Abstract
A colorimetric assay, exploiting the combination of loop-mediated isothermal amplification (LAMP) with DNA barcoding, was developed to address the authentication of some cephalopod species, a relevant group in the context of seafood traceability, due to the intensive processing from the fishing sites to the shelf. The discriminating strategy relies on accurate design of species-specific LAMP primers within the conventional 5’ end of the mitochondrial COI DNA barcode region and allows for the identification of Loligo vulgaris among two closely related and less valuable species. The assay, coupled to rapid genomic DNA extraction, is suitable for large-scale screenings and on-site applications due to its easy procedures, with fast (30 min) and visual readout.
Keywords: colorimetric test, on-site test, LAMP, food authentication, genetic traceability, cephalopod species
1. Introduction
Food adulteration has become a concern of great interest worldwide. This issue is strictly connected to the phenomenon of trade globalization and overexploitation of biodiversity, which are causing profound changes in the whole food supply chain and market systems. Seafood is among the most adulterated food categories [1]. Its long and complex production chain from catching to filleting includes several steps, making fish, crustacean and mollusk products highly exposed to the risk of fraud events [2]. To ensure transparent information to the consumers, food authentication and traceability along the food chain have become essential issues to be addressed. In this framework, the development of rapid, low-cost, and easy-to-use biosensing technologies for on-site diagnostics would be of great and urgent importance. Currently, DNA-based techniques are the most widely used and have proven very effective, especially in combination with the DNA barcoding approach [3,4,5]. DNA barcoding is a method of species identification that is based on the variability analysis of short genomic sequences called DNA barcodes (e.g., the 658 bp 5’ end portion of the mitochondrial gene cytochrome c oxidase subunit I - COI, used for metazoans identification [6,7,8]) that are reported in a unique database called the Barcode of Life Data (BOLD) System [9]. PCR-based methods targeting a DNA barcode sequence have provided several advantages, due to their high discriminating power and sensitivity [10,11,12]. Nevertheless, their requirements for long procedures and complex instrumentation make them unsuitable for rapid and portable testing. In an attempt to simplify the overall procedures, nanotechnologies have offered new perspectives in developing DNA barcoding-based hybrid techniques with naked-eye colorimetric readout, taking advantage of the plasmonic properties of gold nanoparticles [13,14]. A possible advancement toward the development of faster and portable technologies for on-site testing is represented by isothermal amplification techniques. Generally, such reactions work in isothermal conditions (usually 55–65 °C), are very rapid (less than 1 h), and yield amplification efficiencies comparable to the standard PCR [15]. Loop-mediated isothermal amplification (LAMP) was first introduced by Notomi [16] and, due to its rapid and specific DNA amplification in isothermal conditions, is currently one of the most applied techniques in several fields [17,18,19,20,21], including food authentication [22,23,24,25]. This robust methodology uses a DNA polymerase enzyme with strand displacement activity and four to six primers that recognize six to eight regions on the nucleic acid target [16,26]. Recently, the classical analysis of LAMP products by gel electrophoresis has been substituted by real-time [27,28] and naked-eye readout [29,30,31], including the colorimetric method based on pH-sensitive dyes [20,32]. Therefore, LAMP reaction, if performed with portable simple instrumentation such as a heating block, is suitable to realize rapid, one-step, and on-site applications. Here, we present the development of a rapid colorimetric assay combining LAMP with DNA barcoding to address naked-eye authentication of food species. In particular, we applied this approach to a real scenario of seafood fraud, i.e., the authentication of the cephalopod mollusk Loligo vulgaris and its discrimination from two genetically similar and often mislabeled species, Loligo reynaudii and Loligo forbesii [14]. This one-step assay enables the visualization of the colorimetric result within 30 min, also when applied to crude extracts by rapid DNA extraction protocols.
2. Materials and Methods
2.1. Sampling, Dataset Assembling for Primers Design, and DNA Extraction
Loligo spp. tissues were obtained from a previous study assessing the mislabeling rate of cephalopod seafood items in the Italian market using a DNA barcoding approach [14]. COI sequences of the analyzed samples, together with other sequences of Loligo vulgaris, L. forbesii, L. reynaudii, and closely related species (total number of sequences = 140), were downloaded from GenBank and aligned using MUSCLE online [33] to perform LAMP primer design. Similarly, 16S ribosomal RNA sequences of the same species (total number of sequences = 23) were downloaded from GenBank and aligned to design LAMP primers for positive controls.
To perform LAMP experiments, we homogenized squid tissues (≤5 mg) with a sterilized plastic pestle, and DNA was extracted using the InstaGene Matrix (Bio-Rad, München, Germany), following the manufacturer’s instructions. With this protocol, DNA was rapidly extracted (approximately 15 min) and required no further purification steps prior to perform LAMP experiments.
2.2. Colorimetric LAMP Reaction
Colorimetric LAMP reactions were performed in a Bio-Rad T100 Thermal Cycler. Specifically, 4 µL of genomic DNA (≈25 ng/µL) was used as a template and mixed to 21 µL of reaction mix, containing 0.2 µM of forward outer primer (F3) and backward outer primer (B3), 1.6 µM of forward inner primer (FIP) and backward inner primer (BIP), 1.2 µM of forward loop primer (LF) and backward loop primer (LB) (Integrated DNA Technologies, Coralville, IA, USA), 0.6 M betaine (VWR International S.R.L., Milano, Italy), 1x LAMP reaction buffer (New England BioLabs, Ipswich, MA, USA), 6 mM MgSO4, 1.4 mM of each deoxyribonuclotide triphosphate (dNTP) (Promega, Madison, WI, USA), 0.32 U/µL Bst 2.0 WarmStart DNA polymerase (New England BioLabs, Ipswich, MA, USA), and 0.06 mM Xylenol Orange (Sigma-Aldrich-Merck, St. Louis, MO, USA). The colorimetric reaction was performed at 63 °C and the result was visualized after 30 min.
2.3. Fluorimetric LAMP Reaction
Fluorimetric LAMP reactions were performed in an Applied Biosystems StepOne Real-Time PCR System. Specifically, 4 µL of genomic DNA (≈25 ng/µL) were used as a template and mixed to 21 µL of reaction mix, containing 0.2 µM of F3/B3, 1.6 µM of FIP/BIP, 1.2 µM of LF/LB (Integrated DNA Technologies, Coralville, IA, USA), 0.6 M betaine (VWR International S.R.L., Milano, Italy), 1x LAMP reaction buffer (New England BioLabs, Ipswich, MA, USA), 6 mM MgSO4, 1.4 mM of each dNTP (Promega, Madison, WI, USA), 0.32 U/µL Bst 2.0 WarmStart DNA polymerase (New England BioLabs, Ipswich, MA, USA), and 0.2x SYBR Green (Thermo Fisher Scientific, Waltham, MA, USA). The reaction was performed at 63 °C for 60 min.
2.4. LAMP-AGE Procedure
Colorimetric LAMP products were electrophoresed on a 1% agarose gel to assess the amplification reactions. In detail, 500 mg of ultra-pure agarose (Thermo Fisher Scientific, Waltham, MA, USA) was dissolved in 50 mL of 1x tris-borate-EDTA buffer (TBE) (Sigma-Aldrich-Merck, St. Louis, MO, USA) using a microwave oven. A total of 5 µL of SYBR Safe DNA gel stain (Thermo Fisher Scientific, Waltham, MA, USA) was added to the solution, which then was poured into the plate with a spacer for gel polymerization. Each product of reaction was analyzed by mixing 5 µL of LAMP products to 1 µL of gel loading dye. A 0.1–10 Kb DNA ladder (New England BioLabs, Ipswich, MA, USA) was used as a molecular weight marker. The electrophoresis was performed at 80 V, and the result of the run was visualized in a Bio-Rad Gel Doc XR system.
3. Results and Discussion
The species substitution of labelled Loligo vulgaris products is representative of an important issue in the field of seafood since the global demand for cephalopods has shown a positive trend in the last decades, with the main importers and consumers being Italy, Spain, and Japan ((Food and Agriculture Organization, FAO 2016). Given this growing market importance, the correct identification and labelling of traded cephalopods is of primary importance for food safety issues due to the possible occurrence of direct or indirect toxicity phenomena in different species [34,35].
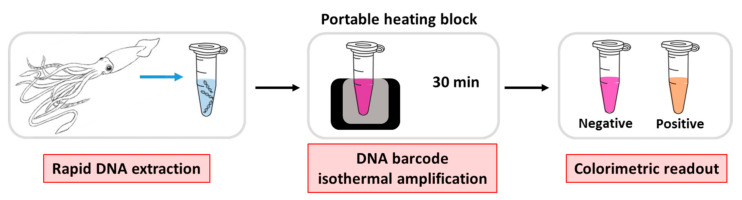
The schematic procedure of the entire method is displayed in Figure 1.
Figure 1.
Schematic procedure of colorimetric loop-mediated isothermal amplification (LAMP) for cephalopod authentication. The proposed method consists of three steps: a rapid DNA extraction; the DNA barcode isothermal amplification process, performed at 63 °C for 30 min; and the colorimetric readout, immediately visible at the end of the reaction.
A rapid DNA extraction from the squid tissue was performed using the InstaGene Matrix (Bio-Rad, München, Germany) following the manufacturer’s instructions. Less than 5 mg of Loligo tissue was used as starting material prior to homogenization with a sterilized plastic pestle. Then, without any purification steps, a few microliters of the extracted target were directly added to the reaction tube, containing all reagents for the amplification reaction. The six primers were designed to be highly specific to the authentic species (unlike the polymorphic variants), and annealed to the selected regions in the presence of the target, triggering the amplification process. The colorimetric result is directly visualized by the naked-eye, due to a color change from purple to pink/orange when amplicons are produced. Remarkably, the overall colorimetric method, including the DNA extraction, the amplification reaction, and the colorimetric readout of the result, requires no instrumentation, apart from a simple heating block.
The key point of the entire strategy relies on the accurate design of species-specific LAMP primers, which target the mitochondrial COI gene of L. vulgaris, and allow its discrimination from the two closely related species. In detail, all the available DNA barcodes of each Loligo species and close relatives (Table S1) were aligned using MUSCLE online [33]. Thus, six LAMP primers (Table 1) were designed to fulfil some general requirements (such as G+C content, annealing temperature, absence of dimers, etc.) and two additional criteria, in order to improve specificity and speed up the amplification mechanism. The six specific primers were designed to target the largest number of interspecies polymorphisms. In particular, the specificity was enhanced by including at least two mismatches per primer, possibly purine/pyrimidine, to increase the melting temperature gap. In addition, where possible, one mismatch was placed at the 3′ end of the primer to hamper the DNA polymerase recognition. Such rules were revealed to be essential for the two inner primers (FIP and BIP), which are responsible for triggering the linear phase of the reaction, as well as for the two loop primers, which accelerate the amplification process [26]. Furthermore, primers were designed on the shortest and highly polymorphic region of the barcode sequence to accelerate the production of the shortest “dumbbell” amplicon, which represents the starting material for the exponential phase of the reaction.
Table 1.
LAMP primer sequences for Loligo vulgaris authentication and its discrimination from Loligo reynaudii and Loligo forbesii.
| Name | Sequence |
|---|---|
| F3 | 5′-ACTGTTAAATGACGATCAACTATAC-3′ |
| B3 | 5′-GGCTAAATCTACAGACGGT-3′ |
| FIP | 5′-AGCGAAGGGGGAAGTAATCAAACGGAAACTGATTAGTGCCTA-3′ |
| BIP | 5′-TCTGCTGTAGAAAGAGGAGCTGCTGCGTGAGAAAGATTTCTAGA-3′ |
| LF | 5′-CTATATCTGGCGCACCTAGTATTA-3′ |
| LB | 5′-GTACAGGATGAACAGTCTACCC-3′ |
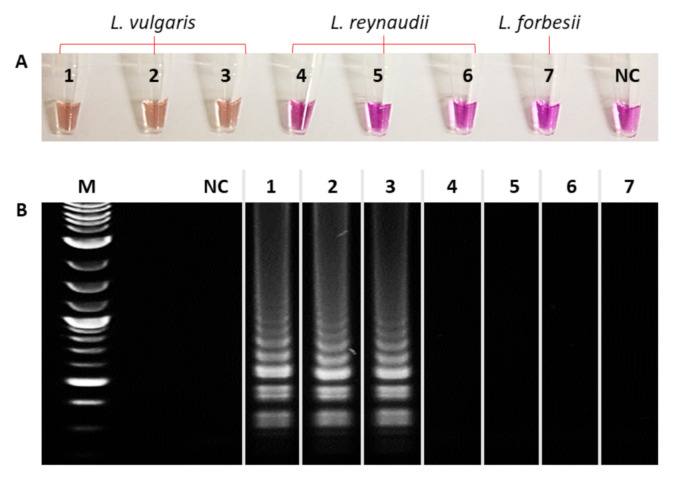
The one-step colorimetric LAMP was applied to various genomic DNA samples, rapidly extracted from the tissues of L. vulgaris, L. reynaudii, and L. forbesii specimens, using 100 ng/µL of each crude DNA extract. The isothermal amplification process was performed in a simple heating block, and the colorimetric result was rapidly and easily visible within 30 min (Figure 2A).
Figure 2.
Representative colorimetric LAMP results for L. vulgaris authentication, applied on samples. 1–3: L. vulgaris, 4–6: L. reynaudii, 7: L. forbesii, NC: pure water. (A) Colorimetric result. (B) 1% agarose gel; M: molecular weight marker.
As is shown, the colorimetric assay was able to correctly identify the target species, which exhibited a clear purple-to-orange color change, whilst the other species maintained the same initial color as the control sample. The reaction was further controlled by agarose gel electrophoresis, confirming that the amplification occurred specifically for L. vulgaris, as shown by the typical profile of LAMP products, unlike the closely related species that were not amplified (Figure 2B). In addition, a set of primers specific for a conserved region of 16S ribosomal RNA were designed as a positive control of the colorimetric LAMP (Table 2) by using the 16S rRNA sequences listed in Table S1.
Table 2.
LAMP 16S rRNA primer sequences for positive control of L. vulgaris.
| Name | Sequence |
|---|---|
| F3-pc | 5′-TCCCTATGGTAACTATATTATAAGCA-3′ |
| B3-pc | 5′-ACAGCTGCGGTATTTTAAC-3′ |
| FIP-pc | 5′-ATTTTCATAGTGAAAAAGCTTGAATTTTTTAAAGGTCCTTAATCACCCCAATTAAAATTTATATAT-3′ |
| BIP-pc | 5′-TTTCTAAAAAATAAAATAGAGACAGATTAACCTTCGTGTACTAAGGTAGCATAATAATTTGCC-3′ |
| LF-pc | 5′-GACGAGAAGACCCTACTGAG-3′ |
| LB-pc | 5′-CAAACCATTCATTCTAGCCTCAAATTAT-3′ |
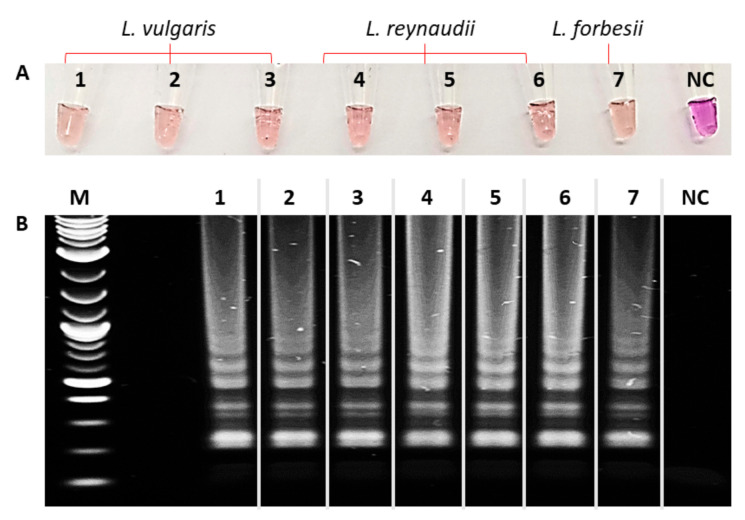
Thus, the positive control reaction was tested on each sample under the same conditions. The visual inspection of the results showed that all samples turned pink/orange (Figure 3), demonstrating the good performance of the reaction, as well as the efficiency of the extraction protocol.
Figure 3.
Colorimetric LAMP positive control applied on samples. 1–3: L. vulgaris, 4–6: L. reynaudii, 7: L. forbesii, NC: pure water. (A) Colorimetric result. (B) 1% agarose gel; M: molecular weight marker.
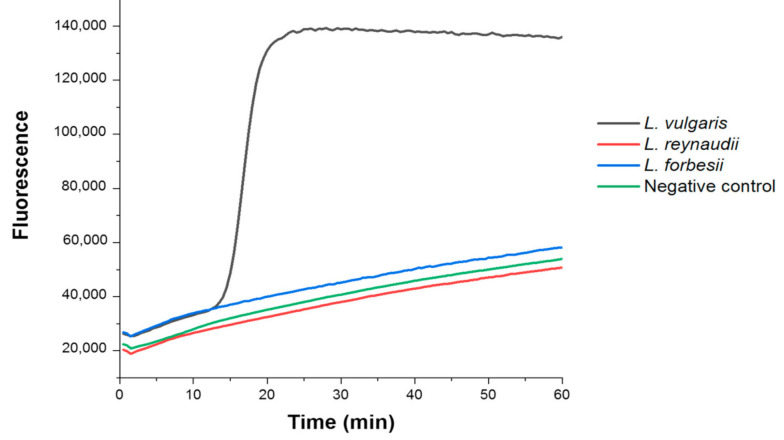
Besides on-site assessments, this reaction may also represent an interesting laboratory-based methodology due to its simplicity and rapidity. Actually, when coupled to a real-time fluorescence reader, using a classical fluorescent intercalating reporter instead of a colorimetric dye, the assay exhibited high discrimination efficiency and ultra-rapidity, being capable of providing clear authentication results in only 15 min (Figure 4).
Figure 4.
Fluorimetric LAMP for L. vulgaris authentication, applied on samples L. vulgaris (black line), L. reynaudii (red line), and L. forbesii (blue line).
Specifically, we analyzed three genomic DNA extracts from L. vulgaris, L. reynaudii, and L. forbesii specimens by fluorimetric LAMP, observing fast amplification after 15 min of reaction only in the case of the target species L. vulgaris. Hence, the optimized design of the LAMP primers, combined with the DNA barcoding approach, enabled us to address the authentication of L. vulgaris. Notably, both the colorimetric and the fluorimetric LAMP were also proven effective when associated with a rapid DNA extraction procedure.
4. Conclusions
In this work, we reported the development of a fast, on-site assay for the authentication of squid species, addressing the specific discrimination of one species from two genetically similar and less valuable ones. By coupling the LAMP technique with DNA barcoding and rapid DNA extraction procedures for a one-step readout, we showed the possibility of achieving fast, naked-eye readout of the species discrimination. Such features make these methodologies suitable for large-scale screenings and on-site industrial quality controls in several fields.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6374/10/12/190/s1: Table S1: Loligo spp. and close relatives cytochrome c oxidase subunit I (COI) and 16S ribosomal RNA sequences used to design LAMP primers.
Author Contributions
Conceptualization, P.P.P.; methodology, G.T., P.C., D.M., A.G. and P.P.P.; validation, G.T. and P.C.; formal analysis, G.T., P.C., D.M., A.G. and P.P.P.; resources, P.P.P.; data curation, G.T., P.C., D.M., A.G. and P.P.P.; writing—original draft preparation, G.T. and P.P.P.; writing—review and editing, G.T., P.C., D.M., A.G. and P.P.P.; supervision, A.G. and P.P.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Naaum A.M., Warner K., Mariani S., Hanner R.H., Carolin C.D. Seafood Mislabeling Incidence and Impacts. In: Naaum A.M., Hanner R., editors. Seafood Authenticity and Traceability. 1st ed. Academic Press; Cambridge, MA, USA: 2016. pp. 3–26. [Google Scholar]
- 2.Fox M., Mitchell M., Dean M., Elliott C., Campbell K. The seafood supply chain from a fraudulent perspective. Food Sec. 2018;10:939–963. doi: 10.1007/s12571-018-0826-z. [DOI] [Google Scholar]
- 3.Hebert P.D.N., Cywinska A., Ball S.L., deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galimberti A., De Mattia F., Losa A., Bruni I., Federici S., Casiraghi M., Martellos S., Labra M. DNA barcoding as a new tool for food traceability. Food Res. Int. 2013;50:55–63. doi: 10.1016/j.foodres.2012.09.036. [DOI] [Google Scholar]
- 5.Galimberti A., Casiraghi M., Bruni I., Guzzetti L., Cortis P., Berterame N.M., Labra M. From DNA barcoding to personalized nutrition: The evolution of food traceability. Curr. Opin. Food Sci. 2019;28:41–48. doi: 10.1016/j.cofs.2019.07.008. [DOI] [Google Scholar]
- 6.Yancy H.F., Zemlak T.S., Mason J.A., Washington J.D., Tenge B.J., Nguyen N.-L.T., Barnett J.D., Savary W.E., Hill W.E., Moore M.M., et al. Potential use of DNA barcodes in regulatory science: Applications of the Regulatory Fish Encyclopedia. J. Food Prot. 2008;71:210–217. doi: 10.4315/0362-028X-71.1.210. [DOI] [PubMed] [Google Scholar]
- 7.Handy S.M., Deeds J.R., Ivanova N.V., Hebert P.D.N., Hanner R.H., Ormnos A., Weigt L.A., Moore M.M., Yancy H.F. A single-laboratory validated method for the generation of DNA barcodes for the identification of fish for regulatory compliance. J. AOAC Int. 2011;94:201–210. doi: 10.1093/jaoac/94.1.201. [DOI] [PubMed] [Google Scholar]
- 8.Deeds J.R., Handy S.M., Fry F., Jr., Granade H., Williams J., Powers M., Shipp R., Weigt L.A. Protocol for building a reference standard sequence library for DNA-based seafood identification. J. AOAC Int. 2014;97:1626–1633. doi: 10.5740/jaoacint.14-111. [DOI] [PubMed] [Google Scholar]
- 9.Ratnasingham S., Hebert P.D.N. BOLD: The barcode of life data system (https://www.barcodinglife.org. ) Mol. Ecol. Resour. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colombo F., Cerioli M., Colombo M.M., Marchisio E., Malandra R., Renon P. A simple polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method for the differentiation of cephalopod mollusc families Loliginidae from Ommastrephidae, to avoid substitutions in fishery field. Food Control. 2002;13:185–190. doi: 10.1016/S0956-7135(01)00101-3. [DOI] [Google Scholar]
- 11.Barbuto M., Galimberti A., Ferri E., Labra M., Malandra R., Galli P., Casiraghi M. DNA barcoding reveals fraudulent substitutions in shark seafood products: The Italian case of “palombo” (Mustelus spp.) Food Res. Int. 2010;43:376–381. doi: 10.1016/j.foodres.2009.10.009. [DOI] [Google Scholar]
- 12.Velasco A., Ramilo-Fernández G., Sotelo C.G. A real-time PCR method for the authentication of common cuttlefish (Sepia officinalis) in food products. Foods. 2020;9:286. doi: 10.3390/foods9030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valentini P., Galimberti A., Mezzasalma V., De Mattia F., Casiraghi M., Labra M., Pompa P.P. DNA barcoding meets nanotechnology: Development of a smart universal tool for food authentication. Angew. Chem. 2017;56:8094–8098. doi: 10.1002/anie.201702120. [DOI] [PubMed] [Google Scholar]
- 14.Maggioni D., Tatulli G., Montalbetti E., Tommasi N., Galli P., Labra M., Pompa P.P., Galimberti A. From DNA barcoding to nanoparticle-based colorimetric testing: A new frontier in cephalopod authentication. Appl. Nanosci. 2020;10:1053–1060. doi: 10.1007/s13204-020-01249-6. [DOI] [Google Scholar]
- 15.Zhao Y., Chen F., Li Q., Wang L., Fan C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015;115:12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 16.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parida M., Horioke K., Ishida H., Dash P.K., Saxena P., Jana A.M., Islam M.A., Inoue S., Hosaka N., Morita K. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 2005;43:2895–2903. doi: 10.1128/JCM.43.6.2895-2903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehme C.C., Nabeta P., Henostroza G., Raqib R., Rahim Z., Gerhardt M., Sanga E., Hoelscher M., Notomi T., Hase T., et al. Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J. Clin. Microbiol. 2007;45:1936–1940. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaroenram W., Cecere P., Pompa P.P. Xylenol orange-based loop-mediated DNA isothermal amplification for sensitive naked-eye detection of Escherichia coli. J. Microbiol. Methods. 2019;156:9–14. doi: 10.1016/j.mimet.2018.11.020. [DOI] [PubMed] [Google Scholar]
- 21.Huang W.E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaagt F., Haase I., Fischer M. Loop-Mediated Isothermal Amplification (LAMP)-based method for rapid mushroom species identification. J. Agric. Food Chem. 2013;61:1833–1840. doi: 10.1021/jf304824b. [DOI] [PubMed] [Google Scholar]
- 23.Abdulmawjood A., Grabowski N., Fohler S., Kittler S., Nagengast H., Klein G. Development of Loop-Mediated Isothermal Amplification (LAMP) assay for rapid and sensitive identification of ostrich meat. PLoS ONE. 2014;9:e100717. doi: 10.1371/journal.pone.0100717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S.-Y., Kim M.-J., Hong Y., Kim H.-Y. Development of a rapid on-site detection method for pork in processed meat products using real-time loop-mediated isothermal amplification. Food Control. 2016;66:53–61. doi: 10.1016/j.foodcont.2016.01.041. [DOI] [Google Scholar]
- 25.Ye J., Feng J., Dai Z., Meng L., Zhang Y., Jiang X. Application of Loop-Mediated Isothermal Amplification (LAMP) for rapid detection of Jumbo Flying Squid Dosidicus gigas (D’Orbigny, 1835) Food Anal. Methods. 2017;10:1452–1459. doi: 10.1007/s12161-016-0700-6. [DOI] [Google Scholar]
- 26.Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 27.Li S., Wang Y., Li Y., Jiang J., Yu R., Xiang M., Xia G. Loop-mediated isothermal amplification (LAMP): Real-time methods for the detection of the survivin gene in cancer cells. Anal. Methods. 2016;8:6277–6283. doi: 10.1039/C6AY01943A. [DOI] [Google Scholar]
- 28.Wang D., Yu J., Wang Y., Zhang M., Li P., Liu M., Liu Y. Development of a real-time loop-mediated isothermal amplification (LAMP) assay and visual LAMP assay for detection of African swine fever virus (ASFV) J. Virol. Methods. 2020;276:113775–113782. doi: 10.1016/j.jviromet.2019.113775. [DOI] [PubMed] [Google Scholar]
- 29.Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 30.Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 31.Goto M., Honda E., Ogura A., Nomoto A., Hanaki K. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 32.Tanner N.A., Zhang Y., Evans T.C. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. BioTechniques. 2015;58:59–68. doi: 10.2144/000114253. [DOI] [PubMed] [Google Scholar]
- 33.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopes V.M., Lopes A.R., Costa P., Rosa R. Cephalopods as vectors of harmful algal bloom toxins in marine food webs. Mar. Drugs. 2013;11:3381–3409. doi: 10.3390/md11093381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y.-J., Lin C.-L., Chen C.-H., Hsieh C.-H., Jen H.-C., Jian S.-J., Hwang D.-F. Toxin and species identification of toxic octopus implicated into food poisoning in Taiwan. Toxicon. 2014;91:96–102. doi: 10.1016/j.toxicon.2014.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.