Abstract
Background
The proportion of infections among young children that are antimicrobial-resistant is increasing across the globe. Newborns may be colonized with enteric antimicrobial-resistant pathogens early in life, which is a risk factor for infection-related morbidity and mortality. Breastfeeding is actively promoted worldwide for its beneficial impacts on newborn health and gut health. However, the role of breastfeeding and human milk components in mitigating young children’s carriage of antimicrobial-resistant pathogens and antibiotic resistance genes has not been comprehensively explored.
Main body
Here, we review how the act of breastfeeding, early breastfeeding, and/or human milk components, such as the milk microbiota, secretory IgA, human milk oligosaccharides, antimicrobial peptides, and microRNA -bearing extracellular vesicles, could play a role in preventing the establishment of antimicrobial-resistant pathogens in young children’s developing gut microbiomes. We describe findings from recent human studies that support this concept.
Conclusion
Given the projected rise in global morbidity and mortality that will stem from antimicrobial-resistant infections, identifying behavioral or nutritional interventions that could decrease children’s susceptibility to colonization with antimicrobial-resistant pathogens may be one strategy for protecting their health. We suggest that breastfeeding and human milk supplements deserve greater attention as potential preventive measures in the global effort to combat antimicrobial resistance, particularly in low- and middle-income settings.
Keywords: Breastfeeding, Human milk, Microbiome, Antimicrobial resistance, Child health, Low- and middle-income countries
Background
Antimicrobial-resistant (AMR) bacterial infections are becoming increasingly common across the globe [1]. As many as 162,000 US adults currently die from multidrug-resistant bacterial infections each year, making resistant infections the third leading cause of death [2]. A 2019 UN report described antimicrobial resistance as a global crisis, with potential for knock-on financial shocks due to increasing healthcare costs, poverty, and inequality comparable to those experienced during the 2008–2009 global financial crisis [3]. Morbidity and mortality stemming from antimicrobial resistance are expected to be highest in low- and middle-income countries (LMICs), where bacterial diseases like lower respiratory infections, diarrheal disease, and tuberculosis remain common.
Infants are particularly susceptible to the potential negative consequences of AMR infections [4]. Evidence suggests that young children may be colonized or infected with enteric AMR pathogens early in life [5–8]. Presence and elevated abundance of AMR pathogens in the intestinal tract are risk factors for transmission to non-enteric sites (e.g., blood, surgical sites) [9–12], horizontal transfer to other individuals, and vertical transmission from mothers to their infants. Neonatal infections with highly resistant pathogens like extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae, which are considered an urgent health threat by the US Centers for Disease Control and Prevention [13], have been documented across the globe and have become increasingly prevalent over the past decade [14, 15]. Children under two are also one of the likeliest age groups to receive antibiotics [16, 17]. Frequent antibiotic exposures may increase the relative abundance of antibiotic resistance genes (ARGs) harbored by gut bacteria, and foster a gut environment supportive of their transfer to pathogens [4, 18–20].
Breastfeeding has long been recognized for its beneficial impacts on newborn health, including provision of maternal immune cells [21] and antibodies to protect against infection [22] and decreased risk of exposure to diarrheal pathogens from non-breastmilk food-sources [23]. However, to date, few studies have examined how early breastfeeding practices and breastmilk constituents could specifically mitigate young children’s colonization with AMR bacteria and ARGs. In general, breastfed children experience fewer diarrhea-related hospitalizations [23, 24] and may consume fewer antibiotics [24], which helps to prevent exposure to, colonization with, and selection for AMR pathogens and their ARGs [18]. In addition, early and exclusive breastfeeding is increasingly recognized as an integral step in the development of healthy infant microbiomes. While the developing infant gut microbiome is shaped by transmission from multiple maternal body sites [25], human milk induces intestinal tolerance to the commensal microbiome through the simultaneous provision of nutrition for the infant and the bacteria themselves [26]. Furthermore, human milk contains maternal immune effectors (e.g., antibodies, immune cells, and antimicrobial peptides) that enhance pathogen exclusion and elimination, and food and microbial antigens that promote infant immune development. Healthy microbiomes provide benefits into adulthood [27] and may also be better able to resist colonization with pathogens, including AMR pathogens, a phenomenon known as “colonization resistance” [28]. However, the potential links between specific breastfeeding practices or human milk components and children’s reduced susceptibility to colonization or infection with AMR bacteria have not been comprehensively explored.
Here, we argue that breastfeeding may be an important strategy for combatting the global antimicrobial resistance crisis. We begin by reviewing how early breastfeeding, exclusivity of breastfeeding, and components of human milk could provide protection against the establishment, proliferation, and/or selection of enteric pathogens in general and AMR bacteria and ARGs, in particular. We describe the findings of recent observational studies and intervention trials that have explored such associations. Finally, we comment on how escalating antimicrobial resistance and changing breastfeeding trends in LMICs make these settings particularly important for exploring breastfeeding as a way to mitigate antimicrobial resistance. Given the scale and scope of the impending antimicrobial resistance crisis, identifying scalable, cost-effective behavioral or nutritional interventions that could decrease children’s susceptibility to colonization by enteric AMR pathogens and ARGs will be critical to protecting their health.
Potential benefits of early breastfeeding
The first form of human milk produced by the mammary glands, called “colostrum,” could be especially important in preventing the establishment and proliferation of antibiotic-resistant bacteria in very young children’s intestines. Colostrum contains concentrations of maternal immune cells and secretory IgA (sIgA) that are up to 6× and 12× higher than subsequent human milk, respectively [22, 29]. Maternal leukocytes provide active immunity against gut pathogens by engulfing and killing them, and may provide protection beyond the gut by crossing the intestinal epithelium and entering the bloodstream [21]. IgA antibodies are secreted by lymphocytes in the mammary gland. These lymphocytes have migrated from the mother’s gut, meaning they target antigens and enteric microorganisms to which mothers have previously been exposed. This includes antibodies to pathogens that can be antibiotic-resistant, such as Shigella and Campylobacter [22, 30]. Once in the gut, sIgA promotes the clearance of targeted pathogens by blocking pathogen access to intestinal epithelial receptors, binding to bacterial adhesion sites like pili, trapping them in mucus, opsonizing them for detection by Fc-receptor-bearing immune cells, and promoting their removal [31, 32]. In addition, sIgA can directly neutralize bacterial virulence factors and toxins, including Escherichia coli heat-labile enterotoxin [31, 33]. Higher concentrations of sIgA antibodies in colostrum and human milk are associated with greater protection against pathogen-mediated diarrhea [33, 34], and could clearly provide early protection against children’s colonization or infection with AMR bacteria. Additionally, early breastfeeding increases the likelihood of exclusive breastfeeding [35], which could confer distinct protection against young children’s colonization with AMR bacteria and ARGs, as outlined below.
Potential benefits of exclusive or predominant breastfeeding for first 6 months
The World Health Organization (WHO) recommends exclusive breastfeeding through the first 6 months of life. While this is not practiced by the majority of mothers in high-income countries (~ 25% of infants in the USA and Europe are exclusively breastfed through 6 months) [36, 37], rates of exclusive and predominant breastfeeding are higher and increasing in countries with the lowest income status [38], although substantial variability exists (Fig. 1). Exclusive or predominant breastfeeding for an extended duration could indirectly protect children against colonization with AMR pathogens and ARGs in several ways (Fig. 2).
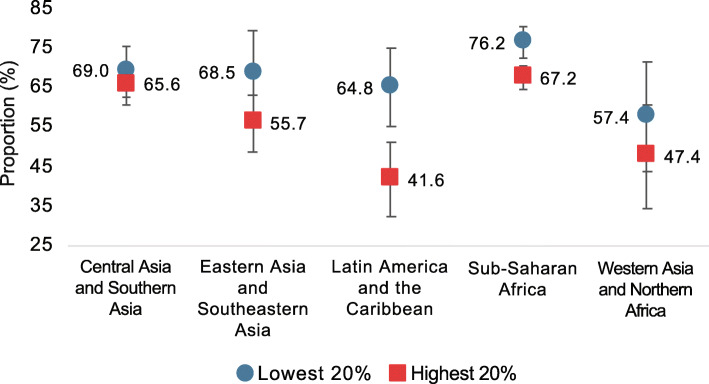
Fig. 1.
Average proportion of children in low- and lower middle-income countries (LMICs) predominately receiving human milk at 0–5 months, by wealth quintile and region, 2010–2017. In LMICs in Latin America, the Caribbean, and Sub-Saharan Africa in particular, babies from the wealthiest families are substantially less likely to be predominately breastfed than babies from the poorest families. Analysis is based on a subset of 66 countries with recent (2010–2017) data for predominant breastfeeding at 0–5 months. Country income classifications provided in source dataset. Error bars show the 90% confidence interval around the mean. Source: United Nations Children’s Fund, Division of Data Research and Policy. Global UNICEF Global Databases: Infant and Young Child Feeding: Predominant breastfeeding (2019)
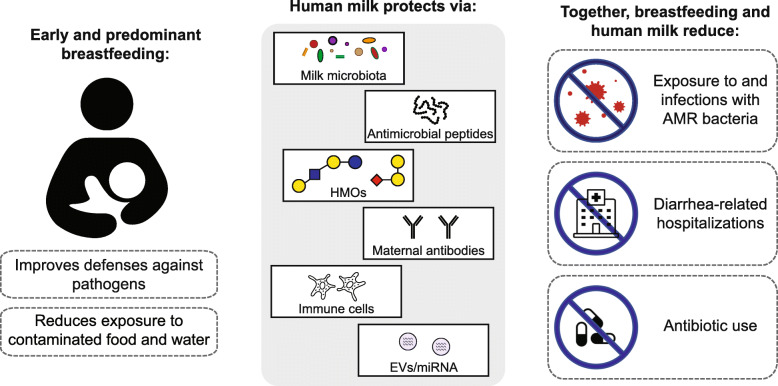
Fig. 2.
Ways breastfeeding and human milk could prevent the establishment, proliferation, and/or selection of antimicrobial-resistant bacteria among young children. Exclusive or predominant breastfeeding for an extended duration improves children’s defenses against pathogens and reduces their food- and waterborne exposures to antimicrobial-resistant bacteria. Human milk contains multiple components that could protect against antimicrobial-resistant pathogens and reduce the lateral transfer of antibiotic resistance genes. The concentrations of some of these components are highest in colostrum, the earliest form of human milk. HMOs, human milk oligosaccharides; EVs, extracellular vesicles; miRNA, microRNA; AMR, antimicrobial resistance
Reduced exposure to antibiotics
Breastfeeding may reduce antibiotic use among children. Recent trials in Finland [39] and France [24] have identified strong inverse relationships between the duration of breastfeeding and frequency of children’s antibiotic use. In France, any breastfeeding at all in the first 6 months was associated with less frequent antibiotic use over the first 2 years of life compared with those who were never breastfed, and longer duration of exclusive or predominant breastfeeding was associated with even lower risks [24]. However, the authors could not exclude that lower antibiotic use may have been due to differences in health-seeking behaviors between breastfeeding versus non-breastfeeding parents. In settings where early-life enteric pathogen exposure is common, breastfeeding may reduce antibiotic use by reducing children’s risk of diarrhea [40]. In LMICs, children’s diarrhea is frequently treated with antibiotics. A review of children’s antibiotic use over the first 2 years of life in eight LMICs found that children were given a median of 3.92 antibiotic courses per child-year, with especially high rates among children in South Asia [19]. By the age of 5, children are estimated to have received an average of 24.5 antibiotic prescriptions in total for treating infections, a rate 5 times higher than observed in high-income countries [20]. At a population level, chronic antibiotic use accelerates the acquisition of ARGs among pathogenic bacteria [41]. Among individuals, antibiotic use can increase gut susceptibility to colonization with virulent pathogens, especially in the hospital setting [42]. By protecting children against diarrhea and improving overall health, breastfeeding may reduce total antibiotic use, reduce diarrhea-related hospitalizations, and prevent associated harms to children’s gut microbiomes during this critical window of development [4].
Reduced exposure to water- and food-borne AMR bacteria
Children who receive their hydration and nutrition via human milk have reduced early-life exposures to water and foods that may be contaminated with AMR bacteria. Potential bacterial contamination of drinking water and feeding bottles is one of many reasons why formula feeding is discouraged by the WHO. A recent study found that formula feeding may in fact increase the relative abundance of ARG-carrying pathogens, including Staphylococcus aureus, Staphyloccoccus epidermidis, Klebsiella pneumoniae, Klebsiella oxytoca, and Clostridioides difficile in the gut of preterm infants, compared to preterm infants receiving human milk or fortified human milk [43]. Once children begin eating complementary foods, they can also be exposed to AMR fecal bacteria that contaminate vegetables and meat, especially in LMIC settings [44, 45]. Preventing children’s food- and waterborne exposures to AMR bacteria is critically important during their first few months of life, particularly in regions where the burden of enteric pathogen exposure is high. Impaired gut microbiome development in these settings may compromise the development of colonization resistance against the establishment and proliferation of pathogens [26, 40]. Exclusive or predominant breastfeeding for an extended duration likely confers the greatest protection, though any level of breastfeeding could theoretically reduce children’s environmental exposures to water- and food-borne AMR bacteria.
Protection by human milk
Human milk constituents can directly protect against the acquisition, establishment, and proliferation of AMR pathogens and ARGs in children’s guts by seeding the gut with non-pathogenic commensals that competitively exclude pathogens and reduce their negative impacts on the host [28, 46–50]; targeting and killing AMR pathogens [21, 48, 51–53]; suppressing gut inflammation which is conducive to pathogen colonization and the lateral transfer of ARGs to unrelated, neighboring bacteria (i.e., via horizontal gene transfer) [47, 54–56]; improving intestinal barrier function to prevent pathogen establishment and translocation [22, 50, 57, 58]; and priming the infant immune system to develop robust responses against exogenous bacteria, including AMR pathogens [50, 59, 60]. Some key protective components beyond sIgA (described above) are reviewed below.
Milk microbiota
Human milk contains approximately 106 bacteria/mL and is a continuous source of commensal, mutualistic, and/or potentially probiotic bacteria to the developing infant’s gut [50]. Dominant genera include Staphylococcus and Streptococcus with hundreds of other taxa identified through 16S rRNA sequencing [61]. Human milk microbiota communities vary between women and over time, and likely comprise both maternal-origin bacteria and infant oral cavity bacteria that enter the milk duct via suckling [62]. Human milk and infant gut microbiomes share a number of commensal organisms by the first week of life [63, 64], with breastfeeding duration and exclusivity influencing these associations in a dose-dependent manner [65]. However, human milk-derived species become less dominant in infants’ guts by the first month [64], suggesting that these initial colonizers could have a specific, early role in infant immune development [25, 66]. Human milk bacteria may protect newborns from AMR pathogen colonization and infection through competitive exclusion of exogenous pathogens, production of antimicrobial compounds, and by enhancing intestinal cell mucin production, which reduces intestinal permeability and improves intestinal barrier function [50, 67]. For example, strains of Lactobacillus rhamnosus isolated from human milk have been demonstrated to be effective in inhibiting a wide range of pathogenic bacteria in vivo [68], including multidrug-resistant E. coli [69]. Finally, it is hypothesized that human milk bacteria prime the infant immune system by training intestinal cells to respond appropriately to commensals versus harmful pathogens [50]. Development of a robust immune response is key to preventing AMR pathogen proliferation and infection in the developing child.
A substantial portion of the human milk microbiota is thought to comprise non-pathogenic maternal gut bacteria that have been endogenously transported from the mother’s intestinal lumen to the mammary glands during late pregnancy and lactation [61]. Because maternal-origin bacteria are particularly stable in the infant gut [25], it has been proposed that modulating the maternal gut microbiome during pregnancy and/or lactation could positively impact the development of the infant gut microbiome [61]. To our knowledge, associations between maternal gut microbiome modifications and increased infant resilience against AMR pathogen colonization or infection have not been directly explored.
Human milk oligosaccharides (HMOs)
Human milk contains high levels of HMOs, a family of structurally diverse unconjugated glycans that are uniquely found in human milk and thought to shape the composition of the developing infant gut microbiome. HMO levels vary between mothers and over time, depending on the stage of lactation [70]. A high concentration of HMOs in the early infant gut could reduce children’s susceptibility to gut colonization by AMR pathogens and ARGs through multiple mechanisms.
First, HMOs are an ideal substrate for Bifidobacterium species, which can dominate the infant gut and may prevent the establishment and proliferation of AMR bacteria and enteric pathogens [27, 46, 48]. Bifidobacterium species like Bifidobacterium longum subsp. infantis efficiently consume HMOs [47], creating acidic by-products which reduce intestinal pH. This lower pH environment is unfavorable for the growth of Enterobacteriaceae, which commonly encode ARGs [46]. Multiple Enterobacteriaceae genera, including all ESBL-producing Enterobacteriaceae and drug-resistant Salmonella and Shigella, are considered critical antibiotic resistance threats by the US Centers for Disease Control and Prevention [71]. Bifidobacteria, on the other hand, much less commonly encode transferable ARGs [27, 72]. Bifidobacteria also produce bacteriocins, which exhibit antimicrobial activity against enteric pathogens and bacteria frequently resistant to antibiotics, such as Escherichia coli [48, 53]. In addition to suppressing their growth, specific Bifidobacterium species have been demonstrated to prevent the lateral exchange of ARGs between neighboring Enterobacteriaceae in mouse models [73]. Previous research among a cohort of Bangladeshi children has demonstrated inverse, dose-response relationships between infants’ gut levels of Bifidobacteria and concentrations of both ARGs and Enterobacteriaceae species [27].
Second, HMOs may directly neutralize invading AMR pathogens and reduce gut inflammation, which otherwise causes a favorable environment for Enterobacteriaceae growth and ARG transfer [55]. The structure of HMOs is similar to cell surface glycoconjugates used by enteric pathogens to bind and enter target cells. Thus, HMOs can prevent the adhesion of enteric pathogens to the intestinal mucosa by serving as alternate binding sites [51]. Further, there is some evidence that HMOs can directly inhibit the expression of genes involved in inflammation, although the mechanisms remain unclear [56]. B. infantis, which thrives in gut environments with high HMO levels, has also been shown to suppress the production of proinflammatory cytokines and is associated with lower concentrations of fecal calprotectin—a marker of intestinal inflammation—throughout the first 2 months of life [47, 54] The capacity for HMOs and breastfed gut environments to suppress gut inflammation is critical in preventing growth blooms of harmful AMR pathogens, including AMR Enterobacteriaceae [74].
Antimicrobial factors
In addition to sIgA and HMOs described above (see the “Potential benefits of early breastfeeding” section and previous section, respectively), human milk contains many other factors that directly inhibit AMR pathogens and/or their adhesion to epithelial cell surfaces. These include lactoferrin, an iron-binding protein with antimicrobial properties that is present in high concentrations in human milk, especially colostrum. Lactoferrin can be extremely effective in preventing the proliferation of E. coli [52], and has also been demonstrated to prevent the growth, impair the virulence, and prevent the epithelial attachment of many other bacterial pathogens via multiple modes of action [75], including Shigella and Salmonella [51, 57]. Recent randomized control trials among infants in LMICs have suggested that lactoferrin supplementation of children’s feeds could reduce risks of neonatal sepsis [76] and reduce the duration of diarrheal illness [77]. In addition, other proteins present in human milk, once they bind to free-floating oligosaccharides, can prevent the adhesion of enteric pathogens and/or their toxins to the intestinal lumen via similar mechanisms to sIgA, HMOs, and lactoferrin. The glycosylated human milk proteins MUC1 and MUC4 are effective against frequently AMR enteric pathogens like Salmonella, for example [51]. β-casein, a common glycosylated human milk protein, has also been shown in mouse models to also stimulate the expression of MUC2 genes and increase the number of mucus-producing goblet cells and Paneth cells in the small intestine. The development of a healthy intestinal mucus layer is critical in preventing the establishment of AMR pathogens, protecting the gut epithelium against pathogen-mediated damage, and promoting pathogen clearance [75]. Lactoferrin and other antimicrobial factors in human milk are present at the highest concentrations directly post-partum [78].
Other antimicrobial proteins may enhance the effectiveness of antibiotics against AMR pathogens. For example, the human milk factor HAMLET (human α-lactalbumin made lethal to tumor cells) has been shown to directly target as well as enhance the effectiveness of antibiotics against drug-resistant Staphylococcus aureus and pathogenic streptococci [79, 80], which could reduce the duration of antibiotic therapy needed to treat an infection and thereby minimize associated harms to infants’ developing microbiomes [4]. Recent experimental studies suggest that other antimicrobial peptides present in human milk, such as β-defensins, may also be able to work in synergy with antibiotics to lower the minimum inhibitory concentrations of intracellular pathogens [81].
There is great interest in developing human milk glycoconjugate supplements that could help support or reinforce natural mechanisms of protection against neonatal infection [51]. Whether these compounds could also protect children against colonization or infection with AMR bacteria merits further investigation.
Extracellular vesicles (EVs)
The potential role of human milk EVs, particularly exosomes, in regulating infants’ gut maturation has only recently gained attention [82, 83]. EVs are non-replicating cell membrane-bound microvesicles (20–2000 nm) naturally produced by almost every mammalian cell. They are believed to be critical in cell-to-cell signaling as they can bear surface receptors or ligands and encapsulate and transport cell contents. EVs secreted into human milk are capable of surviving harsh conditions, including digestion, and appear to be a stable transport mechanism for the transfer of maternal-origin microRNA (miRNA) to infant intestinal cells [83]. miRNAs are small (22 nucleotide length) non-coding RNAs that control gene expression, largely by binding to and accelerating the degradation of messenger RNA. A handful of miRNAs may account for the majority of miRNAs delivered by human milk EVs [83], and recent transcriptomic studies indicate these miRNAs may help protect gut epithelial cells from enteric infection while the immune system is still maturing [58]. Cell surface receptors harbored by human milk EVs may also modulate the developing infant immune system and direct EVs to certain cell types [84]; recent studies have demonstrated that these EVs promote intestinal epithelial cell growth and appear to protect intestinal epithelial cells from oxidative stress [85, 86]. Although human milk EV research is still in its infancy, there is already great interest in determining whether EV supplementation could help protect newborns against colonization or infection with enteric pathogens [82], including AMR pathogens.
Evidence from human studies for the protective benefits of breastfeeding against infants’ colonization by AMR pathogens
Several randomized controlled trials, and observational and mouse model studies have explored the effectiveness of human milk components (e.g., sIgA [30, 31, 33, 34], lactoferrin [51, 52, 57], other glycoconjugates [51]) and human milk-supported taxa (e.g., Bifidobacteria [48, 53]) against enteric pathogens in general. However, to date, fewer studies have examined the potential protective effects of breastfeeding and human milk supplements against AMR bacteria and ARGs specifically [27, 54, 87–91].
Several observational studies have identified a potential protective effect of current breastfeeding against healthy children’s acquisition and/or colonization with enteric AMR bacteria. A longitudinal study of 100 healthy newborns in Spain found that current breastfeeding was protective against first acquisition of ESBL-producing Enterobacteriaceae (adjusted hazard ratio (aHR), 0.29; 95% confidence interval (CI), 0.11, 0.80) [89]. Consumption of any human milk and exclusive breastfeeding were also found to be protective against enteric colonization with ESBL-producing Enterobacteriaceae in relatively small cross-sectional studies conducted in rural Venezuela (n = 78) [90] and Lebanon (n = 117) [91], respectively. Other epidemiological studies have identified a protective effect of exclusive or predominant breastfeeding on children’s infections with enteric pathogens that are commonly drug-resistant, including nontyphoidal Salmonella [45, 92] and Campylobacter [30, 93].
Whether breastfeeding is protective against AMR pathogen colonization among already hospitalized children remains unclear. A cross-sectional study of infants admitted to a hospital in rural Kenya found that current breastfeeding was not protective against gut colonization with ESBL-producing E. coli at the time of admission nor during hospitalization [94]. A 5-year study of infants hospitalized in a neonatal intensive care unit in Italy also found that breastfeeding was not protective against ESBL-producing E. coli acquisition [95]. However, breastfed infants were less likely to become colonized with multidrug-resistant gram-negative bacteria during their stay. Further, among children who did acquire multidrug-resistant gram-negative bacteria, more frequent breastfeeding appeared to be protective against their colonization by multiple species (versus only one) [95].
Recent metagenomic sequencing studies have described associations between breastfeeding and children’s developing gut microbiomes and resistomes, defined as all ARGs present across bacterial species. In Europe, a mother-infant paired study by Pärnänen et al. found that early termination of exclusive breastfeeding was associated with higher relative gut abundance of multiple ARGs and the bacterial taxa that typically encode them (e.g., Gammaproteobacteria) in infants’ guts, compared to infants who were exclusively breastfed through 6 months of age [87]. Specifically, the authors reported that early termination of all breastfeeding (i.e., prior to 6 months) was associated with the increased relative abundance of ARGs conferring resistance to aminoglycoside, sulfonamide, and tetracycline in infants’ resistomes, as well as higher relative abundance of mobile genetic elements that could allow these ARGs to be transferred to pathogens via horizontal gene transfer. Findings from a separate sequencing study of infants’ stool suggest that HMO-supported Bifidobacteria growth could mediate these associations [27]. The authors found that a gut microbiome predominantly made up of Bifidobacteria spp. was associated with significant reductions in both the number and relative abundance of ARGs present in children’s guts by 1 year of age, compared to infant microbiomes with low abundance of Bifidobacteria spp. The authors observed that Bifidobacteria dominance likely suppressed the growth of common ARG-encoding taxa (an observation that has been demonstrated in mouse models) [46]. As described above, HMOs in human milk provide an ideal substrate for Bifidobacterium species.
A recent randomized control trial among healthy newborns in the USA has provided the most robust evidence to date of the potential for human milk supplements to protect infants against AMR pathogen colonization [88]. The authors demonstrated that daily supplementation during the first month of life with a B. infantis strain that relies on HMOs for colonization significantly reduced the relative abundance of ARGs (by 90%) and the bacterial genera commonly associated with them (i.e., Escherichia, Staphylococcus, Bacteroides, and Clostridioides), strongly indicating that HMO-supported growth of Bifidobacteria could be critical to promoting breastfed infants’ gut resilience to AMR pathogens and ARGs [88]. Because HMO diversity and concentrations in human milk can vary between mothers and over time [70], daily Bifidobacteria supplementation could be a way to ensure high Bifidobacteria abundance in the guts of breastfed children, and possibly a way to extend health benefits among children who are only partially breastfed during the first months of life. To our knowledge, no randomized trials have explored whether improved breastfeeding practices or early-life supplementation of other human milk components could directly protect young children from colonization with ARGs or AMR pathogens of critical concern.
Human milk as a source of antimicrobial resistance
Although breastfeeding likely cultivates a gut environment that is ultimately protective against children’s colonization and infection with AMR pathogens, human milk may also be source of AMR bacteria and ARGs to the developing infant [87, 96]. In a recent study of 16 mother-infant pairs by Pärnänen et al., approximately 70% of ARGs and 76% of bacterial species detected in mothers’ human milk were also detected in their infants’ guts, suggesting vertical transmission [87]. The same study observed that maternal antibiotic prophylaxis during delivery was associated with both a higher number and abundance of ARGs in breastfed infants’ guts for up to 6 months, indicating that maternal antibiotic exposures may have lasting impacts on breastfed infants’ gut resistomes [87]. AMR Staphylococcus, Streptococcus, Acinetobacter, Enterococcus, and Corynebacterium strains have also been cultured from human milk [96]. AMR bacteria and ARGs detected in human milk likely do not originate solely from the mammary glands, as bacteria from mothers’ skin and infants’ mouths (through retrograde inoculation during suckling) are also thought to contribute to the milk microbiota [50]. Nevertheless, human milk appears to be one of many sources of AMR bacteria and ARGs to the developing infant. This could be consequential if antibiotic use during delivery becomes more common to prevent maternal and neonatal infections, particularly in LMIC settings [97, 98].
Exploring breastfeeding as a strategy to combat antimicrobial resistance
The potential benefits outlined here suggest that breastfeeding should be actively explored as a scalable, cost-effective strategy to address antimicrobial resistance, in addition to the many other health benefits that breastfeeding confers [23]. In high-income countries, where less than half of newborns receive any human milk at 6 months [23], improving optimal breastfeeding practices could reduce early-life antibiotic use and improve gut colonization resistance against AMR pathogens, which are becoming more common in the community. If breastfeeding or human milk constituents are shown to be protective against colonization or infection with AMR bacteria, this would be one of many important reasons to accelerate efforts to promote breastfeeding at the policy level.
Breastfeeding may be an especially important strategy to explore in LMICs. Due to a higher prevalence of bacterial disease, LMICs are expected to be disproportionately burdened by the antimicrobial resistance crisis [99]. Although absolute antibiotic use has historically been higher in high-income settings due to healthcare access [100], antibiotic consumption is increasing in LMICs and appropriate targeting of antibiotics is limited by widespread informal antibiotic use and restricted resources for identifying, treating, and preventing AMR infections [99, 101]. By 2050, up to 90% of AMR-related mortality is predicted to occur in countries in Asia and Africa [1], most of which are low- or middle-income. Although systematic investigations are sparse, several studies of healthy children in LMICs [5, 7, 102] report colonization with enteric multidrug-resistant gram-negative bacteria at rates that far exceed what is observed in the USA and Europe. For example, 82% of 1–2-year-old children in Bangladesh [103] were recently reported to be colonized with third-generation cephalosporin-resistant Enterobacteriaceae, compared to 4.5% of similarly aged children in the USA [104] and ~ 14% in Switzerland [105]. Household members’ hands, household soil, drinking water, flies, and food have all previously been implicated as sources of both human- and animal-origin fecal bacteria [44, 45, 106–109].
Globally, breastfeeding rates continue to be the highest in countries with the lowest income status [23]. However, within LMICs, differences exist based on mothers’ location (urban versus rural), education status, and household income. If these differences persist, future breastfeeding rates could be impacted. For example, multiple studies have demonstrated that mothers in urban settings exclusively breastfeed their children for a shorter duration of time than rural mothers [110–112]. A recent study from India, a rapidly developing LMIC, suggests that this difference is most pronounced among urban mothers who have at least a primary school education (compared to urban mothers with no formal education), suggesting that the increased employment opportunities afforded by education, and/or greater access to formula and the money to purchase it, may contribute to these differences [110]. Further, fewer wealthy mothers living in LMIC exclusively or predominantly breastfeed their children during their first 6 months of life compared to mothers belonging to the lowest wealth quintiles (Fig. 1) [113]. These discrepancies widen as children grow; 20–23-month-old babies from the poorest families continue to receive at least some breast milk at rates that are 1.3–2.8 times higher than those from the wealthiest families [114]. Collectively, these trends suggest that breastfeeding practices could worsen in LMICs with strong breastfeeding traditions as they urbanize and transition to greater per capita wealth [23, 110]. This may be a particularly important public health concern in settings where reductions in environmental pathogen exposure lag behind wealth- and education-associated reductions in breastfeeding, as a higher fraction of pathogens will be AMR in the coming decades. Policies aimed at promoting breastfeeding in LMICs should find ways to support mothers, such that the benefits for infant health are maximized while mothers’ access to new educational and economic opportunities afforded by economic transition is maintained.
Conclusion
Multiple strategies have been proposed to prevent the coming global antimicrobial resistance emergency, including the development of new antibiotics and vaccines, improved surveillance and education, and improved hospital infection control [1]. Given that AMR pathogens are increasingly prevalent in community settings, early-life interventions like breastfeeding that could protect children against community-acquired gut colonization and infection should also be actively explored. Recent evidence supports the role of breastfeeding in preventing the acquisition, establishment, and proliferation of enteric pathogens, including AMR bacteria, in young children’s guts by reducing children’s early-life exposures to contaminated foods and inappropriate antibiotic use. The emerging mechanistic pathways through which human milk components including the milk microbiota, maternal antibodies, HMOs, antimicrobial peptides, and EVs may protect against early-life colonization by AMR pathogens and ARGs warrant further exploration as novel targets for intervention, and/or supplementation where optimal breastfeeding practices are not possible. The evidence presented here suggests that breastfeeding and human milk supplements deserve greater attention as potential preventive measures in the global effort to combat AMR.
Acknowledgements
None to declare.
Abbreviations
- AMR
Antimicrobial-resistant
- ARG
Antibiotic resistance gene
- HMO
Human milk oligosaccharide
- LMIC
Low- and middle-income country
- EV
Extracellular vesicle
- miRNA
MicroRNA
- sIgA
Secretory IgA
- WHO
World Health Organization
Authors’ contributions
M.L.N. conceptualized the manuscript. M.L.N, C.D.B., R.C.R., E.D.A., A.R.M., and A.J.P. contributed to the literature search, writing, and review. All authors read and approved the final manuscript.
Funding
M.L.N. is supported by the National Center for Advancing Translational Sciences, National Institutes of Health (KL2TR002545). C.D.B. is supported by a Sir Henry Dale Postdoctoral Research Fellowship from Wellcome and the Royal Society (206225/Z/17/Z). R.C.R. is supported by a Sir Henry Wellcome Postdoctoral Fellowship from the Wellcome Trust (206455/Z/17/Z). The funders had no role in manuscript preparation or decision to publish. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Availability of data and materials
The datasets analyzed to generate Fig. 1 of the current study are available from the United Nations Children’s Fund, Division of Data Research and Policy: https://data.unicef.org/resources/dataset/infant-young-child-feeding/.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
None to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jim O’Neill. Tackling Drug-resistant infections globally: final report and recommendations. 2016. p. 84. http://amr-review.org/sites/default/files/160525_Final_paper_withcover.pdf. Accessed 1 Oct 2019.
- 2.Burnham JP, Olsen MA, Kollef MH. Re-estimating annual deaths due to multidrug-resistant organism infections. Infect Control Hosp Epidemiol. 2019;40:112–113. doi: 10.1017/ice.2018.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Interagency Coordination Group on Antimicrobial Resistance. No time to wait: securing the future from drug-resistant infections. 2019. p. 28. https://www.who.int/antimicrobial-resistance/interagency-coordination-group/IACG_final_report_EN.pdf?ua=1. Accessed 27 Oct 2020.
- 4.Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smit PW, et al. Transmission dynamics of hyper-endemic multi-drug resistant Klebsiella pneumoniae in a Southeast Asian neonatal unit: a longitudinal study with whole genome sequencing. Front Microbiol. 2018;9:1197. doi: 10.3389/fmicb.2018.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talbert AWA, Mwaniki M, Mwarumba S, Newton CRJC, Berkley JA. Invasive bacterial infections in neonates and young infants born outside hospital admitted to a rural hospital in Kenya. Pediatr Infect Dis J. 2010;29:945–949. doi: 10.1097/INF.0b013e3181dfca8c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kothari C, et al. Community acquisition of β-lactamase producing Enterobacteriaceae in neonatal gut. BMC Microbiol. 2013;13:136. doi: 10.1186/1471-2180-13-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herindrainy P, et al. Acquisition of extended spectrum beta-lactamase-producing enterobacteriaceae in neonates: a community based cohort in Madagascar. PLoS One. 2018;13:1–17. doi: 10.1371/journal.pone.0193325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Ami R, et al. Influx of extended-spectrum beta-lactamase-producing enterobacteriaceae into the hospital. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2006;42:925–934. doi: 10.1086/500936. [DOI] [PubMed] [Google Scholar]
- 10.Cheikh A, et al. Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) colonization as a risk factor for developing ESBL infections in pediatric cardiac surgery patients: ‘retrospective cohort study’. BMC Infect Dis. 2017;17:237. doi: 10.1186/s12879-017-2346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denis B, et al. Prevalence, risk factors, and impact on clinical outcome of extended-spectrum beta-lactamase-producing Escherichia coli bacteraemia: a five-year study. Int J Infect Dis IJID Off Publ Int Soc Infect Dis. 2015;39:1–6. doi: 10.1016/j.ijid.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Ruppé E, et al. Relative fecal abundance of extended-spectrum-β-lactamase-producing Escherichia coli strains and their occurrence in urinary tract infections in women. Antimicrob Agents Chemother. 2013;57:4512–4517. doi: 10.1128/AAC.00238-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.U.S. Department of Health and Human Services, Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States 2019. 148 (2019).
- 14.Stapleton PJ, et al. Outbreaks of extended spectrum beta-lactamase-producing Enterobacteriaceae in neonatal intensive care units: a systematic review. Arch Dis Child - Fetal Neonatal Ed. 2016;101:72–78. doi: 10.1136/archdischild-2015-308707. [DOI] [PubMed] [Google Scholar]
- 15.Lukac PJ, Bonomo RA, Logan LK. Extended-spectrum β-lactamase–producing Enterobacteriaceae in children: old foe, emerging threat. Clin Infect Dis Off Publ Infect Dis Soc Am. 2015;60:1389–1397. doi: 10.1093/cid/civ020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson C, et al. Estimating global trends in total and childhood antibiotic consumption, 2011-2015. BMJ Glob Health. 2019;4:e001241. doi: 10.1136/bmjgh-2018-001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaz LE, et al. Recent trends in outpatient antibiotic use in children. Pediatrics. 2014;133:375–385. doi: 10.1542/peds.2013-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson MK, Crofts TS, Dantas G. Antibiotics and the developing infant gut microbiota and resistome. Curr Opin Microbiol. 2015;27:51–56. doi: 10.1016/j.mib.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogawski ET, et al. Use of antibiotics in children younger than two years in eight countries: a prospective cohort study. Bull World Health Organ. 2017;95:49–61. doi: 10.2471/BLT.16.176123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fink G, D’Acremont V, Leslie HH, Cohen J. Antibiotic exposure among children younger than 5 years in low-income and middle-income countries: a cross-sectional study of nationally representative facility-based and household-based surveys. Lancet Infect Dis. 2019. 10.1016/S1473-3099(19)30572-9. [DOI] [PubMed]
- 21.Hassiotou F, Geddes DT, Hartmann PE. Cells in human milk: state of the science. J Hum Lact. 2013;29:171–182. doi: 10.1177/0890334413477242. [DOI] [PubMed] [Google Scholar]
- 22.Andreas NJ, Kampmann B, Mehring Le-Doare K. Human breast milk: a review on its composition and bioactivity. Early Hum Dev. 2015;91:629–635. doi: 10.1016/j.earlhumdev.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Victora CG, et al. Breastfeeding in the 21st century: epidemiology, mechanisms, and lifelong effect. Lancet. 2016;387:475–490. doi: 10.1016/S0140-6736(15)01024-7. [DOI] [PubMed] [Google Scholar]
- 24.Davisse-Paturet C, Adel-Patient K, Divaret-Chauveau A, Pierson J, Lioret S, Cheminat M, Dufourg MN, Charles MA, de Lauzon-Guillain B. Breastfeeding Status and Duration and Infections, Hospitalizations for Infections, and Antibiotic Use in the First Two Years of Life in the ELFE Cohort. Nutrients. 2019;11(7):1607. 10.3390/nu11071607. [DOI] [PMC free article] [PubMed]
- 25.Ferretti P, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–145. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson RC, Manges AR, Finlay BB, Prendergast AJ. The human microbiome and child growth – first 1000 days and beyond. Trends Microbiol. 2019;27:131–147. doi: 10.1016/j.tim.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Taft DH, Liu J, Maldonado-Gomez MX, Akre S, Huda MN, Ahmad SM, Stephensen CB, Mills DA. Bifidobacterial Dominance of the Gut in Early Life and Acquisition of Antimicrobial Resistance. mSphere. 2018;3(5):e00441–18. 10.1128/mSphere.00441-18. [DOI] [PMC free article] [PubMed]
- 28.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trend S, de Jong E, Lloyd ML, Kok CH, Richmond P, Doherty DA, Simmer K, Kakulas F, Strunk T, Currie A. Leukocyte Populations in Human Preterm and Term Breast Milk Identified by Multicolour Flow Cytometry. PloS one. 2015;10(8):e0135580. 10.1371/journal.pone.0135580. [DOI] [PMC free article] [PubMed]
- 30.Ruiz-Palacios GM, et al. Protection of breast-fed infants against Campylobacter diarrhea by antibodies in human milk. J Pediatr. 1990;116:707–713. doi: 10.1016/S0022-3476(05)82652-6. [DOI] [PubMed] [Google Scholar]
- 31.Mantis NJ, Rol N, Corthésy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson LA, Korotkova M. The role of breastfeeding in prevention of neonatal infection. Semin Neonatol SN. 2002;7:275–281. doi: 10.1053/siny.2002.0124. [DOI] [PubMed] [Google Scholar]
- 33.Cruz JR, Gil L, Cano F, Caceres P, Pareja G. Breast milk anti-Escherichia coli heat-labile toxin IgA antibodies protect against toxin-induced infantile diarrhea. Acta Paediatr Scand. 1988;77:658–662. doi: 10.1111/j.1651-2227.1988.tb10726.x. [DOI] [PubMed] [Google Scholar]
- 34.Glass RI, et al. Protection against cholera in breast-fed children by antibodies in breast milk. N Engl J Med. 1983;308:1389–1392. doi: 10.1056/NEJM198306093082304. [DOI] [PubMed] [Google Scholar]
- 35.NEOVITA Study Group Timing of initiation, patterns of breastfeeding, and infant survival: prospective analysis of pooled data from three randomised trials. Lancet Glob. Health. 2016;4:e266–e275. doi: 10.1016/S2214-109X(16)00040-1. [DOI] [PubMed] [Google Scholar]
- 36.Division of Nutrition, Physical Activity, and Obesity, National Center for Chronic Disease Prevention and Health Promotion. Breastfeeding Report Card: United States. 2020. https://www.cdc.gov/breastfeeding/pdf/2020-Breastfeeding-Report-Card-H.pdf. Accessed 27 Oct 2020.
- 37.Bagci Bosi AT, Eriksen KG, Sobko T, Wijnhoven TMA, Breda J. Breastfeeding practices and policies in WHO European Region Member States. Public Health Nutr. 2016;19:753–764. doi: 10.1017/S1368980015001767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai X, Wardlaw T, Brown DW. Global trends in exclusive breastfeeding. Int Breastfeed J. 2012;7:12. doi: 10.1186/1746-4358-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korpela K, Salonen A, Virta LJ, Kekkonen RA, de Vos WM. Association of early-life antibiotic use and protective effects of breastfeeding: role of the intestinal microbiota. JAMA Pediatr. 2016;170:750. doi: 10.1001/jamapediatrics.2016.0585. [DOI] [PubMed] [Google Scholar]
- 40.Popkin BM, et al. Breast-feeding and diarrheal morbidity. Pediatrics. 1990;86:874–882. [PubMed] [Google Scholar]
- 41.Lipsitch M, Samore MH. Antimicrobial use and antimicrobial resistance: a population perspective. Emerg Infect Dis. 2002;8:347–354. doi: 10.3201/eid0804.010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buffie CG, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pärnänen, K. et al. Formula alters preterm infant gut microbiota and increases its antibiotic resistance load. bioRxiv 782441 (2019) doi:10.1101/782441.
- 44.Zurfluh K, et al. Extended-spectrum-β-lactamase-producing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl Environ Microbiol. 2015;81:3115–3120. doi: 10.1128/AEM.00258-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nadimpalli M, et al. CTX-M-55-type ESBL-producing Salmonella enterica are emerging among retail meats in Phnom Penh, Cambodia. J Antimicrob Chemother. 2019;74:342–348. doi: 10.1093/jac/dky451. [DOI] [PubMed] [Google Scholar]
- 46.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 47.Underwood MA, German JB, Lebrilla CB, Mills DA. Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr Res. 2015;77:229–235. doi: 10.1038/pr.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez FAC, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv. 2013;31:482–488. doi: 10.1016/j.biotechadv.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 49.Wagner VE, et al. Effects of a gut pathobiont in a gnotobiotic mouse model of childhood undernutrition. Sci. Transl. Med. 2016;8:366ra164. doi: 10.1126/scitranslmed.aah4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boix-Amorós A, et al. Reviewing the evidence on breast milk composition and immunological outcomes. Nutr Rev. 2019;77:541–556. doi: 10.1093/nutrit/nuz019. [DOI] [PubMed] [Google Scholar]
- 51.Peterson R, Cheah WY, Grinyer J, Packer N. Glycoconjugates in human milk: protecting infants from disease. Glycobiology. 2013;23:1425–1438. doi: 10.1093/glycob/cwt072. [DOI] [PubMed] [Google Scholar]
- 52.Bullen JJ, Rogers HJ, Leigh L. Iron-binding proteins in milk and resistance to Escherichia coli infection in infants. Br Med J. 1972;1:69–75. doi: 10.1136/bmj.1.5792.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Touré R, Kheadr E, Lacroix C, Moroni O, Fliss I. Production of antibacterial substances by bifidobacterial isolates from infant stool active against Listeria monocytogenes. J Appl Microbiol. 2003;95:1058–1069. doi: 10.1046/j.1365-2672.2003.02085.x. [DOI] [PubMed] [Google Scholar]
- 54.Henrick BM, et al. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr. Res. 2019;86:749–757. doi: 10.1038/s41390-019-0533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stecher B, et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci U S A. 2012;109:1269–1274. doi: 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He Y, Liu S, Leone S, Newburg DS. Human colostrum oligosaccharides modulate major immunologic pathways of immature human intestine. Mucosal Immunol. 2014;7:1326–1339. doi: 10.1038/mi.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ochoa TJ, Cleary TG. Effect of lactoferrin on enteric pathogens. Biochimie. 2009;91:30–34. doi: 10.1016/j.biochi.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1:7. doi: 10.1186/1758-907X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Field CJ. The immunological components of human milk and their effect on immune development in infants. J Nutr. 2005;135:1–4. doi: 10.1093/jn/135.1.1. [DOI] [PubMed] [Google Scholar]
- 60.Belderbos ME, et al. Breastfeeding modulates neonatal innate immune responses: a prospective birth cohort study. Pediatr Allergy Immunol. 2012;23:65–74. doi: 10.1111/j.1399-3038.2011.01230.x. [DOI] [PubMed] [Google Scholar]
- 61.Fitzstevens JL, et al. Systematic review of the human milk microbiota. Nutr Clin Pract Off Publ Am Soc Parenter Enter Nutr. 2017;32:354–364. doi: 10.1177/0884533616670150. [DOI] [PubMed] [Google Scholar]
- 62.Moossavi S, et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. 2019;25:324–335. doi: 10.1016/j.chom.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 63.Murphy K, et al. The composition of human milk and infant faecal microbiota over the first three months of life: a pilot study. Sci Rep. 2017;7:40597. doi: 10.1038/srep40597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pannaraj PS, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171:647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fehr K, et al. Breastmilk feeding practices are associated with the co-occurrence of bacteria in mothers’ milk and the infant gut: the CHILD cohort study. Cell Host Microbe. 2020;28:285–297. doi: 10.1016/j.chom.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 66.Le Doare K, Holder B, Bassett A, Pannaraj PS. Mother’s milk: a purposeful contribution to the development of the infant microbiota and immunity. Front Immunol. 2018;9:361. doi: 10.3389/fimmu.2018.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olivares M, Díaz-Ropero MP, Martín R, Rodríguez JM, Xaus J. Antimicrobial potential of four Lactobacillus strains isolated from breast milk. J Appl Microbiol. 2006;101:72–79. doi: 10.1111/j.1365-2672.2006.02981.x. [DOI] [PubMed] [Google Scholar]
- 68.Coman MM, et al. In vitro evaluation of antimicrobial activity of Lactobacillus rhamnosus IMC 501(®) , Lactobacillus paracasei IMC 502(®) and SYNBIO(®) against pathogens. J Appl Microbiol. 2014;117:518–527. doi: 10.1111/jam.12544. [DOI] [PubMed] [Google Scholar]
- 69.Li N, et al. Lactobacillus rhamnosus from human breast milk shows therapeutic function against foodborne infection by multi-drug resistant Escherichia coli in mice. Food Funct. 2020;11:435–447. doi: 10.1039/C9FO01698H. [DOI] [PubMed] [Google Scholar]
- 70.Thurl S, Munzert M, Boehm G, Matthews C, Stahl B. Systematic review of the concentrations of oligosaccharides in human milk. Nutr Rev. 2017;75:920–933. doi: 10.1093/nutrit/nux044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.U.S. Department of Health and Human Services, Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States. 2019. p. 148. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. Accessed 15 Jan 2020.
- 72.Ammor MS, et al. Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. J Mol Microbiol Biotechnol. 2008;14:6–15. doi: 10.1159/000106077. [DOI] [PubMed] [Google Scholar]
- 73.Moubareck C, Lecso M, Pinloche E, Butel MJ, Doucet-Populaire F. Inhibitory impact of bifidobacteria on the transfer of beta-lactam resistance among Enterobacteriaceae in the gnotobiotic mouse digestive tract. Appl Environ Microbiol. 2007;73:855–860. doi: 10.1128/AEM.02001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jakaitis BM, Denning PW. Human breast milk and the gastrointestinal innate immune system. Clin Perinatol. 2014;41:423–435. doi: 10.1016/j.clp.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pammi M, Suresh G. Enteral lactoferrin supplementation for prevention of sepsis and necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev. 2020. 10.1002/14651858.CD007137.pub6. [DOI] [PMC free article] [PubMed]
- 77.Ochoa TJ, et al. Randomized double-blind controlled trial of bovine lactoferrin for prevention of diarrhea in children. J Pediatr. 2013;162:349–356. doi: 10.1016/j.jpeds.2012.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trend S, et al. Antimicrobial protein and peptide concentrations and activity in human breast milk consumed by preterm infants at risk of late-onset neonatal sepsis. PLoS One. 2015;10:1–20. doi: 10.1371/journal.pone.0117038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marks LR, Clementi EA, Hakansson AP. Sensitization of Staphylococcus aureus to methicillin and other antibiotics in vitro and in vivo in the presence of HAMLET. PLoS One. 2013;8:e63158. doi: 10.1371/journal.pone.0063158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alamiri F, Riesbeck K, Hakansson AP. HAMLET, a protein complex from human milk, has bactericidal activity and enhances the activity of antibiotics against pathogenic Streptococci. Antimicrob Agents Chemother. 2019;63:e01193–e01119. doi: 10.1128/AAC.01193-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zharkova MS, et al. Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics-a novel way to combat antibiotic resistance? Front Cell Infect Microbiol. 2019;9:128. doi: 10.3389/fcimb.2019.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Galley JD, Besner GE. The therapeutic potential of breast milk-derived extracellular vesicles. Nutrients. 2020;12:745. doi: 10.3390/nu12030745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Reilly D, et al. Perspective: the role of human breast-milk extracellular vesicles in child health and disease. Adv Nutr. 10.1093/advances/nmaa094. [DOI] [PMC free article] [PubMed]
- 84.Admyre C, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol Baltim Md. 2007;1950(179):1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 85.Martin C, et al. Human breast milk-derived exosomes attenuate cell death in intestinal epithelial cells. Innate Immun. 2018;24:278–284. doi: 10.1177/1753425918785715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hock A, et al. Breast milk-derived exosomes promote intestinal epithelial cell growth. J Pediatr Surg. 2017;52:755–759. doi: 10.1016/j.jpedsurg.2017.01.032. [DOI] [PubMed] [Google Scholar]
- 87.Pärnänen K, et al. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat Commun. 2018;9:3891. doi: 10.1038/s41467-018-06393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Casaburi G, et al. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob Resist Infect Control. 2019;8:131. doi: 10.1186/s13756-019-0583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rodríguez-Revuelta MJ, et al. Incidence and risk factors for acquisition of extended-spectrum β-lactamase-producing Enterobacteriaceae in newborns in Seville, Spain: a prospective cohort study. Int J Antimicrob Agents. 2018;52:835–841. doi: 10.1016/j.ijantimicag.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 90.Araque M, Labrador I. Prevalence of fecal carriage of CTX-M-15 beta-lactamase-producing Escherichia coli in healthy children from a rural Andean village in Venezuela. Osong Public Health Res Perspect. 2018;9:9–15. doi: 10.24171/j.phrp.2018.9.1.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hijazi SM, Fawzi MA, Ali FM, Abd El Galil KH. Multidrug-resistant ESBL-producing Enterobacteriaceae and associated risk factors in community infants in Lebanon. J. Infect. Dev. Ctries. 2016;10:947–955. doi: 10.3855/jidc.7593. [DOI] [PubMed] [Google Scholar]
- 92.Rowe SY, et al. Breast-feeding decreases the risk of sporadic salmonellosis among infants in FoodNet sites. Clin Infect Dis Off Publ Infect Dis Soc Am. 2004;38(Suppl 3):S262–S270. doi: 10.1086/381595. [DOI] [PubMed] [Google Scholar]
- 93.Schiaffino F, Colston JM, Paredes-Olortegui M, François R, Pisanic N, Burga R, Peñataro-Yori P, Kosek MN. Antibiotic Resistance of Campylobacter Species in a Pediatric Cohort Study. Antimicrob Agents Chemother. 2019;63(2):e01911–18. 10.1128/AAC.01911-18. [DOI] [PMC free article] [PubMed]
- 94.Kagia N, et al. Carriage and acquisition of extended-spectrum β-lactamase–producing Enterobacterales among neonates admitted to hospital in Kilifi, Kenya. Clin Infect Dis. 2019;69:751–759. doi: 10.1093/cid/ciy976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giuffrè M, Geraci DM, Bonura C, Saporito L, Graziano G, Insinga V, Aleo A, Vecchio D, Mammina C. The Increasing Challenge of Multidrug-Resistant Gram-Negative Bacilli: Results of a 5-Year Active Surveillance Program in a Neonatal Intensive Care Unit. Medicine. 2016;95(10):e3016. 10.1097/MD.0000000000003016. [DOI] [PMC free article] [PubMed]
- 96.Huang M-S, et al. Most commensally bacterial strains in human milk of healthy mothers display multiple antibiotic resistance. MicrobiologyOpen. 2019;8:e00618. doi: 10.1002/mbo3.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oluwalana C, Camara B, Bottomley C, Goodier S, Bojang A, Kampmann B, Ceesay S, D'Alessandro U, Roca A. Azithromycin in Labor Lowers Clinical Infections in Mothers and Newborns: A Double-Blind Trial. Pediatrics. 2017;139(2):e20162281. 10.1542/peds.2016-2281. [DOI] [PubMed]
- 98.Roca A, et al. Oral azithromycin given during labour decreases bacterial carriage in the mothers and their offspring: a double-blind randomized trial. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2016;22:565. doi: 10.1016/j.cmi.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bebell L, Muiru A. Antibiotic use and emerging resistance—how can resource-limited countries turn the tide? Glob Heart. 2014;9:347–358. doi: 10.1016/j.gheart.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Klein EY, et al. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci. 2018;115:E3463. doi: 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.WHO Report on Surveillance of Antibiotic Consumption: 2016–2018 Early Implementation. 2018. https://www.who.int/medicines/areas/rational_use/who-amr-amc-report-20181109.pdf. Accessed 27 Oct 2020.
- 102.Saleem AF, et al. The gut of healthy infants in the community as a reservoir of ESBL and carbapenemase-producing bacteria. Antibiotics. 2020;9:286. doi: 10.3390/antibiotics9060286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Islam MA, et al. Fecal colonization with multidrug-resistant E. coli among healthy infants in rural Bangladesh. Front. Microbiol. 2019;10:640. doi: 10.3389/fmicb.2019.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Islam S, et al. Intestinal carriage of third-generation cephalosporin-resistant and extended-spectrum β-lactamase-producing Enterobacteriaceae in healthy US children. J Pediatr Infect Dis Soc. 2018;7:234–240. doi: 10.1093/jpids/pix045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kaarme J, et al. Rapid increase in carriage rates of Enterobacteriaceae producing extended-spectrum β-lactamases in healthy preschool children, Sweden. Emerg Infect Dis. 2018;24:1874–1881. doi: 10.3201/eid2410.171842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ercumen A, et al. Animal feces contribute to domestic fecal contamination: evidence from E. coli measured in water, hands, food, flies, and soil in Bangladesh. Environ. Sci. Technol. 2017;51:8725–8734. doi: 10.1021/acs.est.7b01710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Odagiri M, et al. Human fecal and pathogen exposure pathways in rural Indian villages and the effect of increased latrine coverage. Water Res. 2016;100:232–244. doi: 10.1016/j.watres.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Boehm AB, et al. Occurrence of host-associated fecal markers on child hands, household soil, and drinking water in rural Bangladeshi households. Environ Sci Technol Lett. 2016;3:393–398. doi: 10.1021/acs.estlett.6b00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Harris AR, et al. Ruminants contribute fecal contamination to the urban household environment in Dhaka, Bangladesh. Environ Sci Technol. 2016;50:4642–4649. doi: 10.1021/acs.est.5b06282. [DOI] [PubMed] [Google Scholar]
- 110.Oakley L, et al. Is increasing urbanicity associated with changes in breastfeeding duration in rural India? An analysis of cross-sectional household data from the Andhra Pradesh children and parents study. BMJ Open. 2017;7:e016331. doi: 10.1136/bmjopen-2017-016331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dahly DL, Adair LS. Quantifying the urban environment: a scale measure of urbanicity outperforms the urban-rural dichotomy. Soc Sci Med. 2007;1982(64):1407–1419. doi: 10.1016/j.socscimed.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thu HN, et al. Breastfeeding practices in urban and rural Vietnam. BMC Public Health. 2012;12:964. doi: 10.1186/1471-2458-12-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.United Nations Children’s Fund, Division of Data Research and Policy . Global UNICEF Global Databases: Infant and Young Child Feeding: Exclusive breastfeeding, Predominant breastfeeding. 2019. [Google Scholar]
- 114.United Nations Children’s Fund, Division of data research and policy . Global UNICEF Global Databases: Infant and Young Child Feeding: Continued breastfeeding. 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed to generate Fig. 1 of the current study are available from the United Nations Children’s Fund, Division of Data Research and Policy: https://data.unicef.org/resources/dataset/infant-young-child-feeding/.