Abstract
The current pandemic situation caused by the Betacoronavirus SARS-CoV-2 (SCoV2) highlights the need for coordinated research to combat COVID-19. A particularly important aspect is the development of medication. In addition to viral proteins, structured RNA elements represent a potent alternative as drug targets. The search for drugs that target RNA requires their high-resolution structural characterization. Using nuclear magnetic resonance (NMR) spectroscopy, a worldwide consortium of NMR researchers aims to characterize potential RNA drug targets of SCoV2. Here, we report the characterization of 15 conserved RNA elements located at the 5′ end, the ribosomal frameshift segment and the 3′-untranslated region (3′-UTR) of the SCoV2 genome, their large-scale production and NMR-based secondary structure determination. The NMR data are corroborated with secondary structure probing by DMS footprinting experiments. The close agreement of NMR secondary structure determination of isolated RNA elements with DMS footprinting and NMR performed on larger RNA regions shows that the secondary structure elements fold independently. The NMR data reported here provide the basis for NMR investigations of RNA function, RNA interactions with viral and host proteins and screening campaigns to identify potential RNA binders for pharmaceutical intervention.
INTRODUCTION
Betacoronaviruses contain a large single-stranded RNA genome of ∼30 000 nucleotides (nts). SCoV2 causing the COVID-19 disease contains multiple regions with very high sequence or secondary structure conservation relative to the 2002 SARS-Coronavirus (SCoV) and other Betacoronaviruses (1,2). The function of these putative cis-acting RNA elements have been characterized in different Betacoronaviruses and are associated with regulation of replication, subgenomic mRNA (sg mRNA) production and translation (3–5). Structural models for many of these RNA elements conserved in SCoV2 have been provided by in silico methods using RNA structure prediction, mutational covariance analysis or homology models (1,6,7).
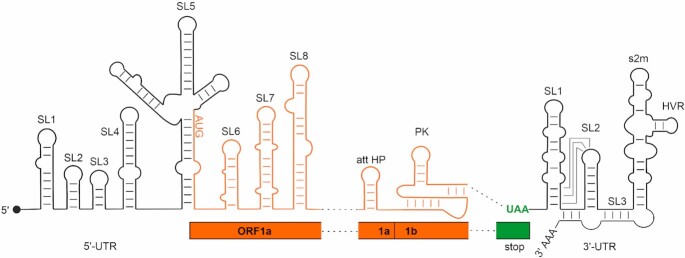
Here, we systematically characterize the secondary structures of all conserved cis-acting RNA elements of SCoV2 by nuclear magnetic resonance (NMR) spectroscopy, providing high-resolution experimental secondary structure models. We analyzed nine RNA constructs representing the eight stem-loop (SL) domains present at the genomic 5′-end, two RNA constructs corresponding to cis-acting elements from the ORF1a/b frameshifting region and four RNA constructs representing functional SLs within the viral 3′-UTR (Figure 1). These 15 RNA elements were chosen based on their conservation within the Betacoronavirus family and their importance for viral propagation. A detailed comparison regarding their structural conservation between SCoV2 and SCoV is shown in Supplementary Figure S1.
Figure 1.
Overview of cis-acting RNA elements of the SCoV2 genome. Black: untranslated regions; orange: coding regions. The SL structures investigated in this study and their relative positions within the SCoV2 genome are shown schematically. ‘stop’ represents the end of ORF9 coding for the Nucleocapsid protein.
We provide a streamlined pipeline for the fast and unambiguous NMR-based assignment of 2D structures in high-throughput. In a coordinated approach involving laboratories at the Goethe-University Frankfurt, the Technical University of Darmstadt, Case Western Reserve University (CWRU), and the Catholic University of Valencia, we produced 20 RNAs in isotopically labeled form representing 15 cis-acting RNA elements of the SCoV2 genome. Here, we report the NMR spectroscopic investigation to experimentally determine the secondary structure for SL1, SL2+3, SL4, three substructures of SL5 (SL5stem, SL5a, SL5b+c), SL6, SL7 and SL8, of the 5′-genomic region (subsequently denoted ‘5_’), the attenuator hairpin (att HP) and the three-stemmed pseudoknot (PK) linked to programmed ribosomal frameshifting (PRF), and SL1, SL2, SL3 and the isolated s2m motif from the 3′-UTR (subsequently denoted ‘3_’)
In addition, we prepared four larger constructs: two constructs representing SLs SL1 to SL8 (5′-geRNA, the first 472 nt of the SCoV2 genome) and SL1 to SL4 (5_SL1-4, nts 6–125) from the 5′-genomic end and two constructs from the 3′-end, the hypervariable region (3_HVR, nts 29 697 to 29 805) and the full-length 337 nt 3′-UTR (3′-UTR, nts 29 534 to 29 870) in 15N-labeled form for NMR investigations.
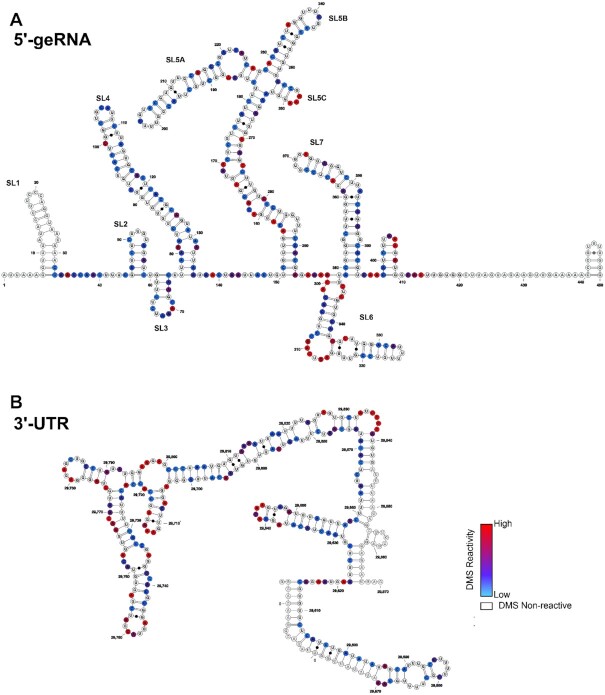
The NMR structural analyses were conducted at Weizmann Institute Rehovot, Karolinska Institute Stockholm, Catholic University of Valencia, CWRU and Goethe University Frankfurt (BMRZ). In addition, a DMS footprinting analysis was performed for the 5′- and 3′-genomic ends of the SCoV2 genomic RNA at CWRU and compared to the NMR results. Our data will facilitate research efforts aiming to map interactions with viral and host proteins or characterize the binding mode of small-molecule ligands targeting the viral RNA.
We continuously update and report results on the webpage http://covid19-nmr.de and NMR chemical shift assignment in the BMRB (8) (http://www.bmrb.wisc.edu/).
MATERIALS AND METHODS
DMS footprinting
A total of 5 μl of 200 ng/μl purified SCoV2 5′-end and 3′-UTR RNA were heated to 95°C for 15 s and flash cooled on ice for 2 min. A total of 95 μl DMS modification buffer (100 mM sodium cacodylate, 140 mM KCl, 3 mM MgCl2, pH 7.5) was added to the RNA sample and incubated for 30 min at room temperature. Two microliter of DMS was added, and the mixture was incubated at 37°C with 500 rpm shaking for 10 min. The methylation reaction was terminated by adding 60 μl of neat β-mercaptoethanol (Sigma-Aldrich). The modified RNA samples were purified and desalted using the RNA-cleanup-and-concentrator-5 column kit (Zymo Research) to recover RNAs containing more than 200 nts.
Methylated RNAs were used for reverse transcription to generate DNA with thermostable group II intron reverse transcriptase, third generation (TGIRT-III, InGex), following the manufacturer's protocol. The reverse primers 5R2 and 3R2 (Supplementary Table S1) for SCoV2 5′-end and 3′-UTR, respectively, were commercially synthesized (IDT). The RNA templates were digested using RNase H (NEB) for 20 min at 37°C following the reverse transcription. The synthesized DNA was sequentially polymerase chain reaction (PCR) amplified using Phusion DNA polymerase (NEB) with the specific primer sets (5F1/5R1,5F2/5R2;3F1/3R1,3F2/3R2, see Supplementary Table S1). The following PCR protocol was applied: denaturing for 30 s at 98°C, followed by 30 PCR cycles, including denaturing for 5 s at 98°C, annealing for 10 s at optimal temperature and extension for 15 s at 72°C; final extension continued for 5 min at 72°C. PCR products were desalted using the DNA-cleanup-and-concentrator-5 column kit (Zymo Research). The homogeneity of the amplified samples was verified using agarose gel electrophoresis prior to sending out for sequencing.
The sequencing was performed on an Illumina HiSeq 2000 system, which used cluster generation and sequencing by synthesis (SBS) chemistry. The sequence results of SCoV2 5′-genomic region and 3′-UTR RNA were aligned against index files and then used to generate the population average and read coverage quality control files (9). The mutational signal of 5′ and 3′ primer overlapping regions and signal from T and G nucleotides in the sequence were determined to be below background. Raw data showing the signal intensities for DMS treated samples and untreated controls are shown in Supplementary Figure S2. The signals from A and C nucleotides were normalized to the highest reactivity following 95% Winsorization. The DMS restraint files for each RNA were used as input to guide folding of full-length SCoV2 5′-end and 3′-UTR with the fold algorithm from the RNAStructure (10) webserver and visualized by VARNA (11).
RNA synthesis for NMR experiments
Fast RNA production was achieved by distribution and parallelization of individual synthesis steps. In general, all RNAs were produced by T7 polymerase-based in-vitro transcription (12). In the DNA template production step, the sequences of 5_SL1, 5_SL2+3, 5_SL4, 5_SL5stem, 5_SL5a, 5_SL5b+c, 5_SL6, 5_SL7, 5_SL8, 5_SL8loop, PK, 3_SL1, 3_SL2, 3_SL3base and 3_s2m together with the T7 promoter were generated by hybridization of complementary oligonucleotides and introduced into the EcoRI and NcoI sites of an HDV ribozyme encoding plasmid based on the pSP64 vector (Promega). The DNA template for 5_SL1-4 was produced by PCR amplification from the full-length 5′-genomic region. RNAs were transcribed as HDV ribozyme fusions to obtain 3′ homogeneity (13). The DNA sequences of the full-length 5’-genomic region, the 3′-UTR and the hypervariable region (3_HVR) were purchased from Eurofins Genomics and introduced into the EcoRI and HindIII sites of the pSP64 vector (Promega). The DNA template for att HP together with the T7 promoter was generated by hybridization of complementary oligonucleotides and directly used for run-off transcription. Isolated RNA hairpins 5_SL5b and 5_SL5c were purchased from Dharmacon and purified according to the manufacturer's instructions. All RNA sequences are summarized in Supplementary Table S2. The complete vector sequences are available upon request.
The recombinant vectors were transformed and amplified in Escherichia coli strain DH5α. Plasmid-DNA (2–10 mg plasmid per liter SB medium) was purified by Gigaprep (Qiagen) according to the manufacturer's instructions and linearized with HindIII (or SmaI in case of 3_HVR) prior to in-vitro transcription by T7 RNA polymerase (P266L mutant, prepared as described in (14)). Four preparative-scale transcriptions (10–15 ml each) were routinely performed in parallel. The RNAs were purified as follows: preparative transcription reactions (6 h at 37°C) were terminated by addition of ethylenediaminetetraacetic acid (EDTA) and RNAs were precipitated with 2-propanol. RNA fragments were separated on 10–12% denaturing polyacrylamide (PAA) gels and visualized by UV shadowing at 254 nm. Desired RNA fragments were excised from the gel and RNA was eluted by passive diffusion into 0.3 M NaOAc, precipitated with EtOH or EtOH/acetone and desalted via PD10 columns (GE Healthcare). Except for the att HP RNA, residual PAA was removed by reversed-phase HPLC using a Kromasil RP 18 column and a gradient of 0–40% 0.1 M acetonitrile/triethylammonium acetate. After freeze-drying of RNA-containing fractions and cation exchange by LiClO4 precipitation (2% in acetone), each RNA was folded in water by heating to 80°C followed by rapid cooling on ice except for the pseudoknot RNA, which was kept below 40°C to optimize folding into monomeric form. Buffer exchange to NMR buffer (25 mM potassium phosphate buffer, pH 6.2, 50 mM potassium chloride) was performed using centrifugal concentrators with a suitable molecular weight cut-off membrane. RNA purity was verified by denaturing PAA gel electrophoresis and homogenous folding was monitored by native PAA gel electrophoresis, loading the same RNA concentration as used for NMR experiments (Supplementary Figures S3 and 4).
The RNA constructs representing the entire 5′-genomic end and the entire 3′-UTR were purified without denaturing steps in order to maintain the native fold. Thus, transcription reactions for 5′-geRNA and 3′-UTR were terminated by addition of EDTA and RNA was extracted by aqueous phenolic extraction at pH 4. Phenol was removed by three cycles of chloroform extraction and subsequent buffer exchange via PD10 columns equilibrated with NMR buffer. The RNAs were purified by size exclusion chromatography using a HiLoad 26/60 Superdex 200 pg column. Fractions containing RNAs of the proper size were identified by denaturing and native gel electrophoresis, pooled and concentrated by centrifugal concentrators (MWCO: 50 kDa).
One production round of four NMR samples in parallel typically took 10 days from plasmid transformation to NMR tube filling. Table 1 provides a summary of the produced RNAs with their isotope labeling scheme, concentrations and site of conducting NMR assignment experiments.
Table 1.
RNA constructs corresponding to individual SCoV2 cis-acting elements, designated according to their location position in the viral genome, together with the isotope labeling scheme and concentration of the samples, and site where the NMR experiments were conducted
| RNA | BMRB ID* | Isotope labeling | Concentration [μM] | NMR experiments |
|---|---|---|---|---|
|
5_SL1234
119 nts |
15N | 446 | BMRZ | |
|
5_SL1
29 nts |
50 349 |
15N 13C,15N |
155 650 |
BMRZ |
|
5_SL2+3
32 nts |
50 344 |
15N 13C,15N |
400 652 |
BMRZ, Catholic University of Valencia |
|
5_SL4
44 nts |
50 347 |
15N (13C,15N G, U) (13C,15N A, C) |
775 400 500 250 |
BMRZ BMRZ BMRZ CWRU |
|
5_SL5stem
69 nts |
50 340 | 15N | 700 | BMRZ |
|
5_SL5a
33 nts |
50 346 |
15N 13C,15N |
807 680 |
BMRZ, Karolinska Institute |
|
5_SL5b+c
37 nts |
50 339 |
15N 13C,15N |
351 728 |
BMRZ, Weizmann Institute |
|
5_SL5b
25 nts |
unlabeled | 1200 | BMRZ | |
|
5_SL5c
12 nts |
unlabeled | 1500 | BMRZ | |
|
5_SL6
46 nts |
50 351 | (15N A, C) (13C,15N G, U) (13C,15N G, U) |
600 250 |
BMRZ CWRU |
|
5_SL7
50 nts |
** | (15N A, C, U) (13C,15N G) |
550 | BMRZ |
|
5_SL8
63 nts |
50 352 | 15N | 830 | BMRZ, Weizmann Institute |
|
5_SL8loop
31 nts |
13C,15N G, U | 411 | BMRZ | |
|
att HP
26 nts |
** | 15N,13C | 90 | Catholic University of Valencia |
|
PK
69 nts |
50 348 | 15N | 700 | BMRZ |
|
3_SL1
72 nts |
50 342 | 15N | 766 | BMRZ |
|
3_SL2
31 nts |
50 343 |
15N 13C,15N |
80 405 |
BMRZ |
|
3_SL3base
90 nts |
50 350 | 15N | 230 | BMRZ |
|
3_HVR
115 nts |
15N,13C G, U | 841 | BMRZ | |
|
3_s2m
45 nts |
50 341 |
15N 13C,15N |
388 596 |
BMRZ |
|
5′-geRNA 472 nts |
15N | 130 | BMRZ | |
|
3′-UTR 337 nts |
15N | 180 | BMRZ |
*Chemical shift assignment will be continuously updated in BMRB (8).
**Deposition in progress/additional experiments required.
At CWRU, transcription reactions were optimized in individual trials (15). The RNA constructs were purified by 8–10% denaturing polyacrylamide gel electrophoresis depending on the size of the RNA and eluted in Tris-borate-EDTA (TBE) buffer. The RNA samples were desalted and adjusted to <20 μM concentration using Nanodrop (ThermoFisher Scientific). All desalted RNA samples were annealed in RNase-free water by heating the sample to 95°C for 2 min followed by snap cooling on ice for a minimum 15 min. The annealed samples were concentrated using a centrifugation filtration system (Amicon) and exchanged into NMR buffer. For isotope labeled samples, uniformly 13C,15N-labeled uridine (rUTP) and guanosine (rGTP) (Cambridge Isotope Laboratories), and unlabeled adenosine (rATP) and cytidine (rCTP) (Sigma-Aldrich) were used in the preparative transcriptions. Comparison of RNAs prepared in different laboratories yielded close-to-identical NMR spectra, demonstrating the value of the coordinated research approach within the NMR consortium Covid19-NMR.
NMR experiments
At BMRZ, NMR measurements were carried out on Bruker 600, 800, 900 and 950 MHz AVIIIHD or AV NEO spectrometers equipped with cryogenic triple-resonance HCN probes, on a Bruker 700 MHz AVIIIHD spectrometer equipped with a cryogenic quadruple-resonance QCI-P probe and a Bruker 800 MHz AVIII spectrometer equipped with a carbon optimized TXO cryogenic probe. All probes at BMRZ have z-axis pulsed-field gradient accessory. At CWRU, NMR experiments were carried out on Bruker AVIII 900/800 MHz NMR spectrometers equipped with cryogenically cooled HCN triple resonance probes and a z-axis pulsed-field gradient accessory. At Valencia, NMR spectra were acquired using a Bruker AVII 600 MHz NMR spectrometer equipped with a triple resonance TCI cryogenic probe. NMR experiments at the Weizmann Institute, were conducted on Bruker AVANCE NEO 1000 and 600 MHz NMR spectrometers equipped with 5-mm cryogenic triple-resonance HCN TCI probes and x, y and z-axis pulsed-field gradient accessory. NMR experiments in Stockholm were carried out on a Bruker 600 MHz AVIII spectrometer equipped with a cryogenic quadruple-resonance QCI probe and z-axis pulsed-field gradient accessory. Data were processed and analyzed using TOPSPIN 4.0 (Bruker BioSpin, Germany) or NMRpipe/NMRDraw. NMRFAM-SPARKY (16), CccpNmr (17) or CARA (http://www.cara.nmr-software.org/downloads/3-85600-112-3.pdf) were used for chemical shift assignment. Table 2 summarizes the NMR experiments conducted for all RNAs.
Table 2.
A set of NMR experiments to delineate secondary structure
| # | NMR experiments* | Sample utilized Solvents Groups detected | Experiment-specific parameter settings | MT | References |
|---|---|---|---|---|---|
| 1 | 1H,1H-NOESY with jump-return water suppression | unlabeled or 15N labeled Homonuclear NOEs between iminos and all other sites |
NOE mixing time: 150 ms | 30 h | (56) |
| 2 | 1H,15N-BEST-TROSY |
15N labeled J-based heteronuclear H,N correlation for imino groups |
Transfer delay Δ = 5.4 ms to match 1J(H,N) ∼ 90 Hz | 1 h | (57,58) |
| 3 | 1H,15N-HSQC |
15N labeled H,N correlation for amino groups |
Relaxation and exchange optimized transfer delay Δ = 4.6 ms | 1 h | (59) |
| 4 | 1H,15N-CPMG-NOESY |
15N labeled 15N-edited NOESY correlation to detect NOEs to fast exchanging protons |
NOE mixing time: 150 ms | 22 h | (60) |
| 5 | 2D-BEST-TROSY HNN-COSY experiments |
15N labeled Through space J-based correlation of nitrogen atoms acting hydrogen bond donor and acceptor nitrogen |
NN-Transfer of 30 ms | 3 h | (58,61–63); in-house optimization |
| 6 | Long range 1H,15N-sfHMQC* |
15N labeled J-based correlation of purine N7/N9 nitrogens with H8 and adenine N1/N3 nitrogens with H2 |
Relaxation optimized transfer delay Δ = 20 ms | 2 h | (64) |
| 7 | Hadamard-encoded NOESY | unlabeled or 15N labeled Homonuclear NOEs between imino sites and all other sites Emphasis on fast exchanging sites |
NOE mixing time: 200 ms | 2 h | (65) |
| 8 | 13C,15N-filter NOESY with WATERGATE water suppression |
13C, 15N labeled X-filter in ω1 and ω2 to selecting 12C and 14N bound protons, e.g. H2, H6, H8 |
NOE mixing time: 250 ms | 24 h | (66–68) |
| 9 | 1H,1H-TOCSY with Excitation sculpting water suppression | unlabeled or 15N labeled J-based correlation of H5 and H6 in pyrimidine nucleobases |
TOCSY mixing time: 30 ms | 4 h | (69,70) |
| 10 | BEST-long range HNN-COSY* | 15N labeled | Long range correlation adenine H2 to base-paired uridine | 8 h | in-house improvements** |
| 11 | 1H,13C sfHMQC | unlabeled or 15N labeled | For aromatic C’s at natural abundance | 5 h-24 h | (64) |
MT is measurement time.
*marks experiments that can be conducted in D2O, but were conducted in 95% H2O, 5% (v/v) D2O to minimize the need for the production of additional samples.
**pulse sequence and parameter sets for in-house optimized experiments can be obtained upon request and data sets can be downloaded at covid19-nmr.de.
The first eight experiments have been conducted for all RNA constructs, additional experiments collected for a subset of RNAs are listed.
The conducted NMR experiments yielded the following information (18): 1H,1H-NOESY experiments (Table 2, #1, #7) correlate signals from RNA protons through space within up to 5.0 Å spatial proximity. In A-form RNA helices, sequential imino protons are within this distance and NOESY spectra thus provide correlation between consecutive base pairs within A-helical regions of the RNA, both to the next nucleotide within the sequence as well as across strands. Uridine and guanosine imino protons can be distinguished by their characteristic 15N chemical shifts in the 1H,15N-TROSY experiment (Table 2, #2), where guanosine imino protons in canonical G-C base pairs have a chemical shift between 145 and 150 ppm (δ 15N) and uridine imino protons in canonical A-U base pairs between 160 and 164 ppm (δ 15N). Non-canonical U-U or G-U base-pairs result in upfield shifts of the corresponding imino proton resonances to 10–11.5 ppm/10.5–12.5 ppm (δ 1H) and 141–145 ppm/155–160 ppm (δ 15N) for guanosine and uridine residues, respectively. Further evidence for canonical base-pairing is provided by the HNN-COSY experiment (Table 2, #5), which correlates the donor nitrogen of the guanosine or uridine imino group with the acceptor nitrogen atom of the corresponding cytidine or adenosine residue via through-space scalar 2hJ-NN coupling. Non-canonical U-U or G-U base-pairs do not give rise to cross peaks in HNN-COSY spectra and can thus be distinguished from involvement into canonical base pairing. For unstable A-U base pairs, for which imino resonances are broadened beyond detection due to solvent exchange, we recorded long-range (lr) HNN-COSY experiments correlating the adenosine H2 proton to the uridine imino group across the hydrogen bond (Table 2, #10). The cytidine amino groups involved in base pairing can be detected in the 1H,1H-NOESY experiment due to their strong cross peaks to the partner guanosine imino protons and can be correlated to their respective N4 nitrogen via an exchange-optimized 1H,15N-HSQC (Table 2, #3). The 1H,15N-CPMG-NOESY experiment (Table 2, #4) complements imino-to-amino correlations in A-form helical structural regions of RNAs. With this standard set of experiments, secondary structure predictions for the abovementioned SCoV2 RNAs were probed. Experimental secondary structures were thus determined for all 15 individual RNA elements and are described in the ‘Results’ section.
For sufficiently small RNAs (up to ∼35 nt), natural abundance 1H,13C HMQC experiments (Table 2, #11) were measured and analyzed to further assign aromatic H6/H8 and H5 protons and anomeric H1′ ribose protons. Assignment of these NMR signals is essential for the NMR sequential assignment procedure via 15N-filtered NOESY experiments in regular A-form conformations (Table 2, #8). The 1H,1H-TOCSY experiment (Table 2, #9) provides a quick identification of intra-nucleobase H5-H6 cross peaks for pyrimidines in this region, reducing ambiguities resulting from poor signal dispersion observed for RNAs larger than ∼30 nts in general. Line shapes of these TOCSY cross peaks are also a good indication for dynamics exhibited by the respective nucleotide. The strongest signals are usually observed for highly flexible pyrimidine residues in loops. In addition, ribose H1′-H2′ cross peaks can be observed for the non-A-form 2′-endo-puckered ribose conformations in the spectral region of 4–6 ppm (δ 1H).
Guanosine H8 protons and adenosine H8 and H2 protons are correlated with N7/N9 or N1/N3 nitrogens by a long-range (lr) 1H,15N-HSQC experiment (Table 2, #6). H2 protons of base-paired adenosines identified in the 1H,1H-NOESY experiment due to their strong cross peak to the partner uridine imino proton can thus be used to assign adenosine N1 and N3 atoms.
NMR experimental data were stored centrally and managed using the scientific data management system LOGS (19) (https://logs-repository.com). All experimental data can be downloaded at www.covid19-nmr.de.
RESULTS
All 20 RNA samples for NMR experiments exhibited high purity and adopted one homogeneous conformation without addition of Mg2+, with the exception of the pseudoknot RNA, which formed dimeric species to a significant extent, as described previously for the PK of SCoV (3). However, in the absence of Mg2+ and at 283 K, the monomeric species dominated (Supplementary Figure S3) and NMR spectra were sufficiently resolved to analyze the imino protons (Figure 13). Thus, secondary structure could be characterized for the entire set of cis-acting RNAs chosen for this study. The NMR spectra of the isolated RNA elements were compared to TROSY spectra for the large 5′-geRNA and 3′-UTR and to DMS footprinting data obtained from both regions, respectively. DMS footprinting confirmed the presence of well-defined SLs in both regions, corroborating the validity of our approach to analyze the structure of cis-regulatory elements in isolation (Figure 2A and B). NMR spectra of both large RNA constructs show extensive stable secondary structure formation in the imino proton region, with differential dynamic behavior of several structural motifs (Supplementary Figure S24A and B), similarly demonstrating independent folding of single SLs. In the following, the secondary structure determination by NMR of individual SLs is described and compared to the obtained DMS data. Where possible, SL constructs were stabilized by terminal G-C base pairs to facilitate in-vitro transcription and structure determination and designed to give one defined structure in prior in-silico predictions in accordance with phylogenetic conservation. Structure predictions using RNAstructure for the individual SLs (20) and pKiss for the PK (21) are shown in Supplementary Figure S5.
Figure 13.
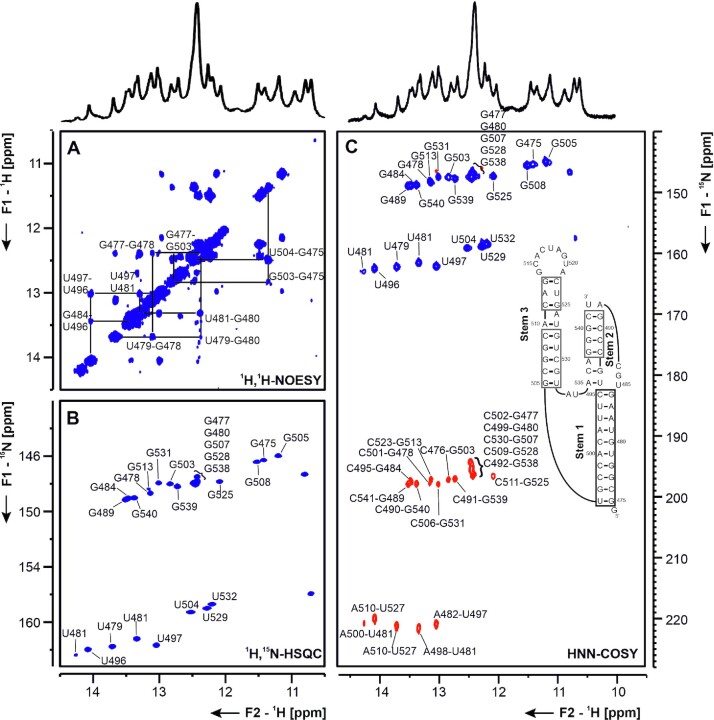
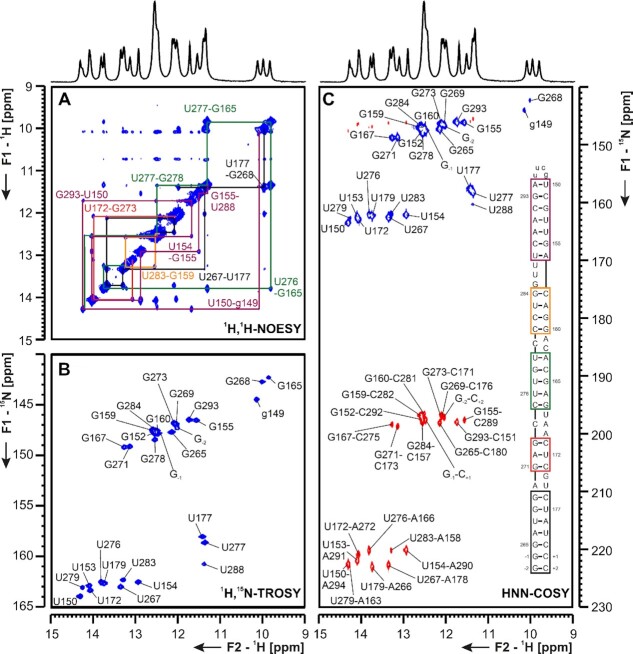
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the genomic-central located PK construct encompassing nts 13 475–13 542. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering, shifted for convenience by 13 000 nts from 5′. Imino-proton correlations in (A) between consecutive base pairs are shown in different colors. Included in (C) is the experimentally observed secondary structures of PK with genomic numbering, shortened by 13k nts from 5′. The secondary structure according to (C) is shown with genomic numbering from SCov2. Colors of boxes are according to the correlations in (A). Gray boxes mark base pairs, which were not assigned based on imino-to-imino correlations.
Figure 2.
Secondary structures of the SCoV2 5′-genomic end and 3′-UTR as determined by DMS-MaPseq. Normalized DMS reactivity indices used to fold the secondary structures of (A) SCoV2 5′-genomic end and (B) SCoV2 3′-UTR are superimposed with color codes scaled from the highest reactivity set to 1 (red) to the lowest set to 0 (blue). G and U nucleotides are rendered as white and sites that overlap with sequencing primers are rendered transparent.
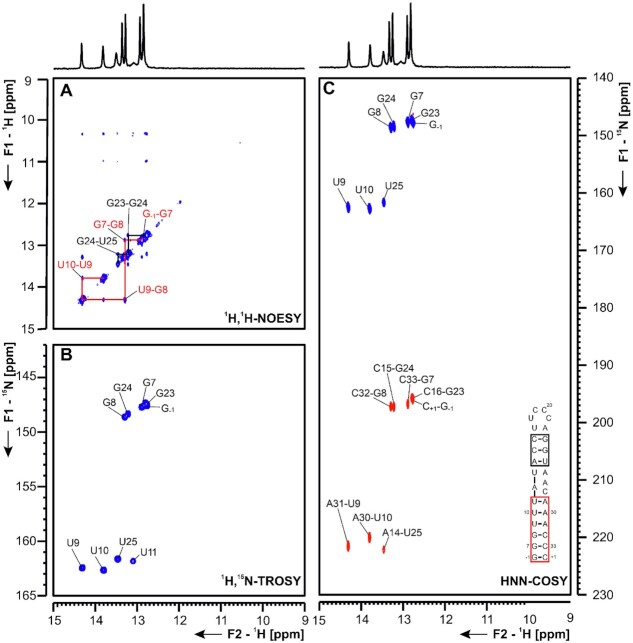
5_SL1 5_SL1 spans nucleotides (nts) 7–33 in the 5′-UTR of the SCoV2 genome. In SCoV, interaction of nsp1 with SL1 has been shown to be crucial to protect viral RNA from degradation (22). NMR studies on SL1 derived from MHV (mouse hepatitis virus) identified a bulged-out adenine corresponding to adenine 27 in SCoV2. Furthermore, the lower-part of SL1 has been shown to be structurally labile, which seems to be crucial for virus propagation in MHV, where SL1 might mediate the cross-talk between the 5′- and the 3′-UTR during viral replication (23). Computational predictions of the secondary structure of 5_SL1 yielded a single lowest-energy conformation with an asymmetrical bulge formed by nucleotides 12, 27 and 28 (Supplementary Figure S6). By assignment of imino resonances, the structural predictions could be confirmed and all observable slow exchanging imino resonances could be assigned from analysis of a 1H,1H-NOESY at 283 K (Figure 3A). Generally, we observe less intense NOE diagonal peaks for residues neighboring the bulge region on the loop helix (U13, U25) compared to those neighboring it on the terminal helix (U10, U11). The terminal helix was stabilized by one additional G-C base pair to facilitate in-vitro transcription. Thus, for instance, the imino resonance of U11 is assignable, while the U13 imino resonance cannot be detected at all. The imino proton-based secondary structure determination was consistent with results from 15N-correlated experiments (Figure 3B and C). Additionally, aromatic and amino resonances of base-paired A and C residues (H2 and H5/H6 resonances) could be assigned (Supplementary Figure S6). Most of the nucleotides forming 5_SL1 are within the primer binding region in DMS footprinting experiments, rendering this SL invisible for DMS analysis (Supplementary Figure S6E).
Figure 3.
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the 5′-genomic end construct 5_SL1 encompassing nts 7–33. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering. Imino-proton correlations in (A) between consecutive base pairs are shown in different colors. Included in (C) is the experimentally observed secondary structures of 5_SL1 with genomic numbering. Additional closing base pairs are annotated with ‘±x’. Colors of boxes are according to the correlations in (A).
5_SL2+3 5_SL2 is the most conserved structure in the 5′-UTR (24). Notably, SL2s from different coronaviruses can functionally replace each other, even across different genera (25). SL2 (nts 45–59) of the 5′-UTR is identical to SCoV SL2. It is thus very likely that it forms the same CUYG-looped short hairpin structure (26). SL3 contains the transcription regulatory sequence (TRS) essential for the synthesis of sg mRNAs by discontinuous transcription (25). Among Betacoronaviruses, SL3 (nts 61–65 in SCoV2) is predicted to be stably formed only for a subset of species, SCoV among them (4). Invariably, whether as part of a hairpin or not, the core sequence (CS-L, CUAAAC) is single-stranded (4). Unwinding of SL3 by the N protein might be necessary for TRS function (27). As SL2, SL3 is identical in sequence between SCoV and SCoV2 (Supplementary Figure S1). In the present study, we investigated a sequence spanning nts 45–75 that contained SLs 2 and 3, the secondary structure of which is shown in Figure 4. The presence of the CUYGU pentaloop in SL2, including a C50:G53 pair and an extrahelical U54 was confirmed by the NMR data, which were also consistent with the presence of a UCUAAAC heptaloop closing the SL3 hairpin and encompassing the CS-L (Figure 4 and Supplementary Figure S7). Interestingly, the two stems in this construct, 5_SL2 and 5_SL3 show different stabilities at 283 K. This difference in stability can be deduced from the difference in imino resonance linewidths for the base pairs of SL2 versus SL3, in line with the requirement of the viral transcription machinery to unfold the leader TRS located in SL3. This differential stability is even more pronounced at 298 K, where the U62 and U63 imino proton resonances of SL3 are broadened beyond detection (Supplementary Figure S8). In this regard, the leader TRS of the Alphacoronavirus TGEV (transmissible gastroenteritis coronavirus) was also found to occupy the apical loop of a pseudo-stable hairpin by NMR spectroscopy (25,28). In agreement with the NMR data, DMS footprinting showed stable base pairs in the stem of SL2 and a loop G-C base pair indicated by the very low reactivity of C50. However, the lower stability of SL3, evident from NMR, could not be detected by DMS footprinting (Supplementary Figure S7). Thus, we tested if Mg2+ ions, present during DMS treatment, but absent in the NMR buffer, could modulate the stability of SL3. Strikingly, in the presence of 3 mM Mg2+, the SL3 is significantly stabilized, illustrated by an increase of imino proton intensities for U62 and U63 (Supplementary Figure S8). This stabilizing effect of Mg2+ is even evident at 298 K, where the U62 and U63 imino proton resonances are broadened beyond detection in the absence of Mg2+, but become clearly observable in the presence of Mg2+ (Supplementary Figure S8). Thus, ionic conditions strongly influence the stability of SL3 and might thereby affect TRS function in vivo.
Figure 4.
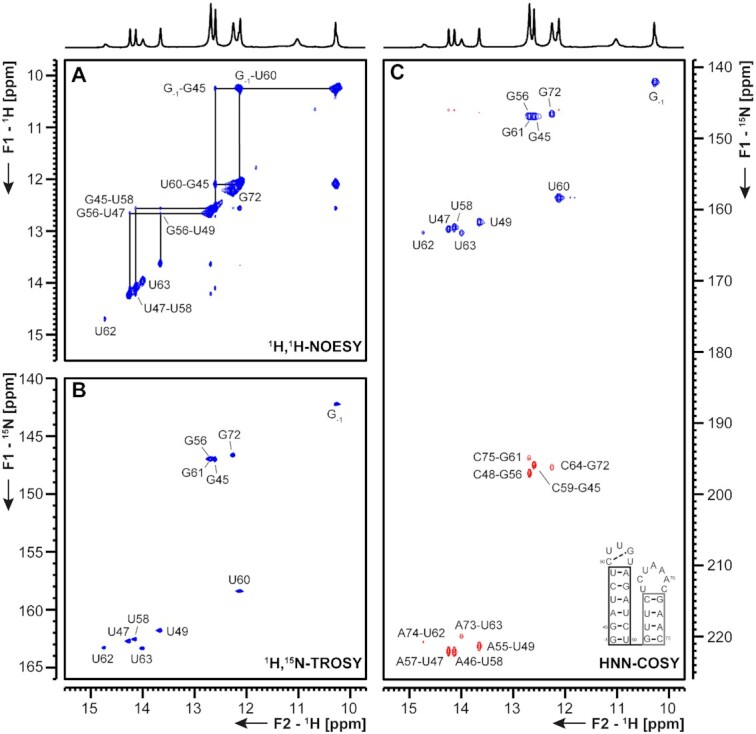
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the 5′-genomic end construct 5_SL2+3 encompassing nts 45–75. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using genomic numbering. Imino-proton correlations in (A) between consecutive base pairs are shown in black. Included in (C) is the experimentally observed secondary structure of 5_SL2+3 with genomic numbering. Additional closing base pairs are annotated with ‘±x’. The black box represents the correlations in (A). The gray boxes marks base pairs which were not assigned based on imino-to-imino correlations.
5_SL4 5_SL4 (nts 86–125) is structurally conserved among Betacoronaviruses and might function as a spacer element required for sg mRNA synthesis, mediating proper relative orientation of SLs 1–3 (29). SL4 carries a short upstream ORF (uORF), which is also conserved among Betacoronaviruses. Its function is still subject to speculation, since genetic pressure to select for the presence of a uORF could be observed, yet manipulations of this uORF that retain the SL4 RNA structure all yielded viable viruses (30). Interestingly, an additional short downstream hairpin structure predicted for MERS-CoV2 might also be present in SCoV2 (4,6), and is currently under investigation at BMRZ. Here, we present our results for 5_SL4 depicted in Figure 5. The NMR data showed the secondary structure of an SL interrupted by mismatches and capped by a non-canonical five nucleotide loop (Figure 5). All predicted base pairs could be detected, including a G-U base pair formed by residues G91 and U120 by the presence of imino signals in the non-canonical region (Figure 5B). The imino NOESY walk is in agreement with a continuous stem for SL4 with a looped-out U95 and a so far structurally incompletely characterized, but A-form helix compatible arrangement of the opposing residues C92 and C119 as well as C100 and U112. So far, the absence of imino resonances for residues of the apical loop, which contains two of the three nucleotides of the start codon of the uORF, precludes any conclusions about the structure of the loop. Starting from the assigned imino protons (Figure 5B), 12 of 14 cytidine amino groups, three out of five adenine amino groups and eight of 14 guanine amino groups were assigned (Supplementary Figure S9D). The cytidine amino groups served as starting point to assign the H5 resonances in a 15N-filtered NOESY spectrum together with a 1H,1H-TOCSY experiment (Supplementary Figure S9B), which also allowed the connection of the H5 to the H6 of the same pyrimidine residue. Starting from the H5-H6 cross peaks and the adenine H2 resonances identified in the 1H,1H-NOESY and the lr-15N-HSQC spectrum (Supplementary Figure S9A), the non-exchangeable aromatic C-H groups of all residues but the two uridine residues in the loop could be assigned in the aromatic-aliphatic walk in the 1H,1H-NOESY spectrum (Supplementary Figure S9C). DMS footprinting data suggest an elongated SL for SL4, ranging from nt 77 to 136.
Figure 5.
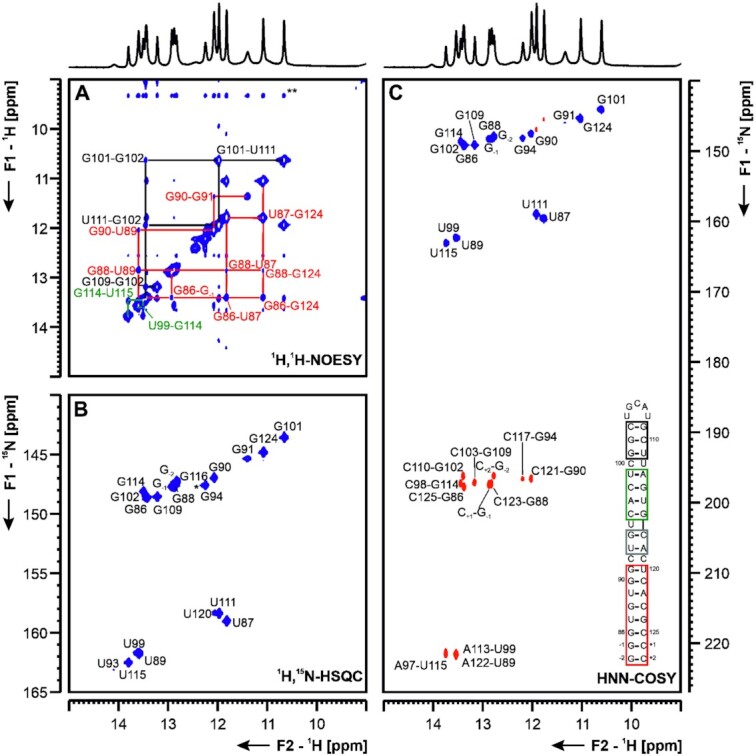
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the 5′-genomic end construct 5_SL4 encompassing nts 86–125. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering. Imino-proton correlations in (A) between consecutive base pairs are shown in different colors. Included in (C) is the experimentally observed secondary structure of 5_SL4 with genomic numbering. Additional closing base pairs are annotated with ‘±x’. Colors of boxes are according to the correlations in (A). Gray boxes mark base pairs, which were not assigned based on imino-to-imino correlations. The asterisk in (B) marks a signal which is visible at lower contour levels.
SL5 (nts 149–294) displays more pronounced structural divergence among the Betacoronaviruses in comparison to the upstream elements, but some features are recurring: a four-helix junction connects the sub-structures SL5a, b, and c to a long-range base-pairing region forming an extended helical stem (31). The AUG start codon for ORF1a is integrated into a stable SL5 helical section, resulting in the 3′-part of SL5 being the N-terminal coding sequence of nsp1 (Figure 1). Sub-structures of SL5 have been associated with viral replication in MHV and BCoV (Bovine coronavirus) (32), but no conclusive model for a conserved functional role of SL5 in Betacoronaviruses is available. We divided SCoV2 SL5 into the three sub-structures 5_SL5stem (nts 150–180 and 265–294), 5_SL5a (nts 188–218) and 5_SL5b+c (nts 227–263), shown in Figures 6–8, respectively.
Figure 6.
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlations of the 5′-genomic end construct 5_SL5stem encompassing nts 150–180 and 265–294. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering. Sequential imino-proton correlations in (A) between neighboring base pairs are shown in colors according to the experimentally determined 5_SL5stem RNA secondary structure schematized as an inset in panel (C). NMR resonances arising from the tetraloop nucleotides are annotated with small letters and the two additional GC closing base pairs at the 5′- and 3′-termini are numbered with ±x, respectively. Colors of boxes are according to the correlations in (A). Note that the stem is depicted upside down with respect to its orientation as presented in the overall scheme of Figure 1.
Figure 8.
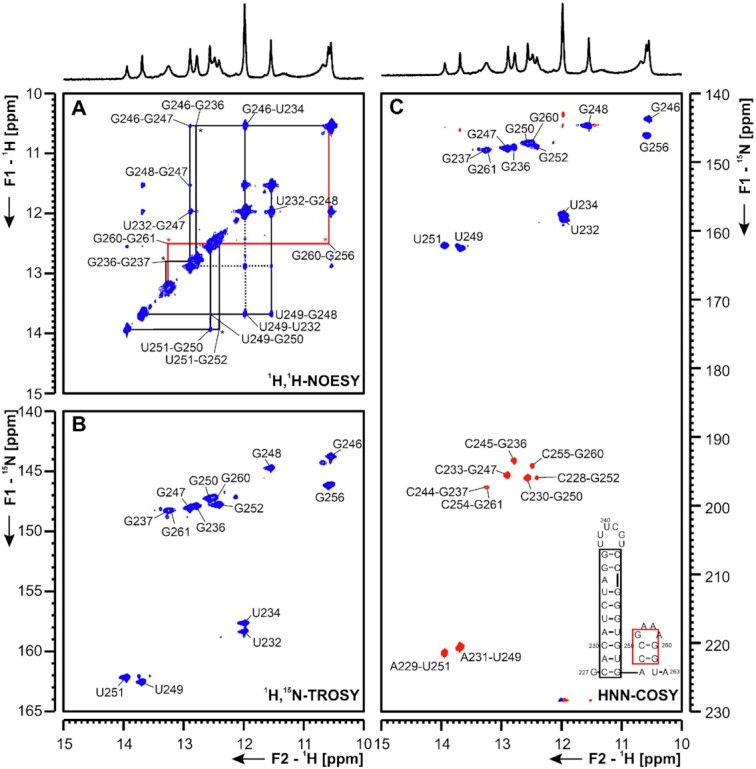
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the 5′-genomic end construct 5_SL5b+c encompassing nucleotides 227 to 263 at 283K. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering. Imino-proton correlations in (A) between consecutive base pairs are shown in different colors, * denote crosspeaks visible at 275K, but not at 283K. Included in (C) is the experimentally observed secondary structure of 5_SL5b+c with genomic numbering. Additional closing base pairs are annotated with ‘±x’. The boxes are according to the correlations in (A).
5_SL5stem For the SL5 core stem RNA element, we used the following construct design: after removal of all predicted SL sub-parts in the SL5 tree (i.e. a, b and c) the remaining core stem encompasses nts 150–180 on its 5′-end and 265–294 on its 3′-end, respectively. This design is fully in line with the suggested arrangements of the SL5 arms and, in particular, the stem structured detected in the DMS footprinting experiments (Figure 2A). Notably, the subgenomic AUG start codon is positioned directly downstream of SL5c, starting from nucleotide A266 (Figures 2 and 6). We decided to stabilize the resulting apical end of 5_SL5stem with two additional G-C base pairs in order to mimick the expected structural rigidity of the nearby four-way-junction and facilitate assignments of the initial base pairs. The 5_SL5stem basal end was stabilized with a bona fide UUCG tetraloop (33), which allowed transcription of the RNA from one DNA template and resulted in a sufficiently compact and homogeneously folded RNA, despite its relatively large size of 69 nt. For convenience, we display the 5_SL5stem in an upside-down arrangement with respect to the scheme in Figure 1 (Figure 6 and Supplementary Figure S10). Using NMR experiments #1 to #6 and #9 on a uniformly 15N-labeled sample (Tables 1 and 2), we obtained assignments for all, except for one (U175), H-bonded imino 1H,15N pairs within the native sequence in agreement with the predicted secondary structure (Figure 6A and B; Supplementary Figures S10 and 11). We found NOEs that show the tight interaction of the tetraloop-stabilized g149 with the closing base pair A294-U150. However, we were only able to see the stabilizing effect of the UUCG cap at 283 K, but not at higher temperatures.
Apart from some spectral overlap in the central guanosine region (around 12.5 ppm), the 5_SL5stem RNA yielded spectra of excellent quality for initial assignments beyond the G/U iminos. We were also able to assign imino nitrogens of all base-pairing adenines (Figure 6C), supporting the underlying base pair pattern and RNA secondary structure. The well-resolved H2 proton chemical shift dispersion allowed us to obtain assignments for the majority of the adenine N3 resonances (Supplementary Figure S10). We could also assign a significant fraction of H-bonded adenine and cytidine amino groups, although a number of amino NMR signals overlap, which suggests that the secondary structure primarily adopts A-form conformation within consecutive stretches, typical for double-stranded RNAs.
The NMR-derived base-pairing pattern is confirmed by the reactivities obtained for the respective RNA stem in the DMS footprinting experiment (Figure 2 and Supplementary Figure S10). The latter data reflect the increase in reactivity along with the bulges compared to the H-bonding within duplexed stretches (see direct comparison of data from both methods in Supplementary Figure S10D). Notably, while an increase in DMS reactivity at A-U closing base pairs adjacent to bulges is explained by a lower average stability of the H-bonds, we also found the innerhelical adenosines 157 and 166 to exhibit relatively high DMS reactivity, which is in contrast to their apparently stable duplex character shown by the NMR data. To examine the role of different experimental conditions between NMR (283 K and no Mg2+) and DMS footprinting (310 K and 3 mM Mg2+), we undertook both an NMR-based titration of 5_SL5stem with Mg2+ as well as a temperature comparison of nitrogen correlation spectra between 283 K and 298 K (Supplementary Figure S11). Our data show that the locally confined increase in DMS reactivity can well be explained by the higher temperature. Of note, the overall comparison between the two buffer conditions reveals identical spectral quality and thus underlines the complementary strength of the two methods to fully probe RNA secondary structure. Consequently, we found the 5_SL5stem part of the SCoV2 RNA to adopt the global secondary structure as suggested in Figures 1 and 2.
5_SL5a based on secondary structure prediction, the SL5a construct is composed of two helical stems that are separated by a three nucleotide U-rich bulge (Figure 7). By NMR spectroscopy, we were able to confirm the presence of both stem regions, while the resonances of the two U nucleotides participating in canonical A-U base pairs (U191 and U209) served as starting points for the sequential imino-walk indicated in Figure 7A. Additionally, we detected a weak U-imino resonance (CS 1H: 11.3 ppm and 15N: 157.8 ppm, Figure 7A and B), which is involved in a non-canonical nucleobase interaction. This U shows correlations to the C193-G213 base pair and is thus either part of the U-rich bulge or represents U195 if the other uridines of the bulge are flipped out and solvent exposed. However, at this point an unambiguous assignment is not possible and also in line with the available SHAPE data (34,35), no imino proton signal for G210 is detected at room temperature. Furthermore, the stem is closed by a UUUCGU loop. While imino resonances remain elusive for this loop, the 1H,15N-HSQC spectrum of the amino groups show characteristic resonances for a UUCG-tetraloop (Supplementary Figure S12) (33). This suggests that the UUUCGU loop might adopt a conformation similar to the UUCG tetraloop. All observations by NMR spectroscopy are in good agreement with the DMS footprinting data.
Figure 7.
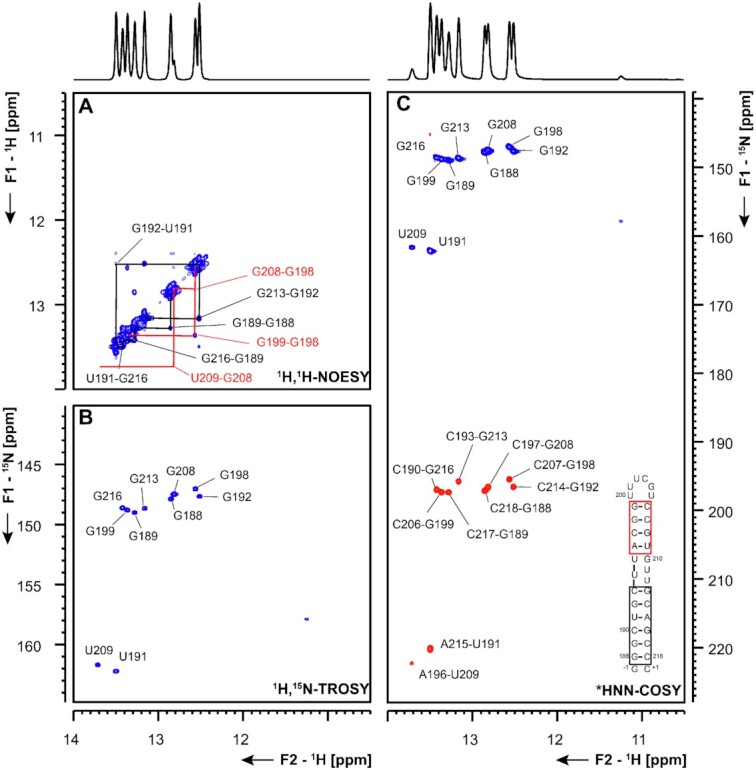
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlations of the 5′-genomic end construct 5_SL5a encompassing nts 188–218. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering. Imino-proton correlations in (A) between consecutive base pairs are shown in different colors. Included in (C) is the experimentally observed secondary structure of 5_SL5a with genomic numbering. The HNN-COSY spectrum in (C) was recorded at 298 K. Additional closing base pairs are annotated with ‘±x’. Colors of boxes are according to the correlations in (A).
5_SL5b+c key to the assignment of 5_SL5b+c (Figure 8 and Supplementary Figures S13-14) was the use of low temperature (275 K) and the comparison of the construct to the single hairpins 5_SL5b and 5_SL5c. In the double hairpin construct 5_SL5b+c, only the lower part of the SL5b stem (nts 229–234, 246–251) gave rise to imino-imino cross peaks in the NOESY at 283 K, while imino-imino correlations for the upper two base pairs closing the UUUCGU-loop, the closing base pair for the stem of 5_SL5b and for the base pairs of 5_SL5c (nts 253-263) only appear at 275 K (Supplementary Figure S14). Stem SL5c is closed by a GAAA-tetraloop (36). In these tetraloops, the H1 of the G is protected by hydrogen bonding to the phosphate of the last adenosine in the loop. In line with this, we observe an additional cross peak in the TROSY (Figure 8B) in the non-canonical region, which arises from G256, however, due to overlap with G246 and ambiguities in the amino region, an assignment was only feasible by comparison of the 5_SL5b+c construct to the single hairpins of 5_SL5b and 5_SL5c (Supplementary Figure S14). Data from DMS footprinting are in line with our observations at 275 K, while we observe more dynamics at higher temperature, suggesting stabilization of SL5b and c within the full length SL5.
5_SL6 5_SL6 comprises nts 302–343 of SCoV2 and is located in the 5′-region of ORF1a coding for nsp1. For a number of members of the Betacoronavirus class, including BCoV, MHV and several human coronaviruses (HCoVs), SL6 was shown to be conserved, but its precise function in viral replication remains unclear (37). Mutational studies leading to destabilization of SL6 resulted in reduced virus viability in MHV. However, this effect was attributed to a reduction of nsp1 protein levels (4,38). A recent study identified the asymmetrically bulged region of SCoV2 SL6 as one major binding site for the N protein (34). Here, we study SCoV2 5_SL6 that comprises nts 302–344 of SCoV2 extended by two G-C base pairs. (Figure 9). We conducted resonance assignment on a sample containing 15N-labeled A and C nucleotides and 13C,15N- labeled G and U nucleotides. We could assign the observable resonances of all exchangeable imino- and C amino-protons in this RNA (Figure 9 and Supplementary Figure S15). Based on these assignments, we can delineate two base-paired regions in 5_SL6. The first paired region is comprised of a stem of eight consecutive canonical Watson–Crick base pairs including the 5′- and 3′-termini. The second base-paired region consists of six base pairs including a terminal G-U base pair, spanning from G319-U324 and A328-U333. This second paired region is capped by a triple-U loop. Both base-paired regions are separated by an asymmetric bulge of 11 and 4 nts. These results were in good agreement with DMS footprinting data. Here, the previously observed asymmetric A-rich bulge showed a high reactivity, whereas both stem regions were well protected from DMS (Supplementary Figure S15).
Figure 9.
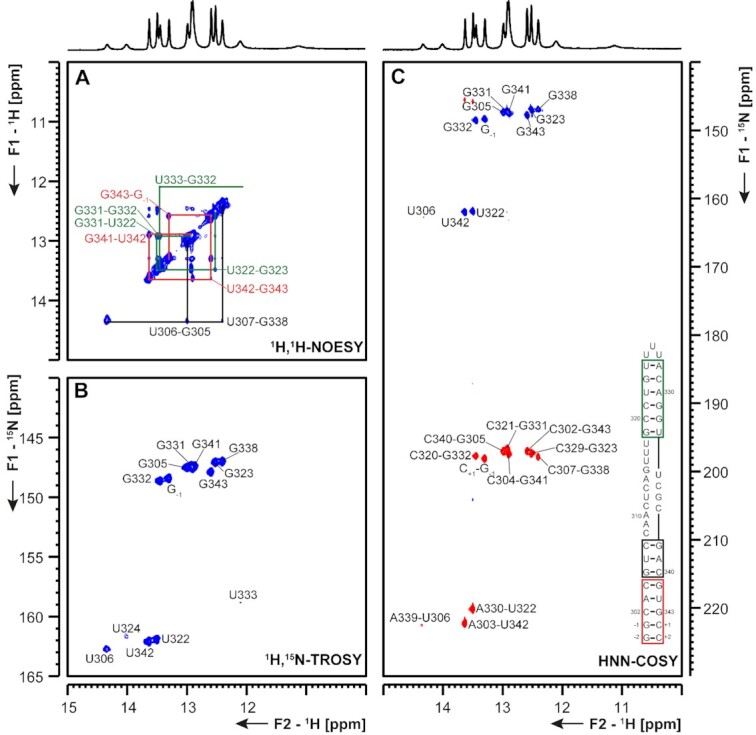
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the 5′-genomic end construct 5_SL6 encompassing nts 302–343. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering. Imino-proton correlations in (A) between consecutive base pairs are shown in different colors. Included in (C) is the experimentally observed secondary structure of 5_SL6 with genomic numbering. Additional closing base pairs are annotated with ‘±x’. Colors of boxes are according to the correlations in (A).
5_SL7 and 5_SL8 The presence of SL7 (nts 349–394) was confirmed by RNA structure probing for MHV, and similar motifs were found by RNA structure prediction in BCoV and SCoV (37). For SCoV and SCoV2, SL8 (nts 413–471) is predicted as a strikingly large hairpin-structure compared to other Coronaviruses, where it is mostly absent (4). We investigated 5_SL7 and 5_SL8 separately as shown in Figures 10 and 11.
Figure 10.
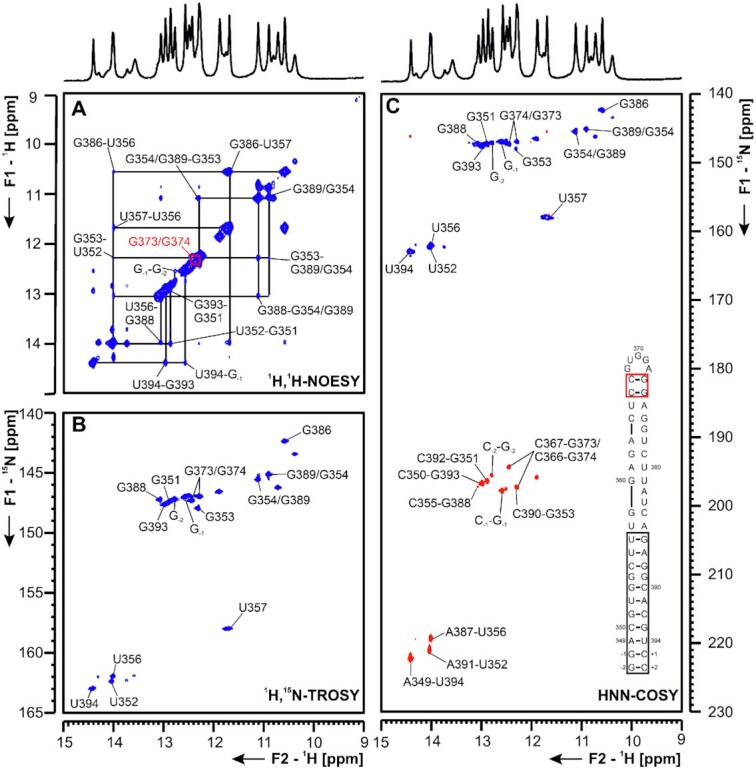
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the 5′-genomic end construct 5_SL7 encompassing nts 349–394. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering. Imino-proton correlations in (A) between consecutive base pairs are shown in different colors. Included in (C) is the experimentally observed secondary structure of 5_SL7 with genomic numbering. Additional closing base pairs are annotated with ‘±x’. Colors of boxes are according to the correlations in (A).
Figure 11.
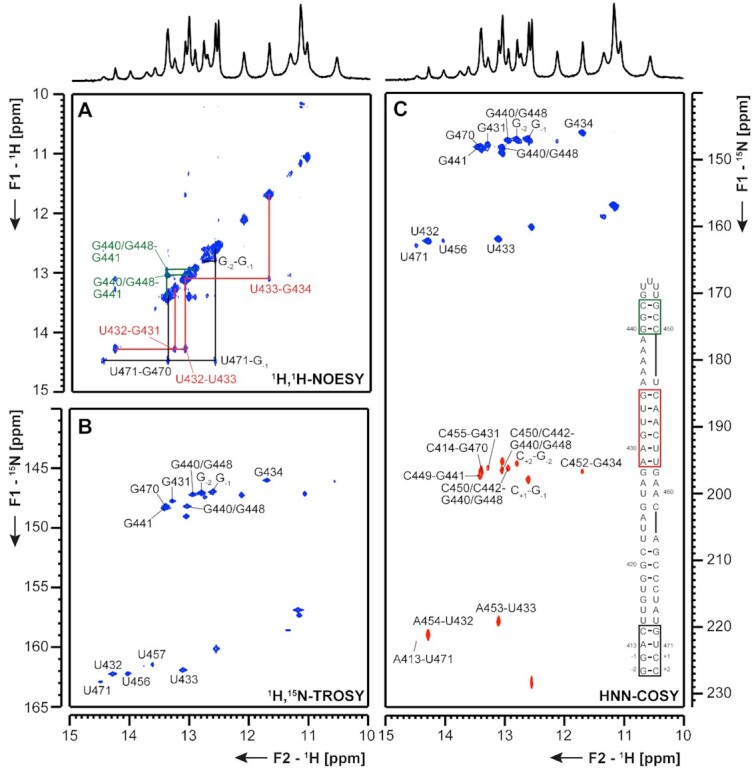
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the 5′-genomic end construct 5_SL8 encompassing nts 413–471. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering. Imino-proton correlations in (A) between consecutive base pairs are shown in different colors. Included in (C) is the experimentally observed secondary structures of 5_SL8 with genomic numbering. Additional closing base pairs are annotated with ‘±x’. Colors of boxes are according to the correlations in (A).
5_SL7 The 50 nt RNA 5_SL7 is subdivided into four different helical sections, bridged by a G-G mismatch and two bulges of one (G377) or two (A382, U383) nucleotides. This general secondary structure, as predicted by RNAstructure and mfold [Supplementary Figure S5 and (39)], could be confirmed by assignment of imino proton resonances in 1H,1H-NOESY spectra (Figure 10). The terminal stem between G-2 and the characteristic G386-U357 wobble base pair at 1H chemical shifts of 11.68 and 10.57 ppm (δ 1H) was unambiguously assigned. For all assigned U imino protons, their corresponding A H2-aromatic protons were assigned (Supplementary Figure S16). The terminal helical stretch comprising two G-C base pairs following one A-U and an additional G-C base-pair was assigned by comparison to the same element present in 5_SL8 (Figure 11). The central unusual G-G-motif gives rise to two non-canonical imino proton resonances, which cannot be unambiguously assigned to one of the two Gs and may result either from stacking or formation of an unusual base pair. In the middle part of 5_SL7, no imino protons could be assigned in the 1H,1H-NOESY spectra, most likely due to an increased instability of this section compared to the remainder of the base pairs. However, two additional, yet unassigned G-C base pairs are detected in the HNN-COSY (Figure 10). The two loop-closing G-C-base pairs were assigned due to their uniqueness. Its unreactive nature was proposed in DMS footprinting. According to DMS footprinting data these base pairs are stable. Further, DMS footprinting data did not detect a higher flexibility of the central region of the SL. Additional insights into the conformation and dynamics of this part of the RNA will include the investigation of shorter RNA constructs.
5_SL8 For 5_SL8, overall 11 imino resonances were assigned unambiguously (Figure 11 and Supplementary Figure S17). With a shortened construct of the upper part of the RNA (5_SL8loop, Table 1) the three consecutive G-C base pairs were assigned (Figure 11, green), while the assignment of the two flanking guanosines, G440 and G448 remains ambiguous. Furthermore, it can be concluded from the HNN-COSY spectrum that a Hoogsteen A-U base pair might be present as the hydrogen bond acceptor nitrogen chemical shift is at ∼230 ppm, which is characteristic for an adenine N7 resonance. Moreover, we detected at least six additional imino protons that are involved in non-canonical structural elements. This NMR experimental fingerprint is not consistent with predictions of the predicted lowest energy structure (Supplementary Figure S5) and will be subject to further studies. As nearly the complete sequence of 5_SL8 constitutes the primer site for DMS footprinting no further DMS analysis was possible.
ORF1a/b frameshifting region
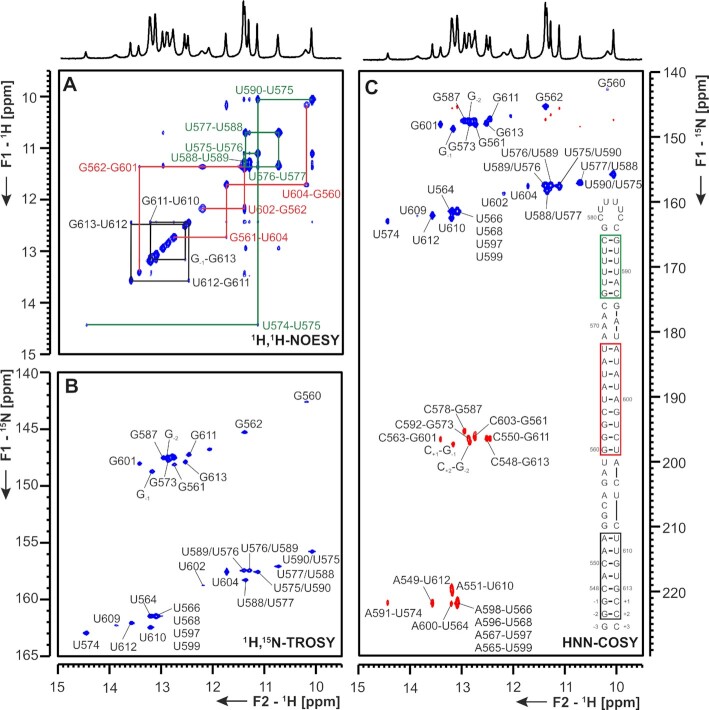
Att HP An attenuator hairpin (att HP) located immediately upstream of the slippery site and the PK responsible for programmed ribosomal frameshifting (PRF) has been reported to regulate PRF function by reducing the activity of the PRF PK (40). Similar to the attenuator hairpin of SCoV, the att HP of SCoV2 contains a 10-nt palindrome of unknown function comprising nts 13 441–13 450 (Figure 12 and Supplementary Figure S18). Indeed, we detected homodimerization for this construct in native polyacrylamide gels containing Mg2+. The SCoV2 att HP exhibits five nt variations relative to SCoV that predict it to be significantly less stable (41). The NMR data indicate that the folding of the att HP of SCoV2 is substantially different to that proposed for the SCoV attenuator, as it contains an eight base pair stem including an intrahelical U-U mismatch, instead of the 10 bp stem predicted for the SCoV hairpin. In addition, the SCoV2 att HP does not expose the palindrome sequence in its apical region (Figure 12). The relative instability of the SCoV2 attenuator is reflected by the detection of possible alternative conformations in the central part of the SL in the NMR spectra, containing either the predicted, adjacent U13 437:U13 450 and U13 438-A13 449 bp, or adjacent U13 437-A13 449 and U13 438-G13 448 bp, with likely extrahelical U13 450 and C13 439 (Figure 12). At the current state of investigation, the assignments of these alternative conformation remains tentative and further constructs will be investigated to clarify the topology of these interesting RNA dynamics.
Figure 12.
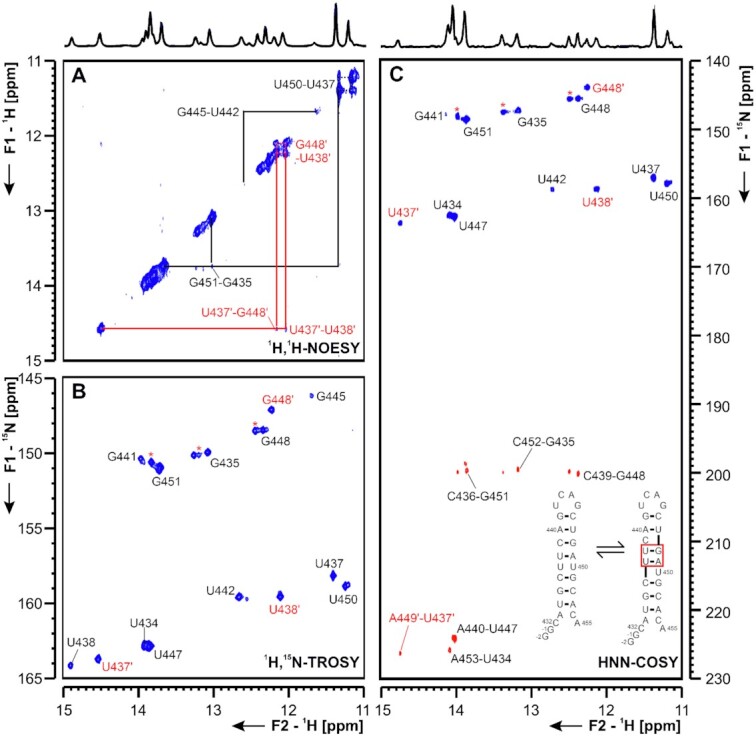
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectrum of the SARS-CoV-2 attenuator hairpin encompassing nts 13 432 to 13 455. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using genomic numbering, shifted for convenience by 13k nts from 5′. Imino-proton correlations in (A) between consecutive base pairs are shown in black and red for the two present exchanging conformations. Included in (A) is the experimentally observed equilibrium between secondary structures of the attenuator. Additional closing basepairs are annotated with ‘±x’. Colors boxes are according to the correlations in (A). Asterisks indicate secondary shifts due to conformational exchange.
PK The three-stemmed pseudoknot (PK, nts 13 475–13 542, Figure 13) structure controlling the ribosomal frameshifting during ORF1a/b translation (42) has a unique fold in SCoV and SCoV2 (43,44). While in other Coronaviruses, and in many ssRNA viruses in general, this RNA element forms a canonical H-type pseudoknot fold, SCoV and SCoV2 PKs include a third SL. This SL harbors a 6-nts palindromic sequence allowing for homodimerization. Mutation of this sequence resulted in lower frameshifting efficiency and altered SCoV growth kinetics in vivo (3). The structure of SCoV PK has been thoroughly investigated before (3,45) and with only one nucleotide changed from SCoV to SCoV2 (C13 533A), assignment of the monomeric form of the PK, once prepared in NMR-suitable purity grade and quantity (∼70% monomer, 8 mg), was straightforward. The presence of the three predicted stem regions could be confirmed in 1H,1H-NOESY spectra combined with the 1H,15N-HSQC spectrum of the imino region and the HNN-COSY spectrum (Figure 13).
3′-UTR
The conserved regulatory RNA sequences necessary for virus replication at the 3′-genomic end of Coronaviruses are exclusively found in the untranslated region. The 5′-most cis-regulatory element is a large, bulged SL (3_SL1, Figure 1), containing a 3′-sequence that forms either the lower part of the SL or base-pairs with a single-stranded sequence within the loop of a downstream, smaller SL (3_SL2, Figure 1). Thus, the SL1-SL2 element is a putative RNA switch, associated with regulation of replication (46,47). Furthermore, single-stranded regions flanking 3_SL2 form long-range interactions with the 3′-most part of the genome in all Betacoronaviruses, associated with nsp8 binding in replication initiation (46).
The downstream part of the 3′-UTR is called hypervariable region (HVR) and affects viral pathogenicity (48). The HVR contains the highly conserved s2m motif in Betacoronaviruses, including SCoV, and also the non-related Astroviruses (Rfam ID: RF00164; (49)). Precisely within this RNA element, a single-nucleotide substitution from SCoV to SCoV2 results in dramatic changes of the predicted secondary structure (Supplementary Figure S5), rendering s2m a very interesting subject for high-resolution structural characterization by NMR spectroscopy.
Thus, we divided the 3′-UTR into the single elements 3_SL1 (nts 29, 548-29, 614), 3_SL2 (nts 29, 630-29, 656), 3_SL3base (nts 29, 620–29, 671 fused to 29 849–29 870) and 3_s2m (nts 29, 728–29, 768), reflecting the most important structural elements in the 3′-UTR (Figure 1).
3_SL1 For 3_SL1, secondary structure prediction tools consistently proposed a long helical stem, interrupted by several small bulges and a larger symmetrical bulge composed of six uridine residues in the upper part of the stem. For the 3_SL1 RNA construct, we could assign the imino protons by analyzing cross peaks in a 1H,1H-NOESY experiment conducted on an unlabeled RNA and 1H,15N-TROSY-and HNN-COSY experiments (Figure 14). In addition to the predicted A-helical parts, we could observe three consecutive non-canonical U-U base pairs in the upper part of 3_SL1. These three base pairs give rise to six well-resolved imino proton resonances in the spectral area typical for U imino protons involved in G-U and U-U base pairs (δ 1H: 10–12.5 ppm, δ 15N: 155–160 ppm) (50). Base pairs U29 564-A29 600 to A29 569-U29 695 are symmetrical, resulting in nearly complete chemical shift degeneracy for the middle four base pairs of this helical stretch. DMS footprinting analysis confirms the presence of these base pairs, showing low reactivity of the respective adenosine residues (Supplementary Figure S20).
Figure 14.
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the 3′-genomic end construct 3_SL1 encompassing nts 29 548–29 613. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering, shifted for convenience by 29 000 nts from 5′. Imino-proton correlations in (A) between consecutive base pairs are shown in different colors. Included in (C) is the experimentally observed secondary structure of 3_SL1 with genomic numbering, shortened by 29k nts from 5′. Additional closing base pairs are annotated with ‘±x’. Colors of boxes are according to the correlations in (A).
3_SL2 3_SL2 is the second hairpin element in the 3′-UTR with a rather large loop sequence (11 nts) that is complementary to the upstream 3′-most sequence of the bulged SL (3_SL1). Imino proton spectra show that the A-helical part of 3_SL2 is composed of nine base pairs (Figure 15 and Supplementary Figure S20). The loop-closing U-A base pair is invisible, likely due to an enhanced life-time of the open state that promotes fast solvent exchange of the imino proton. In contrast to computational secondary structure predictions, no stable base pairs between loop nucleotides are observed (Supplementary Figure S20). In line with NMR data, DMS footprinting showed a high reactivity of the adenosine of the loop-closing U-A base pair. In contrast, DMS data showed only partial reactivity in the loop nucleotides suggestive of a confined conformation (Supplementary Figure S20).
Figure 15.
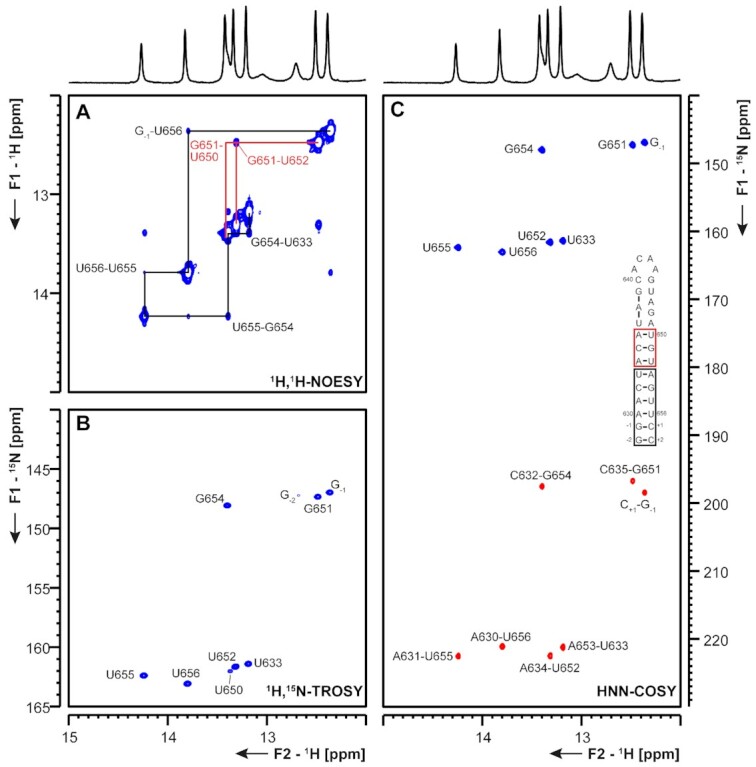
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the 3′-genomic end construct 3_SL2 encompassing nts 29 630–29 656. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering, shifted for convenience by 29 000 nts from 5′. Imino-proton correlations in (A) between consecutive base pairs are shown in different colors. Included in (C) is the experimentally observed secondary structure of 3_SL2. Additional closing base pairs are annotated with ‘±x’. Colors of boxes are according to the correlations in (A).
3_SL3base 3_SL3base represents the long-range RNA interactions between the 3′-end of the genome and the single-stranded regions flanking 3_SL2, with the complete HVR deleted and replaced by a stable UUCG tetraloop. In the NMR spectra of this construct we find three distinct base paired regions arranged in a three-way junction. The first paired region corresponds to a stretch of 4 bp closing the 5′- and 3′-end of the construct. At both sides five additional nucleotides are found that do not form persistent interactions that would lead to exchange protected imino proton signals (Figure 16). The second paired region is found in the SL part of the molecule that is identical to 3_SL2, except for the stabilizing G-C base pairs at the end of the construct introduced for in-vitro transcription. In 3_SL3base a stretch of three Watson–Crick base pairs from A29 634-A29 636 and U29 650-U29 652 plus the additional base pair between C29 632 and G29 654 is formed, the other interactions that were mapped in the smaller and more stabilized 3_SL2 could not be detected (compare Figure 15 and Figure 16). In line with 3_SL2, in the 3_SL3base construct neither the loop nucleotides nor the A-U closing base pair exhibited detectable imino-proton resonances and can therefore most probably be regarded as unpaired. The third paired region in 3_SL3base shows a network of 10 consecutive base pairs including a pair between U29 845 and C29 666 as well as between U29 846 and U29 665, in the middle of the stem (Figure 16). That the stability is an intrinsic feature of the sequence and not induced by the high stability cUUCGg-loop that caps this helical region, is confirmed by DMS footprinting. Here the entire stem shows a low DMS reactivity validating the introduced mutations (Supplementary Figure S21). Although part of the construct constitutes the primer binding site, the DMS data are in good agreement with the entire NMR secondary structure, except for the loop of SL2. To investigate if magnesium ions present during DMS footprinting are able to induce the predicted base pair formation (Supplementary Figure S21), which could explain the lower reactivity of C29 640, we performed Mg2+ titrations with 15N-labeled 3_SL3base. No additional imino proton resonances could be detected at 3 mM Mg2+ (Supplementary Figure S22). Thus, the observed differences between NMR and DMS reactivity is not due to altered base pairing patterns in different ionic conditions, but more likely the result of a restricted conformational flexibility within the loop of 3_SL2 (see ‘Discussion’ section).
Figure 16.
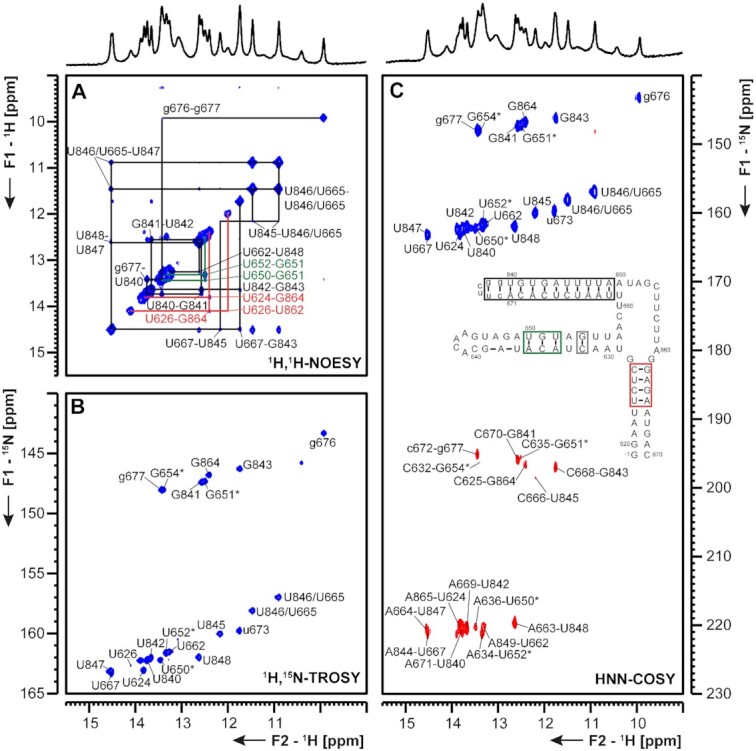
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the 3′-genomic end construct 3_SL3base encompassing nts 29 620–29 671 and 29 840–29 870. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering, shifted for convenience by 29 000 nts from 5′. Assignments that are marked with an asterisk are tentative and might be subject to change. Imino-proton correlations in (A) between consecutive base pairs are shown in different colors. Included in (C) is the experimentally observed secondary structure of 3_SL3base. Additional closing base pairs are annotated with ‘±x’. The colors of boxes are according to the correlations in (A). Gray boxes mark base pairs, which were not assigned based on imino-to-imino correlations.
3_s2m For 3_s2m, we performed assignment of the imino protons by analyzing cross peaks in a 1H,1H-NOESY, 1H,15N-TROSY, HNN-COSY, 1H,15N-CPMG-NOESY and long range 1H,15N-sfHMQC experiments (Figure 17 and Supplementary Figure S23). The assigned base pair pattern unambiguously reveals a secondary structure that consists of two stem regions separated by an internal asymmetric loop. This secondary structure proposal is in line with the protection pattern we observed in DMS-footprinting (Supplementary Figure S23C). In addition, the imino proton resonances of 3_s2m almost perfectly overlay with those of 3_HVR in 1H,15N-TROSY spectra (Figure 18), with 3_HVR representing the native sequence context of 3_s2m.
Figure 17.
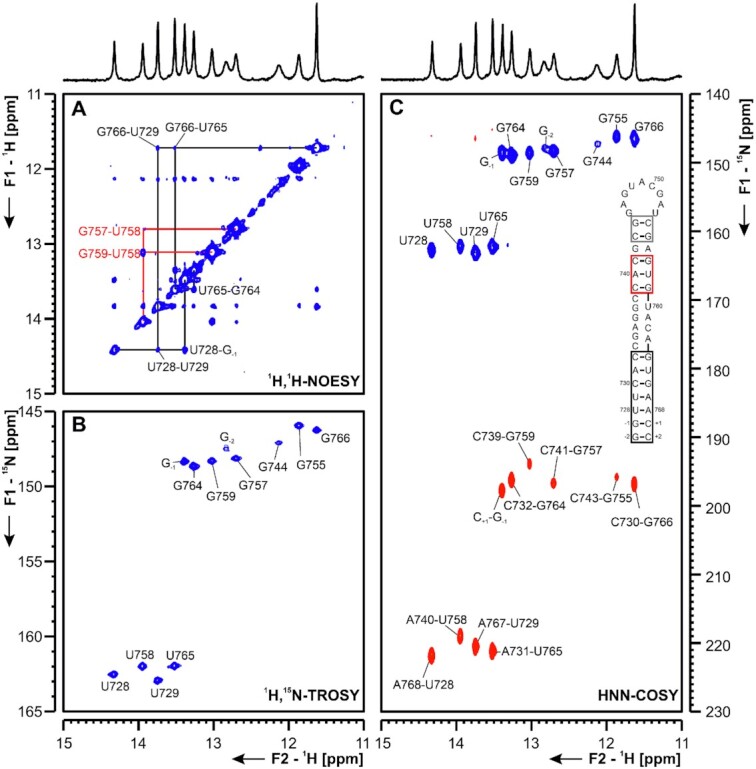
(A) 1H,1H-NOESY, (B) 1H,15N-TROSY and (C) HNN-COSY spectra for imino-proton correlation of the 3′-genomic end construct 3_s2m encompassing nts 29 728–29 768. Positive contours are given in blue, negative contours in red. The imino-proton correlations are annotated using the genomic numbering, shifted for convenience by 29 000 nts from 5′. Imino proton correlations in (A) between consecutive base pairs are shown in different colors. Included in (C) is the experimentally observed secondary structure of 3_s2m. Assignments that are marked with an asterisk are tentative and might be subject to change. Additional closing base pairs are annotated with ‘±x’. Colors of boxes are according to the correlations in (A).
Figure 18.
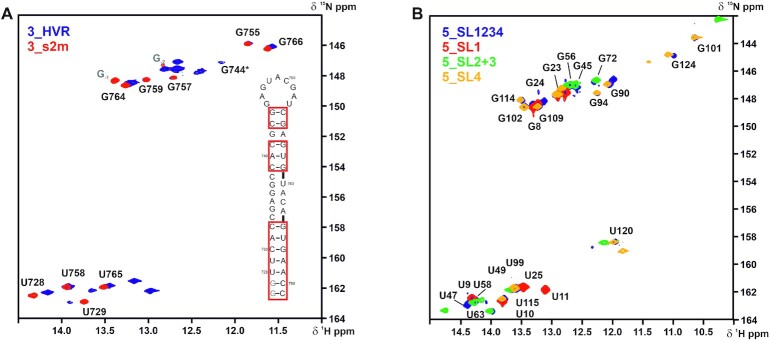
Overlay of 1H,15N-TROSY spectra for the imino-proton correlations of (A) 3_HVR (blue) and 3_s2m (red) and (B). 5_SL1-4 (blue), 5_SL1 (red), 5_SL2+3 (green) and 5_SL4 (yellow). 5_SL1-4 encompasses nts 7–125 and 3_HVR consists of nts 29 698–29 806. Given assignments were derived from the spectra of the single SLs (see Figures 4, 5 and 6 for (B) and Figure 18 for (A) with genomic numbering shifted for convenience by 29,000 nts from 5′. Spectra in (A) were recorded at 298 K and spectra in (B) were recorded at 283 K. In (A), the isolated 3_s2m is depicted for illustration of assignments. The additional nucleotides G-1 and G-2 are colored in gray. The asterisk denotes a signal which shows up at lower contour levels for 3_s2m. In (B), only resonances which are present both in 5_SL1234 and the respective isolated RNA are annotated. Secondary structures are omitted for clarity.
Within the SCoV2 genome, a G29,758U mutation is observed for the s2m element, which seems to cause a register-shifted base-pairing in the upper hairpin stem. Key for the assignment of the symmetric motif of G-C base pairs neighboring A29 740-U29 758 and the two loop closing C-G base pairs was the identification of an imino H2 cross peak to A29 756. The loop-closing G-C base pairs C29 743-G29 755 and G29 744-C29 754 show very weak signals in 1H,15N-HNN-COSY experiments and in 1H,1H-NOESY experiments no imino-imino cross peaks are detected for these residues at 298 K, while they could be observed at 283 K (data not shown). We therefore assume only weak or respectively transient base-pairing interactions at room temperature, despite the fact that C29 743 and C29 754 remain unmodified in DMS footprinting. To understand how this affects the flexibility of the relatively large loop of 9 nts will require further investigation, in particular as—in contrast to SCoV-2—the SCoV 3_s2m element features a rigid loop-geometry (49). The observed secondary structure for SCoV2, thus strongly deviates from the structure of SCoV.
In order to further substantiate the validity of our approach to investigate the cis-acting RNA elements in isolation, we analyzed the SLs SL1 to SL4 from the 5′-UTR and the 3_s2m motif within their native sequence context. We recorded 1H,15N-TROSY spectra of 5_SL1-4 (nts 7–125) and 3_HVR (nts 29 698-29 806) and observed very good agreement with the TROSY spectra of the isolated SLs (Figure 18). Major differences for imino proton resonances are limited to the signals of the additional G/C nucleotides introduced to the isolated RNA elements to enable T7 transcription and stem stabilization. Importantly, these additional nucleotides did not introduce artificial structural changes other than stabilizing the corresponding helical part. The 3_s2m motif forms a stable structural unit within the 3_HVR RNA, demonstrating that its unique fold adopted in SCoV2 is maintained in the longer sequence context. Moreover, it appears to exist as an independent structural unit, since no indications for any additional long-range interactions are observed. This important observation also holds true when comparing the TROSY spectra of SL1-4 with those of the isolated RNAs: no additional resonances appear in the non-canonical spectral regions, which would be indicative for tertiary structure formation or long-range interactions between the individual SLs, exclusively possible within the longer sequence context. Rather, the almost perfect matching of imino proton resonances of 5_SL1, 5_SL2+3 and 5_SL4 onto those of 5_SL1-4, clearly demonstrates that these hairpins form independent structural units within the longer sequence context. Thus, they most likely represent the respective predicted functional units also in the context of the entire genome as well as in the context of the sg mRNA leader sequences (see paragraphs 5_SL1, 5_SL2+3 and 5_SL4).
In summary, by combining reactivity profiles by DMS footprinting and extensive NMR-spectroscopic analysis of a total of 22 RNA constructs representing regulatory relevant regions of the SCoV2 genome, we present conclusive experimental validation of 15 RNA secondary structure models: SLs 1 to 8 of the 5′-genomic end; the attenuator hairpin and the pseudoknot of the frameshift regulating region, and SLs 1 to 3 and s2m of the 3′ genomic end.
DISCUSSION
We analyzed the secondary structures of 15 individual cis-acting RNA elements from SCoV2 by NMR spectroscopy, and complemented these data with DMS footprinting analyses of 11 of these RNAs as well as NMR analyses of four 119/472- and 115/337-nt long sequences representing (parts of) the 5′-and 3′-ends of the SCoV2 RNA genome, respectively. All NMR data have been deposited in the BMRB (8). While secondary structure characterization by NMR relies on detecting protons that are stably involved in base-pairing interactions (first of all G and U imino protons), DMS detects highly flexible A and C residues in non-structured regions of the RNA. By combining these two complementary methods, we were able to structurally characterize all 15 RNA elements at single base pair resolution. We find an excellent overall agreement of RNA secondary structures obtained from DMS footprinting within their native, full-length sequence context (top-down approach) with RNA secondary structures determined by NMR spectroscopy for isolated RNA elements (bottom-up approach). Notably, no conflicting secondary structures are suggested by the two methods, except for the low reactivity of some of the loop nucleotides from 3_SL2 observed by DMS probing (see ‘Discussion’ section below). Recently, preliminary SHAPE and DMS footprinting data have been made available in bioRxiv for the 5′-genomic end, the frameshifting region and even the full-length SCoV2 genome, which overall confirm our secondary structural models (34–35,51–53). Figure 19 shows an overview of the structure of the 15 analyzed cis-elements. Regions showing differences in between the available NMR and structural probing data are depicted in gray and discussed below.
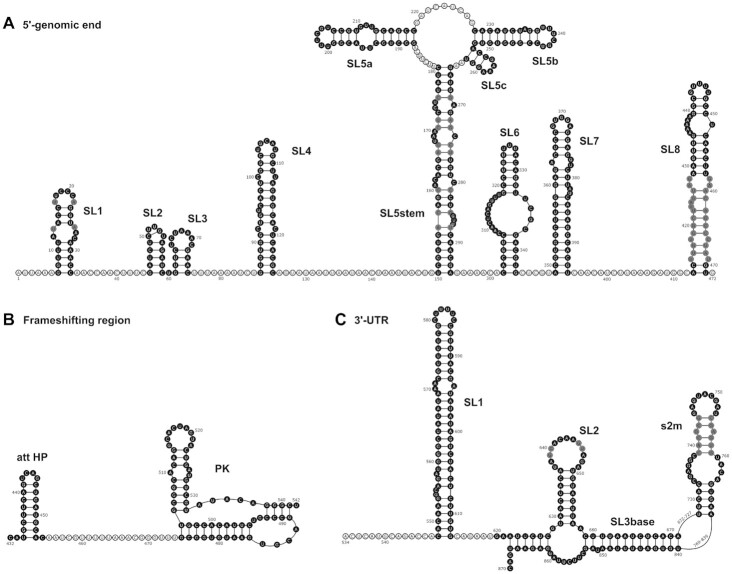
Figure 19.
Overview of the experimentally derived structures for the cis-elements of the 5′-genomic end (A), the frameshifting region (B) and the 3′-UTR (C). Genomic numbering is shifted for convenience by 13 000 from 5′ for the frameshifting region and 29 000 for the 3′-UTR. Cis-elements analyzed by NMR spectroscopy are highlighted in black. Regions with unclear base pairing patterns or high reactivity in structural probing data are shown in gray and discussed in the main text.
5_SL1 NMR analyses indicate open A-U base pairs flanking the upper helix of SL1. Since SL1 is located at the extreme 5'-end of the genome, it is not resolved in most structural studies. In the datasets where SL1 is probed, either the upper or the lower A-U base pair is non-reactive (34,35). This flexibility might affect recognition of SL1 by Nsp1 and should be addressed in future studies of this RNA–protein complex.
5_SL5 Small deviations between DMS- and NMR-based secondary structure models were observed in 5_SL5, where some residues detected to be base-paired in NMR experiments showed high reactivity in DMS footprinting (see Supplementary Figure S10). As DMS experiments were performed at 3 mM Mg2+ and higher temperature, we repeated the NMR experiments for 5_SL5stem at 1.5 and 3 mM Mg2+ as well as at 298 K. While Mg2+ addition mainly induces small chemical shift perturbations (CSPs), the increase in temperature results in dramatic line broadening for imino protons U288, U283, U279 and to a lesser extent U276. These uridine residues are precisely the ones base-pairing with the adenosine residues showing high reactivity in DMS probing, thus demonstrating temperature to be a key effector for stability of this long-range interaction in-vitro. Accordingly, other structural probing data likewise showed enhanced reactivity within the stem region of SL5 (35,51,52). A possible reason might be an enhanced conformational heterogeneity within this larger substructure. Further, division of 5_SL5 into three subparts along with terminal stabilization for NMR spectroscopy abolishes the possibility for tertiary contacts between the four helices that may be expected to form within the entire SL5 RNA. More detailed high-resolution analysis of full-length 5_SL5 should be performed in the future, in order to clarify its exact folding details and intrinsic dynamics.
5_SL8 NMR spectra show several non-canonical interactions within the middle part of SL8, which could not be unambiguously assigned so far. A comparison of the available probing data likewise shows differing reactivities across the various datasets, necessitating further detailed high-resolution analyses (34–35,51).
3_SL2 DMS footprinting data showed only intermediate reactivity for residues in the large loop of 3_SL2, while NMR spectroscopy revealed an entirely open structure, without any stable base-pairing interactions. Addition of up to 3 mM Mg2+ did not show any influence on the loop stability of SL2 in the 3_SL3base construct. Conversely to DMS probing, SHAPE data suggest that the majority of loop residues within SL2 are flexible (35,51,54), suggesting that the lower reactivity found in DMS footprinting analysis reports on a restricted conformational flexibility of the loop of SL2, rather than base pairing interactions. These variations in 3_SL2 loop conformations are highly interesting, as SL2 is supposed to be part of a molecular switch crucial in regulating genome synthesis (40). The loop of SL2 is either free or forms a pseudoknot interaction with the basal stem of 3_SL1. Strikingly, both the two-hairpin and the pseudoknot structures are essential for viral replication (47). While an open loop structure will facilitate pseudoknot formation, a defined folding of the loop might be required for protein interaction in the two-hairpin conformation. Thus, the loop conformation of 3_SL2 needs to be further investigated.
3_s2m Compared to SCoV, the s2m motif from SCoV2 harbors two mutations (Supplementary Figure S1). One compensatory mutation in the lower helix and one mutation that leads to destabilization of the base pairing of the upper helix. This overall instability of the SCoV2 s2m also detected by NMR, is reflected by high reactivity of this motif in several of the available probing datasets (35,51,54).
In addition to the analysis of individual cis-elements, experimental data on the full-length RNA will guide further detailed structural investigations. For example, DMS footprinting suggested an additional helical part for 5_SL4, which is conserved in SCoV. Interestingly, an alternative model for SCoV2 is proposed by computational predictions, where a small SL forms directly downstream of SL4 (45). NMR will be the method of choice to delineate which of the two secondary structures actually form in an RNA construct covering the complete sequence in question. Further, a recent study investigated RNA–RNA long-range interactions within the SCoV2 genome (55). In line with our initial analysis of larger regions of the 5′-genomic end and the 3′-UTR, no long-range interactions between the cis-elements within the two regions were identified other than the SL5stem and SL3base. Notably, long-range interactions of the 5′-genomic end and the 3′-UTR with each other as well as coding regions of the genome were identified, which provide interesting new starting points for the investigation of RNA structures important for virus propagation.
In general, we provide a thorough experimental validation of phylogenetic-based in silico models for the RNA elements characterized in this study. Differences found between prediction and experimental data show a trend toward fewer stable base pairs in the experimentally determined structures, i.e. A-U base pairs next to bulges or two base pair helices including one G-U predicted to be paired are found to be open in the experimental data (both NMR and DMS). In addition, several non-canonical base pairs could be identified that are not considered in the in silico predictions (Supplementary Figure S5). Comparing with the sequence to SCoV, the overall sequence identity of the analyzed RNA elements is 91%, with 5_SL2, 5_SL3 and 5_SL5c showing complete identity, and att HP exhibiting the largest divergence with only 79% identity. In terms of structure, all RNAs are highly conserved although several quite notable exceptions have been detected in this regard. This includes the 5_SL1 element, the s2m element and the att HP. The 24 nt att HP, located immediately upstream of the slippery site and the PRF PK, exhibits five nt variations relative to SCoV that predict it to be significantly less stable (36). In line with this observation, the NMR data indicate that the folding of this motif is substantially different to that proposed for the att HP of SCoV, and involves at least two main conformations (Figure 12). In this last respect, a distinctive advantage of our NMR approach relative to other methodologies is that it allows specific detection of RNA dynamics. Conformational exchange processes have been detected in particular for the att HP and potentially also for other SCoV2 cis-acting elements, including 5_SL7 and 5_SL8. These dynamic processes may be relevant for small-molecule or protein recognition.
Overall, however, it is reasonable to expect very similar functional properties for cis-acting RNAs in SCoV and SCoV2, which will certainly accelerate progress in elucidating virus biology. Further, 3D modeling efforts such as FARFAR, aiming at providing structures of potential drug targets will benefit from independent, high-resolution experimental validation (6). We provide here an extensive set of chemical shift data covering the entire 5′- and large parts of the 3′-genomic ends as well as two elements from the frameshifting region as a conclusive and reliable basis for further structure-based investigations of the SCoV2 RNA genome. This will greatly facilitate research progress toward defeating the virus and fighting Covid-19.
DATA AVAILABILITY
Chemical shifts are deposited at BMRB. All experimental data are deposited and can be downloaded at http://covid19-nmr.de.
Supplementary Material
ACKNOWLEDGEMENTS
Dr L. Frydman holds the Bertha and Isadore Gudelsky Professorial Chair and heads the Helen and Martin Kimmel Institute for Magnetic Resonance Research, the Clore Institute for High-Field Magnetic Resonance Imaging and Spectroscopy and the Fritz Haber Center for Physical Chemistry—whose collective support is also acknowledged. Dr T. Scherf is the incumbent of the Monroy-Marks Research Fellow Chair. We thank Prof. Eric Westhof for helpful discussions. Special thanks goes to Klara R. Mertinkus and Robbin Schnieders for the figure design.
Contributor Information
Anna Wacker, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Julia E Weigand, Department of Biology, Technical University of Darmstadt, Schnittspahnstrasse 10, 64287 Darmstadt, Germany.
Sabine R Akabayov, Faculty of Chemistry, Weizmann Institute of Science, 7610001 Rehovot, Israel.
Nadide Altincekic, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Jasleen Kaur Bains, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Elnaz Banijamali, Department of Medical Biochemistry and Biophysics, Karolinska Institute, Biomedicum 9B, Solnavägen 9, 17177 Stockholm, Sweden.
Oliver Binas, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Jesus Castillo-Martinez, School of Medicine, Catholic University of Valencia, C/Quevedo 2, 46001 Valencia, Spain.
Erhan Cetiner, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Betül Ceylan, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Liang-Yuan Chiu, Department of Chemistry, Case Western Reserve University, Cleveland, OH 44106, USA.
Jesse Davila-Calderon, Department of Chemistry, Case Western Reserve University, Cleveland, OH 44106, USA.
Karthikeyan Dhamotharan, Institute for Molecular Biosciences.
Elke Duchardt-Ferner, Institute for Molecular Biosciences.
Jan Ferner, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Lucio Frydman, Faculty of Chemistry, Weizmann Institute of Science, 7610001 Rehovot, Israel.
Boris Fürtig, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
José Gallego, School of Medicine, Catholic University of Valencia, C/Quevedo 2, 46001 Valencia, Spain.
J Tassilo Grün, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Carolin Hacker, Signals GmbH & Co. KG, Graf-von-Stauffenberg-Allee 83, 60438 Frankfurt/M, Germany.
Christina Haddad, Department of Chemistry, Case Western Reserve University, Cleveland, OH 44106, USA.
Martin Hähnke, Signals GmbH & Co. KG, Graf-von-Stauffenberg-Allee 83, 60438 Frankfurt/M, Germany.
Martin Hengesbach, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Fabian Hiller, Signals GmbH & Co. KG, Graf-von-Stauffenberg-Allee 83, 60438 Frankfurt/M, Germany.
Katharina F Hohmann, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Daniel Hymon, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Vanessa de Jesus, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Henry Jonker, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Heiko Keller, Institute for Molecular Biosciences.
Bozana Knezic, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Tom Landgraf, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Frank Löhr, Institute of Biophysical Chemistry, Center for Biomolecular Magnetic Resonance (BMRZ), Johann Wolfgang Goethe-University Frankfurt, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Le Luo, Department of Medical Biochemistry and Biophysics, Karolinska Institute, Biomedicum 9B, Solnavägen 9, 17177 Stockholm, Sweden.
Klara R Mertinkus, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Christina Muhs, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Mihajlo Novakovic, Faculty of Chemistry, Weizmann Institute of Science, 7610001 Rehovot, Israel.
Andreas Oxenfarth, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Martina Palomino-Schätzlein, School of Medicine, Catholic University of Valencia, C/Quevedo 2, 46001 Valencia, Spain.
Katja Petzold, Department of Medical Biochemistry and Biophysics, Karolinska Institute, Biomedicum 9B, Solnavägen 9, 17177 Stockholm, Sweden.
Stephen A Peter, Department of Biology, Technical University of Darmstadt, Schnittspahnstrasse 10, 64287 Darmstadt, Germany.
Dennis J Pyper, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Nusrat S Qureshi, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Magdalena Riad, Department of Medical Biochemistry and Biophysics, Karolinska Institute, Biomedicum 9B, Solnavägen 9, 17177 Stockholm, Sweden.
Christian Richter, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Krishna Saxena, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Tatjana Schamber, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Tali Scherf, Faculty of Chemistry, Weizmann Institute of Science, 7610001 Rehovot, Israel.
Judith Schlagnitweit, Department of Medical Biochemistry and Biophysics, Karolinska Institute, Biomedicum 9B, Solnavägen 9, 17177 Stockholm, Sweden.
Andreas Schlundt, Institute for Molecular Biosciences.
Robbin Schnieders, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Harald Schwalbe, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Alvaro Simba-Lahuasi, School of Medicine, Catholic University of Valencia, C/Quevedo 2, 46001 Valencia, Spain.
Sridhar Sreeramulu, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Elke Stirnal, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Alexey Sudakov, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Jan-Niklas Tants, Institute for Molecular Biosciences.
Blanton S Tolbert, Department of Chemistry, Case Western Reserve University, Cleveland, OH 44106, USA.
Jennifer Vögele, Institute for Molecular Biosciences.
Lena Weiß, Institute for Molecular Biosciences.
Julia Wirmer-Bartoschek, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Maria A Wirtz Martin, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
Jens Wöhnert, Institute for Molecular Biosciences.
Heidi Zetzsche, Institute for Organic Chemistry and Chemical Biology, Max-von-Laue-Strasse 7, 60438 Frankfurt/M., Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Goethe University; Deutsche Forschungsgemeinschaft (DFG) [CRC902]; National Institutes of Health [U54AI50470, R01GM126833 B.S.T.]; EU Horizon 2020 [828946]; Weizmann's Internal Coronavirus Fund; German-Israel Foundation [G-1501-302]; Spanish Ministerio de Economía y Competitividad [RTI2018-093935-B-I00]; la Caixa Banking Foundation; Catholic University of Valencia; Hessisches Ministerium für Wissenschaft und Kunst [BMRZ]; iNEXT-Discovery [871037]. Funding for open access charge: DFG.
Conflict of interest statement. None declared.
This paper is linked to: doi:10.1093/nar/gkaa1053.
REFERENCES
- 1. Rangan R., Zheludev I.N., Hagey R.J., Pham E.A., Wayment-Steele H.K., Glenn J.S., Das R.. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA. 2020; 26:937–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y.. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020; 9:221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ishimaru D., Plant E.P., Sims A.C., Yount B.L., Roth B.M., Eldho N. V., Pérez-Alvarado G.C., Armbruster D.W., Baric R.S., Dinman J.D.et al.. RNA dimerization plays a role in ribosomal frameshifting of the SARS coronavirus. Nucleic Acids Res. 2013; 41:2594–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang D., Leibowitz J.L.. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015; 206:120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Madhugiri R., Fricke M., Marz M., Ziebuhr J.. Coronavirus cis-acting RNA elements. Adv. Virus Res. 2016; 96:127–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rangan R., Watkins A., Kladwang W., Das R.. De novo 3D models of SARS-CoV-2 RNA elements and small-molecule-binding RNAs to guide drug discovery. 2020; bioRxiv doi:15 April 2020, preprint: not peer reviewed 10.1101/2020.04.14.041962. [DOI]
- 7. Andrews R., Peterson J., Haniff H., Chen J., Williams C., Grefe M., Disney M., Moss W.. An in silico map of the SARS-CoV-2 RNA Structurome. 2020; bioRxiv doi:18 April 2020, preprint: not peer reviewed 10.1101/2020.04.17.045161. [DOI] [PMC free article] [PubMed]
- 8. Ulrich E.L., Akutsu H., Doreleijers J.F., Harano Y., Ioannidis Y.E., Lin J., Livny M., Mading S., Maziuk D., Miller Z.et al.. BioMagResBank. Nucleic Acids Res. 2008; 36:D402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tomezsko P., Swaminathan H., Rouskin S.. Viral RNA structure analysis using DMS-MaPseq. Methods. 2020; 10.1016/j.ymeth.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bellaousov S., Reuter J.S., Seetin M.G., Mathews D.H.. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013; 41:W471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Darty K., Denise A., Ponty Y.. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Milligan J.F., Uhlenbeck O.C.. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989; 180:51–62. [DOI] [PubMed] [Google Scholar]
- 13. Ferré-D¢Amaré A.R., Doudna J.A.. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996; 24:977–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guilleres J., Lopez P.J., Proux F., Launay H., Dreyfus M.. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5958–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chillón I., Marcia M., Legiewicz M., Liu F., Somarowthu S., Pyle A.M.. woodson S.A., Allain F.H.T.. Native purification and analysis of long RNAs. Methods in Enzymology. 2015; 558:Academic Press Inc; 3–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee W., Tonelli M., Markley J.L.. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015; 31:1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D.. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct. Funct. Genet. 2005; 59:687–696. [DOI] [PubMed] [Google Scholar]
- 18. Fürtig B., Richter C., Wöhnert J., Schwalbe H.. NMR spectroscopy of RNA. Chembiochem. 2003; 4:936–962. [DOI] [PubMed] [Google Scholar]
- 19. Lopez J.J. Forschungsdaten: Alle Zahlen mit wenigen Klicks. Nachr. Chem. 2020; 68:27–28. [Google Scholar]
- 20. Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Janssen S., Giegerich R.. The RNA shapes studio. Bioinformatics. 2015; 31:423–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tanaka T., Kamitani W., DeDiego M.L., Enjuanes L., Matsuura Y.. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 2012; 86:11128–11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li L., Kang H., Liu P., Makkinje N., Williamson S.T., Leibowitz J.L., Giedroc D.P.. Structural lability in SL 1 drives a 5′ UTR-3′ UTR interaction in coronavirus replication. J. Mol. Biol. 2008; 377:790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen S.C., Olsthoorn R.C.L.. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology. 2010; 401:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Madhugiri R., Karl N., Petersen D., Lamkiewicz K., Fricke M., Wend U., Scheuer R., Marz M., Ziebuhr J.. Structural and functional conservation of cis-acting RNA elements in coronavirus 5′-terminal genome regions. Virology. 2018; 517:44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee C.W., Li L., Giedroc D.P.. The solution structure of coronaviral SL 2 (SL2) reveals a canonical CUYG tetraloop fold. FEBS Lett. 2011; 585:1049–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., Leibowitz J.L., Giedroc D.P.. Coronavirus N protein N-Terminal Domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 2009; 394:544–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dufour D., Mateos-Gomez P.A., Enjuanes L., Gallego J., Sola I.. Structure and functional relevance of a transcription-regulating sequence involved in coronavirus discontinuous RNA synthesis. J. Virol. 2011; 85:4963–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang D., Liu P., Giedroc D.P., Leibowitz J.. Mouse hepatitis virus SL 4 functions as a spacer element required to drive subgenomic RNA synthesis. J. Virol. 2011; 85:9199–9209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu H.-Y., Guan B.-J., Su Y.-P., Fan Y.-H., Brian D.A.. Reselection of a genomic upstream open reading frame in mouse hepatitis coronavirus 5′-Untranslated-Region mutants. J. Virol. 2014; 88:846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guan B.-J., Su Y.-P., Wu H.-Y., Brian D.A.. Genetic evidence of a long-range RNA-RNA interaction between the genomic 5 = untranslated region and the nonstructural protein 1 coding region in murine and bovine coronaviruses. J. Virol. 2012; 86:4631–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guan B.-J., Wu H.-Y., Brian D.A.. An optimal cis-Replication SL IV in the 5′ untranslated region of the mouse coronavirus genome extends 16 nucleotides into open reading frame 1. J. Virol. 2011; 85:5593–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nozinovic S., Fürtig B., Jonker H.R.A., Richter C., Schwalbe H.. High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acids Res. 2010; 38:683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Iserman C., Roden C., Boerneke M., Sealfon R., McLaughlin G., Jungreis I., Park C., Boppana A., Fritch E., Hou Y.J.et al.. Specific viral RNA drives the SARS CoV-2 nucleocapsid to phase separate. 2020; bioRxiv doi:12 June 2020, preprint: not peer reviewed 10.1101/2020.06.11.147199. [DOI]
- 35. Lan T.C.T.T., Allan M.F., Malsick L.E., Khandwala S., Nyeo S.S.Y.Y., Bathe M., Griffiths A., Rouskin S.. Structure of the full SARS-CoV-2 RNA genome in infected cells. 2020; bioRxiv doi:26 October 2020, preprint: not peer reviewed 10.1101/2020.06.29.178343. [DOI]
- 36. Jucker F.M., Heus H.A., Yip P.F., Moors E.H.M.M., Pardi A.. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996; 264:968–980. [DOI] [PubMed] [Google Scholar]
- 37. Yang D., Liu P., Wudeck E. V., Giedroc D.P., Leibowitz J.L.. Shape analysis of the rna secondary structure of the mouse hepatitis virus 5′ untranslated region and n-terminal nsp1 coding sequences. Virology. 2015; 475:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brockway S.M., Denison M.R.. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology. 2005; 340:209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cho C.P., Lin S.C., Chou M.Y., Hsu H.T., Chang K.Y.. Regulation of programmed ribosomal frameshifting by Co-Translational refolding RNA hairpins. PLoS One. 2013; 8:e62283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kelly J.A., Olson A.N., Neupane K., Munshi S., San Emeterio J., Pollack L., Woodside M.T., Dinman J.D.. Structural and functional conservation of the programmed -1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). J. Biol. Chem. 2020; 295:10741–10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baranov P. V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T.. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005; 332:498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Su M.C., Chang C. Te, Chu C.H., Tsai C.H., Chang K.Y.. An atypical RNA pseudoknot stimulator and an upstream attenuation signal for -1 ribosomal frameshifting of SARS coronavirus. Nucleic Acids Res. 2005; 33:4265–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Plant E.P., Pérez-Alvarado G.C., Jacobs J.L., Mukhopadhyay B., Hennig M., Dinman J.D.. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 2005; 3:1012–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Plant E.P., Perez-Alvarado G.C., Jacobs J.L., Mukhopadhyay B., Hennig M., Dinman J.D., Pérez-Alvarado G.C., Jacobs J.L., Mukhopadhyay B., Hennig M.et al.. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 2005; 3:e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zust R., Miller T.B., Goebel S.J., Thiel V., Masters P.S.. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J. Virol. 2008; 82:1214–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goebel S.J., Hsue B., Dombrowski T.F., Masters P.S.. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2004; 78:669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goebel S.J., Miller T.B., Bennett C.J., Bernard K.A., Masters P.S.. A hypervariable region within the 3′ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J. Virol. 2007; 81:1274–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robertson M.P., Igel H., Baertsch R., Haussler D., Ares M., Scott W.G.. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 2005; 3:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ohlenschläger O., Wöhnert J., Bucci E., Seitz S., Häfner S., Ramachandran R., Zell R., Görlach M.. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure. 2004; 12:237–248. [DOI] [PubMed] [Google Scholar]
- 51. Manfredonia I., Nithin C., Ponce-Salvatierra A., Ghosh P., Wirecki T.K., Marinus T., Ogando N.S., Snijder E.J., van Hemert M.J., Bujnicki J.M.et al.. Genome-wide mapping of therapeutically-relevant SARS-CoV-2 RNA structures. 2020; bioRxiv doi:15 June 2020, preprint: not peer reviewed 10.1101/2020.06.15.151647. [DOI] [PMC free article] [PubMed]
- 52. Sanders W., Fritch E.J., Madden E.A., Graham R.L., Vincent H.A., Heise M.T., Baric R.S., Moorman N.J.. Comparative analysis of coronavirus genomic RNA structure reveals conservation in SARS-like coronaviruses. 2020; bioRxiv doi:16 June 2020, preprint: not peer reviewed 10.1101/2020.06.15.153197. [DOI]
- 53. Tavares R.C.A., Mahadeshwar G., Pyle A.M.. The global and local distribution of RNA structure throughout the SARS-CoV-2 genome. 2020; bioRxiv doi:07 July 2020, preprint:not peer reviewed 10.1101/2020.07.06.190660. [DOI] [PMC free article] [PubMed]
- 54. Huston N., Wan H., Araujo Tavares R., de C., Wilen C., Pyle A.M.. Comprehensive in-vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. 2020; bioRxiv doi:10 July 2020, preprint: not peer reviewed 10.1101/2020.07.10.197079. [DOI] [PMC free article] [PubMed]
- 55. Ziv O., Price J., Shalamova L., Kamenova T., Goodfellow I., Weber F., Miska E.A.. The short-and long-range RNA-RNA Interactome of SARS-CoV-2. 2020; bioRxiv doi:20 July 2020, preprint: not peer reviewed 10.1101/2020.07.19.211110. [DOI] [PMC free article] [PubMed]
- 56. Sklenar V. Suppression of radiation damping in multidimensional NMR experiments using magnetic field gradients. J. Magn. Reson. Ser. A. 1995; 114:132–135. [Google Scholar]
- 57. Solyom Z., Schwarten M., Geist L., Konrat R., Willbold D., Brutscher B.. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J. Biomol. NMR. 2013; 55:311–321. [DOI] [PubMed] [Google Scholar]
- 58. Favier A., Brutscher B.. Recovering lost magnetization: polarization enhancement in biomolecular NMR. J. Biomol. NMR. 2011; 49:9–15. [DOI] [PubMed] [Google Scholar]
- 59. Mori S., Abeygunawardana C., Johnson M.O., Van Zljl P.C.M.. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. Ser. B. 1995; 108:94–98. [DOI] [PubMed] [Google Scholar]
- 60. Mueller L., Legault P., Pardi A.. Improved RNA structure determination by detection of NOE contacts to Exchange-Broadened amino protons. J. Am. Chem. Soc. 1995; 117:11043–11048. [Google Scholar]
- 61. Dingley A.J., Grzesiek S.. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2J(NN) couplings. J. Am. Chem. Soc. 1998; 120:8293–8297. [Google Scholar]
- 62. Lescop E., Kern T., Brutscher B.. Guidelines for the use of band-selective radiofrequency pulses in hetero-nuclear NMR: example of longitudinal-relaxation-enhanced BEST-type 1H-15N correlation experiments. J. Magn. Reson. 2010; 203:190–198. [DOI] [PubMed] [Google Scholar]
- 63. Schulte-Herbrüggen T., Sørensen O.W.. Clean TROSY: compensation for relaxation-induced artifacts. J. Magn. Reson. 2000; 144:123–128. [DOI] [PubMed] [Google Scholar]
- 64. Schanda P., Brutscher B.. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 2005; 127:8014–8015. [DOI] [PubMed] [Google Scholar]
- 65. Novakovic M., Olsen G.L., Pintér G., Hymon D., Fürtig B., Schwalbe H., Frydman L.. A 300-fold enhancement of imino nucleic acid resonances by hyperpolarized water provides a new window for probing RNA refolding by 1D and 2D NMR. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:2449–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ikura M., Bax A.. Isotope-filtered 2D NMR of a protein-peptide complex: study of a skeletal muscle myosin light chain kinase fragment bound to calmodulin. J. Am. Chem. Soc. 1992; 114:2433–2440. [Google Scholar]
- 67. Piotto M., Saudek V., Sklenár V.. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992; 2:661–665. [DOI] [PubMed] [Google Scholar]
- 68. Sklenář V., Piotto M., Leppik R., Saudek V.. Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. Journal of Magnetic Resonance - Series A. 1993; 102:241–245. [Google Scholar]
- 69. Shaka A., Lee C., Pines A.. Iterative schemes for bilinear operators; application to spin decoupling. J. Magn. Reson. 1988; 77:274–293. [Google Scholar]
- 70. Hwang T.L., Shaka A.J.. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. Ser. A. 1995; 112:275–279. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Chemical shifts are deposited at BMRB. All experimental data are deposited and can be downloaded at http://covid19-nmr.de.