Abstract
Clinical development of bromodomain and extra‐terminal (BET) protein inhibitors differs from the traditional course of drug development. These drugs are simultaneously being evaluated for treating a wide spectrum of human diseases due to their novel mechanism of action. BET proteins are epigenetic “readers,” which play a primary role in transcription. Here, we briefly describe the BET family of proteins, of which BRD4 has been studied most extensively. We discuss BRD4 activity at latent enhancers as an example of BET protein function. We examine BRD4 redistribution and enhancer reprogramming in embryonic development, cancer, cardiovascular, autoimmune, and metabolic diseases, presenting hallmark studies that highlight BET proteins as attractive targets for therapeutic intervention. We review the currently available approaches to targeting BET proteins, methods of selectively targeting individual bromodomains, and review studies that compare the effects of selective BET inhibition to those of pan‐BET inhibition. Lastly, we examine the current clinical landscape of BET inhibitor development.
Keywords: BD‐selective, BET, BETi, bromodomain (BD), enhancer
1. INTRODUCTION
A new paradigm is emerging in drug development, centered on the regulation of epigenetic processes for treating a wide spectrum of human diseases. Epigenetic processes involve alterations to gene expression without altering the genetic code. Bromodomain and extra‐terminal (BET) proteins are epigenetic “readers” that recognize and bind posttranslational modifications on histones and other transcriptional machinery to facilitate gene expression. BET proteins have received an increasing amount of attention in recent years, 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 and BET inhibitor(s) (BETi) that target the BET bromodomains (BD) are unique in drug development in that they are consecutively undergoing clinical investigation in vastly different disease areas. Evidence is mounting to support the use of BETi in treating cancer, metabolic, inflammatory, neurologic, cardiovascular, and musculoskeletal diseases. 1 , 2 , 3 , 14 , 19 , 20 , 29 , 31 , 44 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 Investigation of BETi potential as an antiviral has also recently garnered interest. 63 , 64 , 65 , 66 , 67
Unlike other reviews in the field, we focus this review on explaining why targeting BET proteins shows potential benefit in seemingly unrelated disease states. 3 , 9 , 13 , 29 , 33 , 39 , 68 , 69 , 70 , 71 , 72 , 73 , 74 We begin with a brief description of BET protein structure and function. A significant amount of what is currently known about BET proteins was initially discovered in the fields of embryonic development and cancer. This study has led to the identification of BET proteins as key components of “superenhancers” (SE); chromatin structures comprised of clusters of enhancer regions designed to drive transcription. 75 , 76 An understanding of SEs leads into description of similar structures, termed “latent enhancers.” These chromatin structures function in terminally differentiated cells to alter gene expression in disease. BET proteins, BRD4 being the best understood, play prominent roles in the development and progression of cardiovascular, metabolic, and inflammatory diseases. We present the landmark studies in these areas which demonstrate how BET proteins perpetuate disease at the transcriptional level, and provide the preliminary evidence for therapeutic potential of BETi in these areas. These studies support our explanation of why BETi benefit broad therapeutic categories, based on the intrinsic ability of cells from different lineages to respond to disease through evolutionarily conserved programs. We also describe various approaches to BET inhibition, what is known about the differential effects of pan‐ versus selective BD binding, and evidence supporting the use of BD‐selective BETi, particularly in areas outside of cancer. BD‐selective BET inhibition and its potential as a novel therapeutic approach is gaining ground within epigenetic drug development.
2. BROMODOMAIN AND EXTRA‐TERMINAL (BET) PROTEINS
BET proteins belong to a superfamily of bromodomain‐containing proteins (46 members containing 61 BDs), within which they comprise a subfamily of 4 members; BRD2, BRD3, BRD4, and testes‐specific BRDT. 77 The basic structure of BET proteins is comprised of two tandem ~110 amino acid bromodomains (BD1 and BD2) and an extra‐terminal domain, with BRD4 and BRDT including a C‐terminal motif (CTM) (Figure 1). As epigenetic “readers” BET proteins bind to their natural ligand, acetylated lysine, a posttranslational modification found on histone tails and transcription factors. 78 Exceptionally high concentrations of acetylated lysines are found on active chromatin; open regions of DNA containing genes that are accessible for transcription. 69 , 72 , 78 , 79 , 80 , 81 , 82 The binding of BET proteins to acetylated lysines on active chromatin plays an important role in gene transcription, as the BET proteins form docking scaffolds for transcription factors and other transcriptional machinery. In this way, BET proteins directly link epigenetic modifications to gene expression. Here, we describe some of the better‐known functions of each BET protein. Little is known about BRD2, BRD3, and BRDT, while the most well‐studied BET protein is BRD4.
Figure 1.
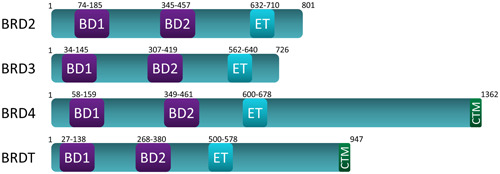
Schematic representation of basic domain organization of human BET (bromodomain and extra‐terminal domain) family proteins; BRD2, BRD3, BRD4, and BRDT. BET proteins contain two ~110 amino acid bromodomains (BD1 and BD2) and an extra‐terminal domain (ET). Only BRD4 and BRDT have a C‐terminal motif (CTM) [Color figure can be viewed at wileyonlinelibrary.com]
BRD2 modulates gene transcription in connection with cell cycle activity of proliferating cells in embryonic development, neural development, and in cancers/tumors at later stages of development. 83 , 84 During embryogenesis, Brd2‐deficient mice exhibit delayed fibroblast proliferation, neural tube closure defects, and overall delayed growth. 83 , 84 Though all BET proteins are highly structurally conserved and associate with numerous common proteins, BRD2's interactome displays the least overlap with other BET proteins. 30 For example, studies have shown that BRD2 primarily associates with E2F transcription factors. 22 , 85 , 86 In fibroblasts transfected with BRD2, interruption of BRD2 interactions with E2F negatively regulated E2F‐dependent cell differentiation and proliferation genes, such as cyclins A, D1, and E, which are required for the G1/S transition. 86 , 87 In addition to E2F, BRD2 interactions with RNA polymerase II (RNA Pol II), a complex that transcribes DNA into messenger, small nuclear and microRNA, are involved in transcription elongation in embryonic kidney cells. 87 BRD2's lack of a CTM indicates that its interactions with RNA Pol II are independent of positive transcription elongation factor b (PTEF‐b), and thus differ from those of BRDT and BRD4 (described below). LeRoy et al. showed that BRD2 acts by chaperoning RNA Pol II through hyperacetylated nucleosomes that typically act as barriers to elongation, thus allowing for transcription. 71 , 87 In later stages of development, overexpression of BRD2 in adult mice has been shown to promote B‐cell expansion and mitogenesis through interactions at the cyclin A promoter, leading to the development of B‐cell lymphoma and leukemia. 88 , 89
Known functions of BRD3, like BRD2, are related primarily to regulating the transcription of genes necessary for embryonic stem cell (ESC) development. BRD3 plays a role in erythroid cell differentiation through interactions with master erythroid transcription factor GATA‐binding factor 1 (GATA1). 30 , 90 , 91 Disruption of BRD3‐GATA1 binding in erythroid progenitor cells results in impaired GATA1‐mediated cell maturation. 90 Additionally, a number of negative transcriptional regulators and chromatin remodelers preferentially associate with BRD3, such as the nucleosome remodeling and deacetylase (NuRD) complex. 30 BRD3 interactions with NuRD are closely linked to interactions with GATA‐1, and thus are implicated in erythroid cell maturation. 30 , 92 BRD3 also interacts with RNA Pol II where it promotes transcription in a P‐TEFb independent manner, similar to BRD2. 30 BRD3 in cancer cell lines may be antiproliferative, in contrast to commonly noted functions of other BET proteins in these cell types. 30 However, further research is needed to fully understand potential BRD3 antiproliferative effects in various cell types and stages of development.
BRDT expression is restricted to the testis. BRDT knockout or mutation in mice leads to lower sperm count and abnormal sperm morphology, indicating its role in spermatogenesis. 93 , 94 , 95 BRDT exhibits the least number of binding interactions with transcriptional regulators of all the BET proteins, associating primarily with the negative elongation factor (NELF) complex through its CTM binding with PTEF‐b. 30 These findings are consistent with BRDT's organ specific localization, and its role in both facilitation of spermatogenic gene expression such as cyclin A1, Y‐box protein 2, serine/threonine‐protein kinase Plk1, aurora kinase C, and A‐kinase anchor protein 4, and repression of genes that require silencing in this process, including nuclear RNA export factor 2 and testes‐expressed gene 11. 93 , 94 , 96 , 97
Among the BET protein family members, the BRD4 protein interactome and functionality have been most extensively studied. One of BRD4's many roles, which may be shared with BRD2, 98 lies in maintaining “mitotic memory” during cell cycle progression by remaining bound to chromatin at essential genes during mitosis. This allows transcriptional activity of these genes to be quickly reestablished as the cell cycle progresses to late‐ and post‐mitotic phases. 69 , 82 , 99 , 100 The critical role of BRD4 in fundamental cellular processes early in development is also apparent in homozygous BRD4 knockout mice, where BRD4 knockout is embryonically lethal before implantation. 101 BRD4, and likely other BET proteins, interact with histone acetyltransferases, deacetylases, and other chromatin remodelers. 102 , 103 , 104 Additionally, and possibly most importantly, BRD4 is involved in the recruitment of transcriptional machinery to actively transcribed genes through its interactions with RNA Pol II and PTEF‐b (via its CTM, similar to BRDT), and numerous transcription factors and transcriptional coactivators. 30 , 105 , 106 , 107 , 108 BRD4 becomes the scaffold holding transcriptional complexes together at open chromatin, thereby facilitating transcription. 99 The central role of BRD4 in transcriptional regulation sets the stage for our discussion of its importance in driving disease‐related transcriptional modifications.
Though we will focus the following discussion on BRD4, the BET protein for which the most is currently known, other BET proteins may play similar or opposing roles in these processes and are likely more important than currently understood. It is important to keep in mind that all four BET proteins exhibit overlap in interactors and in functionality in various cell types. 30
2.1. The role of BRD4 in transcription
Through its primary interactions with epigenetic coregulators, BRD4 plays a number of important roles in gene transcription. First, BRD4 is thought to assist chromatin de‐compaction through histone acetyltransferase activity and association with other histone acetyltransferases, deacetylases, and chromatin remodelers. 82 , 102 , 103 , 104 For example, at enhancer regions in ESCs, BRD4 associates with the histone acetyltransferases P300 and CBP to enhance H3K27 acetylation. 102 Next, with access to the active chromatin, BRD4 binds to acetylated histones at enhancers, promoters, and transcriptional start sites. Binding to its natural ligand, acetylated lysine, on histones and transcription factors creates a scaffold for transcriptional machinery to come together. Mediator, a complex that transduces signals from transcription factors and activators at enhancers to promoters, is recruited by BRD4, which brings together all components of the active transcriptional complex; BRD4, Mediator, transcription factors, and RNA Pol II. 109 , 110 , 111 With the active transcriptional complex in place, BRD4's CTM binding with PTEF‐b results in the phosphorylation of RNA Pol II (at serine 5) to initiate transcription 108 , 112 , 113 , 114 , 115 , 116 (Figure 2A). Following transcription initiation, BRD4 helps guide the complex along an active gene. Approximately 100 base pairs downstream of the transcription start site, where RNA Pol II pauses, 113 , 117 phosphorylation of RNA Pol II serine 2, permitted by maintained interaction between PTEF‐b and BRD4, releases RNA Pol II pausing and advances elongation of the RNA transcript. 117 , 118 Therefore, BRD4's involvement in chromatin decompaction, recruitment of transcriptional complex components, as well as in initiation, pause release, and elongation stages of transcription makes it crucial in regulating gene expression. 61 , 72 , 99 , 119 The central role of BRD4 in these basic yet fundamental cellular processes underscores its importance in many aspects of cellular function.
Figure 2.
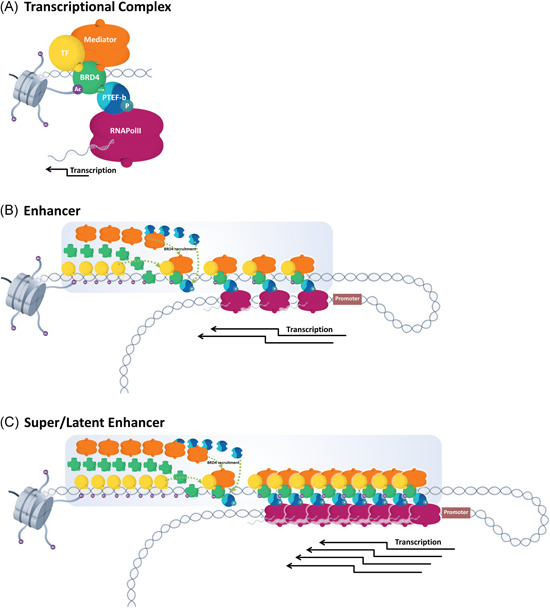
A, Transcriptional complex components. Ac, acetylated lysine; BRD4, bromodomain and extraterminal protein 4; P, phosphorylation; PTEFb, positive transcription elongation factor b; RNA Pol II, RNA polymerase II; TF, transcription factor. B, At typical enhancers, BRD4 binds its ligand, Ac, and becomes a scaffold for recruiting Mediator, transcription factors, and PTEFb to phosphorylate RNA Pol II at the target gene promoter, thereby initiating transcription. C, Latent and super‐enhancers are densely packed with transcriptional complex components. The abundance of transcriptional machinery and BRD4 result in elevated levels of gene transcription [Color figure can be viewed at wileyonlinelibrary.com]
2.2. BRD4 at super‐enhancers
Super‐enhancers (SEs) are chromatin structures comprised of clusters of enhancers that are densely packed with BRD4, Mediator, transcription factors, acetylated histones, and coactivators that span regions of actively transcribed DNA. 75 , 111 , 120 While typical enhancers are approximately 100 base pairs in length, SEs can span as much as 50 kb pairs and contain significantly larger amounts of enhancer elements 111 (Figure 2B,C). SEs are located proximal to their target gene promoters, typically within 100 kb, and ensure that the transcriptional tool kit stays in close proximity to pre‐selected gene promoter regions, priming them for gene responsiveness. 121 SEs were first described by Whyte et al. 111 in areas of the genome that are essential for lineage commitment in pluripotent ESCs. This developmental differentiation process involves a coordinated program requiring abundant expression of a well‐defined, but limited panel of genes, including lineage‐determining genes octamer‐binding transcription factor 4 (OCT4), sex determining region Y‐box transcription factor 2 (SOX2) and homeobox protein NANOG. 122 In ESCs, SEs boost the expression of these lineage‐determining genes to levels needed to define cell identity. 75 , 111 BRD4 aids in the formation of ESC SE sites through its roles in chromatin decompaction, neutralizing positive charges on histones and other acetylated lysines, and recruiting the transcriptional machinery needed for rapid and abundant transcription. 75 , 109 , 123 Hnisz et al. 75 soon after expanded on Whyte's definition of SEs by examining 86 human cell and tissue types, and determining that SEs are involved in cellular processes beyond development, namely those involved in cancer. For example, elevated levels of histone acetylation (particularly H3K27ac), BRD4, and Mediator are present at oncogenic SEs and are often used for SE identification. As at lineage determining SEs, BRD4 and Mediator aid in chromatin remodeling and the aggregation of transcriptional machinery, and BRD4's CTM binding with PTEF‐b releases RNA Pol II pausing, advancing the elongation of oncogene transcripts. 76 These SEs form in proximity to well‐known oncogenes like the MYC proto‐oncogene (MYC), Immunoglobulin Lambda Like Polypeptide 5 (IGLL5), Interferon Regulatory Factor 4/Multiple Myeloma Oncogene 1 (IRF4), and X‐Box Binding Protein 1 (XBP1) in multiple myeloma cells leading to their overexpression. In contrast to lineage determining SEs, oncogenic SEs facilitate maladaptive gene expression, resulting in tumor expansion and disease progression. With the loss of BRD4 function or availability comes the preferential loss of other key transcriptional coregulators from SEs, including Mediator and PTEF‐b, repressing the transcriptional overexpression of oncogenes. 13 , 76 , 124 This finding further underscores the central role of BRD4 in SE‐driven transcriptional upregulation, without it, overexpression does not occur.
2.3. Latent enhancers
Both cellular differentiation and carcinogenesis rely on extensive transcriptional reprogramming that requires recruitment of massive amounts of transcriptional machinery to significantly alter expression of the specific genes which underlie these processes. 75 , 76 , 111 In metabolic, inflammatory, and cardiovascular diseases, transcriptional reprogramming also occurs through similar underlying processes. 4 , 14 , 53 , 56 , 75 When the cellular environment of terminally differentiated cells is disturbed by disease or other noxious stimuli, the cells must respond to the stressor. This response mechanism is the cells' intrinsic ability to alter chromatin structure and form new enhancers in proximity to response genes. 75 , 120 , 125 This “cellular plasticity” affords terminally differentiated cells the ability to facilitate their own transcriptional reprogramming in response to stimuli. Newly created enhancers form at genes that are then quickly expressed as the cell's response to the perturbation.
In response to a novel stimulus, the cell forms a new enhancer in proximity to the response gene promoter. With a single stimulus, enhancer elements at this genomic location may dissolve over time; however, a memory for that event is created speeding the response process for future insults. These primed enhancer regions are referred to as “latent” enhancers and are likely to play a key role in response to repeated cellular insults associated with disease. 120 , 126 , 127 Ostuni et al. 126 have outlined the characteristics of latent enhancers in terminally differentiated cells, distinguishing them from typical enhancers and SEs. Latent enhancers were defined in terminally differentiated cells as genomic regions that acquire the histone marks associated with typical enhancers or SEs in response to stimulation. In mouse bone marrow–derived macrophages, Ostuni et al. found that upon stimulation with lipopolysaccharide (LPS) and various other noxious stimuli, genomic regions that were unmarked in basal conditions acquired these marks, and thus were termed latent enhancers. In fact, identifying features of latent enhancers are similar to those for SEs, such as H3K27ac, 75 , 126 Mediator, 76 , 111 , 120 and BRD4. 75 The response to LPS was time‐dependent in that the H3K27ac mark was present with 4 h LPS stimulation, but by 24 h stimulation H3K27ac had significantly reduced to near basal levels, suggesting that H3K27ac acts as an indicator of active enhancer regions. Additionally, H3K27ac appeared at both overlapping and unique genomic locations in response to various stimuli (LPS vs. TGFβ vs. IL‐4, etc.). Ostuni et al. also noted that each stimulus induced a signature memory of histone methylation (H3K4me1) that persisted for days, and instilled a hyperresponsive state, in that subsequent stimulation resulted in faster and often greater histone acetylation than at previously unstimulated and unmarked genomic regions. Genes adjacent to the latent enhancer regions were also induced faster. Latent enhancers, in effect, afford the cell an opportunity to respond to a stimulus that it would be otherwise unable to respond to. Histone “marks” form a memory or “epigenomic signature” of the stimuli the cell has previously encountered, thus enabling it to respond quicker to the same and often similar stimuli later on. 126 Similar findings in CD4+ T cells showed that BRD4 occupancy at genomic regions activated by stimulation correlated with several different histone acetylation marks (H4K5, H4K8, H3K9) including the enhancer identifier H3K27ac. 108 The pattern of BRD4 co‐localization with various histone acetylation marks and other enhancer identifying factors such as Mediator, overlaps with regions identified as latent enhancers.
Evidence shows that BRD4 maintains its role in recruiting transcriptional machinery to these latent enhancers, perpetuating aberrant gene expression in response to disease stimuli. 82 Though the stimuli, transcription factors, and other coactivators may differ based on the disease conditions and cell type, BRD4 (or other BET proteins) are consistently found at latent enhancers across cell types. In terminally differentiated cells, latent enhancer formation can lead to maladaptive gene overexpression, particularly in disease states where there is recurring or sustained noxious stimuli within the cellular environment. This knowledge leads us to next focus on the studies that support the role of latent enhancers and BRD4's contribution to upregulating maladaptive gene expression at these regions in diseases outside of cancer. These studies are also some of the first to show the potential of BETi in diseases such as atherosclerosis, heart failure (HF), arthritis, and metabolic disorders through examination of disease‐relevant cell types: endothelial cells, cardiac myocytes, immune cells, and adipose tissue. 4 , 14 , 29 , 53 , 56 , 128 , 129 , 130 , 131 , 132
3. BET INHIBITORS IN CARDIOVASCULAR, AUTOIMMUNE, AND METABOLIC DISEASES
In a hallmark study, Brown et al. 56 showed that BRD4 and latent enhancer formation play a significant role in atherosclerotic processes in vascular endothelial cells, a cell type that contributes tremendously to both the development and progression of the disease. 44 , 133 , 134 In these cells, inflammatory stimuli result in translocation of transcriptional regulator nuclear factor kappa‐light‐chain‐enhancer of activated B cell (NF‐κB) from the cytoplasm to the nucleus. NF‐κB engages in cross‐talk with chromatin remodeling machinery, and its subunit v‐rel avian reticuloendotheliosis viral oncogene homolog A (RELA or p65) binds to its consensus sequence near the transcription start site of proinflammatory genes. RELA binding, and subsequent acetylation, mark sites for latent enhancer formation. BRD4 is redistributed from genes associated with resting endothelial cell state, such as tyrosine kinase (TEK), to the latent enhancer regions. These latent enhancers are formed within 50 kb pairs of inflammation‐responsive genes, such as the cytokine C‐C motif chemokine ligand 2 (CCL2), as well as cell adhesion genes that drive atherosclerotic plaque formation such as E‐selectin (SELE) and vascular cell adhesion molecule (VCAM1). These regions then become flooded with transcriptional machinery, increasing expression of these and other response genes. 56 Treatment of stimulated cells with BETi interrupts BRD4 redistribution to the CCL2 and other response gene promoter regions, which ultimately reduces their expression compared to that of untreated, stimulated cells. Brown et al. 56 demonstrate that disease‐driven master transcription factors, such as NF‐κB, induce rapid transcriptional responses through redistribution of BRD4 and other latent enhancer‐binding factors to promote transcription of a new set of inflammatory response genes. 56 Their data also show that inhibition of BRD4 binding to the latent enhancer region of inflammatory and cell adhesion gene promoters in vitro is sufficient to interrupt aberrant transcription. In vivo, BETi also blocks the development of atherosclerosis in mouse models, showing the functional aspect of transcription regulation by BETi. 44 , 56 These groundbreaking discoveries are some of the first to show the prominence of BET proteins in inflammation‐related vascular disease, and allude to BETi potential in therapeutic areas outside of cancer.
Other studies in the field of cardiovascular disease have focused on the role of BRD4 in HF. Though HF and atherosclerosis are both designated as cardiovascular diseases, the underlying pathophysiology is very different. In HF, prolonged stress causes pathological cardiac dysfunction and remodeling, which relies partially on alteration to cardiomyocyte cell state. 14 , 135 , 136 , 137 As has been shown in mouse hypertrophic cardiomyocytes and in agonist‐induced hypertrophy in human‐induced pluripotent stem cell–derived cardiomyocytes (iPSC‐CMs), these cell state changes arise from altered gene expression; enhancer reprogramming, BRD4 redistribution, and overall global changes in histone acetylation and DNA methylation. 53 , 138 BRD4 has been shown to function as a critical coactivator of pathologic gene transactivation in these models. 53 Mirroring what occurs in vascular endothelial cells in atherosclerosis, inflammatory signaling in cardiomyocytes is upregulated through activation of the master transcription factor NF‐κB in HF. 56 Additionally, transforming growth factor beta (TGF‐β) signaling is increased resulting in BRD4 and latent enhancer redistribution around profibrotic myocardial genes, such as connective tissue growth factor (CTGF/CCN2) and serpine1 (Serpin E1/PAI‐1). 14 , 53 , 139 These alterations to the abundance of enhancer elements at new genomic locations (ie, latent enhancer formation) in cardiomyocytes ultimately lead to hypertrophy and contribute to HF. Using a number of currently available and structurally dissimilar BETi, the ability of BET inhibition to halt cardiomyocyte hypertrophy and HF was examined in various HF cell and animal models. 14 , 53 In these experiments, BETi consistently attenuated cardiac remodeling and pathologic hypertrophy, and suppressed key disease‐related gene expression both in vivo and in vitro 14 , 53 . These studies further validate BET inhibition as a viable potential therapeutic option for cardiovascular diseases.
Importantly, however, the development of juvenile idiopathic arthritis, a debilitating disease of the joints affecting children under the age of 16, has also shown a dependency on latent enhancer formation and BRD4 redistribution. In this disease, defects in immune cell function result in a loss of immunological tolerance, which cannot be accounted for by genetic heritability in a large portion of affected individuals. It has now been shown that epigenetic alterations to chromatin and latent enhancer structures result in CD4+ T cell dysfunction. The gene C‐X‐C chemokine receptor type 4 (CXCR4), for example, was found proximal to latent enhancer regions in CD4+ T cells of juvenile idiopathic arthritis patients. The abundance of BRD4 at these latent enhancers and the disease‐driven epigenetic changes in CXCR4 expression were reversed by BETi. 29 , 129 Overall, BET inhibition was shown to preferentially alter an extensive list of juvenile idiopathic arthritis‐specific genes. Though the functional consequences of these findings have yet to be explored, at the cellular level, juvenile idiopathic arthritis provides another example emphasizing the similarities in BRD4′s effect in terminally differentiated cell function across tissues, cell types, and disease states.
In metabolic disorders and obesity, latent enhancers enriched in BRD4 are involved in both adipocyte differentiation as well as chronic inflammation, both of which are key components of metabolic disease. 4 Adipocytes exhibit exceptional plasticity in response to environmental and metabolic changes, at least partially achieved by latent enhancer formation and BRD4 redistribution. Active enhancers placed near the promoter of the master adipogenic transcription factor peroxisome proliferator‐activated receptor gamma (PPARγ) result in BRD4 binding and upregulated PPARγ gene expression, which then activates a number of adipocyte‐specific differentiation genes. This process has been shown in white, and browning (or beige) adipocytes, and inhibition of BRD4 binding with BETi interrupts PPARγ‐mediated adipogenesis. 15 , 19 , 130 , 131 , 140 Though BRD4 activity at the PPARγ promoter is involved in adipocyte differentiation, it is not essential to maintain adipocyte identity. 140 However, BRD4 has been shown to play a role during fat storage and utilization, and in response to insulin resistance‐inducing stimuli. 141 , 142 That is, obesity is characterized not only by accelerated adipocyte differentiation but also by increased accumulation of immune cells, primarily macrophages, within adipose tissue. Inflammatory cytokines produced by these macrophages trigger functional defects in differentiation, insulin resistance, lipolysis, and lipid storage, as well as a cascade of events leading to the activation of NF‐κB. 25 , 40 , 130 As noted earlier in vascular endothelial cells and cardiomyocytes, NF‐κB induces rapid redistribution of BRD4, transcription factors, and latent enhancer formation. NF‐κB has a similar inflammation‐inducing effect on both mouse and human adipocytes. 25 , 40 , 130 , 132 Remarkably, BETi suppreses the expression of inflammatory response genes associated with these latent enhancer regions in adipose tissue, as well as a number of genes involved in insulin sensitivity and lipolysis. 15 , 19 , 142 Not only do these studies show BRD4's involvement in both adipocyte‐related differentiation and macrophage‐driven inflammation, illustrating BRD4's breadth of effect in adipose tissue‐related disorders, but they also show strong potential for the therapeutic use of BETi in metabolic disease and obesity.
Though latent enhancer formation is a beneficial response in acute attacks on the cellular environment, in disease states such as those described here, activation of this natural response becomes constant and maladaptive. In chronic diseases, latent enhancers contribute to sustained overexpression of response genes, which over time contributes to disease pathology and facilitates disease progression. Both the high levels of expression and the chronic activation of these genes become harmful. The latent enhancer regions are flooded with the tools to maintain the overexpression of genes that are now detrimental to the cell and the organism. As key components of latent enhancers, and recruiters of other pertinent transcriptional machinery, BET proteins are pivotal in maintaining the disease‐driven maladaptive activation of genes associated with chronic latent enhancer activation. As shown in the examples provided here, both in vitro and in vivo, in different cell types affected by different stimuli, the central role of BRD4 and likely other BET proteins as a component of latent enhancers puts these proteins at the cross‐roads of controlling gene activity in different disease states. Depending on the situation, the specific HATs, acetylation marks, transcription factors, and coregulators may differ, but BRD4 holds its place in bringing together the transcriptional complex and exacerbating the expression of disease‐driving genes. Inhibition of BET proteins, therefore, is an attractive target for broad therapeutic areas.
4. TARGETING BET PROTEINS
Since BRD4 is a key mediator of transcription at latent enhancers and SEs, which can drive the expression of disease‐causing genes, interruption of BRD4 activity has the potential to alter disease progression. In malignant cells, the cell assembles SEs surrounding oncogenes to then enable cancer cell immortality. In nonmalignant cells, overexpression of disease‐specific genes perpetuates the disease phenotype. A truly remarkable feature of both malignant and nonmalignant disease states is that enhancer formation and location is chosen by cell signaling through evolutionarily conserved response programs. Since BRD4 is redistributed to latent enhancers to respond to a noxious cellular environment, it is thought that BETi treatment in these conditions will attenuate transcription primarily at the abnormally active genes. Controversy remains over the extent to which BET inhibition affects housekeeping genes and what these effects may mean functionally and phenotypically. However, it is generally thought, given current knowledge, that BETi are a means of targeting the altered transcription that occurs at latent enhancers called upon by the cell in responding to and perpetuating the disease state. The attractiveness of BETi is enhanced by the current availability and ongoing development of small molecules that compete for binding of BDs to the natural ligand, acetylated lysine.
4.1. Approaches to targeting BET proteins
Various methods of targeting BET BDs, thus inhibiting their interaction with acetylated histones, have been developed. Proteolytic targeting chimera (PROTAC) compounds not only bind BET proteins, but they can induce BET degradation. These small‐molecule BETi are linked to a ubiquitin ligase recognition module via a flexible linker. Thus, not only do the BETi displace BET proteins from acetylated lysines, they also target the BET protein for ubiquitination and degradation. ARV‐825, ARV‐771, dBETi, and MZ1 are examples of currently available PROTACs that have been tested in various cancer models 7 , 11 , 12 , 39 , 143 , 144 , 145 , 146 , 147 , 148 , 149 , 150 while others continue to be developed. 26 However, BET‐targeting PROTAC research is currently in preclinical stages and thus the therapeutic potential is currently unknown.
Bivalent BETi, which bind both BDs of a BET protein simultaneously, are currently being developed. 151 , 152 , 153 Bivalent BD binding results in a stronger bond and greater potency than the monovalent binding of the more commonly known BETi, discussed below. The furthest bivalent BETi in development is AZD5153; an orally available BRD4 inhibitor. 153 Clinical development of AZD5153 is currently in phase 1, with ongoing studies focused on the treatment of lymphomas. 154 , 155
Another class of BET inhibiting compounds currently in the early stages of development are covalent BETi. These molecules, such as those designed by Kharenko et al., 28 form covalent bonds with residues within the BD binding pocket, creating a stronger interaction than noncovalent binding. Covalent BETi offer a longer lasting effect and potential for pharmacological efficacy at lower concentrations compared to their noncovalent counterparts. 28 , 156 Currently, covalent BETi have yet to reach the clinical investigation stages.
The greatest number of currently available BETi belong to the noncovalent BETi class, which form monovalent interactions with individual BET BDs. Description of the first noncovalent BETi, JQ1 157 has lead the way for IBET‐762, IBET‐151, OTX015, ZEN‐3694, and many others. 1 , 55 These BETi are commonly known for their ability to elicit antitumor activity in cancer cell lines as well as various murine cancer models. 57 , 58 , 158 , 159 , 160 For example, JQ1 has been proposed as a treatment for NUT‐midline carcinoma because this disease stems from an oncoprotein arising from the fusion of BRD4 to NUT. 54 , 58 , 61 , 161 , 162 , 163 Unfortunately, JQ1 is not a clinical candidate due to unfavorable pharmacokinetic properties, 164 however, many other noncovalent BETi are being clinically evaluated for the treatment of cancer and cardiovascular disease. 52 , 59 , 165 , 166 , 167 , 168 , 169 , 170 , 171 , 172 , 173 , 174 , 175 , 176 , 177 , 178 , 179 , 180 , 181 , 182 , 183 , 184 , 185 , 186 , 187 , 188 , 189 , 190 , 191 , 192 , 193 , 194 , 195 , 196 , 197 , 198 , 199 , 200
4.2. Bromodomain‐selective binding
BET bromodomains are comprised of a four‐helix bundle: helices KZ, KA, KB, and KC. The long ZA loop between helices KZ and KA is connected to a BC loop between helices B and C. The binding pockets of both BD1 and BD2 are comprised of a WPF shelf, a ZA channel, and a peptide channel. Amino acid differences between the two BD binding pockets exist; BD1‐specific residues of the binding pocket include Asp144, Ile146, Lys141, and Gln85. BD2‐specific histidine (His433), Val435, Pro430, and Lys374 replace Asp144, Ile146, Lys141, and Gln85, respectively, leading to a narrower binding pocket as well as altered polarity and hydrophobicity compared to BD1 72 , 80 , 157 , 201 (Figure 3).
Figure 3.
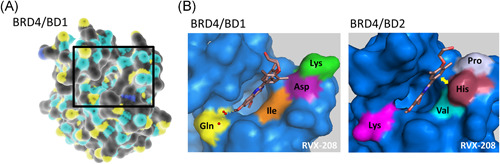
A, X‐ray crystal structure of BRD4/BD1, outlined. B, X‐ray crystal structure of BRD4/BD1 and BRD4/BD2 showing key residue replacements between bromodomains, with the example of apabetalone's (RVX‐208) BD2‐specific His433 binding (yellow arrows) [Color figure can be viewed at wileyonlinelibrary.com]
Most available BETi are pan‐selective, in that they bind both BDs with near equal affinity. This is due to the highly conserved structure and shape of BDs. The chemical structure of pan‐BETi JQ1, for example, allows for interactions with residues in the ZA‐ and BC‐loops of both BD1 and BD2. 77 , 157 There are many pan‐BETi currently available, such as JQ1, I‐BET151, ZEN‐3694, many of which are under clinical investigation for the treatment of cancer. 1 , 3 , 26 , 31 , 34 , 48 , 52 , 59 , 165 , 166 , 167 , 168 , 169 , 170 , 171 , 172 , 173 , 174 , 175 , 176 , 177 , 178 , 179 , 180 , 181 , 182 , 183 , 184 , 185 , 186 , 187 , 188 , 189 , 190 , 191 , 192 , 193 , 194 , 195 , 196 , 197 , 198 , 199 , 200
There are, however, a limited number of BETi compounds that claim to exhibit BD selectivity. The most recognized example of a BD1 selective compound, Olinone, exhibits over 100‐fold higher binding affinity to BD1 than BD2 in all BET proteins. The configuration of Olinone within the BD1‐binding pocket, its specific contact with five key residue replacements between BD1 and BD2, and the number and mobility of hydrogen bonds in the ZA and BC loops drive its BD1 selectivity. 37 , 202 Though Olinone is the most well‐known BD1 selective BETi, other scaffolds, configurations, and binding to unique amino acid residues within a given BD pocket can also be selective for BD1 over BD2. 156 , 203 , 204 A recent evaluation of GSK778, for example, shows >130‐fold selectivity for BRD4 BD1 over BD2 through specific binding with the Asp144/His433 replacement in BD1 versus BD2, respectively. 205 Other examples of BD1 selective compounds, such as MS611 and MS436 and MS402, are derived from a unique chemical scaffold. 202 , 206 , 207 , 208 However, BD1 selective compounds have yet to be investigated in clinical trials.
Examples of BD2‐selective compounds are increasing in number, and their unique therapeutic potential is gaining recognition. Available BD2‐selective BETi in preclinical stages of development include BY27, which binds BD2 with 5–38‐fold selectivity over BD1, 6 GSK620, and GSK046, each of which have been shown to have BD2‐binding affinities >300‐fold over BD1. 205 Two BD2‐selective BETi currently in clinical development include ABBV‐744, which has approximately 300‐fold greater binding affinity to BD2 of BRD4 than BD1, 209 and apabetalone, which selectively binds BD2 of BET proteins with 20–30‐fold greater affinity over BD1. 60 , 210 The first phase 1 clinical trial of ABBV‐744 is in the recruiting stage, enrolling subjects with acute myeloid leukemia. 211 ABBV‐744 was designed after the pan‐BETi ABBV‐075 to target the Asp144/His 437 and Ile146/Val439 sequence differences between BD1 and BD2. 5 , 212 Apabetalone, on the other hand, is the furthest BETi in clinical development, having recently been evaluated in a phase 3 trial in cardiovascular disease. 36 , 213 , 214 Apabetalone is unique in its interactions with BD2‐specific His433, which causes the compound to maintain a single conformation in the narrow BD2‐binding pocket 210 (Figure 3). This specific interaction prevents the multiple rotational positions that commonly occur in the BD1 pocket and highlights the specificity permissible by targeting unique BD features. 210 Another unique feature of apabetalone's binding in BD2 is the lack of interaction with the WPF shelf. Interestingly, initial findings from in vitro and early clinical investigations of both ABBV‐744 and apabetalone suggest increased tolerability in humans compared to pan‐selective agents, 212 , 215 suggesting that BD2‐specificity may be linked to reduced toxicity.
4.3. Pan‐BETi versus selective BETi
There is now a considerable amount of data, generated by various groups, demonstrating differential outcomes of pan‐ versus selective BD inhibition. 16 , 38 , 45 , 157 , 210 In one investigational study, gene expression in hepatocellular carcinoma cell line (HepG2 cells) following treatment with the pan‐BETi JQ1 was contrasted with that of BD2‐selective apabetalone 210 ; JQ1 exhibits equal binding affinity for both BDs, while apabetalone preferentially binds BD2. 157 , 210 JQ1 significantly affected the expression of at least 16 times more genes than apabetalone. More than 750 genes showed changes in expression of at least 1.5‐fold with JQ1, while only 46 genes were altered by apabetalone to the same extent. 210 Genes modified by both apabetalone and JQ1 (42 genes) showed a much larger magnitude of change with JQ1, which ultimately resulted in limited overlap between the top genes affected by each compound. Tyler et al. 45 extended these findings with their discovery that binding BD2 with apabetalone, while subsequently adding JQ1, reduced the ability of JQ1 to engage BRD4 at chromatin in MV4‐11 cells. These novel findings imply that dual BD binding is required for JQ1 to exhibit its full effect, and highlight the complex and dynamic interactions involved in pan‐ versus selective BET inhibition. Runcie et al. 38 also showed variation in the role of BD2 across BET proteins in human osteosarcoma U2OS cells. Compared to other BET proteins, BRD4‐BD2 was essential for regulating gene expression, which suggests that BRD4‐BD2 does, in fact, play a larger role in enhancer‐driven transcriptional regulation than both BRD4‐BD1 and other BET proteins in this cell type. These differential actions may prove to be of pivotal importance in therapeutic BETi use and development.
These and other studies confirm that pan‐BETi lead to vast effects on gene expression and activity while BD2‐selective binding often has less extensive effects. 45 , 157 , 210 They also suggest that the benefits of BD2‐selective BETi may be more specific than pan‐BETi in inhibiting those transcriptional changes associated with enhancer reprogramming and BRD4 redistribution to latent enhancer‐driven response genes. That is, BD2‐selective BETi may benefit disease states in which responses to disease‐driving stimuli are less extensive, for example, in diseases where inflammatory and profibrotic signaling are activated. On the other hand, disease states that result in more extensive alteration to cellular programming, such as cancer where fundamental cellular processes such as proliferation and cell survival signaling are triggered in combination with inflammatory signaling, may exhibit greater benefit from BD1 or pan‐selective BET inhibition. 38 , 45 , 210 Preclinical support for the functional delineation of therapeutic BD‐selectivity is indeed beginning to appear in the literature. The most promising example to date was provided by Gilan and colleagues, who have shown in K562 cells stimulated with interferon‐gamma (IFNγ), THP‐1 cells stimulated with phorbol 12‐myristate 13‐acetate, and CD4+ T cells stimulated with anti‐CD3/CD28, that BD2‐selective BET inhibition specifically targeted stimulus‐induced gene expression, leaving basal gene expression largely unaffected. 205 This finding was corroborated by reduced recruitment of BET proteins to response genes associated with BD2 inhibition. While BD1‐selective inhibition showed similar effects on proliferation, cell cycle arrest and apoptosis to that of pan‐BETi in cancer models (both in vitro and in vivo), BD2‐selective inhibition was effective in inflammation, metabolic disease, and profibrotic models. 205 Other studies have previously shown that BD2‐selective compounds can directly alter BRD4 abundance specifically at latent enhancers. 18 , 44 In these studies, the effects of BD2‐selective BETi on BRD4 redistribution and chromatin occupancy was examined in vascular smooth muscle cells subject to osteogenic (or calcifying) conditions, 18 and in endothelial cells stimulated with TNF‐α. 44 In both studies, BD2‐selective apabetalone reduced BRD4 occupancy at latent enhancers, transcription of genes proximal to latent enhancers, abundance, and activity of related proteins. These studies confirm that BD2‐selective BETi can diminish BET protein redistribution and therefore latent enhancer formation and function in disease areas outside of cancer.
This preliminary evidence, combined with future research and clinical developments, will undoubtedly provide continued support for BD2‐selective BETi in therapeutic research and development in diseases such as metabolic, inflammatory, and cardiovascular diseases. 4 , 14 , 19 , 53 , 56 , 129 , 205 By decreasing the negative effects of enhancer reprogramming and minimizing maladaptive transcription of disease‐driving genes at latent enhancers, BETi with BD2‐selectivity could be specifically beneficial in these areas.
4.4. BD‐selective BETi in the clinic: What human data do we have?
Though evidence of differential effects of pan‐ versus BD‐selective BETi from the clinic is limited due to the early stage of BETi development, some preliminary patterns are beginning to emerge. Clinical trials of the BD2‐selective BETi apabetalone provide the greatest repository of clinical data on BD2‐selective BETi available, and allude to potential benefits and possible differential effects of BD2‐selective BETi versus pan‐selective BETi in humans. First, of all data collected to date from thousands of patients, some of the most important may be related to safety and tolerability. The safety and tolerability profiles of apabetalone far exceed those of pan‐BETi in the clinic. 215 Whereas apabetalone‐treated patients generally experience mild adverse events, pan‐BETi–treated patients frequently experience more severe adverse events including increases in bilirubin, thrombocytopenia, and drug resistance. 3 , 32 , 35 , 52 , 215 , 216 Moreover, clinical data collected to date show that safety is not a limiting factor for chronic dosing of apabetalone, supporting the use of BD2‐selective BETi in diseases outside of cancer. 32 , 214 , 215 Second, apabetalone trials have identified key pathways that benefit from BD2‐selective BETi in cardiovascular and diabetes mellitus patients, 18 , 20 , 32 , 43 , 44 , 46 as well as chronic kidney disease patients. 47 , 217 This further underscores the role of BET proteins in driving the chronic maladaptive overexpression of genes and proteins which participate in networks and pathways that directly contribute to these disease states. Additional analysis of the recently completed phase 3 BETonMACE study will help to reveal more details of the unique biological effects of BD2‐selectivity on BET protein function in diabetic, renal, and cardiovascular disease patients. 36 , 214 Overall, though the number of clinical trials is currently limited, existing evidence supports the use of BD2‐selective BETi as a safe and efficacious therapy for the treatment of various chronic disease states.
4.5. Current clinical landscape of BET inhibitors
The majority of current clinical advancement of BETi remains in the early stages. Generally, clinical trials of BETi focus on treating various forms of cancer. Trials of the pan‐BETi CPI‐0610 have included patients with multiple myeloma, lymphoma, myelofibrosis, myelocytic leukemia, myelodysplastic/myeloproliferative neoplasm, and myelodysplastic syndrome, 171 , 172 , 173 , 197 while the pan‐BETi GSK525762 is being evaluated in ongoing clinical trials for solid tumors, brain tumors, and midline carcinoma. 176 , 177 , 178 , 218 , 219 , 220 , 221 , 222 , 223 , 224 , 225 ZEN‐3694 is being tested in a clinical trial in castration‐resistant prostate cancer and triple‐negative breast cancer. 198 , 199 , 200 ABBV‐744, a potential BD2‐selective BETi, 16 is also in cancer trials, currently being evaluated in a phase 1 trial in acute myeloid leukemia. 211 Apabetalone, the furthest clinically advanced BETi, which is also BD2‐selective, is to date the only BETi being evaluated in therapeutic areas outside of cancer. Apabetalone has been investigated in clinical trials in cardiovascular disease, cardiovascular disease combined with diabetes, chronic kidney disease, and planned clinical trials in pulmonary arterial hypertension are set to begin soon. 193 , 194 , 213 , 226 , 227 , 228 , 229 , 230 , 231 This brief list of examples (for a full list see www.clinicaltrials.gov) of clinical trials completed or currently underway shows that there is strong preclinical evidence that targeting one small protein family; BET proteins, can have the potential for therapeutic action on numerous cellular drivers of disease. 52 , 59 , 165 , 166 , 167 , 168 , 169 , 170 , 171 , 172 , 173 , 174 , 175 , 176 , 177 , 178 , 179 , 180 , 181 , 182 , 183 , 184 , 185 , 186 , 187 , 188 , 189 , 190 , 191 , 192 , 193 , 194 , 195 , 196 , 197 , 198 , 199 , 200 , 211 , 213 , 218 , 219 , 220 , 221 , 222 , 224 , 225 , 226 , 227 , 228 , 229 , 230 , 231 , 232 , 233 At the same time, the landscape of clinical trials suggests there is significant room for future development, particularly for BD‐selective BETi.
5. OUTLOOK
An understanding of the potential benefit of BETi in a wide spectrum of human diseases arises from their ability to target a single family of proteins which play a primary role in conserved cellular responses. BET proteins have a profound impact on transcription in both development and disease. We have shown that the breadth of BETi potential stems from the central role of BET proteins in super and latent enhancer formation and function. In many different diseases with different underlying pathologies, evolutionarily conserved cell signaling culminates in the activation of genes to respond to an insult. BET proteins and other transcriptional machinery are redistributed to enhancers to drive expression of these response genes. Evidence from studies focusing on BRD4 show that it is pivotal in the upregulation of disease‐driving genes and contributes to their overexpression. Inhibition of BRD4 consistently counters the transcriptional upregulation associated with enhancer reprogramming. Thus, the central premise behind therapeutic BETi is based not on the ability of these compounds to target every BET occupied gene, but instead on the cell's response to a stimulus. In other words, the cell sets the stage for BETi sensitivity by activating genes required to respond to disease through redistributing BET proteins to these genetic locations. The genes, transcription factors, and other elements may differ across cell types and disease states, but the underlying processes of redistribution of these elements remains the same. BET proteins are a consistent requirement for the cell's response to be appropriately activated. In chronic disease conditions, such as cardiovascular disease or inflammatory disease, even mild alterations to gene expression can be compounded by sustained activation. That is, though the response to an insult may be protective initially, the constant activation of these pathways and the sustained overexpression of these genes become detrimental. The ability of BETi to target a key component of latent enhancers provides a potential for therapeutic benefit in a variety of diseases, all driven by these same processes.
The current state of BETi research suggests that BD2‐selective BETi may be an effective way to safely target disease‐driven transcriptional alterations in terminally differentiated cells in areas outside of cancer. There are still many questions remaining, but the future of BETi development shows significant promise, and the development of BD‐selective BETi opens new possibilities. Focused development of BD‐selective compounds is certain to alter and expand the clinical landscape for years to come.
CONFLICT OF INTERESTS
Norman C. W. Wong, Brooke D. Rakai, and Ewelina Kulikowski are employees of Resverlogix Corp. and hold shares.
AUTHOR CONTRIBUTIONS
Norman C. W. Wong and Brooke D. Rakai researched and wrote the content; Brooke D. Rakai completed final editing for submission; Ewelina Kulikowski reviewed, edited, and approved content.
ACKNOWLEDGMENTS
The authors would like to thank Olesya Kharenko and Laura Tsujikawa for contributing valueable discussions, comments, and suggestions on the content, and to Resverlogix Corp. for providing funding.
Biographies
Ewelina Kulikowski has over 15 years of experience in the biotechnology industry. She currently leads all research and development activities at Resverlogix Corp. Dr. Kulikowski has contributed to epigenetic drug development from the discovery through to the clinical investigation of apabetalone the first BETi in clinical development for cardiovascular disease and type 2 diabetes mellitus. She has been involved in various aspects of pipeline development including market, reimbursement and pharmacoeconomics, regulatory affairs, commercial and lifecycle management. Dr. Kulikowski has led projects in oncology, ophthalmology, vascular inflammation, and neurodegeneration. In 2004, she received her Doctorate in Oncology from the University of Calgary, AB.
Brooke Rakai has contributed to epigenetic drug development as the Associate Director of Scientific Affairs at Resverlogix Corp. since 2016. She completed her Masters Degree at the Canadian Centre for Behavioural Neuroscience at the University of Lethbridge, AB, and in 2013, received her Doctorate in Experimental Psychology/Neuroscience from the University of Calgary, AB. She continued with her Post‐doctorate studies at the Hotchkiss Brain Institute, University of Calgary, AB, while simultaneously contracting in the pharmaceutical, regulatory, and public health fields. Dr. Rakai has lead and participated in projects in circadian rhythmicity, visual development, stroke, neurogenesis, cardiovascular disease, Fabry disease, neuroinflammation, and vascular inflammation.
Norman C. Wong is Co‐Founder and Chief Scientific Officer of Resverlogix Corp. Dr. Wong serves as Professor of Medicine, University of Calgary, a clinician scientist who specializes in endocrinology, internal medicine, molecular biology, gene/cell therapy, and regulation. His numerous academic accomplishments include over 300 published works, including extensive research in the field of epigenetics. Dr. Wong has served as a medical consultant to many pharmaceutical companies in the areas of diabetes mellitus, thyroid disease, and in the development of cholesterol‐lowering drugs. He is a Director of Zenith Epigenetics, and is or has been a member of the Scientific Advisory Board at Sernova Corp. Dr. Wong's academic record includes an Undergraduate B.Sc., Masters Degree in medical biochemistry, and a Medical Degree from the University of Calgary. https://www.ucalgary.ca/jmdrc/Wong.
Kulikowski E, Rakai BD, Wong NCW. Inhibitors of bromodomain and extra‐terminal proteins for treating multiple human diseases. Med Res Rev. 2021;41:223–245. 10.1002/med.21730
REFERENCES
- 1. Baldan F, Allegri L, Lazarevic M, et al. Biological and molecular effects of bromodomain and extra‐terminal (BET) inhibitors JQ1, IBET‐151, and IBET‐762 in OSCC cells. J Oral Pathol Med. 2019;48(3):214‐221. [DOI] [PubMed] [Google Scholar]
- 2. Bauer K, Berger D, Zielinski CC, Valent P, Grunt TW. Hitting two oncogenic machineries in cancer cells: cooperative effects of the multi‐kinase inhibitor ponatinib and the BET bromodomain blockers JQ1 or dBET1 on human carcinoma cells. Oncotarget. 2018;9(41):26491‐26506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bechter O, Schoffski P. Make your best BET: the emerging role of BET inhibitor treatment in malignant tumors. Pharmacol Ther. 2020;208:107479. [DOI] [PubMed] [Google Scholar]
- 4. Brown JD, Feldman ZB, Doherty SP, et al. BET bromodomain proteins regulate enhancer function during adipogenesis. Proc Natl Acad Sci U S A. 2018;115(9):2144‐2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bui MH, Lin X, Albert DH, et al. Preclinical characterization of BET family bromodomain inhibitor ABBV‐075 suggests combination therapeutic strategies. Cancer Res. 2017;77(11):2976‐2989. [DOI] [PubMed] [Google Scholar]
- 6. Chen D, Lu T, Yan Z, et al. Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins. Eur J Med Chem. 2019;182:111633. [DOI] [PubMed] [Google Scholar]
- 7. Chen P, Yang Y, Yang L, et al. 3‐Hydroxyisoindolin‐1‐one derivates: synthesis by palladium‐catalyzed CH activation as BRD4 inhibitors against human acute myeloid leukemia (AML) cells. Bioorg Chem. 2019;86:119‐125. [DOI] [PubMed] [Google Scholar]
- 8. Chen Y, Xu L, Mayakonda A, et al. Bromodomain and extraterminal proteins foster the core transcriptional regulatory programs and confer vulnerability in liposarcoma. Nat Commun. 2019;10(1):1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cochran AG, Conery AR, Sims RJ 3rd. Bromodomains: a new target class for drug development. Nat Rev Drug Discov. 2019;18(8):609‐628. [DOI] [PubMed] [Google Scholar]
- 10. Deepak V, Wang B, Koot D, et al. In silico design and bioevaluation of selective benzotriazepine BRD4 inhibitors with potent antiosteoclastogenic activity. Chem Biol Drug Des. 2017;90(1):97‐111. [DOI] [PubMed] [Google Scholar]
- 11. DeMars KM, Yang C, Candelario‐Jalil E. Neuroprotective effects of targeting BET proteins for degradation with dBET1 in aged mice subjected to ischemic stroke. Neurochem Int. 2019;127:94‐102. [DOI] [PubMed] [Google Scholar]
- 12. DeMars KM, Yang C, Castro‐Rivera CI, Candelario‐Jalil E. Selective degradation of BET proteins with dBET1, a proteolysis‐targeting chimera, potently reduces pro‐inflammatory responses in lipopolysaccharide‐activated microglia. Biochem Biophys Res Commun. 2018;497(1):410‐415. [DOI] [PubMed] [Google Scholar]
- 13. Donati B, Lorenzini E, Ciarrocchi A. BRD4 and cancer: going beyond transcriptional regulation. Mol Cancer. 2018;17(1):164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duan Q, McMahon S, Anand P, et al. BET bromodomain inhibition suppresses innate inflammatory and profibrotic transcriptional networks in heart failure. Sci Transl Med. 2017;9:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duan Q, Wu P, Liu Z, et al. BET bromodomain inhibition suppresses adipogenesis in mice. Endocrine. 2020;67(1):264‐267. [DOI] [PubMed] [Google Scholar]
- 16. Faivre EJ, McDaniel KF, Albert DH, et al. Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer. Nature. 2020;578:306‐310. [DOI] [PubMed] [Google Scholar]
- 17. Genta S, Pirosa MC, Stathis A. BET and EZH2 inhibitors: novel approaches for targeting cancer. Curr Oncol Rep. 2019;21(2):13. [DOI] [PubMed] [Google Scholar]
- 18. Gilham D., Tsujikawa LM, Sarsons CD, et al. Apabetalone downregulates factors and pathways associated with vascular calcification. Atherosclerosis. 2019;280:75‐84. [DOI] [PubMed] [Google Scholar]
- 19. Goupille O, Penglong T, Kadri Z, et al. Inhibition of the acetyl lysine‐binding pocket of bromodomain and extraterminal domain proteins interferes with adipogenesis. Biochem Biophys Res Commun. 2016;472(4):624‐630. [DOI] [PubMed] [Google Scholar]
- 20. Haarhaus M, Ray KK, Nicholls SJ, et al. Apabetalone lowers serum alkaline phosphatase and improves cardiovascular risk in patients with cardiovascular disease. Atherosclerosis. 2019;290:59‐65. [DOI] [PubMed] [Google Scholar]
- 21. Hajmirza A, Emadali A, Gauthier A, Casasnovas O, Gressin R, Callanan MB. BET family protein BRD4: an emerging actor in NFkappaB signaling in inflammation and cancer. Biomedicines. 2018;6(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsu SC, Gilgenast TG, Bartman CR, et al. The BET protein BRD2 cooperates with CTCF to enforce transcriptional and architectural boundaries. Mol Cell. 2017;66(1):102‐116.e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu J, Wang Y, Li Y, et al. Structure‐based optimization of a series of selective BET inhibitors containing aniline or indoline groups. Eur J Med Chem. 2018;150:156‐175. [DOI] [PubMed] [Google Scholar]
- 24. Huang W, Haynes AC, Mukherjee R, et al. Selective inhibition of BET proteins reduces pancreatic damage and systemic inflammation in bile acid‐ and fatty acid ethyl ester‐ but not caerulein‐induced acute pancreatitis. Pancreatology. 2017;17(5):689‐697. [DOI] [PubMed] [Google Scholar]
- 25. Inoue T, Hariya N, Imamochi Y, et al. Epigenetic regulation of lipoprotein lipase gene via BRD4, which is potentially associated with adipocyte differentiation and insulin resistance. Eur J Pharmacol. 2019;858:172492. [DOI] [PubMed] [Google Scholar]
- 26. Jiang F, Wei Q, Li H, et al. Discovery of novel small molecule induced selective degradation of the bromodomain and extra‐terminal (BET) bromodomain protein BRD4 and BRD2 with cellular potencies. Bioorg Med Chem. 2020;28(1):115181. [DOI] [PubMed] [Google Scholar]
- 27. Jiang G, Deng W, Liu Y, Wang C. General mechanism of JQ1 in inhibiting various types of cancer. Mol Med Rep. 2020;21(3):1021‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kharenko OA, Patel RG, Brown SD, et al. Design and characterization of novel covalent bromodomain and extra‐terminal domain (BET) inhibitors targeting a methionine. J Med Chem. 2018;61(18):8202‐8211. [DOI] [PubMed] [Google Scholar]
- 29. Klein K. Bromodomain protein inhibition: a novel therapeutic strategy in rheumatic diseases. RMD Open. 2018;4(2):e000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lambert JP, Picaud S, Fujisawa T, et al. Interactome rewiring following pharmacological targeting of BET bromodomains. Mol Cell. 2019;73(3):621‐638.e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leal AS, Williams CR, Royce DB, Pioli PA, Sporn MB, Liby KT. Bromodomain inhibitors, JQ1 and I‐BET 762, as potential therapies for pancreatic cancer. Cancer Lett. 2017;394:76‐87. [DOI] [PubMed] [Google Scholar]
- 32. Nicholls SJ, Ray KK, Johansson JO, et al. Selective BET protein inhibition with apabetalone and cardiovascular events: a pooled analysis of trials in patients with coronary artery disease. Am J Cardiovasc Drugs. 2018;18(2):109‐115. [DOI] [PubMed] [Google Scholar]
- 33. Pervaiz M, Mishra P, Gunther S. Bromodomain drug discovery—the past, the present, and the future. Chem Rec. 2018;18(12):1808‐1817. [DOI] [PubMed] [Google Scholar]
- 34. Piha‐Paul SA, Sachdev JC, Barve M, et al. First‐in‐human study of mivebresib (ABBV‐075), an oral pan‐inhibitor of bromodomain and extra terminal proteins, in patients with relapsed/refractory solid tumors. Clin Cancer Res. 2019;25:6309‐6319. [DOI] [PubMed] [Google Scholar]
- 35. Postel‐Vinay S, Herbschleb K, Massard C, et al. First‐in‐human phase I study of the bromodomain and extraterminal motif inhibitor BAY 1238097: emerging pharmacokinetic/pharmacodynamic relationship and early termination due to unexpected toxicity. Eur J Cancer. 2019;109:103‐110. [DOI] [PubMed] [Google Scholar]
- 36. Ray KK, Nicholls SJ, Ginsberg HD, et al. Effect of selective BET protein inhibitor apabetalone on cardiovascular outcomes in patients with acute coronary syndrome and diabetes: rationale, design, and baseline characteristics of the BETonMACE trial. Am Heart J. 2019;217:72‐83. [DOI] [PubMed] [Google Scholar]
- 37. Rodriguez Y, Gerona‐Navarro G, Osman R, Zhou MM. In silico design and molecular basis for the selectivity of Olinone toward the first over the second bromodomain of BRD4. Proteins. 2020;88(3):414‐430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Runcie AC, Zengerle M, Chan KH, et al. Optimization of a "bump‐and‐hole" approach to allele‐selective BET bromodomain inhibition. Chem Sci. 2018;9(9):2452‐2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sahni JM, Keri RA. Targeting bromodomain and extraterminal proteins in breast cancer. Pharmacol Res. 2018;129:156‐176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sakurai N, Inamochi Y, Inoue T, et al. BRD4 regulates adiponectin gene induction by recruiting the P‐TEFb complex to the transcribed region of the gene. Sci Rep. 2017;7(1):11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Segatto M, Fittipaldi R, Pin F, et al. Epigenetic targeting of bromodomain protein BRD4 counteracts cancer cachexia and prolongs survival. Nat Commun. 2017;8(1):1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shadrick WR, Slavish PJ, Chai SC, et al. Exploiting a water network to achieve enthalpy‐driven, bromodomain‐selective BET inhibitors. Bioorg Med Chem. 2018;26(1):25‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shishikura D, Kataoka Y, Honda S, et al. The effect of bromodomain and extra‐terminal inhibitor apabetalone on attenuated coronary atherosclerotic plaque: insights from the ASSURE trial. Am J Cardiovasc Drugs. 2019;19(1):49‐57. [DOI] [PubMed] [Google Scholar]
- 44. Tsujikawa LM, Fu L, Das S, et al. Apabetalone (RVX‐208) reduces vascular inflammation in vitro and in CVD patients by a BET‐dependent epigenetic mechanism. Clin Epigenetics. 2019;11(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tyler DS, Vappiani J, Cañeque T, et al. Click chemistry enables preclinical evaluation of targeted epigenetic therapies. Science. 2017;356(6345):1397‐1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wasiak S, Gilham D, Tsujikawa LM, et al. Downregulation of the complement cascade in vitro, in mice and in patients with cardiovascular disease by the BET protein inhibitor apabetalone (RVX‐208). J Cardiovasc Transl Res. 2017;10(4):337‐347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wasiak S, Tsujikawa LM, Halliday C, et al. Benefit of apabetalone on plasma proteins in renal disease. Kidney Int Rep. 2018;3(3):711‐721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilson AJ, Stubbs M, Liu P, Ruggeri B, Khabele D. The BET inhibitor INCB054329 reduces homologous recombination efficiency and augments PARP inhibitor activity in ovarian cancer. Gynecol Oncol. 2018;149(3):575‐584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang X, Lin J, Liang T, et al. The BET bromodomain inhibitor apabetalone induces apoptosis of latent HIV‐1 reservoir cells following viral reactivation. Acta Pharmacol Sin. 2019;40(1):98‐110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Korb E, Herre M, Zucker‐Scharff I, Darnell RB, Allis CD. BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat Neurosci. 2015;18(10):1464‐1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aguirre‐Portoles C, Feliu J, Reglero G, Ramirez de Molina A. ABCA1 overexpression worsens colorectal cancer prognosis by facilitating tumour growth and caveolin‐1‐dependent invasiveness, and these effects can be ameliorated using the BET inhibitor apabetalone. Mol Oncol. 2018;12(10):1735‐1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Amorim S, Stathis A, Gleeson M, et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose‐escalation, open‐label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016;3(4):e196‐204. [DOI] [PubMed] [Google Scholar]
- 53. Anand P, Brown JD, Lin CY, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154(3):569‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bandopadhayay P, Bergthold G, Nguyen B, et al. BET bromodomain inhibition of MYC‐amplified medulloblastoma. Clin Cancer Res. 2014;20(4):912‐925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boi M, Gaudio E, Bonetti P, et al. The BET bromodomain inhibitor OTX015 affects pathogenetic pathways in preclinical B‐cell tumor models and synergizes with targeted drugs. Clin Cancer Res. 2015;21(7):1628‐1638. [DOI] [PubMed] [Google Scholar]
- 56. Brown JD, Lin CY, Duan Q, et al. NF‐kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56(2):219‐231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dawson MA, Prinjha RK, Dittmann A, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL‐fusion leukaemia. Nature. 2011;478(7370):529‐533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c‐Myc. Cell. 2011;146(6):904‐917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hoffmann‐La R. Study of bromodomain and extra‐terminal protein (BET) inhibitor RO6870810 as mono‐ and combination therapy in advanced multiple myeloma; 2019. https://ClinicalTrials.gov/show/NCT03068351. Accessed August 1, 2019.
- 60. McLure KG, Gesner EM, Tsujikawa L, et al. RVX‐208, an inducer of ApoA‐I in humans, is a BET bromodomain antagonist. PLOS One. 2013;8(12):e83190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mertz JA, Conery AR, Bryant BM, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669‐16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang QG, Qian J, Zhu YC. Targeting bromodomain‐containing protein 4 (BRD4) benefits rheumatoid arthritis. Immunol Lett. 2015;166(2):103‐108. [DOI] [PubMed] [Google Scholar]
- 63. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boehm D, Calvanese V, Dar RD, et al. BET bromodomain‐targeting compounds reactivate HIV from latency via a Tat‐independent mechanism. Cell Cycle. 2013;12(3):452‐462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Boehm D, Conrad RJ, Ott M. Bromodomain proteins in HIV infection. Viruses. 2013;5(6):1571‐1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li Z, Guo J, Wu Y, Zhou Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat‐transactivation. Nucleic Acids Res. 2013;41(1):277‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Banerjee C, Archin N, Michaels D, et al. BET bromodomain inhibition as a novel strategy for reactivation of HIV‐1. J Leukoc Biol. 2012;92(6):1147‐1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Andrieu G, Belkina AC, Denis GV. Clinical trials for BET inhibitors run ahead of the science. Drug Discov Today Technol. 2016;19:45‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Devaiah BN, Gegonne A, Singer DS. Bromodomain 4: a cellular Swiss army knife. J Leukoc Biol. 2016;100(4):679‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ferri E, Petosa C, McKenna CE. Bromodomains: structure, function and pharmacology of inhibition. Biochem Pharmacol. 2016;106:1‐18. [DOI] [PubMed] [Google Scholar]
- 71. Taniguchi Y. The bromodomain and extra‐terminal domain (BET) family: functional anatomy of BET paralogous proteins. Int J Mol Sci. 2016;17(11):1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zeng L, Zhou MM. Bromodomain: an acetyl‐lysine binding domain. FEBS Lett. 2002;513(1):124‐128. [DOI] [PubMed] [Google Scholar]
- 73. Reyes‐Garau D, Ribeiro ML, Roue G. Pharmacological targeting of BET bromodomain proteins in acute myeloid leukemia and malignant lymphomas: from molecular characterization to clinical applications. Cancers(Basel). 2019;11(10):1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Khandekar D, Tiriveedhi V. Role of BET inhibitors in triple negative breast cancers. Cancers (Basel). 2020;12(4):784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hnisz D, Abraham BJ, Lee TI, et al. Super‐enhancers in the control of cell identity and disease. Cell. 2013;155(4):934‐947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lovén J, Hoke HA, Lin CY, et al. Selective inhibition of tumor oncogenes by disruption of super‐enhancers. Cell. 2013;153(2):320‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. 2012;586(17):2692‐2704. [DOI] [PubMed] [Google Scholar]
- 78. Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. BioEssays. 2004;26(10):1076‐1087. [DOI] [PubMed] [Google Scholar]
- 79. Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292(5514):110‐113. [DOI] [PubMed] [Google Scholar]
- 80. Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491‐496. [DOI] [PubMed] [Google Scholar]
- 81. Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074‐1080. [DOI] [PubMed] [Google Scholar]
- 82. Devaiah BN, Case‐Borden C, Gegonne A, et al. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol. 2016;23(6):540‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain‐containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn. 2009;238(4):908‐917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gyuris A, Donovan DJ, Seymour KA, et al. The chromatin‐targeting protein Brd2 is required for neural tube closure and embryogenesis. Biochim Biophys Acta. 2009;1789(5):413‐421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J Proteome Res. 2006;5(3):502‐511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Denis GV, Vaziri C, Guo N, Faller DV. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 2000;11(8):417‐424. [PMC free article] [PubMed] [Google Scholar]
- 87. LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30(1):51‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Belkina AC, Blanton WP, Nikolajczyk BS, Denis GV. The double bromodomain protein Brd2 promotes B cell expansion and mitogenesis. J Leukoc Biol. 2014;95(3):451‐460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Greenwald RJ, Tumang JR, Sinha A, et al. E mu‐BRD2 transgenic mice develop B‐cell lymphoma and leukemia. Blood. 2004;103(4):1475‐1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lamonica JM, Deng W, Kadauke S, et al. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci U S A. 2011;108(22):E159‐E168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gamsjaeger R, Webb SR, Lamonica JM, Billin A, Blobel GA, Mackay JP. Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3. Mol Cell Biol. 2011;31(13):2632‐2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wai DCC, Szyszka TN, Campbell AE, et al. The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators. J Biol Chem. 2018;293(19):7160‐7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gaucher J, Boussouar F, Montellier E, et al. Bromodomain‐dependent stage‐specific male genome programming by Brdt. EMBO J. 2012;31(19):3809‐3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Matzuk MM, McKeown MR, Filippakopoulos P, et al. Small‐molecule inhibition of BRDT for male contraception. Cell. 2012;150(4):673‐684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Berkovits BD, Wolgemuth DJ. The first bromodomain of the testis‐specific double bromodomain protein Brdt is required for chromocenter organization that is modulated by genetic background. Dev Biol. 2011;360(2):358‐368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang L, Wolgemuth DJ. BET protein BRDT complexes with HDAC1, PRMT5, and TRIM28 and functions in transcriptional repression during spermatogenesis. J Cell Biochem. 2016;117(6):1429‐1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Manterola M, Brown TM, Oh MY, Garyn C, Gonzalez BJ, Wolgemuth DJ. BRDT is an essential epigenetic regulator for proper chromatin organization, silencing of sex chromosomes and crossover formation in male meiosis. PLOS Genet. 2018;14(3):e1007209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell. 2004;13(1):33‐43. [DOI] [PubMed] [Google Scholar]
- 99. Devaiah BN, Singer DS. Two faces of BRD4: mitotic bookmark and transcriptional lynchpin. Transcription. 2013;4(1):13‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20(23):4899‐4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS. Growth and early postimplantation defects in mice deficient for the bromodomain‐containing protein Brd4. Mol Cell Biol. 2002;22(11):3794‐3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wu T, Kamikawa YF, Donohoe ME. Brd4's bromodomains mediate histone H3 acetylation and chromatin remodeling in pluripotent cells through P300 and Brg1. Cell Rep. 2018;25(7):1756‐1771. [DOI] [PubMed] [Google Scholar]
- 103. Greer CB, Tanaka Y, Kim YJ, et al. Histone deacetylases positively regulate transcription through the elongation machinery. Cell Rep. 2015;13(7):1444‐1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Shen C, Ipsaro JJ, Shi J, et al. NSD3‐short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell. 2015;60(6):847‐859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Devaiah BN, Lewis BA, Cherman N, et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy‐terminal domain. Proc Natl Acad Sci U S A. 2012;109(18):6927‐6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bhagwat AS, Roe JS, Mok BYL, Hohmann AF, Shi J, Vakoc CR. BET bromodomain inhibition releases the mediator complex from select cis‐regulatory elements. Cell Rep. 2016;15(3):519‐530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wu SY, Chiang CM. The double bromodomain‐containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282(18):13141‐13145. [DOI] [PubMed] [Google Scholar]
- 108. Zhang W, Prakash C, Sum C, et al. Bromodomain‐containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012;287(51):43137‐43155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P‐TEFb and stimulates RNA polymerase II‐dependent transcription. Mol Cell. 2005;19(4):523‐534. [DOI] [PubMed] [Google Scholar]
- 110. Yang Z, Yik JHN, Chen R, et al. Recruitment of P‐TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19(4):535‐545. [DOI] [PubMed] [Google Scholar]
- 111. Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super‐enhancers at key cell identity genes. Cell. 2013;153(2):307‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sims RJ 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18(20):2437‐2468. [DOI] [PubMed] [Google Scholar]
- 113. Itzen F, Greifenberg AK, Bosken CA, Geyer M. Brd4 activates P‐TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014;42(12):7577‐7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Nechaev S, Adelman K. Promoter‐proximal Pol II: when stalling speeds things up. Cell Cycle. 2008;7(11):1539‐1544. [DOI] [PubMed] [Google Scholar]
- 115. Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118(1):1‐10. [DOI] [PubMed] [Google Scholar]
- 116. Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci U S A. 1993;90(17):7923‐7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal‐dependent transcriptional elongation. Cell. 2009;138(1):129‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kanno T, Kanno Y, LeRoy G, et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol. 2014;21(12):1047‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Pott S, Lieb JD. What are super‐enhancers? Nat Genet. 2015;47(1):8‐12. [DOI] [PubMed] [Google Scholar]
- 121. Sabari BR, Dall'Agnese A, Boija A, et al. Coactivator condensation at super‐enhancers links phase separation and gene control. Science. 2018;361:6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Di Micco R, Fontanals‐Cirera B, Low V, et al. Control of embryonic stem cell identity by BRD4‐dependent transcriptional elongation of super‐enhancer‐associated pluripotency genes. Cell Rep. 2014;9(1):234‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A. 2003;100(15):8758‐8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tsang FHC, Law CT, Tang TCC, et al. Aberrant super‐enhancer landscape in human hepatocellular carcinoma. Hepatology. 2019:hep.30544. [DOI] [PubMed] [Google Scholar]
- 125. Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type‐specific enhancers. Nat Rev Mol Cell Biol. 2015;16(3):144‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ostuni R, Piccolo V, Barozzi I, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152(1–2):157‐171. [DOI] [PubMed] [Google Scholar]
- 127. Parker SCJ, Stitzel ML, Taylor DL, et al. Chromatin stretch enhancer states drive cell‐specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110(44):17921‐17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Stratton MS, Lin CY, Anand P, et al. Signal‐dependent recruitment of BRD4 to cardiomyocyte super‐enhancers is suppressed by a microRNA. Cell Rep. 2016;16(5):1366‐1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Peeters JGC, Vervoort SJ, Tan SC, et al. Inhibition of super‐enhancer activity in autoinflammatory site‐derived T cells reduces disease‐associated gene expression. Cell Rep. 2015;12(12):1986‐1996. [DOI] [PubMed] [Google Scholar]
- 130. Loft A, Forss I, Siersbæk MS, et al. Browning of human adipocytes requires KLF11 and reprogramming of PPARgamma superenhancers. Genes Dev. 2015;29(1):7‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Loft A, Schmidt SF, Mandrup S. Modulating the genomic programming of adipocytes. Cold Spring Harb Symp Quant Biol. 2015;80:239‐248. [DOI] [PubMed] [Google Scholar]
- 132. Schmidt SF, Larsen BD, Loft A, Nielsen R, Madsen JG, Mandrup S. Acute TNF‐induced repression of cell identity genes is mediated by NFkappaB‐directed redistribution of cofactors from super‐enhancers. Genome Res. 2015;25(9):1281‐1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Tajbakhsh A, Rezaee M, Kovanen PT, Sahebkar A. Efferocytosis in atherosclerotic lesions: malfunctioning regulatory pathways and control mechanisms. Pharmacol Ther. 2018;188:12‐25. [DOI] [PubMed] [Google Scholar]
- 134. Chistiakov DA, Melnichenko AA, Myasoedova VA, Grechko AV, Orekhov AN. Mechanisms of foam cell formation in atherosclerosis. J Mol Med (Berl). 2017;95(11):1153‐1165. [DOI] [PubMed] [Google Scholar]
- 135. Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358(13):1370‐1380. [DOI] [PubMed] [Google Scholar]
- 136. Sano M, Schneider MD. Still stressed out but doing fine: normalization of wall stress is superfluous to maintaining cardiac function in chronic pressure overload. Circulation. 2002;105(1):8‐10. [PubMed] [Google Scholar]
- 137. van Berlo JH, Maillet M, Molkentin JD. Signaling effectors underlying pathologic growth and remodeling of the heart. J Clin Invest. 2013;123(1):37‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Papait R, Cattaneo P, Kunderfranco P, et al. Genome‐wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc Natl Acad Sci U S A. 2013;110(50):20164‐20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Phanish MK, Winn SK, Dockrell ME. Connective tissue growth factor‐(CTGF, CCN2)—a marker, mediator and therapeutic target for renal fibrosis. Nephron Exp Nephrol. 2010;114(3):e83‐92. [DOI] [PubMed] [Google Scholar]
- 140. Lee JE, Park YK, Park S, et al. Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis. Nat Commun. 2017;8(1):2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Kang S, Tsai LT, Zhou Y, et al. Identification of nuclear hormone receptor pathways causing insulin resistance by transcriptional and epigenomic analysis. Nat Cell Biol. 2015;17(1):44‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Peng X, Zhang Q, Liao C, Han W, Xu F. Epigenomic control of thermogenic adipocyte differentiation and function. Int J Mol Sci. 2018;19(6):1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Piya S, Mu H, Bhattacharya S, et al. BETP degradation simultaneously targets acute myelogenous leukemia stem cells and the microenvironment. J Clin Invest. 2019;129(5):1878–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Lim SL, Damnernsawad A, Shyamsunder P, et al. Proteolysis targeting chimeric molecules as therapy for multiple myeloma: efficacy, biomarker and drug combinations. Haematologica. 2019;104:1209‐1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Sun B, Fiskus W, Qian Y, et al. BET protein proteolysis targeting chimera (PROTAC) exerts potent lethal activity against mantle cell lymphoma cells. Leukemia. 2018;32(2):343‐352. [DOI] [PubMed] [Google Scholar]
- 146. Saenz DT, Fiskus W, Manshouri T, et al. Targeting nuclear beta‐catenin as therapy for post‐myeloproliferative neoplasm secondary AML. Leukemia. 2019;33(6):1373‐1386. [DOI] [PubMed] [Google Scholar]
- 147. Abruzzese MP, Bilotta MT, Fionda C, et al. Inhibition of bromodomain and extra‐terminal (BET) proteins increases NKG2D ligand MICA expression and sensitivity to NK cell‐mediated cytotoxicity in multiple myeloma cells: role of cMYC‐IRF4‐miR‐125b interplay. J Hematol Oncol. 2016;9(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Lu J, Qian Y, Altieri M, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol. 2015;22(6):755‐763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Dai X, Gan W, Li X, et al. Prostate cancer‐associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med. 2017;23(9):1063‐1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Zengerle M, Chan KH, Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol. 2015;10(8):1770‐1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Waring MJ, Chen H, Rabow AA, et al. Potent and selective bivalent inhibitors of BET bromodomains. Nat Chem Biol. 2016;12(12):1097‐1104. [DOI] [PubMed] [Google Scholar]
- 152. Tanaka M, Roberts JM, Seo HS, et al. Design and characterization of bivalent BET inhibitors. Nat Chem Biol. 2016;12(12):1089‐1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Rhyasen GW, Hattersley MM, Yao Y, et al. AZD5153: a novel bivalent BET bromodomain inhibitor highly active against hematologic malignancies. Mol Cancer Ther. 2016;15(11):2563‐2574. [DOI] [PubMed] [Google Scholar]
- 154.AstraZeneca. AZD5153 in Patients With Relapsed or Refractory Solid Tumors, Including Lymphomas. ClinicalTrials.gov.
- 155.CenterWatch. Platform Study for the Treatment of Relapsed or Refractory Aggressive Non‐Hodgkin's Lymphoma (PRISM Study).
- 156. Daguer JP, Zambaldo C, Abegg D, et al. Identification of covalent bromodomain binders through DNA display of small molecules. Angew Chem Int Ed Engl. 2015;54(20):6057‐6061. [DOI] [PubMed] [Google Scholar]
- 157. Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067‐1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Dawson MA, Gudgin EJ, Horton SJ, et al. Recurrent mutations, including NPM1c, activate a BRD4‐dependent core transcriptional program in acute myeloid leukemia. Leukemia. 2014;28(2):311‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367(7):647‐657. [DOI] [PubMed] [Google Scholar]
- 160. Mirguet O, Gosmini R, Toum J, et al. Discovery of epigenetic regulator I‐BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J Med Chem. 2013;56(19):7501‐7515. [DOI] [PubMed] [Google Scholar]
- 161. Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524‐528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Grayson AR, Walsh EM, Cameron MJ, et al. MYC, a downstream target of BRD‐NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. Oncogene. 2014;33(13):1736‐1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Wang CY, Filippakopoulos P. Beating the odds: BETs in disease. Trends Biochem Sci. 2015;40(8):468‐479. [DOI] [PubMed] [Google Scholar]
- 164. Li F, MacKenzie KR, Jain P, Santini C, Young DW, Matzuk MM. Metabolism of JQ1, an inhibitor of BET bromodomain proteins, in human and mouse liver microsomesdagger. Biol Reprod. 2020;103:427‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.AbbVie. A study evaluating the safety and pharmacokinetics of ABBV‐075 in subjects with cancer; 2019. https://ClinicalTrials.gov/show/NCT02391480. Accessed 5 July 2019.
- 166.Bayer. BAY1238097, First in Man; 2016. https://ClinicalTrials.gov/show/NCT02369029. Accessed January 2016.
- 167. Bristol‐Myers Squibb . Study of BMS‐986158 in subjects with select advanced cancers; 2023. https://ClinicalTrials.gov/show/NCT02419417. Accessed 12 July.
- 168.Celgene. Study to Evaluate the Safety, Tolerability, Pharmacokinetic and Pharmacodynamic of CC‐95775 in Subjects With Advanced Solid Tumors and Relapsed/Refractory Non‐Hodgkin Lymphoma; 2023. https://ClinicalTrials.gov/show/NCT04089527. Accessed 18 June.
- 169. Celgene. Study to Assess the Safety, Tolerability , Pharmacokinetics and Preliminary Efficacy of CC‐90010 in Subjects With Advanced Solid Tumors and Relapsed/Refractory Non‐Hodgkin's Lymphomas; 2021. https://ClinicalTrials.gov/show/NCT03220347. Accessed 23 August.
- 170.M.D. Anderson Cancer Center, National Cancer Institute (NCI). PLX51107 and Azacitidine in Treating Patients With Acute Myeloid Leukemia or Myelodysplastic Syndrome; 2022. https://ClinicalTrials.gov/show/NCT04022785. Accessed 31 December.
- 171.Constellation Pharmaceuticals, The Leukemia and Lymphoma Society. A Phase 1 Study Evaluating CPI‐0610 in Patients With Previously Treated Multiple Myeloma; 2017. https://ClinicalTrials.gov/show/NCT02157636. Accessed November.
- 172.Constellation Pharmaceuticals, The Leukemia and Lymphoma Society. A Phase 1 Study Evaluating CPI‐0610 in Patients With Progressive Lymphoma; 2017. https://ClinicalTrials.gov/show/NCT01949883. Accessed December.
- 173.Constellation Pharmaceuticals, The Leukemia and Lymphoma Society. A Phase 2 Study of CPI‐0610 With and Without Ruxolitinib in Patients With Myelofibrosis; 2021. https://ClinicalTrials.gov/show/NCT02158858. Accessed 31 August.
- 174.Dana‐Farber Cancer Institute, Stand Up To Cancer. Study of the Bromodomain (BRD) and Extra‐Terminal Domain (BET) Inhibitor BMS‐986158 in Pediatric Cancer; 2022. https://ClinicalTrials.gov/show/NCT03936465. Accessed 10 July.
- 175.Forma Therapeutics, Inc. Study of a Novel BET Inhibitor FT‐1101 in Patients With Relapsed or Refractory Hematologic Malignancies; 2019. https://ClinicalTrials.gov/show/NCT02543879. Accessed March.
- 176.GlaxoSmithKline. Dose Escalation Study to Investigate the Safety, Pharmacokinetics (PK), Pharmacodynamics (PD), and Clinical Activity of GSK525762 Plus Trametinib in Subjects With Solid Tumors; 2020. https://ClinicalTrials.gov/show/NCT03266159. Accessed 19 August.
- 177.GlaxoSmithKline. Dose Escalation Study to Investigate the Safety, Pharmacokinetics (PK), Pharmacodynamics (PD) and Clinical Activity of GSK525762 in Subjects With Relapsed, Refractory Hematologic Malignancies; 2023. https://ClinicalTrials.gov/show/NCT01943851. Accessed 12 January.
- 178.GlaxoSmithKline. A Study to Investigate the Safety, Pharmacokinetics, Pharmacodynamics, and Clinical Activity of GSK525762 in Subjects With NUT Midline Carcinoma (NMC) and Other Cancers; 2018. https://ClinicalTrials.gov/show/NCT01587703. Accessed 13 April.
- 179.GlaxoSmithKline. Dose Escalation Study of GSK2820151 in Subjects With Advanced or Recurrent Solid Tumors; 2018. https://ClinicalTrials.gov/show/NCT02630251. Accessed 13 December.
- 180.Hoffmann‐La Roche. A Study of RO6870810/TEN‐010 in Participants With Acute Myeloid Leukemia and Myelodysplastic Syndrome; 2017. https://ClinicalTrials.gov/show/NCT02308761. Accessed 9 August.
- 181.Hoffmann‐La Roche. A Two Part Study of RO6870810. Dose‐Escalation Study in Participants With Advanced Solid Tumors and Expansion Study in Participants With Selected Malignancies; 2017. https://ClinicalTrials.gov/show/NCT01987362. Accessed 11 October.
- 182.Incyte Corporation. An Open‐Label, Dose‐Escalation Study of INCB054329 in Patients With Advanced Malignancies; 2018. https://ClinicalTrials.gov/show/NCT02431260. Accessed 31 January.
- 183.Incyte Corporation. Azacitidine Combined With Pembrolizumab and Epacadostat in Subjects With Advanced Solid Tumors (ECHO‐206); 2019. https://ClinicalTrials.gov/show/NCT02959437. Accessed 15 February.
- 184.Incyte Corporation. A Study of INCB059872 in Relapsed or Refractory Ewing Sarcoma; 2020. https://ClinicalTrials.gov/show/NCT03514407. Accessed December.
- 185.Incyte Corporation. Open‐Label Safety and Tolerability Study of INCB057643 in Subjects With Advanced Malignancies; 2019. https://ClinicalTrials.gov/show/NCT02711137. Accessed 13 February.
- 186.Merck Sharp & Dohme Corp. A Dose Exploration Study With MK‐8628 in Participants With Selected Hematologic Malignancies (MK‐8628‐005); 2018. https://ClinicalTrials.gov/show/NCT02698189. Accessed 18 January.
- 187.Merck Sharp & Dohme Corp. A Dose Exploration Study With MK‐8628 in Participants With Selected Advanced Solid Tumors (MK‐8628‐006); 2017. https://ClinicalTrials.gov/show/NCT02698176. Accessed 26 April.
- 188.National Cancer Institute (NCI). Testing of the Addition of a New Anti‐cancer Drug, Molibresib, to Chemotherapy Treatment (Etoposide and Cisplatin) for Patients With NUT Carcinoma; 2021. https://ClinicalTrials.gov/show/NCT04116359. Accessed 20 April.
- 189.Oncoethix GmbH. A Dose‐Finding Study of MK‐8628, a Small Molecule Inhibitor of the Bromodomain and Extra‐Terminal (BET) Proteins, in Adults With Selected Advanced Solid Tumors (MK‐8628‐003); 2017. https://ClinicalTrials.gov/show/NCT02259114. Accessed 3 March.
- 190.Oncoethix GmbH. A Dose‐finding Study of MK‐8628 in Participants With Recurrent Glioblastoma Multiforme (MK‐8628‐002); 2015. https://ClinicalTrials.gov/show/NCT02296476. Accessed 20 October.
- 191.Oncoethix GmbH. A Study Assessing tOTX015 in Combination With Azacitidine (AZA) or AZA Single Agent in Patients With Newly‐diagnosed Acute Myeloid Leukemia (AML) Not Candidate for Standard Intensive Induction Therapy (SIIT); 2016. https://ClinicalTrials.gov/show/NCT02303782. Accessed March.
- 192.Oncoethix GmbH A Dose‐finding Study of the Bromodomain (Brd) Inhibitor OTX015/MK‐8628 in Hematologic Malignancies (MK‐8628‐001); 2017. https://ClinicalTrials.gov/show/NCT01713582. Accessed 20 January.
- 193.Resverlogix Corp. Safety and Effect of Oral RVX000222 in Subjects With Fabry Disease; 2020. https://ClinicalTrials.gov/show/NCT03228940. Accessed 30 September.
- 194.Resverlogix Corp, PPD, ICON plc, Medidata Solutions. Effect of RVX000222 on Time to Major Adverse Cardiovascular Events in High‐Risk T2DM Subjects With CAD (BETonMACE); 2019. https://ClinicalTrials.gov/show/NCT02586155. Accessed 30 November.
- 195.SignalRX Pharmaceuticals, Inc., University of California, San Diego. Phase 1 Study of SF1126 in Combination With Nivolumab in Patients With Advanced Hepatocellular Carcinoma; 2019. https://ClinicalTrials.gov/show/NCT03059147. Accessed 22 April.
- 196.Steeve Provencher, Resverlogix Corp, Steeve Provencher, Laval University. Apabetalone for Pulmonary Arterial Hypertension: a Pilot Study (APPRoAcH‐p); 2020. https://ClinicalTrials.gov/show/NCT03655704. Accessed June.
- 197.University of Texas Southwestern Medical Center. Study of CPI‐0610 in Patients With Malignant Peripheral Nerve Sheath Tumors; 2018. https://ClinicalTrials.gov/show/NCT02986919. Accessed 17 May.
- 198.Zenith Epigenetics. A Study of ZEN003694 in Combination With Enzalutamide in Patients With Metastatic Castration‐Resistant Prostate Cancer; 2019. https://ClinicalTrials.gov/show/NCT02711956. Accessed October.
- 199.Zenith Epigenetics. A Study of ZEN003694 in Patients With Metastatic Castration‐Resistant Prostate Cancer (TNBC); 2017. https://ClinicalTrials.gov/show/NCT02705469. Accessed October.
- 200.Zenith Epigenetics, Pfizer. Study of ZEN003694 and Talazoparib in Patients With Triple Negative Breast Cancer (TNBC); 2020. https://ClinicalTrials.gov/show/NCT03901469. Accessed September.
- 201. Meslamani J, Smith SG, Sanchez R, Zhou MM. Structural features and inhibitors of bromodomains. Drug Discov Today Technol. 2016;19:3‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Gacias M, Gerona‐Navarro G, Plotnikov AN, et al. Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression. Chem Biol. 2014;21(7):841‐854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Zhong HJ, Lu L, Leung KH, et al. An iridium(iii)‐based irreversible protein–protein interaction inhibitor of BRD4 as a potent anticancer agent. Chem Sci. 2015;6(10):5400‐5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Watson RJ, Bamborough P, Barnett H., et al. GSK789: a selective inhibitor of the first bromodomains (BD1) of the bromo and extra terminal domain (BET) proteins. J Med Chem. 2020. 10.1021/acs.jmedchem.0c00614 [DOI] [PubMed] [Google Scholar]
- 205. Gilan O, Rioja I, Knezevic K, et al. Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation. Science. 2020;368(6489):387‐394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Zhang G, Plotnikov AN, Rusinova E, et al. Structure‐guided design of potent diazobenzene inhibitors for the BET bromodomains. J Med Chem. 2013;56(22):9251‐9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Cheung K, Lu G, Sharma R, et al. BET N‐terminal bromodomain inhibition selectively blocks Th17 cell differentiation and ameliorates colitis in mice. Proc Natl Acad Sci U S A. 2017;114(11):2952‐2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Chiang CM. Nonequivalent response to bromodomain‐targeting BET inhibitors in oligodendrocyte cell fate decision. Chem Biol. 2014;21(7):804‐806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Lin X, Huang X, Bellin R, et al. Abstract 800: ABBV‐744, a first‐in‐class and highly selective inhibitor of the second bromodomain of BET family proteins, displays robust activities in preclinical models of acute myelogenous leukemia. Paper presented at the AACR Annual Meeting 2018, Chicago, IL, 14–18 April 2018.
- 210. Picaud S, Wells C, Felletar I, et al. RVX‐208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci U S A. 2013;110(49):19754‐19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.AbbVie. A Study Evaluating the Safety and Pharmacokinetics of ABBV‐744 in Subjects With Advanced Prostate Cancer (CRPC) and Relapsed/Refractory Acute Myeloid Leukemia (AML) Cancer. https://ClinicalTrials.gov/show/NCT03360006
- 212. Kati W. ABBV‐744: A first‐in‐class highly BDII‐selective BET bromodomain inhibitor. AACR Annual Meeting, Chicago, IL, 14‐18 April 2018.
- 213.Resverlogix Corp. Effect of RVX000222 on Time to Major Adverse Cardiovascular Events in High‐Risk T2DM Subjects With CAD (BETonMACE). https://ClinicalTrials.gov/show/NCT02586155
- 214. Ray KK, Nicholls SJ, Buhr KA, et al. Effect of apabetalone added to standard therapy on major adverse cardiovascular events in patients with recent acute coronary syndrome and type 2 diabetes: a randomized clinical trial. JAMA. 2020, 323, 1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Nicholls SJ, Gordon A, Johansson J, et al. Efficacy and safety of a novel oral inducer of apolipoprotein a‐I synthesis in statin‐treated patients with stable coronary artery disease a randomized controlled trial. J Am Coll Cardiol. 2011;57(9):1111‐1119. [DOI] [PubMed] [Google Scholar]
- 216. Berthon C, Raffoux E, Thomas X, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose‐escalation, phase 1 study. Lancet Haematol. 2016;3(4):e186‐195. [DOI] [PubMed] [Google Scholar]
- 217. Kulikowski E, Halliday C, Johansson J, et al. Apabetalone mediated epigenetic modulation is associated with favorable kidney function and alkaline phosphatase profile in patients with chronic kidney disease. Kidney Blood Press Res. 2018;43(2):449‐457. [DOI] [PubMed] [Google Scholar]
- 218.GlaxoSmithKline. Dose Escalation and Expansion Study of GSK525762 in Combination With Fulvestrant in Subjects With Hormone Receptor‐positive (HR+)/Human Epidermal Growth Factor Receptor 2 Negative (HER2‐) Advanced or Metastatic Breast Cancer. https://ClinicalTrials.gov/show/NCT02964507
- 219. GlaxoSmithKline . A Dose Escalation Study to Investigate the Safety, Pharmacokinetics (PK), Pharmacodynamics (PD), and Clinical Activity of GSK525762 Plus Trametinib in Subjects With Solid Tumors. https://ClinicalTrials.gov/show/NCT03266159
- 220. GlaxoSmithKline . A Dose Escalation Study to Investigate the Safety, Pharmacokinetics (PK), Pharmacodynamics (PD) and Clinical Activity of GSK525762 in Subjects With Relapsed, Refractory Hematologic Malignancies. https://ClinicalTrials.gov/show/NCT01943851
- 221. GlaxoSmithKline . A Study to Investigate the Safety, Pharmacokinetics, Pharmacodynamics, and Clinical Activity of GSK525762 in Subjects With NUT Midline Carcinoma (NMC) and Other Cancers. https://ClinicalTrials.gov/show/NCT01587703
- 222. GlaxoSmithKline . Dose Escalation and Dose Expansion Study of GSK525762 in Combination With Androgen Deprivation Therapy and Other Agents in Subjects With Castrate‐resistant Prostate Cancer. https://ClinicalTrials.gov/show/NCT03150056
- 223. GlaxoSmithKline . A Cross‐over Study to Evaluate the Effect of Itraconazole and Rifampicin on the Pharmacokinetics (PK) of GSK525762 in Healthy Female Subjects of Non Child Bearing Potential. https://ClinicalTrials.gov/show/NCT02706535
- 224. National Cancer Institute (NCI) . Testing a New Anti‐cancer Drug Combination, Entinostat and GSK525762C, for Advanced and Refractory Solid Tumors and Lymphomas. https://ClinicalTrials.gov/show/NCT03925428
- 225. GlaxoSmithKline. Compassionate Use Individual Request Program for GSK525762 in NUT Midline Carcinoma . https://ClinicalTrials.gov/show/NCT03702036
- 226. Resverlogix Corp . A Two‐Part Phase 2a Study of RVX000222 in Patients With End‐Stage Renal Disease Treated With Hemodialysis; 2021. https://ClinicalTrials.gov/show/NCT03160430. Accessed 15 March.
- 227. Resverlogix Corp . Safety, Pharmacokinetic Study of RVX000222 in Healthy Subjects and Subjects With Low HDL Cholesterol; 2009. https://ClinicalTrials.gov/show/NCT03160430. Accessed August.
- 228. Resverlogix Corp . Clinical Trial for Dose Finding and Safety of RVX000222 in Subjects With Stable Coronary Artery Disease (ASSERT); 2010. https://ClinicalTrials.gov/show/NCT01058018. Accessed June.
- 229. Resverlogix Corp, Baker IDI Heart and Diabetes Institute, Nucleus Network Ltd . The Effects of RVX000222 on Glucose Metabolism in Individuals With Pre‐diabetes; 2014. https://ClinicalTrials.gov/show/NCT01728467. Accessed March.
- 230. Resverlogix Corp, The Cleveland Clinic. ApoA‐I Synthesis Stimulation and Intravascular Ultrasound for Coronary Atheroma Regression Evaluation (ASSURE I) ; 2013 https://ClinicalTrials.gov/show/NCT01067820. Accessed May.
- 231. Resverlogix Corp, The Cleveland Clinic . The Study of Quantitative Serial Trends in Lipids With ApolpoproteinA‐I Stimulation; 2012. https://ClinicalTrials.gov/show/NCT01423188. Accessed June.
- 232.National Cancer Institute (NCI). Testing of the Addition of a New Anti‐cancer Drug, Molibresib, to Chemotherapy Treatment (Etoposide and Cisplatin) for Patients With NUT Carcinoma. https://ClinicalTrials.gov/show/NCT04116359
- 233. Resverlogix Corp, South Australian Health and Medical Research Institute . Characterization of Multi‐dose RVX000222 in Combination With Statin Treatment in Dyslipidemia; 2013. https://ClinicalTrials.gov/show/NCT01863225. Accessed June.


