Abstract
The nucleus contains diverse phase-separated condensates that compartmentalize and concentrate biomolecules with distinct physicochemical properties. Here we consider whether condensates concentrate small molecule cancer therapeutics such that their pharmacodynamic properties are altered. We found that antineoplastic drugs become concentrated in specific protein condensates in vitro and that this occurs through physicochemical properties independent of the drug target. This behavior was also observed in tumor cells, where drug partitioning influenced drug activity. Altering the properties of the condensate was found to impact the concentration and activity of drugs. These results suggest that selective partitioning and concentration of small molecules within condensates contributes to drug pharmacodynamics and that further understanding of this phenomenon may facilitate advances in disease therapy.
One Sentence Summary:
Phase-separated condensates concentrate small molecule drugs impacting their on-target activity and pharmacodynamics.
The 5–10 billion protein molecules of cells are compartmentalized into both membrane-and non-membrane-bound organelles (1–3). Many non-membrane-bound organelles are phase-separated biomolecular condensates with distinct physicochemical properties that can absorb and concentrate specific proteins and nucleic acids (4–17). We reasoned that selective condensate partitioning might also occur with small molecule drugs whose targets occur within condensates (Figure 1A), and that the therapeutic index and efficacy of such compounds might therefore relate to their ability to partition into condensates that harbor their target. To test this idea, we focused our study on a collection of nuclear condensates previously reported in cell lines, demonstrated that they all occur in normal human cells and in tumor cells, and then developed in vitro condensate droplet assays with key components of each of the nuclear condensates to enable testing of small molecules.
Fig. 1.
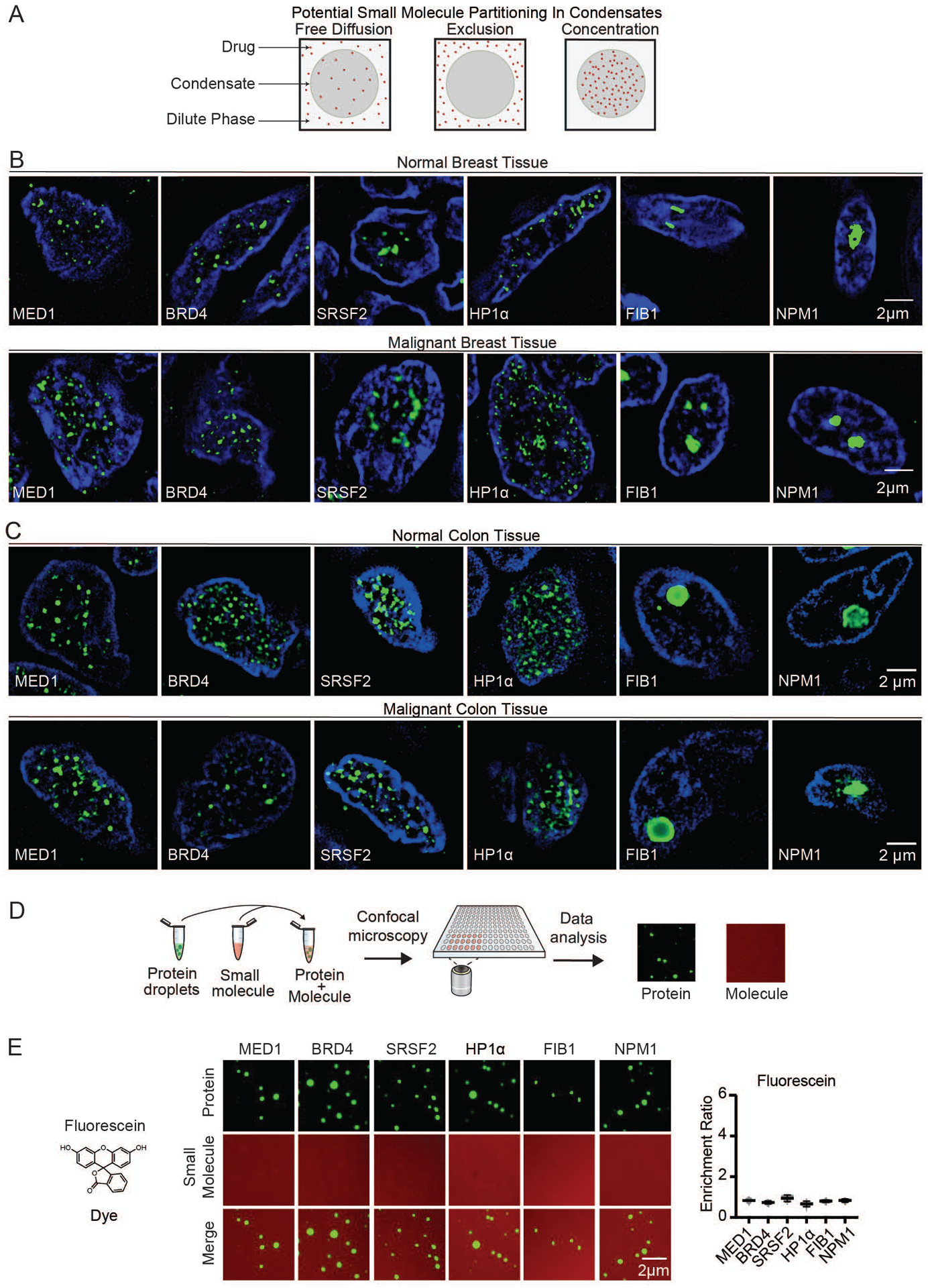
Nuclear condensates in human tissue and in vitro. (A) Model illustrating potential behaviors of small molecules in nuclear condensates. (B–C) Immunofluorescence of scaffold proteins of various nuclear condensates in tissue biopsies from benign and malignant human breast (B), and benign and malignant colon tissue (C), in nuclei stained with Hoechst, imaged at 100× on a fluorescent confocal microscope (see also Figures S1, S2). (D) Schematic of in vitro droplet formation assay to measure small molecule partitioning into nuclear condensates. (E) In vitro droplet assay showing the behavior of fluorescein dye in the presence of six protein condensates formed in 125mM NaCl and 10% PEG, with 10μM protein and 5μM fluorescein, imaged at 150× on a confocal fluorescent microscope (see also Figures S3–S6). Quantification of enrichment of the drug is shown to the right, error bars represent SEM.
Nuclear condensates have been described in diverse cultured cell lines and contain one or more proteins that can serve both as markers of the condensate and as a scaffold for condensate formation in droplet assays in vitro (10–12, 18–32). Specifically, transcriptional condensates are marked by the condensate forming proteins MED1 and BRD4 (10, 12, 19), splicing speckles by SRSF2 (11, 20), heterochromatin by HP1α (21, 22) and nucleoli by FIB1 and NPM1 (23–25) (Figure S1A). To determine whether such condensates can also be observed in the cells of healthy and malignant human tissue, we obtained biopsies of breast ductal epithelium, invasive ductal carcinoma, normal colon, and colon cancer (Figures S1B, S1C). Immunofluorescence revealed nuclear bodies containing these marker proteins in both normal and transformed tissue (Figures 1B, 1C). There was a broad distribution of nuclear body sizes and numbers, as expected for dynamic biomolecular condensates, and no significant differences were observed between benign and malignant tissue (Figures S2A–C). However, tumor cells acquire large super-enhancers (SEs) at driver oncogenes (33) and these can form tumor-specific transcriptional condensates.
We developed an assay to model these nuclear condensates and study the behavior of small molecules within these droplets (Figure 1D). We produced and purified recombinant fluorescently-labeled versions of MED1, BRD4, SRSF2, HP1α, FIB1, and NPM1 (Figure S3), and confirmed the ability of these proteins to form droplets in an in vitro assay (Figures S4A, S4B). To investigate the partitioning behavior of small molecules, we added the dyes Fluorescein (332Da) and Hoechst (452Da), as well as fluorescently-labeled dextrans (4.4 kilodaltons (kDa), to solutions containing each of the six protein condensates. The dyes and dextrans appeared to diffuse through all the condensates without substantial partitioning (Figures 1E, S5, S6A–D). Small molecule drugs are generally smaller than 1 kDa, so these results suggested that small molecules can freely diffuse through these nuclear condensates unless there are factors other than size that influence partitioning.
We next sought to determine whether diverse clinically important drugs with targets that reside in nuclear condensates also exhibit free diffusion across these condensates, or display a different behavior. Cisplatin and mitoxantrone, members of a class of antineoplastic compounds that modify DNA through platination or intercalation, can be modified to have fluorescent properties (cisplatin) (34) or are inherently fluorescent (mitoxantrone). When added to droplet formation buffer with purified MED1, BRD4, SRSF2, HP1α, FIB1, or NPM1, cisplatin was found to be selectively concentrated in MED1 droplets (Figures 2A, S7A), with a partition coefficient of up to 600 (Figures S8A–C). Fluorescent modification of cisplatin did not appear to contribute to this behavior in vitro, as the modified drug could be chased out of the condensate with unmodified cisplatin, and an isomer of cisplatin did not exhibit the same behavior (Figures S7B–D). Mitoxantrone was also concentrated in MED1 condensates, as well as in FIB1 and NPM1 condensates (Figures 2B, S7A, S8A–D). Consistent with these results, mitoxantrone is known to concentrate in the nucleolus where FIB1 and NPM1 reside (35, 36). These results show that in contrast to the dyes tested above, small molecule drugs may concentrate in certain condensates, even in the absence of the drug target.
Fig. 2.
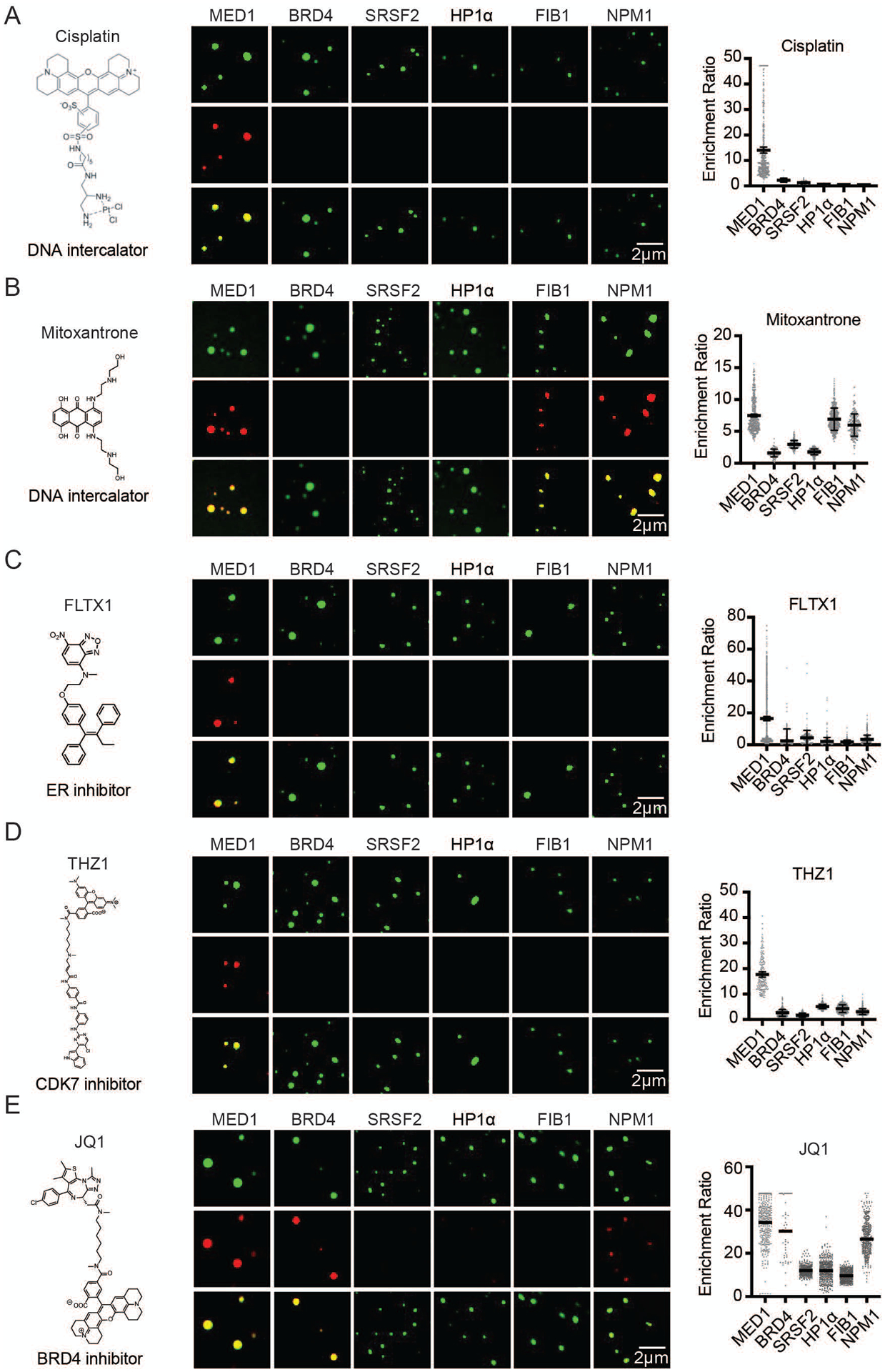
The partitioning behavior of small molecule drugs in nuclear condensates in a droplet assay. Six nuclear condensates formed in 125mM NaCl and 10% PEG, with 10μM protein treated with either (A) 5μM Cisplatin-TMR, (B) 50 μM Mitoxantrone, (C) 100μM FLTX1, (D) 5μM THZ1-TMR, or (D) 1μM JQ1-ROX imaged at 150× on a confocal fluorescent microscope (see also Figures S7–S11). Quantification of enrichment of the drug within droplets is shown to the right of each panel, error bars represent SEM (see also S12–S14).
We selected for further study antineoplastic drugs that target transcriptional regulators expected to be contained within transcriptional condensates in cells. These targets include: a) the estrogen receptor (ER), a transcription factor and nuclear hormone receptor, b) CDK7, a cyclin-dependent kinase that functions in transcription initiation and cell cycle control, and c) BRD4, a bromodomain protein and coactivator involved in oncogene regulation (Figures S9A, S9B). To monitor drug behavior with a confocal fluorescent microscope, we used a fluorescent tamoxifen analog (FLTX1), which targets ER, and modified fluorescent THZ1 and JQ1, which target CDK7 and BRD4, respectively (37, 38). FLTX1 and THZ1 concentrated preferentially in MED1 droplets (Figures 2C, 2D, S7A), and this behavior was not attributable to the fluorescent moiety (Figures S7B, S7D). JQ1 concentration presented a different pattern, being concentrated in MED1, BRD4, and NPM1 droplets (Figures 2E, S7A, S7B). Reinforcing these results, we found that the small molecules that concentrate in MED1 condensates were also concentrated in condensates formed from purified whole Mediator complexes (Figure S10A) and in MED1 condensates formed in an alternative crowding agent (Figure S11A). The targets of these three compounds (ERα, CDK7, and the bromodomains of BRD4) are not present in these in vitro condensates, but are present in the SEs that form condensates with transcription factors and Mediator in vivo (10, 12, 39) (Figures S9A, S9B), suggesting that the ability of some small molecules to concentrate preferentially in the same condensate as their protein target may contribute to the pharmacological properties of these drugs.
To gain additional insight into the nature of interactions governing small molecule enrichment in condensates, we focused on the MED1-IDR condensate. Fluorescence recovery after photobleaching (FRAP) experiments showed that cisplatin molecules are highly mobile in this condensate (Figures S12A, S12B), suggesting that the condensate produces a physiochemical environment that facilitates drug concentration in a state of high dynamic mobility. To gain insights into the chemical features of small molecules that may contribute to selective association with MED1 in condensates, we used a fluorescent boron-dipyrromethene (BODIPY) library of 81 compounds with various combinations of chemical side groups (Figure S13A). Molecules that contained aromatic rings were found to preferentially concentrate in MED1 condensates (Figures S13A–D, S14A). These data suggest that pi-pi or pi-cation interactions are among the physicochemical properties that favor small molecule partitioning into MED1 condensates. Aromatic amino acids participate in pi-system interactions, and are overrepresented in the MED1 IDR relative to the other condensate forming proteins studied (Figure S3B). We generated a MED1 aromatic mutant protein (all 30 aromatic amino acids mutated to alanine) which retained the ability to form droplets in vitro, indicating that the aromatic amino acids are not required for droplet formation (Figure S14B, S14C), but small molecule probes containing aromatic rings and the polar molecule cisplatin no longer partitioned into condensates formed by the MED1 aromatic mutant protein (Figures S14D–F). These results suggest that the aromatic residues of MED1 condensates contribute to the physicochemical properties that selectively concentrate these small molecules.
We anticipated that the ability of small molecules to concentrate in specific condensates would influence target engagement and thus drug pharmacodynamics. To investigate this, we took advantage of the ability of condensates to incorporate DNA (Figure 3A, S15A), and measured the relative efficiency of DNA platination by cisplatin in MED1 condensates, where cisplatin is concentrated, versus HP1α condensates, where cisplatin freely diffuses (Figure 2A). DNA platination, visualized by size-shift on a bioanalyzer, was more prevalent in MED1 condensates than in HP1α condensates (Figure 3B), consistent with the expectation that elevated concentrations of cisplatin in the MED1 condensates yield enhanced target engagement. If cisplatin becomes concentrated in Mediator condensates in cells, we would expect that DNA colocalized within Mediator condensates would be preferentially platinated. To test this idea, we performed co-immunofluorescence in cisplatin-treated HCT116 colon cancer cells using an antibody that specifically recognizes platinated DNA (Figure S16A) (40) together with antibodies specific for MED1, HP1α, or FIB1. Consistent with cisplatin’s preference for MED1 condensates in vitro, we found that platinated DNA frequently colocalized with MED1 condensates, but not with HP1α or FIB1 condensates (Figure 3C). To determine whether the ability of cisplatin to engage DNA is dependent on the presence of a MED1 condensate we treated cells with JQ1, which caused a loss of MED1 condensates (Figure S16B), and observed a concomitant reduction in platinated DNA at the MYC oncogene (Figures S16C, S16D). These results are consistent with the idea that concentration of small molecules in specific condensates can influence the efficiency of target engagement.
Fig. 3.
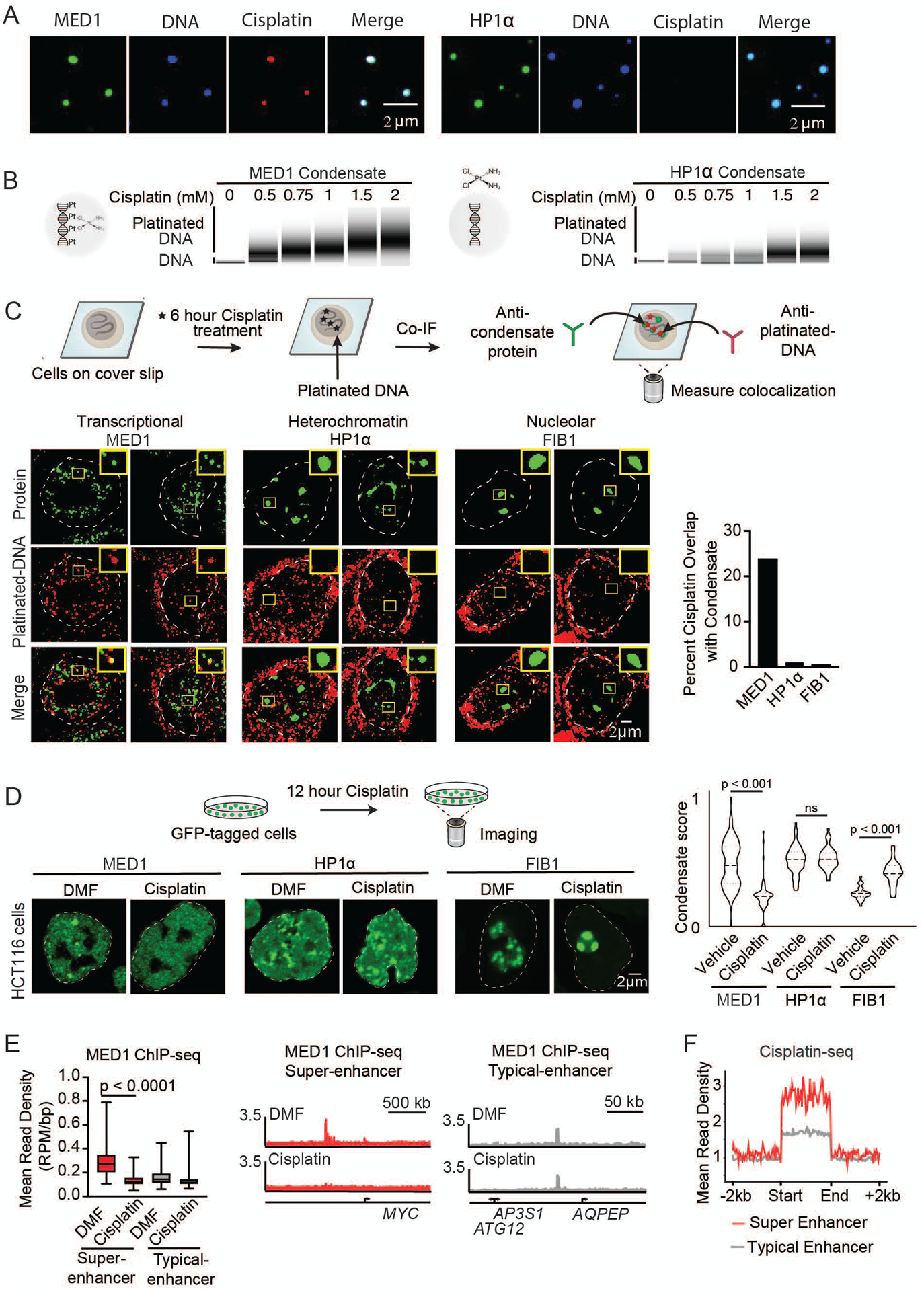
Small molecule concentration within condensates influences drug activity. (A) In vitro droplet assay of MED1 and HP1α condensates formed in 125mM NaCl and 10% PEG, 5nM of 450bp DNA, 10μM MED1, and 5μM cisplatin-TR, imaged at 150× on a confocal fluorescent microscope (see also Figure S15). (B) Bioanalyzer tracings of DNA contained within either MED1 or HP1α droplets exposed to the indicated concentration of cisplatin. (C) (Top) Schematic of an assay to determine the location of platinated DNA relative to various nuclear condensates. (Bottom) Co-immunofluorescence of platinated DNA and the indicated protein in HCT116 cells treated with 50μM cisplatin for 6 hours. Imaged at 100× on a confocal fluorescent microscope. Quantification of overlap shown to the right. (D) (Top) Schematic of a live cell condensate dissolution assay. (Bottom) HCT116 cells bearing endogenously mEGFP-tagged MED1, HP1α, or FIB1 treated with 50μM cisplatin for 12 hours. Quantification of MED1, HP1α, or FIB1 condensate score is shown to the right. (E) MED1 ChIP-seq in HCT116 cells treated with vehicle or 50μM cisplatin for 6 hours. (Left) Plotted are mean read density of MED1 at super-enhancers and typical-enhancers (error bars show min and max) and (Right) gene tracks of MED1 ChIP-Seq at the MYC super-enhancer and AQPEP typical-enhancer. (F) Metaplot of cisplatin-DNA-Seq in cisplatin treated Hela cells comparing super-enhancers and typical enhancers (41) (see also Figures S16–S21).
In cells, the preferential modification of DNA in MED1-containing condensates might be expected to selectively disrupt these condensates with prolonged treatment. To test this, HCT116 colon cancer cells were engineered to express GFP-tagged marker proteins for each of the 6 nuclear condensates (Figures S17A–G, S18A, S18B). When exposed to cisplatin, a selective and progressive reduction in MED1 condensates was observed (Figures 3D, S19A, S19B, S20A). Consistent with this, cisplatin treatment led to a preferential loss of MED1 ChIP-seq signal at SEs (Figures 3E S21A). Furthermore, high throughput sequencing data from platinated-DNA pull-down (41) revealed that cisplatin-modified DNA preferentially occurs at SEs, where MED1 is concentrated (42) (Figure 3F). These results are consistent with reports that cisplatin preferentially modifies transcribed genes (41, 43), and argue that this effect is due to preferential condensate partitioning. Taken together, these results suggest a model where cisplatin preferentially modifies SE DNA, which in turn leads to dissolution of these condensates. Previous studies have shown that diverse tumor cells become highly dependent on SE driven oncogene expression (44–48), which might explain why platinum drugs, which are capable of general DNA modification, are effective therapeutics in diverse cancers (49).
We explored the behavior of another clinically important antineoplastic drug, tamoxifen, to assess whether drug response and resistance are associated with partitioning in condensates (Figure 4A). ERα incorporates into MED1 condensates in an estrogen-dependent manner in vitro (12); droplet assays confirmed this and revealed that the addition of tamoxifen leads to eviction of ERα from the MED1 condensates (Figure 4B). We further investigated the effects of estrogen and tamoxifen on MED1 condensates in breast cancer cells, focusing on the MYC oncogene due to its prominent oncogenic role and responsiveness to estrogen (50). MED1 condensates were observed on the MYC oncogene in the ER+ breast cancer cell line MCF7 (S9A, S22A–D). DNA FISH with MED1 IF revealed that estrogen enhances formation of MED1 condensates at the MYC oncogene and tamoxifen treatment reduces these (Figures S23A, S23B). Artificial MED1 condensates without ER concentrated FLTX1 at the site of the condensate (Figure S24A), indicating that ER is not required for the partitioning of FLTX1 into MED1 condensates in cells. These results are consistent with the model that ERα interacts with MED1 condensates in an estrogen-dependent, tamoxifen-sensitive manner to drive oncogene expression in breast cancer cells.
Fig. 4.
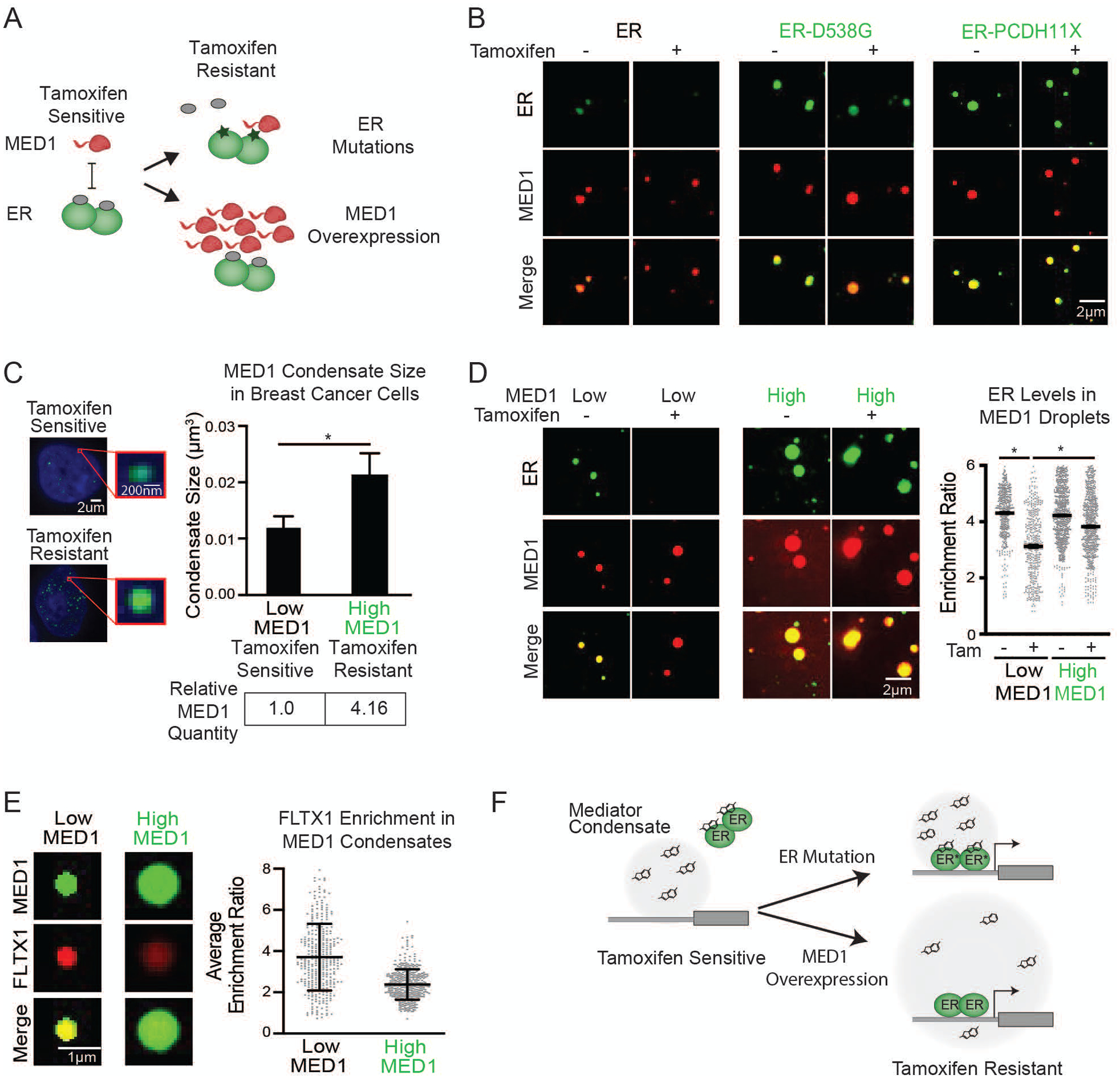
Tamoxifen action and resistance in MED1 condensates. (A) Schematic showing tamoxifen resistance by ER mutation and MED1 overexpression in breast cancer. (B) In vitro droplets assay of the indicated form of GFP-tagged ER in the presence of estrogen, +/− 100μM tamoxifen. Droplets are formed in 125mM NaCl and 10% PEG with 10μM each protein and 100μM estrogen. (C) (Left) Immunofluorescence of MED1 in tamoxifen sensitive (MCF7) and resistant (TAMR7) ER+ breast cancer cell lines imaged at 100× on a confocal fluorescent microscope. (Top right) Quantification of MED1 condensate size in breast cancer cells. (Bottom right) Relative quantities of MED1 in the indicated breast cancer cell line by western blot, error bars show SEM. (D) In vitro droplets assays of ER in the presence of 100μM estrogen, +/− 100μM tamoxifen with either 5μM (Low) or 20μM (High) MED1. Droplets are formed with 5μM ER in 125mM NaCl and 10% PEG, imaged at 150× on a confocal fluorescent microscope, error bars are SEM. (E) In vitro droplet assay with either 5μM (Low) or 20μM (High) MED1 with 100μM FLTX1 in 125mM NaCl and 10% PEG, error bars are SD. (F) Models for tamoxifen resistance due to altered drug affinity (via ER mutation) or concentration (via MED1 overexpression) (see also Figures S22–S30).
The mechanisms that produce drug resistance can provide clues to drug activity in the clinical setting. Endocrine therapy and tamoxifen resistance is an enduring clinical challenge and is associated with multiple mechanisms including ERα mutation and MED1 overexpression (Figure 4A, S25) (51–55). To investigate whether ERα mutations alter ERα behavior in condensates, we produced 4 patient-derived ERα mutant proteins and tested their partitioning in the presence of tamoxifen. In contrast to WT ERα, condensates composed of patient-derived ERα mutants and MED1 were not disrupted upon tamoxifen treatment (Figures 4B, S26A, S26B). The ERα point mutations reduce the affinity for tamoxifen approximately 10-fold (52), indicating that the drug concentration in the droplet is inadequate to evict these ER mutant proteins when this affinity is reduced.
MED1 overexpression is associated with tamoxifen resistance and poor prognosis in breast cancer (51), but it is not clear why overexpression of one subunit of the Mediator complex produces resistance. We considered the possibility that overexpressed MED1 is incorporated into transcriptional condensates, which contain clusters of Mediator molecules (39), thereby expanding their volumes and diluting the available tamoxifen (Figure S27A). We found that the tamoxifen-resistant breast cancer cell line TAMR7 (56), which was derived from the tamoxifen-sensitive cell line MCF7, produces 4-fold elevated levels of MED1 protein (Figure S27B). The volume of MED1-containing condensates is 2-fold larger in these cells (Figures 4C, S27C). When modeled in an in vitro droplet assay, we found that a 4-fold increase in MED1 levels led to a commensurate increase in droplet size (Figures S28A, S28B). Furthermore, we found that 100μM tamoxifen prevented ERα incorporation into MED1 condensates (Figures 4B, 4D), but was much less effective in preventing ERα incorporation into the larger MED1 condensates produced with higher MED1 levels (Figure 4D). To confirm that the levels of tamoxifen in the larger droplets are more dilute, we measured the enrichment of the fluorescent tamoxifen analog FLTX1 in MED1 droplets, and found that the larger condensates have lower concentrations of the drug (Figure 4E). These results were mirrored in cells, where a collection of tethered ERα molecules form a MED1 condensate that is eliminated by tamoxifen, but when MED1 is overexpressed tamoxifen is unable to dissociate the ERα-MED1 condensate (Figure S29A). Similarly, knockdown of MED1 in tamoxifen resistant breast cancer cells sensitizes cells to tamoxifen (51, 55). These results support a model of tamoxifen resistance where MED1 overexpression causes the formation of larger transcriptional condensates, in which tamoxifen is diluted and thereby less effective in dissociating ER from the condensate (Figure 4F).
Our results show that drugs partition selectively into condensates, that this can occur through physicochemical properties that exist independent of their molecular targets, and that cells can develop resistance to drugs through condensate altering mechanisms. This may explain the surprising observation that inhibition of global gene regulators such as BRD4 or CDK7 can have selective effects on oncogenes that have acquired large SEs (46); selective partitioning of inhibitors like JQ1 and THZ1 into SE condensates will preferentially disrupt transcription at those loci. These results also have implications for future development of efficacious disease therapeutics; effective target-engagement will depend on measurable factors such as drug partitioning in condensates (Figures S30A–D). Condensate assays of the type described here may thus help optimize condensate partitioning, target engagement, and the therapeutic index of small molecule drugs.
Supplementary Material
Acknowledgments:
We thank Chris Glinkerman for helpful comments, Wendy Salmon of the W.M Keck Microscopy Facility, and Tom Volkert, Jennifer Love, Stephen Mraz, and Sumeet Gupta of the Whitehead Genome Technologies Core for technical assistance. We thank the light microscopy facility at the MPI-CBG in Dresden for extensive help and support.
Funding: This work was supported by NIH grants GM123511, CA213333, CA155258 (RAY), NSF grant PHY1743900 (RAY), NIH grant GM117370 (DJT), American Society of Clinical Oncology Young Investigator Award (IAK), American Cancer Society Postdoctoral Fellowship (IAK), Ovarian Cancer Research Alliance Mentored Investigator Award (IAK), Swedish Research Council Postdoctoral Fellowship (VR 2017-00372) (AB), Hope Funds for Cancer Research (AD), Gruss-Lipper Postdoctoral Fellowship and by the Rothschild Postdoctoral Fellowship (IS), NIH grant T32:5T32DK007191-45 (JMP), German Research Foundation DFG postdoctoral fellowship DE 3069/1-1 (T-MD), Cancer Research Institute Irvington Fellowship (YEG), Damon Runyon Cancer Research Foundation Fellowship (2309-17) (BRS), ELBE postdoctoral fellowship (PM).
Footnotes
Data and materials availability: All data is available in the main text or the supplementary materials. High-throughput sequencing data sets will be made publicly available in GEO.
Supplementary Materials:
Materials and Methods
Figures S1–S30
References:
- 1.Shin Y, Brangwynne CP, Liquid phase condensation in cell physiology and disease. Science (80-.) 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Banani SF, Lee HO, Hyman AA, Rosen MK, Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hyman AA, Weber CA, Jülicher F, Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol 30, 39–58 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, Jaffrey SR, m6A enhances the phase separation potential of mRNA. Nature. 571, 424–428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langdon EM, Gladfelter AS, A New Lens for RNA Localization: Liquid-Liquid Phase Separation. Annu. Rev. Microbiol 72, 255–271 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, Myong S, Gladfelter AS, mRNA structure determines specificity of a polyQ-driven phase separation. Science (80-.) 360, 922–927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei M-T, Chang Y-C, Shimobayashi SF, Shin Y, Brangwynne CP, Nucleated transcriptional condensates amplify gene expression. bioRxiv, 737387 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Nott TJJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DPP, Pawson T, Forman-Kay JDD, Baldwin AJJ, Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Mol. Cell 57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nott TJ, Craggs TD, Baldwin AJ, Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat. Chem 8, 569–575 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Tong IL, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA, Coactivator condensation at super-enhancers links phase separation and gene control. Science (80-.) 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall’Agnese A, Hannett NM, Spille J-H, Afeyan LK, Zamudio AV, Shrinivas K, Abraham BJ, Boija A, Decker T-M, Rimel JK, Fant CB, Lee TI, Cisse II, Sharp PA, Taatjes DJ, Young RA, Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 572, 543–548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, V Zamudio A, Li CH, Shrinivas K, Manteiga JC, Hannett NM, Abraham BJ, Afeyan LK, Guo YE, Rimel JK, Fant CB, Schuijers J, Lee TI, Taatjes DJ, Young RA, Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell. 175, 1842–1855.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N, Granata D, Marzahn MR, Lindorff-Larsen K, Salvatella X, Schulman BA, Mittag T, Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol. Cell 72, 19–36.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrinivas K, Sabari BR, Coffey EL, Klein IA, Boija A, V Zamudio A, Schuijers J, Hannett NM, Sharp PA, Young RA, Chakraborty AK, Enhancer Features that Drive Formation of Transcriptional Condensates. Mol. Cell 75, 549–561.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bentley EP, Frey BB, Deniz AA, Physical Chemistry of Cellular Liquid-Phase Separation. Chemistry. 25, 5600–5610 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, McSwiggen DT, Kokic G, Dailey GM, Cramer P, Darzacq X, Zweckstetter M, RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol 25, 833–840 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Cramer P, Organization and regulation of gene transcription. Nature. 573, 45–54 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Alberti S, Saha S, Woodruff JB, Franzmann TM, Wang J, Hyman AA, A User’s Guide for Phase Separation Assays with Purified Proteins. J. Mol. Biol 430, 4806–4820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA, Perspective A Phase Separation Model for Transcriptional Control. Cell. 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Belmont AS, Genome organization around nuclear speckles. Curr. Opin. Genet. Dev 55, 91–99 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ, Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 547, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH, Phase separation drives heterochromatin domain formation. Nat. Publ. Gr 547, 241–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP, Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW, Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife. 5 (2016), doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitrea DM, Cika JA, Stanley CB, Nourse A, Onuchic PL, Banerjee PR, Phillips AH, Park C-G, Deniz AA, Kriwacki RW, Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun 9, 842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK, Compositional Control of Phase-Separated Cellular Bodies. Cell. 166, 651–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel A, Lee HOO, Jawerth L, Maharana S, Jahnel M, Hein MYY, Stoynov S, Mahamid J, Saha S, Franzmann TMM, Pozniakovski A, Poser I, Maghelli N, Royer LAA, Weigert M, Myers EWW, Grill S, Drechsel D, Hyman AAA, Alberti S, A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Alberti S, The wisdom of crowds: regulating cell function through condensed states of living matter. J. Cell Sci 130, 2789–2796 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Posey AE, Holehouse AS, Pappu RV, in Methods in enzymology (2018; https://linkinghub.elsevier.com/retrieve/pii/S0076687918304099), vol. 611, pp. 1–30. [DOI] [PubMed] [Google Scholar]
- 30.Shin Y, Brangwynne CP, Liquid phase condensation in cell physiology and disease. Science (80-.) 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Uversky VN, Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol 44, 18–30 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Mao YS, Zhang B, Spector DL, Biogenesis and function of nuclear bodies. Trends Genet. 27, 295–306 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA, XSuper-enhancers in the control of cell identity and disease. Cell. 155, 934–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu YH, Sibrian-Vazquez M, Escobedo JO, Phillips AR, Dickey DT, Wang Q, Ralle M, Steyger PS, Strongin RM, Systemic Delivery and Biodistribution of Cisplatin in Vivo. Mol. Pharm 13, 2677–2682 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vibet S, Mahéo K, Goré J, Dubois P, Bougnoux P, Chourpa I, Differential subcellular distribution of mitoxantrone in relation to chemosensitization in two human breast cancer cell lines. Drug Metab. Dispos 35, 822–8 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Smith PJ, Sykes HR, Fox ME, Furlong IJ, Subcellular distribution of the anticancer drug mitoxantrone in human and drug-resistant murine cells analyzed by flow cytometry and confocal microscopy and its relationship to the induction of DNA damage. Cancer Res. 52, 4000–8 (1992). [PubMed] [Google Scholar]
- 37.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, Dastur A, Amzallag A, Ramaswamy S, Tesar B, Jenkins CE, Hannett NM, McMillin D, Sanda T, Sim T, Kim ND, Look T, Mitsiades CS, Weng AP, Brown JR, Benes CH, Marto JA, Young RA, Gray NS, Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 511, 616–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marrero-Alonso J, Morales A, García Marrero B, Boto A, Marín R, Cury D, Gómez T, Fernández-Pérez L, Lahoz F, Díaz M, Unique SERM-like properties of the novel fluorescent tamoxifen derivative FLTX1. Eur. J. Pharm. Biopharm 85, 898–910 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, Cisse II, Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tilby MJ, Johnson C, Knox RJ, Cordell J, Roberts JJ, Dean CJ, Sensitive detection of DNA modifications induced by cisplatin and carboplatin in vitro and in vivo using a monoclonal antibody. Cancer Res. 51, 123–9 (1991). [PubMed] [Google Scholar]
- 41.Shu X, Xiong X, Song J, He C, Yi C, Base-Resolution Analysis of Cisplatin-DNA Adducts at the Genome Scale. Angew. Chem. Int. Ed. Engl 55, 14246–14249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA, Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rovira-Clave X, Jiang S, Bai Y, Barlow GL, Bhate S, Coskun A, Han G, Zhu B, Ho C-M, Hitzman C, Chen S-Y, Bava F-A, Nolan G, Subcellular localization of drug distribution by super-resolution ion beam imaging. bioRxiv, 557603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Zhang T, Kwiatkowski N, Abraham BJ, Lee TI, Xie S, Yuzugullu H, Von T, Li H, Lin Z, Stover DG, Lim E, Wang ZC, Iglehart JD, Young RA, Gray NS, Zhao JJ, CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell. 163, 174–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansour MR, Abraham JM, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, Loh ML, Hunger SP, Sanda T, Young RA, Look AT, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, Loh ML, Hunger SP, Sanda T, Young RA, Look AT, An Oncogenic Super-Enhancer Formed Through Somatic Mutation of a Noncoding Intergenic Element. Science (80-.) 346, 1373–1377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradner JE, Hnisz D, Young RA, Transcriptional Addiction in Cancer. Cell. 168, 629–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA, Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, Zhao K, Levens D, c-Myc Is a Universal Amplifier of Expressed Genes in Lymphocytes and Embryonic Stem Cells. Cell. 151, 68–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dasari S, Bernard Tchounwou P, Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol 740 (2014), pp. 364–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dubik D, Dembinski TC, R. P. C. Shiu4, “Stimulation of c-myc Oncogene Expression Associated with Estrogen-induced Proliferation of Human Breast Cancer Cells1” (1987). [PubMed] [Google Scholar]
- 51.Nagalingam A, Tighiouart M, Ryden L, Joseph L, Landberg G, Saxena NK, Sharma D, Med1 plays a critical role in the development of tamoxifen resistance. Carcinogenesis. 33, 918–30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fanning SW, Mayne CG, Dharmarajan V, Carlson KE, Martin TA, Novick SJ, Toy W, Green B, Panchamukhi S, Katzenellenbogen BS, Tajkhorshid E, Griffin PR, Shen Y, Chandarlapaty S, Katzenellenbogen JA, Greene GL, Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. Elife. 5 (2016), doi: 10.7554/eLife.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, Ali S, Chin SF, Palmieri C, Caldas C, Carroll JS, Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 481, 389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murtaza M, Dawson SJ, Tsui DWY, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong ASC, Marass F, Humphray S, Hadfield J, Bentley D, Chin TM, Brenton JD, Caldas C, Rosenfeld N, Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 497, 108–112 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Cui J, Germer K, Wu T, Wang J, Luo J, Wang SC, Wang Q, Zhang X, Cross-talk between HER2 and MED1 regulates tamoxifen resistance of human breast cancer cells. Cancer Res. 72, 5625–5634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hole S, Pedersen AM, Hansen SK, Lundqvist J, Yde CW, Lykkesfeldt AE, New cell culture model for aromatase inhibitor-resistant breast cancer shows sensitivity to fulvestrant treatment and cross-resistance between letrozole and exemestane. Int. J. Oncol 46, 1481–1490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, Er JC, Jiang H, Li X, Luo Z, Ramezani T, Feng Y, Tang MK, Chang YT, Vendrell M, A highly selective fluorogenic probe for the detection and: In vivo imaging of Cu/Zn superoxide dismutase. Chem. Commun 52, 9093–9096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meyer KD, Donner AJ, Knuesel MT, York AG, Espinosa JM, and Taatjes DJ, Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J. 27, 1447–57 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA, Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 482, 221–225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rubinstein M, Colby RH, Polymer Phyics (Oxford University Press, 2003). [Google Scholar]
- 62.Mittasch M, Gross P, Nestler M, Fritsch AW, Iserman C, Kar M, Munder M, Voigt A, Alberti S, Grill SW, Kreysing M, Non-invasive perturbations of intracellular flow reveal physical principles of cell organization. Nat. Cell Biol 20, 344–351 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Zhao H, Brown PH, Schuck P, On the distribution of protein refractive index increments. Biophys. J 100, 2309–2317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, Ali R, Yunus AA, Liu DR, V Pappu R, Rosen MK, Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol. Cell 63, 72–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holehouse AS, Das RK, Ahad JN, Richardson MOG, V Pappu R, CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophys. J 112, 16–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei M-T, Elbaum-Garfinkle S, Holehouse AS, Chih-Hsiung Chen C, Feric M, Arnold CB, Priestley RD, V Pappu R, Brangwynne CP, Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. nature.com (2017), doi: 10.1038/NCHEM.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delage-Mourroux R, Martini PGV, Choi I, Kraichely DM, Hoeksema J, Katzenellenbogen BS, Analysis of estrogen receptor interaction with a repressor of estrogen receptor activity (REA) and the regulation of estrogen receptor transcriptional activity by REA. J. Biol. Chem 275, 35848–35856 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Broad Institute Cancer Cell Line Encyclopedia (CCLE), (available at https://portals.broadinstitute.org/ccle).
- 69.cBioPortal for Cancer Genomics, (available at https://www.cbioportal.org/).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


