Abstract
Background
Gene fusion events are significant sources of somatic variation across adult and pediatric cancers and are some of the most clinically-effective therapeutic targets, yet low consensus of RNA-Seq fusion prediction algorithms makes therapeutic prioritization difficult. In addition, events such as polymerase read-throughs, mis-mapping due to gene homology, and fusions occurring in healthy normal tissue require informed filtering, making it difficult for researchers and clinicians to rapidly discern gene fusions that might be true underlying oncogenic drivers of a tumor and in some cases, appropriate targets for therapy.
Results
We developed annoFuse, an R package, and shinyFuse, a companion web application, to annotate, prioritize, and explore biologically-relevant expressed gene fusions, downstream of fusion calling. We validated annoFuse using a random cohort of TCGA RNA-Seq samples (N = 160) and achieved a 96% sensitivity for retention of high-confidence fusions (N = 603). annoFuse uses FusionAnnotator annotations to filter non-oncogenic and/or artifactual fusions. Then, fusions are prioritized if previously reported in TCGA and/or fusions containing gene partners that are known oncogenes, tumor suppressor genes, COSMIC genes, and/or transcription factors. We applied annoFuse to fusion calls from pediatric brain tumor RNA-Seq samples (N = 1028) provided as part of the Open Pediatric Brain Tumor Atlas (OpenPBTA) Project to determine recurrent fusions and recurrently-fused genes within different brain tumor histologies. annoFuse annotates protein domains using the PFAM database, assesses reciprocality, and annotates gene partners for kinase domain retention. As a standard function, reportFuse enables generation of a reproducible R Markdown report to summarize filtered fusions, visualize breakpoints and protein domains by transcript, and plot recurrent fusions within cohorts. Finally, we created shinyFuse for algorithm-agnostic interactive exploration and plotting of gene fusions.
Conclusions
annoFuse provides standardized filtering and annotation for gene fusion calls from STAR-Fusion and Arriba by merging, filtering, and prioritizing putative oncogenic fusions across large cancer datasets, as demonstrated here with data from the OpenPBTA project. We are expanding the package to be widely-applicable to other fusion algorithms and expect annoFuse to provide researchers a method for rapidly evaluating, prioritizing, and translating fusion findings in patient tumors.
Keywords: RNA-seq, Gene fusions, Annotation tool, Oncogenes, Cancer, Shiny web application
Background
Gene fusions arise in cancer as a result of aberrant chromosomal rearrangements or defective splicing, bringing together two unrelated genes that are then expressed as a novel fusion transcript [1]. Detection of therapeutically-targetable fusion calls is of clinical importance and computational methods are constantly being developed to detect these events in real-time. Recent comparative studies show low concordance of fusion predictions across methods [2], suggesting that many predictions may not represent true events. Additionally, transcriptional read-throughs [3], in which the polymerase machinery skips a stop codon and reads through a neighbouring gene, as well as fusions that involve non-canonical transcripts or gene-homologs, are prevalent in disease datasets, yet the biological relevance of such events is still unclear. This makes it difficult for both researchers and clinicians to prioritize disease-relevant fusions and discern the underlying biological mechanisms and thus, appropriate fusion-directed therapy. Gene fusion events leading to gain-of-function or loss-of-function in kinases and putative tumor suppressor genes, respectively, have been shown to be oncogenic drivers with therapeutic potential, especially in pediatric tumors [4–6]. For example, the recurrent fusion KIAA1549-BRAF is found across 66–80% of low grade gliomas and results in a fusion transcript that has constitutive BRAF kinase activity [7]. EWSR1-FLI1 is found in nearly 100% of Ewing’s sarcoma and forms an oncogenic RNA complex, driving tumorigenesis [8]. To capture highly recurrent and validated fusions such as these, the fusion databases ChimerDB [9] and TumorFusions [10] were developed from RNA fusions called in The Cancer Genome Atlas (TCGA) [11, 12] samples. In such large-scale cancer studies, a single algorithm was routinely used to detect fusion calls because using multiple callers often adds complexity of annotation and integration. However, it is now common practice to incorporate data from multiple algorithms to reliably define the fusion landscape of cancers. Recent efforts have reported the importance of using systematic filtering and aggregation of multiple fusion callers to expand the number of biologically-relevant fusions in adult cancers [12, 13]. However, to our knowledge there are no tools or packages developed to filter, aggregate, and detect recurrent and putative oncogenic fusions in a systematic, flexible, and reproducible manner. Despite the existence of a few tools with working open-source code which can assist in fusion annotation or prioritization, only three are algorithm-agnostic with the remaining tools relying on outdated fusion algorithms, rendering them unusable on current gold standard tools to date, such as STAR-Fusion [14] and Arriba [15] (Table 1).
Table 1.
Available fusion annotation and prioritization tools
| Tools | Software implementation | Compatible fusion algorithms | Annotations | Prioritization | Visualization | Code availability | Last Updated |
|---|---|---|---|---|---|---|---|
| annoFuse | R | Arriba, STAR-Fusion for annoFuse; agnostic for shinyFuse | Protein Domains, Kinase, Transcription Factor, Oncogene, Tumor Suppressor Gene, Kinase Domain Retention, Reciprocality, CTAT_HumanFusionLib, Gene Homolog, Expression | Based on quality/artifact filtering, functional, and oncogenic annotation | Genomic view of single or multiple fusion events() with domains, Recurrent fusions and recurrently fused genes barplots | https://github.com/d3b-center/annoFuse/ | September 2020+ |
| AGFusion | Python | Algorithm agnostic; requires two fusion gene partners (gene symbol, Ensembl ID, or Entrez gene ID), predicted fusion junctions in genomic coordinates, genome build | Protein domains | Amino acid structure of fusion | https://github.com/murphycj/AGFusion | September 2019 | |
| chimera | R package | chimeraScan, bellerophontes, deFuse, FusionFinder, FusionHunter, mapSplice, tophat-fusion, FusionMap, STAR, Rsubread, fusionCatcher | Uses Oncofuse to prioritize oncogenic fusions | https://rdrr.io/bioc/chimera/ | March 2015 | ||
| chimeraviz | R package | deFuse, EricScript, InFusion, JAFFA, FusionCatcher, FusionMap, PRADA, SOAPFuse, STAR-Fusion | Genomic view of fusion event (Breakpoints required) | https://github.com/stianlagstad/chimeraviz | October 2019 | ||
| co-Fuse | R | FusionCatcher, SoapFuse, TopHat, DeFuse | Recurrence detection | https://github.com/sakrapee/co-fuse | May 2017 | ||
| FusionAnnotator | perl | Algorithm agnostic; requires fusion gene partners | CTAT_HumanFusionLib, Gene Homolog | https://github.com/FusionAnnotator/FusionAnnotator | July 2019 | ||
| FusionHub | web-based | Algorithm agnostic; requires fusion gene partners to search databases for fusion and uses breakpoints from ChimerSeq to visualize | Utilizes oncofuse for oncogenicity | Circos, domain, network view of fusions | https://fusionhub.persistent.co.in/ | June 2018 | |
| FusionPathway | R | Algorithm agnostic; Fusion gene Entrez IDs and gene symbols, Pfam protein domain IDs of parental genes | Oncogenic pathway association using GSEA | https://github.com/perwu/FusionPathway/ | September 2019 | ||
| Oncofuse | Java/Groovy | Tophat-fusion and TopHat2, FusionCatcher, RNASTAR | Protein domains, expression | Bayesian probability of oncogenicity | https://github.com/mikessh/oncofuse | February 2016 | |
| Pegasus | Java/Perl/Python/Bash | chimerascan 0.4.5, defuse 0.4.3, bellerophontes 0.4.0 | Kinase, Protein domains | Driver Score for oncogenicity | https://github.com/RabadanLab/Pegasus | February 2016 | |
| confFuse | Python | deFuse | Feature dependent scoring | https://github.com/Zhiqin-HUANG/confFuse | November 2017 |
List of ten openly-available fusion annotation and prioritization software tools, compared to annoFuse. Only AGFusion, FusionAnnotator, Fusion Pathway, and certain functions of FusionHub are algorithm agnostic, and most algorithms require outdated fusion algorithm input
Here, we developed annoFuse for annotation, prioritization, and exploration of putative oncogenic gene fusions. We performed technical validation using two independent RNA-Sequencing datasets (TCGA and Pediatric Preclinical Testing Consortium), and finally, applied annoFuse to gene fusion calls from STAR-Fusion and Arriba for 1028 pediatric brain tumor samples provided as part of the OpenPBTA Project [16]. To achieve this, we used FusionAnnotator on raw fusion calls to identify and filter red flag fusions, that is, fusions found in healthy tissues or in gene homology databases. Using annoFuse, we remove fusions known or predicted to be artifactual and retain high-quality fusion calls. Second, fusions that pass quality checks are annotated (Additional file 2: Table S1) as previously found within TCGA [10] and each gene partner is annotated as an oncogene, tumor suppressor [17, 18], kinase [19], transcription factor [20], and whether it has been reported in the Catalogue of Somatic Mutations in Cancer (COSMIC) Cancer Gene Census [21]. In addition, we added the following genes specific to pediatric cancer from literature review: MYBL1 [22], SNCAIP [23], FOXR2 [24], TTYH1 [25], and TERT [26–29] as oncogenes and BCOR [30] and QKI [4] as tumor suppressors. Finally, we determined the recurrence pattern for fusions across the cohort and also recurrently-fused genes within each cancer histology.
Implementation
We implemented annoFuse using the R programming language R version 4.0.2 (2020-08-13). The R packages required to install and run annoFuse are reshape2, dplyr, tidyr, ggplot2, qdapRegex, ggpubr, tibble, ggthemes, EnsDb.Hsapiens.v86, grid, readr, grDevices, stats, utils, stringr, shiny, shinydashboard, rintrojs, shinythemes, DT, rmarkdown, and methods, with the optional package: knitr. We also created an interactive web-based application of annoFuse called shinyFuse using the R/Shiny framework.
R package overview
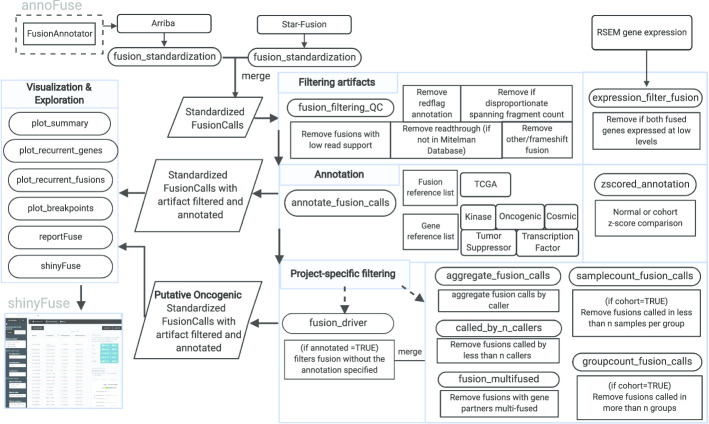
The annoFuse package was developed to provide a standardized filtering and annotation method for fusion calls from Arriba and STAR-Fusion, first and second place winners of the 2017 DREAM SMC-RNA Challenge, respectively [31]. In a 2019 assessment of 23 fusion algorithms for cancer biology, both Arriba and STAR-Fusion ranked in the top three fastest and most accurate tools [32]. annoFuse utilizes a four-step process (Fig. 1) that is available with flexible functions to perform downstream functions such as merging, filtering, and prioritization of fusion calls from multiple fusion calling algorithms on single or batch samples.
Fig. 1.
Graphical representation of the annoFuse pipeline. RNA-seq data processed through STAR-RSEM and fusion calls generated by Arriba v1.1.0 and/or STAR-Fusion 1.5.0 are inputs for the pipeline. The fusion_standardization function standardizes calls from fusion callers to retain information regarding fused genes, breakpoints, reading frame information, as well as annotation from FusionAnnotator. Standardized fusion calls use fusion_filtering_QC to remove false positives such as fusions with low read support, annotated as read-throughs, found in normal and gene homolog databases and remove non-expressed fusions using expression_filter_fusion. Calls are annotated with annotate_fusion_calls to include useful biological features of interest (eg. Kinase, Tumor suppressor etc.) Project-specific filtering captures recurrent fused genes using functions to filter (shown in boxes) as well as putative driver fusion. Outputs available from annoFuse include TSV files of annotated and prioritized fusions, a PDF summary of fusions, recurrently-fused gene/fusion plots, and an HTML report. Finally, users can explore fusion data interactively using shinyFuse. (Created with BioRender.com)
RNA expression and fusion calls
Currently, annoFuse is compatible with fusion calls generated from Arriba v1.1.0 [15] and/or STAR-Fusion 1.5.0 [14]. Both tools utilize aligned BAM and chimeric SAM files from STAR as inputs and STAR-Fusion calls are annotated with GRCh38_v27_CTAT_lib_Feb092018.plug-n-play.tar.gz, which is provided in the STAR-fusion release. Arriba should be provided with strandedness information, or set to auto-detection for poly-A enriched libraries. Additionally, the blacklist file, blacklist_hg38_GRCh38_2018-11-04.tsv.gz contained in the Arriba release tarballs, should be used to remove recurrent fusion artifacts and transcripts present in healthy tissue. An expression matrix with FPKM or TPM values is also required; the matrix should have a column “GeneSymbol” following the same gene naming convention as found in fusion calls.
Fusion call preprocessing
We leveraged the fact that STAR-Fusion uses FusionAnnotator as its final step and thus, require all fusion calls be annotated with FusionAnnotator v. 0.2.0 to contain the additional column, “annots”. Fusion calls for all samples from both algorithms should be merged into a single TSV. Users will also provide a sample ID column (named “Sample” or other character vector specified by the user) for each merged file, which will be used to denote fusion calls per sample as well as identify recurrent fusions in cohorts of samples.
annoFuse steps
Step 1: fusion standardization
To obtain a standardized format for fusion calls from multiple fusion calls we use the fusion_standardization function to format caller specific output files to a standardizedFusionCalls format defined in the package README. fusion_standardization allows users to standardize fusion calls from multiple callers.
Step 2: fusion filtering
Events such as polymerase read-throughs, mis-mapping due to gene homology, and fusions occurring in healthy normal tissue confound detection for true recurrent fusion calls and false positives for genes considered as oncogenic, tumor suppressor or kinases in some cases. In this step, we filter the standardized fusion calls to remove artifacts and false positives (Table 2) using the function fusion_filtering_QC. The parameters are flexible to allow users to annotate and filter the fusions with a priori knowledge of their call set. For example, since the calls are pre-annotated with FusionAnnotator, the user can remove fusions known to be red-flags as annotated with any of the following databases GTEx_recurrent_STARF2019, HGNC_GENEFAM, DGD_PARALOGS, Greger_Normal, Babiceanu_Normal, BodyMap, and ConjoinG. This is done using the parameter, artifact_filter = "GTEx_recurrent_STARF2019|DGD_PARALOGS |Normal| BodyMap". Of note, we decided not to remove genes annotated in HGNC_GENEFAM, as this database contains multiple oncogenes and their removal resulted in missed true fusions using our validation truth set. Likewise, we retained fusions annotated with ConjoinG, as these may represent true chimeric RNA and protein products from adjacent genes, but are a separate class from read-through events [33]. Read-throughs annotated by any algorithm can also be removed at this step by using the parameter “readthroughFilter = TRUE”. During validation, we observed the real oncogenic fusion, P2RY8-CRLF2 [34, 35], annotated as a read-through in acute lymphoblastic leukemia samples, therefore, we implemented a condition such that if a fusion is annotated as a read-through, but is present in the Mitelman cancer fusion database, we recover these fusions as true positive calls.
Table 2.
Fusion filtering and annotation criteria
| Order | Description | Filtering type | Rationale | Output type | Filtering criteria |
|---|---|---|---|---|---|
| 1 | Artifact filtering for readthroughs (readthrough fusions in mittelman database are not filtered) | QC | To filter artifacts | All | General |
| 2 | Artifact filtering for fusions found in normal datasets and gene homologs (red flag database from FusionAnnotator) | QC | To filter artifacts | All | General |
| 3 | JunctionReadCount == 0 | QC | To filter out false calls | All | General |
| 4 | SpanningFragCount-JunctionReadCount >= 100 | QC | To filter false calls from low mapping | All | General |
| 5 | Both gene partners with FPKM < 1 | QC | To filter out not expressed fused genes | All | General |
| 6 | Fused genes with either gene in TSGs,Cosmic,Oncogenic,TCGA fusion list | Gene-list | To capture cancer-specific fusions | Putative-driver | Project-specific |
| 7 | Local Rearrangement | QC | To remove local rearrangement within neighbouring genes | Filtered-fusion | Project-specific |
| 8 | Fusion is called by both callers | QC | To filter out calls from only 1 caller | Filtered-fusion | Project-specific |
| 9 | Fusion is called in atleast 2 samples per histology | Recurrence | To gather recurrent fusion calls | Filtered-fusion | Project-specific |
| 10 | Fusion in Filtered-fusions found in more than 1 histology | QC | To remove fusions from Filered-fusion list that are found in more than1 histology | Filtered-fusion | Project-specific |
| 11 | Fused genes in Filtered-fusion fusions found to be multi-fused more than 5 times in a sample | QC | To remove fusions from Filtered-fusion list that are found to be multi-fused | Filtered-fusion | Project-specific |
| 12 | Add recurrent fusions that pass QC from steps 7–11 | Recurrence | To add non-oncogenic fusions to putative-driver fusion list | Putative-driver + recurrent non-oncogenic fusion | Project-specific |
| Description | Annotation type | Rationale | Output type | Annotation source |
|---|---|---|---|---|
| genelistreference.txt | Gene-list | To annotate filtered and unfiltered fusion calls with gene list of interest saved in reference folder | All | Oncogene, tsgs, kinase, cosmic census, curated tf and predicted tf |
| fusionreference.txt | Gene-list | To annotate filtered and unfiltered fusion calls with fusion list of interest saved in reference folder | All | TCGA |
| GTEx zscored comparison | Expression | To annotate filtered and unfiltered fusion calls zscore and comparison with GTEx | All | GTEx |
| Cohort comparison | Expression | To annotate filtered and unfiltered fusion calls zscore and comparison with cohort level expression | All | Cohort |
Fusion filtering criteria were developed to gather high quality recurrent fusion calls while retaining fusions containing oncogenes and/or tumor suppressor genes. Filtering is divided into 3 types (1) QC: filters known causes of false positives. (2) Gene-list: retains additional fusions in genes and fusions of interest list. (3) Recurrence: filters out non-recurrent fusions in genes not annotated as putative oncogenic. Annotation lists are also described
This function also allows users to flexibly filter out fusions predicted to be artifactual while retaining high-quality fusion calls using junction read support of ≥ 1 (default) and spanning fragment support of < 100 (default) reads compared to the junction read count, as disproportionate spanning fragment support indicates false positive calls [15]. Finally, if both genes of the fusion are deemed not expressed < 1 FPKM or TPM (default), the fusion transcript calls can be removed using function expression_filter_fusion.
Step 3: fusion annotation
The annotate_fusion_calls function annotates standardized fusion calls and performs customizable fusion annotation based on user gene lists as input. If checkReciprocal == TRUE reciprocal status of fusions per sample_id is also provided. As a default setting, we provide lists of, and annotate gene partners as, oncogenes, tumor suppressor genes, and oncogenic fusions (Additional file 2: Table S1).
The optional zscored_annotation function provides z-scored expression values from a user-supplied matrix such as GTEx or within cohort to compare samples with and without the fusion to look for over or under expression of fused genes compared to normal using a zscoreFilter. A cutoff of 2 (default) is set to annotate any score > 2 standard deviations away from the median as differentially-expressed. Researchers can then use this information to decide whether to perform additional downstream filtering.
Single sample run
For single samples, we developed the annoFuse_single_sample function which performs fusion standardization of Arriba and STAR-Fusion calls, fusion filtering, and fusion annotation with user-provided gene and fusion reference lists.
Project-specific filtering
Each study often requires additional downstream analyses to be performed once high-quality annotated fusion calls are obtained. We developed functions to enable analyses at a cohort (or project-level) and/or group-level (eg: histologies) designed to remove cohort-specific artifactual calls while retaining high-confidence fusion calls. The function called_by_n_callers annotates the number of algorithms that detected each fusion. We retained fusions with genes not annotated with the gene lists above (eg: oncogene, etc.) that were detected by both algorithms as inframe or frameshift but not annotated as LOCAL_INVERSION or LOCAL_REARRANGEMENT by FusionAnnotator, as these could represent novel fusions. Additionally, samplecount_fusion_call identifies fusions recurrently called in (default ≥ 2) samples within each group. At the group-level, we add groupcount_fusion_calls (default ≥ 1) to remove fusions that are present in more than one type of cancer. At the sample level, fusion_multifused detects fusions in which one gene partner is detected with multiple partners (default ≥ 5), and we remove these as potential false positives. This enables annoFuse to scavenge back potential oncogenic fusions which may have otherwise been filtered. Separately, the function fusion_driver retains only fusions in which a gene partner was annotated as a tumor suppressor gene, oncogene, kinase, transcription factor, and/or the fusion was previously found in TCGA. Domain retention status for Gene1A (5′ gene) and Gene1B (3′ gene) for the given pfamIDs is also annotated and by default, we assess kinase domain retention status fusion-directed therapy often targets kinases. To further reduce the false positives and fusions containing pseudogenes from the cohort, we next filtered fusions using a cutoff of present in > 4 broad histologies after reviewing the fusion distributions within the OpenPBTA Project (Additional file 1: Figure S1). Finally, potential driver fusions and scavenged back recurrent fusion sets are merged into a final set of putative oncogenic fusions.
Fusion domain annotation
The get_Pfam_domain function in annoFuse provides domain annotation for each fused gene in standardized fusion calls. We used the UCSC pfamID Description database and domain location database (Additional file 2: Table S1), along with bioMart [36, 37] gene coordinates to get genomic locations of each domain in a gene. By identifying the breakpoint within the gene coordinate, we annotate each domain as described in (Additional file 3: Table S2). This annotation provides domain retention information which enables prioritization to generate new hypotheses, validate fusion transcript functional impact, and or identify targeted therapeutic options.
Visualization
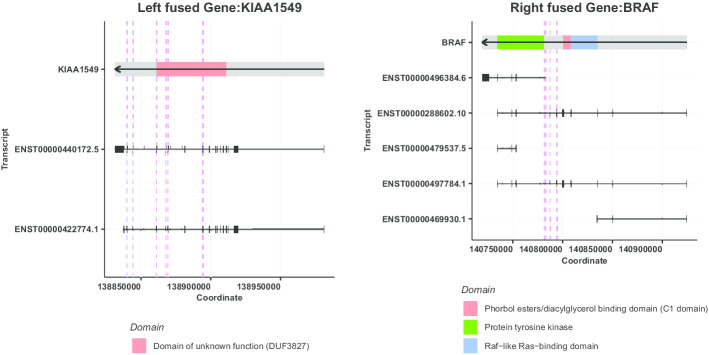
Quick visualization of filtered and annotated fusion calls can provide information useful for review and downstream analysis. We provide the function plot_summary, which provides distribution of intra-chromosomal and inter-chromosomal fusions, number of in-frame and frameshift calls per algorithm, and distribution of gene biotypes, kinase group, and oncogenic annotation. If project-specific filtering is utilized, barplots displaying recurrent fusion and recurrently-fused genes can be generated using plot_recurrent_fusions and plot_recurrent_genes, respectively. Finally, plot_breakpoints can be used to generate all transcripts and breakpoints per gene to visualize the exon and domain retention resulting from the fusion (Fig. 4).
Fig. 4.
Breakpoint distribution for KIAA1549-BRAF fusion. Displayed are each fusion gene, all known transcripts, and their genomic coordinates. Red dotted lines in each gene panel are the locations of breakpoints detected for KIAA1549 (3′) and BRAF (5′) compiled from both Arriba and STAR-Fusion. Strand information is depicted with an arrow in the gene and domain as colored boxes. Black boxes represent exons for each transcript
Interactive fusion exploration using shinyFuse
Depending on the size of the dataset, the prioritized fusions from annoFuse may still contain a considerable amount of information in need of further processing to efficiently extract biological insights. To facilitate this, we developed a web-based application, shinyFuse, in the R/Shiny framework, to assist users in performing drill-down operations while interacting with their fusion results. This feature is included in both the annoFuse package as well as a standalone server (http://shiny.imbei.uni-mainz.de:3838/shinyFuse/) to explore the results table, PutativeDriverAnnoFuse.tsv. Within the web interface, users can easily upload fusion calls from annoFuse or format a file from any fusion algorithm that generates minimal breakpoint location information. Prior to upload, users can choose to append additional columns containing sample descriptors and group information to take advantage of the interactive recurrence analysis feature. There are two major features of shinyFuse: FusionExplorer and FusionSummary.
FusionExplorer
FusionExplorer allows users to interactively search, filter, visualize, and export the output of annoFuse through analysis of PutativeDriverAnnoFuse.tsv. Users can select specific fusion calls according to flexible combinations of filters (e.g. fusion type, caller count, spanning/junction fragments, spanning delta, confidence, and caller). Additionally, selecting a single fusion event (row) in the FusionExplorer tab generates breakpoint, exon, and protein domain plots tailored to transcripts for a specific fusion, sample, or for all samples. Full or filtered data, as well as plots, can be quickly downloaded from the application.
FusionSummary
If a user has more than one cohort of samples and opts to utilize the project-specific filtering (eg: multiple samples from different types of cancer) or has multiple samples with factors by which they would like to group the data (eg: molecular subtype), these data can be explored using FusionSummary. The entire data table can be used for plotting, or it can first be filtered. The user can select a grouping column (factor), a counting column (usually patient-level), and the number of recurrent fusions to display in the plots. Recurrent fusions and recurrently-fused genes will be plotted and figures can be easily exported.
Reproducible fusion analysis with reportFuse
Making results accessible and easier to interpret can play an essential role in reducing time and iterations required to extract actionable knowledge of large datasets, empowering a wide spectrum of collaboration partners. We acknowledge the importance of computational reproducibility [38, 39] when generating analyses, and thus have created reportFuse with functionality to compile a full HTML report (using R Markdown). reportFuse is implemented using a template analysis containing multiple summary functions side by side with the code used to generate them. The report can be a valuable means for persistent storage and sharing of results with colleagues.
Results and discussion
Technical validation of annoFuse
To assess our filtering strategy, we analyzed a subset of samples from TCGA and compared fusions retained with annoFuse filtering and prioritization to those deemed the final call set in a previously published analysis by The Fusion Analysis Working Group [13]. A group of 160 samples were randomly selected BLCA (N = 10), BRCA (N = 11), CESC (N = 5), COAD (N = 11), ESCA (N = 5), GBM (N = 7), HNSC (N = 10), KIRP (N = 9), LGG (N = 9), LIHC (N = 9), LUAD (N = 5), LUSC (N = 11), OV (N = 9), PAAD (N = 8), PCPG (N = 2), PRAD (N = 14), SARC (N = 6), SKCM (N = 9), TGCT (N = 6), THCA (N = 4). We first ran STAR-Fusion, Arriba, and RSEM to generate fusion calls and gene expression values as described in OpenPBTA [16]. Next, we standardized STAR-Fusion and Arriba fusion calls using fusion_standardization, then performed artifact and QC filtering using fusion_filtering_QC. Using a default spanningDelta (spanningFragCount–JunctionReadCount) of 10, annoFuse retained only 70% of the fusions in the final call set (Table 3). Therefore, we visualized the distribution of spanningDelta (spanningFragCount–JunctionReadCount) across fusions called from the TCGA and PBTA cohorts to assess a cutoff for spanningDelta (Additional file 1: Figure S2). We found that sensitivity reaches 96% at cutoff of 100 (Additional file 1: Figure S3 and Table 3) for TCGA 50 to 76 bp read length RNAseq data. Therefore, we have implemented a default spanningDelta of 100 and made this a customizable input parameter.
Table 3.
Sensitivity of TCGA fusion calls
| spanningDelta cutoff | annoFuse Sensitivity |
|---|---|
| 10 | 0.709784411 |
| 20 | 0.807628524 |
| 30 | 0.870646766 |
| 40 | 0.893864013 |
| 50 | 0.922056385 |
| 100 | 0.963515755 |
| 150 | 0.973466003 |
| 200 | 0.983416252 |
Fusion standardization and fusion artifact filtering was conducted on a subset of TCGA samples and compared to published filtered fusion calls from The Fusion Analysis Working Group. SpannigFragCountFilter cutoffs of 10, 20, 30, 40, 50, 100, 150, and 200 were assessed to determine sensitivity of annoFuse prioritized fusion calls
Few gene fusion “truth” sets exist and those that do consist of simulated data or synthetic fusions spiked into breast cancer cell lines or total RNA [31, 32, 40]. We therefore utilized a recent study in which high-confidence fusions were reported in 244 patient-derived xenograft models from the Pediatric Preclinical Testing Consortium (PPTC) [41]. A set of 27 fusions were molecularly validated from acute lymphoblastic leukemia (ALL) models in the PPTC dataset and we deemed this our “truth” set. We first ran Arriba on the PPTC dataset and determined that 23 of the 27 truth fusions were detected using only STAR-Fusion and/or Arriba. Next, we used annoFuse to filter and prioritize putative oncogenic fusions. Table 4 shows the performance of annoFuse, which retained all 23 true positive ALL fusions (100%). Interestingly, only 114 of 166 previously defined as high-confidence (putative oncogenic fusions) in [41] fusions were detected using STAR-Fusion and Arriba (23/27 within the “truth” set), implying gold standard algorithms alone still fail to capture the full landscape of gene fusions, reflecting that additional algorithms should be integrated into our workflow. Of the 114 fusions we detected, 110 (96%) were retained as putative oncogenic fusions using annoFuse. The four fusions annoFuse did not retain were removed with default the “read-through” filter, which can be turned off as an option.
Table 4.
Validation of annoFuse prioritization using PPTC PDX fusion calls
| Histology | PPTC STAR-Fusion/FusionCatcher/SOAPFuse/ deFuse (n detected) | PPTC STAR-Fusion/Arriba (n detected) | annoFuse STAR-Fusion/Arriba (n retained) | % Retained with annoFuse |
|---|---|---|---|---|
| ALL | 117 | 75 | 72 | 96 |
| CNS Embryonal | 4 | 3 | 3 | 100 |
| Ependymoma | 2 | 0 | 0 | NA |
| Ewing Sarcoma | 11 | 10 | 10 | 100 |
| Glioblastoma | 1 | 1 | 0 | 0 |
| Osteosarcoma | 18 | 15 | 15 | 100 |
| Other Brain | 1 | 1 | 1 | 100 |
| Other Sarcoma | 4 | 3 | 3 | 100 |
| Rhabdomyosarcoma | 7 | 6 | 6 | 100 |
| Wilms | 1 | 0 | 0 | NA |
| Total | 166 | 114 | 110 | 96 |
| ALL truth set | 27 | 23 | 23 | 100 |
Retention of high-confidence, putative oncogenic calls averaged 96% across the entire PPTC PDX dataset and was 100% for the ALL truth set (ALL = acute lymphoblastic leukemia). Column 1 = PPTC histology, Column 2 = fusion calls from STAR-Fusion, FusionCatcher, deFuse, and SOAPFuse which were filtered and reported as high-confidence in the PPTC dataset, Column 3 = PPTC reported fusions detected from STAR-Fusion and Arriba, Column 4 = Fusions retained following annoFuse filtering, Column 5 = Percent of fusions retained after applying annoFuse
Case study with annoFuse, shinyFuse, and reportFuse using OpenPBTA
As proof of concept, we utilized RNA expression generated by STAR-RSEM [42] and fusion calls generated by Arriba v1.1.0 [15] and/or STAR-Fusion 1.5.0 [14] which were released as part of the Pediatric Brain Tumor Atlas [43]. Briefly, RNA from fresh-frozen tissue was extracted and libraries were prepped and sequenced at 2 × 100 or 2 × 150 bp to an average of 200 M+ total reads, and at least 60% of reads were required to map to the human genome for fusion analysis to proceed. Additional details can be accessed in the OpenPBTA manuscript [16]. The algorithms were run as described in RNA Expression and Fusion Calls. The RNA expression and fusion workflows are publicly available within the Gabriella Miller KidsFirst GitHub repository [44].
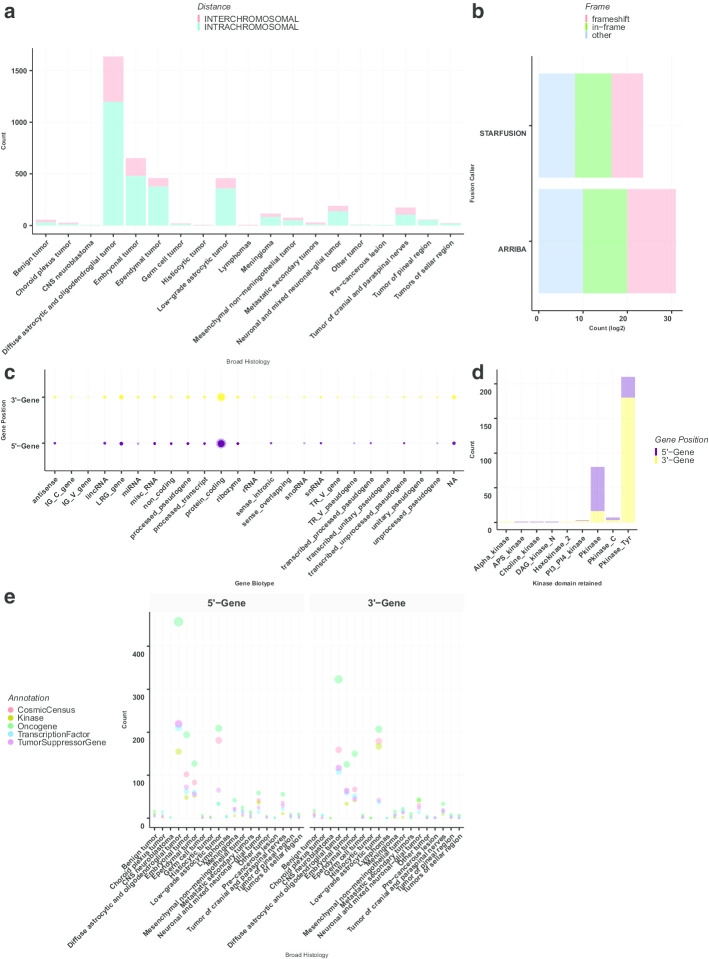
Following fusion standardization, annotation, and filtration, we applied project-specific filtering to the OpenPBTA RNA-Seq cohort (n = 1028 biospecimens from n = 943 patients). Figure 2 is a sample summary PDF designed to give the user an overall glance of the fusion annotations and fusion characteristics within the cohort. From the OpenPBTA cohort, it is clear that there were predominantly more intra-chromosomal fusions called than inter-chromosomal fusions, even after filtering for read-through events (Fig. 2a). While a low-grade astrocytic tumors are the major pediatric brain tumor subtype known to be enriched for gene fusions, it was surprising to observe a large number of fusions in diffuse astrocytic and oligodendroglial tumors and the project-specific utility of annoFuse allows researchers to further prioritize fusions. Histologies within the OpenPBTA project were classified according to broad WHO 2016 subtypes [45].
Fig. 2.
Fusion annotations generated by annoFuse a Distribution of intra- and inter-chromosomal fusions across histologies. b Transcript frame distribution of fusions detected by Arriba and STAR-Fusion algorithms. c Bubble plot of gene partner distribution with respect to ENSEMBL biotype annotation (Size of circle proportional to number of genes). d Barplots representing the distribution of kinase groups represented in the PBTA cohort annotated by gene partner. (Alpha_kinase = Alpha-kinase family, Hexokinase_2 = Hexokinase, PI3_PI4_kinase = Phosphatidylinositol 3- and 4-kinase, Pkinase = Protein kinase domain, Pkinase_C = Protein kinase C terminal domain, Pkinase_Tyr = Protein tyrosine kinase). e Bubble plot representing the distribution of fused genes as oncogenes, tumor suppressor genes, kinases, COSMIC, predicted and curated transcription factors (Size of circle proportional to number of genes). Genes belonging to more than one category are represented in each. In all panels except for B, fusion calls were merged from both STAR-Fusion and Arriba
The number of in-frame and frameshift fusions per algorithm were roughly equivalent within each STAR-Fusion and Arriba fusion calls (Fig. 2b). Figure 2c depicts the density of genes categorized by gene biotype (biological type), and as expected, the filtered and annotated calls were enriched for biologically-functional fusions; the majority of gene partners are classified as protein-coding. The majority of gene partners were annotated as tyrosine kinase (TK) or tyrosine kinase-like (TKL) (Fig. 2d). In Fig. 2e, the user can explore the biological and oncogenic relevance of the fusions across histologies. Of the fusions harboring kinase domains, we found that the majority of 3′ partners retained kinase domains, supporting these fusions as functionally relevant (Additional file 1: Figure S4).
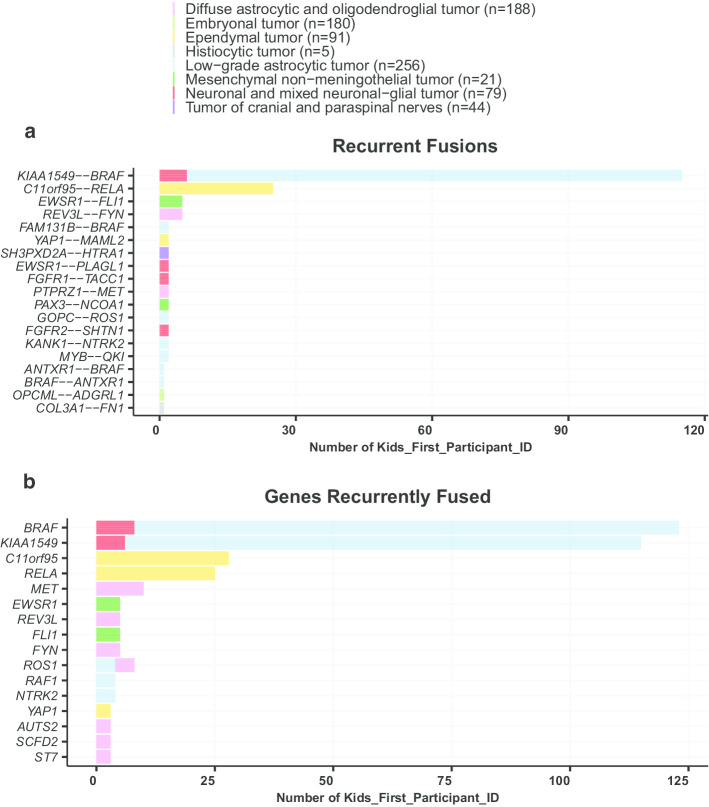
Following project-specific filtering, we observed KIAA1549-BRAF fusions as the most recurrent in-frame fusion in our cohort (n = 109/898), since KIAA1549-BRAF expressing low-grade astrocytic tumors comprise the largest representative histology in the OpenPBTA cohort (n = 236/898). C11orf95-RELA was predominant in ependymal tumors (n = 25/80), as expected in supratentorial ependymomas [46]. Other expected recurrent oncogenic fusions obtained through annoFuse were EWSR1-FLI1 in CNS Ewing sarcomas [47], and KANK1-NTRK2, MYB-QKI, and FAM131B-BRAF in low-grade astrocytic tumors [4, 48] (Fig. 3a). In addition to recurrent fusions, we also detect recurrently-fused genes to account for partner promiscuity. This enables us to see a broader picture of gene fusions, specifically within diffuse astrocytic and oligodendroglial tumors, in which we see fusions prevalent in ST7, MET, FYN, REV3L, AUTS2, and ROS1, and meningiomas, in which NF2 fusions are common (Fig. 3b). Next, we added functionality to visualize domain information (Fig. 4) to quickly scan for domains retained and lost across the dataset. The putative oncogenic fusion table from this analysis is both available in the vignette and on the standalone Shiny server as a demo dataset for shinyFuse, enabling both free exploration of the data as well as reproducible generation of Figs. 3 and 4. Finally, we use reportFuse to include a reproducible HTML report which generates all output tables and figures for this manuscript.
Fig. 3.
Recurrent fusion plots generated by annoFuse. Bar plots as representative of histology showing recurrent fusion calls by number of patients (a) and recurrently-fused genes by number of patients (b) after filtering and annotation
The few openly-available fusion annotation and/or prioritization tools summarized in Table 1 perform only a subset of the functionalities provided in annoFuse and shinyFuse. In addition, the majority are no longer maintained, nor are directly compatible with Arriba, the tool that won the SMC-RNA DREAM challenge for speed and accuracy in 2019. For example, Oncofuse [49], Pegasus [50], chimera [51], and co-Fuse [52] have not been updated in two or more years, and as a result, these tools lack compatibility with newer and improved fusion algorithms. The chimeraviz R package [53] is well-maintained and compatible with nine fusion algorithms, but only performs visualizations of fusions, thus prioritization using database annotation, readthrough and expression is not possible using this tool. Four tools are algorithm agnostic, yet perform only specific aspects of annotation and prioritization. Oncofuse [49] and Pegasus [50] analyze oncogenicity of fusions, and chimera [51] and FusionHub [54] require Oncofuse [49] to be run as a first step. FusionHub [54] is a web-based tool which enables annotation of fusions with 28 databases, however, is not programmatically scalable. FusionAnnotator [55] determines the presence of fusions in 15 cancer-associated databases, oncogene lists, and seven databases for fusions not relevant in cancer, but does not perform prioritization. ConFuse performs filtering and prioritization, but is not algorithm-agnostic; it only works on fusion calls from deFuse [56]. AGFusion [57] annotates protein domains, and Fusion Pathway [58] utilizes fusion and protein domain annotations in gene set enrichment analysis (GSEA) to infer oncogenic pathway association. When used exclusively, none of these tools flexibly perform fusion annotation and prioritization. Furthermore, none enable the interactive exploration and visualization capabilities that we provide with shinyFuse. Instead, we leveraged the algorithm agnostic capabilities of FusionAnnotator to pre-annotate fusion input from STAR-Fusion and Arriba to create annoFuse, an all-in-one tool that performs fusion annotation, oncogenic prioritization, recurrence analysis, visualization, and exploratory fusion analysis.
By integrating FusionAnnotator with functionality of the current gold standard algorithms STAR-Fusion and Arriba, we were able to improve the aforementioned tools’ capabilities by meeting the current demands of the research community. We provide the user with flexible filtering parameters and envision annoFuse will be used to quickly filter sequencing artifacts and false positives, as well as further annotate fusions for additional biological functionality (eg: kinases, transcription factors, oncogenes, tumor suppressor genes) to prioritize fusion calls in a cancer cohort. Additionally, users can opt to simply annotate and filter artifacts or use annoFuse to functionally prioritize fusions as putative oncogenic drivers. During the prioritization steps, we filter based on genes with cancer relevance (see biological functionality list above) and perform analysis of fusion and fused-gene recurrence to create a stringently filtered, prioritized list of fusions likely to have oncogenic potential.
Limitations and future directions
It is worth noting that annoFuse cannot correct fusion calls derived from low-quality RNA-Seq data, thus onus is on the user to design the extraction, library preparation, and sequencing portions of the experiment to enable production of high-quality data. We prioritized Arriba and STAR-Fusion calls as input to annoFuse due to their performance in the SMC-RNA DREAM challenge and we recommend utilizing both algorithms prior to analysis with annoFuse. In addition, users can provide their own informed cutoffs for read support and annotation filters to enable appropriate prioritization of oncogenic fusions. We are actively adding compatibility with additional fusion algorithms currently used by the community, such as deFuse, FusionCatcher, SOAPfuse, and Jaffa to further increase the applicability of annoFuse. As an additional feature, we plan to add expression-based comparison of genes between fused samples, normal, and within a histology or cohort. Future features could link domain retention to drug databases to predict fusion-directed targeting strategies.
Conclusions
Gene fusions provide a unique mutational context in cancer in which two functionally-distinct genes could be combined to function as a new biological entity. Despite showing great promise as diagnostic, prognostic, and therapeutic targets, translation in the oncology clinic is not yet accelerated for gene fusions. This has been partly due to limited translation of the large number of computationally-derived fusion results into biologically meaningful information. In our efforts to address this, we introduce annoFuse, an R Package to annotate and prioritize putative oncogenic RNA fusions and shinyFuse, an algorithm-agnostic web application for interactive fusion exploration and plotting. We include a cancer-specific workflow to find recurrent, oncogenic fusions from large cohorts containing multiple cancer histologies. The filtering and annotation steps within annoFuse enable users to integrate calls from multiple algorithms to improve high-confidence, consensus fusion calling. The lack of concordance among algorithms as well as variable accuracy with fusion truth sets [2, 32] adds analytical complexity for researchers and clinicians aiming to prioritize research or therapies based on fusion findings. Through annoFuse, we add algorithm flexibility and integration to identify recurrent fusions and/or recurrently-fused genes as novel oncogenic drivers. Within the package, shinyFuse and reportFuse deliver interactive and reproducible analysis options to efficiently extract knowledge from the outputs of the annoFuse workflow. We expect annoFuse to be broadly applicable to cancer datasets and empower researchers and clinicians to better inform preclinical studies targeting novel, putative oncogenic fusions and ultimately, aid in the rational design of therapeutic modulators of gene fusions in cancer.
Availability and requirements
Project name: annoFuse: an R Package to annotate, prioritize, and interactively explore putative oncogenic RNA fusions
Project home page: https://github.com/d3b-center/annoFuse
Project web application (shinyFuse): http://shiny.imbei.uni-mainz.de:3838/shinyFuse/
Operating system(s: Platform independent
Programming language: R (> = 4.0.0)
License: MIT
Any restrictions to use by non-academics: None
Supplementary Information
Additional file 1: Figure S1. Fusions found in more than 1 histology. Barplots represent the number of histologies in which each fusion was observed. Dotted line represents the cut off (> 4 counts) used to remove potential false positives and fusions containing pseudogenes. Figure S2. Distribution of spanningDelta for annoFuse prioritized fusions from TCGA and PBTA cohorts. The spanningFragCountFilter and mean are plotted for each filtering cutoff of 10, 20, 30, 40, 50, 100, 150, and 200 spanningDelta for TCGA (N = 160 samples) (A) and PBTA (N = 1028 samples) (B) fusions. Figure S3. Sensitivity of TCGA fusions retained by annoFuse. At a spanningDelta of 100, annoFuse achieved a sensitivity of 96.35% for fusions in the TCGA final call set. The x-axis represents the cutoff for spanningFragCountFilter used and y-axis represents the sensitivity of the fusions retained after fusion_filtering_QC. Figure S4. Distribution of kinase genes fused in 5' and 3' genes per histologies. For each broad histology, pie charts represent the percentage of fusions in which Gene1A (5’) or Gene1B (3’) retain their kinase domains.
Additional file 2: Table S1. Sources for annotation of fusions and gene partners. Listed are annotations utilized by annoFuse for prioritization of oncogenic gene fusions, along with reference files and their sources.
Additional file 3: Table S2. Domain retention conditions for annotation. Listed are conditions for domain retention status for Gene1A (5` gene) and Gene1B (3` gene). Domain retention status for Gene1A depends on the direction of the left breakpoint with respect to the domain and the gene body and domain retention status for Gene1B depends on the direction of the right breakpoint with respect to domain and gene body.
Acknowledgements
We would like to thank Brian J. Haas (Broad Institute), Alexander Dobin (Cold Spring Harbor Laboratory) and Sebastian Uhrig (German Cancer Research Center) for helpful discussions throughout the development of annoFuse. We acknowledge the Children’s Brain Tumor Network and the Pacific Pediatric Neuro-Oncology Consortium for generating RNA-Seq data from pediatric brain tumor tissue and the Gabriella Miller Kids First Data Resource Center for processing and hosting the data. Finally, we graciously thank the patients and families for donating tumor tissue, without which, this research would not be possible.
Abbreviations
- ALL
Acute lymphoblastic leukemia
- BAM
Binary alignment map
- COSMIC
Catalogue of somatic mutations in cancer
- CNS
Central nervous system
- DGD_PARALOGS
Duplicated Genes Database annotated paralogs
- GSEA
Gene set enrichment analysis
- HGNC_GENEFAM
HGNC annotated gene family
- FPKM
Fragments per kilobase million
- OpenPBTA
Open Pediatric Brain Tumor Atlas
- PI3_PI4_kinase
Phosphatidylinositol 3- and 4-kinase
- Pkinase
Protein kinase domain
- Pkinase_C
Protein kinase C terminal domain
- Pkinase_Tyr
Protein tyrosine kinase
- PPTC
Pediatric Preclinical Testing Consortium
- RNA
Ribonucleic acid
- SAM
Sequence alignment map
- SMC-RNA
Somatic Mutation Calling RNA DREAM Challenge (SMC-RNA)
- TCGA
The Cancer Genome Atlas
- TSV
Tab separated value
- TPM
Transcripts per kilobase per million
- WHO
World Health Organization
Authors’ contributions
Conceptualization: KSG, JMM, KSR, PR, JLR, JNT, FM. Methodology: KSG, KSR, PR, JLR, JNT, MAB, BZ, YZ, NAC. Software: KSG, JLR, JNT, FM. Validation: KSG, JLR, JNT, FM. Formal Analysis: KSG, JNT. Investigation: KSG, JLR, FM. Resources: JLR, PBS, ACR, KS. Data Curation: KSG, YZ, JLR, MAB, BZ. Writing—Original Draft: KSG, JLR. Writing—Review and Editing: KSG, JLR, PJ, ASN, JNT, FM. Visualization: KSG, JLR, FM. Supervision: JLR, JNT. Funding Acquisition: JLR, ACR, PBS, JMM, JNT, FM. All authors have read and approved the final manuscript.
Funding
This research was funded by Children’s Hospital of Philadelphia Division of Neurosurgery (PBS and ACR), NIH Grant U2C HL138346-03 (ACR), NIH Grant U24 CA220457-03 (ACR), NIH Grant R35 CA220500 (JMM), Alex’s Lemonade Stand (ALSF) Young Investigator Award (JLR), ALSF Catalyst Award (JLR), ALSF Childhood Cancer Data Lab (JNT), and a German Federal Ministry of Education and Research Grant BMBF 01EO1003 (FM). The funding bodies did not play a role in the design of the study, collection/analysis/interpretation of data, nor manuscript writing.
Availability of data and materials
All brain tumor raw data are available by download from the Gabriella Miller Kids First Data Resource Center with a data access agreement through the Children’s Brain Tumor Network and processed data (release-v16-20200320) available by download through the OpenPBTA project’s GitHub repository: https://github.com/AlexsLemonade/OpenPBTA-analysis. PPTC (Accession Number phs001437.v1.p1) and TCGA (Accession Number phs000178.v1.p1) RNA-Sequencing data are available by download from dbGAP with a data access agreement.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Krutika S. Gaonkar and Federico Marini have contributed equally
Supplementary Information
The online version contains supplementary material available at 10.1186/s12859-020-03922-7.
References
- 1.Wu H, Li X, Li H. Gene fusions and chimeric RNAs, and their implications in cancer. Genes Dis. 2019;6(4):385–390. doi: 10.1016/j.gendis.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar S, Vo AD, Qin F, Li H. Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Sci Rep. 2016 doi: 10.1038/srep21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y, Yuan C, Chen L, Lei M, Zellmer L, Huang H, et al. Transcriptional-Readthrough RNAs Reflect the Phenomenon of “A Gene Contains Gene(s)” or “Gene(s) within a Gene” in the Human Genome, and Thus Are Not Chimeric RNAs. Genes. 2018;9(1):40. doi: 10.3390/genes9010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandopadhayay P, Ramkissoon LA, Jain P, Bergthold G, Wala J, Zeid R, et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat Genet. 2016;48(3):273–282. doi: 10.1038/ng.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain P, Fierst TM, Han HJ, Smith TE, Vakil A, Storm PB, et al. CRAF gene fusions in pediatric low-grade gliomas define a distinct drug response based on dimerization profiles. Oncogene. 2017;36(45):6348–6358. doi: 10.1038/onc.2017.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain P, Surrey LF, Straka J, Luo M, Lin F, Harding B, et al. Novel FGFR2-INA fusion identified in two low-grade mixed neuronal-glial tumors drives oncogenesis via MAPK and PI3K/mTOR pathway activation. Acta Neuropathol. 2018;136(1):167–169. doi: 10.1007/s00401-018-1864-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones DTW, Kocialkowski S, Liu L, Pearson DM, Magnus Backlund L, Ichimura K, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008 doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aurias A, Rimbaut C, Buffe D, Zucker JM, Mazabraud A. Translocation involving chromosome 22 in Ewing’s sarcoma. A cytogenetic study of four fresh tumors. Cancer Genet Cytogenet. 1984;12(1):21–25. doi: 10.1016/0165-4608(84)90003-7. [DOI] [PubMed] [Google Scholar]
- 9.Lee M, Lee K, Yu N, Jang I, Choi I, Kim P, et al. ChimerDB 3.0: an enhanced database for fusion genes from cancer transcriptome and literature data mining. Nucleic Acids Res. 2017;45(1):D784–D789. doi: 10.1093/nar/gkw1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu X, Wang Q, Tang M, Barthel F, Amin S, Yoshihara K, et al. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res. 2018;46(D1):D1144–D1149. doi: 10.1093/nar/gkx1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173(2):321–37.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Q, Liang W-W, Foltz SM, Mutharasu G, Jayasinghe RG, Cao S, et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018;23(1):227–38.e3. doi: 10.1016/j.celrep.2018.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas BJ, Dobin A, Stransky N, Li B, Yang X, Tickle T, et al. STAR-fusion: fast and accurate fusion transcript detection from RNA-Seq. bioRxiv. 2017. p. 120295. 10.1101/120295v1.
- 15.arriba [Internet]. Github. https://github.com/suhrig/arriba. Accessed 30 Sept 2019.
- 16.Shapiro JA, Savonen CL, Bethell CJ, Gaonkar KS, Zhu Y, Brown MA, et al. An Open Pediatric Brain Tumor Atlas. Manubot. 2020. https://alexslemonade.github.io/OpenPBTA-manuscript/v/4a49eff86f96f82f5966a120b9be21c3570389f2/. Accessed 15 Sept 2020.
- 17.Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41(Database issue):D970–D976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao M, Kim P, Mitra R, Zhao J, Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016;44:D1023–D1031. doi: 10.1093/nar/gkv1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 20.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The human transcription factors. Cell. 2018;175(2):598–599. doi: 10.1016/j.cell.2018.09.045. [DOI] [PubMed] [Google Scholar]
- 21.Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer. 2018;18(11):696–705. doi: 10.1038/s41568-018-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramkissoon LA, Horowitz PM, Craig JM, Ramkissoon SH, Rich BE, Schumacher SE, et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci. 2013;110:8188–8193. doi: 10.1073/pnas.1300252110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Northcott PA, Shih DJH, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488(7409):49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sturm D, Orr B, Toprak U, Hovestadt V, Jones D, Capper D, et al. PNR-21FOUR NEW Brain tumor entities emerge from molecular classification of CNS PNETs. Neuro-Oncology. 2016;18:iii11.1–iii11. doi: 10.1093/neuonc/now067.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleinman CL, Gerges N, Papillon-Cavanagh S, Sin-Chan P, Pramatarova A, Quang D-AK, et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nat Genet. 2014;46(1):39–44. doi: 10.1038/ng.2849. [DOI] [PubMed] [Google Scholar]
- 26.Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015;47(12):1411–1414. doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 27.Cobrinik D, Ostrovnaya I, Hassimi M, Tickoo SK, Cheung IY, Cheung N-KV. Recurrent pre-existing and acquired DNA copy number alterations, including focal TERT gains, in neuroblastoma central nervous system metastases. Genes Chromosomes Cancer. 2013;52(12):1150–1166. doi: 10.1002/gcc.22110. [DOI] [PubMed] [Google Scholar]
- 28.Karlsson J, Lilljebjörn H, Holmquist Mengelbier L, Valind A, Rissler M, Øra I, et al. Activation of human telomerase reverse transcriptase through gene fusion in clear cell sarcoma of the kidney. Cancer Lett. 2015;357(2):498–501. doi: 10.1016/j.canlet.2014.11.057. [DOI] [PubMed] [Google Scholar]
- 29.Karsy M, Guan J, Cohen AL, Jensen RL, Colman H. New molecular considerations for glioma: IDH, ATRX, BRAF, TERT, H3 K27M. Curr Neurol Neurosci Rep. 2017;17(2):19. doi: 10.1007/s11910-017-0722-5. [DOI] [PubMed] [Google Scholar]
- 30.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164(5):1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bionetworks S. Synapse|Sage Bionetworks. https://www.synapse.org/. Accessed 12 May 2020.
- 32.Haas BJ, Dobin A, Li B, Stransky N, Pochet N, Regev A. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019;20(1):213. doi: 10.1186/s13059-019-1842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prakash T, Sharma VK, Adati N, Ozawa R, Kumar N, Nishida Y, et al. Expression of conjoined genes: another mechanism for gene regulation in eukaryotes. PLoS ONE. 2010;5(10):e13284. doi: 10.1371/journal.pone.0013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cario G, Zimmermann M, Romey R, Gesk S, Vater I, Harbott J, et al. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non–high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 2010;115(26):5393–5397. doi: 10.1182/blood-2009-11-256131. [DOI] [PubMed] [Google Scholar]
- 35.Panzer-Grümayer R, Köhrer S, Haas OA. The enigmatic role(s) of P2RY8-CRLF2. Oncotarget. 2017;8(57):96466–96467. doi: 10.18632/oncotarget.22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21(16):3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 37.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4(8):1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stodden V, McNutt M, Bailey DH, Deelman E, Gil Y, Hanson B, et al. Enhancing reproducibility for computational methods. Science. 2016;354(6317):1240–1241. doi: 10.1126/science.aah6168. [DOI] [PubMed] [Google Scholar]
- 39.Peng RD. Reproducible research in computational science. Science. 2011;334(6060):1226–1227. doi: 10.1126/science.1213847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tembe WD, Pond SJK, Legendre C, Chuang H-Y, Liang WS, Kim NE, et al. Open-access synthetic spike-in mRNA-seq data for cancer gene fusions. BMC Genomics. 2014;30(15):824. doi: 10.1186/1471-2164-15-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rokita JL, Rathi KS, Cardenas MF, Upton KA, Jayaseelan J, Cross KL, et al. Genomic profiling of childhood tumor patient-derived Xenograft models to enable rational clinical trial design. Cell Rep. 2019;29(6):1675–89.e9. doi: 10.1016/j.celrep.2019.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vivian J, Rao AA, Nothaft FA, Ketchum C, Armstrong J, Novak A, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35(4):314–316. doi: 10.1038/nbt.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ijaz H, Koptyra M, Gaonkar KS, Rokita JL, Baubet VP, Tauhid L, et al. Pediatric high grade glioma resources from the Children’s Brain Tumor Tissue Consortium (Cbttc) Neuro Oncol. 2019 doi: 10.1093/neuonc/noz192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.kf-rnaseq-workflow. Github. https://github.com/kids-first/kf-rnaseq-workflow. Accessed 30 Sept 2019.
- 45.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 46.Fukuoka K, Kanemura Y, Shofuda T, Fukushima S, Yamashita S, Narushima D, et al. Significance of molecular classification of ependymomas: C11orf95-RELA fusion-negative supratentorial ependymomas are a heterogeneous group of tumors. Acta Neuropathol Commun. 2018;6(1):134. doi: 10.1186/s40478-018-0630-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10(7):e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venneti S, Huse JT. The evolving molecular genetics of low-grade glioma. Adv Anat Pathol. 2015;22(2):94–101. doi: 10.1097/PAP.0000000000000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shugay M, Ortiz de Mendíbil I, Vizmanos JL, Novo FJ. Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics. 2013;29(20):2539–2546. doi: 10.1093/bioinformatics/btt445. [DOI] [PubMed] [Google Scholar]
- 50.Abate F, Zairis S, Ficarra E, Acquaviva A, Wiggins CH, Frattini V, et al. Pegasus: a comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst Biol. 2014;4(8):97. doi: 10.1186/s12918-014-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calogero RA, Carrara M, Beccuti M, Cordero F. chimera: A package for secondary analysis of fusion products version 1.28.0 from Bioconductor. 2019. https://rdrr.io/bioc/chimera/. Accessed 12 May 2020. [DOI] [PMC free article] [PubMed]
- 52.co-fuse. Github. https://github.com/sakrapee/co-fuse. Accessed 12 May 2020.
- 53.Lågstad S, Zhao S, Hoff AM, Johannessen B, Lingjærde OC, Skotheim RI. chimeraviz: a tool for visualizing chimeric RNA. Bioinformatics. 2017;33(18):2954–2956. doi: 10.1093/bioinformatics/btx329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panigrahi P, Jere A, Anamika K. FusionHub: a unified web platform for annotation and visualization of gene fusion events in human cancer. PLoS ONE. 2018;13(5):e0196588. doi: 10.1371/journal.pone.0196588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.FusionAnnotator. Github. https://github.com/FusionAnnotator. Accessed 30 Sept 2019.
- 56.Huang Z, Jones DTW, Wu Y, Lichter P, Zapatka M. confFuse: high-confidence fusion gene detection across tumor entities. Front Genet. 2017;8:137. doi: 10.3389/fgene.2017.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy C, Elemento O. AGFusion: annotate and visualize gene fusions. bioRxiv. 2016. p. 080903. 10.1101/080903v1.abstract
- 58.Wu C-C, Beird HC, Zhang J, Andrew Futreal P. FusionPathway: prediction of pathways and therapeutic targets associated with gene fusions in cancer. PLOS Comput Biol. 2018;14:e1006266. doi: 10.1371/journal.pcbi.1006266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Fusions found in more than 1 histology. Barplots represent the number of histologies in which each fusion was observed. Dotted line represents the cut off (> 4 counts) used to remove potential false positives and fusions containing pseudogenes. Figure S2. Distribution of spanningDelta for annoFuse prioritized fusions from TCGA and PBTA cohorts. The spanningFragCountFilter and mean are plotted for each filtering cutoff of 10, 20, 30, 40, 50, 100, 150, and 200 spanningDelta for TCGA (N = 160 samples) (A) and PBTA (N = 1028 samples) (B) fusions. Figure S3. Sensitivity of TCGA fusions retained by annoFuse. At a spanningDelta of 100, annoFuse achieved a sensitivity of 96.35% for fusions in the TCGA final call set. The x-axis represents the cutoff for spanningFragCountFilter used and y-axis represents the sensitivity of the fusions retained after fusion_filtering_QC. Figure S4. Distribution of kinase genes fused in 5' and 3' genes per histologies. For each broad histology, pie charts represent the percentage of fusions in which Gene1A (5’) or Gene1B (3’) retain their kinase domains.
Additional file 2: Table S1. Sources for annotation of fusions and gene partners. Listed are annotations utilized by annoFuse for prioritization of oncogenic gene fusions, along with reference files and their sources.
Additional file 3: Table S2. Domain retention conditions for annotation. Listed are conditions for domain retention status for Gene1A (5` gene) and Gene1B (3` gene). Domain retention status for Gene1A depends on the direction of the left breakpoint with respect to the domain and the gene body and domain retention status for Gene1B depends on the direction of the right breakpoint with respect to domain and gene body.
Data Availability Statement
All brain tumor raw data are available by download from the Gabriella Miller Kids First Data Resource Center with a data access agreement through the Children’s Brain Tumor Network and processed data (release-v16-20200320) available by download through the OpenPBTA project’s GitHub repository: https://github.com/AlexsLemonade/OpenPBTA-analysis. PPTC (Accession Number phs001437.v1.p1) and TCGA (Accession Number phs000178.v1.p1) RNA-Sequencing data are available by download from dbGAP with a data access agreement.