ABSTRACT
Guanine-rich DNA strands can form secondary structures known as G-quadruplexes (G4-DNA or G4s). G4-DNA is important for the regulation of replication and transcription. We recently showed that the expression of Atg7, a gene that is critical for macroautophagy/autophagy, is controlled by G4-DNA in neurons. We demonstrated that the transcription factor SUB1/PC4 and the G4-DNA-specific antibody HF2 bind to a putative G4-DNA motif located in the Atg7 gene. Stabilizing G4-DNA with the G4-ligand pyridostatin (PDS) downregulates Atg7 expression in neurons. Here, we further investigated how G4-DNA in the Atg7 gene is stabilized by PDS. We show that PDS can form 1:1 and 2:1 complexes with the Atg7’s G4. We also demonstrate that PDS downregulates the ATG7 protein and the expression of Atg7 in astrocytes as well as in neurons. Together with our previous findings, these data establish a novel G4-DNA-associated mechanism of autophagy regulation at a transcriptional level in neurons and astrocytes.
KEYWORDS: Aging, astrocytes, autophagy, G-quadruplex, neurodegeneration, neurons
Manuscript
G-quadruplex-DNA (G4-DNA or G4) is a secondary DNA structure formed within guanine (G)-rich strands via self-association of four guanines to form G-quartets. Stacking and stabilization of these G-quartets are promoted by the coordination of certain cations (e.g., by potassium). G4-DNA regulates DNA replication, recombination, transcription, and telomere maintenance [1,2]. Putative G4-DNA motifs occur approximately once per 10 kb in the human genome and are frequently present in promoters of oncogenes and regulatory genes regions [3–5]. G4-DNA-binding proteins and G4-DNA unwinding helicases associate with and regulate G4-DNA formation [6–10]. Evidence indicates that these proteins regulate transcription of genes that contain G4-DNA [11,12]. In yeast, Sub1, a homolog of the mammalian transcription factor SUB1/PC4, interacts with the G4 helicase Pif1 to suppress G4-induced genomic instability by facilitating G4-DNA unfolding and enhancing transcription [13].
G4-DNA is also implicated in neurological diseases. An expansion of a non-coding GGGGCC repeat in the C9orf72 gene is associated with 10% of familial cases of frontotemporal dementia and amyotrophic lateral sclerosis [14]. The repeats form DNA, RNA, and DNA-RNA G4s, leading to an accumulation of ribonucleoprotein complexes that hamper nucleocytoplasmic trafficking and increase intranuclear stress [15,16]. G4-DNA is also implicated in Fragile X syndrome, which is caused by trinucleotide CGG repeat expansion. These abnormally expanded CGG repeats fold into a G4 structure and silence the fragile X mental retardation 1 (FMR1) gene [17]. Moreover, mutations in genes encoding DNA helicases are linked to other G4-DNA-related disorders, such as Werner syndrome with premature aging, Bloom syndrome with skin rash, developmental abnormalities, and predisposition to cancer, and Cockayne syndrome characterized by neurodegeneration and premature aging [18,19].
We recently demonstrated that G4-DNA plays a role in neuronal macroautophagy/autophagy. Autophagy is a fundamental intracellular process that removes aggregated proteins, senescent organelles, and parasites [20]. Autophagy is critical for cell survival and maintenance, proteostasis, organelle quality control, and prevention of cellular senescence and aging, among other processes [20]. The autophagic pathway is orchestrated by the autophagy-related (ATG) proteins, which nucleate the autophagosomal precursor phagophore and elongate the autophagosome, engulf a cytoplasmic cargo, and fuse the autophagosome with the lysosome [20]. Autophagy is regulated by transcription and translation, as well as by protein post-translational modifications [21]. A number of studies demonstrated that autophagic genes are subject to epigenetic silencing [22,23]. Аutophagy plays a positive role in longevity and deceleration of the aging process [24]. Recent studies have shown that autophagy-related genes are critical for longer healthspan and lifespan in worms, flies, and mice [24,25]. The expression of important autophagic genes such as Becn1 (beclin 1), Atg5 and Atg7, which are epigenetically regulated [26], diminishes with aging [27].
ATG7 is an E1-like enzyme that couples LC3-I to the E2-like enzyme ATG3 and leads to the E3-like complex ATG12–ATG5-ATG16L1. This E3-like complex then conjugates LC3-I to phosphatidylethanolamine in phagophore membranes, which is a critical step in autophagosome biogenesis [20]. Strong evidence supports the role of Atg7 in neurodegeneration. Atg7 and ATG7 are found to be downregulated in pre-clinical models of alpha-synucleinopathy and brain samples from patients with Lewy Body disease, suggesting a possible contribution of defective autophagy in the pathogenesis of this disorder. Increasing Atg7 via lentiviral delivery decreases the levels of synuclein alpha (SNCA) and mitigates neurodegeneration [28]. Deletion of Atg7 in Purkinje cells results in neurodegeneration [29]. Degeneration of Atg7-deficient neurons in the midbrain of conditional Atg7 knockout mice is accompanied by the formation of ubiquitinated inclusion bodies [30]. The expression of ATG7 is reduced in the human brain during normal aging [27]. Atg7 and its homologs are regulated by histone and chromatin modifications in flies, worms, and human non-neuronal cells [31,32].
The Atg7 gene (human, mouse, rat) contains numerous putative G4-forming motifs [33]. We recently investigated whether pharmacologically stabilizing G4-DNA with pyridostatin (PDS) [33], a selective G4-binding small molecule (referred to as a G4-ligand [34]), affects neuronal autophagy. Using circular dichroism spectroscopy (CD), thermal difference spectra (TDS) and nuclear magnetic resonance (NMR) analyses, we demonstrated that a putative G4-DNA forming sequence (PQFS) identified in the Atg7 gene folds into a stable G4 structure in vitro, which PDS strongly binds to [35]. We next showed that the G4-specific antibody (HF2) and the G4-binding protein SUB1/PC4 bind to the Atg7’s PQFS (Figure 1A; a scheme of the Atg7 gene) [35]. Cultured cortical neurons treated with PDS exhibited diminished Atg7 expression and reduced autophagy [35]. Mice treated with PDS exhibited memory deficits and accumulation of lipofuscin [35]. We also discovered that G4-DNA is abundantly present in aged mouse brain, but not in the brains of young mice [35].
Figure 1.
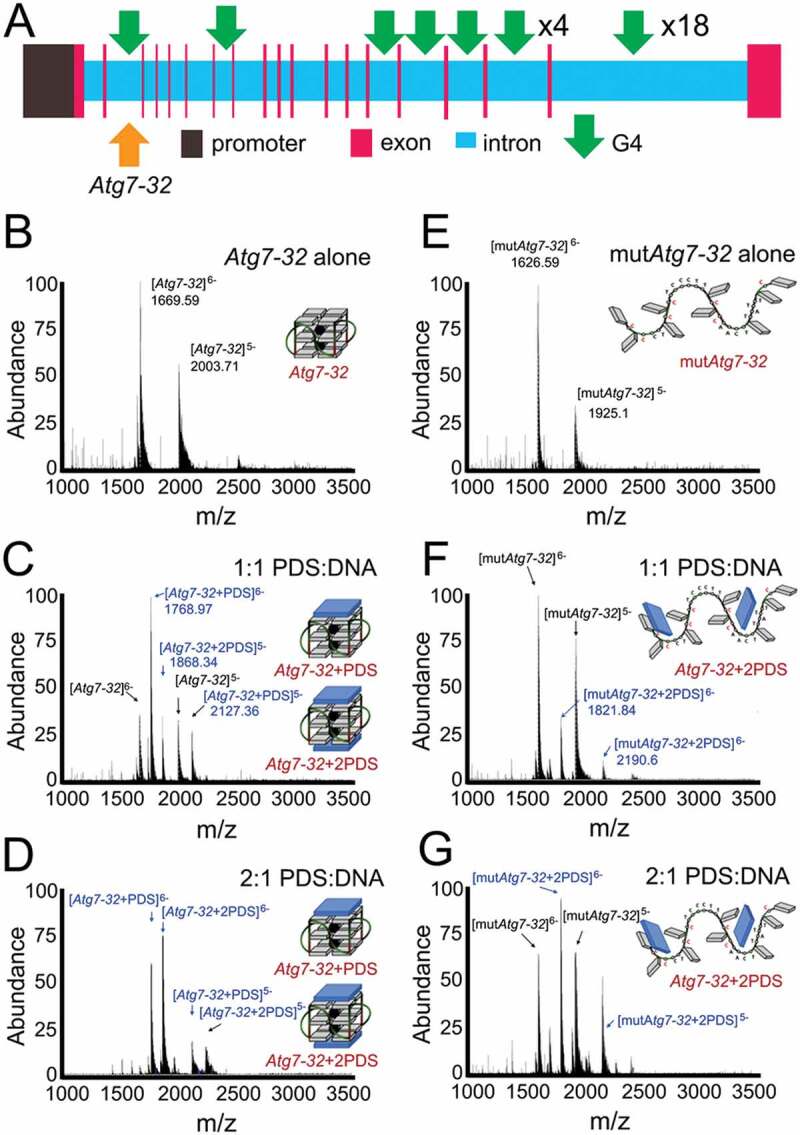
The stoichiometry and equilibrium binding constant of the non-covalent Atg7-32/PDS complexes. (A) A scheme of the rat Atg7 gene and its promoter showing putative G4-DNA locations. PQFSes in Atg7 and in its promoter were analyzed by using the QGRS mapper (http://bioinformatics.ramapo.edu/QGRS/index.php). 5,000 nt upstream the start codon were analyzed. (B-D) Electrospray ionization mass spectrometry (ESI-MS) experiments performed with Atg7-32 alone (10 µM) (B) or in presence of 1 and 2 mol. equiv. PDS (C and D, respectively). Mixtures were prepared in 100 mM ammonium acetate buffer and equilibrated at 25°C for 1 h prior to the experiments (20% of methanol added to the solution for the injection, performed at a flow rate of 10 μL/min). K = 4.82x105 M−1 (C), K = 1.47x107 M−1 (D). (E-G) ESI-MS experiments performed with mutAtg7-32 alone (10 µM) (E) or in presence of 1 and 2 mol. equiv. PDS (F and G, respectively). Mixtures were prepared in 100 mM ammonium acetate buffer and equilibrated at 25°C for 1 h prior to the experiments (20% of methanol added to the solution for the injection, performed at a flow rate of 10 µL/min). K = 2.19x104 M−1 (F), K = 7.73x104 M−1 (G)
Our findings indicate that an age-associated change in DNA conformation via the formation of stable G4-DNA could be a novel mechanism of autophagy regulation in aging neurons. Our data also suggest that anti-neoplastic agents that target G4-DNA [36] (i.e., G4-ligands) may accelerate brain aging and lead to neurodegenerative diseases.
Importantly, our previous findings indicate that endogenous G4-DNA ligands, small molecules or proteins, may regulate neuronal transcription in general and the expression of Atg7 in particular, thereby modulating autophagy. To go a step further, we investigated a mechanism of PDS interaction with a G4-DNA motif from the Atg7 gene, so called Atg7-32, that we discovered previously (Figure 1A). The Atg7-32 motif (d[5ʹG4CTG4TC3T2G4A2CTGTAT2G33']), a 32-nt G-rich sequence, was demonstrated to adopt a stable G4-DNA structure in vitro by CD, TDS and NMR. Here, we used the electrospray ionization mass spectrometry (ESI-MS) technique as it allows measuring both the stoichiometry and equilibrium binding constant of the non-covalent G4-DNA/PDS complexes [37]. With ESI-MS, we showed that a 1:1 mixture of PDS:DNA results in the 1:1 PDS:G4 complex (54.6%), along with unbound DNA (36.4%) and a fraction of the 2:1 PDS:G4 complex (9%) (Figure 1B and Figure 1C). Increasing amount of PDS to a 2:1 mixture of ligand:DNA results in the 2:1 PDS:G4 complex (60.8%), the 1:1 PDS:G4 complex (38.5%), and some residual unbound DNA (0.7%) (Figure 1D). Our data indicate that PDS has a high affinity for the Atg7-32 G4 (K = 1.47x107 M−1), and that both external G-quartets provide equivalent binding sites for PDS. We then performed a series of control experiments with mutated Atg7-32 (mutAtg7-32; d[5ʹGCGCCTGCGCTC3T2GCGCA2CTGTAT2GCG3ʹ]), a DNA sequence that cannot fold into a G4 structure due to the G-to-C mutations (underlined) in Atg7-32. We discovered a low affinity, electrostatically driven association of PDS and mutAtg7-32 (K = 7.73x104 M−1) as a 2:1 mixture PDS:mutAtg7-32, which is ~2 orders of a magnitude lower than of Atg7-32 (Figure 1E-G). We, therefore, confirmed the interaction specificity between PDS and a G4-DNA motif from the Atg7 gene.
Since neurons are highly specialized post-mitotic cells, we speculated that neuronal G4 pathways may be drastically different from G4-based mechanisms in their dividing symbiotic cells, namely in astrocytes. We first confirmed that our astrocytic cultures are pure astrocytes [38]. Astrocytes were grown in culture in DMEM + serum for 3 weeks, fixed, and stained with fluorescent phalloidin and antibodies against GFAP (Figure 2A). Expectedly, neurons, microglia, and oligodendrocytes, which all require specific medium condition, died off and remaining cells were homogeneous cultures of astrocytes (Figure 2A). We then tested whether stabilizing G4s downregulates Atg7 in astrocytes, as in neurons, using qRT-PCR. We found that Atg7’s mRNA levels were 2-fold lower in PDS-treated compared to vehicle-treated astrocytes (p = 0.017) (Figure 2B). Tbp (TATA-binding protein) mRNA was used as loading control as neither Tbp nor its promoter contains a PQFS [39]. Intriguingly, we had previously shown that expression of Atg7 was 7-fold lower in PDS-treated compared to vehicle-treated neurons, suggesting that neurons are more sensitive to PDS treatment than astrocytes. Since the levels of Atg7’s mRNA are reduced by PDS treatment, we tested if the ATG7 protein is similarly affected by PDS. We found that ATG7 was about 30% lower in astrocytes exposed to PDS than in vehicle-treated astrocytes (p = 0.019) (Figure 2C and Figure 2D). As astrocytes basally have low levels of autophagy [38,40], G4 stabilization still leads to downregulating ATG7 levels in these cells. We also looked at the levels of SQSTM1, which is commonly used as an autophagic reporter. PDS promoted accumulation of SQSTM1 by 2-fold, confirming slowed autophagy (p = 0.018) (Figure 2C and Figure 2D).
Figure 2.
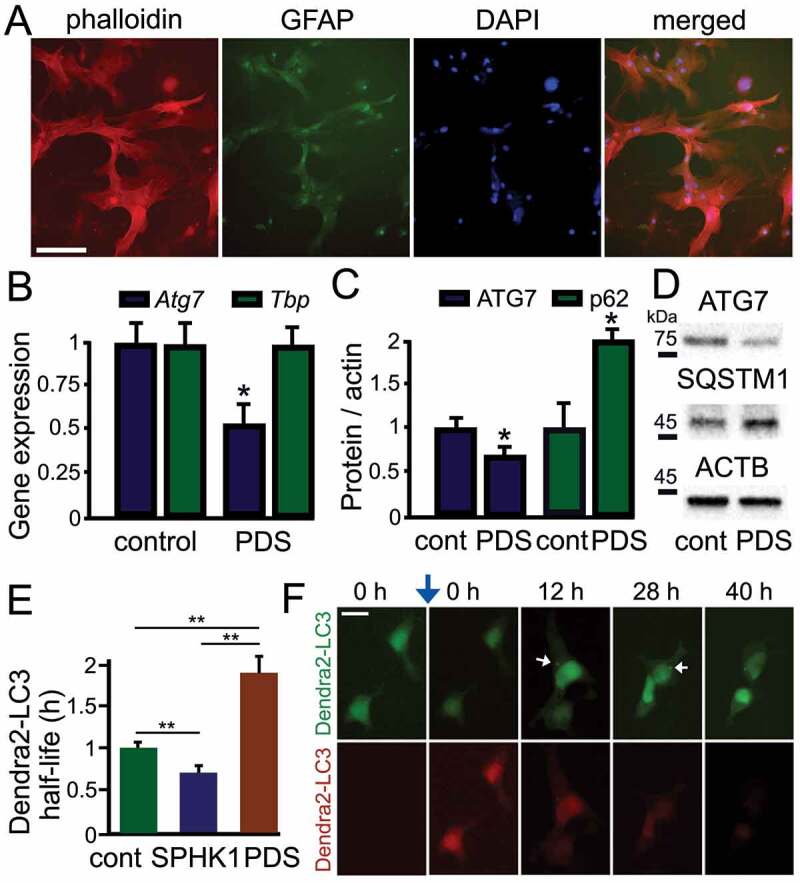
PDS downregulates the ATG7 protein and the expression of Atg7 in astrocytes. (A) Primary astrocytes were cultured for 3 weeks from cortical tissue isolated from rat embryos. Cultures were fixed and stained with Alexa Fluor 633 phalloidin (red), DAPI (blue), and an antibody against GFAP (green). Most cells stained with phalloidin were positive for GFAP (94 ± 6%). Over 200 cells were analyzed, and results were pooled from three independent experiments. Scale bar: 40 µm. (B) Cultured primary astrocytes were treated with a vehicle (control) or with PDS (2 μM) overnight. Astrocytes were then processed to measure mRNA levels. Expression levels of Atg7 and Tbp (housekeeping gene as control) were determined by qRT-PCR. Atg7: p = 0.017 (t-test), *p < 0.05; Tbp: p = 0.7949 (t-test), n.s., non-significant. Results were pooled from six experiments. (C) Cultured primary astrocytes were treated with a vehicle (control) or with PDS (2 μM) overnight. Astrocytes were then processed to measure the levels of ATG7 and SQSTM1. ATG7: *p = 0.019 (t-test), SQSTM1: *p = 0.018 (t-test). Results were pooled from six experiments. (D) An example of western blotting experiments in (C). (E) Dendra2-LC3 was used to measure autophagy flux. Two cohorts of astrocytes were co-transfected with Dendra2-LC3 and an empty plasmid, or with Dendra2-LC3 and untagged SPHK1 (sphingosine kinase 1, a positive control). Astrocytes co-transfected with Dendra2-LC3 and an empty plasmid were treated with a vehicle (control, cont), or with 0.5 µM PDS overnight. After treatment, cells were photoswitched and longitudinally imaged, and the decay of the red fluorescence over time was used to calculate the half-life of Dendra2-LC3. The half-life of Dendra2-LC3 is normalized to one with respect to control astrocytes. cont vs SK1: *p = 0.001, cont vs PDS: **p = 0.001, SK1 vs PDS: **p = 0.001 (one-way ANOVA followed by Tukey’s multiple comparisons testing). Fifty cells per group were analyzed from two independent experiments. (F) An example of a photoswitching experiment with Dendra2-LC3 and SPHK1 (untagged). Astrocytes co-transfected with Dendra2-LC3 and SPHK1 were treated with a vehicle overnight. Cells were photoswitched and longitudinally imaged. Autophagosomes are depicted with white arrows. Blue arrow depicts photoswitching. Scale bar: 25 µm
We confirmed that the flux through autophagy is reduced by PDS with an optical pulse-chase method. The Dendra2-LC3 reporter was used to study autophagic flux in astrocytes. Astrocytic cultures were transfected with Dendra2-LC3 and photoswitched 24 h thereafter. Co-transfection with the Sphk1 (sphingosine kinase 1) gene, a genetic enhancer of autophagy [38,41,42], was used as a positive control. The decay of “red” Dendra2 was analyzed, and the flux via autophagy was measured. PDS inhibited the flux through autophagy (Figure 2E and Figure 2F).
Next, cultured primary astrocytes were treated with either a vehicle or PDS then stained with a G4-DNA fluorophore, N-TASQ that has been used to investigate G4-DNA landscapes in cancer cells [43–45] and neurons [35]. Astrocytes were treated, fixed, permeabilized, and then stained with both N-TASQ (G4s) and DAPI (nuclear DNA). The nuclear puncta index, which is a standard deviation of the intensities measured among pixels within the region of interest (here, the nucleus), was measured. A low index indicates a diffuse N-TASQ nuclear staining while a high index indicates the presence of N-TASQ nuclear foci. We found that PDS significantly promotes G4-DNA formation in the astrocytic nuclei (p < 0.0001) (Figure 3A-C). Intriguingly, we also found that, unlike neuronal cells, both vehicle- and PDS-treated astrocytes contain N-TASQ foci in the cytoplasm. Previously, it was demonstrated that cancerous cells frequently contain N-TASQ foci in the cytoplasm 43–46] (G4-RNA [47]), as cultured astrocytes in our samples. These N-TASQ foci mostly consisted of ribosomal RNA and long non-coding RNA in cancer cells [46]. RNase treatment was thus performed to enhance the contrast of the nuclear staining. We discovered that, in RNase-treated cells, PDS-treated astrocytes exhibit higher levels of N-TASQ fluorescence than vehicle-treated cells, indicating that PDS modulates a G4 landscape in astrocytes, as in neurons (Figure 3A-C). Together, our data indicate that stabilizing G4-DNA with PDS downregulates autophagy in neurons and in astrocytes, suggesting a common mechanism in these symbiotic brain cells.
Figure 3.
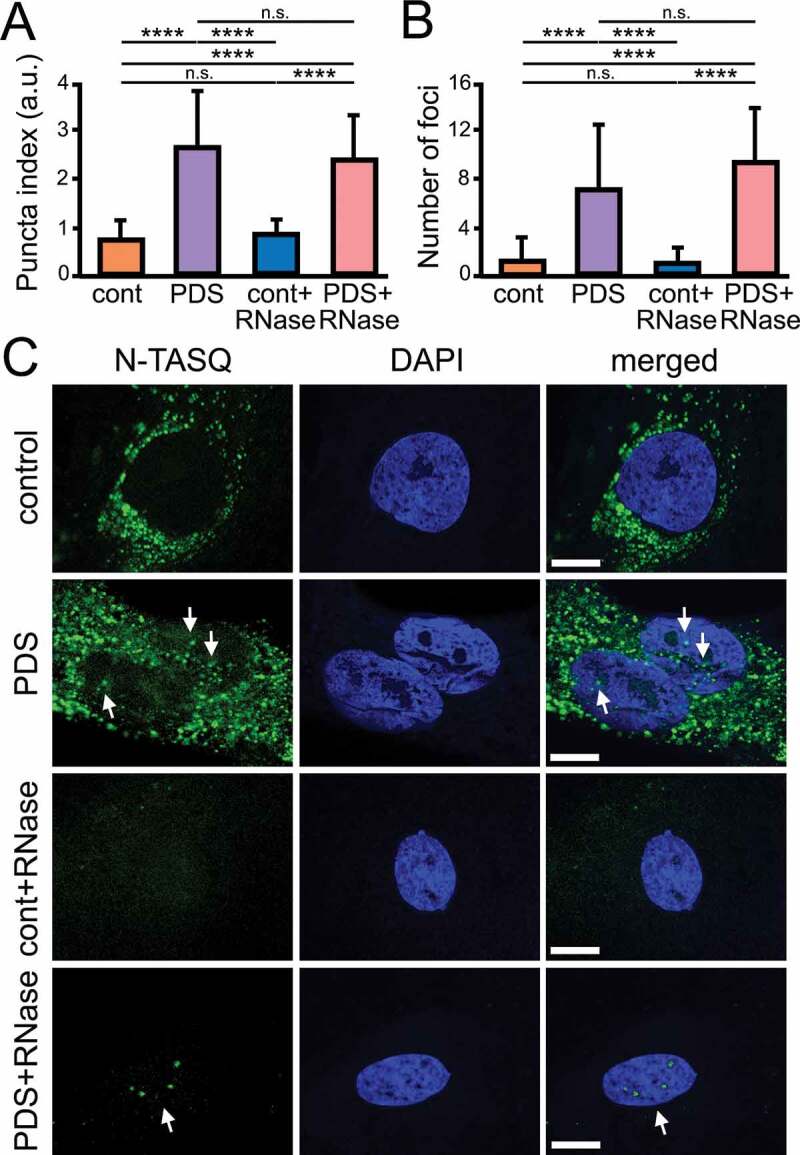
PDS alters G4 landscapes in astrocytes. (A) Cultured primary astrocytes were treated with a vehicle (control) or with PDS (2 μM) overnight. Cells were fixed and stained with N-TASQ (50 µM) and with the nuclear dye Hoechst (DAPI). Some samples were treated with RNase to digest RNA before staining. Fixed cells were automatically imaged with the EVOS microscope (N-TASQ and DAPI) and the nuclear puncta index was analyzed in all conditions. Two-way ANOVA followed by Tukey’s multiple comparisons test were used. ****p < 0.0001, n.s., non-significant. For each experiment, 80 cells were blindly analyzed, and results were pooled from two independent experiments. (B) The number of puncta was analyzed in cells from (A) with ImageJ. Two-way ANOVA followed by Tukey’s multiple comparisons test were used. ****p < 0.0001, n.s., non-significant. (C) Cultured primary astrocytes were treated with a vehicle (control) or with PDS (2 μM) overnight. Cells were fixed and stained with N-TASQ (50 µM) and with the nuclear dye Hoechst (DAPI). Some samples were treated with RNase to digest RNA before staining. Fixed cells were analyzed with a confocal microscope (N-TASQ and DAPI). Scale bar: 1 µm
During aging and age-associated diseases, astrocytic chromatin undergoes epigenetic modifications, including DNA methylation and histone modifications [48]. Our findings demonstrate that stabilized G4-DNA might represent an additional epigenetic-like mechanism that regulates gene expression in astrocytes. Investigating G4-DNA-associated mechanisms in astrocytes and in other cell types in the central nervous system may lead to novel therapies for neurodegenerative disorders.
Materials and methods
Chemicals, plasmids, and antibodies
N-TASQ was synthesized as described [43]. PDS was from Sigma (SML0678) and from Selleck Chemicals (S7444). The DAPI dye was from Thermo Fisher Scientific (D1306). Ribonuclease A (RNase) from bovine pancreas was from Sigma (R4642). pGW1-Dendra2-LC3 and pGW1-SK1 were cloned by us and were described previously [49]. pGW1 (British Biotech, discontinued) was described previously [49]. Antibodies against ACTB/β-actin were from Cell Signaling Technology (clone 8H10D10; 3700; 1:2000). Antibodies against LC3 were from MBL (PD014; 1:1000). Antibodies against SQSTM1/p62 were from BD Biosciences (610833; 1:2000). Antibodies against GFAP were from Santa Cruz Biotechnology (sc-9065; 1:100). Alexa Fluor 633 phalloidin was a gift from Dr. Taeyeop Park (Carmen Dessauer’s lab, Integrative Biology and Pharmacology, the University of Texas McGovern Medical School) (Thermo Fisher Scientific, A22284). Antibodies against rabbit IgG(H + L) conjugated with horseradish peroxidase (HRP; AP307P; 1:3000), and mouse IgG(H + L) conjugated with horseradish peroxidase (AP308P; 1:3000) were from EMD Millipore.
G4-DNA analyses
The QGRS mapper (http://bioinformatics.ramapo.edu/QGRS/index.php) was used to determine the potential G4-DNA structures contained in genes of interest and their G-scores. Search parameters: maximal length: 45; minimal G-group size: 3; loop size: from 0 to 10 [2].
Electrospray ionization mass spectrometry
Electrospray ionization mass spectrometry (ESI-MS) experiments were performed on an LTQ Orbitrap XL (Thermo Fisher Scientific) spectrometer equipped with Ion Max source and HESI-II probe in the negative ion mode. Both Atg7-32 (10 µM) and mutAtg7-32 (10 µM) alone, as well as the corresponding DNA:PDS mixtures (1:1 and 1:2 mol. equiv.), were prepared in 100 mM ammonium acetate buffer (Sigma, 73594–25G-F) and equilibrated at 25°C for 1 h. To obtain a stable electrospray signal, 20% of methanol were added to the solution just before injection. The solutions were injected with syringe pump at a flow rate of 5 µL/min. The full scan mass was recorded in 600–4000 m/z range. The following tuning parameters were used: heater temperature = 50°C, spray voltage = 4.0 kV, capillary temperature = 275°C, tube lens = −160.00 (negative ion mode) and the capillary voltage varied between −35.00 V and −60.00 V. Quantification of the equilibrium affinity constants (K) of PDS for Atg7-32 and mutAtg7-32 was done according to Rosu et al., via the equation K = [DNA:PDS]/([DNAfree][PDSfree]) [50].
Astrocytic Cultures and Treatments
Cortices from rat embryos (E17–18) were dissected, dissociated, and plated on 24-well tissue-culture plates (4x105/well) coated with poly-D-lysine (BD Biosciences, A-003-E). Primary cortical astrocytes were grown in Dulbecco´s Modified Eagle Medium (HyClone, SH3024301) supplemented with 10% heat-inactivated fetal bovine serum (Sigma, F4135) and penicillin-streptomycin as described at least for 3 weeks before experiments [38]. Some cultures were treated with vehicle or PDS (2 μM, overnight). Some astrocytic cultures were fixed with 4% PFA, permeabilized with 0.1% Triton X-100 (Santa Cruz Biotechnology, sc-29112) in PBS (Bio-Rad Laboratories, 161–0780), washed 3 times with PBS, incubated with 1 mg/ml RNase for 1 h at 37°C. Samples were then washed 3 times with PBS and then labeled with N-TASQ (50 µM, overnight, room temperature). Some cultures were transfected with Lipofectamine 2000 (Thermo Fisher Scientific, 11668027) and a total of 1–2 μg of plasmid DNA per well, as described [38].
Western blotting
Western blotting was performed as described [38]. Cleared cellular lysates were analyzed by SDS/PAGE and proteins were transferred onto PVDF membranes (Thermo Fisher Scientific, IB401031) by the iBlot2 system (Thermo Fisher Scientific). Membranes were blocked and incubated overnight with antibodies against ACTB, LC3 or SQSTM1. Membranes were washed and probed for 1 h with anti-rabbit or anti-mouse antibodies conjugated with horseradish peroxidase. Signals were detected using ProSignal Pico (Genesee Scientific, 20–300B) on Medical X-Ray Film (Kodak, 7400).
RNA Extraction and qRT-PCR
Total RNA was extracted from primary culture using the RNeasy Mini kit (Qiagen, 74104), and then reverse transcribed using iScript Reverse Transcription SuperMix (Bio-Rad, 1708840), according to the manufacturer’s protocol and as described [39]. RT-qPCR was performed using a Bio-Rad CFX96 Touch machine using SSoAdvanced Universal SYBR Green (Bio-Rad, 1725275) for visualization and quantification according to the manufacturer’s instructions. Primer sequences were: Atg7, forward: d[5′TC2TGAGAGCATC3TCTA2TC3′], reverse: d[5′CT2CAGT2CGACACAG2TCATC3′]; Tbp, forward: d[5′AGTGC3AGCATCACTGT33′], reverse: d[5′G2TC2ATGACTCTCACT3CT23′]. The PCR conditions were 95°C for 3 min, followed by 40 cycles of 95°C for 10 s and 55°C for 30 s. Relative expression levels were calculated from the average threshold cycle number using the delta-delta Ct method.
Fluorescence microscopy and image analysis
Cell imaging was performed with the EVOS FL Auto Imaging System (Thermo Fisher Scientific). Puncta formation and puncta indexes were analyzed as described [38]. Briefly, the redistribution of green fluorescence (N-TASQ) into punctate structures was reflected by the puncta index, which is the standard deviation of the intensities measured among pixels within the cellular region of interest (the nucleus, the DAPI staining). Diffuse localization corresponds to a low puncta index, and punctate localization corresponds to a high puncta index.
Photoswitching of Dendra2-LC3 was performed with the EVOS automated microscope (Thermo Fisher Scientific) as previously described [49]. Upon irradiation with blue visible light, Dendra2 undergoes an irreversible conformational change. Dendra2’s spectral properties then change from that of a protein that absorbs blue light and emits green fluorescence to that of one that absorbs green light and emits red fluorescence. Photoswitched Dendra2 maintains these spectral properties until the cell degrades the protein. The red fluorescence intensities from a region of interest in individual astrocytes were measured at different time points with the EVOS microscope. The decays of red fluorescence were plotted against time, transformed into log values, and individual half-life (t1/2) was analyzed. t1/2 = (Ln(2)/λ), were λ is the decay value.
Astrocytes were imaged with the Nikon A1 R confocal laser microscope (Nikon Corporation) with the 100X Plan-Apo/1.4 NA oil lens. N-TASQ was imaged with the 488 nm laser and the DAPI dye was imaged with the 405 nm laser. Analyses were done blinded to treatment group.
Ethics statement
Rats were maintained in accordance with guidelines and regulations of the University of Texas McGovern Medical School at Houston. All experimental procedures were approved by the University of Texas McGovern Medical School at Houston.
Statistical analysis
GraphPad Prism was used to perform statistical analyses. Statistical tests are denoted within each figure legend.
Acknowledgments
We thank members of the AST and LDM laboratories for useful discussions. We apologize to all authors whose work we could not cite due to reference number constraints.
Funding Statement
This work was supported by the University of Texas McGovern Medical School at Houston (A.S.T.); Agence Nationale de la Recherche [ANR-17-CE17-0010-01] (D.M.); National Institute of General Medical Sciences [R01GM116007] (N.K.); National Institute of Neurological Disorders and Stroke [R01NS094543] (L.D.M.); National Institute on Aging [RF1AG057576] (A.U.) and [R21AG067204] (A.S.T); Welch Foundation [AU1875] (N.K.).
Disclosure statement
No potential conflicts of interest are disclosed.
References
- [1].Rhodes D, Lipps HJ.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Maizels N, Gray LT.. The G4 genome. PLoS Genet. 2013;9:e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Raiber EA, Kranaster R, Lam E, et al. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012;40:1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gray LT, Vallur AC, Eddy J, et al. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat Chem Biol. 2014;10:313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Paeschke K, Bochman ML, Garcia PD, et al. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sauer M, Paeschke K. G-quadruplex unwinding helicases and their function in vivo. Biochem Soc Trans. 2017. DOI: 10.1042/BST20170097 [DOI] [PubMed] [Google Scholar]
- [10].Chen MC, Tippana R, Demeshkina NA, et al. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature. 2018;558:465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nguyen GH, Tang W, Robles AI, et al. Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc Natl Acad Sci U S A. 2014;111:9905–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tang W, Robles AI, Beyer RP, et al. The Werner syndrome RECQ helicase targets G4 DNA in human cells to modulate transcription. Hum Mol Genet. 2016;25:2060–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lopez CR, Singh S, Hambarde S, et al. Yeast Sub1 and human PC4 are G-quadruplex binding proteins that suppress genome instability at co-transcriptionally formed G4 DNA. Nucleic Acids Res. 2017;45:5850–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Maizels N. G4-associated human diseases. EMBO Rep. 2015;16:910–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Haeusler AR, Donnelly CJ, Rothstein JD. The expanding biology of the C9orf72 nucleotide repeat expansion in neurodegenerative disease. Nat Rev Neurosci. 2016;17:383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol. 2018;14:544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274:12797–12802. [DOI] [PubMed] [Google Scholar]
- [18].Karikkineth AC, Scheibye-Knudsen M, Fivenson E, et al. Cockayne syndrome: clinical features, model systems and pathways. Ageing Res Rev. 2017 Jan;33:3–17. doi:10.1016/j.arr.2016.08.002. https://pubmed.ncbi.nlm.nih.gov/27507608/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mendoza O, Bourdoncle A, Boule JB, et al. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44:1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. Embo J. 2017;36:1811–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lubas M, Harder LM, Kumsta C, et al. eIF5A is required for autophagy by mediating ATG3 translation. EMBO Rep. 2018 Jun;19(6):e46072. doi:10.15252/embr.201846072. https://pubmed.ncbi.nlm.nih.gov/29712776/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Artal-martinez de Narvajas A, Gomez TS, Zhang JS, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013;33:3983–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Baek SH, Kim KI. Epigenetic control of autophagy: nuclear events gain more attention. Mol Cell. 2017;65:781–785. [DOI] [PubMed] [Google Scholar]
- [24].Hansen M, Rubinsztein DC, Walker DW. Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol. 2018 Sep;19(9):579–593. doi:10.1038/s41580-018-0033-y. https://pubmed.ncbi.nlm.nih.gov/30006559/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fernandez AF, Sebti S, Wei Y, et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature. 2018;558:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lapierre LR, Kumsta C, Sandri M, et al. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy. 2015;11:867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lipinski MM, Zheng B, Lu T, et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2010;107:14164–14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Crews L, Spencer B, Desplats P, et al. Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of alpha-synucleinopathy. PLoS One. 2010;5:e9313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [29].Komatsu M, Wang QJ, Holstein GR, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104:14489–14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Friedman LG, Lachenmayer ML, Wang J, et al. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of alpha-synuclein and LRRK2 in the brain. J Neurosci. 2012;32:7585–7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Eisenberg T, Knauer H, Schauer A, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. [DOI] [PubMed] [Google Scholar]
- [32].Eisenberg T, Schroeder S, Andryushkova A, et al. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab. 2014;19:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rodriguez R, Miller KM, Forment JV, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8:301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Neidle S. Quadruplex nucleic acids as novel therapeutic targets. J Med Chem. 2016;59:5987–6011. [DOI] [PubMed] [Google Scholar]
- [35].Moruno-Manchon JF, Lejault P, Wang Y, et al. Small-molecule G-quadruplex stabilizers reveal a novel pathway of autophagy regulation in neurons. eLife. 2020;9:e52283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cimino-Reale G, Zaffaroni N, Folini M. Emerging role of G-quadruplex DNA as target in anticancer therapy. Curr Pharm Des. 2016;22:6612–6624. [DOI] [PubMed] [Google Scholar]
- [37].Rosu F, De Pauw E, Gabelica V. Electrospray mass spectrometry to study drug-nucleic acids interactions. Biochimie. 2008;90:1074–1087. [DOI] [PubMed] [Google Scholar]
- [38].Moruno-Manchon JF, Uzor NE, Ambati CR, et al. Sphingosine kinase 1-associated autophagy differs between neurons and astrocytes. Cell Death Dis. 2018;9:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Moruno-Manchon JF, Koellhoffer EC, Gopakumar J, et al. The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging (Albany NY). 2017;9:1957–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kulkarni A, Dong A, Kulkarni VV, et al. Differential regulation of autophagy during metabolic stress in astrocytes and neurons. 2019. p. 1–17. https://pubmed.ncbi.nlm.nih.gov/31876243/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lavieu G, Scarlatti F, Sala G, et al. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem. 2006;281:8518–8527. [DOI] [PubMed] [Google Scholar]
- [42].Moruno Manchon JF, Uzor NE, Finkbeiner S, et al. SPHK1/sphingosine kinase 1-mediated autophagy differs between neurons and SH-SY5Y neuroblastoma cells. Autophagy. 2016;12:1418–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Laguerre A, Hukezalie K, Winckler P, et al. Visualization of RNA-Quadruplexes in Live Cells. J Am Chem Soc. 2015;137:8521–8525. [DOI] [PubMed] [Google Scholar]
- [44].Laguerre A, Wong JM, Monchaud D. Direct visualization of both DNA and RNA quadruplexes in human cells via an uncommon spectroscopic method. Sci Rep. 2016;6:32141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stefan L, Monchaud D. Applications of guanine quartets in nanotechnology and chemical biology. Nat Rev Chem. 2019;3:650–668. [Google Scholar]
- [46].Yang SY, Lejault P, Chevrier S, et al. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat Commun. 2018;9:4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kamura T, Katsuda Y, Kitamura Y, et al. G-quadruplexes in mRNA: A key structure for biological function. Biochem Biophys Res Commun. 2020;526:261–266. [DOI] [PubMed] [Google Scholar]
- [48].Neal M, Richardson JR. Epigenetic regulation of astrocyte function in neuroinflammation and neurodegeneration. Biochim Biophys Acta (BBA) - Mol Basis Dis. 2018;1864:432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tsvetkov AS, Arrasate M, Barmada S, et al. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol. 2013;9:586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rosu F, Gabelica V, Poncelet H, et al. Tetramolecular G-quadruplex formation pathways studied by electrospray mass spectrometry. Nucleic Acids Res. 2010;38:5217–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]


