Not every gene nominated as a cause of human disease stands the test of time. As additional data become available, the evidence supporting the pathogenicity of a particular variant within a gene can be enhanced or diminished.1 The amyotrophic lateral sclerosis (ALS) field, as much as any other, has been hesitant to address these controversies, leading to uncertainty among the research community.
In 2013, we published a study reporting that mutations in the MATR3 gene were a cause of familial ALS.2 That study was based, in part, on a pedigree in which we described p.Phe115Cys as the pathogenic variant based on exome sequence data obtained from 4 affected individuals. An additional member of this kindred (known as USALS#3, member III:10) was recently diagnosed as having ALS. Clinical genetic testing of this individual showed that they did not carry the MATR3 mutation. Although this individual may be a phenocopy, a more parsimonious explanation was that a different mutation was responsible.
To address this issue, we performed whole-genome sequencing of this amyotrophic lateral sclerosis-frontotemporal dementia (ALS-FTD) family on an Illumina NovaSeq6000 sequencer to identify their true causative mutation (figure 1 and table). The participating institutions' institutional review boards approved the study (clinicaltrials.gov/ct2/show/NCT02014246), and informed consent was obtained from all subjects or their surrogate decision makers, according to the Declaration of Helsinki.
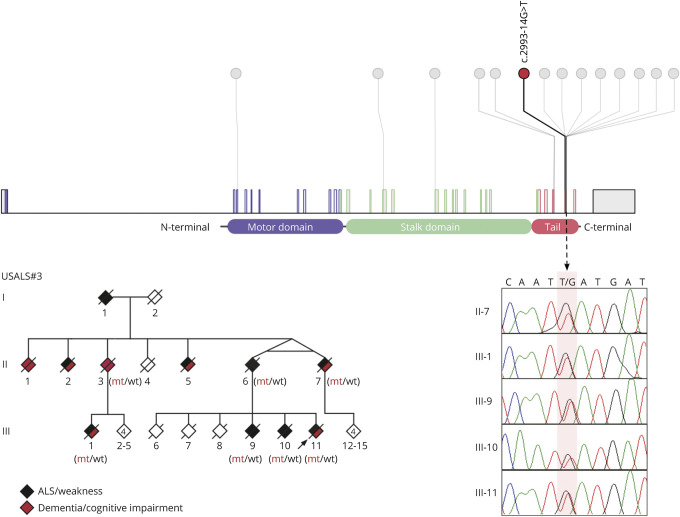
Figure 1. Intronic mutation in KIF5A identified in an ALS-FTD kindred.
The upper panel depicts a lollipop plot of KIF5A depicting the location of the intronic mutation identified in the USALS#3 kindred (red) and known mutations (gray). The chromatograms show mutant alleles from the indicated individuals (mutation is highlighted in pale pink). The pedigree shows the ALS-FTD kindred carrying the KIF5A intronic mutation (updated from ref 5). Individuals II:6, II:7, III:1, III:9, III:10, III:11 were diagnosed with ALS by neurologists. In addition, III:7 had executive dysfunction consistent with behavioral FTD based on formal neuropsychological testing, whereas III:1 and III:11 were observed to have mild cognitive impairment. The phenomenology of other individuals was reconstructed based on history from family members. Mt = mutant alleles; wt = wild-type alleles. Genotypes of presumed obligate carriers are in brackets. Arrow denotes proband. ALS = amyotrophic lateral sclerosis.
Table.
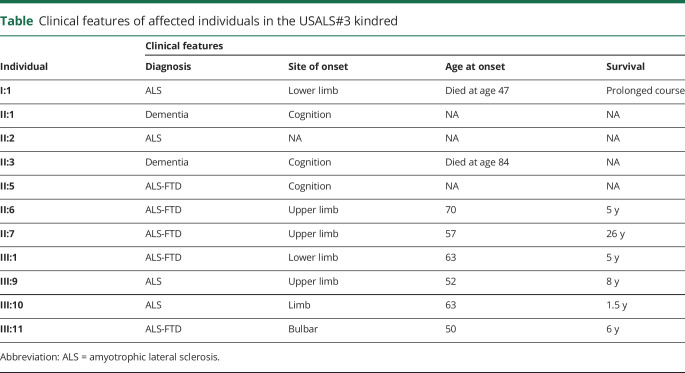
Clinical features of affected individuals in the USALS#3 kindred
Analysis of the sequence data identified 218 variants that were rare and shared across the 5 affected individuals. One variant was located within intron 26 of the KIF5A gene, 14 base pairs from the start of exon 27 (chr12:57582588G>T, build hg38). Exon 27 within KIF5A is a known mutational hotspot underlying familial ALS.3 Exon trap experiments on cDNA obtained from our proband confirmed that this intronic mutation led to aberrant splicing of the KIF5A mRNA transcript. The altered transcript sequence was identical to that produced by other mutations in this intronic region because of skipping of exon 27 (figure 2).3 This family represents the most extensive kindred ascribed to a KIF5A mutation to date, and the affected individuals display both the short survival typically associated with ALS and the prolonged survival previously observed among some patients carrying mutations in this gene (table).3
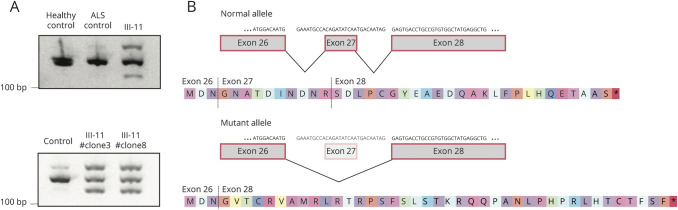
Figure 2. The intronic mutation alters the splicing of KIF5A.
(A) RNA derived from blood (upper panel) from a healthy individual, an ALS patient not carrying the mutation, and individual III-11 carrying the KIF5A intronic mutation. RT-PCR was performed using RNA and previously described primers to amplify a wild-type (155 bp) splice form extending from exon 26 to exon 28.3 An extra band was observed at 127 base pairs indicating aberrant splicing in individual III-11 that was not present in the healthy and disease control subjects. RNA obtained from an IPS cell line (lower panel) derived from fibroblasts of individual III-11 and a control IPS cell line (A18945) showed the same pattern. (B) Sanger sequence analysis of the 127bp transcript/band observed in the patient confirmed the skipping of exon 27 of KIF5A yielding an out of frame and extended disrupted C-terminal peptide sequence.3 ALS = amyotrophic lateral sclerosis.
Discussion
Our previous publication erroneously nominated the p.Phe115Cys variant in MATR3 as the cause of disease within the USALS#3 kindred based on exome sequencing of affected individuals.2 Here, we correct the record to show that an intronic mutation within the known mutational hotspot of KIF5A is the actual cause of disease within this ALS-FTD family. The availability of DNA from an additional affected member within this pedigree was vital to identifying the causative mutation correctly. However, advancements within the genomics field and our understanding of ALS genetics were similarly crucial to resolving this family. In particular, our preexisting knowledge concerning KIF5A allowed us to single out that variant from the list of shared variants.3
Seven members of the kindred (figure 1 and table) developed executive dysfunction during their ALS illness, demonstrating a link between mutations in KIF5A and FTD. Mutations in KIF5A have now been linked to a wide variety of neurodegenerative conditions, including hereditary spastic paraparesis,4 Charcot-Marie-Tooth disease,4 ALS,3 and, more recently, the KIF5A protein has been implicated as having a role in Alzheimer disease.5 These discoveries show the importance of the kinesin protein complex and axonal transport within neurons. Aside from being a striking example of pleiotropy within a single gene, it also suggests that, cumulatively, mutations within KIF5A may be a significant cause of neurologic disease.
Despite our recent findings, we maintain that mutations in MATR3 are a cause of familial ALS. The p.Ser85Cys variant in MATR3 remains the cause of neurologic disease within the other pedigree (USALS#4), segregating with disease among 11 affected members across multiple generations.2 Although there is clear muscle involvement within this family, there is clinical evidence of upper and lower motor neuron involvement.2 MATR3 protein is present within neuronal cytoplasmic inclusions of more than half of sporadic ALS patients,6 and pathogenic MATR3 mutants display neurotoxicity that is mitigated by cytoplasmic redistribution.7 Motor neuron loss and gliosis have been observed within the spinal cords of transgenic mice overexpressing mutant p.Ser85Cys MATR3.8 Finally, there are reports of other MATR3 mutations in patients diagnosed with ALS.9
In conclusion, we identified an intronic mutation in KIF5A that segregated with disease in a large, multigenerational pedigree. Our efforts highlight the rapid advancements that are taking place in our understanding of the genetic architecture of ALS and link mutations in KIF5A to cognitive impairment/frontotemporal dementia.
Appendix. Authors
Study funding
This work was supported by the Intramural Research Programs of the NIH, National Institute on Aging (Z01-AG000949-02). The work was also funded by the Packard Center for ALS Research at Hopkins, the ALS Association, the Muscular Dystrophy Association, the Italian Ministry of Health (RF-2016-02362405), the Italian Ministry of Education, University and Research (Progetti di Ricerca di Rilevante Interesse Nazionale, PRIN, grant n. 2017SNW5MB), the Joint Programme - Neurodegenerative Disease Research (JPND, Brain-Mend projects) granted by Italian Ministry of Education, University and Research, and by the European Community's Health Seventh Framework Programme (FP7/2007–2013, grant agreements no. 259867 and 278611), by the National Institute of Neurologic Disorders and Stroke (NIH grant number R35 NS097261), and by the Collaborative Health Initiative Research Program. This study was performed under the Department of Excellence grant of the Italian Ministry of Education, University and Research to the “Rita Levi Montalcini” Department of Neuroscience, University of Torino, Italy. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH, Bethesda, Maryland (hpc.nih.gov). The institutional review boards of participating institutions approved the study (NIH, 03-AG-N329), and informed consent was obtained from all subjects or their surrogate decision-makers, according to the Declaration of Helsinki.
Disclosure
S. Saez-Atienzar—Reports no disclosures. C.L. Dalgard—Reports no disclosures. J. Ding—Reports no disclosures. A. Chiò—Scientific Advisory Board Member for: Biogen Idec, Cytokinetics, Italfarmaco and Mitsubishi Tanabe, and editorial board member for the journal Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. C. Alba—Reports no disclosures. D.N. Hupalo—Reports no disclosures. M.D. Wilkerson—Reports no disclosures. R. Bowser—founder of Iron Horse Diagnostics, Inc, a company commercialising assays for ALS and other neurologic diseases. E.P. Pioro—Scientific Advisory Board Member for: Biohaven Pharmaceuticals, Cytokinetics, Inc., ITF Pharma, Inc., MT Pharma America, Inc., and Consultant to: Avanir Pharmaceuticals, Inc., Cytokinetics, Inc., ITF Pharma, Inc., MT Pharma America, Inc., Neurotherapia, Inc., and Otsuka America, Inc. R. Bedlack—Scientific Advisory Board Member for: Brainstorm Cell DSMB, Mallinkrodt DSMB, Biogen DSMB, editorial board member for the journal Amyotrophic Lateral Sclerosis and Frontotemporal Dementia, and consultant for the ALS Association, Biohaven, ITF Pharma, MT Pharma America, New Biotic Inc, Woolsey Pharma, Leerink Swan and Company, Easton Associates, and the Gerson Lehman Group. B.J. Traynor—holds US, EU and Canadian patents on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72, and editorial board member for the journals JAMA Neurology, Journal of Neurology, Neurosurgery and Psychiatry, and Neurobiology of Aging. Go to Neurology.org/N for full disclosures.
References
- 1.Wang J, Shen Y. When a “disease-causing mutation” is not a pathogenic variant. Clin Chem 2014;60:711–713. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JO, Pioro EP, Boehringer A, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci 2014;17:664–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicolas A, Kenna KP, Renton AE, et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 2018;97:1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu YT, Laura M, Hersheson J, et al. Extended phenotypic spectrum of KIF5A mutations: from spastic paraplegia to axonal neuropathy. Neurology 2014;83:612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Tian J, Chen H, Du H, Guo L. Amyloid beta-mediated KIF5A deficiency disrupts anterograde axonal mitochondrial movement. Neurobiol Dis 2019;127:410–418. [DOI] [PubMed] [Google Scholar]
- 6.Tada M, Doi H, Koyano S, et al. Matrin 3 is a component of neuronal cytoplasmic inclusions of motor neurons in sporadic amyotrophic lateral sclerosis. Am J Pathol 2018;188:507–514. [DOI] [PubMed] [Google Scholar]
- 7.Malik AM, Miguez RA, Li X, Ho YS, Feldman EL, Barmada SJ. Matrin 3-dependent neurotoxicity is modified by nucleic acid binding and nucleocytoplasmic localization. Elife 2018;7:e35977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Yamashita S, Hara K, et al. Mutant MATR3 mouse model to explain multisystem proteinopathy. J Pathol 2019;249:182–192. [DOI] [PubMed] [Google Scholar]
- 9.Leblond CS, Gan-Or Z, Spiegelman D, et al. Replication study of MATR3 in familial and sporadic amyotrophic lateral sclerosis. Neurobiol Aging 2016;37:209.e17–e209.e21. [DOI] [PubMed] [Google Scholar]