Abstract
To avoid replicative senescence or telomere-induced apoptosis, cancers employ telomere maintenance mechanisms (TMMs) involving either the upregulation of telomerase or the acquisition of recombination-based alternative telomere lengthening (ALT). The choice of TMM may differentially influence cancer evolution and be exploitable in targeted therapies. Here, we examine TMMs in a panel of 17 osteosarcoma-derived cell lines, defining three separate groups according to TMM and the length of telomeres maintained. Eight were ALT-positive, including the previously uncharacterized lines, KPD and LM7. While ALT-positive lines all showed excessive telomere length, ALT-negative cell lines fell into two groups according to their telomere length: HOS-MNNG, OHSN, SJSA-1, HAL, 143b, and HOS displayed subnormally short telomere length, while MG-63, MHM, and HuO-3N1 displayed long telomeres. Hence, we further subcategorized ALT-negative TMM into long-telomere (LT) and short-telomere (ST) maintenance groups. Importantly, subnormally short telomeres were significantly associated with hypersensitivity to three different therapeutics targeting the protein kinase ataxia telangiectasia and Rad3-related (ATR) (AZD-6738/Ceralasertib, VE-822/Berzoserib, and BAY-1895344) compared to long telomeres maintained via ALT or telomerase. Within 24 h of ATR inhibition, cells with short but not long telomeres displayed chromosome bridges and underwent cell death, indicating a selective dependency on ATR for chromosome stability. Collectively, our work provides a resource to identify links between the mode of telomere maintenance and drug sensitivity in osteosarcoma and indicates that telomere length predicts ATR inhibitor sensitivity in cancer.
Keywords: short telomere, telomerase, alternative lengthening of telomeres, osteosarcoma, ATR inhibitor, chromosome bridge
Telomeres are the protective ends of chromosomes, essential for all dividing cells.1 While telomeres shorten with every chromosome replication cycle due to the end-replication problem, critically shortened telomeres elicit DNA damage checkpoint activation, leading to replicative senescence or apoptosis.2 The vast majority of cancer cells maintain the ends of their chromosomes through a telomere maintenance mechanism (TMM), thereby avoiding senescence or death induced by critically shortened telomeres.3 In about 90% of cancers, the reverse transcriptase enzyme telomerase, which replenishes telomeric DNA, is reactivated, permitting indefinite cell divisions.4,5 TMMs may be bypassed only in exceptional circumstances where cancer cells retain substantially long telomeres.6−8
Interestingly, the activity or expression levels of telomerase in cancers does not seem to correlate with the length of telomeres that are maintained.6,9,10 A substantive fraction of telomerase-positive cancers display abnormally short telomeres compared to adjacent noncancerous tissue.6,11−15 Furthermore, shortness of telomeres in tumors is correlated with poor prognosis in many types of cancer.14−16 Whereas the origin of short telomeres in tumors has not been clearly demonstrated, shortness or loss of telomeres is thought to originate from excessive divisions during the benign stages.1,2,17 In telomerase-expressing normal cells, telomerase preferentially elongates the shortest telomeres to extend overall telomere length.18,19 However, such telomerase action appears to be compromised in cancer cells, resulting in retention of critically short telomeres and inefficient telomere lengthening.20−22 These cancer cells evade activation of the DNA repair machinery or senescence pathways caused by shortened telomeres; therefore, the mechanisms to maintain such short telomeres by telomerase are thought to be unique to cancer.2,21
Telomerase activity has been targeted for many types of cancer. However, the cytotoxic effects of telomerase inhibition are not immediate as time is required to achieve telomere attrition and damage accumulation, and cancer cells able to propagate and evolve until their telomeres significantly shorten and become deprotected.7,8,23,24
In the absence of telomerase activity, some tumors gain the ability to undertake telomerase-independent, alternative maintenance of telomeres. Alternative lengthening of telomeres (ALT) is dependent on homologous recombination or homology-directed repair-based DNA replication.25,26 ALT-type TMM is often found in cancers of a mesenchymal origin, including central nervous system cancers as well as in many sarcomas, particularly osteosarcoma, which is potentially explained by the fact that telomerase expression in cells of mesenchymal origin is more strictly regulated compared to that in cells of epithelial origin.5,27−29 To date, ALT activity has not been detected in normal human body cells, highlighting a unique cancer-selective therapeutic opportunity. However, despite this selectively cancer-associated feature, effective drugs that kill ALT-positive cancer cells are yet to be discovered.
Osteosarcoma is a rare cancer, commonly found in young adults. The overall 5 year survival rate has not significantly improved over the past two decades, and therapy-associated toxicities pose a significant problem in the treatment of this cancer.30,31 Therefore, new, less toxic strategies are needed. Children and young adults have significantly longer telomeres in their somatic cells.32 Hence, targeting the mechanisms of short-telomere (ST) maintenance may pose unique opportunities for treatments that effectively eradicate cancer cells, yet minimize normal tissue toxicity and side effects in such patients. To this end, we characterized TMM and telomere length within a panel of 17 osteosarcoma cell lines33 (Table S-1), sorting them into three separate categories based on their mechanism of telomere lengthening and observed telomere length. We provide evidence of different telomere anatomies in telomerase-expressing ALT-negative osteosarcomas, evidenced by either overall long or short telomere length and report synthetic lethality of telomerase-expressing cells with short telomeres to clinical inhibitors of the Ser/Thr protein kinase ataxia telangiectasia and Rad3-telated (ATR; inhibitors of ATR, ATRi). Our data highlight the use of telomere length as a biomarker to identify ATRi sensitivity in osteosarcoma and potentially other cancers.
Results
Categorization of Osteosarcoma Cell Lines by Mode of Telomere Maintenance Mechanism
Of the osteosarcoma cell lines in our panel, six (G292, HuO-9, CAL72, U2OS, NY, and SAOS-2) have previously been characterized as ALT-positive,34,35 while five cell lines (HOS-MNNG, SJSA-1, HOS, MG-63, and HuO-3N1) had previously been characterized as telomerase-positive and ALT-negative35−39 (Table 1). However, TMM in a further six cell lines (OHSN, HAL, 143b, MHM, KPD, and LM7) had not previously been characterized. Therefore, we first sought to assess the TMM used in these remaining lines alongside those previously assessed to confirm the respective published results.
Table 1. Telomere Status of the Osteosarcoma Cell Lines.
| cell line | telomere statusa | qPCR T/S ratiob | median telomere length (kb)c | CC assay level | TERC leveld | TERT mRNA leveld |
|---|---|---|---|---|---|---|
| HOS-MNNG | ST | 0.813 | 3.0 | –0.006 | 19.80% | 56.54% |
| OHSN | ST | 0.837 | 3.2 | –0.011 | 23.17% | 107.37% |
| SJSA-1 | ST | 0.840 | 3.2 | –0.002 | 6.36% | 695.03% |
| HAL | ST | 0.853 | 2.7 | 0.007 | 1.48% | 170.34% |
| 143b | ST | 0.855 | 3.0 | 0.002 | 26.48% | 288.96% |
| HOS | ST | 0.902 | 3.6 | 0.001 | 21.03% | 323.13% |
| MG-63 | LT | 0.966 | 5.8 | –0.003 | 11.91% | 58.47% |
| MHM | LT | 1.086 | 6.4 | 0.001 | 29.03% | 390.11% |
| HuO-3N1 | LT | 1.129 | 6.7 | 0.029 | 35.41% | 7.69% |
| G292 | ALT | 1.415 | 10.7 | 4.156 | 3.98% | 0.06% |
| HuO-9 | ALT | 1.539 | 9.7 | 1.066 | 3.69% | 1.33% |
| CAL72 | ALT | 2.853 | 10.2 | 5.422 | 5.30% | 2.22% |
| U2OS | ALT | 4.221 | 13.7 | 0.835 | 0.00% | 4.21% |
| KPD | ALT | 1.210 | 10.0 | 2.944 | 0.01% | 4.42% |
| NY | ALT | 1.552 | 9.9 | 0.651 | 0.77% | 14.54% |
| SAOS-2 | ALT | 1.230 | 7.6 | 1.975 | 21.87% | 19.36% |
| LM7 | ALT | 1.170 | 8.2 | 1.227 | 12.27% | 0.29% |
ST, ALT-negative, short-telomere. LT, ALT-negative, long-telomere. ALT, ALT-positive.
T/S, telomere repeats to single-copy genes.
Calculated using Telometric software.
Relative to HEK293T expression, determined as expression fold change versus 7SK.
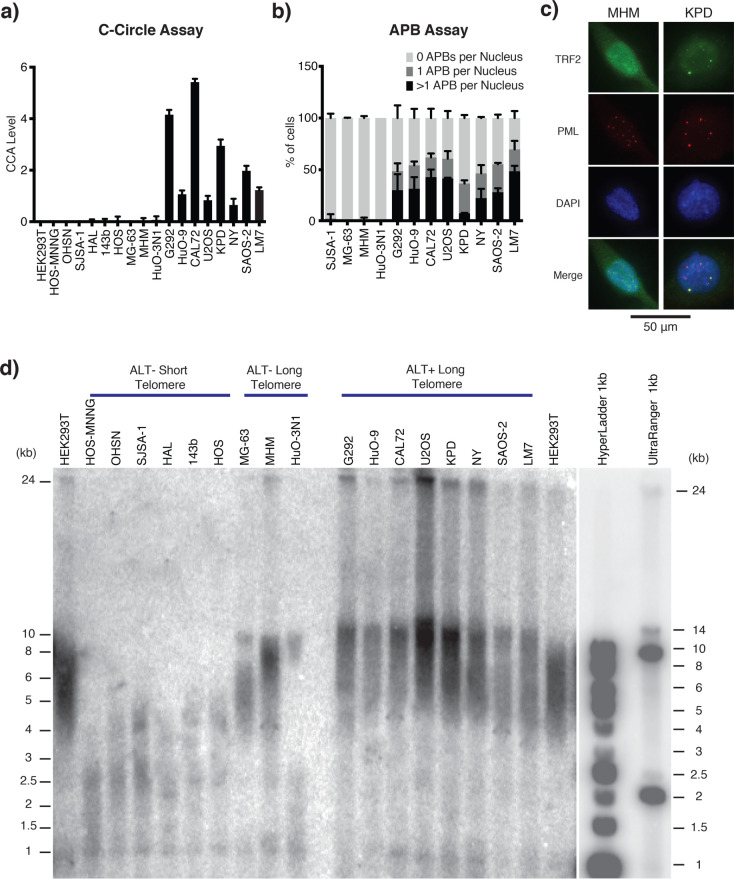
ALT-positive cells are characterized by the presence of C-circles, a byproduct of telomere recombination in ALT, consisting of partially double-stranded extra-chromosomal telomeric DNA circles. C-circles were detected through rolling circle amplification (RCA) followed by quantification of telomeric sequences using either dot-blot hybridization or monochrome-multiplex quantitative polymerase chain reaction (MM-qPCR) (Figures 1a and S-1a). Another hallmark of ALT-based telomere maintenance is the colocalization of telomere clusters with promyelocytic leukemia (PML) bodies, forming structures termed ALT-associated PML bodies (APBs).29 The C-circle positive cell lines, along with three cell lines that were C-circle negative, were subjected to the APB assay (Figures 1b,c and S-2). This combined analysis defined the previously uncharacterized lines KPD and LM7 as ALT-positive, along with previously reported ALT-positive lines G292, HuO-9, CAL72, U2OS, NY, and SAOS-2. We also defined the previously uncharacterized lines OHSN, HAL, 143b, and MHM as ALT-negative, along with the previously ALT-negative lines HOS-MNNG, SJSA-1, HOS, MG-63, and HuO-3N1 (Table 1).
Figure 1.
Characterization of telomere status in osteosarcoma cell lines. (a) C-Circle assay (CCA) Level. Graph shows the CCA level calculated by subtracting the phi-29 polymerase NTC by the phi-29 polymerase free control NTC (see the “Material and Methods” section). A sample is then considered ALT+ if its CCA level lower error bar is >0. Data is represented as a mean of three independent experiments. Error bars show standard deviation. Dot blot result of phi-29 polymerase is shown in Figure S-1a. (b) APB assay for the osteosarcoma cell lines with long telomeres. APBs were scored as a direct colocalization between TRF2 and PML foci. The graph shows the mean percentage of cells containing 0, 1, or >1 APBs in their nuclei for three independently prepared samples of >50 cells. Error bars show standard deviation. (c) Representative immunofluorescence images showing the presence/absence of APBs in the KPD and MHM cell lines, respectively. TRF2 was stained green, and PML bodies were stained red. Yellow foci (colocalization of green and red foci) in the merge panel represent APBs. A 50 μm scale bar is shown. Representative images of all tested cell lines are shown in Figure S-2. (d) Telomere Southern blot of osteosarcoma cell lines. Terminal restriction fragment length analysis of genomic DNA digested with HinfI and RsaI and hybridized with a telomeric TTAGGG probe. A less exposed image of the DNA ladders is shown on the right. ALT-negative cell lines tend to have shorter telomeres than those of HEK293T, while ALT-positive cell lines tend to have longer telomeres than those of HEK293T, which are extremely heterogeneous in length. Digital analysis of telomere length is shown in Table 1.
The ALT pathway involves recombination-mediated robust amplification of telomeric DNA repeats through break-induced replication, generating heterogeneous telomeres that are significantly longer compared to those in telomerase-positive cancer cells. To determine the median length of telomeres in each of the various cell lines, we used telomere restriction fragment (TRF) assays where telomere length is determined by Southern blotting (Figures 1d and S-1b and Table S-2). Previous work proposed that ALT status may be predicted based on excessively long telomeres, with mean, variance, and semi-interquartile ranges of telomere length distribution greater than 16, 200,2 and 4 kb, respectively.29 Consistent with this view, we observed that all of the ALT-positive cell lines exhibited heterogeneous telomere signals reaching over 24 kb in length. Three of the ALT-negative lines, MG-63, MHM and HuO-3N1, harbored telomere length comparable to the telomerase-positive noncancerous cell line HEK293T, serving as a control. Notably, the remaining six ALT-negative cell lines (HOS-MNNG, OHSN, SJSA, HAL, 143b, and HOS) had significantly shorter telomeres and hence may represent situations of telomere maintenance present in cancers with subnormally short telomeres.
To independently confirm the telomere length predictions based on TRF assessment, we used MM-qPCR, which determines telomeric repeat number relative to the repeat number of a reference gene.40 In addition, MM-qPCR is suitable to determine telomere status in patient-derived tumor samples.14,41,42 To reduce errors caused by potential aneuploidy that may variably exist in the cancer lines, we used two different single-copy genes (SCGs) present on different chromosomes, ALB encoding albumin (4q13.3) and HBB encoding beta-globin (11q15.4). The ratio of telomere repeats to single-copy genes (T/S) were calculated (Figure S-1c). More uniform cycle threshold (CT) values were obtained using ALB, suggesting a more reliable SCG for MM-qPCR analysis (Figure S-1d). Using the HEK293T cell line as a reference, the MM-qPCR-based telomere length analysis correlates with the TRF-based analysis and clearly distinguished the cell lines into three categories (Figure S-1e,f).
Together, the above results put the cell lines into three clearly defined categories: (1) ALT-negative cell lines with short telomeres, (2) ALT-negative cell lines with long telomeres, and (3) ALT-positive cell lines with long telomeres (Table 1).
Molecular Profiling of Osteosarcoma Cell Lines
To catalogue the molecular drivers involved in maintaining telomeres in the various osteosarcoma cell lines, we surveyed candidate molecular events known to be associated with different forms of telomere maintenance.
ALT is strongly associated with dysfunction of the alpha-thalassemia/mental retardation syndrome X-linked (ATRX) and the death-domain-associated protein 6 (DAXX) chromatin remodelling complex.34 In our panel, the ALT-positive cell lines, with the exception of G292 and KPD, lacked detectable ATRX expression (Figure 2a). Interestingly, the ALT-negative group of the osteosarcoma lines with short telomeres, except for HOS, expressed remarkably high levels of ATRX, while those with long telomeres had either low or undetectable levels of ATRX. All cell lines expressed DAXX at the expected molecular weight, except G292, where DAXX is known to be fused to the kinesin family member KIFC3.35 The gene encoding SMARCAL1 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1) was recently found to be mutated in a number of ALT-positive glioblastomas.43 In our osteosarcoma panel, only NY lacked SMARCAL1 expression, as was previously reported.35 We did not find a loss of expression in the genes associated with ALT in the newly identified ALT-positive cell line KPD (Figure 2a). Further investigation will be required to elucidate the genetic basis leading to ALT activation in this cell line.
Figure 2.
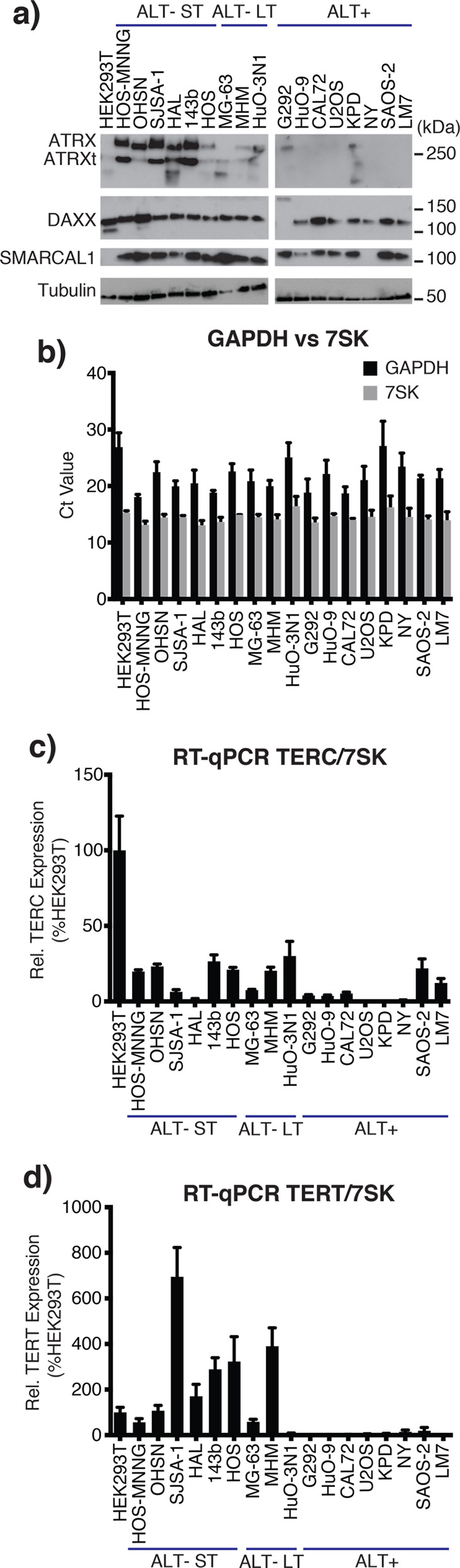
ALT-specific mutations and expression of telomerase. (a) ATRX, DAXX, and SMARCAL1 Western blot. Western blot analysis of the osteosarcoma cell lines for ATRX, DAXX, and SMARCAL1, three previously reported genes with mutations associated with ALT. Alpha-tubulin was used as a loading control. ATRXt = truncated isoform of ATRX. (b) RT-qPCR: 7SK RNA is a more reliable reference than GAPDH mRNA. The graph shows the CT values of GAPDH (black) and 7SK (gray) from normalized mRNA in RT-qPCR. Data is represented as a mean of three independent experiments, each run in triplicate. Error bars show standard deviation. (c) RT-qPCR: TERC/7SK. TERC was quantified using RT-qPCR and normalized first to 7SK and then to expression in the HEK293T cell line. There is no correlation between TERC expression and telomere length. Data are represented as a mean of three independent experiments. Error bars show standard error of the mean. (d) RT-qPCR: TERT/7SK. TERT mRNA was quantified using RT-qPCR and normalized first to 7SK and then to expression in the HEK293T cell line. There is no correlation between TERT expression and telomere length. Data represented as a mean of three independent experiments. Error bars show the standard error of the mean.
Previous studies showed that the ALT-negative cell lines HOS-MNNG, SJSA-1, 143b, HOS, and MG-63 express telomerase.36−39,44,45 In contrast, the ALT-positive cell lines G292, HuO-9, Cal72, U2OS, and NY do not express telomerase, while SAOS-2 expresses telomerase at low levels.37,38,45 To confirm these published results and to determine telomerase status in the newly characterized osteosarcoma lines, we assessed the transcriptional levels of the telomerase subunits, telomerase reverse transcriptase (TERT), and its RNA component (TERC).
Because cancer cells have variable transcription patterns, reliable reference RNA is needed for quantitative RT-PCR (RT-qPCR). We first investigated the expression level of two housekeeping genes, GAPDH and 7SK. We found that 7SK was a more reliable reference RNA as the standard deviation, standard error of the mean, and coefficient of variation of the calculated CT values across the cell lines were all smaller for 7SK than for GAPDH (0.91 vs 2.65%, 0.21 vs 0.63%, and 6.28 vs 12.27%, respectively), indicating that 7SK has smaller relative variability (Figure 2b).
Using 7SK RNA as the reference, TERC and TERT mRNA expression were measured. In general, ALT-negative cell lines expressed TERC at higher levels than the ALT-positive cell lines, with the exception of SAOS-2 and LM7 (Figure 2c). Most of the ALT-negative cell lines had TERT expression comparable to or greater than that in the HEK293T control cell line (Figure 2d). Exceptionally, TERT expression was notably low (7.7% of HEK293T) in the ALT-negative long telomere cell line HuO-3N1. In the ALT-positive cell lines, the TERT expression level was consistently negligible/undetectable (Figure 2d). Overall, our analysis documents telomerase expression almost exclusively in ALT-negative osteosarcoma lines and confirms that there was no correlation between TERC or TERT expression and telomere length in these lines (Figure 2c,d).
Short Telomeres Predict Hypersensitivity to the ATR Inhibitors in Osteosarcoma
Our telomere length analysis categorized the ALT-negative osteosarcoma cell lines into two types: short-telomere (ST) and long-telomere (LT) maintenance. We sought to identify therapeutic options that could synergise with short telomeres and hence may be beneficial in the treatment of osteosarcoma with this feature. Short telomeres are presumed to yield increased vulnerability to events that perturb telomere replication. The DNA damage/replication checkpoint kinase ATR is essential for facilitating replication fork progression,46 and it counteracts telomere shortening in yeasts.47−49 Importantly, ATR contributes to the maintenance of shortened telomeres in mice.19
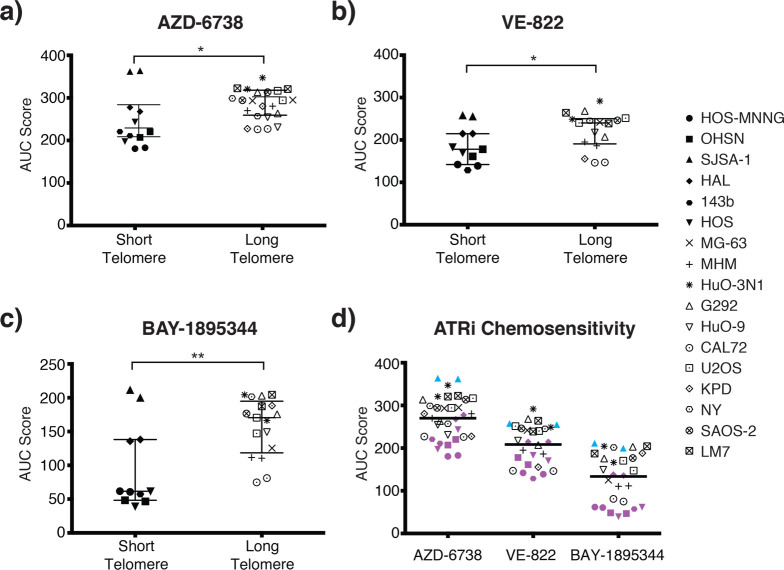
We therefore assessed the response of the various osteosarcoma cell lines against three small-molecule inhibitors of ATR (ATRi): AZD-6738/Ceralasertib, VE-822/Berzoserib, and BAY-1895344, all of which are currently in early phase clinical evaluation, either alone or in combination with DNA-damaging chemotherapies.50−52 In order to compare the responses, we deduced the area under the curve (AUC) based on nine-point concentration response curves established for each line within the panel (Table 2, Figure S-3). Intriguingly, we found that ALT-negative osteosarcoma cell lines with short telomeres were significantly more sensitive to all three of the small-molecule ATRi compared to the ALT-negative lines with long telomeres or the ALT-positive lines (Figure 3). Among the three ATRi, the most recently developed BAY-1895344 was the most potent, yielding the greatest differential between groups (Figure 3d). The half maximal inhibitory concentration (IC50) also support this notion (Table S-3). We observed no significant correlation between telomere length and methotrexate sensitivity (Figure S-4), a drug commonly used to treat osteosarcoma that disturbs DNA replication and induces DNA damage through the inhibition of dihydrofolate reductase. Thus, association of the drug sensitivity with short telomeres is selective for ATRi.
Table 2. Area Under the Curve (AUC) Analysis of ATRi.
| cell line | telomere statusa | AZD-6738 | VE-822 | BAY-1895344 |
|---|---|---|---|---|
| HOS-MNNG | ST | 181.8 ± 1.5 | 140.5 ± 2.2 | 61.2 ± 0.8 |
| OHSN | ST | 213.7 ± 9.3 | 169.7 ± 12.0 | 47.5 ± 1.1 |
| SJSA-1 | ST | 363.0 ± 1.3 | 256.4 ± 1.8 | 205.6 ± 8.2 |
| HAL | ST | 272.7 ± 6.9 | 214.7 ± 0 | 136.9 ± 1.7 |
| 143b | ST | 215.9 ± 6.8 | 128.6 | 57.4 |
| HOS | ST | 220.4 ± 32.4 | 176.8 ± 9.1 | 50.4 ± 15.8 |
| MG-63 | LT | 294.2 ± 1.2 | 241.8 | 125.4 |
| MHM | LT | 275.4 ± 7.4 | 190.7 ± 6.3 | 111.1 ± 0.8 |
| HuO-3N1 | LT | 334.0 ± 18.7 | 270.4 ± 30.3 | 185.4 ± 26.8 |
| G292 | ALT | 288.5 ± 35.4 | 237.7 ± 43.2 | 189.3 ± 19.4 |
| HuO-9 | ALT | 242.8 ± 16.5 | 217.5 | 149.2 |
| CAL72 | ALT | 226.6 ± 0.7 | 146.6 ± 0.4 | 77.9 ± 4.4 |
| U2OS | ALT | 305.6 ± 16.0 | 245.8 ± 8.1 | 158.9 ± 16.5 |
| KPD | ALT | 254.0 ± 37.5 | 155.5 | 188.3 |
| NY | ALT | 278.0 ± 30.0 | 245.2 | 201.5 |
| SAOS-2 | ALT | 304.0 ± 13.5 | 246.2 | 176.6 |
| LM7 | ALT | 321.7 ± 1.3 | 251.5 ± 17.9 | 195.9 ± 12.1 |
ST, ALT-negative, short telomere. LT, ALT-negative, long telomere. ALT, ALT-positive.
Figure 3.
ATRi sensitivity in osteosarcoma cell lines. (a–c) Sensitivity of osteosarcoma lines to ATRi VE-822 (a), AZD-6738 (b), and BAY-1895344 (c). Data represent AUC deduced from concentration response survival curves. Osteosarcoma lines grouped according to telomere length. Individual lines represented by symbols on the right (short telomere lines n = 6, long telomere lines n = 11). Values summarize outcome from two repeats for each cell line. Bars indicate median AUC and 95% confidence interval for each group based on the Mann–Whitney U test. *, p < 0.05; **, p < 0.01. (d) Summary of ATRi sensitivity. Short telomere cell lines are highlighted in purple. An outlier, ATRi-unresponsive short telomere line, SJSA-1, is marked in cyan.
When we compared ATRi sensitivity in the 17 cell lines based on their ALT status, we found no significant difference with two of the inhibitors, AZD-6738/Ceralasertib and VE-822/Berzoserib, although a significant association between ALT-negative status and sensitivity of this inhibitor was detected for BAY-1895344, the most potent of the three ATRi (Figure S-5). These results are consistent with increased presence of ATRi sensitivity within the ALT-negative group and further support the conclusion that telomere length as opposed to telomerase-based maintenance. Collectively, our data indicate that ATRi preferentially eliminate osteosarcoma with short telomeres.
ATRi-Induced Chromosome Segregation Failure and Cell Death in Osteosarcoma with Short Telomeres
To investigate the cause of the increased ATRi sensitivity seen in the osteosarcoma lines with short telomeres, we monitored cell fate in the presence of VE-822 using live cell imaging. We collected phase contrast images and concurrently scored for the incorporation of the live-cell-impermeable SYTOX DNA-binding dye, which indicates cell death. This analysis revealed a strong cytopathic response concurrent with cell death, observed within 24–36 h of ATRi addition in OHSN, HOS-MNNG, and HAL, three osteosarcoma cell lines with short telomeres (Figure 4a–d). In contrast, MG-63 and MHM, two cell lines with long telomeres, did not respond with death or evidence of cytopathic response within the 48 h observation period (Figure 4a,e,f). Similar results were obtained when cells were treated with the ATRi BAY-1895344 (Figure S-4b–f). Notably, an overt cytopathic response remained observable in short telomere cells where SYTOX dye and/or fluorescence imaging was omitted, indicative that the response is not linked to intercalation of the dye into DNA or exposure of the cultures to fluorescent light (Figure S-4a). Thus, our data indicate that ATR activity is required for the survival of osteosarcoma with short telomeres.
Figure 4.
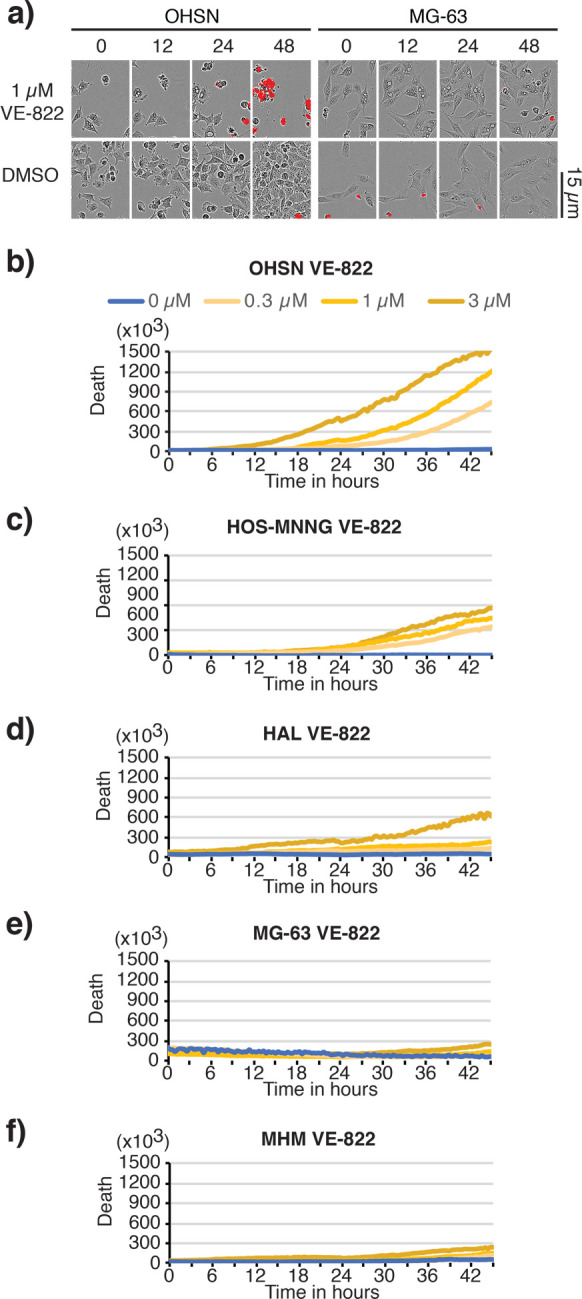
Selective death of osteosarcoma with short telomeres following treatment with ATR inhibitor VE-822. Short telomere cell lines, OHSN, HOS-MNNG, and HAL, and long telomere cell lines, MG-63 and MHM, treated with VE-822 were assessed using IncuCyte live cell analysis. Medium was supplemented with the live-cell-impermeable dye SYTOX to quantify cell death. (a) Representative images of OHSN and MG-63 treated with vehicle (DMSO) or 1 μM VE-822 at the indicated time. A 15 μm scale bar is shown. Phase contrast images are overlaid with SYTOX death dye signal (red) marking dead cells. (b–f) Representative graphs depicting net death over time, reflected by SYTOX death dye incorporation normalized to cell density for OHSN (b), HOS-MNNG (c), HAL (d), MG-63 (e), and MHM (f). Data representative of two or more experiments.
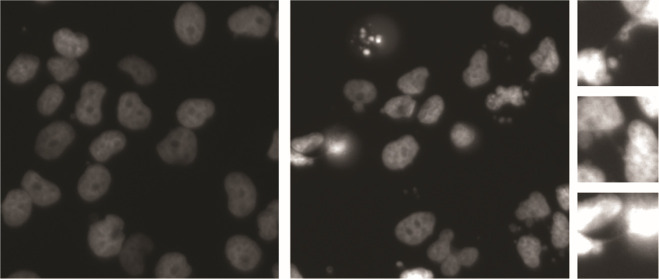
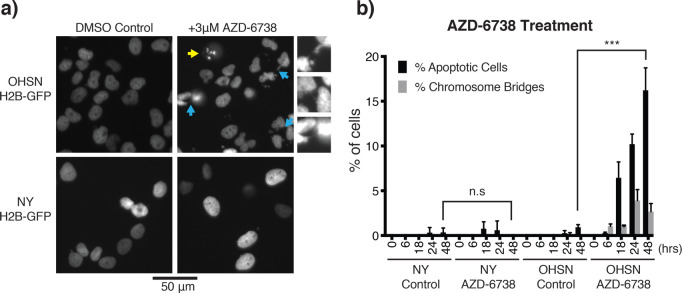
Lack of telomere replication is expected to lead to the loss of telomere protection, in turn eliciting chromosomal fusions and, as a result, defective mitosis with peri- and postmitotic death. To probe for such events, we made use of GFP-fused histone H2B (GFP-H2B), which we expressed in OHSN cells featuring short telomeres and, for comparison, NY cells featuring long telomeres. Longitudinal live-cell analysis revealed a prominent increase in mitotic DNA bridges detectable as early as 6 h following ATRi addition in the short-telomere OHSN cells but not long-telomere NY cells (Figure 5a, cyan arrows). We also observed chromatin fragmentation and apoptotic DNA bodies in OHSN cells but not NY cells (Figure 5a, yellow arrows) with a dramatic increase in prevalence over time (Figure 5b). Thus, we concluded that ATR inhibition causes selective cell death preceded by prominent mitotic aberrations consistent with chromosome fusions in osteosarcoma with short telomeres.
Figure 5.
ATRi-induced chromosome bridges and apoptotic cell death in osteosarcoma with short telomeres. (a) Representative images of OHSN and NY cell lines tagged with H2B-GFP. Yellow arrow shows apoptotic cell. Cyan arrows show chromosome bridges, which are enlarged and shown on the right of the panel. A 50 μm scale bar is shown. (b) Quantification of apoptotic cells and chromosome bridges over 48 h. Cells were treated with vehicle or AZD-6738 and were imaged at 6, 18, 24, and 48 h to count number of apoptotic cells and chromosome bridges from randomly selected samples of at least 200 cells. Graph shows mean percentage of cells classified as apoptotic or containing chromosome bridges for three independent experiments. Error bars show standard deviation. Short telomere cell line OHSN has significantly more apoptotic cells after AZD-6738 treatment compared to control cells, while no significant difference is seen for the long telomere ALT-positive cell line NY (Student’s t test; ***, p < 0.001).
Discussion
In this study, we demonstrated that the mode of telomere maintenance can influence ATRi sensitivity and addressed the reasons for efficacy of ATRi in the case of osteosarcoma. Our study using a panel of 17 osteosarcoma-derived cancer cell lines revealed that ALT-negative lines with short telomeres are hypersensitive to ATRi. Although ATR is an essential factor for genome stability and cell survival, our cytological analysis showed that only osteosarcoma cell lines with short telomeres died within 1–2 days in the presence of the ATR inhibitors. Inactivation of ATR resulted in chromosome bridges in mitosis and cell death. Thus, ATRi selectively eliminates osteosarcoma with short telomeres. Our results highlight the possibility of selectively exploiting the mechanisms of short-telomere maintenance for cancer therapy.
We report comprehensive parallel characterization of TMM covering an extensive panel of osteosarcoma derived lines. TMM for ALT-negative cell lines can be further subcategorized into long- and short-telomere maintenance. Our study agrees with the view that the level of telomerase expression does not strongly correlate with the length of telomeres that are maintained. Of note, we describe one cell line, HuO-3N1, which is categorized as ALT-negative with long telomeres and had low or undetectable levels of TERT. Such cancers were recently reported,6−8 and HuO-3N1 may reflect a model with this molecular feature. Together, our panel of osteosarcoma cell lines covers distinct TMMs and would be a useful tool to study cancer-specific telomere biology and TMM and telomere-length-based cancer therapeutics.
Necessity of ATR Activity at Short Telomeres in Osteosarcoma
A past report suggested that one ATRi, VE-822, selectively kills ALT-positive cells.44 Other reports have failed to detect this association.53−56 These studies focused on ALT status but not telomere length and did not include sufficient numbers of osteosarcoma cell lines that maintain short telomeres, except SJSA-1, which exhibited significant resistance to a wide range of kinase inhibitors in a recent large-scale profiling study.33 Our data provides evidence for the first time that cancer cells that maintain naturally subnormally short telomeres, as occurs in human cancers, require ATR activity for their survival. We speculate that ATR facilitates replication of short telomeres. Telomere replication is inherently difficult due to obstacles encountered by the replication fork such as the repetitive nature of the DNA sequence and the propensity to form secondary structures such as G-quadruplexes.57 A requirement for ATR in telomere replication has been suggested in studies using cell lines derived from patients with Seckel syndrome who have a hypomorphic mutation in ATR (OMIM 210600).58 Similar telomere instability has been recaptured in the Seckel syndrome mouse model.19 Notably, frequent chromosome fusions were observed in the Seckel mouse cells where short telomeres were experimentally induced by deletion of telomerase RNA. This observation is consistent with our finding, documenting elevated chromosome separation defects and cell death in osteosarcoma with short telomeres in the presence of ATR inhibitors.
How short telomeres activate ATR remains to be established. However, various lines of evidence predict a role for ATR in this context. The telomeric DNA binding protein, TRF1, is a component of the shelterin complex that facilitates telomere replication, and reduced TRF1 binding is known to induce telomere replication defects, ATR activation and telomeric ultrafine bridges.59−61 Another shelterin component, Pot1, directly represses ATR activation at the telomeric DNA.62 Loss of TRF1 can be found in short telomeres of aged cells.63 Therefore, we speculate that shortened telomeres with reduced TRF1 and/or Pot1 density regularly activate ATR in order to facilitate telomere replication. ATR, together with the related kinase, ataxia telangiectasia mutated (ATM), is thought to have a role in navigating telomerase to the telomere to extend it.64 Thus, localized ATR activation, mediated by fork-stalling at telomeric sequences, may enhance telomerase recruitment, as emphasized in the replication-mediated telomerase recruitment model.65
Telomere Diagnostic As a Potential Tool for Cancer Stratification
Although Southern blot based TRF analysis has been a gold standard for telomere length analysis, it is labor-intensive and requires a large quantity of intact genomic DNA, making it unsuitable in a clinical setting. Our MM-qPCR analysis for the panel of osteosarcoma cell lines highly correlated with the TRF results. Although cancer cells can be aneuploid or polyploid, the recent development of MM-qPCR made it possible to estimate relative telomere length of cancer.41,42 We were able to clearly identify a group of short-telomere-type TMM in our panel using this technique by comparing telomere repeat number in these cells to those in the reference cell line, HEK293T. As this PCR-based assay is simple, rapid, and high-throughput and needs only a small quantity of sample for diagnosis,40,66,67 we believe this assay is robust and can be used for the primary assessments of telomere repeat number and ALT-status.
In summary, our data demonstrates that short telomeres predict hypersensitivity to ATRi in osteosarcoma. We propose the use of telomere length as a biomarker to predict high efficacy of ATRi.
Material and Methods
Cell Lines
Cell lines and media used in this study are listed in Table S-1. Cells were sourced from the Sanger Centre cell line project.68 H2B-GFP expressing lines were generated as described in.69 All cells were maintained at 37 °C in a humidified incubator with 5% CO2.
DNA/RNA Extraction
Cell pellets were collected from actively proliferating cells grown in 10 cm plates to 70–80% confluency. Genomic DNA and total cellular RNA were extracted using commercially available kits (Invitrogen).
Telomere Length Measurements
Telomere restriction fragment (TRF) analysis and monochrome-multiplex quantitative polymerase chain reaction (MM-qPCR) were performed, as described previously.70
Briefly, TRF assays used 4 μg of genomic DNA digested with HinfI and RsaI and separated on a 30 cm 0.8% agarose gel. Genomic DNA fragments were probed with an alpha-32P-labeled telomeric DNA fragment, released using EcoRI from pKazu-hTelo, containing 55 TTAGGG repeats, cloned into pCR4 TOPO-blunt vector (Invitrogen). DNA weight markers Hyperladder 1-kb (BIoline) and UltraRanger 1kb (Norgen Biotek) were probed with a S. pombe telomeric DNA, released from plasmid p1742.71 The blot image was analyzed using Telometric72 or ImageJ software (NIH).
MM-qPCR was carried out as described in ref (40) with minor alterations. Genomic DNA samples were diluted to 5 ng/μL. Primer sets hTeloG/C (T), AlbuminF/R and GlobinF/R (S) are listed in Table S-4. Five concentrations of reference genomic DNA purified from HEK293T (ATCC CRL-3216) were prepared by 3-fold serial dilution (from 150 ng to 1.85 ng) to generate standard curves for relative quantitation of T/S ratios. For each sample, 20 ng of genomic DNA was mixed with 0.75× PowerUp SYBR Green Master Mix (Thermo Scientific), the primers (300 nM), and water to a final volume of 20 μL per well and analyzed using a Thermo Fisher QuantStudio 5, involving denaturation for 15 min at 95 °C, followed by two cycles of 15 s at 94 °C/15 s at 49 °C and 32 cycles of 15 s at 94 °C/10 s at 62 °C/15 s at 74 °C with signal acquisition and 10 s at 84 °C/15 s at 88 °C with signal acquisition. Samples were analyzed in triplicate, and analysis was repeated at least three times using independent runs. Data with more than 1% of a coefficient of variance for average CT were rejected. Telomere copy number was expressed as fold-enrichment against that in the reference HEK293T genomic DNA.
C-Circle Assay
CC-assays were carried out as previously described.66,67 Briefly, 20 μL CC reactions containing 4 mM dithiothreitol (DTT), 0.1% Tween, 1 mM dNTP mix, 0.2 μg/mL bovine serum albumin (BSA), 7.5 U phi-29 polymerase (New England Bioscience), 1× phi-29 buffer, and 32 ng of gDNA were incubated at 30 °C for 8 h followed by incubation at 65 °C for 20 min. For each sample tested, parallel reactions were carried out without addition of phi-29 polymerase.
For MM-qPCR based quantification, 10 μL of the CC-assay products were diluted with 30 μL of Tris-EDTA buffer and 5 μL of diluted products were used to quantify the C-circles. Standard curves were generated using serial 3-fold dilutions of U2OS DNA, ranging from 3.2 to 0.013 ng/μL. qPCR reaction was run in triplicate and contained 5 μL of DNA, 1× PowerUp SYBR Green Master Mix, 10 mM DTT, 2% DMSO, and 500 nM of the primer sets. Primer sets used were CC-TeloF/R (T) and AlbuminF/R (S). The following PCR conditions was used: 15 min at 95 °C, 33 cycles of 15 s at 95°, and 120 s at 54 °C. CT values were then extracted for analysis and expressed as T/S. CC assay (CCA) level were calculated by subtracting T/S by that of the respective no phi-29 polymerase controls. A sample is then considered to be ALT-positive if its CCA level lower error bar is greater than zero.
For dot blot quantification, CC-assay products were diluted with 200 μL of 2× SSC and transferred to a nitrocellulose membrane using a dot-blot vacuum manifold. DNA was UV-cross-linked on the membrane and probed using human telomere probe, as described for TRF.
Protein Extraction and Immunoblotting
Cell were lysed into 2% w/v SDS (50 mM Tris, pH 6.8). Lysates were separated using discontinuous SDS-PAGE to resolve proteins of molecular weight < 200 kDa or tris-acetate gels (NuPAGE, Invitrogen) to resolve proteins of molecular weight > 200 kDa. Antibodies used for immunoblotting were anti-SMARCAL1 (1/100, D3P5I; Cell Signaling), anti-DAXX (1/1000, 25C12; Cell Signaling), anti-ATRX (1/1000, D1N2E; Cell Signaling), anti-ATRX (1/100, sc-55584; Santa Cruz Biotechnology), anti-alpha-tubulin (1/5000, DM1A; Sigma-Aldrich), and anti-GAPDH (1/5000, ab9485; Abcam).
RNA Quantification by Reverse-Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
RNA quantification using RT-qPCR was carried out as described previously.71 Briefly, 1 μg of total RNA was reverse-transcribed to cDNA using 200 U of SuperScript IV reverse transcriptase (Thermo Scientific), and 50 μM random hexamer oligos. Reverse-transcribed cDNA was diluted to 200 μL with UltraPure DEPC-treated water, and the remaining RNA was removed with RNase A. Samples were analyzed using PowerUp SYBR Green based real-time PCR. Primers sets used were hTERT F1579/R1616, hTERC F27/R163, 7SK F7/R112, and GAPDH F6/R231 (Table S-4). qPCR cycling conditions were 95 °C for 10 min followed by 40 cycles of 10 s at 95 °C, 20 s at 60 °C, and 10 s at 72 °C. Melting curve analysis was performed immediately thereafter. TERT and TERC expression was normalized to 7SK mRNA expression using the ΔCT method. All data are expressed as ΔΔCT compared with HEK293T or as percentage expression compared to the HEK293T cell line. Averages were calculated from three biological replicates.
Fluorescence Microscopy-Based Detection of ALT-Associated Promyelocytic Leukemia (PML) Bodies (APB)
Cells were grown on ethanol-treated glass coverslips until they reached 50–80% confluency, then fixed with −20 °C methanol for 10 min. To detect telomeres and PML bodies, Alexa Fluor conjugated primary antibodies were used: anti-TRF2 [Alexa Fluor 488] (1/100, ab198595; Abcam) and anti-PML [Alexa Fluor 647] (1/50, ab209955; Abcam). Coverslips were mounted into 4′,6-diamidino-2-phenylindole (DAPI) to counterstain the DNA. Imaging was carried out using a DeltaVision Elite (GE Healthcare).
A cell line was considered APB-positive if APBs (a focus of TRF2 localized within a PML body) were detected in more than 10% of nuclei. At least 50 cells were analyzed in each of three separate biological samples.
Cell Viability Assay
Cell viability was assessed using Resazurin reduction. Cells were seeded at densities optimized for each line into 96-well plates. Cells were exposed to nine concentrations of 3-fold serially diluted inhibitors 24 h post seeding. Assays were run in triplicate. Cell viability was assessed 96 h post inhibitor addition. Area under the curve (AUC) metrics were deduced based on concentration–response curves normalized to vehicle for each line.
IncuCyte Live Cell Analysis
Time-lapse observations were carried out using an IncuCyte ZOOM live-cell analysis system (Essen Bioscience), as described previously.69 Cells were seeded into 96-well plates 24 h prior to ATRi addition followed by imaging every 2 h at 37 °C with 5% CO2. SYTOX TM green death dye (Molecular Probes) was added at the time of ATRi addition in some experiments and monitored using fluorescence image acquisition. Data presented are typical for at least two independent assessments, with conditions assessed in triplicate.
Statistical Analysis
Microsoft Excel and GraphPad Prism software were used to carry out statistical analysis for qPCR, chemosensitivity experiments and the extraction of AUC values from cell viability assay data. Mann–Whitney U tests were carried out to determine statistical significance in the chemosensitivity data (*, p < 0.05; **, p < 0.01; and ***, p < 0.001).
Acknowledgments
K.T. and L.C.C. were supported by Cancer Research UK (C36439/A12097) and European research council (281722-HRMCB). G.Z. and C.A.M. were supported by Children with Cancer UK (17-244). C.M. was supported through a Cancer Research UK studentship (C416/A23233). T.G. and K.T. were supported by College of Health, Medicine and Life Sciences, Brunel University London. S.J.S. is supported in part by National Institute for Health Research, UCLH Biomedical Research Centre.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00125.
Origin and characteristics of the osteosarcoma cell lines, telomere length measurement from TRF, IC50 drug response in osteosarcoma cell lines, oligonucleotides used for this study, characterization of telomere status in osteosarcoma cell lines, APB assay, ATRi concentration–survival curves in osteosarcoma lines, relationship between sensitivity to methotrexate and telomere length, comparison of ATRi sensitivities between ALT-negative and positive lines, selective death of osteosarcoma with short telomeres exposed to ATR inhibitor BAY-1895344 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Tomita K., and Collopy L. C. (2019) Telomeres, Telomerase, and Cancer, in Encyclopedia of Cancer (Boffetta P., and Hainaut P., Eds.) 3rd ed., pp 437–454, Academic Press, Oxford, U.K. [Google Scholar]
- Shay J. W.; Wright W. E. (2010) Telomeres and telomerase in normal and cancer stem cells. FEBS Lett. 584, 3819–3825. 10.1016/j.febslet.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddel R. R. (2014) Telomere maintenance mechanisms in cancer: clinical implications. Curr. Pharm. Des. 20, 6361–6374. 10.2174/1381612820666140630101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J. W.; Bacchetti S. (1997) A survey of telomerase activity in human cancer. Eur. J. Cancer 33, 787–791. 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Henson J. D.; Reddel R. R. (2010) Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. 584, 3800–3811. 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Barthel F. P.; Wei W.; Tang M.; Martinez-Ledesma E.; Hu X.; Amin S. B.; Akdemir K. C.; Seth S.; Song X.; Wang Q.; Lichtenberg T.; Hu J.; Zhang J.; Zheng S.; Verhaak R. G. (2017) Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 49, 349–357. 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viceconte N.; Dheur M. S.; Majerova E.; Pierreux C. E.; Baurain J. F.; van Baren N.; Decottignies A. (2017) Highly Aggressive Metastatic Melanoma Cells Unable to Maintain Telomere Length. Cell Rep. 19, 2529–2543. 10.1016/j.celrep.2017.05.046. [DOI] [PubMed] [Google Scholar]
- Dagg R. A.; Pickett H. A.; Neumann A. A.; Napier C. E.; Henson J. D.; Teber E. T.; Arthur J. W.; Reynolds C. P.; Murray J.; Haber M.; Sobinoff A. P.; Lau L. M. S.; Reddel R. R. (2017) Extensive Proliferation of Human Cancer Cells with Ever-Shorter Telomeres. Cell Rep. 19, 2544–2556. 10.1016/j.celrep.2017.05.087. [DOI] [PubMed] [Google Scholar]
- Okamoto K.; Seimiya H. (2019) Revisiting Telomere Shortening in Cancer. Cells 8, 107. 10.3390/cells8020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceja-Rangel H. A.; Sánchez-Suárez P.; Castellanos-Juárez E.; Peñaroja-Flores R.; Arenas-Aranda D. J.; Gariglio P.; Benítez-Bribiesca L. (2016) Shorter telomeres and high telomerase activity correlate with a highly aggressive phenotype in breast cancer cell lines. Tumor Biol. 37, 11917–11926. 10.1007/s13277-016-5045-7. [DOI] [PubMed] [Google Scholar]
- Hastie N. D.; Dempster M.; Dunlop M. G.; Thompson A. M.; Green D. K.; Allshire R. C. (1990) Telomere reduction in human colorectal carcinoma and with ageing. Nature 346, 866–868. 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- de Lange T.; Shiue L.; Myers R. M.; Cox D. R.; Naylor S. L.; Killery A. M.; Varmus H. E. (1990) Structure and variability of human chromosome ends. Mol. Cell. Biol. 10, 518–527. 10.1128/MCB.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaka S.; Davis C.; Lin S. F.; Wang T. L.; Heaphy C. M.; Shih I. M. (2019) Analysis of Telomere Lengths in p53 Signatures and Incidental Serous Tubal Intraepithelial Carcinomas Without Concurrent Ovarian Cancer. Am. J. Surg Pathol 43, 1083–1091. 10.1097/PAS.0000000000001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroupa M.; Rachakonda S. K.; Liska V.; Srinivas N.; Urbanova M.; Jiraskova K.; Schneiderova M.; Vycital O.; Vymetalkova V.; Vodickova L.; Kumar R.; Vodicka P. (2019) Relationship of telomere length in colorectal cancer patients with cancer phenotype and patient prognosis. Br. J. Cancer 121, 344–350. 10.1038/s41416-019-0525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammori M.; Sugishita Y.; Okamoto T.; Kobayashi M.; Yamazaki K.; Yamada E.; Yamada T. (2015) Telomere shortening in breast cancer correlates with the pathological features of tumor progression. Oncol. Rep. 34, 627–632. 10.3892/or.2015.4063. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Chen X.; Li L.; Zhou Y.; Wang C.; Hou S. (2015) The Association between Telomere Length and Cancer Prognosis: Evidence from a Meta-Analysis. PLoS One 10, e0133174 10.1371/journal.pone.0133174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado R. T.; Cooper J. N.; Padilla-Nash H. M.; Sloand E. M.; Wu C. O.; Scheinberg P.; Ried T.; Young N. S. (2012) Short telomeres result in chromosomal instability in hematopoietic cells and precede malignant evolution in human aplastic anemia. Leukemia 26, 700–707. 10.1038/leu.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Kha H.; Ungrin M.; Robinson M. O.; Harrington L. (2002) Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc. Natl. Acad. Sci. U. S. A. 99, 3597–3602. 10.1073/pnas.062549199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNees C. J.; Tejera A. M.; Martinez P.; Murga M.; Mulero F.; Fernandez-Capetillo O.; Blasco M. A. (2010) ATR suppresses telomere fragility and recombination but is dispensable for elongation of short telomeres by telomerase. J. Cell Biol. 188, 639–652. 10.1083/jcb.200908136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.; Blackburn E. H. (2007) Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol. Cell 28, 315–327. 10.1016/j.molcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C. A.; Tomita K. (2017) Fundamental mechanisms of telomerase action in yeasts and mammals: understanding telomeres and telomerase in cancer cells. Open Biol. 7, 160338. 10.1098/rsob.160338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Sfeir A. J.; Zou Y.; Buseman C. M.; Chow T. T.; Shay J. W.; Wright W. E. (2009) Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138, 463–475. 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Mar V.; Zhou W.; Harrington L.; Robinson M. O. (1999) Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 13, 2388–2399. 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekaran V.; Soares J.; Jarstfer M. B. (2014) Telomere maintenance as a target for drug discovery. J. Med. Chem. 57, 521–538. 10.1021/jm400528t. [DOI] [PubMed] [Google Scholar]
- Cesare A. J.; Reddel R. R. (2010) Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 11, 319–330. 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- Zhang J. M.; Yadav T.; Ouyang J.; Lan L.; Zou L. (2019) Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep. 26, 955–968. 10.1016/j.celrep.2018.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy C. M.; Subhawong A. P.; Hong S. M.; Goggins M. G.; Montgomery E. A.; Gabrielson E.; Netto G. J.; Epstein J. I.; Lotan T. L.; Westra W. H.; et al. (2011) Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 179, 1608–1615. 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakin-Smith V.; Jellinek D. A.; Levy D.; Carroll T.; Teo M.; Timperley W. R.; McKay M. J.; Reddel R. R.; Royds J. A. (2003) Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet 361, 836–838. 10.1016/S0140-6736(03)12681-5. [DOI] [PubMed] [Google Scholar]
- Henson J. D.; Hannay J. A.; McCarthy S. W.; Royds J. A.; Yeager T. R.; Robinson R. A.; Wharton S. B.; Jellinek D. A.; Arbuckle S. M.; Yoo J.; et al. (2005) A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin. Cancer Res. 11, 217–225. [PubMed] [Google Scholar]
- Bajpai J.; Chandrasekharan A.; Simha V.; Mandal T.; Shah K.; Hingmare S.; Rangarajan B.; Shetty N.; Vora T.; Ghosh J.; et al. (2019) Osteosarcoma journey over two decades in India: Small steps, big changes. Pediatr. Blood Cancer 66, e27877 10.1002/pbc.27877. [DOI] [PubMed] [Google Scholar]
- Mirabello L.; Troisi R. J.; Savage S. A. (2009) Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer 115, 1531–1543. 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert G.; Lansdorp P. M. (2008) Telomeres and aging. Physiol. Rev. 88, 557–579. 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- Holme H.; Gulati A.; Brough R.; Fleuren E. D. G.; Bajrami I.; Campbell J.; Chong I. Y.; Costa-Cabral S.; Elliott R.; Fenton T.; Frankum J.; Jones S. E.; Menon M.; Miller R.; Pemberton H. N.; Postel-Vinay S.; Rafiq R.; Selfe J. L.; von Kriegsheim A.; Munoz A. G.; Rodriguez J.; Shipley J.; van der Graaf W. T. A.; Williamson C. T.; Ryan C. J.; Pettitt S.; Ashworth A.; Strauss S. J.; Lord C. J. (2018) Chemosensitivity profiling of osteosarcoma tumour cell lines identifies a model of BRCAness. Sci. Rep. 8, 10614. 10.1038/s41598-018-29043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy C. A.; Li W.; Reisenweber S.; Thongthip S.; Bruno J.; de Lange T.; De S.; Petrini J. H.; Sung P. A.; Jasin M.; et al. (2012) Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 8, e1002772 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason-Osann E.; Dai A.; Floro J.; Lock Y. J.; Reiss M.; Gali H.; Matschulat A.; Labadorf A.; Flynn R. L. (2018) Identification of a novel gene fusion in ALT positive osteosarcoma. Oncotarget 9, 32868–32880. 10.18632/oncotarget.26029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; Bobb D.; He J.; Hill D. A.; Dome J. S. (2015) The HSP90 inhibitor alvespimycin enhances the potency of telomerase inhibition by imetelstat in human osteosarcoma. Cancer Biol. Ther. 16, 949–957. 10.1080/15384047.2015.1040964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C.; Schaefer K. L.; Jauch A.; Keller M.; Wai D.; Brinkschmidt C.; van Valen F.; Boecker W.; Dockhorn-Dworniczak B.; Poremba C. (2001) Alternative lengthening of telomeres is associated with chromosomal instability in osteosarcomas. Oncogene 20, 3835–3844. 10.1038/sj.onc.1204493. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y.; Yamamoto N.; Tajima K.; Ohno A.; Washimi Y.; Ishimura D.; Washimi O.; Yamada H. (2011) Characterization of human multicentric osteosarcoma using newly established cells derived from multicentric osteosarcoma. J. Cancer Res. Clin. Oncol. 137, 423–433. 10.1007/s00432-010-0885-9. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Yu L.; Dai G.; Xia K.; Liu G.; Song Q.; Tao C.; Gao T.; Guo W. (2017) Telomerase reverse transcriptase promotes chemoresistance by suppressing cisplatin-dependent apoptosis in osteosarcoma cells. Sci. Rep. 7, 1–11. 10.1038/s41598-017-07204-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon R. M. (2009) Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 37, e21 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropio J.; Chebly A.; Ferrer J.; Prochazkova-Carlotti M.; Idrissi Y.; Azzi-Martin L.; Cappellen D.; Pham-Ledard A.; Soares P.; Merlio J. P.; Chevret E. (2020) Reliable blood cancer cells’ telomere length evaluation by qPCR. Cancer Med. 9, 3153–3162. 10.1002/cam4.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren P. N.; Bishop K.; Dey S.; Herbert B. S.; Tanaka H. (2018) Development of a New Monochrome Multiplex qPCR Method for Relative Telomere Length Measurement in Cancer. Neoplasia 20, 425–431. 10.1016/j.neo.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diplas B. H.; He X.; Brosnan-Cashman J. A.; Liu H.; Chen L. H.; Wang Z.; Moure C. J.; Killela P. J.; Loriaux D. B.; Lipp E. S.; Greer P. K.; Yang R.; Rizzo A. J.; Rodriguez F. J.; Friedman A. H.; Friedman H. S.; Wang S.; He Y.; McLendon R. E.; Bigner D. D.; Jiao Y.; Waitkus M. S.; Meeker A. K.; Yan H. (2018) The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat. Commun. 9, 2087. 10.1038/s41467-018-04448-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R. L.; Cox K. E.; Jeitany M.; Wakimoto H.; Bryll A. R.; Ganem N. J.; Bersani F.; Pineda J. R.; Suva M. L.; Benes C. H.; Haber D. A.; Boussin F. D.; Zou L. (2015) Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 347, 273–277. 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I.; Cai L.; Wei R.-X.; Hu H.; Jin W.; Zhu X.-B. (2011) Different expression of alternative lengthening of telomere (ALT)-associated proteins/mRNAs in osteosarcoma cell lines. Oncol. Lett. 2, 1327–1332. 10.3892/ol.2011.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K. A.; Cortez D. (2008) ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627. 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser B. A.; Chang Y. T.; Kosti J.; Nakamura T. M. (2011) Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat. Struct. Mol. Biol. 18, 1408–1413. 10.1038/nsmb.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H.; Tarumoto Y.; Ishikawa F. (2012) Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 26, 241–246. 10.1101/gad.177873.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener R.; Connelly C. J.; Greider C. W. (2019) Tel1 Activation by the MRX Complex Is Sufficient for Telomere Length Regulation but Not for the DNA Damage Response in Saccharomyces cerevisiae. Genetics 213, 1271–1288. 10.1534/genetics.119.302713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon M. T.; Boylan Z.; Smith D.; Guevara J.; Mohammed K.; Peckitt C.; Saunders M.; Banerji U.; Clack G.; Smith S. A.; Spicer J. F.; Forster M. D.; Harrington K. J. (2018) PATRIOT: A phase I study to assess the tolerability, safety and biological effects of a specific ataxia telangiectasia and Rad3-related (ATR) inhibitor (AZD6738) as a single agent and in combination with palliative radiation therapy in patients with solid tumours. Clin Transl Radiat Oncol 12, 16–20. 10.1016/j.ctro.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carrigan B.; de Miguel Luken M. J.; Papadatos-Pastos D.; Brown J.; Tunariu N.; Perez Lopez R.; Ganegoda M.; Riisnaes R.; Figueiredo I.; Carreira S.; et al. (2016) Phase I trial of a first-in-class ATR inhibitor VX-970 as monotherapy (mono) or in combination (combo) with carboplatin (CP) incorporating pharmacodynamics (PD) studies. J. Clin. Oncol. 34, 2504–2504. 10.1200/JCO.2016.34.15_suppl.2504. [DOI] [Google Scholar]
- Mei L.; Zhang J.; He K.; Zhang J. (2019) Ataxia telangiectasia and Rad3-related inhibitors and cancer therapy: where we stand. J. Hematol. Oncol. 12, 43. 10.1186/s13045-019-0733-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeg K. I.; Chung I.; Bauer C.; Rippe K. (2016) Cancer Cells with Alternative Lengthening of Telomeres Do Not Display a General Hypersensitivity to ATR Inhibition. Front. Oncol. 6, 27–27. 10.3389/fonc.2016.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Soler M.; Morgado-Palacin I.; Lafarga V.; Lecona E.; Murga M.; Callen E.; Azorin D.; Alonso J.; Lopez-Contreras A. J.; Nussenzweig A.; Fernandez-Capetillo O. (2016) Efficacy of ATR inhibitors as single agents in Ewing sarcoma. Oncotarget 7, 58759–58767. 10.18632/oncotarget.11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S. L.; Parmar V.; Lorenzi F.; Marshall L. V.; Jamin Y.; Poon E.; Angelini P.; Chesler L. (2020) Novel therapeutic strategies targeting telomere maintenance mechanisms in high-risk neuroblastoma. J. Exp. Clin. Cancer Res. 39, 78. 10.1186/s13046-020-01582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche-Clary A.; Chaire V.; Verbeke S.; Algeo M. P.; Malykh A.; Le Loarer F.; Italiano A. (2020) ATR Inhibition Broadly Sensitizes Soft-Tissue Sarcoma Cells to Chemotherapy Independent of Alternative Lengthening Telomere (ALT) Status. Sci. Rep. 10, 7488. 10.1038/s41598-020-63294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer O.; Hickson I. D. (2018) Pathways for maintenance of telomeres and common fragile sites during DNA replication stress. Open Biol. 8, 180018. 10.1098/rsob.180018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennarun G.; Hoffschir F.; Revaud D.; Granotier C.; Gauthier L. R.; Mailliet P.; Biard D. S.; Boussin F. D. (2010) ATR contributes to telomere maintenance in human cells. Nucleic Acids Res. 38, 2955–2963. 10.1093/nar/gkp1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A.; Kosiyatrakul S. T.; Hockemeyer D.; MacRae S. L.; Karlseder J.; Schildkraut C. L.; de Lange T. (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138, 90–103. 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P.; Thanasoula M.; Munoz P.; Liao C.; Tejera A.; McNees C.; Flores J. M.; Fernandez-Capetillo O.; Tarsounas M.; Blasco M. A. (2009) Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 23, 2060–2075. 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagno d’Alcontres M. S.; Palacios J. A.; Mejias D.; Blasco M. A. (2014) TopoIIalpha prevents telomere fragility and formation of ultra thin DNA bridges during mitosis through TRF1-dependent binding to telomeres. Cell Cycle 13, 1463–1481. 10.4161/cc.28419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi E. L.; de Lange T. (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071. 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Hohensinner P. J.; Kaun C.; Buchberger E.; Ebenbauer B.; Demyanets S.; Huk I.; Eppel W.; Maurer G.; Huber K.; Wojta J. (2016) Age intrinsic loss of telomere protection via TRF1 reduction in endothelial cells. Biochim. Biophys. Acta, Mol. Cell Res. 1863, 360–367. 10.1016/j.bbamcr.2015.11.034. [DOI] [PubMed] [Google Scholar]
- Tong A. S.; Stern J. L.; Sfeir A.; Kartawinata M.; de Lange T.; Zhu X. D.; Bryan T. M. (2015) ATM and ATR Signaling Regulate the Recruitment of Human Telomerase to Telomeres. Cell Rep. 13, 1633–1646. 10.1016/j.celrep.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C. W. (2016) Regulating telomere length from the inside out: the replication fork model. Genes Dev. 30, 1483–1491. 10.1101/gad.280578.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L. M.; Dagg R. A.; Henson J. D.; Au A. Y.; Royds J. A.; Reddel R. R. (2013) Detection of alternative lengthening of telomeres by telomere quantitative PCR. Nucleic Acids Res. 41, e34 10.1093/nar/gks781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J. D.; Lau L. M.; Koch S.; Martin La Rotta N.; Dagg R. A.; Reddel R. R. (2017) The C-Circle Assay for alternative-lengthening-of-telomeres activity. Methods 114, 74–84. 10.1016/j.ymeth.2016.08.016. [DOI] [PubMed] [Google Scholar]
- Tate J. G.; Bamford S.; Jubb H. C.; Sondka Z.; Beare D. M.; Bindal N.; Boutselakis H.; Cole C. G.; Creatore C.; Dawson E.; Fish P.; Harsha B.; Hathaway C.; Jupe S. C.; Kok C. Y.; Noble K.; Ponting L.; Ramshaw C. C.; Rye C. E.; Speedy H. E.; Stefancsik R.; Thompson S. L.; Wang S.; Ward S.; Campbell P. J.; Forbes S. A. (2019) COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47, D941–D947. 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Stockwell S. R.; Elbanna M.; Ketteler R.; Freeman J.; Al-Lazikani B.; Eccles S.; De Haven Brandon A.; Raynaud F.; Hayes A.; Clarke P. A.; Workman P.; Mittnacht S. (2019) Signalling involving MET and FAK supports cell division independent of the activity of the cell cycle-regulating CDK4/6 kinases. Oncogene 38, 5905–5920. 10.1038/s41388-019-0850-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balogh E.; Chandler J. C.; Varga M.; Tahoun M.; Menyhárd D. K.; Schay G.; Goncalves T.; Hamar R.; Légrádi R.; Szekeres Á.; et al. (2020) Pseudouridylation defect due to DKC1 and NOP10 mutations causes nephrotic syndrome with cataracts, hearing impairment, and enterocolitis. Proc. Natl. Acad. Sci. U. S. A. 117, 15137. 10.1073/pnas.2002328117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collopy L. C.; Ware T. L.; Goncalves T.; í Kongsstovu S.; Yang Q.; Amelina H.; Pinder C.; Alenazi A.; Moiseeva V.; Pearson S. R.; et al. (2018) LARP7 family proteins have conserved function in telomerase assembly. Nat. Commun. 9, 557. 10.1038/s41467-017-02296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant J. D.; Broccoli D.; Muquit M.; Manion F. J.; Tisdall J.; Ochs M. F. (2001) Telometric: a tool providing simplified, reproducible measurements of telomeric DNA from constant field agarose gels. BioTechniques 31, 1314–1316. 10.2144/01316bc02. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.