Abstract
The neurotoxic effects of the chemotherapeutic agent bortezomib on dorsal root ganglia sensory neurons are well documented, yet the mechanistic underpinnings that govern these cellular processes remain incompletely understood. In this study, system-wide proteomic changes were identified in human induced pluripotent stem cell-derived sensory neurons (iSNs) exposed to a clinically relevant dose of bortezomib. Label-free mass spectrometry facilitated the identification of approximately 2,800 iSN proteins that exhibited differential levels in the setting of bortezomib. A significant proportion of these proteins affect the cellular processes of microtubule dynamics, cytoskeletal and cytoplasmic organization, and molecular transport, and pathway analysis revealed an enrichment of proteins in signaling pathways attributable to the unfolded protein response and the integrated stress response. Alterations in microtubule-associated proteins suggest a multifaceted relationship exists between bortezomib-induced proteotoxicity and microtubule cytoskeletal architecture, and MAP2 was prioritized as a topmost influential candidate. We observed a significant reduction in the overall levels of MAP2c in somata without discernable changes in neurites. As MAP2 is known to affect cellular processes including axonogenesis, neurite extension and branching, and neurite morphology, its altered levels are suggestive of a prominent role in bortezomib-induced neurotoxicity.
Keywords: bortezomib, neuropathy, neurotoxicity, UPR, ISR, cytoskeletal dynamics, proteomics, sensory neurons, MAP2
1. Introduction
Chemotherapy-induced peripheral neuropathy (CIPN) affects over 1/3 of the patient population undergoing treatment for cancer and its prevalence is anticipated to rise with improvements in survival rates (Shah et al., 2018). Symptoms of CIPN typically begin during the first 2 months of treatment and become more severe throughout its duration. Countermeasures are limited to dose reduction or a discontinuation of chemotherapy treatment, underscoring the need both to better understand the central pathobiology and to improve our repertoire of therapeutic agents (Staff et al., 2017).
The majority of CIPN signs and symptoms arise from damage to dorsal root ganglion (DRG) neurons or their axons in the form of a dying-back neuropathy, seemingly initiated through defects in axon transport, altered mitochondrial function, or altered calcium homeostasis (Staff et al., 2017), (Meregalli, 2015). Although there are different mechanisms by which chemotherapeutic agents cause this effect, it is not entirely clear how proteasome inhibitors such as bortezomib facilitate axonal degeneration. Bortezomib inhibits the activity of the 26S proteasome, which recognizes ubiquinated proteins and selectively targets them for degradation in order to maintain cellular homeostasis. In tumorigenic cells, proteasome inhibitors lead to the accumulation of damaged and unfolded proteins, which ultimately results in cell cycle arrest and the upregulation of pro-apoptotic signaling proteins (Dou and Zonder, 2014). Nontumorigenic cells rely on protein kinase B (Akt) upregulation, the endoplasmic reticulum (ER) stress response, the unfolded protein response (UPR), and autophagy to mitigate bortezomib’s cytotoxic effects, and the degree to which any of these mechanisms are applicable to bortezomib neurotoxicity remains to be established.
Preclinical studies that yielded encouraging results for the prevention of CIPN failed in clinical trials and reinforced the need to refine our understanding of chemotherapeutic response (Fukuda et al., 2017). Towards that end, technological advancements in the stem cell field now allow for us to create iPSCs (induced pluripotent stem cells) from patients, and the directed differentiation of these cells into various tissue-specific subtypes has reliably recapitulated many disease phenotypes as well as drug intolerances that are not as readily detected in rodents or other species.
Due to the multifaceted impact of proteasome inhibition on cellular function, we employed a proteomics study to identify key processes responsible for bortezomib-induced neurotoxicity. We opted to investigate the proteomics signature due the discordance between RNA and protein levels (Wiita et al., 2013), (Rendleman et al., 2018). The study was performed on human iPSC-derived sensory neurons (iSNs) exposed to a clinically relevant dose of bortezomib to create a proteomics dataset that resembles the in vivo proteotoxicity of bortezomib.
2. Material and Methods
2.1. Human iPSC cell culture and maintenance, iSN differentiation, and bortezomib treatment
Three iPSC lines created from the skin biopsies of individuals with no known health concerns were acquired from the Mayo Clinic Biotrust according to IRB approved guidelines. Fibroblasts were reprogrammed using Cytotune-iPS 2.0 (courtesy of Regen Theranostics) and karyotypically normal iPSC clones for each line that passed quality control testing for pluripotency and germ layer differentiation were selected. The lines used in this study were from 2 females (21-year-old, 6BT1; 77-year-old, 1BT1) and one male (18-year-old, 100BT8), and a single iPSC clone was used to represent each iPSC line. iPSCs were maintained on LDEV-free Geltrex in mTeSR™ 1 and manually passaged every 4 days.
The differentiation of iPSCs into sensory neurons followed a well-established approach based on dual-SMAD inhibition (Chambers et al., 2012) with modification as described previously (Ankam et al., 2019). Briefly, iPSCs were dissociated into single cells using Accutase and plated as a monolayer (2 × 105 cells/cm2) in mTeSR™ 1 supplemented with Y-27632. Differentiation was initiated 24h post-plating with a surface density of 80% confluence. The differentiation medium consisted of KSR medium, N2 medium, or a mixture of both. Days 0 – 4 included 100 nM LDN193189 and 10 μM SB431542 in the culture medium, and for days 2 – 10 the medium was supplemented with 3μM CHIR99021, 10μM DAPT, and 5μM SU5402. Eleven days post-induction, neural crest derivatives were dissociated with Accutase and plated in neuronal media for subsequent iSN maturation and maintenance (N2 supplemented with growth factors: 25 ng/ml NGF, BDNF, GDNF, and NT3, 0.5 mM cAMP, and 200 μM L-ascorbic acid). Typical iSN yields were in the range of 60-70%. The dissociation of cells by Accutase consisted of incubation for an appropriate duration (6 min for iPSCs, and up to 30 min for iSN precursors) followed by trituration to dissociate cell aggregates, and centrifugation at 300 x g for 5 min.
We have previously reported an assessment of iSN maturity for the iPSC lines used in this study using RNA profiling, an analysis of methylation patterns throughout the differentiation process, and the electrophysiological behavior at d30 and d60 (Ankam et al 2019). Proteins representative of neuroectoderm (PAX6, DLK1, CUZD1, OTX2, DKK1) or neural crest (SOX10, MSK1, ID2, AP2B, ETS1, FOXO3, NGN1) were not detected by LC-MS/MS in the samples generated for this study. Proteins indicative of neural progenitors and nociceptors were detectable, and the total protein abundance based on total protein intensities are presented in Supplemental Figure 1. Levels of ion channels, cell surface receptors, and synaptic vesicle proteins are suggestive of a functional nociceptor phenotype.
Bortezomib (PS-341) was dissolved in DMSO for a stock concentration of 3.5mg/ml and diluted in neuronal media to a final concentration of 100nM for all experiments. The amount of DMSO vehicle in the media was 0.0001% for bortezomib-treated iSNs and DMSO controls. An equivalent volume of neuronal media was used for the blank controls.
A complete list of materials and reagents used in this study is located in Supplemental Table 1.
2.2. Study design and sample generation
iSNs were allowed to mature to day 21 prior to the exposure of a clinically relevant dose of bortezomib based on blood plasma levels (Staff et al., 2013), (Judge et al., 2017), (Geisler et al., 2019), (Karademir et al., 2018). Since proteasome inhibitors are known to prolong the half-lives of short-lived proteins (Hakim et al., 2016), a timeframe of 48h was chosen for the duration of exposure. Blank and DMSO controls were included as a baseline for comparison. Samples were plated in triplicate for each condition and 4 experiments were completed for each iPSC line. At the time of harvest, technical replicates were combined into a single representative sample prior to cell lysate extraction. Cells were dissociated using Accutase and washed in PBS containing protease and phosphatase inhibitors, followed by centrifugation at 200 x g for 5 minutes. Cell pellets were flash frozen and stored at −80°C.
2.3. Proteomics methodology & bioinformatic analyses
A label-free shotgun approach guided subsequent sample preparation and has been described previously (Ayers-Ringler et al., 2016). Cell pellets were lysed in buffer comprised of 1.2% SDS, 50 mM Tris, pH 8.2, HALT Protease and Phosphatase Inhibitors, and Benzonase Nuclease, and then cleared by centrifugation at 10,000 x g for 5 minutes. Protein concentrations were measured by BCA assay. 20 ug protein from each sample was denatured and reduced with 2x Laemmli buffer containing 5% βME for 10 minutes at 85°C. Proteins were separated by SDS-PAGE (10.5-14% Criterion Tris-HCl gel, Bio-rad) and visualized with Bio-Safe Coomassie Brilliant Blue G-250 stain (Bio-rad). Samples were subdivided into 7 different fractions that represented different ranges of molecular weights. Proteins were digested into peptide fragments using trypsin prior to LC-MS/MS analysis, and MaxQuant software (version 1.5.2) was used in the identification and quantification of peptide fragments and proteins (Zhang et al., 2013), (Tyanova et al., 2016). For groups of proteins indistinguishable by LC-MS/MS, a minimum of two identifying peptides and a false discovery rate (FDR) of < 1% was required for inclusion in the differential protein expression profile. Protein level changes were based on quantifying differences in protein abundance. Protein intensities were log-transformed and normalized for batch effects. The normalization of protein intensities was modeled using a Gaussian-linked generalized linear model that used study parameters (collection time and experimental group) as independent variables. A separate background model was generated based on collection time as independent variable. An ANOVA test was used to detect significant changes in protein abundances between groups. Differential expression p-values were FDR corrected using Benjamini-Hochberg-Yekutieli procedure. The complete list of protein groups is included in Supplemental Table 2. Protein groups with an adjusted p < 0.05 and an absolute log2 fold change ≥ 0.5 were considered significantly differentially expressed. DMSO exposure appeared to have minimal effects on the levels of most proteins, with 33 proteins that met significance thresholds with 0.05 < p < 0.001 (Supplemental Table 3). The complete list of proteins was analyzed by Ingenuity Pathway Analysis (IPA) using default settings for appropriate cell and tissue sources associated with the nervous system (QIAGEN Redwood City, CA, USA; www.qiagen.com/ingenuity) (Kramer et al., 2014). Additional analysis of gene lists created by IPA was completed using the ClueGO application (version 2.5.4) within Cytoscape (version 3.7.1) (Mlecnik et al., 2018).
2.4. Microscopy & immunofluorescence staining
2.4.1. Immunocytochemistry:
Cells were fixed with 4% paraformaldehyde for 20 minutes at room temperature, permeabilized using 0.5% Triton X-100 for 15 minutes, incubated in SuperBlock Blocking Buffer for 2-4h, and stained overnight with primary antibodies. Antibody information is listed in Supplemental Table 1. The manufacturers’ recommendations were followed for secondary (Alexa Fluor) antibody and DAPI staining. Cells were mounted with ProLong Gold antifade mountant.
2.4.2. Mitochondrial tracking
Experimental:
iSNs plated on Geltrex coated glass bottom dishes were treated with bortezomib or a DMSO vehicle for 24h, and then stained with tetramethylrhodamine methyl ester (TMRM, 20nM) for 30min prior to image acquisition. Each video consisted of 200 frames that were taken at intervals of 1.2s. Each frame was 212.5μm x 212.5μm with a resolution of 2.4 pixels/um. In each experiment, 2 – 3 plates were prepared per condition, and 5 – 7 videos were acquired per plate, all at 37°C. Three independent experiments were completed.
Analysis:
Videos were imported into Fiji (Schindelin et al., 2012) and edited to exclude mitochondria contained outside of iSN neurites. Mitochondrial mobility was analyzed using the automated particle tracker TrackMate with an extension for the inclusion of additional track parameters (Tinevez et al., 2017). The Laplacian of Gaussian (LoG) detector was used to identify mitochondria according to a blob diameter of 3 microns, and the simple LAP (Linear Assignment Problem) algorithm was selected to model track parameters associated with each mitochondrion. Tracks < 5s in duration were eliminated prior subsequent analysis in R.
Analysis of track parameters for mobile mitochondria:
In order to determine the effects of bortezomib on the mitochondria that exhibit characteristics of axonal transport, mitochondria were subdivided into mobile and stationary populations. 15 videos collected for all three iSN lines in which > 98% mitochondria appeared to be stationary on the basis of displacement were used to determine thresholds that appropriately define the stationary fraction. The independent distance-related parameters of confinement ratio and maximum distance traveled provided the most accurate stratification between both populations (Supplementary Figure 2A). The confinement ratio (CR) is defined as the distance traveled from the initial position divided by the total distance traveled. The maximum distance traveled is the furthest distance traveled from the initial position throughout the duration of video acquisition. The mobile fraction of mitochondria was defined by a single threshold of CR > 0.25, and the track parameters for these mitochondria were compared to assess the dose-dependent effects of bortezomib. Analysis of the subset of mitochondria with CR > 0.5, and presumably more representative of axonal transport, indicated a greater reduction in the fraction of mitochondria at lower doses of bortezomib without significant effects on displacement or velocity (Supplemental figure 2C, 2D).
2.4.3. Analysis of MAP2, Tuj-1, and Rab6 staining:
fluorescence intensities for MAP2 and neuronal specific Tuj-1 were determined by encircling whole cell bodies to obtain the WCB measurement and the interior portion of cell bodies for the ICB measurement using Zen 2.0 (blue edition) (Carl Zeiss AG). The interior portion was defined as the region of distinctively lower MAP2 and Tuj-1 fluorescence intensities in bortezomib-treated iSNs. The width between the ICB and WCB was approximately 1μm, and this measurement was used to define the corresponding ICB in control iSNs. Rab6 fluorescence intensities were based on the measurements determined for the ICB. Rab6 fluorescence intensities in the region exterior to the ICB but within the WCB were negligible.
2.4.4. Microscopy:
All immunofluorescent images were acquired on a Zeiss LSM 780 laser scanning confocal microscope using a C-Apochromat 40x/1.20 W Korr FCS M27 water immersion objective (Carl Zeiss (Oberkochen, Germany)). Images were processed and analyzed for immunofluorescent intensities with ZEN software (Carl Zeiss AG, Version ZEN black 2.3, Oberkochen, Germany). For live cell imaging, cells were incubated at 37°C and 5% CO2 in an LCI microscope top-stage incubation system (Live Cell Instrument, Model Chamlide TC-W system, CU-501 controller, and Model FC-9 (O2/CO2/N2 mixer), Seoul, Korea). Laser power was set to 1-2% to minimize photobleaching.
2.5. Westerns
Quantification of specific proteins in cell lysates was determined by a Simple Western immunoassay using a Wes system and chemiluminescent detection (ProteinSimple, San Jose, California). Approximately 3ug cell lysate was loaded per capillary. Antibodies and other materials and reagents used for these assays are included those in Supplementary Table 1. Assays were visualized and analyzed in Compass software (version 3.1.7). Protein abundance was determined from the signal intensity (peak area). Unless indicated otherwise, GAPDH was used for the normalization of protein levels within each sample. Discrepancies in protein mobility in blot images arose from incompatibilities between the protein lysis and assay buffers and were accentuated in assays for the 2-40kDa size range.
2.6. Statistical analyses (R)
Statistical analyses were completed using R (version 3.6.1) (www.R-project.org). Statistical significance was determined from a Student's t-test for pairwise comparisons. An ANOVA test was used to detect significant differences between groups. Tukey's multiple comparison test was used to determine significance among the conditions tested for the mitochondrial tracking experiments. P < 0.05 was considered significant.
3. Results
3.1. Bortezomib induces neurotoxic effects in iSNs
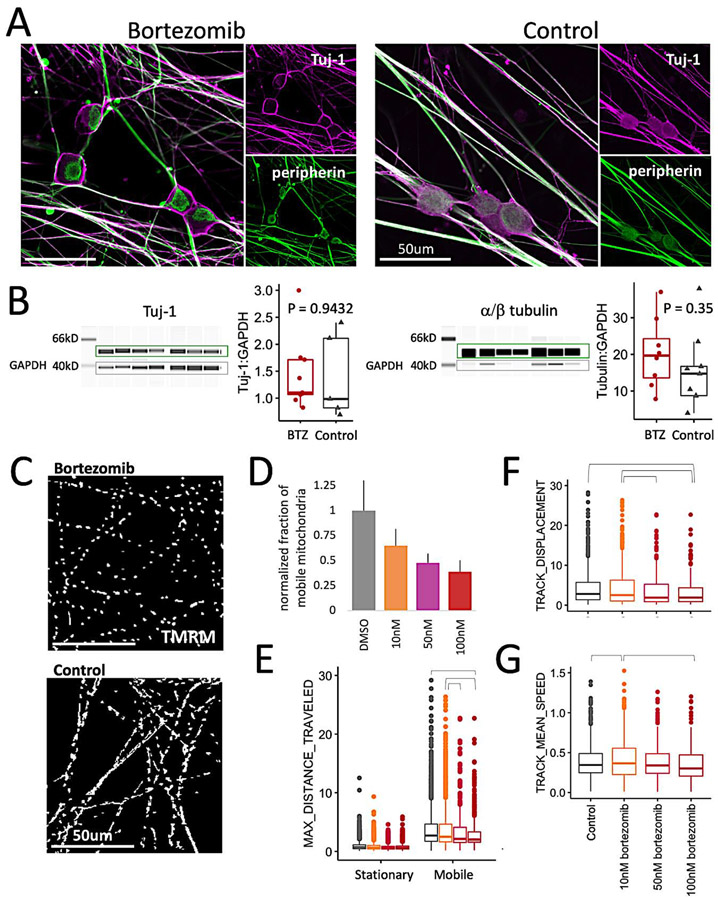
To ensure that the dose and duration of bortezomib exposure was appropriate for this model system, we compared the iSN model to our previous studies in rat E15 DRG neurons. A 24h bortezomib exposure led to the accumulation of β3-tubulin (Tuj-1) proximal to the plasma membrane in the somata of rat E15 DRG neurons (Staff et al., 2013). This same effect was observed in iSNs at both 24h and 48h (Figure 1A). Another similarity shared by both models was that bortezomib exposure had no effect on Tuj-1 protein levels (Figure 1B), nor on overall tubulin levels (Figure 1C). In the iSN model, we also observed an alteration in peripherin localization. While peripherin appears diffuse throughout the soma in control iSNs, in bortezomib, peripherin appears to be compartmentalized within the cytoplasm (Figure 1A).
Figure 1: Bortezomib causes neurotoxicity in iSNs.
a) iSNs exposed to 100nM bortezomib (BTZ) or DMSO for 24h and stained for peripherin and Tuj-1. Scale bar is 50μm.
b) Western blots for Tuj-1 and total α- and β-tubulin (α/β tubulin) for iSNs exposed to bortezomib or DMSO for 48h. Samples represent pooled technical replicates obtained from different experiments using iSNs obtained from 3 different iPSC lines. Intensities for tubulin were normalized to GAPDH levels to determine statistical significance between both conditions. Quantitative measurements were based on band intensities measured in the boxed regions. Significance was determined using a Student’s t-test.
c) Axonal transport of mitochondria was impaired after 24h of bortezomib exposure. Kymograph comparison of TMRM-stained mitochondria in neurites for 300s based on iSN exposure to bortezomib or DMSO.
d) Dose dependence of bortezomib on the fraction of mobile mitochondria. Data shown for panels (d) through (g) were pooled from 3 independent experiments using iSNs obtained from 2 different iPSC lines. The fraction of mobile mitochondria was defined by total distance traveled > 2.5μm from the initial position. Mitochondria that traveled < 2.5μm were designated as the stationary fraction. Significance for all dose-dependent parameters was determined using a Tukey pairwise comparison that corrects for multiple comparisons. All treated conditions were significantly lower than the control condition, and the difference between 10nM and 100nM bortezomib was significant.
e) Dose dependence of the maximum distance traveled by the stationary and mobile populations. Brackets indicate statistically significant comparisons (control vs 100nM, p = 0.036; 10nM vs 100nM, p = 0.003; 10nM vs 50nM, p = 0.02).
f) Dose dependence of the distance traveled by the mobile mitochondrial populations. Brackets indicate statistically significant comparisons (control vs 100nM, p = 0.046; 10nM vs 100nM, p = 0.004; 10nM vs 50nM, p = 0.018).
g) Dose dependence of mean speed velocities of the mobile fraction of mitochondria. Brackets indicate statistically significant comparisons (control vs 10nM, p = 0.006; 10nM vs 100nM, p = 0.001).
Due to mitochondrial axonal transport disruption in the rat model of bortezomib-induced neurotoxicity (Staff et al., 2013), we explored this effect in the iSN model. Here, iSNs were exposed to bortezomib for 24h prior to live cell imaging of TMRM-stained mitochondria. To capture general changes in the bulk population of mitochondria, videos were analyzed using an automated tracking algorithm that provided measurements of velocities and distances traveled (Tinevez et al., 2017). In control conditions < 30% of axonal mitochondria were mobile (Smith and Gallo, 2018), and the use of a distance-based criterion selected for a mobile population of 12% that was further reduced in bortezomib-treated iSNs (Figure 1C-D). Within this mobile population, distance-related parameters of track displacement and maximum distance traveled were inversely proportional to bortezomib dose (Figure 1E-F), but velocity parameters of mean speed, median speed, and max speed were not substantially different (Figure 1G, Supplemental figure 2B). Altogether, the changes in mitochondrial transport and morphological characteristics in a 24-48h timeframe affirm bortezomib-induced iSN neurotoxicity, and changes in the proteome at this stage are sufficient to provide further mechanistic insights.
3.2. Bortezomib toxicity causes system-wide changes in the proteome
Overview of changes to the proteome
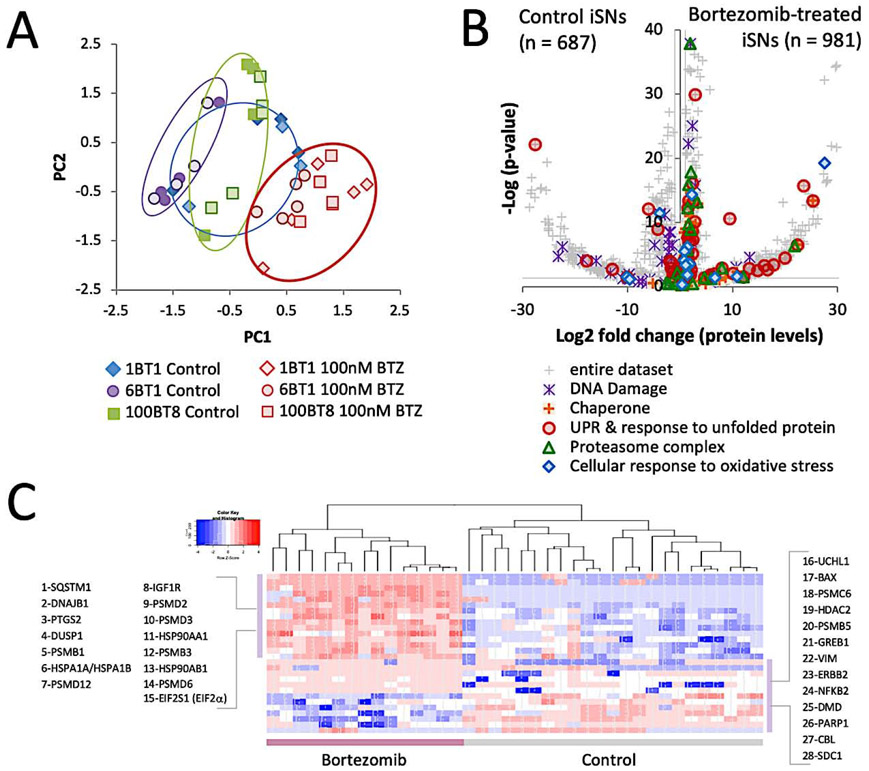
The experimental framework was based on the premise that changes to the proteome would enable the identification of key processes responsible for bortezomib-induced neurotoxicity and the proteins that facilitate axonal degeneration. Approximately 2800 proteins were identified and the global compositional changes of proteins allowed for the unbiased clustering of samples into two distinct categories according to bortezomib exposure (Figure 2A). Exposure of iSNs to the DMSO vehicle had no clear effect on the protein profile since the DMSO controls were intermixed with untreated iSNs (Supplemental figure 3). Batch effects were somewhat more prominent than line-to-line variability. In the analysis that follows, comparisons were drawn from bortezomib-treated iSNs and DMSO controls and were based on 1668 proteins that met a threshold of p < 0.05 (FDR < 0.0872). Among this collection of proteins, 981 increased in levels and 687 were decreased in levels (Figure 2B).
Figure 2: Bortezomib exposure alters the cellular protein profile.
a) Principle component analysis (PCA) of the protein profiles of iSNs treated with bortezomib, DMSO, or a blank vehicle. Samples represent pooled technical replicates obtained from different experiments using iSNs obtained from 3 different iPSC lines.
b) Differential protein levels of all identified proteins. 1599 proteins in total increased in levels and 981 were significant (p < 0.05). 1173 proteins in total decreased in levels and 687 were significant. Lines adjacent to axes represent significance thresholds between bortezomib-treated and control conditions. Proteins associated with protein quality control, DNA damage, and oxidative stress are highlighted.
c) Heatmap of proteins known to change in levels based on bortezomib exposure. All bortezomib-treated iSN samples are clustered as one group and all iSNs from control conditions are clustered as a second group.
Expected changes from bortezomib exposure
It has been well documented that numerous proteins change levels following bortezomib exposure (Obeng et al., 2006), (Karademir et al., 2018), (Sherman and Li, 2020), and a cross-comparison of the proteins identified in this study with those known to change in levels from bortezomib based on the Ingenuity Knowledge Base yielded a list of 28 proteins that met the criteria for significance (Figure 2C). The β1 and β5 subunits of the 26S proteasome are the primary targets of bortezomib (Besse et al., 2019), and in bortezomib-treated iSNs, a significant increase occurred for the β1, β3, and β5 subunits. Bortezomib induces ER stress and increases the levels of chaperones and proteins involved in ER-associated degradation (ERAD) and nonsense-mediated decay (Wiita et al., 2013), (Karagoz et al., 2019), and a similar response was observed in iSNs. Several members of the HSP40, HSP70, and HSP90 were elevated in levels, as were proteins associated with the UPR, as seen in other systems (Kambhampati and Wiita, 2020). Other reported effects of bortezomib include oxidative stress (Karademir et al., 2018), (Yin et al., 2019) and DNA damage (Palanca et al., 2014), and several proteins involved in the mitigation of these stresses were increased in levels (HIF1A, HSPA1A/HSPA2A, BRF2, TMEM161A, and others).
3.3. Cellular stress is a central component to bortezomib neurotoxicity
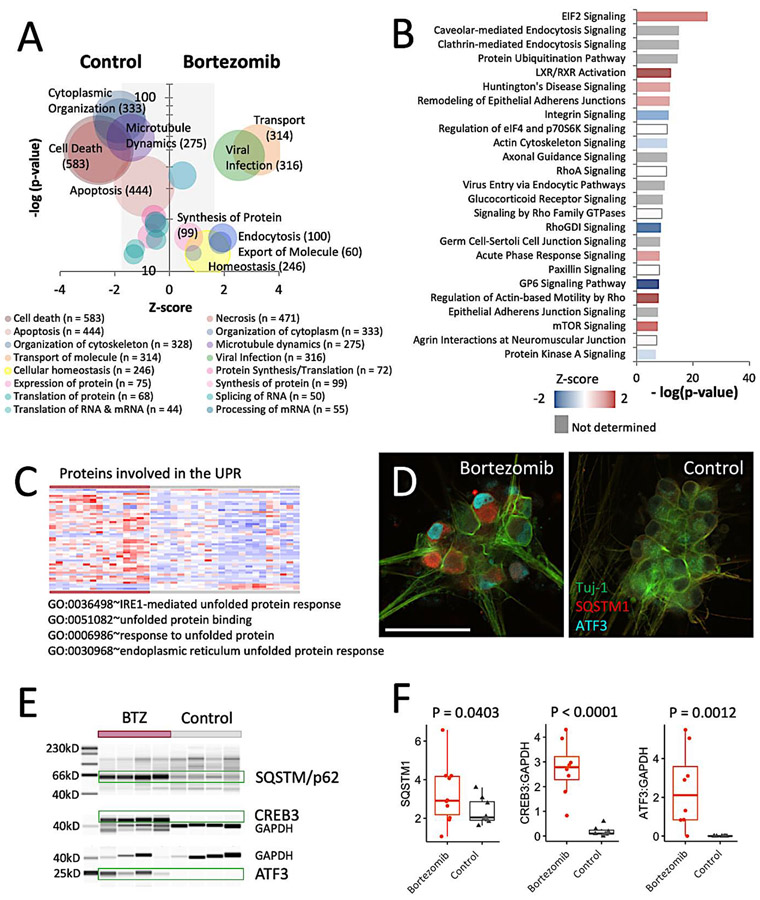
In order to create a broad picture of the impacts that bortezomib treatment has on iSN cellular processes and to prioritize their relative importance, the profile of differential protein levels was analyzed using Ingenuity Pathway Analysis (IPA). Many cellular functions are affected by bortezomib, but those with greatest significance included the IPA categories of cell death, microtubule dynamics, molecular transport, and viral infection. Subcategorization of the proteins comprising these processes in the context of the most significantly affected signaling pathways further supports the interpretation that many the observed changes are attributable to UPR-induced ER stress.
Biological effects of bortezomib indicate a state of cell stress
The general effects of ER stress and the UPR include discordance in translation, changes in amino acid flux, increased cholesterol biosynthesis, and ER membrane remodeling to adjust the capacity of the ER to restore proteostasis (Karagoz et al., 2019). The group of proteins involved in RNA Expression and RNA Translation indicate variation within these cellular functions for iSNs (Figure 3A). To further specify the affected biological processes within these broad IPA categories of cellular effects, each protein group was analyzed for representative gene ontology (GO) terms using ClueGO (Supplemental figure 4A). iSN proteins that changed in levels occurred for RNA Processing (CRNKL1, WDR75, HEATR1), Posttranscriptional Regulation of Gene Expression (ENC1, AGO2, TRIM71), and Translation (IGF2BP2, IGF2BP3, EIF4A1), which may be affected by increased levels of proteins associated with RNA Splicing (hnRNP family members) and RNA Helicase Activity (DEAD-box proteins). The effects of bortezomib on the IPA categories of Protein Expression and Translation appear to be nearly indistinguishable between the two conditions, although Synthesis of Protein is mildly elevated in bortezomib-treated iSNs partly due to the inclusion of proteins involved in the EIF2 and mTOR signaling pathways (Figure 3B).
Figure 3: Pathway analysis prioritizes topmost affected downstream effects and canonical signaling pathways.
a) IPA categories of cellular effects. Bubble size corresponds to number of proteins included within each category. Z-score is based on whether the change in levels of each protein had a positive or negative effect on an associated cellular effect.
b) Topmost affected canonical signaling pathways obtained using IPA. Z-score indicates whether the changes in protein levels for each protein had a positive or negative effect on the signaling pathway.
c) Heatmap of the proteins associated with the GO categories of response of unfolded protein, unfolded protein response, and unfolded protein binding. All iSN samples treated with bortezomib clustered as one group and all iSNs from control conditions clustered as a second group. Samples represent pooled technical replicates obtained from different experiments using iSNs obtained from 3 different iPSC lines.
d) Expression of ATF3 and SQSTM1 in iSNs exposed to bortezomib detected by immunofluorescence.
e) Levels of SQSTM1, ATF3, & CREB3 in bortezomib-treated iSNs after 48h measured by western blotting. Samples submitted for MS analysis are shown.
f) Quantification of SQSTM1, ATF3, & CREB3 levels. Average total intensities for SQSTM1 were used as the basis for statistical analysis. CREB3 and ATF3 were normalized to GAPDH levels prior to statistical assessments. Significance was determined using a Student’s t-test.
The category of Viral Infection is comprised of proteins that have been implicated the cascade of effects that arise from virally-induced cell stress, but are not exclusive to viral infection (e.g., DDX3X, DDX6, PCBP1). The proteins within this category are highly interconnected with proteins involved in other signaling pathways and biological effects. Over half of these proteins are shared with the categories of Cell Death, Cytoplasmic Organization, Cellular Homeostasis, and Molecular Transport; but the more significant effects of this group are associated with altered Gene Expression and Transport. Several signaling pathways are potentially implicated with this cellular effect with many shared proteins that represented > 40% of the proteins contained in the pathways of Caveolar- and Clathrin-Mediated Endocytosis (22 and 45 proteins, respectively), LXR/RXR Activation (21 proteins), Virus Entry via Endocytic Pathways (23 proteins), Huntington’s Disease (31 proteins), mTOR (22 proteins), Actin Cytoskeleton (23 proteins), and EIF2 Signaling (29 proteins). GO biological processes that contained proteins that were increased in levels included Ribonucleoprotein Complex Assembly, Regulation of Gene Silencing by miRNA, Small RNA Loading onto RISC, and de novo Protein Folding (Supplemental figure 4B). GO biological processes with decreased protein levels included Viral Genome Replication, Receptor-mediated Endocytosis, and Protein Localization to Nucleus. The functions of mRNA Processing, Viral Process, and Gene Silencing within this category contained an equal balance of proteins with increased and decreased levels and provides further evidence for the presence of an alteration in protein translation consistent with cellular stress.
Stress-responsive alterations in transport related processes are elevated
Proteins within Transport of Molecule include membrane transporters of proteins (SLC38A1, TIMM44), ions (SLC7A1, PTGS2), and lipids (LDLR, NPC1) and those with central roles in their regulation and trafficking (HIF1A, NDFIP1, SQSTM1). Many of these proteins are associated with endosomes, lysosomes, and the plasma membrane region, and the composition of protein changes within these organelles may reflect an adaptive mechanism for maintaining homeostasis (Supplemental figure 4C). The subcategory Export of Molecule contains several proteins that increased in levels with roles in Cholesterol Homeostasis, mRNA and Protein Export from the Nucleus, and Organic Anion Transmembrane Transport (Supplemental figure 4D). The subcategory of Endocytosis includes additional proteins involved in Cholesterol Transport and Homeostasis that increased in levels, as well as proteins involved in Phagosome Acidification and Transferrin Transport (Supplemental figure 4E). The general importance of cholesterol-related processes is further accentuated by an overlap of proteins involved in LXR/RXR signaling (Figure 3B). Other Endocytosis-related proteins that increased or decreased in levels are associated with the surface membrane of endocytic and synaptic vesicles with roles in trafficking dynamics.
Biological processes with reduced levels of proteins in iSN exposed to bortezomib included Post-Golgi Vesicle Transport, Cytosolic Transport, and Neuron Projection Morphogenesis. The primary vesicle subgroup most affected are Clathrin-Coated Vesicles, although reduced levels of proteins involved in Golgi and Post-Golgi Vesicle Transport could affect the trafficking of a broader group of vesicles. Proteins within the Clathrin-Mediated Endocytosis signaling pathway indicate that both clathrin-dependent and receptor-mediated endocytosis are affected, in part due to a significant decrease in the levels of EPS15 that is involved in clathrin-coated pit formation and the internalization of receptor tyrosine kinases (RTKs), and significantly reduced levels of the levels of the AP-2 adapter complexes (AP2A1, AP2A2, AP2B1). In addition, the AP-1 (AP1B1, AP1G1) and AP-3 (AP3B1, AP3D1) adapter complexes important for endosomal and synaptic vesicle dynamics were significantly reduced in levels. Proteins involved in Neuron Projection Morphogenesis that significantly decreased in levels are important for electrolyte balance (WNK1, NEDD4L), regulating actin polymerization at the plasma membrane (IQGAP1, MAP1S), and in synaptic vesicle transport and secretion (RIMS1, SYT1).
Proteins supporting homeostasis processes are elevated
The IPA category of Homeostasis includes proteins involved in a broad range of processes such as Ion Homeostasis, Autophagy, and Mitochondrial Organization and Transport (Supplemental figure 4F). Within the Ion Homeostasis group, Calcium Homeostasis was the most significant subgroup with an enrichment of iSN proteins following bortezomib exposure (ATP2C1, TMTC2, ATP1B1), and could be related to an increased calcium demand in the ER to accommodate cell stress. Several proteins involved in Autophagy and its regulation (PLK2, WDR6, SQSTM1, TMBIM6) were increased in levels, as were proteins that are shared with the mTOR pathway (EIF4G2, MTOR, and TSC2). Mitochondrial-associated proteins were increased in iSNs for processes associated with Mitochondrion Localization (MFN1, MFN2), Organization (LONP1, OPA1, HSP90AA1), and Microtubule-dependent Transport (HIF1A, RHOT1).
Bortezomib-induced cellular effects are attributable to UPR regulators
The originating source of cell stress presumably commenced with the accumulation of ubiquitinated proteins and subsequent activation of the pro-survival UPR, and the associated protein level changes are likely to be neuroprotective. Regulators of the UPR have been reported to rapidly increase in levels in response to bortezomib and other chemotherapeutic agents and typically subside within 24h (Wiita et al., 2013), (Hakim et al., 2016), (Obeng et al., 2006), so we hypothesize that the presence of downstream effectors within the dataset indicates prior activation of these regulators. GO analysis of the entire dataset identified 50 differentially expressed proteins associated with the GO term Response to Topologically Incorrect Protein and an additional 24 affiliated with Endoplasmic Reticulum Unfolded Protein Response (p = 1.1E-10; Figure 3C, Supplemental figure 4G), which is suggestive of the presence of UPR-related cellular effects. Since the downstream effectors of UPR regulators include a more extensive collection of proteins than those involved with misfolded proteins, the IPA upstream regulator analysis (Kramer et al., 2014) was filtered for the UPR regulators XBP1, NFE2L2, ATF4, HSP90B1, and ATF6, and 112 of their downstream effectors were altered in iSNs in response to bortezomib and accounts for 7.5% of the proteins contained within this dataset. This group of proteins was enriched for Response to ER Stress, Amino Acid Transport, and Cellular Response to Hypoxia (Supplemental figure 4H). To further substantiate the presence of a UPR signature, the protein levels of the UPR effectors ATF3 and CREB3 were assessed by western as both are important signals associated with cell and axonal injury (Figure 3) (Carozzi et al., 2013), (Grandjean et al., 2019), (Ying et al., 2015). Higher levels of both proteins were present in the bortezomib-treated iSNs and support the interpretation that the UPR contributes to a fraction of the observed protein changes and a subset of the biological effects at this stage of bortezomib-induced stress.
3.4. Bortezomib exposure predicts impairment of cytoskeletal dynamics
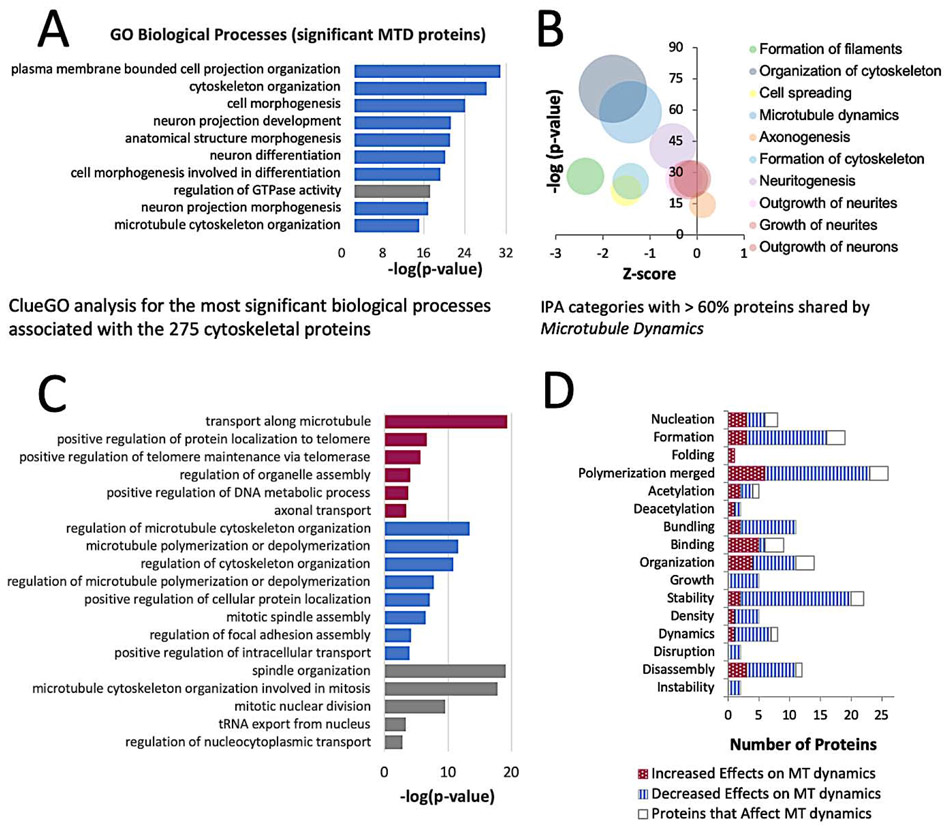
Bortezomib-induced cell stress is accompanied by diminishment of cytoskeletal dynamics markers
The IPA category of Microtubule Dynamics is comprised of 275 proteins that are involved in all aspects of the cytoskeleton, and the changes in the levels of these proteins were suggestive of attenuation in many cytoskeletal-dependent processes. Some of the larger protein groups within this category are involved in the GO biological processes of Cytoskeleton Organization, Neuron Projection Development, Cell Morphogenesis, and all of these cellular processes were enriched for proteins that declined in levels (Figure 4A). The Actin Cytoskeleton, Axonal Guidance, and RhoA signaling pathways were nearly as significant as those associated with a cellular stress response (Figure 3B). Within these pathways, cell surface proteins such as NGFR, NTRK3, and integrin decreased in levels, as did some of the proteins that facilitate signaling events (multiple ARHGEF proteins, IQGAP1/2, CDC42BPA/B, MPRIP). In contrast, the ARP2/3 proteins involved in actin nucleation were increased. The actin cytoskeletal organization proteins spectrin (SPTAN1, SPTBN1), adducin (ADD1), and ankyrin (ANK2, ANK3) that form the membrane-associated periodic skeleton (MPS) were reduced in levels, which limits the effects of extracellular signal–regulated kinase (ERK) signaling events in neurite growth or repair processes (Zhou et al., 2019). Interestingly, protein changes within neuronal subcategories suggest that, at the time point sampled in our experiment, bortezomib does not have a strong impact on Neuritogenesis, Neuronal Morphogenesis, Axonogenesis, or Shape Changes in Neurites (Figure 4B) owing to the increased levels of proteins involved in the cellular stress response.
Figure 4: Bortezomib decreases the levels of Proteins Affecting the Dynamics of Microtubules.
a) Biological processes associated with proteins in the IPA category of Microtubule Dynamics. Blue bars indicate processes predominated by a loss of proteins. Grey bars indicate the process contained a balance of proteins that increased or decreased in levels.
b) IPA categories of cellular effects that contain > 60% overlap of proteins with Microtubule Dynamics.
c) Cellular Compartment and Molecular Function categorization of 84 microtubule-associated proteins that changed in levels from bortezomib exposure.
d) Numbers of proteins that increase, decrease, or affect microtubule-related processes.
The related IPA category Formation of Filaments also was compromised by bortezomib. Nearly 60% of the proteins within this category are associated with the actin and microtubule cytoskeletons and are shared with the category of Microtubule Dynamics. The most significant GO biological processes affected by decreased levels of proteins involved the Organization of the Actin and Microtubule Cytoskeletons and the Regulation of Supramolecular Fibers, primarily through Actin Filament Bundle Assembly, and Actin and Microtubule Polymerization. Several intermediate filament cytoskeleton proteins were significantly decreased in levels (NEFL, NEFM, NES, PHLDB2, PKN2, SYNM, VIM, LMNA, LMNB1, & LMNB2), which increases the susceptibility of iSNs to morphological changes.
Bortezomib broadly impairs microtubule-related processes
Embryonic and adult DRG neurons exposed to bortezomib for 24h show an increase in tubulin polymerization, and similar findings have been reported for in vivo rat models of chronic bortezomib administration (Meregalli et al., 2014). To clarify the effects of bortezomib on the microtubule cytoskeleton, proteins with known associations to microtubules were distilled from this dataset. 84 microtubule-associated proteins within the category of Microtubule Dynamics affiliated with a biological process (GO:0007017~microtubule-based process), cellular component (GO:0015630~microtubule cytoskeleton), or molecular function (GO:0008017~microtubule binding) were differentially expressed at p<0.05 (Table 1). Many of the proteins that changed in levels possess multifunctional roles that regulate the microtubule cytoskeleton, its molecular architecture, as well as transport processes. Some of these influential proteins include CLASP2, MAP2, CKAP5, CLASP1, and MAP4, though the overall list emphasizes that the effects of bortezomib on microtubule dynamics are system-wide and most appropriately considered through a network-centric view given that the levels of half of these proteins are well correlated (Supplemental figure 5A). Cellular processes with significant reductions of proteins include the Regulation of Microtubule Cytoskeleton Organization and Microtubule Polymerization or Depolymerization (Figure 4C). To further assess the bortezomib-induced changes to the microtubule cytoskeleton, proteins associated with different microtubule-dependent processes were curated using the IPA Knowledge Base and yielded an abbreviated list of 72 proteins. The changes in levels for most proteins indicate a decline in all of the categories that concern the dynamics of microtubules (Figure 4D). The proteins were further prioritized based on the extent of functional overlap among the different microtubule related cellular processes (Supplemental figure 5B). MAP2 emerged as the most prominent candidate for further inquiry because of the magnitude of protein loss relative to all of the other cytoskeletal proteins, the number of processes that it is known to affect, and for its somewhat undefined role in axons.
Table 1.
84 microtubule-associated proteins that change in levels in iSNs from bortezomib exposure.
| GO Classification | Gene ID Protein level changes of higher significance (p < 0.0001) |
|---|---|
| Biological Process Cellular Component Molecular Function −log(p-value) > 4 | Increased: PAFAH1B1 Decreased: CLASP1, CLASP2, CKAP5, MAP2, MAP4, DST, MAP1B, MAP1S, ARHGEF2, KIF2A |
| Biological Process −log(p-value) > 4 | Increased: UBB,
OPA1,
TBCD, IFT140, WDR35, Decreased: DOCK7, SRGAP2, KTN1, OBSL1, GCC2, SLK, CEP131, PHLDB2, SON, MYH9, PHLDB1, FLNA |
| Cellular Component −log(p-value) > 4 | Increased: OBSL1, TBCD, IFT140, MFN2, KLHL42, WDR35, Decreased: CC2D1A, IQGAP1, CEP131, RANBP2, MYH9, TAPT1, AKAP11, CHAMP1 |
| Molecular Function −log(p-value) > 4 | Increased: OPA1 Decreased: NEFM |
| Protein level changes of lower significance (0.0001 < p < 0.05) | |
| Biological Process Cellular Component Molecular Function −log(p-value) < 4 | Increased: KIF5A, CENPE Decreased: EML4, NUMA1, KIF5B, MAP1A, GOLGA2, APC, HDAC6, CAMSAP2, CLIP1, TUBGCP6, TRPV4, KIF13B |
| Biological Process −log(p-value) < 4 | Increased: IFT122, TUBB, CUL9 Decreased: BCAS3, EPHA3, UCHL1, PTK2, ROCK2, TACC1, NEFL, KLC1, KPNB1, ABL1, PCM1DLG1 |
| Cellular Component −log(p-value) < 4 | Increased: RAN, IFT80, SARM1, IFT122, TUBB, CENPF, ALS2, RAB8A, KLC1, PRKCI Decreased: TLN1, BCAS3, DAAM1, PTPN23, CEP170, MARCKS, DIAPH1, PTK2, ARHGAP35, ROCK2, TACC1, PCM1, DLG1, NEK1 |
underlined proteins are well correlated (r > ∣0.8∣ to at least one other protein in this list)
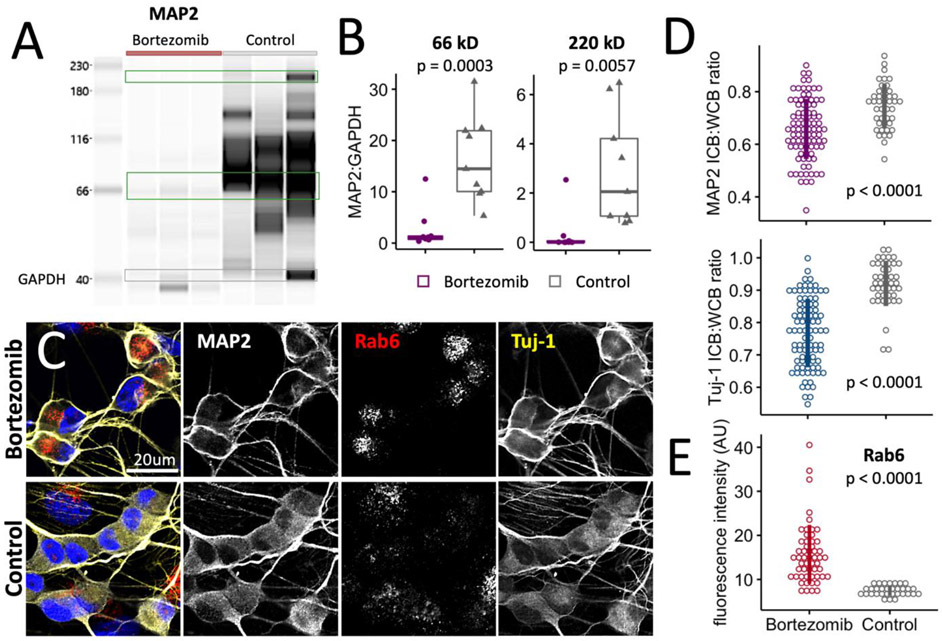
3.5. Bortezomib neurotoxicity drives changes in the cellular levels of MAP2 and its localization
MAP2 is implicated in numerous roles but its function is not entirely understood. MAP2 exerts a stabilizing effect on microtubules through inhibition of catastrophe (Pryer et al., 1992), increased polymer stiffness (Dye et al., 1993), (Edson et al., 1993)), and increased tubule interspatial arrangements (Harada et al., 2002), (Tang et al., 2014). It crosslinks microtubules with actin and neurofilaments (Ozer and Halpain, 2000), (Hirokawa et al., 1988), (Papasozomenos et al., 1985), and it associates with the ER membrane (Farah et al., 2005). The substantial loss of MAP2 was confirmed by western, and in our iSN model this loss is primarily due to the lower molecular weight isoform MAP2c (Figure 5A-B). Alternative splicing of MAP2 yields high or low molecular weight isoforms and the abundance of these isoforms is related to cell maturity (Ramkumar et al., 2018), (Jansen et al., 2017). The higher molecular weight isoforms were expressed in iSNs, and these isoforms also were significantly reduced from bortezomib exposure. Immunostaining for MAP2 indicated the presence of residual protein proximal to the plasma membrane with no obvious differences in localization patterning within neurites (Figure 5C). As a measure to verify this observation within somata, the fluorescence intensities of MAP2 and Tuj-1 were determined for whole cell bodies and the region interior to the subplasma membrane and ratio intensities were compared between control and bortezomib-treated iSNs. Both proteins were significantly concentrated at the subplasma membrane with bortezomib exposure (Figure 5D). MAP2 has been shown to affect the trafficking of Rab6+ dense core vesicles (Gumy et al., 2017). Based on the fluorescence intensities of intracellular Rab6, bortezomib exposure enhanced Rab6 levels and potentially the number of Rab6+ vesicles retained in the soma (Figure 5C, 5E).
Figure 5: MAP2 & TUBB localization is altered after 24h and reduced after 48h in Bortezomib.
a) Western blot for MAP2 in bortezomib-treated and control iSNs. Samples shown represent those submitted for MS analysis.
b) Levels of low molecular weight and high molecular weight isoforms of MAP2 were normalized to GAPDH for statistical comparison. Significance was determined using a Student’s t-test.
c) Localization patterns of Tuj-1 and MAP2 and Rab6 vesicle accumulation in iSNs treated with bortezomib or DMSO for 24h.
d) Localization patterns of Tuj-1 and MAP2 are altered in the presence of bortezomib based on a change in the ratio of fluorescence intensities between the inner cell bodies (ICB) and whole cell bodies (WCB) (p < 0.0001). Measurements were pooled from 4 independent experiments with iSNs obtained from 2 different iPSC lines. Bars represent the median ± 1σ. Significance was determined using a Student’s t-test.
e) Fluorescence intensities for Rab6 in somata. Bars represent the median ± 1σ. Significance was determined using a Student’s t-test.
4. Discussion
The central focus of this study was to better characterize the molecular underpinnings that predispose sensory neurons to bortezomib-induced neurotoxicity. Our approach was to use a model system based on iPSCs to recapitulate bortezomib’s effects on human iSNs. Although iPSC-derived models possess immature characteristics, they have proven to be a useful model system for studying chemotherapeutic drug responses. Our iSN model reproduced a cascade of effects that have been observed in other cellular models. Bortezomib exposure is known to induce the UPR, ER and oxidative cellular stresses, reduced mitochondrial transport, and alterations to the cytoskeleton. The time point at which iSNs were examined was at a stage in which neurotoxic effects are prominent and was accompanied by a robust response in the levels of stress adaptive proteins associated with protein quality control processes (UPR, ERAD, autophagy), the integrated stress response (ISR), and the maintenance of homeostasis; but it also revealed an extensive decline in proteins involved in cytoskeletal dynamics as evidenced by significant reduction in the levels of MAP2. The potent effects of bortezomib on cytoskeletal processes are thought to provide the underlying basis for neurotoxic responses and ultimately are a critical determinant for (iSN) cell survival. This study adds support to this hypothesis and provides some insight that could improve our understanding of bortezomib-induced peripheral neuropathy.
4.1. Bortezomib induces UPR-related cellular stress in iSNs
Translational regulation by the UPR signals that its capacity has been exhausted and is recognized as one of the earliest stages of stress through PERK-mediated phosphorylation of EIF2α to reduce global protein synthesis. Studies based on cell culture models typically observe this event within a few hours of exposure to a proteasome inhibitor and it is accompanied by an increase in the protein levels of PERK-P, EIF2-P, and stress-responsive proteins such as ATF4 that typically return to baseline levels or lower within 12-24h (Wiita et al., 2013), (Hakim et al., 2016), (Obeng et al., 2006). After 48h of bortezomib exposure, the effects of the UPR remained significant in iSNs owing to 61 downstream effectors comprised of several folding proteins and chaperones directly involved in the UPR and response to ER stress. An additional 112 proteins were recognized as downstream effectors of the UPR regulators XBP1, ATF4, ATF6, and NFE2L2, with roles primarily in the GO biological processes related to Response to ER stress, Amino acid transport, Cholesterol and Amino acid metabolism.
Since the measurable timeframe of UPR sensor activation is short-lived, the presence of UPR-induced cell stress was confirmed by the presence of ATF3 and CREB3. ATF3 has been described as a downstream effector of ATF4 that acts in concert with GADD34 in a feedback loop to dephosphorylate EIF2-P and inhibit the effects of the UPR (Jiang et al., 2004). It is important to axonal regeneration (Holland et al., 2019), but it has also been correlated with neuropathic pain (Tsujino et al., 2000), (Linda et al., 2011) and in CIPN based on in vitro and in vivo models of murine DRG sensory neurons (Wang et al., 2020), (Carozzi et al., 2013), (Yin et al., 2019). CREB3/Luman induces the UPR in adult rat DRG neurons injured by nerve crush or axotomy, it is expressed during acute and chronic stages of stress, and it facilitates regenerative axon growth by increasing cholesterol biosynthesis (Ying et al., 2015), (Hasmatali et al., 2019). The presence of ATF3 and CREB3 suggests that multiple branches of the UPR are active and that many of the observed biological effects are a consequence of prolonged bortezomib exposure that are not necessarily observed with other chemotherapeutic agents (e.g. paclitaxel; Supplemental Figure 6).
Nearly half of the proteins that were elevated in iSNs exposed to bortezomib are associated with different aspects of the ISR. Its function is to repress cap-dependent translation in favor of cap-independent translation, and it is responsive to the UPR, viral infection, mitochondrial stress, and chemotherapeutics (Pakos-Zebrucka et al., 2016), (Costa-Mattioli and Walter, 2020). While short-term translational inhibition by the ISR is protective, chronic translational inhibition is considered pathogenic (Bosco, 2018); (Bond et al., 2020). Protein synthesis under conditions of stress is essential for cell survival, but only a limited set of proteins involved in this process have been well characterized. Upon phosphorylation of EIF2, mRNAs with internal ribosome entry sites are translated which generally leads to the expression of ATF4 as well as other stress-induced proteins (Rendleman et al., 2018), (Costa-Mattioli and Walter, 2020). A recent study that examined protein synthesis in human neuroblastoma cells under arsenite-induced translational repression reported > 300 newly synthesized proteins, and 107 of the proteins significantly elevated in levels in bortezomib-treated iSNs overlapped with those findings (Baron et al., 2019). The most significantly increased levels of proteins shared by both cell models included WDR6, ENC1, SQSTM1, DDX3X, HMGCR, POLR2B, and DDX6, and these proteins alone highlight the importance of growth inhibition, curtailment of oxidative stress, cholesterol synthesis, modulation of translation, and the expression of cytoprotective proteins as multifaceted effects of the ISR.
Bortezomib exposure increased the abundance of iSN proteins involved in posttranscriptional gene silencing, mRNA stabilization, DNA damage repair, nonsense mediated decay, ERAD, and was accompanied by a reduction in proteins that facilitate transport across the nuclear pore membrane, all which further indicate a modality of stress-induced regulatory control of translation. The resumption of cap-dependent translation is critical for the synthesis of proteins essential for cell survival and requires dephosphorylation of the EIF2α complex. However, this signal may be decoupled after prolonged bortezomib exposure since it has been observed that translational repression is sustained in MM1.S multiple myeloma cells irrespective of the phosphorylation status of EIF2 (Wiita et al., 2013), (Huang et al., 2020). Insufficient mTOR signaling and broad-scale intron retention have been offered as potential mechanisms, although the reason(s) for this effect remains to be established. For iSNs, changes in the composition of proteins in the EIF2 and mTOR/p70S6K pathways suggest that a stimulatory signal to induce protein synthesis may be present based on decreased levels of the negative mTOR regulator TSC1, but limitations could remain imposed on translation due to a reduced availability of different translational machinery proteins (eIF3, eIF4, eIF5, 40S ribosome, and 60S ribosome subunits) or the presence of less well-characterized proteins such as PAQR3 that have been reported to interfere with mTOR signaling (Wang et al., 2019). It remains to be determined whether translational repression is similarly sustained in sensory neurons or if other mechanisms are central to bortezomib-induced neurotoxicity.
ATF3, CREB3, EIF2 phosphorylation (Khoutorsky et al., 2016), and mTOR signaling (Khoutorsky and Price, 2018) have established roles in the pathobiology of neuronal injury or persistent pain, and some proteins that are altered in levels from bortezomib exposure are associated with peripheral or sensory neuropathy. Bortezomib significantly increased the levels of the ATF4 effectors PTSG2 and MARS, both which are related to the ISR, as well as UPR-induced effectors OPA1, MFN2, and HSPD1. An additional 14 neuropathy-related proteins changed in levels, of which 5 possess functions that involve actin dynamics (ARHGEF10, INF2, DST, SPTAN1, WNK1), and collectively may enhance the susceptibility of iSNs to neurotoxicity.
4.2. Bortezomib affects regulators of cytoskeletal dynamics
The deleterious effects of bortezomib on neuronal cytoskeletal elements were observed in our earlier rat DRG model, neural progenitor cells (Saito and Imaizumi, 2018), rat cortical neurons (von Brzezinski et al., 2017), and murine peripheral nerves in vivo (Carozzi et al., 2013), (Meregalli et al., 2014); and its effects are exacerbated in iPSC-derived cardiomyocytes deficient of the BAG3 chaperone (Judge et al., 2017). Bortezomib induces a collapse in actin dynamics, alters neurofilament protein distribution (Ale et al., 2015), and promotes tubulin polymerization; and the broad decrease in cytoskeletal protein abundance in iSNs highlights the extensive system-wide effects despite insignificant changes in the levels in actin or tubulin. The majority of these proteins regulate cytoskeletal formation, maintenance, organization, and molecular architecture (Figure 4). Some of the changes are anticipated as one of the effects of UPR signaling is to remodel and expand the ER membrane, which is mediated by interactions of FLNA with either PERK or IRE1 and is followed by alterations in the polymerization dynamics of F-actin (Urra et al., 2018), (van Vliet et al., 2017), (Saito and Imaizumi, 2018). Decreased levels of proteins that organize components of the cytoskeletal system in the soma may further accommodate this process and allow for the modulation of inter-organelle communication by altering protein localization and increasing the pool of freely available actin monomers as a feedback signal that controls UPR-related signaling pathways. An additional effect is attributable to reduced levels of the actin-spectrin MPS proteins beta-II spectrin, adducin, and ankyrin that localize to the subplasma membrane along the axonal shaft and in the soma. In the axonal compartment, these proteins form a 2D lattice structure in neurites that organizes ion channels, adhesion molecules, and RTK-dependent signaling processes (Zhou et al., 2019); and contributes to the regulation of microtubule organization (Qu et al., 2017). Under trophic depletion or acute destabilization of actin, the MPS is rapidly disassembled as a prelude to axonal degeneration and fragmentation (Wang et al., 2019), (Unsain et al., 2018). Depletion of these MPS-associated proteins coupled with alteration in actin dynamics suggests the dynamic equilibrium of the MPS is compromised in favor of its disassembly. An added consequence is perturbed intracellular trafficking since beta-II spectrin and ankyrin B are adapters that connect molecular motors with synaptic cargo (Lorenzo et al., 2019). Despite the extent of changes within the cytoskeletal compartment, preservation of Arp2/3-dependent actin nucleation in iSNs could enable ongoing repair of DNA damage (Lambert, 2019) and predispose iSNs to the rapid initiation of MPS reassembly upon reversal of bortezomib-induced cell stress.
Intermediate filament proteins interlink the actin and microtubule cytoskeletons and provide overall mechanical resilience in addition to overall cytoskeletal organization. Intermediate filaments serve as scaffolding proteins that govern the positioning and trafficking of endosomes, lysosomes, synaptic vesicles, and the ER; and decreased levels are indicative of injury (Yuan et al., 2017). Several of these proteins were significantly decreased in levels, including NEFM, NEFL, and NES, which was accompanied by an altered localization of PRPH. Although it is possible that the decreased levels of neurofilament proteins alone could explain previous observations of increased polymerization of microtubules (Yuan et al., 2017), the basis for this effect is likely interdependent on additional proteins (e.g., MAPRE2) that regulate the microtubule cytoskeleton.
The collective decline or loss of 60+ microtubule associated proteins indicates a diminished capacity to assemble, maintain, or reorganize components of the microtubule cytoskeleton. A correlation analysis based on the levels of each protein among all of the samples revealed 40+ proteins with a Pearson correlation coefficient of r > ∣0.8∣ to at least one other protein within this group. The majority of these proteins exhibit co-expression patterns or interact through binding events, and altogether they predominantly affect processes that involve microtubule polymerization and organization, and more specifically microtubule nucleation, intracellular transport, and the regulation of adherens junctions. Many of these proteins are essential to neurite outgrowth by supporting the growth of microtubules from the Golgi and anchoring microtubules at the cell cortex.
In addition to the cytoskeletal protein changes that govern the trafficking of essential axonal and synaptic components, multiple subunits of the AP-1, AP-2, and AP-3 adapter complexes were decreased in levels. These proteins possess important roles in post-Golgi cargo sorting and transport, the axonal trafficking of proteins, and in processes that govern synaptic vesicle dynamics (Guardia et al., 2018). Although the loss of AP3B1 or AP3D1 leads to neurodevelopmental delay, reduced levels are suggestive of a mechanism to preclude the axonal entry of vesicles and introduce another regulatory layer that affects intracellular protein delivery.
One of the proteins that exhibited the highest magnitude of decline in levels was MAP2. The distribution of MAP2 in unperturbed DRG neurons or iSNs is concentrated in the soma and its levels decline in a gradient along neurites. It is primarily regarded to have stabilizing effects on microtubules, but its interactions with multiple proteins involved with cytoskeletal architecture suggest it provides a significant contribution to cellular morphology. In DRG neurons, it has been shown to have an important role in the trafficking of dense core vesicles and lysosomes in the proximal axon (Gumy et al., 2017) through its interactions with the slow kinesin-1 motor (KIF5). Bortezomib exposure led to reduced levels of both a higher molecular weight isoform (MAP2b) and a lower molecular weight isoform (MAP2c), and this loss may be associated with an accumulation of Rab6+ vesicles within the soma. Although the depletion of MAP2 induces the outgrowth of new neurites from the cell body (Gumy et al., 2017), it is unlikely for this to occur in the presence of bortezomib considering that multiple proteins that inhibit growth processes are highly elevated in levels.
Overall, these findings leave us with several questions for future investigation. Are the effects of bortezomib from proteotoxicity or from unknown targets that promote neurotoxicity? Are iSNs self-autonomous in their ability to recover from the severe effects of bortezomib? Do they continue along the path of translational repression at this stage of stress? Could overexpression of MAP2 or other cytoskeletal proteins attenuate the severity of the stress response? In order to address potential avenues of mitigating bortezomib-induced peripheral neuropathy, further elucidation of the mechanistic underpinnings is needed.
Supplementary Material
Supplemental figure 1: Protein abundances indicative of iSN maturity.
a) Total protein levels for markers of neuronal progenitors.
b) Total protein levels for markers of nociceptors.
c) Levels of proteins involved in neuronal maturation identified by GO (GO: 0042551).
d) Levels of proteins involved in action potential and for cell receptors involved in mechanosensation
e) Levels of synaptic vesicle proteins.
Supplemental Figure 2: Tracking of mitochondria
a) The fraction of mitochondria exhibiting mobile characteristics was decreased from exposure to 100nM bortezomib.
b) Additional track parameters provided by the analysis of the mobile mitochondrial subpopulation with different bortezomib doses. Data shown represent 3 independent experiments using iSNs obtained from 2 different iPSC lines. All statistical comparisons were determined using a Tukey’s multiple comparison test. Brackets indicate statistically significant comparisons. Confinement ratio (CR) comparisons: control vs 10nM, p = 0.04; control vs 50nM, p = 0.01; control vs 100nM, p = 0.04. Track median speed comparisons: control vs 10nM, p = 0.03; 10nM vs 50nM, p = 0.046; 10nM vs 100nM, p = 0.0026.
c) Subdivision of the mobile mitochondria into 2 groups illustrates that bortezomib decreased the population of mitochondria that exhibit unidirectional (CR > 0.5) and bidirectional (0.25 < CR < 0.5) mobility patterns. (Mitochondria with CR > 0.5 were more sensitive to the effects of bortezomib.)
d) Track displacement and track mean speed were affected by bortezomib in the CR > 0.5 subgroup, but not in the 0.25 < CR < 0.5 subgroup. Track displacement comparisons (CR > 0.5): control vs 10nM, p = 0.0014; 10nM vs 100nM, p = 0.014. Track mean speed comparisons (CR > 0.5): control vs 10nM, p < 0.000001; 10nM vs 50nM, p = 0.000048; 10nM vs 100nM, p = 0.000014.
Supplemental Figure 3: Heatmap of protein level changes
Supplemental Figure 4: ClueGO results of the IPA cellular effects
a) RNA expression, protein synthesis & translation: biological processes, cellular components, and molecular function.
b) Viral infection: biological processes, cellular components, and molecular function.
c) Transport of molecule: biological processes, cellular components, and molecular function.
d) Export of molecule: biological processes, cellular components, and molecular function.
e) Endocytosis: biological processes, cellular components, and molecular function.
f) Homeostasis: biological processes, cellular components, and molecular function.
g) UPR-related proteins related to the heatmap in Figure 3C.
h) Biological processes of the UPR downstream effectors of XBP1, NFE2L2, ATF4, HSP90B1, and ATF6.
Supplemental Figure 5: microtubule-associated proteins are affected as a network in bortezomib-induced neurotoxicity
a) Heat map based on the Pearson’s correlation coefficient among the most significantly affected microtubule-associated proteins.
b) Illustration of the interconnectivity of microtubule-associated proteins with microtubule-dependent processes. Ribbon width represents the log2-fold change in protein levels.
Supplemental Table 1. Materials and Reagents associated with methodology
Supplemental Table 2. Protein level changes in iSNs for Bortezomib vs DMSO
Supplemental Table 3. Protein level changes in iSNs for DMSO vs Blank controls
Highlights.
Bortezomib exposure strongly induced the UPR and ISR in iSNs
Prolonged bortezomib exposure impairs axonal trafficking of mitochondria in iSNs
Bortezomib caused a system-wide decline in iSN cytoskeletal proteins and processes
Bortezomib promoted Rab6+ vesicle accumulation within iSN somata
Prolonged bortezomib exposure is associated with dramatic loss of MAP2 in iSNs
Acknowledgements
This publication was made possible by support from the Medical Genome Facility Proteomics Core, Mayo Clinic. The core is a shared resource of the Mayo Clinic Cancer Center (NCI P30 CA15083). We thank Dr. Cristine Charlesworth for her guidance on sample preparation for MS analysis.
NPS and SA conceived the study. NPS, SA, SCLH, and RFH contributed to experimental design. Experiments were completed by SA, RFH, SCLH, BA, JPK, BN, and IS. SCLH, RAM, NPS, and SD completed analyses of the bioinformatic and experimental data. RAM and SD developed software for analysis of the proteomics data. SCLH and NPS wrote the manuscript. All co-authors read and approved the submitted version.
Funding: This work was supported by the National Institutes of Health [NPS: R01 CA211887] and the Mayo Foundation for Medical Education and Research.
Abbreviations
- iPSC
induced pluripotent stem cell
- iSN
induced pluripotent stem cell-derived sensory neuron
- CIPN
Chemotherapy-induced peripheral neuropathy
- DRG
dorsal root ganglion neurons
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- UPR
unfolded protein response
- ISR
integrated stress response
- RTK
receptor tyrosine kinase
- IPA
Ingenuity Pathway Analysis
- GO
Gene ontology
- MPS
membrane-associated periodic skeleton
- ERK
extracellular signal–regulated kinase
- BTZ
Bortezomib
- Tuj-1
β3-tubulin
- CR
Confinement ratio
- TMRM
tetramethylrhodamine methyl ester
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
The authors have no conflicting interests.
References
- Ale A, Bruna J, Herrando M, Navarro X, Udina E, 2015. Toxic effects of bortezomib on primary sensory neurons and Schwann cells of adult mice. Neurotox Res 27, 430–440. [DOI] [PubMed] [Google Scholar]
- Ankam S, Rovini A, Baheti S, Hrstka R, Wu Y, Schmidt K, Wang H, Madigan N, Koenig LS, Stelzig K, Resch Z, Klein CJ, Sun Z, Staff NP, 2019. DNA methylation patterns in human iPSC-derived sensory neuronal differentiation. Epigenetics 14, 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers-Ringler JR, Oliveros A, Qiu Y, Lindberg DM, Hinton DJ, Moore RM, Dasari S, Choi DS, 2016. Label-Free Proteomic Analysis of Protein Changes in the Striatum during Chronic Ethanol Use and Early Withdrawal. Front Behav Neurosci 10, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron DM, Matheny T, Lin YC, Leszyk JD, Kenna K, Gall KV, Santos DP, Tischbein M, Funes S, Hayward LJ, Kiskinis E, Landers JE, Parker R, Shaffer SA, Bosco DA, 2019. Quantitative proteomics identifies proteins that resist translational repression and become dysregulated in ALS-FUS. Hum Mol Genet 28, 2143–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse A, Besse L, Kraus M, Mendez-Lopez M, Bader J, Xin BT, de Bruin G, Maurits E, Overkleeft HS, Driessen C, 2019. Proteasome Inhibition in Multiple Myeloma: Head-to-Head Comparison of Currently Available Proteasome Inhibitors. Cell Chem Biol 26, 340–351 e343. [DOI] [PubMed] [Google Scholar]
- Bond S, Lopez-Lloreda C, Gannon PJ, Akay-Espinoza C, Jordan-Sciutto KL, 2020. The Integrated Stress Response and Phosphorylated Eukaryotic Initiation Factor 2alpha in Neurodegeneration. J Neuropathol Exp Neurol 79, 123–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, 2018. Translation dysregulation in neurodegenerative disorders. Proc Natl Acad Sci U S A 115, 12842–12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carozzi VA, Renn CL, Bardini M, Fazio G, Chiorazzi A, Meregalli C, Oggioni N, Shanks K, Quartu M, Serra MP, Sala B, Cavaletti G, Dorsey SG, 2013. Bortezomib-induced painful peripheral neuropathy: an electrophysiological, behavioral, morphological and mechanistic study in the mouse. PLoS One 8, e72995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Qi Y, Mica Y, Lee G, Zhang XJ, Niu L, Bilsland J, Cao L, Stevens E, Whiting P, Shi SH, Studer L, 2012. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol 30, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Walter P, 2020. The integrated stress response: From mechanism to disease. Science 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou QP, Zonder JA, 2014. Overview of proteasome inhibitor-based anti-cancer therapies: perspective on bortezomib and second generation proteasome inhibitors versus future generation inhibitors of ubiquitin-proteasome system. Curr Cancer Drug Targets 14, 517–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye RB, Fink SP, Williams RC Jr., 1993. Taxol-induced flexibility of microtubules and its reversal by MAP-2 and Tau. J Biol Chem 268, 6847–6850. [PubMed] [Google Scholar]
- Edson K, Weisshaar B, Matus A, 1993. Actin depolymerisation induces process formation on MAP2-transfected non-neuronal cells. Development 117, 689–700. [DOI] [PubMed] [Google Scholar]
- Farah CA, Liazoghli D, Perreault S, Desjardins M, Guimont A, Anton A, Lauzon M, Kreibich G, Paiement J, Leclerc N, 2005. Interaction of microtubule-associated protein-2 and p63: a new link between microtubules and rough endoplasmic reticulum membranes in neurons. J Biol Chem 280, 9439–9449. [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Li Y, Segal RA, 2017. A Mechanistic Understanding of Axon Degeneration in Chemotherapy-Induced Peripheral Neuropathy. Front Neurosci 11, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Doan RA, Cheng GC, Cetinkaya-Fisgin A, Huang SX, Hoke A, Milbrandt J, DiAntonio A, 2019. Vincristine and bortezomib use distinct upstream mechanisms to activate a common SARM1-dependent axon degeneration program. JCI Insight 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean JMD, Plate L, Morimoto RI, Bollong MJ, Powers ET, Wiseman RL, 2019. Deconvoluting Stress-Responsive Proteostasis Signaling Pathways for Pharmacologic Activation Using Targeted RNA Sequencing. ACS Chem Biol 14, 784–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardia CM, De Pace R, Mattera R, Bonifacino JS, 2018. Neuronal functions of adaptor complexes involved in protein sorting. Curr Opin Neurobiol 51, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Katrukha EA, Grigoriev I, Jaarsma D, Kapitein LC, Akhmanova A, Hoogenraad CC, 2017. MAP2 Defines a Pre-axonal Filtering Zone to Regulate KIF1- versus KIF5-Dependent Cargo Transport in Sensory Neurons. Neuron 94, 347–362 e347. [DOI] [PubMed] [Google Scholar]
- Hakim V, Cohen LD, Zuchman R, Ziv T, Ziv NE, 2016. The effects of proteasomal inhibition on synaptic proteostasis. EMBO J 35, 2238–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Teng J, Takei Y, Oguchi K, Hirokawa N, 2002. MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J Cell Biol 158, 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasmatali JCD, De Guzman J, Zhai R, Yang L, McLean NA, Hutchinson C, Johnston JM, Misra V, Verge VMK, 2019. Axotomy Induces Phasic Alterations in Luman/CREB3 Expression and Nuclear Localization in Injured and Contralateral Uninjured Sensory Neurons: Correlation With Intrinsic Axon Growth Capacity. J Neuropathol Exp Neurol 78, 348–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Hisanaga S, Shiomura Y, 1988. MAP2 is a component of crossbridges between microtubules and neurofilaments in the neuronal cytoskeleton: quick-freeze, deep-etch immunoelectron microscopy and reconstitution studies. J Neurosci 8, 2769–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland SD, Ramer LM, McMahon SB, Denk F, Ramer MS, 2019. An ATF3-CreERT2 Knock-In Mouse for Axotomy-Induced Genetic Editing: Proof of Principle. eNeuro 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HH, Ferguson ID, Thornton AM, Bastola P, Lam C, Lin YT, Choudhry P, Mariano MC, Marcoulis MD, Teo CF, Malato J, Phojanakong PJ, Martin TG 3rd, Wolf JL, Wong SW, Shah N, Hann B, Brooks AN, Wiita AP, 2020. Proteasome inhibitor-induced modulation reveals the spliceosome as a specific therapeutic vulnerability in multiple myeloma. Nat Commun 11, 1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen S, Melkova K, Trosanova Z, Hanakova K, Zachrdla M, Novacek J, Zupa E, Zdrahal Z, Hritz J, Zidek L, 2017. Quantitative mapping of microtubule-associated protein 2c (MAP2c) phosphorylation and regulatory protein 14-3-3zeta-binding sites reveals key differences between MAP2c and its homolog Tau. J Biol Chem 292, 6715–6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HY, Wek SA, McGrath BC, Lu D, Hai T, Harding HP, Wang X, Ron D, Cavener DR, Wek RC, 2004. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol Cell Biol 24, 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge LM, Perez-Bermejo JA, Truong A, Ribeiro AJ, Yoo JC, Jensen CL, Mandegar MA, Huebsch N, Kaake RM, So PL, Srivastava D, Pruitt BL, Krogan NJ, Conklin BR, 2017. A BAG3 chaperone complex maintains cardiomyocyte function during proteotoxic stress. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambhampati S, Wiita AP, 2020. Lessons Learned from Proteasome Inhibitors, the Paradigm for Targeting Protein Homeostasis in Cancer. Adv Exp Med Biol 1243, 147–162. [DOI] [PubMed] [Google Scholar]
- Karademir B, Sari G, Jannuzzi AT, Musunuri S, Wicher G, Grune T, Mi J, Hacioglu-Bay H, Forsberg-Nilsson K, Bergquist J, Jung T, 2018. Proteomic approach for understanding milder neurotoxicity of Carfilzomib against Bortezomib. Sci Rep 8, 16318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagoz GE, Acosta-Alvear D, Walter P, 2019. The Unfolded Protein Response: Detecting and Responding to Fluctuations in the Protein-Folding Capacity of the Endoplasmic Reticulum. Cold Spring Harb Perspect Biol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoutorsky A, Price TJ, 2018. Translational Control Mechanisms in Persistent Pain. Trends Neurosci 41, 100–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoutorsky A, Sorge RE, Prager-Khoutorsky M, Pawlowski SA, Longo G, Jafarnejad SM, Tahmasebi S, Martin LJ, Pitcher MH, Gkogkas CG, Sharif-Naeini R, Ribeiro-da-Silva A, Bourque CW, Cervero F, Mogil JS, Sonenberg N, 2016. eIF2alpha phosphorylation controls thermal nociception. Proc Natl Acad Sci U S A 113, 11949–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J Jr., Tugendreich S, 2014. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert MW, 2019. The functional importance of lamins, actin, myosin, spectrin and the LINC complex in DNA repair. Exp Biol Med (Maywood) 244, 1382–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linda H, Skold MK, Ochsmann T, 2011. Activating transcription factor 3, a useful marker for regenerative response after nerve root injury. Front Neurol 2, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo DN, Badea A, Zhou R, Mohler PJ, Zhuang X, Bennett V, 2019. betall-spectrin promotes mouse brain connectivity through stabilizing axonal plasma membranes and enabling axonal organelle transport. Proc Natl Acad Sci U S A 116, 15686–15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meregalli C, 2015. An Overview of Bortezomib-Induced Neurotoxicity. Toxics 3, 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meregalli C, Chiorazzi A, Carozzi VA, Canta A, Sala B, Colombo M, Oggioni N, Ceresa C, Foudah D, La Russa F, Miloso M, Nicolini G, Marmiroli P, Bennett DL, Cavaletti G, 2014. Evaluation of tubulin polymerization and chronic inhibition of proteasome as citotoxicity mechanisms in bortezomib-induced peripheral neuropathy. Cell Cycle 13, 612–621. [DOI] [PubMed] [Google Scholar]
- Mlecnik B, Galon J, Bindea G, 2018. Comprehensive functional analysis of large lists of genes and proteins. J Proteomics 171, 2–10. [DOI] [PubMed] [Google Scholar]
- Obeng EA, Carlson LM, Gutman DM, Harrington WJ Jr., Lee KP, Boise LH, 2006. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 107, 4907–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer RS, Halpain S, 2000. Phosphorylation-dependent localization of microtubule-associated protein MAP2c to the actin cytoskeleton. Mol Biol Cell 11, 3573–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM, 2016. The integrated stress response. EMBO Rep 17, 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanca A, Casafont I, Berciano MT, Lafarga M, 2014. Proteasome inhibition induces DNA damage and reorganizes nuclear architecture and protein synthesis machinery in sensory ganglion neurons. Cell Mol Life Sci 71, 1961–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papasozomenos SC, Binder LI, Bender PK, Payne MR, 1985. Microtubule-associated protein 2 within axons of spinal motor neurons: associations with microtubules and neurofilaments in normal and beta,beta'-iminodipropionitrile-treated axons. J Cell Biol 100, 74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer NK, Walker RA, Skeen VP, Bourns BD, Soboeiro MF, Salmon ED, 1992. Brain microtubule-associated proteins modulate microtubule dynamic instability in vitro. Real-time observations using video microscopy. J Cell Sci 103 ( Pt 4), 965–976. [DOI] [PubMed] [Google Scholar]
- Qu Y, Hahn I, Webb SE, Pearce SP, Prokop A, 2017. Periodic actin structures in neuronal axons are required to maintain microtubules. Mol Biol Cell 28, 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramkumar A, Jong BY, Ori-McKenney KM, 2018. ReMAPping the microtubule landscape: How phosphorylation dictates the activities of microtubule-associated proteins. Dev Dyn 247, 138–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendleman J, Cheng Z, Maity S, Kastelic N, Munschauer M, Allgoewer K, Teo G, Zhang YBM, Lei A, Parker B, Landthaler M, Freeberg L, Kuersten S, Choi H, Vogel C 2018. New insights into the cellular temporal response to proteostatic stress. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Imaizumi K, 2018. Unfolded Protein Response-Dependent Communication and Contact among Endoplasmic Reticulum, Mitochondria, and Plasma Membrane. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A, Hoffman EM, Mauermann ML, Loprinzi CL, Windebank AJ, Klein CJ, Staff NP, 2018. Incidence and disease burden of chemotherapy-induced peripheral neuropathy in a population-based cohort. J Neurol Neurosurg Psychiatry 89, 636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman DJ, Li J, 2020. Proteasome Inhibitors: Harnessing Proteostasis to Combat Disease. Molecules 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GM, Gallo G, 2018. The role of mitochondria in axon development and regeneration. Dev Neurobiol 78, 221–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staff NP, Grisold A, Grisold W, Windebank AJ, 2017. Chemotherapy-induced peripheral neuropathy: A current review. Ann Neurol 81, 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staff NP, Podratz JL, Grassner L, Bader M, Paz J, Knight AM, Loprinzi CL, Trushina E, Windebank AJ, 2013. Bortezomib alters microtubule polymerization and axonal transport in rat dorsal root ganglion neurons. Neurotoxicology 39, 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Lu Y, Zheng W, Li Y, 2014. Overexpression of MAP-2 via formation of microtubules plays an important role in the sprouting of mossy fibers in epileptic rats. J Mol Neurosci 53, 103–108. [DOI] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW, 2017. TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90. [DOI] [PubMed] [Google Scholar]
- Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, Yonenobu K, Ochi T, Noguchi K, 2000. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: A novel neuronal marker of nerve injury. Mol Cell Neurosci 15, 170–182. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J, 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11, 2301–2319. [DOI] [PubMed] [Google Scholar]
- Unsain N, Bordenave MD, Martinez GF, Jalil S, von Bilderling C, Barabas FM, Masullo LA, Johnstone AD, Barker PA, Bisbal M, Stefani FD, Caceres AO, 2018. Remodeling of the Actin/Spectrin Membrane-associated Periodic Skeleton, Growth Cone Collapse and F-Actin Decrease during Axonal Degeneration. Sci Rep 8, 3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urra H, Henriquez DR, Canovas J, Villarroel-Campos D, Carreras-Sureda A, Pulgar E, Molina E, Hazari YM, Limia CM, Alvarez-Rojas S, Figueroa R, Vidal RL, Rodriguez DA, Rivera CA, Court FA, Couve A, Qi L, Chevet E, Akai R, Iwawaki T, Concha ML, Glavic A, Gonzalez-Billault C, Hetz C, 2018. IRE1alpha governs cytoskeleton remodelling and cell migration through a direct interaction with filamin A. Nat Cell Biol 20, 942–953. [DOI] [PubMed] [Google Scholar]
- van Vliet AR, Giordano F, Gerlo S, Segura I, Van Eygen S, Molenberghs G, Rocha S, Houcine A, Derua R, Verfaillie T, Vangindertael J, De Keersmaecker H, Waelkens E, Tavernier J, Hofkens J, Annaert W, Carmeliet P, Samali A, Mizuno H, Agostinis P, 2017. The ER Stress Sensor PERK Coordinates ER-Plasma Membrane Contact Site Formation through Interaction with Filamin-A and F-Actin Remodeling. Mol Cell 65, 885–899 e886. [DOI] [PubMed] [Google Scholar]
- von Brzezinski L, Saring P, Landgraf P, Cammann C, Seifert U, Dieterich DC, 2017. Low Neurotoxicity of ONX-0914 Supports the Idea of Specific Immunoproteasome Inhibition as a Side-Effect-Limiting, Therapeutic Strategy. Eur J Microbiol Immunol (Bp) 7, 234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Simon DJ, Wu Z, Belsky DM, Heller E, O'Rourke MK, Hertz NT, Molina H, Zhong G, Tessier-Lavigne M, Zhuang X, 2019. Structural plasticity of actin-spectrin membrane skeleton and functional role of actin and spectrin in axon degeneration. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xiang P, Chew WS, Torta F, Bandla A, Lopez V, Seow WL, Lam BWS, Chang JK, Wong P, Chayaburakul K, Ong WY, Wenk MR, Sundar R, Herr DR, 2020. Activation of sphingosine 1-phosphate receptor 2 attenuates chemotherapy-induced neuropathy. J Biol Chem 295, 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiita AP, Ziv E, Wiita PJ, Urisman A, Julien O, Burlingame AL, Weissman JS, Wells JA, 2013. Global cellular response to chemotherapy-induced apoptosis. Elife 2, e01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Qi X, Qiao Y, Liu H, Yan Z, Li H, Liu Z, 2019. The Association of Neuronal Stress with Activating Transcription Factor 3 in Dorsal Root Ganglion of in vivo and in vitro Models of Bortezomib- Induced Neuropathy. Curr Cancer Drug Targets 19, 50–64. [DOI] [PubMed] [Google Scholar]
- Ying Z, Zhai R, McLean NA, Johnston JM, Misra V, Verge VM, 2015. The Unfolded Protein Response and Cholesterol Biosynthesis Link Luman/CREB3 to Regenerative Axon Growth in Sensory Neurons. J Neurosci 35, 14557–14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Rao MV, Veeranna, Nixon RA, 2017. Neurofilaments and Neurofilament Proteins in Health and Disease. Cold Spring Harb Perspect Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR 3rd, 2013. Protein analysis by shotgun/bottom-up proteomics. Chem Rev 113, 2343–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Han B, Xia C, Zhuang X, 2019. Membrane-associated periodic skeleton is a signaling platform for RTK transactivation in neurons. Science 365, 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 1: Protein abundances indicative of iSN maturity.
a) Total protein levels for markers of neuronal progenitors.
b) Total protein levels for markers of nociceptors.
c) Levels of proteins involved in neuronal maturation identified by GO (GO: 0042551).
d) Levels of proteins involved in action potential and for cell receptors involved in mechanosensation
e) Levels of synaptic vesicle proteins.
Supplemental Figure 2: Tracking of mitochondria
a) The fraction of mitochondria exhibiting mobile characteristics was decreased from exposure to 100nM bortezomib.
b) Additional track parameters provided by the analysis of the mobile mitochondrial subpopulation with different bortezomib doses. Data shown represent 3 independent experiments using iSNs obtained from 2 different iPSC lines. All statistical comparisons were determined using a Tukey’s multiple comparison test. Brackets indicate statistically significant comparisons. Confinement ratio (CR) comparisons: control vs 10nM, p = 0.04; control vs 50nM, p = 0.01; control vs 100nM, p = 0.04. Track median speed comparisons: control vs 10nM, p = 0.03; 10nM vs 50nM, p = 0.046; 10nM vs 100nM, p = 0.0026.
c) Subdivision of the mobile mitochondria into 2 groups illustrates that bortezomib decreased the population of mitochondria that exhibit unidirectional (CR > 0.5) and bidirectional (0.25 < CR < 0.5) mobility patterns. (Mitochondria with CR > 0.5 were more sensitive to the effects of bortezomib.)
d) Track displacement and track mean speed were affected by bortezomib in the CR > 0.5 subgroup, but not in the 0.25 < CR < 0.5 subgroup. Track displacement comparisons (CR > 0.5): control vs 10nM, p = 0.0014; 10nM vs 100nM, p = 0.014. Track mean speed comparisons (CR > 0.5): control vs 10nM, p < 0.000001; 10nM vs 50nM, p = 0.000048; 10nM vs 100nM, p = 0.000014.
Supplemental Figure 3: Heatmap of protein level changes
Supplemental Figure 4: ClueGO results of the IPA cellular effects
a) RNA expression, protein synthesis & translation: biological processes, cellular components, and molecular function.
b) Viral infection: biological processes, cellular components, and molecular function.
c) Transport of molecule: biological processes, cellular components, and molecular function.
d) Export of molecule: biological processes, cellular components, and molecular function.
e) Endocytosis: biological processes, cellular components, and molecular function.
f) Homeostasis: biological processes, cellular components, and molecular function.
g) UPR-related proteins related to the heatmap in Figure 3C.
h) Biological processes of the UPR downstream effectors of XBP1, NFE2L2, ATF4, HSP90B1, and ATF6.
Supplemental Figure 5: microtubule-associated proteins are affected as a network in bortezomib-induced neurotoxicity
a) Heat map based on the Pearson’s correlation coefficient among the most significantly affected microtubule-associated proteins.
b) Illustration of the interconnectivity of microtubule-associated proteins with microtubule-dependent processes. Ribbon width represents the log2-fold change in protein levels.
Supplemental Table 1. Materials and Reagents associated with methodology
Supplemental Table 2. Protein level changes in iSNs for Bortezomib vs DMSO
Supplemental Table 3. Protein level changes in iSNs for DMSO vs Blank controls