Abstract
Shoot branching, determining plant architecture and crop yield, is critically controlled by strigolactones (SLs). However, how SLs inhibit shoot branching after its perception by the receptor complex remains largely obscure. In this study, using the transcriptomic and genetic analyss as well as biochemical studies, we reveal the key role of BES1 in the SL-regulated shoot branching. We demonstrate that BES1 and D53-like SMXLs, the substrates of SL receptor complex D14–MAX2, interact with each other to inhibit BRC1 expression, which specifically triggers the SL-regulated transcriptional network in shoot branching. BES1 directly binds the BRC1 promoter and recruits SMXLs to inhibit BRC1 expression. Interestingly, despite being the shared component by SL and brassinosteroid (BR) signaling, BES1 gains signal specificity through different mechanisms in response to BR and SL signals.
Key words: strigolactones, shoot branching, signaling, D53-like SMXLs, BES1, BRC1
Strigolactones are crucial in shoot branching, but little is known about how SL signals transduce to inhibit shoot branching downstream of the receptor complex D14–MAX2 inducing the degradation of substrates such as D53-like SMXLs and BES1. This study reveals that, in Arabidopsis, BES1 interacts with D53-like SMXLs to inhibit BRC1 expression, which depends on direct binding of BES1 to the BRC1 promoter and subsequent recruitment of D53 to affix DNA and repress BRC1 expression.
Introduction
Strigolactones (SLs), a class of the terpenoid phytohormones (Gomez-Roldan et al., 2008, Umehara et al., 2008), are firstly recognized as symbiotic signals responsible for induction of seed germination of root parasite plants and as branching factors for symbiotic arbuscular mycorrhizal fungi (Cook et al., 1966, Akiyama et al., 2005). Although SLs have been recently found to regulate many plant developmental processes, including root hair elongation, primary root growth, adventitious and lateral root formation, secondary vascular growth, internode growth, and leaf senescence, inhibiting bud outgrowth in shoot branching regulation is one of their well-known functions in plants (Al-Babili and Bouwmeester, 2015). Mutants deficient in SL biosynthesis or signaling in Arabidopsis thaliana (more axillary growth, max), Pisum sativum (ramosus, rms), Oryza sativa (dwarf, d, or high tillering dwarf, htd), and Petunia hybrida (decreased apical dominance, dad), all exhibit enhanced branching phenotypes (Beveridge and Kyozuka, 2010, Domagalska and Leyser, 2011).
SL signaling is initiated when the α/β-hydrolase enzyme DWARF14 (D14) binds SLs and generates a covalently linked intermediate molecule. In turn, this triggers a conformational change in the structure of D14 to facilitate its interaction with an F-box protein DWARF3 (D3)/MAX2 (Nakamura et al., 2013, De Saint Germain et al., 2016, Yao et al., 2016). Recently, it was reported that D3 adopts a conformational state with a dislodged CTH (C-terminal α helix) to bind and inhibit D14 (Shabek et al., 2018). In an SL-dependent manner, D3/MAX2 induces the ubiquitination and degradation of its substrates to transduce SL signals, including D53/D53-like SMXLs (SUPPRESSOR OF MAX2-1 LIKEs, SMXL6, SMXL7, and SMXL8, three orthologs of D53 in Arabidopsis involved in shoot branching) (Jiang et al., 2013, Zhou et al., 2013, Soundappan et al., 2015, Wang et al., 2015), and a basic-helix-loop-helix transcription factor BES1 (bri1-EMS-SUPPRESSOR 1) (Wang et al., 2013). D53/D53-like SMXLs proteins, with ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motifs, act as putative transcriptional repressors to recruit TOPLESS-related proteins. Recently, the crystal structure study demonstrates that D53 promotes assembly of a corepressor–nucleosome complex with TPR2 through the EAR motif, which strongly suggests that the transcriptional regulation is key to transduce SL signaling (Ma et al., 2017). However, D53/D53-like SMXLs are transcription regulators without direct DNA-binding ability (Ma et al., 2017, Song et al., 2017), indicating that they need adaptors to affix DNA to mediate the SL-regulated transcription and shoot branching. BES1 is a transcription factor with DNA-binding activity that directly promotes or inhibits gene expression (Yin et al., 2005). Although BES1 is involved in the SL signaling by the D14–MAX2-mediated degradation in Arabidopsis (Wang et al., 2013), how BES1 mediates the transcriptional regulation in SL signaling is still unknown. In addition, the transcription factor BRC1 (BRANCHED 1) has been reported to be a key switch for inhibiting shoot branching and is regulated by multiple environments and phytohormones, including SLs, in many plant species (Doebley et al., 1995, Aguilar-Martinez et al., 2007, Lewis et al., 2008, Martíntrillo et al., 2011, Choi et al., 2012, Dun et al., 2013, Gonzalez-Grandio et al., 2013). Although BRC1 has been reported to regulate shoot branching genetically downstream of SL signaling (Aguilar-Martinez et al., 2007, Braun et al., 2012, Dun et al., 2012, Guan et al., 2012, Lu et al., 2013), the molecular mechanism of how SL signaling regulates BRC1 is still unknown in Arabidopsis. It is known that transcriptional networks tightly orchestrate the growth and development of mammals and plants, and these networks are triggered by various developmental and environmental cues. Therefore, to complete a signaling pathway, key steps are to identify its essential transcription factors, and reveal that how those transcription factors are regulated by upstream signaling to trigger the signal-specific transcription networks (Hwang and Sheen, 2001, Valverde et al., 2004, Smit et al., 2005, Yin et al., 2005, Pinkston-Gosse and Kenyon, 2007). However, how SL signaling initiates the downstream transcriptional network after SL perception is still unknown.
In addition, BES1 has been initially identified as a primary signaling component in the brassinosteroid (BR) signaling pathway. It is tightly regulated mainly through the dynamic alteration of its phosphorylation status to transduce BR signal by the BR early signaling components, BIN2 (BRASSINOSTEROID INSENSITIVE 2) (Yin et al., 2002, Yin et al., 2005) and PP2A (PROTEIN PHOSPHATASE 2A) (Tang et al., 2011). In BR signaling, the non-phosphorylated and phosphorylated BES1s have different DNA-binding activities to regulate the BR-responsive genes (He et al., 2002). However, in the SL signaling pathway, both phosphorylated and non-phosphorylated BES1s are the direct substrates of SL receptor complex D14–MAX2 to control shoot branching (Wang et al., 2013). Interestingly, the BR signaling components upstream of BES1 display no function in shoot branching in Arabidopsis (Wang et al., 2013). This raises the question of how BES1 differentially functions in the BR and SL signaling to regulate signal-specific developmental processes.
Our transcriptomic and genetic analyss indicate that D53-like SMXLs and BES1 genetically depend on each other to regulate shoot branching through BRC1. This is further supported by the biochemical results thatBES1 physically interacts with D53-like SMXLs to inhibit BRC1 expression, which depends on direct binding of BES1 to the BRC1 promoter, and the EAR motif of D53-like SMXLs that represses the transcription of BRC1. In addition, we demonstrate that BRs treatment has no effect on the interaction of SMXLs with BES1 and the BRC1 expression, and the altered phosphorylation status of BES1 cannot affect its DNA-binding ability with the BRC1 promoter. Together, these findings reveal the mechanisms of how the BES1- D53-like SMXLs complexes transduce SL signals in shoot branching, and how BES1 differentially functions in SL and BR signaling pathways to control signal-specific developmental processes.
Results
BES1- and D53-like SMXLs Genetically Depend on Each Other in Shoot Branching
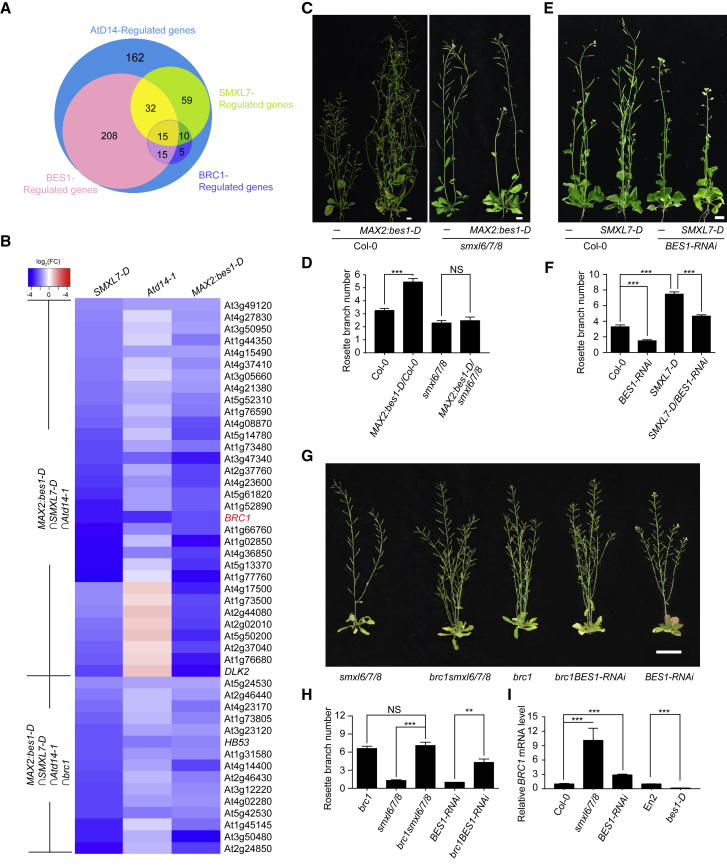
To explore how SL signaling was involved in the transcriptional regulation in shoot branching, we detected the transcriptional profiles in the young buds (bud length ≤3 mm) of the SL signaling-related plant materials, including MAX2:bes1-D-FLAG/Columbia-0 (Col-0) (a gain-of-function form of BES1, which was stable under GR24-induced degradation, defined as MAX2:bes1-D below) (Wang et al., 2013), SMXL7-D-GFP/Col-0 (a gain-of-function form of genomic SMXL7, which was stable under GR24-induced degradation, defined as SMXL7-D below) (Jiang et al., 2013, Soundappan et al., 2015, Wang et al., 2015, Zhou et al., 2013), Atd14-1, and their wild-type Col-0. These materials were reported to exhibit the increased branching number compared with the wild-type Col-0 (Arite et al., 2009, Soundappan et al., 2015, Wang et al., 2013, Wang et al., 2015). First, 506 differentially expressed genes were identified from the comparison between Atd14-1 and the wild-type Col-0, including 42 induced and 464 repressed genes (Supplemental Figure 1A and Supplemental Table 1). These genes were defined as SL-regulated genes because of the high specificity of the receptor AtD14 in SL signaling. There were 516 genes differentially expressed in buds from the comparison of MAX2:bes1-D versus the wild-type Col-0, including 15 upregulated and 501 downregulated genes (Supplemental Figure 1A and Supplemental Table 2). Significantly, 52.33% of the BES1-regulated genes were co-regulated by the SL receptor AtD14 (Figure 1A and Supplemental Figure 1A), and all of them were downregulated in both the MAX2:bes1-D and Atd14-1 (Supplemental Figure 1B and Supplemental Table 3), suggesting that BES1 was a major transcription factor involved in the SL-regulated shoot branching. More independent bes1-D transgenic lines driven by the promoters of MAX2 or BES1, and the bes1-L-D (BES1-L, the long form of BES1) (Jiang et al., 2015) transgenic lines driven by the 35S promoter further confirmed the function of BES1 in promoting shoot branching (Supplemental Figure 2A–2F). In addition, another published independent BES1-RNAi line with reduced expression of BES1 and its close homologs (Yin et al., 2005) also showed decreased branch number compared with the wild type (Supplemental Figure 2G and 2H). Furthermore, BES1 was highly expressed in the axillary buds as indicated by the pBES1-L:GUS and pBES1-S:GUS reporters (Supplemental Figure 3), supporting its key role in shoot branching. Second, there were 116 differentially expressed genes co-regulated by AtD14 and SMXL7 (Figure 1A, Supplemental Figure 1C, Supplemental Tables 4 and 5), all of which showed similar regulatory mode in the Atd14-1 and SMXL7-D (Supplemental Figure 1D). Significantly, 40.52% of them, 47 genes, were also regulated by BES1 (Figure 1A and 1B). Thus, we predicted that SMXL7 might be a major partner for the function of BES1 in SL-regulated shoot branching. Notably, there were still 223 genes co-regulated by BES1 and AtD14, but not by SMXL7 (Figure 1A), suggesting that BES1 was highly specific in the SL-regulated bud outgrowth, and that homologs of SMXL7 were also needed in SL signaling (Stanga et al., 2013, Stanga et al., 2016, Wallner et al., 2017). Several key genes that have been reported to be involved in shoot branching were regulated by both BES1 and SMXL7 in the SL-regulated genes (Figure 1B). For example, HB53 (HOMEOBOX PROTEIN 53), which encodes an HD-ZIP protein in axillary buds, inhibits shoot branching in response to abscisic acid (Gonzalez-Grandio et al., 2017). Importantly, the transcription factor BRC1, a key inhibitor for shoot branching (Aguilar-Martinez et al., 2007, Choi et al., 2012, Doebley et al., 1995, Dun et al., 2013, Gonzalez-Grandio et al., 2013, Lewis et al., 2008, Martíntrillo et al., 2011), was strongly co-regulated by BES1, SMXL7, and AtD14 (Figure 1B), which was consistent to its function in the downstream of SL signaling. Significantly, 53.57% of the BRC1-regulated genes were regulated by AtD14 (Figure 1A, Supplemental Figure 1A and Supplemental Table 6); and 89% of the genes co-regulated by BRC1 and AtD14 were regulated by BES1 and SMXL7 with similar regulatory modes in their buds (Figure 1B, Supplemental Figure 1E and 1F). Some genes that were reported to be involved in bud development, including HB40 (HOMEOBOX PROTEIN 40), NCED3 (9-CIS-EPOXICAROTENOID. DIOXIGENASE 3), NAP (NAC-LIKE, ACTIVATED BY AP3/PI), and UGT74E2 (UDP-glycosyltransferase 74E2), were also found to be under BRC1 regulation (Figure 1B and Supplemental Figure 1E and 1F) (Dong et al., 2008, Tognetti et al., 2010, Gonzalez-Grandio et al., 2013, Gonzalez-Grandio et al., 2017, Holalu and Finlayson, 2017). Therefore, our transcriptome analysis suggests that the SL-regulated transcriptional network in shoot branching is largely dependent on the SMXLs–BES1–BRC1 module.
Figure 1.
BES1- and D53-like SMXLs Genetically Depend on Each Other to Regulate BRC1-Mediated Shoot Branching through BRC1.
(A) Venn diagram of the number of differentially expressed genes in buds of Atd14-1, SMXL7-D, MAX2:bes1-D, and brc1, compared with Col-0, and co-regulated by AtD14. Differentially expressed genes in buds were obtained from cuffdiff analysis with q value <0.05.
(B) Heatmap of the 47 co-regulated genes by AtD14, SMXL7, and BES1 in (A). Original fold change values were transformed by log2 regression for the heatmap shown in the colored bar.
(C) Phenotypes of Col-0, MAX2:bes1-D/Col-0, smxl6/7/8, and MAX2:bes1-D/smxl6/7/8 plants. Scale bar corresponds to 1 cm.
(D) Quantification of rosette branch number of the plants in (C). Data are means ± SE, Col-0 (n = 20), MAX2:bes1-D/Col-0 (n = 27), smxl6/7/8 (n = 27), and MAX2:bes1-Dsmxl6/7/8 (n = 21).
(E) Phenotypes of Col-0, BES1-RNAi, SMXL7-D, and SMXL7-DBES1-RNAi plants. Scale bar corresponds to 1 cm.
(F) Quantification of rosette branch number of the plants in (E). Data are means ± SE, the sample number was Col-0 (n = 17), BES1-RNAi (n = 26), SMXL7-D (n = 19), and SMXL7-DBES1-RNAi (n = 15).
(G) Genetic analysis of BRC1, SMXLs, and BES1 in shoot branching. Scale bar corresponds to 5 cm.
(H) Quantification of rosette branch number of the plants in (G). Data are means ± SE, the sample number was brc1 (n = 20), smxl6/7/8 (n = 20), brc1smxl6/7/8 (n = 20), BES1-RNAi (n = 26), and brc1BES1-RNAi (n = 17).
(I) Relative expression of BRC1 in the buds of Col-0, smxl6/7/8, BES1-RNAi, En2, and bes1-D plants.
Data are means ± SD (n = 6) and P values in (D), (F), (H), and (I) were determined by Student's t-test; ***P < 0.001, **P < 0.01, non-significant (NS), P > 0.05.
See also Supplemental Figures 1 and 2.
To further reveal the relationship among SMXLs, BES1, and BRC1 in SL-inhibited shoot branching, we performed a set of genetic analyses, and found that MAX2:bes1-D could not rescue the branching phenotype of the smxl6/7/8 as indicated by the MAX2:bes1-D/smxl6/7/8 line (Figure 1C and 1D), suggesting that BES1 required SMXLs to promote branching; similarly, the branch number of the SMXL7-D/BES1-RNAi was significantly decreased compared with the SMXL7-D/Col-0 line (Figure 1E and 1F), suggesting that SMXL7 also depended on BES1 to promote branching. Therefore, BES1- and D53-like SMXLs are likely dependent on each other to regulate shoot branching. Furthermore, brc1 was able to rescue the branching phenotypes of either smxl6/7/8 (Seale and Bennett, 2017) or the BES1-RNAi line (Figure 1G and 1H), which indicated that BRC1 acted downstream of both D53-like SMXLs and BES1 to control shoot branching. In addition, the BRC1 expression was lower in the buds of bes1-D, MAX2:bes1-D/Col-0, and SMXL7-D-GFP/Col-0 lines, but higher in the BES1-RNAi lines, smxl6/7/8 (Wang et al., 2015), the MAX2:bes1-D-FLAG/smx6/7/8 lines, and the SMXL7-D-GFP/BES1-RNAi lines than in the wild type (Figure 1I and Supplemental Figure 4A–4C), indicating that knockdown of either BES1 or SMXLs could reduce the inhibitory effect on BRC1 expression. Taken together, we conclude that the D53-like SMXLs and BES1 genetically depend on each other to induce the SL-regulated transcriptional network mainly via BRC1 for Arabidopsis shoot branching.
BES1 Interacts with D53-like SMXLs to Directly Inhibit BRC1 Expression
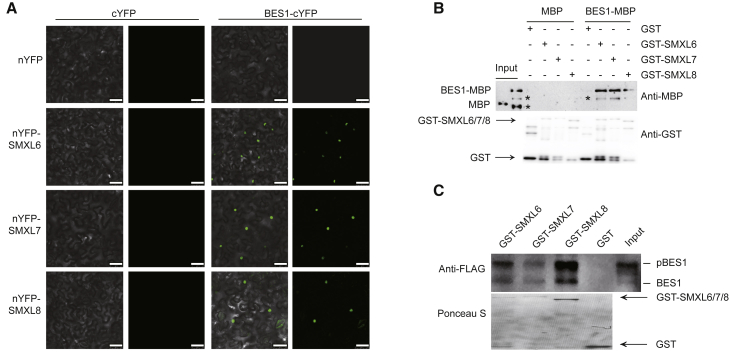
Our further biochemical experiments demonstrated that SMXLs directly interacted with BES1 in pull-down assay, and also with BES1 and its homologs in bimolecular fluorescence complementation (BiFC) assays(Figure 2A and 2B and Supplemental Figure 5). In addition, both the phosphorylated and dephosphorylated BES1s were able to interact with SMXLs (Figure 2C), which was consistent with a previous report that both phosphorylated and dephosphorylated BES1s were able to interact with and be induced to be degraded by MAX2 (Wang et al., 2013), suggesting that both phosphorylated and dephosphorylated BES1s participated in SL signaling. Furthermore, BES1 interacted with D53-like SMXLs with or without additional SLs, BRs, or SLs plus BRs (Supplemental Figure 6).
Figure 2.
The D53-like SMXLs Interact with BES1.
(A) D53-like SMXLs interacted with BES1 in bimolecular fluorescence complementation assays. Scale bars correspond to 50 μm.
(B) D53-like SMXLs interacted with BES1 in a GST pull-down assay. Asterisks (*) indicated a nonspecific band. Anti-GST was used to show the amounts of the loaded GST and GST-SMXLs proteins.
(C) Both the phosphorylated and dephosphorylated forms of BES1 interacted with D53-like SMXLs in a semi-in vivo pull-down assay using 35S:BES1-FLAG plants. Ponceau S staining showed the loaded GST and GST-SMXLs proteins.
See also Supplemental Figures 5 and 6.
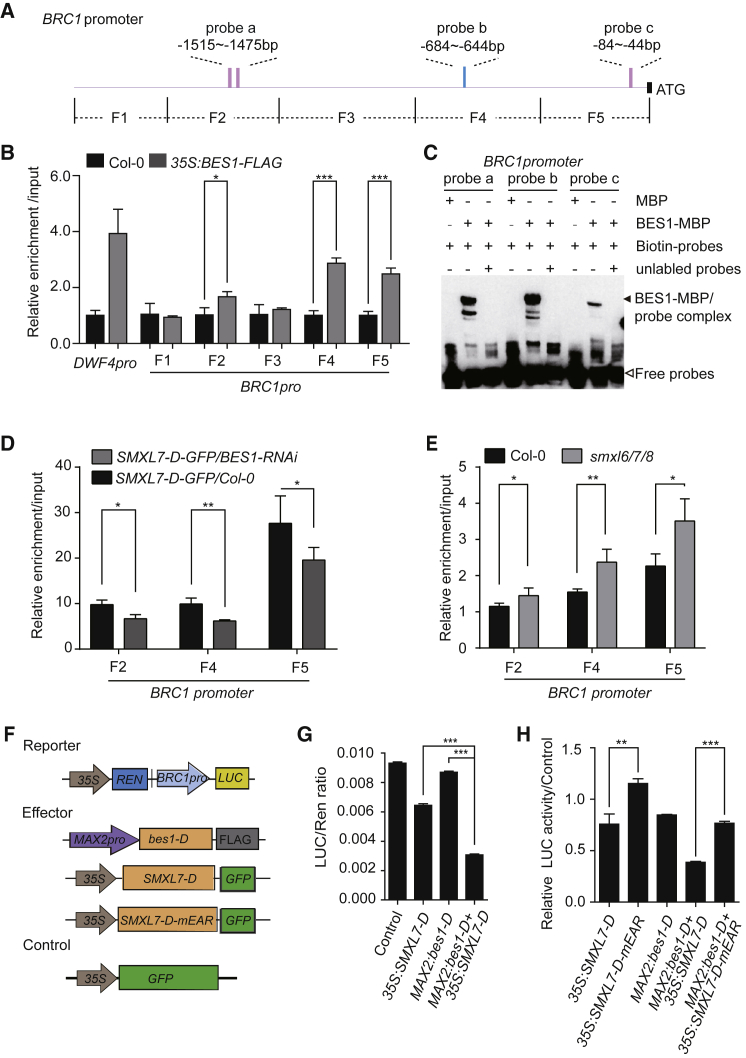
D53/D53-like SMXLs have been reported to induce the oligomerization of TPL tetramer through linking tetramer–tetramer interaction and stabilize the TOPLESS corepressor–nucleosome interaction, which subsequently leads to the formation of repressive chromatin structures to inhibit transcription (Ke et al., 2015, Ma et al., 2017). Due to lacking direct DNA-binding ability, D53 requires an adaptor to specifically target promoters for transcriptional inhibition via chromatin modification (Ma et al., 2017, Song et al., 2017). Therefore, the interaction between BES1 and SMXLs raises the possibility that BES1 and its homologs likely serve as adaptors for SMXLs to proximate DNA and inhibit gene expression. To test this hypothesis, we detected whether BES1 could bind to the BRC1 promoter. Chromatin immunoprecipitation (ChIP)-qPCR and electrophoretic mobility shift assay (EMSA) assays showed that BES1 directly bound to the BRC1 promoter fragments F2, F4, and F5, which contain the E-box and GGTCC elements (BES1 binding cites reported in a previous study [Sun et al., 2010]) (Figure 3A–3C). Furthermore, to investigate the interdependency between BES1 and SMXLs to inhibit BRC1 expression, we performed ChIP assays using the buds in the junction between shoots and roots of different plant materials (Supplemental Figure 7A). We detected the enrichment of BRC1 promoter by SMXL7 in the buds of SMXL7-D-GFP/Col-0 and the SMXL7-D-GFP/BES1-RNAi lines using anti-GFP beads. The results showed the enrichment of BRC1 promoter by SMXL7-D-GFP was much less in the SMXL7-D-GFP/BES1-RNAi plants than in the SMXL7-D-GFP/Col-0 plants (Figure 3D), resulting in a decreased inhibition of BRC1 expression in buds of the SMXL7-D-GFP/BES1-RNAi line compared with that in the SMXL7-D-GFP/Col-0 line (Supplemental Figure 4B). It is indicated that the inhibition of SMXL7 on BRC1 expression requires BES1 binding to the BRC1 promoter. On the other hand, we detected the enrichment of the BRC1 promoter by BES1 in buds of the smxl6/7/8 and the Col-0 plants, and found that although the fragments of the BRC1 promoter enriched by BES1 were significantly higher in the smxl6/7/8 plant than in the Col-0 (Figure 3E), the BRC1 expression level was still higher in the smxl6/7/8 plant than that in Col-0 (Figure 1I and Supplemental Figure 4A), which meant that the inhibition of BES1 on BRC1 expression required SMXLs. In addition, we also tested whether the interdependency between D53-like SMXLs and BES1 directly affected BRC1 expression using the BRC1:LUC reporter in a transient expression assay in N. benthamiana leaves. The SMXL7-D and bes1-D were constructed as effectors, 35S:GFP was used as the control effector, and BRC1:LUC linking 35S controlling Renilla luciferase (REN) was the reporter (Figure 3F). The LUC/REN ratio was significantly reduced in SMXL7-D/bes1-D co-expressed lines compared with the lines expressing SMXL7-D or bes1-D, respectively (Figure 3G). We further measured the effect of D53-like SMXLs and BES1 on BRC1 expression using a direct LUC reporter system in N. benthamiana leaves with 35S:LUC as the reporter (Supplemental Figure 8A). The LUC intensity showed similar results that BRC1 expression was largely inhibited by the co-expression of SMXLs and BES1 (Supplemental Figure 8B–8D). Therefore, the interdependency between BES1 and SMXL7 directly affects BRC1 expression in shoot branching.
Figure 3.
The D53-like SMXLs and BES1 Depend on Each Other to Directly Inhibit BRC1 Expression in Shoot Branching.
(A) Schematic representation showed fragments and probes of the BRC1 promoter in (B)–(E). Pink bars indicated the cis-E-box. Blue bars show the GGTCC element.
(B and C) BES1-MBP directly bound to the BRC1 promoter in ChIP–qPCR (B) and EMSA (C) assays. Solid and open triangles indicate BES1–MBP–DNA bands and free probe, respectively.
(D) The relative enrichment of BRC1 promoter by SMXL7-GFP used anti-GFP beads in buds of SMX7-D-GFP/Col-0 and SMX7-D-GFP/BES1-RNAi plants.
(E) The relative enrichment of BRC1 promoter used anti-BES1 antibody in buds of Col-0 and smxl6/7/8 plants.
(F) Schematic diagrams of the luciferase reporter and effector constructs used in N. benthamiana transient assays.
(G) SMXL7 and BES1 corporately inhibited the expression of BRC1:LUC.
(H) Mutation of the EAR motif in SMXL7 reduced the inhibition of BRC1 expression by the SMXLs–BES1 complex. LUC/REN ratio was normalized to the corresponding control defined as the relative LUC activity.
Data are means ± SD (n = 3) and P values in (B)–(E), (G), and (H) were determined by Student's t-test; ***P < 0.001, **P < 0.01, *P < 0.05.
See also Supplemental Figures 4, 7, 8, and 9.
Because the transcriptional repression by the EAR-contained proteins was highly conserved and general in many signaling pathways among diverse plant species (Kagale and Rozwadowski, 2011), and that the EAR motif in SMXL7 was required for branching (Liang et al., 2016), we next asked whether the EAR motif in SMXL7 was also required by the BES1–SMXLs complex to inhibit BRC1 expression. The SMXL7-D-mEAR-GFP was constructed to detect the function of the EAR motif of SMXL7 in regulating BRC1 expression (Wang et al., 2015). We first tested and confirmed that SMXL7-D-mEAR showed a similar ability to interact with BES1 as SMXL7 and SMXL7-D (Supplemental Figure 9). When using either the dual bioluminescence or the BRC1:LUC reporter system in N. benthamiana, the activities of BRC1:LUC were significantly higher in the SMXL7-D-mEAR/MAX2:bes1-D than in the SMXL7-D/MAX2:bes1-D co-expressing leaves, and were significantly higher in the SMXL7-D-mEAR than in the SMXL7-D expressing leaves (Figure 3H and Supplemental Figure 8E–8G). To further investigate the function of the EAR motif of SMXL7 in shoot branching in planta, SMXL7-D-GFP and SMXL7-D-mEAR-GFP transgenic lines were generated. The SMXL7-D-GFP lines showed an increased number of rosette shoot branches, but the shoot branch number of the SMXL7-D-mEAR-GFP line was similar to that of the wild type (Supplemental Figure 7B and 7C), which was consistent with the results reported in a previous study (Liang et al., 2016). Furthermore, the transcription level of BRC1 in the buds of the SMXL7-D-mEAR-GFP line showed no obvious difference from that of the wild type, but was remarkably higher than that in the SMXL7-D-GFP line (Supplemental Figure 7D). Therefore, the EAR motif of SMXLs is required by the SMXLs–BES1 complex to inhibit BRC1 expression.
BES1 Differentially Functions in SL and BR Signaling in Arabidopsis
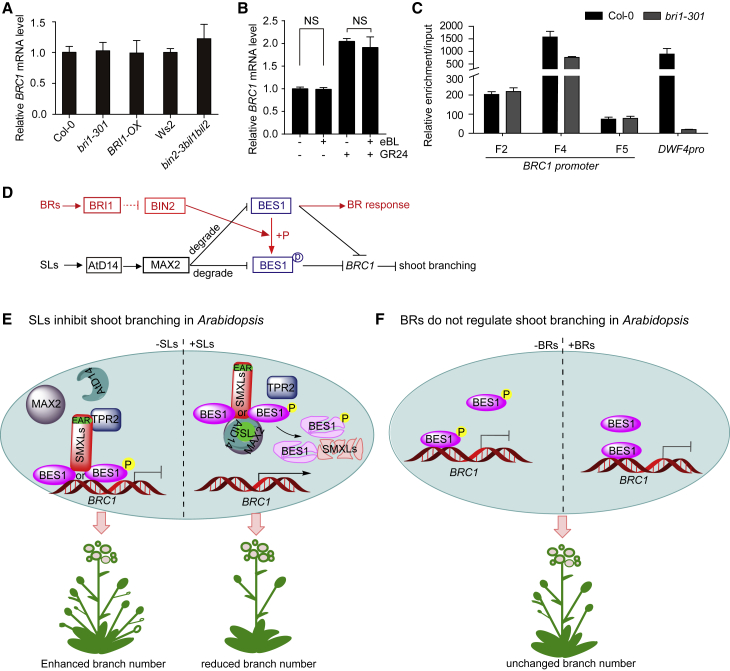
Significantly, BES1 is differently regulated by SL and BR signaling in Arabidopsis. In BR signaling, BES1 is regulated through alteration of its phosphorylation status (Yang et al., 2017), the stability of BES1 is not primarily regulated by BR signaling in Arabidopsis (Jiang et al., 2015, Yang and Wang, 2017); while in SL signaling both the phosphorylated and dephosphorylated BES1s are induced to be degraded by MAX2 (Wang et al., 2013) and interact with D53-like SMXLs (Figure 2C and Supplemental Figure 6). Furthermore, mutants of the BR signaling components upstream of BES1 did not alter branch number (Wang et al., 2013) and BRC1 expression (Figure 4A); and BR treatments had no effects on BRC1 expression, while SLs effectively induced the BRC1 expression with or without BRs (Figure 4B and Supplemental Figure 10). To further reveal the underlying reasons, we performed ChIP–qPCR assays using the BES1 antibody to detect the enrichment of BRC1 promoter by BES1 in Col-0 and the BR receptor mutant bril-301, in which BES1 was mainly in phosphorylated status (Supplemental Figure 4D). Interestingly, BES1 in the BR receptor mutant bril-301 had a similar ability to enrich the BRC1 promoter, but had a largely reduced ability to enrich the DWF4 promoter (Figure 4C), a well-known BR/BES1-targeted gene (He et al., 2005), which well explained the similar BRC1 expression in the BR-related mutants and the wild type (Figure 4A), as well as the unchanged BRC1 expression under BR treatments (Figure 4B). Taken together, these results demonstrate that the alteration of the BES1 phosphorylation status by BR signaling has no effect on BRC1 expression and shoot branching, and that the function of BES1 in SL signaling is independent of that in BR signaling in Arabidopsis (Figure 4D). Therefore, when both SLs and BRs are present, BRs cannot change the SL-controlled shoot branching by altering the phosphorylated status of BES1 in Arabidopsis (Figure 4D).
Figure 4.
BES1 Functions Independently in SL and BR Signaling in Arabidopsis.
(A) Relative BRC1 expression levels in the BR-related materials bri301, BRI-OX, and bin2-3bil1bil2.
(B) The transcription level of BRC1 in isolated buds of Col-0 treated with mock, 5 μM eBL, 5 μM GR24, and 5 μM eBL plus 5 μM GR24 for 3 h, respectively.
(C) The ChIP–qPCR assays of the BRC1 and DWF4 promoters precipitated by BES1 in Col-0 and bri1-301 plants using anti-BES1 antibody. Fold enrichment compared with the ACTIN promoter was normalized to their input.
(D) The model indicates that BES1 functions independently in SL and BR signaling in Arabidopsis. The alteration in the BES1 phosphorylation status by BR signaling has no effect on BRC1 expression in shoot branching, while SL signaling regulates BRC1 expression to inhibit shoot branching through degrading both phosphorylated and dephosphorylated BES1.
(E and F) Working models of the SL- and BR-mediated regulation of shoot branching in Arabidopsis. In Arabidopsis, when SLs are absent, D53-like SMXLs interact with phosphorylated or unphosphorylated BES1 to inhibit BRC1 expression, via direct binding of BES1 to its promoter, and the EAR motif of D53-like SMXLs recruiting TPR2, leading to enhanced shoot branch number. When SLs are present, the D53-like SMXLs–BES1 complex is degraded by AtD14–MAX2 after SLs perception, resulting in the expression of BRC1 to inhibit shoot branching (E). In Arabidopsis, the phosphorylation status change caused by BRs has no effect on BRC1 expression or shoot branching (F).
Data are means ± SD (n = 3) and P values in (B) were determined by Student's t-test; non-significant (NS), P > 0.05.
See also Supplemental Figures 4 and 10.
Discussion
In this study, we reveal that BES1 acts as the adaptor of D53-like SMXLs to trigger the SL-regulated transcriptional network in the buds through the local transcription factor BRC1 for shoot branching in Arabidopsis. First, the genome-wide transcriptomes and genetic analysis using the SL signaling-related plant materials, suggest that BES1- and D53-like SMXLs interdependently trigger an SL-induced transcriptional network for shoot branching mainly through BRC1. Second, we demonstrate that BES1 interacts with D53-like SMXLs to inhibit BRC1 expression, which is dependent on both the direct DNA binding by BES1 and the transcriptional inhibition by the EAR motif of D53-like SMXLs. Third, we reveal that BES1 functions independently in SL and BR signaling in Arabidopsis. Therefore, these data reveal a transcriptional regulation mechanism in the SL-controlled shoot branching via AtD14–MAX2–D53-like SMXLs–BES1–BRC1.
Our genetic and molecular results support the mechanism of how SL signaling directly inhibits BRC1 expression to specifically inhibit bud outgrowth in Arabidopsis. In many species, the TCP transcription factor BRC1 and its homologs are proposed to be key switches to regulate bud outgrowth by coordinating diverse environmental and developmental cues (Doebley et al., 1995, Lewis et al., 2008, Martíntrillo et al., 2011, Dun et al., 2012, Gonzalez-Grandio et al., 2013, Mason et al., 2014, Holalu and Finlayson, 2017). However, the lack of a molecular mechanism by which BRC1 regulates shoot branching means that it has long been controversial whether BRC1 expression is necessary and sufficient for the inhibition of bud outgrowth (Seale and Bennett, 2017). A few studies support the important roles of BRC1 in SL-regulated shoot branching. For instance, the branching number of brc1 is ascribed to rosette branching, but not cauline branching (Aguilar-Martinez et al., 2007), which is consistent with the branching phenotype of the SL-related mutants (Liang et al., 2016); and the expression of BRC1 is also altered in the SL signaling mutants (Zhou et al., 2013, Chevalier et al., 2014, Wang et al., 2015). In addition, in pea and rice, Psbrc1/Osfc1 mutants are insensitive to GR24 treatment and genetically function downstream of SL signaling to inhibit branching (or tillering in rice) (Aguilar-Martinez et al., 2007, Braun et al., 2012, Dun et al., 2012, Guan et al., 2012, Lu et al., 2013). In this study, first, our genetic and molecular results support that BES1 may directly control BRC1 expression depending on D53-like SMXLs to promote bud outgrowth in Arabidopsis (Figure 1). Second, biochemical studies demonstrate that the D53-like SMXLs–BES1 module directly regulates BRC1 expression via DNA binding by BES1 and transcriptional inhibition by the EAR motif of D53-like SMXLs (Figures 1 and 3 and Supplemental Figures 4, 7, and 8). Therefore, our study demonstrates that BRC1, as the SL signaling target, is directly regulated by BES1–SMXLs to inhibit bud outgrowth in Arabidopsis.
We also provide significant insights into how BES1, a component shared by SL and BR signaling pathways, differentially regulates signaling-specific biological processes in Arabidopsis. As a positive component in BR signaling and a key transcription factor directly regulating BR-responsive gene expression (Yin et al., 2005), BES1 regulates BR signaling outputs in Arabidopsis through switching between phosphorylated and dephosphorylated forms to alter its DNA binding and transcription activity (Yang and Wang, 2017, Yin et al., 2002). Recent studies demonstrate that the stability of BES1 is not primarily regulated by BR signaling in Arabidopsis (Jiang et al., 2015, Yang et al., 2017, Yang and Wang, 2017). While in the SL signaling pathway, both the phosphorylated and dephosphorylated forms of BES1 can interact with D53-like SMXLs (Figure 2), and can be induced to be degraded by MAX2 in response to SLs (Wang et al., 2013), indicating that the regulation of BES1 stability is a major mechanism in SL signaling. The differential regulation of BES1 by the two signals indicates that BES1 independently functions in the BR and SL signaling pathways to control different development processes in Arabidopsis (Figure 4D). Consistent with this hypothesis, mutants of the BR signaling components upstream of BES1 have similar branching number (Wang et al., 2013) and similar expression level of BRC1 (Figure 4A) compared with wild type in Arabidopsis; and BR treatment has no effect on BRC1 expression in buds and on the interaction between BES1- and D53-like SMXLs (Figure 4B and Supplemental Figure 6). Furthermore, the altered phosphorylation status of BES1 cannot affect its ability binding to the BRC1 promoter (Figure 4C). These results all support the conclusion that BES1 independently functions in SL and BR signaling to trigger the signal-specific gene expression (Figure 4D).
In addition, a number of genetic data strengthen our conclusion that BES1 and its homologs play an important role in regulating shoot branching in Arabidopsis. In Arabidopsis, BES1 has five homologous genes, BZR1 and BEH1-4. BES1 and its homologs have been reported to work redundantly in BR signaling (Chen et al., 2019; Yin et al., 2005). In addition, it is also known that the homologous genes of BES1 in Arabidopsis are redundant in SL signaling, because BES1 and its homologs are able to interact with MAX2 (Wang et al., 2013) and the D53-like SMXLs (Figure 2 and Supplemental Figure 5). Thus, the BES1-RNAi line, with the reduced expression of BES1 and its homologous genes, displays reduced rosette branching number (Wang et al., 2013) (Figure 1E and 1F and Supplemental Figure 2), and suppresses the branching phenotype of max2-1 (Wang et al., 2013), which well explained why a T-DNA-insertion line, bes1-1, which has abolishes BES1 expression, exhibiting a slightly reduced rosette branches and similar cauline branches compared with that in Col-0 (Bennett et al., 2016). In addition, the bes1-D mutant line in En2 background and the transgenic lines, by expressing bes1-D in the Col-0 background, all exhibited the BR-enhanced phenotypes similar to plants overproducing BRs or BRI1 (Yin et al., 2002), also presented the more branching number than wild-type control in Arabidopsis (Figure 1C, Supplemental Figure 2 and Wang et al., 2013). In this study, the branching phenotype of more independent transgenic lines, including bes1-D and BES1-RNAi (Supplemental Figure 2) further supported the function of BES1 in shoot branching. In addition, in a parallel study, we demonstrated that the OsBZR1-RNAi line (the homolog of BES1 in rice) also exhibits the reduced tiller number, and rescues the tillering phenotype of d14, d3, and d53 in rice; we also demonstrated that the OsBZR1:Osbzr1-D transgenic rice had more tillers than the wild-type Nipponbare. Taken together, these results suggest that the function of AtBES1/OsBZR1 in shoot branching is general and conserved in Arabidopsis and rice.
Therefore, we propose a molecular mechanism how the SL signal is transduced to trigger the transcriptional network in Arabidopsis buds (Figure 4E and 4F). When SLs are insufficient, D53-like SMXLs and BES1, the direct substrates of D14–MAX2, are accumulated, and interact with each other to bind the BRC1 promoter via BES1, which inhibits BRC1 expression by the EAR motif of D53-like SMXLs to increase shoot branching; when SLs are sufficient, BES1 and D53-like SMXLs are all ubiquitinated and induced to be degraded by AtD14–MAX2 complex in buds, which relieves the inhibition of BRC1 expression to inhibit bud outgrowth (Figure 4E). Whereas, the alteration between phosphorylated and dephosphorylated BES1s induced by BR signaling has no effect on the BRC1expression, and does not change the branch number in Arabidopsis (Figure 4F). Therefore, multiple mechanisms have been evolved in regulating BES1 for decoding distinct developmental and environmental cues in plants.
Methods
Plant Materials
The Arabidopsis thaliana mutant alleles used in this study were: brc1 (SALK_091920C) (Aguilar-Martinez et al., 2007), BES-RNAi (Yin et al., 2005), smxl6 (CS847925/SAIL_1285_H05), smxl7 (SALK_082032), smxl8 (SALK_126406) (described in Wang et al., 2015), Atd14-1 mutant (isolated from the Wisconsin DsLox T-DNA insertion collection [CS913109 (N913109)]) (Waters et al., 2012, Vegh et al., 2017), and max2-1 (SALK_092836). All were in the Col-0 background, as well as the brc1smxl6/7/8, brc1BES1-RNAi mutants, and the 35S:BES1-FLAG, SMXL7-D-GFP, SMXL7-D-GFPBES1-RNAi, SMXL7-D-mEAR-GFP, and MAX2:bes1-D smxl6/7/8 transgenic plants. Surface-sterilized seeds were sown on 0.8% agar plates containing Murashige and Skoog (MS) medium. Plates were kept in darkness for 2–3days, and then placed at 22°C under light conditions (16-h light/8-h dark long-day). Primers used for genotyping of these mutants were listed in Supplemental Table 7.
Construction of Transgenic Lines
The Arabidopsis quadruple mutant brc1smxl6/7/8 was generated from a cross between homozygous brc1 and the triple mutant smxl6/7/8, and identified from F2 lines. brc1BES1-RNAi was also obtained from their F2 progeny. Genotyping of the brc1, smxl6, smxl7, and smxl8 mutants was performed by PCR. For Arabidopsis, constructs used to generate transgenic plants were pCAMBIA 1300 with different tags, including SMXL7-D-GFP, SMXL7-D-GFP, SMXL7-D-mEAR-GFP, and MAX2:bes1-D. The genomic DNA fragment of SMXL7, including the promoter region and the transcription region without the stop codon by overlapping PCR (using primer SMXL7pro and SMXL7-R listed in Supplemental Table 7), was fused in-frame to the 5′ end of GFP. SMXL7-D was constructed by overlapping PCR (using primer overlapping-SMXL7-D-F2/R2 listed in Supplemental Table 7) according to the 15-bp deletion of D53 in rice and SMXL7 in Arabidopsis, and resulted in substitution of the amino acids RGKTGI with a single threonine residue (Jiang et al., 2013, Soundappan et al., 2015, Wang et al., 2015, Zhou et al., 2013), which was also fused to the 5′ end of GFP with its promoter. SMXL7-D-mEAR was constructed based on plasmid SMXL7-D by overlapping PCR (using primer overlapping-SMXL7-mEAR-F2/R2 listed in Supplemental Table 7) according to the previous study (Liang et al., 2016). BES1 was amplified using primer BES1-F/R (Supplemental Table 7) to constructed MAX2:bes1-D-FLAG and 35S-BES1-cYFP, and primer BES1-L-F/BES1-R (Supplemental Table 7) to construct 35S:BES1-L-D-mCherry. Genomic BES1 was amplified using BES1pro-F and BES1-R (Supplemental Table 7). Mutated-form bes1-D was obtained by overlapping PCR (using primer overlapping-bes1-D-F/R listed in Supplemental Table 7) according to a previous study (Yin et al., 2002). pBES1-L:GUS and pBES1-S:GUS transgenic lines were used in this paper (Jiang et al., 2015). Constructs were then transfected into Col-0, BES1-RNAi, or smxl6/7/8 by agroinfiltration using the floral dip method (Clough and Bent, 1998). T3 homozygous lines were generated and analyzed for each construct. Primers are listed in Supplemental Table 7.
BiFC, LUC Reporter Assay, and Dual Bioluminescence Assay
For BiFC assays, the full-length coding sequence of each D53-like SMXLs, fused with N-terminal YFP, was cloned into PXY106 vectors. BES1 and its homologous genes, fused with C-terminal YFP, were constructed into PXY104 using the Seamless cloning/in-fusion cloning system. For the LUC reporter assay, the BRC1 promoter (2067 bp length upstream from ATG) and its first exon was constructed into pCAMBIA1300, with LUC as the reporter, and 35S promoter-linked LUC genes as the control reporter. For the effector SMXL7-D, SMXL7-mEAR was constructed into pCAMBIA1300 under the control of a 35S promoter and fused to the 5′ end of the GFP gene, mutated-form SMXL7-D and SMXL7-D-mEAR were constructed as above (described in part construction of transgenic lines), based on the coding sequence of SMXL7 which was amplified using primer SMXL7-F/R (Supplemental Table 7). MAX2:bes1-D was same as the plasmid used to construct the transgenic plant MAX2:bes1-D-FLAG/Col-0. Empty plasmid pCAMBIA1300 with GFP genes under a 35S promoter was used as the control effector.
For dual bioluminescence assays, the BRC1 promoter (2067 bp length with ATG) controlling the LUC reporter gene was constructed into pGreenII 0800-LUC, linked to a 35S promoter regulating the renilla (REN) reporter gene, which was used as the reference. The effectors 35S:SMXL7-D-GFP, 35S:SMXL7-mEAR-GFP, and MAX2:bes1-D-FLAG were constructed in the same way as in the LUC reporter assay. Primers are listed in Supplemental Table 7. Agrobacterium strain GV3101 was transformed with the above vector, then injected into young leaves of N. benthamiana. Plants were grown in the dark for 1 day, then transferred to long-day conditions (16 h light/8 h dark) for 2 days. Fluorescence signals in pavement cells were observed with confocal microscopy (Leica SP8). For the luciferase reporter assay, 2 mM luciferin was used to observe the fluorescence using a CCD system (LUMAZONE PYLON2048B). For dual bioluminescence assay, the fluorescence of LUC and REN were detected using the Dual-Luciferase Report Assay System by Mithras LB940.
In Vitro Pull-Down Assay
The coding sequence of each gene in the D53-like SMXLs family was cloned into pGEX-4T-1 to obtain GST-SMXLs recombinant proteins. Primers are listed in Supplemental Table 7. GST fusion proteins and MBP fusion proteins were purified using glutathione beads (GenScript), and amylose resin (NEB), respectively. Glutathione beads containing GST or GST-SMXLs were incubated with MBP, MBP-BES1 in 1× PBS at 4°C for 2 h. Beads were washed 8–10 times with wash buffer (1× PBS, 0.1% Triton X-100) and boiled with 1× SDS loading buffer at 95°C for 5–10 min, separated by SDS–PAGE, and immunoblotted with anti-MBP antibodies (produced in our lab by rabbits immunized with full-length MBP protein).
Semi-in Vivo Pull-Down Assay
Semi-in vivo pull-down assays were performed using 35S:BES1-FLAG transgenic plants, which were grown on ½ MS medium for 15 days. Plant materials were ground to powder in liquid nitrogen and solubilized with 2× protein extraction buffer (100 mM Tris–HCl [pH 7.5], 300 mM NaCl, 2 mM EDTA [pH 8.0], 1% Triton X-100, 10% glycerol, and protease inhibitor). Extracts were centrifuged twice at 12 000 rpm for 10 min, and the resulting supernatants were collected and incubated with either GST or GST-SMXLs pre-incubated GST beads at 4°C for 2 h. Beads were washed about five times with wash buffer, and then boiled with 1× SDS loading buffer at 95°C for 5–10 min, separated by SDS–PAGE, and immunoblotted with anti-FLAG antibodies.
RT–PCR and RNA Sequencing
Rosette buds ≤3 mm were excised from different plants, which were about 5–10 cm high with only one main branch. Excised buds were immediately put into liquid nitrogen, then collected for RNA extraction. Total RNA was prepared using a plant total RNA extraction kit (TIANGEN), according to the users' manual. For qRT–PCR, RNA samples were reverse transcribed using a first-strand cDNA synthesize kit (Takara) and oligo(dT). Real-time PCR experiments were performed using gene-specific primers (Supplemental Table 7) on a CFX 96 real-time PCR detection system (Bio-Rad) in a total volume of 10 μl containing 2 μl diluted cDNA, 0.3 mM gene-specific primers, and 5 μl SYBR Green Supermix (Bio-Rad). The Arabidopsis U-box gene was used as the internal control. RNA samples were sent to the Beijing Genomics Institute for RNA sequencing (RNA-seq). The RNA-seq data that support the findings of this study are available.
ChIP
Using the method published by Fiil et al. (2008), Col-0 and BES1-FLAG seedlings of about 2–3-weeks-old or buds with junction of shoot and root of Col-0, smxl6/7/8, SMXL7-D-GFP/Col-0, and SMXL7-D-GFP/BES1-RNAi lines were harvested with Fix Buffer (0.4 M sucrose, 10 mM Tris–HCl [pH 8.0], 1 mM EDTA, 1 mM PMSF, 1.0% formaldehyde). Seedlings were vacuum-infiltrated for 30 min for crosslinking. Anti-FLAG gels, anti-GFP gels (40 μl) or endogenous anti-BES1 antibody (needed to pre-clear the chromatin sample using 100–200 μl protein A resin) was used for immunoprecipitation of BES1–DNA complex. Regarding anti-FLAG and anti-GFP gels, chromatin was incubated with gels at 4°C overnight using a rotating mixer wheel before collected. While, as for anti-AtBES1 antibody, after being rotated at 4°C overnight, 40 μl of protein A resin was added and rotated at 4°C for 3 h to collected the BES1–DNA complex. Finally, DNA was isolated by phenol:chloroform. Finally, 50 μl of Milli-Q water was added to dissolve the pellet DNA.
EMSA
The amplified coding sequences of BES1 were fused in-frame with MBP tags and transformed into Escherichia coli. BES1-MBP recombinant proteins were purified. MBP was purified as the control. Recombinant proteins were then incubated with biotin-labeled probes, or with corresponding unlabeled probes for 30 min in EMSA-binding buffer (Thermo Fisher Scientific). Reaction mixtures were separated by non-denaturing polyacrylamide. DNA signals were detected by chemiluminescence.
Quantification and Statistical Analysis
qRT–PCR data were collected using Bio-Rad real-time PCR detection systems. These data were assumed to follow normal distributions and were subjected to one-tailed or two-tailed Student's t-tests according to F-test results. Statistical tests were performed in Microsoft Excel 2016. Statistical parameters, including the exact value of n, the precision measures (mean ± SD) or (mean ± SE) and statistical significance, can be found in the figure legends. Here, n means number of plants for phenotypic analysis, or numbers of technical replicates for qRT–PCR. In Figures, asterisks denote statistical significance test (***P < 0.001, **P < 0.01, *P < 0.05, non-significant [NS], P > 0.05) compared with the corresponding controls, unless otherwise specified by lines connecting the compared pieces of data.
Funding
Supported by NSFC 31430046 (to X.W), 31661143024 (to X.W.), National Key Research and Development Plan 2016YFD0100403 (to S.S.), the Ministry of Agriculture Innovation team plan (0120150092 to X.W.), the School Independent Scientific and Technological Innovation Foundation and Research Startup Foundation of Huazhong Agricultural University (2662015PY020 and 2014RC002 to X.W.).
Author Contributions
X.W., S.S., and J.H. designed the experiments and wrote the manuscript. J.H., Y.J., and X.H. performed the experiments and analyzed the data.
Acknowledgments
We thank J.Y. Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the mutants smxl6/7/8. No conflict of interest declared.
Published: December 12, 2019
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Contributor Information
Shiyong Sun, Email: sunshiyong@mail.hzau.edu.cn.
Xuelu Wang, Email: xlwang@mail.hzau.edu.cn.
Supplemental Information
References
- Aguilar-Martinez J.A., Poza-Carrion C., Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell. 2007;19:458–472. doi: 10.1105/tpc.106.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K., Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435:824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- Al-Babili S., Bouwmeester H.J. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015;66:161–186. doi: 10.1146/annurev-arplant-043014-114759. [DOI] [PubMed] [Google Scholar]
- Arite T., Umehara M., Ishikawa S., Hanada A., Maekawa M., Yamaguchi S., Kyozuka J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009;50:1416–1424. doi: 10.1093/pcp/pcp091. [DOI] [PubMed] [Google Scholar]
- Bennett T., Liang Y., Seale M., Ward S., Muller D., Leyser O. Strigolactone regulates shoot development through a core signalling pathway. Biol. Open. 2016;5:1806–1820. doi: 10.1242/bio.021402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge C.A., Kyozuka J. New genes in the strigolactone-related shoot branching pathway. Curr. Opin. Plant Biol. 2010;13:34–39. doi: 10.1016/j.pbi.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Braun N., de Saint Germain A., Pillot J.P., Boutet-Mercey S., Dalmais M., Antoniadi I., Li X., Maia-Grondard A., Le Signor C., Bouteiller N. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 2012;158:225–238. doi: 10.1104/pp.111.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-G., Gao Z., Zhao Z. BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mol. Plant. 2019;12:1408–1415. doi: 10.1016/j.molp.2019.06.006. [DOI] [PubMed] [Google Scholar]
- Chevalier F., Nieminen K., Sanchez-Ferrero J.C., Rodriguez M.L., Chagoyen M., Hardtke C.S., Cubas P. Strigolactone promotes degradation of DWARF14, an alpha/beta hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell. 2014;26:1134–1150. doi: 10.1105/tpc.114.122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M.S., Woo M.O., Koh E.B., Lee J., Ham T.H., Seo H.S., Koh H.J. Teosinte Branched 1 modulates tillering in rice plants. Plant Cell Rep. 2012;31:57–65. doi: 10.1007/s00299-011-1139-2. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cook C.E., Whichard L.P., Turner B., Wall M.E., Egley G.H. Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science. 1966;154:1189–1190. doi: 10.1126/science.154.3753.1189. [DOI] [PubMed] [Google Scholar]
- De Saint Germain A., Clave G., Badet-Denisot M.A., Pillot J.P., Cornu D., Le Caer J.P., Burger M., Pelissier F., Retailleau P., Turnbull C. An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat. Chem. Biol. 2016;12:787–794. doi: 10.1038/nchembio.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Gustus C. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska M.A., Leyser O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 2011;12:211–221. doi: 10.1038/nrm3088. [DOI] [PubMed] [Google Scholar]
- Dong G., Ma D.P., Li J. The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem. Biophys. Res. Commun. 2008;373:659–664. doi: 10.1016/j.bbrc.2008.06.096. [DOI] [PubMed] [Google Scholar]
- Dun E.A., de Saint Germain A., Rameau C., Beveridge C.A. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 2012;158:487–498. doi: 10.1104/pp.111.186783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun E.A., de Saint Germain A., Rameau C., Beveridge C.A. Dynamics of strigolactone function and shoot branching responses in Pisum sativum. Mol. Plant. 2013;6:128–140. doi: 10.1093/mp/sss131. [DOI] [PubMed] [Google Scholar]
- Fiil B.K., Qiu J.L., Petersen K., Petersen M., Mundy J. Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot5049. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., Fermas S., Brewer P.B., Puech-Pages V., Dun E.A., Pillot J.P., Letisse F., Matusova R., Danoun S., Portais J.C. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Grandio E., Poza-Carrion C., Sorzano C.O., Cubas P. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell. 2013;25:834–850. doi: 10.1105/tpc.112.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Grandio E., Pajoro A., Franco-Zorrilla J.M., Tarancon C., Immink R.G., Cubas P. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. U S A. 2017;114:E245–E254. doi: 10.1073/pnas.1613199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J.C., Koch K.E., Suzuki M., Wu S., Latshaw S., Petruff T., Goulet C., Klee H.J., McCarty D.R. Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol. 2012;160:1303–1317. doi: 10.1104/pp.112.204503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Yang Y., Li J., Wang Z.Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holalu S.V., Finlayson S.A. The ratio of red light to far red light alters Arabidopsis axillary bud growth and abscisic acid signalling before stem auxin changes. J. Exp. Bot. 2017;68:943–952. doi: 10.1093/jxb/erw479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- Jiang L., Liu X., Xiong G., Liu H., Chen F., Wang L., Meng X., Liu G., Yu H., Yuan Y. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature. 2013;504:401–405. doi: 10.1038/nature12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Zhang C., Wang X. A recently evolved isoform of the transcription factor BES1 promotes brassinosteroid signaling and development in Arabidopsis thaliana. Plant Cell. 2015;27:361–374. doi: 10.1105/tpc.114.133678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Rozwadowski K. EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics. 2011;6:141–146. doi: 10.4161/epi.6.2.13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J., Ma H., Gu X., Thelen A., Brunzelle J.S., Li J., Xu H.E., Melcher K. Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci. Adv. 2015;1:e1500107. doi: 10.1126/sciadv.1500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.M., Mackintosh C.A., Shin S., Gilding E., Kravchenko S., Baldridge G., Zeyen R., Muehlbauer G.J. Overexpression of the maize Teosinte Branched1 gene in wheat suppresses tiller development. Plant Cell Rep. 2008;27:1217–1225. doi: 10.1007/s00299-008-0543-8. [DOI] [PubMed] [Google Scholar]
- Liang Y., Ward S., Li P., Bennett T., Leyser O. SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell. 2016;28:1581–1601. doi: 10.1105/tpc.16.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Yu H., Xiong G., Wang J., Jiao Y., Liu G., Jing Y., Meng X., Hu X., Qian Q. Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell. 2013;25:3743–3759. doi: 10.1105/tpc.113.113639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Duan J., Ke J., He Y., Gu X., Xu T.H., Yu H., Wang Y., Brunzelle J.S., Jiang Y. A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martíntrillo M., Grandío E.G., Serra F., Marcel F., Rodríguezbuey M.L., Schmitz G., Theres K., Bendahmane A., Dopazo H., Cubas P. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 2011;67:701–714. doi: 10.1111/j.1365-313X.2011.04629.x. [DOI] [PubMed] [Google Scholar]
- Mason M.G., Ross J.J., Babst B.A., Wienclaw B.N., Beveridge C.A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. U S A. 2014;111:6092–6097. doi: 10.1073/pnas.1322045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Xue Y.L., Miyakawa T., Hou F., Qin H.M., Fukui K., Shi X., Ito E., Ito S., Park S.H. Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 2013;4:2613. doi: 10.1038/ncomms3613. [DOI] [PubMed] [Google Scholar]
- Pinkston-Gosse J., Kenyon C. DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat. Genet. 2007;39:1403–1409. doi: 10.1038/ng.2007.1. [DOI] [PubMed] [Google Scholar]
- Seale M., Bennett T. BRC1 expression regulates bud activation potential but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development. 2017;144:1661–1673. doi: 10.1242/dev.145649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N., Ticchiarelli F., Mao H., Hinds T.R., Leyser O., Zheng N. Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling. Nature. 2018;563:652–656. doi: 10.1038/s41586-018-0743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debelle F., Gough C., Bisseling T., Geurts R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- Song X., Lu Z., Hong Y., Shao G., Xiong J., Meng X., Jing Y., Liu G., Xiong G., Duan J. IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 2017;27:1128–1141. doi: 10.1038/cr.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I., Bennett T., Morffy N., Liang Y., Stanga J.P., Abbas A., Leyser O., Nelson D.C. SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell. 2015;27:3143–3159. doi: 10.1105/tpc.15.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga J.P., Smith S.M., Briggs W.R., Nelson D.C. SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 2013;163:318–330. doi: 10.1104/pp.113.221259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga J.P., Morffy N., Nelson D.C. Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta. 2016;243:1397–1406. doi: 10.1007/s00425-015-2458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Fan X.Y., Cao D.M., Tang W., He K., Zhu J.Y., He J.X., Bai M.Y., Zhu S., Oh E. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Yuan M., Wang R., Yang Y., Wang C., Oses-Prieto J.A., Kim T.W., Zhou H.W., Deng Z., Gampala S.S. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011;13:124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti V.B., Van Aken O., Morreel K., Vandenbroucke K., van de Cotte B., De Clercq I., Chiwocha S., Fenske R., Prinsen E., Boerjan W. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell. 2010;22:2660–2679. doi: 10.1105/tpc.109.071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Vegh A., Incze N., Fabian A., Huo H., Bradford K.J., Balazs E., Soos V. Comprehensive analysis of DWARF14-LIKE2 (DLK2) reveals its functional divergence from strigolactone-related paralogs. Front. Plant Sci. 2017;8:1641. doi: 10.3389/fpls.2017.01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner E.S., Lopez-Salmeron V., Belevich I., Poschet G., Jung I., Grunwald K., Sevilem I., Jokitalo E., Hell R., Helariutta Y. Strigolactone- and karrikin-independent SMXL proteins are central regulators of phloem formation. Curr. Biol. 2017;27:1241–1247. doi: 10.1016/j.cub.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sun S., Zhu W., Jia K., Yang H., Wang X. Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev. Cell. 2013;27:681–688. doi: 10.1016/j.devcel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Wang L., Wang B., Jiang L., Liu X., Li X., Lu Z., Meng X., Wang Y., Smith S.M., Li J. Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell. 2015;27:3128–3142. doi: 10.1105/tpc.15.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Nelson D.C., Scaffidi A., Flematti G.R., Sun Y.K., Dixon K.W., Smith S.M. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development. 2012;139:1285–1295. doi: 10.1242/dev.074567. [DOI] [PubMed] [Google Scholar]
- Yang M., Wang X. Multiple ways of BES1/BZR1 degradation to decode distinct developmental and environmental cues in plants. Mol. Plant. 2017;10:915–917. doi: 10.1016/j.molp.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Yang M., Li C., Cai Z., Hu Y., Nolan T., Yu F., Yin Y., Xie Q., Tang G., Wang X. SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev. Cell. 2017;41:47–58.e44. doi: 10.1016/j.devcel.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R., Ming Z., Yan L., Li S., Wang F., Ma S., Yu C., Yang M., Chen L., Chen L. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature. 2016;536:469. doi: 10.1038/nature19073. [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Moragarcia S., Li J., Yoshida S., Asami T., Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Zhou F., Lin Q., Zhu L., Ren Y., Zhou K., Shabek N., Wu F., Mao H., Dong W., Gan L. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature. 2013;504:406–410. doi: 10.1038/nature12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.