Abstract
Hypertrophy of chondrocytes is a crucial step in the endochondral bone formation process that drives bone lengthening and the transition to endochondral bone formation. Both Parathyroid hormone-related protein (PTHrP) and Histone deacetylase 4 (HDAC4) inhibit chondrocyte hypertrophy. Use of multiple mouse genetics models reveals how PTHrP and HDAC4 participate in a pathway that regulates chondrocyte hypertrophy. PTHrP/cAMP/Protein Kinase A (PKA) signaling pathway phosphorylates the PKA-target sites on salt-inducible kinase 3 (Sik3), which leads to inhibition of Sik3 kinase activity. Inhibition of Sik3 kinase activity decreases phosphorylation of HDAC4 by Sik3 at binding sites for 14-3-3; lower levels of HDAC4 phosphorylation then allow HDAC4 nuclear translocation. In the nucleus, the transcription factor, Myocyte Enhancer Factor 2 (Mef2), activates Runt-related transcription factor 2 (Runx2), and together these two transcription factors drive the hypertrophic process. HDAC4 binds both Mef2 and Runx2 and blocks their activities. There are genetic redundancies in this pathway. Sik1 and Sik2 also mediate PTHrP/cAMP/PKA signaling when Sik3 activity is low. HDAC5 also mediates PTHrP signaling when HDAC4 expression is low. Thus, PTHrP triggers a kinase cascade that leads to inhibition of the key transcription factors (Mef2 and Runx2) that promote chondrocyte hypertrophy.
Keywords: Parathyroid hormone-related protein (PTHrP), Histone deacetylase 4 (HDAC4), Myocyte Enhancer Factor 2 (Mef2), Salt-inducible kinase 3 (Sik3), Protein Kinase A (PKA), Chondrocyte hypertrophy
Introduction
Most of the vertebrate skeleton is generated by endochondral bone formation, in which growth plate chondrocytes direct subsequent bone formation [1]. The fetal growth plate has three morphologically distinct groups of chondrocytes: round chondrocytes, flat chondrocytes, and hypertrophic chondrocytes. Round chondrocytes proliferate and differentiate into flat chondrocytes. Flat chondrocytes proliferate further and form orderly columns of chondrocytes. Then, flat chondrocytes stop proliferating and enlarge many-fold to become hypertrophic chondrocytes [1]. The fetal growth plate is surrounded by perichondrium in which immature osteoblast precursors respond to signals from prehypertrophic and hypertrophic chondrocytes to become osteoblasts, while also signaling back to the growth plate [1].
During endochondral bone formation, chondrocyte hypertrophy is a crucial turning point in which the differentiation program is switched from chondrocyte differentiation to bone formation. Hypertrophic chondrocytes provide crucial signals for vascular invasion and osteoblast differentiation. Chondrocyte hypertrophy is also an important component of bone lengthening. In this review, we discuss the molecular mechanisms of chondrocyte hypertrophy revealed by mouse genetic studies [2, 3].
Parathyroid hormone-related protein (PTHrP)
In 1941, Fuller Albright first recognized humoral hypercalcemia of malignancy (HHM) in a renal cancer patient [4]. He suspected that the tumor might be producing parathyroid hormone (PTH), but PTH bioactivity was not found in the tumor [4] by a PTH bioassay [5]. Even after PTH radioimmunoassay was established in 1963 [6], PTH immunoactivity was not detected in tumor or plasma of patients with HHM [7]. Finally, in 1987, the isolation of full-length complementary DNA (cDNA) clones of a putative hypercalcemia factor from a human lung cancer cell line revealed a previously unrecognized protein, PTH-related protein (PTHrP) [8]. PTHrP shares homology with PTH only within the first 13 amino acids [8], but PTHrP and PTH share a common receptor [9], explaining hypercalcemia by PTHrP excess. These findings raised the question of the normal functions of PTHrP.
Physiological role of PTHrP in the growth plate
Knockout of the gene encoding PTHrP allowed exploration of the physiological roles of PTHrP. PTHrP-knockout (KO) mice die immediately after birth from respiratory failure caused by chondrocyte hypertrophy and bone formation on the anterior rib cartilage, where normally PTHrP represses chondrocyte hypertrophy in wildtype (WT) mice [10]. PTH/PTHrP receptor-KO mice die around embryonic day 18.5 (E18.5), showing the similar phenotype to that of PTHrP-KO mice [11]. In contrast, chondrocyte-specific PTHrP-transgenic (Tg) mice that overexpress human PTHrP protein using a mouse Type 2 collagen promoter exhibit severely delayed chondrocyte hypertrophy [12]. Thus, PTHrP, synthesized by and secreted from round chondrocytes, suppresses chondrocyte hypertrophy in the growth plate.
Histone deacetylases (HDACs)
Unexpected lethal phenotype in chondrocytes in Histone deacetylase 4 (HDAC4)-KO mice revealed a new mediator in the skeletal biology [13]. In mammalian genomes, eleven HDAC proteins have been identified. These proteins are classified into four families (Class I, IIa, IIb, IV) [14]. In 1996, HDAC1 was isolated as a Trapoxin (HDAC inhibitor [15]) binding protein from bovine thymus extracts [16] and HDAC2 was identified as a YY1 (mammalian zinc-finger transcription factor) binding protein by a yeast two-hybrid screen [17]. Within the next six years, HDAC3 to HDAC11 were identified by homology search with HDAC1 or existing HDAC proteins using the GenBank sequence database. Mammalian genomes encode another group of deacetylases, Sirtuin 1 to 7 which are referred as class III HDACs [14].
Class I HDAC proteins (HDAC 1, 2, 3, and 8) are ubiquitously expressed nuclear enzymes that potently deacetylate lysine residues on histone tails. Histone deacetylation leads to chromatin compaction and subsequent transcriptional repression [14]. In contrast, class IIa HDAC proteins are expressed in only specific tissues (HDAC5 and 9 in muscles, heart, and brain, HDAC4 in brain and growth plates, HDAC7 in endothelial cells and thymocytes) and exhibit only minimum histone deacetylase activity due to conserved changes in catalytic domain sequence[14].
In addition to their conserved deacetylase domain, class IIa HDAC proteins (HDAC 4, 5, 7 and 9) have characteristic N-terminal extensions [14], which contain binding sites for 14-3-3 chaperone proteins [18] and for the transcription factor, Myocyte enhancer factor 2 (Mef2) [19]. When bound by 14-3-3 proteins in the cytoplasm, class IIa HDAC proteins do not block the nuclear transcriptional activity of Mef2. In contrast, when released by cytoplasmic 14-3-3 chaperones, class IIa HDAC proteins can translocate into the nucleus and negatively regulate Mef2 activity [18].
KO mice for HDAC proteins
Between 2002 and 2009, all KO mice for Class I and Class IIa HDAC proteins were generated which defined the functions of HDACs in vivo [14, 20] . The ubiquitous expression, high deacetylase activity, and high homology among Class I HDAC proteins suggested functional redundancy in vivo. However, each Class I HDAC KO mouse leads to embryonic or perinatal lethality, demonstrating the unique roles of each Class I HDAC protein [14].
As expected from the crucial actions of Mef2 in skeletal and cardiac muscles, KO mice for class IIa HDAC proteins exhibited cardiovascular phenotypes in HDAC5-KO mice (normal life span) [21], HDAC7-KO mice (embryonic lethality before E11.5) [22], and HDAC9-KO mice (normal life span) [23]. However, HDAC4-KO mouse unexpectedly exhibited a lethal phenotype characterized by abnormally accelerated chondrocyte hypertrophy and bone formation in the anterior rib cartilage (lethality around postnatal day 12 (P12) )[13].
PTHrP regulation of the HDAC4/Mef2 interaction in vitro
A subsequent mouse genetic study revealed that Mef2 also drives chondrocyte hypertrophy [24]. This result demonstrated that the accelerated chondrocyte hypertrophy in HDAC4-KO mice is induced by increased Mef2 action. Then, an in vitro study using cultured chick primary chondrocytes demonstrated that PTHrP treatment leads to lower HDAC4 phosphorylation at the first 14-3-3 binding site and induces subsequent nuclear translocation of HDAC4, proposing that PTHrP signaling pathway regulates the HDAC4/Mef2 interaction [25]. In humans, the haploinsufficiency of PTHrP [26] or of HDAC4 [27] causes the skeletal dysplasia, brachydactyly type E, suggesting a functional linkage between PTHrP and HDAC4.
Genetic evidences in vivo that HDAC4 mediates PTHrP signaling
Using multiple mouse genetics models, we demonstrated in vivo that HDAC4 is a mediator of the PTHrP signaling pathway during chondrocyte differentiation [2]. Here, we summarize the main results of our mouse genetic studies.
(1). PTHrP-KO mouse vs. HDAC4-KO mouse
PTHrP-KO mice die immediately after birth due to abnormal chondrocyte hypertrophy and bone formation on the anterior rib cartilage[10]. In contrast, HDAC4-KO mice have normal rib cartilage at birth. However, progressive rib chondrocyte hypertrophy starts at P2 and HDAC4-KO mice die around P12[2, 13]. In PTHrP-KO mice, the distance from the top of the growth plate to the beginning of the hypertrophic layer is extremely short due to the accelerated chondrocyte hypertrophy35 % of the WT controls)[2]. HDAC4-KO mice show similar but milder phenotype64 % of the WT controls [2].
The similar phenotype between PTHrP-KO mouse and HDAC4-KO mouse supports the hypothesis that HDAC4 mediates PTHrP signaling. However, the milder phenotype of HDAC4-KO mouse also suggests the possible existence of other mediators of PTHrP signaling in HDAC4-KO mouse.
(2). HDAC4-KO mouse vs. Chondrocyte-specific HDAC4 cKO mouse
To exclude the possibility that systemic effects caused by HDAC4 deletion in other organs contribute to the chondrocyte phenotype in universal HDAC4-KO mice, we generated HDAC4 conditional KO (cKO) mice using HDAC4-floxed mice [28] with either Type 2 collagen (Col2)-Cre driver that is active in chondrocytes and perichondrial cells [29] or Osterix (Osx)-Cre driver that is active in hypertrophic chondrocytes, perichondrium, and osteoblasts [30]. Col2-Cre: HDAC4-cKO mice show similar phenotype with universal HDAC4-KO mice, but Osx-Cre: HDAC4-cKO mice show normal phenotype [2].
This result demonstrates that the chondrocyte phenotype in universal HDAC4-KO mouse depends only on HDAC4 deletion in proliferating chondrocytes (round chondrocytes and flat chondrocytes).
(3). PTHrP and HDAC4 double heterozygous (HET) mouse
Both PTHrP-HET and HDAC4-HET mice have normal growth plates. If PTHrP and HDAC4 work independently, then the double HET mouse should also have a normal growth plate. However, the double HET mouse shows a significantly shorter proliferating region (85 % of the WT controls in the proximal tibial growth plate at birth) [2], which could result from lower PTHrP action in the growth plate.
The phenotype of the double HET mouse is mild, but nevertheless similar to that of the individual KO mouse. This experiment indicates that PTHrP and HDAC4 repress chondrocyte hypertrophy though a common pathway.
(4). HDAC4 deletion in the PTHrP-Tg mouse
As a more stringent genetic test of whether HDAC4 is a mediator of PTHrP signaling, we deleted the HDAC4 gene in PTHrP-Tg mouse. If HDAC4 mediates PTHrP signaling, the HDAC4 deletion should modify the PTHrP transgenic phenotype. Heterozygous PTHrP-Tg (PTHrP-Tg/+) mice have only round chondrocytes in all the long bones up to one week after birth, and subsequent bone formation is also severely delayed [12]. Even the heterozygous deletion of HDAC4 gene partially blocks the PTHrP transgenic phenotype at birth. Strikingly, the homozygous deletion of HDAC4 gene almost completely reversed the PTHrP transgenic phenotype [2].
Molecular evidences in vivo that HDAC4 mediates PTHrP signaling
To support in vivo genetic evidences that PTHrP is only working when HDAC4 is present, we investigated the molecular evidences in vivo that HDAC4 mediates PTHrP signaling.
(1). HDAC4 Phosphorylation in vivo
We manually micro-dissected proliferating chondrocytes from the proximal tibial growth plates at birth, and then we compared the phosphorylation of three 14-3-3 binding sites of HDAC4 among PTHrP-Tg/+ mouse, PTHrP-KO mouse, and WT controls by Western blots using antibodies that detect the site-specific phosphorylation. Either PTHrP overexpression or PTHrP deletion leads to reduced or increased levels of HDAC4 phosphorylation at three 14-3-3 binding sites compared to WT controls, respectively. We found the most significant changes at the first 14-3-3 binding site, but the changes at the other two sites are also significant [2].
(2). HDAC4 nuclear translocation in vivo
When intracellular signals decrease the level of phosphorylation at the 14-3-3 binding sites, class IIa HDAC proteins are released from 14-3-3 proteins and then move into the nucleus [18]. To demonstrate HDAC4 nuclear translocation induced by PTHrP signaling in vivo, we compared the intracellular HDAC4 localization between PTHrP-Tg/+ mouse and PTHrP-KO mouse by immunohistochemistry (IHC), focusing on round chondrocytes in the proximal tibial growth plate at birth. Using confocal microscopy, we observed intense nuclear HDAC4 signals in PTHrP-Tg/+ mouse. In contrast, HDAC4 signals appear lower in nuclei, but higher in the cytoplasm in PTHrP-KO mouse [2].
We calculated HDAC4 signal intensities in the nuclei and those in the whole cells by three-color confocal images (for HDAC4 IHC, nuclear stain, and whole cell stain). The average ratio of total HDAC4 signal intensities in the nuclei to those in the whole cells was significantly higher in PTHrP-Tg/+ mouse (PTHrP-Tg/+: PTHrP-KO =51 % vs. 38 %, p<0.0001) [2], supporting the hypothesis that PTHrP increases HDAC4 nuclear translocation.
Identification of HDAC5 as an additional mediator of PTHrP signaling
To identify additional mediators of PTHrP signaling that might work in HDAC4-KO mouse, we focused on other class IIa HDAC proteins, namely HDAC5, HDAC7, and HDAC9. We hypothesized that the common N-terminal extension among class IIa HDAC proteins might be regulated by PTHrP signaling, as we observed with HDAC4.
(1). Expression analysis
In microarray analysis using manually micro-dissected proximal tibial growth plates of newborn WT mice, HDAC5 mRNA and HDAC7 mRNA showed similar expression levels to those of HDAC4 mRNA, but the expression levels of HDAC9 mRNA were at background levels [2]. Thus, we narrowed down the candidates to HDAC5 and HDAC7.
(2). Homology analysis
Then, we analyzed the homology in protein sequence among HDAC4, HDAC5, and HDAC7. We focused on the homology in the N-terminal extension domain that is characteristic for class II HDAC family proteins. We found higher homology between HDAC4 and HDAC5 than between HDAC4 and HDAC7 (50 % vs. 27 %) in contrast to the similarity in the C-terminal histone deacetylase domain (74 % vs. 68 %) [2]. Taken together, we investigated HDAC5 first.
(3). HDAC4 and HDAC5 double KO mouse
We first analyzed HDAC5-KO mouse at birth, but found a normal phenotype in the rib chondrocytes, tibial growth plate, and whole tibia. Next, we analyzed mice with mutations in both HDAC4 and HDAC5. Surprisingly, HDAC4-KO: HDAC5-HET mouse died immediately after birth. The rib phenotype of this mouse was similar to that of PTHrP-KO mouse with dramatically accelerated chondrocyte hypertrophy [2]. This result is consistent with the notion that HDAC5 mediates PTHrP signaling to suppress chondrocyte hypertrophy in HDAC4-KO mouse. HDAC4 and HDAC5 double KO mouse died at birth with a more severe rib phenotype than that of PTHrP-KO mouse [2], and thus had a phenotype similar to that of PTH/PTHrP receptor-KO mouse [31]. The phenotype in HDAC4 and HDAC5 double KO mouse supports the hypothesis that both of these class II HDAC proteins are necessary for PTHrP signaling to efficiently block the initiation of chondrocyte hypertrophy.
We did not analyze the double mutant mice of HDAC4 and HDAC7 to examine if HDAC7 also mediates PTHrP signaling. HDAC7 alone cannot block accelerated chondrocyte hypertrophy in HDAC4 and HDAC5 double KO mouse at birth. In contrast, HDAC5 can block accelerated chondrocyte hypertrophy in HDAC4-KO mouse at birth. Though we cannot exclude a role of HDAC7, we concluded that HDAC4 and HDAC5 are the main mediators of PTHrP signaling in chondrocytes.
(4). HDAC5 deletion in the PTHrP-Tg mouse
To explore the relative actions of HDAC4 and HDAC5 in the setting of PTHrP overexpression in the growth plates, we deleted the HDAC5 gene in PTHrP-Tg/+ mouse. The heterozygous or homozygous deletion of HDAC5 had no effects on the PTHrP-Tg/+ phenotype, in contrast to the findings with the HDAC4 deletion inPTHrP-Tg/+ mouse [2].
Next, we deleted the HDAC5 gene in HDAC4-HET: PTHrP-Tg/+ mouse. With HDAC4 expression limited, we found a stepwise acceleration of chondrocyte differentiation when the HDAC4-HET was combined with HDAC5-HET and thenHDAC5-KO [2]. These results demonstrate that HDAC5 mediates PTHrP signaling, when the expression level of HDAC4 is low.
Downstream of the PTHrP/HDAC4/Mef2 pathway
Next, we investigated mediators working downstream of PTHrP/HDAC4/Mef2 pathway. A previous study showed that Runx2 mRNA expression is reduced in the prehypertrophic region of the growth plate when Mef2C was knocked out of chondrocytes and osteoblasts using Twist2-Cre [24]. This finding is consistent with the idea that, in chondrocytes, Runx2 expression reflects Mef2 action. To test if Runx2 drives chondrocyte hypertrophy downstream of Mef2, we utilized HDAC4 deficient animals.
In HDAC4-KO mice, HDAC4 deletion leads to high Mef2C activity that subsequently induces rib chondrocyte hypertrophy and mineralization anteriorly, where this does not normally occur [13, 24]. The heterozygous deletion of Runx2 in HDAC4-KO mouse abrogates chondrocyte hypertrophy, which enables this mouse to survive longer with a normal rib phenotype [2]. The heterozygous deletion of Runx2 itself had no effects on the rib phenotype. This result indicates that Mef2 fails to induce chondrocyte hypertrophy when Runx2 expression is limited. Taken together, Runx2 works downstream of HDAC4/Mef2 and HDAC5/Mef2 to drive chondrocyte hypertrophy.
PTHrP regulates Sik3 activity in vitro
In the discussions above, how PTHrP action leads to lower phosphorylation of HDAC4 was unknown. In our next paper [3], we investigated molecules that phosphorylate HDAC4. We first focused on a kinase named Salt-inducible kinase (Sik) 3, because a previous study reported that Sik3-KO animals exhibits delay in chondrocyte hypertrophy [32] reminiscent to what is observed in PTHrP-Tg mice [12]. Salt-inducible kinases are AMPK family kinases that are known to phosphorylate N-terminal 14-3-3 binding sites on class IIa HDACs [33]. Moreover, SIK activity is regulated by cAMP/PKA signaling in multiple cell types [33,34]. Cyclic AMP signaling leads to protein kinase A (PKA)-mediated phosphorylation of Sik3, a modification that allosterically inhibits cellular Sik3 kinase activity [35]. Taken together, we hypothesized that Sik3 might phosphorylate the 14-3-3 binding sites of HDAC4 in chondrocytes and that PTHrP might reduce HDAC4 phosphorylation by inhibiting Sik3 activity through cAMP/PKA pathway.
To test this hypothesis, we treated primary rib chondrocytes with PTH and examined phosphorylation status of a 14-3-3 binding site of HDAC4 and that of a PKA target site of Sik3 using Western blot. In these cells, activation of PTH/PTHrP signaling pathway leads to increased Sik3 phosphorylation at T441, one of three PKA-target phosphorylation sites on Sik3 protein [36]. In turn, reduced Sik3 activity leads to reduced HDAC4 phosphorylation at the first 14-3-3 binding site [3]. These results indicate that PTHrP suppresses Sik3 activity, leading to lower phosphorylation of HDAC4.
PTHrP and Sik3 double KO mouse
To identify the link between PTHrP signaling and Sik3 in vivo, we asked if the Sik3 deletion might lower the high HDAC4 phosphorylation status that is characterized in PTHrP-KO mouse. If HDAC4 is phosphorylated by Sik3 in chondrocytes, the lethal phenotype of PTHrP-KO mice should be rescued by knocking out Sik3 activity. To test this hypothesis, we generated PTHrP-KO: Sik3-cKO (Col2-Cre) double KO mice. Strikingly, the perinatal lethality of PTHrP-KO mouse is completely rescued by Sik3 deletion [3]. Consistent with rescued rib chondrocyte hypertrophy, the double KO mice can survive up to one month after birth. The cause of death in the double KO mice is not abnormal rib chondrocyte hypertrophy and mineralization. One possibility is that a complete failure of tooth eruption in the double KO mice might cause the death after weaning [3].
Sik2/Sik3 or Sik1/Sik3 double KO mouse
Sik3-KO mice and chondrocyte specific Sik3 cKO mice show delayed chondrocyte hypertrophy compared to WT mouse [3, 32], but it is not as severe as the delay of chondrocyte hypertrophy in chondrocyte-specific PTHrP-Tg mice [12]. The more severe phenotype in PTHrP-Tg/+ mice suggests that other Sik family proteins (Sik1 or Sik2) may control chondrocyte hypertrophy in addition to Sik3. To confirm this hypothesis, we generated double cKO mice of Sik1 and Sik3 or Sik2 and Sik3.
By additional deletion of the Sik1 [36] or Sik2 [37] gene in Sik3-cKO mice, we observed longer growth plate and delayed separation of proximal and distal growth plates at birth. These results suggest that Sik1 and Sik2 also regulate chondrocyte differentiation when Sik3 is absent, but that Sik2 deletion exhibits more obvious effects [3]. However, Sik2 and Sik3 double cKO mice are still not as severe as the delay of PTHrP-Tg mice. We have not been able to generate Sik1 and Sik2 and Sik3 triple cKO mice.
Sik1-cKO mice, Sik2-cKO mice, and Sik1 and Sik2 double cKO mice exhibited normal growth plate phenotypes, highlighting that Sik3 is the predominant Sik isoform in regulating chondrocyte differentiation [3].
Sik3 and HDAC4 double KO mouse
To confirm that Sik3 exerts its effects on chondrocyte differentiation via the PTHrP/HDAC4 signaling pathway, we generated chondrocyte-specific Sik3 and HDAC4 double mutant mice. By heterozygous or homozygous deletion of the HDAC4 gene in Sik3-cKO mouse, stepwise shorter proliferating chondrocyte regions were observed at birth, consistent with the idea that HDAC4 acts downstream of Sik3 to delay chondrocyte hypertrophy. Delayed chondrocyte hypertrophy observed inSik3-cKO mice is completely abrogated by homozygous deletion of HDAC4 gene. Sik3 and HDAC4 double cKO mice appear phenotypically similar toHDAC4-cKO mice, indicating that HDAC4 functions downstream as a key mediator of Sik3 signaling.
Other pathways whereby Sik3 might regulate chondrocyte hypertrophy
It is reported that Sik3 induces chondrocyte hypertrophy by increasing acetyl-CoA, a substrate for the TCA cycle [38]. The same group identified a new Sik3 inhibitor, Pterosin B by screening ~2,500 compounds. Pterosin B inhibits chondrocyte hypertrophy in articular cartilage, leading to protection of cartilage from osteoarthritis [39]. This result suggests that Sik3 regulates the homeostasis of articular cartilage. Another group demonstrated that Sik3 is an essential positive regulator of mTOR (mammalian target of rapamycin) signaling by inducing the phosphorylation and subsequent proteosomal degradation of DEPTOR (DEP domain containing mTOR Interacting Protein), an inhibitor of mTOR, in response to PTH/PTHrP signaling [40]. Our studies with Sik3/HDAC4 double mutants suggest that class IIa HDACs are likely the Sik3 substrates that dominate in regulation of the differentiation of chondrocytes in the growth plate. The role of acetyl-CoA or DEPTOR in chondrocyte hypertrophy and their regulation by SIKs remains to be studied using mouse genetics.
Summary
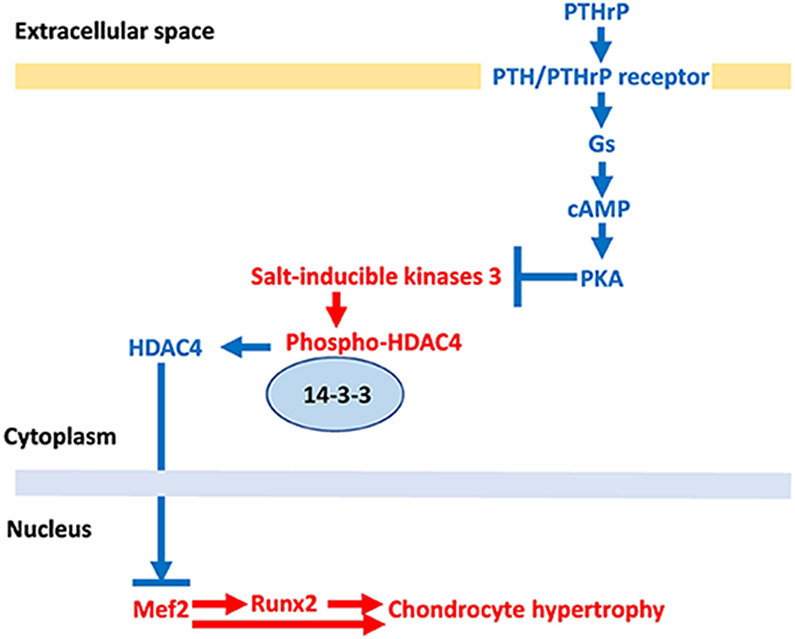
Figure 1 summarizes the model resulting from our data and the previous manuscripts [24, 25]. PTHrP/cAMP/PKA signaling pathway phosphorylates the PKA-target sites on Sik3 molecule, leading to inhibition of Sik3 kinase activity. Inhibition of Sik3 kinase activity decreases phosphorylation of HDAC4 at the 14-3-3 binding sites, which induces HDAC4 nuclear translocation by releasing HDAC4 from 14-3-3 proteins. In the nucleus, HDAC4 blocks the transcriptional activity of Mef2 through its Mef2 binding sites. Mef2 drives chondrocyte hypertrophy both by direct action and by activating=Runx2 expression. Runx2 works downstream of HDAC4&5/Mef2 to drive chondrocyte hypertrophy. Sik1 and Sik2 also mediate the PTHrP/cAMP/PKA signaling pathway when Sik3 activity is low. HDAC5 also mediates PTHrP signaling when HDAC4 expression is low.
Figure 1. Proposed model for regulation of chondrocyte hypertrophy.
The pathways in red letters drive chondrocyte hypertrophy. The pathways in blue letters inhibit chondrocyte hypertrophy. Chondrocyte hypertrophy is determined by the balance between the actions of Mef2 and HDAC4. Mef2 drives chondrocyte hypertrophy both by direct action and by activating Runx2 expression. Sik3 phosphorylates the 14-3-3 binding sites of HDAC4, which keeps HDAC4 in cytoplasm and subsequently inhibits HDAC4 to access Mef2 in nucleus. PTHrP inhibits the Mef2 action by releasing HDAC4 from 14-3-3. PTHrP reduces Sik3 activity through phosphorylation of PKA target sites on Sik3.
Highlights:
Hypertrophy of chondrocytes is a crucial step in endochondral bone formation.
Parathyroid hormone-related protein (PTHrP) inhibits chondrocyte hypertrophy.
Myocyte enhancer factor 2 (Mef2) drives chondrocyte hypertrophy.
Histone deacetylase 4 (HDAC4) blocks this Mef2 action upon nuclear translocation.
PTHrP induces HDAC4 translocation by inactivating salt-inducible kinase 3 (Sik3).
Acknowledgments
Funding:
This work was supported by the National Institutes of Health (DK116716 (MNW), DK011794 (MNW and HMK), and AR066261 (HMK).
Footnotes
Conflict of interest statement
MNW and HMK receive research funding from Galapagos NV. MNW receives research funding from Radius Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kronenberg HM, Developmental regulation of the growth plate, Nature. 423 (2003) 332–336. 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- [2].Nishimori S et al. , PTHrP Targets HDAC4 and HDAC5 to Repress Chondrocyte Hypertrophy, JCI Insight. 4(2019) e97903 10.1172/jci.insight.97903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nishimori S, et al. , Salt-inducible Kinases Dictate Parathyroid Hormone 1 Receptor Action in Bone Development and Remodeling, J Clin Invest. 129 (2019) 5187–5203. 10.1172/JCI130126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Albright F F, et al. , Case records of the Massachusetts General Hospital-Case 27461. N Engl J Med. 225(1941) 789–791. 10.1056/NEJM194111132252007. [DOI] [Google Scholar]
- [5].Collip JB, The extraction of a parathyroid hormone which will prevent or control parathyroid tetany and which regulates the level of blood calcium, J. Biol. Chem 63(1925) 395–438. [Google Scholar]
- [6].Berson SA, Yalow RS, Aurbach GD, Potts JT Jr., Immunoassay of bovine and human parathyroid hormone, ProcNatl Acad Sci U S A. 49(1963) 613–617. 10.1073/pnas.49.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Powell D, Singer FR, Murray TM, Minkin C, Potts JT Jr, Nonparathyroid Humoral Hypercalcemia in Patients with Neoplastic Diseases, N Engl J Med. 289(1973) 176–181. 10.1056/NEJM197307262890403. [DOI] [PubMed] [Google Scholar]
- [8].Suva LJ, et al A parathyroid hormone-related protein implicated in malignant hypercalcemia: cloning and expression, Science. 237 (1987) 893–896. 10.1126/science.3616618. [DOI] [PubMed] [Google Scholar]
- [9].Jiippner H, Abou-Samra AB, Uneno S, Gu WX, Potts JT Jr, Segre GV, The parathyroid hormone-like peptide associated with humoral hypercalcemia of malignancy and parathyroid hormone bind to the same receptor on the plasma membrane of ROS 17/2.8 cells, J Biol Chem. 263 (1988) 8557–60. PMID: 2837457. [PubMed] [Google Scholar]
- [10].Karaplis AC, et al. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene, Genes Dev. 8 (1994) 277–289. 10.1101/gad.8.3.277. [DOI] [PubMed] [Google Scholar]
- [11].Lanske B, et al. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth, Science. 273 (1996) 663–666. 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
- [12].Weir EC, Philbrick WM, Amling M, Neff LA, Baron R, Broadus AE, Targeted overexpression of parathyroid hormone-related peptide in chondrocytes causes chondrodysplasia and delayed endochondral bone formation, ProcNatl Acad Sci U S A. 93 (1996) 10240–10245. 10.1073/pnas.93.19.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Vega RB, et al. , Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis, Cell. 119 (2004) 555–566. 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- [14].Haberland M, Montgomery RL, Olson EN, The many roles of histone deacetylases in development and physiology: implications for disease and therapy, Nat Rev Genet. 10 (2009) 32–42. 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kijima M, Yoshida M, Sugita K, Horinouchi S, Beppu T, Trapoxin, an Antitumor Cyclic Tetrapeptide, Is an Irreversible Inhibitor of Mammalian Histone Deacetylase, J Biol Chem. 268 (1993) 22429–35. PMID: 8226751. [PubMed] [Google Scholar]
- [16].Taunton J, Hassig CA, Schreiber SL, A Mammalian Histone Deacetylase Related to the Yeast Transcriptional Regulator Rpd3p, Science. 1996. (272) 408–411. 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- [17].Yang WM, Inouye C, Zeng Y, Bearss D, Seto E, Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3, Proc Natl Acad Sci U S A 93(1996) 12845–12850. 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Grozinger CM, Schreiber SL, Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization, Proc Natl Acad Sci U S A. 97 (2000) 7835–7840. 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T, HDAC4 Deacetylase Associates with and Represses the MEF2 Transcription Factor, EMBO J. 18 (1999) 5099–5107. 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang JS, S-H. Yoon, M.N. Wein, Role of histone deacetylases in bone development and skeletal disorders, Bone, in press (2020) 10.1016/j.bone.2020.115606. [DOI] [PMC free article] [PubMed]
- [21].Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN, Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development, Mol Cell Biol. 24 (2004) 8467–8476. 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN, Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10, Cell. 126 (2006) 321–334. 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- [23].Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN, Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy, Cell. 110 (2002) 479–488. 10.1016/S0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Arnold MA, et al. , MEF2C transcription factor controls chondrocyte hypertrophy and bone development, Dev Cell. 12 (2007) 377–389. 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- [25].Kozhemyakina E, Cohen T, Yao TP, Lassar AB, Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway, Mol Cell Biol. 29 (2009) 5751–5762. 10.1128/MCB.00415-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Klopocki E, et al. , Deletion and point mutations of PTHLH cause brachydactyly type E, Am J Hum Genet. 86 (2010) 434–439. 10.1016/j.ajhg.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Williams SR, et al. , Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems, Am J Hum Genet. 87 (2010) 219–228. 10.1016/j.ajhg.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Potthoff MJ, et al. , Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers, J Clin Invest. 117 (2007) 2459–2467. 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR, Col2a1-Directed Expression of Cre Recombinase in Differentiating Chondrocytes in Transgenic Mice, Genetics. 26(2000) 145–146. PMID: 10686612 [PubMed] [Google Scholar]
- [30].Rodda SJ, McMahon AP, Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors, Development. 133(2006) 3231–3244. 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- [31].Lanske B, et al. , The parathyroid hormone (PTH)/PTH-related peptide receptor mediates actions of both ligands in murine bone, Endocrinology. 139 (1998) 5194–5204. 10.1210/endo.139.12.6361. [DOI] [PubMed] [Google Scholar]
- [32].Sasagawa S, et al. , SIK3 is essential for chondrocyte hypertrophy during skeletal development in mice, Development. 139 (2012) 1153–1163. 10.1242/dev.072652. [DOI] [PubMed] [Google Scholar]
- [33].Walkinshaw DR, et al. , The tumor suppressor kinase LKB1 activates the downstream kinases SIK2 and SIK3 to stimulate nuclear export of Class IIa histone deacetylases, J Biol Chem. 288 (2013) 9345–9362. 10.1074/jbc.M113.456996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wein MN, Foretz M, Fisher DE, Xavier RJ, Kronenberg HM, Salt-Inducible Kinases: Physiology, Regulation by cAMP, and Therapeutic Potential, Trends Endocrinol Metab. 29 (2018) 723–735. 10.1016/j.tem.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sonntag T, Vaughan JM, Montminy M, 14-3-3 proteins mediate inhibitory effects of cAMP on salt-inducible kinases (SIKs), FEBS J. 285 (2018) 467–480. 10.1111/febs.14351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nixon M, et al. , Skeletal muscle salt inducible kinase 1 promotes insulin resistance in obesity, Mol Metab. 5 (2015) 34–46. 10.1016/j.molmet.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Patel K, et al. , The LKB1-salt-inducible kinase pathway functions as a key gluconeogenic suppressor in the liver, Nat Commun. 5 (2014) 4535 10.1038/ncomms5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kosai A, et al. , Changes in acetyl-CoA mediate Sik3-induced maturation of chondrocytes in endochondral bone formation, Biochem Biophys Res Commun. 516 (2019) 1097–1102. 10.1016/j.bbrc.2019.06.139. [DOI] [PubMed] [Google Scholar]
- [39].Yahara Y, et al. , Pterosin B prevents chondrocyte hypertrophy and osteoarthritis in mice by inhibiting Sik3, Nat Commun. 7 (2016) 10959 10.1038/ncomms10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Csukasi F, et al. , The PTH/PTHrP-SIK3 pathway affects skeletogenesis through altered mTOR signaling, Sci Transl Med. 10 (2018) eaat9356 10.1126/scitranslmed.aat9356. [DOI] [PMC free article] [PubMed] [Google Scholar]