Abstract
Background
Chemo-resistance is still considered one of the key factors in the mortality of ovarian cancer. In this work, we found that ubiquitin-conjugating enzyme E2 N (UBE2N) is downregulated in paclitaxel-resistant ovarian cancer cells. It suggests UBE2N to be critical in the regulation of paclitaxel sensitivity in ovarian cancer.
Materials and Methods
Ovarian cancer cells with stably overexpressed UBE2N were injected into nude mice to assess tumor growth and paclitaxel sensitivity in vivo. The MTT assay was applied to observe the effect of UBE2N expression on paclitaxel sensitivity. A real-time PCR array, specific for human cancer drug resistance, was used to examine the potential downstream target genes of UBE2N. The expression of UBE2N and potential downstream target genes was determined by Western blotting. The analysis of Gene Ontology and protein–protein interactions of these differentially expressed genes (DEGs) was performed using online tools. To evaluate the prognostic value of hub genes expression for ovarian cancer patients treated with paclitaxel, we applied the online survival analysis tool.
Results
Overexpressed UBE2N enhanced the paclitaxel sensitivity of ovarian cancer cells in vitro and in vivo. Thirteen upregulated DEGs and 11 downregulated DEGs were identified when we knockdown UBE2N. Meanwhile, 9 hub genes with a high degree of connectivity were selected. Only Fos proto-oncogene, AP-1 transcription factor subunit (Fos), was overexpressed upon decreasing UBE2N levels, indicating a poor outcome for patients treated with paclitaxel. Moreover, reduced UBE2N could increase Fos expression and reduce P53. Furthermore, reversed regulation of Fos and P53 based on UBE2N reduction could reverse paclitaxel sensitivity, respectively.
Conclusion
Our study suggests that UBE2N could be used as a therapeutic agent for paclitaxel-resistant ovarian cancer through Fos/P53 pathway. Further studies are needed to elucidate the specific mechanism.
Keywords: ovarian cancer, UBE2N, paclitaxel sensitivity, Fos, P53
Introduction
Ovarian cancer is the most devastating form of all gynecologic cancers. Despite the improvement of surgical techniques and chemotherapeutics, the overall 5-year survival rate for ovarian cancer is as low as 47%.1 The primary strategy of the treatment for ovarian cancer consists of primary cytoreductive surgery combined with paclitaxel and carboplatin chemotherapy. According to the data in recent years, more than 80% of the patients have shown response to standard chemotherapy. However, most of them eventually succumb to relapse due to the resistance to chemotherapy.2 It remains the principal factor for treatment failure of ovarian cancer. Evidence has shown that multiple factors could contribute to paclitaxel resistance, which might originate from a series of modifications and regulatory mechanisms.3–7 However, the principal mechanism of chemoresistance in ovarian cancer remains unresolved.
In our previous study, we applied a 2-dimensional fluorescence difference in gel electrophoresis (DIGE) quantitative proteomic analysis to determine differentially expressed proteins between the ovarian cancer cell line (SKOV3) and its corresponding paclitaxel-resistant cell line (SKOV3-TR30). We found that ubiquitin-conjugating enzyme E2 N (UBE2N) was decreased remarkably in SKOV3-TR30 cells.8 As an important post-translational modification, ubiquitination is reported to be involved in various cellular processes.9,10 Additionally, recent findings reveal that ubiquitination participates in the regulation of the sensitivity of tumor cells to some chemotherapeutic agents.11–13 UBE2N, a ubiquitin-conjugating enzyme, plays a central role in ubiquitin-mediated cellular activities, such as signal transduction.14 UBE2N is involved in the development of various cancers.15–18 Additionally, UBE2N mediates non-canonical ubiquitination to regulate DNA damage repair,19–21 and ultimately modulates drug resistance.22,23 However, the link between paclitaxel and UBE2N expression is still unclear.
Here, we first verified the enhanced effect of paclitaxel sensitivity of UBE2N in ovarian cancer cells in vivo. Moreover, using PCR array and bioinformatic analysis, we demonstrated that Fos/P53 pathway participated in regulating paclitaxel sensitivity of UBE2N. Overall, our study reveals that UBE2N is a potential therapeutic agent for ovarian cancer treatment.
Materials and Methods
In vivo Xenograft Experiments
Female BALB/C nude mice of 2–4 weeks age were purchased from the Zhejiang Laboratory Animal Center (Hangzhou, China) and housed within a dedicated SPF facility at the Laboratory Animal Center of Zhejiang University. We harvested the A2780 cells with UBE2N stably overexpressed or in negative expression. They are, after being washed with PBS, resuspended in serum-free medium (Gibco, Grand Island, New York, USA), and injected subcutaneously into the right axilla of each mouse (2 × 106 cells per injection). Once the mice developed detectable tumors, they were divided to the following treatment groups: (1) overexpressing UBE2N, received paclitaxel (Bristol-Myers Squibb Company, New York, USA) (n = 6); (2) negative control, received paclitaxel (n = 5); (3) overexpressing UBE2N, received saline (n = 6); (4) negative control, received saline (n = 5). Paclitaxel (15 mg/kg) and the same volume of saline were intraperitoneally injected once a week for 3 weeks. The health of the mice and tumor growth were examined every 3 days. All mice were sacrificed on day 7 after the completion of drug treatment. The tumors were excised, weighed, and photographed. Tumor volume (mm3) was calculated as width2/length. All experiments were performed following Animal Research Reporting in vivo Experiments (ARRIVE) guidelines for the use of laboratory animals and were approved by the ethics committee of Zhejiang University. The study was performed in accordance with the Chinese Animal Welfare Guideline GB/T 35,892–2018.
Cell Culture
The human ovarian cancer cell lines A2780 and SKOV3 were purchased from Sigma-Aldrich (St. Louis, Missouri, USA) and American Type Culture Collection (ATCC), respectively. Cell lines were maintained in RPMI-1640 (Gibco, Grand Island, New York, USA) or MyCoy’5A (Gibco, Grand Island, New York, USA) with 10% fetal bovine serum (Gibco, Grand Island, New York, USA) in 5% CO2 at 37°C. For more information see Supplementary Materials and Methods.
Cell Viability Assay
A2780 and SKOV3 cells were transfected with siRNA (GenePharma, Shanghai, China) or plasmid (Genscript, Nanjing, China) for 24 hours. The cells were then resuspended in culture medium and seeded in 96-well plates (4000 cells/well). Cells were treated with various concentrations of paclitaxel (2, 5, 10, 20, 50, 100, and 200 nM) for 48 hours after cells adhered onto plates. Cell viability was determined using the CellTiter 96 AQueous One Solution Cell proliferation kit (Promega, Madison, Wisconsin, USA).
Plasmid and siRNA Transfection
Full-length UBE2N was cloned into the pEGFP-C1 vector, while UBE2N-specific shRNA was cloned into the pGPH1/Neo vector. The sequence of UBE2N-shRNA was identical to UBE2N-siRNA1. The X-treme GENE HP DNA Transfection Reagent (Roche, Basel, Switzerland) was used for the transfection of constructs. For G418 (Sigma, USA) selection, cells were transfected with pEGFP-UBE2N or pGPH1-shUBE2N plasmid for 24h, and treated with 400 μg/mL G418 for 14 days. siRNAs against UBE2N (siRNA1, 5ꞌ-GGCUAUAUGCCAUGAAUAA-3ꞌ; siRNA2, 5ꞌ-CCAGAUGAUCCAUUAGCAA-3ꞌ) and Fos (siRNA1, 5ꞌ-GGGAUAGCCUCUCUUACUA-3ꞌ; siRNA2, 5ꞌ-GCAAGGUGGAACAGUUAUC-3ꞌ) were transfected into cells using the DharmaFECT Transfection Reagent (GE healthcare, Boston, Massachusetts, USA).
Real-Time PCR (RT-PCR) Array
SKOV3 cells were transfected with UBE2N siRNA1 (n = 2) and control siRNA (n = 2) for 72 hours and were then harvested. Total RNA was extracted, of which 1.5 µg was reverse-transcribed according to the manufacturer’s instructions (Qiagen, Hilden, Germany). RT-PCR array was performed using the RT Profiler PCR Array Human Cancer Drug Resistance kit (Qiagen, Hilden, Germany). ΔΔCt values were used for quantification. For more information see Supplementary Materials and Methods.
Western Blotting
Ovarian cancer cells were collected and lyzed in 50 mL cell lysis buffer containing protease inhibitors. The cell lysates were separated on 12% SDS–PAGE. After the proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, USA), we blocked the proteins with Tris-buffered saline (TBS) and 0.1% Tween 20 (TBS/T) which containing 5% bovine serum albumin. The PVDF membranes were incubated with specific primary antibodies (Anti-UBE2N antibody: Abcam, 1:1000; Anti-Fos antibody: Cell Signaling Technology, 1:1000; Anti-P53 antibody: Cell Signaling Technology, 1:1000; Anti-GAPDH antibody: Cell Signaling Technology, 1:1000) at room temperature for 1 hour or at 4°C overnight. The membranes were washed three times with TBS/T and then incubated with the appropriate HRP-conjugated secondary antibodies for 1 h at room temperature. The protein concentration was quantified using the BCA Protein Kit (Applygen, Beijing, China). The protein GAPDH was used as an internal control. The relative expression of other proteins was normalized against GAPDH.
Gene Ontology (GO) Analysis
GO annotation analysis of differentially expressed genes (DEGs) was performed with the database for Annotation, Visualization and Integrated Discovery (DAVID 6.8, www.david.ncifcrf.gov). P-value < 0.05 was considered as statistically significant.
PPI Network Construction and Hub Gene Identification
DEGs were mapped to the Search Tool for the Retrieval of Interacting Genes (STRING, www.string-db.org) database to evaluate the potential PPI relationships. The interactions with a combined score > 0.4 were considered as significant. The PPI network was visualized with the Cytoscape software (version 3.7.0).
Survival Data from the Kaplan–Meier Plotter Database
The prognostic values of hub genes were analyzed using the Kaplan–Meier plotter (KM plotter, www.kmplot.com) database for ovarian cancer. In our study, ovarian cancer patients were screened based on the chemotherapy using the administration of paclitaxel. P-value < 0.05 was regarded as statistically significant.
Statistical Analysis
Results are shown as mean ± SEM. Differences between two groups were evaluated using the two-tailed Student’s t-test unless otherwise indicated. Statistical tests were carried out using SPSS (version 21.0, SPSS Inc.) or GraphPad Prism (version 7.0, GraphPad Software Inc.). The level of statistical significance was set at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Results
UBE2N Increases Paclitaxel Sensitivity in Ovarian Cancer in vivo
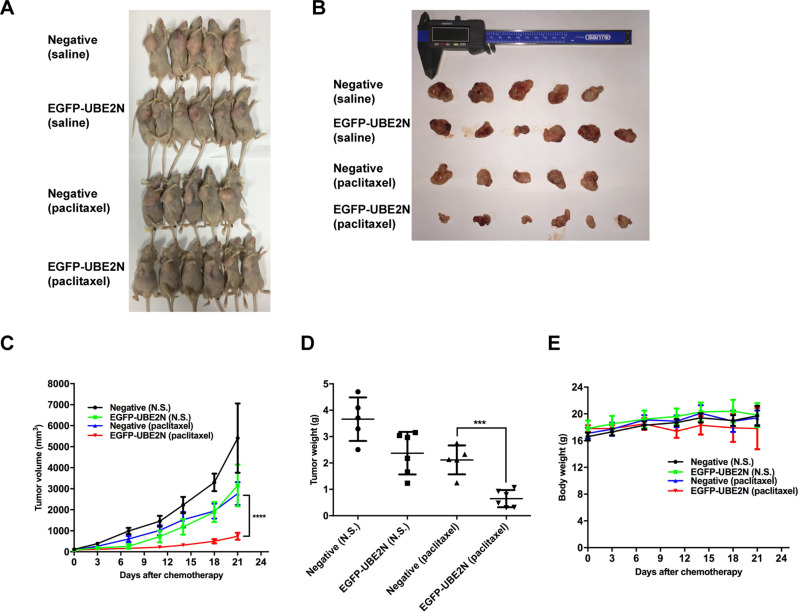
In our previous study, we found that the expression of UBE2N decreased in paclitaxel-resistant ovarian cancer cells (shown in Figure S1A). Thus, we developed A2780 cells which stably expressing increased amounts of UBE2N which were confirmed by qRT-PCR and Western blotting (shown in Figure S1B and D). These cells, along with negative control cells, were injected subcutaneously into female BALB/C nude mice. The mice developed visible tumors were then grouped for the administration of paclitaxel or saline. Paclitaxel (15 mg/kg) or saline was intraperitoneally injected once a week for three weeks. As shown in Figure 1A–C, tumor growth was significantly suppressed in the UBE2N overexpression group than that in the negative control group after the treatment of paclitaxel. Upon the completion of paclitaxel treatment, tumors were removed and weighed. In the paclitaxel treatment group, the weight of excised tumors overexpressing UBE2N was less than that of negative control tumors (Figure 1D). However, the differences in body weight among different groups had no statistical significance (Figure 1E). Therefore, our in vivo findings suggest that UBE2N enhances paclitaxel sensitivity in ovarian cancer.
Figure 1.
UBE2N mediates paclitaxel sensitivity in mouse models. BALB/C nude mice were subcutaneously inoculated with 2*106 A2780 cells with overexpressed UBE2N (n=6) or negative control (n=5). Once the tumors developed obviously, paclitaxel (15mg/kg) and the same volume of saline were then intraperitoneally injected once a week for 3 weeks. All mice were sacrificed on day 7 after complete drug treatment and tumors were excised, weight, and photographed. Tumor volume (mm3) was calculated as Width2/Length. (A–D) Impaired paclitaxel sensitivity in transplanted tumors in mice inoculated by UBE2N-overexpressing A2780 cells was observed. (E) There were no differences of mice weight between groups during the experiment. The level of significance is indicated by ***P < 0.001, ****P < 0.0001.
Exogenous UBE2N Modulated Paclitaxel Sensitivity in Ovarian Cancer Cells in vitro
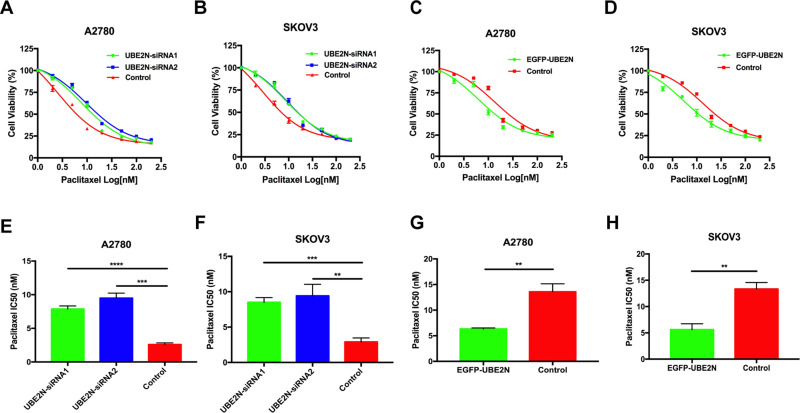
To further evaluate the involvement of UBE2N in regulating paclitaxel sensitivity in ovarian cancer cells, we downregulated or upregulated UBE2N expression exogenously using siRNA transfection and plasmid transfection, respectively (shown in Figure S1C and D). The effects of these treatments on paclitaxel sensitivity were determined using the cell viability assay. As shown in Figure 2A and B, we found that UBE2N knockdown significantly protected ovarian cancer cells from paclitaxel. The IC50 values of paclitaxel in UBE2N knockdown groups were higher than those in control groups in both A2780 and SKOV3 cells (Figure 2E and F). Conversely, UBE2N overexpression significantly increased paclitaxel sensitivity (Figure 2C and D) and reduced IC50 values of paclitaxel in both cell types (Figure 2G and H). Our in vitro findings support that UBE2N could increase paclitaxel sensitivity in ovarian cancer cells.
Figure 2.
UBE2N regulated paclitaxel sensitivity in vitro. (A–D) Cell viability assays in A2780 and SKOV3 cells with UBE2N knockdown and UBE2N overexpression that were treated with paclitaxel at the indicated concentrations. (E–H) IC50 of paclitaxel in A2780 and SKOV3 cells with UBE2N knockdown and UBE2N overexpression. Results are shown as means±SEM for at least 3 separate experiments. The level of significance is indicated by **P < 0.01, ***P < 0.001, ****P < 0.0001.
Identification of DEGs and Functional Enrichment Analyses
To investigate the regulation of downstream genes of UBE2N, we used an RT-PCR array that is specific for genes involved in drug resistance in human cancers. This array contained 84 genes shown to be involved in the regulation of resistance to chemotherapeutic agents. Twenty-four DEGs were identified, including 13 upregulated genes and 11 downregulated genes based on fold-change values ≥ 1.5 (Table 1, all array data are shown in the Supplementary Table).
Table 1.
The Differentially Expressed Genes of RT-PCR Array
| Gene Symbol | Gene Description | Fold Change (UBE2N Knockdown vs Control) |
|---|---|---|
| XPC | Xeroderma pigmentosum, complementation group C | 5.02 |
| Fos | Fos proto-oncogene, AP-1 transcription factor subunit | 4.09 |
| BCL2L1 | BCL2-like 1 | 2.40 |
| CYP2D6 | Cytochrome P450 family 2 subfamily D member 6 | 2.33 |
| RARG | Retinoic acid receptor, gamma | 2.04 |
| RXRB | Retinoid X receptor, beta | 1.87 |
| ABCC2 | ATP-binding cassette subfamily C member 2 | 1.85 |
| NFκBIB | NFκB inhibitor beta | 1.79 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B | 1.60 |
| RB1 | Retinoblastoma 1 | 1.58 |
| CDK4 | Cyclin-dependent kinase 4 | 1.57 |
| AHR | Aryl hydrocarbon receptor | 1.53 |
| IGF2R | Insulin-like growth factor 2 receptor | 1.50 |
| UGCG | UDP-glucose ceramide glucosyltransferase | −1.50 |
| TPMT | Thiopurine S-methyltransferase | −1.59 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A | −1.62 |
| EPHX1 | Epoxide hydrolase 1 | −1.68 |
| APC | Adenomatous polyposis coli | −1.73 |
| EGFR | Epidermal growth factor receptor | −1.94 |
| BLMH | Bleomycin hydrolase | −2.05 |
| CCND1 | Cyclin D1 | −2.29 |
| NFκBIE | NFκB inhibitor epsilon | −3.09 |
| ERBB3 | erb-b2 receptor tyrosine kinase 3 | −3.64 |
| CYP2E1 | Cytochrome P450 family 2 subfamily E member 1 | −4.94 |
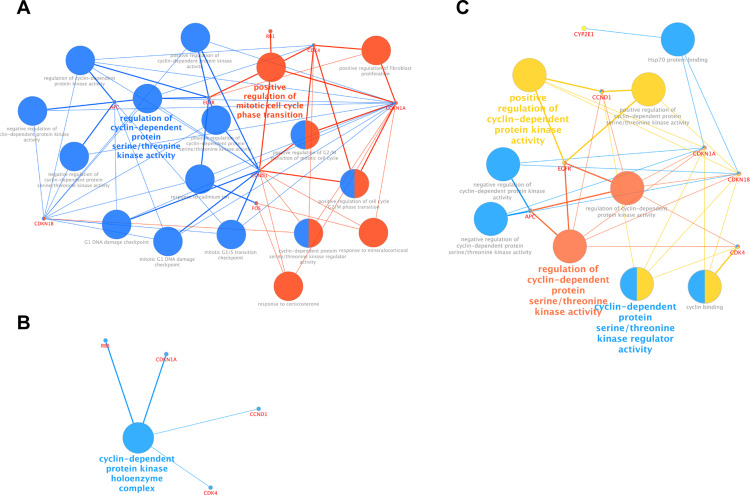
Furthermore, we performed GO function analysis for DEGs with DAVID. Enriched GO terms were divided into biological process (BP), cellular component (CC), and molecular function (MF) ontologies. Biological process analysis indicated that the DEGs were significantly enriched in regulation of cyclin-dependent protein serine/threonine kinase activity, and in positive regulation of mitotic cell cycle phase transition (Figure 3A). Cell component analysis indicated that the DEGs showed enrichment for the members of the cyclin-dependent protein kinase holoenzyme complex (Figure 3B). The DEGs were enriched in positive regulation of cyclin-dependent protein kinase activity, and in cyclin-dependent protein serine/threonine kinase regulator activity with respect to molecular function (Figure 3C).
Figure 3.
GO functions analysis for the DEGs. (A) Enriched biological process of DEGs. (B) Enriched molecular function of DEGs. (C) Enriched cellular component of DEGs.
Construction of PPI Networks
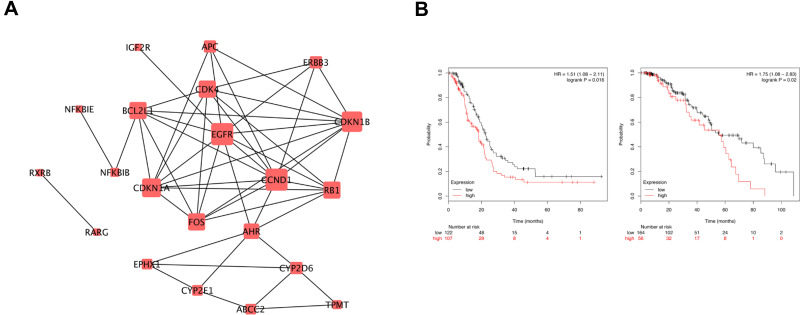
Interactions among proteins expressed by the DEGs were predicted using the STRING tool. In total, 100 nodes and 814 edges were involved in the PPI network (Figure 4A). The top nine genes (degree ≥ 5) in the PPI network are showed in Table 2. Results showed that cyclin D1 (CCND1) and epidermal growth factor receptor (EGFR) were significant genes with connectivity degree D1 = 10, which were downregulated upon the decrease of UBE2N levels, followed by cyclin-dependent kinase inhibitor 1A (CDKN1A; degree = 8). Cyclin-dependent kinase inhibitor 1B (CDKN1B; degree = 9), retinoblastoma 1 (RB1; degree = 7), cyclin-dependent kinase 4 (CDK4; degree = 7), Fos proto-oncogene, AP-1 transcription factor subunit (Fos; degree = 7), BCL2-like 1 (BCL2L1; degree = 7), and aryl hydrocarbon receptor (AHR; degree = 6) were upregulated upon decrease in UBE2N levels.
Figure 4.
Protein-protein interaction network and survival analysis. (A) PPI network constructed with the differentially expressed genes. (B) Kaplan–Meier survival analysis for Fos expression in ovarian cancer patients with paclitaxel treatment.
Table 2.
Top Nine Hub Genes with Higher Degree of Connectivity
| Gene Symbol | Gene Description | Degree |
|---|---|---|
| CCND1 | Cyclin D1 | 10 |
| EGFR | Epidermal growth factor receptor | 10 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B | 9 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A | 8 |
| RB1 | Retinoblastoma 1 | 7 |
| CDK4 | Cyclin-dependent kinase 4 | 7 |
| Fos | Fos proto-oncogene, AP-1 transcription factor subunit | 7 |
| BCL2L1 | BCL-like 1 | 7 |
| AHR | Aryl hydrocarbon receptor | 6 |
Survival Analysis
The Kaplan–Meier plotter bioinformatics analysis platform was used to investigate the prognostic values of these hub genes. To study the relationship between these hub genes and chemotherapy, we restricted the analysis to treatment groups administered with paclitaxel. Two hundred and twenty ovarian cancer patients were enrolled for the analysis of overall survival, while 229 patients for analysis of relapse-free survival (shown in Figure S2). However, we found that only Fos was associated with unfavorable overall survival and relapse-free survival in ovarian cancer patients applied paclitaxel (Figure 4B).
UBE2N Regulated Paclitaxel Sensitivity via Fos/P53 in Ovarian Cancer Cells
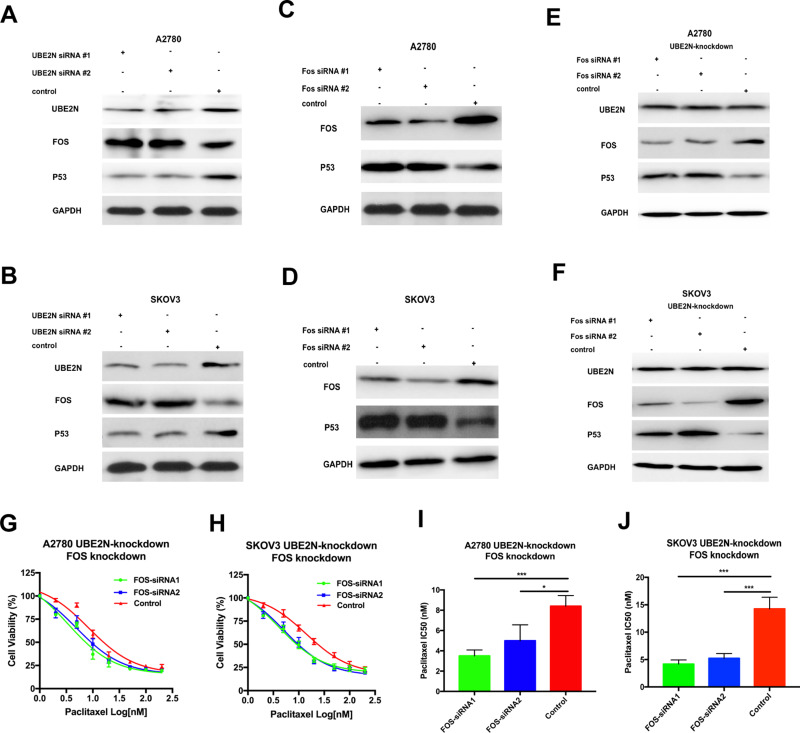
To verify the relationship between UBE2N and Fos, we decreased UBE2N expression and found that Fos was increased in A2780 and SKOV3 cells (Figure 5A and B). Furthermore, we inspected the effect of Fos when UBE2N regulated the paclitaxel sensitivity in ovarian cancer cells. After both cells with UBE2N knockdown selected by G418 for 14 days, siRNAs specific against Fos were transfected into cells. The cytoprotection to the paclitaxel owing to UBE2N down-regulation was reversed by Fos knockdown (Figure 5G and H). Moreover, Fos knockdown by particular siRNAs enhanced the sensitivity to paclitaxel (Figure 5I and J). Our findings together suggest that Fos participates in UBE2N’s regulation of paclitaxel sensitivity in ovarian cancer cells in the opposite direction to UBE2N. To explore the mechanism of regulating paclitaxel sensitivity via UBE2N, we detected P53, an antioncogene. We observed that the expression of P53 was decreased when UBE2N was knocked down (Figure 5A and B). On the contrary, the expression of P53 was increased following the reduction of Fos in both two ovarian cells (Figure 5C and D). Furthermore, after cells with UBE2N down-expression, we reversely knocked down Fos and observed that the expression of P53 was restored (Figure 5E and F). Moreover, the cytoprotection to paclitaxel owing to UBE2N knockdown was reversed by P53 up-regulation, but the expression of UBE2N and Fos was not affected (shown in Figure S3). Our findings suggest that Fos may participate in UBE2N regulation of paclitaxel sensitivity via P53 in ovarian cancer cells.
Figure 5.
Fos regulated paclitaxel sensitivity in vitro and participated in UBE2N regulation of the paclitaxel sensitivity via P53. (A and B) Western blotting of UBE2N, Fos and P53 in A2780 and SKOV3 cells with UBE2N-knockdown. (C and D) Western blotting of Fos, P53 in A2780 and SKOV3 cells with Fos-knockdown. (E and F) Western blotting of Fos, P53 in A2780 and SKOV3 cells with Fos-knockdown, which were transfected in advance with UBE2N-specific shRNA and selected with G418 (400 μg/mL) for 14 days, and treated with paclitaxel at the indicated concentrations. (G and H) Cell viability assays in A2780 and SKOV3 cells with Fos-knockdown, which were transfected in advance with UBE2N-specific shRNA and selected with G418 (400 μg/mL) for 14 days, and treated with paclitaxel at the indicated concentrations. (I and J) IC50 of paclitaxel in A2780 and SKOV3 cells with Fos-knockdown, which were transfected in advance with UBE2N-specific shRNA and selected with G418 (400 μg/mL) for 14 days. Results are shown as means±SEM for at least 3 separate experiments. The level of significance is indicated by *P < 0.05, ***P < 0.001.
Discussion
Resistance to chemotherapy is by far one of the most important constraint on ovarian cancer treatment. As a traditional chemotherapeutic agent, paclitaxel has been used for the treatment of ovarian cancer for decades. However, the key mechanism underlying resistance to chemotherapy is still unclear. In our previous study, using quantitative proteomic analysis and immunoblotting, we have confirmed that UBE2N levels were reduced in paclitaxel-resistant ovarian cancer cells,8 indicating that UBE2N might be involved in regulating paclitaxel resistance of ovarian cancer cells.
To verify our hypothesis, we examined the effect of elevated UBE2N expression on paclitaxel in vivo. We observed that the growth of transplanted tumors with overexpressed UBE2N was significantly restricted in the mouse model. The tumors presented a remarkable sensitivity to paclitaxel. We also found that the upregulation of UBE2N could enhance the sensitivity of ovarian cancer cells to paclitaxel, while the downregulation of UBE2N induced the paclitaxel resistance in ovarian cancer cells in vitro. These findings are consistent with our previous report.
UBE2N plays important roles in various biological processes like protein degradation, cell cycle, apoptosis, and DNA repair.14,20,24 It is also associated with the origin and development of some diseases,25–29 and might be involved in the modulation of chemotherapeutic sensitivity of cancers.23,30 To investigate the downstream factors of UBE2N involved in the regulation of paclitaxel-resistance, we used an RT-PCR array that was specific for human cancer drug resistance. This array contains 84 genes shown to be involved in chemoresistance. We identified 13 upregulated DEGs and 11 downregulated DEGs when UBE2N was knocked down in ovarian cancer cells. Moreover, GO analysis showed that these DEGs were associated with cyclin-dependent protein kinase regulation. A PPI network was constructed to investigate the interrelationship between the DEGs, and nine hub genes identified, including CCND1, EGFR, CDKN1A, CDKN1B, RB1, CDK4, Fos, BCL2L1, and AHR. Finally, the Kaplan–Meier plotter online tool was used to predict the relationship between the expression of hub genes and prognosis of ovarian cancer patients treated using paclitaxel. However, only overexpression of Fos was found to be an unfavorable prognostic factor for ovarian cancer patients treated using paclitaxel.
Fos, also known as c-Fos, is a proto-oncogene involved in various types of cancers. Fos encodes leucine zipper proteins that dimerize with proteins of the JUN family, thereby forming the transcription factor complex AP-1. As a member of AP-1, Fos protein plays important roles in several cellular events, including signal transduction, cellular differentiation and proliferation.31 A few studies have suggested that Fos might be correlated with poor prognosis of some cancers.32–34 Mar et al reported that Fos enhances IL6 and VEGF-A expression in colon cancer.35 Fos can also lead to loss of cell polarity and epithelial–mesenchymal transition (EMT), as a result of invasion and metastasis.36 Moreover, increased Fos expression has been shown to phenotypically promote drug resistance.37 Takahashi et al reported that high levels of Fos led to paclitaxel resistance.38 Although studies have demonstrated that Fos is associated with drug resistance against some chemotherapeutics,39–41 the mechanism underlying paclitaxel resistance due to Fos is still unclear. Here, we observed that the knockdown of UBE2N elevated Fos mRNA and protein expression and induced paclitaxel resistance. GO function analysis showed that these DEGs, including Fos, were enriched in the network involving cyclin-dependent proteins. UBE2N might regulate paclitaxel sensitivity in ovarian cancer by modulation of Fos through the cyclin-dependent protein kinase pathway.
In our study, the reduction of Fos could improve paclitaxel sensitivity in ovarian cancer cells. Moreover, reduced Fos could recover the cytotoxic effect of paclitaxel, which is opposite to the down-regulation of UBE2N. Thus, it suggests that Fos participates in the process of UBE2N regulation of the paclitaxel sensitivity of ovarian cancer cells.
The mechanism responsible for Fos involved in regulating paclitaxel sensitivity is still unclear. Here we found that Fos could regulate P53 under UBE2N’s modulation. The P53 protein is widely reported in human cancers. It is a 53 kDa nuclear phosphoprotein, which regulates the cell cycle by inhibiting DNA synthesis. Protein P53 is coded by tumor-suppressor gene P53, which is located on the short arm of chromosome 17 and exerts an inhibitory effect on inducing cell cycle arrest or apoptosis in response to DNA damage.42,43 Some studies have shown that the overexpression of c-Jun in cancer cells leads to a decrease in P53 levels and accelerates cell proliferation, whereas the absence of c-Jun resulted in elevated expression of P53 and its target gene, the CDK inhibitor P21.44–46 In our study, we observed that P53 decrease was consistent with Fos increase in ovarian cancer cells. In advance with UBE2N-specific siRNA, we found a reverse regulation of P53 induced by Fos-knockdown in ovarian cancer cells. It has been reported that inhibition of P53 transcriptional activity is associated with paclitaxel resistance in ovarian cancer cells, and restore P53 transcriptional activity can reverse paclitaxel sensitivity.47 Hailing Yang et al reported that P53 expression was upregulated in ovarian cancer cells after paclitaxel stimulation, and the overexpression of TAP63, a P53 family member, led to apoptosis in paclitaxel-resistant ovarian cancer cells after treatment with paclitaxel.48 According to these studies, we speculate that Fos is a potential target for the regulation of paclitaxel sensitivity, and maybe participates in the regulation via suppressing P53’s apoptotic contribution to drug response in ovarian cancer cells. However, further works for the specific mechanisms are still essential.
Besides, we detected other hub genes associated with ovarian cancer, including CCND1, EGFR, CDKN1A, CDKN1B, RB1, CDK4, BCL2L1, and AHR. All of them were reported as key factors involved in the development of some cancers and in resistance to some chemotherapeutic. However, in our study, results for survival analysis based on the expression of these hub genes in ovarian cancer patients treated with paclitaxel were not consistent with those from the RT-PCR array. The role of these hub genes in paclitaxel resistance induced upon reduction in UBE2N expression is not clear, demanding further elucidation.
In conclusion, UBE2N was verified to regulate paclitaxel resistance in ovarian cancer in vivo and in vitro, which suggests UBE2N might be used as a therapeutic agent for regulating paclitaxel-resistance in ovarian cancer. RT-PCR array identified 24 DEGs between UBE2N knockdown cells and negative controls. Bioinformatic analysis was used to screen nine hub genes that might be the principal genes, including CCND1, EGFR, CDKN1A, CDKN1B, RB1, CDK4, BCL2L1, Fos, and AHR. Only Fos overexpression is an unfavorable prognostic factor of ovarian cancer upon paclitaxel treatment. According to our experimental results, we speculate that UBE2N regulates paclitaxel sensitivity via Fos/P53 in ovarian cancer cells, our findings offer a new approach to reverse chemoresistance. Further studies are required to elucidate the specific mechanisms.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (Grant No. 81803781). Part of our work was performed at the Women’s Reproductive Health Laboratory of Women’s Hospital, School of Medicine, Zhejiang University. We thank Professor Xing Xie for the help at the facility.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 2.Vargas-Hernandez VM, Moreno-Eutimio MA, Acosta-Altamirano G, Vargas-Aguilar VM. Management of recurrent epithelial ovarian cancer. Gland Surg. 2014;3(3):198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen D, Lin X, Zhang C, et al. Dual PI3K/mTOR inhibitor BEZ235 as a promising therapeutic strategy against paclitaxel-resistant gastric cancer via targeting PI3K/Akt/mTOR pathway. Cell Death Dis. 2018;9(2):123. doi: 10.1038/s41419-017-0132-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janakiraman H, House RP, Talwar S, et al. Repression of caspase-3 and RNA-binding protein HuR cleavage by cyclooxygenase-2 promotes drug resistance in oral squamous cell carcinoma. Oncogene. 2017;36(22):3137–3148. doi: 10.1038/onc.2016.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, Chen S, Yu J, et al. Single-cell transcriptome analyses reveal molecular signals to intrinsic and acquired paclitaxel resistance in esophageal squamous cancer cells. Cancer Lett. 2018;420:156–167. doi: 10.1016/j.canlet.2018.01.059 [DOI] [PubMed] [Google Scholar]
- 6.Yu Y, Gaillard S, Phillip JM, et al. Inhibition of spleen tyrosine kinase potentiates paclitaxel-induced cytotoxicity in ovarian cancer cells by stabilizing microtubules. Cancer Cell. 2015;28(1):82–96. doi: 10.1016/j.ccell.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang SF, Wang XY, Fu ZQ, et al. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy. 2015;11(2):225–238. doi: 10.1080/15548627.2014.998931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Feng Y, Wang XY, et al. The inhibition of UBC13 expression and blockage of the DNMT1-CHFR-Aurora A pathway contribute to paclitaxel resistance in ovarian cancer. Cell Death Dis. 2018;9(2):93. doi: 10.1038/s41419-017-0137-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hershko A, Ciechanover A, Varshavsky A. Basic medical research award. Nat Med. 2000;6(10):1073–1081. doi: 10.1038/80384 [DOI] [PubMed] [Google Scholar]
- 10.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22(1):159–180. doi: 10.1146/annurev.cellbio.22.010605.093503 [DOI] [PubMed] [Google Scholar]
- 11.Jeon YJ, Khelifa S, Ratnikov B, et al. Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell. 2015;27(3):354–369. doi: 10.1016/j.ccell.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomas A, Vaughan SO, Burgoyne T, et al. WASH and Tsg101/ALIX-dependent diversion of stress-internalized EGFR from the canonical endocytic pathway. Nat Commun. 2015;6(1):7324. doi: 10.1038/ncomms8324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galanos P, Vougas K, Walter D, et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat Cell Biol. 2016;18(7):777–789. doi: 10.1038/ncb3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67(1):425–479. doi: 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 15.Habelhah H, Takahashi S, Cho SG, Kadoya T, Watanabe T, Ronai Z. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB. EMBO J. 2004;23(2):322–332. doi: 10.1038/sj.emboj.7600044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unk I, Hajdu I, Fatyol K, et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2006;103(48):18107–18112. doi: 10.1073/pnas.0608595103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu ZH, Wong ET, Shi Y, et al. ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol Cell. 2010;40(1):75–86. doi: 10.1016/j.molcel.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodge CD, Ismail IH, Edwards RA, et al. RNF8 E3 ubiquitin ligase stimulates Ubc13 E2 conjugating activity that is essential for DNA double strand break signaling and BRCA1 tumor suppressor recruitment. J Biol Chem. 2016;291(18):9396–9410. doi: 10.1074/jbc.M116.715698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–351. doi: 10.1038/35085597 [DOI] [PubMed] [Google Scholar]
- 20.Zhang M, Windheim M, Roe SM, et al. Chaperoned ubiquitylation–crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20(4):525–538. doi: 10.1016/j.molcel.2005.09.023 [DOI] [PubMed] [Google Scholar]
- 21.Thorslund T, Ripplinger A, Hoffmann S, et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015;527(7578):389–393. doi: 10.1038/nature15401 [DOI] [PubMed] [Google Scholar]
- 22.Saviozzi S, Ceppi P, Novello S, et al. Non-small cell lung cancer exhibits transcript overexpression of genes associated with homologous recombination and DNA replication pathways. Cancer Res. 2009;69(8):3390–3396. doi: 10.1158/0008-5472.CAN-08-2981 [DOI] [PubMed] [Google Scholar]
- 23.Cheng J, Fan YH, Xu X, et al. A small-molecule inhibitor of UBE2N induces neuroblastoma cell death via activation of p53 and JNK pathways. Cell Death Dis. 2014;5(2):e1079. doi: 10.1038/cddis.2014.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolas NK, Chapman JR, Nakada S, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318(5856):1637–1640. doi: 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukushima T, Matsuzawa S, Kress CL, et al. Ubiquitin-conjugating enzyme Ubc13 is a critical component of TNF receptor-associated factor (TRAF)-mediated inflammatory responses. Proc Natl Acad Sci U S A. 2007;104(15):6371–6376. doi: 10.1073/pnas.0700548104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto M, Sato S, Saitoh T, et al. Cutting edge: pivotal function of Ubc13 in thymocyte TCR signaling. J Immunol. 2006;177(11):7520–7524. doi: 10.4049/jimmunol.177.11.7520 [DOI] [PubMed] [Google Scholar]
- 27.Yan Z, Guo R, Paramasivam M, et al. A ubiquitin-binding protein, FAAP20, links RNF8-mediated ubiquitination to the Fanconi anemia DNA repair network. Mol Cell. 2012;47(1):61–75. doi: 10.1016/j.molcel.2012.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin P, Tu Z, Yin A, et al. Aged monkey brains reveal the role of ubiquitin-conjugating enzyme UBE2N in the synaptosomal accumulation of mutant huntingtin. Hum Mol Genet. 2015;24(5):1350–1362. doi: 10.1093/hmg/ddu544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norris KL, Hao R, Chen LF, et al. Convergence of parkin, PINK1, and alpha-synuclein on stress-induced mitochondrial morphological remodeling. J Biol Chem. 2015;290(22):13862–13874. doi: 10.1074/jbc.M114.634063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Lei Z, Huang Z, et al. Epigallocatechin-3-gallate(EGCG) suppresses melanoma cell growth and metastasis by targeting TRAF6 activity. Oncotarget. 2016;7(48):79557–79571. doi: 10.18632/oncotarget.12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer. 2005;41(16):2449–2461. doi: 10.1016/j.ejca.2005.08.008 [DOI] [PubMed] [Google Scholar]
- 32.Qu X, Yan X, Kong C, et al. c-Myb promotes growth and metastasis of colorectal cancer through c-Fos-induced epithelial-mesenchymal transition. Cancer Sci. 2019;110(10):3183–3196. doi: 10.1111/cas.14141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao Y, Zhu L, Yan L, et al. c-Fos mediates alpha1, 2-fucosyltransferase 1 and Lewis y expression in response to TGF-beta1 in ovarian cancer. Oncol Rep. 2017;38(6):3355–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu ZG, Jiang G, Tang J, et al. c-Fos over-expression promotes radioresistance and predicts poor prognosis in malignant glioma. Oncotarget. 2016;7(40):65946–65956. doi: 10.18632/oncotarget.11779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mar AC, Chu CH, Lee HJ, et al. Interleukin-1 receptor type 2 acts with c-Fos to enhance the expression of interleukin-6 and vascular endothelial growth factor a in colon cancer cells and induce angiogenesis. J Biol Chem. 2015;290(36):22212–22224. doi: 10.1074/jbc.M115.644823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fialka I, Schwarz H, Reichmann E, Oft M, Busslinger M, Beug H. The estrogen-dependent c-JunER protein causes a reversible loss of mammary epithelial cell polarity involving a destabilization of adherens junctions. J Cell Biol. 1996;132(6):1115–1132. doi: 10.1083/jcb.132.6.1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Luo A, Li S, et al. Inhibitor of differentiation/DNA binding 1 (ID1) inhibits etoposide-induced apoptosis in a c-Jun/c-Fos-dependent manner. J Biol Chem. 2016;291(13):6831–6842. doi: 10.1074/jbc.M115.704361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi M, Tsutsui H, Tagawa H, Igarashi-Saito K, Imanaka-Yoshida K, Takeshita A. Microtubules are involved in early hypertrophic responses of myocardium during pressure overload. Am J Physiol. 1998;275(2):H341–348. [DOI] [PubMed] [Google Scholar]
- 39.Li G, Hu X, Sun L, et al. C-fos upregulates P-glycoprotein, contributing to the development of multidrug resistance in HEp-2 laryngeal cancer cells with VCR-induced resistance. Cell Mol Biol Lett. 2018;23(1):6. doi: 10.1186/s11658-017-0067-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shankar E, Song K, Corum SL, et al. A signaling network controlling androgenic repression of c-Fos protein in prostate adenocarcinoma cells. J Biol Chem. 2016;291(11):5512–5526. doi: 10.1074/jbc.M115.694877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi R, Peng H, Yuan X, et al. Down-regulation of c-Fos by shRNA sensitizes adriamycin-resistant MCF-7/ADR cells to chemotherapeutic agents via P-glycoprotein inhibition and apoptosis augmentation. J Cell Biochem. 2013;114(8):1890–1900. doi: 10.1002/jcb.24533 [DOI] [PubMed] [Google Scholar]
- 42.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351(6326):453–456. doi: 10.1038/351453a0 [DOI] [PubMed] [Google Scholar]
- 43.Steele RJ, Thompson AM, Hall PA, Lane DP. The p53 tumour suppressor gene. Br J Surg. 1998;85(11):1460–1467. doi: 10.1046/j.1365-2168.1998.00910.x [DOI] [PubMed] [Google Scholar]
- 44.Schreiber M, Kolbus A, Piu F, et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13(5):607–619. doi: 10.1101/gad.13.5.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eferl R, Ricci R, Kenner L, et al. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112(2):181–192. [DOI] [PubMed] [Google Scholar]
- 46.Yedjou CG, Tchounwou PB. Modulation of p53, c-Fos, RARE, cyclin A, and cyclin D1 expression in human leukemia (HL-60) cells exposed to arsenic trioxide. Mol Cell Biochem. 2009;331(1–2):207–214. doi: 10.1007/s11010-009-0160-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parmakhtiar B, Burger RA, Kim JH, Fruehauf JP. HIF inactivation of p53 in ovarian cancer can be reversed by topotecan, restoring cisplatin and paclitaxel sensitivity. Mol Cancer Res. 2019;17(8):1675–1686. doi: 10.1158/1541-7786.MCR-18-1109 [DOI] [PubMed] [Google Scholar]
- 48.Yang H, Shu Z, Jiang Y, et al. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase-2 regulates TP53-dependent paclitaxel sensitivity in ovarian and breast cancers. Clin Cancer Res. 2019;25(18):5702–5716. doi: 10.1158/1078-0432.CCR-18-3448 [DOI] [PMC free article] [PubMed] [Google Scholar]