Abstract
It has long been recognized that natural selection during the haploid gametophytic phase of the plant life cycle may have widespread importance for rates of evolution and the maintenance of genetic variation. Recent theoretical advances have further highlighted the significance of gametophytic selection for diverse evolutionary processes. Genomic approaches offer exciting opportunities to address key questions about the extent and effects of gametophytic selection on plant evolution and adaptation. Here, we review the progress and prospects for integrating functional and evolutionary genomics to test theoretical predictions, and to examine the importance of gametophytic selection on genetic diversity and rates of evolution. There is growing evidence that selection during the gametophyte phase of the plant life cycle has important effects on both gene and genome evolution and is likely to have important pleiotropic effects on the sporophyte. We discuss the opportunities to integrate comparative population genomics, genome-wide association studies, and experimental approaches to further distinguish how differential selection in the two phases of the plant life cycle contributes to genetic diversity and adaptive evolution.
Key words: gametophyte, pollen, ovule, evolution, genomics, selection
The development of genomic approaches offers fruitful opportunities to investigate key questions concerning the evolutionary consequences of gametophytic selection in plants. This review summarizes the current knowledge about gametophytic selection and highlights opportunities for integrating comparative population genomics, genome-wide association studies, and experimental approaches to better understand the extent and effects of gametophytic selection on genetic diversity and evolution.
Introduction
The gametophytic phase of the plant life cycle is believed to have important implications for genetic variation and plant evolution. The absence of heterozygosity in the haploid phase removes dominance effects, thereby increasing the efficacy of selection on both beneficial and deleterious mutations (Immler, 2019). Furthermore, a variety of selective pressures act only during the gametophytic phase of the life cycle. Competition between male gametophytes can cause variance in fertilization success, creating the potential for strong directional selection (Haldane, 1932). Complex interactions between male gametophytes, female gametophytes, and the sporophyte create opportunities for sexual conflict and coevolution (Higashiyama and Takeuchi, 2015; Lankinen and Green, 2015). Finally, variants favored in gametophytes may have positive or negative pleiotropic effects on sporophyte fitness (Peters and Weis, 2018). A major goal of this review is to explore the prevalence of gametophytic selection and its effects on the sporophyte (Figure 1).
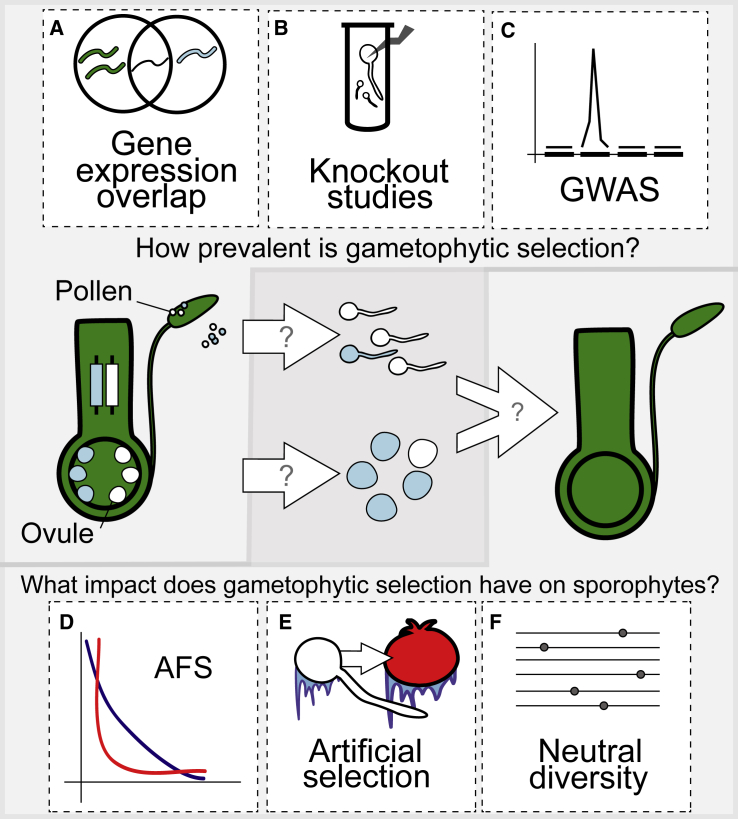
Figure 1.
Two Key Questions Should Guide Future Research on Gametophytic Selection.
Three approaches can be used to determine the prevalence of gametophytic selection:
(A) Investigate gene-expression overlap between the gametophyte and sporophyte.
(B) Reverse genetics: evaluate the impact of genetic knockouts on gametophytes.
(C) Forward genetics: scan genome-wide for associations between alleles and gametophyte phenotypes (GWAS).
To evaluate the effects of gametophytic selection on sporophytes, the following studies will be useful:
(D) Use the allele frequency spectrum (AFS) to compare the frequency of alleles with effects on gametophyte fitness to neutral expectations.
(E) Conduct artificial selection on gametophytes to evaluate the impact on the sporophyte (see, e.g. Domínguez et al., 2005 on selection for cold tolerance in tomatoes).
(F) Use linked neutral genetic diversity to infer signals of past selection.
Despite nearly a century of research, quantifying the extent of gametophytic selection and its fitness effects in sporophytes remains challenging. Pollen has attracted the greatest research effort, and early hypotheses focused on the potential for conflict between pollen and sporophytes (Haldane, 1932). Later researchers proposed that selection during the gametophyte stage in angiosperms could increase sporophyte fitness (Mulcahy, 1979), and early evidence reporting differences in offspring quality due to pollen competition created widespread enthusiasm for this hypothesis (e.g., Stephenson et al., 1986; Davis et al., 1987). The simple life cycle of pollen also attracted the attention of artificial selection programs seeking increased sporophyte yield in crops (Ottaviano et al., 1980). However, convincing evidence for the pervasive genetic variation in gametophyte performance necessary for gametophytic selection was hotly debated, as was the nature of pleiotropic effects of this variation (Charlesworth et al., 1987; Charlesworth, 1988; Walsh and Charlesworth, 1992). The early focus on pollen has continued, although removing dominance effects is as likely in female as in male gametophytes, and similar questions apply to both pollen and ovules regarding pleiotropic expression, the evolutionary dynamics of pollen–pistil interactions, and how these might vary with mating system (Willson and Burley, 1983). If, as comparative studies of pollen limitation suggest (Larson and Barrett 2000; Knight et al., 2005), the number of ovules per flower is often greater than the amount of pollen delivered to stigmas, competition between ovules may also be more prevalent than is generally assumed. Thus, after a century of study, much remains unknown about the evolutionary consequences of gametophytic selection.
Recent theoretical analyses on the consequences of differences in selection during the diploid and haploid phase (Scott and Otto, 2017; Immler and Otto 2018), and a growing resurgence of empirical research on the subject (Williams and Mazer, 2016; Delph, 2019), have reopened discussion on the importance of gametophytic selection for a variety of questions in plant evolution. This, coupled with growing technological advances in functional and evolutionary genomics, leads to novel approaches to further understanding of the evolutionary significance of gametophytic selection and its role in structuring genome-wide diversity and the evolution of genes and genomes. Here, we highlight key open questions and empirical evidence concerning the evolutionary importance of gametophytic selection, with a particular focus on seed plants. We then identify future directions for the field by integrating experimental functional, evolutionary, and quantitative genomics studies of gametophytes.
What Is the Genome-wide Scope for Gametophytic Selection?
The evolutionary importance of the haploid gametophyte phase depends on what proportion of the genome is subject to selection. In addition, the impact of gametophytic selection on evolutionary processes can be constrained or accelerated by pleiotropic effects of gene function shared between the gametophytic and sporophytic stages (Lande and Arnold 1983; Keith and Mitchell-Olds, 2019; and see below). Gene-expression studies, analyses of allelic variation in gametophytic success, and quantitative genetic analyses have all provided useful insights into the number of genes and fraction of mutations that are expressed in gametophytes, how their expression levels compare with sporophytic expression, and which mutations affect the success of both male and female gametophytes.
Gene Expression in the Gametophytic Phase
To characterize the evolutionary importance of the gametophytic phase, it is first necessary to determine whether the phenotype of gametophytes is a result of the genotype of the gametophyte or the sporophyte. For example, if pollen-tube growth rate is determined entirely by the sporophyte that produced the pollen, the phenotype of the gametophyte is a function of the sporophyte's genotype. Studies of gene expression in haploid gametophytes can be used to infer the potential scope for gametophytic selection (Figure 1A). Although initially small in scale and/or of low resolution, evidence from isozyme analysis and early mRNA hybridization studies indicated that significant numbers of genes are expressed in mature pollen in species including Solanum (Lycopersicon) esculentum (Tanksley et al., 1981), Tradescantia paludosa (Willing and Mascarenhas, 1984), Zea mays (Sari-Gorla et al., 1986; Willing et al., 1988), and several other systems (Pedersen et al., 1987). These studies also indicated significant overlap in gene expression between the gametophyte and sporophyte phases; pollen transcriptomes consistently showed evidence for lower complexity (i.e., fewer distinct transcripts) than sporophytic tissue, but the majority of genes expressed in the gametophyte are also expressed in the sporophyte (Figure 2).
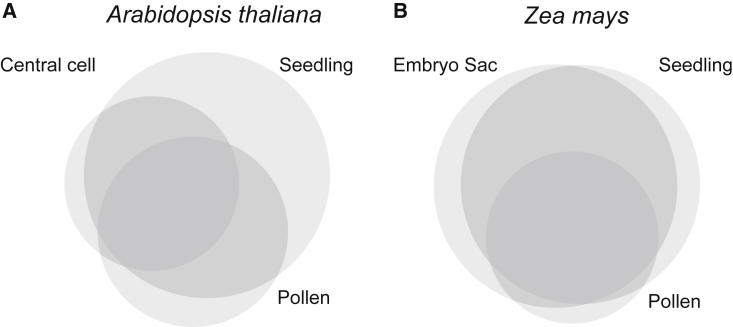
Figure 2.
Gene Expression Overlap between Gametophyte and Sporophyte
The observed overlap in gene expression between the sporophyte and female and male gametophytes in (A)Arabidopsis thaliana (data from Borges et al., 2008 and Wuest et al., 2010) and (B)Zea mays (data from Chettoor et al., 2014). See Supplemental Table 1 for numerical values.
Whole transcriptome-based approaches using microarrays and mRNA sequencing have provided genome-wide confirmation that in male gametophytes, a significant proportion of the genome is transcriptionally active (Figure 2; reviewed in Rutley and Twell, 2015). Male gametophytes grow, exchange nutrients with their surroundings, export and receive signals, and orient themselves in space (reviewed in Bedinger, 1992; Higashiyama and Takeuchi, 2015). This extensive biological function is reflected in transcription: studies across diverse species have revealed thousands of genes transcribed and translated in pollen (e.g., Rutley and Twell, 2015; Sandler et al., 2018; Warman et al., 2020). Furthermore, the transcriptome of mature pollen only represents a fraction of the genes likely to be under selection in male gametophytes; expression varies considerably during the pollen life cycle between growth stages (Honys and Twell, 2004; Wei et al., 2010) and between pre- and post-pollination (Qin et al., 2009; Boavida et al., 2011). Comparisons with expression in sporophytic tissue have indicated a unique transcriptional profile specialized to pollen but also a strong overlap with genes expressed in the sporophyte (Honys and Twell, 2004; Borges et al., 2008; Tang et al., 2010). Whereas male gametophyte expression varies between the species that have been investigated, approximately 40%–60% of genes assayed showed evidence of expression in at least one stage of male gametophyte development, with approximately 90% or more of pollen-expressed transcripts also expressed in sporophytic tissues (Honys and Twell, 2004; Borges et al., 2008; Tang et al., 2010; Whittle et al., 2010; Anderson et al., 2013; Chettoor et al., 2014). This general finding suggests that there is considerable scope for pleiotropic fitness effects between the two life-cycle phases.
Similarly, although it is more technically challenging to isolate female gametophyte tissue, expression studies of the female gametophyte also suggest widespread gene expression that overlaps considerably with the sporophytic phase (Figure 2). Studies identifying genes specific to the gametophyte through comparisons of fertile and infertile ovules (Yu et al., 2005; Yang et al., 2016; Yao et al., 2018), or through directly sequencing female gametophytes following laser microdissection (Wuest et al., 2010; Schmidt et al., 2011; Kubo et al., 2013; Galla et al., 2019), typically report extensive transcriptional activity across a wide variety of systems. Comparisons of pollen and ovule transcriptomes suggest that pollen transcriptomes exhibit the most distinctive expression profiles, but there are some parallel patterns of differential expression in both male and female gametophyte tissue, such as increased expression of repetitive sequences, likely due to regulation of transposable elements (Chettoor et al., 2014).
Taken together, expression studies of gametophyte and sporophyte tissue suggest that (1) a substantial portion of the genome is expressed in the gametophyte and therefore genetic variation is likely visible to selection, and (2) gene expression overlaps incompletely but considerably between sporophyte and male and female gametophytes, which could cause pleiotropic fitness effects.
Experimental Assays of Gametophytic Selection
Although expression studies provide important support for the prediction of widespread phenotypic effects of genes expressed in the gametophyte, and significant overlap with the sporophyte, the connection between gene expression and fitness effects is not always clear. While highly expressed sequences are often under stronger selection (Drummond et al., 2005), gene expression alone does not guarantee a direct effect on fitness (Barrett and Hoekstra, 2011). For example, widespread expression of transposable elements in pollen vegetative cells is likely linked to resetting of epigenetic silencing of transposable elements (Slotkin et al., 2009), but mutations within these transcripts may have little effect on pollen function and competitive ability. To demonstrate fitness effects, experimental assays of differential allelic transmission provide a necessary complement to studies that only describe gene expression.
Early researchers inferred variation in fertilization success among pollen grains from biased segregation patterns and termed this phenomenon “certation” (Correns, 1907; Heribert-Nilsson, 1920; Brink, 1927; Jones, 1928). Non-Mendelian genotype frequencies in the progeny of heterozygous crosses provided the first evidence for heritable variation in pollen competitive ability, suggesting the potential for gametophytic selection. More recently, studies of transmission distortion of genes heterozygous for knockout mutations have helped identify candidate loci with significant effects on male and female gametophyte function (Feldmann et al., 1997; Christensen et al., 1998; Howden et al., 1998; Pagnussat et al., 2005; Boavida et al., 2009). Backcrosses revealed lines where gene knockouts specifically affected pollen transmission, ovule transmission, and in some cases both (Pagnussat et al., 2005; Boavida et al., 2009). Although these screens have revealed significant numbers of genes affecting gametophytic transmission, the percentage of gene knockouts (1%–2%) identified in such screens is considerably smaller than the proportion of genes expressed in gametophytes (Pagnussat et al., 2005). However, these are stringent screens that are meant to survey near-lethal effects on gametophytic fitness. As fitness is a quantitative trait, we expect many more spontaneous mutations to have subtler gametophytic fitness effects. Indeed, earlier studies of induced deletion mutations (reviewed in Walsh and Charlesworth, 1992) indicate a general pattern of biased pollen transmission, suggesting that a high proportion of loci across the genome affect gametophytic performance.
A recent study in maize that combines knockout mutations with data on gene expression is consistent with the expectation that genes overexpressed in pollen are more likely to confer fitness effects on gametophyte transmission (Warman et al., 2020). These authors demonstrated that knockout mutations of genes with high expression in sporophytic tissues showed no evidence of transmission distortion, but 10% of genes highly expressed in sperm cells and 20% of genes highly expressed in pollen vegetative cells caused significant transmission distortion when knocked out. This connection between gene expression and fitness effects provides confirmation that expression levels represent a broadly accurate “search image” for genes with functional effects in pollen. Future work should determine the identity and impact of knockouts that affect transmission in both pollen and ovules, and whether they are biased toward genes that are also expressed in the sporophytes and, thus, may have pleiotropic effects following gametophytic selection.
Standing Variation in Gametophytic Success in Natural Populations
While gene expression and knockout studies highlight the potential for widespread gametophytic selection, they cannot directly address the extent of standing genetic variation for gametophytic fitness in the wild. The amount of heritable variation in gametophyte success in natural populations informs our understanding of the selective forces acting on gametophytic performance and its evolutionary potential (Walsh and Charlesworth, 1992; Mazer et al., 2010). Whether strong selection occurs may also depend on the ecology of pollen deposition on stigmas, the variation in pollen quality, and the intensity of pollen limitation. Variation in ecological context may cause selection on gametophytes to fluctuate and reduce our ability to detect it. Fluctuating selection, in turn, can support the maintenance of genetic variation. Accounting for ecological variation is therefore essential for quantifying and comparing heritable variation in gametophytic performance. Ecological and demographic variation also offers abundant “natural experiments” to compare the effects of pollen competition between populations. To this end, we first review important ecological variables to consider for studies of gametophytes in wild populations, then describe patterns of non-random mating among pollen, among ovules, and between interacting genotypes.
Both the size and the genetic composition of open-pollinated pollen loads on stigmas vary considerably through time and space (Knight et al., 2005; Harder et al., 2016; Ashman et al., 2020). This variation creates heterogeneity in the scope and intensity of selection on gametophyte competition. Genetic heterogeneity in the pollen pool is often highly variable (Dyer and Sork, 2001) and structured across landscapes (Smouse and Robledo-Arnuncio, 2005; Rosas et al., 2011), or even among flowers produced by a single sporophyte (Herrera, 2002, 2004). The abiotic environment also affects reproductive performance in both male and female gametophytes. Poor nutrient conditions (Young and Stanton, 1990; Lau and Stephenson, 1993, 1994) and temperature fluctuations (Hedhly et al., 2004) during sporophyte growth can reduce the competitive ability of pollen, and although ovule number is fixed in some species, in others it can be highly plastic in response to resource availability (Greenway and Harder, 2007). Uniovulate flowers, which are likely to experience the most intense pollen competition, are unevenly distributed across angiosperms and are more common in wind-pollinated taxa (Friedman and Barrett, 2011). Together, the ecological context of mating should significantly influence realized gametophyte competition. Nevertheless, evidence from a wide range of species suggests considerable standing variation in the competitive ability of pollen.
Controlled crosses in diverse angiosperm species have reported variation in siring success in pollinations with mixtures of pollen from different pollen donors. Some of the variation in siring success within populations of angiosperm taxa has been shown to be heritable and at least partially under male gametophytic control (reviewed in Table 1). One important result is that although pollen-tube growth rate often correlates with siring success, this is not a universal finding. For example, Jolivet and Bernasconi (2007) found strong heritability of pollen-tube growth rate based on parent–offspring regressions, but pollen-tube growth rate did not predict fertilization success in a crossing design. Variation in siring success related to pollen-tube growth rate has been functionally linked to specific biological and genetic processes only rarely, and these cases suggest that a complex basis for differential siring success may be common. Further investigation into genetic mechanisms underlying this variation reveals complex patterns (Carlson et al., 2011) and should be pursued in more systems.
Table 1.
Summary of Studies considering the Heritability (h2) of Siring Success (SS) and Pollen-Tube Growth Rate (PTGR), the Effects of PTGR on Traits of the Sporophyte, and whether the Genotype of the Female Parent Interacted with Male Rankings.
| Family | Genus | Species | Method | SS h2 | PTGR h2 | PTGR on SS | PTGR on sporophyte | Interaction with female parent | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Betulaceae | Betula | pendula | vv, vt | Y | Y | Y | – | Mix | Pasonen et al., 1999 |
| Betula | pendula | vv | – | Y | – | Y | Y | Pasonen et al., 2001 | |
| Brassicaceae | Arabidopsis | thaliana | vv | Y | – | – | – | Y | Carlson et al., 2011 |
| Arabidopsis | thaliana | vv | Y | Y | Y | – | – | Swanson et al., 2016 | |
| Raphanus | raphanistrum | vv | N | – | – | – | – | Snow and Mazer, 1988 | |
| Raphanus | sativus | vv | Y | N | N | N | Y | Marshall and Diggle, 2001 | |
| Raphanus | sativus | vv | Y | – | – | – | Y | Marshall and Evans, 2016 | |
| Caryophyllaceae | Silene | latifolia | vt | Y | Y | N | N | Y | Jolivet and Bernasconi, 2007 |
| Silene | latifolia | vv | N | – | – | – | Y | Teixeira et al., 2008 | |
| Convolvulaceae | Ipomoea | purpurea | vv | Y | – | – | – | Y | McCallum and Chang, 2016 |
| Cucurbitaceae | Cucurbita | texana | vt | – | Y | – | Y | – | Jóhannsson and Stephenson, 1997 |
| Cucurbita | pepo | vv | Y | – | – | – | – | Schlichting et al., 1990 | |
| Fabaceae | Chamaecrista | fasciculata | vv | – | – | – | – | Y | Fenster and Sork, 1988 |
| Medicago | sativa | vt | – | Y | – | N | – | Rosellini et al., 1994 | |
| Lobeliaceae | Lobelia | cardinalis | vv | – | – | – | – | Y | Johnston, 1993 |
| Malvaceae | Hibiscus | moscheutos | vv | – | – | Y | N | N | Snow and Spira, 1991a |
| Hibiscus | moscheutos | vv | – | – | Y | N | N | Snow and Spira, 1991b | |
| Hibiscus | moscheutos | vv | – | – | – | – | Y | Snow and Spira, 1993 | |
| Onagraceae | Oenothera | organensis | vt | – | N | – | – | – | Havens, 1994 |
| Pedaliaceae | Sesamum | indicum | vt | – | Y | – | – | – | Pfahler et al., 1997 |
| Pinaceae | Picea | abies | vt, vv | Y | Y | Y | – | Y | Aronen et al., 2002 |
| Plantaginaceae | Collinsia | heterophylla | vt | N | Y | – | N | N | Lankinen et al., 2009 |
| Collinsia | heterophylla | vv | Y | – | – | – | Y | Lankinen et al., 2017 | |
| Poaceae | Zea | mays | vv | Y | – | – | – | – | Sari-Gorla et al., 1992 |
| Zea | mays | vv, vt | Y | Y | Y | – | N | Sari-Gorla et al., 1975 | |
| Zea | mays | vv, vt | Y | Y | Y | – | – | Sari-Gorla et al., 1975 | |
| Zea | mays | vv | – | Y | – | Y | – | Landi et al., 1989 | |
| Zea | mays | vv | Y | – | – | – | – | Ottaviano et al., 1980 | |
| Rosaceae | Prunus | avium | vv, vt | – | Y | – | – | Y | Hedhly et al., 2005 |
| Violaceae | Viola | tricolor | vv, vt | Y | Y | Y | Y | – | Skogsmyr and Lankinen, 2000 |
| Viola | tricolor | vt | – | Y | – | – | – | Lankinen, 2000 |
Heritability or other effects were measured as either in vivo (vv) or in vitro (vt). Y, yes; N, no.
Variation in fertilization success between ovules has been less well documented. Two factors are likely at play in the bias toward studies of pollen: the technical difficulty of accessing developing ovules and the theoretical challenge of differentiating between the activities of ovules, sporophytes, and zygotes. Female mate choice, a core component of sexual selection in animals, may also occur in plants (Willson and Burley, 1983; Skogsmyr and Lankinen 2000, 2002; Gerald et al., 2014; Immler and Otto, 2018) but empirical data remain limited. Several aspects of seed plant reproduction suggest a possible role of ovule genotype in securing cross-fertilization. Sporophytes may selectively abort fertilized ovules in response to pollen limitation (Rosenheim et al., 2016; Huang et al., 2020). Ovules are known to produce signals involved in pollen-tube guidance (Higashiyama and Takeuchi, 2015). Finally, seed size and abortion are often under sporophyte control (Charlesworth, 1988), and it has been hypothesized that early-fertilized ovules are less likely to be aborted (Willson and Burley, 1983). Consistent with this, ovule development success and abortion probability can be affected both by position in a fruit, which affects fertilization timing, and by pollen genotype (Susko and Clubb, 2008). This raises the possibility that ovules compete for early fertilization, and research relating ovule genotype to fertilization and abortion probabilities is likely to be a fertile area of study.
Finally, interactions between male gametophytes and maternal sporophytes may influence reproductive compatibility, leading to variance in mating success. Crossing experiments frequently observe an interaction between maternal and paternal individuals or lines, suggesting that pollen–pistil interactions and style signaling play an important role in siring success (Table 1). Indeed, interactions between pollen and pistil are essential in pollen-tube growth and guidance (Higashiyama and Takeuchi, 2015), and pistils often discriminate among pollen tubes (Swanson et al., 2004). Quantitative trait loci (QTL) analyses of non-random mating between lines in Arabidopsis thaliana (Carlson et al., 2011) and hybrids of Mimulus guttatus and Mimulus nasutus (Fishman et al., 2008) have identified roles for both male gametophytic and female-specific factors and suggest variation in the genetic architecture of compatibility across systems. Interactions between pollen and pistil (whereby different pollen genotypes show differential fertilization success on different pistil genotypes) affect reproductive success in a manner that may not be predictable from pollen genotypes alone, and thus may further alter the selective landscape.
The present state of the field suggests that diverse factors contribute to variation in the fitness of gametophytes. Environmental and ecological context can introduce population structure and reduce heritable variation and signals of selection. Many interactions between gametophytes and sporophytes, including style–pollen mismatches and maternal provisioning, can influence fertilization and subsequent survival of ovule and pollen genotypes. In spite of these complications, evidence has accumulated that the conditions necessary for gametophyte competition are regularly met in natural populations of flowering plants. Moreover, gametophytes express their own genotypes and phenotypes, and, under some circumstances, male gametophytic traits show heritable variation influencing siring success. Female gametophytes often face pollen limitation and signal extensively to growing pollen tubes, and these circumstances suggest competition among ovules for mating opportunities. More thoroughly describing the role of sporophytic and female gametophytic traits should offer further insights into these reproductive processes.
Does Haploid Gametophytic Selection Strengthen the Efficacy of Genome-wide Selection?
If, as it appears, a large fraction of the genome is subject to haploid gametophytic selection and standing variation is present in natural populations, this phase of the life cycle may have important effects on the strength and efficacy of both positive and purifying selection across the genome. This is particularly the case for recessive mutations. Removing dominance effects increases the efficacy of selection because selection depends jointly on selection and dominance (s ・ h). Because there is no dominance coefficient (h) in the effect of selection on haploids, the efficacy of selection on both adaptive and deleterious mutations is increased (Haldane, 1932, Haldane, 1933; Crow and Kimura, 1965). Because recessive mutations are not sheltered in haploids, expression in the haploid phase of the life cycle should lead to more effective purging of recessive deleterious mutations and to a higher probability of fixing recessive beneficial mutations (Charlesworth and Charlesworth, 1992a, 1992b; Immler and Otto, 2018; Immler, 2019).
The direct impacts of gametophytic selection on positive and purifying selection can be inferred experimentally as well as at the population level using patterns of genomic variation and molecular evolution. In this section, we review evidence for the predicted efficacy of selection on haploids, using both experimental and population genomic evidence. Furthermore, as the strength and direction of selection may differ between plant mating systems, we also review the power of mating-system contrasts to further disentangle the role of gametophytic selection and its differential effects across species. Along with the power of directly associating genetic variation with phenotype, population-scale whole-genome sequencing is one of the most promising avenues of the genomics approach to understanding gametophytic selection.
Experimental Tests of the “Unmasking Effect”
Tests of changes in fitness effects caused by decreasing ploidy, called “unmasking,” have been performed in some systems by synthetically increasing ploidy in predominantly haploid species. In synthetic diploids of the predominantly haploid alga Chlamydomonas reinhardtii, most rare mutations with important fitness effects in the haploid phase are recessive in diploids (Orr, 1991). However, in yeast new mutations are more detrimental to rates of cell division in diploids than in haploids, which suggests that the difference between haploids and diploids is more complex than differences in dominance alone, and that differences in the strength of selection may confound simple predictions about differences in dominance (Sharp et al., 2018). Although these results may be specific to systems with predominantly haploid life cycles, they highlight that the role for ploidy differences in selection efficacy varies and should be subject to empirical tests. Additionally, these effects may be taxon specific, and similar analyses in flowering plants may shed light on the relations between ploidy and fitness.
Experimental comparisons of diploid and haploid pollen performance provide one means of testing for a role of haploid selection in flowering plants. Synthetic diploid pollen grows faster than haploid pollen of the same or similar strains in Solanum (Van Breukelen, 1982) and in natural rose hybrids (Gao et al., 2019). Higher competitive ability of diploid pollen is consistent with masking of deleterious recessive mutations, but other mechanisms could be responsible. For example, in Ipomoea diploid pollen is larger than haploid pollen, and larger pollen generally grows faster (McCallum and Chang, 2016). Recent experimental work in Chamerion comparing inbreeding effects on pollen-tube growth rates of diploid and haploid pollen was able to distinguish the effects of ploidy alone on pollen-tube growth rates (Figure 3A), with diploids more able to purge deleterious recessive mutations than polyploids (Husband, 2016). Doubled haploids have been used in breeding for some time and are increasingly important in evolutionary genetics (Ferrie and Möllers 2011; Harkess et al., 2017), and, as they are diploid but entirely homozygous, may offer another valuable tool for distinguishing between effects of ploidy and dominance. These developments highlight the selective importance of exposure of recessive deleterious mutations in the haploid gametophyte, and the power that ploidy variation offers to identify the action of different forms of selection.
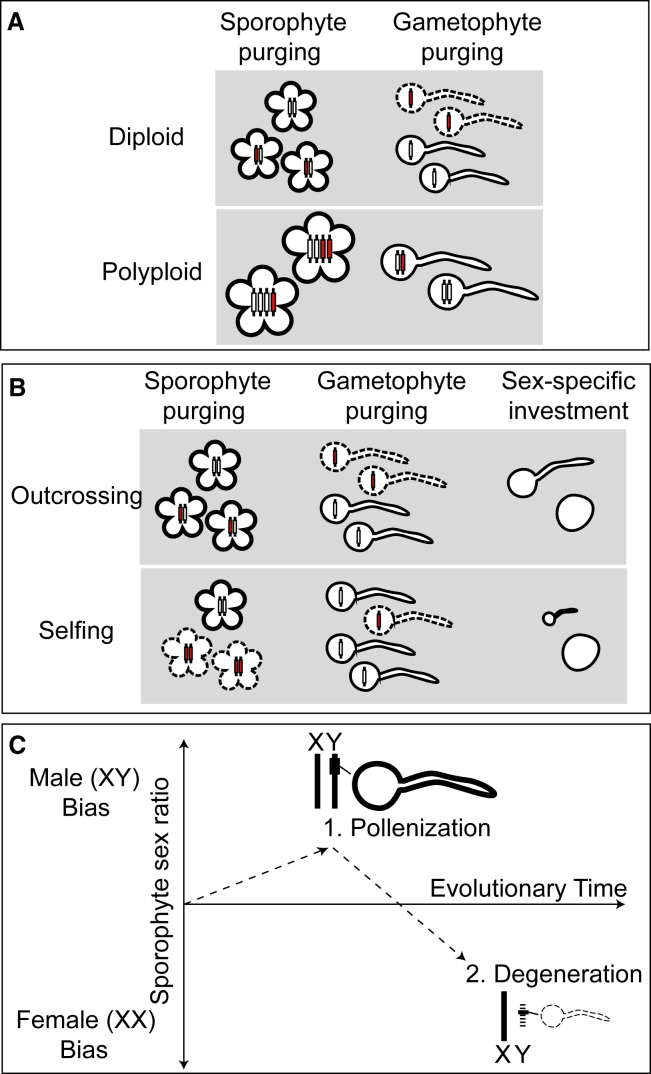
Figure 3.
Ploidy and Mating-System Comparisons Offer Insight into the Influence of Selection on Pollen Competition.
(A) Increased ploidy affects dominance expectations in both the sporophyte and the gametophyte because ploidies greater than 2N can mask deleterious alleles even in the gametophyte stage.
(B) The comparison between outcrossing and selfing mating systems highlights that selfers should experience increased purging in the sporophyte, resulting in more similar selection on recessive alleles between gametophytes and sporophytes. Reduced competition may also decrease investment in pollen compared with ovules.
(C) In dioecious plants, the linkage of pollen-beneficial alleles to the Y chromosome may increase Y-bearing pollen competitive ability and skew sex ratios to male (XY) bias. However, the long-term impact of large linked regions of suppressed recombination can lead to degeneration of Y-linked pollen genes and to an increased competitive ability of X-bearing pollen, resulting in a skew to female-biased (XX) sex ratios.
Genome Evolution and Population Genomic Comparisons
If there is indeed considerable opportunity for selection to act on a large proportion of the genome in the gametophyte phase, we would predict that genes and species more likely to experience gametophytic selection should exhibit signals of stronger purifying selection and faster rates of adaptive evolution.
Recent studies on gene duplicates and on sex chromosomes have highlighted the potential importance of efficient haploid purifying selection in the pollen phase on genome evolution. In the maize genome, pollen-enriched genes throughout the genome are more likely to have been retained as duplicates following the last whole-genome duplication event than other gene-expression categories (Chettoor et al., 2014). Because the retention of gene duplicates is thought to reflect, in part, selection for balanced gene dosage, this is consistent with more efficient purifying selection in the haploid phase, when the effect of a loss of one copy will have a disproportionately greater effect on expression than in the diploid phase. This study did not find similar evidence of differential retention of duplicate genes expressed in the female gametophyte. This may be due to the importance of strong pollen competition in further increasing the efficacy of selection in addition to the effects of haploidy, but could also be explained by ovule function being more supplemented by surrounding sporophyte tissue than pollen. However, research from other plant groups highlights the importance of male-specific evolution and competition. For example, in the predominantly haploid liverworts Marchantia polymorpha and Marchantia inflexa, more rapid divergence on sex chromosomes appears to be driven by male-expressed genes on the sex chromosomes despite the lack of an unmasking effect (Marks et al., 2019).
We expect similar patterns for gene degeneration on Y chromosomes (Haldane, 1932). Genes with pollen-biased expression are less likely to be silenced or deleted from the Y chromosome in Silene latifolia (Chibalina and Filatov, 2011; Krasovec et al., 2018) and in two Rumex species (Crowson et al., 2017; Sandler et al., 2018), presumably because pollen expression unmasks deleterious recessive alleles on the Y chromosome. These lines of evidence suggest that pollen expression influences selection through dosage effects.
Population genomic approaches can allow for model-based comparisons of the efficacy of both positive and purifying selection of genes that function in the gametophyte versus the sporophyte. Several studies in the Brassicaceae involve such comparisons, using gene-expression profiles and the properties of plant systems to predict the scope for gametophytic selection (Arunkumar et al., 2013; Szövényi et al., 2013; Gossmann et al., 2014, 2016; Harrison et al., 2019). Interpreting such comparisons can be challenging because a number of confounding factors also influence evolutionary rates, including the levels and breadth of gene expression across tissues. Thus, controlling for covariates of the distribution of fitness effects can be important when interpreting differences in the strength and direction of selection.
In self-incompatible, outcrossing Capsella grandiflora, population genomic comparisons of genes expressed specifically in the pollen vegetative cell with those expressed only in the sperm and seedlings provided evidence for both more effective purifying and positive selection in pollen-specific genes (Arunkumar et al., 2013). These results are consistent with predictions of an important role for haploid selection and/or gametophytic competition. However, a similar study in A. thaliana (Harrison et al., 2019) showed contrasting results; although rates of adaptive evolution were significantly elevated in pollen-specific genes, there was evidence of weaker purifying selection on pollen-specific compared with sporophyte-specific genes. Furthermore, analyses of polymorphism data indicated a higher burden of deleterious mutations in pollen-specific genes, in contrast to predictions from haploid selection. Similarly, a study identifying pollen-expressed genes in the pistil of A. thaliana showed elevated sequence diversity compared with random genes (Leydon et al., 2017). Thus, comparable studies in related species of Brassicaceae have revealed important differences in their conclusions, with no evidence for greater efficacy of purifying selection in A. thaliana. Expanding these approaches beyond the Brassicaceae and to include female gametophytes as well as male will help determine which patterns predominate and what aspects of plant biology are associated with particular patterns of selection.
Mating-System Contrasts
Although the grounds for generalization are currently limited, a notable aspect of the unexpected findings described above in Arabidopsis and Capsella is the difference in mating system. Self-fertilization is common in flowering plants, with over 70% of angiosperms able to self to some extent (Mable, 2004) and 20% predominantly selfing (Barrett, 2002). Increasing selfing reduces heterozygosity and therefore reduces the impact of unmasking mutations in the haploid stage (Figure 3B). Furthermore, increased rates of self-fertilization will reduce the potential for male–male competition (Mazer et al., 2010; Peters and Weis, 2018), as pollen competes with other pollen of increasingly similar genotypes. This decrease in pollen competition should increase relative investment through female fitness components because less is gained by investing in male components of fitness (Charlesworth and Charlesworth, 1978a, 1978b). Indeed, theoretical studies suggest that the increasing asymmetry between sex-specific fitness gains involving a shift toward female investment should result in decreased maintenance of sexually antagonistic alleles in selfing populations (Jordan and Connallon, 2014; Tazzyman and Abbott, 2015), and this has been explicitly modeled for ploidally antagonistic alleles (Peters and Weis, 2018). Thus, shifts toward high rates of self-fertilization predict relaxed purifying selection on pollen competitive ability (Mazer et al., 2010) or, if gametophytic selection has antagonistic effects on the sporophytic phase (see the next section), directional selection for reduced pollen competitive ability. Therefore, shifts toward increased selfing should influence pollen competition by (1) reducing the benefits of unmasking, (2) lowering standing genetic variation in competitive ability, and (3) decreasing investment in male-fitness components by the hermaphroditic sporophyte. This is reflected in the pattern of self-pollinating plants generally investing less in pollen than outcrossing relatives (Sicard and Lenhard 2011).
Preliminary results from Arabidopsis are consistent with these possibilities (Harrison et al., 2019). A. thaliana has experienced a recent transition to high rates of self-fertilization, so patterns of within-species polymorphism may reflect a recent relaxation of purifying selection or directional selection for reduced investment in pollen. In contrast, estimates of positive selection from between-species divergence may be due to historically higher outcrossing rates because of a signal of high rates of adaptive evolution over longer time scales. Indeed, a recent study reporting evidence of standing genetic variation and recent positive selection on allelic variants reducing pollen number in this species (Tsuchimatsu et al., 2020) highlights the potential for positive directional selection on reduced pollen competitive ability, which may also affect the gametophytic stage. Thus, patterns of polymorphism and divergence in A. thaliana may reflect an important role for mating-system shifts in driving relaxed selection and/or directional selection on pollen competitive ability. As this is the only selfer–outcrosser comparison integrating results on genetic variation of pollen-expressed genes, extending these findings beyond a single comparison is an exciting future research priority. Some of these effects are predicted to be specific to pollen, so comparisons of the evolutionary rates of ovule-expressed genes between species with contrasting mating systems will likely also reveal interesting contrasts.
Phenotypic comparisons and experimental tests on the role of mating-system differences in pollen competition can also test the prediction that pollen competitive ability should be greater in outcrossing species, but empirical results so far have been mixed. In Clarkia tembloriensis, outcrossing populations showed faster pollen-tube growth rates than selfing populations of the same species (Smith-Huerta, 1996; Mazer et al., 2018), whereas there was no pattern of differential siring success between selfing and outcrossing populations of Lobelia cardinalis (Johnston, 1993). In self-compatible Dalechampia scandens, Armbruster and Rogers (2004) manipulated the intensity of competition without varying parent identity or the total number of pollen grains on stigmas by taking advantage of the elongated stigmas of that species. A fixed amount of pollen was either concentrated or dispersed along the elongated stigmatic surface. With pollen identity and quantity held constant, pollen spatial distribution was shown to reliably increase seed mass and offspring growth rate. These results suggest that pollen competition could play a role in compensating for the cost of the reduced efficacy of selection in self-pollinating species, and demonstrate that gametophyte competition alone may be an important selective force. In Turnera ulmifolia, self-pollen showed higher siring success than outcross pollen from other populations, but there was no difference between self and outcross pollen within populations (Baker and Shore, 1995). Combined with the studies of molecular variation, there is growing evidence from comparisons between selfers and outcrossers of relaxed selection on pollen competitive ability and/or directional selection for reduced competitive ability.
Does Gametophytic Selection Have Positive, Negative, or Neutral Effects on Sporophytic Fitness?
The genomic and experimental studies described above suggest that gametophytic selection may play an important role in the efficacy of purifying and positive selection, with implications for rates of molecular evolution and the evolution of genome structure. A central question then concerns how gametophyte selection affects fitness in the sporophyte stage, since overlap between the sporophyte and gametophyte determines the role of gametophyte selection in evolutionary rates and the maintenance of standing genetic variation (Walsh and Charlesworth, 1992).
Genetic covariances can both facilitate and constrain adaptive evolution (Lande and Arnold, 1983), leading to complex patterns of genetic variation in populations (Figure 4). A positive covariance in fitness would enhance rates of adaptive evolution on both life-cycle phases, potentially driving a rapid response to selection. Given positive covariance, we predict low levels of standing variation in gametophyte performance within natural populations, since its maintenance would mostly arise from a balance between deleterious mutation and selection, and the exposure of these mutations to haploid selection should reduce genetic variation (Walsh and Charlesworth, 1992). In contrast, a negative covariance in fitness, representing conflict between the two phases of the life cycle, could play an important role in maintaining standing genetic variation in fitness in natural outcrossing populations, particularly if the deleterious effects of haploid-beneficial mutations are recessive in the sporophyte (Immler et al., 2012; Peters and Weis, 2018). A recent analytical study modeled the invasion potential of an allele that increased pollen performance at the expense of sporophyte fitness (a negative covariance for fitness), and found that the fitness cost, dominance of the allele, and the selfing rate of the population all affected invasion probability (Peters and Weis, 2018). Furthermore, a wide range of conditions allows the alleles to coexist in a stable polymorphism, particularly when outcrossing rates are high (Peters and Weis, 2018). These results suggest that standing variation in populations is possible due to negative fitness covariances between gametophyte and sporophyte even without the fluctuating environmental conditions that plant gametophytes likely experience (see Standing Variation in Gametophytic Success in Natural Populations). Thus, investigations into the pleiotropic effects of genetic variation for gametophytic fitness on the sporophyte are critical for inferring the effect of the gametophytic phase on genetic variation and rates of evolution.
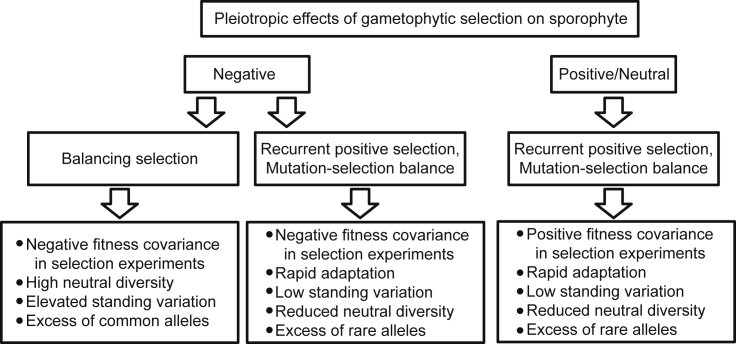
Figure 4.
Pleiotropic Effects of Gametophytic Selection on Sporophyte.
Summary of the population genetic effects of negative/antagonistic and positive or neutral effects of gametophytic selection on sporophyte fitness. Positive covariances can accelerate evolution and give signals of elevated rates of adaptive evolution, and are expected to reduce the amount of standing variation in natural populations. In contrast, negative covariances can have a variety of effects, and may lead to balancing selection, showing population genetic signals of high diversity and an excess of common alleles, or positive and purifying selection, if the conditions for the maintenance of variation are not met.
Shared Genetic Architecture
The possibility of strong covariances in fitness between gametophytes and sporophytes has led to interest in whether selection on pollen competitive ability has effects on sporophytic traits influencing fitness (Mulcahy, 1979). Whereas some aspects of gametophyte performance are unique to the gametophytic life stage, others depend strongly on interaction with the sporophyte, and some depend on traits that covary between gametophytes and sporophytes. Because of complex interactions between sporophytes and gametophytes, neither pollen-tube growth nor the extent of pollen competition is consistently associated with seed quality (reviewed in Baskin and Baskin, 2015). This finding argues against a universally positive covariance in fitness between the life stages. However, some traits shared between sporophyte and gametophyte have a common genetic basis and support the idea that selection on pollen can influence sporophytes.
Artificial selection on specific traits during pollen-tube growth has successfully changed sporophyte trait values under different environmental conditions, including salinity tolerance, toxin tolerance, and metal tolerance (Hormaza and Herrero, 1992). These positive responses suggest a joint genetic basis. More recent investigations involving gametophytic selection have produced novel varieties tolerant to two stressors across three domesticated species: cold tolerance in tomato and chickpea (Clarke et al., 2004; Domínguez et al., 2005), and Alternaria leaf blight resistance in cultivated sunflower (Shobha and Ravikumar, 2006). In cultivated varieties of chickpeas, a combination of genome-wide association study (GWAS) and QTL identified variant haplotypes of a cellulose synthase gene that were strongly associated with increases in pod and seed number and faster pollen-tube growth (Kujur et al., 2015). These chickpea gametophyte QTL had significant effects on sporophyte female fitness, not because the faster gametophytes are generally superior but because the gene is expressed in both pollen and seed pods. Similarly, a major regulator of ovule number, a sporophyte trait identified in a GWAS among A. thaliana accessions, exhibited significant overlap with male gametophyte development loci (Yuan and Kessler, 2019). Evidence for pleiotropy between vegetative and reproductive tissue also appears outside of the seed plants. In the moss Ceratodon purpureus, both negative correlations between vegetative and reproductive traits and weak between-sex trait correlations are consistent with specialized sexual function constrained by antagonistic pleiotropy (McDaniel, 2005). Thus, there is growing evidence for the existence of standing genetic variation with overlapping gametophytic and sporophytic effects.
Combined with the evidence for an intersection in gene expression (see Introduction and Figure 2), pleiotropic effects of alleles in the two phases of the life cycle and between male and female gametophytes seem highly likely. However, the strength and direction of this pleiotropy remains poorly understood, and in Evolutionary Genomics of Gametophytic Selection: The Next Steps we highlight important future directions integrating genomics and experimental approaches to better resolve this question.
Antagonistic Pleiotropy: Evidence from Sex Chromosomes and Sex-Ratio Bias
One of the most well-documented theoretical predictions for negative pleiotropic fitness effects of haploid selection involves plant sex chromosomes (Figure 3C). Recent theory indicates that haploid-specific selection should favor the linkage of pollen-beneficial alleles with the Y chromosome, leading to a male-biased sex ratio in offspring (Scott and Otto, 2017). However, theory also indicates that the accumulation of deleterious mutations on the Y chromosome due to restricted recombination could reduce the haploid competitive ability of the Y chromosome, leading to female-biased sex ratios (Hough et al., 2013; Scott and Otto, 2017).
Recent research in dioecious Rumex species is consistent with these contradictory expectations. Genes with pollen-biased expression are more likely to occur on the Y chromosome, and allele-specific expression of Y alleles is prevalent in pollen-expressed genes (Sandler et al., 2018). The evidence for increased pollen expression following Y linkage is analogous to the “masculinization” effect reported in animal Y chromosomes (e.g., Zhou and Bachtrog, 2012), and accords with the prediction that antagonistic pleiotropy between the gametophytic and sporophytic phases is relaxed following Y linkage, enabling specialization in pollen function.
The combination of selection favoring pollen-beneficial alleles on the Y chromosome and the longer-term degeneration of this chromosome due to inefficient selection may lead to a time-dependent shift in the outcomes of haploid selection between the sex chromosomes, with consequences for the sex ratio of populations (Figure 3C). Initially, the accumulation of pollen-beneficial alleles on the Y may lead to male-biased sex ratios because Y-bearing pollen has enhanced function and can outcompete X-bearing pollen (Scott and Otto, 2017). Indeed, a comparative study of dioecious populations indicated that angiosperm species with less degenerated (homomorphic) sex chromosomes had slightly male-biased sex ratios whereas species with more degenerated (heteromorphic) sex chromosomes exhibited female-biased sex ratios (Stehlik and Barrett, 2005; Field et al., 2013). This apparent incongruity is resolved by the observation that female-biased sex ratios appear to be the result of the inferior competitive ability of Y-bearing pollen (Smith, 1963; Lloyd, 1974; Stehlik and Barrett, 2005; Lenormand et al., 2020), potentially caused by the degeneration of large non-recombining Y chromosomes (Hough et al., 2014; Beaudry et al., 2017). New deleterious mutations can drive evolutionary change to a new stable sex ratio that departs from the expectation of a 1:1 sex ratio (Hough et al., 2013).
Studies in which pollen loads on stigmas of Rumex were experimentally manipulated provided evidence supporting the early proposal by Correns (1907, 1922) that certation favoring X-bearing pollen over Y-bearing pollen through gametophytic competition was the basis of female-biased sex ratios (Field et al., 2012). This pattern of sex-chromosome degeneration causing sex-ratio bias may be unique to angiosperms, because of the combination of a transcriptionally active male gametophyte and a predominantly diploid life cycle. A comparative study of sex ratios in bryophytes and angiosperms found that while population sex ratios vary in both taxa, angiosperms were more likely to exhibit primary sex ratios that deviated significantly from 1:1 (De Jong et al., 2018). As the bryophytes studied also had degenerated sex chromosomes, this suggests that the evolutionary trajectory of sex chromosomes causing first an increase and then a decrease in the proportion of males (Figure 3C) may be specific to angiosperms.
Overall, the results from sex-chromosome evolution and comparative patterns of sex-ratio bias provide evidence of antagonistic pleiotropy between the gametophytic and sporophytic phases. The relative contributions to biased sex ratio of an increased efficacy of selection compared with a high variance in fertilization success of male gametophytes could be further addressed by comparative studies of the effects of pollen expression on W chromosome degeneration in female-heterogametic species such as strawberry (Tennessen et al., 2018) or Datisca (Liston et al., 1990; Wolf et al., 1997, 2001).
Evolutionary Genomics of Gametophytic Selection: The Next Steps
There is clearly considerable scope for future research into the evolutionary genomics of gametophytic selection. First, the mutational variation of the gametophyte stage has yet to be characterized directly. With the growing availability of mutation accumulation lines (e.g., Ossowski et al., 2010), experimental crosses and the tracking of paternal and maternal transmission could be used to infer the distribution of fitness effects of new mutations during the gametophyte stage. Similar experimental approaches were employed to infer the distribution of fitness effects in the primarily haploid C. reinhardtii (Böndel et al., 2019). Such analyses would allow for further understanding of the effects of mutations on gametophytic fitness, especially the proportion and effect sizes of beneficial and deleterious mutations on both phases of the life cycle.
Although studies to date highlight the widespread potential for gametophytic selection, there are a number of specific directions that require further resolution. First, it would be useful to conduct additional QTL mapping and fine-scale characterization of loci influencing gametophytic performance, as conducted by Carlson et al. (2009, 2011), Gerald et al. (2014), and Swanson et al. (2016). These should be conducted on larger population samples and in more species to help clarify the number of loci, their effect sizes, and their population frequencies. By crossing multiple independent accessions, it should be possible to estimate how many genes influence segregating variation in gametophytic competition and the selective forces acting on this variation. With sufficient between-population crosses, this could be scaled up to enable a GWAS of both pollen and ovule fitness. Similarly, GWAS for traits associated with pollen competition (e.g., pollen-tube growth rates, timing of germination) and further exploration of the extent and genetic basis of ovule competition should enable indirect insights into the genetic architecture of standing variation in gametophyte competition. An examination of the overlap between the two sets of GWAS analyses may provide insights into the extent to which gametophyte fitness is determined by known phenotypic effects.
Further population genomic analyses will also be important for providing indirect tests of the relative importance of antagonistic pleiotropy and positive fitness covariance between the gametophytic and sporophytic phases (Figure 4). In particular, ploidally antagonistic selection can more often lead to the maintenance of genetic variation, particularly in outcrossing species (Peters and Weis, 2018). If such alleles are maintained in populations over the long term, they should exhibit signatures of balancing selection on patterns of neutral diversity (Charlesworth, 2006). In contrast, positive fitness covariance is more likely to show molecular signals of high rates of adaptive evolution and recurrent selective sweeps. Although population genomic patterns in pollen-specific genes have been examined in the highly outcrossing C. grandiflora and indicate high rates of positive selection (Arunkumar et al., 2013), no explicit examination of neutral diversity was conducted to test for the action of balancing selection. Furthermore, genes showing pollen-specific expression are unlikely to experience antagonistic pleiotropy (Peters and Weis, 2018); we expect such signals to be more prominent in genes with overlapping expression in the two phases of the life cycle. Indeed, pollen-biased genes with broader expression appear to show different patterns of diversity and molecular evolution compared with pollen-specific genes, suggesting that selective pressures may differ in important ways between these gene sets (Szövényi et al., 2013; Gossmann et al., 2014; Harrison et al., 2019).
Future work examining patterns of neutral diversity in genes with varying levels of overlap in expression between the two phases should provide an important test of the possibility of widespread ploidally antagonistic selection maintaining genetic variation. Similarly, analysis of the population genetic signals of selection surrounding QTL affecting gametophyte expression and performance traits would help distinguish the importance of mutation-selection balance and balancing selection in maintaining genetic variation for these traits (Josephs et al., 2017). Studies that focus on standing variation within single outcrossing populations would be important for disentangling the effects of local directional selection from standing variation within populations. The integration of GWAS and population genomics can provide important connections between gametophytic competition, sporophyte fitness, and the evolutionary forces acting on standing variation.
The genomic location of alleles conferring gametophytic advantage can have a major influence on the chance that they will spread, particularly in the face of deleterious pleiotropic effects on the sporophyte. Regions of recombination suppression can be important in both local adaptation and the development of reproductive barriers between species (Fishman et al., 2013), and the concentration of genes with gametophyte-specific functions on sex chromosomes is both empirically and theoretically supported (Scott and Otto 2017; Sandler et al., 2018). As such, genome assemblies will be useful in further understanding the genomic distribution of gametophyte-expressed genes, and in providing additional tests of the importance of antagonistic pleiotropy between gametophyte and sporophyte and/or between male and female gametophytes.
More extensive research is also necessary to characterize the role of mating-system shifts in changing the selective dynamics of genes expressed during the gametophytic and sporophytic stages of the plant life history. Although the contrast between C. grandiflora and A. thaliana is encouraging and suggests an important role for mating-system differences in the efficacy of selection on gametophyte-expressed genes, large-scale replicate population genomic comparisons across repeated transitions between selfing and outcrossing species will be essential to test whether multiple evolutionary transitions to high selfing rates drive repeated shifts in the efficacy of gametophytic selection on pollen-specific genes. Further analysis of nucleotide diversity of pollen-specialized genes in A. thaliana coupled with GWAS of loci influencing pollen competitive ability would help clarify the potential role of relaxed purifying and positive selection on gametophytic competition in this highly selfing species. In particular, tests examining neutral diversity patterns can help identify signals of recent selective sweeps (Tsuchimatsu et al., 2020).
Another avenue for further exploration is the evolution of female gametophytes. While pollen-tube growth rate has been shown to be a heritable trait in some systems (Table 1), heritable variance in fertilization success for female gametophytes remains underexplored, although mutant screens and between-accession crosses in A. thaliana suggest it is likely to be widespread (Pagnussat et al., 2005; Carlson et al., 2011). One option may be targeted studies of specific genes or gene families, aided by increased understanding of the genes and molecules involved in signaling during fertilization (Higashiyama and Takeuchi, 2015). For example, the reduced competition among gametophytes expected in highly selfing species may also reduce the strength of selection in the signaling peptides used by ovules to communicate with pollen. Similarly, the effects of responses to selection on the female gametophyte on the male gametophyte or the sporophyte are also mostly unknown. Our understanding of pleiotropy can be expanded with more expression (Wuest et al., 2010; Chettoor et al., 2014) and mutation accumulation (Pagnussat et al., 2005) studies. Female gametophytes could also be used to investigate the roles of unmasking compared with high variance in reproductive success on male gametophyte selection. Comparisons of the distribution of fitness effects and rates of adaptive evolution should be made between genes expressed in the male and female gametophyte phase (e.g., Gossmann et al., 2014). The limited understanding of the evolutionary genetics of female gametophytes is in large part due to technical challenges and perhaps the biased assumption that their role in plant mating is largely passive. Yet studies in animals have shown that overlooking the female contribution to mating success can be a major oversight (see Firman et al., 2017). Although mating in plants is obviously fundamentally different from most animals, future study of the evolutionary genomics of plant gametophytes is likely to provide deeper insight by integrating information from both partners in the sexual interaction.
Funding
This project was supported by Discovery Grants from the Natural Sciences and Engineering Research Council of Canada to S.I.W. and S.C.H.B., and a University of Toronto Ecology and Evolutionary Biology postdoctoral fellowship to J.L.R.
Author Contributions
Conceptualization, F.E.G.B., J.L.R., S.C.H.B., and S.I.W.; Investigation, F.E.G.B. and J.L.R.; Writing – Original Draft, F.E.G.B., J.L.R., and S.I.W.; Writing – Review & Editing, F.E.G.B., J.L.R., S.C.H.B., and S.I.W.; Funding Acquisition, S.C.H.B. and S.I.W.; Visualization F.E.G.B., Supervision, S.C.H.B. and S.I.W.; Project Administration, F.E.G.B. and J.L.R.
Acknowledgments
We thank four anonymous reviewers for their thoughtful and insightful comments. We thank Loren Rieseberg for the invitation to participate in this special issue of Plant Communications. No conflict of interest declared.
Published: October 24, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Contributor Information
Felix E.G. Beaudry, Email: felix.beaudry@mail.utoronto.ca.
Stephen I. Wright, Email: stephen.wright@utoronto.ca.
Supplemental Information
References
- Anderson S.N., Johnson C.S., Jones D.S., Conrad L.J., Gou X., Russell S.D., Sundaresan V. Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J. 2013;76:729–741. doi: 10.1111/tpj.12336. [DOI] [PubMed] [Google Scholar]
- Armbruster W.S., Rogers D.G. Does pollen competition reduce the cost of inbreeding? Am. J. Bot. 2004;91:1939–1943. doi: 10.3732/ajb.91.11.1939. [DOI] [PubMed] [Google Scholar]
- Aronen T., Nikkanen T., Harju A., Tiimonen H., Häggman H. Pollen competition and seed-siring success in Picea abies. Theor. Appl. Genet. 2002;104:638–642. doi: 10.1007/s00122-001-0789-9. [DOI] [PubMed] [Google Scholar]
- Arunkumar R., Josephs E.B., Williamson R.J., Wright S.I. Pollen-specific, but not sperm-specific, genes show stronger purifying selection and higher rates of positive selection than sporophytic genes in Capsella grandiflora. Mol. Biol. Evol. 2013;30:2475–2486. doi: 10.1093/molbev/mst149. [DOI] [PubMed] [Google Scholar]
- Ashman T.-L., Alonso C., Parra-Tabla V., Arceo-Gómez G. Pollen on stigmas as proxies of pollinator competition and facilitation: complexities, caveats, and future directions. Ann. Bot. 2020;125:1003–1012. doi: 10.1093/aob/mcaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A.M., Shore J.S. Pollen competition in Turnera ulmifolia (Turneraceae) Am. J. Bot. 1995;82:717–725. [Google Scholar]
- Barrett S.C.H. The evolution of plant sexual diversity. Nat. Rev. Genet. 2002;3:274–284. doi: 10.1038/nrg776. [DOI] [PubMed] [Google Scholar]
- Barrett R.D.H., Hoekstra H.E. Molecular spandrels: tests of adaptation at the genetic level. Nat. Rev. Genet. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- Baskin J.M., Baskin C.C. Pollen (microgametophyte) competition: an assessment of its significance in the evolution of flowering plant diversity, with particular reference to seed germination. Seed Sci. Res. 2015;25:1–11. [Google Scholar]
- Beaudry F.E.G., Barrett S.C.H., Wright S.I. Genomic loss and silencing on the Y chromosomes of Rumex. Genome Biol. Evol. 2017;9:3345–3355. doi: 10.1093/gbe/evx254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger P. The remarkable biology of pollen. Plant Cell. 1992;4:879–887. doi: 10.1105/tpc.4.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boavida L.C., Shuai B., Yu H.-J., Pagnussat G.C., Sundaresan V., McCormick S. A collection of Ds insertional mutants associated with defects in male gametophyte development and function in Arabidopsis thaliana. Genetics. 2009;181:1369–1385. doi: 10.1534/genetics.108.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boavida L.C., Borges F., Becker J.D., Feijó J.A. Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol. 2011;155:2066–2080. doi: 10.1104/pp.110.169813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böndel K.B., Kraemer S.A., Samuels T., McClean D., Lachapelle J., Ness R.W., Colegrave N., Keightley P.D. Inferring the distribution of fitness effects of spontaneous mutations in Chlamydomonas reinhardtii. Plos Biol. 2019;17:e3000192. doi: 10.1371/journal.pbio.3000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F., Gomes G., Gardner R., Moreno N., McCormick S., Feijó J.A., Becker J.D. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 2008;148:1168–1181. doi: 10.1104/pp.108.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink R.A. The sugary gene in maize as a modifier of the waxy ratio. Genetics. 1927;12:461–491. doi: 10.1093/genetics/12.6.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson A.L., Telligman M., Swanson R.J. Incidence and post-pollination mechanisms of non-random mating in Arabidopsis thaliana. Sex. Plant Reprod. 2009;22:257–262. doi: 10.1007/s00497-009-0108-1. [DOI] [PubMed] [Google Scholar]
- Carlson A.L., Gerald J.N.F., Telligman M., Roshanmanesh J., Swanson R.J. Defining the genetic architecture underlying female- and male-mediated non-random mating and seed yield traits in Arabidopsis. Plant Physiol. 2011;157:1956–1964. doi: 10.1104/pp.111.187542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. Evidence for pollen competition in plants and its relationship to progeny fitness: a comment. Am. Nat. 1988;132:298–302. [Google Scholar]
- Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. Plos Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D. A model for the evolution of dioecy and gynodioecy. Am. Nat. 1978;112:975–997. [Google Scholar]
- Charlesworth D., Charlesworth B. Population genetics of partial male-sterility and the evolution of monoecy and dioecy. Heredity. 1978;41:137–153. [Google Scholar]
- Charlesworth D., Charlesworth B. The effects of genetic load of selection in the gametophyte stage. In: Ottaviano E., GorlaMulcahy M.S.D.L., Mulcahu G.B., editors. Angiosperm pollen and ovules. Springer; New York, NY: 1992. [Google Scholar]
- Charlesworth D., Charlesworth B. The effects of selection in the gametophyte stage on mutational load. Evolution. 1992;46:703–720. doi: 10.1111/j.1558-5646.1992.tb02077.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Schemske D.W., Sork V.L. The evolution of plant reproductive characters; sexual versus natural selection. Experientia Suppl. 1987;55:317–335. doi: 10.1007/978-3-0348-6273-8_14. [DOI] [PubMed] [Google Scholar]
- Chettoor A.M., Givan S.A., Cole R.A., Coker C.T., Unger-Wallace E., Vejlupkova Z., Vollbrecht E., Fowler J.E., Evans M.M. Discovery of novel transcripts and gametophytic functions via RNA-seq analysis of maize gametophytic transcriptomes. Genome Biol. 2014;15:414. doi: 10.1186/s13059-014-0414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibalina M.V., Filatov D.A. Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr. Biol. 2011;21:1475–1479. doi: 10.1016/j.cub.2011.07.045. [DOI] [PubMed] [Google Scholar]
- Christensen C.A., Subramanian S., Drews G.N. Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 1998;202:136–151. doi: 10.1006/dbio.1998.8980. [DOI] [PubMed] [Google Scholar]
- Clarke H.J., Khan T.N., Siddique K.H.M. Pollen selection for chilling tolerance at hybridisation leads to improved chickpea cultivars. Euphytica. 2004;139:65–74. [Google Scholar]
- Correns C. Borntrager; Berlin, Germany: 1907. Die Bestimmung und Vererbung des Geschlechtes nach Neuen Versuchen mit Höheren Pflanzen. [Google Scholar]
- Correns C.E. Geschlechtsbestimmung und zahlenverhältnis der geschlechter beim sauerampfer (Rumex acetosa) Biol. Zent. Bl. 1922;42:465–480. [Google Scholar]
- Crow J.F., Kimura M. Evolution in sexual and asexual populations. Am. Nat. 1965;99:439–450. [Google Scholar]
- Crowson D., Barrett S.C.H., Wright S.I. Purifying and positive selection influence patterns of gene loss and gene expression in the evolution of a plant sex chromosome system. Mol. Biol. Evol. 2017;34:1140–1154. doi: 10.1093/molbev/msx064. [DOI] [PubMed] [Google Scholar]
- Davis L.E., Stephenson A.G., Winsor J.A. Pollen competition improves performance and reproductive output of the common zucchini squash under field conditions. J. Am. Soc. Hortic. Sci. 1987;12:712–716. [Google Scholar]
- De Jong T.J., During H.J., Shmida A. Differences and similarities of sex ratios between dioecious angiosperms and dioicous bryophytes. Evol. Ecol. Res. 2018;19:365–386. [Google Scholar]
- Delph L.F. Pollen competition is the mechanism underlying a variety of evolutionary phenomena in dioecious plants. New Phytol. 2019;224:1075–1079. doi: 10.1111/nph.15868. [DOI] [PubMed] [Google Scholar]
- Domínguez E., Cuartero J., Fernández-Muñoz R. Breeding tomato for pollen tolerance to low temperatures by gametophytic selection. Euphytica. 2005;142:253–263. [Google Scholar]
- Drummond D.A., Bloom J.D., Adami C., Wilke C.O., Arnold F.H. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer R.J., Sork V.L. Pollen pool heterogeneity in shortleaf pine, Pinus echinata. Mill. Mol. Ecol. 2001;10:859–866. doi: 10.1046/j.1365-294x.2001.01251.x. [DOI] [PubMed] [Google Scholar]
- Feldmann K.A., Coury D.A., Christianson M.L. Exceptional segregation of a selectable marker (KanR) in Arabidopsis identifies genes important for gametophytic growth and development. Genetics. 1997;147:1411–1422. doi: 10.1093/genetics/147.3.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster C.B., Sork V.L. Effect of crossing distance and male parent on in vivo pollen tube growth in Chamaecrista dasciculata. Am. J. Bot. 1988;75:1898–1903. [Google Scholar]
- Ferrie A.M.R., Möllers C. Haploids and doubled haploids in Brassica spp. for genetic and genomic research. Plant Cell Tissue Organ Cult. 2011;104:375–386. [Google Scholar]
- Field D.L., Pickup M., Barrett S.C.H. The influence of pollination intensity on fertilization success, progeny sex ratio, and fitness in a wind-pollinated, dioecious plant. Int. J. Plant Sci. 2012;173:184–191. [Google Scholar]
- Field D.L., Pickup M., Barrett S.C.H. Comparative analyses of sex-ratio variation in dioecious flowering plants. Evolution. 2013;67:661–672. doi: 10.1111/evo.12001. [DOI] [PubMed] [Google Scholar]
- Firman R.C., Gasparini C., Manier M.K., Pizzari T. Postmating female control: 20 years of cryptic female choice. Trends Ecol. Evol. 2017;32:368–382. doi: 10.1016/j.tree.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., Aagaard J., Tuthill J.C. Toward the evolutionary genomics of gametophytic divergence: patterns of transmission ratio distortion in monkeyflower (Mimulus) hybrids reveals a complex genetic basis for conspecific pollen precedence. Evolution. 2008;62:2958–2970. doi: 10.1111/j.1558-5646.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- Fishman L., Stathos A., Beardsley P.M., Williams C.F., Hill J.P. Chromosomal rearrangements and the genetics of reproductive barriers in Mimulus (monkey flowers) Evolution. 2013;67:2547–2560. doi: 10.1111/evo.12154. [DOI] [PubMed] [Google Scholar]
- Friedman J., Barrett S.C.H. The evolution of ovule number and flower size in wind-pollinated plants. Am. Nat. 2011;177:246–257. doi: 10.1086/657954. [DOI] [PubMed] [Google Scholar]
- Galla G., Basso A., Grisan S., Bellucci M., Pupilli F., Barcaccia G. Ovule gene expression analysis in sexual and aposporous apomictic Hypericum perforatum L. (Hypericaceae) accessions. Front. Plant Sci. 2019;10:654. doi: 10.3389/fpls.2019.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S.-M., Yang M.-H., Zhang F., Fan L.-J., Zhou Y. The strong competitive role of 2n pollen in several polyploidy hybridizations in Rosa hybrida. BMC Plant Biol. 2019;19:127. doi: 10.1186/s12870-019-1696-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald J.N.F., Carlson A.L., Smith E., Maloof J.N., Weigel D., Chory J., Borevitz J.O., Swanson R.J. New Arabidopsis advanced intercross recombinant inbred lines reveal female control of nonrandom mating. Plant Physiol. 2014;165:175–185. doi: 10.1104/pp.113.233213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossmann T.I., Schmid M.W., Grossniklaus U., Schmid K.J. Selection-driven evolution of sex-biased genes is consistent with sexual selection in Arabidopsis thaliana. Mol. Biol. Evol. 2014;31:574–583. doi: 10.1093/molbev/mst226. [DOI] [PubMed] [Google Scholar]
- Gossmann T.I., Saleh D., Schmid M.W., Spence M.A., Schmid K.J. Transcriptomes of plant gametophytes have a higher proportion of rapidly evolving and young genes than sporophytes. Mol. Biol. Evol. 2016;33:1669–1678. doi: 10.1093/molbev/msw044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway C.A., Harder L.D. Variation in ovule and seed size and associated size-number trade-offs in angiosperms. Am. J. Bot. 2007;94:840–846. doi: 10.3732/ajb.94.5.840. [DOI] [PubMed] [Google Scholar]
- Haldane J.B.S. Princeton University Press; Princeton, NJ: 1932. The Causes of Evolution. [Google Scholar]
- Haldane J.B.S. The part played by recurrent mutation in evolution. Am. Nat. 1933;67:5–19. [Google Scholar]
- Harrison M.C., Mallon E.B., Twell D., Hammond R.L. Deleterious mutation accumulation in Arabidopsis thaliana pollen genes: a role for a recent relaxation of selection. Genome Biol. Evol. 2019;11:1939–1951. doi: 10.1093/gbe/evz127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder L.D., Aizen M.A., Richards S.A. The population ecology of male gametophytes: the link between pollination and seed production. Ecol. Lett. 2016;19:497–509. doi: 10.1111/ele.12596. [DOI] [PubMed] [Google Scholar]
- Harkess A., Zhou J., Xu C., Bowers J.E., Van Der Hulst R., Ayyampalayam S., Mercati F., Riccardi P., McKain M.R., Kakrana A. The asparagus genome sheds light on the origin and evolution of a young y chromosome. Nat. Commun. 2017;8:1279. doi: 10.1038/s41467-017-01064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens K. Clonal repeatability of in vitro pollen tube growth rates in Oenothera organensis (Onagraceae) Am. J. Bot. 1994;81:161–165. [Google Scholar]
- Hedhly A., Hormaza J.I., Herrero M. Effect of temperature on pollen tube kinetics and dynamics in sweet cherry, Prunus avium (Rosaceae) Am. J. Bot. 2004;91:558–564. doi: 10.3732/ajb.91.4.558. [DOI] [PubMed] [Google Scholar]
- Hedhly A., Hormaza J.I., Herrero M. Influence of genotype-temperature interaction on pollen performance. J. Evol. Biol. 2005;18:1494–1502. doi: 10.1111/j.1420-9101.2005.00939.x. [DOI] [PubMed] [Google Scholar]
- Heribert-Nilsson N. Zuwachsgeschwindigkeit der pollen-schlauche und gestorte Mendelzahlen bei Oenothera lamarckiana. Hereditas. 1920;1:41–67. [Google Scholar]
- Herrera C.M. Censusing natural microgametophyte populations: variable spatial mosaics and extreme fine-graininess in winter-flowering Helleborus foetidus (Ranunculaceae) Am. J. Bot. 2002;89:1570–1578. doi: 10.3732/ajb.89.10.1570. [DOI] [PubMed] [Google Scholar]
- Herrera C.M. Distribution ecology of pollen tubes: fine-grained, labile spatial mosaics in southern Spanish Lamiaceae. New Phytol. 2004;161:473–484. doi: 10.1111/j.1469-8137.2004.00978.x. [DOI] [PubMed] [Google Scholar]
- Higashiyama T., Takeuchi H. The mechanism and key molecules involved in pollen tube guidance. Annu. Rev. Plant Biol. 2015;66:393–413. doi: 10.1146/annurev-arplant-043014-115635. [DOI] [PubMed] [Google Scholar]
- Honys D., Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormaza J.I., Herrero M. Pollen selection. Theor. Appl. Genet. 1992;83:663–672. doi: 10.1007/BF00226682. [DOI] [PubMed] [Google Scholar]
- Hough J., Immler S., Barrett S.C.H., Otto S.P. Evolutionarily stable sex ratios and mutation load. Evolution. 2013;67:1915–1925. doi: 10.1111/evo.12066. [DOI] [PubMed] [Google Scholar]
- Hough J., Hollister J.D., Wang W., Barrett S.C.H., Wright S.I. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proc. Natl. Acad. Sci. U S A. 2014;111:7713–7718. doi: 10.1073/pnas.1319227111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R., Park S.K., Moore J.M., Orme J., Grossniklaus U., Twell D. Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics. 1998;149:621–631. doi: 10.1093/genetics/149.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Wang W., Barrett S.C.H., Ren M. Plasticity in selective embryo abortion may limit the mating costs of geitonogamy in self-compatible plants: a hypothesis. Am. J. Bot. 2020;107:390–393. doi: 10.1002/ajb2.1448. [DOI] [PubMed] [Google Scholar]
- Husband B.C. Effect of inbreeding on pollen tube growth in diploid and tetraploid Chamerion angustifolium: do polyploids mask mutational load in pollen? Am. J. Bot. 2016;103:532–540. doi: 10.3732/ajb.1500243. [DOI] [PubMed] [Google Scholar]
- Immler S. Haploid selection in “diploid” organisms. Annu. Rev. Ecol. Evol. Syst. 2019;50:219–236. [Google Scholar]
- Immler S., Arnqvist G., Otto S.P. Ploidally antagonistic selection maintains stable genetic polymorphism. Evolution. 2012;66:55–65. doi: 10.1111/j.1558-5646.2011.01399.x. [DOI] [PubMed] [Google Scholar]
- Immler S., Otto S.P. The evolutionary consequences of selection at the haploid gametic stage. Am. Nat. 2018;192:241–249. doi: 10.1086/698483. [DOI] [PubMed] [Google Scholar]
- Jóhannsson M.H., Stephenson A.G. Effects of pollination intensity on the vigor of the sporophytic and gametophytic generation of Cucurbita texana. Sex. Plant Reprod. 1997;10:236–240. [Google Scholar]
- Johnston M.O. Tests of two hypotheses concerning pollen competition in a self-compatible, long-styled species (Lobelia cardinalis: Lobeliaceae) Am. J. Bot. 1993;80:1400–1406. [Google Scholar]
- Jolivet C., Bernasconi G. Within/between population crosses reveal genetic basis for siring success in Silene latifolia (Caryophyllaceae) J. Evol. Biol. 2007;20:1361–1374. doi: 10.1111/j.1420-9101.2007.01344.x. [DOI] [PubMed] [Google Scholar]
- Jones D.A. University of Chicago Press; Chicago, Illinois USA: 1928. Selective Fertilization. [Google Scholar]
- Jordan C.Y., Connallon T. Sexually antagonistic polymorphism in simultaneous hermaphrodites. Evolution. 2014;68:3555–3569. doi: 10.1111/evo.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs E.B., Stinchcombe J.R., Wright S.I. What can genome-wide association studies tell us about the evolutionary forces maintaining genetic variation for quantitative traits. New Phytol. 2017;214:21–33. doi: 10.1111/nph.14410. [DOI] [PubMed] [Google Scholar]
- Keith R.A., Mitchell-Olds S.T. Antagonistic selection and pleiotropy constrain the evolution of plant chemical defenses. Evolution. 2019;73:947–960. doi: 10.1111/evo.13728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight T.M., Steets J.A., Vamosi J.C., Mazer S.J., Burd M., Campbell D.R., Dudash M.R., Johnston M.O., Mitchell R.J., Ashman T.-L. Pollen limitation of plant reproduction: pattern and process. Annu. Rev. Ecol. Evol. Syst. 2005;36:467–497. [Google Scholar]
- Krasovec M., Chester M., Ridout K., Filatov D.A. The mutation rate and the age of the sex chromosomes in Silene latifolia. Curr. Biol. 2018;28:1832–1838. doi: 10.1016/j.cub.2018.04.069. [DOI] [PubMed] [Google Scholar]
- Kubo T., Fujita M., Takahashi H., Nakazono M., Tsutsumi N., Kurata N. Transcriptome analysis of developing ovules in rice isolated by laser microdissection. Plant Cell Physiol. 2013;54:750–765. doi: 10.1093/pcp/pct029. [DOI] [PubMed] [Google Scholar]
- Kujur A., Bajaj D., Upadhyaya H.D., Das S., Ranjan R., Shree T., Saxena M.S., Badoni S., Kumar V., Tripathi S. A genome-wide SNP scan accelerates trait-regulatory genomic loci identification in chickpea. Sci. Rep. 2015;5:11166. doi: 10.1038/srep11166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., Arnold S.J. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Landi P., Frascaroli E., Tuberosa R., Conti S. Comparison between responses to gametophytic and sporophytic recurrent selection in maize (Zea mays L.) Theor. Appl. Genet. 1989;77:761–767. doi: 10.1007/BF00268324. [DOI] [PubMed] [Google Scholar]
- Lankinen Å. Effects of soil pH and phosphorus on in vitro pollen competitive ability and sporophytic traits in clones of Viola tricolor. Int. J. Plant Sci. 2000;161:885–893. [Google Scholar]
- Lankinen Å., Maad J., Armbruster W.S. Pollen-tube growth rates in Collinsia heterophylla (Plantaginaceae): one-donor crosses reveal heritability but no effect on sporophytic-offspring fitness. Ann. Bot. 2009;103:941–950. doi: 10.1093/aob/mcp014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankinen Å., Green K.K. Using theories of sexual selection and sexual conflict to improve our understanding of plant ecology and evolution. AoB Plants. 2015;7:plv008. doi: 10.1093/aobpla/plv008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankinen Å., Hydbom S., Strandh M. Sexually antagonistic evolution caused by male-male competition in the pistil. Evolution. 2017;71:2359–2369. doi: 10.1111/evo.13329. [DOI] [PubMed] [Google Scholar]
- Larson B.M.H., Barrett S.C.H. A comparative analysis of pollen limitation in flowering plants. Biol. J. Linn. Soc. 2000;69:503–520. [Google Scholar]
- Lau T.C., Stephenson A.G. Effects of soil nitrogen on pollen production, pollen grain size, and pollen performance in Cucurbita pepo (Cucurbitaceae) Am. J. Bot. 1993;80:763–768. [Google Scholar]
- Lau T.C., Stephenson A.G. Effects of soil phosphorus on pollen production, pollen size, pollen phosphorus content, and the ability to sire seeds in Cucurbita pepo (Cucurbitaceae) Sex. Plant Reprod. 1994;7:215–220. [Google Scholar]
- Lenormand T., Fyon F., Sun E., Roze D. Sex chromosome degeneration by regulatory evolution. Curr. Biol. 2020;30:3001–3006.e5. doi: 10.1016/j.cub.2020.05.052. [DOI] [PubMed] [Google Scholar]
- Leydon A.R., Weinreb C., Venable E., Reinders A., Ward J.M., Johnson M.A. The molecular dialog between flowering plant reproductive partners defined by SNP-informed RNA-sequencing. Plant Cell. 2017;29:984–1006. doi: 10.1105/tpc.16.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., Rieseberg L.H., Elias T.S. Functional androdioecy in the flowering plant Datisca glomerata. Nature. 1990;343:641–642. [Google Scholar]
- Lloyd D.G. Female-predominant sex ratios in angiosperms. Heredity. 1974;32:35–44. [Google Scholar]
- Mable B.K. Polyploidy and self-compatibility: is there an association? New Phytol. 2004;162:803–811. doi: 10.1111/j.1469-8137.2004.01055.x. [DOI] [PubMed] [Google Scholar]
- Marks R.A., Smith J.J., Cronk Q., Grassa C.J., McLetchie D.N. Genome of the tropical plant Marchantia inflexa: implications for sex chromosome evolution and dehydration tolerance. Sci. Rep. 2019;9:1–13. doi: 10.1038/s41598-019-45039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall D.L., Diggle P.K. Mechanisms of differential pollen donor performance in wild radish, Raphanus sativus (Brassicaceae) Am. J. Bot. 2001;88:242–257. [PubMed] [Google Scholar]
- Marshall D.L., Evans A.S. Can selection on a male mating character result in evolutionary change? A selection experiment on California wild radish, Raphanus sativus. Am. J. Bot. 2016;103:553–567. doi: 10.3732/ajb.1500171. [DOI] [PubMed] [Google Scholar]
- Mazer S.J., Hove A.A., Miller B.S., Barbet-Massin M. The joint evolution of mating system and pollen performance: predictions regarding male gametophytic evolution in selfers vs. outcrossers. Perspect. Plant Ecol. Evol. Syst. 2010;12:31–41. [Google Scholar]
- Mazer S.J., Hendrickson B.T., Chellew J.P., Kim L.J., Liu J.W., Shu J., Sharma M.V. Divergence in pollen performance between Clarkia sister species with contrasting mating systems supports predictions of sexual selection. Evolution. 2018;72:453–472. doi: 10.1111/evo.13429. [DOI] [PubMed] [Google Scholar]
- McCallum B., Chang S.-M. Pollen competition in style: effects of pollen size on siring success in the hermaphroditic common morning glory, Ipomoea purpurea. Am. J. Bot. 2016;103:460–470. doi: 10.3732/ajb.1500211. [DOI] [PubMed] [Google Scholar]
- McDaniel S.F. Genetic correlations do not constrain the evolution of sexual dimorphism in the moss Ceratodon purpureus. Evolution. 2005;59:2353–2361. [PubMed] [Google Scholar]
- Mulcahy D.L. The rise of the angiosperms: a genecological factor. Science. 1979;206:20–23. doi: 10.1126/science.206.4414.20. [DOI] [PubMed] [Google Scholar]
- Orr H.A. A test of Fisher’s theory of dominance. Proc. Natl. Acad. Sci. U S A. 1991;88:11413–11415. doi: 10.1073/pnas.88.24.11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledó J.I., Warthmann N., Clark R.M., Shaw R.G., Weigel D., Lynch M. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviano E., Sari-Gorla M., Mulcahy D.L. Pollen tube growth rates in Zea mays: implications for genetic improvement of crops. Science. 1980;210:437–438. doi: 10.1126/science.210.4468.437. [DOI] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.-J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S., Capron A., Xie L.-F., Ye D., Sundaresan V. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development. 2005;132:603–614. doi: 10.1242/dev.01595. [DOI] [PubMed] [Google Scholar]
- Pasonen H.L., Pulkkinen P., Käpylä M., Blom A. Pollen-tube growth rate and seed-siring success among Betula pendula clones. New Phytol. 1999;143:243–251. [Google Scholar]
- Pasonen H.-L., Pulkkinen P., Käpylä M. Do pollen donors with fastest-growing pollen tubes sire the best offspring in an anemophilous tree, Betula pendula (Betulaceae)? Am. J. Bot. 2001;88:854–860. [PubMed] [Google Scholar]
- Pedersen S., Simonsen V., Loeschcke V. Overlap of gametophytic and sporophytic gene expression in barley. Theor. Appl. Genet. 1987;75:200–206. [Google Scholar]
- Peters M.A.E., Weis A.E. Selection for pollen competitive ability in mixed-mating systems. Evolution. 2018;72:2513–2536. doi: 10.1111/evo.13597. [DOI] [PubMed] [Google Scholar]
- Pfahler P.L., Pereira M.J., Barnett R.D. Genetic variation for in vitro sesame pollen germination and tube growth. Theor. Appl. Genet. 1997;95:1218–1222. [Google Scholar]
- Qin Y., Leydon A.R., Manziello A., Pandey R., Mount D., Denic S., Vasic B., Johnson M.A., Palanivelu R. Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. Plos Genet. 2009;5:e1000621. doi: 10.1371/journal.pgen.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas F., Quesada M., Lobo J.A., Sork V.L. Effects of habitat fragmentation on pollen flow and genetic diversity of the endangered tropical tree Swietenia humilis (Meliaceae) Biol. Conserv. 2011;144:3082–3088. [Google Scholar]
- Rosellini D., Veronesi F., Falcinelli M. Recurrent selection for microgametophytic vigor in alfalfa and correlated responses at the sporophytic level. Crop Sci. 1994;34:933–936. [Google Scholar]
- Rosenheim J.A., Schreiber S.J., Williams N.M. Does an “oversupply” of ovules cause pollen limitation? New Phytol. 2016;210:324–332. doi: 10.1111/nph.13750. [DOI] [PubMed] [Google Scholar]
- Rutley N., Twell D. A decade of pollen transcriptomics. Plant Reprod. 2015;28:73–89. doi: 10.1007/s00497-015-0261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler G., Beaudry F.E.G., Barrett S.C.H., Wright S.I. The effects of haploid selection on Y chromosome evolution in two closely related dioecious plants. Evol. Lett. 2018;2:368–377. doi: 10.1002/evl3.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sari-Gorla M., Ottaviano E., Faini D. Genetic variability of gametophyte growth rate in maize. Theor. Appl. Genet. 1975;46:289–294. doi: 10.1007/BF00281151. [DOI] [PubMed] [Google Scholar]
- Sari-Gorla M., Frova C., Binelli G., Ottaviano E. The extent of gametophytic-sporophytic gene expression in maize. Theor. Appl. Genet. 1986;72:42–47. doi: 10.1007/BF00261452. [DOI] [PubMed] [Google Scholar]
- Sari-Gorla M., Pé M.E., Mulcahy D.L., Ottaviano E. Genetic dissection of pollen competitive ability in maize. Heredity. 1992;69:423–430. [Google Scholar]
- Schlichting C.D., Stephenson A.G., Small L.E., Winsor J.A. Pollen loads and progeny vigor in Cucurbita pepo: the next generation. Evolution. 1990;44:1358–1372. doi: 10.1111/j.1558-5646.1990.tb05238.x. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Wuest S.E., Vijverberg K., Baroux C., Kleen D., Grossniklaus U. Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. Plos Biol. 2011;9:e1001155. doi: 10.1371/journal.pbio.1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M.F., Otto S.P. Haploid selection favors suppressed recombination between sex chromosomes despite causing biased sex ratios. Genetics. 2017;207:1631–1649. doi: 10.1534/genetics.117.300062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp N.P., Sandell L., James C.G., Otto S.P. The genome-wide rate and spectrum of spontaneous mutations differ between haploid and diploid yeast. Proc. Natl. Acad. Sci. U S A. 2018;115:E5046–E5055. doi: 10.1073/pnas.1801040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A., Lenhard M. The selfing syndrome: a model for studying the genetic and evolutionary basis of morphological adaptation in plants. Ann. Bot. 2011;107:1433–1443. doi: 10.1093/aob/mcr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogsmyr I., Lankinen Å. Potential selection for female choice in Viola tricolor. Evol. Ecol. Res. 2000;2:965–979. [Google Scholar]
- Skogsmyr I., Lankinen Å. Sexual selection: an evolutionary force in plants? Biol. Rev. Camb. Philos. Soc. 2002;77:537–562. doi: 10.1017/s1464793102005973. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B.W. The mechanism of sex determination in Rumex hastatulus. Genetics. 1963;48:1265–1288. doi: 10.1093/genetics/48.10.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Huerta N.L. Pollen germination and tube growth in selfing and outcrossing populations of Clarkia tembloriensis (Onagraceae) Int. J. Plant Sci. 1996;157:228–233. [Google Scholar]
- Smouse P.E., Robledo-Arnuncio J.J. Measuring the genetic structure of the pollen pool as the probability of paternal identity. Heredity. 2005;94:640–649. doi: 10.1038/sj.hdy.6800674. [DOI] [PubMed] [Google Scholar]
- Snow A.A., Mazer S.J. Gametophytic selection in Raphanus raphanistrum: a test for heritable variation in pollen competitive ability. Evolution. 1988;42:1065. doi: 10.1111/j.1558-5646.1988.tb02524.x. [DOI] [PubMed] [Google Scholar]
- Snow A.A., Spira T.P. Differential pollen-tube growth rates and nonrandom fertilization in Hibiscus moscheutos (Malvaceae) Am. J. Bot. 1991;78:1419–1426. [Google Scholar]
- Snow A.A., Spira T.P. Pollen vigour and the potential for sexual selection in plants. Nature. 1991;352:796–797. [Google Scholar]
- Snow A.A., Spira T.P. Individual variation in the vigor of self pollen and selfed progeny in Hibiscus moscheutos (Malvaceae) Am. J. Bot. 1993;80:160–164. [Google Scholar]
- Stehlik I., Barrett S.C.H. Mechanisms governing sex-ratio variation in dioecious Rumex nivalis. Evolution. 2005;59:814–825. [PubMed] [Google Scholar]
- Stephenson A.G., Winsor J.A., Davis L.E. Effects of pollen load size on fruit maturation and sporophyte quality in zucchini. In: Mulcahy D.L., Mulcahy G.B., Ottaviano E., editors. Biotechnology and Ecology of Pollen. Springer; New York, NY: 1986. pp. 429–434. [Google Scholar]
- Susko D.J., Clubb M. Pollination effects on patterns of ovule fate in Hesperis matronalis (Brassicaceae) Botany. 2008;86:466–474. [Google Scholar]
- Swanson R., Edlund A.F., Preuss D. Species specificity in pollen-pistil interactions. Annu. Rev. Genet. 2004;38:793–818. doi: 10.1146/annurev.genet.38.072902.092356. [DOI] [PubMed] [Google Scholar]
- Swanson R.J., Hammond A.T., Carlson A.L., Gong H., Donovan T.K. Pollen performance traits reveal prezygotic non-random mating and interference competition in Arabidopsis thaliana. Am. J. Bot. 2016;103:498–513. doi: 10.3732/ajb.1500172. [DOI] [PubMed] [Google Scholar]
- Szövényi P., Ricca M., Hock Z., Shaw J.A., Shimizu K.K., Wagner A. Selection is no more efficient in haploid than in diploid life stages of an angiosperm and a moss. Mol. Biol. Evol. 2013;30:1929–1939. doi: 10.1093/molbev/mst095. [DOI] [PubMed] [Google Scholar]
- Shobha R.T., Ravikumar R.L. Sporophytic and gametophytic recurrent selection for improvement of partial resistance to Alternaria leaf blight in sunflower (Helianthus annuus L.) Euphytica. 2006;147:421–431. [Google Scholar]
- Tang X., Zhang Z.-Y., Zhang W.-J., Zhao X.-M., Li X., Zhang D., Liu Q.-Q., Tang W.-H. Global gene profiling of laser-captured pollen mother cells indicates molecular pathways and gene subfamilies involved in rice meiosis. Plant Physiol. 2010;154:1855–1870. doi: 10.1104/pp.110.161661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley S.D., Zamir D., Rick C.M. Evidence for extensive overlap of sporophytic and gametophytic gene expression in Lycopersicon esculentum. Science. 1981;213:453–455. doi: 10.1126/science.213.4506.453. [DOI] [PubMed] [Google Scholar]
- Tazzyman S.J., Abbott J.K. Self-fertilization and inbreeding limit the scope for sexually antagonistic polymorphism. J. Evol. Biol. 2015;28:723–729. doi: 10.1111/jeb.12592. [DOI] [PubMed] [Google Scholar]
- Tennessen J.A., Wei N., Straub S.C.K., Govindarajulu R., Liston A., Ashman T.-L. Repeated translocation of a gene cassette drives sex-chromosome turnover in strawberries. Plos Biol. 2018;16:e2006062. doi: 10.1371/journal.pbio.2006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira S., Burkhardt A., Bernasconi G. Genetic variation among females affects paternity in a dioecious plant. Oikos. 2008;117:1594–1600. [Google Scholar]
- Tsuchimatsu T., Kakui H., Yamazaki M., Marona C., Tsutsui H., Hedhly A., Meng D., Sato Y., Städler T., Grossniklaus U. Adaptive reduction of male gamete number in the selfing plant Arabidopsis thaliana. Nat. Commun. 2020;11:2885. doi: 10.1038/s41467-020-16679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breukelen E.W.M. Competition between 2x and x pollen in styles of Solanum tuberosum determined by a quick in vivo method. Euphytica. 1982;31:585–590. [Google Scholar]
- Walsh N.E., Charlesworth D. Evolutionary interpretations of differences in pollen tube growth rates. Q. Rev. Biol. 1992;67:19–37. [Google Scholar]
- Warman C., Panda K., Vejlupkova Z., Hokin S., Unger-Wallace E., Cole R.A., Chettoor A.M., Jiang D., Vollbrecht E., Evans M.M.S. High expression in maize pollen correlates with genetic contributions to pollen fitness as well as with coordinated transcription from neighboring transposable elements. Plos Genet. 2020;16:e1008462. doi: 10.1371/journal.pgen.1008462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L.Q., Xu W.Y., Deng Z.Y., Su Z., Xue Y., Wang T. Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics. 2010;11:338. doi: 10.1186/1471-2164-11-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C.A., Malik M.R., Li R., Krochko J.E. Comparative transcript analyses of the ovule, microspore, and mature pollen in Brassica napus. Plant Mol. Biol. 2010;72:279–299. doi: 10.1007/s11103-009-9567-x. [DOI] [PubMed] [Google Scholar]
- Williams J.H., Mazer S.J. Pollen—tiny and ephemeral but not forgotten: new ideas on their ecology and evolution. Am. J. Bot. 2016;103:365–374. doi: 10.3732/ajb.1600074. [DOI] [PubMed] [Google Scholar]
- Willing R.P., Mascarenhas J.P. Analysis of the complexity and diversity of mRNAs from pollen and shoots of Tradescantia. Plant Physiol. 1984;75:865–868. doi: 10.1104/pp.75.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing R.P., Bashe D., Mascarenhas J.P. An analysis of the quantity and diversity of messenger RNAs from pollen and shoots of Zea mays. Theor. Appl. Genet. 1988;75:751–753. [Google Scholar]
- Willson M.F., Burley N. Princeton University Press; Princeton, NJ: 1983. Mate Choice in Plants, Vol. 19: Tactics, Mechanisms, and Consequences. [Google Scholar]
- Wolf D.E., Rieseberg L.H., Spencer S.C. The genetic mechanism of sex determination in the androdioecious flowering plant, Datisca glomerata (Datiscaceae) Heredity. 1997;78:190–204. [Google Scholar]
- Wolf D.E., Satkoski J.A., White K., Rieseberg L.H. Sex determination in the androdioecious plant Datisca glomerata and its dioecious sister species D. cannabina. Genetics. 2001;159:1243–1257. doi: 10.1093/genetics/159.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest S.E., Vijverberg K., Schmidt A., Weiss M., Gheyselinck J., Lohr M., Wellmer F., Rahnenführer J., von Mering C., Grossniklaus U. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 2010;20:506–512. doi: 10.1016/j.cub.2010.01.051. [DOI] [PubMed] [Google Scholar]
- Yang L., Wu Y., Yu M., Mao B., Zhao B., Wang J. Genome-wide transcriptome analysis of female-sterile rice ovule shed light on its abortive mechanism. Planta. 2016;244:1011–1028. doi: 10.1007/s00425-016-2563-x. [DOI] [PubMed] [Google Scholar]
- Yao Y., Han R., Gong Z., Zheng C., Zhao Y. RNA-seq analysis reveals gene expression profiling of female fertile and sterile ovules of Pinus tabulaeformis Carr. during free nuclear mitosis of the female gametophyte. Int. J. Mol. Sci. 2018;19:2246. doi: 10.3390/ijms19082246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young H.J., Stanton M.L. Influences of floral variation on pollen removal and seed production in wild radish. Ecology. 1990;71:536–547. [Google Scholar]
- Yu H.-J., Hogan P., Sundaresan V. Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol. 2005;139:1853–1869. doi: 10.1104/pp.105.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Kessler S.A. A genome-wide association study reveals a novel regulator of ovule number and fertility in Arabidopsis thaliana. Plos Genet. 2019;15:e1007934. doi: 10.1371/journal.pgen.1007934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Bachtrog D. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science. 2012;337:341–345. doi: 10.1126/science.1225385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.