Abstract
The membrane proximal external region (MPER) of HIV-1 gp41 contains epitopes for at least four broadly neutralizing antibodies. Depending on solution conditions and construct design, different structures have been reported for this segment. We show that in aqueous solution the MPER fragment (gp160660–674) exists in a monomer-trimer equilibrium with an association constant in the micro-molar range. Thermodynamic analysis reveals that the association is exothermic, more favorable in D2O than H2O, and increases with ionic strength, indicating hydrophobically driven intermolecular interactions. Circular dichroism, 13Cα chemical shifts, NOE, and hydrogen exchange rates reveal that MPER undergoes a structural transition from predominately unfolded monomer at low concentrations to an α-helical trimer at high concentrations. This result has implications for antibody recognition of MPER prior to and during the process where gp41 switches from a pre-hairpin intermediate to its post-fusion 6-helical bundle state.
Keywords: HIV-1 gp41, MPER, self-association, monomer-trimer equilibrium, structural transition, NMR
Graphical abstract

gp41 MPER structural transition: HIV-1 gp41 undergoes large structural rearrangements to mediate membrane fusion. Its MPER segment, critical to soliciting neutralizing antibodies, is shown to spontaneously assemble into an α-helical trimer in aqueous solution.
Envelope protein (gp160 or gp120/gp41) of HIV-1 mediates viral entry into the host cell. The requisite viral membrane fusion process is initiated by large structural rearrangements of the glycoprotein gp160, triggered by its binding to the host cell receptor and co-receptor.[1–3] These rearrangements expose the gp41 ectodomain and fusion peptide, thereby enabling their interaction with the host-cell membrane and allowing gp41 to transition to the so-called pre-hairpin intermediate state. The membrane proximal external region (MPER) of gp41 connects the N-terminal ectodomain with the single transmembrane helix, anchored in the viral envelope. MPER harbors epitopes for at least four broadly neutralizing antibodies, and its structure has been widely studied.[3–5]
X-ray crystallographic studies in the presence of antibodies (2F5 and 10E8) revealed remarkably different structures of the MPER epitope. It adopts an extended β-turn conformation when bound to 2F5,[6] whereas it folds into two short α-helices when bound to 10E8.[4] Early NMR studies of MPER in the presence of DPC micelles showed a helix-turn-helix conformation.[7] On the other hand, X-ray crystallographic studies in the presence of SDS identified a coiled-coil trimer,[8] when stabilized by an engineered coiled-coil trimeric isoleucine helix zipper motif.[9] Although, a trimeric MPER arrangement was also observed when the peptide was fused at its N-terminus to the trimer-forming foldon domain of T4-fibrittin,[10] the MPER helices splayed apart in the presence of DPC detergent, exposing the C-terminal half of the peptide to the phospholipid surface.[11] By contrast, a recent study in the presence of phospholipid bicelles indicates that MPER forms a stable trimeric structure, with intermolecular contacts at both the N- and C-terminal ends, while the strands in the central region splay apart.[12] Hence, the different structures observed for MPER under varied conditions are indicative of considerable structural plasticity.
While prior studies have provided much structural information about MPER, either bound to antibodies or in the presence of detergents or lipids, structural studies in aqueous solution were limited by its poor solubility. Solubility increases either by truncating the hydrophobic stretch at the C-terminus or by extending the N-terminus with native gp41 residues.[13–16] These studies reported a variety of MPER structures, ranging from a mixture of 310 and α-helical conformations to dynamically disordered states. Here, we report that the 15-residue segment of MPER (gp160 residues 660–674) exists in a concentration-dependent dynamic monomer-trimer equilibrium. Whereas the monomeric state is largely unfolded, MPER self-associates into a homotrimeric α-helical bundle at higher concentrations. The ability of the lipophilic MPER segment to dislodge from the membrane is prerequisite for antibody recognition, and its inherent propensity to adopt an α-helical coiled coil has important implications for understanding viral entry, which may extend to other Class I viral fusion proteins.
A chemically synthesized N-terminal 15-residue fragment (gp160660–674) of MPER was utilized (Figure 1A), representing a. natural variant that contains Asn instead of Asp at position 664,[16] but conclusions drawn apply to both variants (see SI). Analogous to the “host-guest” system previously used for the hemagglutinin fusion peptide,[17] we added a “host” poly-Lys tag at the peptide’s C-terminus, which enhances its solubility to ≥14 mM in the absence of lipids or detergents. At 0.1 mM, the 1H NMR spectrum of MPER shows narrow spectral dispersion for its amide region (Figure S1). However, at higher peptide concentrations (0.35 mM to 14.1 mM) large chemical shift changes are observed, most clearly seen for the resolved Trp indole resonances, pointing to oligomerization (Figure S1). As a single resonance is observed for each indole 1H, the exchange rate is fast on the time scale of the chemical shift difference between monomeric and oligomeric states (ca 1000 rad/s for Trp-672). Natural 15N abundance 1H-15N HSQC NMR spectra also exhibit substantial amide chemical shift changes for nearly all MPER residues upon increasing the peptide concentration from 1.1 to 14 mM (Figure 1).
Figure 1.
Solution NMR study of MPER in H2O. (A) Amino acid sequence of the MPER (residues L660-N674) peptide used in this study, including an N-terminal acetylated Gly and five C-terminal Lys residues. (B) Overlay of the 1H-15N HSQC spectra of 14 mM (green), 3 mM (red) and 1.1 mM (blue) MPER. Assignments are marked for the 1.1 mM sample. (C) Changes in 1H and 15N chemical shifts when increasing MPER concentration from 1.1 mM to 3 mM (red), and from 1.1 mM to 14 mM (green). Spectra were collected at 600 MHz in 50 mM MES buffer, pH 6, 40 °C. The C-terminal poly-Lys residues are shown on a grey background.
The MPER peptide includes four Leu residues, located at the N-terminus (L660, L661 and L663) and at the central region (L669) of the peptide. Taking advantage of the high sensitivity of 1H-13C methyl group HSQC spectra, the Leu CδH3 resonances were used to probe the oligomerization over a wide concentration range. Although the analysis does not require this, tentative stereospecific assignments were made on the basis of the known chemical shifts differences between Leu Cδ1 and Cδ2 in disordered peptides (Figure 2A).[18] Upon increasing the peptide concentration, large chemical shift changes were observed for the methyl groups of L663 and L669.
Figure 2.
Trimerization of MPER peptide. (A) Overlay of excerpts from 1H-13C HSQC spectra obtained at different concentrations (0.1 mM-black, 1.1 mM-blue and 3 mM-red) of MPER in 50 mM MES (pH 6) at 35 °C. Assignments are shown for concentrations of 0.1 mM and 3 mM. (B) Methyl 1H chemical shifts for L663 and L669 as a function of MPER concentration in the absence (green) and presence of salts (blue). Global fitting of the data to a monomer-trimer equilibrium (solid lines) resulted in a ca 8-fold lower χ2 than monomer-dimer (dashed lines). (C) Van’t Hoff analysis of the monomer-trimer equilibrium over the 25 °C to 40 °C temperature range, in H2O buffer (green) and in D2O (purple). ΔH and ΔS were obtained from the linear fit to the data.
NMR measurements were carried out over a wide range of concentrations (0.01–14 mM) in order to distinguish between different possible modes of oligomerization, in particular dimer and trimer. A fit to the monomer-trimer model resulted in an eight-fold lower 𝜒2 value than a monomer-dimer model (Figure 2B). Global fitting for both residues L663 and L669 yields Ka = 1.8×105 M−2 at 35 °C, in MES buffer, increasing to 2.2 ×106 M−2 at moderate ionic strength.
It has long been recognized that hydrophobic intermolecular interactions are stabilized in D2O over H2O solutions.[19–22] For MPER, titrations as a function of peptide concentration in D2O (Figure S2A) showed about a two-fold increase in Ka over measurements in H2O (Figure S2B, Table 1) indicating that trimerization is more favorable by approximately −0.15 kcal.mol−1 (of monomer) in D2O than in H2O. Analysis of the methyl group chemical shifts over a range of temperatures (25 °C to 40 °C) provided access to thermodynamic parameters of the oligomerization process, both in H2O and D2O. The equilibrium is temperature dependent and favors trimeric species at lower temperature, as evidenced by an increase in Ka (Figure 2C). The linear dependence of self-association (ln Ka) on temperature indicates that the heat capacity does not significantly depend on temperature and that the oligomerization process is more exothermic by about −0.6 kcal.mol−1 in H2O than D2O. Enhanced Ka in both D2O and upon increasing ionic strength indicates that the association is largely driven by hydrophobic interactions.
Table 1.
Thermodynamic properties of MPER self-association.
| Buffer | Ka (105 M−2) | ΔG (kcal.mol−1) | ΔH (kcal.mol−1) | ΔS (cal.mol−1) |
|---|---|---|---|---|
| H2Oa | 1.8 ± 0.1 | −2.47 ± 0.01 | −4.7 ± 0.1 | −7.2 ± 0.5 |
| D2Oa | 3.8 ± 0.1 | −2.62 ± 0.01 | −4.13 ± 0.04 | −4.9 ± 0.1 |
| Salts[b] | 22 ± 1 | −2.98 ± 0.02 | NA | NA |
Values for Ka and ΔG values are for 35°C, pH 6. ΔG, ΔH and ΔS are per mole of monomer. ΔH and ΔS were derived from van’t Hoff analysis (Figure 2C).
50 mM MES, pH 6.
20 mM sodium phosphate (pH 6.0), 50 mM NaCl, H2O.
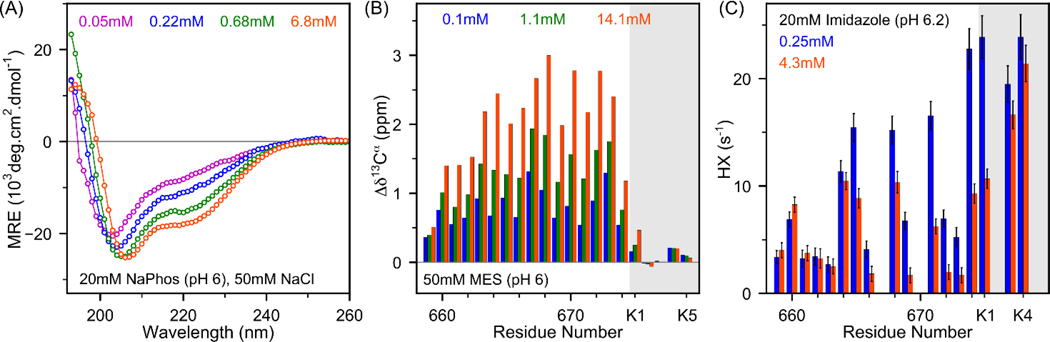
Earlier NMR studies of MPER pointed to the presence of a stable monomeric 310 helix in water (gp41659–671) [13], and substantial helical propensity for a longer fragment (gp41636–677) led to the proposal that this region constitutes an autonomous folding unit that potentially serves as a nucleation site for protein folding.[16] Subsequent NMR, circular dichroism (CD) and molecular dynamics studies of analogous peptides reported a more dynamic, disordered structure.[14–15] To obtain further insights into the origins of our concentration-dependent NMR spectral changes, we also carried out CD measurements as a function of concentration. Analysis of these data confirms that the helical signature strongly increases when the concentration is raised from 0.025 mM ([θ]222 ≈−6350 deg cm2 dmol−1) to 6.8 mM ([θ]222 ≈−17500 deg cm2 dmol−1), corresponding to an increase in α-helicity from ~19% to ~53%, incl. its poly-Lys tag (Figure 3A and S3A-B). A non-linear least-squares fit of [θ]222 as a function of concentration to a monomer-trimer model yields Ka ≈ 1.3 × 107 M−2 at 25 °C, in good agreement with the NMR-derived value (Figure 2B), in particular when taking the temperature dependence of Ka into account (Figure 2C).
Figure 3.
Secondary structure of MPER. (A) Far-UV CD spectra at concentrations ranging from 0.05 to 6.8mM of MPER at 25 °C (see Figure S3 for a complete titration series). (B) Residue specific secondary Δδ13Cα chemical shifts of MPER (0.1 mM – blue; 1.1 mM – green; 14.1 mM – orange) at 35 °C. (C) Hydrogen exchange rates (HX) of 0.25 mM and 4.3 mM MPER at 35 °C. Significant reduction in HX rates at higher concentration (4.3 mM) is indicative of increased α-helical H-bond formation.[23] Buffers used are indicated. C-terminal poly-Lys residues are shown on a grey background.
To obtain residue-specific secondary structure information, 13Cα NMR chemical shifts were analyzed over a wide range of concentrations (0.1 mM to 14 mM) (Figure S4). The deviation in 13Cα chemical shifts (Δδ13Cα) from random coil values provides information about local secondary structure. At 0.1 mM, MPER displays narrow resonances with moderate, positive Δδ13Cα secondary chemical shift values (0.5–1.25 ppm), confirming the previously reported intrinsic propensity of the monomeric peptide to adopt helical structure (Figure 3B).[13–16] At 14 mM, the Δδ13Cα values approximately double, indicative of a strong increase in helicity. The Δδ13Cα values of MPER at intermediate concentrations (~0.5–1 mM, Figure 3B and S11D) agree fairly well with a prior solution NMR study of a biosynthetically prepared, 13C-enriched longer segment that encompassed the entire sequence of the antiviral drug fuzeon.[16] Analysis of HN, N, Cα and Hα chemical shifts by TALOS-N software[24] shows that at high concentration (14.1 mM) the MPER chemical shifts of residues L661-F673 are consistent with α-helical secondary structure (Figure S5).
Increased population of secondary structure was further validated by NMR-based hydrogen exchange (HX) experiments. Amide protons in dynamically disordered regions exchange rapidly with solvent,[25] while amides engaged in H-bonds are protected from HX.[26] Residue-specific HX rates for the strongly overlapping amide resonances commonly are measured from 1H-15N HSQC measurements, which were not feasible for the natural abundance samples used in our study. Instead, we obtained these HX rates by monitoring the amide 1H (F1) cross peaks to aliphatic (F2) cross peaks in a 2D 1H-1H TOCSY spectrum,[27–28] in the absence and presence of inversion of H2O magnetization prior to the TOCSY pulse sequence (Figure S6). Monomer populations of ca. 98% and 43% are expected for sample concentrations of 0.25 mM and 4.3 mM, respectively. Comparison of HX rates measured at 0.25 mM and 4.3 mM concentrations (Figure 3C, Figure S7 and Table S1) shows an average 2.7-fold reduction in HX rates for all amides that donate intramolecular H-bonds when adopting helical structure (W665-N674).
Our results conclusively point to an increase in α-helical structure with concentration. However, differentiating α-helix from 310 helix on the basis of 13Cα chemical shifts or CD spectra can be challenging. To resolve this ambiguity, a 2D NOESY spectrum was recorded which shows dαN(i, i+4) interactions characteristic of α-helix but absent in 310 helix. (Figure S8A). At low concentration (0.25 mM), no NOE/ROE connectivities beyond dαN(i,i+2) were observed, pointing to a dynamic transient population of helical structure[14–15] that may include 310 helix (Figure S8B-C).[13]
To map critical regions essential for oligomerization, shorter versions of the MPER peptide were also examined. A peptide lacking the N-terminal Leu660-Leu661 residues (MPERΔN) showed a 4-fold reduction in Ka (Figure S9). A further exclusion of Trp672-Asn674 (MPERtrunc) yielded NMR spectra that were independent of concentration over 0.1– 2.3 mM range (Figure S10), indicating that these C-terminal residues are essential to trimerization. A MPER peptide that contains Asp at position 664 (MPERD664), as in the antiviral drug fuzeon,[16] also exhibited the monomer-trimer equilibrium but with an 8-fold increase in Ka, accompanied by a small increase in intrinsic helical propensity of the monomeric state, as judged by 13Cα chemical shifts and CD under dilute conditions (Figure S11).
Although, at first sight, the micromolar Ka values we report for MPER may appear high from a biological perspective, it is important to note that other structural elements of gp41, notably the N-terminal heptad repeat region of its ecto domain (residues L544-L582) are generally considered to remain homotrimeric during the entire viral fusion process.[29] Hence, three MPER peptides are physically restrained to remain located in a sphere with a radius of roughly 100 Å, which then corresponds to an effective MPER concentration of ca 1 mM, well within the range relevant for trimerization.
Helical trimer formation has previously been observed for considerably longer “isoleucine-zipper” motifs.[9] The ability of MPER to adopt a helical trimeric arrangement when fused to such a motif was demonstrated by X-ray crystallography.[8] Our observation that the HIV-1 gp41 MPER fragment of only 15 residues spontaneously adopts such an arrangement confirms that this property is of likely significance in gp41’s role in membrane fusion. The precise role of MPER in the all-important structural transition of the gp41 ectodomain from its pre-fusion intermediate to its well-established post-fusion homotrimeric six-helical bundle state remains unknown. Our data clearly indicate a strong propensity for this MPER region to adopt a three-helical bundle motif, which may stabilize the early state of the gp41 during its structural transition.
HIV-1 envelope protein gp160 is a class I viral fusion protein with structural and functional homology to other well-studied viral fusion proteins such as influenza hemagglutinin and Ebola virus envelope protein.[30–31] For both virions, the dynamic properties of their membrane proximal regions have been implicated in antibody binding.[30–31] The SARS-CoV-2 Spike protein is also a member of the class I viral fusion proteins, and it therefore appears likely that similar considerations will apply for its membrane proximal region.
Supplementary Material
Acknowledgements
We thank G. Abdoulaeva, J. Ying, and Y. Shen for technical support and D.A. Torchia and J. Courtney for valuable suggestions. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M, Nature 1995, 373, 123–126. [DOI] [PubMed] [Google Scholar]
- [2].Ryu SE, Kwong PD, Truneh A, Porter TG, Arthos J, Rosenberg M, Dai XP, Xuong NH, Axel R, Sweet RW, Hendrickson WA, Nature 1990, 348, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H, Aids Research and Human Retroviruses 2001, 17, 1757–1765. [DOI] [PubMed] [Google Scholar]
- [4].Huang JH, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang YP, Zhang BS, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M, Nature 2012, 491, 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cerutti N, Loredo-Varela JL, Caillat C, Weissenhorn W, Current Opinion in Hiv and Aids 2017, 12, 250–256. [DOI] [PubMed] [Google Scholar]
- [6].Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD, J. Virol 2004, 78, 10724–10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sun ZYJ, Oh KJ, Kim MY, Yu J, Brusic V, Song LK, Qiao ZS, Wang JH, Wagner G, Reinherz EL, Immunity 2008, 28, 52–63. [DOI] [PubMed] [Google Scholar]
- [8].Liu J, Deng Y, Dey AK, Moore JP, Lu M, Biochemistry 2009, 48, 2915–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Harbury PB, Kim PS, Alber T, Nature 1994, 371, 80–83. [DOI] [PubMed] [Google Scholar]
- [10].Meier S, Guthe S, Kiefhaber T, Grzesiek S, J. Mol. Biol 2004, 344, 1051–1069. [DOI] [PubMed] [Google Scholar]
- [11].Reardon PN, Sage H, Dennison SM, Martin JW, Donald BR, Alam SM, Haynes BF, Spicer LD, Proc. Natl. Acad. Sci. U. S. A 2014, 111, 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fu QS, Shaik MM, Cai YF, Ghantous F, Piai A, Peng HQ, Rits-Volloch S, Liu ZJ, Harrison SC, Seaman MS, Chen B, Chou JJ, Proc. Natl. Acad. Sci. U. S. A 2018, 115, E8892–E8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Biron Z, Khare S, Samson AO, Hayek Y, Naider F, Anglister J, Biochemistry 2002, 41, 12687–12696. [DOI] [PubMed] [Google Scholar]
- [14].Barbato G, Bianchi E, Ingallinella P, Hurni WH, Miller MD, Ciliberto G, Cortese R, Bazzo R, Shiver JW, Pessi A, J. Mol. Biol 2003, 330, 1101–1115. [DOI] [PubMed] [Google Scholar]
- [15].Tulip PR, Gregor CR, Troitzsch RZ, Martyna GJ, Cerasoli E, Tranter G, Crain J, J. Phys. Chem. B 2010, 114, 7942–7950. [DOI] [PubMed] [Google Scholar]
- [16].Biron Z, Khare S, Quadt SR, Hayek Y, Naider F, Anglister J, Biochemistry 2005, 44, 13602–13611. [DOI] [PubMed] [Google Scholar]
- [17].Han X, Tamm LK, Proc. Natl. Acad. Sci. U. S. A 2000, 97, 13097–13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Beck Erlach M, Koehler J, Crusca E, Munte CE, Kainosho M, Kremer W, Kalbitzer HR, J. Biomol. NMR 2017, 69, 53–67. [DOI] [PubMed] [Google Scholar]
- [19].Lee JJ, Berns DS, Biochem. J 1968, 110, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Woodfin BM, Henderson RF, Henderson TR, J. Biol. Chem 1970, 245, 3733–3737. [PubMed] [Google Scholar]
- [21].Aune KC, Goldsmith LC, Timasheff SN, Biochemistry 1971, 10, 1617–1622. [DOI] [PubMed] [Google Scholar]
- [22].Eginton C, Beckett D, Biochemistry 2013, 52, 6595–6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Englander SW, Mayne L, Proc. Natl. Acad. Sci. U. S. A 2017, 114, 8253–8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shen Y, Bax A, J. Biomol. NMR 2013, 56, 227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bai Y, Milne JS, Englander SW, Proteins 1993, 17, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Englander SW, Downer NW, Teitelbaum H, Annu. Rev. Biochem 1972, 41, 903–924. [DOI] [PubMed] [Google Scholar]
- [27].Braunschweiler L, Ernst RR, J. Magn. Reson 1983, 53, 521–528. [Google Scholar]
- [28].Davis DG, Bax A, J. Am. Chem. Soc 1985, 107, 2820–2821. [Google Scholar]
- [29].Harrison SC, Nat. Struct. Mol. Biol 2008, 15, 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Benton DJ, Nans A, Calder LJ, Turner J, Neu U, Lin YP, Ketelaars E, Kallewaard NL, Corti D, Lanzavecchia A, Gamblin SJ, Rosenthal PB, Skehel JJ, Proc. Natl. Acad. Sci. U. S. A 2018, 115, 10112–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lee J, Nyenhuis DA, Nelson EA, Cafiso DS, White JM, Tamm LK, Proc. Natl. Acad. Sci. U. S. A 2017, 114, E7987–E7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.