Abstract
In health research, statistical methods are frequently used to address a wide variety of research questions. For almost every analytical challenge, different methods are available. But how do we choose between different methods and how do we judge whether the chosen method is appropriate for our specific study? Like in any science, in statistics, experiments can be run to find out which methods should be used under which circumstances. The main objective of this paper is to demonstrate that simulation studies, that is, experiments investigating synthetic data with known properties, are an invaluable tool for addressing these questions. We aim to provide a first introduction to simulation studies for data analysts or, more generally, for researchers involved at different levels in the analyses of health data, who (1) may rely on simulation studies published in statistical literature to choose their statistical methods and who, thus, need to understand the criteria of assessing the validity and relevance of simulation results and their interpretation; and/or (2) need to understand the basic principles of designing statistical simulations in order to efficiently collaborate with more experienced colleagues or start learning to conduct their own simulations. We illustrate the implementation of a simulation study and the interpretation of its results through a simple example inspired by recent literature, which is completely reproducible using the R-script available from online supplemental file 1.
Keywords: statistics & research methods, epidemiology, protocols & guidelines
Introduction
In health research, statistical methods are frequently used to address a wide variety of research questions. For almost every analytical challenge, different methods are available. But how do we choose between different methods and how do we judge whether the chosen method is appropriate for our specific study? Most statistical methods are developed under specific assumptions, but these assumptions are often difficult to check in applied settings. Moreover, performance of methods may still be reasonable when some assumptions are violated, such as the linearity of relationships in regression models in the presence of mild non-linear relationships. In real-life studies of human health, some of these formal underlying assumptions may be questionable or definitely violated. For example, frequent problems, such as unusual distributions, missing data, measurement errors, unmeasured confounders or lack of accurate information on event times, may affect the accuracy or even the validity of the proposed analyses. What conditions (eg, what sample size) are needed for a specific method to behave well? Which method is most appropriate in a particular setting?
The main objective of this paper is to demonstrate that simulation studies, that is, evaluation of synthetic data with known properties, are an invaluable tool for addressing these questions. We aim to provide a first introduction to simulation studies for data analysts or, more generally, for researchers involved at different levels in the analyses of health data, for example, data from observational studies or from clinical trials, who (1) may rely on simulation studies published in statistical literature to choose their statistical methods and who, thus, need to understand the criteria of assessing the validity and relevance of simulation results and their interpretation; and/or (2) need to understand the basic principles of designing statistical simulations in order to efficiently collaborate with a more experienced colleague or to start learning to conduct their own simulations. Our paper is intended for an audience that is otherwise not targeted by previous literature on simulation studies and uses a novel approach to introduce the basic principles of simulation studies to clinical researchers and end users of statistical methods. Statisticians interested in more details about statistical simulations are referred to the more technical overviews available in the literature.1–3
More generally, our introduction to simulation studies aims to draw the attention of readers of medical papers, including practitioners, to the importance of the choice of appropriate, validated statistical methods. The use of inappropriate statistical methods contributes to the replication crisis that has drawn increasing attention in recent years; see for example The Lancet series ‘Increasing value, reducing waste’.4 Simulation studies have a role to play in this global process as they are a means of identifying the appropriate methodology for a particular study in a specific context, thus improving research quality. In this context, understanding the principles of simulation studies allows clinical researchers to better use published simulation results. Note that simulation studies themselves also have to be relevant and replicable.
Statistical methodology has seen substantial development in recent times, but many of these developments are largely ignored in the practice of health data analyses. To help bridge the gap between methodological innovation and applications to medical data, the STRengthening Analytical Thinking for Observational Studies (STRATOS) initiative was launched in 2013.5 It aims to provide statistical guidance for key topics in the design and analysis of observational studies. In practice, analyses are sometimes conducted by researchers with limited statistical background. Consequently, STRATOS plans to develop guidance for researchers with different levels of statistical knowledge, including researchers without strong statistical backgrounds (see table 1 in Sauerbrei et al 5). For the analysis of observational studies, typically several approaches are possible, and the properties of each approach should be assessed in comparison with alternative methods. Simulation studies are key instruments for such assessments. Ideally, all data analysts should be familiar with them.
This paper is structured as follows. We first discuss the role of statistical simulation studies in the next section"The role of simulation studies". The section "Examples of statistical methods" outlines four relatively simple examples of statistical methods and then explains how the performance of these methods could be evaluated using simulation studies. The section "Basic principles of simulation studies" sketches out the basic principles of designing and conducting simulations. Finally, the section "An example of a statistical simulation" briefly illustrates the implementation of a simulation study and the interpretation of its results through a simple example inspired by recent literature.
The role of simulation studies
Comparing methods based on theory
During the first half of the 20th century, mathematical theory was the cornerstone of evaluating traditional statistical methods addressing well-defined problems. However, to investigate questions in modern medicine, more complex statistical modelling or the use of machine learning techniques is often required. Only in rare cases of low complexity and often of limited practical relevance mathematics tells us that—given the data satisfy certain properties—the considered method behaves in a particular way. For example, theory tells us that the two-sample t-test has better power to detect a true difference between mean values in two independent groups than the Mann-Whitney test—if the variable of interest is normally distributed within each of the two groups. Most theoretical results of this type are valid only under specific assumptions about the available data. While it may be acceptable to assume normally distributed data in the case of the simple example mentioned above, for more complex problems the required assumptions can be unrealistic; see the second, third and fourth subsections of the section "Examples of statistical methods" for examples beyond this simple case. Moreover, the process of verifying assumptions is often already challenging in practice; see for example Rochon et al 6 for an extensive simulation study of the choice between t-test and Mann-Whitney test, including considerations on normality checks.
Comparing methods using empirical data
Another approach for evaluating statistical methods consists of applying them to representative data sets from the considered field and assessing their performance, or more generally of observing their behaviour when using them in these data sets. Some important characteristics of statistical methods can indeed be derived from real data sets. For example, are results stable if we modify the data set slightly? For many approaches, however, the most important evaluation criteria cannot be assessed for real data, simply because for real data we do not know the true values of the underlying parameters we aim to draw inferences about. For example, if one method estimates a difference of 1 between two groups, and another estimates a difference of 2, we can see that they give us different results (assuming that the confidence intervals are narrow), but we do not know whether 1 or 2 is closer to the correct answer.
Why simulation studies?
A simulation study is useful if theoretical arguments are insufficient to determine whether the method of interest is valid in a specific real-life application or whether violations of the assumptions underlying the available theory (such as normal distribution of residuals, proportional hazards and so on) affect the validity of the results. In methodological research, simulations play a role similar to experiments in basic science.7 The idea of a simulation study is to investigate the behaviour of methods when applied to synthetic data sets with known characteristics. Because the ‘correct’ or ‘true’ answer is known by the researchers, who had full control of how the data were simulated, simulations permit assessment of whether the methods recover this known truth. For example, we may generate data with and without a treatment effect and then assess how often a test correctly or incorrectly rejects the null hypothesis of no treatment effect. Alternatively, we may generate data in which the treatment effect has a certain value and then study how accurately a regression model can estimate this known effect. Notice that such assessment is not possible using real data when the true response or the true effect is not known.
Suppose a scientist is planning a cohort study of the effect of an exposure on time to a clinical event (eg, death) and wants to know what sample size is necessary to achieve a certain power with a given test or a certain precision with a given estimation method. A question that might be explored using a simulation study could be the following: What is the power of the log-rank test (an asymptotic test requiring large sample sizes to ensure validity) in the case of small samples? Here, a simple simulation study, designed to be consistent with the specific settings of the proposed study (sample size, prevalence of the exposure of interest, incidence of events and so on), could provide the necessary answers.
Simulation studies are also helpful to provide objective reproducible answers to more general methodological questions on the behaviour of statistical methods (ie, not necessarily motivated through a specific application). Examples of this type of question, which have been investigated by recent simulation studies, include the following: What is the effect of measurement errors on the estimated exposure-outcome relations in epidemiological studies?8 Does it make sense to check for subgroup-specific treatment effects even if the test for an overall effect is non-significant?9
In addition to the evaluation of individual methods, simulations can also be used to determine which one of several candidate methods will perform best for the application at hand. In the case of simulations reported in statistical literature, candidate methods may include existing methods and may (but do not have to) include new methods proposed by the researchers performing the simulation study. In the latter case, their focus is often on showing in which settings the new method performs better than its existing ‘competitors’.10 11
No matter the context of the simulation study, the objective is to find out if/when methods perform well and when they fail. Regarding the ‘when’ question, simulations provide an ideal setting for a systematic assessment of how variations in the values of relevant parameters and/or assumptions regarding data structure (eg, independence of observations, lack of measurement errors) affect the performance of the methods of interest. The definition of the term ‘good performance’ depends on the context. For example, if we compute a 95% confidence interval (CI), we usually want it to yield 95% coverage (ie, we want 95% of the CIs constructed in this way, using varying data sets, to cover the true value). If we apply a statistical test, we want this test to reject the null hypothesis with high probability if it is false, but to retain it with high probability if it is true. In comparison studies, two or more methods may be compared in this respect. In the case of a simulation performed for sample size calculation, we want to determine the smallest sample size with which a study has a given power to detect clinically important effects.
In practice, nobody can predict with certainty whether a method will yield accurate results for a specific data set, or which of a set of considered methods will perform best on that data set. Simulations can provide systematic evidence regarding how methods perform on average for data sets with similar characteristics to the data set under investigation. In an ideal world, relevant results from simulation studies would be available from previous research to help make rational decisions about which method to use. Data analysts would then use simulation results to verify whether the method they choose is adequate or to pick the most suitable from a range of different methods. Such ‘previous research’ is typically done by statistical researchers working on methods as the focus of research (as opposed to researchers applying methods in health research projects). For a data analyst with little experience and background in statistical methodological research, it is important to be able to interpret the results of such simulation studies. If previous evidence is lacking, or if previous studies do not seem to apply to the specific data setting under consideration, data analysts should conduct a targeted simulation study tailored to their specific data set.
Examples of statistical methods
In this section we present four examples of analyses which help us to explain the basic principles of simulation studies. Key criteria for evaluating the performance of methods related to these examples are summarised in table 1.
Table 1.
Overview of the main criteria for evaluating statistical methods in the four considered examples
| Example | Evaluation criterion | Target value |
| A: testing and CI | Type 1 error | Close to and not greater than nominal value α |
| Type 2 error | Low | |
| Coverage of (1–α) CI | Close to and not lower than nominal value 1–α | |
| B: explaining | Mean coefficient values | Close to true values (low bias) |
| Precision of coefficient estimation | High (low variance) | |
| Coverage of CI | Close to and not lower than nominal value 1–α | |
| Sensitivity of variable selection | High | |
| Specificity of variable selection | High | |
| C: predicting | Prediction error on independent data | Low |
| Accuracy measures | High | |
| D: clustering | Agreement with true cluster structure | High |
| All settings | Stability | High |
| Computational cost | Low | |
| Success of the computation (eg, ‘convergence’) | Yes |
The last column indicates which values the considered evaluation criterion takes if the investigated method is good.
Statistical hypothesis testing and CIs
In most health research projects we perform statistical tests and/or derive CIs. However, their behaviour is often not well characterised in real-world situations. For example, for time-to-event data with censored observations, how do the log-rank test and CI for hazard ratios (HR) behave in relatively small samples? Which technique should be preferred to compute the CI for proportions in a given setting (eg, very small proportions)?12
What is a good test/CI?
A good test is one that yields the correct answer with high probability, that is, one that rejects the null hypothesis with high probability if it is not true and retains it with high probability if it is true. Classical tests are defined in such a way that, in theory, the probability that the null hypothesis is rejected despite being true (called type 1 error) does not exceed a level α chosen by the user (in medicine, often α=0.05)—provided the assumptions are fulfilled. However, it is possible that the actual type 1 error may be larger than α, in which case the results of the test should be interpreted with caution. When evaluating a test, it is thus important to verify that the type 1 error does not exceed the nominal significance level α that was chosen by the researcher. Provided the type 1 error is as it should be (equal to or smaller than α), the most important quantity characterising a statistical test is its power, defined as the probability of correctly rejecting the null hypothesis.
Apart from hypothesis testing, results of statistical analysis are often presented as an estimate with a corresponding CI. A good method for deriving, say, 95% CI, is a method that yields CIs covering the true value with probability of 95%.
Can real data be used for the evaluation?
The main performance criteria cannot simply be assessed based on real data, because the truth (which hypotheses are true or false, or the true value of the parameter being estimated) is generally unknown in practice—we can see that a test has rejected the null hypothesis, but do not know if this was correct or not. If the truth were known, there would be no need to perform the test or compute a CI. Baseline characteristics in correctly randomised trials are a notable exception. Given the randomisation procedure, they are expected to be equally distributed in the two groups by definition.
Model selection for regression models: explaining the effects of covariates on an outcome variable
The second example is regression modelling of an outcome variable of interest, sometimes called ‘dependent’ variable, using several covariates, sometimes denoted as predictor variables or independent variables (often, prognostic or risk factors). In general, such modelling is performed either to explain the outcome variable by determining the effects of the covariates (as considered in this subsection), or to build a model, which will be used later on new patients for prediction purposes (as considered in the next subsection); see Shmueli13 for a discussion of these two related but distinct purposes. In health research, the outcome variable is often of one of the three following types: continuous (eg, amount of cholesterol reduction), categorical (eg, response to therapy) or survival time (eg, disease-free survival in months). Even though for all three cases standard regression modelling is reasonably well understood, the behaviour of regression techniques (including model selection) still raises questions in particular cases; see for example a recent simulation study on the use of resampling techniques for model selection purposes.14
What is a good regression approach?
In principle, a regression technique (including model selection aspects) is expected to (1) correctly distinguish the variables that are related to the outcome variable from those that are not, and (2) correctly fit the regression coefficients of the variables, that is, fit them to provide estimated values close to the true ones (unbiased and low variance). Regarding (1), it is good to have high sensitivity (ie, selecting most/all variables with effects, this is analogous to detecting most/all diseased patients in a diagnostic study) as well as high specificity (ie, not selecting variables without an effect, analogous to correctly identifying participants without disease). Depending on the specific goal, analysts may also aim to eliminate variables with very small effects.
Can real data be used for the evaluation?
In practice, the exact set of variables that have an effect on the outcome variable and the values of these effects are unknown, although previous knowledge from the literature may provide valuable guidance in some cases. Thus, in most cases, real data are of limited use for the evaluation of model selection approaches for regression models.
Model selection for regression models: predicting the values of an outcome using the values of covariates
The third example is related to the second example, but takes a different perspective. While regression models are often used to ‘explain’ the outcome variable (eg, a disease outcome or survival time), in order to understand how different risk factors affect the outcome variable, they can also be used as ‘prediction models’ to predict the outcome of interest for new patients, based on these patients’ values of the covariates. Classical linear regression models can be used for this purpose as well as various more complex alternative procedures, especially algorithms developed in the machine learning community, such as support vector machines or random forests (see Boulesteix et al 15 for a gentle introduction). In this field, simulations can be useful to assess the prediction accuracy of the considered prediction methods in different settings. For example, different penalised regression methods may be compared in simulations with respect to their prediction performance when a small number of clinical covariates are combined with a large number of candidate molecular covariates.16
What is a good prediction model?
A good prediction model is a model that yields accurate predictions in the future patients it will be applied to. For continuous and categorical outcome variables, often predicted and true values are directly compared, and the differences are summarised across patients. For survival times, suitable adjusted scores, like the Brier score, may be used to take into account censoring.17
Can real data be used for the evaluation?
The prediction error can be estimated based on the available data set using a large (possibly external) validation data set if available, or the so-called resampling techniques such as cross-validation.18 Note that this estimation may be unreliable depending on the context (eg, the smaller the sample size, the more unstable the cross-validation estimates).19 What these evaluations tell us about the methods’ accuracy is relevant to the considered specific real data example(s), but may not be relevant to other settings.
Clustering
The last example considered in this paper is clustering, also called cluster analysis. The objective of clustering is to identify clusters, that is, ‘groups’ of patients that behave similarly. For example, clustering methods may be used with the goal of identifying clinically meaningful subgroups of patients, using MRI data and clinical data, among others.20 Clusters should be constructed in such a way that the values of patients within a cluster are more similar (according to the chosen similarity criterion) than the values of patients from different clusters. Many different clustering algorithms have been proposed at the interface between computer science and statistics, for example k-means clustering or hierarchical clustering. Simulation studies may be used to assess the ability of methods to recover a true underlying structure.20 21
What is a good clustering method?
A good clustering procedure is a procedure that correctly recovers a true cluster structure present in the data but does not falsely identify clusters that are not in fact present.
Can real data be used for the evaluation?
In practice, the true cluster structure is often unknown, and even if there is a known cluster structure further sensible cluster structures might exist. The abilities of clustering methods to group similar observations together may be assessed by using data that consist of known subgroups and measuring the degree of overlap between the clustering structure defined by the known subgroups and the clustering structure proposed by the clustering algorithm. However, there might not be only one sensible cluster structure; in fact, the observations may cluster together more strongly according to factors other than the subgroup membership, for example, gene expressions are associated with various phenotypes. Real data may be used to assess aspects such as stability (ie, robustness against small changes in the data) or computational efficiency, but they are of limited use for the evaluation of a clustering method according to the criterion ‘agreement with the true cluster structure’.
Basic principles of simulation studies
Key features of a simulation study
In this section we provide a brief overview of the key features of a simulation study, which are also displayed in table 2, together with the example from the section "An example of a statistical simulation". A more detailed introduction to the concepts of data generating mechanisms and simulation scenarios is given in the subsection "Sampling variability and data generating processes" for interested readers. One may also refer to a recent indepth article on simulation studies addressing an audience of statisticians.3
Table 2.
Overview of the key features of a simulation study (first column) with the NHANES example described in the section "An example of a statistical simulation" (second column)
| Key features of simulation studies | NHANES example |
| Aims | To quantify the impact of measurement error. |
| Data generating mechanism | Take real data, add normally distributed random error to the exposure of interest (HbA1c) and/or the confounder (BMI). |
| Method of analysis | Multivariable linear regression, first on data with no measurement error, then on data with measurement error added. |
| Performance measure | Bias in regression coefficient for exposure of interest (HbA1c). |
| Number of repetitions | 1000 |
This table is inspired by the ‘ADEMP’ system (aims, data generating mechanisms, estimands, methods and performance measures) introduced previously in statistical literature.3
BMI, body mass index; HbA1c, glycated haemoglobin; NHANES, National Health and Nutrition Examination Survey.
The first key feature of a simulation study is its overall objective. Is the simulation study tailored to a specific data set relevant to a particular application or does it address a methodological question of general interest for future applications? Regardless of the overall objective, researchers performing a simulation study should make decisions considering the following key issues.
Aims
What do we want to learn about the method(s) from the simulation study? For example, one may want to assess whether a model selection method selects the right covariates (main aim) and whether it estimates their effects accurately (secondary aim). This point is analogous to the definition of primary and secondary outcomes in clinical trials, for example disease-free survival or side effects.
Data generating mechanism (including choice of relevant parameters)
How do we generate the simulated data sets? From which distribution? Which parameters may affect the results and what values should be considered? Each combination of the relevant assumptions and parameter values defines one simulation scenario (for which several data sets will usually be (randomly) generated, as outlined in the next subsection). There are many ways to generate data sets: by using real data sets as a starting point (see our example later) or by sampling from (possibly multivariate) prespecified distributions, for example the normal distribution. The definition of the scenarios is analogous to the definition of experimental conditions for a lab experiment and should be guided by considerations about clinical plausibility and/or relevance.11 While simulation designs can be made complex, the focus is often on relatively simple properties of the data distributions, such as skewness or outliers. The performance of many widely used basic statistical building blocks, such as the least squares optimisation principle for estimating model parameters, can be severely affected by the type of distribution under consideration. As a result, in order to comprehensively gauge performance, simulation studies should also include the rather innocent looking problems of real data, such as some outlier observations. More insights are given in the subsection "Sampling variability and data generating processes".
Method(s) of analysis to be evaluated/compared
Which method(s)/variant(s) is (are) evaluated? This point is analogous to the definition of the treatments with all necessary details (dose and so on) to be compared in a clinical trial. Further discussion about the analogy between clinical trials and comparisons of statistical methods can be found elsewhere.10
Performance measure(s)
Which criteria are used to assess the performance of the considered data analysis methods? In the example of model selection mentioned above, one may address the main aim by considering the sensitivity of the method for selecting the ‘true effects’ as well as the frequency of ‘false positives’ (ie, selection of variables that have no true associations with the outcome). The secondary aim may be addressed by computing the mean squared deviation or the mean absolute deviation of the coefficient estimates from the true values. This point is analogous to the precise definition of primary and secondary outcomes in a clinical trial: for example, which instruments are used for the assessment of side effects of the therapy, or how do we exactly estimate disease-free survival and compare it across the trial arms?
Number of repetitions
For each considered scenario, how many data sets are randomly drawn? It is necessary to generate several (ideally, ‘many’) data sets in order to average out random fluctuations and ensure sufficiently precise simulation results. The more data sets are generated, the more precise the performance evaluation will be—as can be quantified through, for example, the width of the CI for the selected ‘performance criteria’. The number of repetitions is analogous to the sample size in a clinical trial. In contrast to increasing the sample size in clinical trials, however, it is often easy to extend the number of repetitions in simulation studies. The number of repetitions is chosen as a compromise between precision of the results and computational time.
Sampling variability and data generating processes
This section gives further insights into the data generating process for readers interested in gaining a deeper understanding of the fundamentals of simulation studies, beyond the key points outlined above. To this end we first explain briefly how simulations provide a framework for assessing and accounting for the impact of random sampling error on the results of empirical studies.
Preliminary: sampling variability in real data
Suppose a clinical researcher is interested in the mean difference between the blood pressure of men and women in the population aged 20–60. The true mean difference could only be calculated if we had data on the whole populations of men and women aged 20–60. Of course, in practice, we only have a sample available with a specific (often moderate) size and can only estimate the mean difference using this sample. Different samples will yield different estimates of the same mean difference in the population. Collecting a data sample can be seen as drawing observations from a population of interest that has particular characteristics. In statistical terms, these observations can be seen as random observations generated from the true distribution of the variable(s) of interest in the relevant population. In real-life studies, this distribution and the true values of its parameters (eg, population means) are unknown and we can only estimate them using available sample data.
Simulating data
The principle of simulations is to mimic the process of taking repeated (random) samples from a large population, by repeatedly generating synthetic data (‘virtual observations’) from a virtual population, under prespecified assumptions that can be varied across the considered simulation scenarios. Each synthetic sample is generated from a particular known distribution, with ‘true’ values of all relevant parameters fixed by the researchers. Each simulated sample is then analysed using the method(s) of interest, and its (their) performance is evaluated using prespecified criteria (see table 1 for examples). To give one simple example, we may simulate systolic blood pressure values for a sample of n=100 ‘synthetic subjects’ by generating 100 independent numbers from a normal distribution with, say, mean of 120 and standard deviation (SD) of 15. Doing so, we know that the true population mean is 120 mmHg and that the simulated blood pressure follows the normal distribution. The way in which virtual observations are generated in the context of a simulation (in our example, ‘100 independent numbers from a normal distribution with mean 120 and SD 15’) is termed the data generating mechanism. There is a large number of user-friendly statistical packages that can be used to accomplish this task.
Sampling variability in simulations
Just as random sample-to-sample variability affects real data samples drawn from a population of interest, it also affects the results obtained using simulated data. If we generate two synthetic data sets using the same data generating mechanism and the same parameters, we will get somewhat different results (with the differences decreasing, on average, with increasing size of the generated data sets). It is therefore almost always important to repeat the same data generation and analysis process using many simulated data sets, as outlined above. The variability of the results obtained across the different data sets simulated from the same distribution has to be carefully assessed by, for example, calculating the SD of the individual estimates. Calculating the mean value of the individual estimates provides a more robust estimate of the unknown population-level parameter than a value from a single simulated sample, as averaging over several repetitions reduces the impact of random sampling error.
Choice of data generating mechanisms
When performing a simulation, one has to choose one or several data generating mechanisms that reflect, as closely as possible, the distribution and relevant characteristics of the real data of interest, no matter whether the focus is on a specific application or on a ‘generic’ methodological question, such as evaluation or comparison of specific analytical methods. The difficulty is that, in reality, the true data generating process is unknown as mentioned above in the example of blood pressure. The only possibility is to consider several data generating mechanisms—called simulation scenarios—that, together, will cover the range of situations congruent with the expected structure of real data of interest. Scenarios may differ, among other ways, in the sample size, the true distributions of the considered variables (normal, uniform, exponential and so on), the values of parameters such as means or variances, the correlation structure of the variables or the presence of outliers. For example, we may be interested in the behaviour of a test that assumes a normal distribution in situations where this assumption is not fulfilled. If the variable of interest is expected, based on earlier studies and/or substantive knowledge, to be (approximately) uniformly distributed (meaning that the observations are evenly distributed over a certain interval), priority will be given to corresponding scenarios. However, it may be useful to also consider a few alternative scenarios with other distributions, for example, a positively skewed distribution with most values concentrating below the mean and relatively few high values.
In general, if the focus of the simulation study is on a specific application, the primary goal is essentially to simulate data sets that are as similar as possible to the relevant real data set. This may necessitate making some plausible assumptions and involve some uncertainty if the data have not yet been collected—as is the case when simulations are performed with the aim of calculating the adequate sample size or assessing the expected power and/or precision of future analyses. In contrast, if the focus of the simulation is on the general behaviour of a particular method (or comparison of alternative methods) for a class of applications, the primary goal when choosing scenarios is often to cover a wide spectrum of potentially plausible situations in which the method(s) of interest are likely to be employed. Some scenarios may be unrealistic but are nevertheless helpful in understanding how the method works or when it breaks down (and how it can be improved to cope better with the problematic situations), and thus yield valuable information. The choice of simulation scenarios is thus intrinsically related to the goal of the simulation, but should also account for substantive knowledge in the field of potential real-life applications.
Advantages and drawbacks of simulation studies
To simulate the synthetic data sets, we define the underlying ‘truth’ regarding the research question being explored. For example, in example A in the section "Examples of statistical methods" (testing) we know whether the null hypothesis is true or not. In example B (explaining) we know which variables have independent effects on the outcome variable. In example C (predicting) we know the true values of the outcome variable. In example D (clustering) we know the true cluster structure. To sum up, in all these examples, we know what an accurate method of data analysis is supposed to find. Thus, we can determine how well the method(s) being evaluated perform(s) by comparing their results against this known ‘truth’. This feature is the major advantage of simulations over empirical comparisons of the same methods based on one or few real-life data sets as, in the latter case, the true answers often remain unknown.
Another advantage of simulations is that they allow investigation of a large number of different scenarios, and in particular also scenarios that are not directly observed in real data sets. This means that the analysis can be extended to new or rare scenarios, or scenarios reflecting practically unrealistic settings (eg, randomised trial data or very large sample sizes). A related advantage of simulations is that, by varying the assumptions and the values of relevant parameters used to generate data for different scenarios, one can systematically assess how the performance of different methods depends on these assumptions and parameters. Furthermore, one can also perform, for each considered scenario, as many repetitions as needed to average out random fluctuations. This is in contrast to real data experiments where the quantity of data is often severely limited, which affects the precision of the results.
These advantages, however, come at a cost. First, simulation scenarios are often simplified, that is, they do not reflect the true complexity of the data encountered in real-life data analyses. The lack of complexity of simulated data may lead to a distorted picture of the methods’ performance. For example, an approach that can model data in a very flexible manner might be more severely affected by outliers. Yet simulation designs so far rarely incorporate outliers or skewed distributions. Real-world performance of an approach that has been selected based on simulation study results might be surprisingly bad. Second, large simulation studies can be computationally very expensive, taking days or weeks and even requiring the use of parallel computing, if a large number of scenarios and/or large numbers of repetitions are considered and especially if the analysis also involves large data sets and/or complex statistical methods.
Finally, it is important to note that simulations are not immune to the typical flaws of numerical studies leading to biased results. For example, the effect of single influential points, which are difficult to detect in simulation studies with hundreds or thousands of simulated samples, can be critical. They may be relevant in some of the simulation repetitions, in which they cause unreliable results. If undetected, they can bias the results. Most importantly, selective reporting may be an issue. If a very large number of scenarios are analysed, but only those scenarios that favour one particular method are presented in the paper, the reported results will give a distorted picture of reality. Obviously, this is a serious problem of bad reporting and bad research, which can be easily avoided by being transparent.
An example of a statistical simulation
For illustration, in this section we consider a simple simulation study that investigates the impact of measurement error in linear regression analysis, inspired by a previous study.8 See the overview of its key features in the right column of table 2. Our study is completely reproducible using the R code provided in online supplemental file 1, which uses freely available data. In epidemiological studies of the relation between an exposure and an outcome, this relation is often estimated using regression analysis. As an example, we consider a study of the association between glycated haemoglobin (HbA1c) levels and systolic blood pressure assessed using linear regression. Data from 5092 subjects in the 2015–2016 National Health and Nutrition Examination Survey (NHANES)22 are used to obtain an estimate of the effect of HbA1c on systolic blood pressure, while adjusting for age, gender and body mass index (BMI). Details on the data are described on the NHANES website (https://wwwn.cdc.gov/nchs/nhanes/). After adjustment for age and gender, it was estimated that HbA1c increases systolic blood pressure by 1.13 mmHg (95% CI 0.73 to 1.52) per unit increase in HbA1c. Additional adjustment for BMI resulted in a considerable change in the effect estimate: HbA1c was estimated to increase blood pressure by 0.75 mmHg (95% CI 0.35 to 1.16) per unit increase in HbA1c.
bmjopen-2020-039921supp001.pdf (49.5KB, pdf)
The confounding variable BMI as well as the exposure variable HbA1c may be subject to measurement error. For example, BMI may be self-reported (instead of a standardised measurement using scales) or technical problems in the lab may have affected the HbA1c measurement. Therefore, researchers may want to know the possible impact of measurement error of the exposure and/or confounding variable(s) in terms of bias.23 We are interested both in the direction and magnitude of this bias.
One way to investigate the possible impact of measurement error is through a small simulation study,8 whose steps are schematically represented in figure 1. For the purpose of this example, the original recordings in the NHANES data were assumed to be measured without error (step 1 in figure 1). Then, in addition, new artificial variables were created that represented HbA1c and BMI, but for the situation in which these are measured with error. To create these variables, measurement error was artificially added to the exposure variable (HbA1c) and/or the confounding variable (BMI) (step 2 in figure 1). These errors were drawn from a normal distribution with a mean zero and were independent of all variables considered. This type of measurement error is often referred to as classical measurement error.24 The variance of the normal distribution, defining the amount of measurement error added, was altered for different scenarios. Scenarios ranged from no measurement error on either HbA1c or BMI (reference scenario) to 50% of the variance in HbA1c and/or BMI attributable to measurement error. To minimise the impact of simulation error, each scenario was repeated 1000 times and the results were averaged per scenario over these 1000 repetitions.
Figure 1.
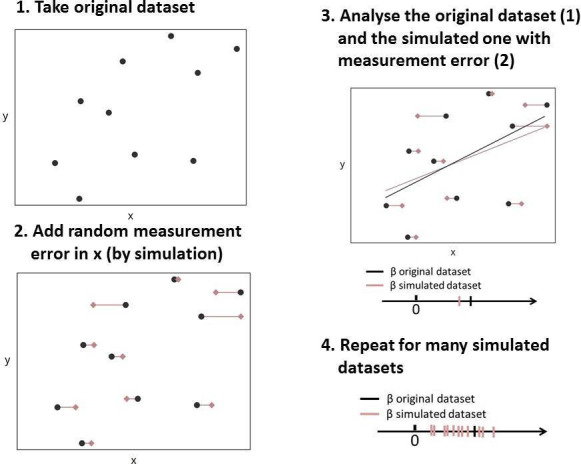
Schematic illustration of the key steps of the example simulation study.
Figure 2 shows the impact of measurement error on HbA1c and/or BMI on the estimate of the regression coefficient of HbA1c (steps 3 and 4 in figure 1). The relation between HbA1c and systolic blood pressure was attenuated when measurement error was added to HbA1c, but not when measurement error was added to BMI. The association became stronger as measurement error was added solely to the confounding variable BMI. The reason for this effect is that, with increasing levels of measurement error on BMI, adjustment for the confounding due to BMI becomes less efficient and the effect estimate gets closer to the unadjusted estimate (1.13 mmHg). Due to measurement error, a type of residual confounding is introduced. In the case of measurement error on HbA1c as well as BMI, both phenomena play a role and may cancel each other out. In this study, measurement error on HbA1c seemed more influential than measurement error on BMI.
Figure 2.
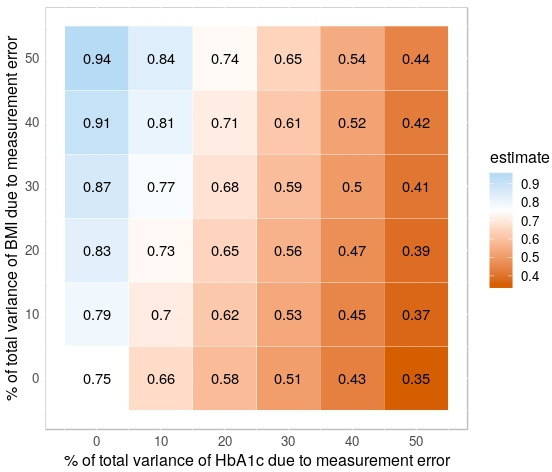
Estimates of the association between HbA1c levels and systolic blood pressure after adjustment for confounding by BMI under various simulation scenarios characterised by different levels of measurement error. Numbers represent effect estimates averaged over 1000 simulation repetitions. Red shading represents low (averaged) estimates. Blue shading represents high (averaged) estimates. CIs are omitted for clarity. See text for details. BMI, body mass index; HbA1c, glycated haemoglobin.
This example illustrates how a simple simulation study could provide insight into an important potential source of bias, namely measurement error. Here, we only considered classical measurement error, but simulations could easily be extended to incorporate more complex forms of measurement error. For example, the errors may not be drawn from a normal distribution with mean zero or may not be independent of all other variables considered. Instead, the mean of the distribution of errors may depend on the value of another variable in the model, for example, error on BMI may depend on gender. Furthermore, non-normal distributions may be considered, or scenarios in which the variance of the errors depends on the true value of the measurement (heteroskedastic errors), among other possible extensions.
Finally, we note that researchers conducting small-scale simulation studies like the one presented here should reflect on the plausibility of the scenarios considered. For example, knowing whether it is realistic to assume that 50% of the total variance of HbA1c and BMI is due to measurement error (top-right scenario in figure 2) requires subject-matter knowledge.
Concluding remarks
Just as randomised clinical trials form part of the evidence base for the choice of therapy in medical practice, simulation studies form part of the evidence base for statistical practice. Large-scale simulation studies allow assessment of the properties of complex estimation and inferential methods, and comparison of complex model building strategies under a variety of alternative assumptions and sample sizes.5 They provide valuable support for decision-making regarding the choice of statistical methods to be used in a given real-life application and they are the cornerstone of the work on guidance for the design and analysis of the STRATOS initiative. They complement—rather than replace—the judgement of a trained expert (a data analyst in the choice of statistical methods, analogous to a physician in the choice of therapies). Increased computational power nowadays makes it possible to examine many possible simulation scenarios with different combinations of distributional parameters and assumptions. This partly addresses the main limitation of simulations, namely that they can never fully reflect the complexity of real data.
Let us again consider our analogy between simulation studies and clinical studies. The design and implementation of clinical studies should be left to teams of trained clinical researchers, but it is crucial for practitioners who want to practise evidence-based medicine to be able to read and understand the results of these clinical studies. Similarly, the design, implementation and reporting of complex simulations are still a subject of debate3 and should be left to methodological statistical experts, but it is important for data analysts to be able to read and understand simulation studies in the literature (or perhaps to implement simple ones themselves). Armed with these skills, they will be better able to identify appropriate data analysis methods for their data and research questions, which will ultimately contribute to improved replicability of research results.
Supplementary Material
Acknowledgments
The authors thank Alethea Charlton for language corrections. The international STRengthening Analytical Thinking for Observational Studies (STRATOS) initiative aims to provide accessible and accurate guidance for relevant topics in the design and analysis of observational studies (http://stratos-initiative.org). Members of simulation panel at the time of first submission were: Michal Abrahamowicz, Harald Binder, Anne-Laure Boulesteix, Rolf Groenwold, Victor Kipnis, Tim Morris, Jessica Myers Franklin, Willi Sauerbrei, Pamela Shaw, Ewout Steyerberg, Ingeborg Waernbaum.
Footnotes
Twitter: @BoulesteixLaure, @tmorris_mrc
Contributors: ALB initiated and coordinated the project and wrote most of the manuscript. RG performed the example analysis and wrote the corresponding section. MA, HB, MB, RH, TPM and JR critically revised the manuscript for important intellectual content. WS initiated and coordinated the project. All authors made substantial contributions to the manuscript’s content and text and approved the final version.
Funding: This project was partly funded by the German Research Foundation (DFG) with grants BO3139/4-3 to ALB and SA580/10-1 to WS. MA is a James McGill Professor at McGill University. His research is supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) (grant 228203) and the Canadian Institutes of Health Research (CIHR) (grant PJT-148946). TPM was funded by the UK MRC (grants MC_UU_12023/21 and MC_UU_12023/29).
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Contributor Information
for the Simulation Panel:
Victor Kipnis, Jessica Myers Franklin, Pamela Shaw, Ewout Steyerberg, and Ingeborg Waernbaum
References
- 1. Burton A, Altman DG, Royston P, et al. . The design of simulation studies in medical statistics. Stat Med 2006;25:4279–92. 10.1002/sim.2673 [DOI] [PubMed] [Google Scholar]
- 2. Sigal MJ, Chalmers RP. Play it again: teaching statistics with Monte Carlo simulation. J Educ Stat 2016;24:136–56. 10.1080/10691898.2016.1246953 [DOI] [Google Scholar]
- 3. Morris TP, White IR, Crowther MJ. Using simulation studies to evaluate statistical methods. Stat Med 2019;38:2074–102. 10.1002/sim.8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Macleod MR, Michie S, Roberts I, et al. . Biomedical research: increasing value, reducing waste. Lancet 2014;383:101–4. 10.1016/S0140-6736(13)62329-6 [DOI] [PubMed] [Google Scholar]
- 5. Sauerbrei W, Abrahamowicz M, Altman DG, et al. . Strengthening analytical thinking for observational studies: the STRATOS initiative. Stat Med 2014;33:5413–32. 10.1002/sim.6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rochon J, Gondan M, Kieser M. To test or not to test: preliminary assessment of normality when comparing two independent samples. BMC Med Res Methodol 2012;12:81. 10.1186/1471-2288-12-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lotterhos KE, Moore JH, Stapleton AE. Analysis validation has been neglected in the age of reproducibility. PLoS Biol 2018;16:e3000070. 10.1371/journal.pbio.3000070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brakenhoff TB, van Smeden M, Visseren FLJ, et al. . Random measurement error: why worry? an example of cardiovascular risk factors. PLoS One 2018;13:e0192298. 10.1371/journal.pone.0192298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abrahamowicz M, Beauchamp M-E, Fournier P, et al. . Evidence of subgroup-specific treatment effect in the absence of an overall effect: is there really a contradiction? Pharmacoepidemiol Drug Saf 2013;22:1178–88. 10.1002/pds.3485 [DOI] [PubMed] [Google Scholar]
- 10. Boulesteix A-L, Wilson R, Hapfelmeier A. Towards evidence-based computational statistics: lessons from clinical research on the role and design of real-data benchmark studies. BMC Med Res Methodol 2017;17:138–38. 10.1186/s12874-017-0417-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boulesteix A-L, Binder H, Abrahamowicz M, et al. . On the necessity and design of studies comparing statistical methods. Biom J 2018;60:216–8. 10.1002/bimj.201700129 [DOI] [PubMed] [Google Scholar]
- 12. Newcombe RG. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med 1998;17:857–72. [DOI] [PubMed] [Google Scholar]
- 13. Shmueli G. To explain or to predict? Stat Sci 2010;25:289–310. 10.1214/10-STS330 [DOI] [Google Scholar]
- 14. De Bin R, Janitza S, Sauerbrei W, et al. . Subsampling versus bootstrapping in resampling-based model selection for multivariable regression. Biometrics 2016;72:272–80. 10.1111/biom.12381 [DOI] [PubMed] [Google Scholar]
- 15. Boulesteix A-L, Wright MN, Hoffmann S, et al. . Statistical learning approaches in the genetic epidemiology of complex diseases. Hum Genet 2020;139:73–84. 10.1007/s00439-019-01996-9 [DOI] [PubMed] [Google Scholar]
- 16. De Bin R, Boulesteix AL, Benner A, et al. . Combining clinical and molecular data in regression prediction models: insights from a simulation study. Brief Bioinformatics 2020. [DOI] [PubMed] [Google Scholar]
- 17. Graf E, Schmoor C, Sauerbrei W, et al. . Assessment and comparison of prognostic classification schemes for survival data. Stat Med 1999;18:2529–45. [DOI] [PubMed] [Google Scholar]
- 18. Boulesteix A-L, Strobl C, Augustin T, et al. . Evaluating microarray-based classifiers: an overview. Cancer Inform 2008;6:77–97. 10.4137/CIN.S408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanczar B, Hua J, Dougherty ER. Decorrelation of the true and estimated classifier errors in high-dimensional settings. EURASIP J Bioinform Syst Biol 2007;38473:1–12. 10.1155/2007/38473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kent P, Jensen RK, Kongsted A. A comparison of three clustering methods for finding subgroups in MRI, SMS or clinical data: SPSS twostep cluster analysis, latent gold and SNOB. BMC Med Res Methodol 2014;14:113–13. 10.1186/1471-2288-14-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coretto P, Hennig C. A simulation study to compare robust clustering methods based on mixtures. Adv Data Anal Classif 2010;4:111–35. 10.1007/s11634-010-0065-4 [DOI] [Google Scholar]
- 22. Zipf G, Chiappa M, Porter KS, et al. . National health and nutrition examination survey: plan and operations, 1999-2010. Vital Health Stat 1 2013;1:1–37. [PubMed] [Google Scholar]
- 23. Brakenhoff TB, Mitroiu M, Keogh RH, et al. . Measurement error is often neglected in medical literature: a systematic review. J Clin Epidemiol 2018;98:89–97. 10.1016/j.jclinepi.2018.02.023 [DOI] [PubMed] [Google Scholar]
- 24. Carroll RJ, Ruppert D, Stefanski LA, et al. . Measurement error in nonlinear models: a modern perspective. 2 edn CRC Press, 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2020-039921supp001.pdf (49.5KB, pdf)


