SUMMARY
The blood vessels in the central nervous system (CNS) have a series of unique properties, termed the blood-brain barrier (BBB), which stringently regulate the entry of molecules into the brain, thus maintaining proper brain homeostasis. We sought to understand whether neuronal activity could regulate BBB properties. Using both chemogenetics and a volitional behavior paradigm, we identified a core set of brain endothelial genes whose expression is regulated by neuronal activity. In particular, neuronal activity regulates BBB efflux transporter expression and function, which is critical for excluding many small lipophilic molecules from the brain parenchyma. Furthermore, we found that neuronal activity regulates the expression of circadian clock genes within brain endothelial cells, which in turn mediate the activity-dependent control of BBB efflux transport. These results have important clinical implications for CNS drug delivery, clearance of CNS waste products including Aβ, and understanding how neuronal activity can modulate diurnal processes.
eTOC Blurb
The authors demonstrate that brain endothelial gene expression and blood-brain barrier efflux exhibit plasticity in response to neuronal activity. This has implications for how the blood-brain barrier regulates the composition of endogenous and exogenous molecules in the central nervous system in relation to circadian time and neuronal activity. This also suggests that neuronal activity can extrinsically modulate otherwise cell-intrinsic oscillatory processes.
INTRODUCTION
The blood vessels that vascularize the CNS exhibit a series of distinct properties compared to peripheral blood vessels (Zlokovic, 2008; Profaci et al., 2020). Many of these properties are possessed by CNS endothelial cells (ECs) which form vessel walls, tightly regulating the chemical microenvironment of the CNS which is important to maintain brain homeostasis and for neural protection. Although BBB properties are predominantly possessed by CNS ECs, they are induced and maintained by cues from the CNS microenvironment, including signals from neural stem cells, pericytes and astrocytes (Stewart and Wiley, 1981; Janzer and Raff, 1987; Abbott, 2002; Weidenfeller, Svendsen and Shusta, 2007; Armulik et al., 2010; Daneman et al., 2010).
The BBB is not a single entity, but a series of properties that together allow CNS ECs to stringently regulate the movement of molecules and cells between the blood and the neural tissue. CNS ECs are held together by tight junctions (TJs) which create a high electrical resistance paracellular barrier and polarize the cells into distinct luminal and abluminal compartments (Furuse, 2010). CNS ECs lack fenestra and exhibit low rates of transcytosis in comparison to peripheral ECs, thus limiting transcellular movement of solutes (Hallmann et al., 1995). CNS ECs also have low expression of leukocyte adhesion molecules (LAMs), thus limiting CNS immune surveillance (Engelhardt, 2008). CNS ECs are also unique in their enriched expression of a wide array of transporters. These include a large breadth of specialized solute carrier (Slc) transporters which have narrow substrate specificities and collectively import a wide range of essential nutrients for the brain (Betz and Goldstein, 1978), and adenosine triphosphate (ATP)-binding cassette (ABC) transporters which are primarily embedded in the luminal side of CNS ECs and utilize ATP to actively efflux many small lipophilic molecules that would otherwise passively diffuse across the EC membrane (Cordon-Cardo et al., 1989). ABC transporters have broad substrate specificities including many xenobiotics such as small molecule drugs, thus presenting a major obstacle for CNS drug delivery (Pardridge, 2007). Although the ABC efflux transporters have predominantly been studied in the context of preventing entry of exogenous molecules into the CNS, they also have a role in the efflux of endogenous molecules, including steroids (Hindle et al., 2017) and neural waste products such as amyloid beta (Aβ) (Lam et al., 2001; Cirrito et al., 2005; Bruckmann et al., 2016). Dysfunction of BBB efflux transport has been hypothesized as a mechanism of Aβ accumulation in Alzheimer’s disease (AD) (Sweeney et al., 2018).
The BBB has largely been studied as a static structure: a wall protecting the CNS from potential toxins. The brain, however, is highly dynamic with constant fluctuations in neuronal activity and energy demand. Therefore, it is plausible that the vasculature could dynamically respond to meet the changing demands of the neuronal circuitry. Neurovascular communication has largely been studied in the context of blood flow where increased neuronal activity leads to an increase in localized blood flow (Roy and Sherrington, 1890; Nersesyan et al., 2004). It is also established that neurovascular signaling is critical for postnatal angiogenesis in the brain (Lacoste et al., 2014; Whiteus, Freitas and Grutzendler, 2014). Much less is known about whether the properties of the BBB are plastic and how these properties respond to changes in neuronal activity. Interestingly, a study utilizing single cell sequencing of the visual cortex in different light paradigms identified that there were changes to vascular cell gene expression in addition to neurons and glia, suggesting that experience-dependent changes in neuronal activity may indeed alter brain EC function (Hrvatin et al., 2018). Because the BBB is essential for regulating the CNS microenvironment, changes in transport, signaling, metabolism or other BBB properties without overt ‘leakiness’ of the BBB to non-specific molecules, could significantly alter the extracellular environment and thus modulate neuronal circuit function and behavior (Kaplan, Chow and Gu, 2020; Profaci et al., 2020). We sought to understand whether neuronal activity can regulate specific barrier properties.
We utilized chemogenetics and a volitional behavior paradigm followed by EC purification and RNA sequencing in order to gain insight into how neuronal activity can modulate brain EC gene expression. These studies identified a core dataset of activity-dependent brain EC genes including key BBB efflux transporters and EC circadian genes. We further found that brain EC expression and function of the major efflux transporter, P-glycoprotein (Pgp), are inversely correlated with neuronal activity across the day and that this diurnal rhythm is attenuated by genetic ablation of EC circadian genes. We further found that EC-specific adult knockout of Arntl (Bmal1) suppressed the activity-dependent changes in Pgp efflux, demonstrating that BBB efflux is regulated by neuronal activity through EC circadian clock genes, and that neuronal activity and behavior can modulate diurnal processes.
RESULTS
DREADDs as a Tool to Manipulate Glutamatergic Activity in vivo
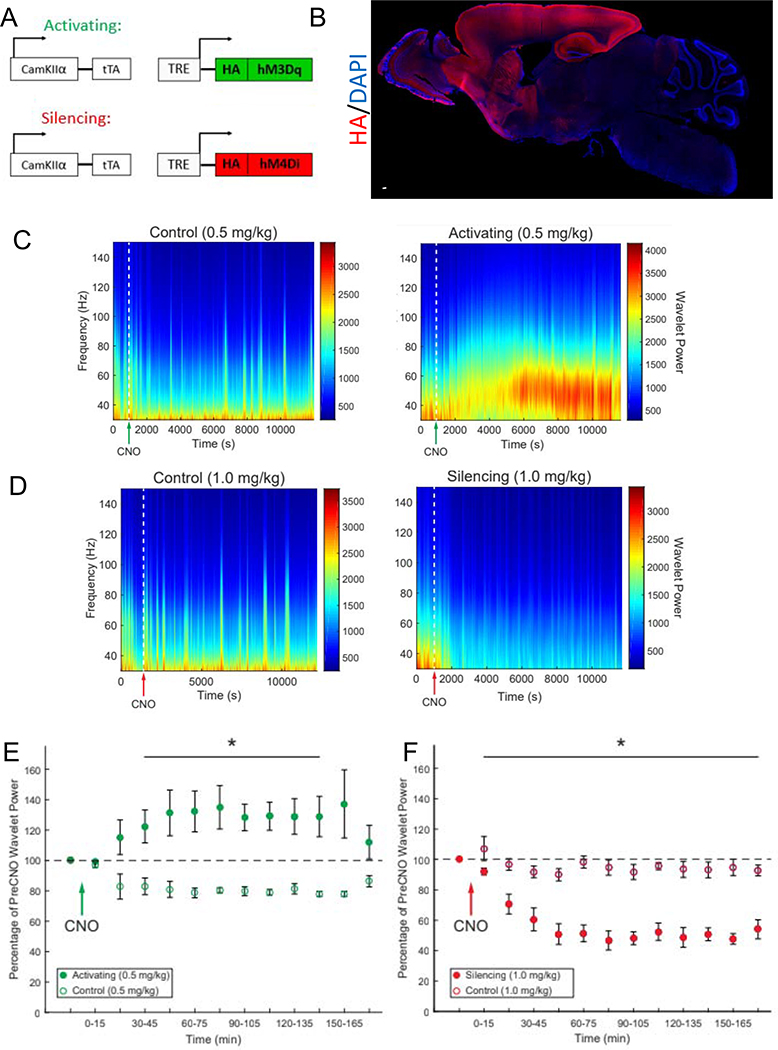
To determine how glutamatergic neuronal activity regulates the brain ECs, we generated two chemogenetic mouse models to express Designer Receptors Exclusively Activated by Designer Drugs (DREADDs) in glutamatergic neurons: CamKIIα-tTA;TRE-hM3Dq (hM3Dq-Activating) mice to activate glutamatergic neurons and CamKIIα-tTA; TRE-hM4Di (hM4Di-Silencing) to silence glutamatergic neurons (Figure 1A) in response to clozapine-N-oxide (CNO) (Alexander et al., 2009). The DREADDs were densely expressed in the cortex and hippocampus (Figure 1B, S1A). To determine the temporal dynamics of chemogenetic neuronal activity modulation we used in vivo multi-electrode recordings to examine neuronal activity in the cortex/hippocampus following injection of CNO. IP injection of 0.5 mg/kg CNO was sufficient to cause a robust increase in gamma local field potential (LFP) power and locomotor activity (Figure S1B) in the hM3Dq-Activating mice, without eliciting seizures (Figure 1C) or excitotoxic cell death (Figure S1D, E, F, G). The increase in neuronal activity was initially observed at 15 minutes post-CNO injection, peaked at 1 hour post-injection and lasted until 3 hours post-injection. No increase in gamma local field potential or locomotor activity was observed in the littermate controls (Figure 1E, S1B). IP injection of 1.0 mg/kg CNO in the hM4Di-Silencing mice robustly attenuated gamma LFP power (Figure 1D, F) and largely did not affect locomotor activity (Figure S1C). Neuronal silencing was initially observed 15 minutes post-CNO, maximum silencing was observed around 1 hour and it lasted the full 3 hours of recording.
Figure 1. DREADDs as a Tool to Manipulate Glutamatergic Activity in vivo.
(A) Schematic representation of the genetic mouse models utilizing DREADDS to activate or silence glutamatergic activity in vivo.
(B) Photomerge of a representative sagittal section of a brain derived from an Activating DREADDs mouse stained for rabbit anti-HA (red) and cell nuclei with DAPI (blue). Color-matched boxes were added to the edges to make a rectangular image. Scale bar is 2 mm.
(C-D) Power density spectrograms depicting gamma local field potential (LFP) power from representative multielectrode array electrophysiological recording sessions from Activating DREADDs mice and controls (C) and Silencing DREADDs mice and control (D) CNO dosages are noted, and delivered when indicated by the green (C) or red (D) arrow on the x-axis.
(E-F) Average gamma local field potential (LFP) power as a percentage of the pre-CNO injection baseline for Activating DREADDs mice and controls (E) and Silencing DREADDs mice and control (F). Data represent mean ± SEM (error bars). n=3 per group, *p<0.05 by unpaired Student’s t-test.
DREADDs-Mediated, Neuronal Activity-Regulated Brain EC Transcriptome
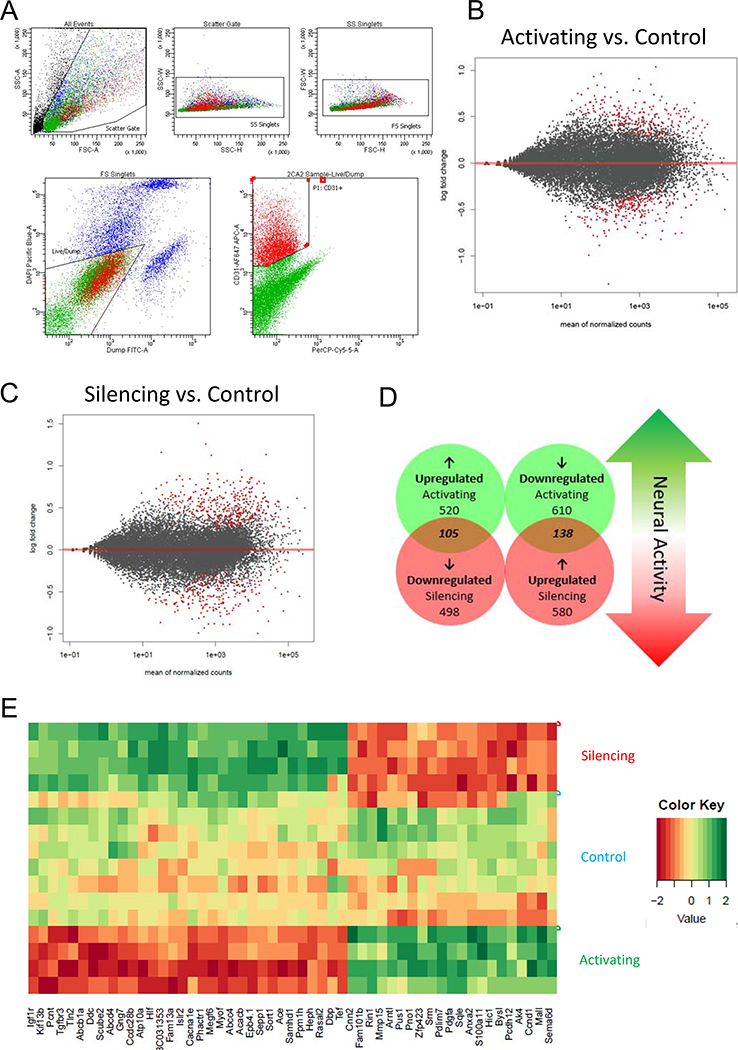
We next determined how neuronal activity regulates brain EC gene expression. 3 hours following CNO injection into hM3Dq-Activating mice and littermate controls (0.5mg/kg CNO) or hM4Di-Silencing mice and littermate controls (1.0mg/kg CNO), we isolated brain ECs via fluorescence-activated cell sorting (FACS, Figure 2A, Figure S2A,B) and examined their gene expression by RNA sequencing (RNA sequencing data can be found in Supplemental Files).
Figure 2. DREADDs-Mediated, Neuronal Activity-Regulated Brain EC Transcriptome.
(A) Representative FACS plot of the gating strategy used to sort brain ECs. First, intact cells were gated using forward and side scatter (top 3 panels). Next, cells were gated against dead cells and (DAPI-positive) and pericytes and immune cells (FITC-positive). Finally, Alexa 647-positive ECs were positively selected.
(B) MA plot representing global gene expression changes in brain ECs after glutamatergic activation vs. control. Red dots signify statistically significant changes by Wald Test. n=4 mice per group.
(C) MA plot for glutamatergic silencing vs. control. n=4 mice per group.
(D) Venn Diagram for statistically significant (p<0.05 by Wald Test) gene expression changes after glutamatergic activation and silencing. “Neuronal activity-dependent genes” are the 243 (105 and 138) that were regulated in opposite directions after glutamatergic activation and silencing.
(E) Clustering heat map of a refined list (at least 0.25 log2 fold change in both directions with Abcd4 added for comparison) of statistically significant (by Wald Test) oppositely regulated neuronal activity-dependent genes. Color scale represents arbitrary units of expression. Red represents lower expression and green represents higher expression.
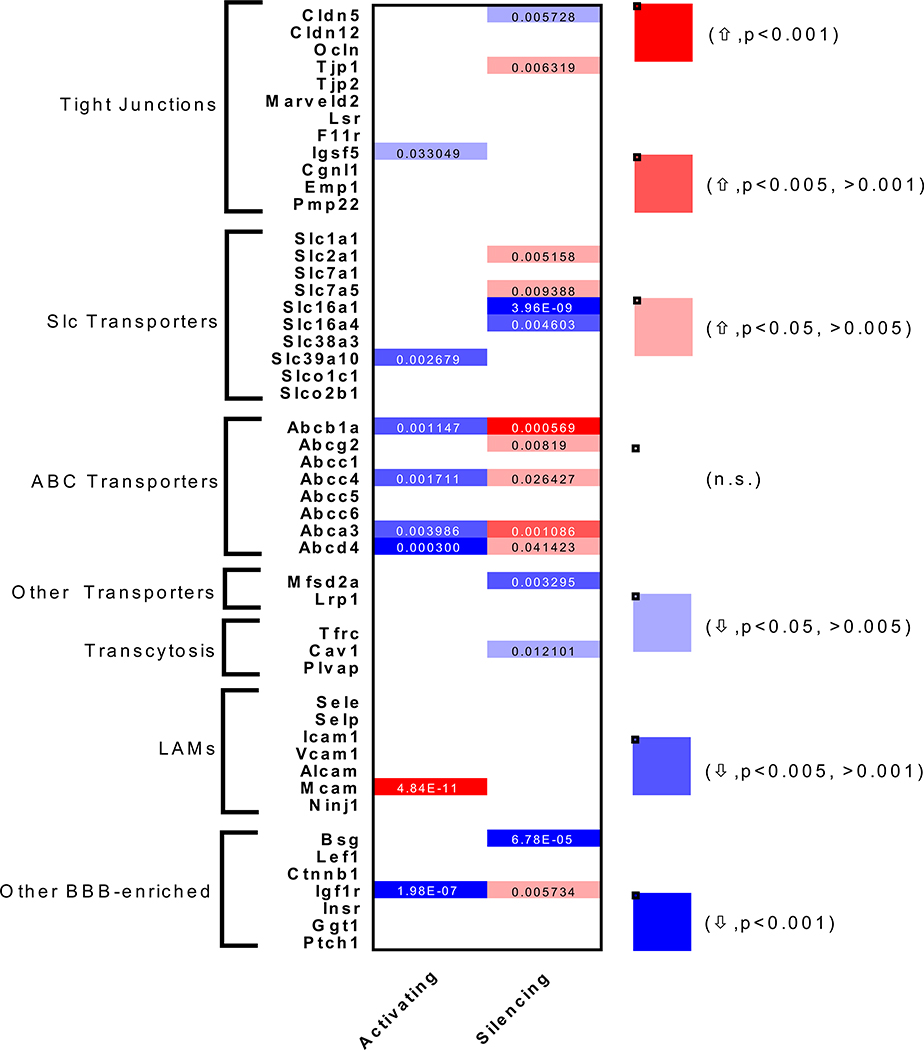
625 genes were significantly upregulated and 748 genes were significantly downregulated by glutamatergic activation (Figure 2B, Table S1, S2, Supplemental File 2). Pathway analysis revealed downregulation of amino acid and lipid metabolic pathways (Table S1). There was a robust upregulation of genes in pathways involving adherens junctions and cytoskeletal remodeling (Table S2). Neuronal activity has recently been shown to cause rapid structural and cytoplasmic changes to arteriolar ECs leading to neurovascular coupling (NVC) (Chow et al., 2020). However, the increases mRNA levels of these structural genes are likely a consequence of increased NVC-mediated blood flow in these brain regions as shear stress-dependent transcriptional regulation of the adherens junctions and consequently the cytoskeleton in ECs is a well-established phenomenon in the vasculature (Galbraith, Skalak and Chien, 1998; Miao et al., 2005; Polacheck et al., 2017). We then examined previously described BBB-enriched genes (Figure 3, Supplemental File 4). TJ gene expression (Cldn5, Ocln, Tjp1, Tjp2, Lsr, Marveld2) was largely unaffected by increased neuronal activity. BBB-enriched Slc transporters were also largely unaffected with the exception of a significant downregulation of Slc39a10, a putative zinc transporter. There were largely no changes to LAMs (Icam1, Vcam1, Alcam, Sele, Selp) with the exception of a significant upregulation of Mcam. Regulators of transcytosis (Mfsd2a, Cav1 and Plvap) were also largely unaffected by glutamatergic activation. Most strikingly, 4 major ABC efflux transporters were significantly downregulated by glutamatergic activation (Pgp/Abcb1a, Mrp4/Abcc4, Abca3, Abcd4).
Figure 3. Neuronal Activity-Regulated BBB Transcriptome.
Heat map for binned p-values and activity-regulated directionality of common BBB genes in Activating vs. Control (left) and Silencing vs. Control (right). Genes were divided into groups for different BBB properties: Tight junctions, Slc transporters, Abc transporters, other transporters, transcytosis, Leukocyte Adhesion Molecules (LAMs), or Other BBB-enriched. Color scale denotes if a given gene was upregulated (↑) (red) or downregulated (↓) (blue) and whether the change was statistically significant by Wald Test (intensity of color). Individual p-values are shown for significantly regulated genes.
718 genes were significantly upregulated and 603 genes were significantly downregulated by glutamatergic silencing (Figure 2C, Table S3, S4, Supplemental File 2). There was downregulation of adherens junction pathways likely indicating a response to decreased local blood flow when neuronal activity is low (Table S3). Interestingly, metabolism-related genes were also downregulated after glutamatergic silencing which included pathways involved in lipid and steroid metabolism (Table S3). When we looked at known BBB-enriched genes (Figure 3, Supplemental File 4), surprisingly Cldn5 and Tjp1 were significantly downregulated and upregulated respectively. There were 4 BBB-enriched Slc transporters involved in glucose (Slc2a1; upregulated), amino acid (Slc7a5; upregulated) and monocarboxylate transport (Slc16a1, Slc16a4; both downregulated) that were significantly changed in response to glutamatergic silencing. Interestingly, transcytosis regulators Mfsd2a and Cav1 were also downregulated after glutamatergic silencing. There were no changes in LAM expression. Most strikingly, 5 major efflux transporters were upregulated by glutamatergic silencing (Pgp/Abcb1a, BCRP/Abcg2, Mrp4/Abcc4, Abca3, Abcd4).
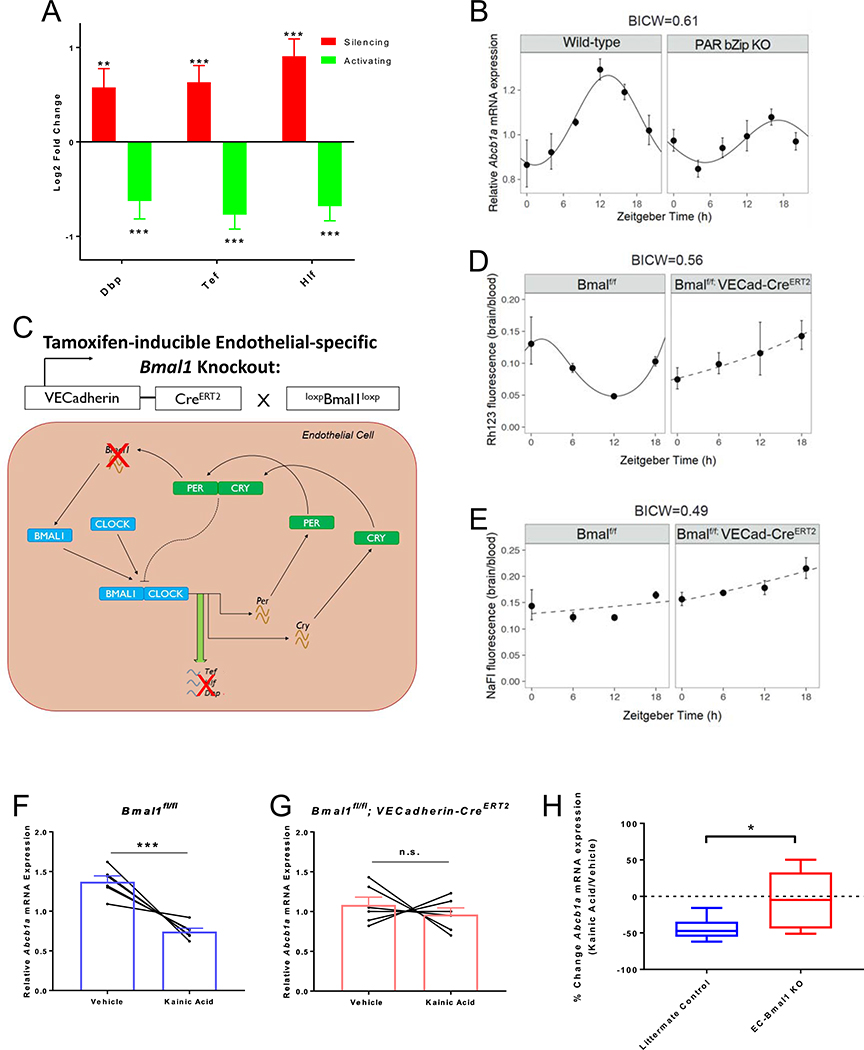
243 genes were regulated in opposing directions by activating and silencing neuronal activity (105 directly correlated and 138 inversely correlated with the amount of neuronal activity), indicating that the absolute mRNA expression level of these brain EC genes are regulated by the total amount of glutamatergic activity (Figure 2D, E, Supplemental File 3). The most enriched pathways that were directly correlated with neuronal activity were adherens junctions and focal adhesion (Table S6), providing further evidence that brain ECs respond to changes in blood flow by undergoing shear stress-induced structural changes. Although neuronal activity has been shown to locally increase permeability of IGF-1 into the CNS (Nishijima et al., 2010), we found that Igf1r expression was inversely correlated with glutamatergic activity (Figure 3). However, this study found that neuronal activity-dependent receptor phosphorylation and cleavage of an IGF binding protein lead to the increased permeability suggesting this phenomenon may be independent of Igf1r transcription. Of particular interest, we found that ABC efflux transporters (Pgp/Abcb1a, Mrp4/Abcc4, Abca3, Abcd4), and PAR bZip circadian transcription factors (Dbp, Tef and Hlf) were both inversely correlated with neuronal activity (Table S5).
Neuronal Activity Regulates ABC Transporter Expression and Function
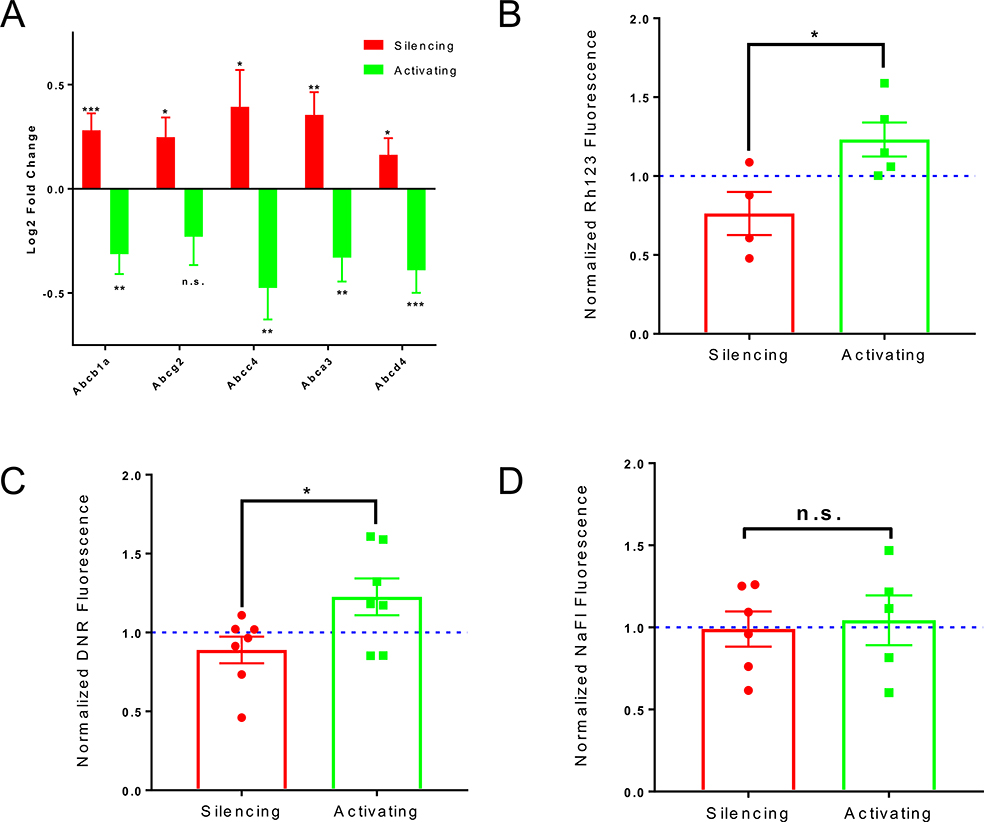
ABC transporters were one of the top pathways inversely correlated with neuronal activity (Table S5, Figure 4A). We observed a similar trend with Pgp protein expression in response to glutamatergic activity modulation (Figure S3A). Interestingly, moesin (Msn) and protein kinase C beta-1 (Prkcb) regulate Pgp function (Hoshi et al., 2019) and the mRNA of both were robustly regulated by DREADDs-mediated changes in glutamatergic activity (Supplemental File 2).
Figure 4. Neuronal Activity Regulates ABC transporter expression and function.
(A) Log2 fold change of mRNA expression in Activating or Silencing groups relative to respective littermate controls of 5 major BBB-specific ABC transporters after DREADDs-mediated manipulation of glutamatergic activity. Data represent mean ± SEM (error bars). n=4 per group. *p<0.05, **p<0.005, ***p<0.001, n.s. (not significant) by Wald Test.
(B) Normalized Rhodamine123 (Rh123) fluorescence of Activating vs. Silencing cortices/hippocampi following CNO administration. Mutants were paired with littermate controls. The rhodamine fluorescence (brain:blood) of each mutant was normalized to the fluorescence of its littermate control (Excitation=505nm, Emission=560nm). Data represent mean ± SEM (error bars). Individual data points are shown. *p=0.0291 by unpaired Student’s t-test.
(C) Normalized Daunorubicin (DNR) fluorescence of Activating vs. Silencing cortices/hippocampi following CNO administration. Mutants were paired with littermate controls The DNR fluorescence (brain:blood) of each mutant was normalized to the fluorescence of its littermate control (Excitation=470, Emission=585). Data represent mean ± SEM (error bars). Individual data points are shown. *p=0.0195 by unpaired Student’s t-test.
(D) Normalized Sodium Fluorescein (NaFl) fluorescence of Activating vs. Silencing cortices/hippocampi following CNO administration. Mutants were paired with littermate controls. The NaFl fluorescence (brain:blood) of each mutant was normalized to fluorescence of its littermate control (Excitation=480nm, Emission=538nm). Data represent mean ± SEM (error bars). Individual data points are shown. n.s. (not significant) by unpaired Student’s t-test.
To determine if there were corresponding activity-dependent changes in BBB efflux function, we injected Rhodamine123 (Rh123), a fluorescent lipophilic substrate of Pgp (Figure S3B) (Wang et al., 2001; Sohet and Daneman, 2013), 2 hours post-CNO injection into hM3Dq-Activating and hM4Di-Silencing mice and their paired littermate controls and measured Rh123 fluorescence in the brain and blood. Significantly more Rh123 entered the brains of the hM3Dq-Activating mice relative to hM4Di-Silencing mice (Figure 4B). Thus Pgp function, as with mRNA, is inversely correlated with glutamatergic activity. Although there was a slight trending activity-dependent increase, there was no significant difference in Rh123 permeability between the two groups in the cerebellum where no DREADDs are expressed. This suggests that this effect is localized to regions directly undergoing robust changes in neuronal activity (Figure S3C), but may be observed to a lesser degree in functionally connected regions. There was no difference in Rh123 uptake in immune-related organs. Surprisingly there was significantly more Rh123 uptake in circulating immune cells in the Activating mice relative to Silencing mice (Figure S3C). However, this peripheral Rh123 effect could not be responsible for the observed CNS Rh123 effect as there was no altered peripheral immune cell infiltration into the brain parenchyma in any of the conditions (Figure S3D). We also analyzed the permeability of daunorubicin (DNR), a fluorescent small molecule chemotherapeutic that is also a Pgp substrate (Egorin et al., 1974; Wang et al., 2001). As with Rh123, there was also significantly more permeability to DNR upon glutamatergic activation than glutamatergic silencing (Figure 4C), suggesting that this activity-dependent regulation of Pgp function is relevant for CNS drug delivery. We observed no differences in brain permeability to sodium fluorescein (NaFl) or Evans Blue, two molecules that are not substrates of Pgp, between Activating mice and Silencing mice (Figure 4D, S3E). This indicates that the permeability changes to Rh123 and DNR are specific to efflux transport, and not non-specific leakiness of the BBB.
Neuronal activity-regulated PAR bZip Transcription Factors Modulate Pgp Expression and Function
We identified that EC expression of the PAR bZip family of circadian transcription factors Dbp, Tef and Hlf are inversely correlated with neuronal activity (Table S5, Figure 5A). Circadian rhythms are intrinsic oscillatory biological processes that are driven by a transcriptional feedback loop and govern the oscillation of a variety of physiological processes such as sleep, cardiovascular function, gastrointestinal motility and metabolism (Richards and Gumz, 2013). The core circadian transcriptional machinery consists of Bmal1 (Arntl) and Clock in the positive loop which drive transcription of genes such as the PAR bZip transcription factors, and Per1/2 and Cry1/2 which act as negative feedback regulators on Bmal1 and Clock (Ripperger and Schibler, 2001, 2006; Mongrain et al., 2011; Curie et al., 2013). The circadian clock has mostly been studied in the context of the suprachiasmatic nucleus (SCN), the central pacemaker which is entrained by light and synchronizes many of the body’s biological rhythms (Welsh et al., 1995; Brancaccio et al., 2019). However, the core clock transcriptional machinery exists in many non-neuronal cells both in the brain and peripheral tissues. Although there has been evidence implicating a significant role of the circadian clock in blood vessel tone, the blood-retinal barrier of vertebrates and the glial-based BBB in flies, the role of an EC-specific circadian clock in the BBB of mammals has not been extensively studied (Curtis et al., 2007; Sandoo et al., 2010; Anea et al., 2013; Richards, Diaz and Gumz, 2014; Savolainen et al., 2016; Durgan, Crossland and Bryan, 2017; Zhang et al., 2018; Hudson et al., 2019). Interestingly, global triple knockouts of the PAR bZip transcription factors have disrupted xenobiotic efflux in the liver, low circulating endogenous steroids and low blood pressure (Gachon et al., 2006; Wang et al., 2010). The PAR bZip transcription factors have recently been further implicated in epilepsy and shown to be modulated by hyperactive neuronal activity, but cell-specific roles have also not yet been explored in this context (Gachon et al., 2004; Rambousek et al., 2020).
Figure 5. Neuronal activity-regulated PAR bZip transcription factors modulate Pgp expression and function.
(A) Log2 fold change of mRNA expression in Activating or Silencing groups relative to respective littermate controls of 3 PAR bZip transcription factors after DREADDs-mediated manipulation of glutamatergic activity. Data represent mean ± SEM (error bars). n=4 mice per group. **p<0.005, ***p<0.001 by Wald Test.
(B) Relative mRNA expression of Abcb1a normalized to GAPDH across a 24 hour day (12:12 hour dark:light) in Wildtype (left) and PAR bZip triple knockout mice (right). Expression levels represent 2−ΔΔct. Data represent mean ± SEM (error bars). n=4 mice per group.
(C) Schematic of genetic strategy to modulate circadian gene oscillation exclusively in ECs in response to tamoxifen. Bmal1 floxed mice were mated to VECadherin-CreERT2 mice and injected with tamoxifen at 5 week of age. Ablation of Bmal1, the master regulator of the positive loop will ablate typical expression of PAR bZip transcription factors.
(D) Rhodamine123 (Rh123) fluorescence (brain:blood) in littermate controls (left) and EC-Bmal1 knockout mice (right) across a day (Excitation=505nm, Emission=560nm). Data represent mean ± SEM (error bars). n=3–7 mice per group.
(E) Sodium Fluorescein (NaFl) fluorescence (brain:blood) in littermate controls (left) and EC-Bmal1 knockout mice (right) across a day (Excitation=480nm, Emission=538nm). Data represent mean ± SEM (error bars). n=3–6 mice per group.
Rhythmicity was assessed by linear regression. Results are represented as solid lines if the statistical model indicates rhythmicity. Non-rhythmic fits are represented by dashed lines. The statistical model was only considered if the BICW > 0.4 (B, D, E).
(F) Relative mRNA expression of EC Abcb1a normalized to Rps20 in FACS-purified brain ECs from littermate control mice (left) and EC-Bmal1 conditional knockout mice (right). Expression levels represent 2−ΔΔct. Data represent mean ± SEM (error bars). Individual pairs are also shown (n=6 mice per group). ***p<0.001, n.s. (not significant) by paired Student’s t-test.
(G) Box and whisker plot depicting percent change of Abcb1a mRNA expression after kainic acid treatment in littermate controls vs. EC-Bmal1 cKOs. n=6 mice per group. *p<0.05 by unpaired Student’s t-test. Data represent median, upper and lower quartiles, and minimum and maximum.
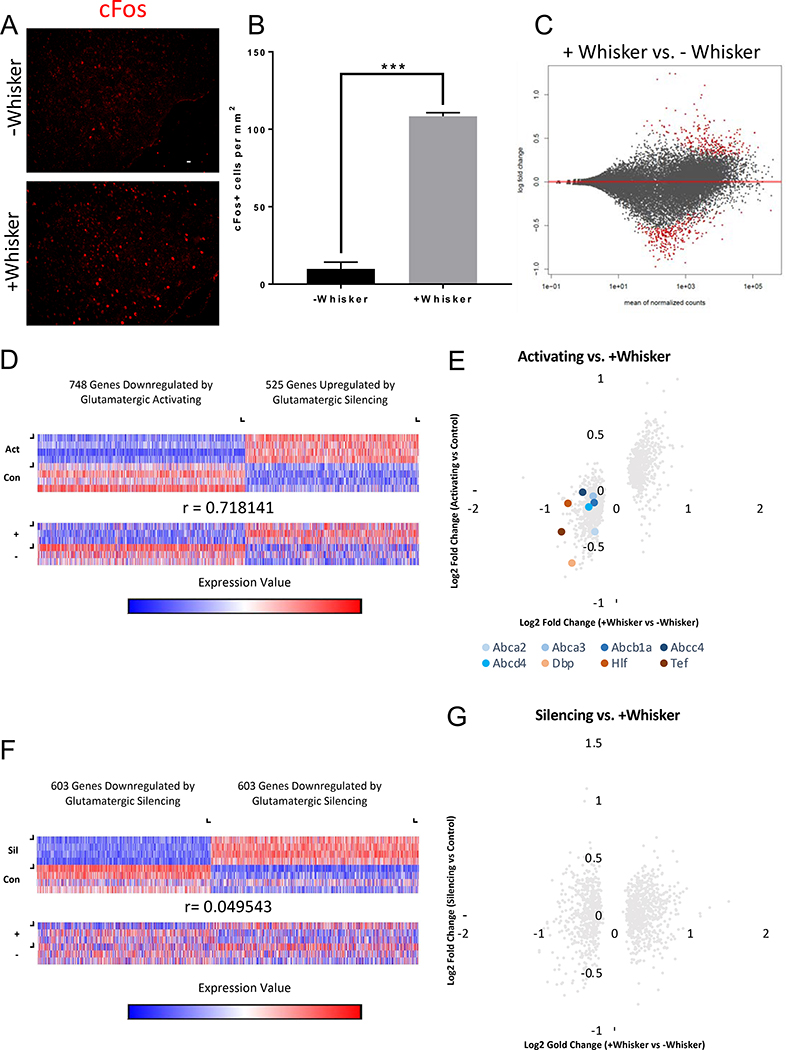
CamKIIα-driven HA-DREADDs are expressed in a subset of neurons in the SCN, as reported previously (Figure S4C, D) (Yokota et al., 2001; Kon et al., 2014). However, DREADDs-mediated activation and silencing did not significantly alter cFos expression in the SCN (albeit a trending increase after activating) (Figure S4E, F, G). Furthermore, as described below, we found similar changes to BBB gene expression in the barrel cortex in response to whisker stimulation, a volitional behavior paradigm not mediated by the SCN (Figure 6), as with DREADDs-mediated activation. This whisker stimulation also led to a decrease in the expression of both efflux transporters and EC circadian genes (Figure 6E). These data suggest that these neuronal activity-dependent EC circadian gene expression changes were not strictly a result of a systemic effect on the core SCN circadian clock.
Figure 6. Behaviorally motivated, Neuronal Activity-Regulated Brain EC Transcriptome.
(A) Representative sections of barrel cortex in whisker-shaven, environmentally-null mice (top) vs. whisker-intact, environmentally-enriched mice (bottom) stained for goat anti-cFos (red). Scale bar is 100 μm.
(B) Quantification of cFos+ cells per mm2 in barrel cortex of “−Whisker” vs. “+Whisker” mice. Data represent mean ± SEM (error bars). n=3 per group. ***p<0.001 by unpaired Student’s t-test.
(C) MA plot for +Whisker vs. −Whisker. Red dots signify statistically significant changes by Wald Test. n=3 mice per group.
(D) Heat maps of genes statistically significantly (by Wald Test) regulated by DREADDs-mediated glutamatergic activation viewed in the Activating replicates (Act) and paired littermate control replicates (Con) (top) and the same genes viewed in the +Whisker replicates (+) and −Whisker replicates (−) (bottom). Color scale represents arbitrary units of expression. Blue represents lower expression and red represents higher expression. Pearson Correlation Coefficient shown.
(E) X/Y scatter plot of the same genes in (D) in the Activating mice and +Whisker mice based on log2 fold change. Each dot represents an individual gene. The majority of genes cluster in the top right (upregulated in both) or bottom left (downregulated in both) quadrants indicating a close correlation of expression changes of these genes between these data sets. ABC transporter genes (blue) and PAR bZip transcription factor genes (red) are shown as colored dots.
(F) Heat maps of genes statistically significantly (by Wald Test) regulated by DREADDs-mediated glutamatergic silencing viewed in the Silencing replicates (Sil) and paired littermate control replicates (Con) (top) and the same genes viewed in the +Whisker replicates (+) and −Whisker replicates (−) (bottom). Color scale represents arbitrary units of expression. Blue represents lower expression and red represents higher expression. Pearson Correlation Coefficient shown.
(G) X/Y scatter plot of the same genes in (F) in the Silencing mice and +Whisker mice based on log2 fold change. Each dot represents an individual gene. The genes do not cluster within specific quadrants indicating a lack of correlation of expression changes of these genes between these data sets.
Similar to the ABC transporters, the expression of Dbp, Tef, and Hlf were inversely correlated with glutamatergic neuronal activity (Table S5, Figure 5A). Both Arntl (Bmal1) of the positive loop and Cry1 of the negative loop were regulated in the opposite manner to that of the PAR bZip transcription factors, and were positively correlated with neuronal activity (Figure S4A). However, modulation of the PAR bZip transcription factors by neuronal activity could be explained by the expression of Nr1d2 (Rev-erbβ), another circadian transcription factor that represses both Bmal1 and Cry1 (Preitner et al., 2002), whose expression was also inversely correlated with neuronal activity (Figure S4A). Cry1 is known to directly repress the expression of Dbp (Ye et al., 2014) and likely plays a role in the regulation of PAR bZip transcription factors by neuronal activity.
It has recently been reported that flies and rodents exhibit diurnal oscillation of BBB efflux transport (Savolainen et al., 2016; Zhang et al., 2018), suggesting that the activity-dependent regulation of BBB efflux may be downstream of EC circadian clock genes. We next determined whether there was a diurnal rhythm of BBB efflux in mice, and whether this was controlled by EC circadian genes. Mice are active during their dark period (ZT12-ZT24) and rest during their light period (ZT0-ZT12), thus exhibiting higher excitatory neuronal activity during the dark period associated with wakefulness and lower excitatory neuronal activity during the light period associated with sleep (Vyazovskiy et al., 2008; Bero et al., 2011). We observed that there was diurnal oscillation of Abcb1a mRNA expression in brains of wildtype mice with a peak occurring at ZT12 (ZT13.3 on the fit curve), the end of the resting light period, and a trough occurring at ZT0, the end of the active dark period, corresponding with its expression after manipulation of neuronal activity (Figure 5B). This rhythmic oscillation exhibited an attenuated amplitude and a substantial phase delay (peak at ZT17.1) in the brains of PAR bZip triple knockout (tKO) mice (Figure 5B).
In order to determine if the circadian clock gene-dependent regulation of BBB efflux transport was specific to its function in ECs, we generated tamoxifen-inducible, EC-specific Bmal1 knockout mice (VECadherin-CREERT2; Bmal1fl/fl) (Figure S5A, B, C). Bmal1 is the master regulator of the positive loop in the circadian transcriptional machinery, and thus deletion of Bmal1 in ECs will disrupt circadian oscillation of Dbp, Tef and Hlf (Figure 5C, S5D) (Ripperger et al., 2000; Ripperger and Schibler, 2006; Mongrain et al., 2011; Curie et al., 2013). EC-specific Bmal1 conditional knockout (EC-Bmal1 cKO) mice and littermate controls (Bmal1fl/fl) both have normal diurnal behavior, displaying a nocturnal eating and drinking schedule (Figure S5E, F), suggesting that inhibition of the EC clock does not interfere with the overall rhythmic activity of the mouse. We also observed that there was diurnal oscillation of Abcb1a transcripts in brains of littermate control mice with a peak occurring at the end of the resting light period (ZT14.6 on the fit curve) and a trough occurring at the end of the active dark period (~ZT0) (Figure S5G). Although this rhythmic oscillation was not completely abolished in the brains of the EC-Bmal1 cKO mice, there was more variability and lower mean expression suggesting that EC circadian genes are partially responsible for rhythmic oscillation of Abcb1a mRNA expression and that non-EC circadian mechanisms are also involved.
We observed a rhythmic oscillation of Rh123 permeability in littermate control animals that was inversely correlated with the rhythmic expression of Abcb1a (Figure 5D), demonstrating that there is indeed diurnal oscillation of BBB efflux function that inversely correlates with mouse diurnal activity and net neuronal activity throughout the day. This rhythmic oscillation of Pgp function was inhibited in EC-Bmal1 cKO mice suggesting that the rhythmic oscillation of BBB efflux transport was dependent on proper function of Bmal1 within ECs (Figure 5D). Therefore, EC Bmal1 appears to regulate Pgp/Abcb1a at two levels: at the transcriptional level as EC deletion led to mild alteration in Abcb1a mRNA levels, and at the functional level as EC deletion abolished the rhythmic BBB efflux of Rh123. We did not observe robust diurnal oscillation of NaFl permeability in either genotype (Figure 5E), suggesting that the diurnal oscillation was specific to BBB efflux, and not passive permeability.
We next determined whether the EC circadian clock genes mediate the neuronal activity-dependent regulation of BBB efflux. We injected vehicle or the glutamatergic receptor agonist kainic acid at a sub-epileptic dose into EC-Bmal1 cKOs and littermate controls, FACS-purified brain ECs from the four groups, and measured Abcb1a mRNA expression. Kainic acid was sufficient to cause a significant decrease in Abcb1a expression in the controls, similar to what we observed with DREADDs mediated glutamatergic activation (Figure 5F). However, kainic acid caused no significant change in Abcb1a expression in the EC-Bmal1 cKOs (Figure 5F, G). This demonstrates that EC circadian clock genes are required for neuronal activity-dependent regulation of BBB efflux gene expression.
Behavioral Regulation of the Brain EC Transcriptome
We sought to ask whether behaviorally motivated changes in neuronal activity would also elicit similar changes in brain EC gene expression as the chemogenetics. We utilized a behavioral paradigm in which whisker somatosensation induces robust firing of the neurons in the barrel cortex (Valles et al., 2011). Mice with whiskers intact were habituated to and permitted to explore a large, environmentally-enriched cage (+Whisker) whereas mice with their whiskers shaved off were habituated to and allowed to explore a large empty cage (−Whisker). The +Whisker group had significantly more cFos+ cells in the barrel cortex in comparison to the −Whisker group (Figure 6A, B), indicating that this behavioral paradigm is sufficient to modulate neuronal activity in the barrel cortex.
We performed RNA sequencing on FACS-purified brain ECs from the barrel cortices of the two groups (+Whisker and −Whisker) and found that 727 genes were significantly upregulated and 508 genes were significantly downregulated in the +Whisker ECs compared to −Whisker ECs (Figure 6C, Figure S2C, Supplemental File 2). We compared the significant changes observed through chemogenetics with those observed through the whisker stimulation paradigm. The brain EC gene expression changes after whisker-mediated increase in neuronal activity were highly correlated with the gene expression changes after DREADDs-mediated glutamatergic activation (r=0.718141), suggesting that the neuronal activity-regulated gene expression changes in brain ECs that we identified by DREADDs-mediated glutamatergic activation are physiologically relevant and represent a robust activity-dependent transcriptomic signature in the brain ECs (Figure 6D, E, S6). There was a large overlap of enriched pathways between the two experiments. Specifically, pathways involving adherens junctions and the cytoskeleton were also upregulated after whisker-mediated increase in neuronal activity (Table S8). In addition, pathways involving metabolism were also downregulated after whisker-mediated increase in neuronal activity (Table S7). Pathways uniquely regulated by whisker stimulation include downregulation of genes involved in the lysosome and glycolysis and upregulation of genes involved in the Src Homology-3 Domain and the Rap1 Signaling Pathway (Table S7, S8). Interestingly, the gene expression changes after whisker stimulation were not inversely correlated with the gene expression changes after DREADDs-mediated glutamatergic silencing (r=0.049543) suggesting that silencing of neuronal activity is likely an active form of signaling to the brain vasculature, rather than a lack of signaling from neuronal activity (Figure 6F, G, S6).
Interestingly, of 155 Slc transporters expressed above a threshold of 10 average CPM across all samples, 65 were significantly changed in at least one of the 3 experiments (Figure S7). Six genes were significantly regulated in the same direction after DREADDs-mediated glutamatergic activation and whisker-mediated increase in neuronal activity (Slc12a2, Slc12a9, Slc20a1, Slc25a24, Slc5a3, Slc9a3r2). These data suggest that brain ECs may plastically change expression of these transporters as a mechanism to meet neuronal circuit-specific energy and nutrient demands.
Most interestingly, we found that whisker stimulation downregulated expression of the same 5 ABC efflux transporters and the PAR bZip circadian clock-regulated transcription factors (Figure 6E) and had an overall similar effect on brain EC core circadian genes (Figure S4B). Thus both DREADDs-mediated manipulation of neuronal activity and the whisker stimulation volitional behavior paradigm had the same effect, in that the expression of the efflux transporter and PAR bZip transcription factors were inversely correlated with neuronal activity. Taken together, physiologically relevant changes in neuronal activity can regulate BBB efflux and EC circadian clock genes.
DISCUSSION
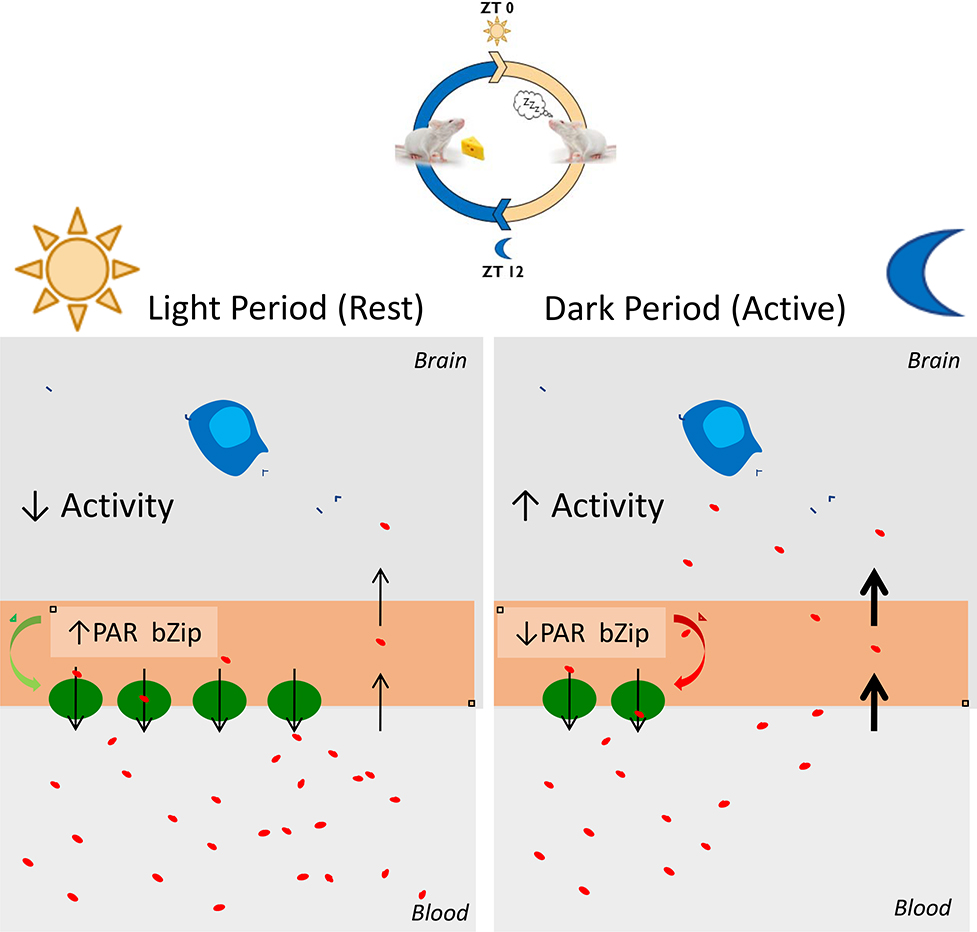
We report that neuronal activity regulates brain EC gene expression, and specifically that neuronal activity inhibits the expression of BBB ABC efflux transporters and PAR bZip circadian clock-regulated transcription factors. We further show that there is a diurnal oscillation to BBB efflux transporter expression and function, which is reduced by global triple knockout of the PAR bZip transcription factors or EC-specific deletion of one of the main circadian clock regulators Bmal1. In particular, we found that there is overall less BBB efflux during the dark period when mice are more active and more BBB efflux during the light period when mice are at rest (Figure 7). Our results also suggest neuronal activity is able to highjack elements of the EC clock molecular machinery to modulate BBB efflux transport.
Figure 7. Model of neuronal activity-dependent expression of EC circadian clock-regulated PAR bZip transcription factors which regulate BBB efflux transport.
Mice are more active during the night vs. day and thus exhibit more neuronal activity during the night. The expression of the PAR bZip transcription factors in brain ECs is inversely regulated by the amount of glutamatergic activity in the brain and their expression regulates the expression and function of BBB efflux transporters. Therefore, there is more BBB efflux during the day/rest period vs. the night/active period in mice. This may be important for maintaining neurochemical balance. Disrupting this system via sleep deprivation or dysfunctional EC circadian function could lead to neurochemical imbalance and impaired waste clearance.
We identified hundreds of brain EC genes, including those involved in transport, metabolism and focal adhesion, that were regulated by neuronal activity and it will be very interesting to determine how these activity-dependent genes may affect the local neural environment and thus circuit function. In addition, it will be interesting to understand the signaling mechanism(s) by which neuronal activity regulate(s) brain EC transcription. These mechanisms could be driven through different possibilities. First, metabolic byproducts from neuronal activity including glutamate, ATP, d-serine and potassium, could directly signal to brain ECs (Anwar et al., 1999; LeMaistre et al., 2012; Hogan-cann and Anderson, 2016; Longden et al., 2017; Mohamed et al., 2019). Second, astrocytes send out polarized processes that ensheathe both synapses and vessels, and thus may mediate activity-dependent changes in brain ECs (Petzold and Murthy, 2011). It is also possible that neurons and/or astrocytes signal to pericytes which in turn regulate brain EC gene expression (Fernández-Klett and Priller, 2015). Third, changes in EC shear stress that result from activity-dependent changes in blood flow may regulate brain EC transcription through luminal mechanosensation of differential shear stress via TRP channels, Piezo1, GPR68 or other EC mechanosensitive mechanisms (Li et al., 2014; Ranade et al., 2014; Baeyens and Schwartz, 2016; Xu et al., 2018). Interestingly, many focal adhesion genes that we found to be increased by neuronal activity are regulated by the mechanosensory ion channel Piezo1 in ECs including Actn1, Actn4, Flna, Flnb, Pls3, Vcl, Tubb2a, Tubb2b, and Tubb4b (Li et al., 2014).
In this study, the brain ECs were analyzed at a single time point after neuronal manipulation, but due to the dynamic nature of transcription and neuronal activity, it would also be interesting to investigate how neuronal activity affects brain EC transcription at multiple time points. The experiments described in this study likely identify “homeostatic” changes to brain ECs in response to altering neuronal activity, as we examined the gene expression changes hours after manipulating neuronal activity. It would be interesting to identify “dynamic” changes to BBB properties that would occur on the order of milliseconds to seconds. Such dynamic changes would likely not involve the alteration of the transcriptional profile of the ECs, but perhaps post-translational regulation including the trafficking of key transporters such as Pgp to and from the surface (McCaffrey et al., 2012; Noack et al., 2014; Tome et al., 2016).
The experiments described specifically analyzed glutamatergic activity since it is the major type of excitatory neurotransmission in the brain, but it would be interesting to see how other types of neuronal activity regulate brain EC gene expression and determine whether there are circuit-specific and regional-specific activity-dependent changes in the brain ECs. It is also worth noting that although we primarily analyzed brain EC transcriptomics and BBB function in the localized areas that we manipulated neuronal activity, based on previous findings and some of our own results, we expect that these changes are specific to these localized areas, and perhaps connected regions, rather than affecting the entire brain vascular network (Figure S3B) (Nishijima et al., 2010; Hrvatin et al., 2018).
Although there is substantial overlap in neuronal activity-dependent brain EC gene expression changes between our study and Hrvatin et al. 2018, we also newly identify many different genes not found in their study, especially many downregulated genes. We think the disparities in data could likely be due to: i) differences in the duration of neuronal activation, ii) our ability to both increase and decrease neuronal activity, iii) differences in sequencing depth between single cell RNA sequences and bulk RNA sequencing, especially when enriching for vascular cells. While we did achieve deep sequencing with presumably primarily microvascular brain ECs, performing scRNAseq in this paradigm would also be beneficial in elucidating how different ECs along the zonation of the brain vascular network respond to neuronal activity (Vanlandewijck et al., 2018).
Our work demonstrating the activity-dependent downregulation of efflux transport and other studies involving increased permeability to IGF-1 and increased transcytosis suggest that the BBB becomes less stringent after high neuronal activity perhaps as a mechanism to deliver more nutrients into the energy-demanding areas of the brain parenchyma (Nishijima et al., 2010; Sadeghian et al., 2018). It’s also possible that BBB efflux transport becomes less stringent during high neuronal activity as an evolutionary mechanism to preserve energy as both neuronal action potentials and active efflux transport require vast amounts of ATP, or two synchronize BBB efflux waste removal with the glymphatic system (Xie et al., 2013).
BBB efflux transporters have a broad range of exogenous substrates and thus act as a major obstacle for delivery of small molecule drugs to the CNS (Cordon-Cardo et al., 1989). These transporters also efflux endogenous substrates and act as a major extrusion route of waste products and metabolites from the brain into the bloodstream (Sweeney et al., 2018). We have previously found that Pgp is important for modulating concentrations of endogenous steroids in the brains of flies and mice and is important for behavior (Hindle et al., 2017). Because our work here shows that the expression and function of Pgp is regulated by neuronal activity, it is possible that BBB efflux and neuronal activity exhibit dynamic crosstalk to maintain the brain concentrations of these important neuromodulators. This would implicate the BBB as a dynamic component of the neuronal circuitry.
We also found that BBB efflux transport exhibits diurnal oscillation in mice which is consistent with the glial-based BBB of fruit flies and previous rodent experiments, suggesting that there is an evolutionarily conserved mechanism to control the amount of efflux throughout the day (Savolainen et al., 2016; Zhang et al., 2018). The diurnal permeability to Pgp substrates in the two organisms are inversely correlated which is consistent with the fact that flies are diurnal and mice are nocturnal when kept on a standard 12:12 hour light:dark cycle. Thus in both cases, the amount of efflux transport decreases with the activity of the organism. This phenomenon has important clinical implications for chronopharmacology, drug treatment that takes the body’s circadian rhythm into consideration as time of day variation may affect the efficiency of drug delivery to the CNS and thus efficacy within the brain. Our work further shows that this diurnal oscillation isn’t entirely controlled by cell intrinsic gene oscillation, as neuronal activity can modulate the levels and function of efflux through regulation of the circadian genes. This identifies that neuronal activity and behavior can modulate diurnal oscillatory functions by regulating the molecular clock machinery.
This study also has substantial clinical implications in the context of waste clearance from the brain and AD. The finding that BBB efflux inversely correlates with neuronal activity is consistent with previous studies that there is increased clearance of waste products such as amyloid beta (Aβ) from the brain during sleep (Xie et al., 2013). Although the results from that study specifically implicated a role of the glymphatic system, BBB transport, and specifically Pgp efflux, is also important in waste clearance and AD (Sweeney et al., 2018). AD patients have been shown to have decreased Pgp function in the brain (Van Assema et al., 2012; Deo et al., 2014), and Aβ has been shown to be cleared from the brain to the blood through abluminal LRP1 and luminal Pgp (Shibata et al., 2000; Cirrito et al., 2005; Pflanzner et al., 2011, 2012; Zhao et al., 2015; Bruckmann et al., 2016). It is also well-established that there is a substantial relationship between aberrant hyperactive neuronal activity and buildup of Aβ plaques in both mouse models of AD and humans with AD, although a mechanistic role for Pgp or the vasculature has not been explored in this context (Dickerson et al., 2005; Palop et al., 2007; Busche et al., 2008; Bero et al., 2011; Elman et al., 2014; Yuan and Grutzendler, 2016; Palmqvist et al., 2017). In addition, circadian rhythm is often disrupted in patients with Alzheimer’s disease (Musiek, Xiong and Holtzman, 2015). We found that increased neuronal activity downregulates expression of both Abcb1a and Lrp1 (Supplemental File 1), and thus, it is plausible that proper circadian balance of activity-dependent regulation of BBB efflux is important for clearing waste products such as Aβ from the brain during sleep when activity is lower and BBB efflux transport is higher. However, it has recently been demonstrated that vasomotion correlates with increased clearance of Aβ via paravascular pathways, highlighting the complexity of the interactions between glymphatic, lymphatic, vascular and phagocytotic pathways of clearance (van Veluw et al., 2020). Taken together, these findings suggest that disruption of circadian-dependent and neuronal activity-modulated BBB efflux may be a component of AD pathogenesis.
STAR METHODS
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Richard Daneman (rdaneman@ucsd.edu).
Materials Availability
This study did not generate new unique reagents. The bZip triple knockout mice are no longer being live bred anywhere to our knowledge. Brain mRNA from the triple knockouts were collected and stored previously independent of this study.
Data and Code Availability
All RNAseq data are provided as Supplemental Files and the raw data can be accessed on Gene Expression Omnibus (GEO) (Accession: GSE156689).
Experimental Models and Subject Details
All animal experiments were performed with national and UCSD IACUC guidelines.
Mouse Strains
CamKIIa-tTA mice (Mayford et al., 1996) were crossed to TRE-hM3Dq mice (Alexander et al., 2009) to generate a tool to activate glutamatergic neurons and TRE-M4Di mice (Alexander et al., 2009) to generate a tool to silence glutamatergic neurons. Littermates not expressing the DREADDs were used as controls. Bmal1 floxed mice (Storch et al., 2007) were crossed to VECadherin-CreERT2 mice (Sörensen, Adams and Gossler, 2009) to generate a tamoxifen-inducible, endothelial-specific, Bmal1 knockout mice. Rosa-lsl-tdTomato mice (Madisen et al., 2010) were crossed to VECadherin-CreERT2 mice to generate a tamoxifen-inducible, endothelial-specific, fluorescent red reporter mouse. Rpl22HA (RiboTag) (Sanz et al., 2009), VECadherin-CreERT2 and Bmal1 floxed mice were crossed to each other to generate tamoxifen-inducible, endothelial-RiboTagged, endothelial-specific, Bmal1 knockout mice. All three sets of mice were administered 3 daily intraperitoneal injection of tamoxifen (100μl of 20 mg/ml) at 5 weeks of age and all experiments involving these mice were performed at least 5 weeks post tamoxifen injection. All experiments involving other strains were performed in adult mice 3–4 months of age. PAR bZip triple knockout mice and corresponding wildtype controls were previously described (Gachon et al., 2004). Other wildtype animals were C57/BL6. All mice were kept on a standard 12:12 hour light:dark cycle. Only male mice were used for the transcriptomic experiments. Both males and females were used for all other experiments.
Method Details
Immunohistochemistry
Mice were anesthetized by i.p. injection of a ketamine/xylazine cocktail and then fixed via transcardial perfusion of D-PBS for 3 minutes, 4% paraformaldehyde (PFA) for 7–10 minutes, and again with D-PBS for 2 minutes using a Dynamax peristaltic pump. Speed was matched to typical cardiac output of a mouse. The brains were then dissected and submersion-fixed in 4% PFA overnight at 4°C. Brains were then submerged in 30% sucrose overnight at 4°C. Brains were then frozen in cryosectioning blocks in a solution consisting of 1:2 30% sucrose: OCT. 10–40μm sagittal sections were obtained using a cryostat.
Sections were stained floating in solution in the wells of a 12-well cell culture plate. They were blocked in a solution consisting of 5% goat serum and 0.1% Triton X-100 in PBS at room temperature for 45 minutes. They were then incubated in the blocking solution with the primary antibody, Rabbit-anti-HA 1/250 overnight at 4°C. They were then incubated in Goat-anti-Rabbit-Alexa 594 secondary antibody at room temperature for 90 minutes and then mounted on slides with DAPI Fluoromount-G for image processing.
For cFos immunostaining, 8 −12 μm coronal sections were obtained with a cryostat and cross referenced with a brain atlas to ensure sections included the barrel cortex or SCN. Sections were put on slides and stained directly on the slides with the same procedure described above using Goat-anti-cFos and 5% donkey serum + 0.1% Triton X-100 as the blocking and staining solution. The same methodology was used for all other staining.
Epifluorescence Imaging
Epifluorescence images of HA-immunostained slides were taken with an Axio Imager D2 (Carl Zeiss) with a 5x, 10x or 20x objective using a digital camera (Axiocam HRm, Carl Zeiss). AxioVision software was used to acquire images. Individual images were stitched together to gain a complete image of a sagittal section of the brain using the photomerge feature of Adobe Photoshop.
cFos Image Analysis
Once the barrel cortex or SCN was identified, a freehand ROI was drawn to exclusively capture the region and cFos-positive cells were quantified using ImageJ. A burned in scale bar was used to calibrate area. The following functions were used in sequential order: “Threshold Adjustment,” “Convert to Mask,” “Fill Holes,” “Watershed,” “Analyze particles”. Then “# cFos+ cells per mm2” was calculated by (Total # cFos-positive)/(Area). Three images (technical replicates) were averaged for each mouse (biological replicates). Similar methodology was used for Caspase-3-positive cell quantification except that positive cells were manually counted. Mice were picked randomly for each group and analyzed blind to genotype/group.
Middle Cerebral Artery Occlusion
To mimic stroke, we used tissue from a permanent focal cerebral ischemia model in mice as described previously (Munji et al., 2019). Briefly, the ischemia was induced by coagulating the distal portion of the left middle cerebral artery (MCA). Mice were anesthetized with 2.5% isoflurane. The left common carotid artery was isolated and temporarily ligated. A 2mm bar hole was made using a dental drill between the left orbit and ear. The distal portion of the left MCA was exposed and coagulated using a small vessel cauterizer (Fine Science Tools). The MCA was then transected. The ligation of the common carotid artery was released after 30 minutes of occlusion. Temperature was monitored rectally and maintained at approximately 37°C with a heating pad.
Multielectrode Array Electrophysiology
Electrophysiological procedures were performed as described previously (Koenig et al., 2011). Briefly, four-tetrode (bundles of four 17 micron platinum-iridium (90/10%) wires) microdrives were implanted stereotaxically (from bregma: −2.0 mm A/P, +1.8mm M/L, 0.00 mm D/V) into the cortex and hippocampus of adult DREADDs mice and littermate controls. Electrode tips were plated with platinum to reduce electrode impedances to between 150–250 kΩ at 1 kHz. The mice were given at least 1 week to recover with additional post-operative care administered as needed. For a given recording session, data were collected for 15 minutes while awake behaving mice were in their home cage to establish an internal control of the baseline level of neuronal activity. The mouse was then injected with the given dose of CNO (0.5 mg/kg or 1.0 mg/kg) i.p. Data were collected for 3 more hours post-injection in the home cage. A preamplifier, tether, and a 32-channel digital data acquisition system (Neuralynx, Inc.) was used. LFP was sampled at 32,000 Hz and filtered between 1 and 1,000 Hz. Wavelet power was calculated by first downsampling by a factor of 25 and filtered between 30–150 Hz. The average gamma power was calculated every 15 minutes beginning with the 15 minutes prior to the CNO injection (“preCNO”), and every 15 minutes following for 3 hours. The percentage of baseline wavelet power was calculated by taking each 15 minute data point and diving by the preCNO gamma power. Locomotion was tracked via head-mounted LEDs and a camera facing down onto the mouse cage. Average velocity was calculated by taking the instantaneous velocity using the x and y coordinates and averaging over every 15 minutes.
After all recordings were completed, mice were perfused with PBS followed by 4% paraformaldehyde in PBS solution. Brains were post-fixed for an additional 24 hours in 4% paraformaldehyde and then cryoprotected using a 30% sucrose solution for an additional 2 days. Brains were frozen and sliced into 40 μm coronal sections on a sliding microtome. Sections were mounted on electrostatic slides and stained with cresyl violet to visualize recording locations for validation of electrode placement.
FACS-purification of Brain Endothelial Cells
For a given experiment, mice were collected in pairs consisting of one DREADDs mouse and one littermate control mouse. Each DREADDs mouse and its littermate control was injected i.p. with the electrophysiologically-verified dose of CNO (0.5mg/kg for Activating mice and 1.0mg/kg for Silencing mice) at approximately ZT23-ZT24. For kainic acid experiments, 5 mg of kainic acid was dissolved in 2 ml of saline to obtain a working concentration of 2.5 mg/ml and saline was used as vehicle. One cKO and one littermate control were injected with 15 mg/kg of kainic acid and another pair of identical genotypes were injected with an equivalent volume of vehicle at approximately ZT23–24. Behavior was closely monitored for 3 hours and the experiment was aborted if behavioral seizures were observed. 3 hours post-injection the mice were live-decapitated using a mouse decapitator (LabScientific, XM-801). Brains were dissected out, the meninges were removed and the cortex and hippocampus were dissected out for further processing. The tissue was then diced using a #10 blade and enzymatically digested in Papain, 1 vial per on a 33° heat-block while being exposed to 95% oxygen, 5% carbon dioxide for 90 minutes. The tissue was then triturated and a second enzymatic digestion was performed in 1.0 mg/ml Collagenase Type 2 and 0.4 mg/ml Neutral Protease on a 33° heat-block while being exposed to 95% oxygen, 5% carbon dioxide for 30 minutes. Myelin was then removed as recommended with myelin removal beads using 30μm filters (MACS Miltenyi Biotec, 130–041-407) and LS columns (MACS Miltenyi Biotec, 130–042-401) on a MidiMACS separator (MACS Miltenyi Biotec, 130–042-302). The remaining single cell suspension was blocked with Rat IgG 1/100 for 20 minutes on ice. The samples were then stained with Rat-anti-CD31-Alexa 647, 1/100, mouse-anti-CD45-FITC 1/150, rat-anti-CD13-FITC 1/100, rat-anti-CD11b-FITC 1/100, rabbit-anti-NG2-Alexa 488 1/150, and DAPI for 20 minutes at 4°. CD31-positive cells were sorted into Trizol using an ARIA II instrument at the Flow Cytometry Core at the VA Hospital in La Jolla, CA. Mice were picked randomly for each group and analyzed blind to genotype/group.
Barrel Cortex Activity Behavior Experiment
The design of this experiment was based off previously described work with minor modifications (Valles et al., 2011). Briefly, for each experiment, 10 C57/BL6 mice were separated into 2 groups, 5 mice in the “+Whisker” group and 5 mice in the “-Whisker” group. All mice underwent “Habituation Period 1” for 7 days in which they were weighed, monitored and handled once daily. The mice then underwent “Habituation Period 2” for 5 days in which each group explored an empty standard rat cage for 30 minutes in the dark daily. Then, on the last day of Habituation Period 2 (Day 11), all mice were transiently anesthetized (2–3 minutes) with 1–3% isoflurane and the whiskers were clipped from the −Whisker mice using an electric hair trimmer. On the Test Day (Day 12), mice were added to their respective habituated rat cages in the dark at approximately ZT23 for a duration of 3 hours. The +Whisker mice had an enriched environment with novel objects which they were encouraged to explore with their whiskers due to the absence of light. Objects were switched and rearranged every 30 minutes to maintain novelty and exploration. −Whisker mice were kept in their empty habituated rat cage. Mice were then live-decapitated using a mouse decapitator.
The barrel cortex was then dissected out using a mouse brain matrix. A single coronal cut was made where the olfactory bulb meets the cortex. Two more posterior coronal cuts were made at 3mm intervals. The barrel cortex was then dissected from the 3mm coronal section by making cuts overlaid on a scaled-down image of the mouse brain atlas coronal section containing the barrel cortex. The meninges were removed from the dissected barrel cortex and the remaining tissue was pooled together from the 5 mice from each group. The pooled tissue was then processed further for FACS-purification as described above.
RNA-sequencing
RNA was harvested from the FACS-purified brain endothelial cells using the Qiagen RNeasy Microkit. RNA samples were then further processed at the UCSD Genomics Core using standard core procedures. The RNA was tested for quality and concentration using a tape station bioanalyzer. Next, cDNA libraries were made using the TruSeq RNA Library Prep Kit v2. Samples were then sequenced on an Illumina HiSeq4000, 100 based, paired ends.
Sequence reads for all samples were mapped to Ensembl mm9 v67 mouse whole genome using Tophat v 2.0.11 and Bowtie 2 v 2.2.1 with parameters no-coverage-search -m 2 -a 5 -p 7. Alignment files were sorted using SAMtools v.0.1.19. Count tables were generated using HTSeq-0.6.1. Differential expression of genes between control and treated samples, log 2-fold changes between control and treated samples, and statistical analysis including p values and FDR was performed using DESeq2 and Excel.
A gene was considered “downregulated” if the average CPMs of the control samples were ≥10 and the Wald Test p-value was <0.05. A gene was considered “upregulated” if the average CPMs of the experimental samples were ≥10 and the Wald Test p-value was <0.05.
Rhodamine123 Permeability Assay
Rhodamine123 (Rh123) was dissolved in DMSO to make a stock solution of 10mg/ml. For each experiment, a working solution of 2 mg/ml was made by diluting the stock solution with saline. For a given DREADDs experiment, mice were collected in pairs and injected with CNO at approximately ZT3-ZT4 as described in the FACS-purification section (Activating mice were injected with 0.45mg/kg CNO instead of 0.5mg/kg as they were more susceptible to seizures when also injected with Rh123). 2 hours post-CNO injection, the mice were then injected i.p. with 25mg/kg Rh123. For a given diurnal experiment, the mice were injected i.p. with Rh123 1.5 hours before the indicated time. 3.75 hours post-CNO injection and 15 minutes before the collection, the mice were then anesthetized by i.p. injection of ketamine/xylazine. Blood was collected via cardiac puncture and kept in an EDTA-coated tube rotating until the end of the tissue collection procedure. For the positive control Pgp inhibitor experiment, mice were injected with 10 mg/kg of Tariquidar, injected 1 hour later with Rh123, then collected 2 hours post-Rh123 injection with ketamine/xylazine anesthesia 15 minutes before collection. The mice were then perfused with D-PBS for 3 minutes using a Dynamax peristaltic pump. Brains were dissected, meninges removed and the cortex and hippocampus combined and cerebellums were then dissected out. Livers, spleens and lymph nodes were also dissected out. The tissue was flash frozen in liquid nitrogen and then stored at −80°C until Rh123 was extracted. The blood samples were then centrifuged. The supernatant (plasma) was collected and stored at −80°C until Rhodamine123 was extracted. For “Immune Cells,” the blood pellet underwent red blood cell lysis with ACK Lysing Buffer, washed with cold PBS and resuspended in cold PBS. The resulting solution was store at −80 until Rh123 was extracted.
Once Rh123 was ready to be extracted from the collected tissue, the brains were weighed and added to tubes with cold PBS (mass/volume-adjusted). They were then homogenized using a bead beater homogenizer. The homogenized tissue was eluted by centrifugation. The Rh123 extraction was performed on the homogenized brain tissue and plasma as described previously with slight modifications to adjust for input mass and volume (Banes et al., 1986). Briefly, n-butanol was added to the samples which were then vortexed. These steps were repeated once and the samples were incubated overnight at 4°C rotating. The samples were then vortexed, spun down and the supernatant was collected. The remaining pellet was then washed with an equivalent volume of n-butanol, the samples vortexed, spun down and the supernatant was collected. The samples were then added to a 96-well plate with 4 technical replicates per sample. A standard curve and blanks were also run to ensure that Rh123 measurements were above the detectable limit. The plate was analyzed on a Tecan Infinite plate reader (Excitation=505nm, Emission=560nm). Brain:Blood ratios were calculated for each sample by taking the average of the technical replicates. To directly compare Activating to Silencing groups, each DREADDs sample’s ratio was then normalized to its paired littermate control sample’s ratio to obtain the “Normalized Rh123 Fluorescence”. For this experiment and all subsequent permeability experiments, mice were picked randomly for each group and analyzed blind to genotype/group.
Daunorubicin Permeability Assay
Daunorubicin (DNR) was dissolved in DMSO to make a stock solution of 10mg/ml. For each experiment, a working solution of 2 mg/ml was made by diluting the stock solution with saline. For a given DREADDs experiment, mice and their corresponding cortices/hippocampi and blood were collected identically to the Rh123 experiment. However, whole blood was collected rather than plasma.
Brain tissue was homogenized identical the Rh123 extraction. The DNR extraction was performed on the homogenized brain tissue and whole blood as described previously with slight modifications (Bachur et al., 1970; Egorin et al., 1974). Briefly, 0.3 N HCl in 50% Ethanol was added to the samples which were then vortexed. These steps were repeated once and the samples were incubated overnight at 4°C rotating. The samples were then vortexed, spun down and the supernatant was collected. The remaining pellet was then washed with an equivalent volume of solvent, the samples vortexed, spun down and the supernatant was collected. Brain samples were evaporated in a fume hood under nitrogen gas. n-butanol was added to the evaporated tubes, vortexed, spun down and the supernatants collected. The brain and blood supernatants were then added to a 96-well plate with 4 technical replicates per sample. A standard curve and blanks were also run to ensure that DNR measurements were above the detectable limit. The plate was analyzed on a Tecan Infinite plate reader (Excitation=470nm, Emission=585nm). Brain:Blood ratios were calculated for each sample by taking the average of the technical replicates. To directly compare Activating to Silencing groups, each DREADDs sample’s ratio was then normalized to its paired littermate control sample’s ratio to obtain the “Normalized DNR Fluorescence”.
Sodium Fluorescein Permeability Assay
Sodium Fluorescein (NaFl) was dissolved in sterile PBS to make 10 mg/ml. For each experiment, a working solution of 2 mg/ml was made by diluting the stock solution with sterile PBS. Mice and their corresponding cortices/hippocampi and plasma were collected identically to Rhodamine123 permeability experiments (dosage was 25 mg/kg).
Brain tissue was homogenized identical the Rh123 extraction. The brain homogenates were then centrifuged and the resulting supernatant was diluted 1:1 in 2% TCA. Plasma samples were diluted 1:400 in sterile PBS, followed by an additional 1:1 dilution in 2% Trichloroacetic Acid (TCA). Brain and plasma samples were incubated overnight rotating at 4°C. Both sets of samples were then centrifuged and the supernatants were diluted 1:1 in borate buffer, pH 11. The samples were then added to a 96-well plate with 4 technical replicates per sample. A standard curve and blanks were also run to ensure that NaFl measurements were above the detectable limit. The plate was analyzed on a Tecan Infinite plate reader (Excitation=480nm, Emission=538nm). Data were analyzed identical to the rhodamine experiment.
Evan’s Blue Permeability Assay
Evan’s Blue was dissolved in sterile PBS to make 2% Evan’s Blue. For each experiment, each mouse was injected with 8.3x body mass of the 2% Evan’s Blue. Tissue collection was carried out identical to NaFl permeability experiments.
Brain tissue was homogenized identical the NaFl extraction. The brain homogenates were then centrifuged and the resulting supernatant was diluted 1:1 in 2% TCA. Plasma samples were diluted 1:400 in sterile PBS, followed by an additional 1:1 dilution in 2% TCA. Brain and plasma samples were incubated overnight rotating at 4°C. Both sets of samples were then centrifuged and the supernatants were diluted 1:1 in borate buffer, pH 11. The samples were then added to a 96-well plate with 4 technical replicates per sample. A standard curve and blanks were also run to ensure that Evan’s Blue measurements were above the detectable limit. The plate was analyzed on a Tecan Infinite plate reader (Excitation=620nm, Emission=680nm).
Pgp Targeted Mass Spectrometry Assay
4 Activating mice were injected with 0.45 mg/kg CNO and 4 Silencing mice were injected with 1.0 mg/kg CNO. They were then perfused and cortices/hippocampi were collected identically to the permeability experiments with the addition of protease inhibitors to the dissection solution. Frozen samples were then shipped to IQ Proteomics in Cambridge, MA for targeted mass spectrometry. 3 heavy synthetic Pgp peptide fragments (GAQLSGGQK, IATEAIENFR and STVVQLLER) were synthesized and acquired from New England Peptide. These peptides were chosen based on abundance of Pgp peptides from previous mouse whole brain shotgun proteomic experiments and availability from New England Peptide. Then a targeted liquid chromatography-mass spectrometry assay was performed on the Activating and Silencing tissue. The 3 heavy synthetic Pgp peptides were spiked into whole cell lysate tryptic digests and the relative light/heavy ratio via targeted parallel reaction monitoring was used to measure the endogenous expression of Pgp. Mice were picked randomly for each group and analyzed blind to genotype/group.
RiboTag Brain Endothelial Cell Purification
RiboTagged EC-Bmal1 cKO mice (VECadherin-CreERT2; Rpl22HA/+; Bmal1fl/fl) and RiboTagged littermate controls (VECadherin-CreERT2; Rpl22HA/+; Bmal1+/+) were live-decapitated at ZT0 and ZT12. Brains were dissected out, meninges were removed and the cortex and hippocampus were dissected out and flash frozen with liquid nitrogen and stored at −80 until further processing. Actively translating mRNA from brain endothelial cells were purified using RiboTag immunoprecipitation as described previously (Sanz et al., 2009). Briefly, thawed brain regions were homogenized using a dounce homogenizer. The brain homogenate was then spun down, Rabbit-HA antibody was added to the supernatant and it was gently rotated at 4° for 4 hours. Protein G magnetic beads were washed on a magnetic column and then added to the antibody-homogenate mix and gently rotated at 4° overnight. Bead-antibody-homogenate mix was washed 3 times on a magnetic column, lysed with RLT Lysis Buffer + 1% 2-mercaptaethanol and spun down to remove any remaining beads. Mice were picked randomly for each group and analyzed blind to genotype/group.
RT-qPCR
Brain homogenate RNA from PAR bZip triple knockout mice and respective littermate controls was purified as described previously (Gachon et al., 2004). Approximately 100 mg of frozen brain was grinded in extraction buffer (3.9 M Guanidium thiocyanate, 0.03 M Sodium citrate, 0.2 M Sodium acetate, 1% (v/v) 2-Mercaptoethanol) using a Polytron PT 2500 E homogenizer. An equal volume of phenol (saturated in H2O) and 0.5 volume of chloroform/isoamylalcohol (49:1 (v/v)) were added to the homogenate. The mixture was vigorously vortexed subsequently. Phase separation was accomplished by a centrifugation step at 4°C 12,000 g for 20 min. RNA of the aqueous phase was precipitated at −20°C during at least 20 min using an equal volume of isopropanol. The precipitate was pelleted by centrifugation at 12,000 g for 15 min at 4°C. Subsequently, the pellet was resuspended in 4M LiCl and subsequently re-pelleted during a centrifugation step of 12,000 g for 15 min at 4°C. The pellet was washed with 75% ethanol with a subsequent centrifugation at 12,000 g for 15 min at 4°C. The washed pellet was then dried at room temperature and dissolved in RNAse/DNAse free water.
RNA was purified from brain homogenates, Ribotag-purified brain endothelial cells and FACS-purified brain endothelial cells from EC-Bmal1 cKOs and littermate controls using the Qiagen RNeasy Microkit. RNA was reverse transcribed to cDNA using iScript Reverse Transcription Supermix. qPCR was performed using SYBR Green Master Mix with Primetime qPCR primers from Integrated DNA Technologies. Relative gene expression of Abcb1a, Dbp, Tef and Hlf was calculated using the ΔΔCt method normalizing to GAPDH for diurnal experiments and Rps20 for the kainic acid neuronal activity experiment.
Validation of Endothelial-Specific Bmal1 Knockout
Endothelial cells and non-endothelial cell populations were FACS-purified from the brains of VECadherin-CreERT2; Bmal1fl/fl mice and littermate controls (Bmal1fl/fl). The cell populations were lysed with Proteinase K in DirectPCR Lysis Reagent, then incubated rotating overnight at 55°C. Lysates were incubated at 85°C for 45 minutes to inhibit the Proteinase K. Multiplex PCR was performed using primers L1, L2 and R4 as described previously (Storch et al., 2007).
Food and Water Intake Rhythmic Behavior
Mice were single-housed with ad libitum access to food and water in a standard 12:12 hour light-dark cycle. Food intake was measured as weight of food in grams to the nearest 0.1 gram. Daily spillage of food has been shown to not exceed 0.1g and therefore considered negligible (Bachmanov et al., 2001). Water intake was measured as the displacement of water in a cylindrical tube fixed with a drinking spout. Measurements were taken at ZT0 and ZT12 every day over a period of 4 days. Mice were picked randomly for each group and analyzed blind to genotype/group.
Rhythmicity Analysis
Rhythmic analysis between different conditions was performed as described previously with minor modifications (Weger et al., 2018). Briefly, we applied multiple linear regressions on the data and subsequently performed a model selection using the Bayesian information criterion (BIC). The function was defined as y(t) = μ + αcos((2π/24 h)t) + βsin((2π/24 h)t) + noise, where y is the log transformed signal, μ is the mean, t is Zeitgeber time and α and β are the coefficients of the cosine and sine functions. As we observed a non-rhythmic increase of signal in the permeability assays, we added a coefficient γ to the function that accounts for non-periodic time-dependent changes when we analyzed these experiments [y(t) = μ + γ⋅t + αcos((2π/24 h)t) + βsin((2π/24 h)t) + noise]. To compare rhythmicity between the wild-type and KO mouse models, we generated five different models with differing α and β accounting for rhythmic and non-rhythmic patterns in each condition. Each of the models were fitted to the data using linear regression and model complexity was subsequently controlled by a BIC based model selection (Kass and Raftery, 1995). Schwarz weight (wj) was used to assess the confidence of all fitted models. The model with the highest BIC weight (BICW) was only considered to reflect the measurements if a threshold of 0.4 was reached.
Quantification and Statistical Analysis
The statistical details of experiments can be found in figure legends. Rhythmicity Analysis was performed using R version 3.5.0. The clustering heat map was generated using R version 3.4.3. Binned heat maps were generated using Graphpad Prism 7. All other heat maps were generated using Morpheus. XY scatter plots were generated using Excel and Student’s t tests were performed using Excel. Wald’s Test was performed using DESeq2.
Supplementary Material
Supplemental File 1. Master File with all Processed RNAseq Data in the 3 Experiments, Related to Figures 2, 3 and 6
Supplemental File 2. Processed RNAseq Data for All Gene Expression Changes in the 3 Experiments, Related to Figures 2, 3 and 6
Supplemental File 3. Processed RNAseq Data for Genes Regulated in Opposing Directions between DREADDs Activating and DREADDs Silencing, Related to Figures 2 and 3
Supplemental File 4. Processed RNAseq Data for Blood-Brain Barrier Genes, Related to Figures 2, 3 and 6
Key Resources Table.
| REAGENT | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-HA | Cell Signaling Technology | Cat. # 3724S |
| Goat anti-Rabbit-Alexa 488 | ThermoFisher | Cat. # R37116 |
| Goat anti-Rabbit-Alexa 594 | ThermoFisher | Cat. # R37117 |
| Goat anti-Rat-Alexa 594 | ThermoFisher | Cat. # A-11007 |
| Goat anti-cFos | Santa Cruz Biotechnology | Cat. # sc-52-G |
| Donkey anti-Goat-Alexa 594 | ThermoFisher | Cat. # A-11058 |
| Rat anti-CD31 | BD Pharminegen | Cat. # 553370 |
| Rat anti-CD31-Alexa 647 | Molecular Probes | Cat. # A14716 |
| Mouse anti-CD45-FITC | eBioscience | Cat. # 11–0451–85 |
| Rat anti-CD13-FITC | BD Pharmingen | Cat. # 558744 |
| Rat anti-CD11b-FITC | eBioscience | Cat. # 11–0112–81 |
| Rabbit anti-NG2-Alexa 488 | Bioss | Cat. # bs-11192R-A488 |
| Mouse anti-NeuN | Abcam | Cat. # ab104224 |
| Rabbit anti-BMAL1 | Novus Biologicals | Cat. # NB100–2288 |
| Rat anti-HA | Roche | Cat. # 11867423001 |
| Rat anti-CD45 | Bio-Rad | Cat. # MCA1031GA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| D-PBS | Gibco | Cat. # 14190–144 |
| DAPI Fluoromount-G | SouthernBiotech | Cat. # 0100–20 |
| Clozapine-N-oxide | Enzo | Cat. # BML NS105–0005 |
| Papain | Worthington Biochemical | Cat. # LK003176 |
| Collagenase Type 2 | Worthington Biochemical | Cat. # LS004176 |
| Neutral Protease | Worthington Biochemical | Cat. # LS02104 |
| Myelin removal beads | MACS Miltenyi Biotec | Cat. # 130–096-433 |
| Rat IgG | Sigma Aldrich | Cat. # I8015 |
| Trizol | Invitrogen | Cat. # 15596026 |
| Rhodamine123 | Sigma Aldrich | Cat. # 83702 |
| Sodium Fluorescein | Sigma Aldrich | Cat. # F6377 |
| Borate buffer, pH 11 | Sigma Aldirch | Cat. # 33650 |
| Proteinase K | Viagen | Cat. # 501-PK |
| DirectPCR Lysis Reagent | Viagen | Cat. # 102-T |
| Daunorubicin | Sigma Aldrich | Cat. # 30450 |
| Kainic Acid | Santa Cruz Biotechnology | Cat. # sc-200454 |
| Tariquidar | ThermoFisher | Cat. # 501365577 |
| Evan’s Blue | Sigma Aldrich | Cat. # E2129 |
| ACK Lysis Buffer | Quality Biological | Cat. # 118–156-101 |
| Pierce™ Protein G Magnetic Beads | ThermoFisher | Cat. # 88847 |
| Fluorescein labeled Griffonia (Bandeiraea) Simplicifolia Lectin I | Vector Laboratories | Cat. # FL-1101 |
| Critical Commercial Assays | ||
| RNeasy Microkit | Qiagen | Cat. # 74004 |
| TruSeq RNA Library Prep Kit v2 | Illumina | Cat. # RS-122–2001 |
| iScript Reverse Transcription Supermix | Bio-Rad | Cat. # 1708841 |
| SYBR Green Master Mix | Applied Biosystems | Cat. # 4367659 |
| Deposited Data | ||
| Raw and analyzed RNAseq data | This paper | GEO: GSE156689 |
| Experimental Models: Organisms/Strains | ||
| Conventionally raised mice (male) (C57BL/6 background) | Envigo | C57BL/6 |
| CamKIIa-tTA mice (male and female) (C57BL/6 background) | The Jackson Laboratory | JAX: 007004 |
| TRE-hM3Dq mice (male and female) (C57BL/6 background) | The Jackson Laboratory | JAX: 014093 |
| TRE-hM4Di mice (male and female) (C57BL/6 background) | The Jackson Laboratory | JAX: 024114 |
| VECadherin-CreERT2 mice (male and female) (mixed background mated 6 generations to C57BL/6) | Dr. Ralf Adams (Max Planck Institute for Molecular Biomedicine) (Sörensen, Adams and Gossler, 2009) | N/A |
| Bmal1fl/fl mice (male and female) (C57BL/6 background) | The Jackson Laboratory | JAX: 007668 |
| Rpl22HA/+ (Ribotag) (female) (C57BL/6NCrl background) | The Jackson Laboratory | JAX: 011029 |
| Rosa-lsl-tdTomato mice (male and female) (C57BL/6 background) | The Jackson Laboratory | JAX: 007909 |
| PAR bZip Triple KO mice and corresponding wildtype mice (male and female) (mixed background) | Brain RNA from Dr. Frédéric Gachon, The University of Queensland, Australia (Gachon et al., 2004) | N/A |
| Oligonucleotides | ||
| Bmal1 L1 Forward: ACTGGAAGTAACTTTATCAAACTG | Weitz Lab (Storch et al., 2007) | N/A |
| Bmal1 L2 Reverse: CTGACCAACTTGCTAACAATTA | Weitz Lab (Storch et al., 2007) | N/A |
| Bmal1 R4 Forward: CTCCTAACTTGGTTTTTGTCTGT | Weitz Lab (Storch et al., 2007) | N/A |
| Abcb1a Primetime® qPCR Primer | Integrated DNA Technology | Mm.PT.56a.33328663 |
| GAPDH Primetime® qPCR Primer | Integrated DNA Technology | Mm.PT.39a.1 |
| Rps20 Primetime® qPCR Primer | Integrated DNA Technology | Mm.PT.58.41623895.g |
| Dbp Primetime® qPCR Primer | Integrated DNA Technology | Mm.PT.58.16911772 |
| Tef Primetime® qPCR Primer | Integrated DNA Technology | Mm.PT.58.6941379 |
| Hlf Primetime® qPCR Primer | Integrated DNA Technology | Mm.PT.58.30817147 |
| Software and Algorithms | ||
| Zeiss | Axiovision 4.8 | https://www.zeiss.com/microscopy/us/products/microscope-software/axiovision.html |
| Photoshop | Adobe | https://www.adobe.com/products/photoshop.html |
| ImageJ | ImageJ | https://imagej.net |
| Neuralynx | Neuralynx, Inc. | https://neuralynx.com/ |
| Tophat v 2.0.11 | John Hopkins University | https://ccb.jhu.edu/software/tophat |
| Bowtie 2 v 2.2.1 | John Hopkins University | http://bowtie-bio.sourceforge.net/bowtie2 |
| SAMtools v.0.1.19 | (Li et al., 2009) | http://samtools.sourceforge.net/ |
| HT-Seq-0.6.1 | (Anders, Pyl and Huber, 2015) | https://htseq.readthedocs.io/en/release_0.11.1/ |
| DESeq2 | (Love, Huber and Anders, 2014) | https://doi.org/10.18129/B9.bioc.DESeq2 |
| Morpheus | Broad Institute | https://software.broadinstitute.org/morpheus/ |
| Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Highlights.
Neuronal activity robustly regulates gene expression in brain endothelial cells
Neuronal activity regulates BBB efflux transport and endothelial circadian genes
Efflux transport rhythmically correlates with neuronal activity across the day
Neuronal activity regulates BBB efflux via modulation of endothelial circadian genes
ACKNOWLEDGEMENTS
This work was in part supported by the UCSD Graduate Training Program in Cellular and Molecular Pharmacology through an institutional training grant from the National Institute of General Medical Sciences, T32 GM007752 and NIH/NINDS R01 NS091281 Diversity Supplement (R.S.P.). This work was also funded by NIH/NINDS R01 NS091281, Rita Allen Foundation, Klingenstein-Simons Foundation, and CureAlz Foundation (R.D.). The authors would like to acknowledge the UCSD Genomics Core and the VA Flow Cytometry Core in La Jolla, CA. We would also like to acknowledge IQ Proteomics in Cambridge, MA for performing the targeted mass spectrometry assay.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abbott NJ (2002). Astrocyte-endothelial interactions and blood-brain barrier permeability. Journal of Anatomy. 200, 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander GM, Rogan SC, Abbas AI, Armbruster BN, Pei Y, Allen JA, Nonneman RJ, Hartmann J, Moy SS, Nicolelis MA, et al. (2009). Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 63(1), 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq – a Phython framework to work with high-throughput sequencing data. Bioinformatics. 31(2), 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anea CB, Zhang M, Chen F, Ali MI, Hart CMM, Stepp DW, Kovalenkov YO, Merloiu A, Pati P, Fulton D, et al. (2013). Circadian Clock Control of Nox4 and Reactive Oxygen Species in the Vasculature, PLoS ONE. 8(10): e78626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar Z, Albert JL, Gubby SE, Boyle JP, Roberts JA, Webb TE, and Boarder MR (1999). Regulation of cyclic AMP by extracellular ATP in cultured brain capillary endothelial cells. British journal of pharmacology. 128(2), 465–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik A, Genové G, Mäe M, Nisancioglu MH, Wallgard E, Niaudet C, He L, Norlin J, Lindblom P, Strittmatter K, et al. (2010). Pericytes regulate the blood-brain barrier. Nature. 468(7323), 557–61. [DOI] [PubMed] [Google Scholar]
- Bachmanov AA, Reed DR, Tordoff MG, Price RA, and Beauchamp GK (2001). Nutrient preference and diet-induced adiposity in C57BL/6ByJ and 129P3/J mice. Physiology and Behavior. 72(4), 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachur NR, Moore AL, Bernstein JG, and Liu A (1970). Tissue distribution and disposition of daunomycin (NCS-82151) in mice: fluorometric and isotopic methods.. Cancer Chemotherapy Reports. 54(2), 89–94. [PubMed] [Google Scholar]
- Baeyens N and Schwartz MA (2016). Biomechanics of vascular mechanosensation and remodeling. Molecular Biology of the Cell. 27(1), 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banes AJ, Link GW, Beckman WC, Camps JL, and Powers SK (1986). High-performance liquid chromatographic quantitation of rhodamines 123 and 110 from tissues and cultured cells. Journal of Chromatography A. 356, 301–309. [DOI] [PubMed] [Google Scholar]
- Bero AW, Yan P, Roh J, Cirrito JR, Stewart FR, Raichle ME, Lee J, and Holtzman DM (2011). Neuronal activity regulates the regional vulnerability to amyloid-β deposition. Nature Neuroscience. 14(6), 750–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz AL and Goldstein GW (1978). Polarity of the blood-brain barrier: neutral amino acid transport into isolated brain capillaries. Science. 202(4364), 225–227. [DOI] [PubMed] [Google Scholar]
- Brancaccio M, Edwards MD, Patton AP, Smyllie NJ, Chesham JE, Maywood ES, and Hastings MH (2019). Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science, 363(6423), 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckmann S, Brenn A, Grube M, Niedrig K, Holtfreter S, von Bohlen und Halbach O, Groschup M, Keller M, and Vogelgesang S (2017). Lack of P-glycoprotein Results in Impairment of Removal of Beta-Amyloid and Increased Intraparenchymal Cerebral Amyloid Angiopathy after Active Immunization in a Transgenic Mouse Model of Alzheimer’s Disease. Current Alzheimer Research. 14(6), 656–667. [DOI] [PubMed] [Google Scholar]
- Busche MA, Eichhoff G, Adelsberger H, Abramowski D, Wiederhold K, Haass C, Staufenbiel M, Konnerth A, and Garaschuk O (2008). Clusters of hyperactive neurons near amyloid plaques in a mouse model of Alzheimer’s disease. Science. 321(5896), 1686–9. [DOI] [PubMed] [Google Scholar]
- Chow BW, Nuñez V, Kaplan L, Granger AJ, Bistrong K, Zucker HL, Kumar P, Sabatini BL, and Gu C (2020). Caveolae in CNS arterioles mediate neurovascular coupling. Nature. 579(7797), 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirrito JR, Deane R, Fagan AM, Spinner ML, Parsadanian M, Finn M, Jiang H, Prior JL, Sagare A, et al. (2005). P-glycoprotein deficiency at the blood-brain barrier increases amyloid-beta deposition in an Alzheimer disease mouse model. Journal of Clinical Investigation. 115(11), 3285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordon-Cardo C, O’Brien JP, Casals D, Rittman-Grauer L, Biedler JL, Melamed MR, and Bertino JR (1989). Multidrug-resistance gene (P-glycoprotein) is expressed by endothelial cells at blood-brain barrier sites. Proceedings of the National Academy of Sciences of the United States of America. 86(2), 695–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie T, Mongrain V, Dorsaz S, Mang GM, Emmenegger Y, and Franken P (2013). Homeostatic and Circadian Contribution to EEG and Molecular State Variables of Sleep Regulation. Sleep. 36(3), 311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis AM, Cheng Y, Kapoor S, Reilly D, Price TS, and Fitzgerald GA (2007). Circadian variation of blood pressure and the vascular response to asynchronous stress. Proceedings of the National Academy of Sciences of the United States of America. 104(9), 3450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman R, Zhou L, Kebede AA, and Barres BA (2010). Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 468(7323), 562–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo AK, Borson S, Link JM, Domino K, Eary JF, Ke B, Richards TL, Mankoff DA, Minoshima S, O’Sullivan F, et al. (2014). Activity of P-Glycoprotein, a β-Amyloid Transporter at the Blood-Brain Barrier, Is Compromised in Patients with Mild Alzheimer Disease. Journal of Nuclear Medicine. 55(7), 1106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Salat DH, Greve DN, Chua EF, Rand-Giovannetti E, Rentz DM, Bertram L, Mullin K, Tanzi RE, Blacker D, et al. (2005). Increased hippocampal activation in mild cognitive impairment compared to normal aging and AD. Neurology. 65(3), 404–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan DJ, Crossland RF and Bryan RM (2017). The rat cerebral vasculature exhibits time-of-day-dependent oscillations in circadian clock genes and vascular function that are attenuated following obstructive sleep apnea. Journal of Cerebral Blood Flow and Metabolism. 37(8), 2806–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorin MJ, Hildebrand RC, Cimino EF, and Bachur NR (1974). Cytofluorescence Localization Of Adriamycin And Daunorubicin. Cancer Research. 34, 2243–2245. [PubMed] [Google Scholar]
- Elman JA, Oh H, Madison CM, Baker SL, Vogel JW, Marks SM, Crowley S, O’Neil JP, and Jagust WJ (2014). Neural compensation in older people with brain amyloid-β deposition. Nature Neuroscience. 17(10), 1316–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt B (2008). Immune cell entry into the central nervous system: involvement of adhesion molecules and chemokines. J Neurol Sci, 274(1–2), 23–6. [DOI] [PubMed] [Google Scholar]
- Fernández-Klett F and Priller J (2015). Diverse functions of pericytes in cerebral blood flow regulation and ischemia. Journal of Cerebral Blood Flow and Metabolism. 35(6), 883–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M (2010). Molecular basis of the core structure of tight junctions. Cold Spring Harbor Perspectives in Biology. 2(1), a002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Fonjallaz P, Damiola F, Gos P, Kodama T, Zakany J, Duboule D, Petit B, Tafti M, and Schibler U (2004). The loss of circadian PAR bZip transcription factors results in epilepsy. Genes and Development. 18(12), 1397–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Olela FF, Schaad O, Descombes P, and Schibler U (2006). The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metabolism. 4(1), 25–36. [DOI] [PubMed] [Google Scholar]
- Galbraith GG, Skalak R and Chien S (1998). Shear stress induces spatial reorganization of the endothelial cell cytoskeleton. Cell Motility and the Cytoskeleton. 40(4), 317–330. [DOI] [PubMed] [Google Scholar]
- Hallmann R, Mayer DN, Berg EL, Broermann R, and Butcher EC (1995). Novel mouse endothelial cell surface marker is suppressed during differentiation of the blood brain barrier. Dev Dyn. 202(4), 325–32. [DOI] [PubMed] [Google Scholar]
- Hindle SJ, Munji RN, Dolghih E, Gaskins G, Orng S, Ishimoto H, Soung A, DeSalvo M, Kitamoto T, Keiser MJ, et al. (2017). Evolutionarily Conserved Roles for Blood-Brain Barrier Xenobiotic Transporters in Endogenous Steroid Partitioning and Behavior. Cell Reports, 21(5), 1304–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan-cann AD and Anderson CM (2016). Physiological Roles of Non- Neuronal NMDA Receptors. Trends in Pharmacological Sciences. 37(9), 750–767. [DOI] [PubMed] [Google Scholar]
- Hoshi Y, Uchida Y, Kuroda T, Tachikawa M, Couraud P, Suzuki, Terasaki T (2019). Distinct roles of ezrin, radixin and moesin in maintaining the plasma membrane localizations and functions of human blood–brain barrier transporters. Journal of Cerebral Blood Flow and Metabolism. 40(7), 1533–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, Zilionis R, Ratner A, Borges-Monroy R, Klein AM, et al. (2018). Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nature Neuroscience. 21, 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson N, Celkova L, Hopkins A, Greene C, Storti F, Ozaki E, Fahey E, Theodoropoulou S, Kenna PF, Humphries MM, et al. (2019). Dysregulated claudin-5 cycling in the inner retina causes retinal pigment epithelial cell atrophy. JCI Insight. 4(15), e130273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzer RC and Raff MC (1987). Astrocytes induce blood-brain barrier properties in endothelial cells. Nature. 325(6101), 253–257. [DOI] [PubMed] [Google Scholar]
- Kaplan L, Chow BW and Gu C (2020). Neuronal regulation of the blood–brain barrier and neurovascular coupling. Nature Reviews Neuroscience. 21(8), 416–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass RE and Raftery AE (1995). Bayes factors. Journal of the American Statistical Association. 90(430), 773–795. [Google Scholar]
- Koenig J, Linder AN, Leutgeb JK, and Leutgeb S (2011). The spatial periodicity of grid cells is not sustained during reduced theta oscillations. Science. 332(6029), 592–5. [DOI] [PubMed] [Google Scholar]
- Kon N, Yoshikawa T, Honma S, Yamagata Y, Yoshitane H, Shimizu K, Sugiyama Y, Hara C, Kaemshita I, Honma K and Fukada Y (2014). CaMKII is essential for the cellular clock and coupling between morning and evening behavioral rhythms. Genes and Development. 28(10), 1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste B, Comin CH, Ben-Zvi A, Kaeser PS, Xu X, Costa LF, Gu C (2014). Sensory-Related Neural Activity Regulates the Structure of Vascular Networks in the Cerebral Cortex. Neuron. 83(5), 117–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25(16), 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hou B, Tumova S, Muraki K, Bruns A, Ludlow MJ, Sedo A, Hyman AJ, McKeown L, Young RS, et al. (2014). Piezo1 integration of vascular architecture with physiological force. Nature. 515(7526), 279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longden TA, Dabertrand F, Koide M, Gonzales AL, Tykocki NR, Brayden JE, Hill-Eubanks D, and Nelson MT (2017). Capillary K+-sensing initiates retrograde hyperpolarization to increase local cerebral blood flow. Nature Neuroscience. 20(5), 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12), 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh S, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neuroscience. 13(1), 133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, and Kandel ER (1996). Control of memory formation through regulated expression of a CaMKII transgene. Science. 274(5293), 1678–83. [DOI] [PubMed] [Google Scholar]
- McCaffrey G, Staatz WD, Sanchez-Covarrubias L, Finch JD, Demarco K, Laracuente M, Ronaldson PT, and Davis TP (2012). P-glycoprotein trafficking at the blood-brain barrier altered by peripheral inflammatory hyperalgesia. Journal of Neurochemistry. 122(5), 962–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H, Hu Y, Shiu Y, Yuan S, Zhao Y, Kaunas R Wang Y, Jin G, Usami S, and Chien S (2005). Effects of flow patterns on the localization and expression of VE-cadherin at vascular endothelial cell junctions: in vivo and in vitro investigations. Journal of Vascular Research. 42(1), 77–89. [DOI] [PubMed] [Google Scholar]
- Mohamed LA, Markandaiah S, Bonanno S, Pasinelli P, and Trotti D (2019). Excess glutamate secreted from astrocytes drives upregulation of P-glycoprotein in endothelial cells in amyotrophic lateral sclerosis. Experimental Neurology. 316, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrain V, La Spada F, Curie T, and Franken P (2011). Sleep loss reduces the DNA-binding of BMAL1, CLOCK, and NPAS2 to specific clock genes in the mouse cerebral cortex. PLoS ONE. 6(10), e26622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munji RN, Soung AL, Weiner GA, Sohet F, Semple BD, Trivedi A, Gimlin K, Kotoda M, Korai M, Aydin S, et al. (2019) Profiling the mouse brain endothelial transcriptome in health and disease models reveals a core blood-brain barrier dysfunction module. Nature Neuroscience. 22(11), 1892–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiek ES, Xiong DD, and Holtzman DM (2015). Sleep, circadian rhythms, and the pathogenesis of Alzheimer disease. Experimental & molecular medicine. 47(3), e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nersesyan H, Herman P, Erdogan E, Hyder F, Blumenfeld. (2004). Relative changes in cerebral blood flow and neuronal activity in local microdomains during generalized seizures. Journal of Cerebral Blood Flow and Metabolism. 24(9), 1057–68. [DOI] [PubMed] [Google Scholar]
- Nishijima T, Piriz J, Duflot S, Fernandez AM, Gaitan G, Gomez-Pinedo U, Garcia Verdugo JM, Leroy F, Soya H, Nuñez A, and Torres-Aleman I (2010). Neuronal Activity Drives Localized Blood-Brain-Barrier Transport of Serum Insulin-like Growth Factor-I into the CNS. Neuron. 67(5), pp. 834–46. [DOI] [PubMed] [Google Scholar]
- Noack A, Noack S, Hoffmann A, Maalouf K, Buettner M, Couraud O, Romero IA, Weksler B, Alms D, Römermann K, et al. (2014). Drug-induced trafficking of P-glycoprotein in human brain capillary endothelial cells as demonstrated by exposure to mitomycin C. PLoS ONE. 9(2), e88154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist S, Schöll M, Strandberg O, Mattsson N, Stromrud E, Zetterberg H, Blennow K, Landau S, Jagust W, and Hansson O (2017). Earliest accumulation of β-amyloid occurs within the default-mode network and concurrently affects brain connectivity. Nature Communications. 8(1), 1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palop JJ, Chin J, Roberson ED, Wang J, Thwin MT, Bien-Ly N, Yoo J, Ho KO, Yu G, Kreitzer A, et al. (2007). Aberrant Excitatory Neuronal Activity and Compensatory Remodeling of Inhibitory Hippocampal Circuits in Mouse Models of Alzheimer’s Disease. Neuron. 55(5), 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardridge WM (2007). Blood-brain barrier delivery Drug Discovery Today. 12(1–2), 54–61. [DOI] [PubMed] [Google Scholar]
- Petzold GC and Murthy VN (2011). Role of astrocytes in neurovascular coupling. Neuron 71, 782–796. [DOI] [PubMed] [Google Scholar]
- Pflanzner T, Janko MC, André-Dohmen B, Reuss S, Weggen S, Roebroek AJM, Kuhlmann RW, and Pietrzik CU (2011). LRP1 mediates bidirectional transcytosis of amyloid-β across the blood-brain barrier. Neurobiology of Aging. 32(12), 2323.e1–11. [DOI] [PubMed] [Google Scholar]
- Pflanzner T, Petsch B, André-Dohmen B, Müller-Sciffmann A, Tschickardt S, Weggen S, Stitz L, Korth C, and Pietrzik CU (2012). Cellular prion protein participates in amyloid-β transcytosis across the blood-brain barrier. Journal of Cerebral Blood Flow and Metabolism. 32(4), 628–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacheck WJ, Kutys ML, Yang J, Eyckmans J, Wu Y, Vasavada H, Hirschi KK, and Chen CS (2017). A non-canonical Notch complex regulates adherens junctions and vascular barrier function. Nature. 552(7684), 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, and Schibler U (2002). The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 110(2), 251–60. [DOI] [PubMed] [Google Scholar]
- Profaci CP, Munji RN, Pulido RS, and Daneman R (2020). The blood–brain barrier in health and disease: Important unanswered questions. Journal of Experimental Medicine. 217(4), e20190062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambousek L, Gschwind T, Lafourcade C, Paterna J, Dib L, Fritschy J, and Fontana A (2020). Aberrant expression of PAR bZIP transcription factors is associated with epileptogenesis, focus on hepatic leukemia factor. Scientific Reports. 10, 3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade SS, Qiu Z, Woo S, Hur SS, Murthy SE, Cahalan SM, Xu J, Mathur J, Bandell M, Coste B, et al. (2014). Piezo1, a mechanically activated ion channel, is required for vascular development in mice. Proceedings of the National Academy of Sciences of the United States of America. 111(28), 10347–10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J, Diaz AN, and Gumz ML (2014). Clock genes in hypertension: Novel insights from rodent models. Blood Pressure Monitoring. 19(5), 249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J and Gumz ML (2013). Invited Review EB 2012: Mechanism of the Circadian Clock in Physiology. American journal of physiology. Regulatory, integrative and comparative physiology. 304(12), R1052–R1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Shearman LP, Reppert SM, and Schibler U (2000). CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes and Development. 14, 679–689. [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, and Schibler U (2001). Circadian regulation of gene expression in animals. Current Opinion in Cell Biology. 13(3), 357–62. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, and Schibler U (2006). Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nature Genetics. 38(3), 369–74. [DOI] [PubMed] [Google Scholar]
- Roy CS, and Sherrington CS (1890). On the Regulation of the Blood-supply of the Brain. The Journal of Physiology. 11(1–2), 85–108, 158–7-158–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghian H, Lacoste B, Qin T, Toussay X, Rosa R, Oka F, Chung DY, Takizawa T, Gu C, and Ayata C (2018). Spreading depolarizations trigger caveolin-1-dependent endothelial transcytosis. Annals of Neurology. 84(3), 409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoo A, van Zanten JJ, Metsios GS, Carroll D, and Kitas GD (2010). The Endothelium and Its Role in Regulating Vascular Tone. The Open Cardiovascular Medicine Journal. 4:302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, and Amieux PS (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America. 106(33), 13939–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen H, Meerlo P, Elsinga PH, Windhorst AD, Dierckx RAJO, Colabufo NA, van Waarde A, and Luurtsema G (2016). P-glycoprotein Function in the Rodent Brain Displays a Daily Rhythm, a Quantitative In Vivo PET Study. The AAPS Journal. 18, 1524–1531. [DOI] [PubMed] [Google Scholar]
- Shibata M, Yamada S, Kumar SM, Calero M, Bading J, Frangione B, Holtzman DM, Miller CA, Strickland DK, Ghiso J, and Zlokovic BV (2000). Clearance of Alzheimer’s amyloid-β1–40 peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier. Journal of Clinical Investigation. 106(12), 1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohet F, and Daneman R (2013). Genetic mouse models to study blood-brain barrier development and function. Fluids and Barriers of the CNS. 10(1), 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sörensen I, Adams RH, and Gossler A (2009). DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood. 113(22), 5680–8. [DOI] [PubMed] [Google Scholar]
- Stewart PA and Wiley MJ (1981). Developing nervous tissue induces formation of blood-brain barrier characteristics in invading endothelial cells: A study using quail-chick transplantation chimeras. Developmental Biology. 84(1),183–192. [DOI] [PubMed] [Google Scholar]
- Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, and Weitz CJ (2007). Intrinsic Circadian Clock of the Mammalian Retina: Importance for Retinal Processing of Visual Information. Cell. 130(4), 730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MD, Kisler K, Montagne A, Toga AW, and Zlokovic BV (2018). The role of brain vasculature in neurodegenerative disorders. Nature Neuroscience. 21(10), 1318–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tome ME, Herndon JM, Schaefer CP, Jacobs LM, Zhang Y, Jarvis CK, and Davis TP (2016). P-glycoprotein traffics from the nucleus to the plasma membrane in rat brain endothelium during inflammatory pain. Journal of Cerebral Blood Flow and Metabolism. 36(11), 1913–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallés A, Boender AJ, Gijsbers S, Haastm RAM, Martens GJM, and de Weerd P (2011). Genomewide analysis of rat barrel cortex reveals time- and layer-specific mRNA expression changes related to experience-dependent plasticity. J Neurosci. 31(16), 6140–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Assema DME, Lubbernik M, Bauer M, Van Der Flier WM, Schuit RC, Windhorst AD, Comans EFI, Hoetjes NJ, Tolboom N, Langer O, et al. (2012). Blood-brain barrier P-glycoprotein function in Alzheimer’s disease. Brain. 135(Pt 1), 181–9. [DOI] [PubMed] [Google Scholar]
- Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature. 554(7693), 475–480. [DOI] [PubMed] [Google Scholar]
- van Veluw SJ, Hou SS, Calvo-Rodriguez M, Arbel-Ornath M, Snyder AC, Frosch MP, Greenberg SM, and Bacskai BJ (2020). Vasomotion as a Driving Force for Paravascular Clearance in the Awake Mouse Brain. Neuron. 105(3), 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyazovskiy VV, Cirelli C, Pfister-Genskow M, Faraguna U, and Tononi G (2008). Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nature Neuroscience. 11(2), 200–8. [DOI] [PubMed] [Google Scholar]
- Wang EJ, Casciano CN, Clement RP, and Johnson WW (2001). Active transport of fluorescent P-glycoprotein substrates: Evaluation as markers and interaction with inhibitors. Biochemical and Biophysical Research Communications. 289(2), 580–5. [DOI] [PubMed] [Google Scholar]
- Wang Q, Maillard M, Schibler U, Burnier M, and Gachon F (2010). Cardiac hypertrophy, low blood pressure, and low aldosterone levels in mice devoid of the three circadian PAR bZip transcription factors DBP, HLF, and TEF. American journal of physiology: Regulatory, integrative and comparative physiology. 299(4), R1013–9. [DOI] [PubMed] [Google Scholar]
- Weger BD, Gobet C, Yeung J, Martin E, Jimenez S, Betrisey B, Foata F, Berger B, Balvay A, Foussier A, et al. (2018). The Mouse Microbiome Is Required for Sex-Specific Diurnal Rhythms of Gene Expression and Metabolism Cell Metabolism. 29(2), 362–382.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidenfeller C, Svendsen CN, and Shusta EV (2007). Differentiating embryonic neural progenitor cells induce blood-brain barrier properties. Journal of Neurochemistry. 101(2), 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, and Reppert SM (1995). Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 14(4), 697–706. [DOI] [PubMed] [Google Scholar]
- Whiteus C, Freitas C, and Grutzendler J (2014). Perturbed neural activity disrupts cerebral angiogenesis during a postnatal critical period. Nature. 505(7483), 407–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O’Donnell JO, Christensen DJ, Nicholson C, Iliff JJ, et al. (2013). Sleep drives metabolite clearance from the adult brain. Science. 342(6156),373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Mathur J, Vessiéres E, Hammack S, Nonomura K, Favre J, Grimaud L, Petrus M, Francisco A, Li J, et al. (2018). GPR68 Senses Flow and Is Essential for Vascular Physiology. Cell. 173(3), 762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Selby CP, Chiou Y, Ozkan-Dagliyan I, Gaddameedhi S, and Sancar A (2014). Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and period proteins in the mammalian circadian clock. Genes and Development. 28(18), 1989–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota SI, Yamamoto M, Moriya T, Akiyama M, Fukunaga K, Miyamoto E, and Shibata S (2001). Involvement of calcium-calmodulin protein kinase but not mitogen-activated protein kinase in light-induced phase delays and Per gene expression in the suprachiasmatic nucleus of the hamster. Journal of Neurochemistry. 77(2), 618–627. [DOI] [PubMed] [Google Scholar]
- Yuan P and Grutzendler J (2016). Attenuation of β-Amyloid Deposition and Neurotoxicity by Chemogenetic Modulation of Neural Activity. Journal of Neuroscience. 36(2), 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Yue Z, Arnold DM, Artiushin G, and Seghal A (2018). A Circadian Clock in the Blood-Brain Barrier Regulates Xenobiotic Efflux. Cell. 173(1), 130–139.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Nelson AR, Betsholtz, and Zlokovic BV (2015). Establishment and Dysfunction of the Blood-Brain Barrier. Cell. 163(5), 1064–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic BV (2008). The Blood-Brain Barrier in Health and Chronic Neurodegenerative Disorders. Neuron. 57(2), 178–201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File 1. Master File with all Processed RNAseq Data in the 3 Experiments, Related to Figures 2, 3 and 6
Supplemental File 2. Processed RNAseq Data for All Gene Expression Changes in the 3 Experiments, Related to Figures 2, 3 and 6
Supplemental File 3. Processed RNAseq Data for Genes Regulated in Opposing Directions between DREADDs Activating and DREADDs Silencing, Related to Figures 2 and 3
Supplemental File 4. Processed RNAseq Data for Blood-Brain Barrier Genes, Related to Figures 2, 3 and 6
Data Availability Statement
All RNAseq data are provided as Supplemental Files and the raw data can be accessed on Gene Expression Omnibus (GEO) (Accession: GSE156689).