Abstract
Ultrasonography (US) is noninvasive and offers real-time, low-cost, and portable imaging that facilitates the rapid and dynamic assessment of musculoskeletal components. Significant technological improvements have contributed to the increasing adoption of US for musculoskeletal assessments, as artificial intelligence (AI)-based computer-aided detection and computer-aided diagnosis are being utilized to improve the quality, efficiency, and cost of US imaging. This review provides an overview of classical machine learning techniques and modern deep learning approaches for musculoskeletal US, with a focus on the key categories of detection and diagnosis of musculoskeletal disorders, predictive analysis with classification and regression, and automated image segmentation. Moreover, we outline challenges and a range of opportunities for AI in musculoskeletal US practice.
Keywords: Ultrasonography, Musculoskeletal system, Artificial intelligence, Machine learning, Deep learning
Introduction
Ultrasound (US) imaging is a useful diagnostic tool for the examination of the musculoskeletal system. It provides real-time imaging without ionizing radiation and is noninvasive. It has also gained popularity for the dynamic imaging of small structures and the evaluation of the ligaments, muscle and tendons, superficial tumors, and peripheral nerves [1]. Progressive advances in US, including refined transducer technology, power Doppler sonography, and real-time US elastography (EUS), have expanded its clinical applications in the field of musculoskeletal imaging. Additionally, new capabilities that enhance spatial resolution and image quality, such as speckle reduction, video capturing, harmonic tissue imaging, compound imaging, and panoramic imaging, have emerged following revolutionary innovations in computing power and algorithms [2]. For example, EUS facilitates the accurate detection of subclinical changes in the muscles and tendons by leveraging the mechanical properties of musculoskeletal tissue for early diagnosis and therapy monitoring [3].
Within the last decade, classical computer-aided diagnosis (CADx) systems emerged as analytic tools that incorporated a selected set of quantitative features (e.g., first-, second- and higher-order statistical features) to detect abnormal regions in US images accurately. Various machine learning (ML) techniques have been developed to support high-performance computer-aided detection (CADe) or CADx systems for the detection of clinically significant regions, automated segmentation, and classification based on extracted radiological features [4,5]. The image processing techniques used with ML algorithms automate the diagnostic process of detection and characterization for numerous diseases. However, they are limited by their inability to generalize different parameters and high dependence on the settings of the US scanner and image acquisition system. For musculoskeletal US imaging, the integration of conventional ML and CADe or CADx techniques has also been limited by small, thin, narrow, or curved anatomical structures such as extensor digitorum tendons, tiny ligaments or retinacula of the hand, and small nerve branches. These structures may have thicknesses ranging between 0.1 and 0.6 mm [6], making them prone to segmentation and classification errors. Additionally, although US has been explored for musculoskeletal imaging, the curation of US musculoskeletal data remains challenging. Concurrently, the identification of the sonographic appearance of specific anatomical structures remains suboptimal and is still evolving [7], and relevant datasets of musculoskeletal US images with expert annotations are limited.
Over the years, deep learning (DL) has become a notable subfield of artificial intelligence (AI) for high-quality image interpretation and acquisition, and offers support to health professionals for objective and accurate US image analysis [8]. DL in musculoskeletal US is receiving significant attention for automated feature engineering with deep neural networks (DNNs). DNNs are expected to be pivotal in the development of next-generation cutting-edge US imaging systems, exploiting the intrinsic complexities of the anatomical structures and tissues to drive effective triaging and rapid diagnosis. This paper provides an overview of definitions and reviews the recent literature on applications of AI in musculoskeletal US. It also evaluates the future of AI in musculoskeletal US imaging, focusing on key categories of AI-based US image enhancement, classification, detection, and automated segmentation. This article aims to provide musculoskeletal professionals with an understanding of AI technology and to outline the implications of its integration in musculoskeletal US for real-world clinical practice.
What Is AI?
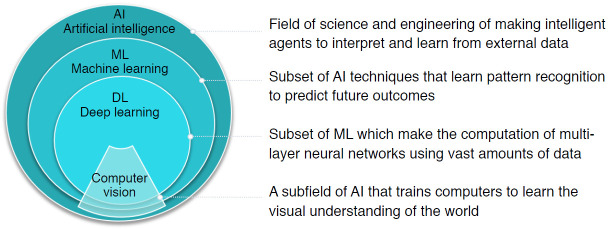
AI is defined as a field of science and engineering that seeks to create intelligent machines to interpret and learn from external data and achieve specific objectives, including natural language processing, robotics, and ML. As a subset of AI, ML is a powerful set of computational tools that have incredible pattern-recognizing abilities, enabling them to automate the reasoning processes of experts (Fig. 1).
Fig. 1. Overview of definitions of artificial intelligence (AI), machine learning (ML), deep learning (DL), and computer vision, as well as their nested relationships.
ML: Feature Extraction and Classification Algorithms
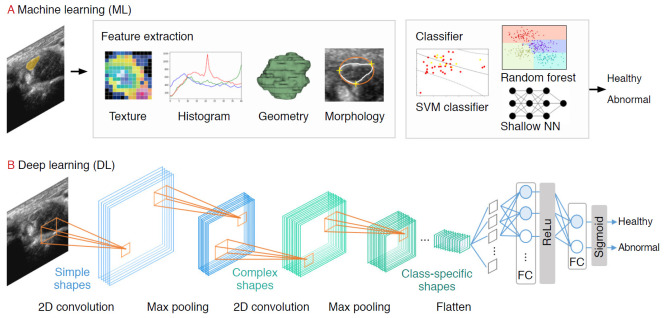
Prior to the explosive growth of DL, ML-based strategies that trained models on descriptive patterns obtained from rules of human inference were dominant [4]. Researchers manually transformed raw data into features, followed by the selection of the best features (e.g., intensity histograms, texture-based features, geometric features, and morphological features). Subsequently, traditional ML classification algorithms (e.g., random forest and support vector machine [SVM] algorithms) were applied to the extracted features [9] (Fig. 2A). A comprehensive review of ML studies shows numerous high-quality techniques to evaluate musculoskeletal disorders based on US images (Table 1) [10-15]. However, a major challenge facing these approaches is that feature selection heavily relies on statistical insights and domain knowledge, and this limitation initiated a paradigm shift from manual feature engineering to DL architectural design.
Fig. 2. A pipeline of ultrasound research using machine learning (ML) and deep learning (DL).
For the ML pipeline, the practitioner extracts the features (e.g., texture, histogram, geometry, morphology) manually before feeding it into the classification model. In the DL pipeline, features are extracted automatically using convolutional filters and pooling. SVM, support vector machine.
Table 1.
Overview of machine learning algorithms and applications used in musculoskeletal ultrasound imaging
| Algorithm | Advantage | Limitation | Example application in musculoskeletal ultrasonography |
|---|---|---|---|
| Logistic regression | Provides probabilistic interpretation of model parameters | Only used to predict discrete function | - |
| Quick model update for incorporating new data | Sensitive to outliers | ||
| K-nearest neighbors | Nonparametric model | Time-consuming and computationally expensive | Nerve identification [10] |
| Used both for classification and regression problems | Number of neighbors must be defined in advance | ||
| Low interpretability | |||
| Naïve Bayes | Suitable for relatively small datasets | Classes must be mutually exclusive | - |
| Handles both binary and multi-class classification problems | Presence of dependency between attributes results in loss of accuracy | ||
| Fast application and high computational efficiency | Assumptions such as the normal distribution might be invalid | ||
| Support vector machines | Good prediction performance in different tasks | Have "black box" characteristics | Lumbar spine classification [11] |
| Can handle multiple feature spaces | Sensitive to manual parameter tuning and kernel choice | Synovitis grading [12] | |
| Nerve identification [10] | |||
| Decision trees | Perform in datasets with large number of features | Only axis-aligned rectangle splits. | Nerve identification [10] |
| Few parameter tuning | Inadequate for regression and continuous value prediction problems | ||
| High representational power and easy to interpret | Mistake in higher labels cause errors in subtrees | ||
| Random forest | Provide estimates of variable or attribute importance in the classification | Complex and computationally expensive | Myositis classification [13] |
| Ensemble-based classifications shows relatively good performance | Number of base classifiers needs to be defined | Hip 2-D US adequacy classification [14] | |
| Overfitting has been observed for noisy data | |||
| Neural networks | Direct image processing | Have "black box" characteristics | Nerve identification [10] |
| Can map complex nonlinear relationships between dependent and independent variables | Have to fine-tune many parameters | ||
| Require a large well-annotated dataset to achieve good performance | |||
| K-means | Can process large datasets | Number of clusters must be defined | Nerve localization [15] |
| Algorithm that is simple to understand and implement |
DL: Convolutional Neural Networks
DL, based on increasing the number of hidden neural network layers, revolutionized the field of end-to-end learning by bypassing the hand-crafted engineering stages that characterize ML pipelines (Fig. 2A) [4]. While conventional radiological assessments are often based on radiologists’ knowledge and experience, DNNs automatically recognize patterns from data and achieve remarkable performance in various applications in radiology [16,17], dermatology [18], and ophthalmology [19], among others.
Several DL applications in radiology are supervised, and most of them are based on convolutional neural networks (CNNs). CNNs comprise a sequence of hidden layers, each responding to unique features that transform images into output class scores (Fig. 2B). CNNs are biologically-inspired neural networks that mimic the physiology of the visual cortex by responding differently to specific features [20]. Simple cells in the visual cortex are the most specific, as they detect lines, edges, and corners in a visual field. Complex features such as colors, shapes, and orientations are captured by complex cells, which show more spatial invariance by pooling the outputs of simple cells. Similar to human visual perception, which is regulated by the visual cortex, two main characteristics make CNNs optimum for image classification: the increasing shape "selectivity" and "the invariance" of the visual representation through feedforward connections.
A CNN is composed of a series of three layers: a convolutional (CONV) layer, followed by a pooling layer, and finally, a fully connected layer (Fig. 2B). The CONV layer forms the basis of the CNN, containing a set of filters with parameters that need to be learned. During the forward pass, the input is convolved by several filters to compute two-dimensional (2D) activation maps of every spatial region. These activations are processed layer by layer to extract high-level features. Repeated convolution of the image results in a map of activations, called feature maps, which represents the location and strength of unique features, pixels, and characters invariant to translation. The pooling layer performs a down-sampling of the spatial dimension and reduces the spatial sizes of the representations. Thus, the number of parameters to be learned and computational complexity is decreased. The fully connected layer maintains full connectivity between the neurons of each preceding and succeeding layer. The CONV and pooling layers perform feature extractions of the given image, and the fully connected layer acts as a classifier that discriminates based on the high-level representation of images.
AI in Musculoskeletal US Imaging
The latest improvements in US imaging technology have been linked to improved accuracy in the diagnosis of musculoskeletal disorders. However, the dependence on subjective assessments of displayed images and the variability in image acquisition and equipment used across studies [21] have delayed the widespread of US compared to that of magnetic resonance imaging (MRI). To help overcome these problems, as well as the ambiguity with which musculoskeletal disorders may present on US imaging, CADe/CADx has become a major solution in radiology.
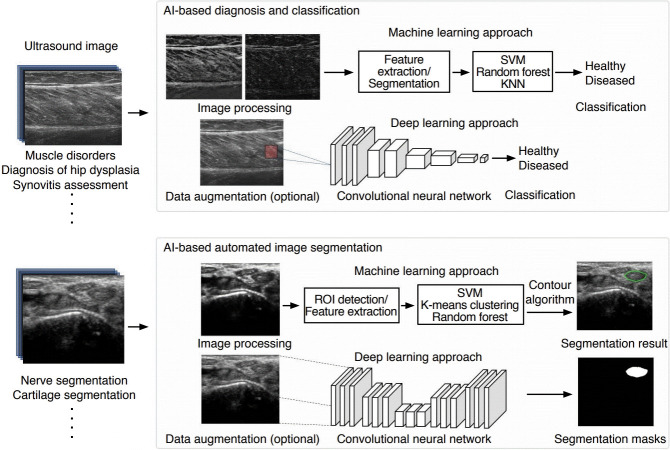
CADe/CADx systems provide quantitative analysis and a complimentary opinion that supports radiologists in making accurate and consistent image assessments quickly. The early versions of CADe/CADx systems in musculoskeletal US imaging were based on hand-crafted ML engineering with multiple processing stages: (1) image preprocessing with segmentation and region of interest (ROI) designation, (2) feature extraction and selection, and (3) classification based on the selected features. However, classical CADe/CADx systems suffer from various algorithmic limitations related to data gathering, preprocessing, filtering, and generalization. Evaluations are also restricted due to bias and variance caused by finite samples collected from different medical centers. DL-based US CADe/CADx systems have been proposed for next-generation AI-powered radiology because trained models can be robust and generalizable given the availability of big data in digitalized radiology. Currently, the most notable application of AI in musculoskeletal imaging is pattern recognition and image classification. Unlike the applications of AI of image reconstruction, enhancement, or synthesis for other modalities (e.g., MRI or computed tomography), AI applied to musculoskeletal US focuses mainly on diagnosis, classification, or segmentation (Fig. 3).
Fig. 3. Schematic of artificial intelligence (AI)-based machine learning and deep learning applications in musculoskeletal ultrasound imaging of AI-based diagnosis and classification and AI-based automated image segmentation.
SVM, support vector machine; KNN, k-Nearest Neighbor; ROI, region of interest.
Automatic Diagnosis and Detection in Musculoskeletal US
Muscle Disorders
Skeletal muscle US is a point-of-care technique for visualizing skeletal muscle structure, fatty atrophy, movement, and function in real-time. The portability and high spatial resolution of US make it an ideal imaging modality for the detection and diagnosis of muscle injuries, myopathy, or myositis [22]. However, due to variations in operator-dependent techniques and the intra- and inter-reader variability in the qualitative assessment of muscle echogenicity [23,24], computer-aided quantitative methods for the detection of muscle pathology and identification of structural abnormalities of muscle have been studied.
In earlier studies of quantitative muscle US, muscle echo intensity were obtained by grayscale analysis of regions of interest using numerous texture descriptors for muscle characterization [25-27]. To identify Duchenne muscular dystrophy (DMD), echo intensity, and muscle thickness were quantified to distinguish myopathic muscles with increased echogenicity [28]. To detect disease progression in DMD patients, the grayscale level and quantitative backscatter analysis have shown to be more sensitive than functional assessments [29]. To further characterize different muscle types in US images according to sex, different combinations of first-order and higher-order texture descriptors were shown to be useful for muscle disorders [30].
Further developments have involved comparative analyses of ML and DL applied to muscle US images to automatically or semi-automatically differentiate among inclusion body myositis, polymyositis, dermatomyositis, and normal presentations [13]. The DL strategy improved accuracy in all scenarios compared to conventional ML algorithms (random forest), and this supports the notion that manually engineered features, while useful, may facilitate suboptimal diagnosis and are inadequate for a full characterization of disease complexity compared to automatically generated feature sets from DL models.
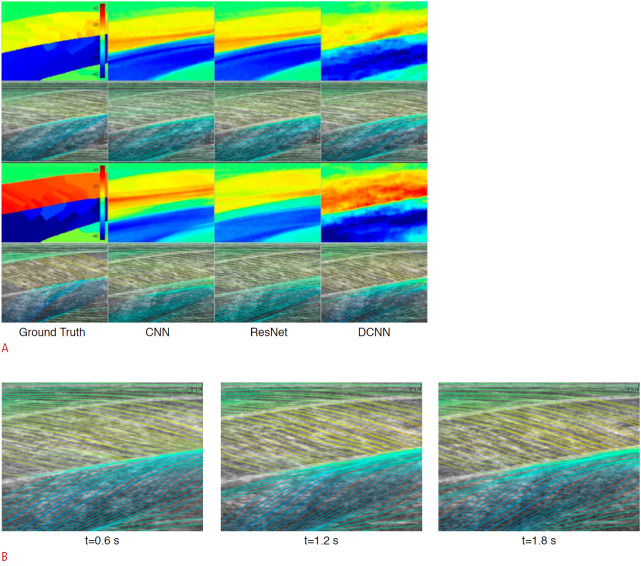
In addition to the detection and classification of muscle pathology, ML and DL approaches have been applied to the objective estimation of muscle fiber orientations from B-mode US images to predict a continuous output. Deep residual networks and deconvolutional CNNs have made better predictions or estimations of full-spatial-resolution muscle fibers than the wavelet method and CNNs (Fig. 4) [31,32]. These objective measurements of muscular length change, thickness, and tendon length are useful for evaluating muscles. They are also useful for understanding, diagnosing, monitoring, and treating muscular disorders.
Fig. 4. Deep convolutional neural network (DCNN)-based fiber orientation.
A. A representation of DCNN predictions for fiber orientation is given. A fiber orientation heatmap is shown in the top image, and a line trace representation overlaid on the ultrasound image is shown in the bottom image. CNN, convolutional neural network. B. The temporal variation in fiber orientation traces of maximum voluntary contraction (starting at 0 second and ending at 2.2 seconds) is given. Reprinted from Cunningham et al. J Imaging 2018;4:29, according to the Creative Commons license.
Diagnosis of Hip Dysplasia
Developmental dysplasia of the hip (DDH) is a developmental deformity of the hip joint characterized by anatomical abnormalities between the head of the femur and the acetabulum [33]. Detecting DDH as early as possible is crucial to avoid the development of residual hip dysplasia or hip osteoarthritis (OA). However, the current US diagnosis of DDH using 2D US is limited by low interobserver reliability [34] and workflow timing of the US evaluation [35]. Thus, AI-based techniques are preferred for the automated interpretation of 2D and three-dimensional (3D) hip US images. The application of AI facilitates a quick estimation of conventional parameters (e.g., alpha angle, acetabular contact angle [ACA]) to provide higher diagnostic accuracy and to minimize interobserver variability with reduced imaging time.
To achieve accurate and rapid semi-automatic delineation of the acetabulum, a semiautomated segmentation technique was used to generate 3D acetabular surface models interpolated from optimal paths passing through user-defined seed points [36]. The ACA delineated from the segmented 3D surface was used to classify the acetabulum as normal, borderline, or dysplastic. Additionally, a fully automated DDH diagnostic technique was proposed, using DNN segmentation with an adversarial network to automatically segment landmarks to estimate Graf's alpha angle in US of infants’ hips [37]. Hareendranathan et al. [38] presented a novel segmentation pipeline, in which US images are first segmented into multiple clusters using a simple linear iterative clustering algorithm. Subsequently, a CNN is trained to classify each segmented cluster, outputting a high-probability region defining the acetabular contour.
Due to advances in segmentation with deep architectures [39-41] and high-level semantic segmentation through complete scene understanding or attentional supervision, it is highly anticipated that DL techniques will replace AI-based applications for DDH assessment. Additionally, the segmentation of 3D inputs using different DL networks [42] to measure the ACA using 3D US may provide a more reliable diagnosis with lower inter-scan variability and shorter processing time.
Automatic Regression and Classification of Musculoskeletal US
Synovitis Assessment
In recent years, US has been used in clinical practice to assist in the assessment and the characterization of patients with synovial proliferative disorders such as inflammatory arthritis, particularly rheumatoid arthritis. The recently published OMERACT-EULAR Synovitis Scoring (OESS) system standardized US scanning for grading synovitis, representing a significant step toward monitoring inflammatory arthritis disease activity [43,44]. Different automated techniques for detection of the synovitis region, quantification of the synovium based on segmentation, and regression models to grade the severity of synovitis have been actively studied.
Synovitis scoring using AI-based computerized techniques may reduce the existing discrepancies between readers [45]. However, designing an automated framework for the quantitative assessment of synovitis in US images is challenging since it is necessary to differentiate synovitis from extra-articular structures, including skin borders and bone regions, under conditions of blurred margins and inhomogeneous echogenicity. The most widely used design of an automated framework for synovitis assessment follows the pipeline of (1) skin border selection, (2) bone location, and (3) synovitis region segmentation. An automated tool for estimation and grading of the synovitis is designed to detect the skin, bone, and joint synovitis through image binarization and image statistics thresholding [46] or in combination with ML classification (SVMs) (Fig. 5) [12].
Fig. 5. Example of synovitis area detection with a machine learning-based pipeline, as suggested by Mielnik et al. [12].
A-D. The detection of synovitis in the proximal interphalangeal joint (A), detection of synovitis in the metacarpophalangeal joint (B), example of underestimated region of synovitis (C), example of error in synovial hypertrophy detection (D) are shown. Reprinted from Mielnik et al. Ultrasound Med Biol 2018;44:489-494, Copyright (2020), with permission from Elsevier [12].
Recently, DL has shown the potential for joint detection and synovitis grading. A hybrid of image processing and an intensity-based algorithm has been proposed for skin border segmentation, while a connectivity algorithm has been proposed for bone region segmentation [47], to grade synovitis. Deep CNNs (DCNNs) have demonstrated the potential to serve as a feasible method for classifying disease activity according to the OESS system. Two DCNN architectures (VGG-16 for classifying healthy or diseased, and Inception-V3 for classifying OESS scores) were applied to Doppler US images, and they achieved high accuracy for low and high-level classification (area under the curve, 0.93) and full-scale OESS classification (quadratically weighted kappa, 0.84) [48]. The applications of DCNNs to the direct quantification of synovitis from entire US images are promising; however, to support an automated DL-based diagnosis, visualization methods such as attention maps that highlight the radiological areas of interest [49] must be integrated into US applications to maintain transparency in the decision process. Given the technological process, dynamic models (e.g., recurrent neural networks) predicting disease progression or quantitative assessment of 3D synovial proliferation and synovitis can be expected.
Spine Level Analysis and Identification
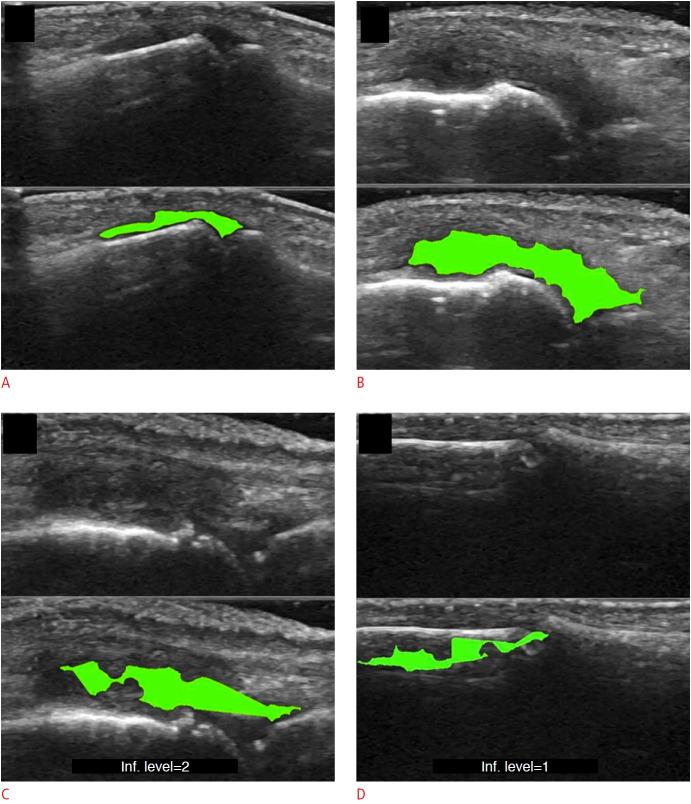
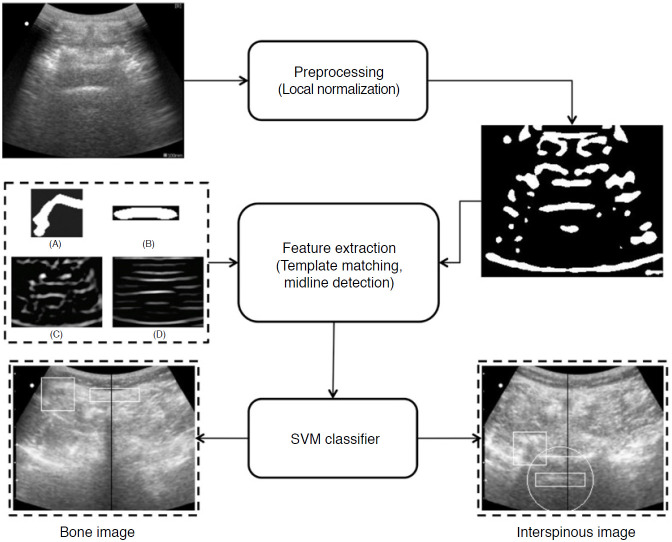
US-guided interventions are gaining popularity for facilitating vascular access, peripheral nerve blocks, and neuraxial anesthesia [50]. Unfortunately, the visualization of spinal US images remains indirect due to inherent speckle noise and the acoustic shadow cast by bone surfaces that hide key anatomical sites, making spinal US technically challenging for a novice who has received limited training in reading US images. Therefore, computer processing algorithms and ML methods have been applied to identify the needle puncture site to aid in US image interpretation. For this, template matching techniques such as lamina and ligamentum flavum (LF) detection [51], midline detection [11], and signatory features detection [52] have been used to identify the skin-to-LF depth and bone structures (Fig. 6). However, the aforementioned template matching-based approaches use a static template stored from a subset of finite subjects for target identification that cannot cover the complete inter-patient variability of spine structures and surroundings.
Fig. 6. Image identification for lumbar ultrasound image.
The pipeline proposed by Yu et al. [11] consists of a feature extraction method to extract important anatomic features and midline detection and classification stage for interspinous region identification. SVM, support vector machine. Reprinted from Yu et al. Ultrasound Med Biol 2015;41:2677-2689, Copyright (2020), with permission from Elsevier [11].
Recently, ML feature extraction and neural network classification techniques were proposed to automate the identification of the optimum plane for epidural steroid and facet joint injections [53]. A method for vertebral localization in the operation room by registering the spinous process shape from radiograph spinous annotations and U-Net segmentation of sagittal US images has been proposed to provide tracker-free 2D US imaging [54]. To visualize the spinal anatomy while performing needle insertion, the SLIDE system [55] has been proposed to classify three characteristic transverse planes, namely the sacrum, intervertebral gaps, and vertebral bones, in real-time. Without the need for predefined features, SLIDE utilizes transfer learning and four different DCNN architectures to learn features of the spine entirely, and a state machine was developed to accurately identify transitions between the planes.
Automated US Image Segmentation Techniques
Nerve Localization and Segmentation
US is the primary diagnostic imaging modality for suspected peripheral neuropathy, particularly when neurological examinations are inconclusive. Nerve conduction velocity or electromyography are commonly analyzed, and morphometric parameters (e.g., cross-sectional area [CSA]) and quantitative measurements of sonographic elastography of peripheral nerves can reflect degrees of peripheral neuropathy. The quantitative evaluation of peripheral nerves using US or sonographic elastography is proceeded by observation of the CSA or assessment of a manually measured ROI. Thus, there is increasing demand for automatic segmentation to extract the ROI of the abnormal signal of the peripheral nerve to reduce the burden of time-consuming and labor-intensive manual measurements. Fully- or semi-automatic AI-based segmentation has shown potential benefits for the segmentation of peripheral nerves due to their near-instantaneous assessment, cost-effectiveness, and high reproducibility.
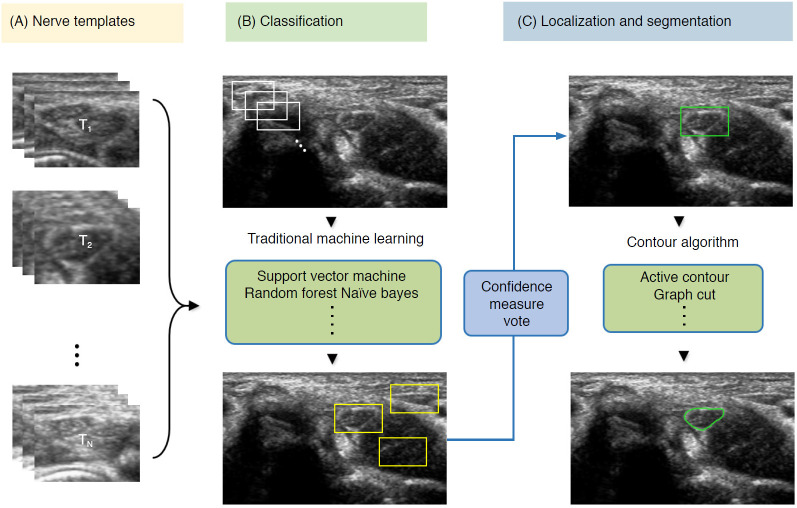
To detect nerve regions to assist in US segmentation, the majority of classical AI methods are based on four main steps: (1) despeckle filtering, (2) template-based ROI detection, (3) feature-based nerve region classification, and (4) segmentation (Fig. 7). The method proposed by Hadjerci et al. [56] uses the median binary pattern and the Gabor filter to the separated hyperechoic tissues, followed by SVM extraction of the nerve region. Furthermore, the authors reported a comparative study of nerve segmentation with a quantitative performance evaluation using 11 despeckling filters, six statistical feature extraction methods, filter- and wrapper-based feature selection, and five ML-based classifiers [10]. However, these classical feature representations require extensive reformulation of numerous frameworks, statistical insights, and expert domain knowledge, and optimality is not guaranteed.
Fig. 7. Conventional machine learning-based segmentation scheme of nerve ultrasonography.
Sliding window template-based classification is applied to generate candidate regions of interest. The nerve region is localized based on a confidence measure vote, and segmentation is applied to obtain nerve boundaries.
To alleviate the time inefficiency of human interventions and to bypass the intermediate stages in conventional ML-based pipeline designs, DL-based segmentation (U-Net architecture) has been applied to identify musculocutaneous, median, ulnar, and radial nerves [57], as well as femoral nerve blocks in US images [58]. DCNN-based nerve segmentation with variants inspired by the original U-Net architecture was applied [59,60] to NERVE datasets (brachial plexus segmentation in US images, available at https://www.kaggle.com/c/ultrasound-nerve-segmentation). Weng et al. [61] employed neural architecture search, which used autoML algorithms that returned the best neural network through the sampling of building blocks to create an end-to-end structure, and achieved encouraging results (mean intersection over union, 0.992; Dice similarity coefficient, 0.881) compared to U-Net segmentation. Additionally, to precisely locate the position while alleviating the resolution reduction of the brachial plexus, Liu et al. [62] developed a deep adversarial network and used dilated convolution to incorporate the global anatomical contextual cues and organ elastic deformation. Since some nerve regions have small areas with indistinguishable characterizations on ultrasonographic images, different DL techniques that optimize the echotexture, incorporate global semantic context, and achieve computationally efficient real-time segmentation [63] may eventually be developed to reduce failed detection of false-positive findings.
Cartilage Segmentation
Knee OA is a common joint disease among older adults, and its prevalence has been increasing; symptomatic OA occurred 10% in men and 13% in women aged 60 years or older [64]. Even though radiography and MRI are standard imaging modalities in clinical practice for the diagnosis of OA, ultrasonography could become a complementary modality for triaging cartilage abnormalities [65] with benefits of non-invasiveness, availability, relative affordability, and safety, as it is performed without ionizing radiation. Therefore, fully automated measurements of cartilage thickness using segmentation-based techniques have been proposed for real-time imaging for knee OA diagnosis and monitoring.
Existing AI-based assessments of cartilage are mainly based on image enhancement and knee cartilage segmentation to represent cartilage thickness with quantitative measures. Hossain et al. [66] used a histogram equalization method of multipurpose beta optimized recursive bi-histogram equalizations to achieve the optimum values of contrast and brightness and to preserve details in the enhancement process of US knee cartilage images. To obtain the true thickness between soft tissue cartilage interface and cartilage-bone interface, quantitative segmentation of the monotonous hypoechoic band was obtained using a locally statistical level set method [67]. Similarly, automated knee-bone surface localization was used to obtain seed points for semi-automatic segmentation methods (random walker, watershed, and graph-cut algorithms), and distance maps are applied to obtain mean cartilage thickness [68]. Recently, the U-Net method has been applied for cartilage segmentation in dynamic, volumetric US images to avoid collision and touching between surgical instruments and anatomical areas during surgery [69] (Fig. 8). Through advances in state-of-art instance segmentation DL models [63], US integrated with DL segmentation might be used to assess of athletic injuries [70], providing accurate real-time feedback.
Fig. 8. Examples of cartilage segmentation based on a U-Net architecture [69].
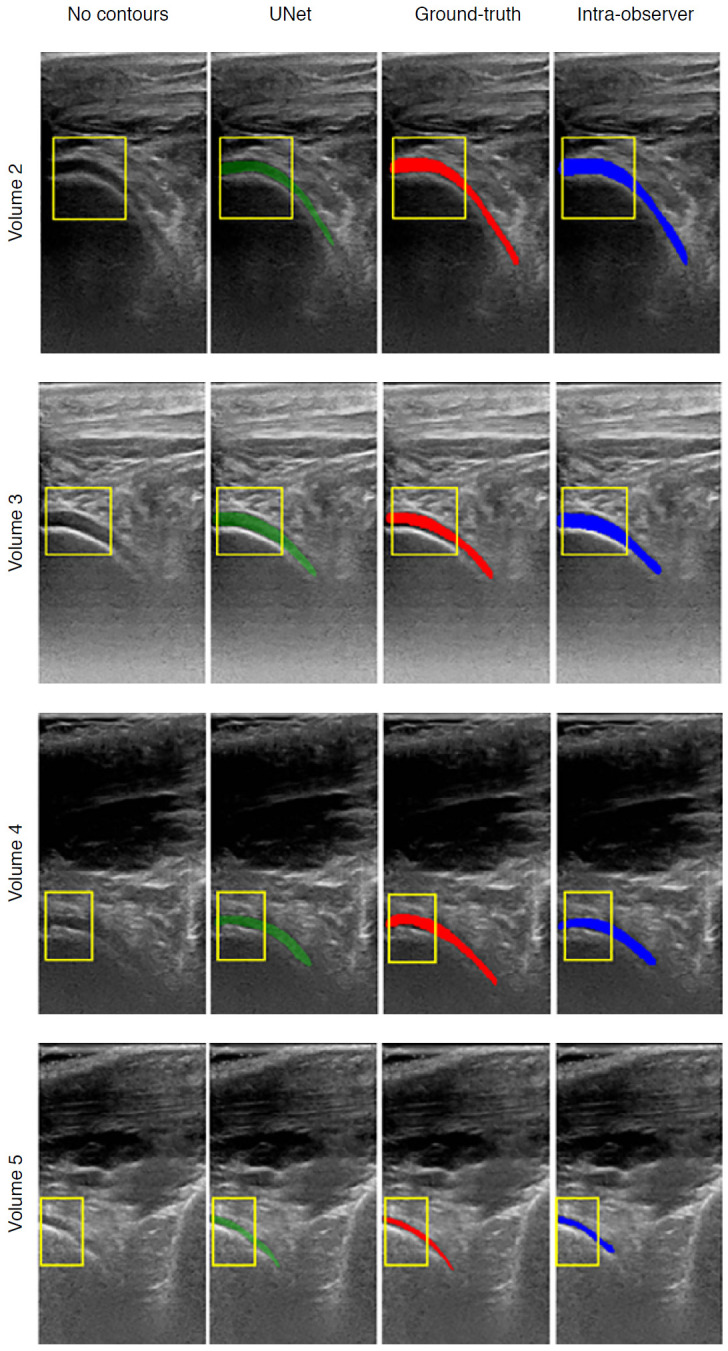
The first column shows examples of images and image regions (yellow box) selected. For each US image in the figure, the segmentations produced by the U-Net (green), by the expert during the ground-truth creation (red), and the intraobserver test (blue) are shown. Reprinted from Antico et al. Ultrasound Med Biol 2020;46:422-435, Copyright (2020), with permission from Elsevier [69].
Challenges and Future Perspectives of AI-Based Musculoskeletal US
Although AI-based musculoskeletal US has shown great potential in overcoming high variability and operator dependency, several limitations must be acknowledged. First, there is a discrepancy between 2D imaging, which is widely utilized in radiology clinics, and 3D imaging. Due to the complexity of musculoskeletal structures and various joints, image preprocessing techniques such as rigid or non-rigid image registration are required for the large-scale application of DL for US. Even for US experts, diagnosis based on 2D US is challenging without a comprehensive understanding of functional anatomy. It is challenging to reproduce and localize the thin 2D US image planes, which is disadvantageous for building a large, standardized medical image dataset. Recent AI-based 3D US imaging techniques may overcome the limitations of 2D US [71,72]. Therefore, the strategies for 3D medical US reconstruction, visualization, and segmentation are promising.
Another challenge is that artifacts, which are frequently encountered, can be mistaken for pathology; furthermore, artifacts can occur together with abnormal conditions in both grayscale and Doppler imaging [73]. Careful curation by healthcare professionals and quality assessment of US training data should be considered beforehand, and appropriate preprocessing and normalization are needed to ensure that the artifacts do not affect AI-based model predictions. Artifact reduction and simultaneous preservation of high resolution via generative adversarial networks or automated quality assessment models may be useful for musculoskeletal US imaging.
Finally, image variability due to motion and differences among US machines and transducers constitutes another fundamental limitation hindering the wide adoption of AI-based musculoskeletal ultrasonography. The sonographer's preferred adjustments and further optimization (e.g., different colormaps for grayscale display, dynamic range, edge enhancements, gamma correction, focal depth) lead to additional high variability and randomness, which limit the accuracy and reproducibility of AI models. Additionally, high-quality musculoskeletal annotations are elusive due to the general paucity of expert musculoskeletal radiologists. Therefore, standardized equipment settings, the use of recent image preprocessing software, and open-source datasets that contain extensive collections of annotations based on a high degree of expert consensus may facilitate the better performance of AI-based musculoskeletal US applications and their increased adoption.
Conclusion
Within the last decade, AI-based musculoskeletal imaging has progressed step by step toward enhancing anatomical structure visualization and automating quantitative measurements. Recent studies on AI-based musculoskeletal US have suggested that DL techniques may become next-generation diagnostic tools for monitoring the condition of joints, bones, cartilage, ligaments, and muscles. The image recognition capability of DL may provide sonographers with real-time diagnostic and decision support. By providing high-quality grayscale images, assessing the appropriateness of US images, and ensuring consistency, AI-based musculoskeletal imaging may facilitate higher-quality patient care.
Acknowledgments
This work was supported by a National Research Foundation (NRF) grant funded by the Korean government, Ministry of Science and ICT (MSIP, 2018R1A2B6009076).
Footnotes
Author Contributions
Conceptualization: Lee YH, Shin Y. Data acquisition: Lee YH, Shin Y. Data analysis interpretation: Lee YH, Shin Y. Drafting of the manuscript: Lee YH, Shin Y. Critical revision of the manuscript: Kim S, Yang J. Approval of the final version of the manuscript: all authors.
No potential conflict of interest relevant to this article was reported.
References
- 1.Nwawka OK. Update in musculoskeletal ultrasound research. Sports Health. 2016;8:429–437. doi: 10.1177/1941738116664326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powers J, Kremkau F. Medical ultrasound systems. Interface Focus. 2011;1:477–489. doi: 10.1098/rsfs.2011.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drakonaki EE, Allen GM, Wilson DJ. Ultrasound elastography for musculoskeletal applications. Br J Radiol. 2012;85:1435–1445. doi: 10.1259/bjr/93042867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18:570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 6.Czyrny Z. Standards for musculoskeletal ultrasound. J Ultrason. 2017;17:182–187. doi: 10.15557/JoU.2017.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douis H, James SL, Davies AM. Musculoskeletal imaging: current and future trends. Eur Radiol. 2011;21:478–484. doi: 10.1007/s00330-010-2024-z. [DOI] [PubMed] [Google Scholar]
- 8.Liu SF, Wang Y, Yang X, Lei B, Liu L, Li SX, et al. Deep learning in medical ultrasound analysis: a review. Engineering. 2019;5:261–275. [Google Scholar]
- 9.Huang Q, Zhang F, Li X. Machine learning in ultrasound computer-aided diagnostic systems: a survey. Biomed Res Int. 2018;2018:5137904. doi: 10.1155/2018/5137904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hadjerci O, Hafiane A, Conte D, Makris P, Vieyres P, Delbos A. Computer-aided detection system for nerve identification using ultrasound images: a comparative study. Inform Med Unlocked. 2016;3:29–43. [Google Scholar]
- 11.Yu S, Tan KK, Sng BL, Li S, Sia AT. Lumbar ultrasound image feature extraction and classification with support vector machine. Ultrasound Med Biol. 2015;41:2677–2689. doi: 10.1016/j.ultrasmedbio.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 12.Mielnik P, Fojcik M, Segen J, Kulbacki M. A novel method of synovitis stratification in ultrasound using machine learning algorithms: results from clinical validation of the MEDUSA project. Ultrasound Med Biol. 2018;44:489–494. doi: 10.1016/j.ultrasmedbio.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Burlina P, Billings S, Joshi N, Albayda J. Automated diagnosis of myositis from muscle ultrasound: exploring the use of machine learning and deep learning methods. PLoS One. 2017;12:e0184059. doi: 10.1371/journal.pone.0184059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quader N, Hodgson AJ, Mulpuri K, Schaeffer E, Abugharbieh R. Automatic evaluation of scan adequacy and dysplasia metrics in 2-D ultrasound images of the neonatal hip. Ultrasound Med Biol. 2017;43:1252–1262. doi: 10.1016/j.ultrasmedbio.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Hadjerci O, Hafiane A, Makris P, Conte D, Vieyres P, Delbos A. Nerve localization by machine learning framework with new feature selection algorithm. In: Murino V, Puppo E, editors. Image Analysis and Processing - International Conference on Image Analysis and Processing 2015. Lecture notes in computer science, Vol. 9279. Cham: Springer; 2015. pp. 246–256. [Google Scholar]
- 16.Liu F, Zhou Z, Samsonov A, Blankenbaker D, Larison W, Kanarek A, et al. Deep learning approach for evaluating knee MR images: achieving high diagnostic performance for cartilage lesion detection. Radiology. 2018;289:160–169. doi: 10.1148/radiol.2018172986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284:574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 18.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arcadu F, Benmansour F, Maunz A, Willis J, Haskova Z, Prunotto M. Deep learning algorithm predicts diabetic retinopathy progression in individual patients. NPJ Digit Med. 2019;2:92. doi: 10.1038/s41746-019-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubel DH, Wiesel TN. Shape and arrangement of columns in cat's striate cortex. J Physiol. 1963;165:559–568. doi: 10.1113/jphysiol.1963.sp007079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mourtzakis M, Parry S, Connolly B, Puthucheary Z. Skeletal muscle ultrasound in critical care: a tool in need of translation. Ann Am Thorac Soc. 2017;14:1495–1503. doi: 10.1513/AnnalsATS.201612-967PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lento PH, Primack S. Advances and utility of diagnostic ultrasound in musculoskeletal medicine. Curr Rev Musculoskelet Med. 2008;1:24–31. doi: 10.1007/s12178-007-9002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandsma R, Verbeek RJ, Maurits NM, van der Hoeven JH, Brouwer OF, den Dunnen WF, et al. Visual screening of muscle ultrasound images in children. Ultrasound Med Biol. 2014;40:2345–2351. doi: 10.1016/j.ultrasmedbio.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Ohrndorf S, Naumann L, Grundey J, Scheel T, Scheel AK, Werner C, et al. Is musculoskeletal ultrasonography an operator-dependent method or a fast and reliably teachable diagnostic tool? Interreader agreements of three ultrasonographers with different training levels. Int J Rheumatol. 2010;2010:164518. doi: 10.1155/2010/164518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Paya JJ, Rios-Diaz J, Del Bano-Aledo ME, Tembl-Ferrairo JI, Vazquez-Costa JF, Medina-Mirapeix F. Quantitative muscle ultrasonography using textural analysis in amyotrophic lateral sclerosis. Ultrason Imaging. 2017;39:357–368. doi: 10.1177/0161734617711370. [DOI] [PubMed] [Google Scholar]
- 26.Wu JS, Darras BT, Rutkove SB. Assessing spinal muscular atrophy with quantitative ultrasound. Neurology. 2010;75:526–531. doi: 10.1212/WNL.0b013e3181eccf8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sogawa K, Nodera H, Takamatsu N, Mori A, Yamazaki H, Shimatani Y, et al. Neurogenic and myogenic siseases: quantitative texture analysis of muscle US data for differentiation. Radiology. 2017;283:492–498. doi: 10.1148/radiol.2016160826. [DOI] [PubMed] [Google Scholar]
- 28.Jansen M, van Alfen N, Nijhuis van der Sanden MW, van Dijk JP, Pillen S, de Groot IJ. Quantitative muscle ultrasound is a promising longitudinal follow-up tool in Duchenne muscular dystrophy. Neuromuscul Disord. 2012;22:306–317. doi: 10.1016/j.nmd.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 29.Zaidman CM, Wu JS, Kapur K, Pasternak A, Madabusi L, Yim S, et al. Quantitative muscle ultrasound detects disease progression in Duchenne muscular dystrophy. Ann Neurol. 2017;81:633–640. doi: 10.1002/ana.24904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molinari F, Caresio C, Acharya UR, Mookiah MR, Minetto MA. Advances in quantitative muscle ultrasonography using texture analysis of ultrasound images. Ultrasound Med Biol. 2015;41:2520–2532. doi: 10.1016/j.ultrasmedbio.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 31.Cunningham R, Harding P, Loram I. Deep residual networks for quantification of muscle fiber orientation and curvature from ultrasound images. In: Valdes Hernandez M, Gonzalez-Castro V, editors. Medical image understanding and analysis. Communications in computer and information science, Vol. 723. Cham: Springer; 2017. pp. 63–73. [Google Scholar]
- 32.Cunningham R, Sanchez MB, May G, Loram I. Estimating full regional skeletal muscle fibre orientation from B-mode ultrasound images using convolutional, residual, and deconvolutional neural networks. J Imaging. 2018;4:29. [Google Scholar]
- 33.Noordin S, Umer M, Hafeez K, Nawaz H. Developmental dysplasia of the hip. Orthop Rev (Pavia) 2010;2:e19. doi: 10.4081/or.2010.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orak MM, Onay T, Cagirmaz T, Elibol C, Elibol FD, Centel T. The reliability of ultrasonography in developmental dysplasia of the hip: How reliable is it in different hands? Indian J Orthop. 2015;49:610–614. doi: 10.4103/0019-5413.168753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel H, Canadian Task Force on Preventive Health Care Preventive health care, 2001 update: screening and management of developmental dysplasia of the hip in newborns. CMAJ. 2001;164:1669–1677. [PMC free article] [PubMed] [Google Scholar]
- 36.Hareendranathan AR, Mabee M, Punithakumar K, Noga M, Jaremko JL. A technique for semiautomatic segmentation of echogenic structures in 3D ultrasound, applied to infant hip dysplasia. Int J Comput Assist Radiol Surg. 2016;11:31–42. doi: 10.1007/s11548-015-1239-5. [DOI] [PubMed] [Google Scholar]
- 37.Golan D, Donner Y, Mansi C, Jaremko J, Ramachandran M. Fully automating Graf’s method for DDH diagnosis using deep convolutional neural networks. In: Carneiro G, editor. Deep learning and data labeling for medical applications. Lecture notes in computer science, Vol. 10008. Cham: Springer; 2016. pp. 130–141. [Google Scholar]
- 38.Hareendranathan A, Zonoobi D, Mabee M, Cobzas D, Punithakumar K, Noga M, et al. Toward automatic diagnosis of hip dysplasia from 2D ultrasound. 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017); 2017 Apr 18-21; Melbourne, VIC, Australia. New York: Institute of Electrical and Electronics Engineers; 2017. pp. 982–985. [Google Scholar]
- 39.Chiao JY, Chen KY, Liao KY, Hsieh PH, Zhang G, Huang TC. Detection and classification the breast tumors using mask R-CNN on sonograms. Medicine (Baltimore) 2019;98:e15200. doi: 10.1097/MD.0000000000015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishra D, Chaudhury S, Sarkar M, Soin AS. Ultrasound image segmentation: a deeply supervised network with attention to boundaries. IEEE Trans Biomed Eng. 2019;66:1637–1648. doi: 10.1109/TBME.2018.2877577. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Ying MT, Yang L, Ahuja AT, Chen DZ. Coarse-to-fine stacked fully convolutional nets for lymph node segmentation in ultrasound images. 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2016 Dec 15-18; Shenzhen, China. New York: Institute of Electrical and Electronics Engineers; 2016. pp. 443–448. [Google Scholar]
- 42.Hesamian MH, Jia W, He X, Kennedy P. Deep learning techniques for medical image segmentation: achievements and challenges. J Digit Imaging. 2019;32:582–596. doi: 10.1007/s10278-019-00227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terslev L, Naredo E, Aegerter P, Wakefield RJ, Backhaus M, Balint P, et al. Scoring ultrasound synovitis in rheumatoid arthritis: a EULAR-OMERACT ultrasound taskforce-Part 2: reliability and application to multiple joints of a standardised consensus-based scoring system. RMD Open. 2017;3:e000427. doi: 10.1136/rmdopen-2016-000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D'Agostino MA, Terslev L, Aegerter P, Backhaus M, Balint P, Bruyn GA, et al. Scoring ultrasound synovitis in rheumatoid arthritis: a EULAR-OMERACT ultrasound taskforce-Part 1: definition and development of a standardised, consensus-based scoring system. RMD Open. 2017;3:e000428. doi: 10.1136/rmdopen-2016-000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D'Agostino MA, Maillefert JF, Said-Nahal R, Breban M, Ravaud P, Dougados M. Detection of small joint synovitis by ultrasonography: the learning curve of rheumatologists. Ann Rheum Dis. 2004;63:1284–1287. doi: 10.1136/ard.2003.012393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nurzynska K, Smolka B. Automatic finger joint synovitis localization in ultrasound images. Real-Time Image and Video Processing 2016; 2016 Apr 6; Brussels, Belgium. Bellingham, WA: International Society for Optics and Photonics; 2016. p. 98970N. [Google Scholar]
- 47.Hemalatha RJ, Vijaybaskar V, Thamizhvani TR. Automatic localization of anatomical regions in medical ultrasound images of rheumatoid arthritis using deep learning. Proc Inst Mech Eng H. 2019;233:657–667. doi: 10.1177/0954411919845747. [DOI] [PubMed] [Google Scholar]
- 48.Andersen JKH, Pedersen JS, Laursen MS, Holtz K, Grauslund J, Savarimuthu TR, et al. Neural networks for automatic scoring of arthritis disease activity on ultrasound images. RMD Open. 2019;5:e000891. doi: 10.1136/rmdopen-2018-000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tiulpin A, Thevenot J, Rahtu E, Lehenkari P, Saarakkala S. Automatic knee osteoarthritis diagnosis from plain radiographs: a deep learning-based approach. Sci Rep. 2018;8:1727. doi: 10.1038/s41598-018-20132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terkawi AS, Karakitsos D, Elbarbary M, Blaivas M, Durieux ME. Ultrasound for the anesthesiologists: present and future. ScientificWorldJournal. 2013;2013:683685. doi: 10.1155/2013/683685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran D, Rohling RN. Automatic detection of lumbar anatomy in ultrasound images of human subjects. IEEE Trans Biomed Eng. 2010;57:2248–2256. doi: 10.1109/TBME.2010.2048709. [DOI] [PubMed] [Google Scholar]
- 52.Yu S, Tan KK, Shen C, Sia AT. Ultrasound guided automatic localization of needle insertion site for epidural anesthesia. 2013 IEEE International Conference on Mechatronics and Automation; 2013 Aug 4-7; Takamatsu, Japan. New York: Institute of Electrical and Electronics Engineers; 2013. pp. 985–990. [Google Scholar]
- 53.Pesteie M, Abolmaesumi P, Ashab HA, Lessoway VA, Massey S, Gunka V, et al. Real-time ultrasound image classification for spine anesthesia using local directional Hadamard features. Int J Comput Assist Radiol Surg. 2015;10:901–912. doi: 10.1007/s11548-015-1202-5. [DOI] [PubMed] [Google Scholar]
- 54.Baka N, Leenstra S, van Walsum T. Ultrasound aided vertebral level localization for lumbar surgery. IEEE Trans Med Imaging. 2017;36:2138–2147. doi: 10.1109/TMI.2017.2738612. [DOI] [PubMed] [Google Scholar]
- 55.Hetherington J, Lessoway V, Gunka V, Abolmaesumi P, Rohling R. SLIDE: automatic spine level identification system using a deep convolutional neural network. Int J Comput Assist Radiol Surg. 2017;12:1189–1198. doi: 10.1007/s11548-017-1575-8. [DOI] [PubMed] [Google Scholar]
- 56.Hadjerci O, Hafiane A, Makris P, Conte D, Vieyres P, Delbos A. Nerve detection in ultrasound images using median Gabor binary pattern. In: Campilho A, Kamel M, editors. Image analysis and recognition. Lecture notes in computer science, Vol. 8815. Cham: Springer; 2014. pp. 132–140. [Google Scholar]
- 57.Smistad E, Johansen KF, Iversen DH, Reinertsen I. Highlighting nerves and blood vessels for ultrasound-guided axillary nerve block procedures using neural networks. J Med Imaging (Bellingham) 2018;5:044004. doi: 10.1117/1.JMI.5.4.044004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang C, Zhou Y, Tan W, Qiu Z, Zhou H, Song Y, et al. Applying deep learning in recognizing the femoral nerve block region on ultrasound images. Ann Transl Med. 2019;7:453. doi: 10.21037/atm.2019.08.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao H, Sun N. Improved U-net model for nerve segmentation. In: Zhao Y, Kong X, Taubman D, editors. International Conference on Image and Graphics 2017. Lecture notes in computer science, Vol. 10667. Cham: Springer; 2017. pp. 496–504. [Google Scholar]
- 60.Baby M, Jereesh A. Automatic nerve segmentation of ultrasound images. 2017 International Conference of Electronics, Communication and Aerospace Technology (ICECA); 2017 Apr 20-22; Coimbatore, India. New York: Institute of Electrical and Electronics Engineers; 2017. pp. 107–112. [Google Scholar]
- 61.Weng Y, Zhou T, Li Y, Qiu X. NAS-Unet: neural architecture search for medical image segmentation. IEEE Access. 2019;7:44247–44257. [Google Scholar]
- 62.Liu C, Liu F, Wang L, Ma L, Lu ZM. Segmentation of nerve on ultrasound images using deep adversarial network. Int J Innov Comput Inform Control. 2018;14:53–64. [Google Scholar]
- 63.He K, Gkioxari G, Dollar P, Girshick R. Mask R-CNN. 2017 IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22-29; Venice, Italy. New York: Institute of Electrical and Electronics Engineers; 2017. pp. 2961–2969. [Google Scholar]
- 64.Zhang Y, Jordan JM. Epidemiology of osteoarthritis. Clin Geriatr Med. 2010;26:355–369. doi: 10.1016/j.cger.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kazam JK, Nazarian LN, Miller TT, Sofka CM, Parker L, Adler RS. Sonographic evaluation of femoral trochlear cartilage in patients with knee pain. J Ultrasound Med. 2011;30:797–802. doi: 10.7863/jum.2011.30.6.797. [DOI] [PubMed] [Google Scholar]
- 66.Hossain MB, Lai KW, Pingguan-Murphy B, Hum YC, Salim MI, Liew YM. Contrast enhancement of ultrasound imaging of the knee joint cartilage for early detection of knee osteoarthritis. Biomed Signal Process Control. 2014;13:157–167. [Google Scholar]
- 67.Faisal A, Ng SC, Goh SL, Lai KW. Knee cartilage segmentation and thickness computation from ultrasound images. Med Biol Eng Comput. 2018;56:657–669. doi: 10.1007/s11517-017-1710-2. [DOI] [PubMed] [Google Scholar]
- 68.Desai P, Hacihaliloglu I. Knee-cartilage segmentation and thickness measurement from 2D ultrasound. J Imaging. 2019;5:43. doi: 10.3390/jimaging5040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Antico M, Sasazawa F, Dunnhofer M, Camps SM, Jaiprakash AT, Pandey AK, et al. Deep learning-based femoral cartilage automatic segmentation in ultrasound imaging for guidance in robotic knee arthroscopy. Ultrasound Med Biol. 2020;46:422–435. doi: 10.1016/j.ultrasmedbio.2019.10.015. [DOI] [PubMed] [Google Scholar]
- 70.Meyer NB, Jacobson JA, Kalia V, Kim SM. Musculoskeletal ultrasound: athletic injuries of the lower extremity. Ultrasonography. 2018;37:175–189. doi: 10.14366/usg.18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prevost R, Salehi M, Jagoda S, Kumar N, Sprung J, Ladikos A, et al. 3D freehand ultrasound without external tracking using deep learning. Med Image Anal. 2018;48:187–202. doi: 10.1016/j.media.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 72.Looney P, Stevenson GN, Nicolaides KH, Plasencia W, Molloholli M, Natsis S, et al. Fully automated, real-time 3D ultrasound segmentation to estimate first trimester placental volume using deep learning. JCI Insight. 2018;3:e120178. doi: 10.1172/jci.insight.120178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taljanovic MS, Melville DM, Scalcione LR, Gimber LH, Lorenz EJ, Witte RS. Artifacts in musculoskeletal ultrasonography. Semin Musculoskelet Radiol. 2014;18:3–11. doi: 10.1055/s-0034-1365830. [DOI] [PubMed] [Google Scholar]