Abstract
Myxobacteria conduct complex social traits that requires populations to be highly related and devoid of exploiters. To enrich for clonal cells in populations, they employ kin discrimination mechanisms. One key system involves a polymorphic cell surface receptor, TraA, which recognizes self by homotypic interactions with neighboring myxobacterial cells. Recent studies revealed that TraA and its partner TraB are fluid outer membrane proteins that coalesce into foci upon recognition of kin. The formation of foci leads to transient membrane fusion junctions and the bidirectional exchange of outer membrane components that facilitates cooperative behaviors. Additionally, expansive suites of polymorphic lipoprotein toxins are exchanged, which act as self-identity barcodes that exquisitely discriminate against nonself to assemble homogenous populations.
Keywords: kin recognition, polymorphic toxins, Myxococcus xanthus, outer membrane exchange, cell surface receptors, membrane fusion, kin discrimination
Graphical Abstract
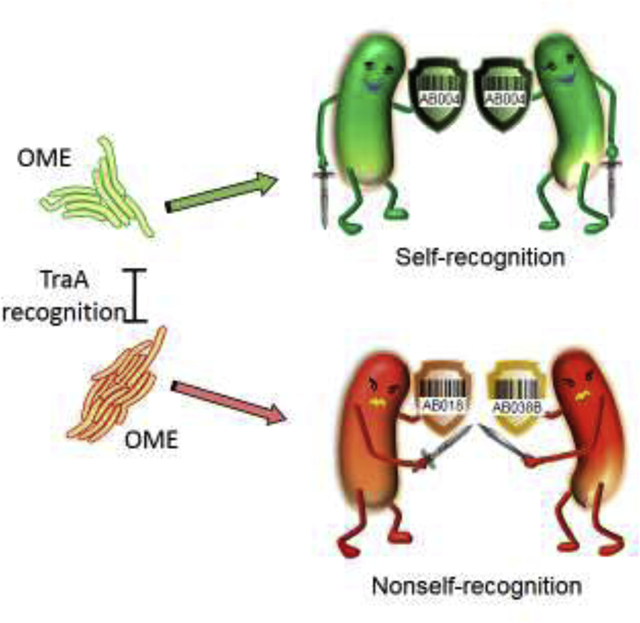
Introduction
Bacteria use molecular recognition tools to recognize the identity of neighboring cells when they interact [1,2]. Cell-cell recognition in turn elicits responses that includes the formation of social groups and initiating programs that leads to development, self-defense, symbiosis or virulence. Among the bacterial kingdom, myxobacteria exhibit some of the most complex social interactions, which have fascinated microbiologists for decades [3,4], and serve as rich models for studying cell-cell recognition [5–8]. Many myxobacterial species are terrestrial where they prey upon other microbes [9,10]. Predation is an active process where cells forage and digest prey cells by secreting a battery of antibiotics and hydrolytic enzymes. In doing so myxobacteria appear to recognize cells as prey [11], but how that occurs is unknown. Upon nutrient exhaustion, they enter a developmental program where kin congregate in large numbers to assemble multicellular fruiting bodies wherein cells differentiate into distinct types, including spores. Because of their complex multicellular lifecycle, when myxobacteria were first discovered they were mistakenly thought to be eukaryotes [12]. Today, strikingly, a leading hypothesis proposes an ancient myxobacterium led to the origin of eukaryotes [13]. This hypothesis suggests a symbiosis relationship evolved into a single cell when a myxobacterium engulfed an Asgard-like archaeon, which evolved into today’s cytoplasm and nucleus, respectively, of eukaryotic cells. This review discusses our recent work on a fascinating myxobacterial behavior that involves kin recognition and the transfer of cellular cargo in a process called outer membrane exchange.
What is outer membrane exchange (OME)?
Myxobacteria move on solid surfaces by gliding motility and in doing so they make frequent contacts with other cells. Upon cell-cell contact, molecular recognition determines whether the adjoining cell is self (clonal) or nonself [14]. Self-recognition is mediated by the polymorphic cell surface receptor, TraA [15–18], and its cohort protein TraB, which leads to the bidirectional transfer of large amounts of outer membrane (OM) proteins and lipids, i.e. OME (Fig. 1). Since lipids are also exchanged [19–21], OME is thought to be driven by transient membrane fusion. In this model, TraA/B function as a fusogen [22]. As the model organism, Myxococcus xanthus, is a Gram-negative bacterium, their OM contains lipopolysaccharide (LPS) and, to our knowledge, TraA/B is the first fusogen suggested for membranes containing LPS. OME is a dynamic process which typically lasts for minutes and occurs while partnering cells are in contact [23]. Gliding motility is critical for OME to ensure membrane fusion/fission occurs [24–26].
Figure 1.
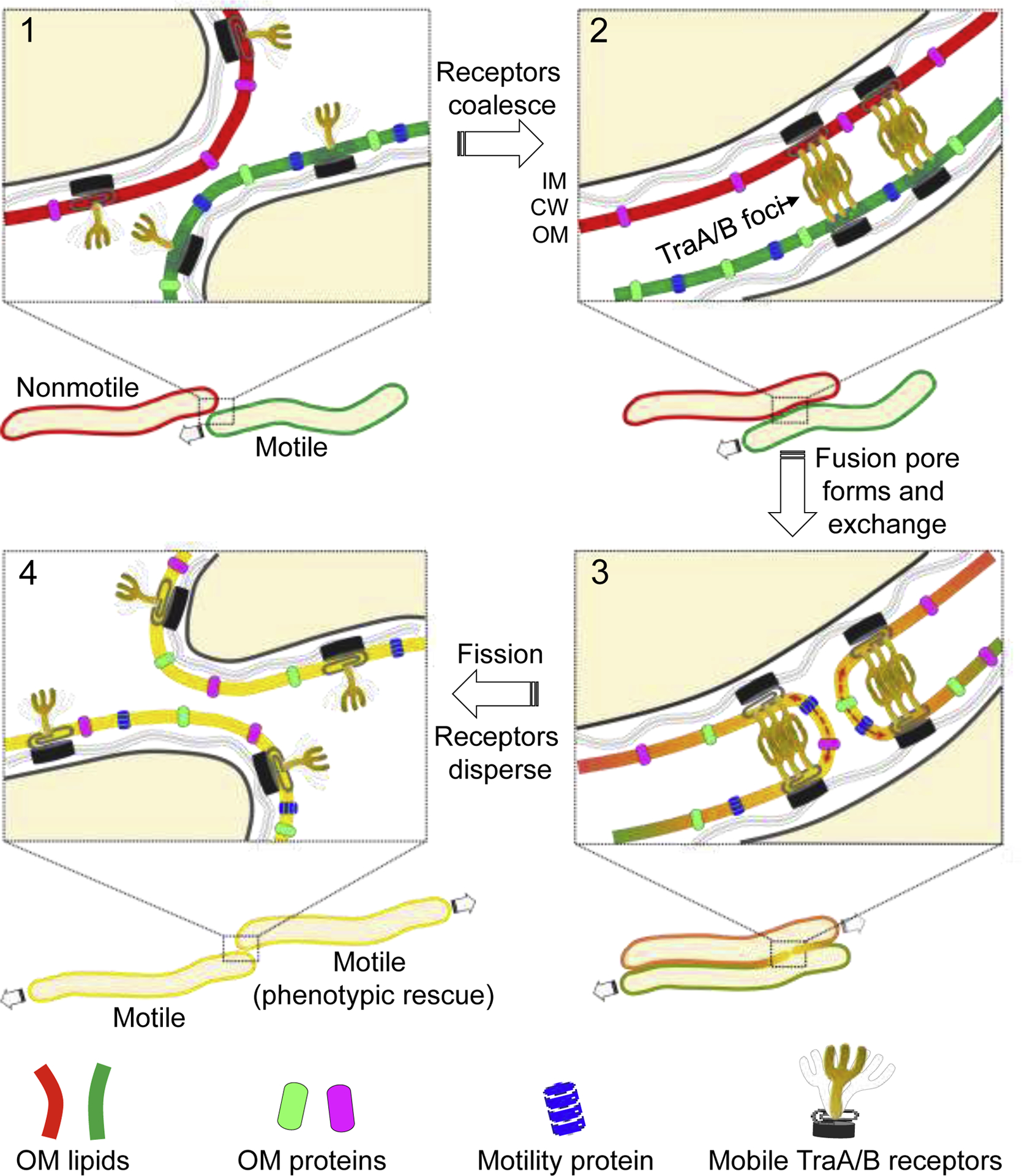
Model of OME. (1) TraA/B are fluid in the OM. (2) After contacting a kin cell TraA/B coalesce (foci) in each cell via homotypic interactions with receptors on adjacent cells. (3) In turn these adhesion junctions lead to OM fusion likely facilitated by membrane stress from gliding motility. During OM fusion lipids and proteins are exchanged bidirectionally. (4) Gliding motility directs OM fission which causes TraA/B complexes to disassemble. Cell that is missing an OM component, e.g. motility protein in nonmotile mutant, is replenished by transfer from partnering cell (phenotypic rescue). Model is based on findings from [23].
OME cargo principally consist of OM components. In particular, many OM lipoproteins are transferred where there is little specificity in cargo recognition. For instance, when a short ~20 amino acid lipoprotein signal sequence is engineered in a foreign protein, e.g. green fluorescent protein (GFP), it localizes to the OM where it is exchanged [24]. There is direct evidence for the transfer of about 30 endogenous lipoproteins, including the motility proteins Tgl and CglC and the diverse families of SitA toxins [27–31]. However, the M. xanthus genome contains >400 lipoproteins [32], and those that are diffusible in the OM we predict are transferred. Diffusible β-barrel OM proteins and peripheral OM proteins are also transferred, e.g. CglE, CglF and particular SitA4 toxins [29,30]. However, OM proteins that are not diffusible in the membrane, for example those anchored to the cell wall or are part of macromolecular complexes, are likely not exchanged. Soluble proteins in the periplasm may also be exchanged, while inner membrane and cytoplasmic proteins and DNA are not exchanged.
Specificity and visualization of TraA kin recognition
Recognition for OME depends on homotypic interactions between identical or highly similar TraA receptors. If the adjacent cell lacks or has a divergent TraA receptor, OME is blocked [15–17]. Among wild myxobacteria, traA alleles are highly polymorphic and therefore this provides a mechanism for kin discrimination. For instance, our study involving 59 wild traA alleles demonstrated or predicted they belong to 42 distinct recognition groups [17]. In a separate study, involving traA alleles from the genus Corallococcus, additional TraA recognition groups were predicted [30]. Figure 2 shows the phylogenetic relationship of the variable domain, involved in specificity, from 96 traA alleles that are predicted to constitute 61 distinct recognition groups. As only a small number of environmental alleles are known, we expect the total number of distinct TraA recognition groups in nature to be much larger.
Figure 2.
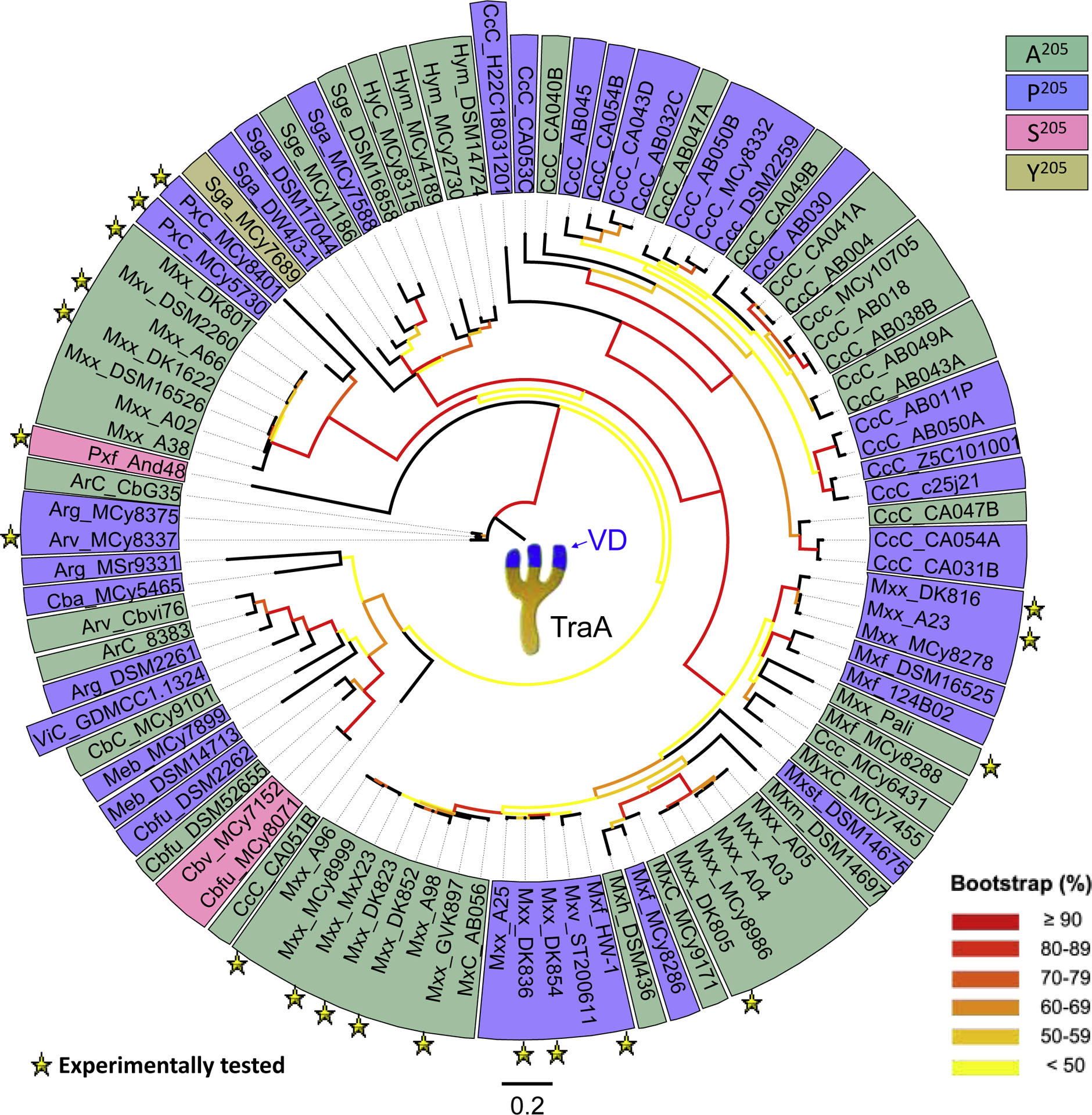
TraA polymorphisms define kin recognition groups. Phylogenetic tree contains 96 sequences of the trimmed variable domain (VD; ~200 amino acids) from TraA proteins. Experimental and bioinformatic criteria define 61 distinct TraA recognition groups as outlined in [17]. Strain names are shown with three letter prefixes that identify species names as listed in [17]. Color shading identifies key reside found at position 205 involved in TraA specificity. All sequences belong to the Cystobacterineae suborder. Although traAB sequences are found in other suborders genomes belonging to the Myxococcales order, they were excluded because their sequences are divergent and their functions are not defined [17].
When TraA/B are overexpressed, they act as adhesins holding cells together in chains for extended periods [16]. Specificity in recognition is determined by TraA and when cells overexpress different alleles of traA they only clump with those that express compatible receptors. Structurally, how homotypic binding between TraA receptors leads to specificity in recognition is an interesting and open question. Nevertheless, to address this topic, a series of chimeras were made between traA alleles with different specificities [16]. Chimeras were within the first 260 amino acids (full length of TraA is ~700 amino acids), which includes the variable domain. Interestingly, many of the chimeras were functional, indicating TraA is malleable to sequence changes, and chimera specificity typically matched one of the parent alleles. Surprisingly, site-directed mutagenesis revealed a particular residue was critical for specificity. That is, a single 205A→P substitution, or vice versa, changed TraA specificity such that the variant no longer recognized the parent receptor and, in one case, formed a novel recognition group. We concluded that this residue acts as a molecular switch where the ‘P configuration’ allows 205P binding, while the ‘A configuration’ allows 205A recognition (Fig. 2). Obviously, other residues also contribute toward specificity.
To observe kin recognition at the molecular level functional fluorescent fusions were made, e.g. TraA-GFP (internal tag) and TraB-mCherry (C-terminal tag) [23]. When cells were isolated these proteins were uniformly distributed around the cell periphery (Fig. 1). In contrast, upon cell-cell contact the TraA/B proteins coalesced to form discrete foci that resemble eukaryotic gap junctions. However, if the adjoining cell lacks receptors or contained a variant traA allele, no foci form and the proteins remain disperse around the OM. Following foci formation, fluorescent cargo is exchanged. TraA/B foci dissipate when cells disengage contact indicating membrane fission occurred. Contact with another sibling results in the cycle repeating itself.
Myxobacteria often travel in groups where dozens of cells remain in contact. Within these packs, cells engage in OME with more than one cell at a time. Topologically this results in multiple cells sharing a connected OM. It also means that cargo exchange can occur serially between cells that are not necessarily in direct contact. Serial transfer was also demonstrated in three-strain mixing experiments [30,33]. Here two strains, a donor and recipient, expressed incompatible TraA receptors that cannot undergo OME. However, when a third strain was added, which contained both traA alleles, it acted as an intermediary where cargo was serially transferred from the donor to intermediary to recipient.
Social consequences: Cooperativity
OME seems to serve two general functions. The first involves resource sharing to form homogenous and fit populations for multicellular functions. The second involves discrimination against nonkin that excludes cheaters and ensures populations remain clonal. Both social consequences are described below.
OME allows cells to act cooperatively and provides a platform to help siblings in need (Fig. 3a). This was shown in phenotypic rescue assays. Here, a donor population rescues a defective or ailing population by replenishing missing or damaged cellular components. For example, a recipient population that lacks an essential motility protein is phenotypically rescued by a donor population that provides that protein [25,26,31,34,35]. However, since no DNA is transferred, rescue is transient. OME also heals damaged membranes. This was demonstrated with a strain that conditionally expressed LpxC, an essential enzyme for LPS biosynthesis [21]. In the absence of LpxC the cells lyses, however, viability is rescued, in a traA-dependent manner, when cells are mixed with siblings that make LPS. Under certain conditions, the healthy donors also benefit because, for example, multicellular development requires a threshold population size [36]. That is, cells cannot form spores when there is not a minimum number of cells. This barrier is relieved by adding lpxC cells. Here, although lpxC cells cannot sporulate on their own, their fitness defect is corrected by OME with healthy donors, and in turn the total population of fit cells rises above the threshold number needed for sporulation.
Figure 3.
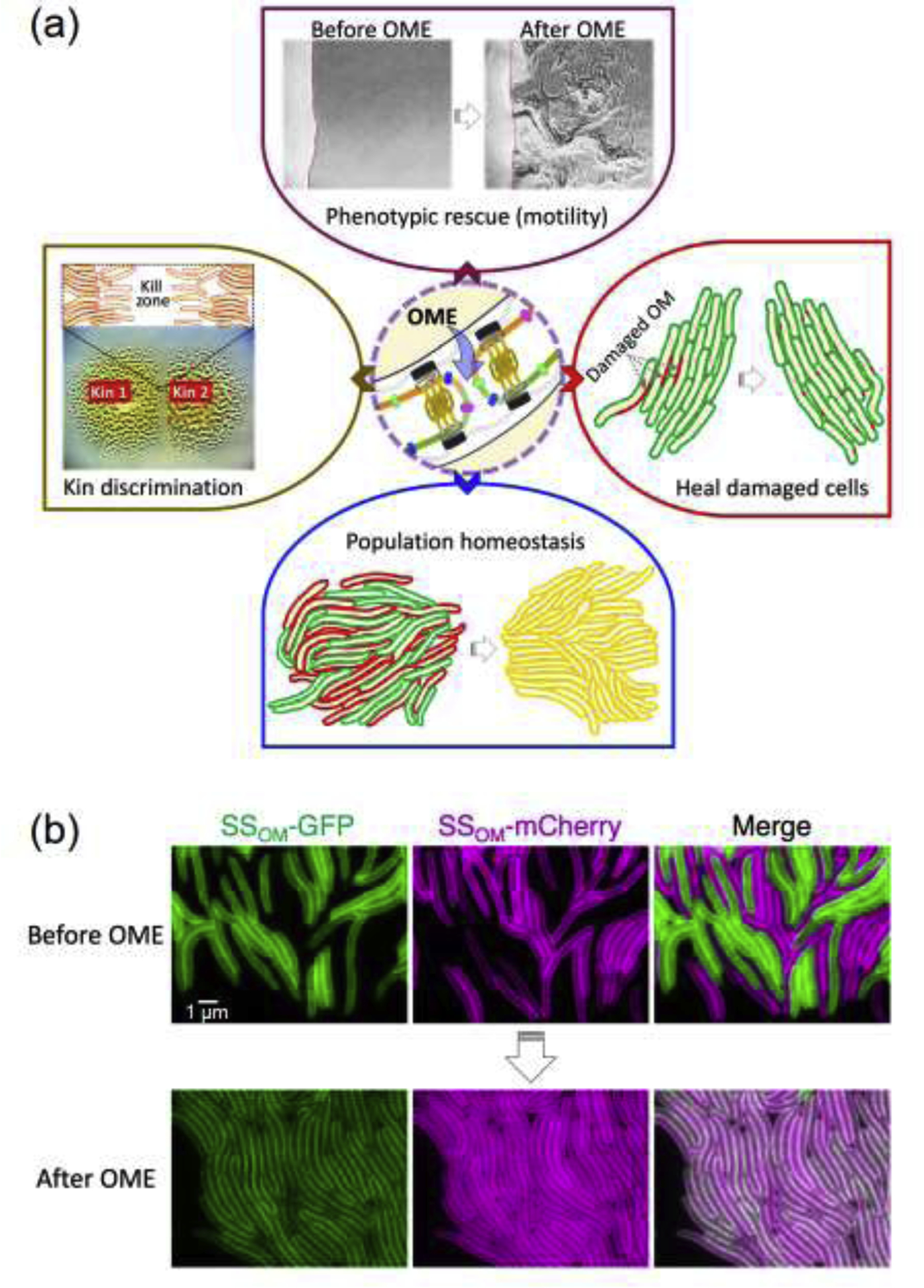
Social consequences of OME. (a) Top panel shows phenotypic rescue of motility defect. A nonmotile strain, lacking an essential OM motility protein, is mixed with a donor nonmotile strain that contains the missing protein. After transfer of the motility protein by OME the recipient cells become motile. Right panel illustrates repair of damaged OM (red) by replenishment of membrane from healthy cells (green) through OME. Bottom panel illustrates homogenization of OM (yellow) in two isogenic populations that originally adapted to different microenvironments (green and red). Left panel shows kin discrimination and formation of demarcation between two populations with identical TraA receptors but different sets of toxins. Nonkin populations are intoxicated and form a killing zone between colonies. (b) Micrographs showing OME between isogenic populations containing identical traA alleles but different OM cargo (GFP and mCherry). Following OME, all cells have equal amounts of both reporters, creating a homogenous population [23]. SSOM = lipoprotein signal sequence for OM localization.
Another striking feature of OME is that it allows populations that are phenotypically distinct, but genotypically congruent, to merge and mix components to create homogenous OMs (Fig. 3b) [23]. We hypothesize that this function helps cells with different life histories and adaptations to transition toward multicellularity by creating harmonious populations, essentially resembling a tissue, wherein cells coherently communicate and behaviors are coordinated and synchronize to conduct multicellular tasks.
Social consequences: Discrimination
OME can occur between nonkin that share compatible TraA receptors. In turn this could lead to exploitation of resources and cooperative behaviors by nonkin. However, in practice, exploitation is blocked by a second layer of discrimination. Here, TraA-TraA recognition is followed by SitA polymorphic toxin exchange (Fig. 3a) [33,37]. In practical terms only true siblings are likely to meet both criteria, with self-identity mediated by toxin repertoire exchange being particularly stringent (Fig. 4). These lipoprotein toxins belong to six distinct families that share domain organization and delivery mechanism. Each SitA family is polymorphic and surprisingly expansive in number; 15 to 83 sitA loci found per genome with the average number being ~38 [30]. Each family contains a unique and conserved ‘escort domain’ thought to facilitate cytoplasmic entry after delivery to the OM by OME. Forty distinct families of toxin domains are known, where they are mostly predicted to function as nucleases. The link between OME and toxin delivery is robust, as all known myxobacterial genomes that contain a bona fide traAB locus also contain sitA loci [30]. In addition, all of these sitA genes are accompanied by a cognate downstream immunity gene (sitI) that is allele specific. In contrast to SitA, SitI proteins are not transferred. Therefore, when the recipient is a clonal cell it is immune to the transfer of SitA proteins by expressing the cognate suite of immunity proteins, while TraA compatible cells that are non-clonal will likely perish, as they will not contain a full complement of immunity proteins. In fact, when examining myxobacterial genomes that contain compatible TraA receptors, dozens of sitAI loci are often found to be unique. For example, when we compared six genomes with compatible TraA receptors, we found 145 sitAI loci predicted to be unique between them [30]. From this pool, if we assume theoretical genomes only contains 20 unique sitAI loci, there is an astronomical number of possible discriminatory combinations (~2 × 1024). For this reason we termed such repertoires of sitAI loci as ‘self-identity barcodes’ that exquisitely discriminate self from nonself (Fig. 4).
Figure 4.
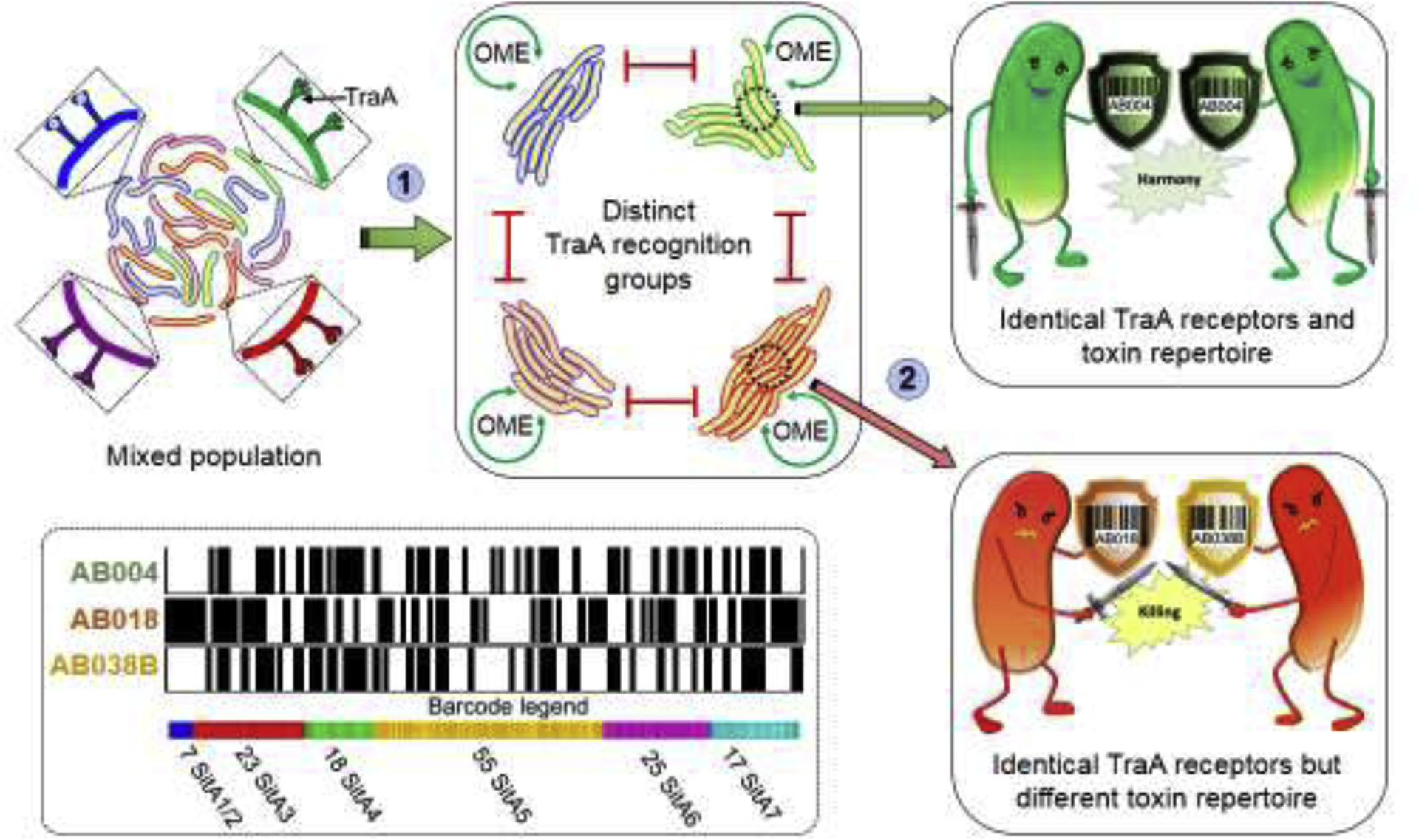
OME of SitA toxins leads to distinct kin groups. Cartoon illustrates two levels of discrimination. (1) Mixed population of myxobacteria discriminate kin from foe by homotypic matching between TraA receptors. Only cells with compatible traA alleles conduct OME. (2) Compatible TraA cells that are clonal contain identical sets of sitAI toxin-immunity loci (barcodes) survive OME and ensures a homogenous population (top). In contrast, cells with compatible TraA receptors that are not clonal contain different toxin-immunity barcodes and annihilate one another (bottom). Bottom left, based on genomic analysis of six wild strains with compatible traA alleles a total of 145 unique sitA loci were identified [30]. Barcodes for three strains shown; black lines indicate the presence of a particular toxin while white lines absence thereof.
Predicting social compatibility
Understanding how myxobacteria recognize self and discriminate against nonself opens the door to testable predictions of social compatibility based on genomic content (Fig. 4). Indeed, in a recent study we predicted social compatibilities of 22 wild M. xanthus isolates based on their repertoire of traAB and sitAI loci as well as toxins delivered by other systems [38]. Strikingly, these 22 isolates consisted of two clades wherein isolates were highly related but nevertheless had diverged into 11 social compatibility groups [39]. We showed that their recent social incompatibilities were caused by the acquisition of mobile elements that contained unique polymorphic toxins, including those delivered by OME. In a predictable manner social incompatibilities were reprogrammed by simply blocking the transfer of unique SitA toxins as well as toxins delivered by type 6 secretion and Rhs systems [38,40]. Additionally, changes in kin discrimination also evolved in these natural populations by the horizontal gene transfer of the traAB loci from one clade to another. We note that another group was unable to correlate traA allele variability with social compatibility groupings [41,42], however this failure was predictable because sitAI loci diversity was not considered, even though it is necessary criteria that was previously established [33].
Kin or greenbeard recognition?
Recognition of related individuals is defined in different evolutionary terms depending on how recognition occurs [43,44]. For example, in kin recognition, individuals identify others when they share many loci involved in social behaviors and they originate from common ancestry. In contrast, so-called greenbeard recognition, instead relies on a single locus to identify others that contain the same molecular (greenbeard) tag or allele [45]. In one mechanism, greenbeard recognition involves homotypic interactions between individuals with identical cell surface receptors [46]. Additionally, in greenbeard recognition other social loci are not necessarily related and individuals do not necessarily share common ancestry. In many ways TraA/B meets the greenbeard definition because the genes overlap in a single operon and TraA polymorphisms allows discrimination between different traA alleles [44]. Moreover, by simply swapping traA alleles cell-cell recognition is reprogrammed [15,16]. However, our recent studies showed that for harmonious OME outcomes to occur the identity of other social loci, i.e. sitAI, matter beyond simple greenbeard recognition. For this reason, TraA/B is also described as kin recognition. Finally, we previously argued that the reason TraA polymorphisms are selected for and maintained in populations is because OME between nonkin leads to antagonism and cell death; hence selectivity in TraA recognition is maintained by natural selection [33].
Conclusions
The external world presents many challenges to microbes and their ability to recognize neighbors as kin, prey or foe provides a strong fitness advantage that involves appropriate responses. Since myxobacteria elicit complex social interactions that transcend into multicellular life, they provide a particularly fruitful model for understanding molecular mechanisms involved in kin recognition. OME is one mechanism, which is specific to the Myxococcales order, but other mechanisms await to be discovered in this group and other groups of bacteria.
Highlights:
Self-recognition mediated by homotypic binding of a dynamic polymorphic cell surface receptor
TraA and TraB function as a fusogen that mediates outer membrane cargo exchange and homeostasis in Myxococcus xanthus
Polymorphic toxin exchange function as self-identity barcodes
Experimental complexities in distinguishing greenbeard recognition from kin recognition
Acknowledgements:
We dedicate this review to the memory of Dale Kaiser. This work was supported by National Institutes of Health Grant GM101449 to DW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
● of special interest
● ● of outstanding interest
- 1.Wall D: Kin recognition in bacteria. Annu Rev Microbiol 2016, 70:143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strassmann JE, Gilbert OM, Queller DC: Kin discrimination and cooperation in microbes. Annu Rev Microbiol 2011, 65:349–367. [DOI] [PubMed] [Google Scholar]
- 3.Kaiser D, Manoil C, Dworkin M: Myxobacteria: Cell interactions, genetics, and development. Annu Rev Microbiol 1979, 33:595–639. [DOI] [PubMed] [Google Scholar]
- 4.Dworkin M: Biology of the myxobacteria. Annu Rev Microbiol 1966, 20:75–106. [DOI] [PubMed] [Google Scholar]
- 5.Bretl DJ, Kirby JR: Molecular mechanisms of signaling in Myxococcus xanthus development. J Mol Biol 2016, 428:3805–3830. [DOI] [PubMed] [Google Scholar]
- 6.Claessen D, Rozen DE, Kuipers OP, Sogaard-Andersen L, van Wezel GP: Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol 2014, 12:115–124. [DOI] [PubMed] [Google Scholar]
- 7.Troselj V, Cao P, Wall D: Cell-cell recognition and social networking in bacteria. Environ Microbiol 2018, 20:923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pathak DT, Wei X, Wall D: Myxobacterial tools for social interactions. Res Microbiol 2012, 163:579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutton D, Livingstone PG, Furness E, Swain MT, Whitworth DE: Genome-wide identification of myxobacterial predation genes and demonstration of formaldehyde secretion as a potentially predation-resistant trait of Pseudomonas aeruginosa. Front Microbiol 2019, 10:2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiery S, Kaimer C: The predation strategy of Myxococcus xanthus. Front Microbiol 2020, 11:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W, Wang Y, Lu H, Liu Q, Wang C, Hu W, Zhao K: Dynamics of solitary predation by Myxococcus xanthus on Escherichia coli observed at the single-cell level. Appl Environ Microbiol 2020, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaiser D: Roland Thaxter’s legacy and the origins of multicellular development. Genetics 1993, 135:249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Garcia P, Moreira D: The Syntrophy hypothesis for the origin of eukaryotes revisited. Nat Microbiol 2020, 5:655–667. [DOI] [PubMed] [Google Scholar]; ● This perspective article provides an up-to-date and in-depth discussion about the origin of eukaryotes and the role an ancient myxobacterium-like cell may have played.
- 14.Wall D: Molecular recognition in myxobacterial outer membrane exchange: Functional, social and evolutionary implications. Mol Microbiol 2014, 91:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pathak DT, Wei X, Dey A, Wall D: Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLoS Genet 2013, 9:e1003891.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao P, Wall D: Self-identity reprogrammed by a single residue switch in a cell surface receptor of a social bacterium. Proc Natl Acad Sci U S A 2017, 114:3732–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao P, Wei X, Awal RP, Muller R, Wall D: A highly polymorphic receptor governs many distinct self-recognition types within the Myxococcales order. mBio 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]; ● This study demonstrates that TraA is polymorphic and that there are many distinct recognition groups among natural isolates.
- 18.Sah GP, Cao P, Wall D: MYXO-CTERM sorting tag directs proteins to the cell surface via the type II secretion system. Mol Microbiol 2020, 113:1039–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D: Cell contact-dependent outer membrane exchange in myxobacteria: Genetic determinants and mechanism. PLoS Genet 2012, 8:e1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ducret A, Fleuchot B, Bergam P, Mignot T: Direct live imaging of cell-cell protein transfer by transient outer membrane fusion in Myxococcus xanthus. eLife 2013, 2:e00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vassallo C, Pathak DT, Cao P, Zuckerman DM, Hoiczyk E, Wall D: Cell rejuvenation and social behaviors promoted by LPS exchange in myxobacteria. Proc Natl Acad Sci U S A 2015, 112:E2939–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao P, Dey A, Vassallo CN, Wall D: How myxobacteria cooperate. J Mol Biol 2015, 427:3709–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao PB, Wall D: Direct visualization of a molecular handshake that governs kin recognition and tissue formation in myxobacteria. Nat Commun 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]; ●● This study created functional fusions of TraA and TraB to fluorescent proteins and showed these fluid OM receptors form foci upon physical contact and kin recognition with neighboring cells. OME was also shown to create homogenous OM between phenotypically divergent populations.
- 24.Wei X, Pathak DT, Wall D: Heterologous protein transfer within structured myxobacteria biofilms. Mol Microbiol 2011, 81:315–326. [DOI] [PubMed] [Google Scholar]
- 25.Wall D, Kaiser D: Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc Natl Acad Sci U S A 1998, 95:3054–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wall D: Social interactions mediated by outer membrane exchange In Myxobacteria: Genomics, Cellular and Molecular Biology. Edited by Yang Z, Higgs PI: Caister Academic Press; 2014. [Google Scholar]
- 27.Rodriguez-Soto JP, Kaiser D: The tgl gene: social motility and stimulation in Myxococcus xanthus. J Bacteriol 1997, 179:4361–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez AM, Spormann AM: Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J Bacteriol 1999, 181:4381–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pathak DT, Wall D: Identification of the cglC, cglD, cglE, and cglF genes and their role in cell contact-dependent gliding motility in Myxococcus xanthus. J Bacteriol 2012, 194:1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassallo CN, Wall D: Self-identity barcodes encoded by six expansive polymorphic toxin families discriminate kin in myxobacteria. Proc Natl Acad Sci U S A 2019, 116:24808–24818. [DOI] [PMC free article] [PubMed] [Google Scholar]; ●● This study discovered six families of lipoprotein toxins delivered by OME. These families are expansive in myxobacterial genomes and act as ‘self-identity barcodes.’
- 31.Nudleman E, Wall D, Kaiser D: Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science 2005, 309:125–127. [DOI] [PubMed] [Google Scholar]
- 32.Bhat S, Zhu X, Patel RP, Orlando R, Shimkets LJ: Identification and localization of Myxococcus xanthus porins and lipoproteins. PLoS One 2011, 6:e27475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vassallo CN, Cao P, Conklin A, Finkelstein H, Hayes CS, Wall D: Infectious polymorphic toxins delivered by outer membrane exchange discriminate kin in myxobacteria. eLife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodgkin J, Kaiser D: Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A 1977, 74:2938–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakobczak B, Keilberg D, Wuichet K, Sogaard-Andersen L: Contact- and protein transfer-dependent stimulation of assembly of the gliding motility machinery in Myxococcus xanthus. PLoS Genet 2015, 11:e1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vassallo CN, Wall D: Tissue repair in myxobacteria: A cooperative strategy to heal cellular damage. BioEssays 2016, 38:306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dey A, Vassallo CN, Conklin AC, Pathak DT, Troselj V, Wall D: Sibling rivalry in Myxococcus xanthus is mediated by kin recognition and a polyploid prophage. J Bacteriol 2016, 198:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vassallo CN, Troselj V, Weltzer ML, Wall D: Rapid diversification of wild social groups driven by toxin-immunity loci on mobile genetic elements. ISME J 2020, 10.1038/s41396-020-0699-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; ●● This study shows how horizontal gene transfer of sitAI and traAB loci led to the social diversification of M. xanthus natural isolates.
- 39.Wielgoss S, Didelot X, Chaudhuri RR, Liu X, Weedall GD, Velicer GJ, Vos M: A barrier to homologous recombination between sympatric strains of the cooperative soil bacterium Myxococcus xanthus. ISME J 2016, 10:2468–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong Y, Zhang Z, Liu Y, Zhou XW, Anwar MN, Li ZS, Hu W, Li YZ: A nuclease-toxin and immunity system for kin discrimination in Myxococcus xanthus. Environ Microbiol 2018, 20:2552–2567. [DOI] [PubMed] [Google Scholar]
- 41.Wielgoss S, Fiegna F, Rendueles O, Yu YN, Velicer GJ: Kin discrimination and outer membrane exchange in Myxococcus xanthus: A comparative analysis among natural isolates. Mol Ecol 2018, 27:3146–3158. [DOI] [PubMed] [Google Scholar]
- 42.Cossey SM, Yu YN, Cossu L, Velicer GJ: Kin discrimination and outer membrane exchange in Myxococcus xanthus: Experimental analysis of a natural population. PLoS One 2019, 14:e0224817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Queller DC: Expanded social fitness and Hamilton’s rule for kin, kith, and kind. Proc Natl Acad Sci U S A 2011, 108 Suppl 2:10792–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madgwick PG, Belcher LJ, Wolf JB: Greenbeard genes: Theory and reality. Trends Ecol Evol 2019, 34:1092–1103. [DOI] [PubMed] [Google Scholar]; ● This opinion article critically reviews the greenbeard concept and examines experimental examples of such genes including the traA/B and sitAI loci.
- 45.Dawkins R: The Selfish Gene. Oxford, UK: Oxford University Press; 1976. [Google Scholar]
- 46.Haig D: Gestational drive and the green-bearded placenta. Proc Natl Acad Sci U S A 1996, 93:6547–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]


