Abstract
Posttraumatic stress disorder (PTSD) is characterized by intrusive thoughts, avoidance, negative alterations in cognitions and mood, and arousal symptoms that adversely affect mental and physical health. Recent evidence links changes in DNA methylation of CpG cites to PTSD. Since clusters of proximal CpGs share similar methylation signatures, identification of PTSD-associated differentially methylated regions (DMRs) may elucidate the pathways defining differential risk and resilience of PTSD. Here we aimed to identify epigenetic differences associated with PTSD. DNA methylation data profiled from blood samples using the MethylationEPIC BeadChip were used to perform a DMR analysis in 187 PTSD cases and 367 trauma-exposed controls from the Grady Trauma Project (GTP). DMRs were assessed with R package bumphunter. We identified two regions that associate with PTSD after multiple test correction. These regions were in the gene body of HLA-DPB1 and in the promoter of SPATC1L. The DMR in HLA-DPB1 was associated with PTSD in an independent cohort. Both DMRs included CpGs whose methylation associated with nearby sequence variation (meQTL) and that associated with expression of their respective genes (eQTM). This study supports an emerging literature linking PTSD risk to genetic and epigenetic variation in the HLA region.
Keywords: EWAS, epigenetics, DNA methylation, eQTM, meQTL
1. Introduction
Posttraumatic stress disorder (PTSD) is a debilitating psychiatric disorder characterized by re-experiencing, avoidance and hyperarousal symptoms that cause negative alterations in cognition, mood and physiologic health (American Psychiatric Association, 2013). Among other psychiatric disorders, PTSD is unique as it requires a trauma exposure to develop. Although 60 to 90% of the population (Benjet et al., 2016; Breslau et al., 1998; Kessler et al., 1995) are exposed to at least one traumatic event during their lifespan, the prevalence of PTSD is 46.2% in highly traumatized US population (Gillespie et al., 2009) and only 3.9% globally (Koenen et al., 2017). This notable difference in prevalence between trauma exposure and PTSD development suggests that some individuals are more vulnerable to trauma and are at higher risk of developing PTSD. Recent evidence suggests that this differential susceptibility may arise from distinct epigenetic changes resulting from trauma (Daskalakis et al., 2018; Morrison et al., 2019; Sheerin et al., 2017).
Epigenetic factors – structural modifications to DNA that regulate gene expression without changing the DNA sequence – link environmental circumstances and experiences (e.g. stress or trauma exposure) to biological response. Identification of epigenetic alterations related to PTSD is a crucial step in characterization of the pathways defining differential risk and resilience of PTSD. Multiple reviews summarize the link between trauma exposure, PTSD and epigenetic alterations (Daskalakis et al., 2018; Kim et al., 2018; Vinkers et al., 2015; Zannas et al., 2015).
To date, a number of epigenome-wide association studies (EWAS) conducted on individual cohorts and meta-analyses reported PTSD-associated differentially methylated CpG sites in genes involved in neurotransmission, neural development and immune pathways (Kuan et al., 2017; Mehta et al., 2013; Smith et al., 2011; Smith et al., 2019; Uddin et al., 2010; Uddin et al., 2018). Since clusters of proximal CpGs often share similar methylation profiles (Martorell-Marugan et al., 2019), joint analysis of multiple CpGs in regions, called differentially methylated regions (DMRs), are more robust with respect to statistical power (Michels et al., 2013) and correlate well with gene expression (Docherty et al., 2014). Therefore, identification of PTSD-associated DMRs may advance our understanding of the molecular mechanisms underlying the disorder. For instance, a recent study conducted on 73 traumatized police officers reported consistently higher methylation of a DMR located at the PAX8 gene (Krzyzewska et al., 2018). A longitudinal study investigating the effect of changes in DNA methylation between postdeployment and pre-deployment samples showed PTSD-associated DMRs in multiple genes, including ZFP57 and SPATC1L (Rutten et al., 2018). A follow-up prospective study reported significant changes in DNA methylation at the DMR located in ZFP57, after successful treatment of PTSD (Vinkers et al., 2019). Recently, a meta-analysis of three longitudinal military cohorts identified 12 novel DMRs associated with PTSD, including MAD1L1 and HLA-DPB1, genes that have previously been implicated in PTSD (Snijders et al., 2020). The SNP rs10235664 in MAD1L1 was reported in the PTSD GWAS of the Million Veteran Program (MVP) (Gelernter et al., 2019), and our group recently identified HLA-DPB1*17:01 as a PTSD risk allele (Katrinli et al., 2019).
In the current study, we performed an EWAS to evaluate the association of current PTSD diagnosis with differentially methylated positions (DMPs) and DMRs. To provide functional context to our findings, we leveraged expression quantitative trait methylation (eQTM) and methylation quantitative trait locus (meQTL) data available for the Grady Trauma Project (GTP) cohort, and also tested the association between the significant DMRs and PTSD in an independent study. Identification of these DMRs may advance our understanding of the genes undergoing regulation in those with PTSD and may provide insights into diagnostic or treatment strategies.
2. Methods
2.1. Grady Trauma Project (GTP)
Participants were recruited as part of the GTP, which investigates the influence of genetic and environmental factors on responses to stressful life events in a predominantly African American, urban population of low socioeconomic status (Gillespie et al., 2009). Briefly, participants were approached in waiting rooms of the general medical clinics of a large, urban, public hospital in Atlanta, GA. Subjects willing to participate provided written informed consent and participated in a verbal interview and blood draw. All procedures in this study were approved by the Institutional Review Board of Emory University School of Medicine and the Grady Health Systems Research Oversight Committee.
Participants provided self-reported demographic information including subject age, sex, and ancestry. For the current study, cases were defined as having current PTSD, and controls were exposed to trauma but had no current or lifetime history of the disorder. PTSD diagnosis was mainly assessed by the Clinician-Administered Post-traumatic Stress Disorder Scale (CAPS) for DSM-IV, a structured diagnostic instrument measuring PTSD that has been demonstrated to have excellent psychometric properties (Blake et al., 1995; Weathers et al., 2001). The CAPS provides a diagnostic status of lifetime and current PTSD. Participants were identified as having PTSD if they met DSM-IV PTSD criteria for current PTSD from the CAPS interview. This resulted in 158 PTSD cases and 332 controls. The CAPS was administered on the same day as the blood draw. For 64 (12%) participants with missing CAPS information, the modified PTSD Symptomatic Scale (PSS) was used as a measure of PTSD symptoms and identified an additional 29 PTSD cases and 35 controls (Foa and Tolin, 2000). PSS frequency items were used to obtain a continuous measure, with values of above 20 considered as clinically significant PTSD symptoms (Falsetti et al., 1993). In addition, to be diagnosed with PTSD, individuals needed to have clinically significant symptoms in three clusters, representing intrusive (B), avoidance/numbing (C), and hyperarousal (C) symptoms on the PSS (Jovanovic et al., 2010). The PSS was only used if it occurred within 30 days of the blood draw (mean = 15, median = 14). Using this PTSD diagnosis algorithm, we defined 187 individuals as PTSD cases and 367 as trauma-exposed controls.
Lifetime exposure to traumatic events was assessed using the Traumatic Events Inventory (TEI) (Bradley et al., 2008; Schwartz et al., 2005), which measures lifetime (childhood and adulthood) exposure to trauma such as natural disaster, serious accident or injury, and physical or sexual assault.
2.2. DNA methylation
DNA was extracted from whole blood using the E.Z.N.A. Blood DNA Midi Kit (Omega Bio-tek, Norcross, GA) and interrogated using the MethylationEPIC BeadChip (Illumina) according to manufacturer’s instructions. Raw methylation beta values were determined via GenomeStudio (Illumina). Internal Illumina controls were used to assess the quality of staining, extension, hybridization, bisulfite conversion, and specificity. Samples with probe detection call rates <90% and those with an average intensity value of either <50% of the experiment-wide sample mean or <2,000 arbitrary units (AU) were removed using R package CpGassoc (Barfield et al., 2012). Probes with detection p-values >0.01 set to missing. CpG sites that cross hybridize between autosomes and sex chromosomes were removed (McCartney et al., 2016). A total of 819,380 probes passed QC and were used in subsequent analyses. Single-sample Noob (ssNoob) normalization method implemented in R package minfi was used for dye bias equalization (Fortin et al., 2017). Following normalization, the ComBat procedure in the R package SVA was used to remove chip and positional batch effects, controlling for age and PTSD status (Leek et al., 2012). Because of the heteroskedastic nature of beta values, logit transformed beta values (M-values) were used for subsequent analysis (Du et al., 2010).
2.3. Analysis of differentially methylated positions
Since DNA methylation varies by cell type, cell proportions can be important confounding factors in DNA methylation analysis of whole blood samples (Houseman et al., 2015). The proportions of CD8+T, CD4+T, natural killer (NK), B cells, monocytes (mono) and neutrophils were estimated using publicly available reference data (GSE110554) and Robust Partial Correlation (RPC) method implemented in R package Epidish (Teschendorff et al., 2017). Estimated proportions of CD8+T, CD4+T, NK, B cells and mono were included as covariates in all analyses.
Logit transformed β values (M-values) were modeled by linear regression as a function of PTSD, adjusting for age, CD8+T, CD4+T, NK, B cell, and monocyte cell proportions, and ancestry using first three principal components (PCs) calculated from GWAS. P-values were adjusted for multiple-testing using the Benjamini-Hochberg FDR procedure at 5% FDR level (Benjamini and Hochberg, 1995).
2.4. Analysis of differentially methylated regions
Identification of DMRs was conducted using bump-hunting method, implemented in R package minfi (Aryee et al., 2014; Jaffe et al., 2012). Probes separated by at least 1000 bp was used to define clusters. For each probe, we estimated the difference in the average logit of beta values (defined as M-values) between PTSD cases and controls, adjusting for sex, age and first three principal components, and smoothed these estimated differences using loess (Cleveland, 1979). For all data, an empirical distribution was created by performing 1000 bootstrap samples. Candidate regions with estimate M-value difference >0.1 and smoothed estimated differences >95% quantile of the empirical distribution were selected. Consistent with the approach of Rhead et al. (Rhead et al., 2018), regions with family-wise error (FWER) <0.2 were identified as significant PTSD-associated DMRs, as controlling the FWER (the probability of making at least one type I error) is more conservative than controlling the false discovery rate (controls the proportion of type I errors). All positions and regions were in reference to GRCh37/hg19.
2.5. Translational Research Center for TBI and Stress Disorders (TRACTS)
Association between PTSD and DMRs identified in the GTP discovery cohort was also tested in an independent replication cohort, using 445 blood samples (309 cases and 136 controls) interrogated using the MethylationEPIC BeadChip (Illumina) from Translational Research Center for TBI and Stress Disorders (TRACTS) cohort (Logue et al., 2020). PTSD was assessed using CAPS and number of different types of trauma was measured by Traumatic Life Events Questionnaire (TLEQ). Cleaning and processing of the methylation data for TRACTS data has been described in detail elsewhere (Logue et al., 2020), using a consortium-based pipeline for processing, similar to that used in the discovery data (Ratanatharathorn et al., 2017).
Bumphunter was used to analyze PTSD in the TRACTS cohort using the same parameters as described above. Analyses included age, sex and cell counts (CD8+T, CD4+T, NK, B cells, and monocytes) as covariates. Any significant region reported in the primary analysis were reported. Based on concerns about the null simulations performed internally by Bumphunter to generate significance values, and to validate the observed evidence of replication, we performed our own null simulations of the DMRs at chr6: 33047185 – 33049505 and chr21: 47604052 – 47605174. As genome-wide null simulations are computationally demanding, we analyzed a reduced set of CpGs in the regions above +/− 50000 bp. First, we tested for association with PTSD as described above in these reduced set of CpGs. Then, we randomly permuted the affection status of TRACTS participants 1000 times, and analyzed the reduced set of CpGs around the two DMRs to generate an empirical p-value distribution of each region under the null. As Bumphunter only evaluates significance of a region when it meets certain criteria, replicates in which Bumphunter did not generate a p-value were recorded as p=1. Our empirical p values for these two regions were then estimated as the percentile of the observed reduced-set p-values in the corresponding region’s null p-value distribution.
2.6. Correlation between genetic effects, gene expression and methylation
Correlation between methylation of CpGs in the DMRs and expression of its annotated gene was evaluated using previously generated expression quantitative trait methylation (eQTM) data from GTP (Kennedy et al., 2018). These eQTMs were identified through analysis of methylation data generated by HumanMethylation450 BeadChip and expression data generated via Illumina HT12v3 and HT12v4 expression BeadChips (Illumina).
To evaluate whether the CpGs in the PTSD-associated DMRs associate by nearby genetic polymorphisms, we have used previously generated meQTL data on 780 subjects from GTP (data not shown). Briefly, 5,971,966 genomic variants (genotyped with Illumina HumanOmni1-Quad, HumanOmniExpress, or Multi-Ethnic Global arrays) were tested for association with 608,245 CpG sites (interrogated with MethylationEPIC BeadChip) using a linear mixed model implemented in matrixeQTL (Shabalin, 2012). The model was adjusted for age, gender, estimated cell proportions and 3 genotype-based principal components (PCs), and treated ethylation (CpG) as the outcome and SNP allele count as an explanatory variable. A stringent Bonferroni threshold of p<8.21E-14 was used to identify meQTLs.
We also tested the association between 24 CpGs in the HLA-DPB1 DMR and HLA-DPB1*17:01 allele, which independently associates with PTSD (Katrinli et al., 2019). To do so, we modeled each CpG as a function of the HLA-DPB1*17:01 allele under a dominant model, adding sex, age and cell proportions as covariates. P-values were adjusted for multiple-testing using a Bonferroni correction for 24 tests.
2.7. Correlation of CpGs in DMRs between blood and brain tissues
The degree of correlation between blood and brain tissue methylation of CpGs in the DMRs were assessed using data from IMAGE-CpG (Iowa Methylation Array Graphing for Experimental Comparison of Peripheral tissue & Gray matter (Brain)) (Braun et al., 2019). This data provides spearman correlation coefficients (rho) and associated p-values for the association of the methylation status of individual CpG sites assayed with MethylationEPIC in blood and the brain.
3. Results
3.1. Demographic characteristics of cohorts
The demographics of the cohorts are presented in Table 1. The GTP cohort (discovery) was predominantly African American (94%) and female (73%). Of 554 participants, 34% had a current PTSD diagnosis, assessed by CAPS and PSS. We observed a moderate correlation between PSS and current CAPS in this dataset (Pearson r = 0.53, p < 2.2E-16). Cases and controls did not differ significantly in terms of age. However, cases were more likely to be female (p = 2.92E-6) and were exposed to more traumatic events as compared to controls (p = 1.10E-15).
Table 1:
Clinical and demographic characteristics of the subjects
| Discovery Study (GTP – EPIC) | Replication Study (TRACTS) | |||||
|---|---|---|---|---|---|---|
| Cases (N=187) | Controls (N=367) | P-value | Cases (N=309) | Controls (N=136) | P-value | |
| Age, mean ± SD | 41.58 ± 11.45 | 42.04 ± 12.86 | 0.668 | 33.10 ± 8.80 | 32.58 ± 9.96 | 0.58 |
| Female (%) | 159 (85.0%) | 245 (66.8%) | 2.92E-06 | 31 (10.03%) | 4 (2.94%) | 0.02 |
| African American, N (%) |
175 (93.6%) | 346 (94.3%) | 1.00 | 25 (8.09%) | 14b (10.37%) | 0.55 |
| Trauma burdena, mean ± SD | 6.94 ± 3.25 | 4.44 ± 2.77 | 1.10E-15 | 2.42 ± 1.93c | 0.89 ± 1.11d | 3.02E-16 |
Abbreviations: PTSD, post-traumatic stress disorder. The p-values were computed from t-test (for Age, PTSD and Trauma burden) and Fisher’s exact test (for sex, ancestry and childhood trauma) comparing PTSD cases to trauma exposed controls.
Based on Traumatic Events Inventory (TEI) measure for GTP and Traumatic Life Events Questionnaire (TLEQ) for TRACTS
Calculated after removing 1 missing data
Calculated after removing 49 missing data
Calculated after removing 32 missing data
The TRACTS cohort (replication), which included 309 cases and 136 controls, was predominantly male (92%) and Caucasian (91%), unlike GTP. However, like GTP, PTSD cases were more likely to be female and had a higher trauma burden relative to controls (Table 1).
3.2. Identification of PTSD-associated DMPs and DMRs
A total of 819,380 CpG sites were evaluated. Of these, 39,597 were nominally associated with PTSD, but no individual DMPs were significant after correction for multiple testing, FDR < 0.05 (Table S1).
Next, we sought to identify DMRs (Lin et al., 2016). In the GTP discovery cohort, 622 potential DMRs with a M-value cut-off of 0.1 were identified and tested for association with PTSD (Table S2). Of these, 110 regions had a p < 0.05, where p is the probability under null distribution, but only two regions remained differentially methylated following correction for multiple comparisons (Table 2). PTSD cases had increased methylation in i) a region (chr6: 33047185 – 33049505) that is annotated to both the promoter of HLA-DPA1 and gene body of HLA-DPB1 and ii) the promoter region of SPATC1L (chr21: 47604052 – 47605174) (Figure 1).
Table 2:
DMRs in discovery cohort with an average M-value difference > 0.1 and FWER < 0.2.
| GTP (Discovery) | TRACTS (Replication) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nearest Gene | Chr | Start | End | Mean ΔM | nCpGs | p-value | Chr | Start | End | Mean ΔM | nCpGs | p-value | Pperm |
| HLA-DPA1, HLA-DPB1 | chr6 | 33047185 | 33049505 | 0.13 | 24 | 2.47E-04 | chr6 | 33047185 | 33050741 | −0.16 | 24 | 2.89E-05* | 0.006 |
| SPATC1L | chr21 | 47604052 | 47605174 | 0.36 | 10 | 6.11E-05 | chr21 | 47604052 | 47605174 | 0.18 | 10 | 0.001 | 0.24 |
Abbreviations: nCpGs, number of CpGs in the DMR; FWER, family-wise error rate; pperm, permutation p-value.
Fig 1: PTSD-associated DMR regions in the discovery cohort.
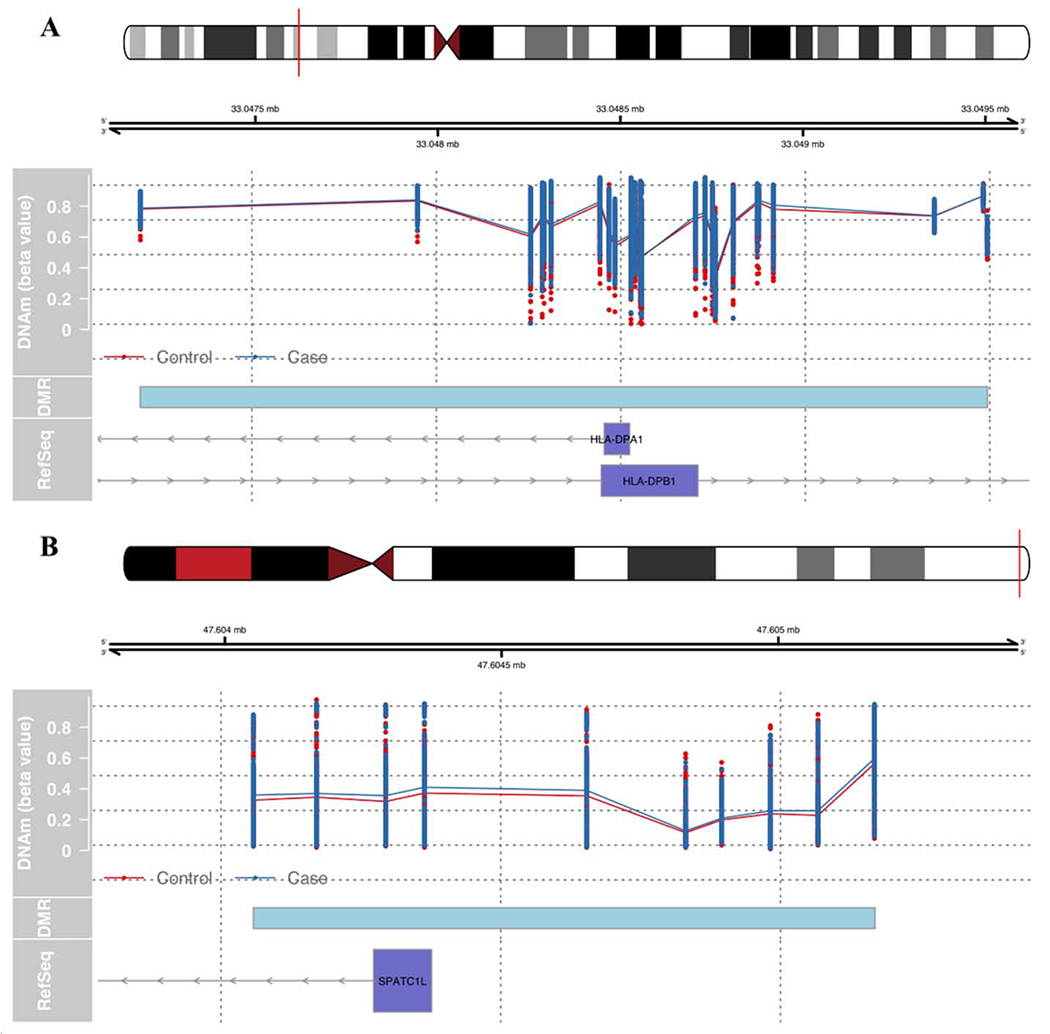
Unadjusted methylation values for the control (red) and PTSD case (blue) groups. Cyan box represents DMR region. The blue axis represents the known RefSeq genes downloaded from UCSC browser: (A) region near HLA-DPA1 and HLA-DPB1L, (B) region near SPATC1.
To evaluate whether DMRs that associated with PTSD in the GTP also associate with PTSD in an independent and demographically different cohort, we performed a DMR analysis in TRACTS cohort. We initially observed nominal associations of the DMRs at chr6: 33047185 – 33050741 (HLA-DPB1 region; p = 2.89E-05) and chr21: 47604052 – 47605174 (SPATC1 region; p = 0.001), but only the DMR at chr6: 33047185 – 33050741 remained associated after generation of an empirical (simulation based) p-value (p = 0.006; Table 2). The HLA-DPB1 DMR identified in TRACTS was wider than that of GTP, and the direction of the effect in TRACTS was opposite of what was observed in GTP, such that the DMR had lower methylation in PTSD cases compared to controls (Mean ΔM = −0.16).
3.3. Gene expression and genetic effects
We evaluated the degree of correlation between methylation of CpGs in DMRs and gene expression, using previously generated eQTM data on GTP cohort. For the 24 CpGs in the DMR that are annotated to both the promoter of HLA-DPA1 and the gene body of HLA-DPB1 we evaluated expression with both genes in GTP. Each of the 18 CpGs in this DMR present on both the HumanMethylation450 array and MethylationEPIC arrays negatively correlated with HLA-DPB1 expression (Table S3) though none correlated with HLA-DPA1 expression. Thus, subsequent analyses focused on HLA-DPB1. Of the eight CpGs in the SPATC1L DMR interrogated on both arrays, each negatively correlated with SPATC1L expression (Table S4).
Next, we investigated genetic-epigenetic correlations in the PTSD-associated DMRs. For the HLA-DPB1 DMR, four of 24 CpGs were associated with 380 unique SNPs (Table S5). It is important to note that the HLA-DPB1*17:01 allele, which independently associated with PTSD (Katrinli et al., 2019), also associated with two CpGs in the HLA-DPB1 DMR (Table S6). For the SPATC1L DMR, methylation of six out of 10 CpGs were associated with 123 unique SNPs (Table S7). None of the meQTL SNPs associated independently with PTSD in the recent Psychiatric Genetics Consortium (PGC) – PTSD meta-analysis (Nievergelt et al., 2019).
3.4. Correlation of DMRs between blood and brain tissues
Using the IMAGE-CpG database, the correlation of CpGs in HLA-DPB1 and SPATC1L DMRs were compared in blood-derived and brain-derived samples (Table S8). The DMR in HLA-DPB1 includes 24 CpGs, but only 21 of those CpGs are available in the IMAGE-CpG database. Out of 21 CpGs tested, five CpGs are correlated between blood and brain tissues (rho > 0.4, p < 0.05). The DMR in SPATC1L includes 10 CpGs, and all CpGs are correlated between blood and brain tissues (rho > 0.88, p < 5e-5).
4. Discussion
Understanding the biological characteristics that distinguish individuals with versus without PTSD is crucial for developing new prevention and intervention strategies following a traumatic event. In the current study, we aimed to identify epigenetic loci associated with PTSD in a predominantly African American cohort of traumatized civilians. We identified two regions, located in HLA-DPB1 and SPATC1L, that were differentially methylated in PTSD. Both DMRs showed increased methylation in PTSD cases compared to trauma-exposed controls in the GTP discovery cohort.
The DMR in HLA-DPB1 supports the increasing evidence of the link between PTSD and immune system dysregulation (Eraly et al., 2014; Michopoulos et al., 2017). Interestingly, HLA-DPB1 region associated with PTSD in a recent longitudinal meta-analysis conducted on three military cohorts (Snijders et al., 2020). This study reported DMRs in HLA-DPB1 body (chr6: 33043976 - 33054001) and CpG island (chr6: 33048416 - 33048814), suggesting that methylation changes throughout HLA locus might be associated with PTSD. However, in this longitudinal study of Snijders et al. (2020), PTSD was associated with decreased HLA-DPB1 DMR methylation. This difference in directions may be based on methodological differences, since their methods reported wider regions than what we identified in our study, or difference in cohort characteristics (e.g. the TRACTS study included military cohorts that are predominantly Caucasian and male, but the GTP cohort consist of predominantly African-American and women civilians). It is important to note that we also observed lower methylation at the HLA-DPB1 DMR in the PTSD cases of TRACTS cohort, which is the opposite of what we observed in GTP. Since our discovery cohort (GTP) is predominantly African American and TRACTS is Caucasian, we hypothesize the opposite direction of association of the DMR in HLA-DPB1 may reflect differences in underlying genetic effects; 96% of the HLA-DPB1 meQTL SNPs differ in minor allele frequencies (MAFs) by more than 40% between those of African versus European decent. The region includes 4 CpGs (out of 24 CpGs) with meQTLs in which the allele frequencies of the underlying SNPs vary by ancestry. For example, the A allele frequency of rs1042117 is 48% in African Americans and 9% in Europeans. Thus, the differences in the direction of association the HLA-DPB1 DMR and PTSD may be attributable to sequence-dependent DNA methylation patterns that are ancestry-specific. Examples of allele-specific methylation, which can vary by ancestry, are well-documented (Hellman and Chess, 2010; Onuchic et al., 2018; Smith et al., 2014; Zhang et al., 2009). Moreover, this DMR may play a role in regulation of HLA-DPB1 gene expression, as 75% of the CpGs in the DMR (18 out of 24) were negatively correlated with HLA-DPB1 expression.
The HLA locus is one of the key elements of the individual’s immunogenetic background, which plays an important role in shaping their response to stress or trauma exposure. HLA-DPB1 belongs to highly polymorphic HLA Class II genes, which are a combination of different SNPs (i.e. haplotypes) that results in functionally distinct proteins. According to the HLA PheWAS database, various HLA alleles associate with neuropsychiatric phenotypes, including substance abuse, schizophrenia, bipolar disorder, anxiety disorder and PTSD (Karnes et al., 2017). In our recent study, we showed that HLA-DPB1*17:01 allele was more frequent in PTSD cases compared to trauma-exposed controls in African Americans (Katrinli et al., 2019). Here, we observed that two CpGs out of 24 CpGs (8%) on HLA-DPB1 DMR were associated with HLA-DPB1*17:01 allele (Bonferroni-adjusted p < 0.05). Interestingly, this PTSD-associated HLA-DPB1*17:01 allele seems to be more prevalent in African Americans, as the HLA-DPB1*17:01 allele frequency is 7 times lower in Caucasians compared to African Americans (0.9% vs 7%, respectively) (Gonzalez-Galarza et al., 2020). Thus, the association between PTSD and the HLA-DPB1 DMR in African Americans may reflect indirect effects of the HLA-DPB1*17:01 allele.
Notably, the DMR in SPATC1L associated with PTSD in a recent longitudinal study in a Caucasian male military cohort (Rutten et al., 2018), where the increased PTSD symptom scores over time were associated with decreased DNA methylation levels at the DMR. The difference in directions between this study and ours may be based on cohort characteristics, since the cohort analyzed in the study of Rutten et al. (2018) was a Caucasian male military cohort, while GTP is a predominantly women and African American civilian cohort, or methodological differences (e.g. Rutten et al. (2018) used continuous PTSD symptom scores and conducted a longitudinal analysis, whereas we used categorical PTSD diagnosis variable and conducted a cross-sectional analysis). It is important to note that SPATC1L DMR did not replicate in TRACTS cohort. This region is under genetic control, with 60% CpGs in the SPATC1L DMR are associated with SNPs, and 78% of those meQTL SNPs differ in MAFs by more than 20% between those of African versus European decent, which again indicating a sequence-dependent allele-specific methylation. Hence, we can speculate that this region may be ancestry specific. We also evaluated if the CpGs in SPATC1L DMR differ by sex, through regressing methylation levels of those CpGs on sex, but we did not find any evidence that this region might be sex-specific. We also demonstrated that SPATC1L DMR has functional implications, as 80% of the CpGs in the DMR (eight out of 10) were negatively correlated with cis gene expression. Therefore, increased methylation on the SPATC1L promoter would be expected to result in decreased SPATC1L expression in PTSD cases.
SPATC1L protects cells from cell death induced by DNA-damaging alkylating agents (Fry et al., 2008), which are byproducts of normal cellular metabolism as well as from oxidative stress and chronic inflammation (Sharma et al., 2017). Increased methylation of SPATC1L promoter in PTSD cases may result in decreased SPATC1L expression, leading to increased sensitivity to DNA alkylating agents (Fry et al., 2008) and cell death, which produces reactive oxygen species. Notably, clinical evidence supports significant differences in blood antioxidant enzyme concentrations and oxidative stress-related gene expression between PTSD cases and controls and supports involvement of oxidative stress in the physiopathology of PTSD (Atli et al., 2016; Borovac Stefanovic et al., 2015; Tylee et al., 2015; Zieker et al., 2007). Moreover, a study conducted on HEK293 cells showed translocation of SPATC1L at cell junctions after neurokinin-2 (NK2) receptor activation, by the neurotransmitter, neurokinin A (NKA) (Lecat et al., 2015). NKA is a member of the tachykinin family of neuropeptides that functions as a neurotransmitter and contributes to inflammation (Onaga, 2014). Animal studies show increased NKA expression following exposure to inflammatory stimuli. For example, administration of bacterial Lipopolysaccharide (LPS) led to increased NKA expression in mouse spinal cord (Bret-Dibat et al., 1994), as well as inflammatory cytokines IL-1 (Kalra et al., 1994) and IL-6 (De Laurentiis et al., 2003) increased NKA expression in rat hypothalamus. Interestingly, a recent study reported increased blood NKA levels in PTSD cases (Oglodek, 2017). In line with the inflammatory physiopathology of PTSD, NK2 receptor antagonists were suggested as novel therapies in the treatment of depression and stress-related disorders. Results from animal trials were promising for NK2 receptor antagonist SR48968 (Saredutant), which has been shown to block NK2 receptors in brain and reduce depressive-like behaviors in mice that undergo forced swimming test, and in guinea pig pups that go through maternal separation (Steinberg et al., 2001). Even though the exact role of SPATC1L in this mechanism is still unknown, it is clear that regulation of SPATC1L expression is important to maintain homeostasis against oxidative stress and inflammation, which are commonly observed in PTSD patients.
Our findings should be considered alongside a number of study limitations. First, our study was limited to CpG sites included on the MethylationEPIC BeadChip, which captures only a fraction of the CpG sites in the genome. Thus, we are likely missing some PTSD-associated CpG sites that can be captured through sequencing methods. However, this array-based technology allowed us to replicate our findings in independent groups of subjects. Second, as this is a cross-sectional study, we cannot ascertain whether the identified epigenetic changes in PTSD cases are a consequence of the disorder or a part of its etiology. Third, our discovery cohort consists of blood samples, which is not the ideal proxy to study psychiatric disorders as brain tissue. However, all CpGs in the SPATC1L DMR and 21% of the CpGs (five out of 24) in the HLA-DPB1 DMR are correlated between blood and brain tissues. Fourth, we used two different PTSD measures (CAPS and PSS), but two measures were moderately correlated in the cohort. Finally, some preliminary simulations by our group reported higher false positive rates while using bumphunter. Hence, we performed null simulations of HLA and SPATC1L to generate emprical p values for replication analysis performed on TRACTS cohort. These simulations supported the association with the HLA-DPB1 DMR, but did not indicate sufficient evidence to conclude that SPATC1L was assocaited in the TRACTS cohort.
Taken together, the findings of the current study support the need for further study of epigenetic regulation of HLA-DPB1 and SPATC1L with regards to PTSD. Longitudinal and functional studies are required to elucidate whether the identified DMRs are consistently and causally associated with PTSD or its treatment.
Supplementary Material
Highlights.
Methylation in HLA-DPB1 and SPATC1L associate with PTSD in African American civilians.
Methylation in HLA-DPB1 replicates in a Caucasian military cohort.
This study links PTSD risk to genetic and epigenetic variation in the HLA region.
5. Acknowledgement
We appreciate the technical support of all of the staff, volunteers and participants from the Grady Trauma Project, supported by the National Institutes of Mental Health (MH096764 to KJR and MH071537 to CFG&KJR). This work was funded by R01MH108826-01, a National Institutes of Mental Health grant to AKS and MWL, I01BX003477, a Department of Veterans Affairs BLR&D grant to MWL, 1R03AG051877, 1R21AG061367-01, and 1I01CX001276-01A2 to EJW, R21MH102834 to MWM, the National Center for PTSD: Behavioral Sciences Division, and the Translational Research Center for TBI and Stress Disorders (TRACTS), a Department of Veterans Affairs Rehabilitation Research and Development (RR&D) Traumatic Brain Injury Center of Excellence (B3001-C) at VA Boston Healthcare System. Genotype and methylation data for the TRACTS Cohort were generated with the support of resources and of facilities at the Pharmacogenomics Analysis Laboratory (Research Service, Central Arkansas Veterans Healthcare System, Little Rock, Arkansas), a core research laboratory funded by the Cooperative Studies Program, Research and Development, Department of Veterans Affairs. The Geneva Foundation contributed to this work and was supported by funds from the Military and Operational Medicine Research Area Directorate III via the US Army Research Office, ARO contracts/grants W911NF-13-1-0376 and W911NF-18-2-0056.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Publisher's Disclaimer: Disclaimer
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70–25.
References
- American Psychiatric Association, 2013. Diagnostic and statistical manual of mental disorders (5th ed.). Arlington, VA. [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA, 2014. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atli A, Bulut M, Bez Y, Kaplan I, Ozdemir PG, Uysal C, Selcuk H, Sir A, 2016. Altered lipid peroxidation markers are related to post-traumatic stress disorder (PTSD) and not trauma itself in earthquake survivors. Eur Arch Psychiatry Clin Neurosci 266, 329–336. [DOI] [PubMed] [Google Scholar]
- Barfield RT, Kilaru V, Smith AK, Conneely KN, 2012. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics 28, 1280–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300. [Google Scholar]
- Benjet C, Bromet E, Karam EG, Kessler RC, McLaughlin KA, Ruscio AM, Shahly V, Stein DJ, Petukhova M, Hill E, Alonso J, Atwoli L, Bunting B, Bruffaerts R, Caldas-de-Almeida JM, de Girolamo G, Florescu S, Gureje O, Huang Y, Lepine JP, Kawakami N, Kovess-Masfety V, Medina-Mora ME, Navarro-Mateu F, Piazza M, Posada-Villa J, Scott KM, Shalev A, Slade T, ten Have M, Torres Y, Viana MC, Zarkov Z, Koenen KC, 2016. The epidemiology of traumatic event exposure worldwide: results from the World Mental Health Survey Consortium. Psychol Med 46, 327–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, Charney DS, Keane TM, 1995. The development of a Clinician-Administered PTSD Scale. J Trauma Stress 8, 75–90. [DOI] [PubMed] [Google Scholar]
- Borovac Stefanovic L, Kalinic D, Mimica N, Beer Ljubic B, Aladrovic J, Mandelsamen Perica M, Curic M, Grosic PF, Delas I, 2015. Oxidative status and the severity of clinical symptoms in patients with post-traumatic stress disorder. Ann Clin Biochem 52, 95–104. [DOI] [PubMed] [Google Scholar]
- Bradley RG, Binder EB, Epstein MP, Tang Y, Nair HP, Liu W, Gillespie CF, Berg T, Evces M, Newport DJ, Stowe ZN, Heim CM, Nemeroff CB, Schwartz A, Cubells JF, Ressler KJ, 2008. Influence of child abuse on adult depression: moderation by the corticotropin-releasing hormone receptor gene. Arch Gen Psychiatry 65, 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun PR, Han S, Hing B, Nagahama Y, Gaul LN, Heinzman JT, Grossbach AJ, Close L, Dlouhy BJ, Howard MA 3rd, Kawasaki H, Potash JB, Shinozaki G, 2019. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl Psychiatry 9, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslau N, Kessler RC, Chilcoat HD, Schultz LR, Davis GC, Andreski P, 1998. Trauma and posttraumatic stress disorder in the community: the 1996 Detroit Area Survey of Trauma. Arch Gen Psychiatry 55, 626–632. [DOI] [PubMed] [Google Scholar]
- Bret-Dibat JL, Kent S, Couraud JY, Creminon C, Dantzer R, 1994. A behaviorally active dose of lipopolysaccharide increases sensory neuropeptides levels in mouse spinal cord. Neurosci Lett 173, 205–209. [DOI] [PubMed] [Google Scholar]
- Cleveland WS, 1979. Robust Locally Weighted Regression and Smoothing Scatterplots. Journal of the American Statistical Association 74, 829–836. [Google Scholar]
- Daskalakis NP, Rijal CM, King C, Huckins LM, Ressler KJ, 2018. Recent Genetics and Epigenetics Approaches to PTSD. Curr Psychiatry Rep 20, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurentiis A, Candolfi M, Pisera D, Seilicovich A, 2003. Effects of lipopolysaccharide on neurokinin A content and release in the hypothalamic-pituitary axis. Regul Pept 111, 91–95. [DOI] [PubMed] [Google Scholar]
- Docherty LE, Rezwan FI, Poole RL, Jagoe H, Lake H, Lockett GA, Arshad H, Wilson DI, Holloway JW, Temple IK, Mackay DJ, 2014. Genome-wide DNA methylation analysis of patients with imprinting disorders identifies differentially methylated regions associated with novel candidate imprinted genes. J Med Genet 51, 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM, 2010. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eraly SA, Nievergelt CM, Maihofer AX, Barkauskas DA, Biswas N, Agorastos A, O’Connor DT, Baker DG, Marine Resiliency Study T, 2014. Assessment of plasma C-reactive protein as a biomarker of posttraumatic stress disorder risk. JAMA Psychiatry 71, 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetti SA, Resnick HS, Resick PA, Kilpatrick DG, 1993. The Modified PTSD Symptom Scale: A brief self-report measure of posttraumatic stress disorder. the Behavior Therapist 16, 161–162. [Google Scholar]
- Foa EB, Tolin DF, 2000. Comparison of the PTSD Symptom Scale-Interview Version and the Clinician-Administered PTSD scale. J Trauma Stress 13, 181–191. [DOI] [PubMed] [Google Scholar]
- Fortin JP, Triche TJ Jr., Hansen KD, 2017. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 33, 558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry RC, Svensson JP, Valiathan C, Wang E, Hogan BJ, Bhattacharya S, Bugni JM, Whittaker CA, Samson LD, 2008. Genomic predictors of interindividual differences in response to DNA damaging agents. Genes Dev 22, 2621–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Sun N, Polimanti R, Pietrzak R, Levey DF, Bryois J, Lu Q, Hu Y, Li B, Radhakrishnan K, Aslan M, Cheung KH, Li Y, Rajeevan N, Sayward F, Harrington K, Chen Q, Cho K, Pyarajan S, Sullivan PF, Quaden R, Shi Y, Hunter-Zinck H, Gaziano JM, Concato J, Zhao H, Stein MB, Department of Veterans Affairs Cooperative Studies, P., Million Veteran P, 2019. Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nat Neurosci 22, 1394–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie CF, Bradley B, Mercer K, Smith AK, Conneely K, Gapen M, Weiss T, Schwartz AC, Cubells JF, Ressler KJ, 2009. Trauma exposure and stress-related disorders in inner city primary care patients. Gen Hosp Psychiatry 31, 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Galarza FF, McCabe A, Santos E, Jones J, Takeshita L, Ortega-Rivera ND, Cid-Pavon GMD, Ramsbottom K, Ghattaoraya G, Alfirevic A, Middleton D, Jones AR, 2020. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res 48, D783–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman A, Chess A, 2010. Extensive sequence-influenced DNA methylation polymorphism in the human genome. Epigenetics Chromatin 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Kim S, Kelsey KT, Wiencke JK, 2015. DNA Methylation in Whole Blood: Uses and Challenges. Curr Environ Health Rep 2, 145–154. [DOI] [PubMed] [Google Scholar]
- Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, Irizarry RA, 2012. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol 41, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic T, Norrholm SD, Blanding NQ, Davis M, Duncan E, Bradley B, Ressler KJ, 2010. Impaired fear inhibition is a biomarker of PTSD but not depression. Depress Anxiety 27, 244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra PS, Dube MG, Kalra SP, 1994. The effects of interleukin 1 beta on the hypothalamic tachykinin, neurokinin A. Brain Res 662, 178–184. [DOI] [PubMed] [Google Scholar]
- Karnes JH, Bastarache L, Shaffer CM, Gaudieri S, Xu Y, Glazer AM, Mosley JD, Zhao S, Raychaudhuri S, Mallal S, Ye Z, Mayer JG, Brilliant MH, Hebbring SJ, Roden DM, Phillips EJ, Denny JC, 2017. Phenome-wide scanning identifies multiple diseases and disease severity phenotypes associated with HLA variants. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katrinli S, Lori A, Kilaru V, Carter S, Powers A, Gillespie CF, Wingo AP, Michopoulos V, Jovanovic T, Ressler KJ, Smith AK, 2019. Association of HLA locus alleles with posttraumatic stress disorder. Brain Behav Immun 81, 655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Goehring GN, Nichols MH, Robins C, Mehta D, Klengel T, Eskin E, Smith AK, Conneely KN, 2018. An integrated -omics analysis of the epigenetic landscape of gene expression in human blood cells. BMC Genomics 19, 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Sonnega A, Bromet E, Hughes M, Nelson CB, 1995. Posttraumatic stress disorder in the National Comorbidity Survey. Arch Gen Psychiatry 52, 1048–1060. [DOI] [PubMed] [Google Scholar]
- Kim GS, Smith AK, Nievergelt CM, Uddin M, 2018. Neuroepigenetics of Post-Traumatic Stress Disorder. Prog Mol Biol Transl Sci 158, 227–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen KC, Ratanatharathorn A, Ng L, McLaughlin KA, Bromet EJ, Stein DJ, Karam EG, Meron Ruscio A, Benjet C, Scott K, Atwoli L, Petukhova M, Lim CCW, Aguilar-Gaxiola S, Al-Hamzawi A, Alonso J, Bunting B, Ciutan M, de Girolamo G, Degenhardt L, Gureje O, Haro JM, Huang Y, Kawakami N, Lee S, Navarro-Mateu F, Pennell BE, Piazza M, Sampson N, Ten Have M, Torres Y, Viana MC, Williams D, Xavier M, Kessler RC, 2017. Posttraumatic stress disorder in the World Mental Health Surveys. Psychol Med 47, 2260–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzewska IM, Ensink JBM, Nawijn L, Mul AN, Koch SB, Venema A, Shankar V, Frijling JL, Veltman DJ, Lindauer RJL, Olff M, Mannens M, van Zuiden M, Henneman P, 2018. Genetic variant in CACNA1C is associated with PTSD in traumatized police officers. Eur J Hum Genet 26, 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan PF, Waszczuk MA, Kotov R, Marsit CJ, Guffanti G, Gonzalez A, Yang X, Koenen K, Bromet E, Luft BJ, 2017. An epigenome-wide DNA methylation study of PTSD and depression in World Trade Center responders. Transl Psychiatry 7, e1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecat S, Matthes HW, Pepperkok R, Simpson JC, Galzi JL, 2015. A Fluorescent Live Imaging Screening Assay Based on Translocation Criteria Identifies Novel Cytoplasmic Proteins Implicated in G Protein-coupled Receptor Signaling Pathways. Mol Cell Proteomics 14, 1385–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD, 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Barton S, Holbrook JD, 2016. How to make DNA methylome wide association studies more powerful. Epigenomics 8, 1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue MW, Miller MW, Wolf EJ, Huber BR, Morrison FG, Zhou Z, Zheng Y, Smith AK, Daskalakis NP, Ratanatharathorn A, Uddin M, Nievergelt CM, Ashley-Koch AE, Baker DG, Beckham JC, Garrett ME, Boks MP, Geuze E, Grant GA, Hauser MA, Kessler RC, Kimbrel NA, Maihofer AX, Marx CE, Qin XJ, Risbrough VB, Rutten BPF, Stein MB, Ursano RJ, Vermetten E, Vinkers CH, Ware EB, Stone A, Schichman SA, McGlinchey RE, Milberg WP, Hayes JP, Verfaellie M, Traumatic Stress Brain Study, G., 2020. An epigenome-wide association study of posttraumatic stress disorder in US veterans implicates several new DNA methylation loci. Clin Epigenetics 12, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorell-Marugan J, Gonzalez-Rumayor V, Carmona-Saez P, 2019. mCSEA: Detecting subtle differentially methylated regions. Bioinformatics. [DOI] [PubMed] [Google Scholar]
- McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL, 2016. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genom Data 9, 22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, Rex-Haffner M, Loeschner A, Gonik M, Mercer KB, Bradley B, Muller-Myhsok B, Ressler KJ, Binder EB, 2013. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A 110, 8302–8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels KB, Binder AM, Dedeurwaerder S, Epstein CB, Greally JM, Gut I, Houseman EA, Izzi B, Kelsey KT, Meissner A, Milosavljevic A, Siegmund KD, Bock C, Irizarry RA, 2013. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods 10, 949–955. [DOI] [PubMed] [Google Scholar]
- Michopoulos V, Powers A, Gillespie CF, Ressler KJ, Jovanovic T, 2017. Inflammation in Fear- and Anxiety-Based Disorders: PTSD, GAD, and Beyond. Neuropsychopharmacology 42, 254–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison FG, Miller MW, Logue MW, Assef M, Wolf EJ, 2019. DNA methylation correlates of PTSD: Recent findings and technical challenges. Prog Neuropsychopharmacol Biol Psychiatry 90, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen CY, Choi KW, Coleman JRI, Dalvie S, Duncan LE, Gelernter J, Levey DF, Logue MW, Polimanti R, Provost AC, Ratanatharathorn A, Stein MB, Torres K, Aiello AE, Almli LM, Amstadter AB, Andersen SB, Andreassen OA, Arbisi PA, Ashley-Koch AE, Austin SB, Avdibegovic E, Babic D, Baekvad-Hansen M, Baker DG, Beckham JC, Bierut LJ, Bisson JI, Boks MP, Bolger EA, Borglum AD, Bradley B, Brashear M, Breen G, Bryant RA, Bustamante AC, Bybjerg-Grauholm J, Calabrese JR, Caldas-de-Almeida JM, Dale AM, Daly MJ, Daskalakis NP, Deckert J, Delahanty DL, Dennis MF, Disner SG, Domschke K, Dzubur-Kulenovic A, Erbes CR, Evans A, Farrer LA, Feeny NC, Flory JD, Forbes D, Franz CE, Galea S, Garrett ME, Gelaye B, Geuze E, Gillespie C, Uka AG, Gordon SD, Guffanti G, Hammamieh R, Harnal S, Hauser MA, Heath AC, Hemmings SMJ, Hougaard DM, Jakovljevic M, Jett M, Johnson EO, Jones I, Jovanovic T, Qin XJ, Junglen AG, Karstoft KI, Kaufman ML, Kessler RC, Khan A, Kimbrel NA, King AP, Koen N, Kranzler HR, Kremen WS, Lawford BR, Lebois LAM, Lewis CE, Linnstaedt SD, Lori A, Lugonja B, Luykx JJ, Lyons MJ, Maples-Keller J, Marmar C, Martin AR, Martin NG, Maurer D, Mavissakalian MR, McFarlane A, McGlinchey RE, McLaughlin KA, McLean SA, McLeay S, Mehta D, Milberg WP, Miller MW, Morey RA, Morris CP, Mors O, Mortensen PB, Neale BM, Nelson EC, Nordentoft M, Norman SB, O’Donnell M, Orcutt HK, Panizzon MS, Peters ES, Peterson AL, Peverill M, Pietrzak RH, Polusny MA, Rice JP, Ripke S, Risbrough VB, Roberts AL, Rothbaum AO, Rothbaum BO, Roy-Byrne P, Ruggiero K, Rung A, Rutten BPF, Saccone NL, Sanchez SE, Schijven D, Seedat S, Seligowski AV, Seng JS, Sheerin CM, Silove D, Smith AK, Smoller JW, Sponheim SR, Stein DJ, Stevens JS, Sumner JA, Teicher MH, Thompson WK, Trapido E, Uddin M, Ursano RJ, van den Heuvel LL, Van Hooff M, Vermetten E, Vinkers CH, Voisey J, Wang Y, Wang Z, Werge T, Williams MA, Williamson DE, Winternitz S, Wolf C, Wolf EJ, Wolff JD, Yehuda R, Young RM, Young KA, Zhao H, Zoellner LA, Liberzon I, Ressler KJ, Haas M, Koenen KC, 2019. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun 10, 4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglodek EA, 2017. Evaluation of ADMA, carbonyl groups, CAT and NKA in depressed patients with and without posttraumatic stress disorder. Pharmacol Rep 69, 730–737. [DOI] [PubMed] [Google Scholar]
- Onaga T, 2014. Tachykinin: recent developments and novel roles in health and disease. Biomol Concepts 5, 225–243. [DOI] [PubMed] [Google Scholar]
- Onuchic V, Lurie E, Carrero I, Pawliczek P, Patel RY, Rozowsky J, Galeev T, Huang Z, Altshuler RC, Zhang Z, Harris RA, Coarfa C, Ashmore L, Bertol JW, Fakhouri WD, Yu F, Kellis M, Gerstein M, Milosavljevic A, 2018. Allele-specific epigenome maps reveal sequence-dependent stochastic switching at regulatory loci. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratanatharathorn A, Boks MP, Maihofer AX, Aiello AE, Amstadter AB, Ashley-Koch AE, Baker DG, Beckham JC, Bromet E, Dennis M, Garrett ME, Geuze E, Guffanti G, Hauser MA, Kilaru V, Kimbrel NA, Koenen KC, Kuan PF, Logue MW, Luft BJ, Miller MW, Mitchell C, Nugent NR, Ressler KJ, Rutten BPF, Stein MB, Vermetten E, Vinkers CH, Youssef NA, Workgroup VAM-AM, Workgroup PPE, Uddin M, Nievergelt CM, Smith AK, 2017. Epigenome-wide association of PTSD from heterogeneous cohorts with a common multi-site analysis pipeline. Am J Med Genet B Neuropsychiatr Genet 174, 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Brorson IS, Berge T, Adams C, Quach H, Moen SM, Berg-Hansen P, Celius EG, Sangurdekar DP, Bronson PG, Lea RA, Burnard S, Maltby VE, Scott RJ, Lechner-Scott J, Harbo HF, Bos SD, Barcellos LF, 2018. Increased DNA methylation of SLFN12 in CD4+ and CD8+ T cells from multiple sclerosis patients. PLoS One 13, e0206511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten BPF, Vermetten E, Vinkers CH, Ursini G, Daskalakis NP, Pishva E, de Nijs L, Houtepen LC, Eijssen L, Jaffe AE, Kenis G, Viechtbauer W, van den Hove D, Schraut KG, Lesch KP, Kleinman JE, Hyde TM, Weinberger DR, Schalkwyk L, Lunnon K, Mill J, Cohen H, Yehuda R, Baker DG, Maihofer AX, Nievergelt CM, Geuze E, Boks MPM, 2018. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Mol Psychiatry 23, 1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz AC, Bradley RL, Sexton M, Sherry A, Ressler KJ, 2005. Posttraumatic stress disorder among African Americans in an inner city mental health clinic. Psychiatr Serv 56, 212–215. [DOI] [PubMed] [Google Scholar]
- Shabalin AA, 2012. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Upton PB, Swenberg J, La D, 2017. Carcinogenic Alkylating Agents. [Google Scholar]
- Sheerin CM, Lind MJ, Bountress KE, Nugent NR, Amstadter AB, 2017. The genetics and epigenetics of PTSD: overview, recent advances, and future directions. Curr Opin Psychol 14,5–11. [DOI] [PubMed] [Google Scholar]
- Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, Bradley B, Tang Y, Gillespie CF, Cubells JF, Ressler KJ, 2011. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet 156B, 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Kilaru V, Kocak M, Almli LM, Mercer KB, Ressler KJ, Tylavsky FA, Conneely KN, 2014. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics 15, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Ratanatharathorn A, Maihofer AX, Naviaux RK, Aiello AE, Amstadter AB, Ashley-Koch AE, Baker DG, Beckham JC, Boks MP, Bromet E, Dennis M, Galea S, Garrett ME, Geuze E, Guffanti G, Hauser MA, Katrinli S, Kilaru V, Kessler RC, Kimbrel NA, Koenen KC, Kuan P-F, Li K, Logue MW, Lori A, Luft BJ, Miller MW, Naviaux JC, Nugent NR, Qin X, Ressler KJ, Risbrough VB, Rutten BPF, Stein MB, Ursano RJ, Vermetten E, Vinkers CH, Wang L, Youssef NA, Uddin M, Nievergelt CM, 2019. Epigenome-wide meta-analysis of PTSD across 10 military and civilian cohorts identifies novel methylation loci. bioRxiv, 585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders C, Maihofer AX, Ratanatharathorn A, Baker DG, Boks MP, Geuze E, Jain S, Kessler RC, Pishva E, Risbrough VB, Stein MB, Ursano RJ, Vermetten E, Vinkers CH, Consortium PPE, Smith AK, Uddin M, Rutten BPF, Nievergelt CM, 2020. Longitudinal epigenome-wide association studies of three male military cohorts reveal multiple CpG sites associated with post-traumatic stress disorder. Clin Epigenetics 12, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg R, Alonso R, Griebel G, Bert L, Jung M, Oury-Donat F, Poncelet M, Gueudet C, Desvignes C, Le Fur G, Soubrie P, 2001. Selective blockade of neurokinin-2 receptors produces antidepressant-like effects associated with reduced corticotropin-releasing factor function. J Pharmacol Exp Ther 299, 449–458. [PubMed] [Google Scholar]
- Teschendorff AE, Breeze CE, Zheng SC, Beck S, 2017. A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinformatics 18, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylee DS, Chandler SD, Nievergelt CM, Liu X, Pazol J, Woelk CH, Lohr JB, Kremen WS, Baker DG, Glatt SJ, Tsuang MT, Marine Resiliency Study I, 2015. Blood-based gene-expression biomarkers of post-traumatic stress disorder among deployed marines: A pilot study. Psychoneuroendocrinology 51, 472–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, Goldmann E, Galea S, 2010. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci U S A 107, 9470–9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Ratanatharathorn A, Armstrong D, Kuan PF, Aiello AE, Bromet EJ, Galea S, Koenen KC, Luft B, Ressler KJ, Wildman DE, Nievergelt CM, Smith A, 2018. Epigenetic meta-analysis across three civilian cohorts identifies NRG1 and HGS as blood-based biomarkers for post-traumatic stress disorder. Epigenomics 10, 1585–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkers CH, Geuze E, van Rooij SJH, Kennis M, Schur RR, Nispeling DM, Smith AK, Nievergelt CM, Uddin M, Rutten BPF, Vermetten E, Boks MP, 2019. Successful treatment of post-traumatic stress disorder reverses DNA methylation marks. Mol Psychiatry. [DOI] [PubMed] [Google Scholar]
- Vinkers CH, Kalafateli AL, Rutten BP, Kas MJ, Kaminsky Z, Turner JD, Boks MP, 2015. Traumatic stress and human DNA methylation: a critical review. Epigenomics 7, 593–608. [DOI] [PubMed] [Google Scholar]
- Weathers FW, Keane TM, Davidson JR, 2001. Clinician-administered PTSD scale: a review of the first ten years of research. Depress Anxiety 13, 132–156. [DOI] [PubMed] [Google Scholar]
- Zannas AS, Provencal N, Binder EB, 2015. Epigenetics of Posttraumatic Stress Disorder: Current Evidence, Challenges, and Future Directions. Biol Psychiatry 78, 327–335. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rohde C, Reinhardt R, Voelcker-Rehage C, Jeltsch A, 2009. Non-imprinted allele-specific DNA methylation on human autosomes. Genome Biol 10, R138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieker J, Zieker D, Jatzko A, Dietzsch J, Nieselt K, Schmitt A, Bertsch T, Fassbender K, Spanagel R, Northoff H, Gebicke-Haerter PJ, 2007. Differential gene expression in peripheral blood of patients suffering from post-traumatic stress disorder. Mol Psychiatry 12, 116–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


