Abstract
Human SH2B3 is involved in growth factor and inflammation signaling. A SH2B3 missense variant (rs3184504) is associated with cardiovascular diseases plus breast, colorectal, and lung cancers, with highly correlated variants across the ATXN2/SH2B3/BRAP locus linked to parental age at death, suggesting a geroscience common mechanism of aging and disease. To better understand the SH2B3-related aging pathway and its potential as an intervention target, we undertook a phenotype-wide association study (PheWAS) of 52 aging traits. Data were obtained from 379,758 European-descent UK Biobank participants, aged 40–70 at baseline: 27% of participants were CC homozygotes and 23% TT at rs3184504. Parental extreme longevity (mothers aged ≥98 years, fathers aged ≥96 years) was more common in CC versus TT (odds ratio [OR] = 1.18, 95% confidence interval [CI]: 1.07 to 1.29) with an additive per allele effect. The C allele associated with better cognitive function and white blood cell counts were more likely to be normal. The C allele reduced risks of coronary heart disease (OR = 0.95, 95% CI: 0.93 to 0.96) but was also associated with a modestly higher cancer rate (OR = 1.03, 95% CI: 1.02 to 1.04), suggesting a trade-off across aging outcomes and limiting its potential as an anti-aging target.
Keywords: Centenarian, Anti-aging, IGF-1, Cancer, UK Biobank
The geroscience hypothesis argues that shared underlying mechanisms drive many diseases of aging (1), but few such mechanisms have been proven in humans (2). A missense variant (rs3184504) in SH2B3 has been linked to many common diseases in genome-wide association studies, including several autoimmune and cardiovascular disorders, hypertension (3), and myeloproliferative cancers (4), plus breast, colorectal, and lung cancers (5). In a genome-wide analysis of parental longevity in UK Biobank (6), we found that 11 highly correlated genetic variants in the wider SH2B3/ATXN2/BRAP locus (including rs3184504) were associated with parent’s attained age, and this longevity association has been replicated in other cohorts (7). Given this involvement in many human diseases and longevity, SH2B3 may shed light on biological mechanisms that contribute to the aging process. Drug targets supported by genetic evidence are twice as likely to succeed in human trials (8). However, the impact of this SH2B3 variant on many aging phenotypes is unknown.
The SH2B3 gene in humans codes for the lymphocyte adaptor protein LNK. Initially characterized as a hematopoiesis and lymphocyte-specific differentiation regulator, LNK is widely expressed in the human body (4) and is involved in transduction and regulation of growth factor and (inflammation-related) cytokine receptor-mediated signaling (9). The rs3184504 T allele missense variant is predicted to disrupt the subcellular localization and functioning of LNK (10).
As noted, the earlier variation in SH2B3 appears highly relevant to geroscience, but little is known about associations with aging-related traits, including muscle weakness, frailty, chronic pain, cognitive measures, blood measured (e.g., of inflammation, with high white cell counts), and several relevant disease diagnoses. We aimed to undertake a phenome-wide association study (PheWAS) of 52 aging traits, to clarify the health outcomes of common variation in SH2B3, as marked by rs3184504. UK Biobank offers a large sample of community volunteers with baseline self-reports of parental age at death, morbidity, and physical measures, plus a follow-up in national hospital, cancer registry, and death certificate data.
Methods
UK Biobank is a volunteer cohort, with 502,642 participants aged 40–70 seen between 2006 and 2010. A range of questionnaire, physiological, and disease data are available, including hospital admissions (up to 10 years follow-up) (11). Genotype information was available on 488,377 participants. Genotype imputation was successful in 487,442 UK Biobank participants and increased the number of available genetic variants to approximately 96 million (12).
Analyses were restricted to European-descent participants (n = 451,433), as numbers from the other ancestry groups in UK Biobank are unfortunately too small to provide sufficiently powered estimates for this PheWAS. European-descent participants were identified based on genetic principal components analysis as described in more detail in Thompson and colleagues (13). To avoid inflated effects from inclusion of closely related family members, only one subject in third-degree or closer pairs were included in analyses (based on the kinship analysis), leaving 379,758 participants aged 40–70 at baseline. None of the included samples were identified by UK Biobank as outliers in heterozygosity and missing rates, which would indicate poor-quality genotypes for these samples (http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=531).
UK Biobank used two Affymetrix microarrays (the BiLEVE array in approximately 50,000 participants, and the Axiom array in approximately 440,000 participants, more than 95% shared marker content) (12). We analyzed the 10 single nucleotide polymorphisms (SNPs) in linkage disequilibrium (LD, R2 > 0.8) with rs3184504, associated with parents’ attained age (p < 5 × 10–8) in our 2017 genome-wide association study (GWAS) (14). See Supplementary Figure 1 for LocusZoom plot of the region (+/–250 kb). None of the 10 SNPs were conditionally associated with parents’ attained age given rs3184504 was in the model (p > 0.05; Supplementary Table 1). The results suggested that the common variation in SH2B3 was marked by rs3184504 for the association with parents’ attained age. We here conducted a PheWAS to study SH2B3 via rs3184504 and aging. rs3184504 (chr12, b37 111,884,608) was directly genotyped on the arrays, and for analyses was coded as 0, 1, or 2 copies of the C (life-span-extending) allele. The C allele frequency was 0.52 and the genotype distribution (23% TT, 50% CT, and 27% CC) was not significantly deviant from Hardy–Weinberg equilibrium (p = 0.642).
Parental Extreme Longevity
Parental age at death was assessed by survey questions, administered to participants, and updated with data from follow-up visits. We derived at least one parent top 1% survival, which was 98 for mothers and 96 for fathers, determined by parental age at death in UK Biobank. Parents who attained the top 1% survival were compared with short-lived parents who both died before the age of 80. Parents who died prematurely (more than 1 standard deviation below the modal age of death: mothers <57 years, fathers <46 years) were excluded from analyses (15).
Baseline Phenotypes
Baseline phenotypes included depression, chronic pain (back, hip, and knee pains), falls, muscle weakness, Fried-defined frailty status, a cumulative morbidity frailty index, cognitive function, and physiological biomarkers.
Depression, chronic pain, and falls in the last year were assessed by the survey questions: “Over the past two weeks, how often have you felt down, depressed or hopeless?” with the responses grouped into several days and longer or not at all; “In the last year, have you had any falls?” with the responses grouped into any fall or no falls”; “Have you had back pains for more than 3 months?” with the responses grouped into yes or no”; “Have you had hip pains for more than 3 months?” with the responses grouped into yes or no”; and “Have you had knee pains for more than 3 months?” with the responses grouped into yes or no.
Low muscle mass (8.87 kg/m2 for men and 6.42 kg/m2 for women) and low handgrip strength (30 kg for men and 20 kg for women) were defined from the European Working Group on Sarcopenia in Older People (16).
For cognitive function, we analyzed reaction time and visual memory errors. “Reaction time” was calculated as the average time (ms) taken to correctly identify a match in a symbol matching game similar to the snap card game. “Visual memory errors” were measured as the number of errors that a participant made to complete a pairs matching task where six pairs of cards were presented for 3 seconds beforehand. Both were log transformed to correct skewness of the distributions. Visual memory errors were right shifted by 1 to avoid infinite values from zero errors.
Fried frailty status (frail or not frail) was frail if meeting three or more of the five conditions: self-reported exhaustion, weight loss, and slow walking pace, plus measured grip strength and physical activity at the lowest 20% where physical activity was measured by the short version of International Physical Activity Questionnaire (17).
Physiological biomarkers included lung function measures of forced expiratory volume in 1 second (FEV1), forced vital capacity (FVC), and FEV1/FVC; heel bone mineral density and blood pressures. Albumin–creatinine ratio at least 3 mg/mmol was used as a biomarker of renal impairment for diagnosing chronic kidney disease, which was associated with hypertension and cardiovascular mortality (18).
We also examined associations with hematological measures, due to previously known links between SH2B3 and bone marrow activation (4). We compared raised or lowered values to reference ranges quoted by the manufacturer (http://biobank.ndph.ox.ac.uk/showcase/showcase/docs/haematology.pdf).
Disease Outcomes
At the baseline assessment, participants self-reported prevalent doctor-diagnosed diseases. These were combined with hospital admission data to identify participants with diagnoses of multiple relevant diseases. The disease status was confirmed regardless of prevalent or incident cases. Common diseases were included such as cancers, diabetes, respiratory diseases, bone diseases, and cardiovascular diseases. We derived “any cancer” excluding non-melanoma skin cancers and “any-cause anemia” that was mild or more severe. In addition, we included the frailty index by Williams and colleagues (19), which scored 49 deficits mostly pains and diseases (range 0–49).
Statistical Analysis
A logistic regression model was performed to associate a binary outcome and the rs3184504 genotype, adjusted for age at measurement (age at baseline or age at the last follow-up), sex, genotyping microarray, assessment center, and principal components 1–5 to account for ancestry composition. A linear regression model was used instead for a continuous outcome, which was z-transformed to standardize the scale. We highlighted associations with p-values smaller than the Bonferroni-corrected level (p < 0.05/52 = 0.00096) and reported 95% confidence intervals. All the statistical analyses were performed in R 3.4.1.
Sensitivity Analyses
We performed subgroup analyses for baseline phenotypes and disease outcomes by sex or using subjects aged 60 and older at measurement (baseline or the last follow-up) to exclude possibly atypical cases. Parental extreme longevity traits are parental traits determined by both parents, therefore, were not included in subgroup analyses. We also linked the 10 SNPs in LD with rs3184504 to traits associated with rs3184504 and tested their effects conditioning on rs3184504.
Power Analysis
Denote by f (=0.52) the C allele frequency of rs3184504. The effect size (ES) defined as 2 × β2 × f (1–f) is the percent of trait variance explained by rs3184504 assuming Hardy–Weinberg equilibrium and an additive polygenic model where β is the standard deviation (SD) change per C allele in a continuous trait (20). Given the sample size of each trait assuming the Bonferroni-corrected significance level (0.05/52 = 0.00096), power to detect a 0.1 or –0.1 standard deviation change (β = 0.1 or –0.1, ES = 0.50%) in a continuous trait was calculated using G*Power (21). Power to detect a relative risk of 1.2 or 0.83 (multiplicative inverse of 1.2) assuming a multiplicative model for a binary trait was calculated using the R package power.ctepd (22) where the case–control ratio and the prevalence were estimated from the sample. Power for all studied phenotypes was more than 80%, except for parental extreme longevity (69% power), type 1 diabetes (44% power), renal failure (75% power), and some hematological measures (see Supplementary Table 2 for details).
Ethical Approval
UK Biobank received an approval from the UK Biobank Research Ethics Committee (REC; REC reference 11/NW/0382). All the participants provided written informed consent to participate in the study and for their data to be used in future research. This research was conducted using UK Biobank resource, under application 14631.
Results
A total of 379,758 European-descent participants in UK Biobank were included in analyses (mean age 56.7, SD = 8.0, range: 40–73), with a mean follow-up of 7.8 years (SD = 1.0). The studied sample (54% women) included n = 168,310 aged 60 plus at baseline interview (n = 261,837 aged 60 and older by the end of follow-up). Overall, 11,014 subjects died during follow-up. A summary of aging traits, overall and by rs3184504 genotypes, was provided in Supplementary Table 2, where traits associated with the SNP (p < 0.00096) were marked by gray background.
Associations with p-values smaller than the Bonferroni-corrected level (p < 0.00096) were discussed later. Complete association results using all mid-age and older adults, 60 or older adults only, and men, or women only are provided in Supplementary Table 3.
Associations With Parental Extreme Longevity
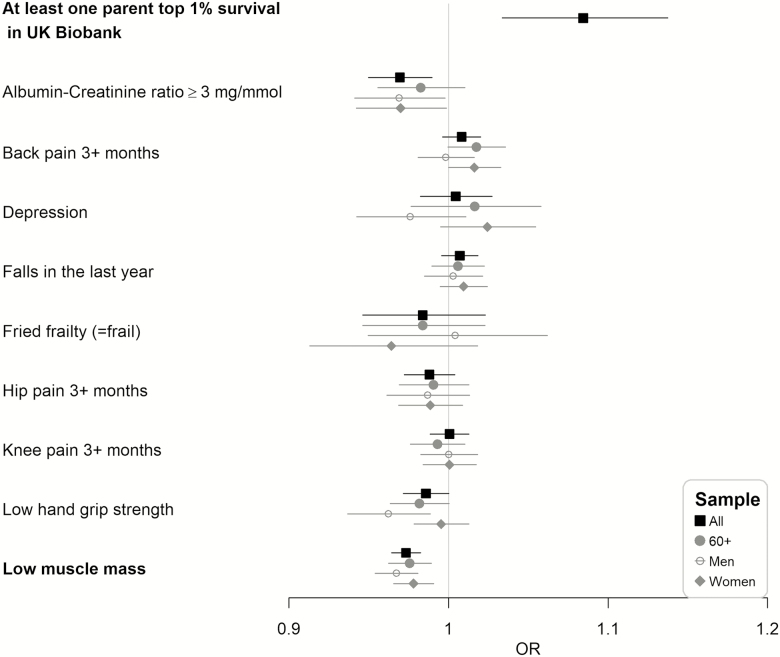
Extreme parental longevity (at least one parents top 1% survival in UK Biobank) was more common for CC compared with TT homozygotes (OR = 1.18, 95% CI: 1.07 to 1.29), with an additive per allele effect (per C allele OR = 1.08, 95% CI: 1.03 to 1.14; Figure 1).
Figure 1.
Odds ratio (OR) per C allele of rs3184504 and 95% confidence interval for parental extreme longevity or a binary baseline phenotype using all samples (All), 60 or older (60+), men only (Men), or women only (Women). Highlighted in bold if the p-value smaller than the Bonferroni-corrected level (0.05/52 = 0.00096).
Associations With Baseline Phenotypes
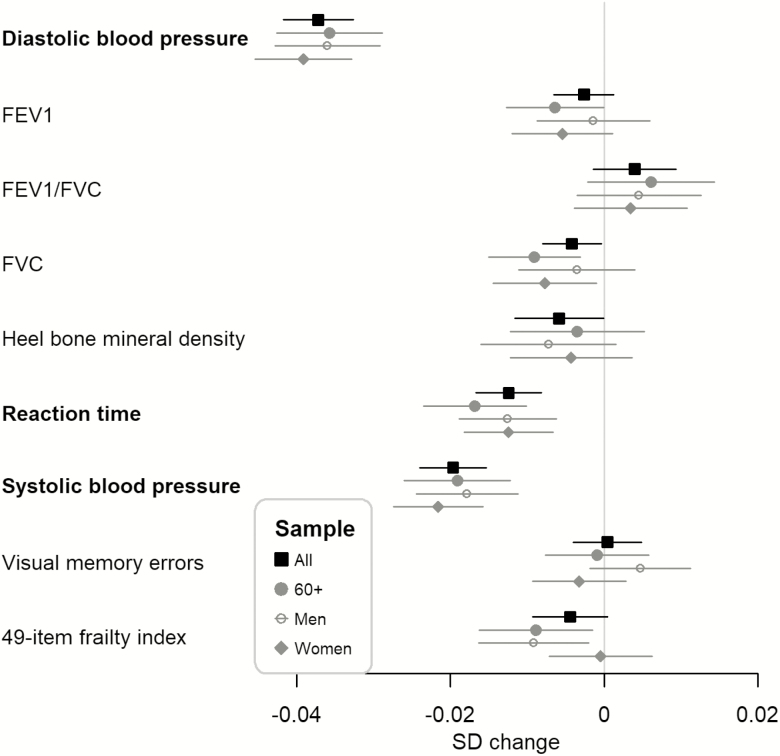
rs3184504 C allele was associated with a reduced prevalence of low muscle mass (per C allele OR = 0.97, 95% CI: 0.96 to 0.98; Figure 1), lower systolic and diastolic blood pressures (systolic SD change = –0.020, 95% CI: –0.024 to –0.015, diastolic SD change = –0.037, 95% CI: –0.042 to –0.033). Also, in cognitive testing, reaction times were shorter (SD change in log scale = –0.012, 95% CI: –0.017 to –0.008; Figure 2). To ensure relevance to aging in men and women, we undertook specific analyses in 60 plus year old, including separate estimates for men and women: effect sizes as odds ratios or SD changes were similar in all mid-age and older adults, 60 or older adults only, and men, or women only (Figures 1 and 2). There was no association (p > 0.00096) with visual memory errors (to measure cognitive function), chronic pain, depression, Fried frailty (comparing frail vs. not frail), falls, grip strength, heel bone mineral density, lung function biomarkers (FEV1, FVC, and FEV1/FVC), and the urinary biomarker, albumin–creatinine ratio (≥3 mg/mmol; Figures 1 and 2).
Figure 2.
Standard deviation (SD) change per C allele of rs3184504 and 95% confidence interval for a continuous baseline phenotype or the 49-item multimorbidity-based frailty index using all samples (All), 60 or older (60+), men only (Men), or women only (Women). Highlighted in bold if the p-value smaller than the Bonferroni-corrected level (0.05/52 = 0.00096).
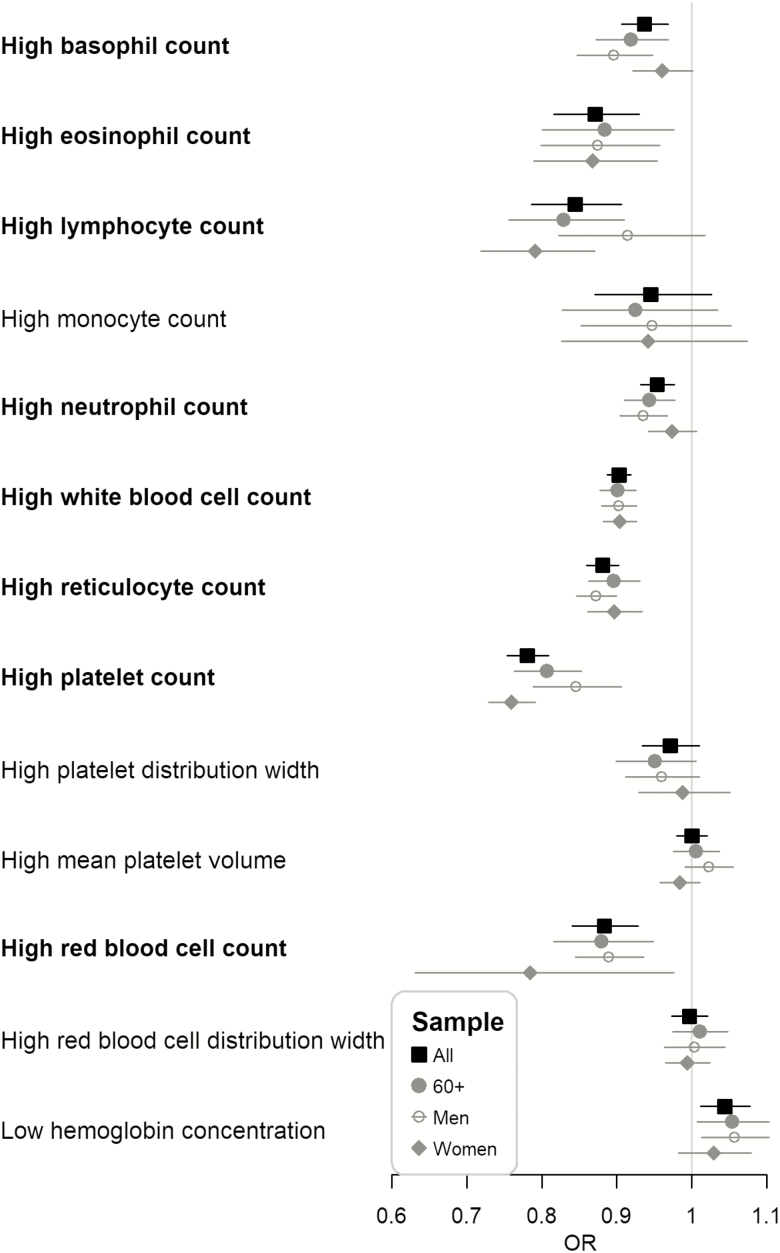
Analyses of baseline hematological measures (Figure 3) showed associations between rs3184504 C allele and lower risks of being above white cell count clinical reference ranges, including total white cell count (OR = 0.90, 95% CI: 0.89 to 0.92), neutrophils (OR = 0.95, 95% CI: 0.93 to 0.98), lymphocytes (OR = 0.84, 95% CI: 0.79 to 0.91), eosinophils (OR = 0.87, 95% CI: 0.82 to 0.93), and basophils (OR = 0.94, 95% CI: 0.91 to 0.97). rs3184504 C allele was also associated with lower risks of being above reference ranges of platelets (OR = 0.78, 95% CI: 0.75 to 0.81), reticulocytes (OR = 0.88, 95% CI: 0.86 to 0.90), and total red blood cell count (OR = 0.88, 95% CI: 0.84 to 0.93). The effect sizes for lymphocytes, platelets, and red cell count were stronger in women than in men. Clinically low hemoglobin concentrations (below sex-specific reference ranges) were slightly less common in C allele carriers, overall, in men, and in women but did not reach statistical significance.
Figure 3.
Odds ratio (OR) per C allele of rs3184504 and 95% confidence interval for being above or below the reference range of a hematological measure using all samples (All), 60 or older (60+), men only (Men), or women only (Women). Highlighted in bold if the p-value smaller than the Bonferroni-corrected level (0.05/52 = 0.00096).
Associations With Disease Outcomes (Binary) and the Multimorbidity-Based Frailty Index
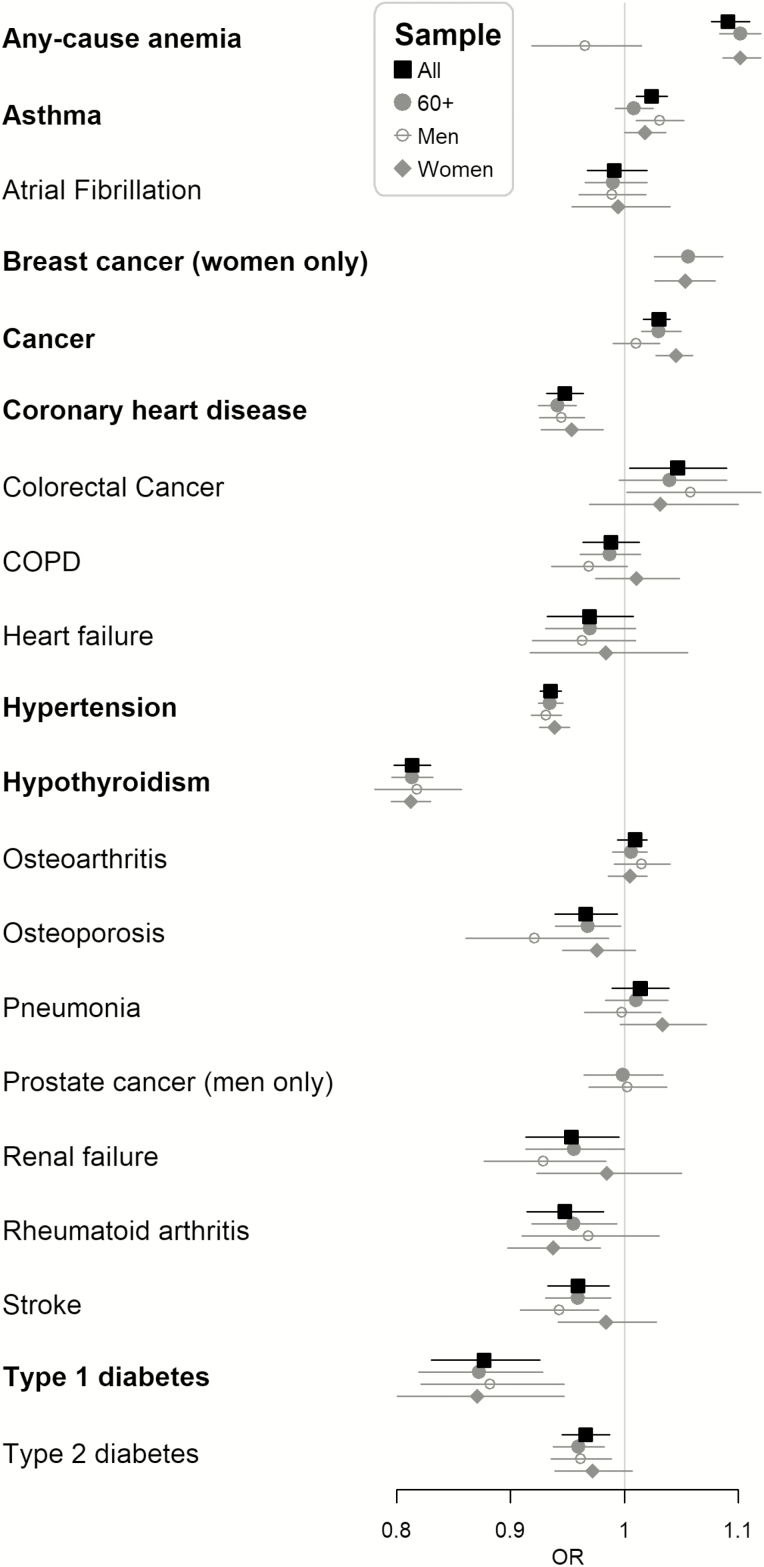
The rs3184504 C allele increased the risk of any-cause anemia (mild or more severe, OR = 1.09, 95% CI: 1.08 to 1.11); however, reduced the risks of hypothyroidism (OR = 0.81, 95% CI: 0.80 to 0.83), and type I diabetes (OR = 0.88, 95% CI: 0.83 to 0.93; Figure 4). Positive associations were found with breast cancer (OR = 1.05, 95% CI: 1.03 to 1.08), and any cancer (excluding non-melanoma skin cancers, OR = 1.03, 95% CI: 1.02 to 1.04) but negative associations with coronary heart disease (OR = 0.95, 95% CI: 0.93 to 0.96), and hypertension (OR = 0.94, 95% CI: 0.93 to 0.94). Except for the association with any-cause anemia (mild or more severe, mainly present in women), the effect sizes overall were similar to those in the subgroups of 60 and older, men only, and women only (Figure 4). Although not highlighted at the Bonferroni-corrected level (p < .00096), the C allele was modestly associated with colorectal cancer (OR = 1.05, 95% CI: 1.00 to 1.09) and protective for renal failure (OR = 0.95, 95% CI: 0.91 to 1.00), rheumatoid arthritis (OR = 0.95, 95% CI: 0.91 to 0.98), stroke (OR = 0.96, 95% CI: 0.93 to 0.99), and type 2 diabetes (OR = 0.97, 95% CI: 0.95 to 0.99; Figure 4). rs3184504, however, was not associated (p > 0.00096) with the 49-item frailty index (Figure 2).
Figure 4.
Odds ratio (OR) per C allele of rs3184504 and 95% confidence interval for a disease outcome using all samples (All), 60 or older (60+), men only (Men), or women only (Women). Highlighted in bold if the p-value smaller than the Bonferroni-corrected level (0.05/52 = 0.00096). All women or women of 60 or older only for breast cancer and all men or men of 60 or older only for prostate cancer.
In additional analysis for each rs3184504-associated trait, we examined the association for the 10 SNPs in LD with rs3184504 at the same locus. Overall, effect sizes and p-values were very similar, and in conditional analyses, none showed significant associations after controlling rs3184504 except an SNP for reaction time (p < 0.00094) but the effect size was minimal (Supplementary Table 4).
Discussion
The SH2B3 missense variant rs3184504 has been linked to several chronic diseases and cancers, and highly correlated variants in the wider SH2B3/ATXN2/BRAP locus (including rs3184504) were associated with parental age at death. Here, we aimed to provide a phenotype scan of 52 aging-related traits, to better understand the SH2B3-related aging pathway and to test the variant’s potential as a target for anti-aging interventions. We showed that rs3184504 is associated with substantially increased chances of having at least one parent top 1% survival in UK Biobank. We also showed that the C allele was associated with lower blood pressures, shorter reaction time (cognitive measure), and healthier muscle mass and hematological measures: that is, lower prevalence of low muscle mass and lower prevalence of abnormally high blood white cell counts. In addition, we found associations between the C allele and reduced rates of hypothyroidism, hypertension, type I diabetes and cardiovascular disease. However, a modest excess in cancer risk was present: rs3184504 was associated with any cancer (excluding non-melanoma skin cancers) and breast cancer, plus the expected association with colorectal cancer, although this appeared to be modest. Our findings thus suggest that modulation of SH2B3-related pathways may be subject to trade-offs between aging outcomes.
Our PheWAS results for those traits that have been previously studied are consistent with the published literature: rs3184504 has been reported to be associated with coronary heart disease risk (23), increased blood pressure (24), type-1 diabetes (25), and platelet counts and leukocytosis (especially neutrophil counts) (26), in the same direction as our results. Similarly, several links have been shown between SH2B3 and cancers, including rs3184504 associations with colorectal cancer (27). Longevity associations were also identified by Fortney and colleagues in the New England Centenarian Study using an “informed GWAS” approach (28), and highly correlated SNPs across the SH2B3/ATXN2/BRAP locus were associated with parental longevity in the LifeGen cohorts (29). We found no association between rs3184504 and a number of age-related traits, including cognitive function (visual memory errors), chronic pain, and frailty. This is consistent with published GWAS (where available in the GWAS catalog) and is unexpected given the association with longevity, although the pleiotropic effect of the variant on cancer and cardiovascular disease may explain this.
This PheWAS cannot address mechanism directly, but there is extensive published evidence suggesting the likely mechanisms driving these associations. The life-span-decreasing T allele of rs3184504 in SH2B3 is predicted to disrupt normal SH2B3/LNK functioning in facilitating transduction and regulation of growth factor and (inflammation-related) cytokine receptor-mediated signaling (9). Inflammation has been suggested as a factor accounting for the association between SH2B3 variants and lung, bowel, and breast cancer in GWAS (5). Several other links have been shown between SH2B3 and cancers, including the presence of activating mutations in acute lymphoblastic leukemia cells (30,31). The rs3184504 C (life-span-increasing) allele is associated with reduced levels of vascular cell adhesion protein 1 (32), which has major roles in development in the spread of cancers (33) plus white cell recruitment in the cellular immune response and in angiogenesis. Similarly, there are several suggested mechanisms linking LNK/SH2B3 with cardiovascular disease and hypertension (10), partly through increased production of IFNy, a pro-inflammatory cytokine. SH2B3 is also a crucial mediator of post-myocardial infarction inflammation and fibrosis (34). A trade-off between chronic diseases and cancer in aging has been suggested (35) based on the hypothesis that more apoptosis of damaged cells might prevent cancers but could also result in less regrowth and repair thus promoting chronic diseases, and vice versa: more regrowth may prevent chronic disease while promoting cancer. More work is clearly needed to fully define SH2B3 aging mechanisms in humans.
Incidentally, in Drosophila, the SH2B gene is involved in insulin-like growth factor (IGF-1) and other energy balance-related signaling, and a Drosophila SH2B loss-of-function mutant had increased life span under starvation conditions through increased carbohydrate stores (36). In mice and humans, several SH2B homologs exist, with SH2B1 and SH2B2 also involved in IGF-1 signaling (37), but no GWAS associations have thus-far been reported for human longevity in these genes. As discussed earlier, SH2B3 is associated with human longevity but is not thought to be an important driver of IGF-1 signaling (37).
There are inevitably limitations to this analysis: UK Biobank is a volunteer study and the sample tended to be healthier at baseline than the general population, but the sample did include a wide range of exposures (38). We have studied only European-ancestry participants, as numbers for other ancestry groups were relatively small. Several phenotype measures are available at baseline only. Follow-up data are limited to discharge hospital records and death certificates, so may underestimate incident diagnoses. The data used from UK Biobank were included in several previous studies of specific diseases and parental age at death, but these associations have been replicated in independent samples, and our focus here on aging phenotypes extends the existing literature.
Conclusion
The human SH2B3 locus harbors common variation associated with human longevity, several chronic diseases and cancers, and may represent a geroscience hypothesized common mechanism of aging. In a large aging PheWAS of 52 relevant traits, common variation marked by rs3184504 was associated with a substantial increase in the chances of having at least one parent top 1% survival. There was also evidence for this variant being associated with better cardiovascular health and better cognition. However, modestly higher cancer rates were found, confirming previous reports. Therefore, despite being associated with parental extreme longevity, common variation in SH2B3 may be subject to trade-offs between aging outcomes, which will limit its potential as an anti-aging intervention target.
Supplementary Material
Supplementary data are available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Funding
This study was partly funded by an award to D.M. by the UK Medical Research Council (MR/M023095/1). C.L.K. was supported by an Intergovernmental Personnel Act agreement (#20170526) with L.F. D.M.. and L.C.P.. are supported by the University of Exeter Medical School and the University of Connecticut School of Medicine. D.M. initiated the project. D.M., M.J., and C.L.K. conducted the literature review. C.L.K., M.J., and L.C.P. were involved in the data analysis. All the authors contributed to the manuscript preparation.
Conflict of Interest
None declared.
Acknowledgments
We wish to thank the UK Biobank participants and coordinators for this unique data set, Dr. Andrew R. Wood for his work identifying the UK Biobank participants of European descent, the group of Dr. Sara Hägg for sharing codes to create the 49-item frailty index and Dr. Janice Atkin’s efforts for the actual implementation.
References
- 1. Pessin JE, Epel ES, Campisi J, Rando TA, Wyss-Coray T, Kennedy BK, et al. Geroscience: linking aging to chronic disease. Cell [Internet]. 2014;159:709–713. doi: 10.1016/j.cell.2014.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferrucci L, Levine ME, Kuo PL, Simonsick EM. Time and the metrics of aging. Circ Res.. 2018;123:740–744. doi: 10.1161/CIRCRESAHA.118.312816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laroumanie F, Humphrey JD, Madhur MS. LNK deficiency promotes acute aortic dissection and rupture. JCI Insight [Internet]. 2018. [cited 2018 Dec 21]; 3 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6237478/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maslah N, Cassinat B, Verger E, Kiladjian JJ, Velazquez L. The role of LNK/SH2B3 genetic alterations in myeloproliferative neoplasms and other hematological disorders [Internet] Vol. 31, Leukemia: Nature Publishing Group; 2017. [cited 2018 Dec 13]. p. 1661–1670. Available from: http://www.nature.com/doifinder/10.1038/leu.2017.139 [DOI] [PubMed] [Google Scholar]
- 5. Hung RJ, Ulrich CM, Goode EL, Brhane Y, Muir K, Chan AT, et al. Cross cancer genomic investigation of inflammation pathway for five common cancers: lung, ovary, prostate, breast, and colorectal cancer. J Natl Cancer Inst [Internet]. 2015. Nov 29 [cited 2019 Jan 29]; 107. doi: 10.1093/jnci/djv246. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26319099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pilling LC, Atkins JL, Bowman K, Jones SE, Tyrrell J, Beaumont RN, et al. Human longevity is influenced by many genetic variants: evidence from 75,000 UK Biobank participants. Aging (Albany NY) [Internet]. 2016. [cited 2016 May 15]; 8:1–24. Available from: http://www.impactaging.com/papers/v8/n3/full/100930.html%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/27015805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Timmers PR, Mounier N, Lall K, Fischer K, Ning Z, Feng X, et al. Genomics of 1 million parent lifespans implicates novel pathways and common diseases and distinguishes survival chances. Elife [Internet]. 2019;8:1–40. doi:10.7554/eLife.39856.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, et al. The support of human genetic evidence for approved drug indications. Nat Genet [Internet]. 2015;47:856–860. doi:10.1038/ng.3314 [DOI] [PubMed] [Google Scholar]
- 9. Devallière J, Charreau B. The adaptor Lnk (SH2B3): an emerging regulator in vascular cells and a link between immune and inflammatory signaling. Biochem Pharmacol.. 2011;82:1391–1402. doi: 10.1016/j.bcp.2011.06.023 [DOI] [PubMed] [Google Scholar]
- 10. Dale BL, Madhur MS. Linking inflammation and hypertension via LNK/SH2B3. Curr Opin Nephrol Hypertens [Internet]. 2016. [cited 2018 Dec 13]; 25:87–93. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25755918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thompson WD, Tyrrell J, Borges MC, et al. Association of maternal circulating 25 (OH) D and calcium with birth weight: a Mendelian randomisation analysis. PLoS Med. 2019;16:e1002828. doi:10.1371/journal.pmed.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pilling LC, Kuo CL, Sicinski K, et al. Human longevity: 25 genetic loci associated in 389,166 UK biobank participants. Aging (Albany NY). 2017;9:2504–2520. doi: 10.18632/aging.101334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dutta A, Henley W, Robine JM, Langa KM, Wallace RB, Melzer D. Longer lived parents: protective associations with cancer incidence and overall mortality. J Gerontol A Biol Sci Med Sci.. 2013;68:1409–1418. doi: 10.1093/gerona/glt061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cruz-Jentoft AJ, Baeyens JP, Bauer JM, et al. ; European Working Group on Sarcopenia in Older People. Sarcopenia: European consensus on definition and diagnosis: report of the European working group on sarcopenia in older people. Age Ageing.. 2010;39:412–423. doi: 10.1093/ageing/afq034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Craig CL, Marshall AL, Sjöström M, et al. International physical activity questionnaire: 12-country reliability and validity. Med Sci Sports Exerc.. 2003;35:1381–1395. doi: 10.1249/01.MSS.0000078924.61453.FB [DOI] [PubMed] [Google Scholar]
- 18. Sung KC, Ryu S, Lee JY, Lee SH, Cheong E, Hyun YY, et al. Urine albumin/creatinine ratio below 30 mg/g is a predictor of incident hypertension and cardiovascular mortality. J Am Heart Assoc. 2016;5:1–11. doi:10.1161/JAHA.116.003245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Williams DM, Jylhävä J, Pedersen NL, Hägg S. A frailty index for UK biobank participants. J Gerontol A Biol Sci Med Sci.. 2019;74:582–587. doi: 10.1093/gerona/gly094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park JH, Wacholder S, Gail MH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet.. 2010;42:570–575. doi: 10.1038/ng.610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Erdfelder E, FAul F, Buchner A, Lang AG. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods.. 2009;41:1149–1160. doi:10.3758/BRM.41.4.1149 [DOI] [PubMed] [Google Scholar]
- 22. Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case-control genetic association analyses. BMC Genet.. 2008;9:36. doi: 10.1186/1471-2156-9-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. ; CARDIoGRAMplusC4D Consortium Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet [Internet]. 2013;45:25–33. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23202125 [DOI] [PMC free article] [PubMed]
- 24. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi:10.1038/nature10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barrett JC, Clayton DG, Concannon P, et al. ; Type 1 Diabetes Genetics Consortium Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet.. 2009;41:703–707. doi: 10.1038/ng.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Astle WJ, Elding H, Jiang T, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schumacher FR, Schmit SL, Jiao S, et al. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat Commun.. 2015;6:7138. doi: 10.1038/ncomms8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fortney K, Dobriban E, Garagnani P, et al. Genome-wide scan informed by age-related disease identifies loci for exceptional human longevity. PLoS Genet.. 2015;11:e1005728. doi: 10.1371/journal.pgen.1005728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Joshi PK, Pirastu N, Kentistou KA, et al. Genome-wide meta-analysis associates HLA-DQA1/DRB1 and LPA and lifestyle factors with human longevity. Nat Commun.. 2017;8:910. doi: 10.1038/s41467-017-00934-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature [Internet]. 2012Jan 12 [cited 2019 Jan 29]; 481:157–63. Available from: http://www.nature.com/articles/nature10725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schlesinger M, Bendas G. Vascular cell adhesion molecule-1 (VCAM-1)–an increasing insight into its role in tumorigenicity and metastasis. Int J Cancer.. 2015;136:2504–2514. doi: 10.1002/ijc.28927 [DOI] [PubMed] [Google Scholar]
- 34. Flister MJ, Hoffman MJ, Lemke A, Prisco SZ, Rudemiller N, O’Meara CC, et al. SH2B3 is a genetic determinant of cardiac inflammation and fibrosis. Circ Cardiovasc Genet [Internet]. 2015April [cited 2019 Jan 29]; 8:294–304. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25628389 [DOI] [PubMed] [Google Scholar]
- 35. Campisi J. Aging, tumor suppression and cancer: high wire-act! Mech Ageing Dev.. 2005;126:51–58. doi: 10.1016/j.mad.2004.09.024 [DOI] [PubMed] [Google Scholar]
- 36. Slack C, Werz C, Wieser D, et al. Regulation of lifespan, metabolism, and stress responses by the Drosophila SH2B protein, Lnk. PLoS Genet.. 2010;6:e1000881. doi: 10.1371/journal.pgen.1000881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Desbuquois B, Carré N, Burnol AF. Regulation of insulin and type 1 insulin-like growth factor signaling and action by the Grb10/14 and SH2B1/B2 adaptor proteins [Internet]. FEBS J.. 2013. [cited 2019 Jan 15];280:794–816. Available from: http://doi.wiley.com/10.1111/febs.12080 [DOI] [PubMed] [Google Scholar]
- 38. Fry A, Littlejohns TJ, Sudlow C, et al. Comparison of sociodemographic and health-related characteristics of UK biobank participants with those of the general population. Am J Epidemiol.. 2017;186:1026–1034. doi: 10.1093/aje/kwx246 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.