Abstract
The stomatal pores of plant leaves control gas exchange with the environment. Stomatal development is prevised regulated by both internal genetic programs and environmental cues. Among various environmental factors, light regulation of stomata formation has been extensively studied in Arabidopsis. In this review, we summarize recent advances in the genetic control of stomata development and its regulation by light. We also present a comparative analysis of the conserved and diverged stomatal regulatory networks between Arabidopsis and cereal grasses. Lastly, we provide our perspectives on manipulation of the stomata density on plant leaves for the purpose of breeding crops that are better adapted to the adverse environment and high-density planting conditions.
Key words: stomatal development, spacing patterns, light signaling, Arabidopsis, cereal crops
This review summarizes recent advances in our understanding of light-regulated stomatal development and patterning in Arabidopsis, with specific emphasis on the comparison of regulatory mechanisms between Arabidopsis and monocotyledonous cereal grasses. The authors provide perspectives about the regulation of stomatal development and patterning in grasses toward ecosystem function and agricultural improvement.
Introduction
Stomata are openings on the leaf surface that control the exchange of water, carbon dioxide, and oxygen with their environment. Stomatal function is essential for photosynthesis and respiration of a plant, and contributes to carbon cycle on a global scale (Hetherington and Woodward, 2003). Remarkable progress has been made in the past two decades toward understanding of the intercellular communication, cell asymmetric division, and stomatal state transitions underlying stomatal development in Arabidopsis thaliana (Facette and Smith, 2012, Pillitteri and Dong, 2013, Torii, 2015, Simmons and Bergmann, 2016, Qu et al., 2017, Hepworth et al., 2018). The formation and opening/closure of stomata is regulated by complex interplays between internal genetic programs and various environmental factors (such as light, humidity, temperature, and concentration of carbon dioxide). Among such environmental factors, the molecular mechanisms by which light regulate stomatal movement, development, and patterning have been best studied thus far. As the signaling mechanisms governing light regulation of stomatal opening/closure has been recently summarized in a few excellent reviews (Chen et al., 2012, Kollist et al., 2014, Inoue and Kinoshita, 2017, Matthews et al., 2019), the scope of this review is limited to highlight recent advances in our understanding of light-regulated stomatal development and patterning in Arabidopsis, and to present a comparison of the shared and distinct regulatory mechanisms between dicotyledenous plants (Arabidopsis as a model) and monocotyledonous cereal grasses. Lastly, we provide a perspective for future research to elucidate the light-mediated regulation of stomatal development and patterning in cereal grasses. Such knowledge may help to facilitate the breeding of climate-resilient crops that can be better adapted to adverse global climate changes and crops suitable for dense planting conditions.
A Core Transcriptional Cascade Regulating Stomata Formation in Arabidopsis
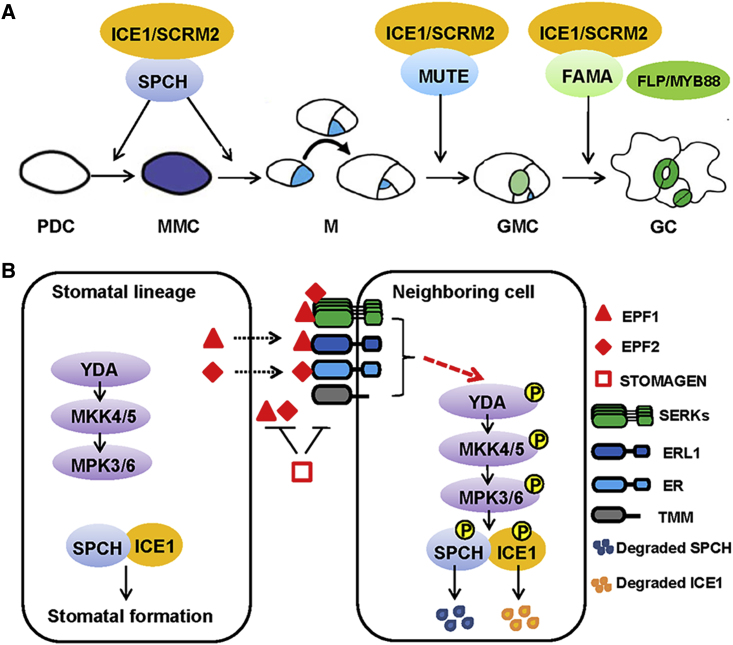
In Arabidopsis, stomata are surrounded by two kidney-shaped guard cells (GCs) and they are scattered throughout the epidermis. The developmental pathway of stomata can be generally divided into four steps (Figure 1A): (1) a subset of leaf protoderm cells (PDCs) differentiate into meristemoid mother cells (MMCs); (2) MMCs divide asymmetrically to produce a smaller meristemoid (M) and a larger stomatal lineage ground cell, which can directly develop into a pavement cell or undergo additional rounds of asymmetrical division; (3) M takes a fate of guard mother cell (GMC); and (4) GMC divides symmetrically to form two GCs followed by GC maturation (Torii, 2015, Simmons and Bergmann, 2016). Collective work from several groups has shown that major transcriptome changes occur during each fate transition and that five basic helix-loop-helix (bHLH) transcription factors act in a sequential manner to form a central transcriptional cascade regulating stomata development. SPCHLESS (SPCH) acts at the entry point and is broadly expressed in the young leaf, but its protein is restricted to M to ensure continued asymmetrical division (MacAlister et al., 2007). How this is achieved still remains unknown and will be an interesting avenue of future research. The decline of SPCH protein level and accompanied increase of MUTE protein in the M cells lead to termination of the amplifying divisions of M cells and enable the differentiation of M into GMC (Pillitteri et al., 2007). Lastly, FAMA protein accumulates in GMCs to promote the symmetric division of GMC and formation of two GCs (Ohashi-Ito and Bergmann, 2006). Two additional bHLH proteins, INDUCER OF CBF1 (ICE1)/SCREAM1 (SCRM1) and SCRM2, being broadly expressed, are able to form heterodimers with SPCH, MUTE, and FAMA and promote their stability, thus modulating the entire stomatal development process (Kanaoka et al., 2008). Two partially redundant paralogous R2R3 MYB transcription factors FOUR LIPS (FLP) and MYB88 also participate in the regulation of GMC symmetrical division and GC differentiation (Lai et al., 2005, Lee et al., 2014).
Figure 1.
Schematic Model of the Signal Transduction Networks Regulating Stomatal Development in Arabidopsis.
(A) Diagram of cell-state transitional steps within stomatal cell lineages in Arabidopsis. The schematic model is modified from Torii (2015). The bHLH transcription factors SPCHLESS (SPCH), MUTE, and FAMA sequentially control the initiation, transition, and differentiation steps, respectively. ICE1/SCRM promotes stomatal development through forming heterodimers with SPCH, MUTE, or FAMA. FOUR LIPS (FLP) and its redundant paralog MYB88 also regulate the GMC symmetric division and GC maturation. PDC, protoderm cell; MMC, meristemoid mother cell; M, meristemoid; GMC, guard mother cell; GC, guard cell.
(B) The negative regulatory pathways involve ligands EPIDERMAL PATTERNING FACTORS (EPF1, EPF2), membrane localized receptor-like kinases including the ERECTA family receptors (ER, ERL1), the kinase-like protein TOO MANY MOUTH (TMM) and SOMATIC EMBRYOGENESIS RECEPTOR KINASEs (SERKs) family receptors (SERK1, SERK2, SERK3/BAK1, SERK4), and the downstream YDA-MKK4/5-MPK3/6 signaling cascade. EPF2 and EPF1 are highly expressed in the stomatal lineage cells, and are secreted from stomatal precursors to restrict the neighboring cells from entering into the stomatal lineage. STOMAGEN/EPF9, expressed in the internal mesophyll cells, competes with EPF2/EPF1 to bind ER/ERL1, thus inhibiting signaling via TMM. SERKs form heterodimers with ER/ERL1 and TMM in a ligand-induced manner. The ligand–receptor–coreceptor complexes activate the YDA-MKK4/5-MPK3/6 signaling cascade by an as yet unidentified mechanism, which leads to phosphorylation and proteasome-mediated degradation of SPCH/ICE1 and eventual arrest of stomatal development in the neighboring cells.
How the identity of stomata lineage is acquired remains an intriguing question. Numerous studies have shown that SPCH acts as the point of signal integration to regulate stomatal patterning. The protein stability of SPCH is modulated by positional cues composed of ligands (EPIDERMAL PATTERNING FACTOR peptides, EPF1, EPF2) and receptors including ERECTA-family kinases (ER, ERL1, and ERL2) and the receptor-like protein TOO MANY MOUTHS (TMM) (Figure 1B). EPF2 is highly expressed in MMC and early M cells in an SPCH-dependent manner, and is secreted from these stomatal precursors to restrict the neighboring cells from entering the stomatal lineage. Application of bioactive EPF2 peptide or overexpression of EPF2 causes a stomataless phenotype as the spch loss-of-function mutants (Hunt and Gray, 2009, Yamamuro et al., 2014). In comparison, EPF1 is expressed in late M, GMCs, and GCs and is emitted to enforce the one-cell spacing rule. The loss-of-function epf1 mutant displays mild clustered stomata with defects in orienting spacing divisions (Hara et al., 2007, Lee et al., 2012). Further studies showed that the EPP2–ERECTA ligand–receptor pair acts primarily to repress asymmetric entry division through inhibiting SPCH whereas the EPF1–ERL1 pair primarily inhibits GMC differentiation, yet the underlying mechanism has not been fully elucidated (Lau and Bergmann, 2012, Torii, 2012). TMM, which lacks a cytoplasmic effector domain, was shown to function as a coreceptor for ER proteins and modulate their activity. Additionally, it was shown that the SOMATIC EMBRYOGENESIS RECEPTOR KINASEs (SERK1, SERK2, SERK3, SERK4) family receptor-like kinases can also form heterodimers with ER and TMM proteins upon ligand binding (Meng et al., 2015). The formation of ligand–receptor–coreceptor complexes leads to the activation of the YDA-MKK4/5-MPK3/6 signaling cascade by an as yet unidentified mechanism, which leads to the phosphorylation and proteasome-mediated degradation of SPCH and eventually the arrest of stomata development in the neighboring cells (Lampard et al., 2008, Lampard et al., 2009).
Intriguingly, ICE1 (SCRM) was recently reported to serve as a scaffold that recruits upstream MPK3/6 to inhibit SPCH. Disruption of the interaction between ICE1 and MPK3/6 results in higher stability and activity of SPCH-SCRM heterodimers and excessive stomatal formation (Putarjunan et al., 2019). On the other hand, EPF9/STOMAGEN, expressed in the internal mesophyll cells, was shown to compete with EPF2 to bind ER (but this binding does not activate the downstream MAP kinase [MAPK]) (Lee et al., 2015, Lin et al., 2017), thus promoting SPCH stabilization and stomata formation. In addition, a polarity factor named BREAKING ASYMMETRY IN THE STOMATAL LINEAGE (BASL) was shown to interact with YDA and cause the distribution of the YDA/BASL complex to the larger daughter cell after an asymmetric division, resulting in lower SPCH protein level and, therefore, loss of stomatal identity of the larger cell (Dong et al., 2009, Zhang et al., 2016).
Light Regulation of Stomata Development and Patterning in Arabidopsis
Light is arguably one of the most important environmental factors regulating plant growth and development. Dark-grown Arabidopsis seedlings undergo skotomorphogenesis (elongated hypocotyls with enclosed cotyledons, and suppressed differentiation of etioplasts). Upon light exposure, Arabidopsis seedlings undergo photomorphogenesis (de-etiolation), characterized by open and expanded cotyledons, suppressed hypocotyl growth, and differentiation of etioplasts into mature green chloroplasts (Whitelam et al., 1998, Dong et al., 2014). Numerous studies have indicated that light regulation of stomata development is another important aspect of plant photomorphogenesis. In dark-grown wild-type plants, only few mature stomata can be observed on the cotyledons, forming occasional stomatal clusters, and many of them are retained in the stomatal precursor stage, such as meristemoids and GMCs (Balcerowicz et al., 2014, Yamamuro et al., 2014, Lee et al., 2017). Light greatly enhances the entry division of MMC to form M and division of GMCs to form mature stomata, as well as ensuring proper stomatal patterning. Consistent with this notion, several studies have shown that the formation of M and stomatal maturation is compromised in various photoreceptor mutants (phyB, phyA, and cry1cry2) or the hy1 mutant that is defective in the assembly of functional holophytochromes due to the lack of heme oxygenase (Kang et al., 2009). These observations are also consistent with the reports that the expression of STOMAGEN, SPCH, MUTE, FAMA, EPF2, and TMM is induced by light (Hronková et al., 2015, Lee et al., 2017). Whether light is involved in post-transcriptional regulation of these proteins is worthy of further investigation.
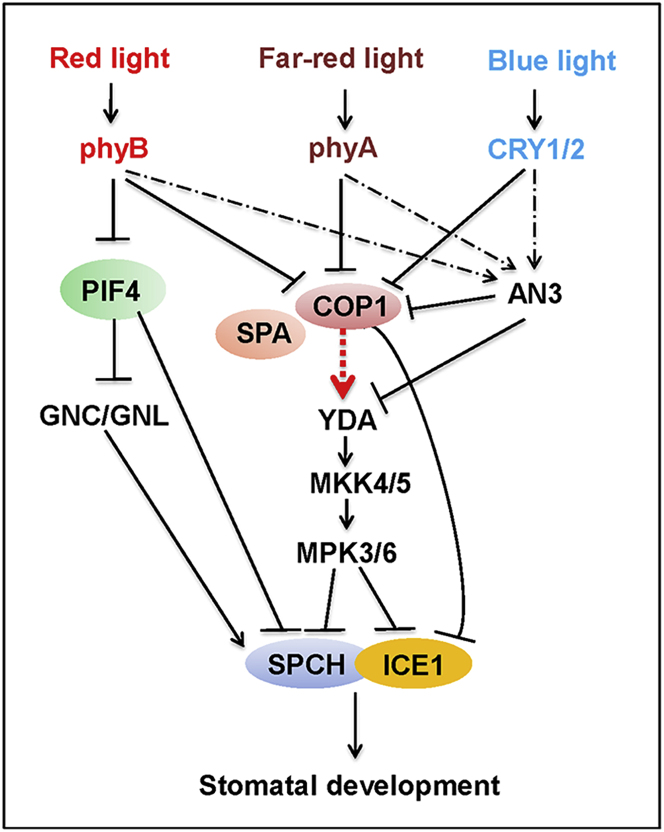
CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) is a central negative regulator of photomorphogenesis acting downstream of the phytochrome and cryptochrome photoreceptors (Lau and Deng, 2012). COP1 acts together with a small family of SPA proteins (SPA1, SPA2, SPA3, SPA4) and the CDD protein complex (formed by COP10, DDB1, and DET1) to form multiple E3 ubiquitin ligases responsible for targeting multiple photomorphogenesis-promoting factors (such as HY5 and HFR1) for 26S-proteasome-mediated degradation in darkness, thus suppressing photomorphogenesis (Chen et al., 2006, Chen et al., 2010). Interestingly, cop1, cop10, and det1 loss-of-function single mutant and spa1spa3spa4 triple mutant exhibited constitutive clustered stomata in the dark (Kang et al., 2009, Delgado et al., 2012, Hoecker, 2017), indicating that these COP/DET/FUS and SPA proteins all act to repress asymmetric cell division and stomatal fate initiation. Recent studies have provided evidence that COP1- and TMM/ER/ERL-mediated signaling converges to YDA to suppress stomata formation by promoting phosphorylation and subsequent degradation of SPCH and ICE1 (Lampard et al., 2008, Zhao et al., 2017) (Figure 2). Moreover, it was shown that COP1 directly interacts with ICE1 protein and mediates its degradation through the ubiquitin pathway (Lee et al., 2017). However, how COP1 regulates YDA remains an intriguing question to be elucidated. Notably, although the protein abundance of SPCH is elevated by light, the degradation of SPCH in darkness in not mediated by the 26S proteasome-mediated pathway (Lee et al., 2017). Thus, it will be interesting to examine in future studies how light stabilizes SPCH protein and whether COP1 is involved in this regulation.
Figure 2.
Model for the Light Regulation of Stomatal Development and Patterning.
COP1 activates the YDA-MKK4/5-MAPK3/6 cascade to mediate protein degradation of SPCH and ICE1, or directly mediates the degradation of ICE1. How COP1 regulates YDA remains an intriguing question to be elucidated. Meanwhile, PIF4 represses transcription of SPCH and GNC/GNL that promotes stomatal development through upregulating the expression of SPCH. In conditions of light, red- and far-red light photoreceptors (phyB and phyA) and blue-light photoreceptors CRY1/2 act in concert to suppress COP1 and PIF4 activity and the associated downstream networks, thus promoting stomatal development. Besides, light promotes transcription and protein abundance of AN3, likely through the photoreceptor-mediated pathways, which then repress the expression of COP1 and YDA. Arrow and bar-ended lines represent activation and inhibition, respectively. Solid lines indicate direct regulation. Dotted lines indicate indirect regulation or through an unidentified mechanism.
Phytochrome-interacting factors (PIFs) are bHLH transcription factors that accumulate in the dark and act in concert with the COP/SPA complex to suppress photomorphogenesis (Pham et al., 2018). Substantial evidence also indicates that PIF proteins act at the nexus of various signaling integration to regulate plant growth and development, as well as responses to various biotic and abiotic stresses (Leivar and Quail, 2011, de Lucas and Prat, 2014). A recent study reported that Arabidopsis PIF4 is expressed in the stomatal precursors and suppresses stomatal development through directly inhibiting the expression of SPCH in response to higher temperature. The pif4 loss-of-function mutants produce more stomata than the wild type and scarcely demonstrate stomatal responses to elevated temperature (Lau et al., 2018). Given the demonstrated central role of PIFs acting as the signaling hub regulating various aspects of plant growth and physiological responses, more detailed studies concerning the roles of different PIF proteins in regulating stomata development are warranted.
In addition, several other light-regulated genes have been implicated in light regulation of stomata development. Two paralogous Arabidopsis genes, GNC (GATA, NITRATE-INDUCIBLE, CARBON METABOLISM-INVOLVED) and GNL (GNC-LIKE), encoding leucine-leucine-methionine (LLM)-domain B-GATA transcription factors, were able to promote cell divisions and stomata formation in cotyledons and hypocotyls during dark-to-light transition, acting upstream of the central stomatal regulators SPCH, MUTE, and SCRM/2 (Klermund et al., 2016). It is noteworthy that PIFs can directly repress the expression of GNC and GNL, implicating a regulatory role of the PIF-GNC/GNL module in light-mediated stomatal development (Richter et al., 2010, Klermund et al., 2016).
The GRF-INTERACTING FACTOR1 (GIF1)/ANGUSTIFOLIA3 (AN3) gene encoding a transcription coactivator was characterized to promote stomatal development. Its loss-of-function mutant produced clusters of stomata, resembling the phenotype of yda or cop1 mutants (Meng and Yao, 2015). AN3 is regulated by light at both the transcription and post-translational levels and in turn directly downregulates the expression of both YDA (Meng and Yao, 2015) and COP1 (Meng et al., 2018), thus allowing the integration of light signaling into the production and patterning of stomata. Moreover, AN3 can associate with its own promoter, forming a light-controlled autopositive feedback loop. Together, the AN3-YDA and AN3-COP1 signaling cascades coordinately modulate light-mediated stomatal development (Figure 2). The possible effects of other identified light-signaling molecules in regulating stomata development, such as FAR-RED ELONGATED HYPOCOTYLS3 (FHY3), FAR-RED IMPAIRED RESPONSE1 (FAR1), ELONGATED HYPOCOTYL (HY5), HY5-HOMOLOG (HYH), LONG AFTER FAR-RED LIGHT1 (LAF1), and B-box zinc finger proteins (BBX4/22/24) remain to be studied in the future.
Conserved and Diverged Stomatal Regulatory Mechanisms in Cereal Grasses
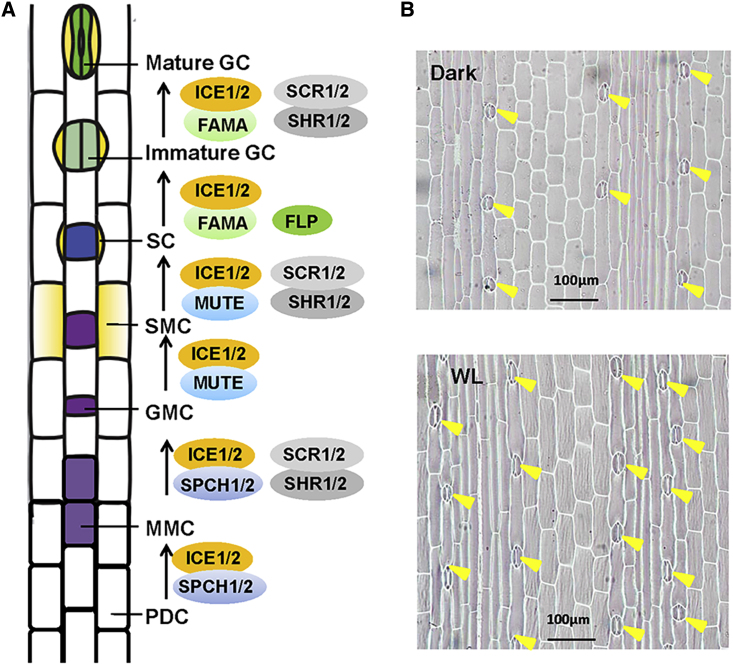
Our understanding of the regulatory mechanisms of stomatal development and patterning in cereal grasses (such as maize, rice, barley, and Brachypodium) still remains rudimentary, despite increased efforts being made in recent years. Monocot stomata contain two dumbbell-shaped GCs flanked by two subsidiary cells (SCs), which are developed in parallel rows within defined epidermal cell files in a base-to-tip gradient (Chen et al., 2017). Based on studies in Brachypodium, wheat, rice, and maize, the developmental process of stomata in grasses can be generally divided into six steps (Figure 3A), namely (1) stomatal file establishment, (2) asymmetric entry division to produce GMC, (3) establishment of subsidiary mother cell (SMC), (4) SMC asymmetric division to form SC, (5) GMC symmetric division to form two immature GCs, and (6) maturation of two dumbbell-shaped GCs (Raissig et al., 2017, Hepworth et al., 2018, Wang et al., 2019).
Figure 3.
Schematic Model for Stomatal Development in Grasses.
(A) A schematic model of stomata development in monocots based on Brachypodium and rice, modified from Raissig et al. (2017). The developmental process of stomata in grasses can be divided into six steps: (1) stomatal file establishment, (2) asymmetric entry division to produce GMC, (3) establishment of SMC, (4) SMC asymmetric division to form SC, (5) GMC symmetric division to form two immature GCs, and (6) maturation of two dumbbell-shaped GCs flanked by two SCs. Grass SPCH1/2 proteins promote the acquisition of stomata file identity and asymmetric entry division to form GMC. The grass MUTE has a unique feature to move from GMCs into SMC where it acts to promote the asymmetric division of SMC to form SCs. FAMA and FLP regulate the symmetric division of GMC and the maturation of GCs. Like the Arabidopsis counterparts, grass ICE1/2 proteins form heterodimers with SPCH1/2, MUTE, or FAMA. In rice, SCR1/2 and SHR1/2, which encode GRAS family proteins, have been shown to act together to control the initiation of stomatal lineage cells, formation of subsidiary cells, and maturation of GCs. PDC, protoderm cells; MMC, meristemoid mother cell; GMC, guard mother cell; SMC, subsidiary mother cell; SC, subsidiary cell; GC, guard cell.
(B) Light promotes stomata formation in maize leaves. Five-day-old maize seedlings grown in darkness were transferred to white light (WL) for 36 h. Light exposure significantly increased the stomata number per unit area of seedling leaf. The stomata are indicated by yellow arrowheads.
Grass stomata are initiated in the leaf base, where asymmetric transverse divisions occur in files with cells that are more close to the square in shape compared with the neighboring cell files. The smaller apical daughter cell of the asymmetrical transverse division is called a GMC, which later divides to form a pair of GCs. Although it has been speculated that internal tissue layers, e.g., the preformed underlying veins, may send a signal to inhibit stomata development in the overlying epidermal cell files (Sylvester et al., 1996), experimental evidence supporting this interesting hypothesis has not been forthcoming, and the exact nature of such a signal still awaits to be determined.
Using a reverse genetic approach, the function of Arabidopsis SPCH, MUTE, FAMA, ICE, and SCRM2 orthologs has been characterized in several grass species. Accumulating evidence suggests that the function of FAMA is conserved between grasses and Arabidopsis, whereas the roles of SPCH and MUTE paralogs show some degree of divergence. Unlike Arabidopsis, rice and Brachypodium have two SPCH genes and maize has three SPCH paralogs, yet the phosphorylation sites by MPK3/6 are conserved in the grass SPCH proteins. It has been shown that BdSPCH1/2 and OsSPCH1/2 are redundantly required for the initiation of the stomata lineage in Brachypodium and rice, respectively, like their Arabidopsis counterpart (Raissig et al., 2016, Wu et al., 2019a, Wu et al., 2019b). Notably, OsSPCH2 driven by the AtSPCH promoter fails to rescue the Arabidopsis spch mutant phenotype, suggesting certain functional divergence between OsSPCH and AtSPCH (Liu et al., 2009).
A unique feature of stomata development in grasses is the formation of SCs that allow swift transport of ions and solutes to GCs. Using maize as a model system, Laura Smith and her colleagues have pioneered the mechanisms of SMC establishment and asymmetric division. SMCs are formed on both sides of a GMC, and divide asymmetrically to produce SCs flanking the GMC on both sides. Earlier studies suggested that the orientation of SMC asymmetrical division is determined by a signal emanating from the GMC (Stebbins and Shah, 1960); however, it is only in recent years that the molecular nature of GMC–SMC cell–cell communication has begun to be unraveled. Two leucine-rich receptor-like kinases PANGLOSS1 (PAN1) and PAN2, were initially identified to be essential for promoting asymmetrical division of SMC (Cartwright et al., 2009, Zhang et al., 2012). Later it was found that PAN1 interacts with small guanosine triphosphatase RHO OF PLANTS (ROP2/9) and that they are colocalized at the GMC contact site, conferring actin patch formation and migration of the SMC nucleus to the GMC–SMC interface, thereby promoting asymmetrical division in SMC (Humphries et al., 2011, Zhang et al., 2012). More recently, it has been shown that the SCAR/WAVE regulatory complex, involved in activation of the actin-nucleating ARP2/3 complex for branched actin formation, is required for the polarization of PAN1 and PAN2 (Facette et al., 2015). Furthermore, two recent studies showed that the Brachypodium, rice, and maize MUTE proteins can move from GMCs, possibly through plasmodesmata, into neighboring SMC where it acts to establish SMC identity (Raissig et al., 2017, Wang et al., 2019). Intriguingly, it was shown that ZmMUTE can directly control the expression of PAN1 and PAN2 in SMC (Raissig et al., 2017, Wang et al., 2019). It has been proposed that OsMUTE and its downstream transcriptional regulators may activate the expression of some peptide signals in GMC (Wu et al., 2019b). Regardless, identification of the signal emanating from GMCs to establish SMC polarity should be a priority for future research.
In comparison with AtMUTE, which is nonmobile, it appears that the grass MUTE proteins (with a grass-specific C terminus) have acquired a novel function to move from GMCs to SMCs and to promote SMC formation. Another interesting feature is that grass MUTE proteins possess multiple predicted MAPK phosphorylation sites, which are absent in the Arabidopsis MUTE protein. Whether the grass MUTE proteins are regulated by phosphorylation, the responsible kinase(s)/phosphatase(s) and the physiological significance of this regulation remain to be unraveled in future studies. In this regard, it is worth mentioning that the maize DCD1 gene, encoding a B″ subunit of the PP2A phosphatase complex, is homologous to Arabidopsis TONNEAU2 and is needed for formation of the preprophase band (PPB) in asymmetrically dividing cells (Camilleri et al., 2002). The discordia1 (dcd1) mutants display misoriented SMC division due to defective PPB formation and phragmoplast expansion (Gallagher and Smith, 1999, Wright et al., 2009). Thus, it will be worth investigating the possible regulatory relationship between ZmMUTE and DCD1 in the future.
Recent studies have generated evidence supporting the notion that grass SCRM (ICE1) orthologs play a conserved function in transcriptional regulation of stomatal development. The Bdice1 mutant showed a stomataless phenotype, indicating that BdICE1 is essential for stomata formation in Brachypodium (Raissig et al., 2016). In line with this, OsICE has been shown to be essential for the initiation of stomatal lineage, the establishment of GMC, asymmetric division of SMC, and stomata maturation in rice. Moreover, the conserved physical interaction between OsICE1/SCRM2 with OsSPCH, OsMUTE, and OsFAMA was also observed (Wu et al., 2019b). Notably, BdICE1 possesses predicted high-fidelity MAPK target sites within the protein degradation associated PEST domain and that YFP-BdICE1 fusion protein driven by the ubiquitin promoter accumulates only in the stomatal lineage cells of the leaf, hinting that this protein might also be subject to post-translational regulation as the Arabidopsis SPCH protein (Raissig et al., 2016). In addition, it was found that OsFLP is required for the regulation of proper orientation of GMC symmetrical division in rice (Wu et al., 2019b).
Intriguingly, recent studies found that OsSCR1 and OsSHR1, which encode two GRAS family proteins, are expressed in stomatal lineage cells and that the transcription of OsSCR1 and OsSCR2 can be activated by both OsSPCH and OsMUTE (Wu et al., 2019b). Moreover, the double mutants of OsSCR1/2 and OsSHR1/2 exhibit an abnormal stomata phenotype with swollen or undivided SCs, arrested meristemoids, and reduced stomatal density (Wu et al., 2019b). These observations suggest that OsSCR and OsSHR act together to control the initiation of stomatal lineage cells and the formation of SCs. Notably, an earlier study showed that ectopic expression of maize ZmSHR1 (orthologous to OsSHR2) causes production of supernumerary stomatal rows in rice (Schuler et al., 2018). This result contradicts the recent observation that rice overexpression lines of OsSHR1 and OsSHR2 do not show a notable effect on stomata development (Wu et al., 2019b). Thus, the roles the SHR proteins in regulating stomata development may have somewhat diverged in different grass species, a point that needs to be further clarified.
Similar to the case in Arabidopsis, the signaling network composed of secretory peptides and downstream MAPK cascade has also been shown to be involved in regulating stomata development and patterning in grasses. Overexpression of the grass EPF1gene has been shown to arrest stomatal production in rice and barley (Hughes et al., 2017, Caine et al., 2019), whereas knockout of a rice ortholog of Arabidopsis EPFL9, OsEPF9a, causes severe reduction in stomata density (Yin et al., 2017), suggesting the coexistence of negative and positive signaling peptides regulating stomata formation in grasses. The loss-of-function Bdyda1 mutant bears a large group of stomata in contact in a single row, breaking the stomatal spacing rule in wild-type plants. No ectopic stomatal rows were found in the Bdyda1 mutant, suggesting that the defect occurred after the stomatal lineage had been determined. Further detailed analysis revealed that clusters in Bdyda1 arise from improper enforcement of alternative cell fates in the epidermis, rather than from default physical asymmetry in cell divisions (Abrash et al., 2018).
Light Regulation of Stomata Development in Grasses?
Despite tremendous advances in understanding the mechanisms of light-regulated stomatal development and behavior in Arabidopsis, only limited evidence has been accumulated to support a role for light in regulating stomata development in grasses. For example, earlier studies showed that the mutations of rice PhyB1 or PhyB2 result in decreased stomata density and smaller stomata (Liu et al., 2012), and that ectopic overexpression of maize ZmPIF1 in rice cause smaller stomata aperture and, therefore, better drought resistance (Gao et al., 2018). Our preliminary analysis showed that the stomatal number per unit area of maize seedling leaves was significantly increased after light exposure for 36 h (Figure 3B). Several recent studies have shown that many key light-signaling components in grasses have conserved functions as do their counterparts in Arabidopsis. For example, heterologous expression of the maize ZmPIFs genes can partially or fully complement the phenotype of the Arabidopsis pifq mutant (Shi et al., 2017, Wu et al., 2019a, Wu et al., 2019b). Similarly, expression of rice OsCOP1 can rescue the phenotype of Arabidopsis cop1 mutant (Ranjan et al., 2014). Whether and how these proteins regulate stomatal development and patterning in grasses will be avenues worthy of future investigation.
Linking the Modulation of Stomatal Development/Patterning with Ecosystem Function and Agricultural Improvement
In the context of global warming, we are encountering increasing temperature, higher atmospheric CO2 concentration, and increased frequency of drought. Changes in climate are bringing about negative effects on many agricultural ecosystems around the world (IPCC, 2018). Also, it has been shown that increases in air vapor pressure deficit, rather than changes in precipitation, is a dominant factor affecting future grassland productivity (Konings et al., 2017). As stomata are fundamental regulators of water use efficiency (WUE, the ratio of carbon gained to water loss), genetic manipulation of stomatal development and physiology may contribute to improving the ecosystem function and agricultural productivity. Stomatal morphology, including stomatal numbers, stomatal size, and presence or absence of SCs, can be adjusted to optimize CO2 uptake for photosynthesis while reducing water loss. Indeed, the size and density of stomata have changed over evolutionary time to facilitate the adaptation of plant species to new environments. For instance, based on the observation of herbarium and fossilized samples, smaller stomatal size and higher stomatal density were formed in response to low atmospheric CO2 concentration, while increased stomatal size and decreased stomatal density were associated with high CO2 levels (Dilcher et al., 2000, Franks and Beerling, 2009). Grass species have improved WUE and are better adapted to arid environments, which is correlated with their unique stomata complex composed of dumbbell-shaped stomata and neighboring SCs. The swift response of grass stomata to environmental cues is believed to confer adaptive advantages of grasses over most of the dicot plants (Chen et al., 2017). Of note, reduced stomatal density and decreased stomatal aperture may also compromise plants' ability to prevent overheating, possibly resulting in photoinhibition and yield penalties (Chaves et al., 2016). Thus it is imperative to better understand the regulatory mechanisms of stomatal size and density in cereal crops to tackle the dilemma between water conservation and evaporative cooling in the scenario of drier and warmer temperature. Significantly, exogenous manipulation of stomatal development can be achieved through synthetic chemistry approaches (Kim et al., 2012, Sakai et al., 2017), which provide an alternative solution for the genetic modification of stomatal development and patterning using genome-editing techniques such as the CRISPR/Cas9 system. It is envisaged that the production of synthetic cyclic peptides, such as EPFs, with high efficiency and low cost, may encourage large-scale agricultural applications in the future, thus bringing about a post-green-revolution era in agriculture (Endo and Torii, 2019).
In modern agricultural practices, increasing planting density has been an effective means of improving crop yields per unit land area. However, a reduction of red to far-red light ratio below the canopy can trigger plants' shade-avoidance responses including increased plant height, accelerated flowering, decreased leaf blade area, and decreased stomata density (Casson and Hetherington, 2014, Xie et al., 2017). Although the key regulators of stomatal development in grasses have been identified, how light signaling integrates with the core regulatory pathways to modulate stomatal development and patterning has not yet been explored. Given that smaller stomatal size allows faster aperture response and contributes to maximizing WUE under fluctuating light conditions (McAusland et al., 2016), we envisage that molecular breeding of crops with smaller stomatal size and increased stomatal density might be beneficial to maintain or improve crop performance under conditions of high-density planting.
Funding
H.W. is supported by a project sponsored by the Education Department of Guangdong Province (2018KQNCX022).
Acknowledgments
No conflict of interest declared.
Published: February 13, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
References
- Abrash E., Anleu G.M.X., Matos J.L., Bergmann D.C. Conservation and divergence of YODA MAPKKK function in regulation of grass epidermal patterning. Development. 2018;145 doi: 10.1242/dev.165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcerowicz M., Ranjan A., Rupprecht L., Fiene G., Hoecker U. Auxin represses stomatal development in dark-grown seedlings via Aux/IAA proteins. Development. 2014;141:3165–3176. doi: 10.1242/dev.109181. [DOI] [PubMed] [Google Scholar]
- Caine R.S., Yin X., Sloan J., Harrison E.L., Mohammed U., Fulton T., Biswal A.K., Dionora J., Chater C.C., Coe R.A. Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 2019;221:371–384. doi: 10.1111/nph.15344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri C., Azimzadeh J., Pastuglia M., Bellini C., Grandjean O., Bouchez D. The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell. 2002;14:833–845. doi: 10.1105/tpc.010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright H.N., Humphries J.A., Smith L.G. PAN1: a receptor-like protein that promotes polarization of an asymmetric cell division in maize. Science. 2009;323:649–651. doi: 10.1126/science.1161686. [DOI] [PubMed] [Google Scholar]
- Casson S.A., Hetherington A.M. Phytochrome B is required for light-mediated systemic control of stomatal development. Curr. Biol. 2014;24:1216–1221. doi: 10.1016/j.cub.2014.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves M.M., Costa J.M., Zarrouk O., Pinheiro C., Lopes C.M., Pereira J.S. Controlling stomatal aperture in semi-arid regions—the dilemma of saving water or being cool? Plant Sci. 2016;251:54–64. doi: 10.1016/j.plantsci.2016.06.015. [DOI] [PubMed] [Google Scholar]
- Chen H., Shen Y., Tang X., Yu L., Wang J., Guo L., Zhang Y., Zhang H., Feng S., Strickland E. Arabidopsis cullin4 forms an e3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell. 2006;18:1991–2004. doi: 10.1105/tpc.106.043224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Huang X., Gusmaroli G., Terzaghi W., Lau O.S., Yanagawa Y., Zhang Y., Li J., Lee J.H., Zhu D. Arabidopsis CULLIN4-damaged DNA binding protein1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22:108–123. doi: 10.1105/tpc.109.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Xiao Y., Li X., Ni M. Light-regulated stomatal aperture in Arabidopsis. Mol. Plant. 2012;5:566–572. doi: 10.1093/mp/sss039. [DOI] [PubMed] [Google Scholar]
- Chen Z.H., Chen G., Dai F., Wang Y., Hills A., Ruan Y.L., Zhang G., Franks P.J., Nevo E., Blatt M.R. Molecular evolution of grass stomata. Trends Plant Sci. 2017;22:124–139. doi: 10.1016/j.tplants.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Delgado D., Ballesteros I., Torres-Contreras J., Mena M., Fenoll C. .Dynamic analysis of epidermal cell divisions identifies specific roles for COP10 in Arabidopsis stomatal lineage development. Planta. 2012;236:447–461. doi: 10.1007/s00425-012-1617-y. [DOI] [PubMed] [Google Scholar]
- Dilcher D., de Boer H.J., Dekker S.C., Dilcher D.L., Lotter A.F., Wagner-Cremer F. Toward a new synthesis: major evolutionary trends in the angiosperm fossil record. Proc. Natl. Acad. Sci. U S A. 2000;97:7030–7036. doi: 10.1073/pnas.97.13.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., MacAlister C.A., Bergmann D.C. BASL controls asymmetric cell division in Arabidopsis. Cell. 2009;137:1320–1330. doi: 10.1016/j.cell.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Tang D., Gao Z., Yu R., Li K., He H., Terzaghi W., Deng X.W., Chen H. Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell. 2014;26:3630–3645. doi: 10.1105/tpc.114.130666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo H., Torii K.U. Stomatal development and perspectives toward agricultural improvement. Cold Spring Harb. Perspect. Biol. 2019;11:a034660. doi: 10.1101/cshperspect.a034660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facette M.R., Smith L.G. Division polarity in developing stomata. Curr. Opin. Plant Biol. 2012;15:585–592. doi: 10.1016/j.pbi.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Facette M.R., Park Y., Sutimantanapi D., Luo A., Cartwright H.N., Yang B., Bennett E.J., Sylvester A.W., Smith L.G. The SCAR/WAVE complex polarizes PAN receptors and promotes division asymmetry in maize. Nat. Plants. 2015;1:14024. doi: 10.1038/nplants.2014.24. [DOI] [PubMed] [Google Scholar]
- Franks P.J., Beerling D.J. Maximum leaf conductance driven by CO2 effects on stomatal size and density over geologic time. Proc. Natl. Acad. Sci. U S A. 2009;106:10343–10347. doi: 10.1073/pnas.0904209106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher K., Smith L.G. Discordia mutations specifically misorient asymmetric cell divisions during development of the maize leaf epidermis. Development. 1999;126:4623–4633. doi: 10.1242/dev.126.20.4623. [DOI] [PubMed] [Google Scholar]
- Gao Y., Wu M., Zhang M., Jiang W., Ren X., Liang E., Zhang D., Zhang C., Xiao N., Li Y. A maize phytochrome-interacting factors protein ZmPIF1 enhances drought tolerance by inducing stomatal closure and improves grain yield in Oryza sativa. Plant Biotechnol. J. 2018;16:1375–1387. doi: 10.1111/pbi.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Kajita R., Torii K.U., Bergmann D.C., Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one- cell-spacing rule. Genes Dev. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth C., Caine R.S., Harrison E.L., Sloan J., Gray J.E. Stomatal development: focusing on the grasses. Curr. Opin. Plant Biol. 2018;41:1–7. doi: 10.1016/j.pbi.2017.07.009. [DOI] [PubMed] [Google Scholar]
- Hetherington A.M., Woodward F.I. The role of stomata in sensing and driving environmental change. Nature. 2003;424:901–908. doi: 10.1038/nature01843. [DOI] [PubMed] [Google Scholar]
- Hoecker U. The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling Ute Hoecker. Curr. Opin. Plant Biol. 2017;37:63–69. doi: 10.1016/j.pbi.2017.03.015. [DOI] [PubMed] [Google Scholar]
- Hronková M., Wiesnerová D., Šimková M., Skůpa P., Dewitte W., Vráblová M., Zažímalová E. Light-induced STOMAGEN-mediated stomatal development in Arabidopsis leaves. J. Exp. Bot. 2015;66:4621–4630. doi: 10.1093/jxb/erv233. [DOI] [PubMed] [Google Scholar]
- Hughes J., Hepworth C., Dutton C., Dunn J.A., Hunt L., Stephens J., Waugh R., Cameron D.D., Gray J.E. Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 2017;174:776–787. doi: 10.1104/pp.16.01844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries J.A., Vejlupkova Z., Luo A., Meeley R.B., Sylvester A.W., Fowler J.E., Smith L.G. ROP GTPases act with the receptor-like protein PAN1 to polarize asymmetric cell division in Maize. Plant Cell. 2011;23:2273–2284. doi: 10.1105/tpc.111.085597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L., Gray J.E. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 2009;19:864–869. doi: 10.1016/j.cub.2009.03.069. [DOI] [PubMed] [Google Scholar]
- Inoue S.I., Kinoshita T. Blue light regulation of stomatal opening and the plasma membrane H+-ATPase. Plant Physiol. 2017;174:531–538. doi: 10.1104/pp.17.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC . Global warming of 1.5°C. In: Masson-Delmotte V., editor. Summary for Policy Makers. An IPCC Special Report on the Impacts of Global Warming of 1.5°C above Pre-industrial Levels and Related Global Greenhouse Gas Emission Pathways, in the Context of Strengthening the Global Response to the Threat of Climate Change, Sustainable Development, and Efforts to Eradicate Poverty. World Meteorological Organization; Geneva, Switzerland: 2018. p. 32. [Google Scholar]
- Kanaoka M.M., Pillitteri L.J., Fujii H., Yoshida Y., Bogenschutz N.L., Takabayashi J., Zhu J.K., Torii K.U. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell. 2008;20:1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.Y., Lian H.L., Wang F.F., Huang J.R., Yang H.Q. Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell. 2009;21:2624–2641. doi: 10.1105/tpc.109.069765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Michniewicz M., Bergmann D.C., Wang Z.Y. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482:419–422. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klermund C., Ranftl Q.L., Diener J., Bastakis E., Richter R., Schwechheimer C. LLM-domain B-GATA transcription factors promote stomatal development downstream of light signaling pathways in Arabidopsis thaliana hypocotyls. Plant Cell. 2016;28:646–660. doi: 10.1105/tpc.15.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollist H., Nuhkat M., Roelfsema M.R.G. Closing gaps: linking elements that control stomatal movement. New Phytol. 2014;203:44–62. doi: 10.1111/nph.12832. [DOI] [PubMed] [Google Scholar]
- Konings A., Williams A., Gentine P. Sensitivity of grassland productivity to aridity controlled by stomatal and xylem regulation. Nat. Geosci. 2017;10:284–288. [Google Scholar]
- Lai L.B., Nadeau J.A., Lucas J., Lee E.K., Nakagawa T., Zhao L., Geisler M., Sack F.D. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell. 2005;17:2754–2767. doi: 10.1105/tpc.105.034116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G.R., MacAlister C.A., Bergmann D.C. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science. 2008;322:1113–1116. doi: 10.1126/science.1162263. [DOI] [PubMed] [Google Scholar]
- Lampard G.R., Lukowitz W., Ellis B.E., Bergmann D.C. Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell. 2009;21:3506–3517. doi: 10.1105/tpc.109.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Bergmann D.C. Stomatal development: a plant’s perspective on cell polarity, cell fate transitions and intercellular communication. Development. 2012;139:3683–3692. doi: 10.1242/dev.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17:584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Lau O.S., Song Z., Zhou Z., Davies K.A., Chang J., Yang X., Wang S., Lucyshyn D., Tay I.H.Z., Wigge P.A. Direct control of SPEECHLESS by PIF4 in the high- temperature response of stomatal development. Curr. Biol. 2018;28:1273–1280. doi: 10.1016/j.cub.2018.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Kuroha T., Hnilova M., Khatayevich D., Kanaoka M.M., McAbee J.M., Sarikaya M., Tamerler C., Torii K.U. Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 2012;26:126–136. doi: 10.1101/gad.179895.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Lucas J.R., Sack F.D. Deep functional redundancy between FAMA and FOUR LIPS in stomatal development. Plant J. 2014;78:555–565. doi: 10.1111/tpj.12489. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Hnilova M., Maes M., Lin Y.C.L., Putarjunan A., Han S.K., Avila J., Torii K.U. Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature. 2015;522:439–443. doi: 10.1038/nature14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Jung J.H., Park C.M. Light inhibits COP1-mediated degradation of ICE transcription factors to induce stomatal development in Arabidopsis. Plant Cell. 2017;29:2817–2830. doi: 10.1105/tpc.17.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Zhang L., Han Z., Yang X., Liu W., Li E., Chang J., Qi Y., Shpak E.D., Chai J. A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev. 2017;31:927–938. doi: 10.1101/gad.297580.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Ohashi-ito K., Bergmann D.C. Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development. 2009;136:2265–2276. doi: 10.1242/dev.032938. [DOI] [PubMed] [Google Scholar]
- Liu J., Zhang F., Zhou J., Chen F., Wang B., Xie X. Phytochrome B control of total leaf area and stomatal density affects drought tolerance in rice. Plant Mol. Biol. 2012;78:289–300. doi: 10.1007/s11103-011-9860-3. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Prat S. PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 2014;202:1126–1141. doi: 10.1111/nph.12725. [DOI] [PubMed] [Google Scholar]
- MacAlister C.A., Ohashi-Ito K., Bergmann D.C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445:537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- Matthews J.S.A., Vialet-Chabrand S., Lawson T. Role of blue and red light in stomatal dynamic behaviour. J. Exp. Bot. 2019 doi: 10.1093/jxb/erz563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAusland L., Vialet-Chabrand S., Davey P., Baker N.R., Brendel O., Lawson T. Effects of kinetics of light-induced stomatal responses on photosynthesis and water-use efficiency. New Phytol. 2016;211:1209–1220. doi: 10.1111/nph.14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L.S., Yao S.Q. Transcription co-activator Arabidopsis ANGUSTIFOLIA3 (AN3) regulates water-use efficiency and drought tolerance by modulating stomatal density and improving root architecture by the transrepression of YODA (YDA) Plant Biotechnol. J. 2015;13:893–902. doi: 10.1111/pbi.12324. [DOI] [PubMed] [Google Scholar]
- Meng X., Chen X., Mang H., Liu C., Yu X., Gao X., Torii K.U., He P., Shan L. Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 2015;25:2361–2372. doi: 10.1016/j.cub.2015.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L.S., Li C., Xu M.K., Sun X.D., Wan W., Cao X.Y., Zhang J.L., Chen K.M. Arabidopsis ANGUSTIFOLIA3 (AN3) is associated with the promoter of CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) to regulate light-mediated stomatal development. Plant Cell Environ. 2018;41:1645–1656. doi: 10.1111/pce.13212. [DOI] [PubMed] [Google Scholar]
- Torii K.U. Mix-and-match: Ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci. 2012;17:711–719. doi: 10.1016/j.tplants.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D.C. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell. 2006;18:2493–2505. doi: 10.1105/tpc.106.046136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham V.N., Kathare P.K., Huq E. Phytochromes and phytochrome interacting factors. Plant Physiol. 2018;176:1025–1038. doi: 10.1104/pp.17.01384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Dong J. Stomatal development in Arabidopsis. Arabidopsis Book. 2013;11:e0162. doi: 10.1199/tab.0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- Putarjunan A., Ruble J., Srivastava A., Zhao C., Rychel A.L., Hofstetter A.K., Tang X., Zhu J., Tama F., Zheng N. Bipartite anchoring of SCREAM enforces stomatal initiation by coupling MAP kinases to SPEECHLESS. Nat. Plants. 2019;5:742–754. doi: 10.1038/s41477-019-0440-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Peterson K.M., Torii K.U. Stomatal development in time: the past and the future. Curr. Opin. Genet. Dev. 2017;45:1–9. doi: 10.1016/j.gde.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Raissig M.T., Abrash E., Bettadapur A., Vogel J.P., Bergmann D.C. Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc. Natl. Acad. Sci. U S A. 2016;113:8326–8331. doi: 10.1073/pnas.1606728113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M.T., Matos J.L., Gil M.X.A., Kornfeld A., Bettadapur A., Abrash E., Allison H.R., Badgley G., Vogel J.P., Berry J.A. Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science. 2017;355:1215–1218. doi: 10.1126/science.aal3254. [DOI] [PubMed] [Google Scholar]
- Ranjan A., Dickopf S., Ullrich K.K., Rensing S.A., Hoecker U. Functional analysis of COP1 and SPA orthologs from Physcomitrella and rice during photomorphogenesis of transgenic Arabidopsis reveals distinct evolutionary conservation. BMC Plant Biol. 2014;14:1–15. doi: 10.1186/1471-2229-14-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K., Haslbeck M., Buchner J. The heat shock response: life on the verge of death. Mol. Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Sakai Y., Sugano S.S., Kawase T., Shirakawa M., Imai Y., Kawa-moto Y., Sugiyama H., Nakagawa T., Hara-Nishimura I., Shimada T. The chemical compound bubblin in- duces stomatal mispatterning in Arabidopsis by disrupting the intrinsic polarity of stomatal lineage cells. Development. 2017;144:499–506. doi: 10.1242/dev.145458. [DOI] [PubMed] [Google Scholar]
- Schuler M.L., Sedelnikova O.V., Walker B.J., Westhoff P., Langdale J.A. SHORTROOT-mediated increase in stomatal density has no impact on photosynthetic efficiency. Plant Physiol. 2018;176:757–772. doi: 10.1104/pp.17.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q., Zhang H., Song X., Jiang Y., Liang R., Li G. Functional characterization of the maize phytochrome-interacting factors PIF4 and PIF5. Front. Plant Sci. 2017;8:2273. doi: 10.3389/fpls.2017.02273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons A.R., Bergmann D.C. Transcriptional control of cell fate in the stomatal lineage. Curr. Opin. Plant Biol. 2016;29:1–8. doi: 10.1016/j.pbi.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G., Shah S. Developmental studies of cell differentiation in the epidermis of monocotyledones. II. Cytological features of stomatal development in the Gramineae. Dev. Biol. 1960;2:477–500. [Google Scholar]
- Sylvester A.W., Smith L., Freeling M. Acquisition of identity in the developing leaf. Annu. Rev. Cell Dev. Biol. 1996;12:257–304. doi: 10.1146/annurev.cellbio.12.1.257. [DOI] [PubMed] [Google Scholar]
- Torii K.U. Stomatal differentiation: the beginning and the end. Curr. Opin. Plant Biol. 2015;28:16–22. doi: 10.1016/j.pbi.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Wang H., Guo S., Qiao X., Guo J., Li Z., Zhou Y., Bai S., Gao Z., Wang D., Galbraith D.W. BZU2/ZmMUTE controls symmetrical division of guard mother cell and specifies neighbor cell fate in maize. PLoS Genet. 2019;15:e1008377. doi: 10.1371/journal.pgen.1008377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam G.C., Patel S., Devlin P.F. Phytochromes and photomorphogenesis in Arabidopsis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1998;353:1445–1453. doi: 10.1098/rstb.1998.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A.J., Gallagher K., Smith L.G. discordia1 and alternative discordia1 function redundantly at the cortical division site to promote preprophase band formation and orient division planes in maize. Plant Cell. 2009;21:234–247. doi: 10.1105/tpc.108.062810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Zhao Y., Shen R. Characterization of maize phytochrome-interacting factors in light signaling and photomorphogenesis. Plant Physiol. 2019;181:789–803. doi: 10.1104/pp.19.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Chen L., Yu Q., Zhou W., Gou X., Li J., Hou S. Multiple transcriptional factors control stomata development in rice. New Phytol. 2019;223:220–232. doi: 10.1111/nph.15766. [DOI] [PubMed] [Google Scholar]
- Xie Y., Liu Y., Wang H., Ma X., Wang B., Wu G., Wang H. Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 2017;8:348. doi: 10.1038/s41467-017-00404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C., Miki D., Zheng Z., Ma J., Wang J., Yang Z., Dong J., Zhu J.K. Overproduction of stomatal lineage cells in Arabidopsis mutants defective in active DNA demethylation. Nat. Commun. 2014;5:4062. doi: 10.1038/ncomms5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Biswal A.K., Dionora J., Perdigon K.M., Balahadia C.P., Mazumdar S., Chater C., Lin H.C., Coe R.A., Kretzschamr T. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 2017;36:745–757. doi: 10.1007/s00299-017-2118-z. [DOI] [PubMed] [Google Scholar]
- Zhang X., Facette M., Humphries J.A., Shen Z., Park Y., Sutimantanapi D., Sylvester A.W., Briggs S.P., Smith L.G. Identification of PAN2 by quantitative proteomics as a leucine-rich repeat- receptor-like kinase acting upstream of PAN1 to polarize cell division in Maize. Plant Cell. 2012;24:4577–4589. doi: 10.1105/tpc.112.104125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Guo X., Dong J. Phosphorylation of the polarity protein BASL differentiates asymmetric cell fate through MAPKs and SPCH. Curr. Biol. 2016;26:2957–2965. doi: 10.1016/j.cub.2016.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Wang P., Si T., Hsu C.C., Wang L., Zayed O., Yu Z., Dong J., Tao W.A., Zhu J.K. MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell. 2017;43:618–629. doi: 10.1016/j.devcel.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]