Abstract
The principle of monoclonality with regard to bacterial infections was considered immutable prior to 30 years ago. This view, espoused by Koch for acute infections, has proven inadequate regarding chronic infections as persistence requires multiple forms of heterogeneity among the bacterial population. This understanding of bacterial plurality emerged from a synthesis of what-were-then novel technologies in molecular biology and imaging science. These technologies demonstrated that bacteria have complex life cycles, polymicrobial ecologies, and evolve in situ via the horizontal exchange of genic characters. Thus, there is an ongoing generation of diversity during infection that results in far more highly complex microbial communities than previously envisioned. This perspective is based on the fundamental tenet that the bacteria within an infecting population display genotypic diversity, including gene possession differences, which result from horizontal gene transfer mechanisms including transformation, conjugation, and transduction. This understanding is embodied in the concepts of the supragenome/pan-genome and the distributed genome hypothesis (DGH). These paradigms have fostered multiple researches in diverse areas of bacterial ecology including host–bacterial interactions covering the gamut of symbiotic relationships including mutualism, commensalism, and parasitism. With regard to the human host, within each of these symbiotic relationships all bacterial species possess attributes that contribute to colonization and persistence; those species/strains that are pathogenic also encode traits for invasion and metastases. Herein we provide an update on our understanding of bacterial plurality and discuss potential applications in diagnostics, therapeutics, and vaccinology based on perspectives provided by the DGH with regard to the evolution of pathogenicity.
Keywords: bacterial pathogenesis, comparative genomics, pan-genome
Introduction
Established paradigms of monoclonality may have adequately described a subset of acute bacterial infections, however, they have been found lacking when dealing with persistent and chronic bacterial infections, including biofilms. Monoclonality implies that colonizing bacteria all belong to the same genotype; however, this is an oversimplified view of most bacterial infections, especially those that are chronic. Traditional diagnostic methods that relied on bacterial cultures helped to entrench these paradigms in medical microbiology. However, newer diagnostic and research tools, such as polymerase chain reaction (PCR), advanced microscopic techniques for metabolic profiling, and whole-genome sequencing (WGS), have demonstrated extensive phenotypic and genotypic diversity among bacterial strains within a population or species [1–3]. Genotypic diversity within infecting bacterial populations suggests these populations are in fact polyclonal, rather than monoclonal; i.e. multiple independent strains of the same bacterial species are present in these populations at the same time [1,3–7]. This diversity also implies that even mono-species bacterial populations are neither phenotypically or genotypically clonal [1].
Genetic diversity within a prokaryotic species incorporates two distinct phenomena: (1) genetic heterogeneity, i.e. individuals in a population possess different alleles (variant forms of the same gene); and (2) genomic plasticity, i.e. individuals in a population possess different genes [1,8]. The rubric of genomic plasticity, thus teaches that the genome of one strain within a species cannot account for the entire set of genes for the species as a whole, and that studies of individual isolates will greatly underestimate the biological properties of the species. Therefore, the genomes of multiple independent isolates need to be fully characterized to even estimate the total biological complexity of a bacterial species [8–16]. Collectively, the genes contained in the genomes of all individuals within a species comprise the supragenome (or pan-genome) of that species [8,9,17]. Multiple mathematical models have been developed to estimate the species-level supragenome based on the sequencing of a limited number of strains chosen to be unclustered with respect to geographic location and phenotypic properties [13,14,18,19]. The concepts of genomic plasticity and the supragenome/pan-genome are integral components of the distributed gene hypothesis (DGH), which posits that not all individuals in a population have the same set of genes, and that no one individual strain has all the genes of a species. Individual members of the population have ‘access to’ a species supragenome from which genetic material can be taken up and recombined as a means of generating genomic diversity [1,2,7]. This genomic plasticity, propagated by horizontal gene transfer (HGT), is highly conserved among bacterial species. There are multiple examples wherein HGT has been shown to be the mechanism that has provided for bacterial adaptation, survival, and evolution through the transfer of genes that provide for adaptation to various environmental pressures including host immune surveillance, antibiotic resistance, and changes in pH [20–26]. Obviously, vertical transmission of the core genome is required to maintain species viability and to provide a framework within which HGT mechanisms operate.
In this review, we provide an updated perspective with regard to bacterial genomic plasticity, and how HGT mechanisms together with a large, species-level, supragenome provides for bacterial adaptation. In addition, we show how these new understandings provide for targeted clinical interventions that have the potential to fundamentally change how we approach microbial infections such that it should be possible to ‘surgically’ eliminate highly pathogenic strains without undue damage to the host's microbiome
Current perspective
Bacterial genomic plasticity
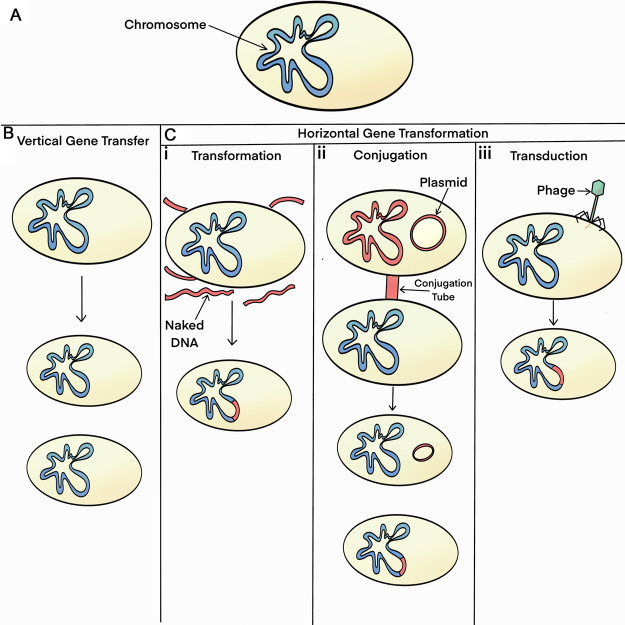
Bacterial genomic plasticity is characterized by each strain in a population possessing a unique set of distributed/accessory genes from the population supragenome [1,7,13,15]. We have demonstrated high degrees of genomic plasticity within multiple species that underlies the observation that these species have both pathogenic and commensal strains, these include: Haemophilus influenzae [10,14,19], Pseudomonas aeruginosa [12,27,28], Streptococcus pneumoniae [11,18–20,29], Moraxella catarrhalis [30,31] among others. Genomic diversity results from a balance between genome expansion (addition of genes) and genome reduction (deletion of genes) [14–15]. Our analyses comparing type strains of both H. influenzae (Rd) and S. pneumoniae (Sp6) to clinical isolates (of each of their respective species) revealed only minor changes in genome size, but more than 200 INDELS per genome-pair comparison (on average) indicating that there is very extensive recombination occurring [14,29]. Genomic plasticity is generated via three types of horizontal gene transfer (HGT) processes: (1) transformation; (2) conjugation; and (3) bacteriophage-mediated transduction (Figure 1). A large proportion of novel bacterial genes arise from DNA duplication and other DNA-modifying processes [15], or via acquisition from mobile genetic elements (e.g. transposons, insertion elements, etc.) and transferable plasmids all of which provide genomic fodder for the evolution of novel functions as they are not associated with gene loss.
Figure 1. Schematic demonstrating the different types of bacterial inheritance.
(A) Bacterial cell with a single circular chromosome in blue; (B) vertical transmission from mother to daughter cells with the daughter cells being genomically identical with each other and the mother cell; (C) horizontal gene transfer methods with transferred or transferrable DNA signified by red shading: (i) transformation, foreign DNA is picked up from the environment by competent bacteria which is then recombined into the bacterial chromosome via homologous recombination; (ii) conjugation, transfers either a plasmid (episome) or chromomosonal DNA via a pilus from a ‘male’—parasitizing bacteria to a recipient; (iii) transduction, transfers foreign bacterial DNA via a phage using nonhomologous recombination.
Vertical gene transfer is the standard mechanism by which a mother cell replicates its entire DNA complement and passes identical (or nearly identical) copies of each chromosome and plasmid/episome (an episome is usually described as a plasmid that is able to integrate into the main bacterial chromosome) to each daughter cell during cell division [7]. In contrast, the HGT processes involve unidirectional gene movement between two, often unrelated, bacterial cells in which one or more blocks of donor chromosomal DNA (range: several hundred bases to >100 Kb) (and/or plasmid and episomal DNAs in the case of conjugation) are transferred into the recipient cell resulting in either the partial replacement of the recipient bacterium's chromosome or the acquisition of a new replicon [1,7]. Mell et al. [46] using high molecular mass DNA for in vitro transformation of H. influenzae found that the mean recombination tract length was 8.1 ± 4.5 Kb. Hiller et al. [20] in a study of S. pneumoniae evolution in situ during a polyclonal pediatric infection found 23 transformation events ranging in size from 0.4 to 235 Kb, with a mean size of 28 Kb and a median size of 13 Kb.
It is important to understand that the cell from which the transferred DNA comes does not necessarily have to be viable at the time of transfer (and never is in the case of transduction). Each of the HGT mechanisms can occur between different strains of the same species and also between related species. Conjugation, however, particularly when it involves episomal transfers, can also occur between very divergent species that exist in different phyla [32,33]. Transformation rates decline rapidly with increased levels of genetic heterogeneity as they rely on homologous recombination machinery within the recipient cell [34–37]. Transduction is based on the host-range of the infecting temperate bacteriophage and is, therefore, usually confined to a single species or closely related sister species.
Some bacterial species use a single HGT process, while others use two or even all three. For example, Staphylococcus aureus principally relies on transduction — and accordingly has a much smaller pan-genome than many other bacterial species [19,38]. In contrast, Escherichia coli uses both mating [39] and transduction [40,41] whereas H. influenzae utilizes all three HGT processes [37,42–44]. Mating and transformation are active processes, requiring that the donor and recipient organisms live in close proximity to one another. Both processes require substantial energetic expenditures by one or the other of the participating bacteria. Thus, either the donor or the recipient bacteria, and indirectly the bacterial population engaging in these processes must receive an evolutionary advantage from these processes, or they would not persist. Any mechanistic process that requires energetic expenditures, by definition must provide an evolutionary advantage for its continued propagation. In the case of mating, the primary energy expenditure is via the male (DNA donating) bacteria. This process has been described as parasitic and an example of selfish genes ensuring their own propagation [45]. This is because the genes for the mating apparatus are parts of transposable elements (either chromosomally or episomally located) that collectively ensure their own horizontal propagation — as well as transferring physically adjacent genes that have been selected for their ability to provide for survival advantages to the host bacteria in stressful environments. Thus, mating often also results in the transfer of genes that enhance the survival of the recipient demonstrating that a singular process can have multiple ecological outcomes. In the mating process, two live bacteria are joined temporarily by a pilus (or similar structure) through which one bacterium sends a copy of its DNA (chromosomal and/or episomal) into the other bacterium. Therefore, the evolutionary advantage accrues to both the donor (selfish gene) as it propagates itself, and the recipient as it gains genes enabling survival such as those encoding resistance for antibiotics and heavy metals. For these reasons, we have referred to these HGT processes as population-level virulence (or survival) traits [3]. Similarly, the benefit of transformation accrues to the recipient and the population.
In the case of competence and transformation, the energy expenditure is via the recipient bacteria as it uptakes DNA from its environment (competence) and homologously recombines the exogenous DNA into its chromosome (transformation). This results in changing the genotype and phenotype of the cell which may provide a selective advantage in a stressful environment. Multiple transfers distributed around the chromosome may occur during a single competence event [20,7,46–48]. Competence, the metabolic state of being able to take up foreign DNA from the environment into the cell, is triggered by nutrient limitation or other stress conditions as part of the bacterial cell's SOS response. One of the molecular triggers of competence among the Streptococci, and related gram-positive bacteria, is the production of a quorum-sensing pheromone, competence stimulating peptide (CSP). CSP is a seventeen amino acid peptide that serves as an intercellular activating signal leading to the expression of the ComABCDE regulon that encodes genes that control and produce the cellular machinery necessary for competence and transformation. In S. pneumoniae there are several CSP alleles that divide the species into specificity groups as each CSP has a corresponding receptor (ComD) which is specific for a given CSP peptide. In some cases, particularly among the Streptococci and Vibrio spp. competence is also an auto-parasitic process, as the first bacteria in a population to become competent kill their neighbors to ensure a source of DNA for transformation [49–52]. Finally, viral transduction results when a lysogenic (temperate) bacterial virus, or bacteriophage, excises itself from the host genome and inadvertently takes some of the host's genes along with its own genes and then reinserts these genes into the recipient's genome when it establishes a lysogenic state [1,7,15]. Through transduction (specialized or generalized), bacteriophages play a major role in HGT, especially regarding antibiotic resistance, virulence factors, and invasion-related functions [13,15]. Viral transduction is a bacterially passive process for both the donor and recipient bacteria (in that it results from viral infection) which can result in bacterial species living in different environments to exchange genetic material [7,13,15].
DNA exchange via these HGT mechanisms provides for diversity generation within a bacterial population; and thus, they provide the same advantages as sex in eukaryotic organisms and can be thought of analogous processes [53,54]. Recently Colnaghi et al. [55] demonstrated that eukaryotic sex developed to provide the same population-level benefits as HGT in prokaryotes, including protection form Muller's ratchet, because as genome size increased there was a need for increases in recombination length. Without the ability to rapidly generate diversity, all individuals within the population would have the same fitness level with respect to environmental challenges which would decrease a population's chance of survival during times of environmental change. Moreover, the lack of HGT would result in the bacterial equivalent of in-breeding as there would be no mechanism to replace mutant loss-of-function alleles; this would, according to Muller's ratchet, result in their eventual extinction caused by the accumulation of slightly deleterious mutations via genetic drift [56]. The universality of the need for genetic recombination was recently reinforced when it was found that there is extensive HGT among metazoan ‘asexual’ bdelloid rotifers. The authors of this work, and those commenting on its implications, have clearly and unequivocally construed HGT as the equivalent of sex among these metazoans, and used it to explain their long-term survival as it creates diversity preventing evolutionary bottlenecks associated with the lack of sex [57,58]. Thus, bacterial HGT mechanisms help to ensure that the population survives both during periods of environmental and nutritional challenges [7] by providing an ‘evolutionary shortcut’ that enables organisms to quickly adapt to a changing or new environment [13] and by providing for replacement of mutant alleles [56,59]. HGT can also induce/produce major lifestyle changes for recipient cells and permit radiation into different ecological niches [15] such as occurs when commensal organisms within a holobiont [60–62] acquire virulence genes and become pathogens. Taken together, these observations suggest that evolutionary pressures select for mechanisms that generate diversity [7].
Bacterial supragenome/pan-genome
A species’ supragenome contains three different sets of genes: (1) the core genes, which are found in all strains of the species; (2) the distributed/accessory genes, which are found in a subset of strains of the species; and (3) the shell/unique genes, which are found in only a very small fraction of the strains of the species. Thus, in addition to the set of core genes, each strain has its own unique set of noncore genes from the supragenome [1,9,63]. The core genome consists of ‘essential’ genes responsible for the basic aspects of a species’ metabolism and major phenotypic traits [13,15], including genes for housekeeping functions, such as energy production, amino acid metabolism, nucleotide metabolism, lipid transport, and translational machinery [48]. In contrast, the noncore (distributed, accessory, or adaptive) genome includes genes encoding for supplementary or modified biochemical functions that may be useful in contexts other than basic survival, such as adaptation to new environments, antibiotic resistance, or colonization of new environments and hosts [13,15]. Being noncore implies that these genes can be deleted from the genome, but such deletions may result in the loss of important phenotypic traits, such as the ability to grow on nontraditional nutrients and substrates, virulence and antibiotic resistance [13,15]. Early studies demonstrated that the majority of noncore genes within a species supragenome have been evolving with the core genes of that species [64] and that many of the unannotated distributed genes are associated with survival in different environments [25,65]. The noncore genes also include those with parasitic functions, the selfish genes. These include those that promote their own transfer and propagation [66] as well as those that run a ‘protection racket’ — the toxin–antitoxin genes [67].
WGS of multiple strains of multiple bacterial species has demonstrated that, with each new strain sequenced, multiple new genes are found. The number of novel genes added per genome, ranges from many hundreds at the start of a pan-genome project to just a handful as the growth becomes asymptotic [9–12,9–14,16,19–20,31,68]. These early observations led to the recognition that in most cases thousands of genomes would be needed to fully describe the supragenomes/pan-genomes of many bacterial species [13]. Recently Park et al. [38] performed pan-genome analyses on seven pathogenic bacterial species using data downloaded from some 27 000 genomes from the NCBI prokaryotic genome database. They calculated pan-genome sizes of 22 000 genes for S. aureus and 128 193 genes for E. coli with the others being intermediate. It is interesting to note, that even the smallest of these bacterial pan-genomes contain as many genes as the human genome based on these recent estimates [69].
Our laboratory's recent sequencing of more than 2000 H. influenzae clinical isolates has demonstrated that we have yet to fully characterize this species’ supragenome, albeit we are getting very close as our recent analyses have not identified any novel MLST types (unpublished observations); moreover, the number of novel genes identified with each additional strain sequenced is now in the single digits. This analysis includes specimens from all six permanently inhabited continents, and from patients with every known disease type that H. influenzae has been associated with; therefore we think the probability is high that there are very few widely divergent genotypes that have not been characterized, and that the number of additional novel genes to be identified is relatively small compared with the number that has been found. With this knowledge, the best means to approximate a bacterial species genome in terms of gene numbers are mathematical models, including the Pan-Genome Model [13,18] and the Finite Supragenome Model [14,16,19] both of which have proven to be quite accurate in predicting the total number of genes within a species using data from WGS of a few independent isolates.
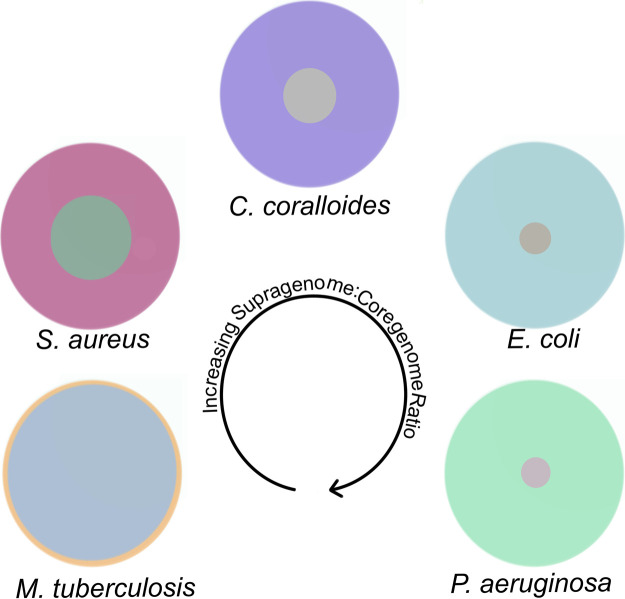
As no two strains in a species contain the same complement of genes (per the DGH), collectively the species’ supragenome/pan-genome is often quite large and can actually exceed the number of genes in mammalian genomes. For example, as noted above, there are over 100 000 genes in the E. coli supragenome. However, it is also easy for the prokaryotic cell to undergo genomic deletions, disposing of unnecessary or deleterious genes [15]. The size of a species’ supragenome/pan-genome, relative to its core genome size, is highly variable among bacterial species. In an analysis of 295 species-specific supragenome projects published from 2005 to 2019, the supragenome was found to be substantially larger than the core genome in essentially all cases with the core genome making up from <20% to >60% of the supragenome [48] (Figure 2). It is interesting to note that both genome and supragenome size are associated with a species biology. Free-living environmental bacterial species tend to have the largest genomes 4–12 megabases (Mb); with commensal and pathogenic bacteria having intermediate size genomes 1.5–4 Mb; and obligate and intracellular pathogens having the smallest genomes 0.6–1.5 Mb. These rules, however, are not hard and fast as there are exceptions as not all pathogens have a reduced genome size [48].
Figure 2. Schematic showing the relationship between the size of the core and supragenomes for different bacterial species.
The area of the inner circle for each species represents the size of the core genome relative to the supragenome which is represented by the entire area of the circle. All areas are representative of the actual percentage differences between the core and supragenomes. Starting at the lower left with Mycobacterium tuberculosis and moving clockwise through Staphylococcus aureus, Corallococcus coralloides, Escherichia coli and Pseudomonas aeruginosa the relative size of the core genome decreases compared with the supragenome.
In summary, the DGH provides a theoretical framework for understanding bacterial genomic plasticity and the supragenome/pan-genome are the functional constructs that embody the genic diversity that the DGH predicts. The DGH states, with respect to chronic bacterial pathogens, that they utilize a survival strategy wherein a majority of their genes are distributed among a population and are not found in all members of a species; thus there exists a supragenome at the population level which is greater than the genome of any one organism and that this distribution of genes among a population serves as a population-level virulence factor [3] that provides for improved population survival through continual HGT mechanisms which provide for rapid adaptation to environmental conditions through the reassortment of genes (and alleles). Moreover, the set of genes in a species’ supragenome can expand through the introduction of genes via inter-species exchange or via gene duplication and the evolution of paralogous functions within a species. This is not to say that all genes and all different gene combinations will lead to correspondingly large phenotypic differences as multiple genes can often provide the same functionality; moreover, it is quite possible that some genes that enter a genome via HGT mechanisms may not be functional in that genome due to the need for other corresponding gene products or signaling pathways that are not present in the new host. With those caveats, the shuffling of noncore genes and alleles (different forms of the same gene) of core genes generates new combinations that are subsequently subjected to the forces of selection. Genomic plasticity represents a successful strategy for bacteria to adapt, survive, and evolve.
Research applications based on the supragenome
In the 15 years since the initial characterizations of bacterial supra/pan-genomes in both Gram-negative and Gram-positive species [9,10] the integration of the DGH with our parallel increases in understanding of microbial ecology has revolutionized thinking about many aspects of bacteriology. These include: (1) basic bacterial biology, e.g. their possession of a life cycle, evolution, population genetics, and taxonomy/phylogeny; (2) host–bacterial interactions both at the species and microbiome levels encompassing bacterial colonization, persistence and invasion into host tissues; and (3) clinical applications, e.g. the diagnosis, treatment, prevention, and epidemiology of bacterial infections [1,7,13,15,63]. It is important in these contexts to make the distinction between the bacterial meta-genome which refers to the collective genomes of multiple species assemblages, i.e. microbiomes, and the supra/pan-genome which refers to the collective genome of a single species, or population of that species within a particular microbiome. Here, we discuss several research applications from the current literature using these perspectives as theoretical bases.
Supragenome-wide association studies
The DGH teaches that each individual strain within a species and even within a polyclonal bacterial population possesses a unique complement of distributed genes. This means that each strain possesses variation with respect to gene possession with respect to all other strains. Thus, it follows in the case of pathogens that each strain has a unique set of heritable traits with regard to antigen presentation to the host; virulence, including antibiotic resistances and serum resistances; and tissue tropisms, all which contribute to a strain's pathogenicity affecting its ability to colonize, persist in the face of antimicrobial therapy, invade cells and tissues, metastasize to distant sites via systemic spread, and evade or disarms various aspects of the host's innate and adaptive immune systems [1,7,64]. Based on the DGH we developed a comparative and functional genomics program that has provided the data establishing the veracity of the postulate that the genetic determinants of virulence and antibiotic resistance are unique to each strain for multiple pathogenic bacterial species (reviewed in [48]). These include: the Gram-negative pathogens, H. influenzae [14,19,62,65,70–73], M. catarrhalis [30,31], P. aeruginosa [12,27,28], and Burkholderia cenocepacia [68]; and the Gram-positive pathogens S. pneumoniae [19–20,29,74–77], S. aureus [19], and Gardnerella vaginalis [16]. This information can be exploited for the development of targeted prevention and treatment strategies [62,65,71].
We first proposed the use of SGWAS (SupraGenome-Wide Association Studies) for the identification of bacterial virulence and tropism genes over a decade ago [62]. SGWAS, as with GWAS, can analyze large numbers of genetic variants (including gene possession variants) to test for a statistical association between each variant and a phenotype of interest [78] being careful to apply methodologies that account for multiple comparisons such as the Bonferroni correction or the Benjamini–Hochsberg method to decrease the false discovery rate. Since that time, we and others have used SGWAS as a valuable tool to identify specific bacterial genes that gives rise to specific phenotypic traits [65,71,78,79]. To date, bacterial GWAS studies have principally focused on identifying genes that are associated with clinically relevant phenotypes, such as virulence and antibiotic resistance [62,65,71,78,79] (Table 1).
Table 1. Examples of Pathogen SGWAS.
| Organism | Sample size | Phenotype | Reference |
|---|---|---|---|
| Haemophilus influenzae | 220 | Virulence | [70,64] |
| Mycobacterium tuberculosis | 498 | Antibiotic resistance | [78] |
| Staphylococcus aureus | 75 | Antibiotic resistance | [167] |
| Streptococcus pneumoniae | 3701 | Antibiotic resistance | [168] |
| Plasmodium falciparum | 1063 | Antibiotic resistance | [169] |
| Staphylococcus aureus | 90 | Virulence | [170] |
| Staphylococcus epidermidis | 83 | Virulence | [171] |
| Burkholderia cenocepacia | 215 | Biofilm formation | [67] |
| Haemophilus influenzae | 220 | Virulence | [71,65] |
| Mycobacterium tuberculosis | 498 | Antibiotic resistance | [79] |
| Staphylococcus aureus | 75 | Antibiotic resistance | [170] |
| Streptococcus pneumoniae | 3701 | Antibiotic resistance | [171] |
| Plasmodium falciparum | 1063 | Antibiotic resistance | [172] |
| Staphylococcus aureus | 90 | Virulence | [173] |
| Staphylococcus epidermidis | 83 | Virulence | [174] |
| Burkholderia cenocepacia | 215 | Biofilm formation | [68] |
Recently, we have conducted SGWAS studies using multiple advanced algorithms including machine-learning approaches. In Lee et al. [68] we conducted a SGWAS using Spearman rank correlation to study 215 B. cenocepacia strains isolated from 16 CF patients and observed recurrent loss-of-function mutations that were associated with decreases in biofilm formation. In two random-forest-based machine-learning SGWAS studies, both involving large numbers of H. influenzae strains (the first with >1600 genomes and the second with >200 genomes) that were performed to associate clinical provenance with gene presence/absence, we identified a preponderance of unannotated genes among the most important classifiers. This finding is of immense theoretical importance as it teaches us two important lessons. First, that these unbiased methods can point us to specific genes within the enormous background of genomic ‘dark matter’ that are relevant clinically, and second that their examination is highly likely to lead to novel biology, as we previously demonstrated [65]. In this study, we applied statistical genetic analysis methods to clinical meta-data on large numbers of H. influenzae strains from which we identified multiple unannotated genes that were associated with virulence. Characterization of one of these genes, which we named msf1, demonstrated that it was a major virulence factor providing for invasion and survival in human macrophages, and also increased trafficking of H. influenzae to the brain in the Chinchilla lanigera model of otitis media leading to increased morbidity and mortality.
Microbiome transplantations
Microbiomes are polykingdom communities, often including bacteria, archaea, fungi, protozoa, and viruses, which colonize particular environments in or on animal bodies (skin, oral cavity, gastrointestinal tract, respiratory tract, urogenital tract, etc.), higher plants, soils and other terrestrial and aquatic environments. The microbiome can also be thought of as the combined genetics and metabolic capabilities of the community of organisms. For the host/holobiont, defined as host-microbiota symbioses [60–62], individual members of the microbiome can display a wide range of symbiotic relationships — from mutualism to commensalism to parasitism to pathogen.
Microbes inhabiting the human body (1) play roles in multiple important physiological functions, such as digestion, metabolism and immunity; (2) vary according to body site; and (3) in most cases establish an equilibrium with healthy hosts [80,81]. When this homeostasis is disrupted by an overgrowth of pathogenic microorganisms or by a lack of sufficient numbers of mutualistic or commensal microorganisms at particular body sites or expression of accessory genes triggered by some environmental stimulus, the consequences of the resulting microbial dysbiosis are malfunctioning physiological processes and perhaps ultimately disease [82–86]. Thus, understanding host–microbiome interactions provides insights into disease diagnoses, treatment, and prevention. Disturbances in microbiomes at various human body sites have been linked to the development of various traits and diseases, including weight gain, obesity, inflammatory bowel disease, diabetes, liver cirrhosis, cardiovascular disease, rheumatoid arthritis, cancer, depression, autism, asthma [80,84], and even premature and still birth [86,87]. Treatments involving the human oral and gut microbiomes include probiotics, which have become popular with the public in recent years. More recently microbiome transplantations (although the Chinese have been using them for thousands of years) have also captured the public imagination, however, they are still largely in the experimental stages at this point, but they have produced promising results as a potential therapy for diseases caused, or exacerbated by, microbial dysbiosis [88]. Transplantation of the intestinal and skin microbiomes have been used to treat a range of diseases. Fecal microbial transplants (FMT) have shown promising results in patients with obesity, C. difficile infections, and ulcerative colitis [89,90]. There are ongoing trials to measure the potential therapeutic effects of FMT in a host of other diseases; however, the benefits have yet to be determined [91]. Though some benefits of FMT have been shown, there have also been adverse effects, such as obesity post-transplant and infection [92] and even infection and death [93]. Hopefully, with a better understanding of strain-specific genetic profiles, donors and mock-laboratory-built fecal microbiomes could be more carefully and strategically selected/constructed to avoid adverse effects owing to pathogenic distributed genes in strains from donor samples. This could be accomplished using a combination of 16S microbiome analyses with metagenomic sequencing [94,95] to provide data on species/strain composition and gene content which could be used to aid in the creation of a safer fecal transplant.
While the gut microbiome has been investigated extensively, the skin microbiome has been the focus of more recent research. The skin is colonized by a large number of diverse microorganisms, of which most are beneficial or harmless [96,97]. The composition of the abundant species is relatively stable over time, although varies with anatomical site. However, skin-associated diseases, such as acne vulgaris, eczema, psoriasis, and dandruff are associated with strong and specific microbiome alterations. Thus, manipulation of the skin microbiome holds promise as a novel therapeutic approach for these diseases (e.g. [98,99]). Paetzold et al. [100] used mixtures of different skin microbiome components to alter the composition of recipient skin microbiomes and showed that, after sequential applications of donor microbiomes, recipient microbiomes became more similar to those of the donors. As the degree of engraftment depended on the recipient and donor microbiome composition, applied bacterial load, and application site, these parameters will need to be explored more fully in future experiments.
Microbiome-sparing therapeutics and vaccines
While HGT drives the evolution of many virulent and drug-resistant bacterial strains that contribute to increasing levels of morbidity and mortality [7,15], studies of pathogen supragenomes/pan-genomes, on the other hand, help to identify distributed genes that could serve as biomarkers of virulence, and perhaps more importantly as targets for precision medicine-based treatments and preventions [15,65]. For species-level diagnostic purposes the core genome can be utilized as it contains genes possessed by every member of the species, but we need to use the noncore (distributed/accessory) genes to identify the strains with specific phenotypes if we wish to target specific populations for intervention (since different genes and gene combinations produce different disease phenotypes and tissue tropisms) [48,65]. For global prevention and treatment, core genes can be used to target an entire species whereas targeting distributed genes allows for selective strain targeting, ensuring that only strains containing the gene of interest are affected (microbiome-sparing approach) [48,65]. As a technical note, the risk of misclassifying genes as core or distributed/accessory (when they are not) has decreased considerably as the field moves to long-read genomic sequencing methodologies such as PacBio and Oxford Nanopore. These methodologies which have been employed for the last half-dozen years routinely provide closed circular genomes directly from the initial sequencing run. Thus, they have eliminated the need to start genome assemblies with alignments to a reference genome which can be problematic when there is extensive genomic plasticity, and instead provide high-quality sequences for de novo assemblies [25,68,73].
Antibiotics have been considered the standard-of-care for the treatment of bacterial infections caused by drug-susceptible organisms since World War II. However, this situation is changing due to the worldwide spread of antibiotic resistance driven by widespread use (and misuse) of antibiotics [101,102]. This emergence of drug-resistant bacterial pathogens has led to a decline in the efficacy of traditional antimicrobial therapies and has greatly limited the repertoire of antibiotics available to effectively treat patients [101,102]. The rapid rise in multi-drug-resistant bacteria is a direct consequence of natural selection operating on HGT-driven mechanisms of gene exchange whereby multiple antibiotic resistance genes have become clustered together with the genes that promote conjugation. Thus, a single gene transfer event can provide for the survival of the recipient in the face of combination antibiotic therapy.
Much of the rise in antibiotic resistances (both individual resistances and multi-drug resistances) could have been avoided, or at least greatly delayed, by the employment of common-sense guidelines for treatment. Easily the greatest mistake made by antibiotic stewards worldwide for most of the last 75 years was due to their slavish adherence to the ‘one antibiotic at a time’ doctrine. On the face of it, this was a statistically flawed approach from the start. If resistance to a given antibiotic were to arise spontaneously in one in 107 bacteria (a widely agreed upon rate), then an infection with 109 bacteria (not an unreasonable number) would produce 100 resistant bacteria that would survive, and could go on to colonize and infect other individuals — which is precisely what has occurred over and over again. If, on the other hand, the patient had been treated with two (or even better three) different antibiotics with non-overlapping mechanisms of action then to produce a doubly resistant organism it would require an infection with 1014 bacteria (107×107 = 1014) — which is a number far greater than any infection, even during sepsis; and to produce a triply resistant organism it would require a starting population of 1021 bacteria which is equivalent to 1000 metric tons of bacteria! Unfortunately, it is now too late to universally adopt such an approach and have a uniform positive patient outcome. This is because our past mistakes, caused largely by an unawareness of the teachings of the DGH, have resulted in the evolution of transferable plasmids and transposons that not only promote HGT of their own core dispersal genetic machinery (selfish genes), but also include multiple genetically encoded resistances to antibiotics. Thus, a single HGT event will result in the formation of a new multiply-drug-resistant bacterial strain [103,104]. However, one need only look at HIV HAART (highly active anti-retroviral therapy) to understand the utility of such a multi-target approach to pathogen treatment. The simultaneous targeting of multiple HIV enzymatic functions, in the late 1990s, changed HIV-1-related disease from a near certain death sentence to a treatable chronic disease.
Historically, antibiotics have been identified by screening natural compounds for their ability to kill bacteria grown in vitro. Recently, genome sequencing and supragenomic characterization of microorganisms have enabled the collection of detailed information regarding the physiological repertoire of entire microbial species (vide supra). This has led to a shift in the discovery of novel anti-microbials from an empirical approach to a knowledge-based approach based on specified targets. In these ‘target-based’ approaches, potential drugs are identified, or preferentially, designed using in silico modeling algorithms [105,106] that take advantage of structural information of the target molecule predefined via its roles in a key metabolic process. Comparative metabolomics [107] combined with comparative genomics [7] can be used to identify genes essential for pathogen survival and pathogenicity, which are then tested as targets of specific compounds derived from large chemical libraries [105,106].
Aside from the antibiotic resistance crisis, research has revealed the harmful effects of broad-spectrum antibiotic therapy on the community structure of beneficial host microbiota, which in turn can have negative effects on long-term host health [102]. To combat these threats, the microbiome-sparing approach aims to modify or replace broad-spectrum antibiotics with precision anti-microbials that selectively target and remove pathogenic strains while leaving the community structure of the surrounding microbiota unchanged [102]. These new therapeutic strategies include the development of anti-virulence compounds that inhibit specific bacterial pathogenesis and persistence traits of targeted strains encoded by distributed genes, which therefore spare other strains of the same species that do not encode such virulence genes. As such these strategies are designed to identify compounds that are bactericidal or bacteriostatic to a minimal number of bacterial strains [102], and thus provide precision treatments, based on druggable small molecules and natural compounds that are bacterial strain specific (Table 2). By preserving patients’ microbiota, this microbiome-sparing strategy of identifying pathogen-specific targets has the potential to improve patient health during and after bacterial infections [48,65,102].
Table 2. Examples of microbiome-sparing treatments for infections.
| Infection | Organism | Model | Treatment | Reference |
|---|---|---|---|---|
| Urinary tract | Escherichia coli | Mouse | Mannosides | [175] |
| Intestinal tract | Escherichia coli | Mouse, human | Mannosides | [176] |
| Respiratory tract | Pseudomonas aeruginosa | Mouse | LecB inhibitors | [177] |
| Nasopharynx | Staphylococcus aureus | Mouse | Natural compound produced by S. lugdunensis (lugdinin) | [178] |
| Skin | Propionibacterium acnes | Human | Natural compound produced by Staphylococcus epidermidis (succinic acid) | [179] |
Whole-genome sequencing in bacterial diagnostics
Identification and characterization of infection-causing microorganisms are crucial for successful treatment, recovery, and safety of patients. Culture fails to detect an organism in 80% of cases in which a patient has signs and/or symptoms of infection. This under-detection has multiple causes, including: antibiotic treatment; bacteria growing as biofilms; and slow-growing, fastidious organisms that cannot be cultured or literally take weeks to culture [2,64,108–129]. Conventional clinical diagnostic culture methods are biased toward the 2% of microorganisms able to grow rapidly in standard culture media. Particularly for chronic or biofilm-related infections, these cultured bacteria are usually presumed to be relevant. However, we now know that in many chronic infections the organisms that grow out rapidly are often not the representative of the species that are driving the infection [120–122,130–141]. In part, this understanding is based on the reduction in time to closure of chronic wounds when patients are treated based on the results of molecular diagnostics and WGS as opposed to culture [130–133]. A meta-analysis currently ongoing in the authors’ laboratories covering multiple studies of multiple infectious diseases over more than a decade, in which multiple molecular diagnostic methods were used to diagnose over 7000 infections across a wide range of clinical presentations and anatomic sites demonstrated that under the best conditions culture would detect S. aureus ∼50% of the times it was actually present, and could detect other staphylococcal and most streptococcal species ∼20% of the time. For nearly all other pathogens the detection rate was less than 20% and for anaerobes such as Cutibacterium acnes (formerly Proprionibacterium acnes), and multiple Treponena spp. Prevatella spp. it was far less. Culture is also inadequate to rapidly and accurately distinguish among multiple strains and sub-strains of even the most common pathogens, in part because they often evolve rapidly through HGT mechanisms during the infectious process [7,16,20,25,28,68,142].
Despite its shortcomings, culture is entrenched as the primary diagnostic technique for the identification and characterization of bacterial pathogens. Nucleic acid amplification techniques (principally PCR) overcame many of these shortcomings, and DNA sequencing methodologies, such as multiple-locus sequence typing (MLST), were a step in the right direction, but they lack the resolution needed for strain-specific diagnostics [63,143–145]. Thus, there was a need for better molecular diagnostic tools to improve the accuracy and efficiency of diagnoses.
WGS promises to provide the ultimate in resolution for strain, and even sub-strain identification, as it takes into account the genomic plasticity and supragenomes of bacterial species [63,142] and can be used to identify novel strains as they appear. The superior resolution of WGS in the identification and characterization of pathogens has great potential for routine use in diagnostic laboratories (e.g. [143,144]). However, despite this promise, WGS has not completely superseded current diagnostic methods in most clinical microbiology laboratories. There are several obstacles to its routine implementation, including the high cost of WGS, a lack of training in bioinformatics among clinical microbiologists, a lack of the necessary computational infrastructure in most hospitals, and the difficulty of establishing the proper bioinformatic protocols [144]. Much of this can be overcome through the use of standardized or centralized ‘cloud-based’ computational systems. The European Society of Clinical Microbiology and Infectious Diseases (ESCMID) has recently published a review covering the need for such systems and a framework for their implementation [143,145]. As with any system of analysis, appropriate technical requirements need to be instituted to prevent the caveats associated with poor quality sequencing that can lead to low-coverage and lost sequences. Doyle et al. [146] have highlighted the need for quality sequence for clinical diagnostics wherein they showed missed WGS-based antibiotic resistance calls due to short read, low-coverage data. This has led to the push to adopt long-read, and even circular consensus sequencing, which obviate these issues [37,147–148].
Reverse and microbiome-sparing approaches to bacterial vaccinology
For over a century, infectious diseases have been controlled by vaccination and the administration of antibiotics [101]. Nevertheless, pathogenic microorganisms are still the most important threat to health worldwide [149]. Conventional vaccinology approaches were successful in conferring protection against some but not all infectious diseases [149]. The vaccine counterpart to antibiotic resistance — ‘vaccine escape’ — is partly responsible for vaccine failure. Vaccine escape implies that the target pathogen has mutated such that it no longer expresses the same form of the antigen as used to prepare the vaccine. This type of vaccine failure is completely different from those associated with host-based ‘immunological windows’ wherein antigen presentation does not induce a memory immune response to the vaccine antigen.
Until very recently essentially all vaccines were based on killed or live-attenuated microorganisms, or their chemically deactivated toxins [101,149]. With the genomic era we and others developed the concept of the supragenome/pan-genome [8–12,14,18] and from this came the recognition that distributed/accessory genes associated with virulence could be used to target specific pathogenic strains in a microbiome-sparing approach that would not result in the elimination of commensal strains [65,71,149]. Conversely, the pan-genome can be used to identify core genes that are universally present among all strains if the wish is to eliminate a bacterial species in its entirety. These potential vaccine antigens, whether core or distributed, are often identified in reverse manner starting from an analysis of the supragenome/pan-genome of a species as opposed to the use of a single whole organism — a process called reverse vaccinology [150,151]. Reverse vaccinology makes use of bioinformatics and for microbiome-sparing approaches, statistical methods using strain meta-data with regard to clinical provenance, i.e. commensal or pathogen [71,65], to utilize the information derived from the supragenome/pan-genome of the target bacterial species to predict potential vaccine candidates ‘in silico’. The first the application of reverse vaccinology was for a vaccine against serotype B Neisseria meningitidis [152]. For a vaccine against the more complex Streptococcus agalactiae, reverse vaccinology using the supragenome/pan-genome was employed for the first time [153]. Somewhat later reverse vaccinology was used to compare pathogenic and nonpathogenic strains of the same species to find antigens that truly affect pathogenesis These successes led to application of reverse vaccinology to other pathogens including N. gonorrheae and Mycoplasma pneumoniae [154,155]. It is important to point out that reverse vaccinology and microbiome-sparing vaccine approaches are still susceptible to the evolution of vaccine escape mutants. Thus, it will always be necessary to constantly surveil the circulating pathogen population for the evolution of mutants.
Bacterial single-cell ‘omics’
The vast majority (>99%) of microbial species in the biosphere cannot be cultured in the laboratory with current culturing methods and thus are contained within what has been referred to as the microbial ‘dark matter’ [156–158]. This cultural inability has limited our ability to understand the biology of these organisms. Microbiologists have traditionally studied populations of bacterial cells, typically using millions to billions of cells for analysis in bulk, rather than individual cells, as it has been assumed that individual cells are representative of the population. However, this assumption neglects any heterogeneity present in the population [159–161]. The individual behavior of single cells, particularly in spatially and taxonomically complex assemblages are substantially different from that of the whole population; thus, conclusions based on average molecular or phenotypic measurements of a population can be biased, as the patterns of subpopulations would not be revealed [162].
The recent development of single-cell meta-omics has greatly enhanced our understanding of the individuality and heterogeneity of microbes in multiple biological systems [162].
Single-cell omic technologies (genomics, transcriptomics, metabolomics) help reveal this hidden information from both unculturable organisms and low-abundance organisms in complex microbial communities [161]. These technologies are providing new perspectives with regard to our understanding of population diversity by bringing the power of meta-omics [163] to the single-cell level for studies of taxonomically and metabolically complex biofilms, microbiomes, and holobionts [60–62]. The ability to comprehensively characterize single cells or small populations of cells within a more complex system is revolutionizing our understanding of bacterial metabolic differentiation and how this contributes to the robustness of the biology of microbiomes and their holobionts [60–62,160]. Single-cell sequencing (SCS) is one of the tools used in single-cell omic studies and complements metagenomic deep-sequencing methods [161]. The three main applications of single-cell omics in relation to bacterial populations are to: (1) investigate the genomes of unculturable microorganisms; (2) delineate cell-to-cell diversity within diverse populations [159–162]; and (3) compare the transcriptional activities of genomically identical cells based on their spatial orientations with respect to nutrient availability and access to the substrate. SCS has been applied in widely diverse biological and environmental contexts, including human microbiomes (e.g. [156,164]), seawater and marine sediments (e.g. [165–167]), and even a hospital sink [168–169].
Summary
The advent of the concepts of the supragenome/pan-genome and the DGH have revolutionized not only our understanding of bacterial genomics, evolution and adaptability, but they have also provided the framework for novel approaches to diagnosis, precision medicine, and vaccinology. As the scientific community continues to expand upon the past 20 years’ accomplishments in this field, humanity stands to reap substantial rewards with regard to personalized medicine and public health.
Perspectives
The DGH and its prediction of the bacterial supragenome/pan-genome together with the biofilm paradigm has resulted in the formation of a new rubric, bacterial plurality. Bacterial plurality encompasses the concepts that persistent bacterial infections require both genotypic and metabolic heterogeneity, as well as evolution in situ, to explain what had been previously paradoxical findings associated with chronic infections.
Horizontal gene transfer (HGT) mechanisms provide the engine for robust and continuous recombination among bacteria that provides for continuous strain evolution during polyclonal colonizations and chronic infections as a means to adapt to changing environmental and host immune pressures.
The realization that there are differences in gene content and gene expression between commensal and pathogenic strains of the same species provides for specific targeting of the pathogenic strains in the design of drugs and vaccines, resulting in microbiome-sparing approaches.
Acknowledgements
The authors would like to thank Ms. Carol Hope, MBA, for administrative support in the preparation and submission of this paper. We are also grateful to Josh Earl for his principal role in the development and implementation of much of the bioinformatic software that underlies so much of the data discussed herein. Finally, we thank the entire faculty and staff of the Center for Genomic Sciences and the Center for Advanced Microbial Processing for generating much of the data that is summarized in this review. This work was supported by Drexel University College of Medicine; the Oskar Fisher Project, a gift from Dr. James Truchard; the Bill and Marion Cook Foundation; and NIH R01 DC-02148 and NIH U01 DK-082316 to GDE.
Abbreviations
- CSP
competence stimulating peptide
- DGH
distributed genome hypothesis
- FMT
fecal microbial transplants
- HGT
horizontal gene transfer
- MLST
multiple-locus sequence typing
- PCR
polymerase chain reaction
- SCS
single-cell sequencing
- WGS
whole-genome sequencing
Competing Interests
The authors declare no competing interests
Author Contributions
J.A.H. together with E.A.G. wrote the first draft; K.M.S. made the figures; J.C.M. provided data necessary for the response to the reviewers and critical reading of the revisions; G.D.E. outlined the manuscript and provided many of the references; edited the entire original submission; and was responsible for all revisions for the subsequent versions. All authors read and approved the final version.
References
- 1.Ehrlich G.D., Hu F.Z., Shen K., Stoodley P. and Post J.C. (2005) Bacterial plurality as a general mechanism driving persistence in chronic infections. Clin. Orthop. Relat. Res. 437, 20–24 10.1097/00003086-200508000-00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrlich G.D., Hu F.Z., Sotereanos N., Sewecke J., Parvizi J., Nara P.L. et al. (2014) What role do periodontal pathogens play in osteoarthritis and periprosthetic joint infections of the knee? J. Appl. Biomater. Funct. Biomater. 12, 13–20 10.5301/jabfm.5000203 [DOI] [PubMed] [Google Scholar]
- 3.Hu F.Z. and Ehrlich G.D. (2008) Population-level virulence factors amongst pathogenic bacteria: relation to infectious outcome. Fut. Med. 3, 31–42 10.2217/17460913.3.1.31 [DOI] [PubMed] [Google Scholar]
- 4.Smith-Vaughan H.C., Leach A.J., Shelby-James T.M., Kemp K., Kemp D.J. and Mathews J.D. (1996) Carriage of multiple ribotypes of non-encapsulated Haemophilus influenzae in aboriginal infants with otitis media. Epidemiol. Infect. 116, 177–183 10.1017/S0950268800052419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith-Vaughan H.C., Sriprakash K.S., Mathews J.D. and Kemp D.J. (1997) Nonencapsulated Haemophilus influenzae in Aboriginal infants with otitis media: prolonged carriage of P2 porin variants and evidence for horizontal P2 gene transfer. Infect. Immun. 65, 1468–1474 10.1128/IAI.65.4.1468-1474.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy T.F., Sethi S., Klingman K.L., Brueggemann A.B. and Doern G.V. (1999) Simultaneous respiratory tract colonization by multiple strains of nontypeable Haemophilus influenzae in chronic obstructive pulmonary disease: implications for antibiotic therapy. J. Infect. Dis. 180, 404–409 10.1086/314870 [DOI] [PubMed] [Google Scholar]
- 7.Ehrlich G.D., Ahmed A., Earl J., Hiller N.L., Costerton J.W., Shen K. et al. (2010) The distributed gene hypothesis as a rubric for understanding evolution in situ during chronic bacterial biofilm infectious processes. FEMS Immunol. Med. Microbiol. 59, 269–279 10.1111/j.1574-695X.2010.00704.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrlich G.D., Hu Z.F. and Post J.C. (2004) Role for biofilms in infectious disease In Microbial Biofilms (Ghannoum M., and O'Toole G.A. eds), pp. 332–358, ASM Press, Washington, DC [Google Scholar]
- 9.Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L. et al. (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.”. Proc. Natl. Acad. Sci. U.S.A. 102, 13950–13955 10.1073/pnas.0506758102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen K., Antalis P., Gladitz J., Sayeed S., Ahmed A., Yu S. et al. (2005) Identification, distribution, and expression of novel genes in 10 clinical isolates of nontypeable Haemophilus influenzae. Infect. Immun. 73, 3479–3491 10.1128/IAI.73.6.3479-3491.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen K., Gladitz J., Antalis P., Dice B., Janto B., Keefe R. et al. (2006) Characterization, distribution, and expression of novel genes among eight clinical isolates of Streptococcus pneumoniae. Infect. Immun. 74, 321–330 10.1128/IAI.74.1.321-330.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen K., Sayeed S., Antalis P., Gladitz J., Ahmed A., Dice B. et al. (2006b) Extensive genomic plasticity in Pseudomonas aeruginosa revealed by identification and distribution studies of novel genes among clinical isolates. Infect. Immun. 74, 5272–5283 10.1128/IAI.00546-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medini D., Donati C., Tettelin H., Masignani V. and Rappuoli R. (2005) The microbial pan-genome. Curr. Opin. Gen. Dev. 15, 589–594 10.1016/j.gde.2005.09.006 [DOI] [PubMed] [Google Scholar]
- 14.Hogg J.S., Hu F.Z., Janto B., Boissy R., Hayes J., Keefe R. et al. (2007) Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains. Genome Biol. 8, R103 10.1186/gb-2007-8-6-r103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mira A., Martin-Cuadrado A.B., D'Auria G. and Rodriguez-Valera F. (2010) The bacterial pan-genome: a new paradigm in microbiology. Int. Microbiol. 13, 45–57 10.2436/20.1501.01.110 [DOI] [PubMed] [Google Scholar]
- 16.Ahmed A., Earl J., Retchless A., Hillier S.L., Rabe L.K., Cherpes T.L. et al. (2012) Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J. Bacteriol. 194, 3922–3937 10.1128/JB.00056-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen K., Wang X., Post J.C. and Ehrlich G.D. (2003) Molecular and translational research approaches for the study of bacterial pathogenesis in otitis media In Evidence-based Otitis Media, 2nd edn (Rosenfeld R. and Bluestone C.D., eds), pp. 91–119, B.C. Decker Inc., Hamilton, Ontario, Canada [Google Scholar]
- 18.Donati C., Hiller N.L., Tettelin H., Muzzi A., Croucher N.J., Angiuoli S.V. et al. (2010) Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 11, R107 10.1186/gb-2010-11-10-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boissy R., Ahmed A., Janto B., Earl J., Hall B.G., Hogg J.S. et al. (2011) Comparative supragenomic analyses among the pathogens Staphylococcus aureus, Streptococcus pneumoniae, and Haemophilus influenzae using a modification of the finite supragenome model. BMC Genom. 12, 187 10.1186/1471-2164-12-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiller N.L., Ahmed A., Powell E., Eutsey R.E., Earl J., Martin D. et al. (2010) Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog. 6, e1001108 10.1371/journal.ppat.1001108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golubchik T., Brueggemann A., Street T., Gertz R.E., Spencer C.C.A., Ho T. et al. (2012) Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat. Genet. 44, 352–355 10.1038/ng.1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chewapreecha C., Marttinen P., Croucher N.J., Salter S.J., Harris S.R., Mather A.E. et al. (2014) Comprehensive identification of single nucleotide polymorphisms associated with β-lactam resistance within pneumococcal mosaic genes. PLoS Genet. 10, e1004547 10.1371/journal.pgen.1004547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillings M.R. (2017) Lateral gene transfer, bacterial genome evolution, and the Anthropocene. Ann. N. Y. Acad. Sci. 1389, 20–36 10.1111/nyas.13213 [DOI] [PubMed] [Google Scholar]
- 24.David S., Sánchez-Busó L., Harris S.R., Marttinen P., Rusniok C., Buchrieser C., et al. (2017) Dynamics and impact of homologous recombination on the evolution of Legionella pneumophila [published correction appears in PLoS Genet. 2017 Dec 1;13(12):e1007116]. PLoS Genet. 13, e1006855 Published 2017 Jun 26. https://doi.org/10.1371/journal.pgen.1006855 10.1371/journal.pgen.1006855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moleres J., Fernández-Calvet A., Ehrlich R.L., Martí S., Pérez-Regidor L., Euba B., et al. (2018) Antagonistic pleiotropy in the bifunctional surface protein fadL (OmpP1) during adaptation of Haemophilus influenzae to chronic lung infection associated with chronic obstructive pulmonary disease. mBio 9, e01176-18 Published 2018 Sep 25. https://doi.org/10.1128/mBio.01176-18 10.1128/mBio.01176-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettigrew M.M., Ahearn C.P., Gent J.F., Kong Y., Gallo M.C., Munro J.B., et al. (2018) Haemophilus influenzae genome evolution during persistence in the human airways in chronic obstructive pulmonary disease. Proc. Natl. Acad. Sci. U.S.A. 115, E3256–E3265 10.1073/pnas.1719654115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erdos G., Sayeed S., Hu F.Z., Antalis P.T., Shen K., Hayes J.D. et al. (2006) Ehrlich GD construction and characterization of a highly redundant Pseudomonas aeruginosa genomic library prepared from 12 clinical isolates: application to studies of gene distribution among populations. Int. J. Pediatr. Otorhinolaryngol. 70, 1891–1900 10.1016/j.ijporl.2006.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rau M.H., Marvig R.L., Ehrlich G.D., Molin S. and Jelsbak L. (2012) Deletion and acquisition of genomic content during early stage adaptation of Pseudomonas aeruginosa to a human host environment. Environ. Microbiol. 14, 2200–2211 10.1111/j.1462-2920.2012.02795.x [DOI] [PubMed] [Google Scholar]
- 29.Hiller N.L., Janto B., Hogg J.S., Boissy R., Yu S., Powell E. et al. (2007) Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J. Bact. 189, 8186–8195 10.1128/JB.00690-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davie J.J., Earl J., de Vries S.P.W., Ahmed A., Hu F.Z., Bootsma H.J. et al. (2011) Comparative analysis and supragenome modeling of twelve Moraxella catarrhalis clinical isolates. BMC Genom. 12, 70 10.1186/1471-2164-12-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Earl J.P., de Vries S.P.W., Ahmed A., Powell E., Schultz M.P., Hermans P.W.M. et al. (2016) Comparative genomic analyses of the Moraxella catarrhalis serosensitive and seroresistant lineages demonstrate their independent evolution. Genome Biol. Evol. 8, 955–974 10.1093/gbe/evw039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahlberg C., Bergström M. and Hermansson M. (1998) In situ detection of high levels of horizontal plasmid transfer in marine bacterial communities. Appl. Environ. Microbiol. 64, 2670–2675 10.1128/AEM.64.7.2670-2675.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahlberg C., Bergström M., Andreasen M., Christensen B.B., Molin S. and Hermansson M. (1998b) Interspecies bacterial conjugation by plasmids from marine environments visualized by gfp expression. Mol. Biol. Evol. 15, 385–390 10.1093/oxfordjournals.molbev.a025935 [DOI] [Google Scholar]
- 34.Majewski J. (2001) Sexual isolation in bacteria. FEMS Microbiol. Lett. 199, 161–169 10.1111/j.1574-6968.2001.tb10668.x [DOI] [PubMed] [Google Scholar]
- 35.Fraser C., Hanage W.P. and Spratt B.G. (2007) Recombination and the nature of bacterial speciation. Science 315, 476–480 10.1126/science.1127573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray J.L., Harms K., Wikmark O.-G., Starikova I., Johnsen P.J. and Nielsen K.M. (2009) Sexual isolation in Acinetobacter baylyi is locus-specific and varies 10,000-Fold over the genome. Genetics 182, 1165–1181 10.1534/genetics.109.103127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mell J.C. and Redfield R.J. (2014) Natural competence and the evolution of DNA uptake specificity. J. Bacteriol. 196, 1471–1483 10.1128/JB.01293-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park S.C., Lee K., Kim Y.O., Won S. and Chun J. (2019) Large-scale genomics reveals the genetic characteristics of seven species and importance of phylogenetic distance for estimating pan-genome size. Front. Microbiol. 10, 834 10.3389/fmicb.2019.00834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lederberg J. and Tatum E.L. (1946) Gene recombination in E. coli. Nature 158, 558 10.1038/158558a0 [DOI] [PubMed] [Google Scholar]
- 40.Morse M.L., Lederberg E.M. and Lederberg J. (1956) Transduction in Escherichia coli K-12. Genetics 41, 142–156 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morse M.L., Lederberg E.M. and Lederberg J. (1956) Transductional heterogenotes in Escherichia coli. Genetics. 41, 758–779 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilsdorf J.R., Marrs C.F. and Foxman B. (2004) Haemophilus influenzae: genetic variability and natural selection to identify virulence factors. Infect. Immun. 72, 2457–2461 10.1128/IAI.72.5.2457-2461.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juhas M., Power P.M., Harding R.M., Ferguson D.J., Dimopoulou I.D., Elamin A.R. et al. (2007) Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 8, R237 10.1186/gb-2007-8-11-r237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juhas M., Crook D.W., Dimopoulou I.D., Lunter G., Harding R.M., Ferguson D.J. et al. (2007) Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 189, 761–771 10.1128/JB.01327-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawkins R. (1976) The Selfish Gene, Oxford University Press, Oxford, UK [Google Scholar]
- 46.Mell J.C., Shumilina S., Hall I.M. and Redfield R.J. (2011) Transformation of natural genetic variation into Haemophilus influenzae genomes. PLoS Pathog. 7, e1002151 10.1371/journal.ppat.1002151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mell J.C., Hall I.M. and Redfield R.J. (2012) Defining the DNA uptake specificity of naturally competent Haemophilus influenzae cells. Nucleic Acids Res. 40, 8536–8549 10.1093/nar/gks640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Innamorati, K.A., Earl, J.P., Aggarwal, S.D., Ehrlich, G.D. and Hiller, N.L. (2020) The bacterial guide to designing a diversified portfolio In The Pangenome (Tettelin H. and Medinie D., eds.), pp. 51–88, Springer, Cham, Switzerland: [PubMed] [Google Scholar]
- 49.Guiral S., Mitchell T.J., Martin B. and Claverys J.P. (2005) Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. U.S.A. 102, 8710–8715 10.1073/pnas.0500879102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Håvarstein L.S., Martin B., Johnsborg O., Granadel C. and Claverys J.P. (2006) New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 59, 1297–1307 10.1111/j.1365-2958.2005.05021.x [DOI] [PubMed] [Google Scholar]
- 51.Claverys J.P., Martin B. and Håvarstein L.S. (2007) Competence-induced fratricide in streptococci. Mol. Microbiol. 64, 1423–1433 Review. Erratum in: Mol Microbiol. 2007;65:230 10.1111/j.1365-2958.2007.05757.x [DOI] [PubMed] [Google Scholar]
- 52.Veening J. and Blokesch M. (2017) Interbacterial predation as a strategy for DNA acquisition in naturally competent bacteria. Nat. Rev. Microbiol. 15, 621–629 10.1038/nrmicro.2017.66 [DOI] [PubMed] [Google Scholar]
- 53.Shapiro B.J., Friedman J., Cordero O.X., Preheim S.P., Timberlake S.C., Szabó G., et al. (2012) Population genomics of early events in the ecological differentiation of bacteria. Science 336, 48–51 10.1126/science.1218198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costa T.R.D., Ilangovan A., Ukleja M., Redzej A., Santini J.M., Smith T.K. et al. (2016) Structure of the bacterial sex F pilus reveals an assembly of a stoichiometric protein-phospholipid complex. Cell 166, 1436–1444.e10 10.1016/j.cell.2016.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colnaghi M., Lane N. and Pomiankowski A. (2020) Genome expansion in early eukaryotes drove the transition from lateral gene transfer to meiotic sex. eLife 9, e58873 Published 2020 Sep 29. https://doi.org/10.7554/eLife.58873 10.7554/eLife.58873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koonin E.V. (2016) Horizontal gene transfer: essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000Res 5, F1000 Faculty Rev-1805. Published 2016 Jul 25. https://doi.org/10.12688/f1000research.8737.1 10.12688/f1000research.8737.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwander T. (2016) Evolution: the end of an ancient asexual scandal. Curr. Biol. 26, R233–R235 10.1016/j.cub.2016.01.034 [DOI] [PubMed] [Google Scholar]
- 58.Debortoli N., Li X., Eyres I., Fontaneto D., Hespeels B., Tang C.Q. et al. (2016) Genetic exchange amongbdelloid rotifers is more likely due to horizontalgene transfer than to meiotic sex. Curr. Biol. 26, 723–732 10.1016/j.cub.2016.01.031 [DOI] [PubMed] [Google Scholar]
- 59.Bryant J., Chewapreecha C. and Bentley S.D. (2012) Developing insights into the mechanisms of evolution of bacterial pathogens from whole-genome sequences. Fut. Microbiol. 7, 1283–1296 10.2217/fmb.12.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meyer-Abich A. (1943) Beiträge zur Theorie der Evolution der Organismen. I. Das typologische Grundgesetz und seine Folgerungen für Phylogenie und Entwicklungsphysiologie [Contributions to the evolutionary theory of organisms: I. The basic typological law and its implications for phylogeny and developmental physiology]. Acta Biotheor. 7, 1–80 10.1007/BF01603792 [DOI] [Google Scholar]
- 61.Margulis L. (1991). Symbiogenesis and symbionticism In Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis (Margulis L. and Fester R. eds), pp. 1–14, MIT Press, Cambridge, MA: [PubMed] [Google Scholar]
- 62.Ehrlich G.D., Hiller N.L. and Hu F.Z. (2008) What makes pathogens pathogenic? Genome Biol. 9, 225 10.1186/gb-2008-9-6-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ehrlich G.D. and Post J.C. (2013) The time is now for gene- and genome-based bacterial diagnostics: “you say you want a revolution.”. JAMA Int. Med. 173, 1405–1406 10.1001/jamainternmed.2013.7042 [DOI] [PubMed] [Google Scholar]
- 64.Gladitz J., Antalis P., Hu F.Z., Post J.C. and Ehrlich G.D. (2005) Codon usage comparison of novel genes in clinical isolates of Haemophilus influenzae. Nucleic Acids Res. 33, 3644–3658 10.1093/nar/gki670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kress-Bennett J.M., Hiller N.L., Eutsey R.A., Powell E., Longwell M.J., Hillman T. et al. (2016) Identification and characterization of msf, a novel virulence factor in Haemophilus influenzae. PLoS One 11, e0149891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lawrence J.G. (1997) Selfish operons and speciation by gene transfer. Trends Microbiol. 5, 355–359 10.1016/S0966-842X(97)01110-4 [DOI] [PubMed] [Google Scholar]
- 67.Van Melderen L. and De Bast M S. (2009) Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5, e1000437 10.1371/journal.pgen.1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee A.H.Y., Flibotte S., Sinha S., Paiero A., Ehrlich R.L., Balashov S. et al. (2017) Phenotypic diversity and genotypic flexibility of Burkholderia cenocepacia during long-term chronic infection of cystic fibrosis lungs. Genome Res. 27, 650–662 10.1101/gr.213363.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willyard C. (2018) New human gene tally reignites debate. Nature 558, 354–355 10.1038/d41586-018-05462-w [DOI] [PubMed] [Google Scholar]
- 70.Janto B.A., Hiller N.L., Eutsey R.A., Dahlgren M.E., Earl J.P., Powell E. et al. (2014) Development and validation of an Haemophilus influenzae supragenome hybridization (SGH) array for transcriptomic analyses. PLoS One 9, e105493 10.1371/journal.pone.0105493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kress-Bennett J.M. (2012) Identificatiuon and Characterization of a Novel Bacterial Virulence Factor in Haemophilus Influenzae (Doctoral dissertation, Drexel University, College of Medicine, Microbiology and Immunology Program)
- 72.Eutsey R.A., Hiller N.L., Earl J.P., Janto B.A., Dahlgren M.E., Ahmed A. et al. (2013) Design and validation of a supragenome array for determination of the genomic content of Haemophilus influenzae isolates. BMC Genom. 14, 484 10.1186/1471-2164-14-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mell J.C., Sinha S., Balashov S., Viadas C., Grassa C.J., Ehrlich G.D. et al. (2014) Complete genome sequence of Haemophilus influenzae strain 375 from the middle ear of a pediatric patient with otitis media. Genome Announc. 2, e01245-14 10.1128/genomeA.01245-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hiller N.L., Eutsey R.A., Powell E., Earl J.P., Janto B., Martin D.P. et al. (2011) Differences in genotype and virulence among four multidrug-resistant Streptococcus pneumoniae isolates belonging to the PMEN1 clone. PLoS One 6, e28850 10.1371/journal.pone.0028850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu F.Z., Eutsey R., Ahmed A., Frazao N., Powell E., Hiller N.L. et al. (2012) In vivo capsular switch in Streptococcus pneumoniae–analysis by whole genome sequencing. PLoS One 7, e47983 10.1371/journal.pone.0047983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kadam A., Janto B., Eutsey R., Earl J.P., Powell E., Dahlgren M.E. et al. (2015) Streptococcus pneumoniae supragenome hybridization arrays for profiling of genetic content and gene expression. Curr. Protoc. Microbiol. 36, 9D.4.1–9D.4.20 10.1002/9780471729259.mc09d04s36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eutsey R.A., Powell E., Dordel J., Salter S.J., Clark T.A., Korlach J. et al. (2015) Genetic stabilization of the drug-resistant PMEN1 pneumococcus lineage by its distinctive DpnIII restriction-modification system. mBio 6, e00173 10.1128/mBio.00173-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Power R.A., Parkhill J. and de Oliveira T. (2017) Microbial genome-wide association studies: lessons from human GWAS. Nat. Rev. Genet. 18, 41–50 10.1038/nrg.2016.132 [DOI] [PubMed] [Google Scholar]
- 79.Chen P.E. and Shapiro B.J. (2015) The advent of genome-wide association studies for bacteria. Curr. Opin. Microbiol. 25, 17–24 10.1016/j.mib.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 80.Awany D., Allali I., Dalvie S., Hemmings S., Mwaikono K.S., Thomford N.E. et al. (2019) Host and microbiome genome-wide association studies: current state and challenges. Front. Genet. 9, 637 10.3389/fgene.2018.00637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chow J., Lee S.M., Shen Y., Khosravi A. and Mazmanian S.K. (2010) Host-bacterial symbiosis in health and disease. Adv. Immunol. 107, 243–274 10.1016/B978-0-12-381300-8.00008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chow J., Tang H. and Mazmanian S.K. (2011) Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr. Opin. Immunol. 23, 473–480 10.1016/j.coi.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jayasinghe T.N., Chiavaroli V., Holland D.J., Cutfield W.S. and O'Sullivan J.M. (2016) The new era of treatment for obesity and metabolic disorders: evidence and expectations for gut microbiome transplantation. Front. Cell Infect. Microbiol. 6, 15 10.3389/fcimb.2016.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang J. and Jia H. (2016) Metagenome-wide association studies: fine-mining the microbiome. Nat. Rev. 14, 508–522 10.1038/nrmicro.2016.83 [DOI] [PubMed] [Google Scholar]
- 85.Desselberger U. (2018) The mammalian intestinal microbiome: composition, interaction with the immune system, significance for vaccine efficacy, and potential for disease therapy. Pathogens 7, 57 10.3390/pathogens7030057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han Y.W., Redline R.W., Li M., Yin L., Hill G.B. and McCormick T.S. (2004) Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect. Immun. 72, 2272–2279 10.1128/IAI.72.4.2272-2279.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han Y.W., Fardini Y., Chen C., Iacampo K.G., Peraino V.A., Shamonki J.M., et al. (2010) Term stillbirth caused by oral Fusobacterium nucleatum. Obstet. Gynecol. 115, 442–445 10.1097/AOG.0b013e3181cb9955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaakoush N.O. (2020) Fecal transplants as a microbiome-based therapeutic [published online ahead of print, 2020 Jun 29]. Curr. Opin. Microbiol. 56, 16–23 10.1016/j.mib.2020.05.008 [DOI] [PubMed] [Google Scholar]
- 89.Hirsch B.E., Saraiya N., Poeth K., Schwartz R.M., Epstein M.E. and Honig G. (2015) Effectiveness of fecal-derived microbiota transfer using orally administered capsules for recurrent Clostridium difficile infection. BMC Infect. Dis. 15, 191 Published 2015 Apr 17 10.1186/s12879-015-0930-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Costello S.P., Hughes P.A., Waters O., Bryant R.V., Vincent A.D., Blatchford P., et al. (2019) Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA 321, 156–164 10.1001/jama.2018.20046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leshem A., Horesh N. and Elinav E. (2019) Fecal microbial transplantation and its potential application in cardiometabolic syndrome. Front. Immunol. 10, 1341 10.3389/fimmu.2019.01341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alang N. and Kelly C.R. (2015) Weight gain after fecal microbiota transplantation. Open Forum Infect. Dis. 2, ofv004 10.1093/ofid/ofv004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.FDA. MedWatch Alert: Fecal microbiota for transplantation: safety communication — risk of serious adverse reactions due to transmission of multidrug-resistant organisms. June 18, 2019. Available at: https://bit.ly/2XDJrAK
- 94.Franzosa E.A., Hsu T., Sirota-Madi A., Shafquat A., Abu-Ali G., Morgan X.C., et al. (2015) Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat. Rev. Microbiol. 13, 360–372 10.1038/nrmicro3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pasolli E., Asnicar F., Manara S., Zolfo M., Karcher N., Armanini F., et al. (2019) Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 10.1016/j.cell.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grice E. and Segre J. (2011) The skin microbiome. Nat. Rev. Microbiol. 9, 244–253 10.1038/nrmicro2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Byrd A.L., Belkaid Y. and Segre J.A. (2018) The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155 10.1038/nrmicro.2017.157 [DOI] [PubMed] [Google Scholar]
- 98.Nodake Y., Matsumoto S., Miura R., Honda H., Ishibashi G., Matsumoto S., et al. (2015) Pilot study on novel skin care method by augmentation with Staphylococcus epidermidis, an autologous skin microbe–a blinded randomized clinical trial. J. Dermatol. Sci. 79, 119–126 10.1016/j.jdermsci.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 99.Myles I.A., Williams K.W., Reckhow J.D., Jammeh M.L., Pincus N.B., Sastalla I., et al. (2016) Transplantation of human skin microbiota in models of atopic dermatitis. JCI Insight 1, e86955 10.1172/jci.insight.86955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Paetzold B., Willis J.R., de Lima J P., Knödlseder N., Brüggemann H., Quist S.R. et al. (2019) Skin microbiome modulation induced by probiotic solutions. Microbiome 7, 95 10.1186/s40168-019-0709-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Muzzi A., Masignani V. and Rappuoli R. (2007) The pan-genome: towards a knowledge-based discovery of novel targets for vaccines and antibacterials. Drug Discov. Today 12, 429–439 10.1016/j.drudis.2007.04.008 [DOI] [PubMed] [Google Scholar]
- 102.Spaulding C.N., Klein R.D., Schreiber H.L., Janetka J.W. and Hultgren S.J. (2018) Precision antimicrobial therapeutics: the path of least resistance? Biofilms Microbiomes 4, 4 10.1038/s41522-018-0048-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Popowska M. and Krawczyk-Balska A. (2013) Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 4, 44 Published 2013 Mar 7 10.3389/fmicb.2013.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hall R.M. (1997) Mobile gene cassettes and integrons: moving antibiotic resistance genes in gram-negative bacteria. Ciba Found. Symp. 207, 192–205 10.1002/9780470515358.ch12 [DOI] [PubMed] [Google Scholar]
- 105.Jr H.D. and Ji H.F. (2020) A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel. Med. Infect. Dis. 35, 101646 10.1016/j.tmaid.2020.101646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jr H.D., Król J.E., Cahill J.P., Ji H.F. and Ehrlich G.D. (2020) The development of a pipeline for the identification and validation of small-molecule RelA inhibitors for use as anti-biofilm drugs. Microorganisms 8, E1310 10.3390/microorganisms8091310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nguyen D., Joshi-Datar A., Lepine F., Bauerle E., Olakanmi O., Beer K., et al. (2011) Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986 10.1126/science.1211037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang Y., Isaacman D., Wadowsky R.M., Rydquist-White J., Post J.C. and Ehrlich G.D. (1995) Detection of Streptococcus pneumoniae in whole blood by PCR. J. Clin. Microbiol. 33, 546–601 10.1128/JCM.33.3.596-601.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Isaacman D.J., Zhang Y., Rydquist-White J., Wadowsky R.M., Post J.C. and Ehrlich G.D. (1995) Identification of a patient with Streptococcus pneumoniaebacteremia and meningitis by the polymerase chain reaction (PCR). Mol. Cell. Probes 9, 157–160 10.1006/mcpr.1995.0026 [DOI] [PubMed] [Google Scholar]
- 110.Post J.C., Preston R.A., Aul J.J., Larkins-Pettigrew M., Rydquist-White J., Anderson K.W. et al. (1995) Molecular analysis of bacterial pathogens in otitis media with effusion. JAMA 273, 1598–1604 10.1001/jama.1995.03520440052036 [DOI] [PubMed] [Google Scholar]
- 111.Post J.C., Aul J.J., White G.J., Wadowsky R.M., Zavoral T., Tabari R. et al. (1996) PCR-based detection of bacterial DNA after antimicrobial treatment is indicative of persistent, viable bacteria in the chinchilla model of otitis media. Am. J. Otolaryngol. 17, 106–111 10.1016/S0196-0709(96)90005-8 [DOI] [PubMed] [Google Scholar]
- 112.Rayner M.G., Zhang Y., Gorry M.C., Chen Y., Post J.C. and Ehrlich G.D. (1998) Evidence of bacterial metabolic activity in culture-negative otitis media with effusion. JAMA 279, 296–299 10.1001/jama.279.4.296 [DOI] [PubMed] [Google Scholar]
- 113.Hall-Stoodley L., Hu F.Z., Stoodley P., Nistico L., Link T.R., Burrows A. et al. (2006) Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296, 202–211 10.1001/jama.296.2.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wolcott R.D. and Ehrlich G.D. (2008) Biofilms and chronic infections. JAMA 299, 2682–2684 10.1001/jama.299.22.2682 [DOI] [PubMed] [Google Scholar]
- 115.Kathju S., Nistico L., Hall-Stoodley L., Post J.C., Ehrlich G.D. and Stoodley P. (2009) Chronic surgical site infection due to suture-associated polymicrobial biofilm. Surg. Infect. 10, 457–461 10.1089/sur.2008.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stoodley P., Braxton E., Nistico L., Hall-Stoodley L., Johnson S., Quigley M. et al. (2010) Direct demonstration of Staphylococcus biofilm in an external ventricular drain in a patient with a history of recurrent ventriculoperitoneal shunt failure. Pediatr. Neurosurg. 46, 127–132 10.1159/000319396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nistico L., Gieseke A., Kreft R., Coticchia J.M., Burrows A., Khampang P. et al. (2011) Pathogenic biofilms in adenoids: a reservoir for persistent bacteria. J. Clin. Microbiol. 49, 1411–1420 10.1128/JCM.00756-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gallo P.H., Melton-Kreft R., Nistico R., Sotereanos N.G., Sewecke J.J., Stoodley P. et al. (2011) Demonstration of Bacillus cereus in Peri-implant infection using a multi-primer PCR-mass spectrometric assay: report of two cases. J. Bone Jt. Surg. 93, e85 10.2106/JBJS.J.01181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stoodley P., Conti S.F., DeMeo P.J., Nistico L., Melton-Kreft R., Johnson S. et al. (2011) Characterization of a mixed MRSA/MRSE biofilm in an explanted total ankle arthroplasty. FEMS Immunol. Med. Microbiol. 62, 66–74 10.1111/j.1574-695X.2011.00793.x [DOI] [PubMed] [Google Scholar]
- 120.Tuttle M.S., Mostow E., Mukherjee P., Hu F.Z., Melton-Kreft R., Ehrlich G.D. et al. (2011) Characterization of bacterial communities in venous insufficiency wounds by use of conventional culture and molecular diagnostic methods. J. Clin. Microbiol. 49, 3812–3819 10.1128/JCM.00847-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jacovides C.L., Kreft R., Adeli B., Hozack B., Ehrlich G.D. and Parvizi J. (2012) Successful identification of pathogens by polymerase chain reaction (PCR)-based electron spray ionization time-of-flight mass spectrometry (ESI-TOF-MS) in culture-negative periprosthetic joint infection. J. Bone Joint Surg. Am. 94, 2247–2254 10.2106/JBJS.L.00210 [DOI] [PubMed] [Google Scholar]
- 122.Yun H.C., Kreft H.E., Castillo M.A., Ehrlich G.D., Guymon C.H., Crouch H.K. et al. (2012) Comparison of PCR/electron spray ionization - time-of-flight - mass spectrometry versus traditional clinical microbiology for detection of organisms contaminating high-use surfaces in a burn unit, an orthopaedic ward and healthcare workers. BMC Infect. Dis. 12, 252 10.1186/1471-2334-12-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ehrlich G.D., DeMeo P., Costerton, J.W. and Winkler, H. (eds) Culture-Negative Orthopedic Biofilm Infections, Springer Verlag Series on Biofilms; 144 pages, 47 illustrations. ISBN: 978-3-642-29553-9 (Print) 978-3-642-29554-6 (Online) 2012. http://dx.doi.org/10.1007/978-3-642-29554-6 [Google Scholar]
- 124.Boase S., Foreman A., Cleland E., Tan L., Melton-Kreft R., Pant H. et al. (2013) The microbiome of chronic rhinosinusitis: culture, molecular diagnostics and biofilm detection. BMC Infect. Dis. 13, 210 10.1186/1471-2334-13-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vento T.J., Mende K., Kreft R.E., Ehrlich G.D., Wenke J.C., Spirk T. et al. (2013) Detection of methicillin-resistant and methicillin-susceptible Staphylococcus aureus colonization of healthy military personnel by traditional culture, PCR, and mass spectrometry. Scand. J. Infect. Dis. 45, 752–759 10.3109/00365548.2013.816439 [DOI] [PubMed] [Google Scholar]
- 126.Melton-Kreft R., Costerton J.W. and Ehrlich G.D. (2013) PCR is changing clinical diagnostics. Microbe 8, 15–20 [Google Scholar]
- 127.Palmer M.P., Altman D., Alman G., Sewecke J., Ehrlich G.D., Hu F.Z. et al. (2014) Can we trust intraoperative culture results in nonunions? J. Orthop. Trauma 28, 384–390 10.1097/BOT.0000000000000043 [DOI] [PubMed] [Google Scholar]
- 128.Nickel J.C., Stephens A., Landis R., Mullins C., Melton-Kreft R. and Ehrlich G.D. (2015), And the MAPP research network. Search for microorganisms in men with urologic chronic pelvic pain syndrome: a culture-independent analysis in the MAPP research network. J. Urol. 194, 127–135 10.1016/j.juro.2015.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Palmer M.P., Hu F.Z., Nistico L., Melton-Kreft R., Altman H.K., Altman G.T. et al. (2016) Synovial fluid analysis using PCR-ESI-TOF-MS for detection of bacterial and fungal pathogens in native knee arthritis. Genet. Test. Mol. Biomark. 20, 721–731 10.1089/gtmb.2016.0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wolcott R.D., Gontcharova V., Sun Y., Zischakau A. and Dowd S.E. (2009) Bacterial diversity in surgical site infections: not just aerobic cocci any more. J. Wound Heal. 18, 317–323 10.12968/jowc.2009.18.8.43630 [DOI] [PubMed] [Google Scholar]
- 131.Wolcott R., Costerton J.W., Raoult D. and Cutler S.J. (2013) The polymicrobial nature of biofilm infection. Clin. Microbiol. Infect. 19, 107–112 10.1111/j.1469-0691.2012.04001.x [DOI] [PubMed] [Google Scholar]
- 132.Wolcott R.D., Hanson J.D., Rees E.J., Koenig L.D., Phillips C.D., Wolcott R.A., et al. (2016) Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 24, 163–174 10.1111/wrr.12370 [DOI] [PubMed] [Google Scholar]
- 133.Wolcott R., Sanford N., Gabrilska R., Oates J.L., Wilkinson J.E. and Rumbaugh K.P. (2016) Microbiota is a primary cause of pathogenesis of chronic wounds. J. Wound Care 25, S33–S43 10.12968/jowc.2016.25.Sup10.S33 [DOI] [PubMed] [Google Scholar]
- 134.Bianchi T., Wolcott R.D., Peghetti A., Leaper D., Cutting K., Polignano R., et al. (2016) Recommendations for the management of biofilm: a consensus document. J. Wound Care 25, 305–317 10.12968/jowc.2016.25.6.305 [DOI] [PubMed] [Google Scholar]
- 135.Malone M., Bjarnsholt T., McBain A.J., James G.A., Stoodley P., Leaper D., et al. (2017) The prevalence of biofilms in chronic wounds: a systematic review and meta-analysis of published data. Journal of Wound Care 26, 20–25 10.12968/jowc.2017.26.1.20 [DOI] [PubMed] [Google Scholar]
- 136.Schultz G., Bjarnsholt T., James G.A., Leaper D.J., McBain A.J., Malone M., et al. (2017) Consensus guidelines for the identification and treatment of biofilms in chronic nonhealing wounds. Wound Repair Regen. 25, 744–757 10.1111/wrr.12590 [DOI] [PubMed] [Google Scholar]
- 137.Snyder R.J., Bohn G., Hanft J., Harkless L., Kim P., Lavery L., et al. (2017) Wound biofilm: current perspectives and strategies on biofilm disruption and treatments. Wounds 29, S1–S17. PMID: [PubMed] [Google Scholar]
- 138.Tipton C.D., Mathew M.E., Wolcott R.A., Wolcott R.D., Kingston T. and Phillips C.D. (2017) Temporal dynamics of relative abundances and bacterial succession in chronic wound communities. Wound Repair Regen. 25, 673–679 10.1111/wrr.12555 [DOI] [PubMed] [Google Scholar]
- 139.Wolcott R.D. (2017) Biofilms cause chronic infections. J. Wound Care 26, 423–425 10.12968/jowc.2017.26.8.423 [DOI] [PubMed] [Google Scholar]
- 140.Murphy C., Atkin L., Dissemond J., Hurlow J., Tan Y.K., Apelqvist J., et al. (2019) Defying hard-to-heal wounds with an early antibiofilm intervention strategy: ‘wound hygiene’. J. Wound Care 28, 818–822 10.12968/jowc.2019.28.12.818 [DOI] [PubMed] [Google Scholar]
- 141.Yu Y., Tsitrin T., Bekele S., Tsitrin T., Bekele S., Thovarai V., et al. (2019) Aerococcus urinae and Globicatella sanguinis persist in polymicrobial urethral catheter biofilms examined in longitudinal profiles at the proteomic level. Biochem Insights. 12, 1178626419875089 10.1177/1178626419875089Published 2019 Sep 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hall H.G., Ehrlich G.D. and Hu F.Z. (2010) Pan-genome analysis provides much higher strain typing resolution than does MLST. Microbiology 156, 1060–1068 10.1099/mic.0.035188-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Deurenberg R.H., Bathoorn E., Chlebowicz M.A., Couto N., Ferdousa M., García-Cobos S. et al. (2017) Application of next generation sequencing in clinical microbiology and infection prevention. J. Biotechnol. 243, 16–24 10.1016/j.jbiotec.2016.12.022 [DOI] [PubMed] [Google Scholar]
- 144.Balloux F., Brønstad Brynildsrud O., van Dorp L., Shaw L.P., Chen H., Harris K.A. et al. (2018) From theory to practice: translating whole-genome sequencing (WGS) into the clinic. Trends Microbiol. 26, 1035–1048 10.1016/j.tim.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rossen J.W.A., Friedrich A.W. and Moran-Gilad J. (2018) ESCMID study group for genomic and molecular diagnostics (ESGMD). practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin. Microbiol. Infect. 24, 355–360 10.1016/j.cmi.2017.11.001 [DOI] [PubMed] [Google Scholar]
- 146.Doyle R.M., O'Sullivan D.M., Aller S.D., Bruchmann S., Clark T., Coello Pelegrin A. et al. (2020) Discordant bioinformatic predictions of antimicrobial resistance from whole-genome sequencing data of bacterial isolates: an inter-laboratory study. Microb. Genom. 6, e000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Earl J.P., Adappa N.D., Krol J., Bhat A.S., Balashov S., Ehrlich R.L. et al. (2018) Species-level bacterial community profiling of the healthy sinonasal microbiome using pacific biosciences sequencing of full-length 16S rRNA genes. Microbiome 6, 190 10.1186/s40168-018-0569-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Greathouse K.L., White J.R., Vargas A.J., Bliskovsky V.V., Beck J.A., von Muhlinen N. et al. (2018) Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 19, 123 10.1186/s13059-018-1501-6. Erratum in: Genome Biol. 2020;21:41. PMID: 30143034; PMCID: PMC6109311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Serruto D., Serino L., Masignani V. and Pizza M. (2009) Genome-based approaches to develop vaccines against bacterial pathogens. Vaccine 27, 3245–3250 10.1016/j.vaccine.2009.01.072 [DOI] [PubMed] [Google Scholar]
- 150.Rappuoli R. (2001) Reverse vaccinology, a genome-based approach to vaccine development. Vaccine 19, 2688–2691 10.1016/S0264-410X(00)00554-5 [DOI] [PubMed] [Google Scholar]
- 151.Rappuoli R., Bottomley M.J., D'Oro U., Finco O. and De Gregorio E. (2016) Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J. Exp. Med. 213, 469–481 10.1084/jem.20151960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pizza M., Scarlato V., Masignani V., Giuliani M.M., Aricò B., Comanducci M., et al. (2000) Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287, 1816–1820 10.1126/science.287.5459.1816 [DOI] [PubMed] [Google Scholar]
- 153.Maione D., Margarit I., Rinaudo C.D., Masignani V., Mora M., Scarselli M., et al. (2005) Identification of a universal group B streptococcus vaccine by multiple genome screen [published correction appears in science. 2013 Jan 11;339:141. Nardi Dei, Vincenzo [corrected to Nardi-Dei, Vincenzo]]. Science 309, 148–150 10.1126/science.1109869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhu T., McClure R., Harrison O.B., Genco C. and Massari P. (2019) Integrated bioinformatic analyses and immune characterization of new Neisseria gonorrhoeae vaccine antigens expressed during natural mucosal infection. Vaccines 7, 153 10.3390/vaccines7040153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vilela Rodrigues T.C., Jaiswal A.K., de Sarom A., de Castro Oliveira L., Freire Oliveira C.J., Ghosh P. et al. (2019) Reverse vaccinology and subtractive genomics reveal new therapeutic targets against Mycoplasma pneumoniae: a causative agent of pneumonia. R. Soc. Open Sci. 6, 190907 Published 2019 Jul 31 10.1098/rsos.190907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Marcy Y., Ouverney C., Bik E.M., Losekann T., Ivanova N., Martin H.G., et al. (2007) Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. U.S.A. 104, 11889–11894 10.1073/pnas.0704662104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., Cheng J.F. et al. (2103) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 10.1038/nature12352 [DOI] [PubMed] [Google Scholar]
- 158.Ehrlich G.D., Multi-informatic Approaches to Identifying OM-specific Virulence Genes from within the Bacterial Genomic Dark Matter. International Society for Otitis Media (ISOM) Biennial Meeting, Los Angeles, CA, June 9–13, 2019 [Google Scholar]
- 159.Wang Y. and Navin N.E. (2015) Advances and applications of single cell sequencing technologies. Mol. Cell 58, 598–609 10.1016/j.molcel.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gawad C K. and Quake S.R. (2016) Single-cell genome sequencing: current state of the science. Nat. Rev. 17, 175 10.1038/nrg.2015.16 [DOI] [PubMed] [Google Scholar]
- 161.Wang J. and Song Y. (2017) Single cell sequencing: a distinct new field. Clin. Trans. Med. 6, 10 10.1186/s40169-017-0139-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen Z., Chen L. and Zhang W. (2017) Tools for genomic and transcriptomic analysis of microbes at the single-cell level. Front. Microbiol. 8, article 1831 10.3389/fmicb.2017.01831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rath C.M., Janto B., Earl J., Ahmed A., Hu F.Z., Hiller L. et al. (2011) Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem. Biol. 6, 1244–1256 10.1021/cb200244t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Browne H.P., Forster S.C., Anonye B.O., Kumar N., Neville B.A., Stares M.D. et al. (2016) Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546 10.1038/nature17645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mason O.U., Hazen T.C., Borglin S., Chain P.S.G., Dubinsky E.A., Fortney J.L., et al. (2012) Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to deepwater horizon oil spill. ISME J. 6, 1715–1727 10.1038/ismej.2012.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Flood B.E., Fliss P., Jones D.S., Dick G.J., Jain S., Kaster A.K. et al. (2016) Single-cell (meta-)genomics of a dimorphic Candidatus Thiomargarita nelsonii reveals genomic plasticity. Front. Microbiol. 7, 603 10.3389/fmicb.2016.00603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Song Y., Kaster A.K., Vollmers J., Song Y., Davison P.A., Frentrup M. et al. (2017) Single-cell genomics based on Raman sorting reveals novel carotenoid-containing bacteria in the Red Sea. Microb. Biotechnol. 10, 125–137 10.1111/1751-7915.12420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.McLean J.S., Lombardo M.J., Ziegler M.G., Novotny M., Yee-Greenbaum J., Badger J.H., et al. (2013) Genome of the pathogen Porphyromonas gingivalis recovered from a biofilm in a hospital sink using a high-throughput single-cell genomics platform. Genome Res. 23, 867–877 10.1101/gr.150433.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.McLean J.S., Lombardo M.J., Badger J.H., Edlund A., Novotny M., Yee-Greenbaum J., et al. (2013b) Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc. Natl. Acad. Sci. USA 110, E2390–E2399 10.1073/pnas.1219809110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Alam M.T., Petit R.A., Crispell E.K., Thornton T.A., Conneely K.N., Jiang Y., et al. (2014) Dissecting vancomycin-intermediate resistance in Staphylococcus aureus using genome-wide association. Genome Biol. Evol. 6, 1174–1185 10.1093/gbe/evu092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chewapreecha C., Harris S., Croucher N.J., Turner C., Marttinen P., Cheng L., et al. (2014) Dense genomic sampling identifies highways of pneumococcal recombination. Nat. Genet. 46, 305–309 10.1038/ng.2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Miotto O., Amato R., Ashley E.A., MacInnis B., Almagro-Garcia J., Amaratunga C., et al. (2015) Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 47, 226–234 10.1038/ng.3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Laabei M., Recker M., Rudkin J.K., Aldeljawi M., Gulay Z., Sloan T.J., et al. (2014) Predicting the virulence of MRSA from its genome sequence. Genome Res. 24, 839–849 10.1101/gr.165415.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Espadinha D., Sobral R.G., Mendes C.I., Méric G., Sheppard S.K., Carriço J.A. et al. (2019) Distinct phenotypic and genomic signatures underlie contrasting pathogenic potential of Staphylococcus epidermidis clonal lineages. Front. Microbiol. 10, 1971 10.3389/fmicb.2019.01971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Guiton P.S., Cusumano C.K., Kline K.A., Dodson K.W., Han Z., Janetka J.W., et al. (2012) Combinatorial small-molecule therapy prevents uropathogenic Escherichia coli catheter-associated urinary tract infections in mice. Antimicrob. Agents Chemother. 56, 4738–4745 10.1128/AAC.00447-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Alvarez Dorta D., Sivignon A., Chalopin T., Dumych T.I., Roos G., Bilyy R.O., et al. (2016) The Antiadhesive strategy in Crohn's disease: orally active mannosides to decolonize pathogenic Escherichia coli from the gut. Chembiochem 17, 936–952 10.1002/cbic.201600018 [DOI] [PubMed] [Google Scholar]
- 177.Sommer R., Wagner S., Rox K., Varrot A., Hauck D., Wamhoff E.-C., et al. (2018) Glycomimetic, orally bioavailable LecB inhibitors block biofilm formation of Pseudomonas aeruginosa. J. Am. Chem. Soc. 140, 2537–2545 10.1021/jacs.7b11133 [DOI] [PubMed] [Google Scholar]
- 178.Zipperer A., Konnerth M.C., Laux C., Berscheid A., Janek D., Weidenmaier C., et al. (2016) Human commensals producing a novel antibiotic impair pathogen colonization [published correction appears in nature. 2016 Nov 10;539:314]. Nature 535, 511–516 10.1038/nature18634 [DOI] [PubMed] [Google Scholar]
- 179.Nakase K., Nakaminami H., Takenaka Y., Hayashi N., Kawashima M. and Noguchi N. (2017) Propionibacterium acnes is developing gradual increase in resistance to oral tetracyclines. J. Med. Microbiol. 66, 8–12 10.1099/jmm.0.000392 [DOI] [PubMed] [Google Scholar]