Abstract
The small heat shock proteins (sHsps) are a ubiquitous family of ATP-independent stress proteins found in all domains of life. Drosophila melanogaster Hsp27 (DmHsp27) is the only known nuclear sHsp in insect. Here analyzing sequences from HMMER, we identified 56 additional insect sHsps with conserved arginine-rich nuclear localization signal (NLS) in the N-terminal region. At this time, the exact role of nuclear sHsps remains unknown. DmHsp27 protein-protein interaction analysis from iRefIndex database suggests that this protein, in addition to a putative role of molecular chaperone, is likely involved in other nuclear processes (i.e., chromatin remodeling and transcription). Identification of DmHsp27 interactors should provide key insights on the cellular and molecular functions of this nuclear chaperone.
Electronic supplementary material
The online version of this article (10.1007/s12192-020-01156-3) contains supplementary material, which is available to authorized users.
Keywords: Small heat shock protein (sHsp), DmHsp27, Chaperone, Alpha-crystallin domain (ACD), Drosophila melanogaster, Insect
Introduction
The nucleus is the cellular organelle that distinguishes eukaryotes from prokaryotes. A large number of biological activities occur in the nucleus, including DNA replication and damage repair, the biogenesis of ribosomal RNA precursors, transcription, and splicing of pre-mRNAs. Many proteins are involved in nuclear transport in processes dependent on Karyopherin-β proteins (both importins and exportins).
The small heat shock proteins (sHsps) are a ubiquitous family of ATP-independent stress proteins found in all domains of life (Caspers et al. 1995; de Jong et al. 1998; Fu et al. 2006; Maaroufi and Tanguay 2013). These proteins are upregulated in response to a variety of stresses that negatively impact protein homeostasis. sHsp sequence analysis indicates a tripartite architecture, with a conserved α-crystallin domain (ACD) flanked by a variable non-conserved N- and C-terminal regions (NTR and CTR) (Kappé et al. 2010; Basha et al. 2012). sHsp number, level and stage of expression, tissue distribution, and intracellular localization can show differences between species, suggesting that sHsps have adapted to their environment and to specific cell needs (Michaud et al. 2002; Morrow and Tanguay 2015).
Relocation of cytosolic sHsps to the nucleus is observed under certain conditions of stress in virtually all mammals (Van De Klundert and De Jong 1999; Borrelli et al. 2002), but this is distinct from nuclear sHsps that are present all the time in the nucleus in Drosophila melanogaster and plant, due to the presence of a nuclear localization signal (NLS) (Beaulieu et al. 1989; Marin and Tanguay 1996; Scharf et al. 2001; Siddique et al. 2003, 2008; Michaud et al. 2008).
Determining the cellular localization of proteins is important for understanding protein functions. Here we focused on the nuclear-localized small Hsp27 of Drosophila melanogaster (DmHsp27). This sHsp was the first reported to be localized in the nucleus (Beaulieu et al. 1989), and since this discovery, other sHsps, especially from plants, have been shown to have a nuclear localization such as Solanum peruvianum LpHsp16.1, Arabidopsis thaliana AtHsp17.4, Gossypium arboreum GaHsp17.3, Medicago truncatula MtHsp17.1, Lactuca sativa LsHsp17.8, Triticum aestivum TaHsp19.0, Hordeum vulgare HvHsp19.1, and Oryza sativa OsHsp18.6 (Scharf et al. 2001; Siddique et al. 2003, 2008). The import of proteins into the nucleus is dependent on a nuclear localization signals (NLS), and members of the nuclear transport receptor (importin β-like) superfamily. In plants, the nuclear localization depends on a short motif (N(G)KRKR) located between β-strands 5 and 6 in the conserved ACD (Siddique et al. 2003). This mode requires importin α to serve as a bridge between the NLS and the import receptor importin β (Marfori et al. 2012). In Drosophila melanogaster DmHsp27, the NLS motif is composed of three arginine (Arg54-Arg55-Arg56) in the NTR (Michaud et al. 2008). Authors suggested that arginine 54 and 55 form a single functional group that acts in concert with arginine 56 to dictate nuclear localization of DmHsp27 (Michaud et al. 2008). Arginine-rich NLS uses importin β for import by a mechanism independent of importin α (Palmeri and Malim 1999). The difference between the nuclear localization of sHsps in mammals, plants, and D. melanogaster suggests that they adapt to their environments and their specific cellular needs. While we know the existence of several sHsps with an NLS in plants, DmHsp27 is the only member reported in insect. NTR in sHsp was reported as a disordered non-conserved region due to the lack of sequence conservation and structure among this family (Kim et al. 1998; van Montfort et al. 2001). Therefore, localization of a nuclear signal in this disordered and non-conserved region is surprising. Here we characterize nuclear insect sHsps containing an arginine-rich NLS and suggest possible functions of this group of sHsps based on DmHsp27 interacting partners.
Materials and methods
Sequence collection of the insect sHsps
All insect protein sequences containing one or more instances of ACD (accession number PF00011) were extracted from HMMER web server (Finn et al. 2011). Proteins containing the term “fragment” in their description were removed. From this data, we extracted only sequences containing arginine-rich NLS (residues XRR and RXR) in NTR as defined in Drosophila melanogaster by Michaud et al. (2008). After alignment using ClustalW (Larkin et al. 2007), only proteins showing a conservation of NLS with DmHsp27 have been kept.
Lengths, amino acid composition, alignment, and logos of insect sHsp
The beginning and end locations ACD of all sequences were extracted from PFAM database (Finn et al. 2014). The sequences were split into the 3 building blocks (ACD, NTR, and CTR). The length frequencies were analyzed using R ggplot2 package (Wickham 2009), and amino acid distribution average was evaluated using R stringi package (https://cran.r-project.org/web/packages/stringi/).
The sequences were aligned with ClustalW default parameters. Secondary structure of the alignment generated by ESPript (Robert and Gouet 2014) according to DmHsp27 ACD predicted structure by Moutaoufik et al. (2017b). UniProt accession numbers of sequences used in the alignments are indicated. To identify conserved regions, we used WebLogo 3 program (Crooks et al. 2004) to create blocks (LOGOS) of conserved amino acid residues from the multiple protein sequences.
Phylogenetic analysis of the insect sHsps
Multiple alignments of insect sHsp sequences were calculated in MEGAX ((Kumar et al. 2018)) using ClustalW default parameters. Maximum likelihood phylogenetic trees were constructed using MEGAX default parameters and 50 bootstrap replicates. Phylogenetic trees were visualized using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Protein-protein interaction data
In order to understand nuclear functions of insect sHsps, we searched protein-protein interaction (PPI) data of insect nuclear sHsps using iRefIndex database (Razick et al. 2008). From all other insect species identified in this study, only Drosophila melanogaster PPI data was available in iRefIndex. DmHsp27 experimentally verified PPI data were downloaded, filtered by nuclear localization, and imported into Cytoscape (Shannon et al. 2003) to construct PPI network.
Results and discussion
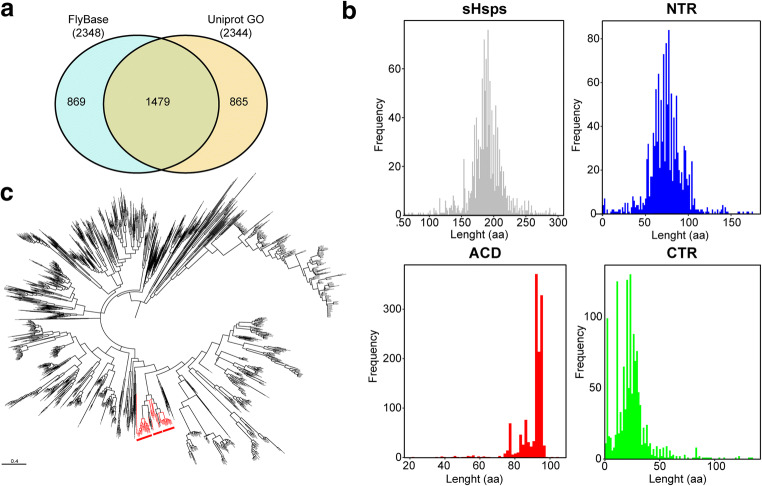
Nucleus plays a central role in eukaryotes. However, its protein content is far from complete. A mouse nuclear proteomic prediction reveals that mouse nucleus is the house of between 4084 and 9122 proteins (Fink et al. 2008). Using FlyBase (Thurmond et al. 2019) and UniProt Gene Ontology (GO) (2019), we identified reported nuclear proteins in Drosophila melanogaster. The total number of nuclear proteins reported in FlyBase and UniProt Go is constant with 2348 and 2344 entries, respectively (Fig. 1a, Supplementary Table 1), although more than half proteins (1479) are shared between two databases.
Fig. 1.
Drosophila melanogaster proteome and insect sHsps. A Venn diagram showing common and specific Drosophila melanogaster nuclear proteins between public FlyBase and UniProt GO databases. B Frequency of nuclear sHsp length. Full length sequence presented in (gray), NTR (blue), ACD (red), and CTR (green). C Nuclear sHsp evolutionary analysis by Maximum Likelihood method. Nuclear sHsps highlighted in red
Collection of nuclear insect sHsp sequences
It is accepted that the presence of the conserved ACD is a sufficient criterion for assigning a sequence to the sHsp family (Caspers et al. 1995; Kappé et al. 2010). To collect the sequences of sHsps, we searched the presence of ACD by using the domain PF00011 in HMMER web server (Finn et al. 2011). We obtained 1321 insect sequences containing ACD (Supplementary Table 2), with an average length of 188.82 (± 26.89) (Fig. 1b). NTR have a length average of 74.50 (± 20.67) and CTR 23.98 (± 16.26); both show a variation in the length and frequency (Fig. 1b). ACD less variable region with a length average of ACD 90.33 (± 7.95).
Analysis of NTR shows that 57 sequences contain an arginine-rich NLS in NTR (Table 1). The potential nuclear localization was confirmed by Euk-mPLoc (Chou and Shen 2007), a powerful tool, developed by hybridizing the functional domain information and sequential evolutionary information through three different modes of pseudo amino acid composition (Eq.4, Eq.6, and Eq.12).
Table 1.
Insect nuclear sHsps
| Protein name | UniProt ID | Species | Length | Mass (Dalton) | Arginine-rich NLS localization |
|---|---|---|---|---|---|
| Hsp 27 | P02518 | Drosophila melanogaster | 213 | 23,617 | Arg 54,55,56 |
| GM25143 | B4HKP4 | Drosophila sechellia | 213 | 23,625 | Arg 54,55,56 |
| GD14177 | A0A0J9UHX1 | Drosophila simulans | 213 | 23,625 | Arg 54,55,56 |
| GD14177 | B4QN53 | Drosophila simulans | 213 | 23,566 | Arg 54,55,56 |
| GG15371 | B3NCF2 | Drosophila erecta | 212 | 23,650 | Arg 54,55,56 |
| GE20833 | B4PEX9 | Drosophila yakuba | 212 | 23,594 | Arg 54,55,56 |
| ND | A0A1I8M7V4 | Musca domestica | 216 | 24,395 | Arg 52–53 |
| Hsp20 | T1PAE8 | Musca domestica | 216 | 24,364 | Arg 52–53 |
| Hsp27 | A0A1L8EIU6 | Haematobia irritans | 203 | 23,200 | Arg 48–50 |
| Hsp27 | A0A1L8EIY3 | Haematobia irritans | 203 | 23,250 | Arg 48–50 |
| Hsp27 | A0A1L8EJ25 | Haematobia irritans | 203 | 23,327 | Arg 48–50 |
| Hsp27 | A0A1L8EJ27 | Haematobia irritans | 203 | 23,223 | Arg 48–50 |
| Hsp27 | A0A1L8EJ69 | Haematobia irritans | 203 | 23,350 | Arg 48–50 |
| Hsp27 | A0A1L8EJ80 | Haematobia irritans | 203 | 23,212 | Arg 48–50 |
| Hsp27 | A0A1X9JRV5 | Musca domestica | 203 | 23,031 | Arg 47–49 |
| ND | A0A1I8M631 | Musca domestica | 203 | 23,040 | Arg 47–49 |
| Hsp27 | A0A1L8EJ49 | Haematobia irritans | 203 | 23,261 | Arg 48–50 |
| Hsp27 | A0A1L8EIP8 | Haematobia irritans | 202 | 23,112 | Arg 49–50–51 |
| Hsp27 | A0A1L8EIV9 | Haematobia irritans | 202 | 23,126 | Arg 49–50–51 |
| Hsp27 | A0A1L8EJ16 | Haematobia irritans | 202 | 23,098 | Arg 49–50–51 |
| Hsp27 | A0A1L8EIS9 | Haematobia irritans | 203 | 22,956 | Arg 48–50 |
| Hsp27 | A0A1L8EIV6 | Haematobia irritans | 203 | 23,117 | Arg 48–50 |
| Hsp27 | A0A1L8EIW9 | Haematobia irritans | 203 | 23,078 | Arg 48–50 |
| Hsp27 | A0A1L8EIX8 | Haematobia irritans | 203 | 23,097 | Arg 48–50 |
| Hsp27 | A0A1L8EIY9 | Haematobia irritans | 203 | 23,022 | Arg 48–50 |
| Hsp27 | A0A1L8EJ10 | Haematobia irritans | 203 | 23,068 | Arg 48–50 |
| Hsp27 | A0A1L8EJ18 | Haematobia irritans | 203 | 23,038 | Arg 48–50 |
| Hsp27 | A0A1L8EJ35 | Haematobia irritans | 203 | 23,123 | Arg 48–50 |
| Hsp27 | A0A1L8EJ40 | Haematobia irritans | 203 | 23,105 | Arg 48–50 |
| Hsp27 | A0A1L8EJ47 | Haematobia irritans | 203 | 23,127 | Arg 48–50 |
| ND | T1PK02 | Musca domestica | 198 | 22,646 | Arg 47–49 |
| Hsp27 | A0A1X9JUH8 | Musca domestica | 198 | 22,628 | Arg 47–49 |
| Hsp27 | A0A0L0CSL4 | Lucilia cuprina | 203 | 23,331 | Arg 47–49–50 |
| GK17637 | B4MN48 | Drosophila willistoni | 227 | 25,285 | Arg 58,59,60 |
| Hsp27 | A9UEZ2 | Drosophila buzzatii | 211 | 23,695 | Arg 51,52,53 |
| ND | A0A1B0G3A3 | Glossina morsitans morsitans | 212 | 24,164 | Arg 55–57 |
| ND | A0A1A9UGE3 | Glossina austeni | 184 | 21,061 | Arg 55–57 |
| Hsp27 | I1T1H1 | Bactrocera dorsalis | 210 | 23,831 | Arg 51–52–53 |
| Hsp27 | A0A0K8UP97 | Bactrocera latifrons | 212 | 24,037 | Arg 53–54–-55 |
| Hsp27 | A0A0A1XCZ4 | Bactrocera cucurbitae | 212 | 23,920 | Arg 53–54–55 |
| Hsp27 | B3GK93 | Ceratitis capitata | 214 | 23,944 | Arg 54–55–56 |
| Hsp27 | A0A0M4EGE5 | Drosophila busckii | 217 | 24,445 | Arg 51–52–53 |
| Hsp27 | A0A3B0JUV4 | Drosophila guanche | 225 | 25,016 | Arg 58–59–60 |
| GL22445 | B4H1L8 | Drosophila persimilis | 225 | 25,041 | Arg 58,59,60 |
| GA18205 | Q29F98 | Drosophila pseudoobscura pseudoobscura | 225 | 25,041 | Arg 58,59,60 |
| GF10692 | B3M6F4 | Drosophila ananassae | 219 | 24,490 | Arg 57,58,59 |
| Hsp27 | F5B960 | Drosophila albomicans | 222 | 24,770 | Arg 52,53,54 |
| Hsp27 | F5B961 | Drosophila sulfurigaster albostrigata | 221 | 24,675 | Arg 52,53,54 |
| GJ13835 | B4LGS9 | Drosophila virilis | 211 | 23,792 | Arg 51,52,53 |
| GH17009 | B4IYY1 | Drosophila grimshawi | 218 | 24,442 | Arg 54,55,56 |
| Hsp25 | A1E385 | Sarcophaga crassipalpis | 221 | 24,982 | Arg 60–61 |
| ND | A0A484BBT9 | Drosophila navojoa | 211 | 23,628 | Arg 51–52–53 |
| GI13087 | B4L112 | Drosophila mojavensis | 209 | 23,459 | Arg 51,52,53 |
| Hsp27 | F5B962 | Drosophila repletoides | 220 | 24,504 | Arg 48,49,50 |
| CSON012237 | A0A336MGY1 | Culicoides sonorensis | 167 | 19,445 | Arg 40–41 |
| CSON11881 | A0A336MMP2 | Culicoides sonorensis | 167 | 19,401 | Arg 40–41 |
| Hsp27 | A0A1W4VN65 | Drosophila ficusphila | 213 | 23,784 | Arg 53–54–55 |
Next, we studied the evolutionary history of insect sHsp sequences (Fig. 1c). It is interesting that insect nuclear sHsps were clustered in one phytogenic group, suggesting that all nuclear insect sHsps evolved from the same ancestral sequence.
Analysis of insect nuclear sHsps
Insect sHsp sequences that presented arginine-rich NLS include drosophila genus: Drosophila simulans (2 sequences) and 1 sequence for Drosophila albomicans, Drosophila ananassae, Drosophila busckii, Drosophila buzzatii, Drosophila erecta, Drosophila ficusphila, Drosophila grimshawi, Drosophila guanche, Drosophila melanogaster, Drosophila mojavensis, Drosophila navojoa, Drosophila persimilis, Drosophila pseudoobscura pseudoobscura, Drosophila repletoides, Drosophila sechellia, Drosophila sulfurigaster albostrigata, Drosophila virilis, Drosophila willistoni, and Drosophila yakuba. In addition to other genera: Haematobia irritans (20 sequences), Musca domestica (6 sequences), Culicoides sonorensis (2 sequences) and 1 sequence for Bactrocera cucurbitae, Bactrocera dorsalis, Bactrocera latifrons, Ceratitis capitate, Glossina austeni, Glossina morsitans morsitans, Lucilia cuprina, and Sarcophaga crassipalpis (Table 1) suggesting that arginine-rich NLS is not specific to drosophila.
The average mass in Dalton is 23,489.00 (± 1086.55). The average amino acid length is 207.77 (± 11.45). The longest sequence is B4MN48 from Drosophila willistoni with 227 amino acids (aa) and the shortest is A0A336MGY1 from Culicoides sonorensis with sequence 167 aa (Table 1).
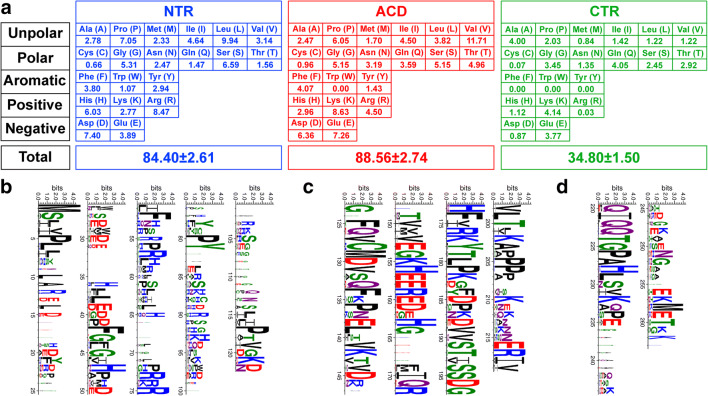
The average length of amino acids in NTR is 84.40 (± 2.61) (Fig. 2a). The analysis of NTR revealed underrepresentation of cysteine and an overrepresentation of leucine, proline, aspartic acid, and arginine (Fig. 2a and b). ACD has an average length of 88.56 (± 2.74) (Fig. 2a). In contrast, within the ACD, a significant enrichment of charged positively (lysine) and negatively (aspartic acid and glutamic acid) residues is observed, whereas tryptophan is absent (Fig. 2a and c). Most sequences present one cysteine in ACD, suggesting that under oxidative conditions, a disulfide crosslink may be formed between ACDs to form a dimer as shown for DmHsp27 (Moutaoufik et al. 2017b). In CTR, the shortest region has an average amino acid number of 34.80 (± 1.50) with absence of tryptophan, phenylalanine, and tyrosine. Additionally, there is underrepresentation of cysteines and arginine (Fig. 2a and d). This is consistent with a previous study from 1935 metazoan proteins which contain ACD by Kriehuber et al. (2010).
Fig. 2.
Insect nuclear sHsps. A Distribution of residues within the nuclear sHsps in insect. Average number of each amino acid within the NTR (blue), CTR (green), and the ACD (red) is presented. B, C, and D Logo presentation using WebLogo 3 program (Crooks et al. 2004). The height of each letter is made proportional to its frequency. Amino acids are colored according to their chemical properties: acidic (D,E) in red, basic (K,R,H) in blue, polar (G,S,T,Y,C) in green and (N,Q) in purple, and hydrophobic (A,V,L,I,P,W,F,M) in black. Logo representation for Logos presentation of NTR (B), ACD (C), and CTR (D) of insect nuclear sHsp sequences
Blocks of the most conserved residues were made as logos to visualize the conserved regions in three regions—NTR, ACD, CTR—of nuclear insect sHsps (Fig. 2c, d, and e) in addition to a complete sequence alignment which (Supplementary Fig. 1) shows that motif characteristics of sHsps were conserved. This is the case of the arginine in the Beta 6 + 7 strand: DmHsp27-R122 equivalent of HspB1-R127, DmHsp27-R131 equivalent of HspB1-R136, and DmHsp27-R135 equivalent of HspB1-R140, HspB5-R120, and HspB4-R116 associated with human pathologies (Vicart et al. 1998; Bera and Abraham 2002; Evgrafov et al. 2004; Inagaki et al. 2006; Gentil and Cooper 2012; Ylikallio et al. 2015). In addition to the conserved I/V/L-X-I/V/L motif in ACD, implicated in sHsps higher assembly by interaction with β4/β8 pocket in ACD of adjacent dimer (de Jong et al. 1998; van Montfort et al. 2001; Stamler et al. 2005; Poulain et al. 2010; Clark et al. 2018) and the FGFG motif in NTR, important for the oligomeric structure and chaperone-like activity (Moutaoufik et al. 2017a). In addition, B4PEX9 (Drosophila yakuba), B4HKP4 (Drosophila sechellia), B3NCF2 (Drosophila erecta), A1E385 (Sarcophaga crassipalpis), and A0A0J9UHX1 and B4QN53 (Drosophila simulans) show a conservation in both phosphorylated serines in DmHsp27 (S58 and S75) reported by Moutaoufik et al. (2017a). T1PAE8 (Musca domestica), B4L112 (Drosophila mojavensis), A9UEZ2 (Drosophila buzzatii), A0A1I8M7V4 (Musca domestica), and A0A484BBT9 (Drosophila navojoa) show only a conservation in phosphorylated serine (S58). However, A0A1L8EIU6, A0A1L8EIY3, A0A1L8EJ25, A0A1L8EJ27, A0A1L8EJ69, A0A1L8EJ80, A0A1L8EJ49, A0A1L8EIP8, A0A1L8EIV9, A0A1L8EJ16, A0A1L8EIS9, A0A1L8EIV6, A0A1L8EIW9, A0A1L8EIX8, A0A1L8EIY9, A0A1L8EJ10, A0A1L8EJ18, A0A1L8EJ35, A0A1L8EJ40 and A0A1L8EJ47 (Haematobia irritans), A0A1X9JRV5, A0A1I8M631, T1PK02 and A0A1X9JUH8 (Musca domestica), A0A3B0JUV4 (Drosophila guanche), B3M6F4 (Drosophila ananassae), A0A1W4VN65 (Drosophila ficusphila), A0A0L0CSL4 (Lucilia cuprina), A0A1B0G3A3 (Glossina morsitans morsitans), A0A1A9UGE3 (Glossina austeni), Q29F98 (Drosophila pseudoobscura pseudoobscura), B4MN48 (Drosophila willistoni), and B4H1L8 (Drosophila persimilis) show only a conservation in phosphorylated serine (S75).
DmHsp27 interactome and nuclear functions
The nuclear functions of insect sHsps remain unknown. A study had reported an association of DmHsp22, DmHsp23, DmHsp26, and DmHsp27 to heterogeneous nuclear ribonucleoprotein (hnRNP) (Kloetzel and Bautz 1983). In addition, under heat shock conditions, DmHsp27 and cytoplasmic DmsHsps were co-purified with the proteasome (Arrigo et al. 1985).
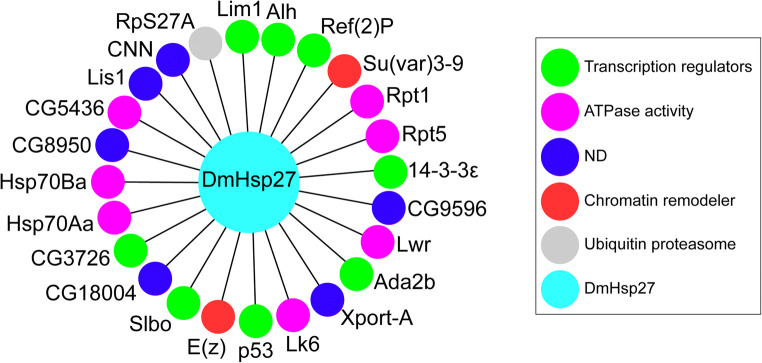
We searched the protein partners of insect sHsps identified in this study, using iRefIndex (Razick et al. 2008). Only Drosophila melanogaster protein-protein interactions are presented in the used database. DmHsp27 shows 46 associations with both nuclear and cytoplasmic proteins. After filtering by subcellular localization, we identified 24 nuclear proteins that tend to interact with DmHsp27 in addition to self-interacting to form oligomeric structures (Fig. 3, Table 2). Third (8, 33%) of DmHsp27 nuclear partners were involved in regulation of transcription, more than one-quarter (7, 29%) in ATPase activity, (2, 0.08%) were implicated in chromatin remodeling, (1, 0.04%) in ubiquitin proteasome complex and quarter (6, 25%) with no known nuclear function (Fig. 3b). These include proteins involved in many biological processes such as fly development (14-3-3ε, Lk6, enhancer of zeste E(Z), centrosomin (cnn), lissencephaly-1 (Lis-1), lesswright (lwr), p53, slow border cells (slbo), and suppressor of variegation 3–9 (Su(var)3–9)) (Ashton-Beaucage et al. 2014; Arquier et al. 2005; Jones et al. 1998; Eisman et al. 2009; Liu et al. 1999; Miles et al. 2008; Bauer et al. 2007; Borghese et al. 2006; Kunert et al. 2003). In addition, LIM homeobox 1 (Lim1) implicated in eye development (Roignant et al. 2010). However, 14-3-3ε, Alhambra (Alh), and transcriptional adaptor 2 Ada2b were associated with locomotor rhythm and muscle and wing development (Bahri et al. 2001; Gause et al. 2006; Bejarano et al. 2008). Finally, 14-3-3ε, p53, and CG5436 (Hsp68) were related to fly viability (Bauer et al. 2005; Nielsen et al. 2008; Biteau et al. 2010), lwr to immune system response (Bhaskar et al. 2002), and CG5436 (Hsp68), Hsp70Aa, and Hsp70Ba to chaperone activity (Gong and Golic 2006; Gaudet et al. 2011). This is consistent with the related expression of DmHsp27 during oogenesis and spermatogenesis (Marin and Tanguay 1996; Michaud et al. 1997), eye development (Chen et al. 2012), lifespan (Hao et al. 2007; Liao et al. 2008), and defense against pathogens (Chen et al. 2010).
Fig. 3.
DmHsp27 and its nuclear-associated proteins. Representation of DmHsp27 and associated protein assemblies. Green color indicates transcription regulators, purple color designate proteins with ATPase activity, blue color indicate proteins with non-defined nuclear functions, red color indicate chromatin remodeler, and gray indicates protein involved in ubiquitin proteasome system
Table 2.
Proteins associated with DmHsp27
| Protein 1 | UniprotID1 | Protein 2 | UniprotID2 | Method | Reference |
|---|---|---|---|---|---|
| 14-3-3ε | P92177 | DmHsp27 | P02518 | Colocalization | Müller et al. (2010) |
| Ada2b | Q8I8V0 | Dmhsp27 | P02518 | Two-hybrid | Giot et al. (2003) |
| Alh | Q9VI61 | DmHsp27 | P02518 | Two-hybrid | Giot et al. (2003) |
| CG18004 | A1Z8D4 | DmHsp27 | P02518 | AP-MS | Guruharsha et al. (2011) |
| CG3726 | X2JDV1 | DmHsp27 | P02518 | AP-MS | Rhee et al. (2014) |
| CG8950 | Q7JVW5 | DmHsp27 | P02518 | AP-MS | Rhee et al. (2014) |
| CG9596 | Q8MS69 | DmHsp27 | P02518 | AP-MS | Guruharsha et al. (2011) |
| CG5436 | O97125 | DmHsp27 | P02518 | AP-MS | Guruharsha et al. (2011) |
| CNN | P54623 | DmHsp27 | P02518 | MS | Habermann et al. (2012) |
| E(z) | P42124 | DmHsp27 | P02518 | AP-MS | Kang et al. (2015) |
| DmHsp27 | P02518 | DmHsp27 | P02518 | BN gel | Moutaoufik et al. (2017a) |
| Hsp70 Aa | P82910 | Dmhsp27 | P02518 | AP-MS | Guruharsha et al. (2011) |
| Hsp70Ba | Q8INI8 | DmHsp27 | P02518 | AP-MS | Guruharsha et al. (2011) |
| Lim1 | M9PJE4 | DmHsp27 | P02518 | Two-hybrid | Giot et al. (2003) |
| Lis1 | Q7KNS3 | DmHsp27 | P02518 | Two-hybrid | Giot et al. (2003) |
| Lk6 | Q9VGI4 | DmHsp27 | P02518 | Two-hybrid | Giot et al. (2003) |
| Lwr | Q7KNM2 | DmHsp27 | P02518 | AP-WB/Two-hybrid | Joanisse et al. (1998) |
| p53 | A0A0B4K7P1 | DmHsp27 | P02518 | AP-WB | Lei et al. (2017) |
| Rpt1 | Q7KMQ0 | DmHsp27 | P02518 | AP-MS | Guruharsha et al. (2011) |
| Ref(2)P | P14199 | DmHsp27 | P02518 | AP-MS | Guruharsha et al. (2011) |
| RpS27A | P15357 | DmHsp27 | P02518 | AP-WB | Lee et al. (2014) |
| Rpt5 | Q9V3V6 | DmHsp27 | P02518 | AP-WB | Cho-Park and Steller (2013) |
| Slbo | Q02637 | DmHsp27 | P02518 | Genetic Interactions | Rørth et al. (1998) |
| Su(var)3–9 | I0DHL3 | DmHsp27 | P02518 | Two-hybrid | Giot et al. (2003) |
| Xport-A | Q9VDS3 | DmHsp27 | P02518 | AP-WB | Rosenbaum et al. (2011) |
Conclusion
Here we have shown that the NLS signal rich in arginine is not a peculiarity of Drosophila melanogaster. The progress of genome sequencing projects should confirm the presence of other insect sHsp sequences with NLS signal, of the same type or not. The exact role of nuclear sHsps remains unknown. The interaction network of DmHsp27 suggests that this protein does not only play the role of molecular chaperone, but it is likely involved in different nuclear processes. Such studies open the perspectives to establish the functional activities of DmHsp27 and associated proteins in different biological processes.
Electronic supplementary material
Sequence alignment of nuclear drosophila small heat shock proteins. Alignment was generated using ClustalW. UniProt accession numbers of sequences used in the alignments are indicated. Secondary structures of the alignment generated by ESPript (Robert and Gouet 2014), according to DmHsp27 ACD predicted structure by Moutaoufik et al. 2017b. NTRs show conservation of FGFG motif and arginine-rich NLS (Black arrow). The I/V/L-X-I/L/V motif in the CTR and conserved arginine R122, R131, and R135 are indicated by black arrow and black star, respectively. (PNG 2876 kb).
(XLS 839 kb).
(XLS 212 kb).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Arquier N, Bourouis M, Colombani J, Léopold P. Drosophila Lk6 kinase controls phosphorylation of eukaryotic translation initiation factor 4E and promotes normal growth and development. Curr Biol. 2005;15:19–23. doi: 10.1016/j.cub.2004.12.037. [DOI] [PubMed] [Google Scholar]
- Arrigo AP, Darlix JL, Khandjian EW, Simon M, Spahr PF. Characterization of the prosome from Drosophila and its similarity to the cytoplasmic structures formed by the low molecular weight heat-shock proteins. EMBO J. 1985;4:399–406. doi: 10.1002/j.1460-2075.1985.tb03642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton-Beaucage D, Udell CM, Gendron P, Sahmi M, Lefrançois M, Baril C, Guenier AS, Duchaine J, Lamarre D, Lemieux S, Therrien M. A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signaling in Drosophila. PLoS Biol. 2014;12:e1001809. doi: 10.1371/journal.pbio.1001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahri SM, Chia W, Yang X. The Drosophila homolog of human AF10/AF17 leukemia fusion genes (Dalf) encodes a zinc finger/leucine zipper nuclear protein required in the nervous system for maintaining EVE expression and normal growth. Mech Dev. 2001;100:291–301. doi: 10.1016/S0925-4773(00)00539-6. [DOI] [PubMed] [Google Scholar]
- Basha E, O’Neill H, Vierling E. Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr Biol. 2005;15:2063–2068. doi: 10.1016/j.cub.2005.10.051. [DOI] [PubMed] [Google Scholar]
- Bauer JH, Chang C, Morris SNS, Hozier S, Andersen S, Waitzman JS, Helfand SL. Expression of dominant-negative Dmp53 in the adult fly brain inhibits insulin signaling. Proc Natl Acad Sci U S A. 2007;104:13355–13360. doi: 10.1073/pnas.0706121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JF, Arrigo AP, Tanguay RM. Interaction of Drosophila 27,000 Mr heat-shock protein with the nucleus of heat-shocked and ecdysone-stimulated culture cells. J Cell Sci. 1989;92:29–36. doi: 10.1242/jcs.92.1.29. [DOI] [PubMed] [Google Scholar]
- Bejarano F, Luque CM, Herranz H, Sorrosal G, Rafel N, Pham TT, Milán M. A gain-of-function suppressor screen for genes involved in dorsal–ventral boundary formation in the Drosophila wing. Genetics. 2008;178:307–323. doi: 10.1534/genetics.107.081869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera S, Abraham EC. The alphaA-crystallin R116C mutant has a higher affinity for forming heteroaggregates with alphaB-crystallin. Biochemistry. 2002;41:297–305. doi: 10.1021/bi011010v. [DOI] [PubMed] [Google Scholar]
- Bhaskar V, Smith M, Courey AJ. Conjugation of Smt3 to dorsal may potentiate the Drosophila immune response. Mol Cell Biol. 2002;22:492–504. doi: 10.1128/MCB.22.2.492-504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, DeGennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 2010;6:e1001159. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese L, Fletcher G, Mathieu J, Atzberger A, Eades WC, Cagan RL, Rørth P. Systematic analysis of the transcriptional switch inducing migration of border cells. Dev Cell. 2006;10:497–508. doi: 10.1016/j.devcel.2006.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli MJ, Bernock LJ, Landry J, Spitz DR, Weber LA, Hickey E, Freeman ML, Corry PM. Stress protection by a fluorescent Hsp27 chimera that is independent of nuclear translocation or multimeric dissociation. Cell Stress Chaperones. 2002;7:281–296. doi: 10.1379/1466-1268(2002)007<0281:SPBAFH>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers GJ, Leunissen JA, de Jong WW. The expanding small heat-shock protein family, and structure predictions of the conserved “alpha-crystallin domain.”. J Mol Evol. 1995;40:238–248. doi: 10.1007/BF00163229. [DOI] [PubMed] [Google Scholar]
- Chen J, Xie C, Tian L, Hong L, Wu X, Han J. Participation of the p38 pathway in Drosophila host defense against pathogenic bacteria and fungi. Proc Natl Acad Sci U S A. 2010;107:20774–20779. doi: 10.1073/pnas.1009223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S-F, Kang M-L, Chen Y-C, Tang HW, Huang CW, Li WH, Lin CP, Wang CY, Wang PY, Chen GC, Wang HD. Autophagy-related gene 7 is downstream of heat shock protein 27 in the regulation of eye morphology, polyglutamine toxicity, and lifespan in Drosophila. J Biomed Sci. 2012;19:52. doi: 10.1186/1423-0127-19-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho-Park PF, Steller H. Proteasome regulation by ADP-ribosylation. Cell. 2013;153:614–627. doi: 10.1016/j.cell.2013.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K-C, Shen H-B. Euk-mPLoc: a fusion classifier for large-scale eukaryotic protein subcellular location prediction by incorporating multiple sites. J Proteome Res. 2007;6:1728–1734. doi: 10.1021/pr060635i. [DOI] [PubMed] [Google Scholar]
- Clark AR, Vree Egberts W, Kondrat FDL, Hilton GR, Ray NJ, Cole AR, Carver JA, Benesch JLP, Keep NH, Boelens WC, Slingsby C. Terminal regions confer plasticity to the tetrameric assembly of human HspB2 and HspB3. J Mol Biol. 2018;430:3297–3310. doi: 10.1016/j.jmb.2018.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong WW, Caspers G-J, Leunissen JAM. Genealogy of the α-crystallin—small heat-shock protein superfamily. Int J Biol Macromol. 1998;22:151–162. doi: 10.1016/S0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- Eisman RC, Phelps MAS, Kaufman TC. Centrosomin: a complex mix of long and short isoforms is required for centrosome function during early development in Drosophila melanogaster. Genetics. 2009;182:979–997. doi: 10.1534/genetics.109.103887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgrafov OV, Mersiyanova I, Irobi J, van den Bosch L, Dierick I, Leung CL, Schagina O, Verpoorten N, van Impe K, Fedotov V, Dadali E, Auer-Grumbach M, Windpassinger C, Wagner K, Mitrovic Z, Hilton-Jones D, Talbot K, Martin JJ, Vasserman N, Tverskaya S, Polyakov A, Liem RKH, Gettemans J, Robberecht W, de Jonghe P, Timmerman V. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- Fink JL, Karunaratne S, Mittal A, Gardiner DM, Hamilton N, Mahony D, Kai C, Suzuki H, Hayashizaki Y, Teasdale RD. Towards defining the nuclear proteome. Genome Biol. 2008;9:R15. doi: 10.1186/gb-2008-9-1-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Jiao W, Chang Z. Phylogenetic and biochemical studies reveal a potential evolutionary origin of small heat shock proteins of animals from bacterial class A. J Mol Evol. 2006;62:257–266. doi: 10.1007/s00239-005-0076-5. [DOI] [PubMed] [Google Scholar]
- Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Eissenberg JC, MacRae AF, et al. Nipped-A, the Tra1/TRRAP subunit of the Drosophila SAGA and Tip60 complexes, has multiple roles in notch signaling during wing development. Mol Cell Biol. 2006;26:2347–2359. doi: 10.1128/MCB.26.6.2347-2359.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentil BJ, Cooper L. Molecular basis of axonal dysfunction and traffic impairments in CMT. Brain Res Bull. 2012;88:444–453. doi: 10.1016/j.brainresbull.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna M, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Loss of Hsp70 in Drosophila is pleiotropic, with effects on thermotolerance, recovery from heat shock and neurodegeneration. Genetics. 2006;172:275–286. doi: 10.1534/genetics.105.048793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Rual J-F, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, McKillip E, Shah S, Stapleton M, Wan KH, Yu C, Parsa B, Carlson JW, Chen X, Kapadia B, VijayRaghavan K, Gygi SP, Celniker SE, Obar RA, Artavanis-Tsakonas S. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann K, Mirgorodskaya E, Gobom J, Lehmann V, Muller H, Blumlein K, Deery MJ, Czogiel I, Erdmann C, Ralser M, von Kries JP, Lange BMH. Functional analysis of centrosomal kinase substrates in Drosophila melanogaster reveals a new function of the nuclear envelope component otefin in cell cycle progression. Mol Cell Biol. 2012;32:3554–3569. doi: 10.1128/MCB.00814-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X, Zhang S, Timakov B, Zhang P. The Hsp27 gene is not required for Drosophila development but its activity is associated with starvation resistance. Cell Stress Chaperones. 2007;12:364–372. doi: 10.1379/CSC-308.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki N, Hayashi T, Arimura T, Koga Y, Takahashi M, Shibata H, Teraoka K, Chikamori T, Yamashina A, Kimura A. Alpha B-crystallin mutation in dilated cardiomyopathy. Biochem Biophys Res Commun. 2006;342:379–386. doi: 10.1016/j.bbrc.2006.01.154. [DOI] [PubMed] [Google Scholar]
- Joanisse DR, Inaguma Y, Tanguay RM. Cloning and developmental expression of a nuclear ubiquitin-conjugating enzyme (DmUbc9) that interacts with small heat shock proteins inDrosophila melanogaster. Biochem Biophys Res Commun. 1998;244:102–109. doi: 10.1006/bbrc.1998.8214. [DOI] [PubMed] [Google Scholar]
- Jones CA, Ng J, Peterson AJ, Morgan K, Simon J, Jones RS. The Drosophila esc and E(z) proteins are direct partners in polycomb group-mediated repression. Mol Cell Biol. 1998;18:2825–2834. doi: 10.1128/MCB.18.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, McElroy KA, Jung YL, et al. Sex comb on midleg (Scm) is a functional link between PcG-repressive complexes in Drosophila. Genes Dev. 2015;29:1136–1150. doi: 10.1101/gad.260562.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappé G, Boelens WC, de Jong WW. Why proteins without an α-crystallin domain should not be included in the human small heat shock protein family HSPB. Cell Stress Chaperones. 2010;15:457–461. doi: 10.1007/s12192-009-0155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- Kloetzel P-M, Bautz EKF. Heat-shock proteins are associated with hnRNA in Drosophila melanogaster tissue culture cells. EMBO J. 1983;2:705–710. doi: 10.1002/j.1460-2075.1983.tb01488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriehuber T, Rattei T, Weinmaier T, Bepperling A, Haslbeck M, Buchner J. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 2010;24:3633–3642. doi: 10.1096/fj.10-156992. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunert N, Marhold J, Stanke J, Stach D, Lyko F. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development. 2003;130:5083–5090. doi: 10.1242/dev.00716. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee SY, Ramirez J, Franco M, Lectez B, Gonzalez M, Barrio R, Mayor U. Ube3a, the E3 ubiquitin ligase causing Angelman syndrome and linked to autism, regulates protein homeostasis through the proteasomal shuttle Rpn10. Cell Mol Life Sci. 2014;71:2747–2758. doi: 10.1007/s00018-013-1526-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Liu K, Hou L, Ding L, Li Y, Liu L. Small chaperons and autophagy protected neurons from necrotic cell death. Sci Rep. 2017;7:5650. doi: 10.1038/s41598-017-05995-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao P-C, Lin H-Y, Yuh C-H, Yu LK, Wang HD. The effect of neuronal expression of heat shock proteins 26 and 27 on lifespan, neurodegeneration, and apoptosis in Drosophila. Biochem Biophys Res Commun. 2008;376:637–641. doi: 10.1016/j.bbrc.2008.08.161. [DOI] [PubMed] [Google Scholar]
- Liu Z, Xie T, Steward R. Lis1, the Drosophila homolog of a human lissencephaly disease gene, is required for germline cell division and oocyte differentiation. Development. 1999;126:4477–4488. doi: 10.1242/dev.126.20.4477. [DOI] [PubMed] [Google Scholar]
- Maaroufi H, Tanguay RM. Analysis and phylogeny of small heat shock proteins from marine viruses and their cyanobacteria host. PLoS One. 2013;8:e81207. doi: 10.1371/journal.pone.0081207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfori M, Lonhienne TG, Forwood JK, Kobe B. Structural basis of high-affinity nuclear localization signal interactions with importin-α. Traffic. 2012;13:532–548. doi: 10.1111/j.1600-0854.2012.01329.x. [DOI] [PubMed] [Google Scholar]
- Marin R, Tanguay RM. Stage-specific localization of the small heat shock protein Hsp27 during oogenesis in Drosophila melanogaster. Chromosoma. 1996;105:142–149. doi: 10.1007/BF02509495. [DOI] [PubMed] [Google Scholar]
- Michaud S, Marin R, Westwood JT, Tanguay RM. Cell-specific expression and heat-shock induction of Hsps during spermatogenesis in Drosophila melanogaster. J Cell Sci. 1997;110(Pt 17):1989–1997. doi: 10.1242/jcs.110.17.1989. [DOI] [PubMed] [Google Scholar]
- Michaud S, Morrow G, Marchand J, Tanguay RM. Drosophila small heat shock proteins: cell and organelle-specific chaperones? In: Arrigo A-P, Müller WEG, editors. Small Stress Proteins. Berlin Heidelberg: Springer; 2002. pp. 79–101. [DOI] [PubMed] [Google Scholar]
- Michaud S, Lavoie S, Guimond M-O, Tanguay RM. The nuclear localization of Drosophila Hsp27 is dependent on a monopartite arginine-rich NLS and is uncoupled from its association to nuclear speckles. Biochim Biophys Acta Mol Cell Res. 2008;1783:1200–1210. doi: 10.1016/j.bbamcr.2008.01.031. [DOI] [PubMed] [Google Scholar]
- Miles WO, Jaffray E, Campbell SG, Takeda S, Bayston LJ, Basu SP, Li M, Raftery LA, Ashe MP, Hay RT, Ashe HL. Medea SUMOylation restricts the signaling range of the Dpp morphogen in the Drosophila embryo. Genes Dev. 2008;22:2578–2590. doi: 10.1101/gad.494808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Tanguay RM. Drosophila small heat shock proteins: an update on their features and functions. In: Tanguay RM, Hightower LE, editors. The big book on small heat shock proteins. Cham: Springer International Publishing; 2015. pp. 579–606. [Google Scholar]
- Moutaoufik MT, Morrow G, Finet S, Tanguay RM. Effect of N-terminal region of nuclear Drosophila melanogaster small heat shock protein DmHsp27 on function and quaternary structure. PLoS One. 2017;12:e0177821. doi: 10.1371/journal.pone.0177821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutaoufik MT, Morrow G, Maaroufi H, Férard C, Finet S, Tanguay RM. Oligomerization and chaperone-like activity of Drosophila melanogaster small heat shock protein DmHsp27 and three arginine mutants in the alpha-crystallin domain. Cell Stress Chaperones. 2017;22:455–466. doi: 10.1007/s12192-016-0748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H, Schmidt D, Steinbrink S, Mirgorodskaya E, Lehmann V, Habermann K, Dreher F, Gustavsson N, Kessler T, Lehrach H, Herwig R, Gobom J, Ploubidou A, Boutros M, Lange BMH. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 2010;29:3344–3357. doi: 10.1038/emboj.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MD, Luo X, Biteau B, Syverson K, Jasper H. 14-3-3 Epsilon antagonizes FoxO to control growth, apoptosis and longevity in Drosophila. Aging Cell. 2008;7:688–699. doi: 10.1111/j.1474-9726.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeri D, Malim MH. Importin β can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin α. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/MCB.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain P, Gelly J-C, Flatters D. Detection and architecture of small heat shock protein monomers. PLoS One. 2010;5:e9990. doi: 10.1371/journal.pone.0009990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee DY, Cho D-Y, Zhai B, Slattery M, Ma L, Mintseris J, Wong CY, White KP, Celniker SE, Przytycka TM, Gygi SP, Obar RA, Artavanis-Tsakonas S. Transcription factor networks in Drosophila melanogaster. Cell Rep. 2014;8:2031–2043. doi: 10.1016/j.celrep.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant J-Y, Legent K, Janody F, Treisman JE. The transcriptional co-factor Chip acts with LIM-homeodomain proteins to set the boundary of the eye field in Drosophila. Development. 2010;137:273–281. doi: 10.1242/dev.041244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P, Szabo K, Bailey A, et al. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Rosenbaum EE, Brehm KS, Vasiljevic E, Liu CH, Hardie RC, Colley NJ. XPORT-dependent transport of TRP and rhodopsin. Neuron. 2011;72:602–615. doi: 10.1016/j.neuron.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K-D, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains (Acd proteins) Cell Stress Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:TEFOAT>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M, Port M, Tripp J, Weber C, Zielinski D, Calligaris R, Winkelhaus S, Scharf KD. Tomato heat stress protein Hsp16.1-CIII represents a member of a new class of nucleocytoplasmic small heat stress proteins in plants. Cell Stress Chaperones. 2003;8:381–394. doi: 10.1379/1466-1268(2003)008<0381:THSPHR>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M, Gernhard S, von Koskull-Döring P, Vierling E, Scharf KD. The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones. 2008;13:183–197. doi: 10.1007/s12192-008-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler R, Kappé G, Boelens W, Slingsby C. Wrapping the alpha-crystallin domain fold in a chaperone assembly. J Mol Biol. 2005;353:68–79. doi: 10.1016/j.jmb.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, Kaufman TC, Calvi BR, the FlyBase Consortium. Perrimon N, Gelbart SR, Agapite J, Broll K, Crosby L, Santos G, Emmert D, Gramates LS, Falls K, Jenkins V, Matthews B, Sutherland C, Tabone C, Zhou P, Zytkovicz M, Brown N, Antonazzo G, Attrill H, Garapati P, Holmes A, Larkin A, Marygold S, Millburn G, Pilgrim C, Trovisco V, Urbano P, Kaufman T, Calvi B, Czoch B, Goodman J, Strelets V, Thurmond J, Cripps R, Baker P. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019;47:D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Gene Ontology (GO) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Klundert FAJM, De Jong WW. The small heat shock proteins Hsp20 and αB-crystallin in cultured cardiac myocytes: differences in cellular localization and solubilization after heat stress. Eur J Cell Biol. 1999;78:567–572. doi: 10.1016/S0171-9335(99)80022-3. [DOI] [PubMed] [Google Scholar]
- van Montfort RL, Basha E, Friedrich KL, et al. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- Vicart P, Caron A, Guicheney P, Li Z, Prévost MC, Faure A, Chateau D, Chapon F, Tomé F, Dupret JM, Paulin D, Fardeau M. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2009. [Google Scholar]
- Ylikallio E, Konovalova S, Dhungana Y, Hilander T, Junna N, Partanen JV, Toppila JP, Auranen M, Tyynismaa H. Truncated HSPB1 causes axonal neuropathy and impairs tolerance to unfolded protein stress. BBA Clin. 2015;3:233–242. doi: 10.1016/j.bbacli.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of nuclear drosophila small heat shock proteins. Alignment was generated using ClustalW. UniProt accession numbers of sequences used in the alignments are indicated. Secondary structures of the alignment generated by ESPript (Robert and Gouet 2014), according to DmHsp27 ACD predicted structure by Moutaoufik et al. 2017b. NTRs show conservation of FGFG motif and arginine-rich NLS (Black arrow). The I/V/L-X-I/L/V motif in the CTR and conserved arginine R122, R131, and R135 are indicated by black arrow and black star, respectively. (PNG 2876 kb).
(XLS 839 kb).
(XLS 212 kb).