, 6762–6770 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Naik S. H., et al. , Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8, 1217–1226 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Eisenbarth S. C., Dendritic cell subsets in T cell programming: Location dictates function. Nat. Rev. Immunol. 19, 89–103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson J. T., et al. , Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages. EMBO J. 30, 2690–2704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashiwada M., Pham N. L., Pewe L. L., Harty J. T., Rothman P. B., NFIL3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development. Blood 117, 6193–6197 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grajales-Reyes G. E., et al. , Batf3 maintains autoactivation of Irf8 for commitment of a CD8α(+) conventional DC clonogenic progenitor. Nat. Immunol. 16, 708–717 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hildner K., et al. , Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sichien D., et al. , IRF8 transcription factor controls survival and function of terminally differentiated conventional and plasmacytoid dendritic cells, respectively. Immunity 45, 626–640 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Aliberti J., et al. , Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood 101, 305–310 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Schlitzer A., et al. , IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 38, 970–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott C. L., et al. , The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J. Exp. Med. 213, 897–911 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Persson E. K., et al. , IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38, 958–969 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Tussiwand R., et al. , Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42, 916–928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis K. L., et al. , Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 35, 780–791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fasnacht N., et al. , Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J. Exp. Med. 211, 2265–2279 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito T., et al. , Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 18, 675–685 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Tanigaki K., et al. , Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 3, 443–450 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Tan J. B., et al. , Lunatic and manic fringe cooperatively enhance marginal zone B cell precursor competition for delta-like 1 in splenic endothelial niches. Immunity 30, 254–263 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Briseño C. G., et al. , Notch2-dependent DC2s mediate splenic germinal center responses. Proc. Natl. Acad. Sci. U.S.A. 115, 10726–10731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satpathy A. T., et al. , Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14, 937–948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caton M. L., Smith-Raska M. R., Reizis B., Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 204, 1653–1664 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balan S., et al. , Large-scale human dendritic cell differentiation revealing notch-dependent lineage bifurcation and heterogeneity. Cell Rep. 24, 1902–1915.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkling M. E., et al. , Notch signaling facilitates in vitro generation of cross-presenting classical dendritic cells. Cell Rep. 23, 3658–3672.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dan I., Watanabe N. M., Kusumi A., The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11, 220–230 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Strange K., Denton J., Nehrke K., Ste20-type kinases: Evolutionarily conserved regulators of ion transport and cell volume. Physiology 21, 61–68 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Delpire E., The mammalian family of sterile 20p-like protein kinases. Pflugers Arch. 458, 953–967 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Hammad H., et al. , Transitional B cells commit to marginal zone B cell fate by Taok3-mediated surface expression of ADAM10. Nat. Immunol. 18, 313–320 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Lambrecht B. N., Vanderkerken M., Hammad H., The emerging role of ADAM metalloproteinases in immunity. Nat. Rev. Immunol. 18, 745–758 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Gibb D. R., et al. , ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J. Exp. Med. 207, 623–635 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Gassen S., et al. , FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Yi T., Cyster J. G., EBI2-mediated bridging channel positioning supports splenic dendritic cell homeostasis and particulate antigen capture. eLife 2, e00757 (2013).23682316 [Google Scholar]
- 38.Witmer M. D., Steinman R. M., The anatomy of peripheral lymphoid organs with emphasis on accessory cells: Light-microscopic immunocytochemical studies of mouse spleen, lymph node, and Peyer’s patch. Am. J. Anat. 170, 465–481 (1984). [DOI] [PubMed] [Google Scholar]
- 39.Schlitzer A., et al. , Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16, 718–728 (2015). [DOI] [PubMed] [Google Scholar]
- 40.De Smedt T., et al. , Regulation of dendritic cell numbers and maturation by lipopolysaccharide in vivo. J. Exp. Med. 184, 1413–1424 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gatto D., et al. , The chemotactic receptor EBI2 regulates the homeostasis, localization and immunological function of splenic dendritic cells. Nat. Immunol. 14, 446–453 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Morelli A. E., et al. , Internalization of circulating apoptotic cells by splenic marginal zone dendritic cells: Dependence on complement receptors and effect on cytokine production. Blood 101, 611–620 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Calabro S., et al. , Bridging channel dendritic cells induce immunity to transfused red blood cells. J. Exp. Med. 213, 887–896 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Briseño C. G., et al. , Deficiency of transcription factor RelB perturbs myeloid and DC development by hematopoietic-extrinsic mechanisms. Proc. Natl. Acad. Sci. U.S.A. 114, 3957–3962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hobeika E., et al. , Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. U.S.A. 103, 13789–13794 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kopan R., Ilagan M. X., The canonical notch signaling pathway: Unfolding the activation mechanism. Cell 137, 216–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murtaugh L. C., Stanger B. Z., Kwan K. M., Melton D. A., Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. U.S.A. 100, 14920–14925 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown C. C., et al. , Transcriptional basis of mouse and human dendritic cell heterogeneity. Cell 179, 846–863.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gentle M. E., Rose A., Bugeon L., Dallman M. J., Noncanonical Notch signaling modulates cytokine responses of dendritic cells to inflammatory stimuli. J. Immunol. 189, 1274–1284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dongre A., et al. , Non-canonical notch signaling drives activation and differentiation of peripheral CD4(+) T cells. Front. Immunol. 5, 54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin H. M., et al. , NOTCH1 can initiate NF-κB activation via cytosolic interactions with components of the T cell signalosome. Front. Immunol. 5, 249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamada Y., et al. , Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development 126, 3415–3424 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Damle S. R., et al. , ADAM10 and Notch1 on murine dendritic cells control the development of type 2 immunity and IgE production. Allergy 73, 125–136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujita K., et al. , Cell-autonomous FLT3L shedding via ADAM10 mediates conventional dendritic cell development in mouse spleen. Proc. Natl. Acad. Sci. U.S.A. 116, 14714–14723 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abram C. L., Roberge G. L., Hu Y., Lowell C. A., Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J. Immunol. Methods 408, 89–100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srinivas S., et al. , Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Czeloth N., et al. , Sphingosine-1 phosphate signaling regulates positioning of dendritic cells within the spleen. J. Immunol. 179, 5855–5863 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Lu E., Dang E. V., McDonald J. G., Cyster J. G., Distinct oxysterol requirements for positioning naïve and activated dendritic cells in the spleen. Sci. Immunol. 2, eaal5237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calabro S., et al. , Differential intrasplenic migration of dendritic cell subsets tailors adaptive immunity. Cell Rep. 16, 2472–2485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arnon T. I., Horton R. M., Grigorova I. L., Cyster J. G., Visualization of splenic marginal zone B-cell shuttling and follicular B-cell egress. Nature 493, 684–688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cinamon G., Zachariah M. A., Lam O. M., Foss F. W. Jr, Cyster J. G., Follicular shuttling of marginal zone B cells facilitates antigen transport. Nat. Immunol. 9, 54–62 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.You Y., et al. , Marginal zone B cells regulate antigen capture by marginal zone macrophages. J. Immunol. 186, 2172–2181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patel S. R., et al. , Marginal zone B cells induce alloantibody formation following RBC transfusion. Front. Immunol. 9, 2516 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quadros R. M., et al. , Easi-CRISPR: A robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 18, 92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Desmarets M., Cadwell C. M., Peterson K. R., Neades R., Zimring J. C., Minor histocompatibility antigens on transfused leukoreduced units of red blood cells induce bone marrow transplant rejection in a mouse model. Blood 114, 2315–2322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.