Abstract
A mixed-aged flock of 130 turkeys in Bangladesh reported the sudden death of 1 bird in September 2017. Highly pathogenic avian influenza A(H5N1) virus was detected in 3 turkeys, and phylogenetic analysis placed the viruses in the reassortant clade 2.3.2.1a. The birds had clinical signs of depression, diarrhea, weakness, closed eyes, and finally death. The mortality rate of the flock was 13% over the 6 d prior to the flock being euthanized. At autopsy, we observed congestion in lungs and brain, hemorrhages in the trachea, pancreas, breast muscle, coronary fat, intestine, bursa of Fabricius, and kidneys. Histopathology revealed hemorrhagic pneumonia, hemorrhages in the liver and kidneys, and hemorrhages and necrosis in the spleen and pancreas. Significant changes in the brain included gliosis, focal encephalomalacia and encephalitis, and neuronophagia.
Keywords: avian influenza A virus, Bangladesh, clade 2.3.2.1a, H5N1, histopathology, turkeys
Avian influenza A viruses (avian IAVs; Orthomyxoviridae, Alphainfluenzavirus, Influenza A virus) can be divided into 2 distinct groups based on their ability to cause disease and certain molecular and biological properties of the virus: highly pathogenic avian influenza virus (HPAIV) and low pathogenicity avian influenza virus (LPAIV). Avian influenza A(H5N1) virus affects multiple domestic and wild bird species and is extremely contagious, causing multi-organ systemic disease of poultry leading to high mortality.18 The ancestor of panzootic avian influenza A(H5N1) viruses, A/Goose/Guangdong/1/1996 (Gs/Gd), was recognized in Guangdong province of China in 1996 and is considered the progenitor of subsequently emerged clades of H5N1 subtype viruses.21 In Bangladesh, HPAI (H5N1) viruses were first detected in February 2007.3 The first introduced viruses belonged to clade 2.2.2, which was later replaced by clade 2.3.2.1a; apart from these clades, there was also brief circulation or sporadic isolation of clade 2.3.4.2 and 2.3.2.1c viruses in Bangladesh.6,7,11,12,14,15 Analyses of surveillance samples from live bird markets (LBMs) and waterfowl in wetland areas revealed the emergence of a new genotype of HPAI (H5N1) virus in domestic ducks in 2015,2 and subsequently detected in clinical outbreaks in commercial and backyard poultry in 2017.12 The newly emerged virus is a segment reassortant with the hemagglutinin (HA), neuraminidase (NA), and matrix (M) genes of circulating 2.3.2.1a Bangladeshi H5N1 subtype viruses and 5 other genes of low pathogenicity non-H9N2 Eurasian-lineage avian IAVs.12 The reassortment event was proposed to happen in the haor wetland areas of Bangladesh.2
Turkey farming is gaining popularity among young farmers in Bangladesh. However, the lack of knowledge of farmers about husbandry and biosecurity practices, the well-entrenched circulation of several subtypes of avian IAVs in multiple species,14 and recent cocirculation of HPAIVs and LPAIVs in commercial poultry flocks in the country13 remain as the main constraints for successful turkey farming in Bangladesh. In comparison with other poultry species, turkeys are highly susceptible to HPAIV infection.8 Herein we report the clinical and pathologic features of an outbreak of HPAI (H5N1) virus in a commercial turkey farm in Bangladesh during 2017.
On 13 September 2017, a small-scale turkey farm in the Mymensingh district of Bangladesh with 130 turkeys of different ages reported the sudden death of 1 bird. All affected turkeys were reared in a single shed with open windows and with minimal biosecurity measures. There were 130 turkeys aged 1 mo to 1 y. On-farm investigation on the first day of the outbreak found 4 sick turkeys. By day 6 of the outbreak, 17 turkeys had died, and 4 birds were sick. Among different age groups, mostly young turkeys (≤ 5 mo old) were affected. The birds that showed clinical signs had depression, diarrhea, weakness, closed eyes, and finally death. The overall morbidity and mortality could not be assessed given that the remaining birds were culled. However, 13% mortality was recorded in the flock over the 6 d prior to the flock being euthanized.
The farm owner brought the dead turkey and a sick 2-mo-old turkey to the Department of Pathology, Bangladesh Agricultural University, Mymensingh. On 16 September 2017, another dead 2-mo-old turkey was brought to the Department. The protocol and procedures employed in our study were ethically reviewed and approved by the Committee for Ethical Standard of Research of Bangladesh Agricultural University, Mymensingh. Before autopsy, sick birds were euthanized by a veterinarian. Autopsies were performed following strict biosafety measures and gross pathologic findings were recorded. Pieces of trachea and lungs were collected aseptically, pooled, and stored at −80°C for virologic study. For histopathology, tissues from various organs were collected in 10% neutral-buffered formalin. Formalin-fixed tissue samples were processed routinely and stained with hematoxylin and eosin.
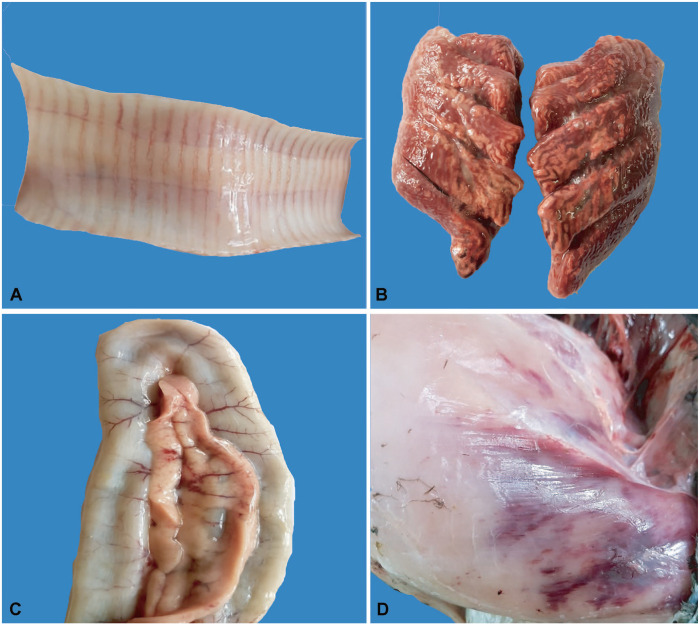
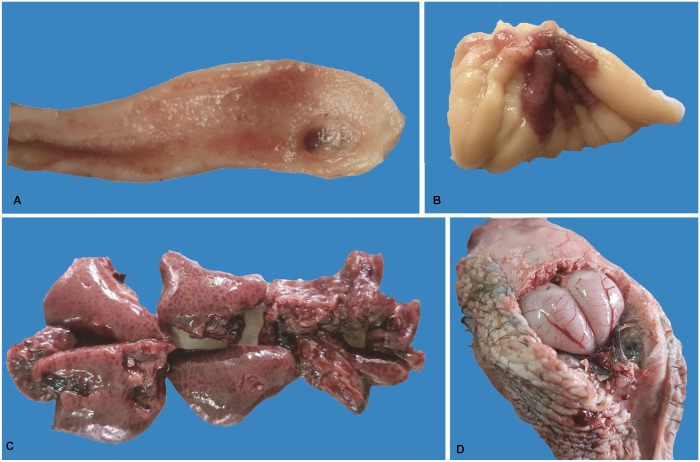
The whole carcass was congested and hemorrhagic. Lesions included hemorrhages in the trachea, congestion and consolidation of lungs, and hemorrhages in the pancreas, muscles, the intestine, cecal tonsils, the bursa of Fabricius, and kidneys (Figs. 1, 2). The brain was congested (Fig. 2D). Hemorrhages and/or congestion were also seen in the liver and spleen.
Figure 1.
Gross lesions in avian influenza A(H5N1) virus–affected turkeys. A. Hemorrhages in the trachea. B. Pulmonary congestion and consolidation. C. Hemorrhages in the pancreas. D. Hemorrhages in breast muscle.
Figure 2.
Gross lesions in avian influenza A(H5N1) virus–affected turkeys. A. Hemorrhages in the intestine. B. Hemorrhages in the bursa of Fabricius. C. Hemorrhages in kidneys. D. Congestion in the brain.
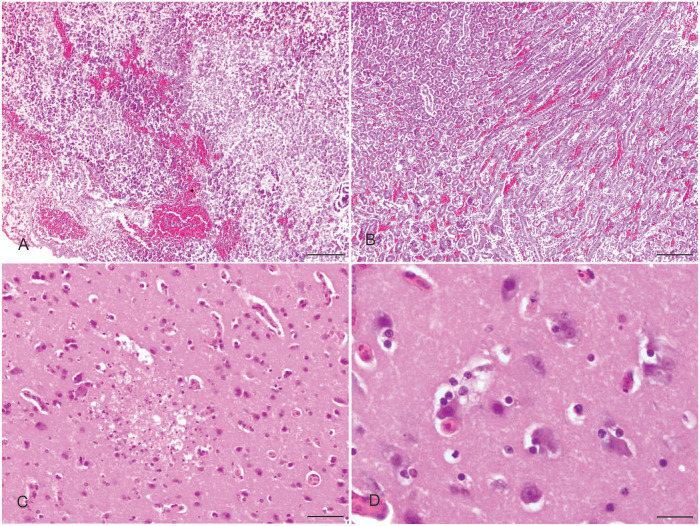
Histologic examination revealed hemorrhagic pneumonia (Suppl. Fig. 1A), necrohemorrhagic pancreatitis (Fig. 3A), and hemorrhagic kidneys (Fig. 3B). In addition, hemorrhages, congestion, and focal necrotizing hepatitis were present, as was multifocal necrosis and hemorrhage in the spleen (Suppl. Fig. 1B, C). In the brain, there was gliosis and focal encephalomalacia with encephalitis (Fig. 3C, Suppl. Fig. 1D). Neuronophagia was also evident in the brain (Fig. 3D).
Figure 3.
Histopathologic changes in avian influenza A(H5N1) virus–affected turkeys. A. Hemorrhages and necrosis in the pancreas. Bar = 50 µm. B. Hemorrhage in the kidney. Bar = 50 µm. C. Gliosis and focal encephalomalacia with encephalitis. Bar = 10 µm. D. Microglial accumulation around a damaged neuron in the brain. Bar = 4 µm. H&E.
A 20% tissue homogenate was prepared from pooled trachea and lung tissues using sterile phosphate-buffered saline (PBS) containing gentamicin (10 mg/mL). Viral RNA was extracted from tissue homogenates (MagMAX viral RNA isolation kit, KingFisher purification system; Thermo Fisher Scientific). All samples were first screened for IAV using universal M gene–specific reverse-transcription real-time PCR17 (RT-rtPCR; Ag path-ID one step RT-PCR kit; Thermo Fisher Scientific). Samples with a cycle quantification (Cq) ≤ 40 and a positive band of appropriate size (100 bp) in agarose gel electrophoresis were considered as positive. The M gene–positive samples were subtyped with H5, H7, H9, and N1–specific primers, as described previously.10,13,20 All samples were also tested for Newcastle disease virus (NDV) using F gene–specific primers.1,19 In all assays, RNA extracted from H5N1, H7N1, H9N2 subtypes of IAVs, and NDV available in the laboratory were used as positive controls.
For virus isolation, the 3 M gene RT-rtPCR–positive tissue samples were inoculated into 10-d-old embryonated chicken eggs by the allantoic route. Following the death of embryos within 48 h post-infection, infected allantoic fluids were harvested and tested using a hemagglutination assay (HA). The HA titer of these allantoic fluids was 16-512 HA units. Testing of the allantoic fluids using IAV M gene–specific RT-rtPCR and subtyping using H5, H7, H9, and N1–specific primers as described above1,10,13,19,20 confirmed the H5N1 subtype of the virus and the absence of coinfections with H7 and H9 subtypes of IAV, and of NDV.
The M gene RT-rtPCR showed positive amplification with Cq values of 23–25 in all 3 tissue samples from turkeys, which was visualized by agarose gel electrophoresis. In RT-PCR, the H5 and N1 subtype–specific primer sets successfully amplified 545 bp and 343 bp fragments, respectively, from all samples, indicating the H5N1 subtype of the virus. RT-PCR for H7 and H9 subtypes of IAV and NDV did not show any amplification, ruling out the coinfections with these pathogens. Phylogenetic analysis based on the complete genome sequences placed the viruses into reassortant clade 2.3.2.1a, which has been reported from poultry of LBMs, aquatic birds, and field outbreaks in Bangladesh.2,12 Details of the phylogeny and deduced amino acid analysis of these isolates have been published.12
In Bangladesh, avian influenza A(H5N1) virus was first identified in 2007. During 2007–2010, clade 2.2.2 viruses were circulating in the field, and most of the outbreaks were reported in chickens.6,7 However, in 2011, clade 2.3.2.1 (later designated as 2.3.2.1a) appeared.7 The new clade 2.3.2.1a affected multiple avian species, including chickens, ducks, quails, feral crows, and migratory birds.6,9,11,15 Most of the outbreaks involving clade 2.3.2.1a viruses in chickens, quails, and crows had sudden onset of high mortality with respiratory distresses. A few outbreaks were recorded in ducks with relatively low mortality and nervous signs.5,11 From 2015 and onward, a new reassortant of clade 2.3.2.1a viruses was reported in LBMs and aquatic birds,2 which was subsequently detected in domestic poultry with clinical outbreaks.12 The 3 H5N1 subtype viruses from turkeys in our outbreak were also segment reassortants containing HA, NA, and M genes of the circulating H5N1 clade 2.3.2.1a viruses and other internal genes of LPAIVs.2,12 Chickens infected with the new reassortant had signs of high mortality with diarrhea and respiratory distress, whereas the affected ducks and geese had nervous signs with low mortality.12 In our study, turkeys that died of new reassortant clade 2.3.2.1a viruses had a sudden onset of mortality with signs of diarrhea and drowsiness, as described previously for chickens.12
The pathologic findings from our turkey samples were similar to those of other HPAIV subtype H5N1 infections. HPAIV often produces hemorrhagic pneumonia, necrohemorrhagic pancreatitis, and multifocal splenic necrosis in chickens.4,16 However, neurologic lesions such as gliosis, encephalomalacia, and encephalitis with neuronophagia were described mainly for ducks infected with clade 2.3.2.1a viruses.5,11 We found similar neurologic lesions in turkeys naturally infected with H5N1 clade 2.3.2.1a viruses in our outbreak in Bangladesh.
Supplemental Material
Supplemental material, Supplemental_material for Pathology of an outbreak of highly pathogenic avian influenza A(H5N1) virus of clade 2.3.2.1a in turkeys in Bangladesh by Tanjin T. Mumu, Mohammed Nooruzzaman, Azmary Hasnat, Rokshana Parvin, Emdadul H. Chowdhury, Abu S. M. Bari and Mohammad R. Islam in Journal of Veterinary Diagnostic Investigation
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The study was partially supported by The World Academy of Sciences (TWAS, RGA 15-083 RG/BIO/AS_I) to Rokshana Parvin, and Ministry of Science and Technology, Bangladesh (grant BS93/2019-20) to Mohammed Nooruzzaman.
ORCID iDs: Mohammed Nooruzzaman  https://orcid.org/0000-0002-9358-1494
https://orcid.org/0000-0002-9358-1494
Mohammad R. Islam  https://orcid.org/0000-0001-6874-0628
https://orcid.org/0000-0001-6874-0628
Supplementary material: Supplementary material for this article is available online.
References
- 1. Barman LR, et al. Phylogenetic analysis of Newcastle disease viruses from Bangladesh suggests continuing evolution of genotype XIII. Arch Virol 2017;162:3177–3182. [DOI] [PubMed] [Google Scholar]
- 2. Barman S, et al. Role of domestic ducks in the emergence of a new genotype of highly pathogenic H5N1 avian influenza A viruses in Bangladesh. Emerg Microbes Infect 2017;6:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biswas PK, et al. Avian influenza outbreaks in chickens, Bangladesh. Emerg Infect Dis 2008;14:1909–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown JD, et al. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg Infect Dis 2006;12:1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haider N, et al. Unusually high mortality in waterfowl caused by highly pathogenic avian influenza A(H5N1) in Bangladesh. Transbound Emerg Dis 2017;64:144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haque ME, et al. Molecular evolution of H5N1 highly pathogenic avian influenza viruses in Bangladesh between 2007 and 2012. Avian Pathol 2014;43:183–194. [DOI] [PubMed] [Google Scholar]
- 7. Islam MR, et al. New introduction of clade 2.3.2.1 avian influenza virus (H5N1) into Bangladesh. Transbound Emerg Dis 2012;59:460–463. [DOI] [PubMed] [Google Scholar]
- 8. Kapczynski DR, et al. Vaccine protection of turkeys against H5N1 highly pathogenic avian influenza virus with a recombinant turkey herpesvirus expressing the hemagglutinin gene of avian influenza. Avian Dis 2016;60:413–417. [DOI] [PubMed] [Google Scholar]
- 9. Khan SU, et al. Investigating a crow die-off in January– February 2011 during the introduction of a new clade of highly pathogenic avian influenza virus H5N1 into Bangladesh. Arch Virol 2014;159:509–518. [DOI] [PubMed] [Google Scholar]
- 10. Lee MS, et al. Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J Virol Methods 2001; 97:13–22. [DOI] [PubMed] [Google Scholar]
- 11. Nooruzzaman M, et al. Pathology of clade 2.3.2.1 avian influenza virus (H5N1) infection in quails and ducks in Bangladesh. Avian Pathol 2019;48:73–79. [DOI] [PubMed] [Google Scholar]
- 12. Nooruzzaman M, et al. A new reassortant clade 2.3.2.1a H5N1 highly pathogenic avian influenza virus causing recent outbreaks in ducks, geese, chickens and turkeys in Bangladesh. Transbound Emerg Dis 2019;66:2120–2133. [DOI] [PubMed] [Google Scholar]
- 13. Parvin R, et al. Co-subsistence of avian influenza virus subtypes of low and high pathogenicity in Bangladesh: challenges for diagnosis, risk assessment and control. Sci Rep 2019;9:8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parvin R, et al. Review analysis and impact of co-circulating H5N1 and H9N2 avian influenza viruses in Bangladesh. Epidemiol Infect 2018;146:1259–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parvin R, et al. Genetic characterization of highly pathogenic H5N1 avian influenza virus from live migratory birds in Bangladesh. Virus Genes 2014;49:438–448. [DOI] [PubMed] [Google Scholar]
- 16. Sims LD, et al. Avian influenza in Hong Kong 1997–2002. Avian Dis 2003;47:832–838. [DOI] [PubMed] [Google Scholar]
- 17. Spackman E, et al. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol 2002;40:3256–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swayne DE, et al. Highly pathogenic avian influenza. Rev Sci Tech 2000;19:463–482. [DOI] [PubMed] [Google Scholar]
- 19. Wang Z, et al. Genotyping of Newcastle disease viruses isolated from 2002 to 2004 in China. Ann N Y Acad Sci 2006;1081: 228–239. [DOI] [PubMed] [Google Scholar]
- 20. World Health Organization (WHO). Recommendations and Laboratory Procedures for Detection of Avian Influenza A (H5N1) Virus in Specimens from Suspected Human Cases. Geneva: WHO, 2007. [cited 2020 Sept 21]. https://www.who.int/influenza/resources/documents/h5n1_laboratory_procedures/en/ [Google Scholar]
- 21. Xu X, et al. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 1999;261:15–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_material for Pathology of an outbreak of highly pathogenic avian influenza A(H5N1) virus of clade 2.3.2.1a in turkeys in Bangladesh by Tanjin T. Mumu, Mohammed Nooruzzaman, Azmary Hasnat, Rokshana Parvin, Emdadul H. Chowdhury, Abu S. M. Bari and Mohammad R. Islam in Journal of Veterinary Diagnostic Investigation