Abstract
Cortical computations are critically reliant on their local circuit, GABAergic cells. In the hippocampus, a large body of work has identified an unprecedented diversity of GABAergic interneurons with pronounced anatomical, molecular and physiological differences. Yet, little is known about the functional properties and activity dynamics of the major hippocampal interneuron classes in behaving animals. Here we use fast, targeted, three-dimensional (3D) two-photon calcium imaging coupled with immunohistochemistry-based molecular identification to retrospectively map in vivo activity onto multiple classes of interneurons in the mouse hippocampal area CA1 during head-fixed exploration and goal-directed learning. We find examples of preferential subtype recruitment with quantitative differences in response properties and feature selectivity during key behavioral tasks and states. These results provide new insights into the collective organization of local inhibitory circuits supporting navigational and mnemonic functions of the hippocampus.
In Brief:
Geiller, Vancura, et al. utilized 3D AOD-based 2-photon functional imaging and post hoc immunohistochemistry in mouse hippocampal region CA1 to simultaneously image and characterize the dynamics of hundreds of molecularly-defined interneurons in vivo during spatial navigation, quiet wakefulness, and goal-oriented spatial learning.
Introduction
Local circuits throughout the mammalian brain are composed of relatively homogenous populations of glutamatergic principal cells and numerically fewer but highly diverse gamma-aminobutyric acid (GABA)-releasing interneurons (INs). While cortical functions are thought to be primarily carried out by excitatory cells, GABAergic inhibition provided by INs has been shown to be indispensable for a range of elementary cortical computations (Isaacson and Scanziani, 2011; Tremblay, Lee and Rudy, 2016; Fishell and Kepecs, 2020).
Much of our current knowledge about cortical INs comes from decades of investigations in the mammalian hippocampus. Pioneering studies have uncovered an astounding anatomical, physiological, developmental and molecular diversity of hippocampal INs and parsed them into distinct cell-types in each hippocampal region (Freund and Buzsaki, 1996; Klausberger and Somogyi, 2008; Pelkey et al., 2017; Booker and Vida, 2018). To date, there has been an unprecedented characterization of the IN populations that comprise hippocampal region CA1, revealing several organizational principles of local circuit inhibition. First, excitatory input-output transformation in CA1 pyramidal cells (CA1PCs) is under strong inhibitory control by feedforward and feedback inhibitory circuits (Pouille and Scanziani, 2001, 2004; Lovett-Barron et al., 2012). Second, subpopulations of CA1 INs inhibit specific subcellular compartments of CA1PCs, such as the axon initial segment, soma, and different dendritic domains (Buhl, Halasy and Somogyi, 1994; Freund and Buzsaki, 1996; Klausberger and Somogyi, 2008; Bloss et al., 2016). Third, these IN subtypes are differentially modulated during hippocampal network oscillations, thus organizing the release of GABA onto PCs in both space and time (Klausberger et al., 2003, 2004; Somogyi et al., 2014). Finally, subpopulations of INs preferentially innervate other INs (Acsády, Görcs and Freund, 1996; Gulyás, Hájos and Freund, 1996; Chamberland and Topolnik, 2012), as well as distant brain regions via long-range projections (Jinno et al., 2007; Caputi et al., 2013; Katona et al., 2017; Francavilla et al., 2018; Wick, Tetzlaff and Krook-Magnuson, 2019).
While these features of inhibitory circuit organization collectively point to specialized behavioral functions for IN subtypes in the hippocampus, this long-standing hypothesis remains largely open as little is known about the in vivo activity dynamics of identified INs in behaving animals. This knowledge gap partly stems from the limited ability of traditional electrophysiological techniques to study the in vivo population-level activity dynamics of identified INs, as they are either too low-throughput or unable to molecularly or morphologically identify the recorded cells (Dupret, O’Neill and Csicsvari, 2013; English et al., 2017, Klausberger et al., 2003; Varga, Golshani and Soltesz, 2012). Similarly, more recent one- or two-photon (2p) imaging approaches in Cre-driver lines are typically limited to one or several imaging planes and broad classes of INs (Arriaga and Han, 2017, 2019; Sheffield, Adoff and Dombeck, 2017; Turi et al., 2019). An optimal strategy would offer monitoring of the collective activity dynamics of all INs in large tissue volumes in vivo in combination with post hoc multiplexed characterization of the recorded cells, as has been implemented in the neocortex (Langer and Helmchen, 2012; Kerlin et al., 2010; Khan et al., 2018). As a step towards a comprehensive characterization of inhibitory circuit dynamics in the behaving mouse hippocampus, here we combine acousto-optic deflection (AOD) microscopy-based 3D calcium imaging (Katona et al., 2012; Szalay et al., 2016) with post hoc, multiplexed fluorescence immunohistochemistry to record simultaneously from populations of molecularly-defined INs throughout all CA1 strata during spatial navigation, learning, and sharp-wave ripple (SWR) events during quiet wakefulness. Our results demonstrate the validity and utility of this approach, as they largely confirm previous findings in cases where similar data is available, extend previous findings in cases where existing data is hampered by small sample sizes or a lack of subtype-specific information, and provide several novel insights about IN subtype dynamics during behavior.
Results
Fast, targeted in vivo calcium imaging in 3D and post hoc molecular identification of CA1 INs
To obtain large-scale functional recordings of CA1 INs, we performed 3-dimensional (3D) AOD-2p calcium imaging following virus injection in dorsal CA1 of VGAT-Cre mice to express the genetically encoded calcium sensor GCaMP6f in all INs. Then, we implanted a cannula window over dorsal CA1 and trained mice to run head-fixed on a treadmill. We used AOD-2p microscopy to rapidly target small frames in 3D surrounding INs located within an 800×800×500 μm volume (total of 3626 INs in 16 mice, 226.6 ± 27.3 INs per mouse, mean ± std) from the CA1 stratum oriens/alveus border to the stratum lacunosum moleculare, and imaged them simultaneously at a rate of 4–5 Hz during behavior (Figure 1A, 1B, 1D, 1E, 1F, S1). Post hoc immunohistochemistry was then performed in fixed brain slices and the molecular identity of imaged INs was retrospectively determined by registering confocal images to in vivo 2p stacks (Figure 1B, 1C, S1). In the first set of experiments we focused on CA1PC-targeting INs by using a combination of 5 molecular markers (Methods, Figure 1C, S1) to identify 6 subsets of INs that account for ~80% of the total IN population in CA1 (Bezaire and Soltesz, 2013). These included three types of parvalbumin (PV)-immunopositive INs: basket cells (PVBC), axo-axonic cells (AAC) and bistratified cells (BiC). In addition, three broader groups included somatostatin (SOM)-expressing cells (SomC), which comprises oriens lacunosum moleculare (OLM) INs and long-range projecting cells; cholecystokinin (CCK)-expressing cells (CCKC), which contain basket and dendrite-targeting INs; and neuropeptide Y (NPY)-immunopositive and SOM-immunonegative ivy/neurogliaform cells (IvC/NGFC) (Freund and Buzsaki, 1996; Klausberger and Somogyi, 2008; Pelkey et al., 2017). A total of 3129 INs were found post hoc in the slices (195.6 ± 42.6 per mouse, mean ± std), and 1936 were assigned to a putative IN subtype identity (121.0 ± 28.4 per mouse, mean ± std). The overall proportion (Figure S1) and layer distribution of each of these subtypes was consistent with previous studies (Figure 1F), which used detailed morphological reconstructions for cell-type identification, demonstrating the utility of our method for high-throughput recordings of distinct IN subpopulations.
Figure 1 -. AOD-based two-photon imaging in 3D and interneuron subtype identification.
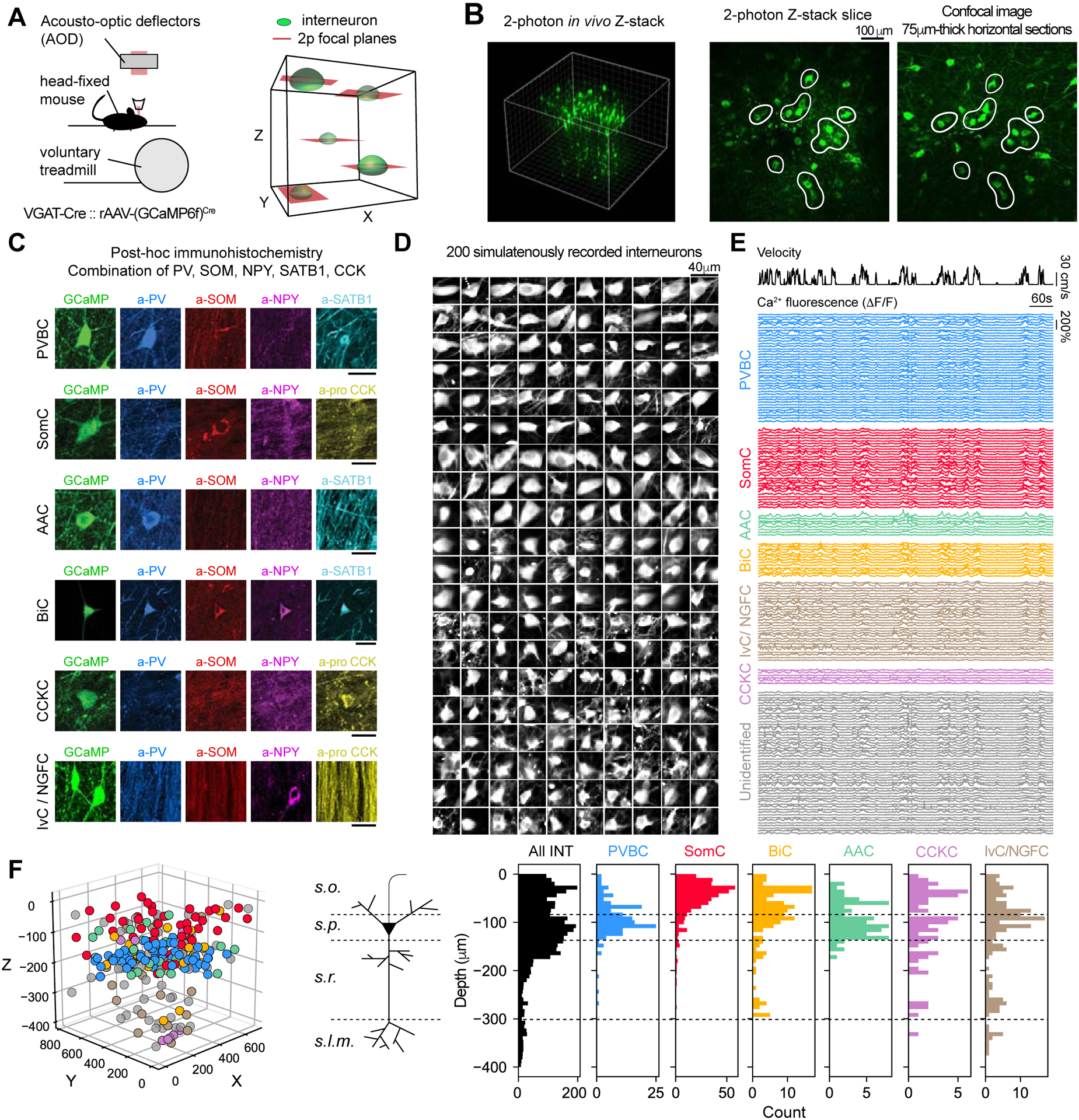
A. Left: Experimental setup. VGAT-Cre mice injected in CA1 with a rAAV to express GCaMP6f in all interneuron run voluntarily on a treadmill. Right: Schematic of 3D chessboard scanning, based on fast AOD imaging.
B. Left: 3D rendering of an in vivo CA1 Z-stack, 800×800×450μm FOV. Green cells are GCaMP6f-expressing interneurons. Middle: XY-orthoslice of the Z-stack. Right: Confocal image of a 75μm-thick horizontal slice from the same brain after fixation. Cells in slices can be found in the in vivo Z-stack (white contours).
C. Post hoc immunohistochemistry is performed on horizontal slices using a combination of markers and cells are assigned a subtype among parvalbumin basket cells (PVBCs), somatostatin-expressing cells (SomC), bistratified cells (BiC), axo-axonic cells (AAC), cholecystokinin-expressing cells (CCKCs), Ivy/Neurogliaform cells (IvC/NGFC) or unidentified cells.
D. Example time-averaged images of 200 simultaneously recorded interneurons.
E. Relative GCaMP fluorescence (ΔF/F) traces from molecularly-identified interneurons from D.
F. Left: X-Y-Z positions of molecularly-identified interneurons from E. Right: Depth distribution for all interneurons (black, all INT) and molecularly-identified subtypes (n = 16 mice). Depth 0 corresponds to the position of the most dorsal interneuron in each mouse, located at the top of stratum oriens (see CA1 pyramidal cell schematic). s.o.: stratum oriens, s.p.: stratum pyramidale, s.r.:stratum radiatum, s.l.m.: stratum lacunosum-moleculare.
Locomotion-state-dependent activity of molecularly identified IN subpopulations during spatial navigation
To characterize the relationship between the ambulatory state of the animal and each IN’s activity, we imaged mice that were water-restricted and trained to run on a 2m-long belt rich in tactile cues during a random foraging task (Figure 2A). We first computed the Pearson’s correlation between the animal’s velocity and each cell’s activity (Figure 2B) and found that the majority of INs were positively correlated with velocity (Varga, Golshani and Soltesz, 2012; Lapray et al., 2012; Varga et al., 2014; Katona et al., 2014; Lee et al., 2014; Somogyi et al., 2014; Fuhrmann et al., 2015; Arriaga and Han, 2017, 2019; Francavilla et al., 2018; Turi et al., 2019) (Figure 2C). However, we also observed that ~10% of cells exhibited negative correlations, present across all subtypes (Figure 2C). Negatively correlated cells were significantly enriched in CCKC, where nearly half of cells showed increased activity during immobility (Figure 2B, 2C). We also found that IvC/NGFC were less modulated by locomotion, consistent with previous results (Fuentealba et al., 2008; Lapray et al., 2012). To examine whether the activity-velocity modulations are reflected in ambulatory state transitions, we calculated the average run-start and run-stop responses for each cell (Figure S2), which further detailed the unicity of immobility-triggered activation of CCKC. Regardless of molecular identity, negatively correlated cells were significantly enriched in stratum oriens, while positively correlated cells were predominantly in stratum pyramidale (Figure 2D), with small but noticeable differences in multiple subtypes (Figure S2). In addition, while the activity of positively correlated cells generally followed changes in velocity by up to several seconds, the activity of negatively correlated cells was more tightly linked in time to changes in velocity (Figure S2). Together, these results demonstrate that locomotion positively modulates the activity of most INs across molecularly defined subtypes, while identifying CCKC as comprising a significant population of negatively modulated cells.
Figure 2 -. Locomotion-state-dependent activity of IN subtypes.
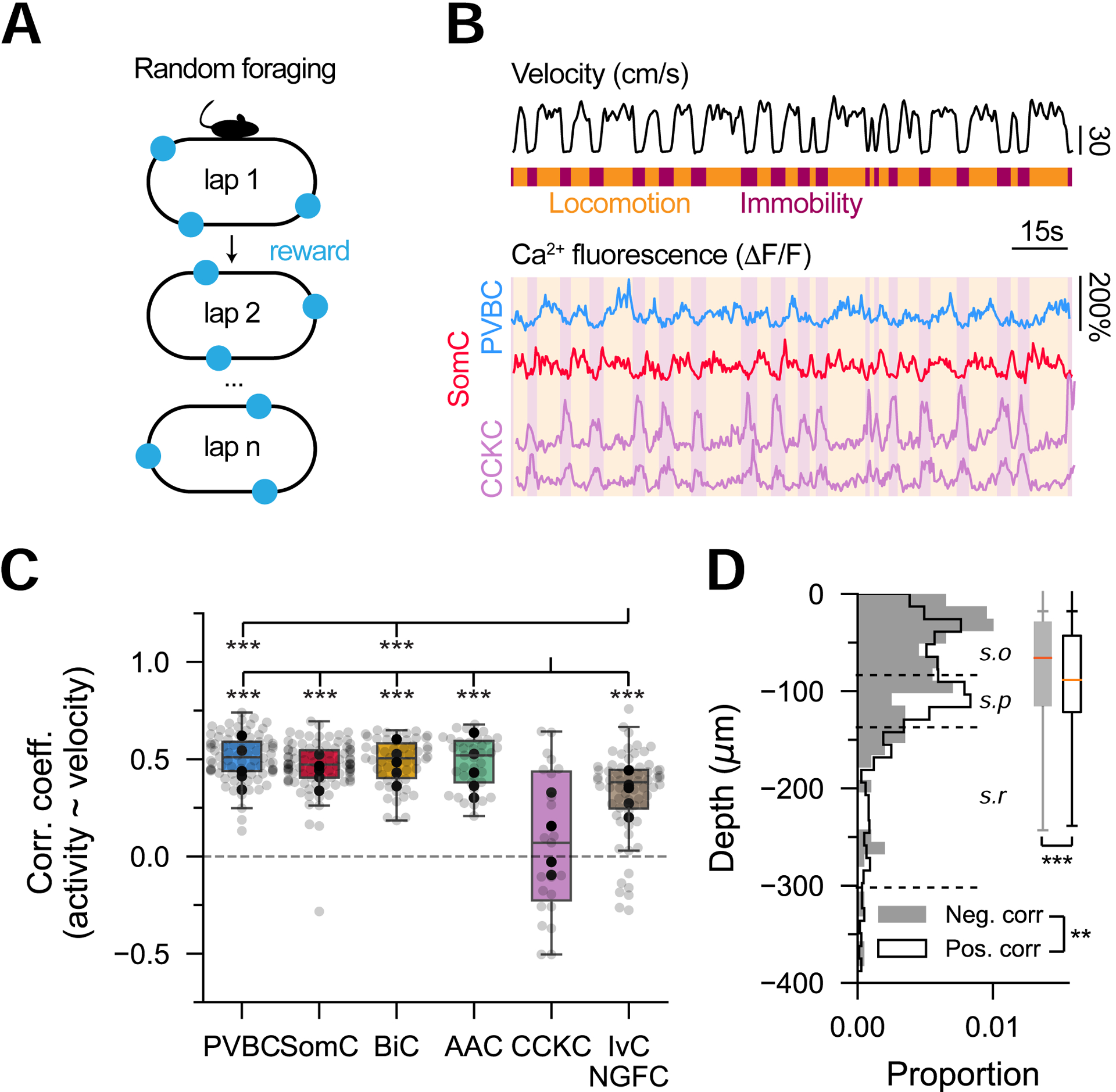
A. Schematic of the random foraging task.
B. Representative ΔF/F traces from PVBC, SomC and CCKC. Locomotion and immobility epochs are overlaid in orange and purple, respectively.
C. Pearson’s correlation coefficients of ΔF/F traces and velocity (n = 134 PVBC, 180 SomC, 75 BiC, 56 AAC, 44 CCKC, 119 IvC/NGFC from n = 7 mice). One-way ANOVA (p < 10−10) with post hoc Tukey’s range test corrected for multiple testing. Mouse averages are indicated by the black dots.
D. Depth distribution of all negatively (filled gray, n = 156) and positively (empty black, n = 1429) correlated cells with velocity, regardless of subtype (n = 7 mice). Negatively correlated cells are more dorsal in stratum oriens than positively correlated cells, which are closer to stratum pyramidale. Two-sample Kolmogorov-Smirnov test on distributions (p = 0.02) and Mann-Whitney U test on populations (p < 0.001).
Decoding of IN molecular identity with a machine-learning approach
The ability to infer the subtype of an imaged interneuron from its fluorescence trace and other easily measured behavioral or anatomical variables, without the need for genetic targeting or post hoc identification, would represent a significant advance in the ability to study interneuron subtypes at high throughput during behavior. As a first step towards this goal, we developed a machine learning-based classification tool to ask how well the molecular identity of a given IN can be predicted from its relative fluorescence calcium signal (ΔF/F), the animal’s velocity, and the depth of the cell (Figure S3) (Troullinou et al., 2019). While prediction accuracy was well above chance for all 6 molecularly defined IN subtypes, it remained suboptimal as an automated classification approach. We found that many decoding errors resulted from misclassification between PV-expressing INs (PVBC and AAC) and between SOM-expressing INs (SomC and BiC). Given these findings, we next asked whether discrimination would improve by merging the similar categories to create a 4-class problem consisting of, PV-expressing INs, SOM-expressing INs, CCKC, and IvC/NGFC. The classification showed higher accuracy (64%, 71% 60% and 59%, respectively) (Figure S3). Overall, our findings suggest that the broader groups of molecularly defined INs have general signatures in their dynamics that can be recognized at above chance-level accuracy by a machine learning algorithm, although the error rates still remain too high for general use as an automatic classification approach.
Subtype-specific modulation of CA1 INs during sharp-wave ripple oscillations
CA1 INs have been shown to have highly stereotyped, subtype-specific activity patterns during SWRs, fast oscillatory events associated with memory consolidation (Buzsáki, 2015). However, only small numbers of identified INs have been recorded during SWRs in awake mice (Varga et al., 2014), making it unclear how generally these conclusions apply at the population level. To monitor hippocampal network oscillations during imaging, mice were chronically implanted with an extracellular electrode placed in the CA1 region on the contralateral side to record the local field potential (LFP) while simultaneously performing large-scale AOD-2p imaging at an imaging rate of 40Hz (Figure 3A, 3B, 3C, S4). To quantify the SWR-triggered activation in different subtypes, we developed a measure of modulation to assess the percentage of SWRs during which an IN increased its activity above baseline levels during immobility (Figure 3D). We observed subtype-specific modulation patterns that are largely consistent with previous reports. PVBC were generally strongly modulated (Klausberger et al., 2003; Lapray et al., 2012; Varga et al., 2014), while most SomC were not (Katona et al., 2014, 2017)(Figure 3D). AAC and BiC showed prominent bimodal profiles (Figure S4), with some cells strongly modulated during SWRs but others remaining silent (Viney et al., 2013; Katona et al., 2014; Varga et al., 2014)(Figure 3D), with no correlation with the anatomical depth of each cell (Figure S4). To investigate the differences between modulated and non-modulated INs within each subtype, we used a clustering method to infer the number of functionally distinct groups from the data. All subtypes but CCKC and NGFC separated into 2 distinct groups, suggesting within-subtype dissociations in SWR modulation (Figure 3E). Across subtypes, modulation probability values of modulated INs were similar, with slightly lower values for AAC (Figure 3E). The fraction of modulated INs in each group indicate a large proportion of previously unsuspected SWR-modulated AAC and non-SWR-modulated BiC (Figure 3F). Finally, we found differences in the axial location of modulated and non-modulated cells for SomC and BiC, as modulated cells were generally closer to stratum pyramidale (Figure 3G). These results reveal that although general trends do exist for the responses of molecularly defined IN subtypes during awake SWRs, there is also a previously underestimated within-subtype heterogeneity at the population level.
Figure 3: Between- and within-subtype functional heterogeneity during sharp wave ripples.
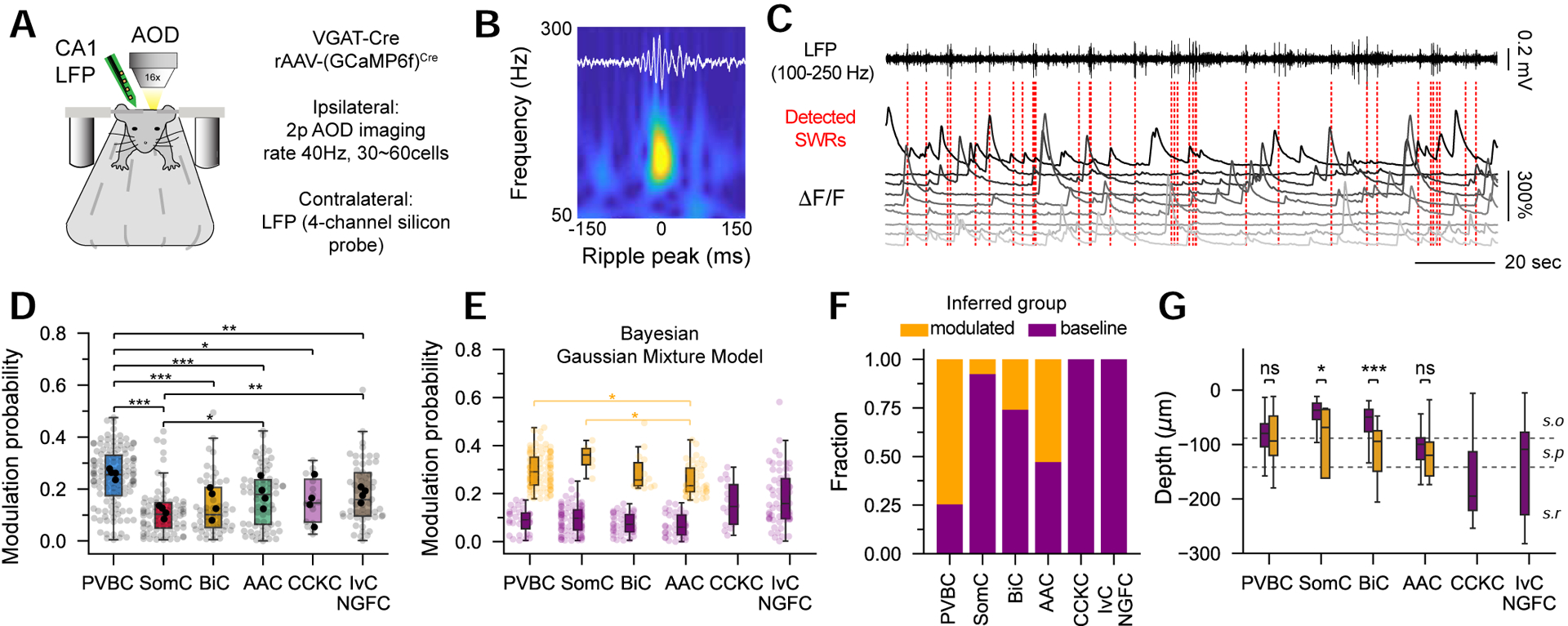
A. Mice are implanted with a 4-channel silicon probe on the contralateral side to record LFP. 3D scanning is performed at higher imaging rates (40Hz) by decreasing the number of simultaneously recorded neurons (~30 cells simultaneously).
B. Representative averaged sharp-wave ripple (SWR)-triggered spectrogram for one session.
C. Representative LFP and ΔF/F traces. Red dashed lines represent the onset of a detected SWR.
D. SWR modulation probability for each subtype (n = 122 PVBC, 79 SomC, 54 BiC, 72 AAC, 16 CCKC, 58 IvC/NGFC from n = 4 mice, one-way ANOVA, p < 0.001).
E. Bayesian Gaussian Mixture Models, in which clusters are inferred from the data, show bimodal distributions for ripple modulation (modulated and non-modulated groups) in PVBC, SomC, BiC and AAC subtypes. Small but significant quantitative differences are seen among the modulated groups (one-way ANOVA, p = 0.004).
F. Fraction of cells in each inferred group (orange: modulated, purple: baseline) for the different subtypes.
G. Comparison of the depth profiles between the modulated and baseline groups for the different subtypes (unpaired t-tests).
ANOVA tests are corrected for multiple testing using post hoc Tukey’s range test. Mouse averages are indicated by the black dots. *p<0.05, **p<0.01, ***p<0.001.
Spatially selective activity dynamics of CA1 INs during head-fixed spatial navigation
While place cells have been traditionally considered to be principal cells (O’Keefe and Dostrovsky, 1971), spatial modulation has also been sparingly reported for INs, although without rigorous subtype identification (Marshall et al., 2002; Ego-Stengel and Wilson, 2007; Wilent and Nitz, 2007; Hangya et al., 2010; Grienberger et al., 2017). Therefore, we sought to examine spatial information content in a large sample of INs during navigation (Figure 4A). We used standard shuffling procedures for the detection of significant peaks in activity at particular regions on the belt (Figure 4A). We found a wide spectrum of spatially tuned INs, with some exhibiting high and stable selectivity across laps (Figure 4B, 4H). To show that our detection method managed to identify spatially modulated cells, we trained a linear classifier to decode the position of the animal based on IN calcium activity dynamics (Figure 4C). While the errors of the decoded position were largely greater than those reported when decoding position from CA1PC activity (Figure S5), inferring the animal’s position from the activity of the small subset of spatially-selective INs (blind of subtype, 17.5 ± 6.4% in total passed the threshold, mean ± sem, n = 6 mice, 3 sessions per mouse) was approximately as effective as doing so from the entire IN population (Figure 4D). Conversely, the decoding error was significantly greater from a subset of randomly chosen INs (matching the number of spatially modulated INs), as well as when the position and activity were randomly shuffled relative to one another (Figure 4D). While spatially selective INs were found in virtually all subtypes, BiC tended to have a significantly higher recurrence probability (Figure 4E). The majority of INs had broad tuning, as measured by the selectivity index, but a subset of SomC and BiC, and to some extent PVBC, displayed sharper fields (Figure 4F). Place field centroids of INs were generally uniformly distributed along the treadmill, with some observable preference in PVBC for the seam of the belt, a location likely providing a very salient sensory cue (Geiller et al., 2017)(Figure 4G). In addition, spatially-selective PVBC displayed a significantly more stable within-session recruitment than spatially-selective SomC (Figure 4H). Previous studies have also reported that some INs selectively decrease their activity in particular regions of an environment (“negative place fields”) (Marshall et al., 2002; Ego-Stengel and Wilson, 2007; Wilent and Nitz, 2007; Hangya et al., 2010). Therefore, we also examined negative spatial selectivity but did not find that it was enriched among a specific subtype. The fraction of negative spatially modulated cells was also slightly lower (blind of subtype, 12.7 ± 8.3%, mean ± sem, n = 6 mice, 3 sessions per mouse) than the fraction of positive spatially modulated cells, as was the recurrence probability across all subtypes (Figure S5), although these reduced numbers may originate from difficulties in observing decreased activity with calcium sensors. These results demonstrate that hippocampal INs can represent spatial information at the population level and that a subset of INs exhibit spatial tuning similar to CA1PCs.
Figure 4: A subset of cells show significant spatial selectivity and drive spatial information content among interneurons at the population level.
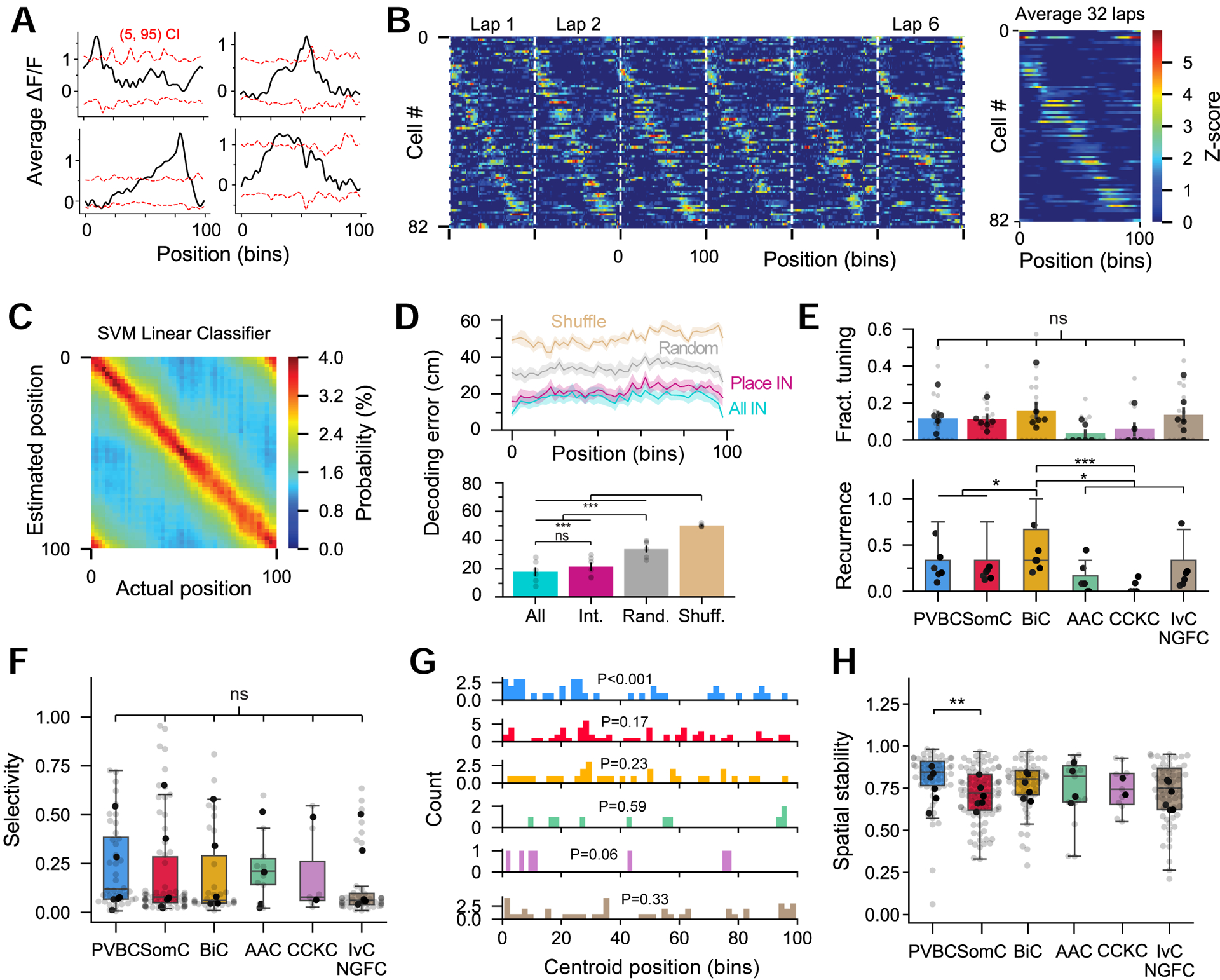
A. Representative examples of spatially modulated interneurons. Red dashed lines represent the 5% and 95% confidence intervals of the shuffled data.
B. Activity of 82 spatially modulated interneurons recorded simultaneously. Each row is a heatmap of one interneuron’s Z-scored ΔF/F trace. Left: Successive laps are concatenated (only the first 6 laps are shown). Right: Average of the entire session (32 laps).
C. Representative example of the posterior probabilities obtained from a Support Vector Machine (SVM) classifier used to decode mouse position from interneuron activity.
D. Top: Decoding error as a function of position for 4 different groups. Cyan, activity from all interneurons in a given session was used for decoding. Magenta, only interneurons detected as spatially modulated were used. Gray, activity from randomly picked interneurons but matching the number of spatially modulated interneurons in the session. Brown, only spatially modulated interneurons but the position of the animal was randomly shuffled (n = 18 sessions from n = 6 mice). Bottom: Average decoding error for each group above. Decoding accuracy originates mainly from spatially modulated cells, although they represent only a small fraction of all recorded interneurons (one-way ANOVA, p < 10−8).
E. Top: Fraction of spatially modulated neurons for each subtype. Light gray dots represent sessions (n = 18 sessions from 6 mice) and black dots represent mouse averages. BiC have higher fractions but the difference is not significant from other subtypes (one-way ANOVA, p = 0.06).
Bottom: Recurrence probability of spatial modulation. BiC have a higher probability to stay spatially modulated on different sessions (one-way ANOVA, p < 0.001; n = 105 PVBC, 145 SomC, 50 BiC, 39 AAC, 41 CCKC, 111 IvC/NGFC from n = 6 mice).
F. Selectivity index, quantifying the sharpness of the tuning curve, for the different subtypes. SomC have a subset of cells with sharp fields but the mean is not different from that of the other subtypes (one-way ANOVA, p = 0.35). Each dot represents one neuron’s selectivity index or its average over multiple sessions if imaged multiple times (n = 40 PVBC, 66 SomC, 32 BiC, 10 AAC, 7 CCKC, 43 IvC/NGFC from n = 6 mice).
G. Distribution of interneuron activity centroids along the belt. P-values are derived from a Kolmogorov-Smirnov uniformity test (same n as in F).
H. Tuning curve correlation coefficients between odd and even laps within the same session for spatially modulated interneurons, depicting the stability of the place field. Mouse averages are indicated by the black dots.
ANOVA tests are corrected for multiple testing using post hoc Tukey’s range test. Data are represented as mean ± sem. *p<0.05, **p<0.01, ***p<0.001.
We next sought to assess the stability of IN spatial representations by comparing the tuning curves of the same recorded neurons at three different time points, regardless of their spatial modulation: after virtually no delay (within-session), after 1 hour, and finally after 24 hours (Figure S6). While there were differences in within session stability across molecularly defined IN subtypes, with PVBC and CCKC showing the highest and lowest within-session correlations, respectively (Figure S6A, S6B), all subtypes had similar tuning curves correlations after 1 hour and 24 hours (Figure S6I). Interestingly, INs located more superficially within CA1, particularly superficially located PVBC and IvC/NGFC, were more stable within sessions, but the same trend did not hold for 1h correlations (Figure S6C, S6F). Tuning curve correlations after both 1 hour and 24 hours were significantly lower than the within-session correlations across all IN subtypes, indicating a general reorganization of IN spatial representations with time.This general decorrelation is likely to reflect small fluctuations in the relatively uniform spatial responses of INs, as only a small subset of cells display significant place tuning. However, correlations at later time points (1h and 24h) were still significantly higher than chance levels (a distribution in which IN subtype identity was shuffled), indicating that INs retain a certain degree of spatial information over time (Figure S6I). The general decorrelation of tuning curves is two-fold lower in INs than in CA1PC, as reported previously under similar experimental conditions (Danielson et al., 2016). While it is not surprising that INs are a highly plastic element of the hippocampal network, further experiments can help dissect the causality of these representational drifts.
CA1 IN activity dynamics rapidly reconfigure in novel contexts
As CA1PC representations have been shown to undergo remapping in response to changes in the environment (Colgin, Moser and Moser, 2008; Kubie, Levy and Fenton, 2019), we next sought to assess the degree to which IN subtypes remap upon exposure to a novel context. To do so, mice performed the random foraging task twice on a familiar belt (context A1 then A2) and were finally exposed to a novel belt, decorated with a distinct set of tactile cues (context B) (Figure 5A). At the population level, INs displayed a significant decorrelation in their spatial tuning upon exposure to belt B (Figure 5B). This remapping could not be explained simply by the passage of time, as A1-A2 correlations were significantly higher than both A1-B and A2-B values, and no difference between A1-B and A2-B was observed (Figure 5B). All subtypes, with the exception of CCKC, displayed a significant decrease in tuning curve correlation after exposure to the novel context (Figure 5D), with no subtype-specific differences in the amplitude of this decrease (Figure 5E). A linear classifier trained on a subset of the IN activity could decode the context in which the held-out test data was imaged at near-perfect levels, further suggesting that IN activity carries strong contextual information at the population level (Figure 5C).
Figure 5: Interneurons encode contextual information at the population level and rapidly modulate their activity in novel contexts.
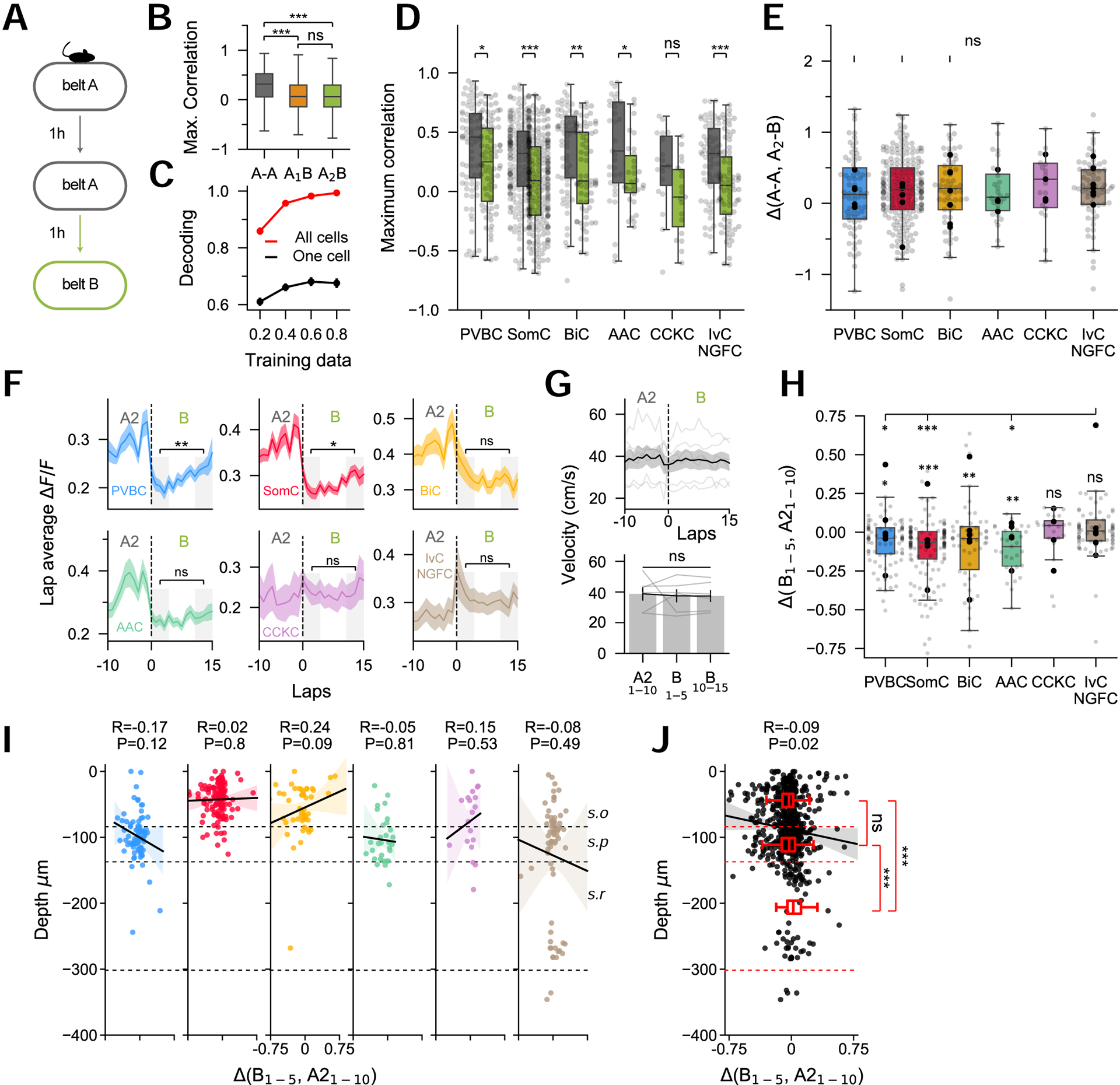
A. Mice ran in a familiar context (belt ‘A’) twice and were then exposed to a new context (belt ‘B’).
B. Tuning curve correlation coefficients between contexts ‘A’ and ‘B’ (n = 1080 cells from 6 mice, one-way ANOVA, p < 10−10).
C. Context identity can be reliably decoded using a SVM classifier (n = 6 mice).
D. Correlations between A1-A2 (gray) and between A2-B (green) for the different subtypes (n = 74 PVBC, 216 SomC, 64 BiC, 29 AAC, 19 CCKC, 73 IvC/NGFC from n = 6 mice, paired t-tests).
E. Difference (delta) between A1-A2 and A2-B correlation coefficients (same n as K, one-way ANOVA, p = 0.96).
F. Average ΔF/F during running on each lap during the last 10 laps of A2 and the first 15 laps of B. In belt B, paired t-tests between the first 5 and last 5 laps (same n as K).
G. Top: Velocity profiles during change of belt for n = 6 mice. Bottom: Average velocity quantification in last 10 laps of A2, first 5 laps in B and last 5 laps in B (one-way ANOVA, p = 0.96).
H. Difference (delta) in ΔF/F between the last 10 laps in A2 and the first 5 laps in B (one-way ANOVA, p < 0.001 between groups). The decrease seen in belt B was also assessed by testing delta to a mean of 0 (one-sample t-test, P-value above each bar).
I. Depth distribution as a function of the change in activity (delta) for each subtype. Shaded areas represent bootstrapped confidence intervals.
J. Depth distribution as a function of delta for all interneurons (n = 660 cells), regardless of subtype identity. Interneurons in s.r. are less affected by the change of context (unpaired t-tests).
ANOVA tests are corrected for multiple testing using post hoc Tukey’s range test. Mouse averages are indicated by the black dots. Data are represented as mean ± sem. *p<0.05, **p<0.01, ***p<0.001.
Several recent studies have leveraged calcium imaging from genetically-defined CA1 INs in virtual reality systems to assess changes in IN dynamics upon rapid exposure to a novel environment (Sheffield, Adoff and Dombeck, 2017; Hainmueller and Bartos, 2018; Arriaga and Han, 2019). These experiments revealed that SOM-expressing INs decrease their activity in novel contexts, while conflicting results have been reported for PV-expressing INs. With our treadmill apparatus, we observed a significant decrease in IN activity for the first several laps in context B for PVBC, SomC, BiC, and AAC subtypes (Figure 5F, H). CCKC were not modulated by the context change, while IvC/NGFC displayed a small but statistically nonsignificant increase in activity during the first 2 laps in the novel context (Figure 5F, H). In PVBC and SomC, activity returned to near-baseline levels later in the session, while this appeared more gradual for AAC and BiC (Figure 5F). These results could not be simply explained by differences in locomotion, as the velocity remained nearly identical across laps and between contexts (Figure 5G). To assess whether the decrease in activity was related to the anatomical location of each cell, we correlated the novel context activity modulation with cell depth for each subtype (Figure 5I). Although no statistically significant correlation was found between modulation and depth for a given subtype (Figure 5I), interneurons located deeper in CA1 were generally less modulated by the context change (Figure 5J), consistent with the enrichment of CCKC and IvC/NGFC in deeper sublayers. Taken together, these results indicate that IN activity is modulated by context and can rapidly reconfigure upon exposure to a novel environment. The majority of INs, including both perisomatic-targeting and dendrite-targeting subtypes, modulate their activity in novel contexts via both an overall decrease in activity levels and a general reorganization of their tuning properties. Both mechanisms could facilitate remapping in downstream pyramidal cell targets by influencing the rate and place reallocation of the fields.
Reward modulation of IN activity during goal-oriented spatial learning
CA1 place cell maps undergo prominent reorganization toward rewarded locations in goal-directed learning tasks (Hollup et al., 2001; Dupret, O’Neill and Csicsvari, 2013; Zaremba et al., 2017; Kaufman, Geiller and Losonczy, 2020). To investigate if CA1 IN activity is modulated over the course of spatial reward learning, we trained mice in a goal-oriented spatial learning task, in which water-restricted mice run on a cue-rich treadmill belt for a water reward delivered at a fixed location on each lap (Figure 6A). When mice learned the task, they displayed selective, anticipatory licking preceding the reward zone, and their velocity decreased within the same location (Figure 6B). We thus compared the average activity profiles for the various subtypes at different locations along the treadmill (Figure 6C). We observed a selective increase in SomC and BiC activity in the area immediately preceding the reward zone, while the activity of the other subtypes decreased in this region, as did the animal’s velocity (Figure 6C). We quantified this increased activity by defining a reward modulation index (RMI), a measure of the activity in the area immediately preceding the reward zone compared to the activity throughout the rest of the treadmill (Figure 6D). On average, SomC and BiC had significantly higher RMI values than the other subtypes, reflecting a preferential reward-related modulation (Figure 6D). More specifically, most subtypes displayed negative RMI values, representing decreased activity in the pre-reward area compared to the rest of the treadmill, consistent with the animal’s lower velocity in the pre-reward area (Figure 6D). However, SomC and BiC displayed RMI values near 0, suggesting the contribution of a specific reward-related modulation to their activity. To more formally disambiguate the effects of velocity and position on interneuron activity during the GOL task, we created a multivariate regression model (Figure S7). This analysis revealed positive weights for the position variable immediately preceding the reward zone for SomC and BiC subtypes, but not other subtypes, confirming their preferential recruitment. To analyze how IN dynamics are reconfigured when the reward location is changed, we then focused on the sessions after translocation of the reward zone. Within these sessions, mice learned to lick in the new reward location, but often also displayed residual licking around the old reward zone (Figure 6E). This residual licking in the old reward zone was accompanied by increased SomC and BiC activity in this area (Figure 6F). To quantify this effect, we directly compared the activity change around the old reward zone and observed a significant increase from chance level only in SomC and BiC (Figure 6G, S7). To show that this increase was the remnant representation of the old reward zone, we split the recording session in half and observed that SomC and BiC activity levels gradually decreased and increased in the old and current reward zones, respectively (Figure 6H). Other IN subtypes did not show such reorganization (Figure S7). Taken together these results demonstrate prominent reward modulation of SOM-immunopositive IN activity during goal-directed spatial learning.
Figure 6: SomC and BiC subtypes are preferentially modulated by reward during goal-oriented learning.
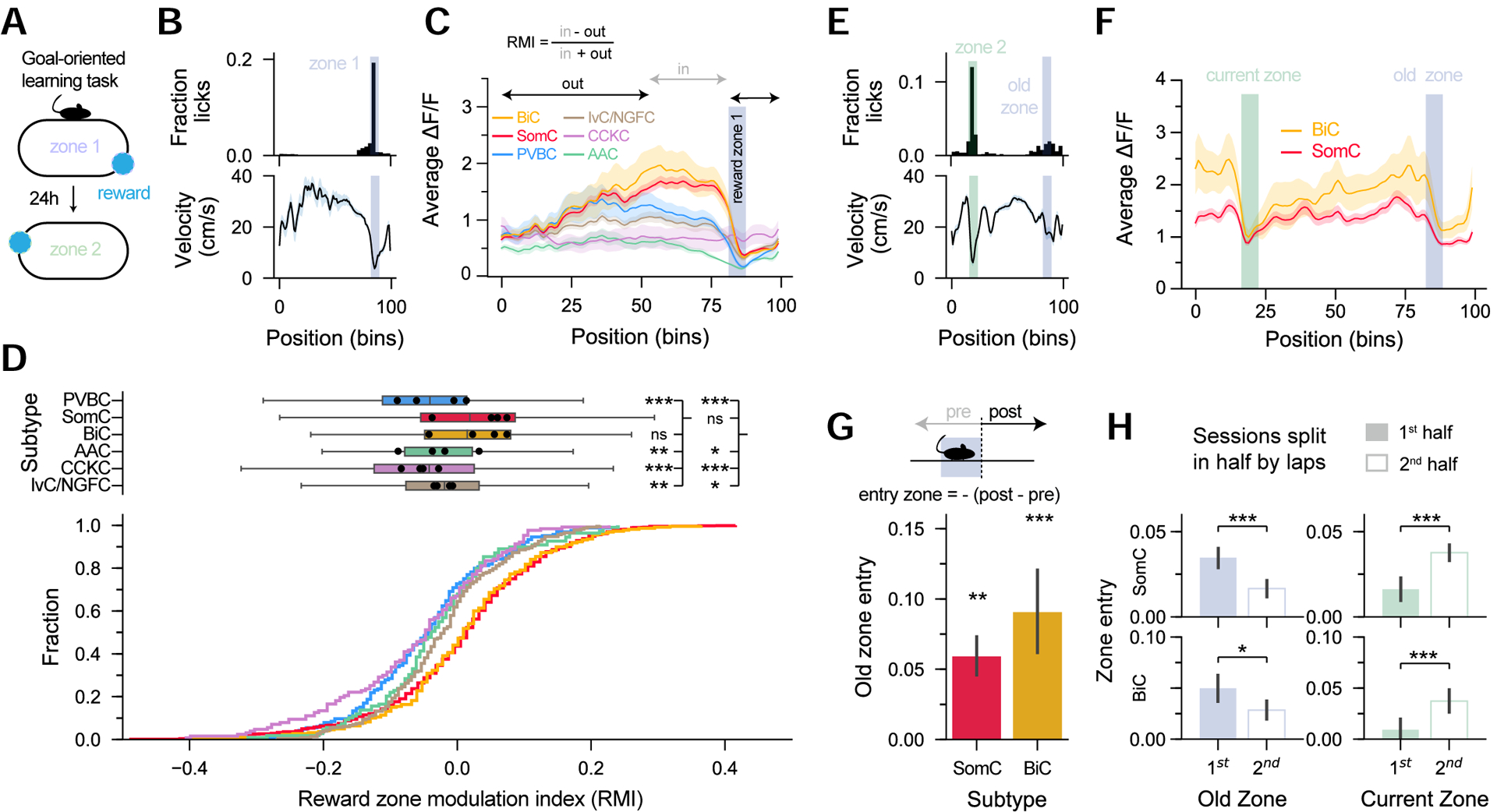
A. Mice ran to find a ‘hidden’ water reward, kept in a fixed location throughout the session, but changed each day (zone1 to zone 2).
B. Top: Representative histogram of licks in each position bin from one session. Blue shaded area represents the rewarded zone. Bottom: Corresponding velocity profile.
C. Representative average ΔF/F in each position bin for each subtype. BiC and SomC show increased activity preceding the reward zone.
D. Reward zone modulation index cumulative fraction (bottom) and box plots (top). BiC and SomC have higher modulations than other subtypes (n = 165 PVBC, 830 SomC, 220 BiC, 109 AAC, 127 CCKC, 228 IvC/NGFC from n = 4 mice, one-way ANOVA, p < 10−10).
E. Top: Representative histogram of licks and (bottom) velocity profile after translocation of the reward zone.
F. Representative BiC and SomC average activity by position bin from one session, showing traces of the old zone location.
G. Quantification of the old zone entry shows significantly higher modulation than expected by chance (n = 314 SomC, 103 BiC from n = 4 mice, one-sample t-test).
H. Quantification of old zone modulation and comparison between the first half and second half of the session. The modulation in the old zone for both BiC and SomC decreases with time while the modulation in the current reward zone increases, showing that BiC and SomC have remnants of the old reward representation (Same n as in G, paired t-tests).
ANOVA tests are corrected for multiple testing using post hoc Tukey’s range test. Mouse averages are indicated by the black dots. Data are represented as mean ± sem. *p<0.05, **p<0.01, ***p<0.001.
Disinhibitory circuit dynamics and interactions in CA1
The experiments described above focus on INs directly targeting CA1PCs. These INs are themselves under inhibitory control from distinct types of interneuron-specific INs (ISIs) which can further be divided into 3 subtypes (Type I, II and III ISI) (Acsády, Arabadzisz and Freund, 1996; Acsády, Görcs and Freund, 1996; Gulyás, Hájos and Freund, 1996; Freund and Buzsaki, 1996; Pelkey et al., 2017). However, little is known regarding the in vivo functional properties of ISIs and how their dynamics relate to those of CA1PC-targeting INs. Given that the molecular diversity of ISIs and their postsynaptic IN targets together exceeds the multiplexing capacity of post hoc immunohistochemistry, in a next series of experiments we sought to use a two-step strategy to address these outstanding questions. In a first cohort of VGAT-Cre mice, we performed post hoc immunostaining for vasoactive intestinal polypeptide (VIP) and calretinin (CR) to identify ISI subtypes in addition to PV, SOM and NPY to identify the major postulated targets of ISIs (Chamberland and Topolnik, 2012; Donato, Rompani and Caroni, 2013; Pi et al., 2013; Tyan et al., 2014; Luo et al., 2020) (Figure S1, 7A, 7B). To characterize ISIs and VIP-expressing populations with greater molecular resolution and at a larger scale, in a second cohort we performed 3D chessboard scanning in VIP-Cre mice (Turi et al., 2019) which allowed us to identify additional VIP-expressing classes: VIP+/CCK+ basket cells and a previously described IN subtype with long-range projection targets, expressing VIP and muscarinic acetylcholine receptor 2 (M2R) (Francavilla et al., 2018)(Figure 7A, 7B). We observed that all ISI subtypes were positively correlated with velocity, with the strongest and most uniform modulation from Type II ISIs (Figure 7C). Interestingly, VIP+/CCK+ INs all exhibited negative velocity correlations and were more active during immobility. These results strongly suggest that the functional dichotomy we observe within CCKC (Figure 2C, S1) reflects differences between perisomatic-targeting CCK+ basket cells and dendrite-targeting CCK+ IN subtypes (Cope et al., 2002; Booker and Vida, 2018). We also found that all VIP+/M2R+ cells were activated during immobility (Figure 7C), in accordance with previous work (Francavilla et al., 2018). Interestingly, we observed further major differences between these two immobility-active VIP+ subpopulations (Figure 7D): VIP+/CCK+ cells transiently responded to run-stop events but their activity subsequently decreased rapidly, while VIP+/M2R+ cell activity remained elevated throughout the duration of the immobility epoch without decay (Figure 7D). This distinction could represent differences in immobility-related inputs between the two subtypes. Finally, Type III ISIs exhibited different response profiles than all other INs, with pyramidal cell-like, phasic bursts of calcium transients (Figure 7E). We next investigated potential interactions between ISI subtypes and other INs in VGAT-Cre mice, where we could additionally image and identify PV+ cells, SomC and BiC. We found that the activity dynamics of VIP+/CR− cells and Type III ISIs, but not Type I ISIs, were significantly less correlated with SomC and BiC than with perisomatic-targeting PV+ INs (PVBCs and AACs) (Figure 7H, 7I), suggesting preferential functional connectivity between IN subtypes in CA1 disinhibitory circuits. Together these results demonstrate major differences in the functional dynamics of ISI and VIP-expressing subtypes and in their interactions with downstream IN targets.
Figure 7: ISIs and VIP+ subpopulations display heterogeneous functional properties and preferential functional connectivity with subtypes of pyramidal cell-targeting interneurons.
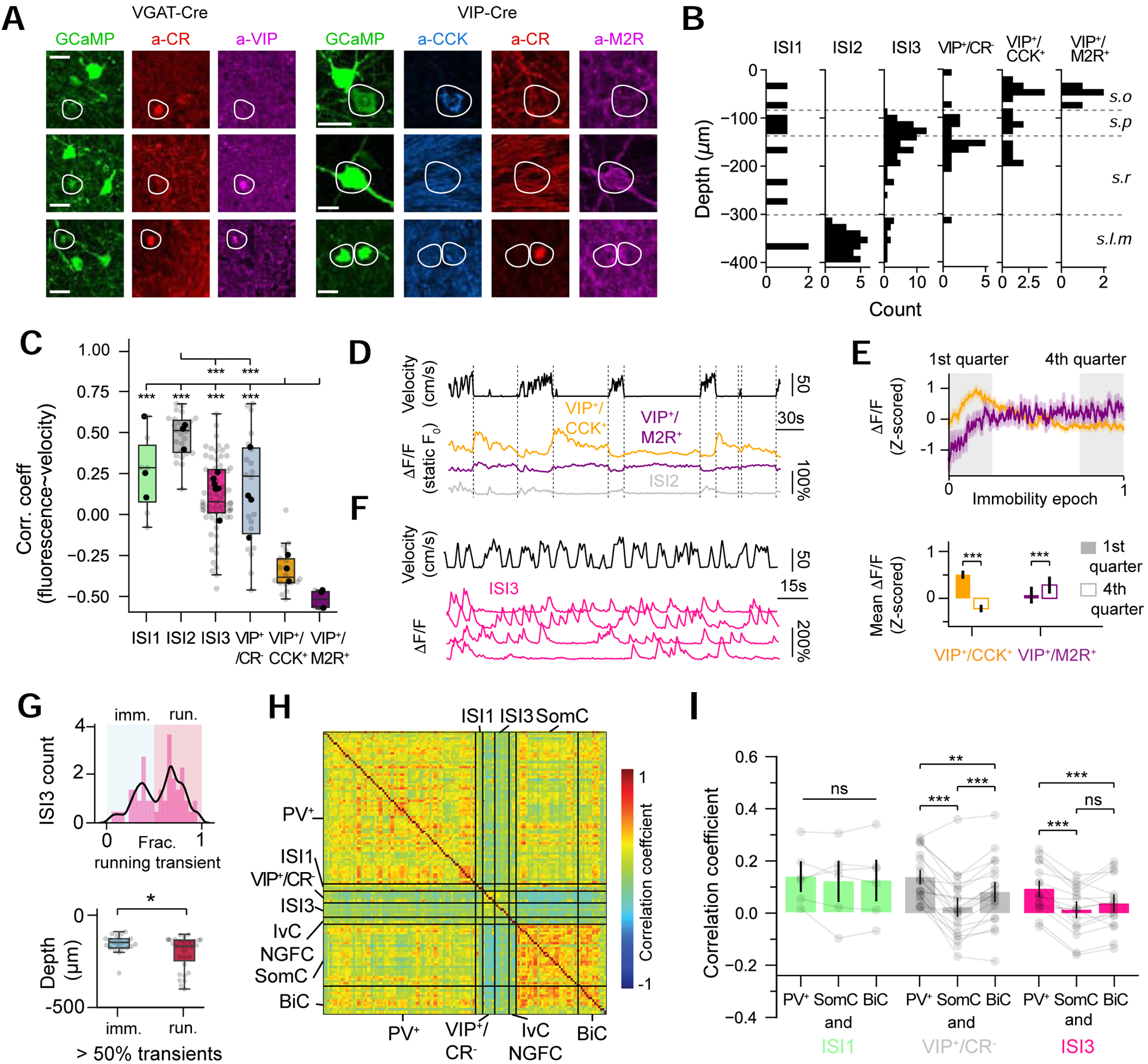
A. Immunohistochemistry in VGAT-Cre and VIP-Cre mice. Slices from VGAT-Cre animals (n = 4 mice) were immunostained for PV, SOM, NPY, VIP and CR. Slices from VIP-Cre animals (n = 3 mice) were immunostained for CCK, CR, M2R. Scale bars: 15 μm.
B. Depth distributions of all ISI subtypes, VIP+/CR−, VIP+/CCK+ and VIP+/M2R+ neurons detected in all mice (n = 7, VGAT-Cre and VIP-Cre pooled).
C. Correlation coefficients between activity and velocity (one-way ANOVA, p < 0.001 with n = 6 ISI1, 27 ISI2, 69 ISI3, 20 VIP+/CR−, 18 VIP+/CCK+ and 4 VIP+/M2R+ from 4 VGAT-Cre and 3 VIP-Cre mice).
D. Representative mouse velocity and ΔF/F traces for 3 simultaneously recorded VIP+/CCK+, VIP+/M2R+ and ISI2 cells. The activity was corrected using a static baseline to show the sustained or decreased activity during immobility.
E. Top: Average VIP+/CCK+ and VIP+/M2R+ response profiles (orange and purple, respectively) for all time-normalized immobility epochs across all cells. Bottom: Quantification of the difference in activity between the first and last quarter of each time-normalized immobility epoch. VIP+/CCK+ slowly decrease their activity while VIP+/M2R+ have a ramping and sustained activity during immobility (n = 18 VIP+/CCK+ cells, n = 4 VIP+/M2R+ cells from 3 mice, paired t-tests).
F. Representative ΔF/F traces for 4 simultaneously recorded ISI3 cells. Unlike other interneuron fluorescence traces, ISI3 cells show calcium transients underlying a potential phasic firing mode such as bursts of action potentials.
G. Top: Distribution of the ISI3 cells as a function of their number of detected transients occurring during locomotion. The distribution is bimodal with cells firing the majority of their transients either during immobility (blue, imm.) or locomotion (red, loc.). Bottom: Comparison of depth profiles for immobility and locomotion-specific groups (unpaired t-test).
H. Representative correlation matrix between all simultaneously recorded interneurons in one imaging session. The PV+ group includes both PVBC and AAC subtypes as we did not perform SATB1 immunostaining in these mice.
I. Average correlation coefficients for all ISI subtypes with PV+, SomC, or BiC neurons (n = 6 ISI1, 20 VIP+/CR− and 16 ISI3 from 4 mice, paired t-tests).
ANOVA tests are corrected for multiple testing using post hoc Tukey’s range test. Mouse averages are indicated by the black dots. Data are represented as mean ± sem. *p<0.05, **p<0.01, ***p<0.001.
Discussion
In this work, we combine cellular-resolution, fast, targeted 3D calcium imaging and post hoc immunolabeling to record simultaneously from populations of molecularly defined subtypes of INs in hippocampal area CA1 during spatial navigation, goal-oriented learning, contextual manipulations, and SWR events during quiet wakefulness. Our results support the fidelity of this approach, as they are largely in agreement with previous findings on CA1 interneuron recruitment in cases where similar data is available. Our results also extend previous studies by separating broad molecular class of PV-expressing and SOM-expressing INs and by providing subtype-specific information or sufficient sample sizes in cases where similar data is available but lacks this information. Finally, our results provide the first population-level description of in vivo response profiles and feature selectivity of some major IN subpopulations implicated in regulating CA1PC input-output transformation, such as CCK-expressing or NPY-expressing cells (Fuentealba et al., 2008; Klausberger, 2009; Tricoire et al., 2010; Katona and Freund, 2012; Milstein et al., 2015). Overall, while our results provide some examples of preferential subtype recruitment during behavior, they also suggest significant similarities in the activation patterns of many subtypes, which argues in favor of a previously underestimated functional homogeneity of INs.
In summary, the group of SOM-expressing, dendrite-targeting INs, which includes OLM (McBain, DiChiara and Kauer, 1994; Sik et al., 1995) and bistratified cells (Buhl, Halasy and Somogyi, 1994; Buhl et al., 1996), are strongly modulated by locomotion, spatial learning, and are themselves under strong disinhibitory control. These results suggest a close, bi-directional interaction of these INs with active CA1PC ensembles during exploration and a major role for these INs in regulating experience and learning-related reorganization of CA1PC dynamics through flexible regulation of synaptic integration and plasticity in CA1PC dendrites (Lovett-Barron et al., 2012, 2014; Royer et al., 2012). The second group of NPY-expressing and SOM-immunonegative, dendrite-targeting INs, which includes Ivy and neurogliaform cells (Fuentealba et al., 2008; Tricoire et al., 2010; Lapray et al., 2012; Milstein et al., 2015; Overstreet-Wadiche and McBain, 2015), shows overall weaker modulation by locomotion and spatial signals as well as less novelty-induced decrease in their activity following a change of context. The third group of perisomatic targeting PV+ INs, which comprises basket and axo-axonic cells (Buhl, Halasy and Somogyi, 1994; Sik et al., 1995; Klausberger et al., 2003), is strongly modulated by locomotion signals, but exhibits less dynamic reward-related modulation and disinhibitory control, suggesting that these INs predominantly regulate behavioral state and network oscillation-related activity dynamics of the overall CA1PC population (Cobb et al., 1995; Bartos, Vida and Jonas, 2007; Hu, Gan and Jonas, 2014). Finally, vis-à-vis these exploration- and locomotion-coupled inhibitory subcircuits, perisomatic targeting CCK-expressing INs seem to primarily regulate immobility-related activity of CA1PCs without carrying significant spatial or contextual information. While immobility-related activity is present across all IN subtypes, CCK-VIP basket cells appear to be a major component of an inhibitory subnetwork within CA1 that controls PC excitability primarily during immobility. This unique behavioral function of perisomatic-targeting CCK-expressing INs could be further aided by long-lasting, asynchronous, and retrograde endocannabinoid signaling-regulated GABA release from their axon terminals (Hefft and Jonas, 2005; Daw et al., 2009; Lee, Földy and Soltesz, 2010). It remains to be determined how subtype-specific differences in excitatory and inhibitory input convergence and integration onto IN types (Gulyás et al., 1999; Martina, Vida and Jonas, 2000; Matyas, Freund and Gulyas, 2004; Lovett-Barron et al., 2012; Chiovini et al., 2014; Milstein et al., 2015; Luo et al., 2020) as well as cell type-specific differences in neuromodulatory regulation (Freund and Katona, 2007; Varga et al., 2009; Wester and McBain, 2014) specifically contribute to the emergence of distinct in vivo response profiles and tuning specificity.
Our results provide a population-level landscape of SWR-related modulation across IN subtypes, by large consistent with previous in vivo electrophysiological studies (Klausberger et al., 2003, 2004, 2005; Lapray et al., 2012; Varga, Golshani and Soltesz, 2012; Katona et al., 2014). Our large-scale imaging approach also reveals considerable within-subtype heterogeneity during SWRs, as suggested by a growing body of studies. For instance, AAC have traditionally been considered to be silenced during SWRs (Klausberger et al., 2003; Viney et al., 2013), although very few cells have been recorded in awake rodents. Our observations are in agreement with a recent study reporting heterogeneous dynamics of AAC during SWRs (Varga et al., 2014). However, this study reported a segregation of AAC activity during SWRs based on axial location that we did not observe in our data: within the AAC subtype, modulated and unmodulated INs were present in both stratum oriens and stratum pyramidale. Finally, while BiC have been previously described as an IN subtype activated during SWRs (Klausberger et al., 2004; Katona et al., 2014), we report here a bimodal distribution of responses. It is possible that further heterogeneity might exist even within canonical inhibitory cell-types, such as axo-axonic cells, related to differential postsynaptic targets (Taniguchi, Lu and Huang, 2013).
Our findings also provide further insights into in vivo activity dynamics of disinhibitory circuits. They are consistent with an interpretation that VIP+ ISIs, and in particular Type III ISIs, exert disinhibitory influence on CA1PCs preferentially through SOM-expressing dendrite targeting INs and less so via PV-expressing perisomatic-targeting INs. This confirms previous anatomical and in vitro electrophysiological results showing that Type III ISIs preferentially innervate and inhibit SOM-expressing OLM cells (Chamberland and Topolnik, 2012; Tyan et al., 2014), similar to neocortical disinhibitory local circuit motifs (Lee et al., 2013; Pfeffer et al., 2013; Pi et al., 2013). We also observe prominent differences in locomotion modulation in CCK-VIP INs and Type II ISIs. While the origin of locomotion and immobility signals remain unclear, the presence of strong and specific anatomical contacts from ISI2s onto CCK-VIP INs suggest that the striking immobility-related activation we observe in CCK-VIP basket cells could result from this inhibitory interaction (Acsády, Arabadzisz and Freund, 1996; Acsády, Görcs and Freund, 1996).
The strategy employed here represents a major advance in our ability to record the activity of multiple molecularly-defined populations during behavior, but there are challenges and limitations associated with both calcium imaging in GABAergic cells and post hoc cell-type identification. While AOD-based 2p calcium imaging allows for recording of neural dynamics and longitudinal tracking of the same cells in 3D with high signal-to-noise and speed, calcium imaging is not ideal for measuring neural activity from fast-spiking populations, as modulations in constantly high firing rates lead to relatively small changes in collected fluorescence. Conversely, it is not known whether single spikes from low firing-rate GABAergic interneurons would be visible in calcium fluorescence traces in vivo, as no simultaneous recordings have been performed in such subtypes, to the best of our knowledge. Thus, heterogeneities in IN recruitment observed with calcium imaging may be due to inherent heterogeneities in the spike-to-calcium relationship for the different subtypes. In addition, as with all imaging approaches in scattering tissues, the ability to collect dynamic signals at cellular resolution degrades with imaging depth, introducing a potential bias for IN subtypes located more superficially in the hippocampus. For this reason, populations of CCKC and IvC/NGFC located below the pyramidal cell layer may have been less sampled than the other subtypes. In the future, the development of photostable, genetically-encoded voltage indicators compatible with 2p population imaging might enable more detailed measurements of IN activity (Lin and Schnitzer, 2016; Lee et al., 2017). Similarly, retrospective, multiplexed immunohistochemistry is a robust approach for identifying proteins within cells while preserving their relative spatial arrangement, allowing for the post hoc registration that we performed here. However, antibody-based probes are inherently constrained in their multiplexing capability and cannot be routinely removed from tissue sections. The development of in situ hybridization and high throughput transcriptomics (Cembrowski et al., 2016; Harris et al., 2018; Yao et al., 2020), preserving spatial information in thick tissue sections (Chen et al., 2015; Wang et al., 2018), could circumvent these issues and enable quantitative molecular profiling of imaged cells. Finally, the improvement of automated classification approaches based on machine learning algorithms that incorporate various behavioral and neural response features could achieve high decoding accuracy scores and enable immunolabeling-free identification of various subtypes. These comprehensive characterization efforts should be coupled with cell-type-specific manipulations to directly test for general organizational principles related to segregated behavioral functions of inhibitory circuits.
STAR METHODS
Resource availability
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Attila Losonczy (al2856@columbia.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The machine-learning approach and the dataset are available on GitHub at https://github.com/losonczylab/Geiller_Vancura_Neuron2020. All other codes generated during this study are available upon request.
Experimental model and subject details:
All experiments were conducted in accordance with NIH guidelines and with the approval of the Columbia University Institutional Animal Care and Use Committee. Experiments were performed with healthy, 3–5 month old, heterozygous adult male and female VGAT-IRES-Cre mice (Jackson Laboratory, Stock No: 016962, referred to as VGAT-Cre mice) and VIP-IRES-Cre mice (Jackson Laboratory, Stock No: 010908, referred to as VIP-Cre mice). Both strains were kept on a C57BL/6J background. Mice were kept in the vivarium on a 12-hour light/dark cycle and housed 3–5 mice in each cage. Experiments were performed during the light portion of the cycle.
Viruses:
Cre-dependent recombinant adeno-associated virus (rAAV) expressing GCaMP6f under the control of the Synapsin promoter (rAAV1-Syn-FLEX-GCaMP6f-WPRE-Sv40, Addgene #100833, titer: 1×1013 vg/mL, referred to as rAAV2/1:Syn(GCaMP6f)Cre) was used to express GCaMP6f either in VGAT+ interneurons (VGAT-Cre mice) or in VIP+ interneurons (VIP-Cre mice). Because of the high specificity of the VIP-Cre mouse line for labeling VIP-immunopositive cells in CA1 (see Figure 1 in Turi et al., 2019), interneurons labeled in VIP-Cre mice with Cre-dependent AAV were considered VIP-immunopositive in the present study.
Virus injections and hippocampal window/headpost implant:
For viral injections, 2 to 4-month-old VGAT-Cre mice were anesthetized with isoflurane and placed into a stereotaxic apparatus. Meloxicam and bupivacaine were administered subcutaneously to minimize discomfort. After the skin was cut in the midline to expose the skull, the skull was leveled, and a craniotomy was made over the right hippocampus using a drill. A sterile glass capillary loaded with rAAV2/1:Syn(GCaMP6f)Cre was attached to a Nanoject syringe (Drummond Scientific) and slowly lowered into the right hippocampus. Dorsal CA1 was targeted at coordinates AP −2.2, ML −1.75, DV −1.8, −1.6, −1,4, −1.2, −1.0, −0.8 relative to Bregma, with 50–64 nL of virus injected at each DV location. After injection, the pipette was left in place for 5–10 minutes and slowly retracted from the brain. The skin was closed with several sutures and the mice were allowed to recover for 4 days before the window/headpost implant.
The surgical procedure for CA1 window/headpost implant has been described previously (Lovett-Barron et al., 2014). Briefly, the injected mice were anesthetized with isoflurane and placed into the stereotaxic apparatus. After subcutaneous administration of meloxicam and bupivacaine, the skull was exposed, leveled, and a 3 mm craniotomy was made over the right hippocampus, centered on coordinates AP −2.2, ML −1.75 relative to Bregma. The dura overlying the cortex was removed, and the cortex overlying the hippocampus was slowly removed with negative pressure while ice-cold cortex buffer was simultaneously applied. This process was performed until the white, horizontal fibers overlying CA1 became visible and any bleeding subsided. A stainless steel, 3 mm circular cannula fitted with a glass window was inserted into the craniotomy and secured in place with Vetbond applied on the skull. Subsequently, dental cement was applied to the entire skull, and a headpost was affixed to the posterior skull with dental cement. The mice received a 1.0 mL subcutaneous injection of PBS and recovered in their home cage while heat was applied. The mice were monitored for 3 days post-operatively until behavioral training began.
Behavioral training and paradigms:
After recovery from surgery, mice were handled for several days and habituated to head-fixation. Mice were subsequently water-restricted to 85–90% of their original weight and trained to run on a single-fabric, cue-free belt. Mice were trained to operantly lick and receive water rewards (water was delivered in response to tongue contact with a capacitive sensor) at random locations along the belt. As performance improved, the number of rewards delivered on each lap decreased. After several days of training on this cue-free belt, the mice were trained for ~1 week on a 2m long, cue-rich belt for randomly delivered water rewards. The belt consisted of three joined fabric ribbons and included some combination of the following tactile cues: colored pom poms, velcro, glue gun spikes, pink foam strips, and silver glitter masking tape (Danielson et al., 2016; Zaremba et al., 2017). For Random Foraging experiments, imaging was started after mice could run approximately 10 laps in 10 minutes (usually after 10–14 days of total training). For context remapping experiments, mice were subsequently imaged for one or two sessions on a novel belt decorated with a distinct set of tactile cues (Danielson et al., 2016; Zaremba et al., 2017). As the familiar belt, this novel belt was composed of three joined fabric ribbons and some combination of additional tactile cues. However, to maximize the novelty of the new belt, both the fabric ribbons and the tactile cues were different, and they were arranged in a unique order along the belt. For Goal-Oriented Learning (GOL) experiments, the mice were then trained for several days in the GOL paradigm, in which a single water reward was delivered at a fixed location each lap. Imaging was started for GOL experiments after several days of training in this paradigm; the location of the reward was switched between each training session. For combined imaging and LFP experiments, data acquisition was started once GCaMP6f expression was optimal, hippocampal windows were clear, and the mice were habituated to head-fixation; these mice did not undergo additional behavioral training.
AOD-based two-photon calcium imaging:
Once behavioral training was complete, and 24 hours before functional imaging, mice underwent a single imaging session consisting of a high-resolution structural scan. This step was necessary to obtain a reference Z-stack and derive the X-Y-Z positions of GCaMP-expressing neurons. The mice were head-fixed under a custom-modified AOD microscope (Femto3D-ATLAS, Femtonics Ltd) and anesthetized with ketamine/xylazine to reduce motion artifacts during the stack. To provide stable transmission parameters during chronic imaging in the entire 3D scanning volume, the AOD microscope was extended with a high speed and precision beam stabilization unit which was directly attached to the AOD scan head, sensitive to input beam misalignment. The beam stabilization unit consisted of two quadrant detectors (PDQ80A and TPA101, Thorlabs) and two broadband dielectric mirrors (Thorlabs) mounted on motorized mirror mounts (Femtonics). The beam alignment was performed by the LaserControl software (Femtonics). A water-immersion objective (16x Nikon CFI75) was placed above the glass window and lowered until the CA1 pyramidal cell layer was in focus. At this stage, the objective was fixed in position and focus was subsequently adjusted using AO crystals (Szalay et al., 2016). The laser (Coherent Ultra II) was tuned to λ=920 nm, and the reference Z-stack was taken from the most dorsal position in stratum oriens/alveus (150–200 μm above the pyramidal cell layer) to the stratum lacunosum-moleculare layer (~300 μm below the pyramidal cell layer) with 800×800 pixel images (X-Y resolution of 1.25 μm/pixel) every 4 μm. Laser power and photomultiplier (PMT) detectors (GaAsP, H10770PA-40 Hamamatsu) were compensated appropriately in Z throughout the stack (power at 20–40 mW and detector gain at 80% in stratum oriens/alveus, power at 120–150 mW and detector gain at 90% in stratum lacunosum-moleculare). After completion, the mice were returned to their home cage and allowed to recover for 24h until the start of functional imaging.
To determine X-Y-Z positions of GCaMP-expressing neurons, the Z-stack was scrolled through, and each visible interneuron was manually selected using the integrated software (MES, Femtonics Ltd) to generate a list of 200–300 X-Y-Z coordinates defined as the center of each cell. These points constituted the center of region of interests (ROI) used on subsequent days for functional imaging. Each ROI was defined as a square of 40 to 50 μm2 (chessboard scan) (Szalay et al., 2016) with a resolution of 1 to 1.5 μm/px. The advantage of the chessboard scanning method is that only neurons and small areas around the pre-selected cells are recorded. Therefore, a high ratio of the total recording time (~20–50%) is spent reading out information from the selected neurons. In contrast, volumetric imaging with the same 2P excitation provides an orders-of-magnitude worse ratio for measurement time utilization as the somata of INs occupy a relatively small ratio of the total scanning volume.
On each day of functional imaging, the same field of view was found using the reference Z-stack and X-Y-Z coordinates were loaded into the software to perform 3D imaging. Once all cells were in focus, 10–15-minute functional imaging sessions were conducted at a frame rate of 3–5 Hz for most experiments (frame rate was dependent on ROI size and resolution). For experiments involving contralateral LFP recordings, imaging was conducted at a higher rate (40 Hz), which restricted imaging to only 30 cells simultaneously. During functional imaging, the laser power and detector gain were compensated based on the reference Z-stack parameters.
Silicon probe implantation, LFP recordings, and sharp-wave ripple identification:
For experiments requiring simultaneous two-photon calcium imaging and LFP recordings, mice were implanted with a glass window over the hippocampus as above, and additionally a chronic, 4-channel silicon probe (Qtrode, Neuronexus) was inserted into the contralateral CA1 at a 45-degree angle. The probe was secured in place with dental acrylic and the mouse was allowed to recover for several days, as above. LFP signals were recorded with a multichannel recording system (Intan Technologies) synchronized with the AOD imaging system. The correct position of the silicon probe was confirmed by the presence of sharp-waves ripples in the data. LFP signals were recorded at 20kHz. To identify putative sharp-wave ripple events, the raw LFP signal was band-pass filtered from 100–250 Hz and thresholded at 3 standard deviations above the mean value with a minimum duration of 50ms. All putative sharp-wave ripple events were then manually inspected to obtain the final set of sharp-wave ripple events used for analysis.
Perfusion and tissue processing:
After the completion of imaging experiments, mice were transcardially perfused with 40 mL of ice-cold Phosphate-Buffered Saline (PBS, Thermo Fisher), followed by 40 mL of ice-cold 4% paraformaldehyde (PFA, Electron Microscopy Sciences). Brains were stored overnight in 4% PFA at 4°C. The next day, the 4% PFA was removed and the brains were rinsed 3×5 min in PBS. 75 μm horizontal sections of the imaged hippocampus were cut on a vibrating microtome (Leica VT1200S) and washed 3×15 minutes in PBS. Subsequently, sections were permeabilized for 2×20 minutes in PBS with 0.3% Triton X-100 (Sigma-Aldrich). Blocking was then performed with 10% Normal Donkey Serum (Jackson ImmunoResearch, Catalog #017-000-121) in PBST (PBS with 0.3% Triton X-100) for 45 minutes. The sections were then incubated in a PBS solution containing 3 primary antibodies (see below for antibody information and dilutions) for one hour at room temperature, followed by 2 days at 4°C. After 2 days, the primary antibody solution was removed from the slices and the slices were washed 3×15 minutes in PBS to remove unbound primary antibodies. The slices were subsequently incubated in a PBS solution containing a mixture of appropriate secondary antibodies conjugated to fluorescent labels (see below for antibody information and dilutions) for 2 hours at room temperature. The sections were then washed 5×15 minutes in PBS at room temperature. Finally, sections were mounted on glass slides in Fluoromount-G aqueous mounting medium (ThermoFisher Scientific) and coverslipped. The slides were allowed to dry at 4°C for at least one hour before confocal imaging (see below). After confocal imaging, the slides were submerged in PBS to remove the coverslip, and the sections were removed from the slides with gentle rocking. After washing 3×15 min in PBS and blocking with 10% Normal Donkey Serum in PBST for 45 minutes, the sections were incubated in an additional 2–3 primary antibodies. The sections were subsequently washed, incubated in secondary antibodies, washed again, and mounted and imaged, as in the first round of staining. We considered antibody combinations to be compatible for sequential imaging in the same fluorescence channel if they were against antigens known to be present only in non-overlapping interneuron populations in CA1 (see below for detailed strategies).
Immunohistochemistry (see Key Resources Table for catalog number):
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rat anti-somatostatin | Millipore | MAB354 |
| sheep anti-neuropeptideY | Abcam | ab6173 |
| rabbit anti-proCCK | Frontier Institute | CCK-pro-Rb-Af350 |
| chicken anti-parvalbumin | Gift from Susan Brenner-Morton | |
| rabbit anti-SATB1 | Abcam | ab70004 |
| guinea pig anti-calretinin | Swant | CRgp7 |
| rabbit anti-VIP | Immunostar | 20077 |
| rat anti-M2R | Millipore | MAB367 |
| donkey anti-chicken DyLight 405 | Jackson Immunoresearch | 703-475-155 |
| donkey anti-rabbit DyLight 405 | Jackson Immunoresearch | 711-475-152 |
| donkey anti-guinea pig Rhodamine Red X | Jackson Immunoresearch | 706-295-148 |
| donkey anti-rat Alexa 568 | Abcam | ab175475 |
| donkey anti-sheep F(ab)2 Alexa 647 | Jackson Immunoresearch | 713-606-147 |
| donkey anti-rabbit Alexa 647 | Jackson Immunoresearch | 711-605-152 |
| donkey anti-rat Alexa 647 | Jackson Immunoresearch | 712-605-153 |
| Bacterial and Virus Strains | ||
| rAAV1-Syn-FLEX-GCaMP6f-WPRE-Sv40 | UPenn Viral Vector Core | N/A |
| Experimental Models: Organisms/Strains | ||
| VGAT-IRES-Cre | The Jackson Laboratory | Jax stock: 016962 |
| VIP-IRES-Cre | The Jackson Laboratory | Jax stock: 010908 |
| Software and Algorithms | ||
| Python 2.7 | Python | https://www.python.org |
| FIJI (ImageJ) | NIH | http://fiji.sc |
| Scikit-Learn | Pedregosa et al., 2011 | http://scikit-learn.org |
| SIMA | Kaifosh et al., 2014 | https://github.com/losonczylab/sima |
| FISSA | Keemink et al., 2018 | https://github.com/rochefort-lab/fissa |
| Machine-learning classification of subtypes | This paper | https://github.com/losonczylab/Geiller_Vancura_Neuron2020 |
In VGAT-Cre mice, three strategies were used for staining.
Strategy 1.
Mice ID: TG16, TG17, TG18, TG19, BV90, TG300, TG302, TG310 (see Supplementary Figure S1A)
First round primary antibodies: rabbit anti-proCCK (1:500), rat anti-somatostatin (1:500), and sheep anti-NPY (1:500)
First round secondary antibodies: donkey anti-rabbit DyLight 405 (1:300), donkey anti-rat Alexa 568 (1:300), and donkey anti-sheep F(ab)2 Alexa 647 (1:300)
Second round primary antibodies: chicken anti-PV (1:5,000) and rabbit anti-SATB1 (1:1,000).
Second round secondary antibodies: donkey anti-chicken DyLight 405 (1:300) and donkey anti-rabbit Alexa 647 (1:300)
Strategy 2.
Mice ID: TG325, TG326, TG327 (see Supplementary Figure S1A)
First round primary antibodies: chicken anti-PV (1:5,000), rat anti-somatostatin (1:500), and rabbit anti-SATB1 (1:1,000)
First round secondary antibodies: donkey anti-chicken DyLight 405 (1:300), donkey anti-rat Alexa 568 (1:300), and donkey anti-rabbit Alexa 647 (1:300)
Second round primary antibodies: rabbit anti-proCCK (1:500) and sheep anti-NPY (1:500)
Second round secondary antibodies: donkey anti-rabbit DyLight 405 (1:300) and donkey anti-sheep F(ab)2 Alexa 647 (1:300)
Strategy 3.
Mice ID: TG276, BV106, BV107, BV109 (see Supplementary Figure S1B).
First round primary antibodies: chicken anti-PV (1:5,000), rat anti-somatostatin (1:500), and rabbit anti-VIP (1:5,000)
First round secondary antibodies: donkey anti-chicken DyLight 405 (1:300), donkey anti-rat Alexa 568 (1:300), and donkey anti-rabbit Alexa 647 (1:300)
Second round primary antibodies: guinea pig anti-calretinin (1:1,000) and sheep anti-NPY (1:500)
Second round secondary antibodies: donkey anti-guinea pig Rhodamine Red X (1:200) and donkey anti-sheep F(ab)2 Alexa 647 (1:300)
In VIP-Cre mice, immunohistochemistry labeling consisted of only one round.
Mice ID: TG338, TG339, TG340 (see Supplementary Figure S1C).
Primary antibodies: rabbit anti-proCCK (1:500), guinea pig anti-calretinin (1:1,000), and rat anti-M2R (1: 2,000)
Secondary antibodies: donkey anti-rabbit DyLight 405 (1:300), donkey anti-guinea pig Rhodamine Red X (1:200), and donkey anti-rat Alexa 647 (1:300)
Assignment of subtype identity based on immunostaining:
Axo-axonic cells (AAC):
AAC were immunopositive for PV, and immunonegative for SATB1, SOM, NPY, and CCK (Klausberger et al., 2003; Viney et al., 2013; Varga et al., 2014); they were thus differentiated from parvalbumin-expressing basket cells on the basis of SATB1 transcription factor immunonegativity. Although only 2 anatomically-verified AAC have been shown to be SATB1-immunonegative in CA1 (Viney et al., 2013; Varga et al., 2014), a larger number of identified AAC have been shown to have these molecular characteristics in CA3 (Viney et al., 2013). In addition, recent single-cell RNA sequencing studies have demonstrated the presence of a significant cluster of PV-immunopositive SATB1-immunonegative cells within mouse CA1, suggesting that the molecular characteristics of CA3 AAC generalize to CA1 (Harris et al., 2018; Qian et al., 2020). To confirm undetectable SATB1 expression, every cell we considered to be an AAC was re-imaged under the confocal microscope at Nyquist resolution and inspected for any evidence of nuclear immunolabeling.
Parvalbumin-expressing basket cells (PVBC):
PVBC were immunopositive for PV and immunonegative for SOM, NPY, and CCK (Sik et al., 1995; Freund and Buzsaki, 1996; Klausberger et al., 2003; Lapray et al., 2012; Viney et al., 2013; Hu, Gan and Jonas, 2014; Pelkey et al., 2017). These cells were also immunopositive for transcription factor SATB1 (Viney et al., 2013; Harris et al., 2018).
Bistratified cells (BiC):
The BiC category included all cells that were immunopositive for PV, SOM, and NPY (Maccaferri et al., 2000; Losonczy et al., 2002; Pawelzik, Hughes and Thomson, 2002; Klausberger et al., 2004; Katona et al., 2014). Although not used as a criteria, all PV, SOM, and NPY triple immunopositive cells with conclusive SATB1 immunoreactivity were also SATB1-immunopositive, in accordance with a previous study (Viney et al., 2013).
Somatostatin-expressing cells (SomC):
Cells in the SomC category were immunopositive for SOM and immunonegative for CCK and PV. Cells within this category include both OLM interneurons (Maccaferri and McBain, 1996; Losonczy et al., 2002; Katona et al., 2014; Forro et al., 2015) and long-range projecting SOM-immunopositive cells (Jinno et al., 2007; Jinno, 2009; Katona et al., 2017). Although previous studies have shown that OLM cells can be weakly PV-immunopositive (Losonczy et al., 2002; Varga, Golshani and Soltesz, 2012; Katona et al., 2014), PV levels are universally significantly lower in OLM cells than in PVBC, AAC, or BiC (Pelkey et al., 2017; Winterer et al., 2019). Thus, cells with barely detectable levels of PV immunoreactivity were considered PV-immunonegative. Although studies in rats have shown OLM cells to be immunonegative for NPY (Katona et al., 2014), a more recent study of anatomically identified OLM cells in mice has shown a significant fraction of them to be NPY-immunopositive (Winterer et al., 2019). In addition, some long-range projecting SOM-immunopositive cells have been shown to be NPY-immunopositive (Jinno et al., 2007). Thus, the presence or absence of NPY immunopositivity was not used as a criterion for this category, and this category includes both NPY-immunopositive and NPY-immunonegative cells. Although more than half of cells within this category were SATB1-immunopositive, SATB1 immunoreactivity was not used as a criteria for this category and it includes some SATB1-immunonegative cells.
Cholecystokinin-expressing cells (CCKC):
Cells in the CCKC category were immunopositive for pro-CCK, immunonegative for SOM, PV, and SATB1, and either positive or negative for NPY (Somogyi et al., 2004; Klausberger et al., 2005; Harris et al., 2018). This category includes both CCK-expressing basket cells and CCK-expressing dendrite-targeting cells (Vida et al., 1998; Cope et al., 2002; Pawelzik, Hughes and Thomson, 2002; Klausberger et al., 2005; Bezaire and Soltesz, 2013; Pelkey et al., 2017).
Ivy cells (IvC) and Neurogliaform cells (NGFC):
Cells in the IvC/NGFC category include both Ivy cells and neurogliaform cells (Vida et al., 1998; Zsiros and Maccaferri, 2005; Price et al., 2005, 2008; Fuentealba et al., 2008; Tricoire et al., 2010; Capogna, 2011; Maccaferri, 2011; Armstrong, Krook-Magnuson and Soltesz, 2012; Lapray et al., 2012; Overstreet-Wadiche and McBain, 2015; Pelkey et al., 2017; Harris et al., 2018), and these cells were always NPY-immunopositive and immunonegative for CCK, SOM, and PV, while they could be either immunopositive or immunonegative for SATB1. Although long-range projecting interneurons have recently been identified that share these molecular characteristics (Wick, Tetzlaff and Krook-Magnuson, 2019), the number of these cells is low compared to estimated numbers of Ivy cells and neurogliaform cells (Fuentealba et al., 2008; Bezaire and Soltesz, 2013). For this reason, we refer to this category as IvC/NGFC.
Type I interneuron-specific interneurons (ISI1):
ISI1 cells were identified as CR-immunopositive and VIP-immunonegative (Gulyás, Hájos and Freund, 1996; Bezaire and Soltesz, 2013; Pelkey et al., 2017). These cells were identified in VGAT-Cre mice.
Type II interneuron-specific interneurons (ISI2):
ISI2 were always VIP-immunopositive and immunonegative for CCK, CR, M2R (Acsády, Arabadzisz and Freund, 1996; Acsády, Görcs and Freund, 1996). We also restricted our definition of ISI2 cells to those located in SR or the SR-SLM border (Acsády, Arabadzisz and Freund, 1996; Acsády, Görcs and Freund, 1996). Because we did not stain for CCK, VIP, and M2R simultaneously in VGAT-Cre mice (see Immunohistochemistry section above), cells matching those criteria could only be identified in VIP-Cre mice and are analyzed in Figure 7.
Type III interneuron-specific interneurons (ISI3):
ISI3 cells were classified on the basis of the co-expression of VIP and CR (Acsády, Arabadzisz and Freund, 1996; Acsády, Görcs and Freund, 1996; Tyan et al., 2014; Pelkey et al., 2017; Luo et al., 2020). All such cells were immunonegative for PV, SOM, and NPY. Cells matching these criteria could be identified in both VGAT-Cre and VIP-Cre mice.
VIP-immunopositive, CR-immunonegative cells (VIP+/CR−):
Cells within the VIP+/CR− category were immunopositive for VIP and immunonegative for CR, PV, SOM, and NPY. This category potentially includes ISI2 cells (Acsády, Arabadzisz and Freund, 1996; Acsády, Görcs and Freund, 1996), VIP+ basket cells (Acsády, Arabadzisz and Freund, 1996; Acsády, Görcs and Freund, 1996), or long-range projecting VIP+ interneurons (Francavilla et al., 2018). In VGAT-Cre mice, our staining strategies (see Immunohistochemistry section above) did not allow us to further separate these subpopulations, so we left them classified by their immunolabel, VIP+/CR−.
VIP-immunopositive, CCK-immunopositive cells (VIP+/CCK+):
VIP/CCK cells were CCK+, CR−, and M2R−. This population represents perisomatic-targeting basket cells (Acsády, Arabadzisz and Freund, 1996; Acsády, Görcs and Freund, 1996).
VIP-immunopositive, M2R-immunopositive cells (VIP+/M2R+):
VIP/M2R cells were immunonegative for CCK and CR, and immunopositive for M2R. These cells represent a long-range projecting VIP-expressing subtype (Francavilla et al., 2018).
Parvalbumin-immunopositive cells (PV+):
In Supplementary Figure 6 and Figure 7H, 7I.
In mice where staining strategy 3 was used (see Immunohistochemistry section above), cells that were PV-immunopositive, SOM-immunonegative, NPY-immunonegative, VIP-immunonegative, and CR-immunonegative were classified as PV+ cells. This category includes both PVBC and AAC, but we could not distinguish between these two populations as we did not stain for SATB1 (see Supplementary Figure 1).
Unidentified cells
All cells not assigned to one of the subtypes described above were classified as unidentified.
Confocal imaging:
A Nikon A1 confocal microscope was used to acquire multi-channel fluorescence images of the immunolabeled tissue sections. 405 nm, 488 nm, 561 nm, and 640 nm laser lines were used for excitation. Each channel was acquired sequentially with a 10x Plan Apo NA 0.45 objective (Nikon) at 1.2–1.3x Zoom. 2048×2048 pixel images were acquired every ~3 microns through the entire depth of the tissue sections, with the pinhole size set to ~1 Airy unit. Fluorescence was collected with 2 GaAsP PMTs (488 nm and 561 nm channels) and 2 multi-alkali PMTs (405 nm and 640 nm channels). The resulting 4-channel Z-stacks were viewed in FIJI.
Registration of confocal images to in vivo Z-stacks and identification of immunopositivity/negativity:
The following steps were performed by an experimenter without the use of any automated methods: First, the confocal stacks were rotated and translated until the cells in the green channel (GCaMP-labeled) matched the cells seen in the in vivo Z stack. Second, each imaged cell was found in the confocal stacks, and it was evaluated for immunopositivity or immunonegativity for the tested molecule. For a cell to be considered positive, the fluorescence intensity inside the cell had to be significantly greater than the background intensity level. A cell was considered positive for a given marker only if clear examples of immunonegative cells could be found on the same tissue section. Similarly, a cell was considered negative for a given marker only if clear examples of immunopositive cells could be found on the same tissue section. In the case of ambiguous immunolabeling, cells were discarded and not grouped into a subtype for further analysis. In experiments in which PV and SATB1 immunoreactivity were assessed together, all cells that were initially identified as axo-axonic cells (PV+, SATB1−) were imaged again under the confocal microscope at Nyquist resolution to confirm the absence of detectable SATB1 immunoreactivity. Overall, all efforts were made to use the most stringent criteria for cell classification prior to analysis.
Pre-processing of Ca2+ imaging data:
The raw movies containing each cell were motion corrected independently using a whole-frame cross-correlation algorithm, as implemented in the SIMA software (Kaifosh et al., 2014). The time-average of each imaged cell was manually inspected and a ROI was hand-drawn over each cell. Fluorescence was extracted from each ROI using the FISSA software (Keemink et al., 2018) package to correct for neuropil contamination. For each resulting raw fluorescence trace, a baseline F was calculated by taking the 1st percentile in a rolling window of 30s and a ΔF/F trace was calculated. The ΔF/F trace for each cell was smoothed using an exponential filter and all further analysis was performed on the resulting ΔF/F traces. In Figure 7D and 7E, a static baseline calculated as the 1st percentile of the whole trace was used instead of an adaptive baseline. This way, we could examine small fluctuations during immobility epochs that would otherwise be suppressed by an adaptive baseline. All further analysis was implemented in Python 2.7 and is detailed below.
Cross-correlation between each cell’s GCaMP Ca2+ activity and velocity:
To calculate the correlation between each cell’s activity and the animal’s velocity, the Pearson’s correlation coefficient was calculated between each cell’s ΔF/F trace and the smoothed velocity trace by shifting each cell’s fluorescence trace one frame at a time within a −5s (activity lagging velocity) to +5s (activity leading velocity) interval. The maximum of the absolute value of all correlation coefficients within this interval was taken as the correlation coefficient for the cell, and the shift at which it occurred was taken as the lag. As most cells were cross-registered across many imaging experiments, the correlation coefficients and lags for each cell were averaged across all sessions in which the cell was imaged.
Responses to run-start and run-stop events:
Run-start and run-stop events were identified in the imaging data as frames during which the animal’s velocity increased above 0.2 cm/s (run-start event) or decreased below 0.2 cm/s (run-stop event). In addition, each run-start/run-stop event had to be separated from the previous run-start/run-stop event by at least several seconds to be considered as a separate event. For each event, the mean of the pre-event ΔF/F was subtracted from the mean of the post-event ΔF/F in a −3s to +3s window to calculate a response magnitude. For each cell, the run-start and run-stop responses were averaged over all run-start and run-stop events in the given imaging experiment. If a cell was imaged across more than one imaging experiment, the average run-start and run-stop responses from each experiment were averaged over all experiments.
Spatial tuning curve:
To calculate a spatial tuning curve for each imaged cell in a given experiment, the 2m treadmill was divided into 100 2cm-long bins. For each bin, we calculated the average ΔF/F from frames where the animal was in locomotion (velocity > 5cm/s) and smoothed the resulting trace with a Gaussian kernel (σ = 3 bins) to obtain the spatial tuning curve
Spatial tuning analysis:
To determine whether a cell was spatially tuned during an imaging session, we generated 1000 shuffled tuning curves by circularly rotating position in relation to ΔF/F tuning curves (restricted to frames during locomotion). A cell was detected spatially selective if it had 10 consecutive bins (20cm) exceeding the 95th percentile of the shuffle distribution (or lower than the 5th percentile for negative fields) and a peak firing in the field exceeding 100% ΔF/F (no threshold for negative fields). Recurrence probability was calculated as the number of times a cell was detected spatially modulated over the number of times it was imaged. Selectivity index was calculated as previously described (Acharya et al., 2016) by computing the spatial sparsity of the rate map given N bins with rn the rate in the nth bin as :
Position decoding:
We used a linear classifier (support vector machine, SVM) to decode the animal’s position from interneuron / pyramidal cell population activity using the spatial tuning curves. For cross-validation, a spatial tuning curve was generated for each lap and we trained the classifier on (n-1) laps and then tested on the held-out lap. The testing lap was then rotated to cover all the n laps of the session.
Stability analysis:
Stability analysis was performed by correlating (Pearson’s R) the spatial tuning curves of the same cells at different time points. For the within-session comparison, each session was first divided between odd and even laps, and stability was calculated as the correlation between the average tuning curve for each split.
Context remapping analysis:
In the context remapping analysis, we correlated the tuning curves in two different belts (A1, A2 and B). To avoid aligning arbitrarily the two belts, we iteratively rotated the tuning curves in B to maximize the population’s average correlation coefficient for each mouse independently, which resulted in reporting the highest possible values.
To analyze the change in activity upon exposure to a new belt, we averaged the ΔF/F values (restricted to frames during locomotion) in each lap and compared values from the 10 last laps of A2 (just preceding the change of belt) to the 15 first laps of B.
Context decoding:
We used a linear classifier (support vector machine, SVM) to decode the context identity based on interneuron population activity. For cross-validation, a spatial tuning curve was generated for each lap and we trained the classifier on blocks of n randomly-chosen laps in the session (20%, 40%, 60% and 80% of the laps used for training and the rest for testing). This training/testing procedure was repeated 100 times.
GOL analysis:
We quantified the change in activity for each cell near the reward zone by defining a reward modulation index (RMI) to normalize activity amplitudes across different IN subtypes. RMI is defined as the difference of the average activity preceding the reward zone (10cm) and the average activity in the rest of the belt, divided by the sum of the two. To quantify the representation of the old reward zone by different subtypes without being affected by the activity in the current zone, we calculated a reward zone entry value as the difference in activity between 10cm after the start of the old zone and 10cm before it (based on the spatial tuning curves).
Multivariate ridge regression:
To more explicitly dissociate the effects of the various behavioral variables on each cell’s activity during the GOL task, we developed a multivariate linear regression model to predict each cell’s spatial tuning curve (the average ΔF/F value in each position bin) from the following behavioral variables: 1) the animal’s velocity, 2) licking, and 3) position. The position variable was itself divided into 5 predictors, corresponding to 5 zones of the treadmill. The model utilized Ridge regression to minimize the effects of potential relationships between the independent variables. The fit values are reported in Supplementary Figure S7, as well as the coefficients for the predictors for each subtype, and the relationship between predictors and fit quality.
Ripple modulation probability:
We used two measures of activity during sharp-wave ripple (SWR) events. The average response was defined as the averaged difference in activity in a 400ms window centered around each SWR onset. The first 200ms (left side of the window) serves as the baseline and the baseline is subtracted from the average value in the 200ms on the right side of the window. Here, the baseline is subtracted in case multiple ripples occur successively and that the GCaMP signal is on the descending phase, which could have potential confounds on the fluorescence values. Therefore, only 200ms is used to compute a modulation value and not the full 400ms. This analysis is similar to a PSTH, but it is not displayed as an average trace for each subtype. This is because the neurons belonging to a given subtype can have different response traces for each ripple event, largely dependent on the number of spikes fired. The traces do not bring useful quantitative information, and so instead, we represented each cell by a single point, whereby the heterogeneity within each subtype can be better appreciated. It is also important to remember that the ripples detected contralaterally can have a small but consequent time jitter with the activity seen in the ipsilateral side, which may further degrade the ability to nicely visualize average responses. This analysis can be seen in Supplementary Figure S4.
In the second analysis (in Figure 3), we reasoned that limitations in calcium sensitivity, baseline levels and ripple amplitude can influence the response activity seen in different subtypes. To circumvent those limitations, we developed a measure of modulation to evaluate whether a cell had an increased activity during a SWR event statistically higher than its baseline fluctuation during immobility. To do so, we computed for each cell a shuffle distribution that consisted of 1000 values. Each value was the average of N randomly chosen ΔF/F points during immobility (N corresponds to the number of SWRs detected in the session). Then, for each SWR, if the difference in activity in a 400ms window centered around the onset was greater than the 95th percentile of the shuffle distribution, the cell was modulated during this event. The modulation probability reports the percentage of SWRs leading to an increase above baseline.
Bayesian gaussian mixture model:
To separate and cluster groups of cells in a given subtype that are modulated during SWR events, we used a Bayesian estimation of a Gaussian model where the number of groups is directly inferred from the data, removing any arbitrary threshold in modulation probability values. The prior for the weight distributions was taken from a finite mixture model, and the weights were initialized using k-means.
Type III ISI transient detection:
Activity in Type III ISIs showed calcium transients akin to pyramidal cells. We detected the onset of each transient by finding peaks in the Z-score traces with a threshold of 0.3 sigma, and a minimum value of 15 frames (~2 seconds) for minimal horizontal distance, with smaller transients being discarded first until the condition is fulfilled.
Interneuron dynamics during immobility epochs:
To compare the dynamics of VIP+/CCK+ and VIP+/M2R+ interneurons during immobility epochs, immobility intervals were first identified during each imaging session as periods when the velocity remained below 0.2 cm/s for at least 15–20 seconds. Immobility intervals within a given imaging experiment were normalized by upsampling each interval to the length of the longest interval. The baseline-corrected fluorescence (ΔF/F, with static baseline F0, see above) of each cell was divided into 1,000 bins within each normalized immobility interval and averaged over all immobility intervals within the imaging session to generate an average immobility response. If a given cell was imaged in more than one experiment, its immobility trace was averaged over all experiments in which it was imaged.
Interactions between different interneuron subtypes:
To analyze the activity relationships between simultaneously recorded interneurons, the pairwise correlation coefficient (Pearson’s R) was calculated between each pair of simultaneously imaged cells.
Machine learning analysis:
To assess whether the six IN subtypes described in Figures 1–6 can be discriminated based on their activity profiles during behavior and/or anatomical features, we employed a machine learning approach. Specifically, we developed a Deep Neural Network (DNN) and tested its discrimination performance on various tasks:
a 4-class problem consisting of PV-expressing, perisomatic-targeting INs (PVBCs and AACs), SOM-expressing, dendrite-targeting INs (SomCs and BiCs), CCK-expressing (CCKCs) and NPY-expressing INs (IvCs/NGFCs) (Supplementary Figure S3)
a 6-class problem consisting of all IN subtypes identified in Figures 1–6 (PVBC, AAC, SomC, BiC, CCKC, and IvC/NGFC) (Supplementary Figure S3)
Data Preprocessing
We used three types of features to predict the subtype identity of each imaged IN: 1) the cell’s ΔF/F signal, 2) the velocity of the animal, and 3) the z position of the imaged cell within CA1 (Figure 1E, Figure 1F). Each one of these features was added incrementally to the network in order to assess its relative contribution to the decoding performance.
Each cell’s ΔF/F signal and the velocity signal (which is the same for all cells within the same imaging session) change over time, while the z position is a single variable for each cell. We considered the ΔF/F and velocity signals during individual laps, defined by respective run-start and run-stop signals measured at the onset and finish of each lap. To normalize the number of measurements within each lap, we resampled the ΔF/F and velocity signals to include 100 measurements for each lap by applying a cubic interpolation to the data. The alignment process is graphically shown in Supplementary Figure 3A. The z position of the cell was converted to a 100-element vector by simply repeating the value 100 times.
This process resulted in three types of input features for each cell that were used incrementally as inputs to the DNN (namely ΔF/F signal alone, ΔF/F signal plus velocity, and ΔF/F signal plus velocity plus z position). Specifically, a ΔF/F lap is a single vector of 100 elements that contains the cell’s interpolated ΔF/F signal between a run-start and run-stop event. Accordingly, a ΔF/F-velocity lap consists of two vectors, the ΔF/F signal and the velocity signal of a lap. A calcium-velocity-z-position lap includes the ΔF/F and velocity vectors plus an additional vector with the z-position of the cell.
DNN architectures
We employed a Convolutional Neural Network architecture previously shown to achieve high accuracy in a cell-type classification task (Troullinou et al., 2019). We first built a 1-Dimensional Convolutional Neural Network (1D-CNN) architecture and used ΔF/F signals as the only input feature. The network architecture consisted of a 1-Dimensional Convolutional layer consisting of 256 filters with a kernel of size 3 followed by a ReLu activation function. This layer is followed by a Batch Normalization layer. Then, the model is built up with the following pattern: a 1-Dimensional Convolutional layer-a Batch Normalization layer-1-Dimensional Max-Pooling layer. This pattern is repeated 3 times. In each pattern each convolutional layer consists of 128, 64 and 32 filters respectively, with a kernel of size 3, followed by a ReLU activation function. All max-pooling layers have a kernel of size 3 and stride is defined to 2. All these layers are followed by a dropout layer, where individual nodes of a specific layer are kept with probability p = 0.5. At the end of the architecture we used a fully connected layer, which has as many nodes as the number of classes and represents the activation maps of high-level features determining which features most correlate to a particular class. As a final classification step, we used the softmax activation function, which assigns decimal probabilities to each class in a multi-class problem. The optimizer that is used in order to train our model is the Adam gradient-descent based algorithm with learning rate 0.001, and the loss we used for this multi-class problem is the categorical cross entropy.
Similarly, when using the combined ΔF/F-velocity laps as input features for each cell, we built a 2-Dimensional Convolutional Neural Network (2D-CNN) architecture. The general architecture of the 2D-CNN model is the same as that of the 1D-CNN model with some modifications. 1-Dimensional Convolutional and Max-Pooling layers are replaced with 2-Dimensional Convolutional and Max-Pooling layers, respectively. Moreover, the kernel of the first convolutional layer is of size [2,3], so that both of our features (ΔF/F and velocity) are convolved with the filter. For the rest of the convolutional layers we used a kernel of size [1,3] and retained the same dimensions for the input and output channels of each layer. This serves implementation purposes, given that each example is now 2-dimensional.
To account for all three input features of our cells, we used the 2D-CNN architecture with the addition of the z position. The kernel of the first convolutional layer is of size [3,3], so that the kernel is also convolved with the z position information. As previously, for the rest of the convolutional layers we used a kernel of size [1,3] and retained the same dimensions for the input and output channels of each layer.
The Deep Learning models that were used in our analysis were implemented with the help of Tensorflow and Keras open-source libraries written in Python. Both TensorFlow and Keras can run calculations on GPU, dramatically decreasing the computational time of the network’s training. For these experiments we used Python 3.7.3™, Tensorflow version 2.1, Keras version 2.3.1 and NVIDIAs GPU model, GeForce GTX 1080 Ti, running on a Linux CentOS 7 operating system.
Prediction analysis
The data set consists of representative examples from each cell class. As the number of cell examples is very different for the 6 classes, we have adopted different ways of splitting the data. For each task, we used a fixed number of test examples for all classes to ensure fair comparison. The training set was constrained by the number of examples in the smallest cell class. We thus used a semi-balanced approach, whereby larger categories were augmented by additional samples on a case-by-case basis (reported separately for each figure). In all cases, 10% of the training set is used for validation and based on its performance progress in every epoch, we select the best model to be the one with the highest validation accuracy. We have also applied an early stopping criterion, so that if there is no improvement for 200 consecutive epochs the algorithm stops. In order to evaluate the performance of our models, we apply 10 random train-test splits and report the mean prediction accuracy for different combinations of input features as well as the confusion matrices for better visibility of misclassified samples.
Statistical analyses:
Statistical details of comparisons are specified in either the main text or figure legends. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications. Box plots represent median and interquartile range (IQR) while whiskers extend to cover the distribution without outliers (defined as points above 1.5 IQR below or above the box edges). Bar plots represent mean and s.e.m. Between-subtype comparisons were tested using a one-way ANOVA followed by a Tukey’s range test with correction for multiple testing if appropriate. For comparisons between two populations, a paired sample or unpaired t-test was applied if the data points followed a normal distribution (confirmed using the Kolmogorov-Smirnov test). To analyze data that was not normally distributed, the Mann-Whitney U test was used. *, p < 0.05, **, p < 0.01, ***, p < 0.001. Data analysis and figures were done using custom made software in Python 2.7.15™.
Supplementary Material
Supplementary Table 1. Means and medians with corresponding SEM or IQR for data distributions presented in the main figures. Related to Figures 1–7.
Supplementary Video 1. 3D calcium imaging of 296 interneurons during running (5-minute session at 20x speed, non-motion corrected). Related to Figure 1.
Highlights:
A subset of CA1 interneurons exhibit spatially tuned activity
Axo-axonic and bistratified cells show bimodal recruitment during sharp-wave ripples
SOM+ cells are preferentially recruited during goal-oriented spatial learning
Disinhibition from VIP+ cells is preferentially mediated through SOM+ cells
Acknowledgements:
A.L. is supported by NIMH-1R01MH100631, NINDS-1U19NS104590, NINDS-1R01NS094668, and the Kavli Foundation. P.P. is supported by the European Commission, FET Open Grant (NEUREKA, 863245), the Fondation Santé, and the Alexander von Humboldt Foundation. B.V. is supported by grant T32 GM007367. E.T. is supported by the program ‘Strengthening Human Resources Research Potential via Doctorate Research’ (MIS-5000432), implemented by the State Scholarships Foundation (IKY). B.R. is supported by ERC 682426, KFI-2018-00097, VKE-2018-00032, NUMBER 871277 — AMPLITUDE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests:
The authors declare no competing interests.
REFERENCES
- Acharya L, Aghajan ZM, Vuong C, Moore JJ and Mehta MR (2016). Causal Influence of Visual Cues on Hippocampal Directional Selectivity. Cell 164, 197–207. [DOI] [PubMed] [Google Scholar]
- Acsády L, Arabadzisz D and Freund TF (1996). Correlated morphological and neurochemical features identify different subsets of vasoactive intestinal polypeptide-immunoreactive interneurons in rat hippocampus. Neuroscience 73, 299–315. [DOI] [PubMed] [Google Scholar]
- Acsády L, Görcs TJ and Freund TF (1996). Different populations of vasoactive intestinal polypeptide-immunoreactive interneurons are specialized to control pyramidal cells or interneurons in the hippocampus. Neuroscience 73, 317–334. [DOI] [PubMed] [Google Scholar]
- Armstrong C, Krook-Magnuson E and Soltesz I (2012). Neurogliaform and Ivy Cells: A Major Family of nNOS Expressing GABAergic Neurons. Front. Neural Circuits 6, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriaga M and Han EB (2017). Dedicated hippocampal inhibitory networks for locomotion and immobility. J. Neurosci 37, 9222–9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriaga M and Han EB (2019). Structured inhibitory activity dynamics in new virtual environments. eLife 8, e47611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartos M, Vida I and Jonas P (2007). Synaptic mechanisms of synchronized gamma oscillations in inhibitory interneuron networks. Nat. Rev. Neurosci 8, 45–56. [DOI] [PubMed] [Google Scholar]
- Bezaire MJ and Soltesz I (2013). Quantitative assessment of CA1 local circuits: Knowledge base for interneuron-pyramidal cell connectivity. Hippocampus 23, 751–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloss EB, Cembrowski MS, Karsh B, Colonell J, Fetter RD and Spruston N (2016). Structured Dendritic Inhibition Supports Branch-Selective Integration in CA1 Pyramidal Cells. Neuron 89, 1016–1030. [DOI] [PubMed] [Google Scholar]
- Booker SA and Vida I (2018). Morphological diversity and connectivity of hippocampal interneurons. Cell Tissue Res. 373, 619–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhl EH, Szilágyi T, Halasy K and Somogyi P (1996). Physiological properties of anatomically identified basket and bistratified cells in the CA1 area of the rat hippocampus in vitro. Hippocampus 6, 294–305. [DOI] [PubMed] [Google Scholar]
- Buhl EH, Halasy K and Somogyi P (1994). Diverse sources of hippocampal unitary inhibitory postsynaptic potentials and the number of synaptic release sites. Nature 368, 823–828. [DOI] [PubMed] [Google Scholar]
- Buzsáki G (2015). Hippocampal sharp wave-ripple: A cognitive biomarker for episodic memory and planning. Hippocampus 25, 1073–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capogna M (2011). Neurogliaform cells and other interneurons of stratum lacunosum-moleculare gate entorhinal-hippocampal dialogue. J. Physiol 589, 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi A, Melzer S, Michael M and Monyer H (2013). The long and short of GABAergic neurons. Curr. Opin. Neurobiol 23, 79–186. [DOI] [PubMed] [Google Scholar]
- Cembrowski MS, Wang L, Sugino K, Shields BC and Spruston N (2016). Hipposeq: A comprehensive RNA-seq database of gene expression in hippocampal principal neurons. eLife 5, e14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberland S and Topolnik L (2012). Inhibitory control of hippocampal inhibitory neurons. Front. Neurosci 6, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S and Zhuang X (2015). Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiovini B, Turi GF, Katona G, Kaszás A, Pálfi D, Maák P, Szalay G, Szabó MF, Szabó G, Szadai Z, et al. (2014). Dendritic Spikes Induce Ripples in Parvalbumin Interneurons during Hippocampal Sharp Waves. Neuron 82, 908–924 [DOI] [PubMed] [Google Scholar]
- Cobb SR, Buhl EH, Halasy K, Paulsen O and Somogyi P (1995). Synchronization of neuronal activity in hippocampus by individual GABAergic interneurons. Nature 378, 75–78. [DOI] [PubMed] [Google Scholar]
- Colgin LL, Moser EI and Moser MB (2008). Understanding memory through hippocampal remapping. Trends Neurosci. 31, 469–477. [DOI] [PubMed] [Google Scholar]
- Cope DW, Maccaferri G, Márton LF, Roberts JD, Cobden PM and Somogyi P (2002). Cholecystokinin-immunopositive basket and Schaffer collateral-associated interneurones target different domains of pyramidal cells in the CA1 area of the rat hippocampus. Neuroscience 109, 63–80. [DOI] [PubMed] [Google Scholar]
- Danielson NB, Zaremba JD, Kaifosh P, Bowler J, Ladow M and Losonczy A (2016). Sublayer-Specific Coding Dynamics during Spatial Navigation and Learning in Hippocampal Area CA1. Neuron 91, 652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw MI, Tricoire L, Erdelyi F, Szabo G and McBain CJ (2009). Asynchronous transmitter release from cholecystokinin-containing inhibitory interneurons is widespread and target-cell independent. J. Neurosci 29, 11112–11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato F, Rompani SB and Caroni P (2013). Parvalbumin-expressing basket-cell network plasticity induced by experience regulates adult learning. Nature 504, 272–276. [DOI] [PubMed] [Google Scholar]
- Dupret D, O’Neill J and Csicsvari J (2013). Dynamic Reconfiguration of Hippocampal Interneuron Circuits during Spatial Learning. Neuron 78, 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ego-Stengel V and Wilson MA (2007). Spatial selectivity and theta phase precession in CA1 interneurons. Hippocampus 17, 161–174. [DOI] [PubMed] [Google Scholar]
- English DF, McKenzie S, Evans T, Kim K, Yoon E and Buzsáki G (2017). Pyramidal Cell-Interneuron Circuit Architecture and Dynamics in Hippocampal Networks. Neuron 96, 505–520.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G and Kepecs A (2020). Interneuron Types as Attractors and Controllers. Annu. Rev. Neurosci 43, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forro T, Valenti O, Lasztoczi B and Klausberger T (2015). Temporal organization of GABAergic interneurons in the intermediate CA1 hippocampus during network oscillations. Cereb. Cortex 25, 1228–1240. [DOI] [PubMed] [Google Scholar]
- Francavilla R, Villette V, Luo X, Chamberland S, Muñoz-Pino E, Camiré O, Wagner K, Kis V, Somogyi P and Topolnik L (2018). Connectivity and network state-dependent recruitment of long-range VIP-GABAergic neurons in the mouse hippocampus. Nat. Commun 9, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund TF and Buzsáki G (1996). Interneurons of the hippocampus. Hippocampus 6, 347–470. [DOI] [PubMed] [Google Scholar]
- Freund TF and Katona I (2007). Perisomatic Inhibition. Neuron 56, 33–42. [DOI] [PubMed] [Google Scholar]
- Fuentealba P, Begum R, Capogna M, Jinno S, Márton LF, Csicsvari J, Thomson A, Somogyi P and Klausberger T (2008). Ivy Cells: A Population of Nitric-Oxide-Producing, Slow-Spiking GABAergic Neurons and Their Involvement in Hippocampal Network Activity. Neuron 57, 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann F, Justus D, Sosulina L, Kaneko H, Beutel T, Friedrichs D, Schoch S, Schwarz MK, Fuhrmann M and Remy S (2015). Locomotion, Theta Oscillations, and the Speed-Correlated Firing of Hippocampal Neurons Are Controlled by a Medial Septal Glutamatergic Circuit. Neuron 86, 1253–1264. [DOI] [PubMed] [Google Scholar]
- Geiller T, Fattahi M, Choi JS and Royer S (2017). Place cells are more strongly tied to landmarks in deep than in superficial CA1. Nat. Commun 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C, Milstein AD, Bittner KC, Romani S and Magee JC (2017). Inhibitory suppression of heterogeneously tuned excitation enhances spatial coding in CA1 place cells. Nat. Neurosci 20, 417–426. [DOI] [PubMed] [Google Scholar]
- Gulyás AI, Megías M, Emri Z and Freund TF (1999). Total number and ratio of excitatory and inhibitory synapses converging onto single interneurons of different types in the CA1 area of the rat hippocampus. J. Neurosci 19, 10082–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyás AI, Hájos N and Freund TF (1996). Interneurons containing calretinin are specialized to control other interneurons in the rat hippocampus. J. Neurosci 16, 3397–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainmueller T and Bartos M (2018). Parallel emergence of stable and dynamic memory engrams in the hippocampus. Nature 558, 292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangya B, Li Y, Muller RU and Czurkó A (2010). Complementary spatial firing in place cell-interneuron pairs. J. Physiol 588, 4165–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Hochgerner H, Skene NG, Magno L, Katona L, Bengtsson Gonzales C, Somogyi P, Kessaris N, Linnarsson S and Hjerling-Leffler J (2018). Classes and continua of hippocampal CA1 inhibitory neurons revealed by single-cell transcriptomics. PLoS Biol. 16, e2006387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefft S and Jonas P (2005). Asynchronous GABA release generates long-lasting inhibition at a hippocampal interneuron-principal neuron synapse. Nat. Neurosci 8, 1319–1328. [DOI] [PubMed] [Google Scholar]
- Hollup SA, Molden S, Donnett JG, Moser MB and Moser EI (2001). Accumulation of hippocampal place fields at the goal location in an annular watermaze task. J. Neurosci 21, 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Gan J and Jonas P (2014). Fast-spiking, parvalbumin+ GABAergic interneurons: From cellular design to microcircuit function. Science 345, 1255263. [DOI] [PubMed] [Google Scholar]
- Isaacson JS and Scanziani M (2011). How inhibition shapes cortical activity. Neuron 72, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinno S, Klausberger T, Marton LF, Dalezios Y, Roberts JD, Fuentealba P, Bushong EA, Henze D, Buzsáki G and Somogyi P (2007). Neuronal diversity in GABAergic long-range projections from the hippocampus. J. Neurosci 27, 8790–8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinno S (2009). Structural organization of long-range GABAergic projection system of the hippocampus. Front. Neuroanat 3, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifosh P, Zaremba JD, Danielson NB and Losonczy A (2014). SIMA: Python software for analysis of dynamic fluorescence imaging data. Front. Neuroinform 8, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona G, Szalay G, Maák P, Kaszás A, Veress M, Hillier D, Chiovini B, Vizi ES, Roska B and Rózsa B (2012). Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat. Methods 9, 201–208. [DOI] [PubMed] [Google Scholar]
- Katona I and Freund TF (2012). Multiple Functions of Endocannabinoid Signaling in the Brain. Annu. Rev. Neurosci 35, 529–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona L, Lapray D, Viney TJ, Oulhaj A, Borhegyi Z, Micklem BR, Klausberger T and Somogyi P (2014). Sleep and Movement Differentiates Actions of Two Types of Somatostatin-Expressing GABAergic Interneuron in Rat Hippocampus. Neuron 82, 872–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona L, Micklem B, Borhegyi Z, Swiejkowski DA, Valenti O, Viney TJ, Kotzadimitriou D, Klausberger T and Somogyi P (2017). Behavior-dependent activity patterns of GABAergic long-range projecting neurons in the rat hippocampus. Hippocampus 27, 359–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman AM, Geiller T and Losonczy A (2020). A Role for the Locus Coeruleus in Hippocampal CA1 Place Cell Reorganization during Spatial Reward Learning. Neuron 105, 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keemink SW, Lowe SC, Pakan J, Dylda E, van Rossum M and Rochefort NL (2018). FISSA: A neuropil decontamination toolbox for calcium imaging signals. Sci. Rep 8, 3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerlin AM, Andermann ML, Berezovskii VK and Reid RC (2010). Broadly Tuned Response Properties of Diverse Inhibitory Neuron Subtypes in Mouse Visual Cortex. Neuron 67, 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AG, Poort J, Chadwick A, Blot A, Sahani M, Mrsic-Flogel TD and Hofer SB (2018). Distinct learning-induced changes in stimulus selectivity and interactions of GABAergic interneuron classes in visual cortex. Nat. Neurosci 21, 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T, Magill PJ, Márton LF, Roberts JD, Cobden PM, Buzsáki G and Somogyi P (2003). Brain-state- and cell-type-specific firing of hippocampal interneurons in vivo. Nature 421, 844–848. [DOI] [PubMed] [Google Scholar]
- Klausberger T, Márton LF, Baude A, Roberts JD, Magill PJ and Somogyi P (2004). Spike timing of dendrite-targeting bistratified cells during hippocampal network oscillations in vivo. Nat. Neurosci 7, 41–47. [DOI] [PubMed] [Google Scholar]
- Klausberger T, Marton LF, O’Neill J, Huck JH, Dalezios Y, Fuentealba P, Suen WY, Papp E, Kaneko T, Watanabe M, et al. (2005). Complementary roles of cholecystokinin- and parvalbumin-expressing GABAergic neurons in hippocampal network oscillations. J. Neurosci 25, 9782–9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T (2009). GABAergic interneurons targeting dendrites of pyramidal cells in the CA1 area of the hippocampus. Eur. J. Neurosci 30, 947–957. [DOI] [PubMed] [Google Scholar]
- Klausberger T and Somogyi P (2008). Neuronal diversity and temporal dynamics: The unity of hippocampal circuit operations. Science 321, 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubie JL, Levy ERJ and Fenton AA (2020). Is hippocampal remapping the physiological basis for context? Hippocampus 30, 851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer D and Helmchen F (2012). Post hoc immunostaining of GABAergic neuronal subtypes following in vivo two-photon calcium imaging in mouse neocortex. Pflugers Arch. 463, 339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapray D, Lasztoczi B, Lagler M, Viney TJ, Katona L, Valenti O, Hartwich K, Borhegyi Z, Somogyi P and Klausberger T (2012). Behavior-dependent specialization of identified hippocampal interneurons. Nat. Neurosci 15, 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kruglikov I, Huang ZJ, Fishell G and Rudy B (2013). A disinhibitory circuit mediates motor integration in the somatosensory cortex. Nat. Neurosci 16, 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Marchionni I, Bezaire M, Varga C, Danielson N, Lovett-Barron M, Losonczy A and Soltesz I (2014). Parvalbumin-positive basket cells differentiate among hippocampal pyramidal cells. Neuron 82, 1129–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Földy C and Soltesz I (2010). Distinct endocannabinoid control of GABA release at perisomatic and dendritic synapses in the hippocampus. J. Neurosci 30, 7993–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Geiller T, Jung A, Nakajima R, Song YK and Baker BJ (2017). Improving a genetically encoded voltage indicator by modifying the cytoplasmic charge composition. Sci Rep 7, 8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MZ and Schnitzer MJ (2016). Genetically encoded indicators of neuronal activity. Nat. Neurosci 19, 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losonczy A, Zhang L, Shigemoto R, Somogyi P and Nusser Z (2002). Cell type dependence and variability in the short-term plasticity of EPSCs in identified mouse hippocampal interneurones. J. Physiol 542, 193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Barron M, Turi GF, Kaifosh P, Lee PH, Bolze F, Sun XH, Nicoud JF, Zemelman BV, Sternson SM and Losonczy A (2012). Regulation of neuronal input transformations by tunable dendritic inhibition. Nat. Neurosci 15, 423–430. [DOI] [PubMed] [Google Scholar]
- Lovett-Barron M, Kaifosh P, Kheirbek MA, Danielson N, Zaremba JD, Reardon TR, Turi GF, Hen R, Zemelman BV and Losonczy A (2014). Dendritic inhibition in the hippocampus supports fear learning. Science 343, 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Guet-McCreight A, Villette V, Francavilla R, Marino B, Chamberland S, Skinner FK and Topolnik L (2020). Synaptic Mechanisms Underlying the Network State-Dependent Recruitment of VIP-Expressing Interneurons in the CA1 Hippocampus. Cereb. Cortex 30, 3667–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri G, Roberts JD, Szucs P, Cottingham CA and Somogyi P (2000). Cell surface domain specific postsynaptic currents evoked by identified GABAergic neurones in rat hippocampus in vitro. J. Physiol 524, 91–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri G (2011). Microcircuit-specific processing in the hippocampus. J. Physiol 589, 1873–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri G and McBain CJ (1996). Long-term potentiation in distinct subtypes of hippocampal nonpyramidal neurons. J. Neurosci 16, 5334–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, Henze DA, Hirase H, Leinekugel X, Dragoi G and Buzsáki G (2002). Hippocampal pyramidal cell-interneuron spike transmission is frequency dependent and responsible for place modulation of interneuron discharge. J. Neurosci 22, RC197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina M, Vida I and Jonas P (2000). Distal initiation and active propagation of action potentials in interneuron dendrites. Science 287, 295–300. [DOI] [PubMed] [Google Scholar]
- Matyas F, Freund TF and Gulyas AI (2004). Convergence of excitatory and inhibitory inputs onto CCK-containing basket cells in the CA1 area of the rat hippocampus. Eur. J. Neurosci 19, 1243–1256. [DOI] [PubMed] [Google Scholar]
- McBain CJ, DiChiara TJ and Kauer JA (1994). Activation of metabotropic glutamate receptors differentially affects two classes of hippocampal interneurons and potentiates excitatory synaptic transmission. J. Neurosci 14, 4433–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein AD, Bloss EB, Apostolides PF, Vaidya SP, Dilly GA, Zemelman BV and Magee JC (2015). Inhibitory Gating of Input Comparison in the CA1 Microcircuit. Neuron 87, 1274–1289. [DOI] [PubMed] [Google Scholar]
- O’Keefe J and Dostrovsky J (1971). The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 34, 171–175. [DOI] [PubMed] [Google Scholar]
- Overstreet-Wadiche L and McBain CJ (2015). Neurogliaform cells in cortical circuits. Nat. Rev. Neurosci 16, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelzik H, Hughes DI and Thomson AM (2002). Physiological and morphological diversity of immunocytochemically defined parvalbumin- and cholecystokinin-positive interneurones in CA1 of the adult rat hippocampus. J. Comp. Neurol 443, 346–367. [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. (2011). Scikit-learn: Machine learning in Python. J. Mach. Learn. Res 12, 2825–2830. [Google Scholar]
- Pelkey KA, Chittajallu R, Craig MT, Tricoire L, Wester JC and McBain CJ (2017). Hippocampal gabaergic inhibitory interneurons. Physiol. Rev 97, 1619–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer CK, Xue M, He M, Huang ZJ and Scanziani M (2013). Inhibition of inhibition in visual cortex: The logic of connections between molecularly distinct interneurons. Nat. Neurosci 16, 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi HJ, Hangya B, Kvitsiani D, Sanders JI, Huang ZJ and Kepecs A (2013). Cortical interneurons that specialize in disinhibitory control. Nature 503, 521–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouille F and Scanziani M (2001). Enforcement of temporal fidelity in pyramidal cells by somatic feed-forward inhibition. Science 293, 1159–1163. [DOI] [PubMed] [Google Scholar]
- Pouille F and Scanziani M (2004). Routing of spike series by dynamic circuits in the hippocampus. Nature 429, 717–723. [DOI] [PubMed] [Google Scholar]
- Price CJ, Cauli B, Kovacs ER, Kulik A, Lambolez B, Shigemoto R and Capogna M (2005). Neurogliaform neurons form a novel inhibitory network in the hippocampal CA1 area. J. Neurosci 25, 6775–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CJ, Scott R, Rusakov DA and Capogna M (2008). GABAB receptor modulation of feedforward inhibition through hippocampal neurogliaform cells. J. Neurosci 28, 6974–6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Harris KD, Hauling T, Nicoloutsopoulos D, Muñoz-Manchado AB, Skene N, Hjerling-Leffler J and Nilsson M (2020). Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat. Methods 17, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer S, Zemelman BV, Losonczy A, Kim J, Chance F, Magee JC and Buzsáki G (2012). Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nat. Neurosci 15, 769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield MEJ, Adoff MD and Dombeck DA (2017). Increased Prevalence of Calcium Transients across the Dendritic Arbor during Place Field Formation. Neuron 96, 490–504.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sik A, Penttonen M, Ylinen A and Buzsáki G (1995). Hippocampal CA1 interneurons: An in vivo intracellular labeling study. J. Neurosci 15, 6651–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi J, Baude A, Omori Y, Shimizu H, El Mestikawy S, Fukaya M, Shigemoto R, Watanabe M and Somogyi P (2004). GABAergic basket cells expressing cholecystokinin contain vesicular glutamate transporter type 3 (VGLUT3) in their synaptic terminals in hippocampus and isocortex of the rat. Eur. J. Neurosci 19, 552–569. [DOI] [PubMed] [Google Scholar]
- Somogyi P, Katona L, Klausberger T, Lasztóczi B and Viney TJ (2014). Temporal redistribution of inhibition over neuronal subcellular domains underlies state-dependent rhythmic change of excitability in the hippocampus. Philos. Trans. R. Soc. Lond. B Biol. Sci 369, 20120518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalay G, Judák L, Katona G, Ócsai K, Juhász G, Veress M, Szadai Z, Fehér A, Tompa T, Chiovini B, et al. (2016). Fast 3D Imaging of Spine, Dendritic, and Neuronal Assemblies in Behaving Animals. Neuron 92, 723–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, Lu J and Huang ZJ (2013). The spatial and temporal origin of chandelier cells in mouse neocortex. Science 339, 70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay R, Lee S and Rudy B (2016). GABAergic Interneurons in the Neocortex: From Cellular Properties to Circuits. Neuron 91, 260–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricoire L, Pelkey KA, Daw MI, Sousa VH, Miyoshi G, Jeffries B, Cauli B, Fishell G and McBain CJ (2010). Common origins of hippocampal ivy and nitric oxide synthase expressing neurogliaform cells. J. Neurosci 30, 2165–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troullinou E, Tsagkatakis G, Chavlis S, Turi GF, Li WK, Losonczy A, Tsakalides P and Poirazi P (2019). Artificial neural networks in action for an automated cell-type classification of biological neural networks. arXiv, http://arxiv.org/abs/1911.09977. [Google Scholar]
- Turi GF, Li WK, Chavlis S, Pandi I, O’Hare J, Priestley JB, Grosmark AD, Liao Z, Ladow M, Zhang JF, et al. (2019). Vasoactive Intestinal Polypeptide-Expressing Interneurons in the Hippocampus Support Goal-Oriented Spatial Learning. Neuron 101, 1150–1165.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyan L, Chamberland S, Magnin E, Camiré O, Francavilla R, David LS, Deisseroth K and Topolnik L (2014). Dendritic inhibition provided by interneuron-specific cells controls the firing rate and timing of the hippocampal feedback inhibitory circuitry. J. Neurosci 34, 4534–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga C, Oijala M, Lish J, Szabo GG, Bezaire M, Marchionni I, Golshani P and Soltesz I (2014). Functional fission of parvalbumin interneuron classes during fast network events. eLife 3, e04006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga C, Golshani P and Soltesz I (2012). Frequency-invariant temporal ordering of interneuronal discharges during hippocampal oscillations in awake mice. Proc. Natl. Acad. Sci. USA 109, E2726–E2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Losonczy A, Zemelman BV, Borhegyi Z, Nyiri G, Domonkos A, Hangya B, Holderith N, Magee JC and Freund TF (2009). Fast synaptic subcortical control of hippocampal circuits. Science 326, 449–453. [DOI] [PubMed] [Google Scholar]
- Vida I, Halasy K, Szinyei C, Somogyi P and Buhl EH (1998). Unitary IPSPs evoked by interneurons at the stratum radiatum-stratum lacunosum-moleculare border in the CA1 area of the rat hippocampus in vitro. J. Physiol 506, 755–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viney TJ, Lasztoczi B, Katona L, Crump MG, Tukker JJ, Klausberger T and Somogyi P (2013). Network state-dependent inhibition of identified hippocampal CA3 axo-axonic cells in vivo. Nat. Neurosci 16, 1802–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al. (2018). Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester JC and McBain CJ (2014). Behavioral state-dependent modulation of distinct interneuron subtypes and consequences for circuit function. Curr. Opin. Neurobiol 29, 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick ZC, Tetzlaff MR and Krook-Magnuson E (2019). Novel long-range inhibitory nNOS-expressing hippocampal cells. eLife 8, e46816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilent WB and Nitz DA (2007). Discrete Place Fields of Hippocampal Formation Interneurons. J. Neurophysiol 97, 4152–4161. [DOI] [PubMed] [Google Scholar]
- Winterer J, Lukacsovich D, Que L, Sartori AM, Luo W and Földy C (2019). Single-cell RNA-Seq characterization of anatomically identified OLM interneurons in different transgenic mouse lines. Eur. J. Neurosci 50, 3750–3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Nguyen TN, van Velthoven CTJ, Goldy J, Sedeno-Cortes A, Baftizadeh F, Bertagnolli D, Casper T, Crichton K, Ding S, et al. (2020). A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. bioRxiv, https://www.biorxiv.org/content/10.1101/2020.03.30.015214v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba JD, Diamantopoulou A, Danielson NB, Grosmark AD, Kaifosh PW, Bowler JC, Liao Z, Sparks FT, Gogos JA and Losonczy A (2017). Impaired hippocampal place cell dynamics in a mouse model of the 22q11.2 deletion. Nat. Neurosci 20, 1612–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsiros V and Maccaferri G (2005). Electrical coupling between interneurons with different excitable properties in the stratum lacunosum-moleculare of the juvenile CA1 rat hippocampus. J. Neurosci 25, 8686–8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Means and medians with corresponding SEM or IQR for data distributions presented in the main figures. Related to Figures 1–7.
Supplementary Video 1. 3D calcium imaging of 296 interneurons during running (5-minute session at 20x speed, non-motion corrected). Related to Figure 1.
Data Availability Statement
The machine-learning approach and the dataset are available on GitHub at https://github.com/losonczylab/Geiller_Vancura_Neuron2020. All other codes generated during this study are available upon request.


