Abstract
Recent studies have demonstrated immunologic dysfunction in severely ill coronavirus disease 2019 (COVID-19) patients. We use single-cell RNA sequencing (scRNA-seq) to analyze the transcriptome of peripheral blood mononuclear cells (PBMCs) from healthy (n = 3) and COVID-19 patients with moderate disease (n = 5), acute respiratory distress syndrome (ARDS, n = 6), or recovering from ARDS (n = 6). Our data reveal transcriptomic profiles indicative of defective antigen presentation and interferon (IFN) responsiveness in monocytes from ARDS patients, which contrasts with higher responsiveness to IFN signaling in lymphocytes. Furthermore, genes involved in cytotoxic activity are suppressed in both natural killer (NK) and CD8 T lymphocytes, and B cell activation is deficient, which is consistent with delayed viral clearance in severely ill COVID-19 patients. Our study demonstrates that COVID-19 patients with ARDS have a state of immune imbalance in which dysregulation of both innate and adaptive immune responses may be contributing to a more severe disease course.
Keywords: acute respiratory distress syndrome, ARDS, coronavirus disease 2019, COVID-19, severe acute respiratory syndrome coronavirus 2, SARS-CoV-2, single-cell RNA sequencing, network analysis
Graphical Abstract
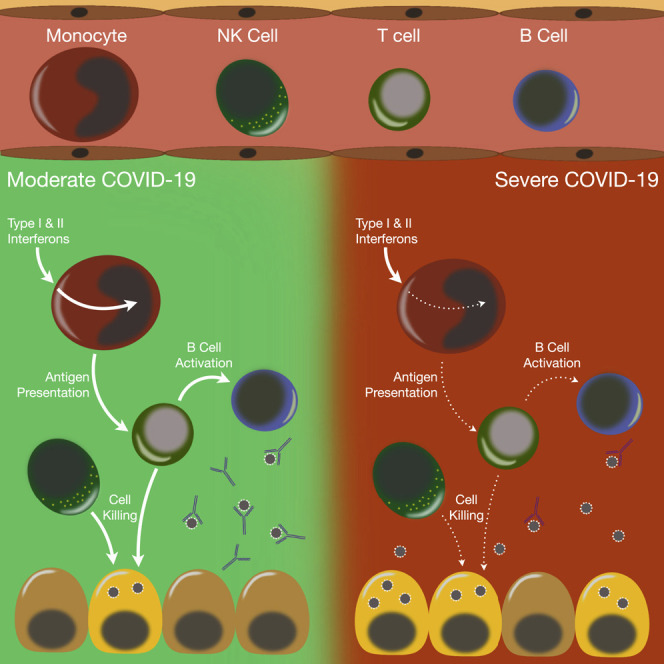
Yao et al. provide evidence that widespread, cell-specific dysregulation of immune responses in COVID-19 patients with acute respiratory distress syndrome may underlie disease severity. Defects include impaired antigen presentation pathways, suppressed monocyte response to early antiviral interferon signals, deficient lymphocyte expression of cytotoxicity genes, and reduced B cell activation.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has quickly spread worldwide to cause the coronavirus disease 2019 (COVID-19) pandemic (Zhu et al., 2020). Coronaviruses are single, positive-stranded RNA viruses that can infect a range of hosts. Some are known to cause seasonal, upper respiratory infections (i.e., common colds), but coronaviruses that cause severe lower respiratory infection have emerged, including those that cause SARS, Middle Eastern respiratory syndrome (MERS), and now COVID-19 (Cui et al., 2019; Drosten et al., 2003; Zaki et al., 2012). SARS-CoV-2 has reached pandemic proportions and is likely to remain a world health emergency until an effective vaccine is widely available due to limited treatments and a high likelihood of recurrent outbreaks. The World Health Organization lists the primary symptoms of COVID-19 as fever, dry cough, and fatigue but other symptoms include diarrhea, loss of taste and smell, and rashes. Those more than 60 years of age and people with obesity, cardiovascular disease, and diabetes have the highest risk for severe COVID-19 (Ebinger et al., 2020; Grasselli et al., 2020). Most COVID-19 patients have mild respiratory illness; however, ~20% become seriously ill and require hospitalization due to pneumonia (Wu and McGoogan, 2020). This can progress into acute respiratory distress syndrome (ARDS) and systemic inflammation referred to as “cytokine storm” (Ye et al., 2020).
Instead of beneficial antiviral immunity in response to infection, severe COVID-19 is characterized by dysregulated immune responses that allow the virus to persist, causing lung damage, ARDS, and systemic inflammation (Giamarellos-Bourboulis et al., 2020). While mechanisms underlying SARS-CoV-2 evasion of antiviral immunity and pathogenic inflammation are not clear at this time, commonalities in the pathogenic response with this novel coronavirus and SARS-CoV-1 and MERS-CoV have become apparent (Felsenstein et al., 2020; Ye et al., 2020). Cells sense RNA viruses using endosomal and cytosolic pattern recognition receptors (PRRs) that signal through other mediators including tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3) and TRAF6 to activate interferon (IFN) regulatory factors (IRFs) and nuclear factor κB (NF-κΒ), resulting in transcription of early antiviral type I IFNs by resident alveolar macrophages (AMs) and epithelial cells in the lungs, which sets up an immune response that clears the virus and resolves inflammation (Lester and Li, 2014). SARS-CoV-1, and likely SARS-CoV-2, inhibits multiple viral sensing PRRs and downstream signals, effectively blocking recognition of virus and early antiviral type I IFN and initiating a dysregulated inflammatory cascade that can lead to ARDS and systemic inflammation (Hu et al., 2017; Li et al., 2016; Siu et al., 2009). Moreover, transcriptomic analysis of peripheral blood mononuclear cells (PBMCs) from COVID-19 patients found upregulated pro-inflammatory pathways in monocytes and CD4 T cells, suggesting that the basic hallmarks of the “cytokine storm” in COVID-19 parallel SARS and MERS (Wen et al., 2020). However, we are now also appreciating immunologic dysfunctions that may be causing a more severe disease course (Hadjadj et al., 2020; Lee et al., 2020; Wilk et al., 2020).
COVID-19 patients have higher circulating levels of interleukin-6 (IL-6), TNF-α, and C-X-C motif chemokine ligand 10 (CXCL10), particularly those with severe disease, and these “early” cytokines are sustained weeks into infection, suggesting an inability to resolve inflammation (Xiong et al., 2020; Yang et al., 2020). Adaptive immune cells recruited from nearby lymph nodes (via circulatory and lymphatic systems) can also contribute to pathogenic inflammation in the lung, particularly if polarized to T helper 1 (Th1) and Th17 responses that contribute to neutrophil recruitment and pro-inflammatory monocyte/macrophage activation (Wong et al., 2019). However, severe lung damage due to pneumonia or sepsis is more often characterized by a lack of adaptive immune cells in the periphery (Bermejo-Martin et al., 2017; Roth et al., 2003). This is due not only to migration of cells to sites of inflammation but also to T cell dysfunction and death (Hotchkiss et al., 2005; Nakajima et al., 2010; Roth et al., 2003). Prolonged antigen stimulation and pro-inflammatory cytokine exposure cause T cell exhaustion and apoptosis, leading to insufficient B cell activation and loss of the immune-resolving functions of T and B cells (De Biasi et al., 2020; Erickson et al., 2015; Kahan et al., 2015). Despite exuberant innate immunity, lymphopenia has been observed in COVID-19 and correlates with poor outcome (De Biasi et al., 2020; Diao et al., 2020). Characterization of T cells from COVID-19 patients found increased surface expression of exhaustion markers T cell immunoglobulin and mucin-domain containing-3 (Tim-3) and programmed cell death protein 1 (PD-1), decreased expression of pathways involved in T cell expansion, and increased expression of apoptotic pathways (Diao et al., 2020; Wen et al., 2020; Xiong et al., 2020). Therefore, dysfunction of circulating T and B cells during ARDS may result in an inability to resolve inflammation and perpetuate the systemic inflammation caused by the cytokine storm.
The innate-driven pathogenic inflammation and suppressed adaptive immunity during ARDS in COVID-19 patients indicate dysfunction in immune regulation and the switch from innate to adaptive immunity. To better understand this dysregulated immune response that drives patients to have more severe illness from SARS-CoV-2 infection, we used single-cell RNA sequencing (scRNA-seq) to analyze the transcriptome of PBMCs collected from hospitalized COVID-19 patients with a moderate course of disease (moderate group) and those that developed ARDS (severe group). Samples were collected within the first 5 days of hospitalization to minimize the potential for therapeutic interventions to confound interpretation of the data. As expected, when we compared the samples from acutely ill COVID-19 patients (moderate and severe groups) with samples from healthy controls, we found enrichment of viral-induced immunologic pathways. We next focused our analysis on the COVID-19 patient groups and investigated the signals that drive lung inflammation in ARDS. We also delineated novel pathways activated during the resolution of lung inflammation by comparing the severe group with the recovering group (i.e., those recovering from ARDS). Severely ill patients had an overall higher activation of inflammatory pathways than moderate and recovering patients. However, we unexpectedly found that patients with severe SARS-CoV-2 infection had monocytes with transcriptomic profiles consistent with a blunted IFN response and dysregulation of antigen presentation as well as defective humoral and lymphocyte cytotoxic activity that could be contributing to the delayed viral clearance found in severely ill COVID-19 patients. The implications of these results are clinically relevant and indicate that treatment of patients with ARDS from SARS-CoV-2 infection may require a targeted approach instead of broad immunosuppressive therapy.
Results
COVID-19 Patients Have a Hyperinflammatory Immune State
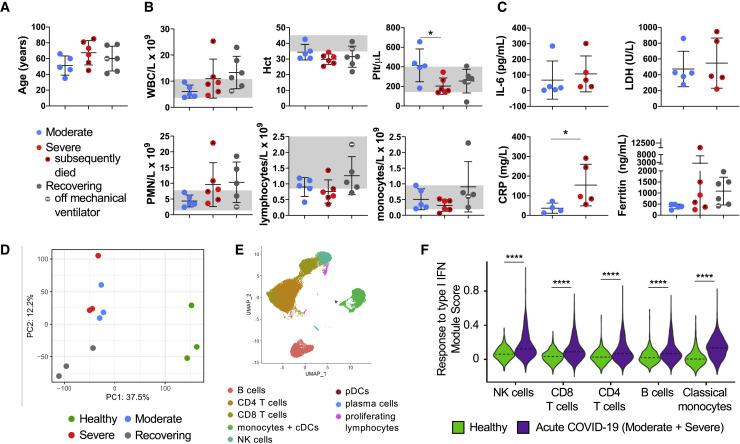
We identified and collected blood for buffy coat samples from hospitalized patients confirmed to have SARS-CoV-2 infection (Table S1). There were no significant differences in age, white blood cell count, cell subsets, and hematocrit between the three groups—acutely ill patients with moderate disease, acutely ill patients with severe disease, and patients recovering from ARDS—but the platelet count was decreased in the severe versus moderate group (Figures 1A and 1B). Using available clinical data, we compared the levels of various inflammatory markers in the blood between groups at the time of sample collection and identified a significantly higher level of C-reactive protein in patients with severe disease, reflecting the hyperinflammatory state relative to patients with moderate disease (Figure 1C).
Figure 1.
Evaluation of Blood Cells Subsets in Moderate, Severe (ARDS), and Recovering (post-ARDS) COVID-19 Patients
(A) Age distribution of hospitalized COVID-19 patients requiring minimal respiratory support (moderate, n = 5), with ARDS (severe, n = 6), and recovering from ARDS (recovering, n = 6). Mean ± SD overlaid on dot plot.
(B) Clinical complete blood count (CBC) with differential cell count for COVID-19 patients. One-way ANOVA with Tukey’s multiple comparisons was used to test significance. Normal ranges are indicated by gray shading. Mean ± SD overlaid on dot plot. ∗p < 0.05.
(C) Inflammatory markers in patients’ peripheral blood samples at admission (IL-6, lactate dehydrogenase [LDH], and C-reactive protein [CRP] were not available for the recovering group). Mann-Whitney test was used for IL-6, LDH, and CRP, and Kruskal-Wallis test with Dunn’s multiple comparison was used for ferritin. ∗p < 0.05.
(D–F) Peripheral blood leukocytes from COVID-19 patients were assessed by scRNA-seq in comparison with healthy controls. Principal component analysis (PCA) of patient pairs from the same group sequenced together demonstrates clustering by disease stage (healthy control, acute COVID-19, recovering ARDS), but does not separate moderately and severely ill patients (D). UMAP visualization reveals the major immune cell subsets (E). Violin plot of response to type I IFN module genes for each cell from healthy versus acute COVID-19 patients (moderate and severe groups combined; F). Kruskal-Wallis test was used to test overall significance in module scores, p < 2.2 × 10−16. Wilcoxon test was used for pairwise comparisons, ∗∗∗∗p < 0.0001.
Using scRNA-seq, we first compared the transcriptome of PBMCs from healthy controls and COVID-19 patients. Principal component analysis (PCA) of the samples demonstrated a clear segregation between healthy controls and COVID-19 patients, indicating that SARS-CoV-2 infection causes a profound perturbation in the circulating immune cell transcriptome (Figure 1D), but it did not clearly separate the moderate and severe COVID-19 patients. To understand the pathways activated by SARS-CoV-2 infection, we identified each of the major immune cell subtypes (natural killer [NK] cells, CD8 and CD4 T cells, B cells, and monocytes; Figure 1E; Figures S1A and S1B; Table S2) and performed Gene Ontology (GO) analysis of the significantly up- and downregulated differentially expressed genes between acutely ill COVID-19 patients (moderate and severe groups) and healthy controls (Figure S2; Table S3). Consistent with their paucity among circulating immune cells, insufficient conventional and plasmacytoid dendritic cells (cDCs and pDCs, respectively) were captured for analysis. Neutrophils were not identified because they, unlike other peripheral blood subsets, do not tolerate the cell freezing process (Wilk et al., 2020).
As expected, the response to type I IFN was activated in peripheral immune cells from COVID-19 patients compared with healthy controls (Figure 1F; Figure S2; Table S3).
Cytotoxic Lymphocytes Have a Higher IFN Response in Patients with Severe Disease but Reduced Expression of Cell-Killing Genes
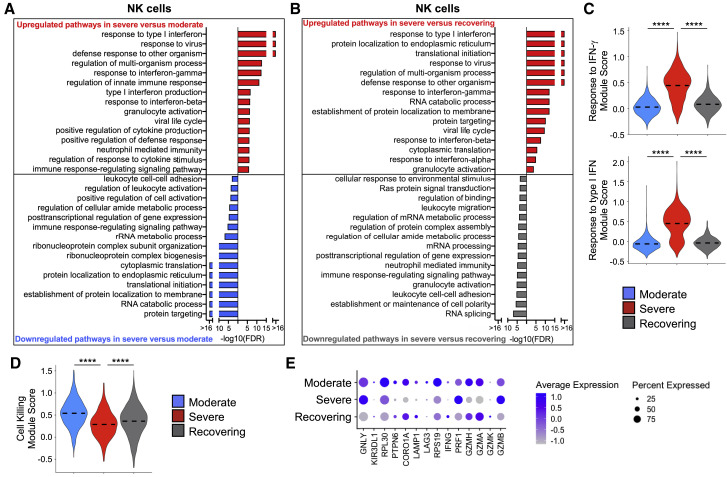
To understand why some patients have a less severe disease course, whereas others develop ARDS, we next analyzed immune cell populations within the scRNA-seq data from moderate, severe, and recovering COVID-19 patients (Figures S1C and S1D). NK cells are an important arm of the innate lymphocytic antiviral response (Abel et al., 2018). We compared the moderate versus severe group to identify differences in NK cell gene expression that may determine why some have a more benign course of disease compared with those that develop ARDS (Figure 2 ; Figures S3A–S3C; Table S4). Pathway analysis revealed differences in the NK cell transcriptome between groups. Biological processes that were highly represented by genes upregulated and downregulated in patients with severe disease compared with patients with moderate disease were identified (Figure 2A; Table S4). Patients with severe disease have higher respiratory viral loads (Zheng et al., 2020). Accordingly, biological processes such as “response to type I IFN,” “response to virus,” “response to IFN-γ,” and “response to IFN-β,” among several others characteristic of a higher viral response, were significantly upregulated in the severe disease group compared with the moderate disease group.
Figure 2.
NK Cells in Severe Patients Have Gene Expression Profiles Indicative of Higher Interferon (IFN) Signaling but Defective Cell Killing
(A and B) Global transcriptome differences between severe and moderate (A) and between severe and recovering (B) were evaluated in all NK cells by overrepresentation analysis of up- and downregulated biological processes.
(C and D) Violin plots of response to IFN-γ and response to type I IFN modules (C) and cell killing module (D) of each cell from patient groups. Kruskal-Wallis test was used to test overall significance in module scores, p < 2.2 × 10−16. Wilcoxon test was used for pairwise comparisons, ∗∗∗∗p < 0.0001.
(E) Average expression of differentially expressed genes (DEGs) involved in cytotoxicity from patient groups.
Comparing critically ill patients with ARDS during their acute illness (i.e., severe group) with those that are recovering from severe disease could provide insight into pathways that drive the non-resolving inflammation in ARDS pathogenesis (Matthay et al., 2012). Thus, we compared the biological processes that were affected by evaluating both up- and downregulated genes between severe and recovering groups (Figure 2B; Table S4). The recovering group exhibited resolution of the antiviral pathways, which would be expected from the waning respiratory viral load over time. The transcriptomic response to both types I and II IFNs was clearly upregulated in NK cells from the severe group in comparison with the moderate and recovering groups (Figure 2C; Figure S3A). Despite this response, which reflects a higher respiratory viral load in the severe group, expression of genes associated with cytotoxic function was lower than in the moderate and recovering groups, suggesting a dysfunctional effector antiviral response by NK cells in ARDS patients (Figures 2D and 2E; Figures S3B and S3C). Evaluating the data as pairs of samples from individual patients from the same group that were sequenced together (Figures S3A and S3B) rather than all cells from the patient groups (Figures 2C and 2D) revealed the same pattern of changes in IFN signaling and cytotoxic function.
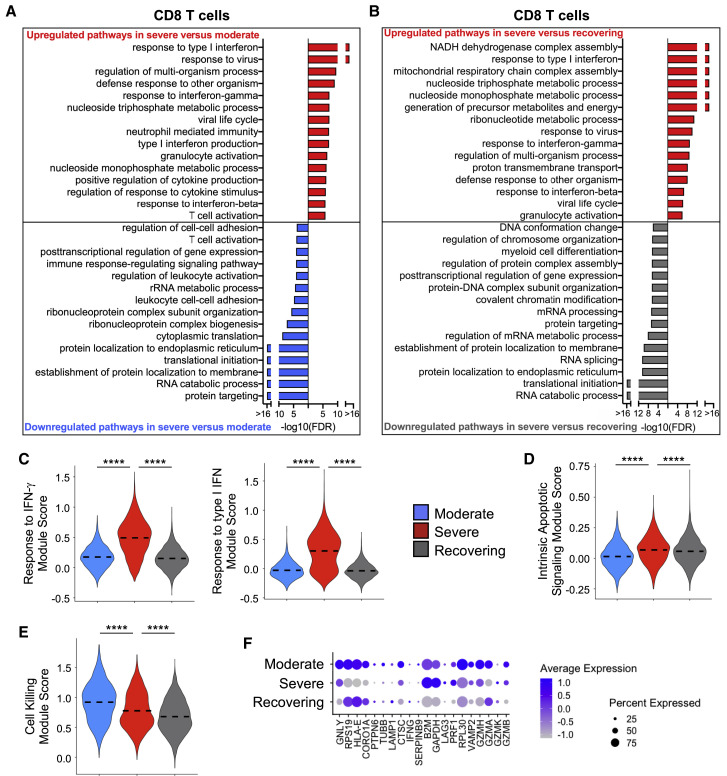
We performed a similar evaluation of CD8 T lymphocytes (Figure 3 ; Figures S3D–S3G; Table S5) to investigate the biological processes that differentiate the severity of disease and those that are activated during the recovery phase. CD8 T cells had appropriate activation of IFN signaling that correlated with the disease severity (Figures 3A–3C; Figure S3D). We also found a gene signature indicating increased apoptosis in CD8 T cells from the severe group, which is consistent with a prior report (Merad and Martin, 2020) and could be driving the lymphopenia associated with COVID-19 illness (Figure 3D; Figure S3E). Interestingly, CD8 T cells had a pattern of gene expression that suggested a deficiency in cytotoxic function in both the severe and recovering groups in comparison with patients with moderate disease (Figures 3E and 3F; Figures S3F and S3G). Although this pattern is expected in recovering patients who have resolving SARS-CoV-2 infection, the altered cytotoxic functional pathways in patients with ARDS mirror the findings of NK cells (Figures 2D and 2E).
Figure 3.
CD8 T Cells in Severe Group Patients Have Gene Expression Profiles Indicative of Higher IFN Signaling, Increased Apoptotic Gene Expression, and Defective Cell Killing
(A and B) Global transcriptome differences between severe and moderate (A) and severe and recovering (B) were evaluated in all CD8 T cells by overrepresentation analysis of up- and downregulated biological processes.
(C–E) Violin plots of response to IFN-γ and response to type I IFN modules (C), intrinsic apoptotic signaling module (D), and cell killing module (E) of each cell from patient groups. Kruskal-Wallis test was used to test overall significance in module scores, p < 2.2 × 10−16. Wilcoxon test was used for pairwise comparisons, ∗∗∗∗p < 0.0001.
(F) Average expression of DEGs involved in cytotoxicity from patient groups.
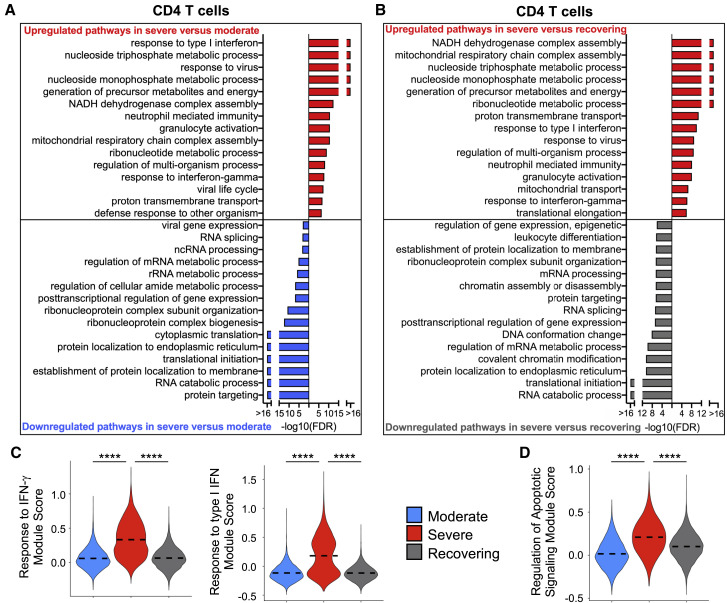
Next, we evaluated CD4 T lymphocytes to determine up- and downregulated biological pathways in moderate versus severe and severe versus recovering groups (Figure 4 ; Figures S3H and S3I; Table S6). Consistent with NK and CD8 T lymphocytes, the severe group had a transcriptome that was indicative of a higher IFN response in CD4 T cells than that of moderate group, and this was appropriately diminished in recovering patients (Figures 4A–4C; Figure S3H). In addition to the antiviral response, expression of gene sets in metabolic and apoptotic pathways was elevated in CD4 T cells in the severe group in comparison with the other groups (Figures 4A, 4B, and 4D; Figure S3I).
Figure 4.
CD4 T Cells in Severe Group Patients Have Gene Expression Profiles Indicative of Higher IFN Signaling, Increased Apoptotic Gene Expression, and Metabolic Activation
(A and B) Global transcriptome differences between severe and moderate (A) and severe and recovering (B) were evaluated in CD4 T cells (all T cells expressing CD4) by overrepresentation analysis of up- and downregulated pathways for biological processes.
(C and D) Violin plots of response to IFN-γ and response to type I IFN (C) and regulation of apoptotic signaling (D) modules of each cell from patient groups. Kruskal-Wallis test was used to test overall significance in module scores, p < 2.2 × 10−16. Wilcoxon test was used for pairwise comparisons, ∗∗∗∗p < 0.0001.
Taken together, these transcriptomic findings indicate that NK and T lymphocytes in severely ill COVID-19 patients, who have higher respiratory viral titers and delayed clearance of SARS-CoV-2 (Merad and Martin, 2020), have an expected stronger response to IFN signaling. Moreover, T lymphocytes have a gene signature consistent with increased activation of metabolic and apoptotic pathways. However, both innate and adaptive cytotoxic lymphocytes have dysfunctional cytotoxic-activity-associated pathways in severely ill COVID-19 patients
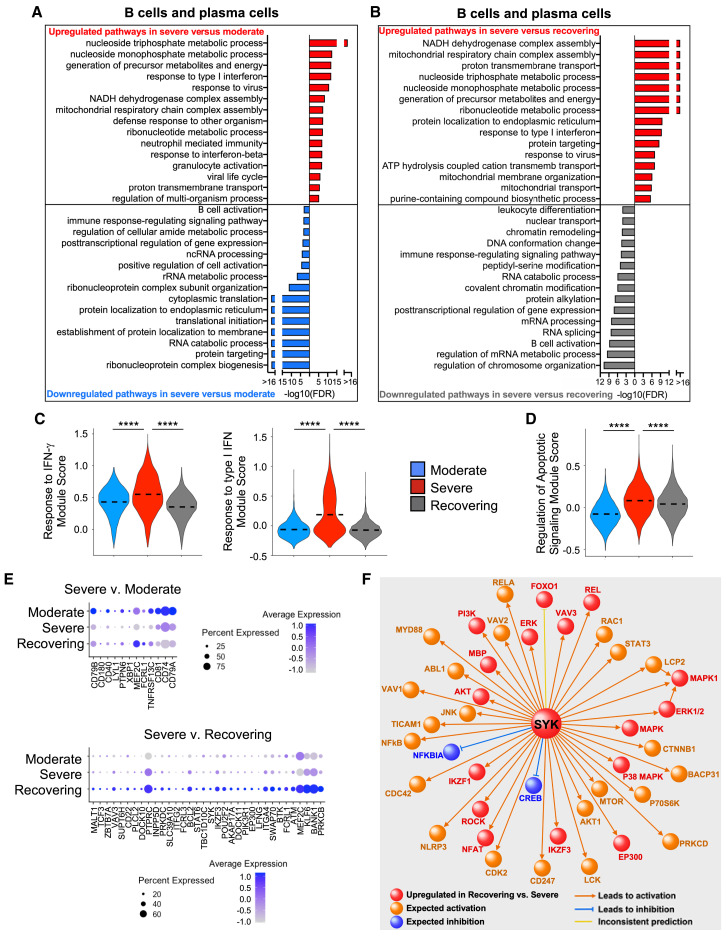
B Cells Have Gene Expression Profiles Indicative of Dysregulated Activation in Patients in the Severe Group
B lymphocytes also play an important role in the antiviral response (Chiu and Openshaw, 2015). We evaluated B cells and plasma cells to determine the differentially expressed genes between patient groups (Figure 5 ; Figures S4A and S4B; Table S7). Similar to NK and T lymphocytes, the B lymphocytes also had changes in their gene signature that were consistent with higher IFN-mediated responses and activation of apoptotic signals in patients in the severe group compared with those in both the moderate and recovering groups (Figures 5A–5D; Figures S4A and S4B). Moreover, transcriptomic changes suggested B cell activation was upregulated in the recovering group in comparison with the severe group, which is the expected response during the recovery phase (Figure 5E). By contrast, B cell activation genes were reduced in the severe group compared with that of patients in the moderate group, supporting the concept that a delayed humoral response could be contributing to the severity of disease (Figures 5B and 5E).
Figure 5.
B Cells in Severe Group Patients Have Gene Expression Profiles Indicative of Higher IFN Signaling, Increased Apoptotic Gene Expression, and Defective Activation
(A and B) Global transcriptome differences between severe and moderate (A) and severe and recovering (B) were evaluated in B cells and plasma cells by overrepresentation analysis of up- and downregulated pathways for biological processes.
(C and D) Violin plots of response to IFN-γ and response to type I IFN (C) and regulation of apoptotic signaling (D) modules of each cell from patient groups. Kruskal-Wallis test was used to test overall significance in module scores, p < 2.2 × 10−16. Wilcoxon test was used for pairwise comparisons, ∗∗∗∗p < 0.0001.
(E) Average expression of DEGs in the B cell activation pathway between moderate and severe and severe and recovering groups.
(F) IPA causal pathway analysis demonstrates that SYK is the primary upstream mediator of the upregulated pathways in B cells from recovering versus severe groups.
In order to understand the pathways that are activated in B cells during the resolution from SARS-CoV-2 infection, we used ingenuity pathway analysis (IPA) to evaluate canonical pathways enriched among the 642 genes that were upregulated in the recovering group compared with the severe group. The top canonical pathways enriched among these genes were B cell receptor signaling (false discovery rate [FDR] of 2.5 × 10−11), IL-3 signaling (FDR of 4.0 × 10−6), and phosphatidylinositol 3-kinase (PI3K) signaling (FDR of 3.9 × 10−7), which are important signals required for B cell memory and the humoral response (Table S7). We performed causal network analysis to understand the upstream signals that regulate the differential upregulation of genes in the patients in the recovering group relative to the severe group and identified spleen tyrpsine kinase (SYK), which is a critical controller of B cell differentiation, maturation, and signal transduction by the B cell receptor, as the top upstream regulator candidate among the differentially expressed genes (Figure 5F) (Mócsai et al., 2010).
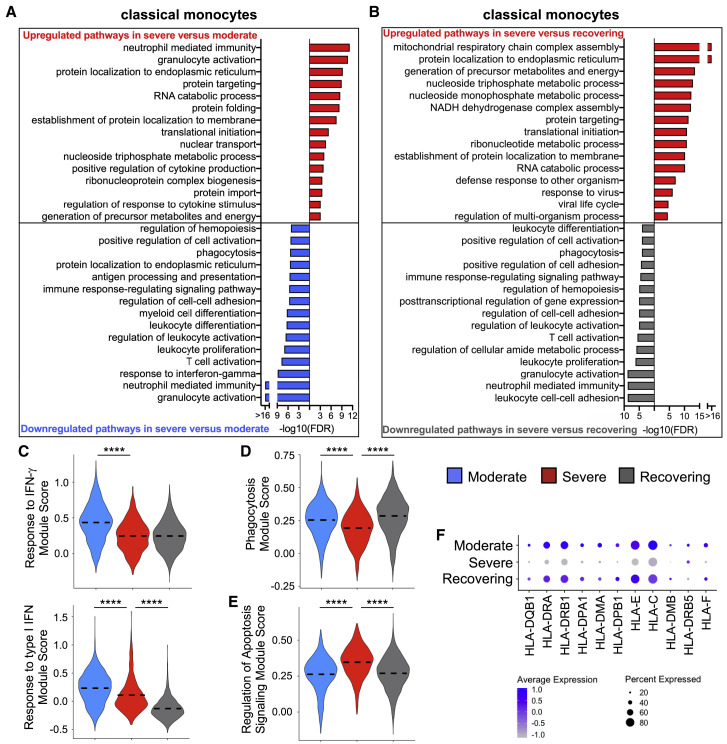
Monocyte Gene Expression Profiles Indicate That They Are Hyporesponsive in Patients in the Severe Group
Emerging evidence demonstrates disruptions to the myeloid compartment in COVID-19 patients (Wilk et al., 2020). As expected, classical monocytes comprised the majority of the cells (Figures S4C and S4D). Surprisingly, classical monocytes had a gene signature consistent with a decreased activation state, impaired phagocytosis, and altered differentiation in the severe compared with moderate group (Figures 6A and 6D; Figure S4E; Table S8). This functional deficiency in classical monocytes appears to recover during the resolution of ARDS, suggesting that improvement in monocyte function could facilitate the resolution of inflammation in COVID-19 patients who develop ARDS (Figures 6B and 6D; Figure S4E). Moreover, monocytes in the severe group had more activation of apoptotic signals (Figure 6E; Figure S4F) and dysfunction in antigen processing and presentation, including lower expression of major histocompatibility complex class I (MHC class I) and MHC class II (Figures 6A and 6F). Additionally, in contrast to the lymphocytes, monocytes in severely ill patients had a deficient transcriptomic response to IFN signaling in comparison with moderately sick subjects that could at least partially explain the hypofunctional state that we identified in this cell compartment (Figure 6C; Figure S4G).
Figure 6.
Classical Monocytes in Severe Group Patients Have Decreased Gene Expression for IFN Signaling, Phagocytosis, and Antigen Presentation
(A and B) Global transcriptome differences between severe and moderate (A) and severe and recovering (B) were evaluated in classical monocytes (CD14+CD16−) by overrepresentation analysis of up- and downregulated pathways for biological processes.
(C–E) Violin plots of response to IFN-γ and response to type I IFN (C), phagocytosis (D), and regulation of apoptotic signaling (E) modules of each cell from patient groups. Kruskal-Wallis test was used to test overall significance in module scores, p < 2.2 × 10−16. Wilcoxon test was used for pairwise comparisons, ∗∗∗∗p < 0.0001.
(F) Average expression of differentially expressed human leukocyte antigen (HLA) genes by classical monocytes from patient groups.
Potential Mechanisms Driving the Immunoparalysis of Specific Immune Cells
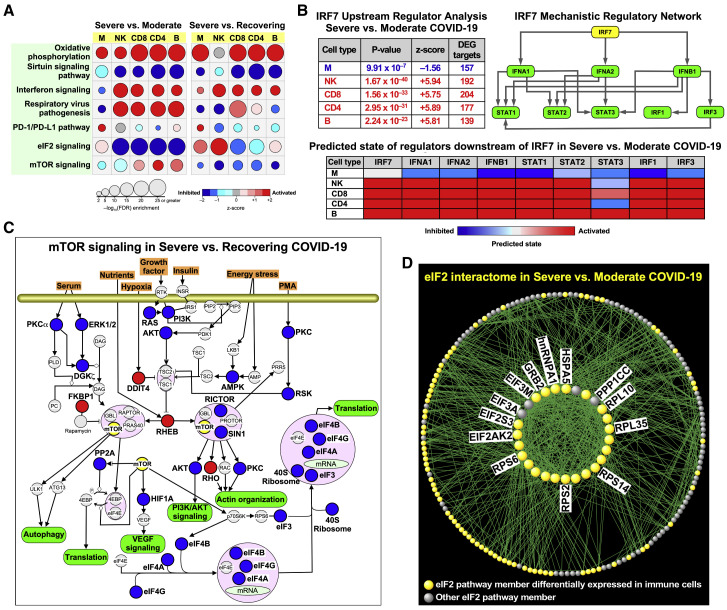
To further evaluate the potential mechanisms that were driving the selective immunodeficiencies identified within various cell compartments, we performed a canonical pathway analysis with IPA. In contrast to standard GO analysis where activation or inhibition of a process is inferred from up- and downregulation of its member genes, respectively, canonical pathway analysis incorporates prior knowledge of the overall consequences of up- and downregulation of individual members of a process in activating or suppressing that pathway (e.g., downregulation of an inhibitor can lead to the activation of a process). In this analysis, we found cell-specific enrichment patterns; for example, CD4 and CD8 T cells had significant activation of the T cell exhaustion pathway in the severe versus the moderate group (Table S9), which is consistent with another report (Wilk et al., 2020).
We next explored patterns of altered signaling pathway changes across all cell compartments (Figure 7 A). Oxidative phosphorylation pathways were uniformly elevated in the severe group compared with the moderate and recovering groups, likely reflecting the increased metabolic state with more severe infection. By contrast, signaling by sirtuins, which are known mediators of antiviral defenses (Budayeva et al., 2015; Koyuncu et al., 2014), was suppressed in severe conditions across all cell compartments. As with our GO evaluation of biological processes, IFN signaling was decreased in monocytes when comparing severe with moderate groups. Similarly, respiratory viral signaling pathways were also uniquely suppressed only within monocytes when comparing severe with moderate groups. Notably, PD-1/programmed death-ligand 1 (PD-L1) signaling, an immunologic checkpoint inhibitory pathway, was higher in monocytes within the severe group, which could be contributing to the suppressed response of this cellular compartment.
Figure 7.
Canonical Pathway Analysis Demonstrates Defective Signaling Programs across All Immune Cells in Severely Ill COVID-19 Patients
(A) Significantly enriched pathways activated or inhibited between severe versus moderate and severe versus recovering groups in all cell types. An FDR < 0.01 was used to designate a pathway as significantly enriched, and z-score was applied to determine the activation/inhibition state of a given pathway in the severe condition.
(B) Upstream regulator analysis of DEGs identified IRF7 as a putative master regulator across all cell types. Top left: the mechanistic regulatory network, with IRF7 as the key orchestrator, was constructed based on the overlap between the patterns of differential gene expression and IPA’s knowledge base across immune cell types. Top right: each member of this network is itself a key regulator of many other DEG targets in each cell type. Bottom: a heatmap summary highlighting whether each regulator is expected to be activated or inhibited for each immune cell population in severe versus moderate groups.
(C) mTOR canonical pathway in severe versus recovering group. The up- (red) and downregulated (blue) nodes are based on composite information across all cell types. Some of the nodes do not represent a single DEG but potentially a family of genes (e.g., 40S ribosome).
(D) Gene product interaction network analysis of the eIF2 pathway, which is downregulated in lymphocytes, but not monocytes, in the severe versus moderate group. A majority of the interactome’s nodes were differentially expressed across cell types, particularly within the densely connected network hubs shown in the center (selectively labeled in the figure and fully detailed in Table S9).
IFN signaling (types I and II) was a prominent pathway that was repeatedly enriched in our comparison of the severe versus moderate groups. We used IPA to perform an upstream regulator analysis for each cell type (severe versus moderate) based on the cell type’s pattern of differential gene expression and also identified IRF7 as one of the most significant regulators consistently enriched in lymphocytes (Figure 7B). We constructed a mechanistic network with IRF7 as the master regulator and found that type I IFN signaling components were enriched as highly interactive nodes (Krämer et al., 2014). Using this framework, we assessed which of the hierarchical components were activated or inhibited in our data and found that lymphocyte subsets had the expected activation, whereas monocytes had inhibition of type I IFN pathways (Figure 7B), which is consistent with our prior analysis. We also compiled the genes identified by GO analysis across all immune cell populations to evaluate common genes that were enriched in response to IFN signaling (Figures S5A–S5C). The lymphocyte subsets from patients with severe disease had similarly elevated type I IFN responses (consistent with enhanced IRF7 expression). By contrast, the classical monocytes from patients in the severe group were no more responsive than those from the moderate group, with lower expression of IRF1 and IRF9, perhaps due to subversion of Toll-like receptor (TLR) and retinoic acid-inducible gene I (RIG-I) signaling by SARS-CoV-2, as was previously reported for SARS-CoV-1 (Hu et al., 2017; Li et al., 2016; Siu et al., 2009). Consistent with this analysis, GO pathway analysis revealed a reduction in the response to IFN-γ compared with healthy controls and a weaker type I IFN response in monocytes from severe patients, which contrasts with the stronger lymphocytic responses (Figures S5D–S5H).
Finally, eukaryotic translation initiation factor 2 (eIF2) signaling, a known target of many respiratory viruses (Groskreutz et al., 2010; Liu et al., 2020; Rabouw et al., 2020), was the most significantly downregulated pathway across all cells (except monocytes) when comparing severe to moderate groups, whereas mammalian target of rapamycin (mTOR) signaling was depressed in the severe versus recovering groups (Figure 7A). We investigated the mTOR canonical pathway in the severe versus recovering groups and noted that several nutrient-sensing and stress response processes were downregulated in severe COVID-19 patients relative to those with moderate disease (Figure 7C). To better define how the components of the eIF2 pathway were altered in immune cells, we performed a gene product interaction analysis using IPA. This network was built by initially including all eIF2-associated members and limiting gene product relationships to those with experimentally validated direct interactions. We observed that many members of this network were differentially expressed in patients with severe versus moderate COVID-19 including eIF1 and eIF2 as well as a number of ribosomal genes such as ribosomal protein S6 (RPS6) that were previously reported to be downregulated in influenza virus infection (Figure 7D; Table S9) (Zhai et al., 2015). This overrepresentation was more pronounced when assessing the most highly connected network nodes—i.e., hubs—suggesting that key drivers of the eIF2 pathway are affected in severe SARS-CoV-2 infection.
Discussion
ARDS is driven by a dysregulated inflammatory response that has been largely described as a “cytokine storm” associated with severely ill patients infected with SARS-CoV-2. A unique feature of the lung injury that perpetuates ARDS is a non-resolving inflammation even after the inciting factor, in this case SARS-CoV-2 infection, has resolved (Matthay et al., 2012). A growing literature has provided a better understanding of how immune cell dysfunction contributes to the inflammatory response in COVID-19 patients (Giamarellos-Bourboulis et al., 2020). Several groups have adopted scRNA-seq as an approach to functionally interrogate distinct immune compartments (Hadjadj et al., 2020; Lee et al., 2020; Wen et al., 2020; Wilk et al., 2020). Our data are largely consistent with those findings, but our study further delineates the profound immune dysregulation in COVID-19 patients with severe illness compared with those with moderate symptoms and those recovering from ARDS. Moreover, our findings were consistent with prior scRNA-seq studies that compared the transcriptomes of PBMCs from healthy controls versus COVID-19 patients and identified an inflammatory signal that increased with disease severity (Hadjadj et al., 2020; Wilk et al., 2020). Focusing our analysis to COVID-19 patients with varying severity allowed us to deeply interrogate the immune cell dysfunction and differentiate those that have moderate disease from patients that develop ARDS. Our analysis revealed that although most immune cellular compartments had an expected hyperinflammatory response in severe patients, several key pathways were dysfunctional in severely ill patients, which could be contributing to their inability to control the viral infection. Indeed, PBMCs from the severe group had a transcriptomic signal suggesting deficiencies in functions necessary to clear the virus: cytotoxic killing in NK and CD8 T cells, B cell activation, and impaired antigen presentation by monocytes. Together, these findings indicate that distinct functional impairments in innate and adaptive immune responses during SARS-CoV-2 infection contribute to the disease severity.
One of the first immunologic features identified in COVID-19 patients was decreased but hyperactive lymphocytes (Song et al., 2020; Xu et al., 2020). Our data suggest that both CD4 and CD8 T lymphocytes have a normal response to the viral pathogen with activation of antiviral pathways and a resultant hypermetabolic state. Notably, we found an increase in mitochondrial respiratory chain activity in the severe group, which could reflect a mitochondrial antiviral signaling (MAVS) response to the SARS-CoV-2 infection (Moreno-Altamirano et al., 2019; Shi et al., 2014). It has also been suggested that lymphocytopenia results from increased apoptosis, which is consistent with our findings (Vabret et al., 2020; Xiong et al., 2020). Additionally, our analysis indicates that CD8 T lymphocytes have a deficiency in cell killing that could be contributing to the pathobiology of ARDS in COVID-19 patients. Similarly, NK cells had a transcriptomic signature that indicated defective effector killing of virally infected cells in patients with severe disease. Clinical data on COVID-19 patients reported high respiratory viral load and impaired clearance in severe disease cases compared with moderate cases (Shi et al., 2020). Additionally, studies in COVID-19 patients have found that SARS-CoV-2 persisted longer in respiratory samples of patients with severe disease than in those with milder symptoms, and this delayed viral clearance coincides with more lung damage from the infection (Chang et al., 2020; Zheng et al., 2020). Collectively, these results demonstrate a functional defect in both the innate and adaptive cytotoxic lymphocytic responses in COVID-19 patients with ARDS that could be contributing to the severity of disease.
Successful viral clearance relies on adaptive T cell and B cell immunity. Whereas cytotoxic T lymphocytes induce apoptosis of virus-infected cells, CD4 T cells activate B cells to differentiate into antibody-producing plasma cells and memory B cells (Liao et al., 2017; Vos et al., 2000). Delayed immunoglobulin M (IgM) has been reported in severely ill COVID-19 patients, suggesting a defect in B cell function, although it is unclear whether this is due to direct defects in B cells or in CD4 Th cells (Shen et al., 2020). Our findings suggest CD4 T lymphocytes have normal activity despite the decreased numbers described in COVID-19 patients. By contrast, pathways involved in B cell activation were downregulated in the severe group, suggesting an inherent dysfunction in the B lymphocyte compartment that limits their activity. Moreover, SYK, which is essential for B cell activation, was identified as the most significant upstream regulator that was downregulated in B cells within the severe group (Cheng et al., 1995; Cornall et al., 2000). Although recovery from SARS-CoV-2 infection is not fully dependent on B cell function and antibody production (Fallet et al., 2020), these immune cells likely still have an important role in controlling the severity of disease, and a dysfunctional humoral response may result in higher viral titers and delayed viral clearance that could contribute to the development of ARDS during SARS-CoV-2 infection.
Much attention has been devoted to lymphocyte dysfunction in COVID-19, particularly to the importance of cytotoxic T cells in overcoming viral infection (De Biasi et al., 2020; Diao et al., 2020; Song et al., 2020); yet, the most dramatic difference we found in the severe group was a marked decline in monocyte function. We identified excessive activation of endoplasmic reticulum (ER) stress in monocytes with a concomitant increase in apoptosis, which could be augmenting death of these immune cells in the severe group. Peripheral blood monocytes also had a deficiency in their response to IFN signaling. Type I IFN activity is important during an appropriate antiviral immune response (Acharya et al., 2020), but a recent study demonstrated that patients with severe COVID-19 have an apparent deficiency in type I IFN signaling (Hadjadj et al., 2020). Our findings are congruous with this finding but further reveal that the deficient type I IFN response emanates from the monocytic compartment in contrast to lymphocytes, which had an appropriate response. Circulating monocytes from the severe group may be exhibiting immunoparalysis (Giamarellos-Bourboulis et al., 2020), as our results imply that they were not responsive to IFN signaling. Sudden loss of monocytes and their MHC expression is an indicator of severe acute illness in other conditions characterized by “cytokine storm” such as sepsis and community acquired infections (Cajander et al., 2016; Lekkou et al., 2004). We also found that monocytes were characterized by decreased expression of class I and II MHC genes in the severe group, consistent with other descriptions (Giamarellos-Bourboulis et al., 2020; Wilk et al., 2020), which may be related to reduced responsiveness to both type I IFN and IFN-γ. A concomitant augmentation of PD-1/PD-L1 signaling was found in monocytes, which could lead to selective immunosuppression in this population. Additionally, we postulate that suppressed protein translation by an augmented unfolded protein response in monocytes may be further contributing to the immunoparalysis phenotype identified in the myeloid compartment. We also identified IRF7 as a master regulator that could be driving the aberrant type I IFN response in severe conditions, perhaps as a consequence of SARS-CoV-2 suppression of TLR/RIG-I signaling (Hu et al., 2017; Li et al., 2016; Siu et al., 2009). Consequently, altered antigen presentation by monocytes can also result in the observed defect in T cell function in the severe group, resulting in inhibition of the downstream adaptive immunity necessary for viral clearance and resolution of systemic inflammation (Jakubzick et al., 2017).
Several cytokines have been found to be dysregulated in COVID-19 patients (Vabret et al., 2020). Type I IFNs are crucial for a successful antiviral immune response, and subverting the early type I IFN response contributes to pathogenesis in MERS-CoV, SARS-CoV-1, and SARS-CoV-2 infections (Hadjadj et al., 2020; Lee et al., 2020; Siu et al., 2009; Yang et al., 2015). Clinical trials with type I IFN therapy are ongoing; however, this treatment may be harmful if patients are already upregulating IFN-stimulated genes. In fact, our pathway analysis suggests adaptive immune cells have a normal IFN response, but monocytes have a selective dysregulation with a depressed IFN response in patients with severe COVID-19. Furthermore, our findings suggest that monocytes in patients in the severe group have an inherent defect in their ability to respond to IFN signaling, and this dysfunction may not be ameliorated by simply treating patients at high risk for developing ARDS from SARS-CoV-2 infection with type I IFN (Acharya et al., 2020).
SARS-CoV-2 infection of immune cells may be causing the dysregulated response in PBMCs. However, immune cells do not express angiotensin-converting enzyme (ACE2), the receptor required for viral entry, suggesting an inability for viral infection (Hamming et al., 2004; Hoffmann et al., 2020; Radzikowska et al., 2020; Zou et al., 2020). Moreover, we did not find any evidence of ACE2 expression or any viral RNA in our scRNA-seq of PBMCs (unpublished data). These observations make it less likely, but do not exclude the possibility, that viral infection of immune cells is antagonizing inflammatory signaling pathways. An alternative explanation is that genetic, environmental, and age-related changes could confer a predisposition to a dysregulated response to pro-inflammatory signals. This concept has been previously demonstrated where certain individuals have attenuated, while others have augmented, inflammatory responses upon LPS stimulation of PBMCs (Wurfel et al., 2005). Recently, a loss-of-function mutation in TLR7 was found to decrease the IFN response and was associated with the development of severe COVID-19 in small group of young men (van der Made et al., 2020). Aging, a risk for more severe COVID-19 illness, could also be contributing to immunoparalysis (Ebinger et al., 2020; Fulop et al., 2018; Grasselli et al., 2020). Indeed, sirtuin signaling was uniformly depressed in all immune cell compartments of the severe group irrespective of comparison with moderate or recovering COVID-19 patients. Sirtuins are histone deacetylases that have diverse effects in antiviral defense and control of longevity, inflammation, and cellular senescence (Budayeva et al., 2015; Imai and Guarente, 2014; Koyuncu et al., 2014). In particular, sirtuin activity decreases with age, and immunosenescence could be driving some of the dysregulated immune response in severe COVID-19 patients (Fulop et al., 2018). Obesity is another risk factor for development of severe COVID-19 (Ebinger et al., 2020), and immunometabolic alterations could be contributing to the dysfunctional immune response in COVID-19 patients. The immunomodulatory effects of obesity can alter both the innate and adaptive arms of the immune system and have profound effects in tissue inflammation (Hotamisligil, 2017). Autophagy has a prominent role in immunometabolism (Rathmell, 2012), and our findings demonstrate disrupted mTOR signaling in multiple cellular compartments of severe patients particularly when compared with recovering patients, suggesting another possible link to the dysregulated immunologic findings in COVID-19 patients.
In summary, our study supports the concept that COVID-19, and especially severe cases that have progressed to ARDS, is characterized by multifaceted immune dysregulation that is not uniformly hyperinflammatory but more accurately described as a state of immune imbalance. Although immune cells have augmented inflammatory signatures in severe COVID-19 patients compared with the moderate and recovering patient groups, transcriptomic analysis suggests distinct patterns of immunoparalysis such as impaired cytotoxic cell killing, attenuated B cell activation, and dysregulated monocyte antigen presentation that could contribute to the severity of illness. It is unclear whether the divergent immunological responses in patients with a moderate-versus-severe disease course are specific to SARS-CoV-2 infection or generalizable to other infectious and non-infectious causes of ARDS, and future research using larger sample sizes is needed to further delineate the transcriptional landscape of immune cells in different ARDS populations. However, our study suggests that therapies should consider a nuanced approach, particularly those involving targeted augmentation of pathways within specific immune compartments to limit disease severity as well as promote resolution of the unrelenting inflammation in COVID-19-associated ARDS.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Human adult PBMC from COVID-19+ patient | Cedars Sinai Medical Center BioBank and Translational Research Core http://www.cedars-sinai.org/research/cores/biobank-translational.html | Patient information is available as Table S1 |
| Critical Commercial Assays | ||
| Single Cell 3′ Next GEM V3.1 | 10X Genomics | PN-1000121 |
| KAPA Library Quantification Kit | Roche | KK4824 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE154567 |
| Healthy PBMC dataset | Chen et al., 2018 | GEO: GSE112845 |
| SARS-COV2 genome | https://www.ncbi.nlm.nih.gov/nuccore/MT246667.1 | GenBank: MT246667.1 |
| Human reference transcriptome GRCh38 | https://www.gencodegenes.org/human/ | GRCh38.p13 |
| Software and Algorithms | ||
| CellRanger v3.0.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| R package Seurat v.3.1.5 | Stuart et al., 2019 | https://github.com/satijalab/seurat |
| R package SoupX | (Young and Behjati, 2018) | https://github.com/constantAmateur/SoupX |
| Harmony | Korsunsky et al., 2019 | https://github.com/immunogenomics/harmony |
| R package MAST | Finak et al., 2015 | https://github.com/RGLab/MAST |
| Webgestalt v2019 | Liao et al., 2019 | http://www.webgestalt.org |
| Ingenuity Pathway Analysis | QIAGEN | N/A |
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| FACS Aria III | BD Biosciences | N/A |
| 10X Chromium Controller | 10X Genomics | Cat.#1000202 |
| Novaseq 6000 | Illumina | N/A |
Resource Availability
Lead Contact
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Peter Chen (peter.chen@cshs.org).
Materials Availability
This study did not generate new unique materials.
Data and Code Availability
The accession number for the COVID-19 patient scRNA-seq data reported in this paper is GEO: GSE154567.
Experimental Model and Subject Details
Ethics statement
This study was approved by the Institutional Review Board at Cedars-Sinai Medical Center (CSMC; IRB# STUDY00000602). Informed consent was obtained from all enrolled patients according to CSMC BioBank’s Phase 2 Protocol (IRB# PRO00043021).
Study design and sample collection
Patients admitted to CSMC and diagnosed with COVID-19 by RT-PCR of nasopharyngeal swabs were stratified into moderate, severe, and recovering groups (n = 6/group). One sample in the moderate group was lost during processing. The patient cohort included 9 females and 8 males between the ages of 35-85 (Refer to Table S1 for details). Analysis by sex was not done due to the small sample size per group. Patients with moderate disease had minimal oxygenation requirements (≤4 l via nasal cannula) at the time of sample collection. Severe disease included critically ill patients admitted to the medical intensive care unit and requiring mechanical ventilation for acute respiratory distress syndrome (ARDS) during their hospitalization. Instead of sample collection based on days of symptoms, which is subjective and liable to variability, we chose to collect samples within 5 days of admission from moderate and severe groups to maintain uniformity of timing for comparison between groups based on disease severity and reduce the potential for therapeutic interventions to confound interpretation of differences in immune cell gene expression between patients and groups. Furthermore, we collected samples from a recovering group comprised of patients 18 to 25 days after admission who were recovering from ARDS secondary to SARS-CoV-2 infection. Venous blood was collected into EDTA coated tubes and centrifuged to separate plasma and buffy coat. Plasma was collected and frozen at −80C, and the buffy coat was collected into cryo-preservation media and frozen at −80C. Our analyses focused on comparing differences between the moderate versus severe and severe versus recovering groups. Because the moderate and recovering groups had different acuity and severity, these two groups were not directly compared with each other.
Method Details
Sample processing and single cell RNA-seq
The frozen buffy coats were thawed and washed with PBS containing 10% FBS. The cells were stained with DAPI (3μM) for 5 min to assess viability. Live cells were sorted using a BD FACS Aria III (BD Biosciences, San Jose, CA, USA) in the CSMC Flow Cytometry Core. Pairs of samples from the same patient group were mixed together at a 1:1 ratio before methanol fixation using the 10X Genomics methanol fixation protocol (Chen et al., 2018). Single cells were captured using a 10X Chromium Controller (10X Genomics) and libraries were prepared according to the Single Cell 3′ Next GEM V3.1 Reagent Kits User Guide (10X Genomics). The barcoded sequencing libraries were quantified by quantitative PCR using the KAPA Library Quantification Kit (KAPA Biosystems, Wilmington, MA). Libraries were sequenced using a Novaseq 6000 (Illumina) with custom sequencing settings of 28bp and 91bp for read 1 and 2, respectively, to obtain a sequencing depth of ~5x104 reads per cell.
Alignment, Demultiplexing, Quality Control and Batch Correction
CellRanger v3.0.0 software was used with the default settings for demultiplexing, aligning reads with STAR software to a custom human GRCh38 transcriptome reference downloaded from https://www.gencodegenes.org/, containing all protein coding and long non-coding RNA genes based on human GENCODE version 33 annotation with SARS-Cov2 virus genome MT246667.1, https://www.ncbi.nlm.nih.gov/nuccore/MT246667.1.
Single cell analysis R package Seurat v3.1.5 was used for data analysis (Stuart et al., 2019). For quality control and filtering out low quality cells, only cells expressing more than 200 genes (defined as genes detected in at least 3 cells) and fewer than 20% mitochondrial genes were selected. To minimize doublet contamination for each dataset, we removed cells with a high number of genes detected using a fit model generated from suggested mulitplet rate over number of cells recovered as in the 10X Genomics user manual. Ambient RNA derived from lysed cells was removed using SoupX package (Young and Behjati, 2018). A total number of 69983 captured single cells (about 4000 cells per patient) passed quality control for further batch correction and unbiased clustering. We used default normalization and data scaling from the Seurat package, which is a log normalization and linear model for data scaling. Batch correction package Harmony with Seurat 3 wrapper was used for data integration (Korsunsky et al., 2019). The batch correction was processed with PCA (Principal Component Analysis) using the 5000 most variable genes, and the first 20 independent components were used for downstream unbiased clustering with a resolution of 0.4. The UMAP (Uniform Manifold Approximation and Projection) method was used for visualization of unsupervised clustering. We used the Seurat RunUMAP function for UMAP reduction using the first 20 harmonized dimensions for immune cell types and 30 harmonized dimensions for immune cell subsets. We applied default setting embedding in Seurat RunUMAP function, with min.dist of 0.3 and n_neighbors of 30. Cell cluster identities were determined using known gene markers of individual cell types. Differentially expressed genes between different clusters and groups were calculated using Mode-based Analysis of Single-cell transcriptomics (MAST) (Finak et al., 2015). Major immune cell types were assigned according their expression of known cell type specific gene signatures (Figure S1; Table S2). Erythrocyte and Platelet clusters were removed from further analysis. Lymphocyte clusters without obvious subset markers and with relatively lower UMI counts were annotated as “undefined lymphocytes” and were excluded from further analysis. Three methanol fixed healthy PBMC datasets generated using the Chromium Single Cell 3′ Reagent v2 Kit were downloaded from GEO: GSE112845 (Chen et al., 2018). Each individual dataset went through the same QC step as the COVID-19 patient samples. To reduce the technical variability caused by different versions of 10x Chromium Single Cell 3′ Reagent kits, when comparing healthy samples with moderate and severe COVID-19 patient samples, ribosomal and mitochondrial genes were removed from the analysis.
Pathway analysis and module scores
To understand the biological processes that were enriched by the differentially expressed genes within each immune compartment, we sorted differentially expressed genes (FDR < 0.01) into those that were upregulated or downregulated between two groups (i.e., moderate versus severe and severe versus recovering) and did further analysis of gene lists separately. Using Webgestalt (Version 2019) (Liao et al., 2019), gene lists were entered to do an over-representation analysis using the Gene Ontology (GO) database to identify the biological processes that were affected by the differentially expressed genes between severity groups in each cell type.
In order to complement our GO analysis, we also performed canonical pathway analysis on all major immune cell types using Ingenuity Pathway Analysis (IPA, QIAGEN). Enrichment analysis on differentially expressed genes (FDR < 0.01) for each cell type (Classical monocytes, NK, CD8 T cell, CD4 T cell, and B cell) across conditions (Moderate, Severe, Recovering) was performed by leveraging over 700 manually curated pathways using Fisher’s exact test with Benjamini Hochberg adjusted P-values (FDR). Additionally, pathway activity analysis was performed to assess whether significantly enriched pathways (FDR < 0.01) were activated or inhibited based on IPA’s knowledgebase of expression or phosphorylation patterns of gene products in a given canonical pathway using a Z score statistic. While each cell type and condition elicited a distinct set of enriched and differentially activated/inhibited pathways, we focused on the most significant programs shared by immune cells under different clinical states.
Upstream regulator analysis was performed by using differentially expressed genes (FDR < 0.01) for each immune cell type as input seeds. The direction of expression of these genes was compared to IPA’s knowledge base using a statistical model (Krämer et al., 2014) to identify key putative regulators and construct a mechanistic regulatory network. An overlap P-value was used to measure enrichment of network-regulated genes in the dataset and an activation Z score was calculated to identify likely regulating molecules based on statistically significant patterns of up- and downregulation as well as expected activation state (activated or inhibited) of each regulator.
Pathway module scores within each immune compartment were determined using the union of differentially expressed genes in all COVID-19 groups returned from GO over-representation analysis for biological processes of interest. Pathways were defined by the GO database. Genes present from the sorted differentially expressed genes that are attributed to a biological process or pathway of interest by the GO database were used to form the modules, and module genes were selected based on actual enriched GO processes. Pathway module scores were calculated using the AddModuleScore function of the Seurat package that calculated the average expression of each gene signature list and subtracted by the aggregated expression of control feature sets. All analyzed features are binned based on averaged expression, and the control features are randomly selected from each bin (Tirosh et al., 2016).
Quantification and Statistical Analysis
Statistical analysis of clinical data was performed using GraphPad Prism software v9. Data were assessed for normal distribution and plotted in the figures as mean ± SEM unless described otherwise. One-way ANOVA with Tukey’s multiple comparisons test was used for normally disturbed data, and Kruskal-Wallis test with Dunn’s multiple comparisons test and Mann-Whitney test were used for data that were not normally distributed. Statistical analysis of scRNaseq data was performed using R Studio v1.3.1 and IPA. Kruskal-Wallis test with Wilcoxon test and Fisher’s exact test with Benjamini Hochberg adjusted P-values were used for module scores and canonical pathway analysis, respectively.
Acknowledgments
We thank the staff of the Biobank and Translational Research Core, the Flow Cytometry Core, and the Applied Genomics, Computation, and Translational Core for their help in this project. This study was funded by the Parker B. Francis Foundation fellowship (C.Y.); the Plum Foundation (P.C.); the Erwin Rautenberg Foundation fellowship (T.P.); and NIH grants T32HL134637 (S.A.B. and J.A.P.), P01HL108793 (B.R.S.), R01HL135163 (B.R.S.), R01AI127406, R01AI103542, R21AI151987 (G.A.M.), R01AI134987 (H.S.G.), R01AI137111 (S.A.G.), R01HL137076 (P.C.) and UCLA CTSI NCATS KL2TR001882 (C.Y. and T.P.).
Author Contributions
Conceptualization: C.Y., S.A.B., S.A.G., H.S.G., and P.C.; Methodology, Formal Analysis, and Investigation: C.Y., S.A.B., J.S., A.A.L., G.A.M., B.R.S., S.A.G., H.S.G., P.C.; Resources: T.P., T.Z., O.A.F., J.A.P., J.S.S., Y.P.M., G.C.C., A.G.K., N.P., and C.E.R.G.; Data Curation: C.Y., S.A.B., A.W.A., H.S., S.A.G., H.S.G., and P.C.; Writing – Original Draft: C.Y., S.A.B., S.A.G., H.S.G., and P.C.; Writing – Review & Editing: C.Y., S.A.B., T.P., G.A.M., S.A.G., H.S.G., and P.C.; Visualization: C.Y., S.A.B., G.A.M., S.A.G., H.S.G., and P.C.; Supervision: S.A.G., H.S.G., and P.C.; Project Administration: S.A.G., H.S.G., and P.C.; Funding Acquisition: P.C.
Declaration of Interests
The authors declare no competing interests.
Published: January 5, 2021; corrected online March 22, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108590.
Supplemental Information
References
- Abel A.M., Yang C., Thakar M.S., Malarkannan S. Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front. Immunol. 2018;9:1869. doi: 10.3389/fimmu.2018.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya D., Liu G., Gack M.U. Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 2020;20:397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo-Martin J.F., Cilloniz C., Mendez R., Almansa R., Gabarrus A., Ceccato A., Torres A., Menendez R., NEUMONAC group Lymphopenic Community Acquired Pneumonia (L-CAP), an Immunological Phenotype Associated with Higher Risk of Mortality. EBioMedicine. 2017;24:231–236. doi: 10.1016/j.ebiom.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budayeva H.G., Rowland E.A., Cristea I.M. Intricate Roles of Mammalian Sirtuins in Defense against Viral Pathogens. J. Virol. 2015;90:5–8. doi: 10.1128/JVI.03220-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajander S., Tina E., Bäckman A., Magnuson A., Strålin K., Söderquist B., Källman J. Quantitative Real-Time Polymerase Chain Reaction Measurement of HLA-DRA Gene Expression in Whole Blood Is Highly Reproducible and Shows Changes That Reflect Dynamic Shifts in Monocyte Surface HLA-DR Expression during the Course of Sepsis. PLoS ONE. 2016;11:e0154690. doi: 10.1371/journal.pone.0154690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Zhao P., Zhang D., Dong J.H., Xu Z., Yang G., Li B.Y., Liu H.X., Li B.A., Qin C.F. Persistent Viral Presence Determines the Clinical Course of the Disease in COVID-19. J. Allergy Clin. Immunol. Pract. 2020;8:2585–2591.e1. doi: 10.1016/j.jaip.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Cheung F., Shi R., Zhou H., Lu W., Consortium C.H.I., CHI Consortium PBMC fixation and processing for Chromium single-cell RNA sequencing. J. Transl. Med. 2018;16:198. doi: 10.1186/s12967-018-1578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A.M., Rowley B., Pao W., Hayday A., Bolen J.B., Pawson T. Syk tyrosine kinase required for mouse viability and B-cell development. Nature. 1995;378:303–306. doi: 10.1038/378303a0. [DOI] [PubMed] [Google Scholar]
- Chiu C., Openshaw P.J. Antiviral B cell and T cell immunity in the lungs. Nat. Immunol. 2015;16:18–26. doi: 10.1038/ni.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornall R.J., Cheng A.M., Pawson T., Goodnow C.C. Role of Syk in B-cell development and antigen-receptor signaling. Proc. Natl. Acad. Sci. USA. 2000;97:1713–1718. doi: 10.1073/pnas.97.4.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Gozzi L., Iannone A., Lo Tartaro D., Mattioli M. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Ebinger J.E., Achamallah N., Ji H., Claggett B.L., Sun N., Botting P., Nguyen T.T., Luong E., Kim E.H., Park E. Pre-existing traits associated with Covid-19 illness severity. PLoS ONE. 2020;15:e0236240. doi: 10.1371/journal.pone.0236240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J.J., Lu P., Wen S., Hastings A.K., Gilchuk P., Joyce S., Shyr Y., Williams J.V. Acute Viral Respiratory Infection Rapidly Induces a CD8+ T Cell Exhaustion-like Phenotype. J. Immunol. 2015;195:4319–4330. doi: 10.4049/jimmunol.1403004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallet B., Kyburz D., Walker U.A. Mild course of Coronavirus disease 2019 and spontaneous severe acute respiratory syndrome coronavirus 2 clearance in a patient with depleted peripheral blood B-cells due to treatment with rituximab. Arthritis Rheumatol. 2020;72:1581–1582. doi: 10.1002/art.41380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein S., Herbert J.A., McNamara P.S., Hedrich C.M. COVID-19: Immunology and treatment options. Clin. Immunol. 2020;215:108448. doi: 10.1016/j.clim.2020.108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop T., Larbi A., Dupuis G., Le Page A., Frost E.H., Cohen A.A., Witkowski J.M., Franceschi C. Immunosenescence and Inflamm-Aging As Two Sides of the Same Coin: Friends or Foes? Front. Immunol. 2018;8:1960. doi: 10.3389/fimmu.2017.01960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasselli G., Greco M., Zanella A., Albano G., Antonelli M., Bellani G., Bonanomi E., Cabrini L., Carlesso E., Castelli G. Risk Factors Associated With Mortality Among Patients With COVID-19 in Intensive Care Units in Lombardy, Italy. JAMA Intern. Med. 2020;180:1345–1355. doi: 10.1001/jamainternmed.2020.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groskreutz D.J., Babor E.C., Monick M.M., Varga S.M., Hunninghake G.W. Respiratory syncytial virus limits alpha subunit of eukaryotic translation initiation factor 2 (eIF2alpha) phosphorylation to maintain translation and viral replication. J. Biol. Chem. 2010;285:24023–24031. doi: 10.1074/jbc.M109.077321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil G.S. Foundations of Immunometabolism and Implications for Metabolic Health and Disease. Immunity. 2017;47:406–420. doi: 10.1016/j.immuni.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss R.S., Osmon S.B., Chang K.C., Wagner T.H., Coopersmith C.M., Karl I.E. Accelerated lymphocyte death in sepsis occurs by both the death receptor and mitochondrial pathways. J. Immunol. 2005;174:5110–5118. doi: 10.4049/jimmunol.174.8.5110. [DOI] [PubMed] [Google Scholar]
- Hu Y., Li W., Gao T., Cui Y., Jin Y., Li P., Ma Q., Liu X., Cao C. The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Inhibits Type I Interferon Production by Interfering with TRIM25-Mediated RIG-I Ubiquitination. J. Virol. 2017;91:e02143-16. doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C.V., Randolph G.J., Henson P.M. Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 2017;17:349–362. doi: 10.1038/nri.2017.28. [DOI] [PubMed] [Google Scholar]
- Kahan S.M., Wherry E.J., Zajac A.J. T cell exhaustion during persistent viral infections. Virology. 2015;479-480:180–193. doi: 10.1016/j.virol.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyuncu E., Budayeva H.G., Miteva Y.V., Ricci D.P., Silhavy T.J., Shenk T., Cristea I.M. Sirtuins are evolutionarily conserved viral restriction factors. MBio. 2014;5:e02249-14. doi: 10.1128/mBio.02249-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.S. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekkou A., Karakantza M., Mouzaki A., Kalfarentzos F., Gogos C.A. Cytokine production and monocyte HLA-DR expression as predictors of outcome for patients with community-acquired severe infections. Clin. Diagn. Lab. Immunol. 2004;11:161–167. doi: 10.1128/CDLI.11.1.161-167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester S.N., Li K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 2014;426:1246–1264. doi: 10.1016/j.jmb.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.W., Wang C.Y., Jou Y.J., Huang S.H., Hsiao L.H., Wan L., Lin Y.J., Kung S.H., Lin C.W. SARS Coronavirus Papain-Like Protease Inhibits the TLR7 Signaling Pathway through Removing Lys63-Linked Polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 2016;17:678. doi: 10.3390/ijms17050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Hua Z., Liu C., Lin L., Chen R., Hou B. Characterization of T-Dependent and T-Independent B Cell Responses to a Virus-like Particle. J. Immunol. 2017;198:3846–3856. doi: 10.4049/jimmunol.1601852. [DOI] [PubMed] [Google Scholar]
- Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang M., Cheng A., Yang Q., Wu Y., Jia R., Liu M., Zhu D., Chen S., Zhang S. The role of host eIF2α in viral infection. Virol. J. 2020;17:112. doi: 10.1186/s12985-020-01362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay M.A., Ware L.B., Zimmerman G.A. The acute respiratory distress syndrome. J. Clin. Invest. 2012;122:2731–2740. doi: 10.1172/JCI60331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mócsai A., Ruland J., Tybulewicz V.L. The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat. Rev. Immunol. 2010;10:387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Altamirano M.M.B., Kolstoe S.E., Sánchez-García F.J. Virus Control of Cell Metabolism for Replication and Evasion of Host Immune Responses. Front. Cell. Infect. Microbiol. 2019;9:95. doi: 10.3389/fcimb.2019.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T., Suarez C.J., Lin K.W., Jen K.Y., Schnitzer J.E., Makani S.S., Parker N., Perkins D.L., Finn P.W. T cell pathways involving CTLA4 contribute to a model of acute lung injury. J. Immunol. 2010;184:5835–5841. doi: 10.4049/jimmunol.0903238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouw H.H., Visser L.J., Passchier T.C., Langereis M.A., Liu F., Giansanti P., van Vliet A.L.W., Dekker J.G., van der Grein S.G., Saucedo J.G. Inhibition of the integrated stress response by viral proteins that block p-eIF2-eIF2B association. Nat. Microbiol. 2020;5:1361–1373. doi: 10.1038/s41564-020-0759-0. [DOI] [PubMed] [Google Scholar]
- Radzikowska U., Ding M., Tan G., Zhakparov D., Peng Y., Wawrzyniak P., Wang M., Li S., Morita H., Altunbulakli C. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy. 2020;75:2829–2845. doi: 10.1111/all.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell J.C. Metabolism and autophagy in the immune system: immunometabolism comes of age. Immunol. Rev. 2012;249:5–13. doi: 10.1111/j.1600-065X.2012.01158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth G., Moser B., Krenn C., Brunner M., Haisjackl M., Almer G., Gerlitz S., Wolner E., Boltz-Nitulescu G., Ankersmit H.J. Susceptibility to programmed cell death in T-lymphocytes from septic patients: a mechanism for lymphopenia and Th2 predominance. Biochem. Biophys. Res. Commun. 2003;308:840–846. doi: 10.1016/s0006-291x(03)01482-7. [DOI] [PubMed] [Google Scholar]
- Shen L., Wang C., Zhao J., Tang X., Shen Y., Lu M., Ding Z., Huang C., Zhang J., Li S. Delayed specific IgM antibody responses observed among COVID-19 patients with severe progression. Emerg. Microbes Infect. 2020;9:1096–1101. doi: 10.1080/22221751.2020.1766382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.S., Qi H.Y., Boularan C., Huang N.N., Abu-Asab M., Shelhamer J.H., Kehrl J.H. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D., Wu W., Wang Q., Xu K., Xie J., Wu J., Lv L., Sheng J., Guo J., Wang K. Clinical characteristics and factors associated with long-term viral excretion in patients with SARS-CoV-2 infection: a single center 28-day study. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.L., Kok K.H., Ng M.H., Poon V.K., Yuen K.Y., Zheng B.J., Jin D.Y. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.W., Zhang C., Fan X., Meng F.P., Xu Z., Xia P., Cao W.J., Yang T., Dai X.P., Wang S.Y. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020;11:3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., 2nd, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., Sinai Immunology Review Project Immunology of COVID-19: Current State of the Science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Made C.I., Simons A., Schuurs-Hoeijmakers J., van den Heuvel G., Mantere T., Kersten S., van Deuren R.C., Steehouwer M., van Reijmersdal S.V., Jaeger M. Presence of Genetic Variants Among Young Men With Severe COVID-19. JAMA. 2020;324:1–11. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos Q., Lees A., Wu Z.Q., Snapper C.M., Mond J.J. B-cell activation by T-cell-independent type 2 antigens as an integral part of the humoral immune response to pathogenic microorganisms. Immunol. Rev. 2000;176:154–170. doi: 10.1034/j.1600-065x.2000.00607.x. [DOI] [PubMed] [Google Scholar]
- Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.J.M., Leong J.Y., Lee J.H., Albani S., Yeo J.G. Insights into the immuno-pathogenesis of acute respiratory distress syndrome. Ann. Transl. Med. 2019;7:504. doi: 10.21037/atm.2019.09.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wurfel M.M., Park W.Y., Radella F., Ruzinski J., Sandstrom A., Strout J., Bumgarner R.E., Martin T.R. Identification of high and low responders to lipopolysaccharide in normal subjects: an unbiased approach to identify modulators of innate immunity. J. Immunol. 2005;175:2570–2578. doi: 10.4049/jimmunol.175.4.2570. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z., Tan W. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015;5:17554. doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shen C., Li J., Yuan J., Wei J., Huang F., Wang F., Li G., Li Y., Xing L. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020;146:119–127.e4. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Wang B., Mao J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Behjati S. SoupX removes ambient RNA contamination from droplet based single cell RNA sequencing data. bioRxiv. 2018 doi: 10.1101/303727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhai Y., Franco L.M., Atmar R.L., Quarles J.M., Arden N., Bucasas K.L., Wells J.M., Niño D., Wang X., Zapata G.E. Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections--A Prospective Cohort Study. PLoS Pathog. 2015;11:e1004869. doi: 10.1371/journal.ppat.1004869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the COVID-19 patient scRNA-seq data reported in this paper is GEO: GSE154567.