Abstract
Purpose of Review
Close to two million individuals globally become infected with human immunodeficiency virus (HIV-1) each year and just over two thirds will have access to life-prolonging antivirals. However, the rapid development of drug resistance creates challenges, such that generation of more effective therapies is not only warranted but a necessary endeavour. This review discusses a group of HIV-1 entry inhibitors known as CD4 mimics which exploit the highly conserved relationship between the HIV-1 envelope glycoprotein (Env) and the receptor, CD4.
Recent Findings
We review the structure/function guided evolution of these inhibitors, vital mechanistic insights that underpin broad and potent functional antagonism, recent evidence of utility demonstrated in animal and physiologically relevant in vitro models, and current progress towards effective new-generation inhibitors.
Summary
This review highlights the promising potential of CD4 mimetics as multifunctional therapeutics.
Keywords: HIV entry inhibitors, CD4 mimics, CD4 mimetics, HIV Env, ADCC, Phe43 cavity, gp120
INTRODUCTION
Developing a therapeutic effective against human immunodeficiency virus (HIV-1) remains the goal for successfully halting an epidemic that has spanned multiple decades, killing more than 770 000 individuals in 2018 (1). Vaccines represent the central strategy for combating a wide range of pathogens; however despite ongoing efforts, the recent premature termination of the HIV vaccine clinical trial, HVTN 702 in South Africa due to futility (2, 3) highlights the ongoing challenges faced by the field. Though Highly Active Antiretroviral Therapy (HAART) targeting viral replication has helped prolong the life of HIV-1-infected individuals, emergence of resistance and the inability of HAART to purge the latent viral reservoir is an unresolved problem, calling for approaches that are aimed at eradicating or functionally curing HIV infection. Entry inhibitors, in particular CD4 mimics, have gained significant traction due to their potential utility as multifunctional therapeutics that can be employed for both prevention and treatment.
HIV-1 entry is mediated by the interaction of gp120, the surface subunit of its Envelope glycoprotein complex (Env), with the primary receptor CD4 and one of two chemokine coreceptors (CXCR4 or CCR5) (Reviewed in (4)). In its unliganded form, the Env trimer exists in a structurally constrained ‘closed’ conformation. Receptor engagement initiates a series of structural rearrangements within gp120 that result in the sequential opening of the trimer into an asymmetric intermediate and finally into an “open” conformation (5–7). CD4 engagement is a major driving force for Env structural rearrangements. CD4 is comprised of four extracellular immunoglobulin-like domains (D1-D4) linked to a short cytoplasmic tail through its transmembrane domain. Early work identified D1 and D2 as the active regions of CD4 that interacted with gp120 (8), with key structural analysis clearly demonstrating the critical interaction between gp120 and D1 (9). Importantly, this showed that the CD4 binding site (CD4bs) formed by the inner domain, outer domain and bridging sheet of gp120 accommodated a large hydrophobic cavity known as the Phe43 cavity. The side chain of Phe43 of D1 of CD4 is inserted into the opening of this cavity, allowing for a critical electrostatic interaction between the cavity lining residue Asp368 of gp120 and Arg59 of CD4 (9). This and other interactions within and surrounding the cavity modulate infection (10, 11) and as discussed by a detailed study conducted by Prevost et al,.(12), influence recognition by CD4bs antibodies (12, 13). Importantly in the context of this review, changes in the Phe43 cavity affect the binding and potency of small-molecule CD4 mimetic compounds (CD4mc)(12). Furthermore, Env-CD4 interaction at the surface of HIV-producing cells is critical for exposure of epitopes for CD4-induced (CD4i) antibodies that mediate Antibody-dependent cellular cytotoxicity (ADCC) (14, 15). Incidentally, these epitopes are recognized by non-neutralizing antibodies (nnAbs) present in natural infection (7, 16). However since the accessory proteins Nef and Vpu play active roles in downregulating membrane-bound CD4 as part of HIV’s evasion mechanism (Reviewed in (17–19), CD4 mimics can act as a surrogate to ‘open’ cell-surface Env (Figure 1 and Table 1). All these reasons provide the impetus for targeting the Phe43 cavity with rationally designed CD4 mimics.
Figure 1: Functional attributes of CD4 mimetics.
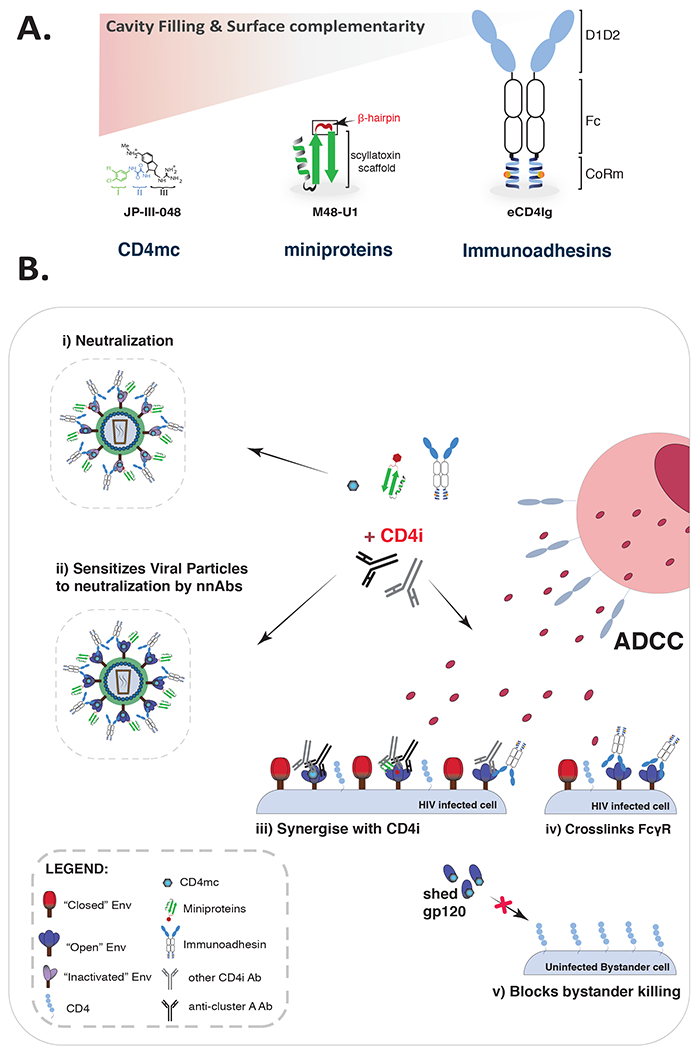
(A) Comparison of cavity filling and surface complementarity relative to CD4 size. (B) Env-CD4mimetic ligation elicits distinct responses. With differing potency all CD4 mimetics are able to (i) inactive viral particles, (ii) facilitate neutralization of viral particles by non-neutralizing antibodies (nnAbs) and (iii) synergize with CD4i antibodies to mediate ADCC. However, only eCD4Ig (immunoadhesin)(iv) is able to independently cross-link FcγR. Only small molecule CD4mc (vi) are able to prevent bystander killing.
Table 1:
Functional characteristics of lead/potent CD4 mimics
| Class | Compound | Description | Functional properties | Therapeutic potential | References |
|---|---|---|---|---|---|
| Immunoadhesins | |||||
| CD4Ig (PRO542) | D1D2 of CD4 fused to Fc portion of Ig |
i, ii |
Prevention Passive immunization |
Jacobsen JM et al, 2000 Jacobsen JM et al, 2004 |
|
| eCD4Ig | CD4Ig fused with CoR mimic | i, ii, iii, iv, v |
Treatment Antiviral |
Davis-Gardner ME et al, 2017 Fellinger CH et al, 2019 Gardner MR et al, 2019 |
|
|
Cure Kill part of ‘Shock and kill’ |
|||||
| Miniproteins | |||||
| M48U1 | Select CD4 β-hairpin residues transplanted on scyllatoxin scaffold | ii, iv, v |
Prevention Topical microbicide |
Acharya P et al, 2015 | |
| gp120-S-S-M46U1 | M46U1 fused with gp120 | i, ii, iii, iv, vi |
Treatment Antiviral |
Martin G et al, 2011 Dey AK et al, 2012 |
|
| gp140-S-S-M46U1 | M46U1 fused with gp140 | i, ii, iii, iv, vi |
Cure Kill part of ‘Shock and kill’ |
||
| CD4mc | |||||
|
NBD Series NBD14110 |
Analog of NBD556 modified to contain bulky benzodioxole moiety in region I | ii, iv, v | Curelli F et al, 2018 | ||
|
DMJ-II-121 Analogs BNM-III-170 |
Analog of NBD556 modified in region III to an indane core with guanidinium moiety | ii, iv, v, vi |
Prevention As part of vaccination |
Melillo B et al, 2016; Madani N et al, 2018; Prevost J et al, 2020 |
|
|
Treatment Antiviral |
|||||
|
YYA-021 Analogs YIR-821 |
Analog of NBD556 structurally modified in region III to consist of a mono-cyclohexy with long guanidino extension attached to piperidine core |
ii |
Cure Kill part of ‘Shock and kill’ |
Ohashi N et al, 2016 | |
|
MCG Series MCG-IV-210 |
Novel molecule containing a short amide linker between piperidine core and halogenated aromatic ring |
ii, iv, v, vi |
Ding S et al, 2019; Grenier MC et al, 2018 Prevost J et al, 2020 |
||
High avidity binding to gp120
Inactivates viral particle
Mediates ADCC of infected cells
Sensitizes infected cells to ADCC
Sensitizes viral particle to neutralisation by nnAbs
Protects against bystander killing
Brief history of CD4 mimics
The earliest evidence of CD4 mimics as potential HIV therapies was the discovery that soluble forms of CD4 (sCD4) were able to neutralize HIV-1 (20–24). Unfortunately, these early promising in vitro studies yielded disappointing results in clinical studies. Despite showing anti-viral activity, administration of sCD4 resulted in rapid viral rebound; alarmingly, suboptimal concentrations were observed to enhance infection (25, 26). Although widespread clinical application of sCD4 was promptly abandoned, it was evident from these early studies that sCD4 preserved high affinity binding and conformational and functional characteristics similar to that of native membrane-bound CD4 (reviewed in (27)), encouraging the development of CD4 mimics with more favourable qualities. Currently CD4 mimics fall into the broad categories of; (i) CD4 immunoadhesins, (ii) miniproteins, and (iii) small-molecule CD4 mimetics (CD4mc) (Fig 1; Table 1). This review discusses the functional evolution of these inhibitors and current progress.
CD4 IMMUNOADHESINS
CD4 immunoadhesins (CD4-Ig), are antibody-like chimeric proteins typically comprising the immunoglobulin (Ig) constant domain fused with the D1 and D2 domains of CD4. A first-generation tetrameric CD4 immunoadhesin, CD4-IgG2, demonstrated that replacement of the variable fragment (Fv) portions of IgG2 with D1 and D2 afforded a longer half-life than sCD4, exhibited cross-clade neutralization (28), and blocked HIV-1 Env-mediated syncytium formation (29). Importantly both tetrameric (28, 29) and dodecameric CD4-Ig (D1D2-Igαtp) (30) lacked the unfavourable feature of viral enhancement inherent to sCD4 derivatives. The tetrameric immunoadhesin PRO542, the first of this class to be approved for clinical trials, was shown to be safe and effectively reduced plasma viremia (31), with especially pronounced effects in patients with advanced disease (32). Additionally, the unique ability to crosslink multiple gp120s afforded immunoadhesins potent avidity, which translated to heightened viral clearance in vitro (30). Fusion of sulfated peptide sequences corresponding to the amino terminus of the CCR5 coreceptor to the carboxy termini of tetrameric CD4-Ig (33), generated the bi-specific immunoadhesin, eCD4-Ig (34). This molecule demonstrated unmatched breadth and potency, neutralizing all HIV-1, HIV-2 and SHIV strains tested with IC50 lower than some broadly neutralizing CD4bs antibodies (34, 35). While CD4-Ig proved inefficient at eliciting antibody-mediated antiviral effector functions, the inclusion of the sulfated peptide afforded eCD4-Ig the capability to promote ADCC (34–36), in particular by enhancing recognition by otherwise occluded CD4i V3 antibodies; eCD4Ig decreased the binding of CD4bs, V2-apex and interface antibodies (35). Davis-Gardner et al. (35) showed that eCD4-Ig synergised with patient sera to kill reactivated latently infected primary cells (35). Interestingly, eCD4-Ig has minimal propensity to allow the generation of escape mutants (37), highlighting that in addition to its potential to purge the latent reservoir it may also be potentially utilised as a long-acting antiviral.
Further variations of eCD4-Ig, excluding D2, were developed to improve specificity for the Phe43 cavity (36); however, were shown to have reduced stability and often enhanced CD4-independent infection (36). Others have also explored conjugation of D1D2 with single-chain Fv domains of CD4i antibodies such as the co-receptor binding site (CoRBs) antibody 17b. These proteins showed potent neutralization; however, data on their ADCC potential is currently lacking (38), though the absence of the Fc portion of the antibody likely precludes efficient effector cell engagement.
Finally, current eCD4-Ig approaches include passive immunization through gene therapy utilising Adeno-associated viral (AAV) vectors following proof of concept studies highlighted by Gardner et al.,(39) in a macaque model. These studies showed durable antibody production, and unlike AAV-delivered broadly neutralizing antibodies (bNAbs) (40), did not induce anti-drug antibodies. Immunoadhesins such as eCD4-Ig hold promise as multifunctional antivirals.
MINIPROTEINS
In the early 1990s, toxicology research highlighted the intriguing therapeutic potential of the cysteine stabilized alpha-beta motif (CSα/β) of scorpion toxins (41). Work showed that these highly stable biologically active structures could accommodate large sequence substitutions from unrelated proteins, with little to no impact on overall structure while adopting the functionality of the transplanted protein (41, 42). The scorpion toxins could therefore act as appropriate molecular scaffolds onto which the binding surfaces of proteins could be transplanted. Interestingly, the complementarity determining region (CDR)-2 loop of CD4 D1 bore a striking structural similarity to the scorpion toxin’s β-hairpin protrusion, consequently inspiring the generation of CD4-miniproteins.
CD4 miniproteins are typically ~27-31 amino acids in length comprising the critical gp120-interacting residues of CD4 D1 grafted onto the β-hairpin region of a short scorpion toxin, scyllatoxin (43). Through a series of rational engineering approaches, Vita and colleagues generated the M-series of miniproteins, of which CD4M9 (M9) was the most potent; CD4M9 retained a native-like conformation, bound gp120 as well as CD4, and exhibited antagonism of HIV-1 infection at μM concentrations. The generation of bivalent (44) and trivalent (45) reconstructions of M9 designed to target multiple CD4 binding sites effectively enhanced inhibitor potency. Further critical improvements of M9 designed for deeper Phe43 cavity penetration, through the replacement of residue 23 (analogous to CD4 Phe43) with a non-natural amino acid, biphenylalanine (Bip) (M33) (46), and later with more flexible groups (47), generated the M48 series of peptides. Amongst these peptides, M48U1 showed the most potent antiviral activity, with neutralization of almost all HIV-1 strains tested; only those HIV-1 strains with atypical Phe43 cavities occluded by larger residues, such as the CRF_01 A/E strains, were resistant to M48U1 (48). Nonetheless, M48U1 has been shown to retain its potent antiviral activity in hydrogels in macaque models of transmission expanding its utility as a topical microbicide (49).
More recent approaches include vaccination with chimeric immunogens of CD4 miniproteins crosslinked to gp120 or gp140. However, despite eliciting high titres of CD4i Abs in rabbits (50, 51) and strong ADCC responses in macaques (52, 53), the high avidity due to multivalent antibody binding was short-lived (51, 53). Additional structure-guided engineering could one day see the promises of this approach achieved.
SMALL MOLECULES (CD4mc)
A screen of chemical libraries looking for inhibitors of the gp120-CD4 interaction by Debnath and colleagues led to the discovery of NBD-556 and NBD-557; these two lead N-phenyl-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamide analogues bound within the gp120 Phe43 cavity with specific, μM activity against the gp120-CD4 interaction, for both lab-adapted and primary isolates (54). The NBD series of compounds were subsequently characterized as comprising 3 distinct regions: a para-halide substituted aromatic ring (Region I) connected to a tetramethyl piperidine heterocyclic ring moiety (Region III) by an oxalamide linker (Region II) (Fig 1). Subsequent structure-guided re-engineering by the Sodroski/Smith and Matsushita/Tamamura groups (55, 56) aimed at deeper penetration of the compound into the Phe43 cavity showed that region I modifications incorporating additional halide groups on the phenyl ring improved gp120 binding compared to the miniprotein M33 and native CD4 (55), while methyl additions demonstrated favourable toxicity profiles in rodents and non-human primates (57).
Further structural analysis by Debnath’s group (58, 59) suggested that the lack of region III insertion into the Phe43 cavity by NBD analogues likely reduced binding efficiency. Consequent replacement of the piperidine moiety in Region III showed improved inhibitory activity and potent neutralization against diverse strains, although still lacking efficient cavity penetration and surface complementarity. Importantly, these Region III modifications provided an alternate scaffold to improve on, yielding a new generation of small molecules. In addition to replacing the piperidine group with an indane, inclusion of a guanidinium moiety to the five-membered ring (60–62) altered positioning within the cavity allowing for deeper penetration that facilitated unique binding characterised by additional H-bonding networks spanning residues Met426 and Asp368 (61, 63). This translated to highly potent functional antagonism and diminished CD4-independent infection for compounds such as AWS-I-169, DMJ-I-228 and DMJ-II-121; subsequent studies by Madani and colleagues showed that the activated Env intermediate induced by these compounds is very short lived (64). Further reconstructions by Matsushita (65) and Sodroski/Smith (64) groups took advantage of region III as a scaffold (YIR-821; BNM-III-147, BNM-III-170 and JP-III-048 respectively). Despite failing to replicate the crucial Arg59 (CD4)-Asp368 (gp120) interaction, these compounds exhibited greater potency against primary strains of HIV-1. The Sodroski/Smith groups reported a pivotal discovery that CD4mc strongly induced conformational changes in gp120 that expose otherwise-hidden highly conserved epitopes (62). This translated to sensitization of viral particles to neutralization by otherwise non-neutralizing CD4i antibodies; such antibodies are easily generated through vaccination with CD4-bound stabilised gp120s (62, 66, 67), leading to the concept that CD4mc could be used to improve the vaccine efficacy of weak immunogens.
Further, work by our group (15, 68) showed that, independent of their region III modifications (JP-III-048 and DMJ-I-228 and BNM-III-170) or class (M48U1 or sCD4), CD4 mimics “opened” Env on cells infected with primary isolates and sensitized them to ADCC mediated by antibodies present in biologically relevant body fluids (ie. sera, cervicovaginal lavage and breast milk) from HIV-1-infected individuals. Structural differences in the CD4mc influenced the magnitude of Env opening and thereby potency at activating ADCC, with more superior responses observed for the indane derivative, JP-III-048 against transmitted/founder HIV-1 in primary cells; both JP-III-048 and DMJ-I-228 exhibited better responses compared to the miniprotein M48U1 and sCD4, suggesting that small CD4 mimetics could avoid conformational constraints imposed by primary Envs on larger protein ligands (15, 68).
Importantly, Richard et al,.(15), presented evidence that CD4mc were able to sensitize ex vivo-amplified primary CD4+ T cells to ADCC mediated by autologous sera and effector cells. This sensitization required synergy between CD4mc and CoRBS Abs, which “open” the Env trimer and facilitate recognition by anti-cluster A antibodies; this results in the stabilization of the asymmetric State 2A Env conformation, which is vulnerable to ADCC (7). Subsequent work by Richard et al.,(68) and Anand et al.,(69) confirmed sequential and synergistic action of CoRBS antibodies and anti-cluster A antibodies to promote efficient FcγRIII engagement, enhanced by CD4mc. These studies provided an otherwise unrecognised biological and mechanistic explanation for the potency of CD4mc in influencing ADCC responses. These results suggested that CD4mc might be useful in the “kill” part of the “shock and kill” strategy being pursued to eliminate the HIV-1 reservoir (70). In these approaches, latently infected cells are activated and subsequently killed by host immune responses. Preclinical studies are ongoing to establish the value of the CD4mc BNM-III-170 to decrease the size of the viral reservoir in humanized mice and non-human primates.
By allowing ‘easy to elicit’ nnAbs to target both viral particles and infected cells, CD4mc represent an alternative approach to prevention of HIV-1 transmission. Recently the small CD4mc JP-III-048 was shown to protect BLT-humanized mice from HIV-1 challenge (71). BNM-III-170 was demonstrated to augment the protective efficacy of an otherwise weak gp120 immunogen in non-human primates stringently challenged with a heterologous Tier 2 SHIV (66, 72). On the backdrop of this functional attribute of CD4mc, the Finzi and Smith Groups (73) using a carefully designed high-throughput cell-based ELISA (CBE) aimed at detecting small molecules that exposed vulnerable HIV-1 Env epitopes, identified and developed a novel family of small molecules from a chemical library of over a hundred thousand compounds (73, 74). This family of compounds, though resembling a binding structure similar to that of BNM-III-170, differ in two respects: (i) they exhibit a smaller, more compact structure, comprising an amide bond linking a halogenated aromatic ring to a piperidine core; (ii) they can easily be synthesized. Structurally, these differences translate to deeper cavity filling anchored by the aromatic halogen ring; this in turn allows for a closer distance between the piperidine core and Asp368, particularly members with sulfonamide extensions. Furthermore, positioning of the halogenated aromatic ring also facilitates direct interactions with another important Phe43 cavity residue, Glu370, as well as close packing against residue 375.
The lead compound, (S)MCG-IV-210, from this series was shown to have the most contacts with gp120, is able to expose Env to nnAbs, and can sensitize infected cells to ADCC (73). In addition, it efficiently sensitized viral particles to neutralization by otherwise non-neutralizing antibodies, consistent with previous studies with NBD analogues (15, 68). Furthermore, as reported previously for other CD4mc (75), this new family was able to protect uninfected cells from bystander killing by preventing shed gp120 from interacting with CD4 on uninfected cells. This mechanism potentially reduces the detrimental depletion of uninfected immune cells, particularly CD4+ T cells during natural course of infection.
Interestingly, Prevost et al,.(12) recently showed that natural polymorphisms of residue 375 and a series of six co-evolving gp120 inner domain residues (12, 76) can structurally remodel the Phe43 cavity and directly impact CD4mc affinity. For instance, the presence of Thr375 (in ~17% of clade B HIV-1 strains) in place of the Ser375 found in the majority of M group HIV-1 isolates, increased affinity for all small CD4mc tested. Furthermore, changes in residue 375 and its coevolving gp120 inner domain residues restructure the CD4bs and thus influence the binding mode, occupancy and potency of the different CD4mc classes, which modulates virus neutralization and ADCC. These findings are especially pivotal for the design and synthesis of next-generation compounds with broader activity. As suggested, shaping of the Phe43 cavity by these residues has major implications for efficient targeting of CRF_01.A/E as well as strains with alternate Phe43 cavity residues.
A mechanistic understanding of how CD4mc engage the functional HIV-1 Env trimer has begun to emerge. Indeed, computational prediction suggests that potent functional antagonism mediated by CD4mc can be achieved by i) engagement of two or three protomers; or ii) high-affinity interactions demonstrated by the different conformational states induced by the different CD4 mimics (64). Overall, these studies show that the ability of CD4mc to allosterically transform Env structure to conformations susceptible to the vast majority of antibodies present in sera from HIV-1-infected individuals promises novel prevention, treatment and cure strategies.
CONCLUSION
Major strides have been made in the development of CD4 mimetics, which has been aided immensely by structure-guided bioengineering. Importantly, each category has provided promising candidates, currently undergoing evaluation, or as scaffolds on which rationally designed enhancements are being considered and pursued.
KEY POINTS.
The need for more effective HIV therapies is driven largely by the emergence of resistance and the inability of HAART to target the latent viral reservoir
Among current therapies, CD4 mimetics, which target the highly conserved CD4-gp120 interaction have demonstrated broad functional potency
Several potent CD4 mimetics have been identified that have shown promise as multifunctional therapeutics that could also be utilized to purge the latent reservoir
Acknowledgements
We would like to thank Jonathan Richard for helpful discussion and assistance with generating figure 1. We also thank Shilei Ding and Jérémie Prevost for helpful suggestions and comments.
Financial Support and Sponsorship
This work was supported by a CIHR foundation grant #352417, NIH R01 AI148379 and R01 AI150322 to A.F. This study was also supported by P01-GM56550/AI150741 to A.F., A.B.S., and J.S. A.F. is the recipient of a Canada Research Chair on Retroviral Entry #RCHS0235. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
References and Recommended reading
- 1.World Health Organisation. Global Health Observatory (GHO) Data 2020 [Available from: https://www.who.int/gho/hiv/en/.
- 2.Cohen J Another HIV vaccine strategy fails in a large scale study: Science; [updated 3rd of February 2020 Available from: https://www.sciencemag.org/news/2020/02/another-hiv-vaccine-strategy-fails-large-scale-study.
- 3.National Institutes of Health (NIH). Experimental HIV Vaccine Regimen Ineffective in Preventing HIV [updated 31st of January 2020 Available from: https://www.niaid.nih.gov/news-events/experimental-hiv-vaccine-regimen-ineffective-preventing-hiv.
- 4.Wilen CB, Tilton JC, Doms RW. HIV: cell binding and entry. Cold Spring Harb Perspect Med. 2012;2(8):a006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munro JB, Gorman J, Ma X, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346(6210):759–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma X, Lu M, Gorman J, et al. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.**Alsahafi N, Bakouche N, Kazemi M, et al. An Asymmetric Opening of HIV-1 Envelope Mediates Antibody-Dependent Cellular Cytotoxicity. Cell Host Microbe. 2019;25(4):578–87 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified a new Env conformation, State 2A, which is susceptible to ADCC and that can be stabilized by the combination of CD4mc, CoRBS and cluster A Abs.
- 8.Traunecker A, Luke W, Karjalainen K. Soluble CD4 molecules neutralize human immunodeficiency virus type 1. Nature. 1988;331(6151):84–6. [DOI] [PubMed] [Google Scholar]
- 9.Kwong PD, Wyatt R, Robinson J, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duenas-Decamp MJ, Peters PJ, Burton D, et al. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J Virol. 2009;83(6):2575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herschhorn A, Gu C, Moraca F, et al. The beta20-beta21 of gp120 is a regulatory switch for HIV-1 Env conformational transitions. Nat Commun. 2017;8(1):1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.**Prévost J. TW, Medjahed H, Sherburn RT, et al. The HIV-1 Env gp120 inner domain shapes the Phe43 cavity and the CD4 binding site. mBio. 2020; 11(3):e00280–20. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study performed an in depth analysis of natural polymorphisms of residue 375 in the Phe43 cavity and the impact on susceptibility to CD4 mimetics. Overall results highlight how the gp120 inner domain shapes the Phe43 cavity and the CD4BS.
- 13.Wibmer CK, Bhiman JN, Gray ES, et al. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog. 2013;9(10):e1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veillette M, Coutu M, Richard J, et al. Conformational evaluation of HIV-1 trimeric envelope glycoproteins using a cell-based ELISA assay. J Vis Exp. 2014(91):51995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richard J, Veillette M, Brassard N, et al. CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc Natl Acad Sci U S A. 2015;112(20):E2687–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams KL, Cortez V, Dingens AS, et al. HIV-specific CD4-induced Antibodies Mediate Broad and Potent Antibody-dependent Cellular Cytotoxicity Activity and Are Commonly Detected in Plasma From HIV-infected humans. EBioMedicine. 2015;2(10):1464–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piguet V, Schwartz O, Le Gall S, et al. The downregulation of CD4 and MHC-I by primate lentiviruses: a paradigm for the modulation of cell surface receptors. Immunol Rev. 1999;168:51–63. [DOI] [PubMed] [Google Scholar]
- 18.Forthal DN, Finzi A. Antibody-dependent cellular cytotoxicity in HIV infection. AIDS. 2018;32(17):2439–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richard J, Prevost J, Alsahafi N, et al. Impact of HIV-1 Envelope Conformation on ADCC Responses. Trends Microbiol. 2018;26(4):253–65. [DOI] [PubMed] [Google Scholar]
- 20.Smith DH, Byrn RA, Marsters SA, et al. Blocking of HIV-1 infectivity by a soluble, secreted form of the CD4 antigen. Science. 1987;238(4834):1704–7. [DOI] [PubMed] [Google Scholar]
- 21.Hussey RE, Richardson NE, Kowalski M, et al. A soluble CD4 protein selectively inhibits HIV replication and syncytium formation. Nature. 1988;331(6151):78–81. [DOI] [PubMed] [Google Scholar]
- 22.Fisher RA, Bertonis JM, Meier W, et al. HIV infection is blocked in vitro by recombinant soluble CD4. Nature. 1988;331(6151):76–8. [DOI] [PubMed] [Google Scholar]
- 23.Deen KC, McDougal JS, Inacker R, et al. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature. 1988;331(6151):82–4. [DOI] [PubMed] [Google Scholar]
- 24.Berger EA, Chaudhary VK, Clouse KA, et al. Recombinant CD4-Pseudomonas exotoxin hybrid protein displays HIV-specific cytotoxicity without affecting MHC class II-dependent functions. AIDS Res Hum Retroviruses. 1990;6(6):795–804. [DOI] [PubMed] [Google Scholar]
- 25.Schooley RT, Merigan TC, Gaut P, et al. Recombinant soluble CD4 therapy in patients with the acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. A phase I-II escalating dosage trial. Ann Intern Med. 1990;112(4):247–53. [DOI] [PubMed] [Google Scholar]
- 26.Schutten M, Andeweg AC, Bosch ML, et al. Enhancement of infectivity of a non-syncytium inducing HIV-1 by sCD4 and by human antibodies that neutralize syncytium inducing HIV-1. Scand J Immunol. 1995;41(1):18–22. [DOI] [PubMed] [Google Scholar]
- 27.Sattentau QJ, Moore JP. The role of CD4 in HIV binding and entry. Philos Trans R Soc Lond B Biol Sci. 1993;342(1299):59–66. [DOI] [PubMed] [Google Scholar]
- 28.Trkola A, Pomales AB, Yuan H, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69(11):6609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allaway GP, Davis-Bruno KL, Beaudry GA, et al. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res Hum Retroviruses. 1995;11(5):533–9. [DOI] [PubMed] [Google Scholar]
- 30.Arthos J, Cicala C, Steenbeke TD, et al. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein: implications for therapeutic and vaccine strategies. J Biol Chem. 2002;277(13):11456–64. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson JM, Lowy I, Fletcher CV, et al. Single-dose safety, pharmacology, and antiviral activity of the human immunodeficiency virus (HIV) type 1 entry inhibitor PRO 542 in HIV-infected adults. J Infect Dis. 2000;182(1):326–9. [DOI] [PubMed] [Google Scholar]
- 32.Jacobson JM, Israel RJ, Lowy I, et al. Treatment of advanced human immunodeficiency virus type 1 disease with the viral entry inhibitor PRO 542. Antimicrob Agents Chemother. 2004;48(2):423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farzan M, Vasilieva N, Schnitzler CE, et al. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J Biol Chem. 2000;275(43):33516–21. [DOI] [PubMed] [Google Scholar]
- 34.Gardner MR, Kattenhorn LM, Kondur HR, et al. AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature. 2015;519(7541):87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis-Gardner ME, Gardner MR, Alfant B, et al. eCD4-Ig promotes ADCC activity of sera from HIV-1-infected patients. PLoS Pathog. 2017;13(12):e1006786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fetzer I, Gardner MR, Davis-Gardner ME, et al. eCD4-Ig Variants That More Potently Neutralize HIV-1. J Virol. 2018;92(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fellinger CH, Gardner MR, Weber JA, et al. eCD4-Ig Limits HIV-1 Escape More Effectively than CD4-Ig or a Broadly Neutralizing Antibody. J Virol. 2019;93(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dey B, Del Castillo CS, Berger EA. Neutralization of human immunodeficiency virus type 1 by sCD4-17b, a single-chain chimeric protein, based on sequential interaction of gp120 with CD4 and coreceptor. J Virol. 2003;77(5):2859–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardner MR, Fellinger CH, Kattenhorn LM, et al. AAV-delivered eCD4-Ig protects rhesus macaques from high-dose SIVmac239 challenges. Sci Transl Med. 2019;11(502). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardner MR, Fetzer I, Kattenhorn LM, et al. Anti-drug Antibody Responses Impair Prophylaxis Mediated by AAV-Delivered HIV-1 Broadly Neutralizing Antibodies. Mol Ther. 2019;27(3):650–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vita C, Roumestand C, Toma F, et al. Scorpion toxins as natural scaffolds for protein engineering. Proc Natl Acad Sci U S A. 1995;92(14):6404–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drakopoulou E, Zinn-Justin S, Guenneugues M, et al. Changing the structural context of a functional beta-hairpin. Synthesis and characterization of a chimera containing the curaremimetic loop of a snake toxin in the scorpion alpha/beta scaffold. J Biol Chem. 1996;271(20):11979–87. [DOI] [PubMed] [Google Scholar]
- 43.Vita C, Drakopoulou E, Vizzavona J, et al. Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein. Proc Natl Acad Sci U S A. 1999;96(23):13091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Song H, Heredia A, et al. Synthetic bivalent CD4-mimetic miniproteins show enhanced anti-HIV activity over the monovalent miniprotein. Bioconjug Chem. 2004;15(4):783–9. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Guan Y, Szczepanska A, et al. Synthesis and anti-HIV activity of trivalent CD4-mimetic miniproteins. Bioorg Med Chem. 2007;15(12):4220–8. [DOI] [PubMed] [Google Scholar]
- 46.Martin L, Stricher F, Misse D, et al. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat Biotechnol. 2003;21(1):71–6. [DOI] [PubMed] [Google Scholar]
- 47.Van Herrewege Y, Morellato L, Descours A, et al. CD4 mimetic miniproteins: potent anti-HIV compounds with promising activity as microbicides. J Antimicrob Chemother. 2008;61(4):818–26. [DOI] [PubMed] [Google Scholar]
- 48.Acharya P, Luongo TS, Louder MK, et al. Structural basis for highly effective HIV-1 neutralization by CD4-mimetic miniproteins revealed by 1.5 A cocrystal structure of gp120 and M48U1. Structure. 2013;21(6):1018–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouchemal K, Aka-Any-Grah A, Dereuddre-Bosquet N, et al. Thermosensitive and mucoadhesive pluronic-hydroxypropylmethylcellulose hydrogel containing the mini-CD4 M48U1 is a promising efficient barrier against HIV diffusion through macaque cervicovaginal mucus. Antimicrob Agents Chemother. 2015;59(4):2215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin G, Burke B, Thai R, et al. Stabilization of HIV-1 envelope in the CD4-bound conformation through specific cross-linking of a CD4 mimetic. J Biol Chem. 2011;286(24):21706–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dey AK, Burke B, Sun Y, et al. Elicitation of neutralizing antibodies directed against CD4-induced epitope(s) using a CD4 mimetic cross-linked to a HIV-1 envelope glycoprotein. PLoS One. 2012;7(1):e30233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen X, Bogers WM, Yates NL, et al. Cross-Linking of a CD4-Mimetic Miniprotein with HIV-1 Env gp140 Alters Kinetics and Specificities of Antibody Responses against HIV-1 Env in Macaques. J Virol. 2017;91(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bogers W, Barnett SW, Oostermeijer H, et al. Increased, Durable B-Cell and ADCC Responses Associated with T-Helper Cell Responses to HIV-1 Envelope in Macaques Vaccinated with gp140 Occluded at the CD4 Receptor Binding Site. J Virol. 2017;91(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Q, Ma L, Jiang S, et al. Identification of N-phenyl-N’-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology. 2005;339(2):213–25. [DOI] [PubMed] [Google Scholar]
- 55.Madani N, Schon A, Princiotto AM, et al. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure. 2008;16(11):1689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamada Y, Ochiai C, Yoshimura K, et al. CD4 mimics targeting the mechanism of HIV entry. Bioorg Med Chem Lett. 2010;20(1):354–8. [DOI] [PubMed] [Google Scholar]
- 57.Hashimoto C, Narumi T, Otsuki H, et al. A CD4 mimic as an HIV entry inhibitor: pharmacokinetics. Bioorg Med Chem. 2013;21(24):7884–9. [DOI] [PubMed] [Google Scholar]
- 58.Curreli F, Choudhury S, Pyatkin I, et al. Design, synthesis, and antiviral activity of entry inhibitors that target the CD4-binding site of HIV-1. J Med Chem. 2012;55(10):4764–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Curreli F, Kwon YD, Zhang H, et al. Binding mode characterization of NBD series CD4-mimetic HIV-1 entry inhibitors by X-ray structure and resistance study. Antimicrob Agents Chemother. 2014;58(9):5478–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LaLonde JM, Kwon YD, Jones DM, et al. Structure-based design, synthesis, and characterization of dual hotspot small-molecule HIV-1 entry inhibitors. J Med Chem. 2012;55(9):4382–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lalonde JM, Le-Khac M, Jones DM, et al. Structure-Based Design and Synthesis of an HIV-1 Entry Inhibitor Exploiting X-Ray and Thermodynamic Characterization. ACS Med Chem Lett. 2013;4(3):338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Madani N, Princiotto AM, Schon A, et al. CD4-mimetic small molecules sensitize human immunodeficiency virus to vaccine-elicited antibodies. J Virol. 2014;88(12):6542–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwon YD, LaLonde JM, Yang Y, et al. Crystal structures of HIV-1 gp120 envelope glycoprotein in complex with NBD analogues that target the CD4-binding site. PLoS One. 2014;9(1):e85940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madani N, Princiotto AM, Zhao C, et al. Activation and Inactivation of Primary Human Immunodeficiency Virus Envelope Glycoprotein Trimers by CD4-Mimetic Compounds. J Virol. 2017;91(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohashi N, Harada S, Mizuguchi T, et al. Small-Molecule CD4 Mimics Containing Mono-cyclohexyl Moieties as HIV Entry Inhibitors. ChemMedChem. 2016;11(8):940–6. [DOI] [PubMed] [Google Scholar]
- 66.Madani N, Princiotto AM, Mach L, et al. A CD4-mimetic compound enhances vaccine efficacy against stringent immunodeficiency virus challenge. Nat Commun. 2018;9(1):2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Madani N, Princiotto AM, Easterhoff D, et al. Antibodies Elicited by Multiple Envelope Glycoprotein Immunogens in Primates Neutralize Primary Human Immunodeficiency Viruses (HIV-1) Sensitized by CD4-Mimetic Compounds. J Virol. 2016;90(10):5031–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richard J, Pacheco B, Gohain N, et al. Co-receptor Binding Site Antibodies Enable CD4-Mimetics to Expose Conserved Anti-cluster A ADCC Epitopes on HIV-1 Envelope Glycoproteins. EBioMedicine. 2016;12:208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.**Anand SP, Prevost J, Baril S, et al. Two Families of Env Antibodies Efficiently Engage Fc-Gamma Receptors and Eliminate HIV-1-Infected Cells. J Virol. 2019;93(3). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study highlighted the importance of synergistic action of coreceptor binding site antibodies and anti-cluster A antibodies in mediating ADCC in the presence of CD4mc.
- 70.Finzi A Exposing HIV-1 Env: Implications for therapeutic strategies. Clin Invest Med. 2019;42(4):E2–E6. [DOI] [PubMed] [Google Scholar]
- 71.Princiotto AM, Vrbanac VD, Melillo B, et al. A Small-Molecule CD4-Mimetic Compound Protects Bone Marrow-Liver-Thymus Humanized Mice From HIV-1 Infection. J Infect Dis. 2018;218(3):471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ding S, Verly MM, Princiotto A, et al. Short Communication: Small-Molecule CD4 Mimetics Sensitize HIV-1-Infected Cells to Antibody-Dependent Cellular Cytotoxicity by Antibodies Elicited by Multiple Envelope Glycoprotein Immunogens in Nonhuman Primates. AIDS Res Hum Retroviruses. 2017;33(5):428–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.**Ding S, Grenier MC, Tolbert WD, et al. A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity. J Virol. 2019;93(24). [DOI] [PMC free article] [PubMed] [Google Scholar]; High throughput cell-based ELISA was performed in this study to identify a novel family of small molecule CD4mc that were able to sensitize HIV infected cells to ADCC. Overall the study confirmed rapid synthesis of these novel molecules using structure based design with improved potent functional antagonism.
- 74.Grenier MC, Ding S, Vezina D, et al. Optimization of Small Molecules That Sensitize HIV-1 Infected Cells to Antibody-Dependent Cellular Cytotoxicity. ACS Med Chem Lett. 2020;11(3):371–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richard J, Veillette M, Ding S, et al. Small CD4 Mimetics Prevent HIV-1 Uninfected Bystander CD4 + T Cell Killing Mediated by Antibody-dependent Cell-mediated Cytotoxicity. EBioMedicine. 2016;3:122–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zoubchenok D, Veillette M, Prevost J, et al. Histidine 375 Modulates CD4 Binding in HIV-1 CRF01_AE Envelope Glycoproteins. J Virol. 2017;91(4). [DOI] [PMC free article] [PubMed] [Google Scholar]


