Since our eyes were opened to the microscopic world centuries ago, microbiologists have been dazzled by the incredible diversity of shapes and sizes adopted by bacteria (1). Given this dizzying morphological potential, it is remarkable that most species gravitate to a particular shape. It is tempting to speculate that the “one species, one form” paradigm reflects the selective benefits of the chosen shape in the natural environment of each species. But how is cell shape determined? And does a given shape truly confer a fitness advantage? Although these questions are often asked, definitive answers have remained elusive. Vibrio cholerae, the pathogen responsible for the diarrheal disease cholera, typically adopts a characteristic comma (curved) shape. In PNAS, Fernandez et al. (2) report that cellular curvature in V. cholerae is regulated by the second messenger cyclic dimeric guanosine monophosphate (c-di-GMP), whose levels are known to regulate cell motility (3) and biofilm formation (4). Fernandez et al. further suggest that c-di-GMP regulation of cell shape may provide an advantage during environmental transitions in which motility and formation of a sessile biofilm are alternately beneficial (4).
Unlike most model bacteria that are not typically considered shape shifters, the typical shape of V. cholerae cells changes over the course of a batch culture. In 1928, Arthur Henrici (5) published a tour-de-force quantification of the shapes of thousands of V. cholerae cells sampled from a culture as it grew from lag phase to exponential phase and then to stationary phase; the population transited from approximately straight cells at low density to highly curved cells at high density. A recent study uncovered the molecular driver of V. cholerae cell curvature: the intermediate filament-like protein CrvA introduces curvature by decreasing the rate of peptidoglycan insertion on one side of the cell (6). Mutants lacking crvA grow as straight rods, migrate less far in soft agar, and are less virulent in animal models (6). CrvA likely underlies the mechanism by which V. cholerae curves at high cell density, but any regulatory pathways that actuate this shift, as well as the fitness advantage of altering curvature, have remained unknown. Fernandez et al. (2) close this gap by determining that c-di-GMP, whose levels are controlled by the density-dependent quorum-sensing circuitry (7), promotes cell straightening by activating the transcription factors VpsR and VpsT and posttranscriptionally regulating CrvA messenger RNA (mRNA) and protein levels (2). Intriguingly, c-di-GMP is known to repress motility, and VpsR and VpsT induce synthesis of the major component of the V. cholerae biofilm matrix (3, 4). Consistent with these opposing roles, Fernandez et al. (2) find that curved cells are faster swimmers, while straighter cells form more robust biofilms.
Given the widespread presence of c-di-GMP signaling across the bacterial kingdom, these findings potentially have implications beyond V. cholerae. High levels of c-di-GMP have recently been shown to regulate biofilm formation in several other bacteria, including the major human pathogen Pseudomonas aeruginosa (3, 8). While the connection between c-di-GMP signaling and cell shape remains to be established beyond V. cholerae, many species are known to change size as a function of cell density and growth rate (9). The work of Fernandez et al. (2) arose from a fortuitous observation that V. cholerae mutant cells with high levels of c-di-GMP more frequently adopt straight, rod-shaped morphology; treatment of other species with exogenous c-di-GMP may reveal a menagerie of shape consequences large and small. Moreover, morphological screens of mutant libraries (10, 11), particularly in curved bacteria, may firmly establish the role of c-di-GMP in shape adaptation, just as deletion of vpsR or vpsT abolished the c-di-GMP−mediated decrease in curvature in V. cholerae (2).
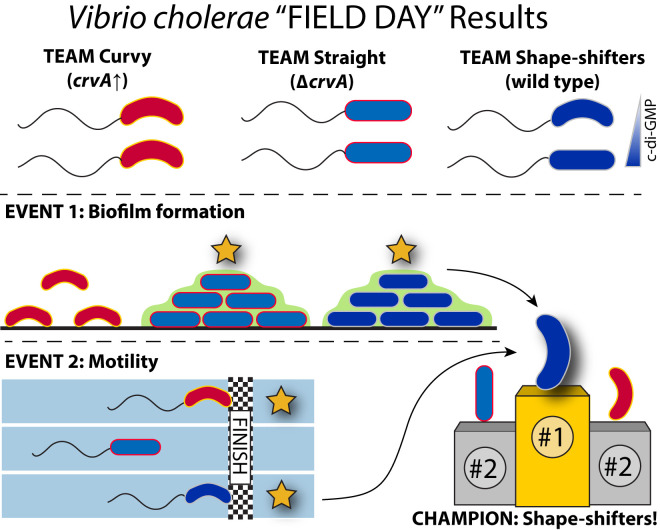
To establish the functional connections between motility and biofilm formation, Fernandez et al. (2) locked V. cholerae cells into straight or curved morphologies by manipulating CrvA levels. When cells were constitutively curved, biofilm mass was reduced, but straight cells swam more slowly, indicating a direct competition between the two phenotypes that suggests a fitness trade-off between straightness and curvature, depending on the environmental context (Fig. 1). The extent to which this fitness trade-off occurs in the wild and in animal hosts remains to be established, but it seems likely that shape modification has been under evolutionary pressure, given the importance of motility and biofilm formation in V. cholerae’s natural habitats.
Fig. 1.
Regulation of cell curvature by c-di-GMP allows V. cholerae to optimize both biofilm formation and motility. Fernandez et al. (2) find that elevated levels of c-di-GMP at high cell density promote straightening of V. cholerae cells through regulation of the transcription factors VpsR and VpsT. Constitutively curved or straight mutants were hampered in biofilm formation or motility, respectively. The ability of wild-type cells to shift their shape from straight to curved depending on environmental conditions may provide an evolutionary advantage for surviving environmental transitions.
Like most studies exploring the links between environment and fitness, the “why” questions remain difficult to answer. Laboratory experiments involving cocultures of wild-type cells and constitutively straight or curved mutants could establish the magnitude of fitness differences due to cell curvature across environments, and long-term laboratory evolution experiments may reveal whether the ability to shape-shift is selected against when the environment is kept constant. Perhaps the most important outstanding question is whether shape-shifting gives V. cholerae an advantage in any of its native environments, which range from chitin-containing shells in brackish water or saltwater, to fresh water sources, to infected human hosts. Given the straightforward nature of quantifying cell shape via imaging, it will be intriguing to probe the cell-density dependence of cell shape in other Vibrio species, for instance, during the quorum sensing-dependent behaviors of Vibrio fischeri when colonizing the Hawaiian bobtail squid. The future discovery of how c-di-GMP posttranscriptionally regulates CrvA mRNA and protein levels could also facilitate comparisons of genomes across currently uncultured bacteria to rapidly determine the prevalence and evolutionary history of shape shifting.
Importantly, Fernandez et al.’s (2) findings for V. cholerae build on several recent studies linking shape to fitness. One investigation demonstrated that rod-shaped Escherichia coli cells colonize basal surface layers better than spherical cells within a colony (12). Another study reported that curved Caulobacter crescentus cells are more able to adhere to surfaces under conditions of high flow (13). A comparative study across motile curved bacteria with a wide range of cell shapes suggested shape-based fitness trade-offs among swimming, chemosensing, and cell growth (14). As we understand more about the advantages of certain morphologies across organisms, the age-old idea that cell shape has been a driver of selection on evolutionary time scales—particularly via motility (15)—gains more credibility. Similar to a quote usually attributed to Oscar Wilde (“I spent all morning putting in a comma, and all afternoon taking it out again”), V. cholerae puts in the work to change its shape in the hope that it will help to survive as motile cells or in biofilms, depending on the changing environment. The work of Fernandez et al. punctuates the flexibility of this remarkable organism.
Acknowledgments
We acknowledge support from NIH RM1 Award GM135102. K.C.H. is a Chan Zuckerberg Biohub Investigator.
Footnotes
The authors declare no competing interest.
See companion article, “Vibrio cholerae adapts to sessile and motile lifestyles by cyclic di-GMP regulation of cell shape,” 10.1073/pnas.2010199117.
References
- 1.Young K. D., The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70, 660–703 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez N. L., et al. , Vibrio cholerae adapts to sessile and motile lifestyles by cyclic di-GMP regulation of cell shape. Proc. Natl. Acad. Sci. U.S.A. 117, 29046–29054 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conner J. G., Zamorano-Sánchez D., Park J. H., Sondermann H., Yildiz F. H., The ins and outs of cyclic di-GMP signaling in Vibrio cholerae. Curr. Opin. Microbiol. 36, 20–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krasteva P. V., et al. , Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327, 866–868 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrici A. T., Morphologic variation and the rate of growth of bacteria. Am. J. Public Health Nations Health 19, 1068–1069 (1929). [Google Scholar]
- 6.Bartlett T. M., et al. , A periplasmic polymer curves Vibrio cholerae and promotes pathogenesis. Cell 168, 172–185.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waters C. M., Lu W., Rabinowitz J. D., Bassler B. L., Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190, 2527–2536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valentini M., Filloux A., Biofilms and cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 291, 12547–12555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaechter M., Maaloe O., Kjeldgaard N. O., Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J. Gen. Microbiol. 19, 592–606 (1958). [DOI] [PubMed] [Google Scholar]
- 10.Ursell T., et al. , Rapid, precise quantification of bacterial cellular dimensions across a genomic-scale knockout library. BMC Biol. 15, 17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campos M., et al. , Genomewide phenotypic analysis of growth, cell morphogenesis, and cell cycle events in Escherichia coli. Mol. Syst. Biol. 14, e7573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith W. P., et al. , Cell morphology drives spatial patterning in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 114, E280–E286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Persat A., Stone H. A., Gitai Z., The curved shape of Caulobacter crescentus enhances surface colonization in flow. Nat. Commun. 5, 3824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuech R., Hoehfurtner T., Smith D. J., Humphries S., Motile curved bacteria are Pareto-optimal. Proc. Natl. Acad. Sci. U.S.A. 116, 14440–14447 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pijper A., Shape and motility of bacteria. J. Pathol. Bacteriol. 58, 325–342 (1946). [DOI] [PubMed] [Google Scholar]