Abstract
Background
Species of the fungal genus Arthrinium (Sordariomycetes, Amphisphaeriales, Apiosporaceae) are often found on bamboo in Asia. They are endophytes, saprobes and important plant pathogens. The genus Arthrinium currently contains 92 species and is widely distributed in North and South America, Europe, Africa, Asia and Oceania.
New information
In this study, a new species, Arthrinium bambusicola sp. nov., is described and illustrated. The new taxon is characterised by oval to broadly or irregularly round, medium brown, multi-guttulate to roughened, granular conidia, with finely pale slits in the outer edges. Arthrinium bambusicola can be distinguished from the closest related species A. gutiae by its conidial characteristics. Phylogenetic analyses of a four-locus dataset (ITS, LSU, TEF1, TUB2) confirm that A. bambusicola is a distinct new species.
Keywords: one new species, Bambusicolous fungi, multi-locus phylogeny, saprobic, Sordariomycetes , taxonomy
Introduction
The genus Arthrinium, with A. caricicola as type species, was established by Schmidt and Kunze (Kunze 1817). Species of Arthrinium are endophytes, saprobes and important plant pathogens of various hosts, particularly grasses and bamboo (Agut and Calvo 2004, Li et al. 2016, Dai et al. 2017, Wang et al. 2018, Jiang et al. 2019, Rashmi et al. 2019). The sexual morph is characterised by black, linear, fusiform ascostromata with a long, slit-like opening at the apex. The ascomata are globose to subglobose, with flattened bases and brown to blackish, with or without setae (Jiang et al. 2019, Pintos et al. 2019, Yang et al. 2019).
Species of Arthrinium produce both hyphomycetous and coelomycetous asexual morphs. The hyphomycetous morph is characterised by septate conidiophores, arising from basal cells or that are reduced to conidiogenous cells. Conidiogenous cells are holoblastic, monoblastic or polyblastic and are hyaline to pale brown, smooth or finely roughened, doliiform, ampulliform or subcylindrical and conidia are dark brown, brown to pale olivaceous and of various shapes (Hyde et al. 2016). The coelomycetous morph is immersed, black, globose to subglobose, septate, hyphoid conidiomata and hyaline to pale brown conidiophores arising from basal cells or that are reduced to conidiogenous cells. The conidiogenous cells are subhyaline to pale brown, smooth-walled or verrucose, holoblastic, monoblastic or polyblastic and cylindrical. The conidia are dark brown, smooth, globose to subglobose, with or without a germ slit or truncate scar at the base (Senanayake et al. 2015, Dai et al. 2017, Jiang et al. 2018, Yang et al. 2019). The presence of both hyphomycetous and coelomycetous asexual morphs has complicated the taxonomy of Arthrinium .
There are 92 species epithets for Arthrinium in Index Fungorum (2020). A total of 63 species have been introduced, based on the combination of morphological and molecular phylogenetic data (Wijayawardene et al. 2017, Jiang et al. 2018, Wang et al. 2018, Zhao et al. 2018, Jiang et al. 2019, Pintos et al. 2019, Yan et al. 2019, Yang et al. 2019). In this study, we propose a new species, based on morphological study and comparison with other species, in combination with phylogenetic analyses of a concatenated dataset of ITS, LSU, TEF1 and TUB2 sequences.
Materials and methods
Sample collection and isolation
Fresh samples of dead culms of Schizostachyum brachycladum (Poales, Poaceae and Bambusoideae) were collected at the campus of Mae Fah Luang University, Chiang Rai, Thailand on 7 May 2019. Single-spore isolation was performed as in Chomnunti et al. (2014). The holotype is deposited at the herbarium of Mae Fah Luang University, Chiang Rai, Thailand (MFLU) and the ex-type living culture is preserved at the Mae Fah Luang University Culture Collection (MFLUCC). Facesoffungi and Index Fungorum numbers for the new taxon were obtained (Jayasiri et al. 2015, Index Fungorum 2020).
Morphological examination
Conidiomata present on the surface of the host were observed using a stereomicroscope (Motic SMZ-171, Wetzlar, Germany). Sections of conidiomata were taken and mounted in water on a microscope slide to observe fungal characters. Photographs were taken using a Nikon ECLIPSE Ni-U compound microscope connected with a Nikon camera series DS-Ri2. Morphological structures (conidiophores, conidiogenous cells, conidia) were measured by Image Frame Work software v. 0.9.7. Adobe Photoshop CC 2019 was used for editing the photographic plate. Colonies were described, based on the colour charts of Rayner (1970).
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from fresh mycelia obtained from living cultures that were grown on potato dextrose agar (PDA) for 15 days at room temperature, using the EZgene Fungal gDNA Kit (GD2416, Biomiga, San Diego, California, USA) following the manufacturer’s instructions. PCR amplification was done for the internal transcribed spacer region (ITS), the large subunit of the ribosomal RNA gene (LSU), translation elongation factor 1-alpha (TEF1) and beta-tubulin (TUB2). The following primers were used: ITS5 and ITS4 for ITS (White et al. 1990); LR0R and LR5 for LSU (Vilgalys and Hester 1990, Hopple 1994); EF1-728F and EF-2 for TEF1 (O’Donnell et al. 1998, Carbone and Kohn 1999).
PCR amplification was done in 50-μl volumes consisting of 2 μl of DNA template, 2 μl of each 10 μM forward and reverse primers, 25 μl of 2 ×Taq PCR Master Mix and 19 μl of deionised water. Cycling conditions were as follows: for ITS: initial denaturation at 94°C for 5 min, then 35 cycles of denaturation at 94°C for 45 s, annealing at 52°C for 50 s and extension at 72°C for 1 min; and final extension at 72°C for 10 min. For LSU: initial denaturation at 94°C for 5 min, then 35 cycles of denaturation at 94°C for 45 s, annealing at 52°C for 50 s and extension at 72°C for 1 min; and final extension at 72°C for 10 min. Lastly, for TEF1: initial denaturation at 94°C for 5 min; then 35 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1 min and extension at 72°C for 90 s; and final extension at 72°C for 10 min.
PCR products were checked in 1% agarose gels and sent to Sangon Biotech (Shanghai) Co. Ltd, China for sequencing, using the same primers.
Phylogenetic analyses
Raw sequence reads were combined using BioEdit v. 7.0.5.3 (Hall 1999) and subjected to BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to find closely-related taxa. To confirm the phylogenetic position of our taxon, sequences of four loci (ITS, LSU, TEF1 and TUB2) were downloaded from NCBI GenBank (Table 1). Note that no TUB2 sequence was generated for the new species, A. bambusicola. Notwithstanding, this locus was included in our phylogenetic analyses to increase phylogenetic resolution. Sequences of individual loci were aligned using MAFFT v. 7 using the 'auto' option (https://mafft.cbrc.jp/alignment/server/index.html) (Katoh et al. 2019) and, where necessary, improved in BioEdit v. 7.0.5.3 (Hall 1999). Multiple loci were combined by SequenceMatrix (Vaidya et al. 2011). The alignment was trimmed using trimAl v 1.2 with the 'gappyout' option (Capella-Gutiérrez et al. 2009). A phylogenetic tree was reconstructed from the concatenated ITS–LSU–TEF1–TUB2 dataset using Maximum Likelihood (ML), Maximum Parsimony (MP) and Bayesian Inference (BI) analyses.
Table 1.
Details of fungal taxa used in this study. Newly-generated sequences are indicated by ▲ after the species name; type materials are in bold.
| Species | Strain numbers | Substrates | Origin | ITS | LSU | TUB2 | TEF 1 |
| Arthrinium aquaticum | S-642 | Submerged wood | China | MK828608 | MK835806 | - | - |
| A. arundinis | CBS 133509 | Aspergillus flavus sclerotium buried in sandy field | USA | KF144886 | KF144930 | KF144976 | KF145018 |
| A. arundinis | CBS 449.92 | Bamboo | Canada | KF144887 | KF144931 | KF144977 | KF145019 |
| A. aureum | CBS 244.83 | - | Japan | AB220251 | KF144935 | KF144981 | KF145023 |
| A. balearicum | CBS 145129 | Undetermined Poaceae | Spain | MK014869 | MK014836 | MK017975 | MK017946 |
| A. bambusae | LC7106 | Leaves of bamboo | China | KY494718 | KY494794 | KY705186 | KY806204 |
| A. bambusae | LC7124 | Leaves of bamboo | China | KY494727 | KY494803 | KY705195 | KY806206 |
| A. bambusicola▲ | MFLUCC 20-0144 | Culms of Schizostachyum brachycladum | Thailand | MW173030 | MW173087 | - | MW183262 |
| A. camelliaesinensis | LC5007 | Camellia sinensis | China | KY494704 | KY494780 | KY705173 | KY705103 |
| A. camelliaesinensis | LC8181 | Brassica rapa | China | KY494761 | KY494837 | KY705229 | KY705157 |
| A. caricicola | CBS 145127 | Dead leaves of Carex ericetorum | China | MK014871 | MK014838 | MK017977 | MK017948 |
| A. chinense | CFCC 53036 | Fargesia qinlingensis | China | MK819291 | - | MK818547 | MK818545 |
| A. chinense | CFCC 53037 | Fargesia qinlingensis | China | MK819292 | - | MK818548 | MK818546 |
| A. chromolaenae | MFLUCC 17-1505 | Chromolaena odorata | Thailand | MT214342 | MT214436 | - | MT235802 |
| A. descalsii | CBS 145130 | Dead culms of Ampelodesmos mauritanicus | Spain | MK014870 | MK014837 | MK017976 | MK017947 |
| A. dichotomanthi | LC4950 | Dichotomanthes tristaniicarpa | China | KY494697 | KY494773 | KY705167 | KY705096 |
| A. dichotomanthi | LC8175 | Dichotomanthes tristaniicarpa | China | KY494755 | KY494831 | KY705223 | KY705151 |
| A. esporlense | CBS 145136 | Dead culms of Phyllostachys aurea | Spain | MK014878 | MK014845 | MK017983 | MK017954 |
| A. euphorbiae | IMI 285638b | Bambusa sp. | Bangladesh | AB220241 | AB220335 | AB220288 | - |
| A. gaoyouense | CFCC 52301 | Living leaves and culms of Phragmites australis | China | MH197124 | - | MH236789 | MH236793 |
| A. gaoyouense | CFCC 52302 | Living leaves and culms of Phragmites australis | China | MH197125 | - | MH236790 | MH236794 |
| A. garethjonesii | KUMCC 16-0202 | Dead culms of bamboo | China | KY356086 | KY356091 | - | - |
| A. guizhouense | LC5318 | Air in karst cave | China | KY494708 | KY494784 | KY705177 | KY705107 |
| A. guizhouense | LC5322 | Air in karst cave | China | KY494709 | KY494785 | KY705178 | KY705108 |
| A. gutiae | CBS 135835 | Gut of a grasshopper | India | KR011352 | MH877577 | KR011350 | KR011351 |
| A. hispanicum | IMI 326877 | Beach sand | Spain | AB220242 | AB220336 | AB220289 | - |
| A. hydei | CBS 114990 | Culms of Bambusa tuldoides | China | KF144890 | KF144936 | KF144982 | KF145024 |
| A. hydei | KUMCC 16-0204 | Dead culms of bamboo | China | KY356087 | KY356092 | - | - |
| A. hyphopodii | MFLUCC 15-0003 | Culms of Bambusa tuldoides | China | KR069110 | - | - | - |
| A. hyphopodii | KUMCC 16-0201 | Culms of bamboo | China | KY356088 | KY356093 | - | - |
| A. hysterinum | CBS 145133 | Phyllostachys aurea | Spain | MK014875 | MK014842 | MK017981 | MK017952 |
| A. hysterinum | ICPM6889 | Bamboo | New Zealand | MK014874 | MK014841 | MK017980 | MK017951 |
| A. ibericum | CBS 145137 | Dead culms of Arundo donax | Portugal | MK014879 | MK014846 | MK017984 | MK017955 |
| A. italicum | CBS 145138 | Dead culms of Arundo donax | Italy | MK014880 | MK014847 | MK017985 | MK017956 |
| A. italicum | CBS 145139 | Dead culms of Phragmites australis | Spain | MK014881 | MK014848 | MK017986 | - |
| A. japonicum | IFO30500 | - | Japan | AB220262 | AB220356 | AB220309 | |
| A. japonicum | IFO 31098 | Leaves of Carex despalata | Japan | AB220264 | AB220358 | AB220311 | - |
| A. jatrophae | AMH-9557 | Jatropha podagrica | India | JQ246355 | - | - | - |
| A. jatrophae | AMH-9556 | Jatropha podagrica | India | HE981191 | - | - | - |
| A. jiangxiense | LC4494 | Phyllostachys sp. | China | KY494690 | KY494766 | KY705160 | KY705089 |
| A. jiangxiense | LC4577 | Maesa sp. | China | KY494693 | KY494769 | KY705163 | KY705092 |
| A. kogelbergense | CBS 113332 | Dead culms of Cannomois virgata | South Africa | KF144891 | KF144937 | KF144983 | KF145025 |
| A. kogelbergense | CBS 113333 | Dead culms of Restionaceae | South Africa | KF144892 | KF144938 | KF144984 | KF145026 |
| A. locuta-pollinis | LC11688 | Bee bread | China | MF939596 | - | MF939623 | MF939618 |
| A. locuta-pollinis | LC11683 | Hive-stored pollen of Brassica campestris | China | MF939595 | - | MF939622 | MF939616 |
| A. longistromum | MFLUCC 11-0479 | Dead culms of bamboo | Thailand | KU940142 | KU863130 | - | - |
| A. longistromum | MFLUCC 11-0481 | Dead culms of bamboo | Thailand | KU940141 | KU863129 | - | - |
| A. malaysianum | CBS 102053 | Macaranga hullettii stems colonised by ants | Malaysia | KF144896 | KF144942 | KF144988 | KF145030 |
| A. marii | CBS 497.90 | Beach sands | Spain | AB220252 | KF144947 | KF144993 | KF145035 |
| A. mediterranei | IMI 326875 | Air | Spain | AB220243 | AB220337 | AB220290 | - |
| A. minus | AP25418 | Leaves of Carex sp. | China | MK014872 | MK014839 | MK017978 | MK017949 |
| A. minus | CBS 145131 | Dead leaves of Carex sp. | Germany | MK014872 | MK014839 | MK017978 | MK017949 |
| A. mytilomorphum | DAOM 214595 | Dead blades of Andropogon sp. | India | KY494685 | - | - | - |
| A. neogarethjonesii | DQD 2019a | Bamboo | China | MK070897 | MK070898 | - | - |
| A. neosubglobosa | JHB006 | Dead culms of bamboo | China | KY356089 | KY356094 | - | - |
| A. neosubglobosa | KUMCC 16-0203 | Bamboo | China | KY356090 | KY356095 | - | - |
| A. obovatum | LC4940 | Lithocarpus sp. | China | KY494696 | KY494772 | KY705166 | KY705095 |
| A. obovatum | LC8177 | Lithocarpus sp. | China | KY494757 | KY494833 | KY705225 | KY705153 |
| A. ovatum | CBS 115042 | Arundinaria hindsii | China | KF144903 | KF144950 | KF144995 | KF145037 |
| A. paraphaeospermum | MFLUCC 13-0644 | Dead culms of bamboo | Thailand | KX822128 | KX822124 | - | - |
| A. phaeospermum | CBS 114317 | Leaves of Hordeum vulgare | Iran | KF144906 | KF144953 | KF144998 | KF145040 |
| A. phaeospermum | CBS 114318 | Leaves of Hordeum vulgare | Iran | KF144907 | KF144954 | KF144999 | KF145041 |
| A. phragmitis | CPC 18900 | Culms of Phragmites australis | Italy | KF144909 | KF144956 | KF145001 | KF145043 |
| A. phyllostachium | MFLUCC 18-1101 | Dead culms of Phyllostachys heteroclada | China | MK351842 | MH368077 | MK291949 | MK340918 |
| A. piptatheri | CBS 145149 | Dead culms of Piptatherum miliaceum | Spain | MK014893 | MK014860 | - | MK017969 |
| A. pseudoparenchymaticum | LC7234 | Leaves of bamboo | China | KY494743 | KY494819 | KY705211 | KY705139 |
| A. pseudoparenchymaticum | LC8173 | Leaves of bamboo | China | KY494753 | KY494829 | KY705221 | KY705149 |
| A. pseudosinense | CPC 21546 | Leaves of bamboo | Netherlands | KF144910 | KF144957 | - | KF145044 |
| A. pseudospegazzinii | CBS 102052 | Macaranga hullettii stem colonised by ants | Malaysia | KF144911 | KF144958 | KF145002 | KF145045 |
| A. pterospermum | CBS 123185 | Leaves lesion of Machaerina sinclairii | New Zealand | KF144912 | KF144959 | KF145003 | - |
| A. pterospermum | CPC 20193 | Leaves of Lepidosperma gladiatum | Australia | KF144913 | KF144960 | KF145004 | KF145046 |
| A. puccinioides | CBS 549.86 | Leaves of Lepidosperma gladiatum | Germany | AB220253 | AB220347 | AB220300 | - |
| A. qinlingense | CFCC 52303 | Dead culms of Fargesia qinlingensis | China | MH197120 | - | MH236791 | MH236795 |
| A. qinlingense | CFCC 52304 | Dead culms of Fargesia qinlingensis | China | MH197121 | - | MH236792 | MH236796 |
| A. rasikravindrae | LC8179 | Brassica rapa | China | KY494759 | KY494835 | KY705227 | KY705155 |
| A. rasikravindrae | NFCCI 2144 | Soil | Norway | JF326454 | - | - | - |
| A. sacchari | CBS 372.67 | Air | - | KF144918 | KF144964 | KF145007 | KF145049 |
| A. sacchari | CBS 664.74 | Soil under Calluna vulgaris | Netherlands | KF144919 | KF144965 | KF145008 | KF145050 |
| A. saccharicola | CBS 191.73 | Air | Netherlands | KF144920 | KF144966 | KF145009 | KF145051 |
| A. saccharicola | CBS 831.71 | - | Netherlands | KF144922 | KF144969 | KF145012 | KF145054 |
| A. serenense | IMI 326869 | Food, pharmaceutical excipients, atmosphere and home dust | Spain | AB220250 | AB220344 | AB220297 | - |
| A. setostromum | KUMCC 19-0217 | Dead branches of bamboo | China | MN528012 | MN528011 | - | MN527357 |
| A. sporophleum | CBS 145154 | Dead leaves of Juncus sp. | Spain | MK014898 | MK014865 | MK018001 | MK017973 |
| A. subglobosum | MFLUCC 11-0397 | Dead culms of bamboo | Thailand | KR069112 | KR069113 | - | - |
| A. subroseum | LC7291 | Leaves of bamboo | China | KY494751 | KY494827 | KY705219 | KY705147 |
| A. subroseum | LC7292 | Leaves of bamboo | China | KY494752 | KY494828 | KY705220 | KY705148 |
| A. thailandicum | MFLUCC 15-0199 | Dead culms of bamboo | Thailand | KU940146 | KU863134 | - | - |
| A. thailandicum | MFLUCC 15-0202 | Dead culms of bamboo | Thailand | KU940145 | KU863133 | - | - |
| A. trachycarpum | CFCC 53038 | Dead branches of Trachycarpus fortune | China | MK301098 | - | MK303394 | MK303396 |
| A. trachycarpum | CFCC 53039 | Dead branches of Trachycarpus fortune | China | MK301099 | - | MK303395 | MK303397 |
| A. urticae | IMI 326344 | - | - | AB220245 | AB220339 | AB220292 | - |
| A. vietnamense | IMI 99670 | Citrus sinensis | Vietnam | KX986096 | KX986111 | KY019466 | - |
| A. xenocordella | CBS 478.86 | Soil from roadway | Zimbabwe | KF144925 | KF144970 | KF145013 | KF145055 |
| A. xenocordella | CBS 595.66 | Soil | Austria | KF144926 | KF144971 | - | - |
| A. yunnanum | DDQ00281 | Dead culms of Phyllostachys nigra | China | KU940148 | KU863136 | - | - |
| A. yunnanum | MFLUCC 15-1002 | Dead culms of Phyllostachys nigra | China | KU940147 | KU863135 | - | - |
| Seiridium phylicae | CPC 19962 | Phylica arborea | UK | LT853092 | KC005807 | LT853239 | LT853189 |
| Seiridium phylicae | CPC 19965 | Phylica arborea | UK | LT853093 | KC005809 | LT853240 | LT853190 |
Notes: Newly-generated sequences are indicated by ▲ after the species name. Ex-type strains are in bold. Abbreviations: AMH: Ajrekar Mycological Herbarium, Pune, Maharashtra, India; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands; CFCC: China Forestry Culture Collection Center, Beijing, China; CPC: Culture collection of Pedro Crous, housed at the Westerdijk Fungal Biodiversity Institute; DAOM: Canadian Collection of Fungal Cultures, Ottawa, Canada; DDQ: D.Q. Dai; ICMP: International Collection of Microorganisms from Plants, New Zealand; IFO: Institute for Fermentation, Osaka, Japan; IMI: Culture collection of CABI Europe UK Centre, Egham, UK; JHB: H.B. Jiang; KUMCC: Culture collection of Kunming Institute of Botany, Yunnan, China; LC: personal culture collection of Lei Cai, housed in the Institute of Microbiology, Chinese Academy of Sciences, China; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; NFCCI: National Fungal Culture Collection of India.
Phylogenetic analyses were performed using the CIPRES Science Gateway web portal (Miller et al. 2010). ML was done using the RAxML-HPC on XSEDE tool under the GTRGAMMA+I-Invar substitution model (Stamatakis et al. 2008). MP analysis was performed using the PAUP on XSEDE tool (Swofford 2002). A heuristic search with 1000 random taxa additions was used to infer MP trees. The value of MaxTrees was set to 5000, with branches of zero length collapsed and all multiple parsimonious trees saved. Parsimony score values for tree length (TL), consistency index (CI), retention index (RI) and homoplasy index (HI) were calculated for trees generated under different optimum criteria. Robustness of branches was estimated by maximum parsimony bootstrap proportions, using 100 bootstrap replicates, with tree bisection-reconnection branch swapping and a re-arrangement limit of 1000.
BI analysis was performed using the MrBayes on XSEDE tool available on the CIPRES Science Gateway (Huelsenbeck and Ronquist 2001, Miller et al. 2010, Ronquist et al. 2012). The best-fit model for each locus was selected by MrModeltest version 2.3, under the Akaike Information Criterion. Four Markov Chain Monte Carlo (MCMC) chains were run, starting from a random tree topology. The operation was stopped automatically when the average standard deviation of split frequencies fell below 0.01. Markov chains were set to run 10,000,000 generations with sampling every 1000 generations. A burn-in set at 25% was discarded. The Maximum Clade Credibility tree was inferred with the highest product of separate clade posterior probabilities (PP). Trees were visualised in FigTree version 1.4.0 and edited with Adobe Illustrator v. 51.1052.0.0 (Adobe Inc., San Jose, California, USA).
Taxon treatments
Arthrinium bambusicola
X. Tang, K.D. Hyde & J.C. Kang, 2020 sp. nov.
FB1F54D9-CA01-58E5-BECB-BC1495B4EE76
Materials
Type status: Holotype. Occurrence: recordedBy: Xia Tang; Taxon: scientificName: Arthrinium bambusicola; kingdom: Fungi; phylum: Ascomycota; class: Sordariomycetes; order: Amphisphaeriales; family: Apiosporaceae; genus: Arthrinium; Location: country: Thailand; countryCode: TH; stateProvince: Chiang Rai; locality: Mae Fah Luang University; Identification: identifiedBy: Xia Tang; dateIdentified: 2019; Event: year: 2019; month: May; day: 7; habitat: terrestrial; fieldNotes: on dead culms of Schizostachyum brachycladum; Record Level: type: Holotype; collectionID: MFLU 20-0528; collectionCode: M19050706
Description
Saprobic on dead culms of Schizostachyum brachycladum (Poales, Poaceae, Bambusoideae). Sexual morph: Undetermined. Asexual morph: Colonies on natural substrate, superficial, gregarious, scattered, irregular, dark brown to black (Fig. 1a-b). Mycelium consisting of branched, septate, hyaline to dark brown (Fig. 1c-e). Conidiophores 0.8–3.5 μm diam. semi-micronematous to macronematous, mononematous, solitary, branched, flexuous, smooth, hyaline, aseptate when immature, becoming brown, septate when mature (Fig. 1d). Conidiogenous cells 1.5–4.5 × 1–4 μm, monoblastic or polyblastic, terminal, determinate, cylindrical, hyaline to light brown, smooth, aggregated, ampulliform, in clusters on aerial mycelium (Fig. 1e-g). Conidia pleurogenous, solitary, oval to broadly round or irregularly round, brown to medium brown, guttulate to roughened, granular, in surface view 6–8 × 6–7.8 μm (x̅ = 6.5 × 7 μm, n = 39), in lateral view 3.5–6 × 3.5–6.5 μm (x̅ = 4.5 × 5 μm, n = 39), with finely pale slit at outer edge (Fig. 1h-l).
Figure 1.
Arthrinium bambusicola (MFLU 20-0528, holotype). a, b. Appearance of the fungus on dead culms of Schizostachyum brachycladum; c. Conidia with mycelia; d. Mycelia; e–f. Mycelia bearing conidiogenous cells and conidia; h–l. Conidia; m. Germinated conidium; n. forward culture; o. reversed culture. Scale bars: b = 500 μm, c–e = 20 μm, f, g = 10 μm, h–m = 5 μm.
Culture characteristics
colonies flat, spreading, with moderate, pale, aerial mycelium. On PDA, surface white, lightly yellow with patches of dirty white, reverse lightly pigmented.
Facesoffungi number
FoF 09162
Etymology
Referring to the host from which the holotype was isolated, a member of the bamboo subfamily (Bambusoideae).
Notes
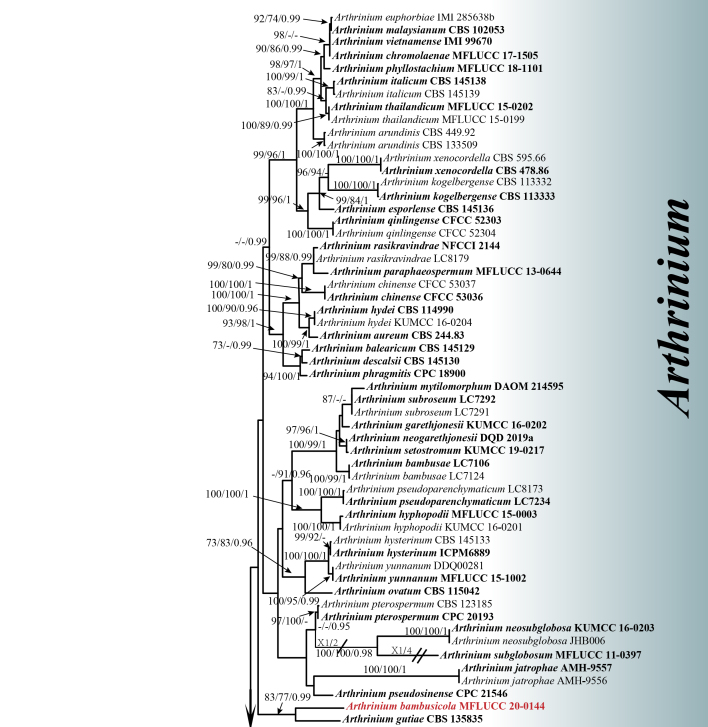
Arthrinium bambusicola forms were retrieved as a sister taxon of A. gutiae, with relatively good support (83 ML, 77 MP, 0.99 PP). Morphologically, A. bambusicola differs from A. gutiae in having larger conidia [surface view: 5.5–8 × 6–8 μm diam., lateral view: 3.5–6 × 3.5–6.5 μm diam. versus surface view: 4.5–6 μm (x̅ = 5.5 μm) diam., lateral view: 2–6 μm (x̅ = 4) diam.] and irregularly rounded, guttulate to roughened conidia (A. gutiae: smooth-walled, globose conidia). The conidiogenous cells of A. bambusicola are smaller (1.5–4.5 × 1–4 μm versus 3–7× 2–4 μm). Based on pairwise nucleotide comparisons, A. bambusicola is different from A. gutiae in 31/ 620 bp (5%) of the ITS, 7/814 (0.98%) of the LSU and 44/342 bp (12%) of TEF1. Based on the combination of morphological characters and sequence data, we consider A. bambusicola as a distinct species.
Analysis
Phylogenetic analyses
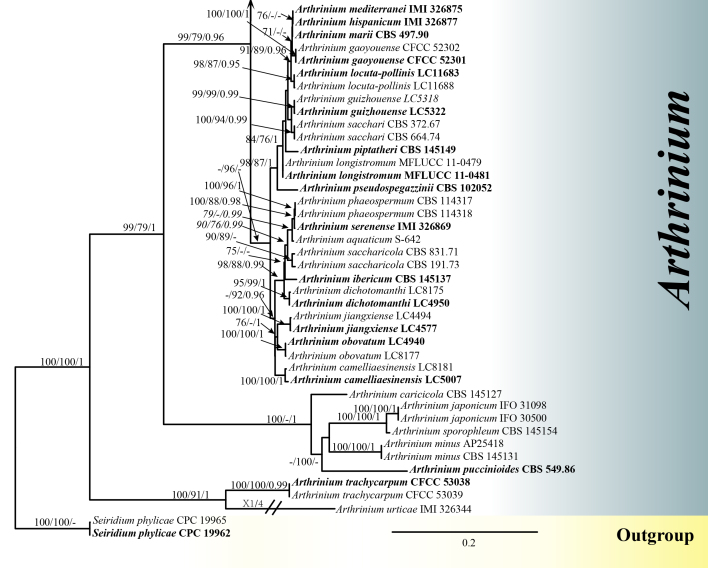
The concatenated ITS–LSU–TEF1–TUB2 dataset consisted of 98 taxa with Seiridium phylicae (Sporocadaceae), isolates CPC 19962 and CPC 19965, as the outgroup. The data matrix consisted of 2414 total characters including gaps, of which 1126 were parsimony-informative (LSU: 1–812 bp, ITS: 813–1264 bp, TEF1: 1265–1688 bp, TUB2: 1689–2414 bp). MP (Suppl. material 3), ML (Suppl. material 4) and BI (Suppl. material 2) analyses of the concatenated dataset resulted in largely similar tree topologies. The best-scoring RaxML tree (-lnL = 25256.155227) is presented in Figs 2, 3. The most parsimonious tree showed the following values: TL = 4653, CI = 0.490, RI = 0.808, RC = 0.396 and HI = 0.510. For the Bayesian posterior probabilities analysis, the best-fit models were selected as GTR+I+G for ITS and LSU and HKY+I+G for TEF1 and TUB2; 2,895,000 generations were run. A total of 2172 trees were maintained after discarding 25% as burn-in. Bayesian PP were evaluated with a final average standard deviation of split frequencies of 0.009965.
Figure 2.
Figs 2, 3 The best-scoring RAxML tree reconstructed from a concatenated ITS–LSU–TEF1–TUB2 dataset. The tree is rooted with Seiridium phylicae (strains CPC 19962 and CPC 19965). ML and MP bootstrap values ≥ 70 and Bayesian PP ≥ 0.95 are shown at the nodes (ML/MP/PP). Ex-type strains are in bold; the newly-described species is highlighted in red.
Figure 3.
Figure 2 Continued.
Discussion
The family Apiosporaceae was introduced by Hyde et al. (1998) to accommodate Apiospora and Appendicospora, based on their unique morphology. Arthrinium is one of the asexual morphs of Apiospora, along with Cordella and Pteroconium (Hyde et al. 1998). Based on molecular evidence, Crous and Groenewald (2013) confirmed that the genus Arthrinium belongs to Apiosporaceae (Hyde et al. 2020b, Wijayawardene et al. 2020). Apiospora was shown to be synonymous with Arthrinium, which is the oldest name (Hawksworth et al. 2011, Crous and Groenewald 2013).
Arthrinium species have a highly-variable morphology (Crous and Groenewald 2013, Dai et al. 2017). They produce hyphomycetous fungal structures in culture or coelomycetous fruiting bodies on their host, depending on the substrate and period of incubation (Crous and Groenewald 2013, Dai et al. 2017). However, as more species of Arthrinium are discovered, identification, based on morphology alone, has become very difficult because some species exhibit similar micro-morphological characters (Jiang et al. 2019).
ITS sequence data provide limited resolution to distinguish species for some Arthrinium species, for example, in the case of A. phyllostachium and A. vietnamensis. The ITS sequences of these species are > 99% similar. However, both species can be distinguished using the secondary barcodes TEF1 and TUB2 (Wang et al. 2018, Yang et al. 2019). In our phylogenetic analyses, A. neogarethjonesii (Hyde et al. 2020b) and A. setostromum (Jiang et al. 2019) cluster together with strong support (97 ML/96 MP/1 PP). Again, the ITS sequences of these species are > 99% similar. However, some morphological characters can be used to separate these two taxa. Whereas A. neogarethjonesii lacks setae, A. setostromum bears setae on the surface of the stromata. The former has larger stromata [1000–2000 μm × 175–250 μm versus 250–600 μm × 140–180 μm] and asci [95–125 μm × 20–25 μm versus 82.5–102.5 μm × 20–30 μm], smaller ascomata [120–230 μm × 125–230 μm versus 210–260 μm × 100–170 μm] and conidiogenous cells [10–48 μm × 4–5.5 μm versus 42–66 μm × 1.5–2.7 μm]. In their asexual morphs, A. setostromum has micronematous, holoblastic and monoblastic conidiogenous cells, but A. neogarethjonesii has basauxic conidiogenous cells. For A. neogarethjonesii, only ITS and LSU sequences are available; for A. setostromum, also a TEF1 sequence is available. Fresh collections of A. neogarethjonesii are necessary to generate sequences of the protein-coding genes for improved species delimitation (Hyde et al. 2020c).
Arthrinium species have been reported from soil debris, plants, lichens, marine algae and hive-stored pollen (Senanayake et al. 2015, Wijayawardene et al. 2017, Zhao et al. 2018), in the gut of insects (Crous et al. 2015), in nodules of human skin (Sharma et al. 2014) and especially associated with bamboo. To date, 24 Arthrinium species have been found in association with the bamboo subfamily Bambusoideae (Dai et al. 2016, Dai et al. 2017, Jiang et al. 2018, Wang et al. 2018, Jiang et al. 2019, Yang et al. 2019, Jiang et al. 2020, https://nt.ars-grin.gov/fungaldatabases/). Arthrinium species have been reported from all continents, except Antarctica (Ellis 1963, Dyko and Sutton 1979, Calvo and Guarro 1980, Von Arx 1981, Crous and Groenewald 2013, Sharma et al. 2014, Wang et al. 2018, Jiang et al. 2020).
To date, seven species of Arthrinium have been reported from Thailand. These are A. bambusicola (this study), A. chromolaenae (Mapook et al. 2020), A. longistromum (Dai et al. 2017), A. paraphaeospermum (Hyde et al. 2016), A. rasikravindrii (Dai et al. 2017), A. subglobosum (Senanayake et al. 2015) and A. thailandicum (Dai et al. 2017). Contrasting morphological features amongst these species are presented in Suppl. material 1. Six of the seven Thai species are found in association with bamboo: Arthrinium bambusicola, A. longistromum, A. paraphaeospermum, A. rasikravindrii , A. subglobosum and A. thailandicum (Senanayake et al. 2015, Hyde et al. 2016, Dai et al. 2017). Only Arthrinium chromolaenae was reported from a non-bamboo host, Chromolaena odorata (Asterales, Asteraceae) (Mapook et al. 2020). Further studies on this genus in Thailand and other countries, as well as from different hosts, are likely to result in the discovery of more new species (Hyde et al. 2018, Hyde et al. 2020a).
Supplementary Material
Morphological comparison of the seven Arthrinium species introduced from Thailand
Xia Tang, Ishani D. Goonasekara, Ruvishika S. Jayawardena, Hong B. Jiang, Jun F. Li, Kevin D. Hyde, Ji C. Kang
Data type
Morphological comparison
Brief description
The morphological comparison of seven Arthrinium species introduced from Thailand.
File: oo_453721.txt
BI output file
Xia Tang
Data type
Phylogenetic output file for BI
Brief description
The output file of BI
File: oo_478675.txt
MP output file
Xia Tang
Data type
The phylogenetic output file of MP
File: oo_478682.txt
The output file of ML
Xia Tang
Data type
The phylogenetic output file of ML
File: oo_478686.txt
Acknowledgements
This work was funded by grants of the National Natural Science Foundation of China (NSFC Grants nos. 31670027 & 31460011). The authors are grateful to the Thailand Research Fund grant “Impact of climate change on fungal diversity and biogeography in the Greater Mekong Sub-region” (RDG6130001). Shaun Pennycook is thanked for his suggestions on naming the new fungus. The authors would also like to thank Mae Fah Luang University.
References
- Agut M, Calvo María Ángeles. In vitro conidial germination in Arthrinium aureum and Arthrinium phaeospermum. Mycopathologia. 2004;157(4):363–367. doi: 10.1023/B:MYCO.0000030432.08860.f3. [DOI] [PubMed] [Google Scholar]
- Calvo A, Guarro J. Arthrinium aureum sp. nov. from Spain. Transactions of the British Mycological Society. 1980;75(1) [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez J M, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn L M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91(3):553–556. doi: 10.2307/3761358. [DOI] [Google Scholar]
- Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MundefinedK, Alias AundefinedS, Xu JundefinedC, Liu XundefinedZ, Stadler M, Hyde KundefinedD. The sooty moulds. Fungal Diversity. 2014;66:1–36. doi: 10.1007/s13225-014-0278-5. [DOI] [Google Scholar]
- Crous P W, Groenewald J Z. A phylogenetic re-evaluation of Arthrinium. https://www.ncbi.nlm.nih.gov/pubmed/23898419. IMA Fungus. 2013;4(1):133–154. doi: 10.5598/imafungus.2013.04.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P W, Wingfield M J, Le Roux J J, Richardson D M, Strasberg D, Shivas R G, Alvarado P, Edwards J, Moreno G, Sharma R, Sonawane M S, Tan Y P, Altés A, Barasubiye T, Barnes C W, Blanchette R A, Boertmann D, Bogo A, Carlavilla J R, Cheewangkoon R, Daniel R, de Beer Z W, de Jesús Yáñez-Morales M, Duong T A, Fernández-Vicente J, Geering A D W, Guest D I, Held B W, Heykoop M, Hubka V, Ismail A M, Kajale S C, Khemmuk W, Kolařík M, Kurli R, Lebeuf R, Lévesque C A, Lombard L, Magista D, Manjón J L, Marincowitz S, Mohedano J M, Nováková A, Oberlies N H, Otto E C, Paguigan N D, Pascoe I G, Pérez-Butrón J L, Perrone G, Rahi P, Raja H A, Rintoul T, Sanhueza R M V, Scarlett K, Shouche Y S, Shuttleworth L A, Taylor P W J, Thorn R G, Vawdrey L L, Solano-Vidal R, Voitk A, Wong P T W, Wood A R, Zamora J C, Groenewald J Z. Fungal Planet description sheets: 371-399. https://www.ncbi.nlm.nih.gov/pubmed/26823636. Persoonia. 2015;35:264–327. doi: 10.3767/003158515X690269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D Q, Jiang H B, Tang L Z, Bhat D J. Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere. 2016;7(9):1332–1345. doi: 10.5943/mycosphere/7/9/7. [DOI] [Google Scholar]
- Dai D Q, Phookamsak R, Wijayawardene N N, Li W J, Bhat D J, Xu J C, Taylor J E, Hyde K D, Chukeatirote E. Bambusicolous fungi. Fungal Diversity. 2017;82:1–105. doi: 10.1007/s13225-016-0367-8. [DOI] [Google Scholar]
- Dyko B J, Sutton B C. New and interesting dematiaceous Hyphomycetes from Florida. Mycotaxon. 1979;8(1):119–124. [Google Scholar]
- Ellis M B. Dematiaceous Hyphomycetes. IV. Mycological Papers. 1963;87 [Google Scholar]
- Hall T A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; Nucleic acids symposium series; [London]: Information Retrieval Ltd., c1979-c2000.; 1999. 3 [Google Scholar]
- Hawksworth D L, Crous P W, Redhead S A, Reynolds D R, Samson R A, Seifert K A, Taylor J W, Wingfield M J, Abaci Özlem, Aime C, Asan A, Bai F Y, Wilhelm de Beer Z, Begerow D, Berikten D, Boekhout T, Buchanan P K, Burgess T, Buzina W, Cai L, Cannon P F, Leland Crane J, Damm U, Daniel H M, van Diepeningen A D, Druzhinina I, Dyer P S, Eberhardt U, Fell J W, Frisvad J C, Geiser D M, Geml J, Glienke C, Gräfenhan T, Groenewald J Z, Groenewald M, de Gruyter J, Guého-Kellermann E, Guo L D, Hibbett D S, Hong S B, Sybren de Hoog G, Houbraken J, Huhndorf S M, Hyde K D, Ismail A, Johnston P R, Kadaifciler D G, Kirk P M, Kõljalg U, Kurtzman C P, Lagneau P E, André Lévesque C, Liu X Z, Lombard L, Meyer W, Miller A, Minter D W, Najafzadeh M J, Norvell L, Ozerskaya S M, Öziç R, Pennycook S R, Peterson S W, Pettersson O V, Quaedvlieg W, Robert V A, Ruibal C, Schnürer J, Schroers H J, Shivas R, Slippers B, Spierenburg H, Takashima M, Taşkoin E, Thines M, Thrane U, Uztan AundefinedH, van Raak M, Varga J, Vasco A, Verkley G, Videira SundefinedIundefinedR, RundefinedP de Vries, Weir BundefinedS, Yilmaz N, Yurkov A, Zhang N. The Amsterdam declaration on fungal nomenclature. IMA Fungus. 2011;2(1):105–111. doi: 10.5598/imafungus.2011.02.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopple J S. Phylogenetic investigations in the genus Coprinus based on morphological and molecular characters. Duke University; Durham, NC: 1994. [Google Scholar]
- Huelsenbeck J P, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hyde K D, Fröhlich J, Taylor J E. Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia. 1998;50(1):21–80. [Google Scholar]
- Hyde K D, Hongsanan S, Jeewon R, Bhat D J, McKenzie E H C, Jones E B G, Phookamsak R, Ariyawansa H A, Boonmee S, Zhao Q, Abdel-Aziz F A, Abdel-Wahab M A, Banmai S, Chomnunti P, Cui B K, Daranagama D A, Das K, Dayarathne M C, de Silva N I, Dissanayake A J, Doilom M, Ekanayaka A H, Gibertoni T B, Góes-Neto A, Huang S K, Jayasiri S C, Jayawardena R S, Konta S, Lee H B, Li W J, Lin C G, Liu J K, Lu Y Z, Luo Z L, Manawasinghe I S, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera R H, Phukhamsakda C, Richter C, C M de A. Santiago, A L, Drechsler-Santos E R, Senanayake I C, Tanaka K, Tennakoon T M D S, Thambugala K M, Tian Q, Tibpromma S, Thongbai B, Vizzini A, Wanasinghe D N, Wijayawardene N N, Wu H X, Yang J, Zeng X Y, Zhang H, Zhang J F, Bulgakov T S, Camporesi E, Bahkali A H, Amoozegar M A, Araujo-Neta L S, Ammirati J F, Baghela A, Bhatt R. P, Bojantchev D, Buyck B, da Silva G A, de Lima C L F, de Oliveira R J V, de Souza C A F, Dai Y C, Dima B, Duong T T, Ercole E, Mafalda-Freire F, Ghosh A, Hashimoto A, Kamolhan S, Kang J C, Karunarathna S C, Kirk P M, Kytövuori I, Lantieri A, Liimatainen K, Liu Z Y, Liu X Z, Lücking R, Medardi G, Mortimer P E, Nguyen T T T, Promputtha I, Raj K N A, Reck M A, Lumyong S, Shahzadeh-Fazeli S A, Stadler M, Soudi M R, Su H Y, Takahashi T, Tangthirasunun N, Uniyal P, Wang Y, Wen T C, Xu J C, Zhang Z K, Zhao Y C, Zhou J L, Zhu L. Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity. 2016;80(1):1–270. doi: 10.1007/s13225-016-0373-x. [DOI] [Google Scholar]
- Hyde K D, Norphanphoun C, Chen J, Dissanayake A J, Doilom M, Hongsanan S, Jayawardena R S, Jeewon R, Perera R H, Thongbai B, Wanasinghe D N, Wisitrassameewong K, Tibpromma S, Stadler M. Thailand’s amazing diversity: up to 96% of fungi in northern Thailand may be novel. Fungal Diversity. 2018;93:215–239. doi: 10.1007/s13225-018-0415-7. [DOI] [Google Scholar]
- Hyde K D, Jeewon R, Chen Y J, Bhunjun C S, Calabon M S, Jiang H B, G Lin C, Norphanphoun C, Sysouphanthong P, Pem D, Tibpromma S, Zhang Q, Doilom M, Jayawardena R S, Liu J K, Maharachchikumbura S S N, Phukhamsakda C, Phookamsak R, Abdullah Al-Sadi A M, Thongklang N, Wang Y, Gafforov Y, Jones E B G, Lumyong S. The numbers of fungi: is the descriptive curve flattening? Fungal Diversity. 2020:1–53. doi: 10.1007/s13225-020-00458-2. [DOI]
- Hyde K D, Norphanphoun C, Maharachchikumbura S S N, Bhat D J, Jones E B G, Bundhun D, Chen Y J, Bao D F, Boonmee S, Calabon M S, Chaiwan N, Chethana K W T, Dai D Q, Dayarathne M C, Devadatha B, Dissanayake A J, Dissanayake L S, Doilom M, Dong W, Fan X L, Goonasekara I D, Hongsanan S, Huang S K, Jayawardena R S, Jeewon R, Karunarathna A, Konta S, Kumar V, Lin C G, Liu J K, Liu N G, Luangsa-ard J, Lumyong S, Luo Z L, Marasinghe D S, McKenzie E H C, Niego A G T, Niranjan M, Perera R H, Phukhamsakda C, Rathnayaka A R, Samarakoon M C, Samarakoon S M B C, Sarma V V, Senanayake I C, Shang Q J, Stadler M, Tibpromma S, Wanasinghe D N, Wei D P, Wijayawardene N N, Xiao Y P, Yang J, Zeng X Y, Zhang S N, Xiang M M. Refined families of Sordariomycetes. Mycosphere. 2020;11(1):305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- Hyde K D, Dong Y, Phookamsak R, Jeewon R, Bhat D J, Jones E B G, Liu N G, Abeywickrama P D, Mapook A, Wei D, Perera R H, Manawasinghe I S, Pem D, Bundhun D, Karunarathna A, Ekanayaka A H, Bao D F, Li J F, Samarakoon M C, Chaiwan N, Lin C G, Phutthacharoen K, Zhang S N, Senanayake I C, Goonasekara I D, Thambugala K M, Phukhamsakda C, Tennakoon D S, Jiang H B, Yang J, Zeng M, Huanraluek N, Liu J K, Wijesinghe S N, Tian Q, Tibpromma S, Brahmanage R S, Boonmee S, Huang S K, Thiyagaraja V, Lu Y Z, Jayawardena R S, Dong W, Yang E F, Singh S K, Singh S M, Rana S, Lad S S, Anand G, Devadatha B, Niranjan M, Sarma V V, Liimatainen K, Aguirre-Hudson B, Niskanen T, Overall A, Alvarenga R L M, Gibertoni T B, Pfliegler W P, Horváth E, Imre A, Alves A L, da Silva Santos A C, Tiago P V, Bulgakov T S, Wanasinghe D N, Bahkali A H, Doilom M, Elgorban A M, Maharachchikumbura S S N, Rajeshkumar K C, Haelewaters D, Mortimer P E, Zhao Q, Lumyong S, Xu J C, Sheng J. Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity. 2020;100:5–277. doi: 10.1007/s13225-020-00439-5. [DOI] [Google Scholar]
- Fungorum Index. https://www.indexfungorum.org/Names/Names.asp. [2020-11-23T00:00:00+02:00];
- Jayasiri S C, Hyde K D, Ariyawansa H A, Bhat J, Buyck B, Cai L, Dai Y C, Abd-Elsalam K A, Ertz D, Hidayat I, Jeewon R, Jones E B G, Bahkali A H, Karunarathna S C, Liu J K, Luangsa-ard J J, Lumbsch H T, Maharachchikumbura S S N, McKenzie E H C, Moncalvo J M, Ghobad-Nejhad M, Nilsson H, Pang K L, Pereira O L, Phillips A J L, Raspé O, Rollins A W, Romero A I, Etayo J, Selçuk F, Stephenson S L, Suetrong S, Taylor J E, Tsui C K M, Vizzini A, Abdel-Wahab M A, Wen T C, Boonmee S, Dai D Q, Daranagama D A, Dissanayake A J, Ekanayaka A H, Fryar S C, Hongsanan S, Jayawardena R S, Li W J, Perera R H, Phookamsak R, de Silva N I, Thambugala K M, Tian Q, Wijayawardene N N, Zhao R L, Zhao Q, Kang J C, Promputtha I. The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Diversity. 2015;74(1):3–18. doi: 10.1007/s13225-015-0351-8. [DOI] [Google Scholar]
- Jiang H B, Hyde K D, Doilom M, Karunarathna S C, Xu J C, Phookamsak R. Arthrinium setostromum (Apiosporaceae, Xylariales), a novel species associated with dead bamboo from Yunnan, China. AJOM. 2019;2(1):254–268. doi: 10.5943/ajom/2/1/16. [DOI] [Google Scholar]
- Jiang N, Li J, Tian C M. Arthrinium species associated with bamboo and reed plants in China. https://www.ncbi.nlm.nih.gov/pubmed/32467884. Fungal Syst Evol. 2018;2:1–9. doi: 10.3114/fuse.2018.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Liang Y M, Tian C M. A novel bambusicolous fungus from China, Arthrinium chinense (Xylariales) Sydowia. 2020;72:77–83. doi: 10.12905/0380.sydowia72-2020-0077. [DOI] [Google Scholar]
- Katoh K, Rozewicki J, Yamada K D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in bioinformatics. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G. Zehn neue Pilzgattungen. Mykologische Hefte. 1817;1:1–18. [Google Scholar]
- Li B J, Liu P Q, Jiang Y, Weng Q Y, Chen Q H. First report of culm rot caused by Arthrinium phaeospermum on Phyllostachys viridis in China. Plant Disease. 2016;100(5) doi: 10.1094/PDIS-08-15-0901-PDN. [DOI] [Google Scholar]
- Mapook A, Hyde K D, McKenzie E H C, Jones E B G, Bhat D J, Jeewon R, Stadler M, Samarakoon M C, Malaithong M, Tanunchai B, Buscot F, Wubet T, Purahong W. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed) Fungal Diversity. 2020;101:1–175. doi: 10.1007/s13225-020-00444-8. [DOI] [Google Scholar]
- Miller M A, Pfeiffer W, Schwartz Te. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; 2010 gateway computing environments workshop (GCE); Ieee; 2010. 7. [DOI] [Google Scholar]
- O’Donnell K, Kistler H C, Cigelnik E, Ploetz R C. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences. 1998;95(5):2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintos Á, Alvarado P, Planas J, Jarling R. Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. hosts. https://www.ncbi.nlm.nih.gov/pubmed/30918449. MycoKeys. 2019;49:15–48. doi: 10.3897/mycokeys.49.32115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashmi M, Kushveer J S, Sarma V V. A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere. 2019;10(1):798–1079. doi: 10.5943/mycosphere/10/1/19. [DOI] [Google Scholar]
- Rayner R W. A mycological colour chart. Commonwealth Mycological Institute, Kew, Surrey.; 1970. [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres D L, Darling A, Höhna S, Larget B, Liu L, Suchard M A, Huelsenbeck J P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and model choice across a large model space. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake I C, Maharachchikumbura S S N, Hyde K D, Bhat J D, Jones E B G, McKenzie E H C, Dai D Q, Daranagama D A, Dayarathne M C, Goonasekara I D, Konta S, Li W J, Shang Q J, Stadler M, Wijayawardene N N, Xiao Y P, Norphanphoun C, Li Q R, Liu X Z, Bahkali A H, Kang J C, Wang Y, Wen T C, Wendt L, Xu J C, Camporesi E. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes) Fungal Diversity. 2015;73(1):73–144. doi: 10.1007/s13225-015-0340-y. [DOI] [Google Scholar]
- Sharma R, Kulkarni G, Sonawane M S, Shouche Y S. A new endophytic species of Arthrinium (Apiosporaceae) from Jatropha podagrica. Mycoscience. 2014;55(2):118–123. doi: 10.1016/j.myc.2013.06.004. [DOI] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. https://www.ncbi.nlm.nih.gov/pubmed/18853362. Systematic Biology. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Swofford D L. PAUP*: Phylogenetic analysis using parsimony (and other methods), version 4.0 b10. MA: Sinauer Associates; Sunderland, UK.: 2002. [Google Scholar]
- Vaidya G, Lohman D J, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics. 2011;27(2):171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990;172(8):4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Arx J A. The genera of fungi sporulating in pure culture. 3. Cramer, Lehre, Germany; 1981. [Google Scholar]
- Wang M, Tan X M, Liu F, Cai L. Eight new Arthrinium species from China. https://www.ncbi.nlm.nih.gov/pubmed/29755262. MycoKeys. 2018;(34):1–24. doi: 10.3897/mycokeys.34.24221. [DOI] [PMC free article] [PubMed]
- White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. Vol. 18. Academic Press; 1990. 7. [DOI] [Google Scholar]
- Wijayawardene N N, Hyde K D, Rajeshkumar K C, Hawksworth D L, Madrid H, Kirk P M, Braun U, Singh R V, Crous P W, Kukwa M, Lücking R, Kurtzman C P, Yurkov A, Haelewaters D, Aptroot A, Lumbsch H T, Timda l E, Ertz D, Etayo J, Phillips A J L, Groenewald J Z, Papizadeh M, Selbmann L, Dayarathne M C, Weerakoon G, Jones E B G, Suetrong S, Tian Q, Castañeda-Ruiz R F, Bahkali AH, Pang K L, Tanaka K, Dai D Q, Sakayaroj J, Hujslová M, Lombard L, Shenoy B D, Suija A, Maharachchikumbura S S N, Thambugala K M, Wanasinghe D N, Sharma B O, Gaikwad S, Pandit G, Zucconi L, Onofri S, Egidi E, Raja H A, Kodsueb R, Cáceres M E S, Pérez-Ortega S, Fiuza P O, Monteiro J S, Vasilyeva L N, Shivas R G, Prieto M, Wedin M, Olariaga I, Lateef A A, Agrawal Y, Fazeli S A S, Amoozegar M A, Zhao G Z, Pfliegler W P, Sharma G, Oset M, Abdel-Wahab M A, Takamatsu S, Bensch K, de Silva N I, De Kese A, Karunarathna A, Boonmee S, Pfister D H, Lu Y Z, Luo Z L, Boonyuen N, Daranagama D A, Senanayake I C, Jayasiri S C, Samarakoon M C, Zeng X Y, Doilom M, Quijada L, Rampadarath S, Heredia G, Dissanayake A J, Jayawardana R S, Perera R H, Tang L Z, Phukhamsakda C, HernándezRestrepo M, Ma X Y, Tibpromma S, Gusmao L F P, Weerahewa D K, Karunarathna S C. Notes for genera: Ascomycota. Fungal Diversity. 2017;86(1):1–594. doi: 10.1007/s13225-017-0386-0. [DOI] [Google Scholar]
- Wijayawardene N N, Hyde K D, Al-Ani L K T, Tedersoo L, Haelewaters D, Rajeshkumar K C, Zhao R L, Aptroot A, Leontyev D V, Saxena R K, Tokarev Y S, Dai D Q, Letcher P M, Stephenson S L, Ertz D, Lumbsch H T, Kukwa M, Issi I V, Madrid H, Phillips A J L, Selbmann L, Pfliegler W P, Horváth E, Bensch K, Kirk P M, Kolaříková K, Raja H A, Radek R, Papp V, Dima V, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal P B, Triebel D, Gautam A K, Avasthi S, S Suetrong, Timdal E, Fryar S C, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska J, Humber RundefinedA, Kodsueb R, Sánchez-Castro I, Goto B T, Silva D K A, de Souza F A, Oehl F, da Silva G A, Silva I R, Błaszkowski J, Jobim K, Maia LundefinedC, Barbosa FundefinedR, Fiuza PundefinedO, Divakar PundefinedK, Shenoy BundefinedD, Castañeda-Ruiz R F, Somrithipol S, Lateef A A, Karunarathna S C, Tibpromma S, Mortimer P E, Wanasinghe D N, Phookamsak R, C Xu J, Wang Y, Tian F, Alvarado P, Li D W, Kušan I, Matočec N, Maharachchikumbura SundefinedSundefinedN, Papizadeh M, Heredia G, Wartchow F, Bakhshi M, Boehm E, Youssef N, Hustad VundefinedP, Lawrey JundefinedD, Santiago AundefinedLundefinedCundefinedMundefinedA, Bezerra JundefinedDundefinedP, Souza-Motta CundefinedM, Firmino AundefinedL, Tian Q, Houbraken J, Hongsanan S, Tanaka K, Dissanayake AundefinedJ, Monteiro JundefinedS, Grossart HundefinedP, Suija A, Weerakoon G, Etayo J, Tsurykau A, Vázquez V, Mungai P, Damm U, Li Q R, Zhang H, Boonmee S, Lu Y Z, Becerra A G, Kendrick B, Brearley F Q, Motiejūnaitė J, Sharma B, Khare R, Gaikwad S, Wijesundara DundefinedSundefinedA, Tang LundefinedZ, He MundefinedQ, Flakus A, Rodriguez-Flakus P, Zhurbenko MundefinedP, McKenzie EundefinedHundefinedC, Stadler M, Bhat DundefinedJ, Liu JundefinedK, Raza M, Jeewon R, Nassonova EundefinedS, Prieto M, Jayalal RundefinedGundefinedU, Erdoğdu M, Yurkov A, Schnittler M, Shchepin OundefinedN, Novozhilov YundefinedK, Silva-Filho AundefinedGundefinedS, Liu P, Cavender JundefinedC, Kang Y, Mohammad S, Zhang LundefinedF, Xu RundefinedF, Li YundefinedM, Dayarathne MundefinedC, Ekanayaka AundefinedH, Wen TundefinedC, Deng CundefinedY, Pereira OundefinedL, Navathe S, Hawksworth DundefinedL, Fan XundefinedL, Dissanayake LundefinedS, Kuhnert E, Grossart HundefinedP, Thines M. Outline of fungi and fungus-like taxa. Mycosphere. 2020;11(1):1060–1456. doi: 10.5943/mycosphere/11/1/8. [DOI] [Google Scholar]
- Yang C L, Xu X L, Dong W, Wanasinghe D N, Liu Y G, Hyde K D. Introducing Arthrinium phyllostachium sp. nov. (Apiosporaceae, Xylariales) on Phyllostachys heteroclada from Sichuan Province, China. Phytotaxa. 2019;406(2):91–110. doi: 10.11646/phytotaxa.406.2.2. [DOI] [Google Scholar]
- Yan H, Jiang N, Liang L Y, Yang Q, Tian C M. Arthrinium trachycarpum sp. nov. from Trachycarpus fortunei in China. Phytotaxa. 2019;400(3):203–210. doi: 10.11646/phytotaxa.400.3.7. [DOI] [Google Scholar]
- Zhao Y Z, Zhang Z F, Cai L, Peng W J, Liu F. Four new filamentous fungal species from newly-collected and hive-stored bee pollen. Mycosphere. 2018;9(6):1089–1116. doi: 10.5943/mycosphere/9/6/3. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphological comparison of the seven Arthrinium species introduced from Thailand
Xia Tang, Ishani D. Goonasekara, Ruvishika S. Jayawardena, Hong B. Jiang, Jun F. Li, Kevin D. Hyde, Ji C. Kang
Data type
Morphological comparison
Brief description
The morphological comparison of seven Arthrinium species introduced from Thailand.
File: oo_453721.txt
BI output file
Xia Tang
Data type
Phylogenetic output file for BI
Brief description
The output file of BI
File: oo_478675.txt
MP output file
Xia Tang
Data type
The phylogenetic output file of MP
File: oo_478682.txt
The output file of ML
Xia Tang
Data type
The phylogenetic output file of ML
File: oo_478686.txt