Abstract
Background
The chromosome 22q11.2 deletion is an extremely high risk genetic factor for various neuropsychiatric disorders; however, the 22q11.2 deletion-related brain pathology in humans at the cellular and molecular levels remains unclear.
Methods
We generated iPS cells from healthy controls (control group) and patients with 22q11.2 deletion (22DS group), and differentiated them into dopaminergic neurons. Semiquantitative proteomic analysis was performed to compare the two groups. Next, we conducted molecular, cell biological and pharmacological assays.
Findings
Semiquantitative proteomic analysis identified ‘protein processing in the endoplasmic reticulum (ER)’ as the most altered pathway in the 22DS group. In particular, we found a severe defect in protein kinase R-like endoplasmic reticulum kinase (PERK) expression and its activity in the 22DS group. The decreased PERK expression was also shown in the midbrain of a 22q11.2 deletion mouse model. The 22DS group showed characteristic phenotypes, including poor tolerance to ER stress, abnormal F-actin dynamics, and decrease in protein synthesis. Some of phenotypes were rescued by the pharmacological manipulation of PERK activity and phenocopied in PERK-deficient dopaminergic neurons. We lastly showed that DGCR14 was associated with reduction in PERK expression.
Interpretation
Our findings led us to conclude that the 22q11.2 deletion causes various vulnerabilities in dopaminergic neurons, dependent on PERK dysfunction.
Funding
This study was supported by the AMED under grant nos JP20dm0107087, JP20dm0207075, JP20ak0101113, JP20dk0307081, and JP18dm0207004h0005; the MEXT KAKENHI under grant nos. 16K19760, 19K08015, 18H04040, and 18K19511; the Uehara Memorial Foundation under grant no. 201810122; and 2019 iPS Academia Japan Grant.
Keywords: 22q11.2 deletion, Neuropsychiatric disorders, iPS cells, Dopaminergic neurons, PERK
Research in Context.
Evidence before this study
The 22q11.2 deletion syndrome (22q11.2DS) is one of the most common microdeletion syndromes in humans. The 22q11.2 deletion is an extremely strong genetic risk factor for various neuropsychiatric disorders, including intellectual disability, schizophrenia and Parkinson's disease. However, the brain pathology in patients with 22q11.2DS at the cellular and molecular levels remains unclear.
Added value of this study
Herein, we compare the dopaminergic neurons derived from healthy controls and 22q11.2DS patients. We found the PERK expression and its activity to be severely decreased in dopaminergic neurons of 22q11.2DS patients. This dysfunction causes various vulnerabilities such as low tolerance to ER stress and abnormal F-actin dynamics in dopaminergic neurons of 22q11.2DS patients.
Implications of all the available evidence
Our findings suggest that PERK is one of the key factors for understanding the 22q11.2DS-related pathology in dopaminergic neurons and may act as a candidate target for the development of therapeutic and preventive strategies for diseases encountered in the future.
Alt-text: Unlabelled box
1. Introduction
The 22q11.2 deletion syndrome (22q11.2DS), which has a frequency of 1:4000 live births, is one of the most common microdeletion syndromes in humans. This chromosomal defect is a deletion of ~3 Mb that includes more than 40 genes, most of which are expressed in the brain [1,2]. The 22q11.2 deletion is an extremely strong genetic risk factor for various neuropsychiatric disorders, including intellectual disability and schizophrenia [3], [4], [5], [6]. Moreover, the remaining characteristic phenotypes of individuals with 22q11.2DS include Parkinson's disease [7,8]. Despite these obvious relationships between this variant and neuropsychiatric phenotypes, as well as several attempts to elucidate the etiopathology using the 22q11.2DS mouse model, the 22q11.2DS-related brain pathology in humans remains to be elucidated.
Human induced pluripotent stem cell (iPSC)-derived neurons are powerful tools that can potentially recapitulate the central nervous system development, including any genetic signatures of the individual from which they were derived. Many studies have used iPSCs for modelling neurodevelopmental impairment [9,10]. Efforts to elucidate the brain cellular and molecular pathology of 22q11.2DS using iPSCs have also been made. Neuronal cells derived from patients with 22q11.2DS exhibit differentially expressed microRNAs [11] and mitochondrial deficits [12]. Therefore, iPSCs can be used to provide novel findings at the cellular and molecular levels in these patients. iPSC-derived neurons likely recapitulate the neurons in the developing brain better than in the adult brain, as mentioned above. However, previous studies have reported that iPSC-derived neurons can also exhibit the pre-onset or early phenotypes of neurodegenerative diseases, such as Parkinson's disease, Alzheimer's disease and Huntington's disease [13], [14], [15]. Hence, we hypothesised that 22q11.2DS iPSCs could be used to extensively characterise the cellular and molecular phenotypes of neurons derived from individuals with 22q11.2DS, including those related to neurodegenerative diseases, such as Parkinson's disease.
This study aimed to identify the cellular and molecular mechanisms underlying the 22q11.2DS-related brain pathology in humans using 22q11.2DS patient-derived iPSCs. To date, brain pathology regarding the 22q11.2 deletion has been studied primarily in psychiatric disorders, especially schizophrenia, focusing on the cortex and hippocampus. For example, the 22q11.2DS mouse model exhibits schizophrenia-like cognitive dysfunction due to impaired hippocampal place cells or prefrontal cortex dysfunction [16,17]. In addition to schizophrenia, a recent study reported that the 22q11.2DS mouse model shows the signatures of Parkinson's disease at the molecular level [18]. Considering that defects in dopaminergic neurons of the midbrain are observed in Parkinson's disease [19], it can be expected that 22q11.2DS patient-derived dopaminergic neurons show the characteristic signatures, regardless of the presence or absence of Parkinson's disease onset.
Here, we generated 22q11.2DS patient-derived iPSCs, which were then homogenously differentiated into dopaminergic neurons using our previously described method [20]. Semiquantitative proteomic analysis revealed that ‘protein processing in the endoplasmic reticulum (ER)’ is the most altered pathway in the patient-derived dopaminergic neurons compared with healthy controls. Based on proteomic results, we identified protein kinase R-like endoplasmic reticulum kinase (PERK) as a key factor that contributes to the 22q11.2DS-related brain pathology in dopaminergic neurons.
2. Methods
2.1. Subjects
The human iPSC line 201B7 (HPS0063) [21] was provided by RIKEN BRC and used as Control1. Three healthy Japanese subjects, including a 41-year-old male (Control2) [20], 30-year-old female (Control3) [22,23] and 65-year-old female (Control4) [23], were selected as additional sources of control iPSCs. We performed psychological examinations and array comparative genomic hybridisation (aCGH) analyses on the healthy subjects. Clinically significant genomic variants were defined according to our previous study [24]. We confirmed that they had no neuropsychiatric disorders. Control4-derived iPSCs were used to generate isogenic iPSC lines. We identified three subjects with a heterozygous 22q11.2 deletion: a 23-year-old Japanese male with intellectual disability (22DS1), a 42-year-old Japanese female with schizophrenia and intellectual disability [22DS2, reported as SCZ1231 in our previous study [4]] and a 23-year-old Japanese female with intellectual disability (22DS3). Detailed information on these individuals is provided in Supplementary Table 1. The given ages of the subjects are those at the time of the blood sampling for iPSC generation.
2.2. iPSC generation and dopaminergic neuron differentiation
All iPSC lines, except for Control1, were generated from mononuclear cells of the peripheral blood of subjects using episomal vectors and the iPSC clones that were used for further analysis were selected as previously described [20]. One iPSC clone derived from each healthy control subject and two iPSC clones derived from each patient with 22q11.2DS were used for further analysis. Dopaminergic neuronal induction was performed as previously reported [20].
2.3. Ethics
All subjects provided written informed consent. The use of human samples and genomic analyses were approved by the Ethics Committee of Nagoya University (Approval Number: 2012-0184).
The animal experiments were approved by the Animal Care and Use Committee of Nagoya University and conducted in accordance with the guidelines of Nagoya University (Approval Number: 20237).
2.4. Animals
Del(3.0 Mb)/+ mice, which reproduce the most common 3.0 Mb deletion in 22q11.2DS, were generated as previously described [25]. The animals used in this study were 6-month-old male mice, which were littermates under the same conditions of breeding and rearing by in vitro fertilization. All animals were housed at a temperature of 23°C ± 1°C, under a 12 h light/dark cycle (light on at 09:00, off at 21:00), with free access to food and water. Collecting brain samples was performed at the laboratory with genetically modified organism diffusion prevention measures (P1A level). Three wild-type and three deletion type mice were used for quantitative analysis of immunoblots. Since the same tendency could be observed in both two groups, there were no excluded animals.
2.5. Mass spectrometry and data processing
For protein extraction, cells were homogenised using a Tenbroeck Grinder (WHEATON, USA) in a pH 7.5 buffer with 15 mM Tris, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 250 mM sucrose and a protease inhibitor (Wako, Japan). After centrifugation, the supernatants were used for mass spectrometry. Proteins (15 μg) were digested by trypsin for 16 h at 37°C after reduction and alkylation. The peptides were analysed by liquid chromatography−mass spectrophotometry (LC−MS) using an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, USA) coupled to an UltiMate3000 RSLCnano LC system (Dionex Co., Amsterdam, The Netherlands) and a nano high-performance liquid chromatography capillary column (150 mm × 75 µm i.d., Nikkyo Technos Co., Japan) via a nanoelectrospray ion source. Reversed-phase chromatography was performed with a linear gradient (0 min, 5% B; 100 min, 40% B) of solvent A (2% acetonitrile with 0.1% formic acid) and solvent B (95% acetonitrile with 0.1% formic acid) at an estimated flow rate of 300 nL/min. A precursor ion scan was performed using a 400–1600 mass-to-charge ratio (m/z) prior to tandem MS/MS analysis. MS/MS was performed by isolation at 0.8 Th with quadrupole high-energy collision dissociation fragmentation with a normalised collision energy of 30% and rapid scan MS analysis in the ion trap. Only precursors with charge states 2–6 were sampled for MS. The dynamic exclusion duration was set to 15 s with a 10 ppm tolerance. The instrument was run in top speed mode with 3 s cycles.
The raw data were processed using Proteome Discoverer 1.4 (Thermo Fisher Scientific) in conjunction with MASCOT search engine version 2.6.0 (Matrix Science Inc., Boston, MA, USA) for protein identification. Peptides and proteins were identified using a human protein database in UniProt (release 2019_11), with a 10 ppm precursor mass tolerance and fragment ion mass tolerance of 0.8 Da. The fixed modification was set to cysteine carbamidomethylation, and variable modifications were set to methionine oxidation. Two missed cleavages by trypsin were allowed.
To identify the differentially expressed proteins in the 22DS group, we initially compared the quantity between the control (n = 3) and 22DS groups (n = 3) using t-test without correction, and then selected candidate proteins with p < 0.05. Next, we narrowed down the candidate proteins based on FC > 10 (control group vs. 22DS group). Using these final candidate proteins, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed with DAVID's functional annotation tool (https://david.ncifcrf.gov/).
2.6. Immunocytochemistry and immunoblotting
For immunocytochemistry, cells were fixed in 4% paraformaldehyde (PFA) for 15 min, permeabilised, blocked in phosphate-buffered saline (PBS) containing 0.3% TritonX-100 and 1% BSA for 60 min and incubated with the indicated primary antibodies overnight at 4°C. For the immunostaining of calreticulin and translocase of outer mitochondrial membrane 20 (TOMM20), cells were incubated in 100% methanol for 5 min before permeabilisation and blocking. After washing with PBS, immunolabeled cells were incubated with appropriate fluorophore-tagged secondary antibodies for 1 h at room temperature. Images were captured using a BZ-9000 fluorescence microscope (Keyence, Japan) or a TiE-A1R confocal microscope (Nikon, Japan). The ImageJ Neuron J plugin was used to manually measure the neurite and filopodia lengths. The primary antibodies used for immunocytochemistry were as follows: anti-TRA-1-60 (ab16288, Abcam), anti-NANOG (ab21624, Abcam), anti-SOX17 (AF1924, R&D Systems), anti-αSMA (A2547, Sigma-Aldrich), anti-βIII-tubulin (T8660, Sigma-Aldrich), anti-tyrosine hydroxylase (TH) (AB152, Millipore), anti-microtubule-associated protein 2 (MAP2) (ab5392, Abcam), anti-cleaved caspase 3 (AF835, R&D systems), anti-activating transcription factor 4 (ATF4) (ab184909, Abcam), anti-phosphorylated eukaryotic translation initiation factor 2α (p-eIF2α) (ab32157, Abcam), anti-calreticulin (ab196158, Abcam) and anti-TOMM20 (ab210665, Abcam). For staining lamellipodia and filopodia, Alexa Fluor 488- or Alexa Fluor 555-Phalloidin (Cytoskeleton or Thermo Fisher Scientific) was used.
The quantification of immunoblots, other than those in Figs. 3b−3h, was performed using ImageJ software. The secondary antibodies included anti-mouse or anti-rabbit horseradish-peroxidase-conjugated antibodies (GE Healthcare, UK). For Figs. 3b−3h, the membrane was probed with the indicated primary antibody, followed by Alexa Fluor 680 and 800 (Thermo Fisher Scientific) secondary antibodies. An infrared imager was used for detection and quantification (Odyssey, LI-COR Biosciences). The primary antibodies used for immunoblotting were as follows: anti-PERK (5683 or 3192, Cell Signaling Technology), anti-phosphorylated-PERK (MA5-15033, ThermoFischer), anti-eIF2α (2103, Cell Signaling Technology), anti-phosphorylated- eIF2α (9721, Cell Signaling Technology), anti-ATF4 (184906, Abcam), anti-GADD34 (10449-1-AP, Proteintech), anti-CHOP (15204-1-AP, Proteintech), anti-ATF6 (24169-1-AP, Proteintech), anti-GRP78 (NBP1-06274SS, NOVUS), anti-IRE1 (27528-1-AP, Proteintech), anti- phosphorylated -IRE1 (NB100-2323SS, NOVUS), anti-TH (AB152, Millipore; 213-004, Synaptic Systems), anti-SNAP29 (11-302, Synaptic Systems), anti-βIII-tubulin (T8660, Sigma-Aldrich), anti-puromycin (MABE343, Millipore), anti-GAPDH (M171-3, MBL; CB1001, Millipore) and anti-β-actin-peroxidase (A3854, Sigma-Aldrich). Full blots were provided in Supplementay Material.
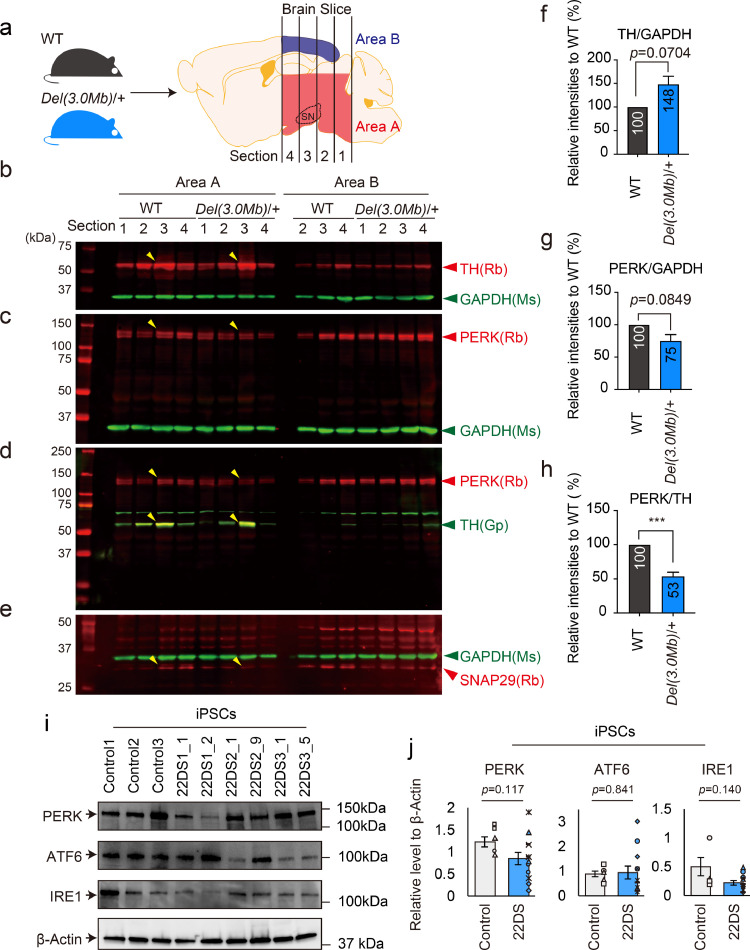
Fig. 3.
Examination of PERK expression in the brain of 22q11.2DS model mouse and iPSCs. (a) Schematic diagram of preparing brain slices from adult WT and Del(3.0Mb)/+ mice. SN = substantial nigra. (b–e) Immunoblot analysis using area A and area B. Yellow arrows indicate the section used for quantification. b and c: PERK and TH were simultaneously detected, with GAPDH as the internal standard. d: Double detection of PERK and TH. e: Decreased expression of SNAP29, one of the factors in the 22q11.2 deletion region. (f–h) Quantification of the TH/GAPDH, PERK/GAPDH and PERK/TH ratios in the area A of section no. 3. f, g and h correspond to b, c and d, respectively. Quantitative detection by immunoblot was performed by Odyssey. Immunoblots were independently performed four (f and g) or six (h) times. The numbers indicated in the bars represent relative values (WT = 100). The bars represent means ± SEs. ***p < 0.001. (i) Immunoblotting for PERK, ATF6, IRE1 and β-actin using proteins extracted from iPSCs. (j) Quantification of the PERK/β-actin, ATF6/β-actin and IRE1/β-actin ratios. Two independent experiments were performed (in summary, Control: n = 6; 22DS: n = 12). The value of Control1 is 1. Each plot represents the value of each line. The bars represent means ± SEs.
2.7. Immunohistochemistry
For immunohistochemistry, animals were deeply anaesthetised using 2,2,2-tribromoethanol (Avertin, 200 mg/kg, i.p., FUJIFILM, Wako Pure Chemical, Japan) and perfused with 4% PFA. The brains were postfixed with 4% PFA overnight, then maintained in 20% sucrose in PBS at 4°C, embedded in Tissue-Tek O.C.T. compound (Sakura Finetek Japan, Japan) and sectioned on a freezing cryostat (CM3000, Leica Microsystems, Germany) at 40 μm thickness. Floating brain slices were incubated with blocking buffer (M.O.M. Blocking Reagent, Vector Laboratories, USA) for 1 h at room temperature, and then incubated with the indicated primary antibodies overnight at 4°C. Labelled cells were visualised with the appropriate fluorescent secondary antibodies. The brain slices were imaged with a fluorescence microscope (BZ-9000, Keyence, Japan) or confocal laser-scanning microscope (LSM880-ELYRA PS.1, Zeiss, Germany). The primary antibodies used were anti-TH (AB152, Millipore) and anti-PERK (sc-377400, Santacruz).
2.8. Establishment of isogenic PERK-deficient iPSCs via targeted genome editing
Isogenic PERK-deficient iPSCs were generated as reported previously [20]. This experiment targeted exon 3 of PERK (NM_004836).
2.9. Cell treatments
To investigate cell death vulnerability, cells were treated with tunicamycin (TCM; T7765, Sigma-Aldrich) for 24 h (from Day 23 to Day 24). Salubrinal (Sal; 14735, Cayman), GSK2656157 (17372, Cayman), CCT020312 (324879, Sigma-Aldrich) and Cycloheximide (01810, Sigma-Aldrich) were added immediately after cell plating. CellTiter96 Aqueous One Solution (Promega) was used for the cell viability assay. Undetectable values (≤ blank) were defined as zero. To evaluate the status of protein synthesis, cells were incubated with puromycin (2 μg/ml) for 30 min on Day 24.
2.10. Knockdown of DGCR14
DiGeorge syndrome critical region 14 (DGCR14) small interfering RNAs (siRNAs) and negative control siRNA were purchased from Sigma-Aldrich as follows: DGCR14 siRNA#1 SASI_HS01_00246741, DGCR14 siRNA#2 SASI_HS01_00246743, DGCR14 siRNA#3 SASI_HS01_00246744 and negative control siRNA SIC-001-10. siRNAs were transfected into HEK293 cells using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer's instructions. After 72 h of transfection, the cells were collected for further experiments.
2.11. Quantitative PCR (qPCR)
Total RNA was extracted using the RNeasy Plus Mini Kit (QIAGEN, Germany). Reverse transcription RT was performed using a High-Capacity cDNA Transcription Kit (Applied Biosystems, USA). qPCR-based gene expression analysis was conducted using QuantStudio5 (Applied Biosystems) and a KAPA SYBR Fast qPCR Kit (KAPA BIOSYSTEMS, USA). The primers used were DGCR14: fw CACTGGGCCCCTTTTAATCGG, rv TTCCAGCAGACCAACTCAAAGTG and RPS18: fw GCGGCGGAAAATAGCCTTTG, rv GATCACACGTTCCACCTCATC.
2.12. Live imaging
Time-lapse movies were obtained using IncuCyte (Essen Bioscience, USA). Sequential neurite dynamics on phase-contrast images were quantified using the NeuroTrack software module. To compare the control and 22DS groups, we used fields in which the areas of cell body clusters were 0.12–0.20 mm2 per 1 mm2 at 24 h after plating.
2.13. Quantification of immunofluorescence
We used the NIS-Elements software (Nikon, Japan) to quantify immunofluorescent images. The same gain and exposure settings were used for the digital capture of images. Image acquisitions were performed on the same day in comparison targets. For fluorescence intensity quantification, the region of interest (ROI) was defined as the entire nucleus. For the co-localisation analysis, the ROI was selected at the peripheral area of the nucleus because the localisation of ER and mitochondria varies according to the subcellular zone. Mander's coefficients were calculated using a module of the NIS-Elements software.
2.14. Auto-detection of F-actin dynamics
To evaluate the effect of PERK activity on F-actin dynamics, we developed an automated detection system using MATLAB (MathWorks, Natick, USA). Images were processed in several steps. First, phalloidin-stained images were adjusted for brightness and contrast before quantitative analysis (Supplementary Figure 1a). Second, filopodia were highlighted using several steps of image processing. The application of unsharp masking and differentiation using a threshold filter was used to generate a binarised image in which filopodia-like structures were highlighted (Supplementary Figure 1b). A morphological operation was conducted to obtain the image skeleton (Supplementary Figure 1c). To identify cell body regions, a combination of contrast imaging and nuclear imaging of DAPI staining was used (Supplementary Figure 1d). A binary image of the cell body was generated by applying a Gaussian smoothing filter and threshold filtering (Supplementary Figure 1e). After two combinations of dilation and erosion (Supplementary Figure 1f), the binary image was used to mask irrelevant skeletal signals in the central part of cells. In the third step, the regionprops function of MATLAB was applied to the skeletal image to extract the properties of the segmented lines (Supplementary Figure 1g). The measured properties of the segmented lines provided the length of filopodia. In the final step, images of the line segments were plotted over the phalloidin-stained images to verify the quality of the analysis. All samples were also evaluated via the naked eye; however, no major contradictions were identified between the measurements obtained and the appearance of the plotted images.
2.15. Statistics
Groupwise binary comparisons were performed using Student's or Welch's t-test (unpaired, two-tailed) unless otherwise noted. For the analysis of three groups, an analysis of variance followed by Dunnett's test for pairwise comparisons was used. Significance was set at p < 0.05.
3. Results
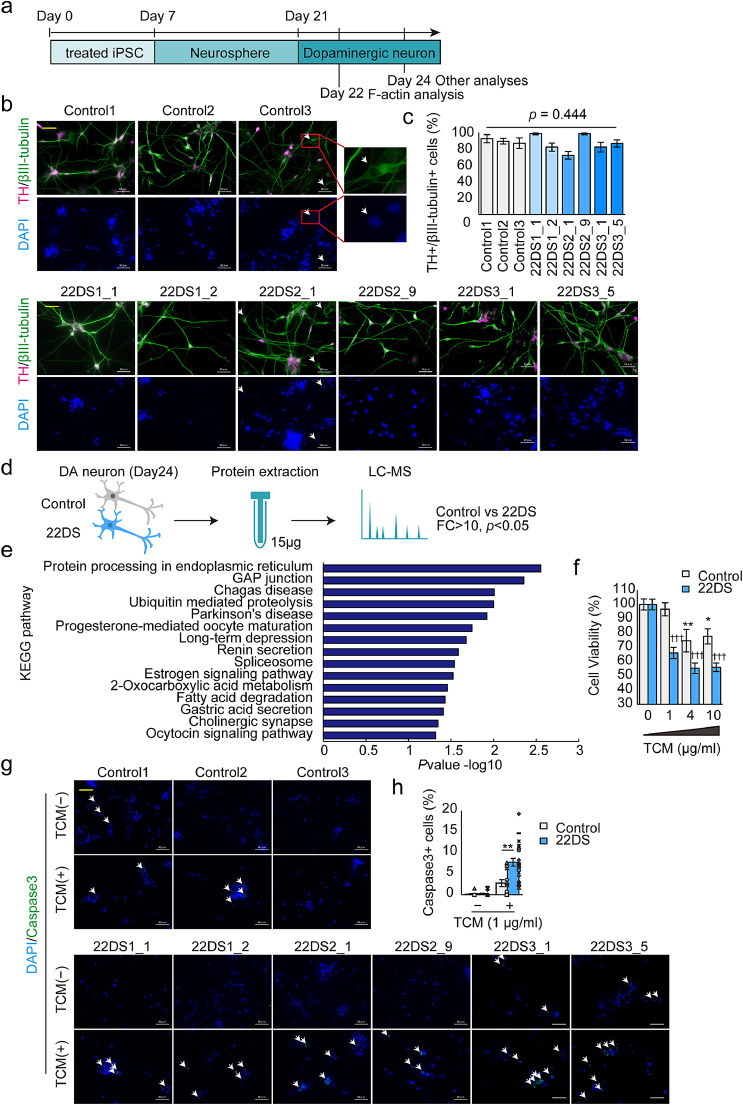
3.1. Characterisation of dopaminergic neuron development in 22q11.2DS iPSCs
We generated iPSCs from heterozygous 22q11.2DS patients (Supplementary Table 1). The all generated iPSCs showed the pluripotent markers (TRA-1-60 and NANOG) and had the ability to differentiate into three germ layers in vitro (Supplementary Figure 2). aCGH analysis confirmed the heterozygous deletion in the 22q11.2 region (Supplementary Figure 3). We differentiated control and patient iPSCs into dopaminergic neurons and examined their development (Fig. 1a). All iPSC lines differentiated into TH-positive neurons by Day 24 with similar efficiency (Figs. 1b and 1c). However, the differentiated neurons derived from the patients with 22q11.2DS (22DS group) exhibited significantly shorter βIII-tubulin-positive neurites compared with those of the healthy controls (control group) (Supplementary Figures 4a and 4b; p = 1.25 × 10–12). On Day 28, the number of MAP2-positive primary dendrites was smaller in the 22DS group compared with the control group (Supplementary Figures 4c and 4d; p = 2.78 × 10–16).
Fig. 1.
Characterisation of 22q11.2DS iPSC-derived dopaminergic neurons. (a) Schematic illustration of the differentiation of dopaminergic neurons. (b) Representative image of dopaminergic neurons (Day 24) immunostained for TH and βIII-tubulin. The yellow scale bar in the images represents 50 μm. White arrows indicate TH-negative, βIII-tubulin-positive (+) neurons. (c) Analysis of dopaminergic neuron differentiation efficiency by quantifying the ratio of TH+ to βIII-tubulin+ cells at Day 24. The bars represent means ± SEs. Five fields were analyzed. (d) Schematic illustration of the proteomic analysis. DA neuron = dopaminergic neurons. (e) KEGG pathway analysis. Proteins used showed a fold change (FC) > 10 and p < 0.05 (Control vs. 22DS). (f) Cell viability in the presence of ER stress. Control: n = 12 (Control1, n = 4; Control2, n = 4; Control3, n = 4); 22DS: n = 30 (22DS1_1, n = 4; 22DS1_2, n = 4; 22DS2_1, n = 4; 22DS2_9, n = 4; 22DS3_1, n = 7; 22DS3_5, n = 7). The plots represent the means ± SEs. *p < 0.05, **p < 0.01 vs. without TCM in the control group, †††p < 0.001 vs. without TCM in the 22DS group. (g) Representative images of immunostained cleaved-caspase3+ cells. White arrows indicate cleaved-caspase3+ cells. The yellow scale bar in the image represents 50 μm. (h) Quantification of the celaved-caspase 3+ cell ratio. Control: n = 12 (each, n = 4 fields); 22DS: n = 24 (each, n = 4 fields). Bars represent means ± SEs. **p < 0.01. Each plot represents the value of each line.
To analyse dynamic neurite elongation during neuronal development, we performed time-lapse image analysis. To minimise bias, we used an automatic detector of neurites (pink) and cell bodies (green) (Supplementary Figure 4e). Shortened neurites were observed in the 22DS group on Day 22, which remained shorter than those in the control group throughout the experiment (Days 23, 24 and 25) (Supplementary Figure 4f). These data confirm the impairment of dopaminergic neuritogenesis in the 22DS group.
3.2. Downregulation of PERK in 22q11.2DS iPSC-derived dopaminergic neurons
Next, we investigated the molecular pathology in the 22DS group. Semiquantitative proteomic analysis using mass spectrometry identified 272 differentially expressed proteins (8 upregulated and 264 downregulated, FC > 10, p < 0.05) in the 22DS group compared with the control group (Fig. 1d and Supplementary Tables 2 and 3). Among proteins encoded by genes in the 22q11.2 region, RANBP1, ARVCF, PI4KA, SEPT5, SLC25A1, TXNRD2 and UFD1L were detected by proteomic analysis, but only ARVCF was included in the 264 significantly downregulated proteins. KEGG pathway analysis revealed that the most altered pathway in the 22DS group was ‘Protein processing in ER’, which is responsible for ER stress response and protein quality control (Fig. 1e). Thus, we predicted that low tolerance to ER stress is observed in the 22DS group.
We exposed dopaminergic neurons to various concentrations of tunicamycin (TCM, an inducer of ER stress). We then analysed cell viability and performed immunostaining of cleaved caspase 3. In the presence of 1 μg/ml TCM, cell viability was significantly low in the 22DS group (Fig. 1f, p = 3.84 × 10–8). Similarly, immunostaining for cleaved caspase 3 was more pronounced in the 22DS group compared with the control group (Figs. 1g and 1h, p = 1.25 × 10–3). As predicted, the 22DS group exhibited a low tolerance to ER stress.
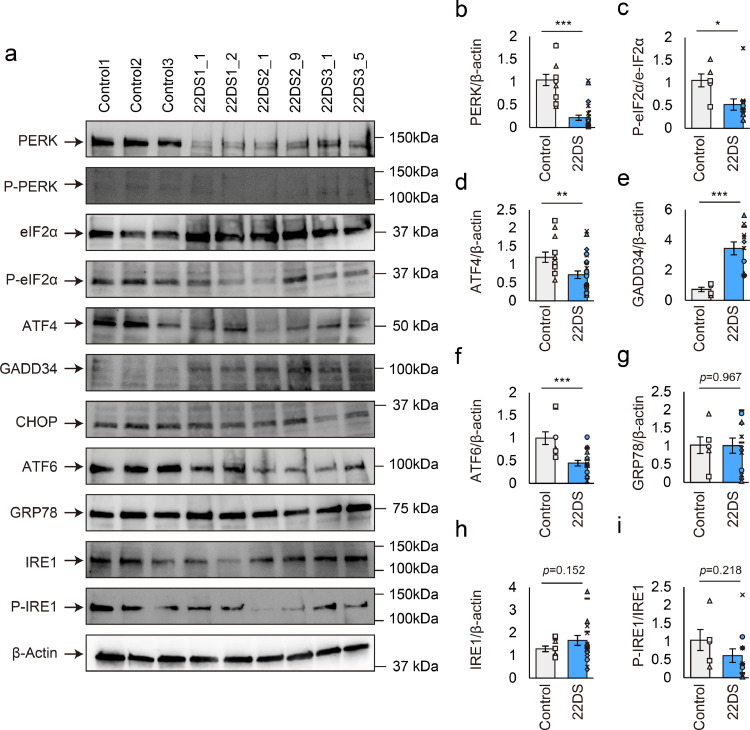
Cellular response to ER stress is regulated mainly by three stress sensors in mammals: PERK, IRE1 and ATF6 [26]. We examined the expression of these sensors and related proteins in the control and 22DS groups. As shown in Figs. 2a and 2b, the protein expression of PERK was significantly lower in the 22DS group compared with the control group (20.7% of the control group, p = 4.45 × 10–8). We also found that phosphorylation of eIF2α and expression of ATF4 decreased in the 22DS group, both of which are target signals of PERK activity (Figs. 2a, 2c and 2d). In agreement with this observation, the expression of GADD34, which promotes eIF2α dephosphorylation, was higher in the 22DS group (Figs. 2a and 2e). These findings indicate that PERK activity was disrupted in the 22DS group. However, although ATF6 expression was reduced in the 22DS group (44.9% of the control group, p = 3.54 × 10–4, Figs. 2a and 2f), the target protein GRP78 expression level was not significantly different between the control group and the 22DS group (Figs. 2a and 2g). Regarding IRE1, no differences in IRE1 expression and phosphorylation were detected (Figs. 2a, 2h and 2i). Hence, we focused on PERK, which exhibited altered expression and activity in the 22DS group, for subsequent analyses.
Fig. 2.
Downregulation of PERK expression and activity in 22q11.2DS iPSC-derived dopaminergic neurons. (a) Immunoblotting for ER stress sensors and their related proteins using dopaminergic neurons (Day 24). P-PERK: phosphorylated PERK; P-eIF2α: phosphorylated eIF2α; P-IRE1: phosphorylated IRE1. (b–i) Quantification of the PERK/β-actin, P-eIF2α/eIF2α, ATF4/β-actin, GADD34/β-actin, ATF6/β-actin, GRP78/β-actin, IRE1/β-actin, P-IRE1/IRE1 ratios. Immunoblots were independently performed four (b and d; control [n = 12], 22DS [n = 24]), three (f and h; control [n = 9], 22DS [n = 18]) or two times (c, e, g and i; control [n = 6], 22DS [n = 12]). The value of Control1 was set to 1. Each plot represents the value of each line. Bars represent means ± SEs. *p < 0.05, **p < 0.01, ***p < 0.001.
3.3. Downregulation of PERK in the midbrain of the 22q11.2DS mouse models
Next, we wondered whether the reduction in PERK expression in the 22DS group is caused by the chromosome 22q11.2 deletion. To address this, we compared PERK protein expression in the midbrain, where dopaminergic neurons are enriched, between wild-type (WT) and 22q11.2DS model [Del(3.0Mb)/+] mice. The Del(3.0Mb)/+ mouse is a novel mouse model with the most common 3.0-Mb deletion in the human 22q11.2 locus as we recently established.[25]
Littermate controls were used for comparison. Immediately after dissection, four thin sections of the whole brain were prepared using a vibratome and the two areas (area A including midbrain and area B including cortex) were cut from each section under a stereoscopic microscope (Fig. 3a). Quantitative immunoblot analysis showed a much higher expression of TH in the area A of both the WT and Del(3.0 Mb)/+ mice compared with that of the area B (Fig. 3b). In addition, immunohistochemistry analysis showed that TH-positive cells expressed PERK (Supplementary Figures. 5a–5f). To ensure the same condition among comparisons, we selected the sections with the highest levels of TH expression (area A of section no. 3, indicated by yellow arrows in Figs. 3b–3e) for further quantification using immunoblot data. Area A of section no. 3 mainly included substantial nigra (SN, Fig. 3a).
The expression levels of TH and PERK normalised to GAPDH were not significantly different between WT and Del(3.0 Mb)/+ mice (Figs. 3b, 3c, 3f and 3g). To focus on only TH-positive neurons, the expression level of PERK normalised to TH was examined. Consistent with the results using iPSC-derived dopaminergic neurons, a significant decrease in PERK was observed in Del(3.0 Mb)/+ mice (53% ± 6.5%, p = 8.00 × 10–4, Figs. 3d and 3h). Of note, we validated that the protein expression of SNAP29 encoded by the Snap29 gene in the deleted region was reduced in Del(3.0 Mb)/+ mice compared with WT mice (Fig. 3e). We also confirmed similar results using iPSC-derived dopaminergic neurons; a significant decrease in the value of PERK/TH and decreased SNAP29 expression level in the 22DS group (Supplementary Figure 6).
These data support that the reduction in PERK protein expression in the 22DS group (= 22q11.2DS patient iPSC-derived dopaminergic neurons) is caused by the chromosome 22q11.2 deletion.
3.4. PERK expression was unaltered in 22q11.2DS patient iPSCs
PERK is a ubiquitously expressed ER protein. It is possible that reduced PERK expression is also observed in other cell types. Thus, we compared PERK expression between healthy control and 22q11.2DS patient iPSCs (before neuronal differentiation) and detected no significant differences (Figs. 3i and 3j). Similarly, ATF6 and IRE1 expression levels were not different between healthy control and 22q11.2DS patient iPSCs (Figs. 3i and 3j).
3.5. Evaluation of PERK-dependent cellular functions: response to ER stress
To determine whether the low tolerance to ER stress in the 22DS group was dependent on PERK dysfunction, we examined the effect of salubrinal (Sal, an activator of this pathway),[27] on cell viability. Exposure of the 22DS group to Sal under ER stress significantly inhibited the effects of TCM on cell viability and cleaved caspase 3 immunoreactivity (Figs. 4a and 4b). Moreover, the 22DS group exhibited a significantly lower protein expression in ATF4 under TCM-induced ER stress compared with the control group (Supplementary Figure 7). These data suggest that PERK dysfunction contributes to the reduced tolerance to ER stress observed in the 22DS group.
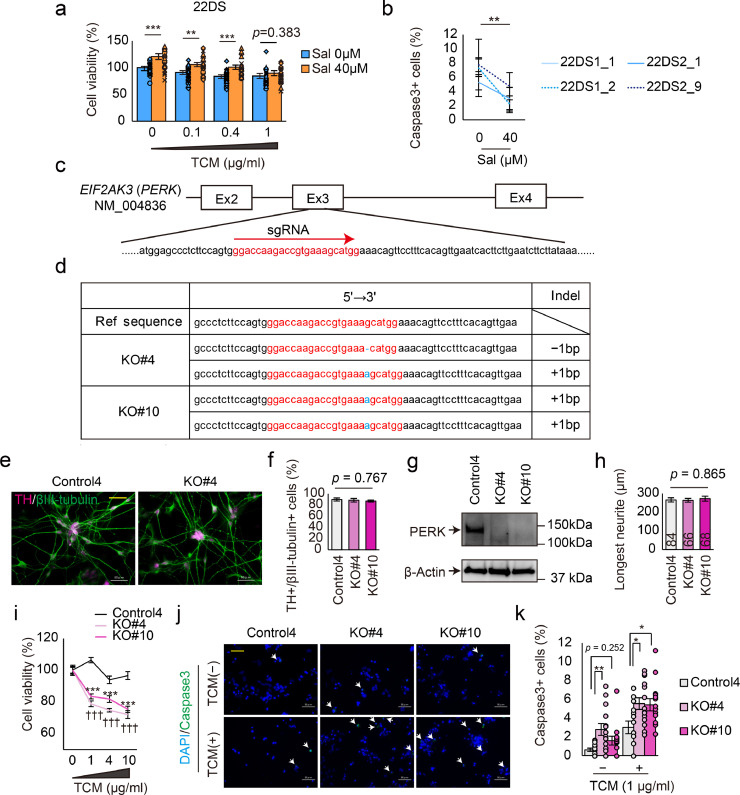
Fig. 4.
Generation of PERK-deficient iPSCs. (a) Cell viability in the presence of TCM with or without salubrinal (Sal). n = 24 (22DS1_1, n = 6; 22DS1_2, n = 6; 22DS2_1, n = 6; 22DS2_9, n = 6). Each plot represents the value of each line. The bars represent means ± SEs. **p < 0.01, ***p < 0.001. (b) Quantification of the caspase 3-positive (+) cell ratio using the 22DS group. TCM treatment (0.4 μg/ml) with or without Sal. Twenty fields were used (each, n = 5). The plots represent means ± SEs. ** p < 0.01. (c) The target site of the CRISPR/Cas9 system used in this study. (d) Indel patterns of the PERK-deficient isogenic lines. (e) Representative images of dopaminergic neurons (Day 24) immunostained for TH and βIII-tubulin. The scale bar represents 50 μm. (f) Analysis of dopaminergic neuron differentiation efficiency via the quantification of the ratio of TH+ to βIII-tubulin+ cells at Day 24. The bars represent means ± SEs. Five fields were used. (g) Immunoblotting of PERK and β-actin using DA neurons at Day 24. (h) Measurement of neurite length. The numbers indicated in the bars represent the number of counted cells. The bars represent means ± SEs. (i) Cell viability in the presence of ER stress (n = 7). The plots represent the means ± SEs. *** p < 0.001, ††† p < 0.001 vs. without TCM. (j) Representative images of immunostaining for cleaved-Caspase3 using dopaminergic neurons. White arrows indicate Caspase3+ cells. The yellow scale bar in the image represents 50 μm. (k) Quantification of the cleaved-caspase 3+ cell ratio (n = 14). The bars represent means ± SEs. * p < 0.05, ** p < 0.01. Each plot represents the value of each line.
Based on the above findings, we hypothesised that PERK is a key factor in the mechanism underlying the 22q11.2DS-related pathology in dopaminergic neurons. To verify this, we established PERK-deficient iPSC lines from healthy control iPSCs (Control4) using CRISPR/Cas9 systems. In comparison with Control 4, the 22DS group showed similar ratios of TH+/βIII-tubulin+ cells (p = 0.640), shorter neurites (p = 1.44 × 10−13), low cell viability (p = 4.64 × 10−11) and a higher ratio of Caspase3+ cells (p = 8.84 × 10−4) under the TCM treatment.
The target site of the single-guide RNA (sgRNA) was exon 3 of PERK (NM_004836), which included a binding site for misfolded proteins (Fig. 4c). For reproducibility and to exclude the possibility of off-target effects, we generated two isogenic iPSC lines, KO#4 and KO#10, both of which were predicted to be a homozygous knockout because of a frameshift event (Fig. 4d). All iPSCs (Control4, KO#4 and KO#10) differentiated into dopaminergic neurons with similar efficiency (Figs. 4e and 4f); however, PERK expression was not detected in KO#4 and KO#10 (Fig. 4g). No differences in the length of neurites were observed among Control4, KO#4 and KO#10 (Fig. 4h). Using these iPSC-derived dopaminergic neurons, we investigated the effect of PERK deficiency on the tolerance to ER stress. As observed in the 22DS group, KO#4 and KO#10 exhibited low tolerance to ER stress compared with Control4 (Figs. 4i, 4j and 4k). The results of pharmacological manipulation and genome editing regarding PERK confirm that PERK dysfunction is one of the underlying mechanisms involved in the low tolerance to ER stress in the 22DS group.
The combined use of the 22DS group and these PERK-deficient dopaminergic neurons may allow us to verify our hypothesis that PERK is a key factor in the mechanism underlying the 22q11.2DS-related pathology in dopaminergic neurons. Hence, we further evaluated other PERK-dependent cellular functions, including F-actin dynamics [28,29], protein synthesis [30,31] and ER-mitochondria contacts.[32]
3.6. Evaluation of PERK-dependent cellular functions: F-actin dynamics
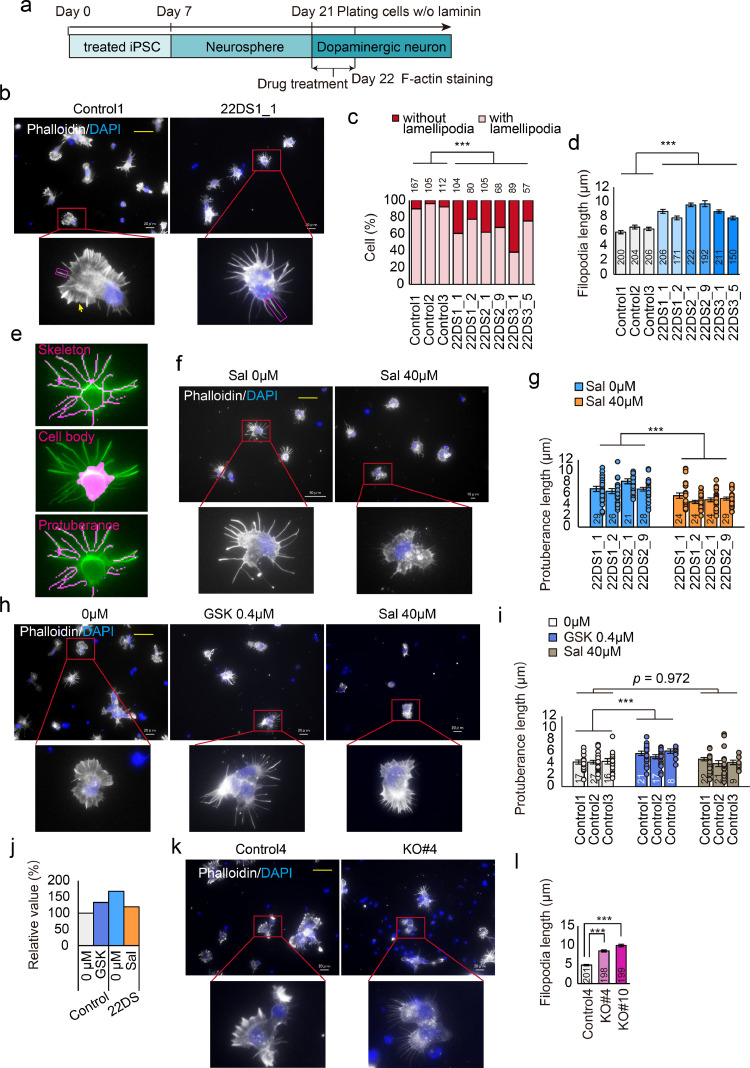
Neuritogenesis requires the formation of filopodia and lamellipodia, both of which rely on dynamic changes in F-actin [33]. Based on our finding that neuritogenesis is impaired in the 22DS group (Supplementary Figure 4), we predicted that this is likely due to abnormal F-actin dynamics. Laminin can partially rescue neuritogenesis through the formation of filopodia-like actin-rich protrusions [34]. Thus, to directly compare F-actin dynamics related to filopodium and lamellipodium formation, we used dopaminergic neurons in the absence of laminin (Fig. 5a). On Day 22, the control group showed typical developing filopodia (magenta box, Fig. 5b) and lamellipodia (yellow arrow, Fig. 5b), whereas the 22DS group exhibited abnormal filopodia that were >1.5-fold longer (p = 7.32 × 10–43) and loss of typical lamellipodia (p = 6.89 × 10–16) (Figs. 5b–5d). Our data indicate that F-actin dynamics are abnormal in the 22DS group.
Fig. 5.
Impairment of F-actin dynamics and PERK dysfunction. (a) Schematic illustration of experimental timing. (b) Representative images of Control1 and 22DS1_1 stained with phalloidin at Day 22. The yellow scale bar in the images represents 50 μm. The magenta box represents filopodia. The yellow arrow represents lamellipodia. (c) Ratio of cells exhibiting typical lamellipodia. The numbers over the bars represent the number of counted cells. ***p < 0.001. (d) Measurement of filopodium length. The numbers indicated in the bars represent the number of counted filopodia. The bars represent means ± SEs. ***p < 0.001. (e) Representative results for the automatic detection system. A skeletonised region (Upper), except for the region detected as a cell body (Middle), is recognised as a filopodia-like protuberance (Lower). (f) Representative images of cells stained with phalloidin at Day 22 with or without Sal (40 μM) in the 22DS group. The yellow scale bar represents 50 μm. (g) Measurement of protuberance length using an automated detection system. The numbers indicated in bars represent the number of counted fields. Each plot represents the value of each line. The bars represent means ± SEs. *** p < 0.001. (h) Representative images of cells stained with phalloidin at Day 22 with or without GSK2656157 (GSK, 0.4 μM) or Sal (40 μM) in the control group. The yellow scale bar represents 50 μm. (i) Measurement of protuberance length using an automated detection system. The numbers indicated in bars represent the number of counted fields. Each plot represents the value of each line. The bars represent means ± SEs. *** p < 0.001. (j) Summary of the effects of pharmacological manipulation (g and i) for PERK activity. The average length of the control group was set as 100. (k) Representative images of Control4 and KO#4 stained with phalloidin at Day 22. The yellow scale bar represents 50 μm. (l) Measurement of filopodium length. The numbers indicated in the bars represent the number of counted filopodia. The bars represent means ± SEs. *** p < 0.001.
To determine whether abnormal F-actin dynamics in the 22DS group were dependent on PERK dysfunction, we activated the PERK pathway using Sal, which increased the expression of p-eIF2α (Supplementary Figures 8a and 8b). To objectively evaluate the effect of Sal, we developed an automated F-actin dynamics detection and analysis system (Fig. 5e and Supplementary Figure 1). The automated system demonstrated that Sal significantly ameliorated the abnormally long filopodium-like protuberances observed in the 22DS group (Figs. 5f and 5g, and Supplementary Figure 9, p = 3.93 × 10–11).
To assess whether the abnormally long filopodia observed in the 22DS group were related to the inactivation of the PERK pathway, we directly inhibited the pathway in the control group using the PERK-selective inhibitor GSK2656157. [35]. After treatment with GSK2656157, the control group exhibited a decrease in the expression of p-eIF2α (Supplementary Figures 8c and 8d) and the presence of abnormal filopodia, which were reminiscent of those observed in the 22DS group (Figs. 5h and 5i, and Supplementary Figure 9, p = 1.78 × 10–4). We also confirmed that Sal at a concentration of 40 μM had no effect on the length of filopodia in the control group (Figs. 5h and 5i, and Supplementary Figure 9, p = 0.972). These results suggest that the abnormal F-actin dynamics detected in the 22DS group were dependent on PERK dysfunction (Fig. 5j). Consistent with this, KO#4 and KO#10 exhibited longer filopodia compared with Control4 (Figs. 5k and 5l) (p = 3.47 × 10–8 and p = 3.68 × 10–4, respectively). Taken together, these results show that PERK dysfunction is one of the underlying mechanisms contributing to abnormal F-actin dynamics in the 22DS group.
Conversely, Sal did not ameliorate neurite length in the 22DS group (Supplementary Figures 10a and 10b), which was consistent with those of KO#4 and KO#10.
We also evaluated the effect of another PERK activator, CCT020312,[36,37] on the 22DS group phenotypes. Similar to salubrinal, CCT020312 ameliorated cell vulnerability to ER stress and F-actin dynamics in the 22DS group with the upregulation of PERK activity (Supplementary Figures 11).
3.7. Evaluation of PERK-dependent cellular functions: protein synthesis
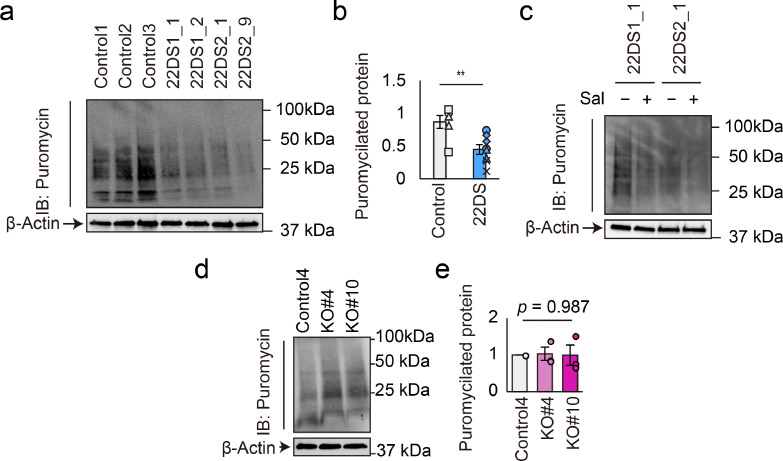
Next, we compared global protein synthesis between the control and 22DS groups using the SUnSET assay, in which newly synthesised proteins are labelled by puromycin.[38] The amount of puromycilated proteins was remarkably decreased by ~50% in the 22DS group compared with the control group, indicating that global protein synthesis is impaired in the 22DS group (Figs. 6a and 6b, p = 4.32 × 10–3). However, this decrease in protein synthesis was not ameliorated by the addition of Sal (Fig. 6c). In agreement with this, PERK-deficient dopaminergic neurons (KO#4 and KO#10) did not show alterations in global protein synthesis (Figs. 6d and 6e, p = 0.987). These findings indicate that the reduced protein synthesis in the 22DS group is independent of PERK.
Fig. 6.
Global protein synthesis and PERK dysfunction. (a) Immunoblotting for puromycin and β-actin in the control and 22DS groups at Day 24. Treatment with puromycin lasted for 30 min. (b) Quantification of puromycilated protein signal intensity in a. The value of Control1 was set to 1. Two independent experiments were performed (in summary, Control, n = 6; 22DS, n = 8). Each plot represents the value of each line. The bars represent means ± SEs. ** p < 0.01. (c) Immunoblotting for puromycin and β-actin using the 22DS group at Day 24 with or without Sal (40 μM). Treatment with puromycin lasted for 30 min. (d) Immunoblotting for puromycin and β-actin using Control4, KO#4 and KO#10 DA neurons at Day 24. Treatment with puromycin lasted for 30 min. (e) Quantification of puromycilated protein signal intensity in d. The value of Control4 was set to 1. Three independent experiments were performed. Each plot represents the value of each line. The bars represent means ± SEs.
3.8. Evaluation of PERK-dependent cellular functions: mitochondria-ER contacts
We evaluated mitochondria-ER contacts, which are thought to represent a signalling hub where Ca2+ and lipid exchange can occur between these two organelles.[39,40] Changes in the extent of mitochondria-ER contacts have been reported in various models of neuropsychiatric disorders, especially neurodegenerative diseases.[41]
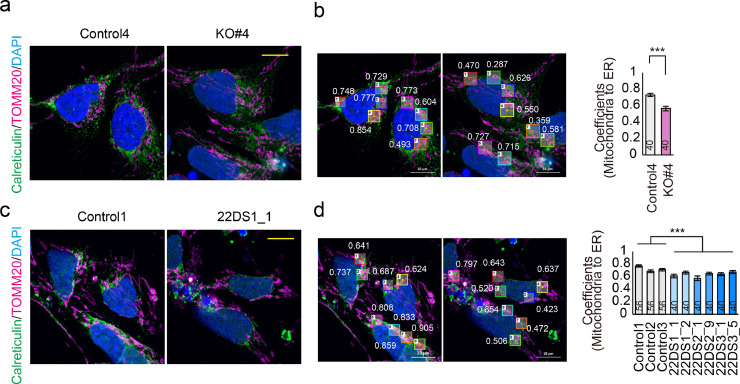
PERK-deficient dopaminergic neurons showed a significantly lower level of mitochondria-ER contacts compared with Control4 (Figs. 7a and 7b, p = 8.12 × 10–6). Similarly, the 22DS group showed reduced co-localisation between mitochondria and the ER (Figs. 7c and 7d, p = 9.38 × 10–8). These data support that PERK dysfunction is involved in the phenotype observed in the 22DS group.
Fig. 7.
ER-mitochondria contacts in the 22DS and PERK-deficient groups. (a) Representative images of dopaminergic neurons from Control4 and KO#4 (Day 24) immunostained for calreticulin (for ER) and TOMM20 (for mitochondria). The yellow scale bar in the image represents 10 μm. (b) Left panel: Representative results of the co-localisation analysis that was performed using Mander's method. Right panel: Mander's co-localisation analysis. The numbers indicated in the bars represent the number of counted fields. The bars represent means ± SEs. *** p < 0.001. (c) Representative images of dopaminergic neurons from Control1 and 22DS1_1 (Day 24) immunostained for calreticulin and TOMM20. (d) Left panel: Representative results of a co-localisation analysis that was performed using Mander's method. The yellow scale bar in the image represents 10 μm. Right panel: Mander's co-localisation analysis. The numbers indicated in the bars represent the number of counted fields. The bars represent means ± SEs. *** p < 0.001.
3.9. DGCR14 is involved in the downregulation of PERK expression
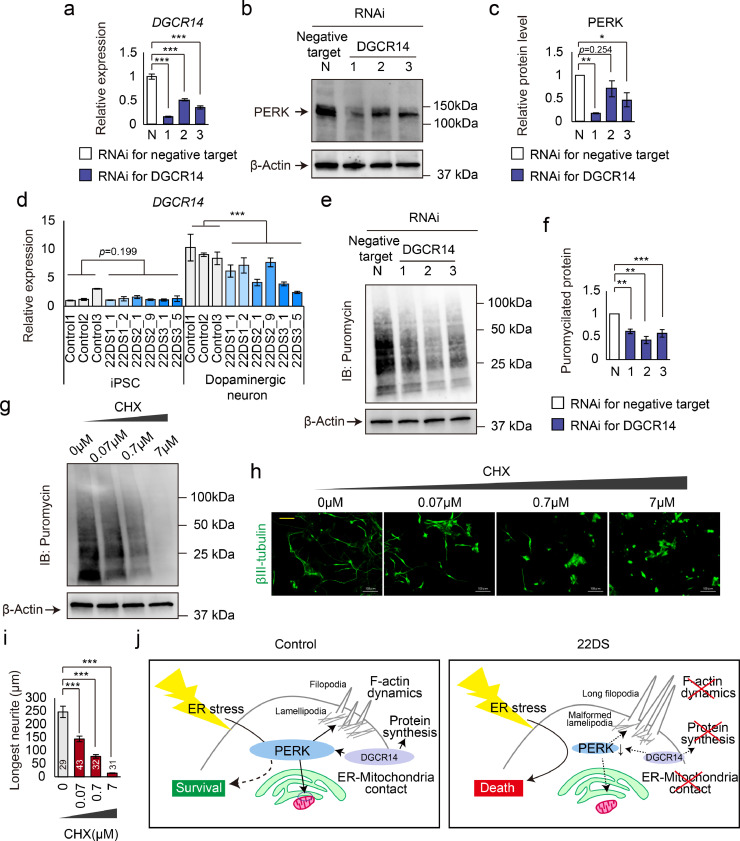
Finally, we assessed the association between reduced PERK expression and the 22q11.2 deletion. Our proteomic analysis showed that the pathway ‘Spliceosome’ is also significantly altered in the 22DS group (Fig. 1e). Among genes in the 22q11.2 region, DGCR14 (also known as ESS2) is reported to be involved in spliceosome formation.[42] Thus, we speculated that focusing on DGCR14 may provide useful information on the association between PERK and 22q11.2 deletion.
We performed RNA interference (RNAi) experiments to knockdown DGCR14 in HEK293 cells. Three different siRNAs targeting DGCR14 (siRNA#1, siRNA#2 and siRNA#3) were used. All siRNAs effectively knocked down DGCR14 expression (Fig. 8a). Compared with cells transfected with negative control RNAi (N), DGCR14 knockdown cells exhibited decreased protein expression of PERK (Figs. 8b and 8c). These data suggest that downregulated DGCR14 is associated with the reduction in PERK expression in the 22DS group.
Fig. 8.
DGCR14 is associated with the downregulation of PERK expression. (a) Relative expression levels of DGCR14 in DGCR14 knockdown cells. *** p < 0.001. (b) Immunoblotting for PERK and β-actin using the negative target and DGCR14 knockdown cells. ‘Negative target’ means the use of the negative control RNAi. (c) Quantification of the PERK/β-actin ratio. Three independent experiments were performed. The bars represent means ± SEs. * p < 0.05, ** p < 0.01. The value of N was set to 1. (d) Quantification of the mRNA expression level of DGCR14 in iPSCs and dopaminergic neurons (Day 24) derived from healthy controls and 22q11.2DS patients. The bars represent means ± SEs. 22DS1_2 and 22DS2_1 in dopaminergic neurons: n = 4; others: n = 3. *** p < 0.001. (e) Immunoblotting for puromycin and β-actin in DGCR14 knockdown cells. (f) Quantification of puromycilated protein signal intensity in e. The value of N was set to 1. The bars represent means ± SEs. Each n = 4. ** p < 0.01, *** p < 0.001 vs N. (g) Immunoblotting for puromycin and β-actin in dopaminergic neurons treated with cycloheximide (CHX). (h) Representative images of immunostaining for βIII-tubulin using CHX-treated dopaminergic neurons. The yellow bar in the image was 100 μm. (i) Measurement of neurite length. The numbers indicated in/over the bars represent the number of counted cells. The bars represent means ± SEs. *** p < 0.001. (j) Graphic abstract.
To determine whether DGCR14 is expressed in dopaminergic neurons, we compared the mRNA expression level of DGCR14 between iPSCs and dopaminergic neurons (Day 24). DGCR14 mRNA expression in dopaminergic neurons was much higher than that in iPSCs (Fig. 8d). Before neuronal differentiation (iPSCs), there were no significant differences in DGCR14 expression between the controls and patients. However, after the differentiation into dopaminergic neurons, the expression level of DGCR14 in the 22DS group was lower than that in the control group (p = 3.03 × 10−4, Fig. 8d). These findings imply that DGCR14 plays several roles in dopaminergic neurons.
Given that DGCR14 is involved in splicing, DGCR14 deficiency may cause a global decrease in protein synthesis. To address this, we examined the protein synthesis of DGCR14 knockdown cells using the SUnSET assay. As shown in Figs. 8e and 8f, knockdown of DGCR14 resulted in a decrease in global protein synthesis. In addition, we asked whether the decrease in protein synthesis leads to short neurites in dopaminergic neurons. Dopaminergic neurons treated with cycloheximide (CHX), an inhibitor of protein synthesis, showed generation of CHX dose-dependent short neurites (Figs. 8g, 8h and 8i). These findings suggest that PERK-independent phenotypes of the 22DS group are involved in DGCR14 deficiency.
4. Discussion
In this study, we showed that PERK dysfunction is one of the underlying causes of the vulnerabilities in dopaminergic neurons carrying the 22q11.2 deletion. Also, our findings suggest that DGCR14, among genes located in the 22q11.2 region, may be involved in the reduction of PERK expression (Fig. 8j).
Our KEGG pathway-based analysis identified that the pathways ‘protein processing in ER’ and ‘Parkinson's disease’ were altered in the 22DS group. This result may reflect the pre-onset or early phenotypes of these patients at the cellular and molecular levels and the high vulnerability to early-onset Parkinson's disease in 22q11.2DS patients[7]. Indeed, the 22q11.2DS patients in this study have not yet shown Parkinson's disease symptoms. Moreover, the analysed dopaminergic neurons are still early and immature. It follows that 22q11.2DS patients have a vulnerability in dopaminergic neurons, even from brain development. The current findings are useful not only for understanding 22q11.2DS-related brain pathology but also for developing strategies to prevent future Parkinson's disease onset in patients with 22q11.2DS.
Along with PERK dysfunction, the 22DS group showed the characteristic phenotypes of low tolerance to ER stress and abnormal F-actin dynamics. These phenotypes prompt us to discuss the association with 22q11.2DS. First, the low tolerance to ER stress probably reflects the high sensitivity to Parkinson's disease in 22q11.2DS patients. In Drosophila, the downregulation of PERK in dopaminergic neurons led to the loss of dopaminergic neurons like Parkinson's disease.[43] Second, normal F-actin dynamics are essential for attractive axon guidance and neural connectivity,[44] and defects in F-actin cause impairment of axonal growth and projection. The dopaminergic neurons used here are considered to be midbrain dopaminergic neurons. Hence, it can be assumed that the projection of midbrain dopaminergic neurons is impaired in 22q11.2DS patients. The projection of midbrain dopaminergic neurons to the prefrontal cortex, hippocampus, amygdala and the ventral striatum are believed to regulate and contribute to various types of learning and memory.[45,46] The impairment of F-actin dynamics in the 22DS dopaminergic neurons may reflect the clinical phenotypes of learning and memory dysfunction in 22q11.2DS patients.[47] Supporting this, PERK signalling in neurons is required for axon guidance and neural connectivity.[48] Moreover, a recent study shows that a specific-deletion of PERK in mouse midbrain dopaminergic neurons drives the cognitive and motor dysfunction.[49] We should certainly validate this hypothesis with in vivo analysis in the future. In addition, we believe that PERK dysfunction is at least partially responsible for the defect in F-actin dynamics based on our pharmacological manipulations and gene-editing analyses; however, we do not exclude the possibility of other genes in the 22q11.2 region directly affecting the F-actin dynamics.
By the proteomic analysis, only ARVCF was included in the significatnly down regulated proteins among proteins encoded by genes located in the 22q11.2 region. Several possible reasons can be considered. One is the detection limit of the proteomic analysis. For example, PERK was not detected in both the control and 22DS group by the proteomic analysis. PERK is one of transmembrane proteins, which are often difficult to detect using this method. Another possible reason is that the deletion of genes does not always lead to changes in their corresponding protein levels. Indeed, in the study of a mouse model of human 3q29 deletion, the transcripts for 18 genes in the interval showed a decrease in expression that was consistent with their deletion; however, the corresponding protein levels did not always vary.[50]
Severe deficits in global protein synthesis were another characteristic phenotype observed in the 22DS group. However, PERK dysfunction was not involved with this phenotype. Similarly, a minimal effect of decreased PERK expression on global protein synthesis was reported in mice with a forebrain-specific disruption of PERK.[51] PERK regulates protein synthesis to restore homeostasis,[52] and its downstream target ATF4 is activated during the ER stress response.[53] Given that the 22DS group does not activate ATF4 expression even under ER stress conditions (Supplementary Figure 7), the severe loss of protein synthesis regulation beyond the PERK pathway may occur in the 22DS group. A possible causal factor of defective protein synthesis in the 22DS group is the haploinsufficiency of DGCR14, one of the genes located in the critical region of 22q11.2DS. In the present study, we found that DGCR14 knockdown leads to decreased protein synthesis. Moreover, the inhibition of protein synthesis resulted in short neurites. Considering that both the decreased protein synthesis and shortened neurites had no connection to PERK dysfunction, it can be assumed that DGCR14 is involved in the PERK-independent phenotypes of the 22DS group. We plan to continue further research, focusing on DGCR14.
Lastly, our study suggested that DGCR14 is associated with reduced PERK expression. Although the detailed function of DGCR14 remains unknown, some studies suggest that DGCR14 associates with transcriptional factors and spliceosomal complexes to modulate mRNA splicing reactions.[42,54] The identification of the pathway ‘Spliceosome’ in the proteomic analysis may reflect the effect of DGCR14 deletion. Because dopaminergic neurons showed much higher expression of DGCR14 than iPSCs, it can be expected that the effect of DGCR14 deletion is larger in dopaminergic neurons than iPSCs. Supporting this idea, reduced PERK expression was observed in 22q11.2DS patient-derived dopaminergic neurons, but not in 22q11.2DS patient-derived iPSCs. On the other hand, it remains unclear why the DGCR14 downregulation or 22q11.2 deletion causes reduced PERK expression. One possibility is that defective spliceosome formation causes abnormal mRNAs, which subsequently promote nonsense-mediated mRNA decay (NMD). Considering that PERK expression is involved in NMD suppression,[55,56] the over-activated NMD possibly induces downregulation of PERK. The underlying mechanism between DGCR14 and PERK will be the next primary question for a future study.
In conclusion, our findings highlight that 22q11.2 deletion causes PERK-dependent vulnerabilities in dopaminergic neurons. In humans, PERK gene mutations cause Wolcott–Rallison syndrome, which is characterised by growth retardation and intellectual disability.[57] In addition, PERK expression is reduced in the postmortem brains of schizophrenia patients.[51,58] Here, we suggest that PERK is one of the key factors for understanding the 22q11.2DS-related pathology in dopaminergic neurons and may represent a candidate target for the development of therapeutic and preventive strategies for future diseases in dopaminergic neurons.
Contributors
Y.A and N.O. conceived the project and designed the experiments; Y.A. performed the experiments using iPSCs; T.S., R.S., A.A. and D.M. performed the experiments using mice; Y.A., E.S., I.K., D.M. analysed the data; and all authors wrote and reviewed the paper.
Declaration of Competing Interest
Dr. N. Ozaki has received research support or speakers’ honoraria from, or has served as a consultant to, Sumitomo Dainippon Pharma Co., Ltd, Otsuka Holdings Co., Ltd., Pfizer Japan Inc., The KAITEKI Institute, Inc., Eisai Co., Ltd., Astellas, Meiji Seika Pharma Co., Ltd., Janssen Pharmaceutical K.K., and Taisho Pharmaceutical Co., Ltd., outside the submitted work.
Acknowledgments
Acknowledgements
We thank the patients and their families for participating in this study. This study was supported by the AMED under grant nos JP20dm0107087, JP20dm0207075, JP 20ak0101113, JP20dk0307081 and JP18dm0207004h0005; the MEXT KAKENHI under grant nos. 16K19760, 19K08015, 18H04040 and 18K19511; and the Uehara Memorial Foundation under grant no. 201810122. The sponsors of this study did not have any role in the study design, data collection or analysis, interpretation of results, or manuscript preparation or submission.
Data sharing statement
The data that support the findings of this study are available from the corresponding authors upon reasonable requests.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103138.
Contributor Information
Yuko Arioka, Email: ariokay@med.nagoya-u.ac.jp.
Norio Ozaki, Email: ozaki-n@med.nagoya-u.ac.jp.
Appendix. Supplementary materials
References
- 1.Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maynard TM, Haskell GT, Peters AZ, Sikich L, Lieberman JA, LaMantia AS. A comprehensive analysis of 22q11 gene expression in the developing and adult brain. Proc Natl Acad Sci U S A. 2003;100:14433–14438. doi: 10.1073/pnas.2235651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butcher NJ, Chow EW, Costain G, Karas D, Ho A, Bassett AS. Functional outcomes of adults with 22q11.2 deletion syndrome. Genet Med. 2012;14:836–843. doi: 10.1038/gim.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kushima I, Aleksic B, Nakatochi M. High-resolution copy number variation analysis of schizophrenia in Japan. Mol Psychiatry. 2017;22:430–440. doi: 10.1038/mp.2016.88. [DOI] [PubMed] [Google Scholar]
- 5.Schneider M, Debbané M, Bassett AS. Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry. 2014;171:627–639. doi: 10.1176/appi.ajp.2013.13070864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun D, Ching CRK, Lin A. Large-scale mapping of cortical alterations in 22q11.2 deletion syndrome: convergence with idiopathic psychosis and effects of deletion size. Mol Psychiatry. 2020;25:1822–1834. doi: 10.1038/s41380-018-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butcher NJ, Kiehl TR, Hazrati LN. Association between early-onset Parkinson disease and 22q11.2 deletion syndrome: identification of a novel genetic form of Parkinson disease and its clinical implications. JAMA Neurol. 2013;70:1359–1366. doi: 10.1001/jamaneurol.2013.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mok KY, Sheerin U, Simón-Sánchez J. Deletions at 22q11.2 in idiopathic Parkinson's disease: a combined analysis of genome-wide association data. Lancet Neurol. 2016;15:585–596. doi: 10.1016/S1474-4422(16)00071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ardhanareeswaran K, Mariani J, Coppola G, Abyzov A, Vaccarino FM. Human induced pluripotent stem cells for modelling neurodevelopmental disorders. Nat Rev Neurol. 2017;13:265–278. doi: 10.1038/nrneurol.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattis VB, Svendsen CN. Induced pluripotent stem cells: a new revolution for clinical neurology. Lancet Neurol. 2011;10:383–394. doi: 10.1016/S1474-4422(11)70022-9. [DOI] [PubMed] [Google Scholar]
- 11.Toyoshima M, Akamatsu W, Okada Y. Analysis of induced pluripotent stem cells carrying 22q11.2 deletion. Transl Psychiatry. 2016;6:e934. doi: 10.1038/tp.2016.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Ryan SK, Deboer E. Mitochondrial deficits in human iPSC-derived neurons from patients with 22q11.2 deletion syndrome and schizophrenia. Transl Psychiatry. 2019;9:302. doi: 10.1038/s41398-019-0643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conforti P, Besusso D, Bocchi VD. Faulty neuronal determination and cell polarization are reverted by modulating HD early phenotypes. Proc Natl Acad Sci U S A. 2018;115 doi: 10.1073/pnas.1715865115. E762–71-e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen HN, Byers B, Cord B. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y, Kirwan P, Smith J, MacLean G, Orkin SH, Livesey FJ. A human stem cell model of early Alzheimer's disease pathology in Down syndrome. Sci Transl Med. 2012;4:124ra29. doi: 10.1126/scitranslmed.3003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sigurdsson T, Stark KL, Karayiorgou M, Gogos JA, Gordon JA. Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature. 2010;464:763–767. doi: 10.1038/nature08855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tripathi A, Spedding M, Schenker E, Didriksen M, Cressant A, Jay TM. Cognition- and circuit-based dysfunction in a mouse model of 22q11.2 microdeletion syndrome: effects of stress. Transl Psychiatry. 2020;10:41. doi: 10.1038/s41398-020-0687-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sumitomo A, Horike K, Hirai K. A mouse model of 22q11.2 deletions: molecular and behavioral signatures of Parkinson's disease and schizophrenia. Sci Adv. 2018;4 doi: 10.1126/sciadv.aar6637. eaar6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein MO, Battagello DS, Cardoso AR, Hauser DN, Bittencourt JC, Correa RG. Dopamine: functions, signaling, and association with neurological diseases. Cell Mol Neurobiol. 2019;39:31–59. doi: 10.1007/s10571-018-0632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arioka Y, Shishido E, Kubo H. Single-cell trajectory analysis of human homogenous neurons carrying a rare RELN variant. Transl Psychiatry. 2018;8:129. doi: 10.1038/s41398-018-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi K, Tanabe K, Ohnuki M. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 22.Arioka Y, Hirata A, Kushima I, Aleksic B, Mori D, Ozaki N. Characterization of a schizophrenia patient with a rare RELN deletion by combining genomic and patient-derived cell analyses. Schizophr Res. 2020;216:511–515. doi: 10.1016/j.schres.2019.10.038. [DOI] [PubMed] [Google Scholar]
- 23.Arioka Y, Shishido E, Kushima I, Mori D, Ozaki N. Cell body shape and directional movement stability in human-induced pluripotent stem cell-derived dopaminergic neurons. Sci Rep. 2020;10:5820. doi: 10.1038/s41598-020-62598-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kushima I, Aleksic B, Masahiro Nakatochi. Comparative analyses of copy-number variation in autism spectrum disorder and schizophrenia reveal etiological overlap and biological insights. Cell Rep. 2018;24:2838–2856. doi: 10.1016/j.celrep.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 25.Saito R, Koebis M, Nagai T. Comprehensive analysis of a novel mouse model of the 22q11.2 deletion syndrome: a model with the most common 3.0-Mb deletion at the human 22q11.2 locus. Transl Psychiatry. 2020;10:35. doi: 10.1038/s41398-020-0723-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyce M, Bryant KF, Jousse C. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 28.van Vliet AR, Agostinis P. PERK and filamin A in actin cytoskeleton remodeling at ER-plasma membrane contact sites. Mol Cell Oncol. 2017;4 doi: 10.1080/23723556.2017.1340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Vliet AR, Giordano F, Gerlo S. The ER stress sensor PERK coordinates ER-plasma membrane contact site formation through interaction with filamin-A and F-actin remodeling. Mol Cell. 2017;65 doi: 10.1016/j.molcel.2017.01.020. 885–-899.e6. [DOI] [PubMed] [Google Scholar]
- 30.Bellato HM, Hajj GN. Translational control by eIF2alpha in neurons: beyond the stress response. Cytoskeleton (Hoboken, NJ) 2016;73:551–565. doi: 10.1002/cm.21294. [DOI] [PubMed] [Google Scholar]
- 31.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verfaillie T, Rubio N, Garg AD. PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 2012;19:1880–1891. doi: 10.1038/cdd.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Da Silva JS, Dotti CG. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci. 2002;3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- 34.Dent EW, Kwiatkowski AV, Mebane LM. Filopodia are required for cortical neurite initiation. Nat Cell Biol. 2007;9:1347–1359. doi: 10.1038/ncb1654. [DOI] [PubMed] [Google Scholar]
- 35.Atkins C, Liu Q, Minthorn E. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2013;73:1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- 36.Ganz J, Shacham T, Kramer M. A novel specific PERK activator reduces toxicity and extends survival in Huntington's disease models. Sci Rep. 2020;10:6875. doi: 10.1038/s41598-020-63899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruch J, Xu H, Rösler TW. PERK activation mitigates tau pathology in vitro and in vivo. EMBO Mol Med. 2017;9:371–384. doi: 10.15252/emmm.201606664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt EK, Clavarino G, Ceppi M, Pierre P. Sunset, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 39.Hirabayashi Y, Kwon SK, Paek H. ER-mitochondria tethering by PDZD8 regulates Ca(2+) dynamics in mammalian neurons. Science. 2017;358:623–630. doi: 10.1126/science.aan6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kornmann B, Currie E, Collins SR. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paillusson S, Stoica R, Gomez-Suaga P. There's something wrong with my MAM; the ER-mitochondria axis and neurodegenerative diseases. Trends Neurosci. 2016;39:146–157. doi: 10.1016/j.tins.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takada I, Tsuchiya M, Yanaka K. Ess2 bridges transcriptional regulators and spliceosomal complexes via distinct interacting domains. Biochem Biophys Res Commun. 2018;497:597–604. doi: 10.1016/j.bbrc.2018.02.110. [DOI] [PubMed] [Google Scholar]
- 43.Elvira R, Cha SJ, Noh GM, Kim K, Han J. PERK-Mediated eIF2alpha Phosphorylation contributes to the protection of dopaminergic neurons from chronic heat stress in Drosophila. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21030845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sainath R, Gallo G. Cytoskeletal and signaling mechanisms of neurite formation. Cell Tissue Res. 2015;359:267–278. doi: 10.1007/s00441-014-1955-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Björklund A, Dunnett SB. Dopamine neuron systems in the brain: an update. Trends Neurosci. 2007;30:194–202. doi: 10.1016/j.tins.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 46.Broussard JI, Yang K, Levine AT. Dopamine regulates aversive contextual learning and associated in vivo synaptic plasticity in the hippocampus. Cell Rep. 2016;14:1930–1939. doi: 10.1016/j.celrep.2016.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lajiness-O'Neill RR, Beaulieu I, Titus JB. Memory and learning in children with 22q11.2 deletion syndrome: evidence for ventral and dorsal stream disruption? Child Neuropsychol. 2005;11:55–71. doi: 10.1080/09297040590911202. [DOI] [PubMed] [Google Scholar]
- 48.Cagnetta R, Wong HH, Frese CK, Mallucci GR, Krijgsveld J, Holt CE. Noncanonical modulation of the eIF2 pathway controls an increase in local translation during neural wiring. Mol Cell. 2019;73:474–489. doi: 10.1016/j.molcel.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Longo F, Mancini M, Ibraheem PL. Genetic reduction of PERK-eIF2α signaling in dopaminergic neurons drives cognitive and age-dependent motor dysfunction. bioRxiv. 2020;07 [Google Scholar]
- 50.Rutkowski TP, Purcell RH, Pollak RM. Behavioral changes and growth deficits in a CRISPR engineered mouse model of the schizophrenia-associated 3q29 deletion. Mol Psychiatry. 2019 doi: 10.1038/s41380-019-0413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trinh MA, Kaphzan H, Wek RC, Pierre P, Cavener DR, Klann E. Brain-specific disruption of the eIF2alpha kinase PERK decreases ATF4 expression and impairs behavioral flexibility. Cell Rep. 2012;1:676–688. doi: 10.1016/j.celrep.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 53.Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J Biol Chem. 2010;285:33165–33174. doi: 10.1074/jbc.M110.167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noma K, Goncharov A, Jin Y. Systematic analyses of rpm-1 suppressors reveal roles for ESS-2 in mRNA splicing in Caenorhabditis elegans. Genetics. 2014;198:1101–1115. doi: 10.1534/genetics.114.167841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z, Vuong JK, Zhang M, Stork C, Zheng S. Inhibition of nonsense-mediated RNA decay by ER stress. RNA. 2017;23:378–394. doi: 10.1261/rna.058040.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Usuki F, Yamashita A, Fujimura M. Environmental stresses suppress nonsense-mediated mRNA decay (NMD) and affect cells by stabilizing NMD-targeted gene expression. Sci Rep. 2019;9:1279. doi: 10.1038/s41598-018-38015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Julier C, Nicolino M. Wolcott-Rallison syndrome. Orphanet J Rare Dis. 2010;5:29. doi: 10.1186/1750-1172-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim P, Scott MR, Meador-Woodruff JH. Dysregulation of the unfolded protein response (UPR) in the dorsolateral prefrontal cortex in elderly patients with schizophrenia. Mol Psychiatry. 2019 doi: 10.1038/s41380-019-0537-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.