Abstract
Nuclear pore complexes (NPCs), which comprise multiple copies of nucleoporins (Nups), are large protein assemblies embedded in the nuclear envelope connecting the nucleus and cytoplasm. Although it has been known that Nups affect flowering in Arabidopsis, the underlying mechanisms are poorly understood. Here, we show that loss of function of Nucleoporin 160 (Nup160) leads to increased abundance of CONSTANS (CO) protein and the resulting upregulation of FLOWERING LOCUS T (FT) specifically in the morning. We demonstrate that Nup160 regulates CO protein stability through affecting NPC localization of an E3-ubiquitin ligase, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1), which destabilizes CO protein in the morning period. Taken together, these results provide a mechanistic understanding of Nup function in the transition from vegetative to reproductive growth, suggesting that deposition of HOS1 at NPCs by Nup160 is essential for preventing precocious flowering in response to photoperiod in Arabidopsis.
Key words: nuclear pore complexes, HOS1, CO, flowering time
Nuclear pore complexes (NPCs) are large protein assemblies embedded in the nuclear envelope. This study reveals that Nup160, a scaffold component in NPCs, interacts with an E3-ubiquitin ligase HOS1 and mediates its localization at NPCs. This facilitates HOS1 function in destabilizing a key flowering promoter CO in the photoperiod pathway, thus preventing precocious flowering in Arabidopsis.
Introduction
Nuclear pore complexes (NPCs) are vast protein assemblies embedded in nuclear pores at the nuclear envelope, mediating communications between nucleus and cytoplasm. Each NPC is composed of multiple copies of around 30 different kinds of nucleoporins (Nups) (Rout et al., 2000, Cronshaw et al., 2002, Tamura et al., 2010). NPCs are of fundamental importance in nucleocytoplasmic transport. Meanwhile, there is mounting evidence to suggest that NPCs contribute to genome integrity, chromatin structure, and gene expression regulation, all of which are exquisitely controlled by different Nups, independently of their transport functions (Capelson and Hetzer, 2009, Ibarra and Hetzer, 2015).
The overall structure and composition of plant NPCs are similar to those in vertebrates and yeast (Fiserova et al., 2009, Tamura et al., 2010). One of the conserved scaffold components in eukaryotic NPCs is the Nup107-160 subcomplex (Nup84 subcomplex in yeast), which normally contains nine Nups (Nup160, Nup107, Nup96, Nup133, Nup85, Nup43, Nup37, SEH1, and SEC13). Studies in yeast and vertebrates have revealed an essential role of the Nup107-160 complex in NPC assembly, kinetochore assembly and function, and DNA damage repair (Walther et al., 2003, Nagai et al., 2008, Ryu et al., 2015). Functional studies in plants have shown that individual Nups are differentially involved in multiple biological processes, including pathogen defense, hormone signaling, abiotic stresses, symbiosis, and flowering time regulation (Zhang and Li, 2005, Dong et al., 2006b, Jacob et al., 2007, Groth et al., 2010, Wiermer et al., 2012). The early-flowering phenotype is the most remarkable developmental defects found in several Nup mutants, including Nup160 and Nup96 in the Nup107-160 subcomplex (Dong et al., 2006b, Parry et al., 2006, Jacob et al., 2007, Tamura et al., 2010). Except for the bulk poly(A)-mRNA export defects found in most of these mutants, the molecular mechanisms underlying these pleiotropic phenotypes in plants are largely uncharacterized (Parry et al., 2006, Wiermer et al., 2012).
Flowering under favorable conditions is essential for successful reproduction of plants. The timing of flowering is precisely controlled by environmental and endogenous signals. Day length is one of the most important environmental cues that affect plant flowering in response to seasonal changes at various latitudes. Studies in Arabidopsis thaliana have identified CONSTANS (CO) as the key player in perceiving the photoperiodic information (Suarez-Lopez et al., 2001). The circadian-clock regulation of CO transcription and photoreceptor regulation of its protein stability restrict CO activity to a narrow window in the late afternoon (Andres and Coupland, 2012, Song et al., 2015). This allows CO to promote flowering through directly activating the expression of the mobile florigen gene FLOWERING LOCUS T (FT) specifically in the long-day (LD) afternoon (Suarez-Lopez et al., 2001, An et al., 2004).
Although CO transcripts are expressed at high levels from afternoon to next early morning under both short days (SDs) and LDs, CO protein only accumulates in the LD afternoon. Destabilization of CO protein in the dark period is due to activity of the CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1)-SUPPRESSOR of PHYA-105 (SPA) complex, which prevents flowering under SDs (Jang et al., 2008, Liu et al., 2008). Under LDs, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1) affects degradation of CO protein in the daylight period possibly through its interaction with a photoreceptor phytochrome B (phyB) (Valverde et al., 2004, Lazaro et al., 2012, Lazaro et al., 2015). Apart from its E3-ubiquitin ligase activity, HOS1 has been reported to be associated either with chromatin to influence gene expression through its nonproteolytic functions (Dong et al., 2006a, Lazaro et al., 2012, Jung et al., 2013, Jung et al., 2014), or with NPCs to affect the mRNA export (Tamura et al., 2010, MacGregor et al., 2013). Despite the above progress in understanding HOS1 function, the molecular mechanism underlying its control of CO abundance during the daylight period remains largely unknown.
In this study, we reveal that a scaffold nucleoporin Nup160 prevents precocious flowering through mediating localization of HOS1 at NPCs, which is required for HOS1 function in degrading CO protein in the morning. Loss of function of Nup160 results in mislocalization of HOS1 from NPCs, which abolishes HOS1’s role in destabilizing CO protein in the morning period. The increased abundance of CO protein causes pre-activation of FT in the morning, thus causing an early-flowering phenotype. Our findings demonstrate that scaffold Nup functions as a docking site to provide spatial control specifically over a key regulator during cell signaling and plant development.
Results
Nup160 Affects Flowering Time in Arabidopsis
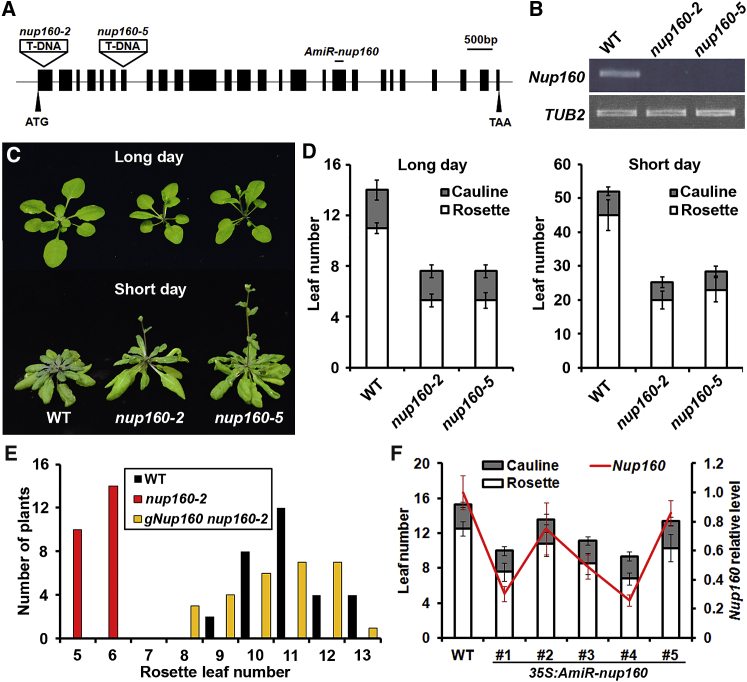
As it has been mentioned that Nup160 affects flowering in Arabidopsis (Cernac et al., 1997, Dong et al., 2006b), we proceeded to understand its underlying mechanism through first examining the flowering phenotypes of two T-DNA insertional mutants, nup160-2 and nup160-5 (also known as sar1-5) (Parry, 2014), in which the N-terminal region of Nup160 was undetectable (Figure 1A and 1B). Both nup160 mutants exhibited much earlier flowering than wild-type plants under LDs and SDs (Figure 1C and 1D), indicating a repressive function of Nup160 in regulating flowering time. To understand how Nup160 influences flowering in response to various flowering signals, we examined nup160-2 phenotype and Nup160 expression in different growth conditions and various flowering mutants. Gibberellic acid, vernalization, and ambient temperature similarly affected flowering of wild-type and nup160-2 (Supplemental Figure 1) and barely changed Nup160 mRNA expression (Supplemental Figure 2B, 2C, and 2E), indicating that Nup160 is not specifically required for flowering responses to these environmental and developmental signals. In addition, Nup160 mRNA expression was only slightly altered in various mutants of the photoperiod and autonomous pathways (Supplemental Figure 2A and 2D), indicating some effects of these two pathways on Nup160 expression.
Figure 1.
Nup160 Regulates Flowering Time in Arabidopsis.
(A) Schematic diagram shows the gene structure of Nup160, the location of T-DNA insertion sites in nup160-2 (SALK_016091) and nup160-5 (SALK_133728), and the target site of the AmiR in 35S:AmiR-nup160. Exons are represented by black boxes, while introns and untranslated regions are indicated by black lines.
(B)Nup160 expression is undetectable in nup160-2 and nup160-5 by semi-quantitative PCR using primers that amplify the 5′ end of the Nup160 transcript. TUB2 was amplified as an internal control.
(C)nup160-2 and nup160-5 exhibit early flowering under both long days and short days.
(D) Flowering time of nup160 mutants under long days and short days (n ≥ 16, ±SD).
(E) Distribution of flowering time in T1 transgenic plants harboring the Nup160 genomic fragment in nup160-2 background grown under long days.
(F) Downregulation of Nup160 in independent 35S:AmiR-nup160 transgenic plants correlates to the degree of early flowering (left axis). Expression levels of Nup160 determined by quantitative real-time PCR in 9-day-old seedlings were normalized against the expression of TUB2 and shown as relative values (red line) to the wild-type level as 1 (right axis). Error bars indicate SD.
To verify that loss of function of Nup160 is responsible for the early-flowering phenotype of nup160-2, an 11.4-kb genomic fragment of Nup160 (gNup160) that includes the 2.1-kb upstream sequence, the 9-kb coding sequence plus introns, and the 0.3-kb downstream sequence (Supplemental Figure 3A), was transformed into nup160-2. Most of the gNup160 nup160-2 T1 transformants displayed similar flowering time to wild-type plants, confirming that Nup160 is required for repressing the floral transition (Figure 1E).
We further created Nup160 knockdown transgenic plants by artificial microRNA (AmiR) interference. Most of the T1 transgenic AmiR-nup160 plants showed an early-flowering phenotype to different extents. The levels of downregulation of Nup160 in five selected 35S:AmiR-nup160 lines were closely related to the degrees of early flowering (Figure 1F), indicating that downregulation of Nup160 has a dosage-dependent effect on the floral transition.
Nup160 Represses FT Expression in the Morning in a CO-Dependent Manner
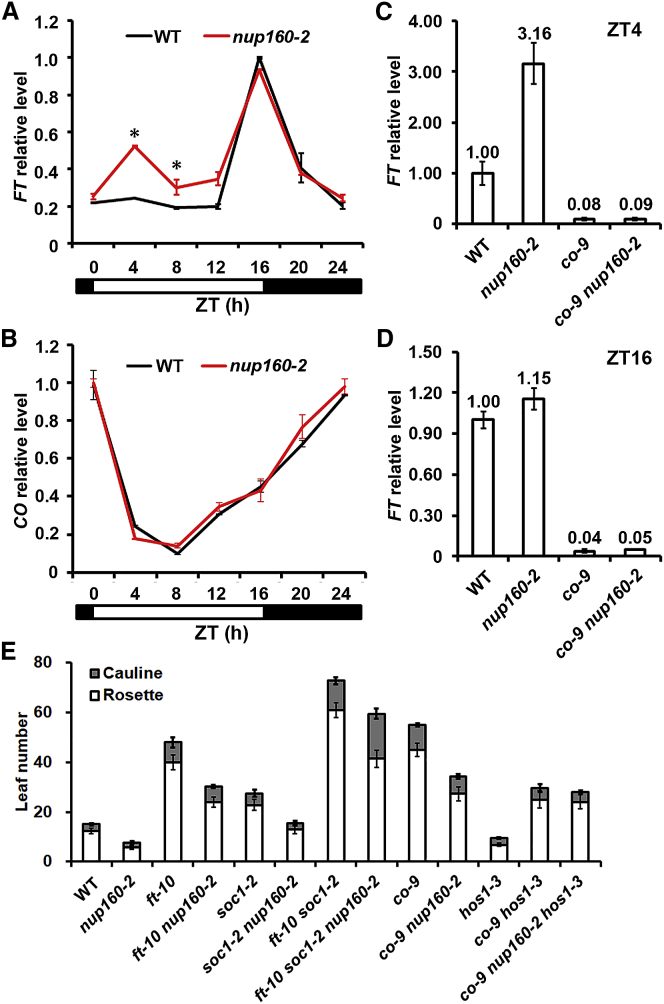
To understand the mechanism through which Nup160 affects flowering, the expression of several key flowering time genes was examined in wild-type and nup160-2 plants. We observed a significant upregulation of FT expression during the morning period (Zeitgeber time [ZT] 4 and 8) in nup160-2 under LDs, while peak expression of FT at ZT16 was not altered (Figure 2A). Because FT activation in response to photoperiod is largely determined by the activity of CO protein (Golembeski and Imaizumi, 2015, Suarez-Lopez et al., 2001), we also examined CO circadian expression pattern and did not observe an obvious change in its expression between wild-type and nup160-2 plants under LDs (Figure 2B). Similarly, there was an obvious increase in FT expression at ZT4 and ZT8 in nup160-2 versus wild-type plants under SDs (Supplemental Figure 4A), while CO circadian expression pattern under SDs was only slightly changed in nup160-2 versus wild-type plants (Supplemental Figure 4B). In addition, the expression of another floral integrator, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), which was activated by FT (Wigge et al., 2005), was consistently upregulated within 24 hours in the nup160-2 under both LDs and SDs (Supplemental Figure 4C and 4D).
Figure 2.
Nup160 Represses FT Expression in the Morning through CO.
(A and B) Diurnal expression of FT(A) and CO(B) determined by quantitative real-time PCR in 9-day-old wild-type (WT) and nup160-2 seedlings grown under long days. Samples were harvested every 4 hours from the onset of illumination, which are shown in hours as Zeitgeber time (ZT). Gene expression levels were normalized against the expression of TUB2 and shown as relative values to the highest level in each panel as 1.0. Asterisks indicate statistically significant differences in FT expression between WT and nup160-2 plants (two-tailed paired Student's t-test, P < 0.005).
(C and D) Expression levels of FT at ZT4 (C) and ZT16 (D) determined by quantitative real-time PCR in 9-day-old WT, nup160-2, co-9, and co-9 nup160-2 seedlings grown under long days. Gene expression levels were normalized against the expression of TUB2 and shown as relative values to the WT level as 1.0. Error bars indicate SD.
(E) Flowering time of various mutants grown under long days (n ≥ 16, ±SD).
To test whether upregulation of FT in nup160-2 depends on CO, we measured FT transcript levels in wild-type, nup160-2, co-9, and co-9 nup160-2 seedlings grown under LDs at ZT4 and ZT16. Loss of function of CO almost completely abolished the activation of FT in both wild-type and nup160-2 backgrounds at ZT4 and ZT16 (Figure 2C and 2D), indicating that upregulation of FT in nup160-2 at ZT4 is still dependent on CO. Further genetic analyses revealed that the crossed progenies between nup160-2 and ft-10, ft-10 soc1-2, or co-9 all displayed much later flowering than wild-type plants (Figure 2E), suggesting that the early-flowering phenotype of nup160-2 is suppressed by these mutants. These observations support that Nup160 suppresses flowering at least partially through the CO-FT regulatory module.
Nup160 Destabilizes CO Protein in the Morning Period
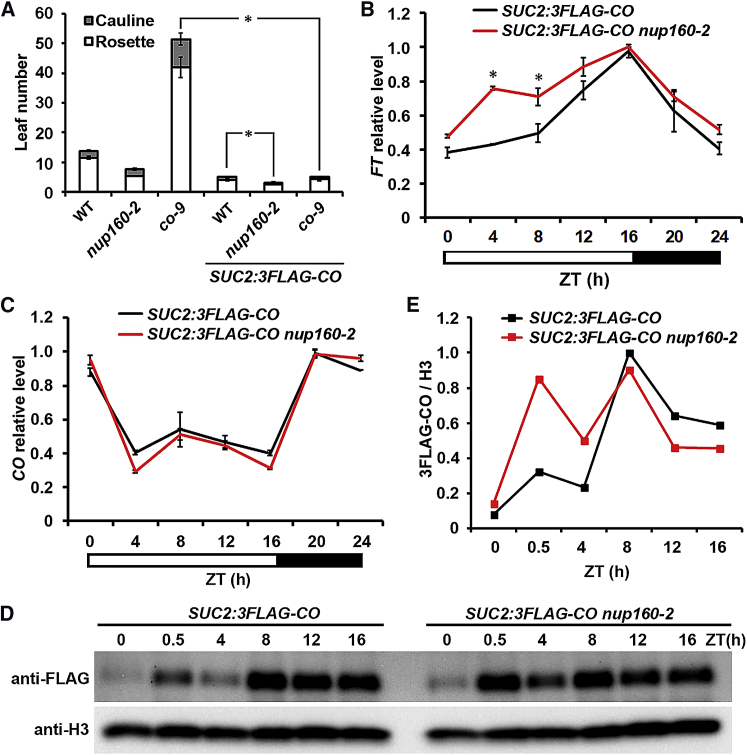
Because CO mRNA expression is not greatly changed in nup160-2 under both LDs and SDs (Figure 2B and Supplemental Figure 4B), we sought to test whether Nup160 affects CO at the protein level. To this end, we generated a SUC2:3FLAG-CO transgenic line, in which FLAG-tagged CO was driven by the promoter of SUCROSE TRANSPORTER 2 (SUC2) that is actively expressed in leaf companion cells where CO promotes FT transcription (Imlau et al., 1999, An et al., 2004). SUC2:3FLAG-CO flowered earlier than wild-type plants and substantially rescued the late-flowering phenotype of co-9 (Figure 3A and Supplemental Figure 5A), indicating that 3FLAG-CO protein retains its biological function in promoting flowering. We further crossed this SUC2:3FLAG-CO allele with nup160-2 (Supplemental Figure 5), and found that nup160-2 further accelerated SUC2:3FLAG-CO flowering as compared with SUC2:3FLAG-CO (Figure 3A). In agreement with this phenotype, FT expression was significantly upregulated at ZT4 and ZT8 in SUC2:3FLAG-CO nup160-2 versus SUC2:3FLAG-CO (Figure 3B). This result is consistent with the observed pattern of upregulation of FT in nup160-2 versus wild-type plants under LDs (Figure 2A). In contrast, circadian expression of CO mRNA, including the endogenous CO and 3FLAG-CO, remained almost at the same levels in SUC2:3FLAG-CO nup160-2 and SUC2:3FLAG-CO (Figure 3C). We then compared the circadian patterns of 3FLAG-CO expression in nuclear extracts of wild-type and nup160-2 plants. CO protein was expressed throughout the daytime with two peaks at ZT0.5 and ZT8 in wild-type plants, while its expression in nup160-2 was much higher at ZT0.5 and ZT4 (Figure 3D and 3E). This increased expression of CO protein in nup160-2 is consistent with an increased FT expression in the morning (Figure 3B), indicating that loss of function of Nup160 results in an increase in CO protein abundance in the morning period, which precociously activates FT expression.
Figure 3.
Nup160 Destabilizes CO Protein during the Morning.
(A) Flowering time of WT, nup160-2, co-9, SUC2:3FLAG-CO, SUC2:3FLAG-CO nup160-2, and SUC2:3FLAG-CO co-9 plants grown under long days (n ≥ 16, ±SD). Asterisks indicate a statistically significant difference between specified genotypes (two-tailed paired Student's t-test, P < 0.005).
(B and C) Diurnal expression of FT(B) and CO(C) mRNA determined by real-time qPCR in 9-day-old SUC2:3FLAG-CO and SUC2:3FLAG-CO nup160-2 seedlings grown under long days. Gene expression levels were normalized against the expression of TUB2 and shown as relative values to the highest level in each panel as 1. Error bars indicate SD. Asterisks indicate statistically significant differences in FT expression between two genotypes (two-tailed paired Student's t-test, P < 0.005).
(D) CO protein expression in 9-day-old SUC2:3FLAG-CO and SUC2:3FLAG-CO nup160-2 seedlings grown under long days. Nuclear protein extracts from seedlings collected at different ZT time points were detected by anti-FLAG antibody. Histone H3 is shown as a loading control.
(E) Quantification of CO protein abundance against the H3 expression levels in (D).
Nup160 Genetically and Physically Interacts with HOS1
Nup160 functions to downregulate CO protein specifically during the morning period. However, there were no obvious changes in circadian expression of Nup160 under LDs (Supplemental Figure 6A), and its protein localization and abundance were also not altered in response to light signals (Supplemental Figure 6B). Thus, it is possible that Nup160 acts through cooperating with other component(s) that confer morning-specific functions. Among the reported CO protein regulators, phyB and HOS1 promote CO degradation during the morning, while COP1 destabilizes CO under dark conditions (Suarez-Lopez et al., 2001, Jang et al., 2008, Lazaro et al., 2015). Although hos1-3, phyB-9, and cop1-4 single mutants all flowered earlier than wild-type plants, only phyB-9 and cop1-4 further enhanced the early-flowering phenotype of nup160-2, while nup160-2 hos1-3 exhibited similar flowering time to nup160-2 (Supplemental Figure 6C). This raises the possibility that Nup160 and HOS1 could function in the same genetic pathway to regulate the floral transition.
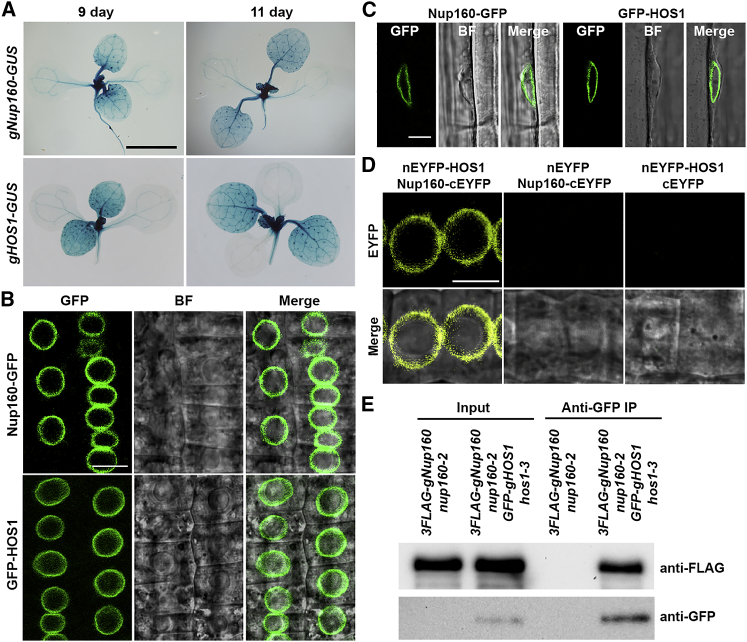
We then proceeded to compare the spatial expression patterns of Nup160 and HOS1 by generating gNup160-GUS and gHOS1-GUS, in which the β-Glucuronidase (GUS) reporter gene was fused in frame at the C terminus of Nup160 and HOS1 in their genomic fragments that were able to rescue their respective mutants (Supplemental Figure 3). Because most of the transgenic lines generated for each construct showed similar expression patterns, we selected one representative line each for gNup160-GUS or gHOS1-GUS for further investigation. GUS staining revealed that both Nup160-GUS and HOS1-GUS were highly expressed in the shoot apex and young rosette leaves, including vasculature tissues in 9- and 11-day-old seedlings (Figure 4A). The similar expression patterns of Nup160 and HOS1 during the floral transition support that they may function in the same genetic pathway.
Figure 4.
Nup160 Interacts with HOS1 at NPCs.
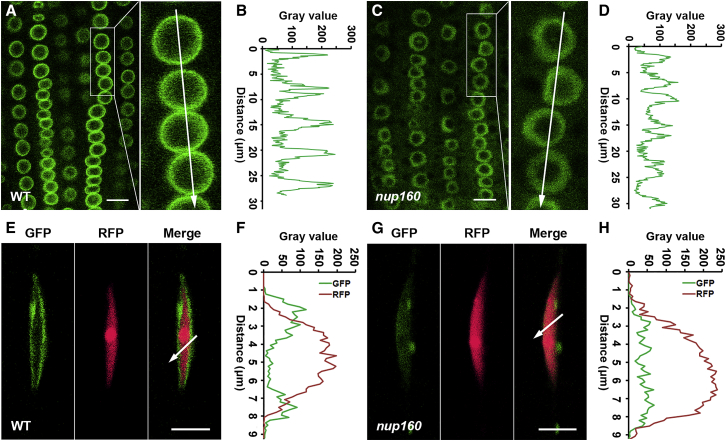
(A) GUS staining of 9- and 11-day-old gHOS1-GUS and gNup160-GUS seedlings grown under long days. Scale bar corresponds to 1 mm.
(B and C) Subcellular localization of Nup160-GFP and GFP-HOS1 in meristematic root cells (B) and mature root cells (C) of 5-day-old gNup160-GFP nup160-2 and GFP-gHOS1 hos1-3 seedlings, respectively. GFP, green fluorescence protein; BF, bright field; Merge, merge of GFP and BF images. Scale bar corresponds to 10 μm.
(D)In vivo BiFC analysis of the interaction between Nup160 and HOS1 in meristematic root cells of 5-day-old nEYFP-gHOS1 gNup160-cEYFP, nEYFP-gHOS1 cEYFP, or nEYFP gNup160-cEYFP seedlings. EYFP, enhanced yellow fluorescence protein; Merge, merge of EYFP and bright-field images. Scale bar corresponds to 10 μm.
(E)In vivo interaction between Nup160 and HOS1 shown by co-immunoprecipitation. Nuclear protein extracts from 9-day-old 3FLAG-gNup160 nup160-2 and 3FLAG-gNup160 nup160-2 GFP-gHOS1 hos1-3 seedlings were incubated with anti-GFP antibody. The coimmunoprecipitated protein was detected by anti-FLAG and anti-GFP antibodies, respectively.
Nup160 is a conserved scaffold Nup found in different eukaryotes (Aitchison et al., 1995, Dong et al., 2006b, Tamura et al., 2010). Although previous studies identified HOS1 as a potential component associated with Arabidopsis NPCs, its localization results obtained through overexpressing GFP-tagged HOS1 were controversial (Lee et al., 2001, Lazaro et al., 2015). To characterize and compare the endogenous subcellular localization of Nup160 and HOS1, we generated gNup160-GFP nup160-2 and GFP-gHOS1 hos1-3 transgenic lines, in which the early-flowering phenotype of nup160-2 and hos1-3 was rescued (Supplemental Figure 3A and 3B). Subcellular localization of Nup160-GFP and GFP-HOS1 was then examined in different tissues of gNup160-GFP nup160-2 and GFP-gHOS1 hos1-3. As expected, Nup160-GFP was localized exclusively at the nuclear periphery of different types of root cells (Figure 4B, upper panel; Figure 4C, left panel). GFP-HOS1 was also enriched at the nuclear rim with detectable signals in the nucleoplasm, but not in the nucleolus of meristematic root cells (Figure 4B, lower panel). Localization of GFP-HOS1 in the nuclear envelope was more prominent in differentiated root cells (Figure 4C, right panel).
The overlapping tissue expression pattern and subcellular localization of Nup160-GFP and GFP-HOS1 prompted us to investigate their protein interaction. To perform in vivo bimolecular fluorescence complementation (BiFC) assay, we created nEYFP-gHOS1 gNup160-cEYFP transgenic plants, in which the coding sequences of the N- and C-terminal halves of the enhanced yellow fluorescence protein (EYFP) were fused in frame with the genomic sequences of HOS1 and Nup160 (Supplemental Figure 3A), respectively. nEYFP-gHOS1 gNup160-cEYFP was further crossed with the transgenic plants bearing cEYFP or nEYFP to obtain the control plants containing either nEYFP-gHOS1 cEYFP or nEYFP gNup160-cEYFP, respectively. We only detected EYFP signals at the nuclear rim of root cells in nEYFP-gHOS1 gNup160-cEYFP, but not in the control plants, indicating an in planta interaction between Nup160 and HOS1 in NPCs (Figure 4D). Furthermore, coimmunoprecipitation analysis confirmed the in vivo interaction of FLAG-Nup160 and GFP-HOS1 in nuclear extracts from 3FLAG-gNup160 GFP-gHOS1 Arabidopsis seedlings, but not in those from 3FLAG-gNup160 seedlings (Figure 4E). Taken together, these results suggest that HOS1 interacts with Nup160 at NPCs to regulate flowering. Notably, localization of GFP-HOS1 was detectable in both nuclear rim and nucleoplasm (Figure 4B, lower panel), implying that HOS1 may not be constitutively associated with NPCs.
Nup160 Is Required for NPC Localization of HOS1
Because Nup160 does not affect HOS1 mRNA expression (Supplemental Figure 6D), we proceeded to investigate whether interaction between Nup160 and HOS1 affects HOS1 protein localization or abundance. We first compared subcellular localization of GFP-HOS1 in GFP-gHOS1 hos1-3 and GFP-gHOS1 nup160-2 hos1-3. In contrast to the prominent NPC localization in the wild-type background (Figure 5A), GFP-HOS1 was less enriched at the nuclear envelope in nup160-2, and diffused to the nucleoplasm and the cytoplasm (Figure 5C). Quantitative analysis of GFP-HOS1 distribution across cells showed that peak signals of GFP-HOS1 at the nuclear envelope was diminished in nup160-2 versus the wild-type background (Figure 5B and 5D). Mislocalization of GFP-HOS1 was consistently observed in root mature cells and leaf petiole cells in nup160-2 (Supplemental Figure 7). These observations suggest that early flowering of nup160-2 is associated with a compromised NPC localization of HOS1.
Figure 5.
Nup160 Is Required for the Nuclear Envelope Localization of HOS1.
(A and C) Subcellular localization of GFP-HOS1 in meristematic root cells of 5-day-old GFP-gHOS1 hos1-3 seedlings in either WT (A) or nup160-2(C) background. The right panels show the magnified views of the boxes indicated in (A) and (C), respectively. Scale bars correspond to 10 μm.
(B and D) Measurement of fluorescence intensity profiles of GFP-HOS1 along the lines indicated in the enlarged figures shown in (A) and (C), respectively. Similar results were observed in meristematic root cells of 20 independent plants for each genotype.
(E and G) Subcellular localization of GFP-HOS1 in leaf phloem companion cells of 9-day-old SUC2:nlsRFP GFP-gHOS1 hos1-3 seedlings in either WT (E) or nup160-2 background (G). GFP, GFP fluorescence; RFP, RFP fluorescence; Merge, merge of GFP and RFP images. Scale bars correspond to 10 μm.
(F and H) Measurement of fluorescence intensity profiles of GFP-HOS1 and nlsRFP along the lines indicated in (E) and (G), respectively. The similar results were observed in leaf phloem companion cells of 10 independent plants for each genotype. Scale bars correspond to 10 μm (A, C, E, and G).
We also generated 35S:GFP-HOS1 hos1-3 transgenic lines, in which the early-flowering phenotype of hos1-3 was partially rescued (Supplemental Figure 8A; Lazaro et al., 2012). This transgenic line was further introduced into the nup160-2 background. Like GFP-HOS1 expressed at the native level (Figure 5A–5D), overexpression of GFP-HOS1 also exhibited relatively prominent localization of GFP-HOS1 in the nuclear envelope of root cells in hos1-3 versus nup160-2 hos1-3 (Supplemental Figure 8B and 8C). This observation again suggests that Nup160 is required for NPC localization of HOS1 even when HOS1 is overexpressed. Notably, 35S:GFP-HOS1 nup160-2 hos1-3 showed a comparable early-flowering phenotype to nup160-2 (Supplemental Figure 8A), indicating that, in the absence of Nup160, compromised NPC localization of overproduced HOS1 is unable to prevent early flowering.
As HOS1 has been shown to function in leaf companion cells to destabilize CO protein (Lazaro et al., 2012), we then investigated whether Nup160 affects NPC localization of HOS1 in these cells. To label the nuclei of leaf companion cells, we generated SUC2:nlsRFP by fusing the red fluorescent protein (RFP) with a nuclear localization signal (nls), which was driven by the SUC2 promoter. The nlsRFP signal was specifically detected in leaf vasculature cells of the SUC2:nlsRFP transgenic lines (Supplemental Figure 9). We further crossed SUC2:nlsRFP with GFP-gHOS1 hos1-3 and GFP-gHOS1 nup160-2 hos1-3, and compared GFP-HOS1 localization in nlsRFP-labeled leaf companion cells between hos1-3 and nup160-2 hos1-3. GFP-HOS1 was clearly detected at nuclear envelope of nlsRFP-labeled companion cells in hos1-3 (with the presence of wild-type Nup160) (Figure 5E and 5F), whereas the NPC localization of GFP-HOS1 largely disappeared in nlsRFP-labeled companion cells in the nup160-2 background (Figure 5G and 5H). These observations substantiate that Nup160 is required for NPC localization of HOS1 in leaf companion cells. In contrast to the effect of Nup160 on subcellular localization of HOS1, Nup160 did not affect GFP-HOS1 mRNA and protein expression levels (Supplemental Figure 10A and 10B) or HOS1-GUS protein expression pattern in seedlings (Supplemental Figure 10C–10F), suggesting that NUP160 affects HOS1 subcellular localization rather than its protein levels.
Nup160-Dependent NPC Localization Is Required for HOS1 to Repress Flowering
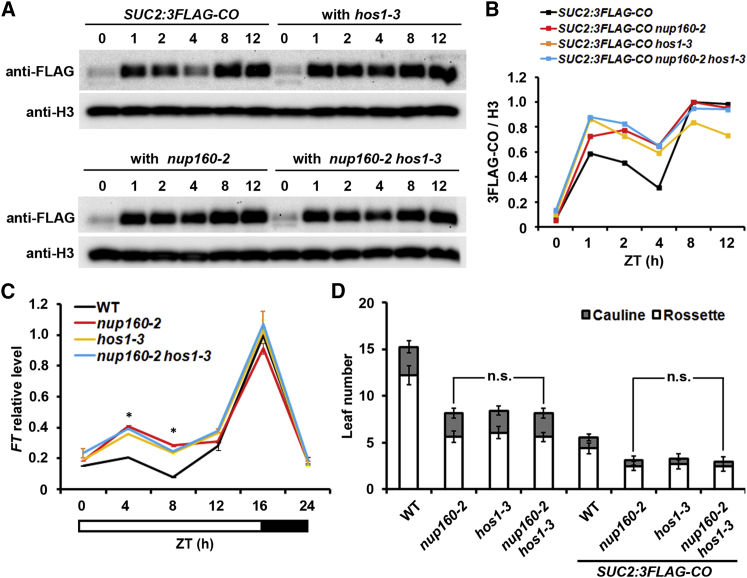
To test whether Nup160-mediated NPC localization of HOS1 affects CO protein degradation, we introduced SUC2:3FLAG-CO into hos1-3 and nup160-2 hos1-3, and compared FLAG-CO protein abundance in different genetic backgrounds. Compared with the expression in the wild-type background, FLAG-CO protein was expressed at comparably higher levels in hos1-3 and nup160-2 hos1-3 only during the morning period, which is similar to the changes observed in nup160-2 (Figure 6A and 6B). Consistent with the higher CO protein abundance, FT mRNA was expressed at higher levels in nup160-2 hos1-3, nup160-2, and hos1-3 versus wild-type plants (Figure 6C). This is in agreement with the earlier flowering time observed in the backgrounds of nup160-2, hos1-3, and nup160-2 hos1-3 compared with wild-type plants regardless of the presence of SUC2:3FLAG-CO (Figure 6D). These results demonstrate that mislocalization of HOS1 caused by loss of Nup160 displays the same flowering defect to loss of HOS1, indicating that Nup160-mediated NPC localization of HOS1 is critical for HOS1 to suppress flowering.
Figure 6.
The Nup160-HOS1 Module Prevents Pre-activation of FT through Destabilizing CO Protein.
(A) Effects of nup160-2 and hos1-3 on CO protein abundance in SUC2:3FLAG-CO. 9-day-old seedlings of SUC2:3FLAG-CO in the WT, nup160-2, hos1-3, and nup160-2 hos1-3 backgrounds were collected at different ZT time points under long days. Nuclear protein was extracted and detected with anti-FLAG antibody. Histone H3 is shown as a loading control.
(B) Quantification of CO protein abundance against the H3 expression levels in (A).
(C) Diurnal expression of FT mRNA determined by real-time qPCR in 9-day-old WT, nup160-2, hos1-3, and nup160-2 hos1-3 seedlings grown under long days. Samples were harvested every 4 hours from ZT0, except ZT20. Gene expression levels were normalized against the expression of TUB2 and shown as relative values to the WT level as 1.0. Error bars indicates SD. Asterisks indicate statistically significant differences in FT expression between WT and nup160-2, hos1-3, or nup160-2 hos1-3 (two-tailed paired Student's t-test, P < 0.005).
(D) Flowering time of nup160-2, hos1-3, nup160-2 hos1-3, and their crossed progenies with SUC2:3FLAG-CO grown under long days (n ≥ 16, ±SD). n.s. indicates no significant difference (two-tailed paired Student's t-test, P > 0.05).
Discussion
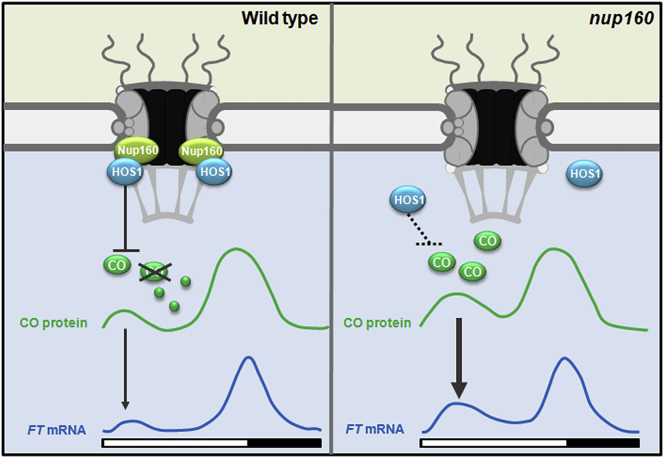
Nups are actively involved in multiple cell signaling processes independently of their trafficking functions in mediating the nucleocytoplasmic transport of macromolecules. Although plant Nups have been found to play important regulatory roles during plant development, the underlying molecular mechanisms are mostly unclear. In this study, we have shown that the Arabidopsis Nup160, a scaffold Nup, is important for regulating the floral transition, a key developmental phase change in flowering plants, through modulating the subcellular localization of HOS1 at NPCs. Knockout of Nup160 causes mislocalization of HOS1 from nuclear pores, which disables HOS1 function in destabilizing CO protein during the morning period, resulting in pre-activation of FT expression and early flowering (Figure 7).
Figure 7.
Nup160 Regulates Flowering through HOS1-Mediated CO Degradation.
Nup160 is required for HOS1 localization at NPCs, which facilitates HOS1 to destabilize CO protein in the morning period. In the absence of Nup160, as HOS1 is no longer associated with NPCs, CO protein accumulates to higher levels in the morning, which activates FT expression, leading to early flowering of nup160.
In this study, we provide several lines of evidence to support an essential role of Nup160 in destabilizing CO protein through anchoring HOS1 at NPCs. First, loss of function of Nup160 results in precocious upregulation of FT during the morning period, which is due to an increased CO protein expression in the same period. Second, Nup160 physically interacts with HOS1 at NPCs, which is necessary for anchoring HOS1 at the nuclear periphery in leaf companion cells. Third, Nup160 affects CO protein levels and flowering time primarily through influencing subcellular localization of HOS1 rather than the abundance of HOS1 mRNA and protein. Last, compromised NPC localization of HOS1 caused by loss of Nup160 results in the same early-flowering defect exhibited by loss of HOS1 function. Taken together, these results suggest that Nup160-mediated NPC localization of HOS1 is critical for HOS1 to destabilize CO protein in the morning to prevent precocious flowering in Arabidopsis.
CO plays a predominant role during the photoperiod-regulated floral transition. Its function and its abundance at the mRNA and protein levels are tightly controlled throughout the day to ensure the induction of FT only in the LD afternoon (Song et al., 2015). It has been found that tight suppression of precocious activation of FT is mediated by several proteins relevant to CO. For example, TOE proteins and BBX19 have been reported to antagonize CO function to suppress FT expression in the morning (Wang et al., 2014, Zhang et al., 2015). Here, we show that the scaffold Nup160 plays a hitherto unknown role in promoting degradation of CO protein through a subtle control of subcellular localization of a CO upstream repressor HOS1 to regulate the expression of FT specifically in the morning in response to photoperiod.
Unlike knockout of Nup160 in the metazoans, Arabidopsis nup160 mutants are still viable, indicating compensatory functions of other plant Nups in maintaining the integrity of NPCs in various plant developmental processes (Galy et al., 2003, Parry et al., 2006, Maehara et al., 2012). The evolutionally conserved molecular function of the components in the Nup107-160 complex is to mediate bulk mRNA export (Vasu et al., 2001, Bai et al., 2004, Parry et al., 2006, Wiermer et al., 2012, Parry, 2014). However, although SEH1 and Nup85 are two members in the Nup107-160 complex, the seh1-1 and nup85-2 mutants, in which poly(A)-mRNA is accumulated to much higher level than wild-type plants, do not show flowering defects (Wiermer et al., 2012, Parry, 2014). This suggests that the defect in mRNA export is not necessarily correlated with the early-flowering phenotype exhibited by nup160 in Arabidopsis. In addition to their primary function in the bulk mRNA export, the human Nup107-160 subcomplex and its ortholog in yeast (Nup84 subcomplex) also function as docking sites for a desumoylating enzyme SENP2 (yeast Ulp1) at NPCs, which is closely associated with their capability in maintaining the cellular sumoylation homeostasis (Palancade et al., 2007, Goeres et al., 2011). Moreover, Nup107-160 (Nup84) subcomplexes also provide anchoring sites for SUMO-dependent E3 ligases, which facilitate the relocation of DNA double-strand break to the nuclear periphery for efficient repair in Drosophila and yeast (Nagai et al., 2008, Ryu et al., 2015). In this study, our findings suggest that the plant Nup160 also functions as a docking site for a plant-specific E3-ubiquitin ligase HOS1, which is essential for HOS1 activity to degrade CO protein specifically in the morning. Association of HOS1 with NPCs may affect HOS1 activity through different mechanisms. For example, NPC-localized HOS1 could permit its quick interaction with other co-regulators, such as phyB that translocates into the nucleus from cytoplasm in response to red light during the morning period (Yamaguchi et al., 1999). This could facilitate HOS1 at NPCs to quickly respond to the light stimulus, thus ensuring swift degradation of CO proteins during the morning period. In addition, HOS1 interaction with the scaffold Nup160 may expose the catalytic surface of HOS1 for its interaction with either CO or phyB. Further analysis of chemical and physical properties of the Nup160-HOS1 interface, the resulting structural changes in HOS1, and the specific domains required for interaction between HOS1 and NUP160 or HOS1 and CO will be helpful to elucidate the biological implication of Nup160-mediated HOS1 deposition at NPCs.
Although our data suggest that Nup160 represses the floral transition by destabilizing CO protein in the morning through spatially confining HOS1 at NPCs, there is evidence indicating that interaction between Nup160 and HOS1 may also mediate the floral transition through other regulators in addition to CO. nup160-2, hos1-3, and hos1-3 nup160-2 all partially suppress the late-flowering phenotype of co-9 (Figure 2E), implying that CO is not the only downstream target of Nup160 and HOS1. One CO-independent target of Nup160 and HOS1 could be FLOWERING LOCUS C (FLC), whose activation has been shown to require HOS1 (Jung et al., 2013). Indeed, the decrease of FLC expression was similarly detected in both nup160-2 and hos1-3 (Supplemental Figure 11A), while flc-3 only slightly enhanced early flowering of nup160-2 and hos1-3 (Supplemental Figure 11B). In addition to Nup160-mediated HOS1 deposition at NPCs, Nups could affect flowering through other regulatory mechanisms. For example, Nup96 has been reported to stabilize HOS1 protein, thus modulating CO protein levels (Cheng et al., 2019), while Nup98 might regulate flowering time in a CO-independent manner (Jiang et al., 2019). These observations suggest that, although Nups are commonly localized in NPCs, they could be involved in various regulatory pathways to control flowering time.
It is noteworthy that, in addition to their regulatory roles in the floral transition, both Nup160 and HOS1 are also required for cold-stress response and ABA signaling (Lee et al., 2001, Dong et al., 2006b, Zhu et al., 2017). Moreover, hos1 mutants display pleiotropic phenotypes, many of which are comparable with those exhibited by loss of function of different components of NPCs (MacGregor and Penfield, 2015), indicating that the interaction between HOS1 and NPCs may affect various plant developmental processes. Further investigation of the dynamic deposition of HOS1 at NPCs and the interacting partners of NPCs and HOS1 will provide additional insights into the mechanisms by which the NPCs-HOS1 module perceives various environmental cues to modulate multiple signaling pathways.
Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana plants were grown on soil or Murashige and Skoog (MS) medium under LDs (16 h light/8 h dark) or SDs (8 h light/16 h dark) at 23°C ± 2°C. All the mutants used in this study, such as nup160-2 (SALK_016091), nup160-5 (SALK_133728), hos1-3 (SALK_069312), ft-10, co-9, soc1-2, flc-3, phyb-9, and cop1-4, are in the Col ecotype.
Plasmid Construction
For the complementation test, an 11.3-kb Nup160 genomic region (gNup160) was amplified and cloned into pENTR/D-TOPO (Invitrogen). Based on this construct, 3FLAG-gNup160 was generated using a modified QuikChange site-directed mutagenesis approach. One KpnI site was introduced before the stop codon of Nup160 in gNup160 (gNup160-KpnI). Based on this construct, gNup160-GFP and gNup160-GUS were generated by translationally fusing GFP and GUS to the C terminus of Nup160. For gHOS1, an 8.08-kb genomic fragment of HOS1 was amplified and cloned into pENTR/D-TOPO (Invitrogen). One KpnI site was further introduced after the start codon of HOS1 using the QuikChange site-directed mutagenesis approach. GFP-gHOS1 was then generated by translationally fusing GFP to the N terminus of HOS1. To construct 35S:GFP-HOS1, the coding region of GFP was first amplified and cloned into pGreen 0229-35S (Yu et al., 2004) to generate pGreen 35S:GFP. The coding sequence of HOS1 was amplified and cloned into pGreen 35S:GFP to obtain 35S:GFP-HOS1.
To construct AmiR-Nup160, a set of fours primers targeting Nup160 were designed on the website (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) based on the published protocol (Schwab et al., 2006). The resulting fragment was cut by EcoRI and XbaI and ligated into the pGreen 0229-35S vector. To construct SUC2:3FLAG-CO, three tandem repeats of FLAG were fused in frame with the full-length CO coding sequence, and the resulting fragment was subsequently inserted into a modified pENTR/D-TOPO vector with the SUC2 promoter. The SUC2:nlsRFP construct was cloned in a similar way. The primers for creating the above constructs are listed in Supplemental Table 1.
Expression Analysis
Total RNA was extracted using the FavorPrep Plant Total RNA Mini Kit (Favorgen) and reverse transcribed using M-MLV reverse transcriptase (Promega) according to the manufacturer’s instructions. Real-time qPCR was performed in triplicate on each of three individually collected samples using the 7900HT Fast Real-Time PCR System (Applied Biosystems) and Maxima SYBR Green/ROX qPCR Master Mix (Fermentas). The difference between the cycle threshold (Ct) of target genes and that of TUB2 was used to calculate the normalized expression of target genes. All primers used for gene expression analysis are listed in Supplemental Table 1.
GUS Staining
Plant tissues were harvested and incubated in cold 90% acetone on ice for 20 min. Tissues were then washed three times with staining buffer without the X-Gluc substrate. New staining buffer was subsequently added together with 2 mM X-Gluc substrate. The tissues were infiltrated in a vacuum chamber for 20 min and subsequently placed in a 37°C oven for an appropriate duration. Samples were washed through an ethanol series until all chlorophyll was removed, after which the tissues were immersed in the clearing reagent and placed on the glass slides for observation.
Confocal Image Analysis
Confocal images used for comparison were taken with the same confocal settings. Quantification of GFP-HOS1 and nlsRFP signal intensity was performed using ImageJ software. A straight line was drawn across the nucleus using the Straight Line Selection Tool in ImageJ. Relative intensity plots for GFP-HOS1 and nlsRFP distribution across the drawn lines were generated using the Plot Profile function in ImageJ.
Coimmunoprecipitation and Western Blot Analysis
Coimmunoprecipitation assay of the interaction between Nup160 and HOS1 was performed with 9-day-old 3FLAG-gNup160 nup160-2 and 3FLAG-gNup160 nup160-2 GFP-gHOS1 hos1-3 seedlings grown under LDs. Whole seedlings were harvested for extraction of nuclear protein as described below. GFP-HOS1 was immunoprecipitated by anti-GFP antibody (Invitrogen) bound to Protein A/G PLUS-Agarose (Santa Cruz). Proteins bound to the beads were resolved by SDS–PAGE and detected by anti-FLAG (Sigma) or anti-GFP (Santa Cruz) antibody.
To examine the abundance of CO protein, seedlings grown on MS plates were harvested at different time points for extraction of nuclear protein using the nuclear isolation buffer (20 mM Tris–HCl [pH 6.8], 40% glycerol, 20 mM MgCl2, 5% sucrose, 0.08% β-mercaptoethanol, 0.8% Triton X-100) with protease inhibitor and 1.3 mM phenylmethylsulfonyl fluoride. After washed three times with the same nuclear isolation buffer, pellets were suspended in the 2× SDS loading buffer and thereafter heated at 95°C for 10 min. The samples were then loaded on the 12% SDS–PAGE gel. For Western blot analysis, anti-FLAG (Sigma) antibody was used to detect FLAG-CO, while anti-histone 3 (Abcam) was used to detect H3 as a loading control. Signals were detected by Bio-Rad ChemiDoc Touch Imaging System and analyzed Bio-Rad Image Lab software.
BiFC Analysis
To detect in vivo interaction between Nup160 and HOS1 in Arabidopsis, the coding sequences of nEYFP and cEYFP were amplified from pSAT vectors and translationally fused with the N terminus of gHOS1 and C terminus of gNup160, respectively. The resulting nEYFP-gHOS1 and gNup160-cEYFP were further subcloned into destination vectors with Hygromycin and Basta selection markers via the Gateway LR reaction, respectively. These two vectors were simultaneously transformed into wild-type plants and screened by both selection markers. The selected transgenic plants were used for confocal analysis, and also crossed with the transgenic plants bearing cEYFP or nEYFP to obtain the control plants containing either nEYFP-gHOS1 cEYFP or nEYFP gNup160-cEYFP, respectively.
Funding
This work was supported by the Singapore National Research Foundation Investigatorship Program (NRF-NRFI2016-02), and the intramural research support from National University of Singapore and Temasek Life Sciences Laboratory.
Author Contributions
C.L., L.L., and H.Y. conceived and designed the experiments. C.L., L.L., L.S., and Z.W.N.T. performed the experiments. C.L., L.L., L.S., and H.Y. analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
Acknowledgments
We thank George Coupland for providing cop1-4 seeds, the Arabidopsis Biological Resource Center for providing T-DNA insertional mutants, and members of the Yu lab for discussion and comments on the manuscript. No conflict of interest declared.
Published: February 19, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental Information
References
- Aitchison J.D., Blobel G., Rout M.P. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J. Cell Biol. 1995;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- Andres F., Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Bai S.W., Rouquette J., Umeda M., Faigle W., Loew D., Sazer S., Doye V. The fission yeast Nup107-120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol. Cell Biol. 2004;24:6379–6392. doi: 10.1128/MCB.24.14.6379-6392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M., Hetzer M.W. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009;10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A., Lincoln C., Lammer D., Estelle M. The SAR1 gene of Arabidopsis acts downstream of the AXR1 gene in auxin response. Development. 1997;124:1583–1591. doi: 10.1242/dev.124.8.1583. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Zhang X., Huang P., Huang G., Zhu J., Chen F., Miao Y., Liu L., Fu Y., Wang X. Nup96 and HOS1 are mutually stabilized and gate CONSTANS protein level, conferring long-day photoperiodic flowering regulation in Arabidopsis. Plant Cell. 2019;32:374–391. doi: 10.1105/tpc.19.00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw J.M., Krutchinsky A.N., Zhang W., Chait B.T., Matunis M.J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U S A. 2006;103:8281–8286. doi: 10.1073/pnas.0602874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.H., Hu X., Tang W., Zheng X., Kim Y.S., Lee B.h., Zhu J.K. A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell Biol. 2006;26:9533–9543. doi: 10.1128/MCB.01063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserova J., Kiseleva E., Goldberg M.W. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 2009;59:243–255. doi: 10.1111/j.1365-313X.2009.03865.x. [DOI] [PubMed] [Google Scholar]
- Galy V., Mattaj I.W., Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol. Biol. Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeres J., Chan P.K., Mukhopadhyay D., Zhang H., Raught B., Matunis M.J. The SUMO-specific isopeptidase SENP2 associates dynamically with nuclear pore complexes through interactions with karyopherins and the Nup107-160 nucleoporin subcomplex. Mol. Biol. Cell. 2011;22:4868–4882. doi: 10.1091/mbc.E10-12-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golembeski G.S., Imaizumi T. Photoperiodic regulation of florigen function in Arabidopsis thaliana. Arabidopsis Book. 2015;24:e0178. doi: 10.1199/tab.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Draxl S., Brachmann A., Sato S., Tabata S., Kawaguchi M., Wang T.L. NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell. 2010;22:2509–2526. doi: 10.1105/tpc.109.069807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A., Hetzer M.W. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015;29:337–349. doi: 10.1101/gad.256495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlau A., Truernit E., Sauer N. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell. 1999;11:309–322. doi: 10.1105/tpc.11.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y., Mongkolsiriwatana C., Veley K.M., Kim S.Y., Michaels S.D. The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol. 2007;144:1383–1390. doi: 10.1104/pp.107.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C.S., Wenkel S., Soppe W., Deng X.-W., Valverde F., Coupland G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008;27:1277–1288. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Xiao L., Huang P., Cheng Z., Chen F., Miao Y., Fu Y.F., Chen Q., Zhang X.M. Nucleoporin Nup98 participates in flowering regulation in a CONSTANS-independent mode. Plant Cell Rep. 2019;38:1263–1271. doi: 10.1007/s00299-019-02442-w. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Lee H.J., Park M.J., Park C.M. Beyond ubiquitination: proteolytic and nonproteolytic roles of HOS1. Trends Plant Sci. 2014;19:538–545. doi: 10.1016/j.tplants.2014.03.012. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Park J.H., Lee S., To T.K., Kim J.M., Seki M., Park C.M. The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell. 2013;25:4378–4390. doi: 10.1105/tpc.113.118364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A., Mouriz A., Piñeiro M., Jarillo J.A. Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis. Plant Cell. 2015;27:2437–2454. doi: 10.1105/tpc.15.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A., Valverde F., Pineiro M., Jarillo J.A. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell. 2012;24:982–999. doi: 10.1105/tpc.110.081885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., Zhu J.K. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo–cytoplasmic partitioning. Genes Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.-J., Zhang Y.-C., Li Q.-H., Sang Y., Mao J., Lian H.-L., Wang L., Yang H.-Q. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell. 2008;20:292–306. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor D.R., Gould P., Foreman J., Griffiths J., Bird S., Page R., Stewart K., Steel G., Young J., Paszkiewicz K. HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 is required for circadian periodicity through the promotion of nucleo-cytoplasmic mRNA export in Arabidopsis. Plant Cell. 2013;25:4391–4404. doi: 10.1105/tpc.113.114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor D.R., Penfield S. Exploring the pleiotropy of hos1. J. Exp. Bot. 2015;66:1661–1671. doi: 10.1093/jxb/erv022. [DOI] [PubMed] [Google Scholar]
- Maehara K., Murata T., Aoyama N., Matsuno K., Sawamura K. Genetic dissection of Nucleoporin 160 (Nup160), a gene involved in multiple phenotypes of reproductive isolation in Drosophila. Genes Genet. Syst. 2012;87:99–106. doi: 10.1266/ggs.87.99. [DOI] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M.B., Roberts T.M., Brown G.W., Varela E., Hediger F., Gasser S.M., Krogan N.J. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., Doye V. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell. 2007;18:2912–2923. doi: 10.1091/mbc.E07-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G. Components of the Arabidopsis nuclear pore complex play multiple diverse roles in control of plant growth. J. Exp. Bot. 2014;65:6057–6067. doi: 10.1093/jxb/eru346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Ward S., Cernac A., Dharmasiri S., Estelle M. The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell. 2006;18:1590–1603. doi: 10.1105/tpc.106.041566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu T., Spatola B., Delabaere L., Bowlin K., Hopp H., Kunitake R., Karpen G.H., Chiolo I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 2015;17:1401–1411. doi: 10.1038/ncb3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Shim J.S., Kinmonth-Schultz H.A., Imaizumi T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015;66:441–464. doi: 10.1146/annurev-arplant-043014-115555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- Tamura K., Fukao Y., Iwamoto M., Haraguchi T., Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2010;22:4084–4097. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Vasu S., Shah S., Orjalo A., Park M., Fischer W.H., Forbes D.J. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Alves A., Pickersgill H., Loïodice I., Hetzer M., Galy V., Hülsmann B.B., Köcher T., Wilm M., Allen T. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Wang C.Q., Guthrie C., Sarmast M.K., Dehesh K. BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell. 2014;26:3589–3602. doi: 10.1105/tpc.114.130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Cheng Y.T., Imkampe J., Li M., Wang D., Lipka V., Li X. Putative members of the Arabidopsis Nup107-160 nuclear pore sub-complex contribute to pathogen defense. Plant J. 2012;70:796–808. doi: 10.1111/j.1365-313X.2012.04928.x. [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Yamaguchi R., Nakamura M., Mochizuki N., Kay S.A., Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Ito T., Wellmer F., Meyerowitz E.M. Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat. Genet. 2004;36:157–161. doi: 10.1038/ng1286. [DOI] [PubMed] [Google Scholar]
- Zhang B., Wang L., Zeng L., Zhang C., Ma H. Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 2015;29:975–987. doi: 10.1101/gad.251520.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li X. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell. 2005;17:1306–1316. doi: 10.1105/tpc.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wang B., Tang K., Hsu C.-C., Xie S., Du H., Yang Y., Tao W.A., Zhu J.-K. An Arabidopsis nucleoporin NUP85 modulates plant responses to ABA and salt stress. PLoS Genet. 2017;13:e1007124. doi: 10.1371/journal.pgen.1007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.