Abstract
Background:
Manual annotation of seizures and interictal-ictal-injury continuum (IIIC) patterns in continuous EEG (cEEG) recorded from critically ill patients is a time-intensive process for clinicians and researchers. In this study, we evaluated the accuracy and efficiency of an automated clustering method to accelerate expert annotation of cEEG.
New method:
We learned a local dictionary from 97 ICU patients by applying k-medoids clustering to 592 features in the time and frequency domains. We utilized changepoint detection (CPD) to segment the cEEG recordings. We then computed a bag-of-words (BoW) representation for each segment. We further clustered the segments by affinity propagation. EEG experts scored the resulting clusters for each patient by labeling only the cluster medoids. We trained a random forest classifier to assess validity of the clusters.
Results:
Mean pairwise agreement of 62.6% using this automated method was not significantly different from interrater agreements using manual labeling (63.8%), demonstrating the validity of the method. We also found that it takes experts using our method 5.31 ± 4.44 min to label the 30.19 ± 3.84 h of cEEG data, more than 45 times faster than unaided manual review, demonstrating efficiency.
Comparison with existing methods:
Previous studies of EEG data labeling have generally yielded similar human expert interrater agreements, and lower agreements with automated methods.
Conclusions:
Our results suggest that long EEG recordings can be rapidly annotated by experts many times faster than unaided manual review through the use of an advanced clustering method.
Keywords: EEG, Critical care, Ictcal-interictal continuum, Clustering, Unsupervised learning
1. Introduction
The electroencephalogram (EEG) is a cornerstone diagnostic modality employed clinically for epilepsy evaluation, sleep studies, and neurocritical care. In all of these settings, clinicians retain a high burden of manually annotating EEG into classes such as seizures, periodic discharges, or sleep stages. Detecting electrographic events like these by manual review of EEG remains a critical bottleneck. In both clinical and research settings, the ability to automatically annotate EEG with high accuracy would greatly improve the efficiency of multiple analyses as compared to present practices.
In the last decade, continuous EEG (cEEG) monitoring of high-risk patients in the intensive care unit (ICU) has become the standard of care (Hirsch, 2004; Friedman et al., 2009). Events of clinical interest in the ICU often presenting with correlates on cEEG include seizures, ischemia, hemorrhage, and elevated intracranial pressure (Friedman et al., 2009; Kennedy and Gerard, 2012). The volume of cEEG data recorded during a typical ICU stay for a single patient imposes a significant annotation burden on clinicians. Automated annotation of cEEG from the ICU specifically could significantly accelerate classification and diagnosis to support clinical decisions for critically ill patients.
Nonconvulsive seizures (NCS) constitute a clinically impactful class of events with prognostic significance (Claassen, 2009). Up to 48% of patients in the ICU may exhibit NCS (Friedman et al., 2009). NCS can cause or exacerbate neuronal injury, inducing worse outcomes including permanent neurologic dysfunction and mortality (Hirsch, 2004). While these events most often have no discernable behavioral or functional correlate, they are detectable on cEEG (Friedman et al., 2009; Shneker and Fountain, 2003).
Due to the clinical significance of these events, NCS is one of the labels used in this study of automated cEEG annotation. While seizure detection studies on convulsive seizures in patients with epilepsy syndromes have yielded performances with sensitivity and specificity over 95%, NCS detection on ICU patient cEEG data has performed considerably worse, and has not been evaluated on large datasets (Bose et al., 2017; Golmohammadi et al., 2017; Sackellares et al., 2011).
In addition to NCS, the IIIC includes several other rhythmic seizure-like patterns not considered to be definite seizures, but still associated with poor outcomes and increased seizure risk (Gaspard et al., 2014). Therefore, in this study, we explore not only automatic labeling of NCS, but also additional IIIC patterns. In particular, lateralized (L) or generalized (G) periodic discharges (PD) and rhythmic delta activity (RDA), as defined by American Clinical Neurophysiology Society (ACNS) ICU EEG terminology, have been shown to correlate with poor neurologic outcomes (Claassen, 2009; Hirsch, 2011; Foreman et al., 2016; Halford et al., 2015). Efficient labeling of voluminous cEEG data will help enable development of better performing diagnostic and prognostic algorithms for clinical use, as well as more accurate models of the functional impact of IIIC events in research use.
In order to divide cEEG data into discrete segments that can be annotated, we employ changepoint detection (CPD), a method that identifies sudden changes in sequential data (Adams and MacKay, 2007). In addition to EEG segmentation, variants of CPD methods have also been used effectively in process control, DNA segmentation, and epidemiology (Adams and MacKay, 2007; Reeves et al., 2007; Barlow et al., 1981; Kaplan and Shishkin, 2000). We then apply a bag-of-words (BoW) model, which summarizes each cEEG segment into a histogram of its composite “words”, which is then used for clustering. This BoW approach is adapted from machine learning methods developed for text and image classification (Zhang et al., 2010).
In the presented analysis, we apply a method we introduced in an earlier pilot study (Jing et al., 2018), BoW-based clustering, to CPD-segmented cEEG data from critically ill patients in the ICU. Our method is designed to facilitate rapid and efficient labeling of cEEG recordings by experts, as compared to manual labeling. In this paper, we evaluate the performance of our method both in terms of the quality of its results, as measured in terms of interrater agreement of experts using the method, and in terms of the mean time required for expert annotation.
2. Materials and methods
2.1. EEG samples and feature extraction
We selected archival data from 97 ICU patients with a variety of IIIC patterns. The local institutional review board (IRB) waived the requirement for informed consent for this retrospective analysis of EEG data. We used the MATLAB R2017 (Natick, MA) Signal Processing Toolbox for signal processing. For each patient, we collected at least 24 h of EEG data. We converted this data to longitudinal bipolar montage and resampled it to 200 Hz. Furthermore, we applied bandpass filtering between 0.5 Hz and 40 Hz to denoise the data. We did not apply additional artifact detection and removal before clustering, so that the clustering method would be robust to real-world clinical signal irregularities.
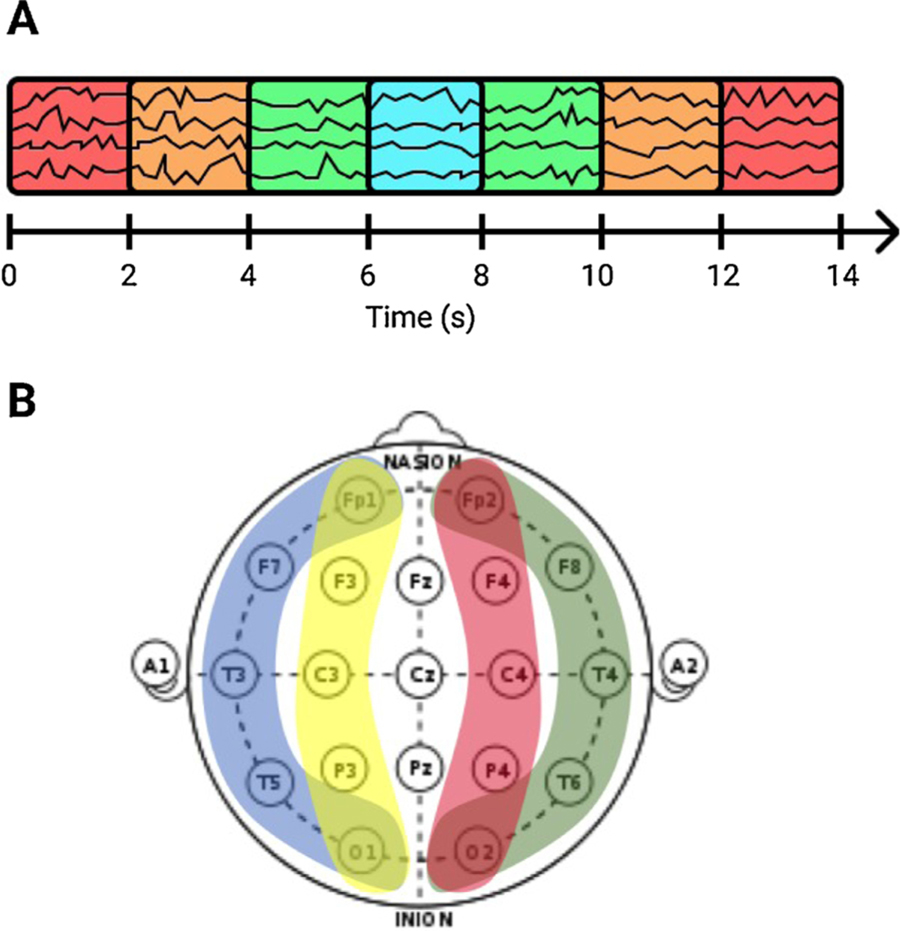
We then divided all cEEG recordings into 2 s segments, and extracted a number of features in the spectral and time domains. These features include classic measures such as line length, kurtosis, entropy, nonlinear energy operator activation, relative power, power ratios, and power kurtosis (see Table 1). The chosen features were for a large part based on prior work, e.g. on automated sleep staging (Sun et al., 2017). The spectral features were calculated with the use of spectrograms, which were estimated with a multitaper (MT) framework (Babadi and Brown, 2014). To include contextual information from the surrounding EEG, we also computed these features within windows of 6, 10 and 14 s centered on each 2 s segment (see Fig. 1A). We divided the scalp into 4 different brain regions for feature construction (LL: Left Lateral, RL: Right Lateral, LP: Left Parasagittal, and RP: Right Parasagittal) in order to represent spatial information (see Fig. 1B).
Table 1.
EEG features.
| Temporal features | Feature calculation | Measurement |
|---|---|---|
| Line length | Total variation | |
| Kurtosis | Extreme values | |
| Shannon entropy | Absolute value | Signal irregularity |
| Nonlinear energy operator | Mean and SD | Changes of stationarity |
| Spectral features | ||
| Absolute δ, θ, α, and β power | Kurtosis | |
| Relative δ, θ, α, and β power | Mean, min, SD, 95th percentile | |
| δ/θ, δ/α, and θ/α power ratios | Mean, min, SD, 95th percentile | |
δ = 1–4 Hz, θ = 4–8 Hz, α = 8–12 Hz, β = 12–18 Hz.
Fig. 1.
Featurization of cEEG data. Features were computed from windows of 2, 6, 10 and 14 s, centered on each 2 s segment (A), and from 4 different brain regions (B), to include contextual and spatial information in the feature representation of the EEG.
The 37 different spectral and temporal features from all 4 temporal scales and all 4 spatial regions resulted in a total of 592 features, which collectively describe each 2 s segment of cEEG. This rich set of features is intended to suffice for differentiating patterns encountered in the cEEGs of ICU patients, including variations of NCS and patterns along the IIIC.
2.2. CPD-BoW based unsupervised clustering
The following steps were applied on each individual subject.
Changepoint detection (CPD) is a general method to find abrupt changes in time series (Guralnik and Srivastava, 1999; Lund et al., 2007). We applied CPD on the averaged spectrograms of each cEEG recording using a parametric global method, implemented in the MATLAB (Natick, MA) Signal Processing Toolbox. This method finds K changepoints in the signal x1, x2, …, xN by minimizing the following objective function for each recording:
| (1) |
Here, k1, …, kK are the indices of the changepoints, with k0 and kK+1 defined as the first and last sample in the signal respectively. is the mean operator and βK represents the penalty term added to avoid overfitting (i.e. introducing too many changepoints). This penalty term had a default of 10 times the variance in power within the segment, but could be manually adjusted by the user in the GUI. For the minimization we applied a recursive optimization algorithm based on dynamic programming with early abandonment (Killick et al., 2012). This breaks each cEEG into variable length segments that are relatively homogeneous between changepoints. The changepoints were rounded up to the future to fit within the 2 s temporal scale. To preempt the possibility of hypo-segmentation by automated CPD, we use a conservative threshold for CPD to attain uniform segments; this threshold is optimized based on iterative user testing and feedback.
Subsequently, we applied a bag-of-words (BoW) model (also known as a “term-frequency counter”) (Zhang et al., 2010). This model is commonly applied in document classification by recording the frequency of occurrence of each word. In this study, we consider each cEEG recording as a special kind of “text,” with the variable length segments between the changepoints as “sentences,” and the consecutive 2 s segments as the elementary “words.”For each patient, feature dimensionality was reduced using principal component analysis (PCA), with 95% variance retained. We learned a dictionary of words by clustering these dimensionality-reduced feature arrays using k-medoids clustering with k = 100 (chosen empirically). Here, each cluster represents one type of word, so that each sentence is represented as a collection of words. We then calculated the normalized histogram of words in each sentence, which is known as the BoW. Finally, we clustered the sentences based on the corresponding BoW by applying χ2-based affinity propagation (AP) (see Fig. 2) (Dueck and Frey, 2007). The APCLUSTER toolbox in MATLAB was applied for this purpose. AP is a clustering algorithm that starts by considering all data points as potential “exemplars” and then updates the availability of each point by recursively transmitting real-valued messages along the edges of the network until the optimal exemplars with corresponding clusters remain. The advantage of this method is that it does not require a pre-defined number of clusters.
Fig. 2.
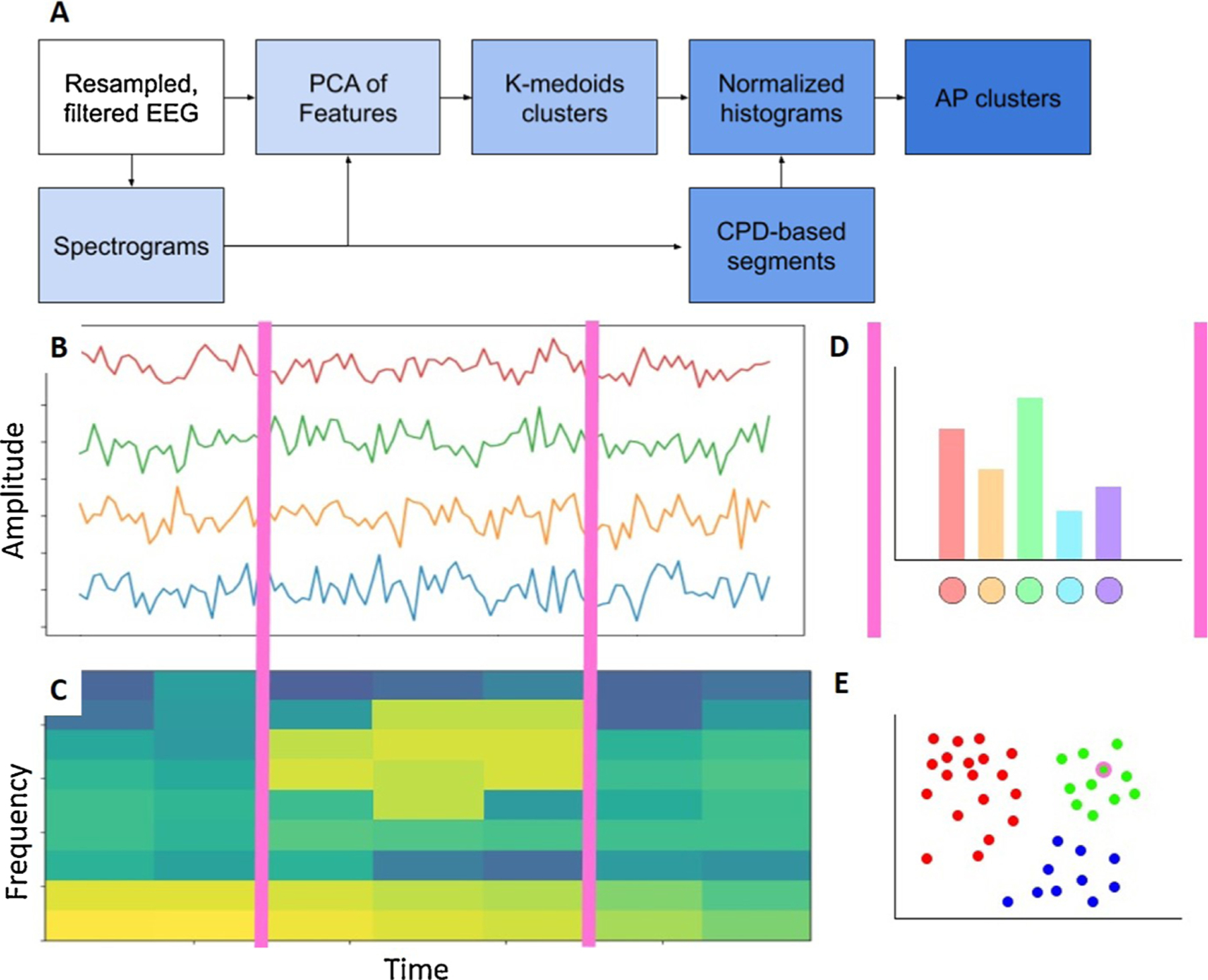
Affinity propagation based clustering of CPD-BoW represented EEG. (A) A step-by-step representation of the proposed method. Filtered EEG (B) and corresponding spectrograms (C) were segmented via changepoint detection (CPD), demarcated with magenta lines. (D) Each segment was then represented as a bag-of-words (BoW) histogram. (E) Chi-squared affinity propagation (AP) clustering then assigned the sample segment, encircled in magenta, to one of several clusters. This figure, intended to illustrate our methodology, is based on synthetic data. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3. NCS and IIIC annotation
Three EEG experts (MT, MN, AH), i.e. fellowship trained epileptologists, independently performed manual scoring of the center of each exemplar sentence, defined as the medoid of each cluster identified by AP. A MATLAB-based graphical user interface (GUI) was developed for this purpose, as shown in Fig. 3. For each 2-second segment, the raw EEG and spectrograms were displayed within a wider temporal context of 14 s (EEG) and 10–60 min (spectrograms). An embedding map showing the clusters in a two-dimensional space (computed via t-SNE) was also displayed. The initial reduced dimensionality and the perplexity of the Gaussian kernel of the t-SNE were both set to 30 and it was implemented in the MATLAB Toolbox for Dimensionality Reduction (v0.8.1b). The GUI presented the medoids sequentially to the experts, and experts annotated each pattern by clicking one of six label buttons.
Fig. 3.
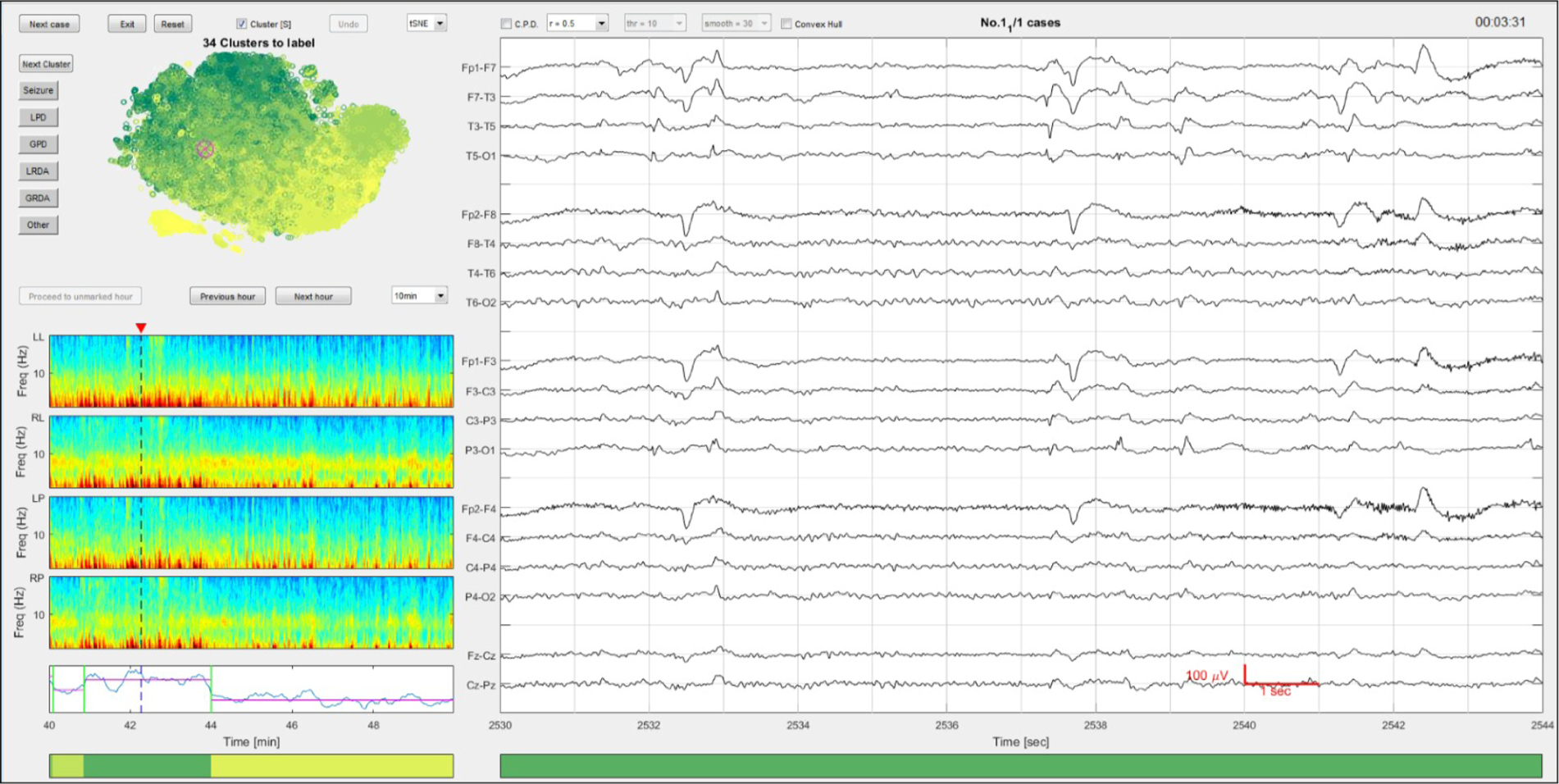
The graphical user interface for rapid annotation. 14 s of EEG are shown on the right. The regional average spectrograms are shown on the left with the changepoint detection results below. The unsupervised clustering membership assignment is illustrated by the horizontal color bar at the bottom, as determined by the CPD-BoW-AP steps. The colors are assigned based on the average total power from all members in that cluster. The higher the power values usually correlate with severity of the EEG patterns (darker colors are more likely to be seizures or IIIC patterns). Above the spectrograms is a 2D embedding map computed using t-SNE (Maaten and Hinton, 2008) for data visualization and exploration. Each scattered point in this map corresponds to a 592-dimensional feature vector extracted from a 2 s EEG interval. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The different EEG patterns that we aimed to distinguish were “Seizure”, and the most common IIIC patterns: “LPD”, “GPD”, “LRDA” and “GRDA”, as defined by the ACNS (Hirsch et al., 2013). An “Other” class was added as well to cover any other patterns, including baseline/background EEG, and major artifacts. We hypothesized that our CPD-BoW based clustering would render the data into relatively uniform groups of EEG patterns that can be accurately labeled as a group, by only inspecting the medoid exemplar of each cluster.
To reduce label ‘noise’ (as distinct from true inter-rater disagreement), we applied a simple label de-noising rule, based on domain knowledge. For each of the following pairs of labels, if two experts agreed on one label and the third expert disagreed, the third expert’s label was changed to agree with the other two: (Seizure, LPD), (Seizure, GPD), (Seizure, LRDA). The justification for this label-denoising rule is as follows: because IIIC patterns lie along a continuum, the classification of some EEG patterns are ambiguous, i.e. there can be more than one ‘correct’ classification. This is particularly true for distinguishing between seizure, LPD, and GPD patterns. In such cases, labels from the three experts such as (seizure, LPD, LPD) do not represent true disagreement. We validated this rule by all three experts reviewing approximately 100 of these cases, confirming that indeed in nearly every case, the third expert conceded no difference in choosing between the label they originally gave versus the label given by their peers. The denoised labels were used throughout the remainder of the analysis.
2.4. Validity of clusters
To assess the validity of the BoW-based clustering, we trained a random forest classifier (RF) using a combination of labeled and pseudolabeled data. The labeled data were the cluster medoids (2 s segments) that were directly annotated by our experts. The pseudolabeled data consisted of all non-medoid 2 s segments. Each of these segments received the label corresponding to that of the medoid of the cluster it belonged to. We reasoned that if this classifier is able to learn from the pseudolabeled data, this would indicate that clusters indeed are suitably uniform in terms of the EEG patterns they encompass. For example, if a medoid of a cluster is labeled as pattern A, but the majority of data points in this cluster have true label pattern B, the classifier would falsely learn to classify a similar data point as pattern A, as all data points received the label of the medoid. On the other hand, if the large majority of points have true labels that match that of the medoid, the classifier should be able to learn to classify correctly.
First, each cluster was labeled according to the majority vote of the medoid of that cluster. If all three experts disagreed on the label of a medoid, its cluster was ignored for training. Thereafter, we pooled all patient data, i.e. the original 592 features of the 2 s segments, and randomly selected 100 samples from 80% of all clusters, excluding the medoid, which was the sample shown to experts for scoring. If a cluster consisted of less than 100 data points, all data points were selected. We applied PCA with 95% variance retained to the selected data and used this as training data for the RF. The center 2 s segments of the medoids of the remaining 20% of clusters, which were all visually scored by the experts, were used as testing data. This way, we made sure that no clusters involved in testing the classifier was also involved in training.
We used the dimensionality-reduced feature arrays as input for a RF containing 1000 trees. We applied balanced class-weights for training the RF. We performed a non-stratified 5-fold cross-validation to assess the performance of this training and testing procedure.
For each fold of cross-validation, we calculated the percentage agreement between the RF and the majority vote, as well as between the RF and each individual expert, and compared this to the agreement between the experts. We hypothesized that the agreement between the model and the experts would be statistically equivalent to the agreement within the experts. This was tested with a two one-sided test for equivalence (TOST) with the limits set to ±SD of the agreement within the experts (TOST toolblox version 1.0.0.0 in MATLAB) (Rogers et al., 1993). A significant equivalence test would constitute evidence that ‘pseudolabeled’ samples inherited from a single manually-annotated cluster center are of sufficiently high quality to justify forgoing the labor-intensive process of manually annotating all individual EEG samples within a cluster.
To assess the added value of the CPD-BoW based clustering over a more straightforward method, we applied the same RF classification method to the k-medoids clusters upon which the BoW model was based. Hereafter, we compared the percentage agreement between this benchmark model and the majority vote of the experts with the percentage agreement between our more advanced model and the majority vote.
3. Results
The cEEG recordings had a mean length of 30.19 h (SD: 3.84), and the BoW-based clustering of each recording resulted in a mean number of clusters of 27 (SD: 11, range: 5–50). Table 2 shows the time per patient taken by the experts to label all cluster medoids of that patient.
Table 2.
Annotation time cost per-patient (in minutes).
| Mean ± SD | Median | IQR | |
|---|---|---|---|
| Expert 1 | 5.61 ± 7.66 | 3.73 | 2.23–5.64 |
| Expert 2 | 2.08 ± 1.38 | 1.55 | 1.12–2.94 |
| Expert 3 | 8.86 ± 3.33 | 8.40 | 6.74–11.34 |
| Overall | 5.52 ± 5.60 | 3.97 | 1.68–7.70 |
IQR: inter quartile range.
As can be seen, the median time taken by the 3 experts to label 24+ h of EEG data is around 4 min. In comparison, conventional review consists of serially reviewing 10–15 s EEG intervals of the 24 h of EEG, which requires visual inspection of between 5760–8640 individual intervals. Annotating 24 h of EEG at a temporal resolution of one label per 2 s, which is the resolution obtained by the proposed annotation scheme, requires applying 43,200 labels per EEG. In practice, some time saving is often possible by “drawing boxes” around events of interest and labeling the entire events at once. Nevertheless, even done this way, manual annotation generally takes 2–4 h per 24 h of EEG (unpublished observations of author MBW), and is thus not scalable. Using 3 h as a conservative lower bound for unaided manual annotation, we estimate that our method provides a speedup of at least 45 times.
Pooling all clusters for training the RF resulted in a total of 2623 clusters (mean of 27 per subject). So in each fold, the test set consisted of 524 or 525 data points (20% of all clusters). 19.75% of the clusters had less than 100 data points. The training sets ranged from 187,320 to 188,799 data points. 61% of the clusters was labeled as ‘other’ based on the majority vote. For ‘Seizure’, ‘LPD’, ‘GPD’, ‘LRDA’ and ‘GRDA’ this was 5%, 14%, 8%, 4% and 8%, respectively. 162 principal components remained after application of PCA with 95% variance retained.
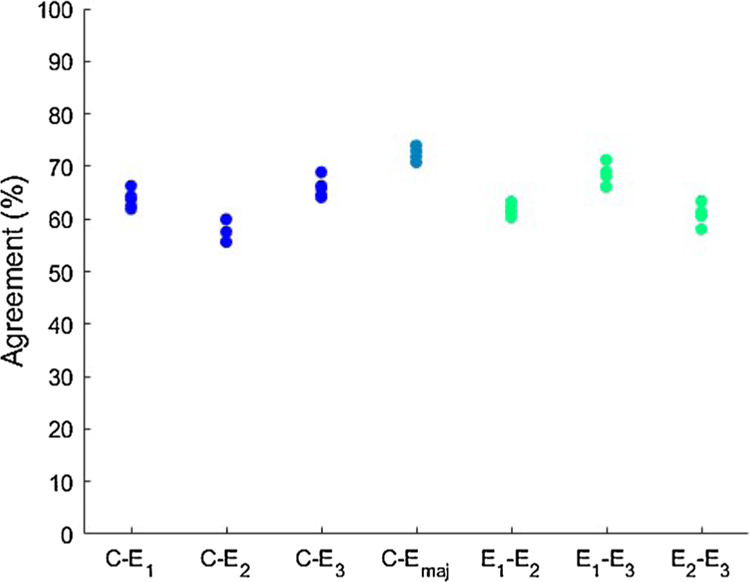
Fig. 4 shows box plots of the pairwise percentage agreements between the model and the experts and within the experts. It also shows the percentage agreements with the majority vote of the experts. In the majority vote, the medoids on which all three experts disagreed were left out in testing, as we could not set a ‘true’ label for these segments. The mean pairwise agreement between the model and the experts was 62.6% (SD: 3.8) and within the experts 63.8% (SD: 4.2). The TOST rendered two significant one-sided t-tests with p < 0.001 and p = 0.0255 (df = 14).
Fig. 4.
Percentage agreements of all 5 folds between the RF classifier (C) and the experts (E1 to E3) and within the experts. Emaj is the majority vote of the experts, with the samples on which all experts disagreed left out.
The mean agreement between the model and the majority vote of the experts was 72.5% (SD: 1.2). Our benchmark model, which was based upon the k-medoids clusters, had a mean agreement of 57.5% (SD: 2.4) with the majority vote. A two-sided t-test shows these results significantly differ (p < 0.001, df = 14).
4. Discussion
We have validated a method to aggregate cEEG data into a small number of clusters, which can rapidly be annotated by EEG experts with an easy-to-use GUI. We did this by applying BoW-based clustering to cEEG, a method we have introduced elsewhere (Jing et al., 2018). Our method allows experts to quickly identify and tag seizures and IIIC patterns in critically ill patients.
4.1. Findings
The validity of the clustering results was assessed by training a RF classifier using randomly selected points within a cluster and assigning them the same label as the center of the cluster, and then comparing the predictions of this model with labels assigned by experts. If the clusters are ‘pure,’ i.e. if the whole cluster indeed belongs to a specific pattern type, the model predictions should agree well with annotations assigned by experts, whereas if there exist a large variety of pattern types within a cluster, agreement should be low. As shown in Fig. 4, agreement between experts and the automatic clustering is comparable with the interrater agreement of the three experts. The lack of significant difference supports the validity of this method: interrater agreements between the automated method and the human experts and those calculated among human experts are comparable in performance. In other words, the samples ‘pseudolabeled’ by inheriting the label of their cluster center were informative; the model trained with these pseudolabels was able to predict the score of an expert as well as the judgment of another expert would. This provides evidence that our methodology creates clusters in such a way that they meaningfully distinguish different pattern types.
Our comparison with a less advanced methodology shows that the CPD-BoW based unsupervised clustering significantly improves the results. Moreover, it shows that it is rather difficult for an algorithm to achieve a level of agreement similar to the interrater agreement, even with the ‘tricky’ samples, where all experts had disagreed, left out.
Previous studies of EEG data labeling have generally yielded similar human expert interrater agreements, and lower agreements with automated methods. The majority of studies conducted using experts labeling EEG data from the ICU or EMU including both seizure and multiple IIIC labels have resulted in kappas in the range of 0.50–0.66 (Hermans et al., 2016; Wusthoff et al., 2017; Halford et al., 2011, 2015; Shellhaas et al., 2008; Foreman et al., 2016; Mani et al., 2012). While Gaspard and colleagues report high agreement for seizures and IIIC patterns (κ> 90 %), their study used carefully curated examples, whereas our study included a wide variety of patterns which were not filtered in any way to fit any given pattern category (Gaspard et al., 2014).
Labeling the cEEG data with our method took the experts at least 45 times less time than manual labeling: the median time per patient was less than 4 min. This time saving effect is comparable to the order of magnitude time reduction we found in our earlier pilot study of this methodology (Jing et al., 2018). This result suggests that the method is fast and easy to use, enabling rapid generation of a large labeled EEG dataset. This dataset can in turn be reliably used for relating the different pattern types to patient outcomes in a supervised manner. Once applied to such large dataset, our interpretable method will enable analysis of which features most strongly define cEEG labels, which will be discussed in a future study.
4.2. Limitations
While this study suggests a valid novel approach for rapidly annotating cEEG data, there are several limitations and caveats. Firstly, the interrater agreement remains relatively low, despite being comparable to the interrater agreement of human experts, which may be due to intrinsic overlap in EEG labeling criteria and resultant indeterminate labels (Hermans et al., 2016; Wusthoff et al., 2017; Halford et al., 2011, 2015; Shellhaas et al., 2008; Foreman et al., 2016; Mani et al., 2012). Secondly, the data ended up having a relative overrepresentation of ‘Other’ labeled cEEG (61%), which was mitigated by using balanced class weights in training the RF. Finally, not every segment of the training data was manually annotated so there could be ‘false’ labels relative to the ground truth labels of human experts. The RF classifier demonstrates scalable performance on the held-out data incorporating pseudolabels, with the caveat that this procedure does not specifically test the equivalence of the pseudolabels to the ground truth labels.
4.3. Future directions
We anticipate three key use cases for our method: (1) categorization and labeling of large EEG datasets for population-level research; (2) creation and curation of labeled EEG databases to train machine learning models; and (3) rapid annotation of a specific EEG recording for patient care in the ICU or epilepsy monitoring unit (e.g. to estimate patient seizure burden for clinical management).
Our work builds on previous work that has demonstrated the clinical utility of an EEG clustering approach (Hassan et al., 2015). We anticipate that this study and future related studies can significantly improve clinical workflows for clinical neurophysiologists, who currently work to manually label large quantities of data. Our clustering method performs robustly enough that clinicians labeling data will only have to label a representative subset of a patient’s data, and can rely on the algorithm to effectively apply the labels across the dataset. In addition to saving clinician time, our approach preserves accuracy by eliminating long labeling sessions, and allowing experts to evaluate exemplary EEG segments in more detail.
5. Conclusion
This work supports the hypothesis that cEEG data can be validly clustered into a small number of distinct patterns. Our results suggest that long EEG recordings can be rapidly annotated by experts many times faster than unaided manual review. Using our system, we are currently in the process of labeling >30TB of EEG data from 2000 ICU subjects. The resulting EEG data will provide sufficient data to train deep neural network models to automatically detect NCS and IIIC patterns. This rich data will also allow us to gain a deeper understanding of the clinical consequences of NCS and IIIC events, and how the consequences depend on the attributes of different NCS and IIIC patterns.
Acknowledgments
MBW was supported by NIH (NIH-NINDS1K23NS090900,1R01NS102190,1R01NS102574,1R01N-S107291). JJ, MBW, and MT received research support from SAGE therapeutics. EdA was supported by ZonMw(The Netherlands Organisation for Health Research and Development, 636310010) and SE was supported by the NIH (F31NS105161, K24NS088568, T32MH020017, T32GM007753), the Harvard Medical Scientist Training Program, and the Paul & Daisy Soros Fellowship.
Footnotes
Declaration of Competing Interest
The authors report no declarations of interest.
References
- Adams RP, MacKay DJC, 2007. Bayesian Online Changepoint Detection.
- Babadi B, Brown EN, 2014. A review of multitaper spectral analysis. IEEE Trans. Biomed. Eng 61 (5), 1555–1564. [DOI] [PubMed] [Google Scholar]
- Barlow JS, Creutzfeldt OD, Michael D, Houchin J, Epelbaum H, 1981. Automatic adaptive segmentation of clinical EEGs. Electroencephalogr. Clin. Neurophysiol 51 (May), 512–525. [DOI] [PubMed] [Google Scholar]
- Bose S, Rama V, Warangal N, Rao CBR, 2017. EEG signal analysis for seizure detection using discrete wavelet transform and random forest. 2017 International Conference on Computer and Applications (ICCA) 369–378 ieeexplore.ieee.org. [Google Scholar]
- Claassen J, 2009. How I treat patients with EEG patterns on the Ictal-Interictal continuum in the neuro ICU. Neurocrit. Care 11 (October), 437. [DOI] [PubMed] [Google Scholar]
- Dueck D, Frey BJ, 2007. Non-metric affinity propagation for unsupervised image categorization. In: ICCV 2007. IEEE 11th International Conference on Computer Vision, 2007 IEEE, pp. 1–8. [Google Scholar]
- Foreman B, Mahulikar A, Tadi P, Claassen J, Szaflarski J, Halford JJ, Dean BC, Kaplan PW, Hirsch LJ, LaRoche S, et al. , 2016. Generalized periodic discharges and ‘triphasic waves’: a blinded evaluation of inter-rater agreement and clinical significance. Clin. Neurophysiol 127 (2), 1073–1080. [DOI] [PubMed] [Google Scholar]
- Friedman D, Claassen J, Hirsch LJ, 2009. Continuous electroencephalogram monitoring in the intensive care unit. Anesth. Analg 109 (2), 506–523. [DOI] [PubMed] [Google Scholar]
- Gaspard N, Hirsch LJ, LaRoche SM, Hahn CD, Westover MB, 2014. Interrater agreement for critical care EEG terminology. Epilepsia 55 (9), 1366–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golmohammadi M, Ziyabari S, Shah V, De Diego SL, Obeid I, Picone J, 2017. Deep Architectures for Automated Seizure Detection in Scalp EEGs.
- Guralnik V, Srivastava J, 1999. Event detection from time series data. Proceedings of the Fifth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 33–42. [Google Scholar]
- Halford JJ, Pressly WB, Benbadis SR, Tatum WO 4th, Turner RP, Arain A, Pritchard PB, Edwards JC, Dean BC, 2011. Web-based collection of expert opinion on routine scalp EEG: software development and interrater reliability. J. Clin. Neurophysiol 28 (April), 178–184. [DOI] [PubMed] [Google Scholar]
- Halford JJ, Shiau D, Desrochers JA, Kolls BJ, Dean BC, Waters CG, Azar NJ, Haas KF, Kutluay E, Martz GU, Sinha SR, Kern RT, Kelly KM, Sackellares JC, LaRoche SM, 2015. Inter-rater agreement on identification of electrographic seizures and periodic discharges in ICU EEG recordings. Clin. Neurophysiol 126 (September), 1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan M, Shamas M, Khalil M, El Falou W, Wendling F, 2015. EEGNET: an open source tool for analyzing and visualizing M/EEG connectome. PLOS ONE 10 (September), e0138297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans MC, Westover MB, van Putten MJAM, Hirsch LJ, Gaspard N, 2016. Quantification of EEG reactivity in comatose patients. Clin. Neurophysiol 127 (January), 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch LJ, 2004. Continuous EEG monitoring in the intensive care unit: an overview. J. Clin. Neurophysiol 21 (5), 332–340. [PubMed] [Google Scholar]
- Hirsch LJ, 2011. Classification of EEG patterns in patients with impaired consciousness. Epilepsia 52 (Suppl. 8 (October)), 21–24. [DOI] [PubMed] [Google Scholar]
- Hirsch L, LaRoche S, Gaspard N, Gerard E, Svoronos A, Herman S, Mani R, Arif H, Jette N, Minazad Y, et al. , 2013. American clinical neurophysiology society’s standardized critical care EEG terminology: 2012 version. J. Clin. Neurophysiol 30 (1), 1–27. [DOI] [PubMed] [Google Scholar]
- Jing J, D’angremont E, Zafar S, Rosenthal ES, Tabaeizadeh M, Ebrahim S, Dauwels J, Westover MB, 2018. Rapid annotation of seizures and interictal-ictal continuum EEG patterns. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, vol. 2018 – July 3394–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan AY, Shishkin SL, 2000. Application of the change-point analysis to the investigation of the brain’s electrical activity. Non-Parametric Statistical Diagnosis. Springer, pp. 333–388. [Google Scholar]
- Kennedy JD, Gerard EE, 2012. Continuous EEG monitoring in the intensive care unit. Curr. Neurol. Neurosci. Rep 12 (August), 419–428. [DOI] [PubMed] [Google Scholar]
- Killick R, Fearnhead P, Eckley IA, 2012. Optimal detection of changepoints with a linear computational cost. J. Am. Stat. Assoc 107 (500), 1590–1598. [Google Scholar]
- Lund R, Wang XL, Lu QQ, Reeves J, Gallagher C, Feng Y, 2007. Changepoint detection in periodic and autocorrelated time series. J. Clim. 20 (20), 5178–5190. [Google Scholar]
- Maaten L.v.d., Hinton G, 2008. Visualizing data using t-SNE. J. Mach. Learn. Res 9 (November), 2579–2605. [Google Scholar]
- Mani R, Arif H, Hirsch LJ, Gerard EE, LaRoche SM, 2012. Interrater reliability of ICU EEG research terminology. J. Clin. Neurophysiol 29 (June), 203–212. [DOI] [PubMed] [Google Scholar]
- Reeves J, Chen J, Wang XL, Lund R, Lu QQ, 2007. A review and comparison of changepoint detection techniques for climate data. J. Appl. Meteorol. Climatol 46 (June), 900–915. [Google Scholar]
- Rogers JL, Howard KI, Vessey JT, 1993. Using significance tests to evaluate equivalence between two experimental groups. Psychol. Bull 113 (3), 553–565. [DOI] [PubMed] [Google Scholar]
- Sackellares JC, Shiau D-S, Halford JJ, LaRoche SM, Kelly KM, 2011. Quantitative EEG analysis for automated detection of nonconvulsive seizures in intensive care units. Epilepsy Behav 22 (Suppl. 1 (December)), S69–S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shellhaas RA, Gallagher PR, Clancy RR, 2008. Assessment of neonatal electroencephalography (EEG) background by conventional and two amplitude-integrated EEG classification systems. J. Pediatr 153 (September), 369–374. [DOI] [PubMed] [Google Scholar]
- Shneker BF, Fountain NB, 2003. Assessment of acute morbidity and mortality in nonconvulsive status epilepticus. Neurology 61 (October), 1066–1073. [DOI] [PubMed] [Google Scholar]
- Sun H, Jia J, Goparaju B, Huang GB, Sourina O, Bianchi MT, Westover MB, 2017. Large-scale automated sleep staging. Sleep 40 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wusthoff CJ, Sullivan J, Glass HC, Shellhaas RA, Abend NS, Chang T, Tsuchida TN, 2017. Interrater agreement in the interpretation of neonatal electroencephalography in hypoxic-ischemic encephalopathy. Epilepsia 58 (March), 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jin R, Zhou Z-H, 2010. Understanding bag-of-words model: a statistical framework. Int. J. Mach. Learn. Cybern 1 (1–4), 43–52. [Google Scholar]