By using a degron system, Zhang and Seemann find that acute depletion of GRASP55 and 65 does not affect Golgi stacking but indirectly compromises the integrity of the Golgi ribbon through loss of vesicle transport components.
Abstract
GRASP55 and GRASP65 have been implicated in stacking of Golgi cisternae and lateral linking of stacks within the Golgi ribbon. However, RNAi or gene knockout approaches to dissect their respective roles have often resulted in conflicting conclusions. Here, we gene-edited GRASP55 and/or GRASP65 with a degron tag in human fibroblasts, allowing for induced rapid degradation by the proteasome. We show that acute depletion of either GRASP55 or GRASP65 does not affect the Golgi ribbon, while chronic degradation of GRASP55 disrupts lateral connectivity of the ribbon. Acute double depletion of both GRASPs coincides with the loss of the vesicle tethering proteins GM130, p115, and Golgin-45 from the Golgi and compromises ribbon linking. Furthermore, GRASP55 and/or GRASP65 is not required for maintaining stacks or de novo assembly of stacked cisternae at the end of mitosis. These results demonstrate that both GRASPs are dispensable for Golgi stacking but are involved in maintaining the integrity of the Golgi ribbon together with GM130 and Golgin-45.
Introduction
The Golgi apparatus is a membrane-bound organelle essential for processing and sorting of secretory and membrane proteins. As the central module of the exocytic pathway, it receives proteins and lipids from the endoplasmic reticulum, post-translationally modifies them, and sorts them to their appropriate cellular destinations (Pantazopoulou and Glick, 2019). This secretory function of the Golgi is highly conserved among eukaryotes and is performed by stacks of flattened, disk-shaped cisternae. Concentrating cisternae into spatially proximate stacks enables efficient cargo sorting as well as post-translational modifications such as glycosylation (Lowe, 2011). Most vertebrate cells contain 100–200 stacks, each comprised of four to six cisternae (Storrie et al., 2012; Sin and Harrison, 2016). In addition, these stacked cisternae are laterally interconnected into a single, contiguous, ribbon-like entity that is positioned next to the centrosomes in the perinuclear region (Lowe, 2019). By joining adjacent cisternae between stacks, the Golgi can accommodate and process large secretory cargos such as collagen or Weibel-Palade bodies that do not fit into individual cisternae (Ferraro et al., 2014; Lavieu et al., 2014). On the other hand, the concentration and polarized positioning of the Golgi ribbon within the cells facilitates directional delivery of cargo and signaling molecules and is thereby indispensable for cell polarization and differentiation (Wei and Seemann, 2017).
Despite the characteristic organization and unique morphological appearance, Golgi membranes are highly dynamic and are constantly remodeled to accommodate extensive vesicular trafficking through the stacks (Lowe, 2011). An imbalanced flux of membranes through the Golgi leads to fragmentation of the Golgi ribbon, which is commonly observed under pathological conditions (Wei and Seemann, 2017; Makhoul et al., 2019). Physiologically, the disassembly process is harnessed during mitosis, when a block of vesicle fusion initiates the conversion of the Golgi ribbon into vesicles. Disassembly of the ribbon is not only required for mitotic progression (Sütterlin et al., 2002; Colanzi et al., 2007; Guizzunti and Seemann, 2016), but it also enables Golgi partitioning by the spindle into the daughter cells, where vesicles fuse to reform a ribbon of stacked cisternae (Shorter and Warren, 2002; Wei and Seemann, 2009a). Such dramatic structural rearrangements raise tremendous interest in investigating the proteins and molecular mechanisms underlying Golgi stacking and lateral linking.
The highly dynamic and unique structure of the Golgi critically depends on structural Golgi proteins, which jointly act as a scaffold or matrix to support its morphology and functions (Ramirez and Lowe, 2009; Gillingham and Munro, 2016). These structural Golgi proteins are peripherally associated with the cytoplasmic face of the Golgi membrane and are categorized into two major protein families: GRASPs (Golgi reassembly and stacking proteins) and Golgins. GRASPs, composed of GRASP55 and GRASP65 (referred to as GRASP55 and 65 for brevity), were first identified by their roles in postmitotic stacking of cisternae in a cell-free system (Barr et al., 1997; Shorter et al., 1999). On the other hand, Golgins such as GM130, Golgin-97, Golgin-84, and Golgin-45, are long rod-like coiled-coil proteins that act as vesicle tethers at distinct cisternae to facilitate cargo transport through the stack (Gillingham and Munro, 2016). Notably, members of the GRASP and Golgin families form distinct subcomplexes with each other. For example, GRASP65 is recruited to the cis-Golgi by GM130 while GRASP55 and Golgin-45 form a complex at medial/trans-Golgi cisternae (Barr et al., 1998; Short et al., 2001). Furthermore, GRASP55 and GRASP65 also directly participate in vesicle transport through the Golgi by binding to p24 cargo receptors (Barr et al., 2001).
The role of GRASP55 and GRASP65 in establishing and maintaining the integrity of the Golgi ribbon is thought to depend on their ability to physically bridge apposing cisternae. Both GRASPs share a highly homologous GRASP domain at their N-terminal domains, which allows each GRASP to assemble into anti-parallel homo-oligomers in trans (Wang et al., 2005; Feng et al., 2013). Oligomerization of GRASP65 is sufficient to hold adjacent membrane compartments together, as demonstrated by clustering of mitochondria induced by expression of GRASP65 fused to a mitochondria targeting sequence (Bachert and Linstedt, 2010). Based on their membrane-linking capacity, GRASP55 and 65 have been implicated in packing cisternae into stacks as well as in lateral linking of adjacent stacks into a ribbon. During mitosis, phosphorylation of GRASP55 and 65 breaks the trans-oligomers, which coincides with lateral unlinking of the ribbon and unstacking of the cisternae (Wang et al., 2003; Tang et al., 2010). However, the precise function of GRASPs in organizing the Golgi structure in vivo is still unclear.
In efforts to dissect the contributions of GRASPs to Golgi stacking and ribbon linking, several RNAi and gene knockout-based studies have been conducted in both cellular and animal models, which led to inconsistent results and conclusions. Down-regulation of either GRASP55 or 65 by RNAi only induced Golgi fragmentation or ribbon unlinking (Sütterlin et al., 2005; Duran et al., 2008; Feinstein and Linstedt, 2008; Puthenveedu et al., 2006), while others reported a loss of cisternae (Xiang and Wang, 2010; Bekier et al., 2017). Similarly, double suppression of GRASP55 and 65 by RNAi or CRISPR-mediated gene inactivation disrupted stacks and vesiculated cisternae (Xiang and Wang, 2010; Bekier et al., 2017), while other studies found no effect on stacking upon RNAi and in knockout mice (Lee et al., 2014; Grond et al., 2020).
One key limitation of these RNAi or gene knockout–based approaches is that protein levels are gradually reduced over an extended period of time. Typically, it takes 2–3 d or sometimes up to weeks to efficiently eliminate protein expression. As such, chronic depletion can lead to complex phenotypes that are indirect and difficult to interpret. In addition, long-term loss of critical functions provides cells ample time to adapt and skew the phenotypes by activating compensatory pathways (Rothman, 2010). To avoid such chronic complications, we set out to determine the immediate effects caused by acute loss of GRASP55 and 65 at the protein level. To this end, we employed the auxin-inducible degron (AID) system (Nishimura et al., 2009; Holland et al., 2012) to acutely degrade GRASP55 and/or 65 within 2 h. Our results show that simultaneous depletion of both GRASPs, but not individual depletion, displaces GM130 and p115 from the Golgi and indirectly affects lateral linking of the Golgi ribbon. Notably, consistent with the ribbon unlinking effect reported for RNAi (Feinstein and Linstedt, 2008; Xiang and Wang, 2010), long-term degradation of GRASP55 also disrupts the Golgi ribbon, while short-term degradation does not. Moreover, acute elimination of GRASP55 and/or GRASP65 does not affect Golgi stacking during interphase or in cells that reassembled a Golgi ribbon at the end of mitosis.
Results
Generation of SV589 cell lines endogenously expressing degron-tagged GRASP55 and/or 65
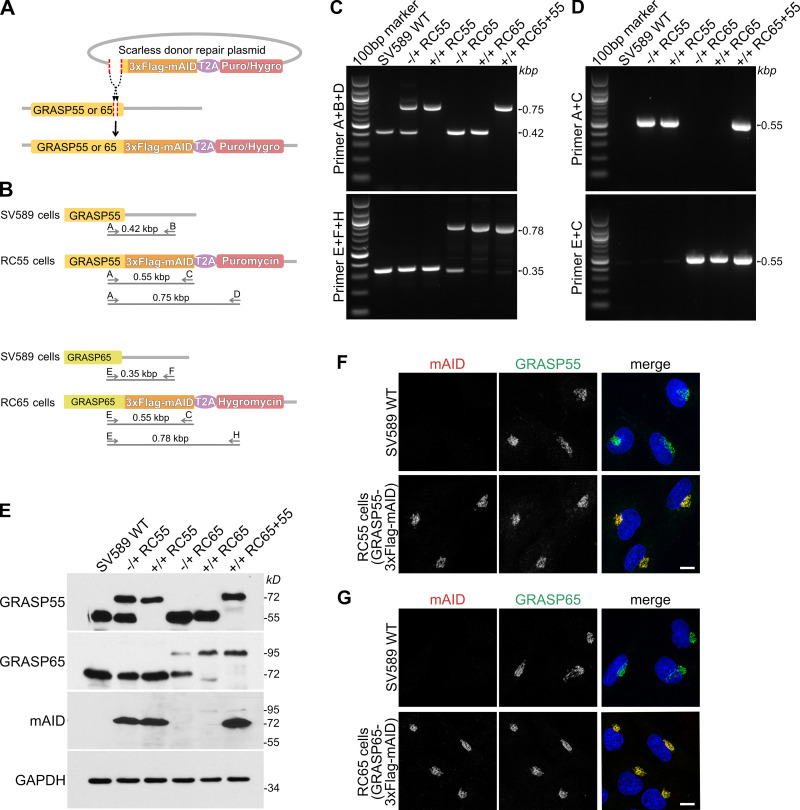
To study the cellular effects caused by acute loss of GRASP55 and 65, we employed AID technology. When cells are incubated with the small molecule auxin, proteins with an AID tag are rapidly degraded by the proteasome (Nishimura et al., 2009; Holland et al., 2012). Degradation of AID-tagged proteins requires expression of the auxin perceptive F-box protein TIR1 from Oryza sativa (OsTIR1), which forms a functional E3 ubiquitin ligase complex with endogenous SCF (Skp1–Cullin–F-box) of the host cells. When added to cells, the auxin indole-3-acetic acid (IAA) binds to TIR1, leading to polyubiquitination and acute proteolysis of target proteins tagged with the minimal AID (mAID) sequence of 68 amino acids (Kubota et al., 2013). To create homozygous cell lines where endogenous GRASP55 and/or 65 are tagged with mAID (hereafter referred to as RC55, RC65, and RC65+55 cell lines), we employed the CRISPaint (CRISPR-assisted insertion tagging) system (Schmid-Burgk et al., 2016). Using this approach, a double-strand break is induced in front of the stop codon, where a repair sequence is then integrated. This exchanges the endogenous stop codon with the 3xFlag epitope tag (to facilitate detection), followed by the mAID degron, a T2A self-cleaving peptide, and an antibiotic resistance gene (Fig. 1, A and B). We successfully established immortalized SV589 human fibroblast cell lines that endogenously express either GRASP55-3xFlag-mAID (RC55), GRASP65-3xFlag-mAID (RC65), or both (RC65+55, generated by endogenous tagging GRASP55 with 3xFlag-mAID in RC65 cells).
Figure 1.
Generation of cell lines expressing endogenous GRASP55 and/or 65 tagged with mAID. (A) Scheme for tagging the endogenous loci of GRASP55 or 65 with 3xFlag and mAID. The DNA sequence encoding 3xFlag-mAID separated by a self-cleaving T2A peptide from the resistance gene (puromycin [Puro] or hygromycin [Hygro]) was inserted by CRISPR-Cas9 gene editing in front of the stop codon of GRASP55 or 65. The amino acids between the insert location and the stop codon were scarlessly repaired by adding the sequence in front of 3xFlag-mAID of the repair donor plasmid. (B) Scheme of the primer sets used for genotyping gene-edited cell lines. The cell line with GRASP55 scarlessly tagged with 3xFlag-mAID and puromycin is referred to as RC55, GRASP65 scarlessly tagged with 3xFlag-mAID and hygromycin as RC65, and both GRASP55 and 65 tagged with 3xFlag-mAID and indicated resistance genes as RC65+55. (C and D) Genomic PCR to genotype heterozygous (−/+) and homozygous (+/+) RC55, RC65, and RC65+55 cell lines for GRASP55 tagged with 3xFlag-mAID or GRASP65-3xFlag-mAID. Parental SV589 cells are shown as negative control (WT). (E) Immunoblot analysis of whole cell lysates of heterozygous (−/+) and homozygous (+/+) RC55, RC65, and RC65+55 cells using antibodies against GRASP55, GRASP65, the degron mAID tag, and GAPDH. (F) SV589 cells and RC55 cells expressing GRASP55-3xFlag-mAID were immunostained for mAID (red) and GRASP55 (green) and labeled for DNA (blue). Scale bar, 10 μm. (G) SV589 cells and RC65 cells stably expressing GRASP65-3xFlag-mAID were immunostained for mAID (red) and GRASP65 (green) and labeled for DNA (blue). Scale bar, 10 μm.
We identified heterozygous and homozygous clones by genomic PCR using forward primers targeting the 3′-region of the GRASP55 or 65 coding sequence (primers A and E) and reverse primers binding to the 3′-UTRs of each GRASP (primers B and F), the mAID sequence (primer C), or the antibiotic resistance gene (primers D and H; Fig. 1, B–D). We then performed Western blotting analysis to confirm that the cell lines expressed mAID-tagged GRASP proteins. Compared with parental SV589 cells, the bands recognized by antibodies against GRASP55 or 65 were upshifted (corresponding to mAID and Flag tag) in RC55 and RC65 cells and were also detected by an anti-mAID antibody (Fig. 1 E). Of note, the mAID band corresponding to GRASP65-3xFlag-mAID at 95 kD was much weaker than the GRASP55-3xFlag-mAID signal at 72 kD, indicating that endogenous protein levels of GRASP55 are much higher than those of GRASP65. This result is consistent with quantitative proteomics showing that the protein copy number of GRASP55 in HeLa cells is 45-fold higher than that of GRASP65 (Kulak et al., 2014). In accordance with the Western blotting results, our immunofluorescence analysis revealed that mAID was only detected in RC55, RC65, or RC65+55 cells, but not in parental SV589 cells (Fig. 1, F and G; and Fig. S1 C). Furthermore, the staining for mAID and GRASP55 or 65 was localized to the Golgi ribbon in the perinuclear region, demonstrating that the C-terminal 3xFlag-mAID-tag does not alter Golgi targeting of the GRASP proteins (Fig. 1, F and G).
Figure S1.
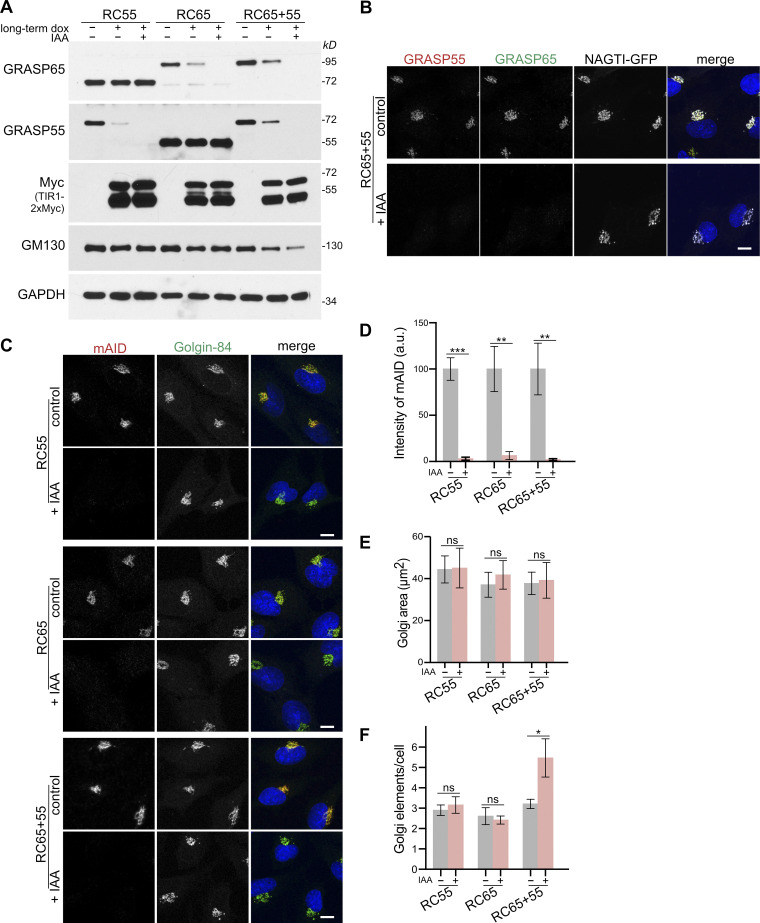
Long-term TIR1 expression in the absence of auxin leads to partial loss of GRASP55 and 65. (A) RC55, RC65, and RC65+55 cells were treated with doxycycline to induce TIR1 expression for 20 h (long-term dox) before IAA was added for a further 2 h. Cell lysates were subjected to immunoblotting with indicated antibodies. (B) RC65+55 cells stably expressing the Golgi enzyme NAGTI-GFP were treated with doxycycline for 6 h and IAA for an additional 2 h. Fixed cells were immunostained for GRASP55 (red) and GRASP65 (green) and labeled for DNA (blue). Scale bar, 10 µm. (C) RC55, RC65, and RC65+55 cells were treated with doxycycline for 6 h and with IAA for a further 2 h as in B and immunolabeled for mAID (red) and Golgin-84 (green) and stained for DNA (blue). Scale bar, 10 µm. (D) Quantification of the fluorescence signal of mAID in the Golgi region marked by Golgin-84 from C. n = 3 independent experiments with >50 cells analyzed per experiment and condition. ** P < 0.01; *** P < 0.001. Error bars represent mean ± SD. (E) Quantitation of the Golgi area marked by Golgin-84 from C. n = 3 independent experiments with an average number of cells analyzed per experiment: RC55 cells: control: n = 58 cells, +IAA: n = 50; RC65 cells: control: n = 66, +IAA: n = 84; RC65+55 cells: control: n = 74, IAA: n = 58. ns, not significant. Error bars represent mean ± SD. (F) Quantitation of the number of Golgi elements labeled by Golgin-84 from C. n = 3 independent experiments with an average cell number analyzed per experiment: RC55 cells: control: n = 60 cells, +IAA: n = 53; RC65 cells: control: n = 86, +IAA: n = 89; RC65+55 cells: control: n = 64, IAA: n = 70. * P < 0.05; ns, not significant. Error bars represent mean ± SD.
GRASP55 and 65 are acutely degraded within 2 h upon auxin induction
To enable acute degradation of mAID-tagged proteins, we stably introduced myc-tagged and codon-optimized OsTIR1 (TIR1-2xMyc) into homozygous RC55, RC65, and RC65+55 cells. In line with previous studies (Natsume et al., 2016), we noticed that constitutive expression of TIR1 reduced the protein levels of mAID-tagged proteins even in the absence of IAA (data not shown). To circumvent potential side effects caused by chronic basal degradation, we generated RC55, RC65, and RC65+55 cell lines expressing TIR1-2xMyc under doxycycline control to precisely induce its timely expression. Since only homozygous RC55, RC65, and RC65+55 cells stably expressing inducible TIR1-2xMyc were used for further experiments, for simplicity we hereafter refer to them as RC55, RC65, and RC65+55 cells. We first compared the stability of mAID-tagged GRASP55 and 65 after short-term 6-h induction or long-term 20-h induction of TIR1. 20-h TIR1 expression in the absence of IAA significantly decreased GRASP55 and 65 levels (Fig. S1 A). By contrast, 6-h TIR1 induction showed only minimal to no basal degradation of GRASP55 and 65 (Fig. 2 A). Subsequent treatment with IAA led to complete degradation of both GRASPs within 2 h but did not change the protein levels of the Golgi resident protein Golgin-84 or the GM130-associated tethering factor p115 (Fig. 2 A and Fig. S1 A).
Figure 2.
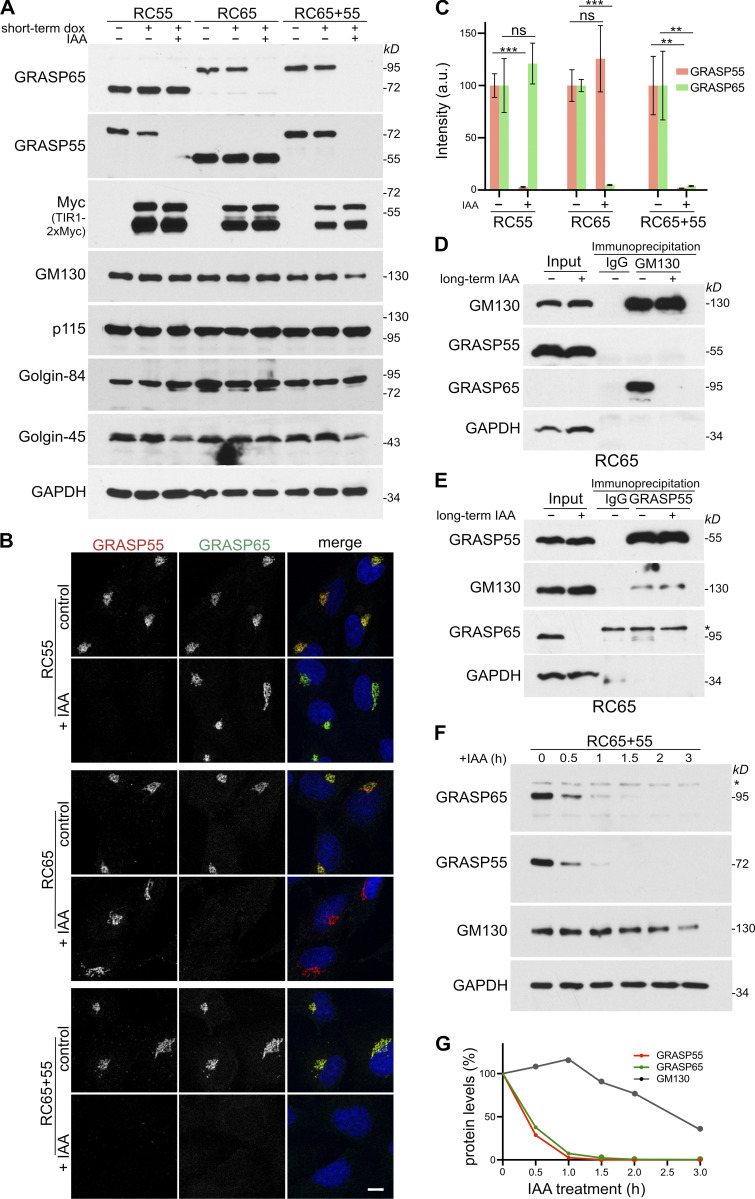
The auxin IAA induces rapid depletion of mAID-tagged GRASP55 and 65. (A) Homozygous RC55, RC65, and RC65+55 cells inducibly expressing TIR1-2xMyc were treated with doxycycline for 6 h (short-term dox) and then with the auxin IAA for a further 2 h to degrade GRASP55 and/or 65. Cell lysates were immunoblotted with the indicated antibodies. (B) RC55, RC65, and RC65+55 cells were treated with doxycycline for 6 h and with IAA for a further 2 h as in A. Cells were then immunolabeled for GRASP55 (red) and GRASP65 (green) and stained for DNA (blue). Scale bar, 10 µm. (C) Quantitation of the fluorescence signal of GRASP55 and 65 in the Golgi region from B for RC55, RC65, and (S1B) for RC65+55. n = 3 independent experiments with >50 cells analyzed per experiment and condition. ** P < 0.01; *** P < 0.001; ns, not significant. Error bars represent mean ± SD. (D and E) GRASP55 does not compensate for the loss of GRASP65 to stabilize GM130. RC65 cells were treated with doxycycline for 6 h before adding IAA for 2 d (long-term IAA) to deplete GRASP65. GM130 (D) and GRASP55 (E) were then immunoprecipitated from the cell lysates and analyzed by Western blotting. * denotes unspecific band. (F and G) Simultaneous degradation of GRASP55 and 65 causes a delayed reduction of GM130 levels. RC65+55 cells expressing TIR1 for 6 h were treated with IAA. Cell lysates were collected at the indicated time points and subjected to Western blotting with the indicated antibodies. * denotes unspecific band. The intensities of each band are shown in G.
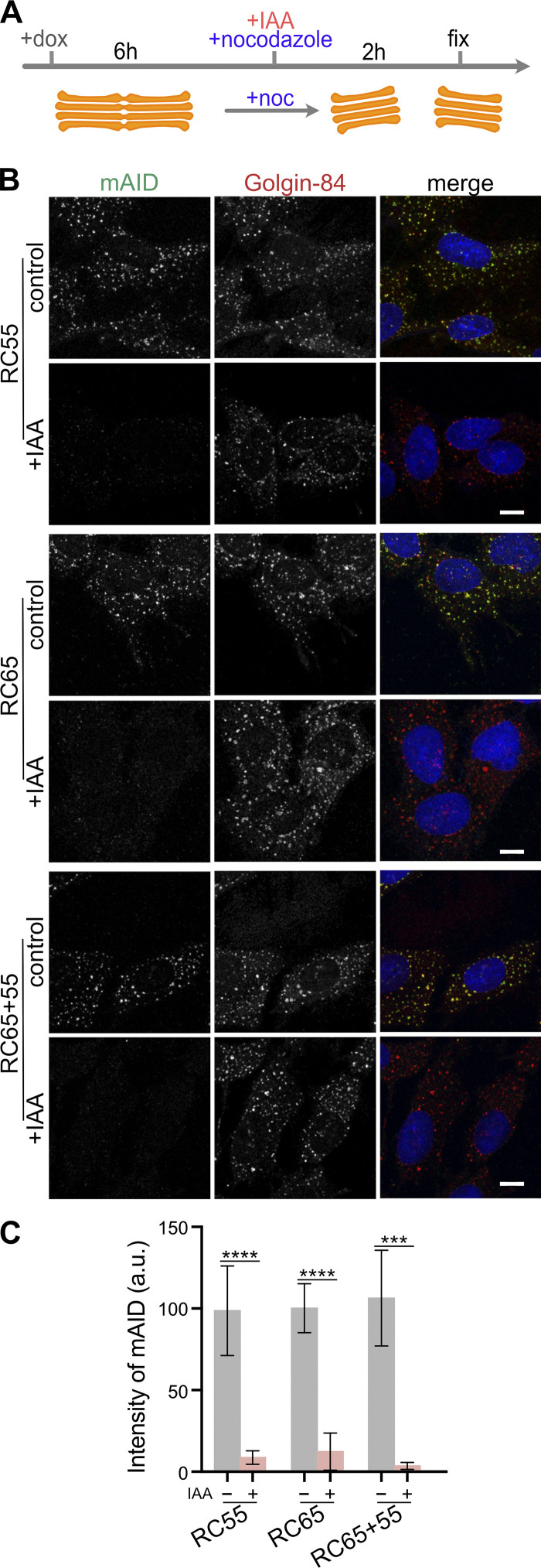
Degradation of GRASP55 and 65 was further corroborated by means of immunofluorescence microscopy. Short-term GRASP55 elimination did not alter the GRASP65 signal on the Golgi and vice versa (Fig. 2, B and C), and depletion of both GRASPs had no effect on the localization of the GFP-tagged Golgi enzyme N-acetylglucosaminyl transferase I (NAGTI-GFP; Fig. S1 B). Furthermore, after 2-h IAA treatment, the mAID signal was lost from the Golgi while the perinuclear localization of Golgin-84 remained (Fig. S1, C and D). GRASP depletion did not change the size of the Golgi area (Fig. S1 E), but double degradation of both GRASPs increased the number of Golgi elements from 3.2 to 5.5 (Fig. S1 F). Together, these results indicate that acute depletion of GRASP55 and 65 does not disperse the Golgi ribbon, suggesting that neither GRASP55 nor 65 is required for maintaining the perinuclear Golgi morphology.
Next, we determined if degradation of GRASP55 and 65 affects the stability of their respective binding partners. Both GRASP proteins form tight complexes via their PDZ domains with members of the Golgin protein family. Specifically, GRASP55 binds to Golgin-45, a medial-Golgin that facilitates secretory protein transport (Short et al., 2001), while GRASP65 interacts with GM130, a cis-Golgi matrix protein that mediates vesicle docking by forming a complex with p115 (Barr et al., 1998). Whether degradation of GRASP55 and 65 affects the stability of Golgin-45 and GM130 was tested by Western blotting. While Golgin-45 was notably reduced by degradation of GRASP55 (Fig. 2 A), GM130 was not affected when either GRASP55 or 65 was depleted, showing that GRASP65 is not degraded in a complex with GM130. However, GM130 levels decreased when both GRASPs were simultaneously eliminated (Fig. 2, A and F), which was also observed after chronic down-regulation by siRNA (Xiang and Wang, 2010; Lee et al., 2014). One possible explanation is that once GRASP65 is degraded, GRASP55 compensates for the loss of GRASP65 and binds via its GRASP domain to stabilize GM130, which was reported for in vitro translated GRASP55 and GM130 (Shorter et al., 1999). To test if GRASP55 interacts with GM130 in cells, we degraded GRASP65 in RC65 cells for 2 d and then immunoprecipitated GM130 or GRASP55. GRASP65 copelleted with GM130 from control lysates as reported (Barr et al., 1997), but GRASP55 was neither pulled down by GM130 from control cells nor from GRASP65-depleted lysates (Fig. 2 D). As previously reported (Short et al., 2001), we also found a minor fraction of GM130 coimmunoprecipitated with GRASP55, but GRASP55 was below the detection limit in GM130 immunoprecipitates (Fig. 2, D and E). However, no increase in the amount of GM130 pulled down by GRASP55 was detected after GRASP65 was depleted, demonstrating that GRASP55 does not compensate for the loss of GRASP65 to stabilize GM130.
To further evaluate GM130 stability upon acute depletion of GRASP55 and 65, we followed the degradation over a time course (Fig. 2, F and G). Protein levels of GRASP55 and 65 in IAA-treated RC65+55 cells were significantly decreased within 30 min, and only trace amounts remained after 1 h. On the other hand, GM130 levels appeared to be unaffected after 1 h but started to decrease by 1.5 h and were significantly diminished after 3 h. The delay of GM130 loss compared with GRASP55 and 65 loss suggests that the stability of GM130 was indirectly compromised by the simultaneous loss of GRASP55 and 65. Considered together, the data shown in Fig. 2 suggest that induced degradation is highly efficient in our system and that degradation of GRASP55 and/or 65 affects the stability of Golgin-45 and GM130 but does not change the overall morphology of the perinuclear Golgi ribbon.
Acute double degradation of GRASP55 and 65 partially displaces GM130 and p115 from the Golgi
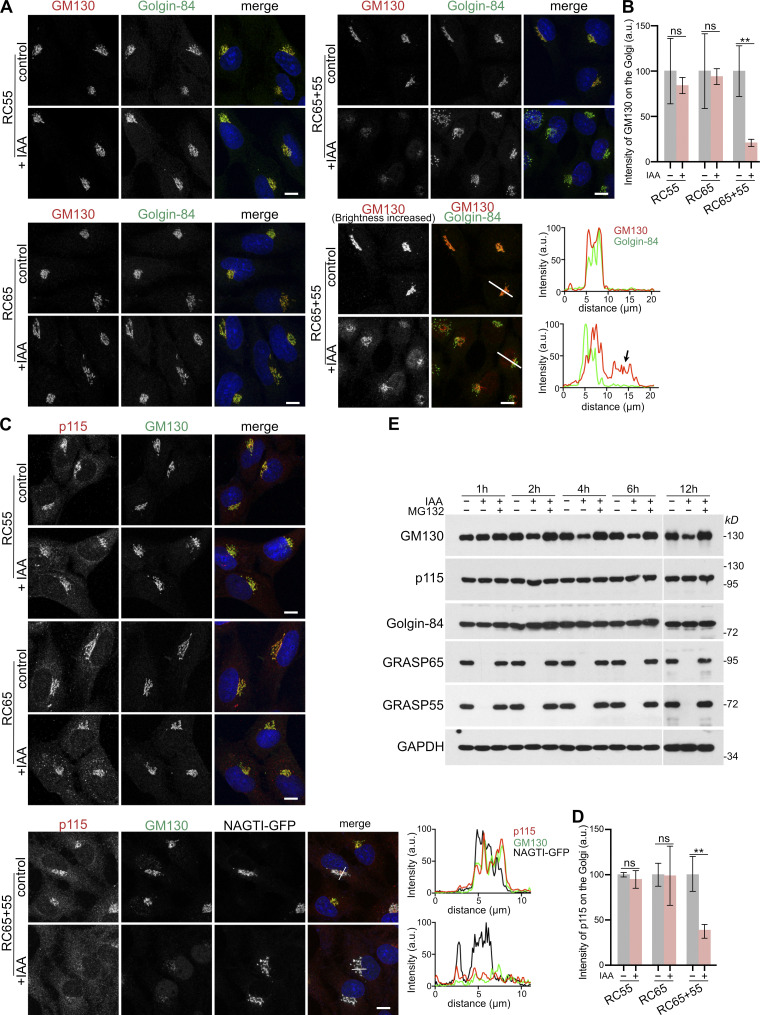
Because depletion of GRASP55 and 65 coincided with a time-delayed reduction of GM130 levels, we analyzed if it also alters the localization of GM130. Cells were treated with IAA for 2 h to degrade GRASP55 and/or 65 and then double-stained for GM130 and another cis-Golgi marker, Golgin-84. GM130 and Golgin-84 colocalized at the perinuclear Golgi ribbon in control cells, which was not changed after the degradation of either GRASP55 or 65 (Fig. 3 A). By contrast, in cells depleted of both GRASPs, the overlapped intensity of GM130 with Golgin-84 was reduced and GM130 was partially displaced from the perinuclear Golgi ribbon (Fig. 3, A and B). In line with our previous report (Wei et al., 2015), Golgi-displaced GM130 relocalized to the nucleus via its N-terminal nuclear localization signal, which became more noticeable after increasing the brightness of the image (Fig. 3 A, arrow). In addition, since p115 can localize to the Golgi by interacting with GM130 (Nakamura et al., 1997; Seemann et al., 2000a), we determined if p115 Golgi localization was affected by GRASP depletion (Fig. 3 C). Co-localization of p115 and GM130 at the Golgi was unaffected when either GRASP55 or 65 was degraded. In marked contrast, in IAA-treated RC65+55 cells, p115 binding to the Golgi was diminished (Fig. 3 D), indicating that p115 was displaced together with GM130 from the perinuclear region where the Golgi-resident enzyme NAGTI-GFP remained. Unlike GM130, p115 was redistributed from the Golgi to the cytosol and was not detected in the nucleus (Fig. 3 C), and its protein level was not reduced either (Fig. 2 A and Fig. 3 E).
Figure 3.
Acute simultaneous depletion of GRASP55 and 65, but not separate depletion, partially dislocates GM130 from the Golgi. (A) GM130 is partially displaced from the Golgi and relocated to the nucleus after degradation of GRASP55 together with GRASP65, but not after individual depletion of GRASP55 or 65. RC55, RC65, or RC65+55 cells were treated with doxycycline for 6 h and then with IAA for a further 2 h. The cells were then fixed and immunolabeled for GM130 (red) and the cis-Golgi protein Golgin-84 (green) and stained for DNA (blue). The brightness of the GM130 image of RC65+55 cells in the bottom panel was increased to better visualize the redistribution of GM130. The arrow indicates the nuclear signal of GM130. White lines show the position of the line-scan used to measure the fluorescence intensity of GM130 (red) and Golgin-84 (green) across the Golgi and nucleus as shown in the graphs. Scale bar, 10 µm. (B) Quantitation of the GM130 fluorescence signal on the Golgi (marked by Golgin-84) from A. n = 3 independent experiments with >50 cells analyzed per experiment and condition. ** P < 0.01; ns, not significant. Error bars represent mean ± SD. (C) Degradation of GRASP55 and 65 partially delocalizes p115 together with GM130 from the Golgi. RC55, RC65, and NAGTI-GFP–expressing RC65+55 cells were treated as in A and immunostained for p115 (red) and GM130 (green) and labeled for DNA (blue). The Golgi is marked by the GFP fluorescence signal of the Golgi enzyme NAGTI-GFP. White lines show the position of the line-scan used to measure the fluorescence intensity of p115 (red), GM130 (green), and NAGTI-GFP across the Golgi as shown in the graphs. Scale bars, 10 µm. (D) Quantitation of the p115 fluorescence signal on the Golgi marked by GM130 or NAGTI-GFP from C. n = 3 independent experiments with >50 cells analyzed per experiment and condition. ** P < 0.01; ns, not significant. Error bars represent mean ± SD. (E) GM130 is partially degraded by the proteasome after acute double depletion of GRASP55 and 65. RC65+55 cells were treated with doxycycline for 6 h, followed by IAA or IAA plus 10 µM MG132. Cell lysates were collected at the indicated time points and subjected to immunoblotting with the indicated antibodies.
Western blotting analysis showed that within 1 h of IAA addition, GRASP55 and 65 amounts dropped below detection levels, while GM130 more gradually decreased within the first 4 h and then persisted at a stable level for the 12-h duration of the experiment (Fig. 3 E). Blocking GRASP55 and 65 degradation with the proteasome inhibitor MG132 also stabilized GM130 and led to its accumulation. By contrast, p115 and Golgin-84 levels were not notably changed after long-term depletion of both GRASPs (Fig. 3 E). Taken together, these results show that acute and simultaneous depletion of GRASP55 and 65, but not individual depletion, partially delocalizes GM130 from the Golgi, leading to its degradation by the proteasome.
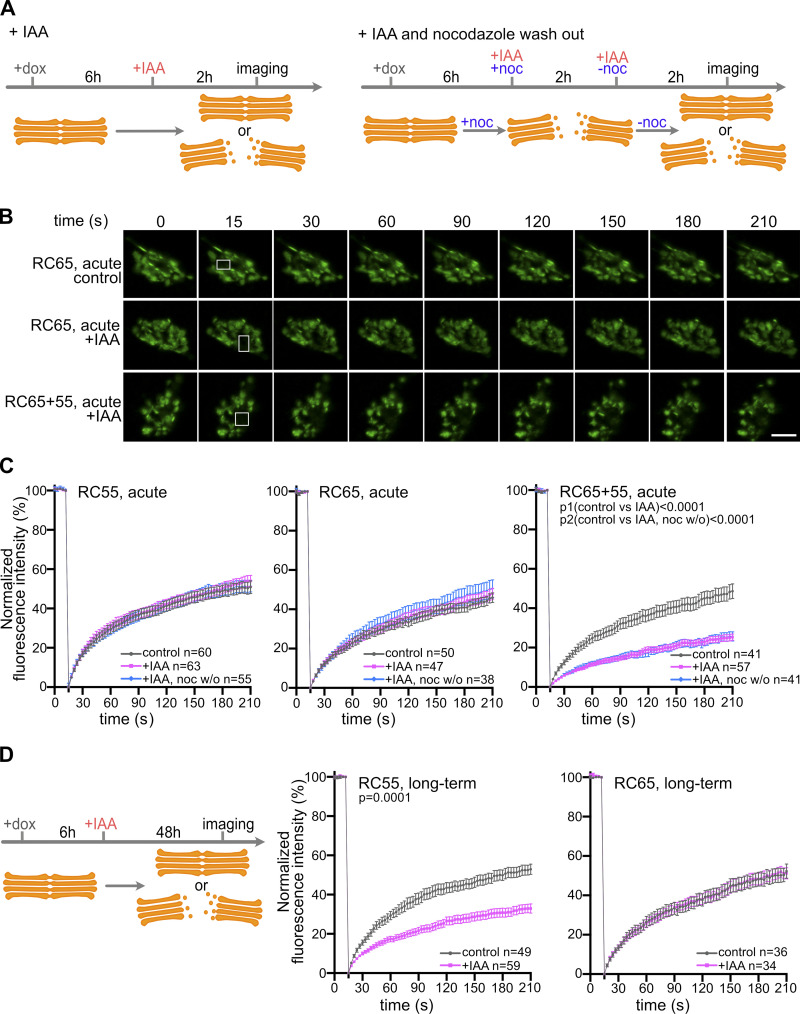
Acute degradation of GRASP55 and 65 disrupts lateral linking of the Golgi ribbon
Previous studies using RNAi and gene knockout have implicated GRASP55 and 65 in laterally connecting adjacent stacks to facilitate proper distribution of Golgi-resident enzymes within the interconnected Golgi ribbon (Puthenveedu et al., 2006; Feinstein and Linstedt, 2008; Xiang and Wang, 2010; Veenendaal et al., 2014). To determine if acute depletion of GRASP55 and/or 65 affects lateral linking of the ribbon, we stably expressed the Golgi enzyme NAGTI-GFP in RC55, RC65, and RC65+55 cells and then performed FRAP to analyze GFP recovery as a measure of lateral mobility within the Golgi ribbon. In contrast to previous reports using RNAi (Puthenveedu et al., 2006; Feinstein and Linstedt, 2008; Xiang and Wang, 2010), acute degradation of either GRASP55 or 65 did not change the rate of recovery, showing that stacks remained laterally linked (Fig. 4, A–C). One possibility is that GRASP55 and 65 are required for the initial connection of the stacks during Golgi ribbon formation but are dispensable once the ribbon is established. To test this idea, cells depleted of GRASPs were first treated with nocodazole to disperse the ribbon into isolated stacks. Nocodazole was then washed out to allow reformation of the perinuclear Golgi ribbon (Fig. 4 A and Fig. S2). The rate of NAGTI-GFP recovery in cells lacking GRASP55 or 65 was indistinguishable from that of control cells, indicating that an interconnected ribbon reformed (Fig. 4, B and C). However, acute and simultaneous degradation of both GRASPs disrupted lateral linking, both in a preexisting ribbon and in a newly reformed Golgi after nocodazole washout (Fig. 4, B and C). In contrast to acute loss of GRASP55, we also noticed that chronically diminished GRASP55 levels disrupted the integrity of the ribbon, which was also observed after RNAi (Xiang and Wang, 2010; Fig. 4 D). Moreover, although RNAi of GRASP65 was reported to compromise lateral connections of stacks (Puthenveedu et al., 2006; Veenendaal et al., 2014), we did not detect a change in FRAP rates after long-term depletion of GRASP65 (Fig. 4 D). Together, the FRAP results show that acute loss of GRASP55 or 65 does not alter the lateral linking of cisternae.
Figure 4.
Acute simultaneous degradation of GRASP55 and 65, but not separate degradation, disrupts the lateral linking of the Golgi ribbon. (A) Schematic illustration of the treatment with doxycycline (dox) and IAA or IAA with nocodazole (noc) washout. (B) FRAP analysis of RC55, RC65, and RC65+55 cells stably expressing the Golgi enzyme NAGTI-GFP. Representative images at the indicated time points are shown, with white boxes indicating the photobleached area of the Golgi. Scale bar, 5 µm. (C) Quantitation of the FRAP results. The recovery rate at each time point was calculated as the ratio of the average intensity of the photobleaching area to that of the adjacent area and then normalized to the closest time point before bleaching. Error bars represent mean ± SEM from the indicated number of cells (n) per condition from three to four independent experiments. (D) Long-term degradation of GRASP55, but not GRASP65, disrupts the integrity of the Golgi ribbon. Scheme of the experiment. RC55 and RC65 cells were treated with doxycycline for 6 h and IAA for a further 48 h before FRAP analysis. FRAP results were quantified as in C. Error bars represent mean ± SEM from the indicated number of cells (n) per condition from three independent experiments.
Figure S2.
GRASP55 and 65 can be degraded in nocodazole-treated cells. (A) Scheme of the experiment. RC55, RC65, and RC65+55 cells were treated with doxycycline (dox) for 6 h. IAA was then added to degrade GRASPs and nocodazole (noc) to depolymerize microtubules and to disperse Golgi stacks. (B) 2 h later, the cells were fixed and immunostained for Golgin-84 (red) and mAID (green) and labeled for DNA (blue). Scale bar, 10 µm. (C) Quantification of the mAID fluorescence signal on the Golgi (labeled by Golgin-84). n ≥ 3 independent experiments with >50 cells analyzed per experiment and condition. *** P < 0.001; **** P < 0.0001. Error bars represent mean ± SD. Scale bar, 10 µm.
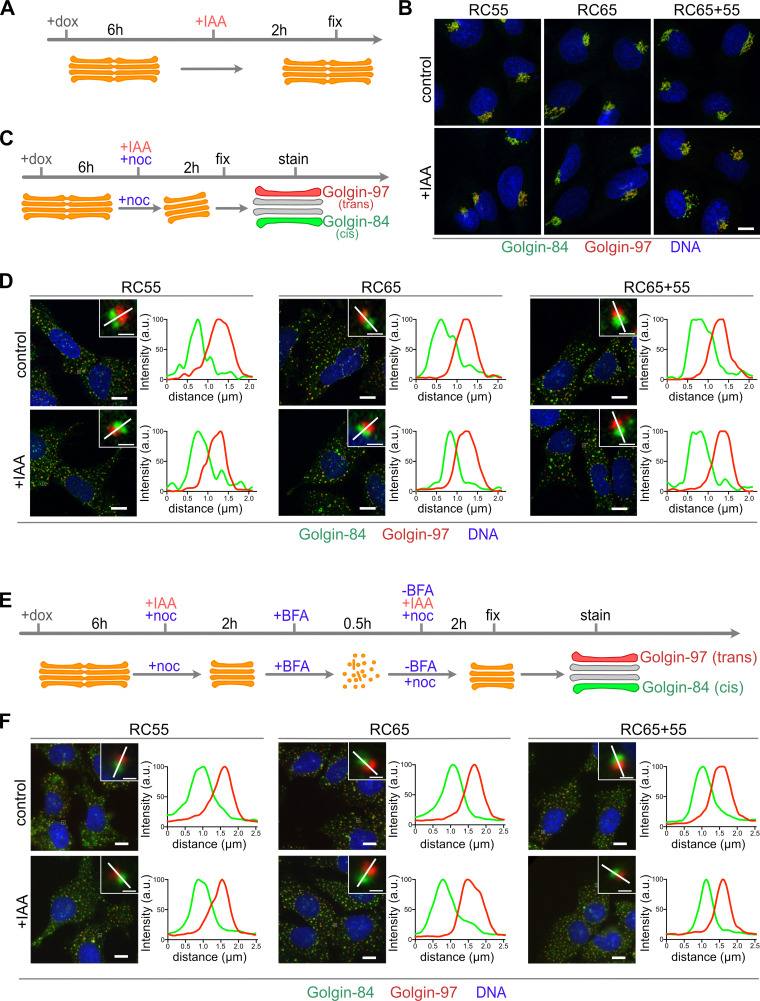
Acute degradation of GRASP55 and 65 does not unstack the Golgi in interphase cells
GRASP55 and 65 were first isolated as factors required to link single cisternae into stacks in vitro (Barr et al., 1997; Shorter et al., 1999), but subsequent knockdown and knockout studies reported conflicting results regarding their ability to stack Golgi cisternae in cells (Kondylis et al., 2005; Xiang and Wang, 2010; Lee et al., 2014). To determine if acute degradation of GRASP55 and 65 affects stacking in cells, we double stained for the cis-Golgi marker Golgin-84 and the trans-Golgi marker Golgin-97 in RC55, RC65, and RC65+55 cells (Fig. 5). Both proteins colocalized at the perinuclear Golgi ribbon in control cells, which was unchanged after acute degradation of GRASP55 and/or 65 (Fig. 5, A and B). To determine if the stacks remained polarized, we treated cells with the microtubule depolymerizing compound nocodazole (Fig. 5 C). Microtubule depolymerization disperses the convoluted perinuclear Golgi ribbon into individual stacks, which allows better discrimination between cis- and trans-Golgi markers by fluorescence microscopy (Shima et al., 1997; Wei and Seemann, 2009a). As shown in Fig. 5 D, the cis Golgin-84 signal was adjacent to the trans-marker Golgin-97 under control conditions and remained so after elimination of GRASPs, indicating that Golgi polarization was maintained with the cis-Golgi attached to the trans-cisternae.
Figure 5.
Acute degradation of GRASP55 and/or 65 during interphase does not affect the cis to trans polarization of Golgi stacks. (A and B) Cells were treated with doxycycline (dox) for 6 h, IAA was added for an additional 2 h, and cells were prepared for immunofluorescence analysis. IAA treatment did not change the localization of the cis-Golgi marker Golgin-84 (green) and the trans-Golgi protein Golgin-97 (red) to the perinuclear Golgi ribbon. Scale bar, 10 μm. (C and D) Cells incubated with doxycycline were treated with IAA or IAA with nocodazole (noc) for an additional 2 h to depolymerize microtubules and to disperse the Golgi ribbon into individual stacks. Cells were then immunostained for Golgin-84 (green), Golgin-97 (red), and DNA (blue). Insets show a magnified Golgi stack with the white line marking the line scan of the fluorescence intensities shown in the graphs. Scale bar, 10 µm. Inset scale bar, 1 µm. (E and F) Polarized Golgi stacks reassemble in cells depleted of GRASPs. Cells treated with doxycycline were incubated with IAA and nocodazole for 2 h, followed by BFA to fragment the Golgi stacks. BFA was then removed, and nocodazole (plus dox and IAA) was maintained to allow reformation of individual Golgi stacks. The cells were fixed and immunostained for Golgin-84 (green) and Golgin-97 (red) and labeled for DNA (blue). Insets show a magnified Golgi stack with the white line marking the line scan of the fluorescence intensities shown in the graphs. Scale bar, 10 µm. Inset scale bar, 1 µm.
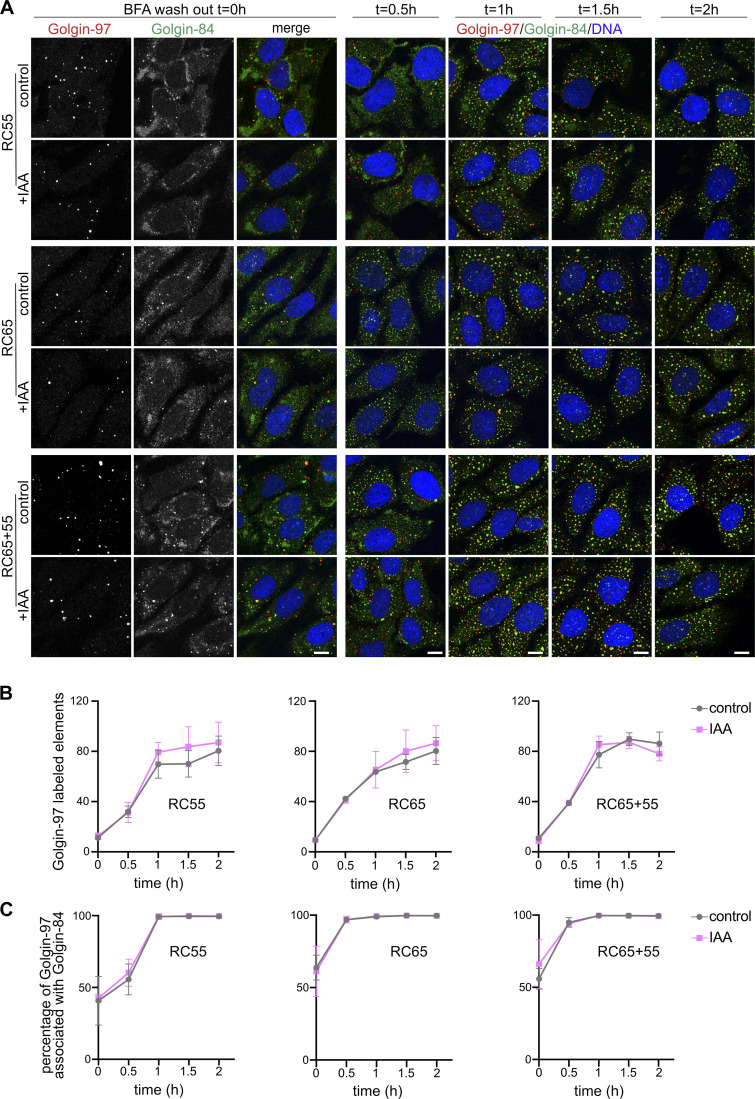
One possibility is that GRASP55 and 65 function during de novo formation of stacked cisternae but are dispensable for maintaining stacks. To test this possibility, we treated cells with the fungal metabolite Brefeldin A (BFA), which unstacks and converts cisternae into a collection of tubules and vesicles. BFA was then removed, and nocodazole was added to allow for the reformation of the Golgi into single stacks (Fig. 5 E and Fig. S3). In control cells and cells depleted of GRASPs, Golgin-84 was associated with Golgi-97 to the same extent as in cells not treated with BFA (Fig. 5 F), suggesting that GRASP55 and 65 are not essential for the assembly of polarized stacks in cells. Furthermore, the rate of stack reformation after BFA washout was not affected by the absence of GRASP55 and/or 65 (Fig. S3, A and B).
Figure S3.
Acute degradation of GRASP55 and/or 65 does not affect Golgi stack reformation after BFA washout. (A) Cells were treated with doxycycline (dox) and then incubated with IAA and nocodazole for an additional 2 h. BFA was then added for 30 min to disassemble Golgi stacks. BFA was removed, and nocodazole (plus dox and IAA) was maintained to allow the reformation of individual Golgi stacks. The cells were fixed at the indicated time points and immunostained for Golgin-97 (red) and Golgin-84 (green) and labeled for DNA (blue). Scale bar, 10 µm. (B) Quantitation of the number of Golgin-97–marked Golgi elements per cell from A. n = 3 with an average of >10 cells per time point. Error bars represent mean ± SD. (C) Quantitation of stack formation from A as the percentage of Golgin-97–labeled elements that associate with Golgin-84. n = 3 with >10 cells per time point per condition per experiment. Error bars represent mean ± SD.
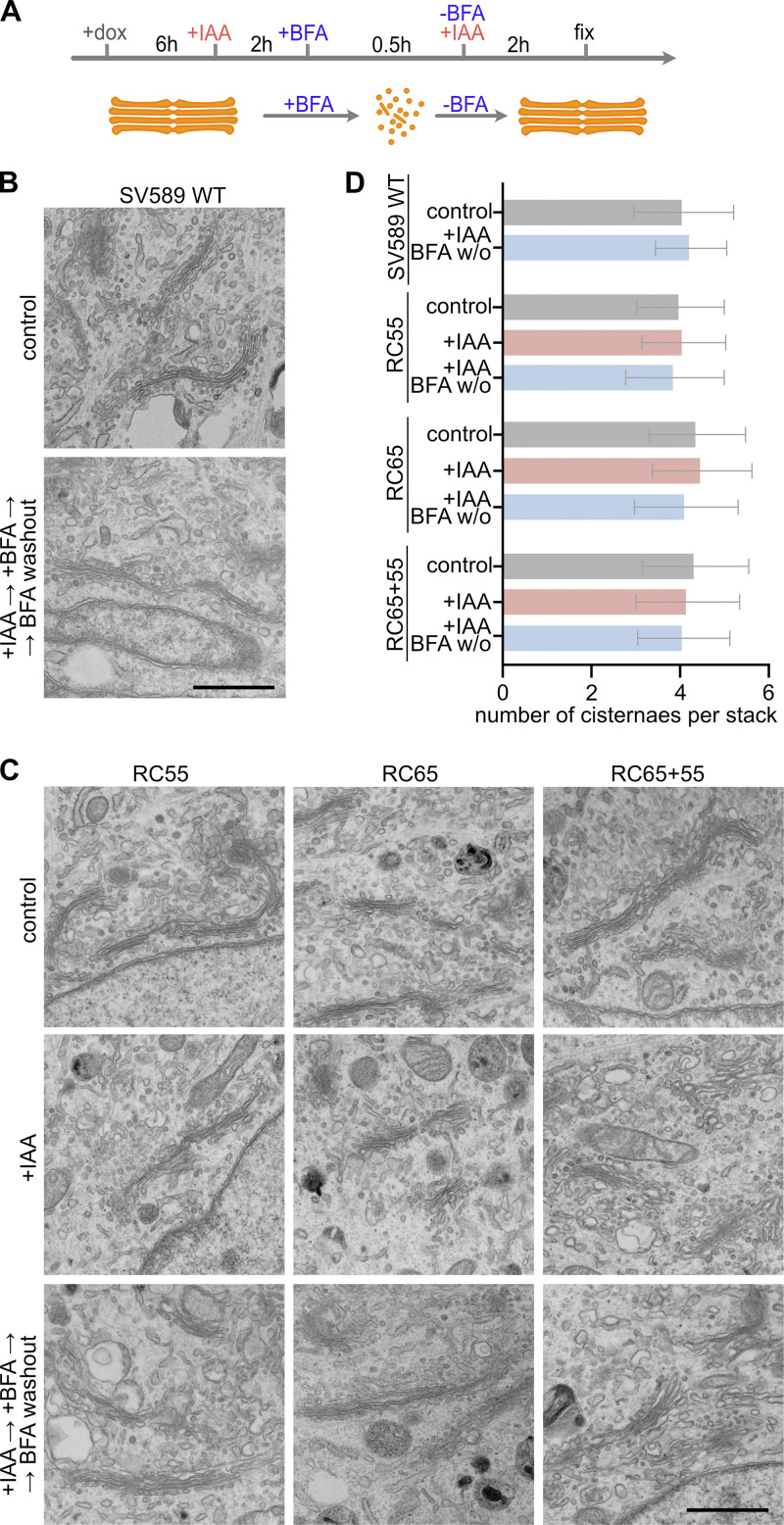
To examine the ultrastructure of the stacks, we performed electron microscopy. As shown in Fig. 6, cells acutely depleted of GRASP55 and/or 65 showed the same extent of stacking as SV589 parental cells. Moreover, there was no difference in the number of cisternae per stack after BFA washout in the absence of GRASPs. The Golgi in RC65+55 cells contained an average of 4.4 ± 1.2 cisternae per stack in controls and 4.1 ± 1.0 cisternae after GRASP55 and 65 depletion followed by BFA washout (Fig. 6, C and D). These results show that acute depletion of GRASP55 and/or 65 does not affect stack formation in interphase cells.
Figure 6.
GRASP55 and 65 are dispensable for Golgi stacking. (A) Experiment scheme. TIR1 expression in RC55, RC65, and RC65+55 cells was induced for 6 h with doxycycline (dox), and IAA was added for 2 h to degrade GRASP55 and/or 65. The cells were then treated for 30 min with BFA to disassemble the Golgi. BFA was removed for 2 h to allow Golgi reformation, and the cells were processed for EM. (B) Representative EM images showing stacked Golgi cisternae in parental SV589 cells and upon Golgi reassembly after BFA washout (+IAA → +BFA → BFA washout). Scale bar, 1 µm. (C) EM images of RC55, RC65, and RC65+55 cells after control treatment, after acute degradation of GRASP55 and/or 65 (+IAA) and after degradation and Golgi reassembly upon BFA washout (+IAA → +BFA → BFA washout). Scale bar, 1 µm. (D) Quantitation of the number of cisternae per stack (RC55 cells: control: n = 15 cells, +IAA: n = 16, +IAA BFA w/o: n = 25; RC65 cells: control: n = 19, +IAA: n = 20, +IAA BFA w/o: n = 30; RC65+55 cells: control: n = 44, +IAA: n = 40, +IAA BFA w/o: n = 44; SV589 cells: control: n = 12, +IAA BFA w/o: n = 10 cells). An average of 4.5 stacks per cell was counted. Error bars represent mean ± SD.
GRASP55 and 65 are dispensable for postmitotic reformation of Golgi stacks
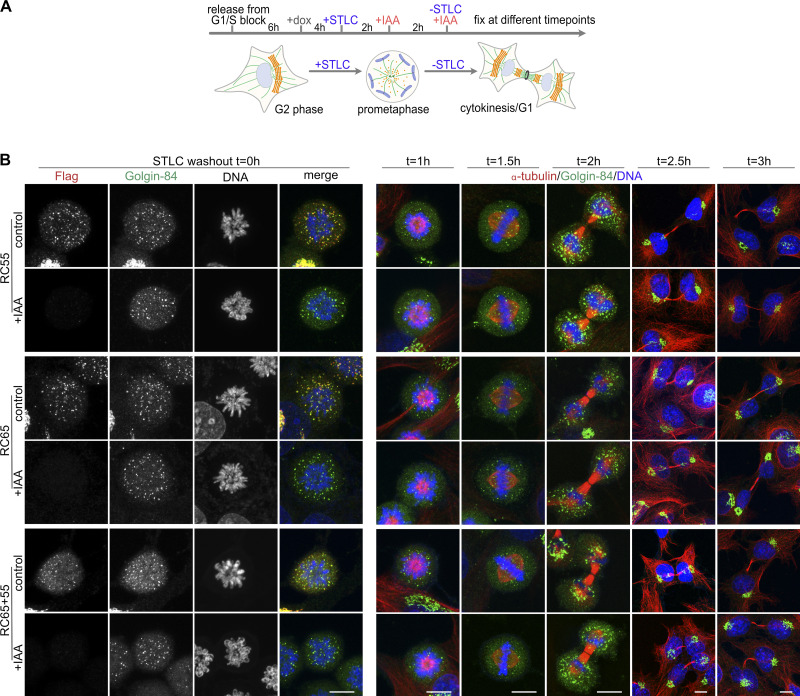
Our data so far indicate that acute loss of GRASP55 and/or 65 does not displace cisternae from preexisting stacks and does not affect de novo assembly of stacks following BFA washout. Next, we tested if acute loss of GRASPs affects the reassembly of Golgi stacks at the end of mitosis. During mitosis, the Golgi unstacks and vesiculates and then reassembles back to stacked cisternae after partitioning by the spindle into the daughter cells (Shima et al., 1997; Wei and Seemann, 2009a). GRASP55 and 65 were first identified in an in vitro postmitotic Golgi reassembly assay as factors required for stacking of cisternae, hence the name GRASPs (Golgi Reassembly Stacking Proteins; Barr et al., 1997; Shorter et al., 1999). We therefore sought to investigate if acute depletion of GRASPs impairs stack reformation at the end of mitosis. To this end, cells were synchronized in prometaphase with the Eg5 kinesin inhibitor S-trityl-L-cysteine (STLC), which prevents centrosome separation and arrests cells with monopolar spindles (Fig. 7 A and Fig. S4 A; Skoufias et al., 2006). Immunofluorescence analysis showed that the addition of IAA while the cells remained arrested in prometaphase also efficiently depleted GRASP55 and/or 65 (Fig. S4 B; t = 0 h). STLC was then washed out to allow bipolar spindle formation and progression through mitosis. Following STLC removal, metaphase spindles formed after 1.5 h, and by 2 h cells entered telophase. After 2.5 h, cells reached cytokinesis/G1 as identified by daughter cells containing reformed nuclei, decondensed chromatin, reassembled Golgi ribbon, and tightly bundled microtubules connecting the daughter cells (Fig. S4 B). There was no indication that depletion of GRASP55 and/or 65 affected the timing of Golgi reformation.
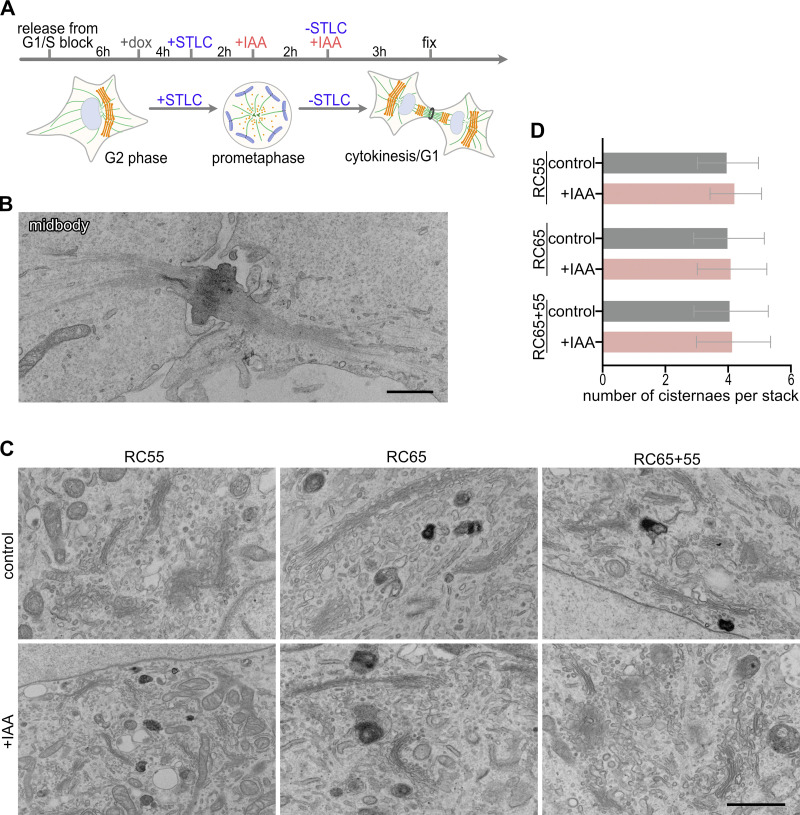
Figure 7.
Golgi stacks reform at the end of mitosis in the absence of GRASP55 and/or 65. (A) Scheme of the experiment. Cells released from double thymidine G1/S block were treated with doxycycline (dox) and arrested in prometaphase with the Eg5 kinesin inhibitor STLC. The mitotic cells were treated with IAA to degrade GRASPs, released from STLC block to allow mitotic progression, and processed for EM analysis. (B) Representative EM images showing a midbody between a pair of daughter cells. Scale bar, 1 µm. (C) Representative EM images of Golgi stacks at the end of mitosis/G1 in control and IAA-treated RC55, RC65, and RC65+55 cells. Scale bar, 1 µm. (D) Quantitation of the number of cisternae per stack (RC55 cells: control: n = 15 pairs of daughter cells, +IAA: n = 17; RC65 cells: control: n = 15, +IAA: n = 15; RC65+55 cells: control: n = 10, +IAA: n = 15). An average of 6.6 stacks per pair of daughter cells was counted. Error bars represent mean ± SD.
Figure S4.
GRASP55 and 65 can be degraded in STLC-arrested mitotic cells. (A) Scheme of the approach. Cells were released from double thymidine G1/S block, treated with doxycycline (dox), and then with STLC to arrest cells at prometaphase. The mitotic cells were then treated with IAA for 2 h to degrade GRASPs followed by STLC washout and the addition of dox and IAA to allow mitotic progression and then fixed at different time points. (B) Cells fixed at t = 0 h after STLC washout were immunostained for Flag-tag (red) and Golgin-84 (green) and labeled for DNA (blue). Cells at later time points were immunostained for α-tubulin (red) and Golgin-84 (green) and labeled for DNA (blue). Scale bar, 10 µm.
To examine the ultrastructure of the Golgi, cells were fixed 3 h after release from STLC and processed for EM. In cells depleted of both GRASP55 and 65, stacks reformed to a comparable extent as those in control cells (Fig. 7 C). Quantitation of the EM images showed that elimination of GRASPs in mitosis did not alter the number of cisternae in reformed stacks at the end of mitosis (Fig. 7, C and D). Together, these results indicate that GRASP55 and 65 are dispensable for stacking of cisternae during postmitotic Golgi reassembly in cells.
Discussion
It has been long debated whether or not GRASP55 and 65 function in stacking Golgi cisternae and/or lateral linking of stacks into a Golgi ribbon. Despite multiple efforts to dissect their respective roles, several loss-of-function studies nevertheless resulted in conflicting conclusions (Duran et al., 2008; Feinstein and Linstedt, 2008; Xiang and Wang, 2010; Bekier et al., 2017; Grond et al., 2020). These approaches employed RNAi or targeted gene disruption in cellular and animal models, which indirectly down-regulate GRASP55 and/or 65 proteins. Although being powerful tools, these genetic approaches can take days to weeks to effectively suppress protein expression. Consequently, the long time period required to eliminate proteins by these approaches can result in complex phenotypes that are indirect and difficult to interpret (Rothman, 2010). In addition, long-term loss of function allows cells to activate compensatory pathways and become adapted, which may mask primary phenotypes. Given conflicting reports for the chronic loss of GRASP proteins, we set out to circumvent these complications by acutely eliminating both proteins and investigate their immediate effects. Our approach was to generate gene-edited stable cell lines that endogenously express GRASP55 and/or 65 tagged with the mAID degron, allowing rapid and inducible degradation of GRASP55 and/or 65 by the proteasome. Both proteins are depleted within 1–2 h after addition of the auxin IAA, enabling us to determine the immediate effects of GRASP55 and/or 65 loss on stacking and lateral linking.
Previous reports showed that GRASP65 down-regulation by RNAi in cells or gene-trap insertion in mice disrupted ribbon interconnectivity (Puthenveedu et al., 2006; Veenendaal et al., 2014). This differs from the findings presented here, showing no such defect following either acute or chronic degradation of GRASP65. Similarly, down-regulation of GRASP55 by RNAi was reported to unlink the Golgi ribbon (Feinstein and Linstedt, 2008; Xiang and Wang, 2010), which was not observed by others (Duran et al., 2008). We also observed defects in the connectivity of the ribbon, but only after long-term and not acute GRASP55 degradation. Our data therefore provide evidence that lateral linking defects are indirectly caused by prolonged GRASP55 depletion. The chronic defect arising after long-term depletion of GRASP55 might be due to partial loss of its binding partner Golgin-45 (Fig. 2 A; Lee et al., 2014; Bekier et al., 2017). GRASP55 and Golgin-45 form a rab2 effector complex that functions in vesicle tethering. Down-regulation of Golgin-45 inhibits protein transport and, as a consequence, disrupts the Golgi ribbon (Short et al., 2001; Feinstein and Linstedt, 2008).
Another important finding here is that short-term ablation of either GRASP55 or 65 did not change the connectivity of the ribbon, while simultaneous depletion of both GRASPs impaired lateral linking of the ribbon without altering the organization of stacks. Defects in lateral linking were also reported for double knockdown of GRASP55 and 65 by RNAi, but in contrast to our data, these treatments caused swollen cisternae (Lee et al., 2014) or vesiculation and disorganization of the stacks (Xiang and Wang, 2010). One possibility is that GRASP55 and 65 have redundant roles in the linking of stacks, where individual elimination of either GRASP has no effect, but depletion of both GRASPs leads to loss of lateral connectivity. Such complementary roles might explain the observation that gene deletion of one GRASP increases the expression levels of the other GRASP protein (Bekier et al., 2017). Another possibility is that our observed lateral connectivity defect upon degradation of both GRASPs is due to the indirect loss of Golgin-45, GM130, and p115 from the Golgi (Fig. 3). Down-regulation of these Golgins by RNAi inhibits cargo transport through the Golgi and, as a consequence, disrupts the dynamic equilibrium of membrane flux through the Golgi, ultimately resulting in ribbon fragmentation (Short et al., 2001; Puthenveedu and Linstedt, 2004; Sohda et al., 2005; Puthenveedu et al., 2006).
In particular, knockdown of GM130 is sufficient to unlink the ribbon and further generates shortened cisternae with accumulated vesicles around the stacks (Puthenveedu et al., 2006). This result is consistent with increased vesiculation and shortened cisternae after acute inhibition of p115 binding to GM130 (Seemann et al., 2000a). Blocking p115-mediated vesicle tethering by microinjection of an N-terminal peptide of GM130 or replacing GM130 with a mutant lacking its p115-binding domain displaced p115 from the Golgi, reduced cargo transport, and concomitantly increased vesicles and shortened the cisternae (Seemann et al., 2000a). The striking similarities of the phenotypes in the two studies support the notion that the lateral unlinking effect upon GM130 depletion is indirectly caused by vesiculation that gradually consumes the cisternae. During mitosis, a comparable but accelerated vesiculation process drives mitotic disassembly of the Golgi. Here, CDK1 phosphorylation of GM130 at Ser25 blocks p115 binding (Lowe et al., 1998, 2000). As a consequence, vesicles continue to bud but fail to fuse, leading to vesicle accumulation and disassembly of the Golgi ribbon (Shorter and Warren, 2002).
Although GM130 is predominantly found in a complex with GRASP65, it can also bind GRASP55 (Fig. 2, D and E; Short et al., 2001). Acute or chronic depletion of either GRASP55 or 65 did not change the localization or protein levels of GM130, demonstrating that the GM130–GRASP complex is dissociated during proteolysis, leaving GM130 behind on the Golgi (Fig. 2 A and Fig. 3 A). In contrast, acute depletion of both GRASPs displaced GM130 from the Golgi, which was also observed in double knockout cells and animals (Bekier et al., 2017; Grond et al., 2020). Although a minor fraction of GM130 can be pulled down by GRASP55 (Fig. 2 E; Short et al., 2001), binding is not increased after long-term GRASP65 elimination, demonstrating that GRASP55 does not compensate for loss of GRASP65 to stabilize GM130 on the Golgi (Fig. 2 D). This result, however, raises the question of why double depletion of both GRASPs expels GM130 together with p115 and Golgin-45 from the Golgi. GM130 together with other Golgins and GRASPs is thought to form a matrix through multimeric interactions to stabilize the Golgi structure (Slusarewicz et al., 1994; Seemann et al., 2000b). Removal of one such component (e.g., GRASP55 or 65) does not affect the overall integrity of the matrix, but displacement of several matrix proteins after double GRASP depletion might be sufficient to destabilize the matrix and weaken the structure, ultimately leading to lateral unlinking of the ribbon. As such, the effect of unlinking is not caused by GRASP55 or 65 depletion alone, arguing that the structural effect is indirect. In support of this notion, GM130 was recently shown to self-organize by liquid-liquid phase separation (Rebane et al., 2020), which might be a mechanism for assembly and maintenance of the matrix by including other Golgi proteins in the condensate.
GRASP55 and 65 were first isolated as proteins required to link single cisterna into stacks in vitro (Barr et al., 1997; Shorter et al., 1999). Further evidence for the role of GRASPs as stacking factors came from RNAi experiments or gene disruption of GRASPs in cells, which showed a reduction in the number of cisternae per stack (Sütterlin et al., 2005; Xiang and Wang, 2010; Bekier et al., 2017). However, tissue-specific genetic deletion of GRASPs in mice did not reveal a noticeable stacking defect (Grond et al., 2020). In our experiments, degradation of GRASP55 and/or 65 did not affect stacking of preexisting stacks, nor did it prevent de novo stack assembly after BFA washout or at the end of mitosis (Fig. 6 and Fig. 7). Further studies will be necessary to determine if stacking is differentially affected in different cell cycle stages or cell types or specific conditions such as stress.
We demonstrated here that indirect phenotypes can arise due to the chronic reduction of GRASP proteins, which might explain previous conflicting reports. We showed by acute degradation that GRASP55 and/or 65 are dispensable for the maintenance and de novo formation of stacked cisternae. Furthermore, short-term depletion of either GRASP55 or 65 had no effect on the lateral connections of the Golgi ribbon, while depletion of both GRASPs coincided in loss of the vesicle tethering proteins GM130, p115, and Golgi-45 from the Golgi and compromised ribbon linking. Because of the high flux of membranes through the Golgi, an imbalance of vesicle transport has been shown to disrupt Golgi ribbon integrity; thus it is reasonable to conclude that double depletion of GRASPs indirectly affects ribbon linking. As such, GRASPs may be described as adaptors or membrane tethers that target the vesicle transport machinery to the Golgi membranes. On the other hand, vesicle transport is required for reformation of Golgi stacks at the end of mitosis or after BFA washout, even though they are not affected by the absence of GRASPs. Moreover, impairment of GRASP55 and 65 expression has been shown to accelerate the rate of cargo transport and to cause underglycosylation (Bekier et al., 2017). However, the precise role of GRASPs in vesicle transport needs further clarification. The ability to acutely disable GRASP function while avoiding chronic side effects will help to dissect the functional contributions of GRASPs to fundamental biological processes.
Materials and methods
Plasmids and cell lines
To generate SV589 cells endogenously expressing either GRASP55-3xFlag-mAID (RC55 cells) or GRASP65-3xFlag-mAID (RC65 cells) or both (RC65+55 cells), we employed the CRISPaint (CRISPR-assisted insertion tagging) approach (Schmid-Burgk et al., 2016) to replace the stop codon with the sequence encoding three tandem copies of the Flag epitope tag (DYKDDDDK), followed by the 7.4 kD mAID sequence (Kubota et al., 2013), a T2A self-cleaving peptide sequence, and a promoterless antibiotic resistance gene. This approach is composed of three plasmids: (1) the single-guide RNA (sgRNA) expression plasmid containing Cas9 to introduce a double-strand break at the targeted genomic location of GRASP55 or GRASP65; (2) the GRASP55- or 65-specific scarless donor plasmid containing short synonymous codons compensating for the lost coding area (but lacking the stop codon) caused by cutting and the 3xFlag-mAID plus resistance gene sequences; and (3) the reading-frame selector, which is an sgRNA expression plasmid aiming to introduce a double-strand break in the scarless donor in the front of the synonymous codons and 3xFlag-mAID sequence. Thereby, the donor plasmid is linearized and can be inserted into the targeted genomic location of GRASP55 or 65.
The sgRNA expression plasmid for GRASP55 was assembled by cloning two annealed oligos (5′-CACCGTTAAGGTGACTCAGAAGCAT-3′ and 5′-AAACATGCTTCTGAGTCACCTTAAC-3′) into pX330-hSpCas9-hGem (Addgene; Shalem et al., 2014). To construct the GRASP55-specific scarless donor plasmid, we first inserted the mAID sequence after 3xFlag in the plasmid (p) pCRISPaint-3xFlag-puro (Addgene; Schmid-Burgk et al., 2016) using the In-Fusion HD Cloning kit (Takara Bio) to generate pCRISPaint-3xFlag-mAID-puro. Then, the scarless donor plasmid for GRASP55 was assembled by inserting two annealed oligos (5′-CTAGCGGGCCAGTACCCAAAAACATCGGAGTCACCTGGGTCTGGTGGCAGTGGAGGGG-3′ and 5′-GATCCCCCTCCACTGCCACCAGACCCAGGTGACTCCGATGTTTTTGGGTACTGGCCCG-3′) into pCRISPaint-3xFlag-mAID-puro with NheI and BamHI. The corresponding reading-frame selector was assembled by cloning two annealed oligos (5′-CACCGGGGCCAGTACCCAAAAACAT-3′ and 5¢-AAACATGTTTTTGGGTACTGGCCCC-3′) into pX330-hSpCas9-hGem. Similarly, the sgRNA plasmid for GRASP65 was assembled by cloning two annealed oligos (5′-CACCGTTATTCTGTGGTAGAGATCT-3′ and 5′-AAACAGATCTCTACCACAGAATAAC-3′) into pX330-hSpCas9-hGem. The GRASP65-specific scarless donor plasmid was generated by replacing the puromycin resistance gene in pCRISPaint-3xFlag-mAID-puro with the hygromycin resistance gene from pQCXIH (Clontech) to yield pCRISPaint-3xFlag-mAID-hygro. Then, two annealed oligos (5′-CTAGCGGGCCAGTACCCAAAAATCTCGGGCACAGAAGGGTCTGGTGGCAGTGGAGGGG-3′ and 5′-GATCCCCCTCCACTGCCACCAGACCCTTCTGTGCCCGAGATTTTTGGGTACTGGCCCG-3′) were cloned into pCRISPaint-3xFlag-mAID-hygro. The reading-frame selector for GRASP65 was assembled by inserting two annealed oligos (5′-CACCGGGGCCAGTACCCAAAAATCT-3′ and 5′-AAACAGATTTTTGGGTACTGGCCCC-3′) into pX330-hSpCas9-hGem.
To generate cell lines expressing endogenously tagged GRASP55 (RC55 cells) or 65 (RC65 cells), SV589 cells were transfected with the sgRNA expression plasmid, the scarless donor plasmid, and the reading-frame selector for GRASP55 or 65 at the ratio of 1:2:1 (micrograms). 48 h after the transfection, 1 µg/ml puromycin (RPI) was added for another 48 h to select RC55 cells or 250 µg/ml hygromycin B (GoldBio) for 7 d to select RC65 cells. Individual colonies were picked, and single clones were isolated by limited dilution. Homozygous gene-edited clones were identified by genomic PCR and immunoblotting with antibodies against mAID, GRASP55, and GRASP65. To establish RC65+55 cells, the plasmid set for GRASP55 was transfected into homozygous RC65 cells, and gene-edited homozygous clones were isolated using puromycin selection as for RC55 cells.
To induce rapid degradation of GRASP55 and 65, the human codon–optimized E3 ubiquitin ligase from OsTIR1 (Addgene; Natsume et al., 2016) was stably introduced into RC55, RC65, and RC65+55 cells. We first exchanged the puromycin resistance gene in pLVX-TetOne-puro (Clontech) with the neomycin gene from pQCXIN (Clontech) to generate pLVX-TetOne-neo. Then OsTIR1 from pMK232(CMV-OsTIR1-PURO) (Addgene; Natsume et al., 2016) was cloned into pLVX-TetOne-neo with EcoRI and BamHI. Finally, we added the 2xMyc sequence generated by two annealed oligos at the 3′-end of OsTIR1. The generated pLVX-TetOne-OsTIR1-2xMyc-neo was cotransfected with psPAX and pVSVG (Addgene) into HEK293T cells to produce lentiviruses, which were then used to transduce RC55, RC65, and RC65+55 cells. After 48 h, 500 µg/ml G418 (Enzo Life Sciences) was added for 2 wk. Individual colonies were picked, and single clones were isolated by limited dilution and verified by immunofluorescence analysis and Western blotting with anti-Myc antibodies.
For FRAP analyses, the sequence of NAGTI-GFP (Shima et al., 1997) was cloned into pLVX-Puro (Clontech). The constructed plasmid was then cotransfected with psPAX and pVSVG into HEK293T cells to produce lentiviruses, which were then used to transduce RC55, RC65, and RC65+55 cells stably expressing OsTIR1-2xMyc. The mixed population of each was used for subsequent FRAP analyses.
Genomic PCR
Genomic DNA was extracted with the Quick-DNA Microprep Plus Kit (Zymo Research) according to the manufacturer’s instruction. PCR amplification was performed with primers targeting the indicated regions (Fig. 1 B and Table S1) using Taq DNA Polymerase (New England Biolabs) for 30 cycles of 95°C for 30 s, 54°C for 30 s, and 68°C for 1 min (Fig. 1 C) or using EmeraldAmp GT PCR Master Mix (Takara Bio) for 30 cycles of 98°C for 10 s, 57°C for 30 s, and 72°C for 1 min (Fig. 1 D).
Cell culture and drug treatments
HEK293T (American Tissue Culture Collection), SV589 (SV40 immortalized human fibroblasts; Yamamoto et al., 1984), RC55, RC65, and RC65+55 cells were cultured at 37°C and 5% CO2 in DMEM (Mediatech) supplemented with 10% cosmic calf serum (HyClone), 100 U/ml penicillin (GoldBio), and 100 µg/ml streptomycin (GoldBio; PenStrep). Expression of OsTIR1-2xMyc was induced with 0.5 µg/ml doxycycline (Sigma) for 6 h or 20 h before inducing degradation of GRASP55 and/or 65 by the addition of 500 µM IAA (Abcam). The Golgi ribbon was disassembled into single stacks by treatment with 3 µg/ml nocodazole (EMD Millipore) for 2 h, while complete disassembly into vesicles was triggered with 5 µg/ml BFA (LC Laboratories) for 30 min. MG132 (Boston Biotech) was added at 10 µM to inhibit protein degradation by the proteasome.
To enrich cells in late M-phase, RC55, RC65, and RC65+55 cells were first synchronized at the G1/S phase transition by double thymidine block. Cells were treated with 2 mM thymidine (Chem-Impex) for 16 h; thymidine was then washed out for 8 h and added again for an additional 16 h. 10 h after the last release from thymidine block, we added 20 µM STLC (Acros) for 4 h to arrest cells in prometaphase. STLC was then removed to allow cells to progress into telophase/cytokinesis, and cells were fixed and processed for immunofluorescence or EM analysis.
Antibodies
We used the following primary antibodies: mouse monoclonal antibody against mAID (clone 1E4, catalog #M214-3; MBL), FLAG-tag (M2, catalog #F3165; Sigma), GAPDH (GA1R, catalog #MA5-15738; Invitrogen), Golgin-45 (E-3, catalog #sc-515193; Santa Cruz Biotechnology), Golgin-97 (CDF4, catalog #A-21270; Invitrogen), GM130 (35/GM130, catalog #610822; BD Transduction Labs), GRASP55 (1C9A3, catalog #66627–1-lg; Proteintech), GRASP65 (D-12, catalog #sc-374423; Santa Cruz Biotechnology), p115 (4H1; Waters et al., 1992), and α-tubulin (DM1A, catalog #14–4502-80; eBioscience) and rabbit polyclonal antibodies against Golgin-84 (Beard et al., 2005), GM130 (Wei et al., 2015), GRASP55 (catalog #PTG10598-1-AP; Proteintech), GRASP65 (UT465; Wei et al., 2015), Myc-tag (A-14, catalog #sc-789; Santa Cruz Biotechnology), and p115 (catalog #130509–1-AP; Proteintech). Secondary antibodies were as follows: Alexa Fluor 488–, Alexa Fluor 555–, Alexa Fluor 594–, or Alexa Fluor 647–conjugated highly cross-absorbed goat anti-mouse IgG (H+L) or goat anti-rabbit IgG (H+L; Invitrogen), HRP-conjugated goat anti-rabbit, or goat anti-mouse IgG (Jackson ImmunoResearch).
Co-immunoprecipitation
IAA- or control-treated RC65 cells were lysed in lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and protease inhibitor cocktail Complete [Roche]) for 30 min on ice. Cell lysates were then cleared by centrifugation for 10 min at 16,000 g at 4°C. For GM130 immunoprecipitation (IP), lysates were mixed with 5 µl rabbit polyclonal anti-GM130 serum or 5 µl preimmune serum for 60 min at 4°C and incubated with protein A-Sepharose (GE Healthcare) for an additional 60 min. For GRASP55 IP, cleared lysates were mixed with 2 µg mouse monoclonal anti-GRASP55 IgG or 2 µg mouse γ-globulin (Jackson ImmunoResearch) for 60 min at 4°C and incubated with protein A/G-Sepharose (Santa Cruz Biotechnology) for an additional 60 min. Beads were then washed with lysis buffer and bound proteins released by boiling in SDS sample buffer.
Immunofluorescence microscopy
Cells grown on glass coverslips were fixed and permeabilized in precooled methanol for 15 min at −20°C, washed in PBS, incubated with indicated primary antibodies for 30 min at 37°C, washed, and incubated with Alexa Fluor–conjugated secondary antibodies for 30 min at 37°C. DNA was stained for 5 min with 1 µg/ml Hoechst 33342 (Invitrogen) in PBS at room temperature, and cells were mounted in Mowiol 4–88 (Calbiochem) embedding solution (Wei and Seemann, 2009b). Confocal images were acquired using an LSM780 inverted microscope (Zeiss) with a Plan-Apochromat 63×/1.4 objective (Zeiss) and Zen imaging software (Zeiss). Z-sections were captured at 0.47-µm intervals. All images are confocal maximum-intensity projections assembled using the Zen software package (Zeiss) with the exception of Fig. 5 F. Images shown in Fig. 5 F were captured using an Axiovert 200M microscope (Zeiss) with a Plan-Neofluar 40×/1.3 differential interference contrast objective (Zeiss), an Orca 285 camera (Hamamatsu), and Openlab 4.0.2 software (Improvision).
FRAP
FRAP assays were performed at 37°C using an LSM780 inverted microscope in combination with a Plan-Apochromat 63×/1.4 objective and Zen imaging software. RC55, RC65, and RC65+55 cells stably expressing NAGTI-GFP grown on 35-mm glass-bottom dishes (MatTek Corporation) were treated with doxycycline and IAA to degrade GRASP55 and/or 65. The medium was then changed to CO2-independent medium (Invitrogen) supplemented with 10% cosmic calf serum, 2 mM GlutaMax (Invitrogen), PenStrep, and the same amount of doxycycline and IAA as for the initial treatment. The Golgi area labeled by NAGTI-GFP was magnified six times to facilitate photobleaching. Time-lapse images were recorded at 3-s intervals, and the average intensity of the photobleached area and the adjacent area at every time point was measured using ImageJ 2.0. The recovery rate at each time point was calculated as the ratio of the average intensity of the photobleached area to that of the adjacent area and finally normalized to prebleach values.
Image analysis and quantification
Image analysis was performed using ImageJ 2.0. Statistical analyses were conducted using Prism 8.4 software (GraphPad). Data shown are from three or more independent experiments. Error bars in Fig. 4 C and Fig. 4 D represent SEM, and error bars in other figures represent SD. The statistical significance was assessed by Student’s t tests.
To measure the fluorescence signal of GRASP55, GRASP65 (Fig. 2 C), mAID (Fig. S1 D and Fig. S2 C), GM130 (Fig. 3 B), and p115 (Fig. 3 D), the Golgi area was first marked by encircling the costained Golgi marker using the Freehand tool (ImageJ). The fluorescence signal of the marked area was then calculated using the Measure function (ImageJ). The average number of cells analyzed per experiment and condition from n ≥ 3 independent experiments was at least 50.
The Golgi area (Fig. S1 E), as defined by the signal distribution of Golgin-84, was encircled using the Freehand tool and analyzed using the Measure function. The average number of cells analyzed per experiment from n = 3 independent experiments was RC55 cells: control: n = 58 cells, +IAA: n = 50; RC65 cells, control: n = 66, +IAA: n = 84; RC65+55 cells, control: n = 74, IAA: n = 58.
To quantify the number of Golgi elements per cell, Golgin-84–labeled (Fig. S1 F) or Golgin-97–labeled (Fig. S3 B) elements were selected by applying a fixed threshold. The objects were then counted using the Analyze Particles function (ImageJ). The average cell number analyzed from n = 3 independent experiments per condition and experiment in Fig. S1 F were RC55 cells: control: n = 60 cells, +IAA: n = 53; RC65 cells: control: n = 86, +IAA: n = 89; RC65+55 cells, control: n = 64, IAA: n = 70. In Fig. S3 B, the average number of cells analyzed for each time point was >10.
Stacks in Fig. S3 (BFA washout) were defined as Golgin-84 structures that associate with Golgin-97 elements by >50% of their length (Rabouille et al., 1995). The length of at least 10 of the largest Golgin-97 elements per cell and the associated Golgin-84 structures were measured using ImageJ. At least 10 cells for each time point per condition per experiment were analyzed in each of three independent experiments.
Transmission EM
Cells grown on glass-bottom dishes were fixed for 30 min with 2.5% (wt/vol) glutaraldehyde in 0.1 M sodium cacodylate, pH 7.4, stained with 1% osmium tetroxide and 0.8% (wt/vol) potassium cyanoferrate in 0.1 M sodium cacodylate, pH 7.4, for 1 h, treated with 2% uranyl acetate overnight, and embedded in Embed-812 resin (Electron Microscope Sciences). The coverslips were removed by hydrofluoric acid. Thin sections were cut and poststained with 2% (wt/vol) uranyl acetate and lead citrate. Images were acquired using a JEOL 1400 Plus Electron Microscope equipped with an LaB6 source using a voltage of 120 kV.
Online supplemental material
Fig. S1 shows that long-term TIR1 expression in the absence of auxin leads to partial loss of GRASP55 and 65. Fig. S2 shows that GRASP55 and 65 can be degraded in nocodazole-treated cells. Fig. S3 shows that acute degradation of GRASP55 and/or 65 does not affect Golgi stack reformation after BFA washout. Fig. S4 shows that GRASP55 and 65 can be degraded in STLC-arrested mitotic cells. Table S1 lists the oligonucleotides used but not listed in Materials and methods.
Supplementary Material
lists the oligonucleotides not listed in Materials and methods.
Acknowledgments
We thank Haijing Guo for suggestions and insightful discussions, Jen-Hsuan Wei for discussions and critical reading of the manuscript, Carlton Adams for comments and technical assistance, and the Live Cell Imaging facility and the Electron Microscopy core facility at the University of Texas Southwestern Medical Center for imaging support and for processing the EM samples.
This work was supported by the National Institutes of Health (grant GM096070) and the Welch Foundation (grant I-1910).
The authors declare no competing financial interests.
Author contributions: Y. Zhang and J. Seemann designed the project, performed the experiments, analyzed the data, prepared the figures, and wrote the manuscript.
References
- Bachert C., and Linstedt A.D.. 2010. Dual anchoring of the GRASP membrane tether promotes trans pairing. J. Biol. Chem. 285:16294–16301. 10.1074/jbc.M110.116129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F.A., Puype M., Vandekerckhove J., and Warren G.. 1997. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 91:253–262. 10.1016/S0092-8674(00)80407-9 [DOI] [PubMed] [Google Scholar]
- Barr F.A., Nakamura N., and Warren G.. 1998. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 17:3258–3268. 10.1093/emboj/17.12.3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F.A., Preisinger C., Kopajtich R., and Körner R.. 2001. Golgi matrix proteins interact with p24 cargo receptors and aid their efficient retention in the Golgi apparatus. J. Cell Biol. 155:885–892. 10.1083/jcb.200108102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard M., Satoh A., Shorter J., and Warren G.. 2005. A cryptic Rab1-binding site in the p115 tethering protein. J. Biol. Chem. 280:25840–25848. 10.1074/jbc.M503925200 [DOI] [PubMed] [Google Scholar]
- Bekier M.E. II, Wang L., Li J., Huang H., Tang D., Zhang X., and Wang Y.. 2017. Knockout of the Golgi stacking proteins GRASP55 and GRASP65 impairs Golgi structure and function. Mol. Biol. Cell. 28:2833–2842. 10.1091/mbc.e17-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanzi A., Hidalgo Carcedo C., Persico A., Cericola C., Turacchio G., Bonazzi M., Luini A., and Corda D.. 2007. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J. 26:2465–2476. 10.1038/sj.emboj.7601686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran J.M., Kinseth M., Bossard C., Rose D.W., Polishchuk R., Wu C.C., Yates J., Zimmerman T., and Malhotra V.. 2008. The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Mol. Biol. Cell. 19:2579–2587. 10.1091/mbc.e07-10-0998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein T.N., and Linstedt A.D.. 2008. GRASP55 regulates Golgi ribbon formation. Mol. Biol. Cell. 19:2696–2707. 10.1091/mbc.e07-11-1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Yu W., Li X., Lin S., Zhou Y., Hu J., and Liu X.. 2013. Structural insight into Golgi membrane stacking by GRASP65 and GRASP55 proteins. J. Biol. Chem. 288:28418–28427. 10.1074/jbc.M113.478024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro F., Kriston-Vizi J., Metcalf D.J., Martin-Martin B., Freeman J., Burden J.J., Westmoreland D., Dyer C.E., Knight A.E., Ketteler R., et al. . 2014. A two-tier Golgi-based control of organelle size underpins the functional plasticity of endothelial cells. Dev. Cell. 29:292–304. 10.1016/j.devcel.2014.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham A.K., and Munro S.. 2016. Finding the Golgi: Golgin Coiled-Coil Proteins Show the Way. Trends Cell Biol. 26:399–408. 10.1016/j.tcb.2016.02.005 [DOI] [PubMed] [Google Scholar]
- Grond R., Veenendaal T., Duran J.M., Raote I., van Es J.H., Corstjens S., Delfgou L., El Haddouti B., Malhotra V., and Rabouille C.. 2020. The function of GORASPs in Golgi apparatus organization in vivo. J. Cell Biol. 219:e202004191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizzunti G., and Seemann J.. 2016. Mitotic Golgi disassembly is required for bipolar spindle formation and mitotic progression. Proc. Natl. Acad. Sci. USA. 113:E6590–E6599. 10.1073/pnas.1610844113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Fachinetti D., Han J.S., and Cleveland D.W.. 2012. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl. Acad. Sci. USA. 109:E3350–E3357. 10.1073/pnas.1216880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., Spoorendonk K.M., and Rabouille C.. 2005. dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol. Biol. Cell. 16:4061–4072. 10.1091/mbc.e04-10-0938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T., Nishimura K., Kanemaki M.T., and Donaldson A.D.. 2013. The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol. Cell. 50:273–280. 10.1016/j.molcel.2013.02.012 [DOI] [PubMed] [Google Scholar]
- Kulak N.A., Pichler G., Paron I., Nagaraj N., and Mann M.. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 11:319–324. 10.1038/nmeth.2834 [DOI] [PubMed] [Google Scholar]
- Lavieu G., Dunlop M.H., Lerich A., Zheng H., Bottanelli F., and Rothman J.E.. 2014. The Golgi ribbon structure facilitates anterograde transport of large cargoes. Mol. Biol. Cell. 25:3028–3036. 10.1091/mbc.e14-04-0931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Tiwari N., Dunlop M.H., Graham M., Liu X., and Rothman J.E.. 2014. Membrane adhesion dictates Golgi stacking and cisternal morphology. Proc. Natl. Acad. Sci. USA. 111:1849–1854. 10.1073/pnas.1323895111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M. 2011. Structural organization of the Golgi apparatus. Curr. Opin. Cell Biol. 23:85–93. 10.1016/j.ceb.2010.10.004 [DOI] [PubMed] [Google Scholar]
- Lowe M. 2019. The Physiological Functions of the Golgin Vesicle Tethering Proteins. Front. Cell Dev. Biol. 7:94 10.3389/fcell.2019.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M., Rabouille C., Nakamura N., Watson R., Jackman M., Jämsä E., Rahman D., Pappin D.J., and Warren G.. 1998. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 94:783–793. 10.1016/S0092-8674(00)81737-7 [DOI] [PubMed] [Google Scholar]
- Lowe M., Gonatas N.K., and Warren G.. 2000. The mitotic phosphorylation cycle of the cis-Golgi matrix protein GM130. J. Cell Biol. 149:341–356. 10.1083/jcb.149.2.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhoul C., Gosavi P., and Gleeson P.A.. 2019. Golgi Dynamics: The Morphology of the Mammalian Golgi Apparatus in Health and Disease. Front. Cell Dev. Biol. 7:112 10.3389/fcell.2019.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Lowe M., Levine T.P., Rabouille C., and Warren G.. 1997. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 89:445–455. 10.1016/S0092-8674(00)80225-1 [DOI] [PubMed] [Google Scholar]
- Natsume T., Kiyomitsu T., Saga Y., and Kanemaki M.T.. 2016. Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep. 15:210–218. 10.1016/j.celrep.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., and Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 6:917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Pantazopoulou A., and Glick B.S.. 2019. A Kinetic View of Membrane Traffic Pathways Can Transcend the Classical View of Golgi Compartments. Front. Cell Dev. Biol. 7:153 10.3389/fcell.2019.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M.A., and Linstedt A.D.. 2004. Gene replacement reveals that p115/SNARE interactions are essential for Golgi biogenesis. Proc. Natl. Acad. Sci. USA. 101:1253–1256. 10.1073/pnas.0306373101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M.A., Bachert C., Puri S., Lanni F., and Linstedt A.D.. 2006. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 8:238–248. 10.1038/ncb1366 [DOI] [PubMed] [Google Scholar]
- Rabouille C., Misteli T., Watson R., and Warren G.. 1995. Reassembly of Golgi stacks from mitotic Golgi fragments in a cell-free system. J. Cell Biol. 129:605–618. 10.1083/jcb.129.3.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez I.B.-R., and Lowe M.. 2009. Golgins and GRASPs: holding the Golgi together. Semin. Cell Dev. Biol. 20:770–779. 10.1016/j.semcdb.2009.03.011 [DOI] [PubMed] [Google Scholar]
- Rebane A.A., Ziltener P., LaMonica L.C., Bauer A.H., Zheng H., López-Montero I., Pincet F., Rothman J.E., and Ernst A.M.. 2020. Liquid-liquid phase separation of the Golgi matrix protein GM130. FEBS Lett. 594:1132–1144. 10.1002/1873-3468.13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.E. 2010. The future of Golgi research. Mol. Biol. Cell. 21:3776–3780. 10.1091/mbc.e10-05-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk J.L., Höning K., Ebert T.S., and Hornung V.. 2016. CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism. Nat. Commun. 7:12338 10.1038/ncomms12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann J., Jokitalo E.J., and Warren G.. 2000a The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol. Biol. Cell. 11:635–645. 10.1091/mbc.11.2.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann J., Jokitalo E., Pypaert M., and Warren G.. 2000b Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature. 407:1022–1026. 10.1038/35039538 [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., et al. . 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima D.T., Haldar K., Pepperkok R., Watson R., and Warren G.. 1997. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J. Cell Biol. 137:1211–1228. 10.1083/jcb.137.6.1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short B., Preisinger C., Körner R., Kopajtich R., Byron O., and Barr F.A.. 2001. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155:877–884. 10.1083/jcb.200108079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., and Warren G.. 2002. Golgi architecture and inheritance. Annu. Rev. Cell Dev. Biol. 18:379–420. 10.1146/annurev.cellbio.18.030602.133733 [DOI] [PubMed] [Google Scholar]
- Shorter J., Watson R., Giannakou M.E., Clarke M., Warren G., and Barr F.A.. 1999. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18:4949–4960. 10.1093/emboj/18.18.4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin A.T.-W., and Harrison R.E.. 2016. Growth of the Mammalian Golgi Apparatus during Interphase. Mol. Cell. Biol. 36:2344–2359. 10.1128/MCB.00046-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D.A., DeBonis S., Saoudi Y., Lebeau L., Crevel I., Cross R., Wade R.H., Hackney D., and Kozielski F.. 2006. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J. Biol. Chem. 281:17559–17569. 10.1074/jbc.M511735200 [DOI] [PubMed] [Google Scholar]
- Slusarewicz P., Nilsson T., Hui N., Watson R., and Warren G.. 1994. Isolation of a matrix that binds medial Golgi enzymes. J. Cell Biol. 124:405–413. 10.1083/jcb.124.4.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohda M., Misumi Y., Yoshimura S., Nakamura N., Fusano T., Sakisaka S., Ogata S., Fujimoto J., Kiyokawa N., and Ikehara Y.. 2005. Depletion of vesicle-tethering factor p115 causes mini-stacked Golgi fragments with delayed protein transport. Biochem. Biophys. Res. Commun. 338:1268–1274. 10.1016/j.bbrc.2005.10.084 [DOI] [PubMed] [Google Scholar]
- Storrie B., Micaroni M., Morgan G.P., Jones N., Kamykowski J.A., Wilkins N., Pan T.H., and Marsh B.J.. 2012. Electron Tomography Reveals Rab6 Is Essential to the Trafficking of trans-Golgi Clathrin and COPI-Coated Vesicles and the Maintenance of Golgi Cisternal Number. Traffic. 13:727–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C., Hsu P., Mallabiabarrena A., and Malhotra V.. 2002. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 109:359–369. 10.1016/S0092-8674(02)00720-1 [DOI] [PubMed] [Google Scholar]
- Sütterlin C., Polishchuk R., Pecot M., and Malhotra V.. 2005. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol. Biol. Cell. 16:3211–3222. 10.1091/mbc.e04-12-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Yuan H., and Wang Y.. 2010. The role of GRASP65 in Golgi cisternal stacking and cell cycle progression. Traffic. 11:827–842. 10.1111/j.1600-0854.2010.01055.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenendaal T., Jarvela T., Grieve A.G., van Es J.H., Linstedt A.D., and Rabouille C.. 2014. GRASP65 controls the cis Golgi integrity in vivo. Biol. Open. 3:431–443. 10.1242/bio.20147757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Seemann J., Pypaert M., Shorter J., and Warren G.. 2003. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 22:3279–3290. 10.1093/emboj/cdg317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Satoh A., and Warren G.. 2005. Mapping the functional domains of the Golgi stacking factor GRASP65. J. Biol. Chem. 280:4921–4928. 10.1074/jbc.M412407200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.G., Clary D.O., and Rothman J.E.. 1992. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J. Cell Biol. 118:1015–1026. 10.1083/jcb.118.5.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J.-H., and Seemann J.. 2009a The mitotic spindle mediates inheritance of the Golgi ribbon structure. J. Cell Biol. 184:391–397. 10.1083/jcb.200809090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J.-H., and Seemann J.. 2009b Induction of asymmetrical cell division to analyze spindle-dependent organelle partitioning using correlative microscopy techniques. Nat. Protoc. 4:1653–1662. 10.1038/nprot.2009.160 [DOI] [PubMed] [Google Scholar]
- Wei J.-H., and Seemann J.. 2017. Golgi ribbon disassembly during mitosis, differentiation and disease progression. Curr. Opin. Cell Biol. 47:43–51. 10.1016/j.ceb.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J.-H., Zhang Z.C., Wynn R.M., and Seemann J.. 2015. GM130 Regulates Golgi-Derived Spindle Assembly by Activating TPX2 and Capturing Microtubules. Cell. 162:287–299. 10.1016/j.cell.2015.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., and Wang Y.. 2010. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J. Cell Biol. 188:237–251. 10.1083/jcb.200907132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Davis C.G., Brown M.S., Schneider W.J., Casey M.L., Goldstein J.L., and Russell D.W.. 1984. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell. 39:27–38. 10.1016/0092-8674(84)90188-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the oligonucleotides not listed in Materials and methods.