Abstract
Deep sea has an extreme environment which leads to biodiversity of microorganisms and their unique physical and biochemical mechanisms. Deep-sea derived microorganisms are more likely to produce novel bioactive substances with special mechanism of action for drug discovery. This article reviews secondary metabolites with biological activities such as anti-tumor, anti-bacterial, anti-viral, and anti-inflammatory isolated from deep-sea fungi and bacteria during 2018–2020. Effective methods for screening and obtaining natural active compounds from deep-sea microorganisms are also summarized, including optimizing the culture conditions, using genome mining technology, biosynthesis and so on. The comprehensive application of these methods makes broader prospects for the development and application of deep sea microbial bioactive substances.
Keywords: deep-sea fungi, deep-sea actinomycetes, secondary metabolites, bioactivity
1. Introduction
Deep sea is one of the latest extreme environments developed on earth. The deep sea is an environment with extreme features including: (1) For every 10 m of increase in depth, the pressure increases by one atmosphere, so the water pressure is higher than 1000 atmospheres in the deep sea trench; (2) The temperature decreases with depth, which is usually around 2 °C on the deep sea bottom; (3) The seawater oxygen concentration mainly depends on the absorption of oxygen at the sea-air interface, the photosynthesis rate of autotrophs in the true light layer, and rate of consumption of marine life respiration; (4) The light intensity is close to zero below the depth of 250 m [1]. In conclusion, deep sea has the characteristics of extreme ecological environment, including high pressure, low temperature, lack of oxygen and darkness. The cold seeps, hydrothermal and seamounts of the world deep-sea locations may worth favoring for bioprospection. Deep-sea microbes have unique biological metabolic pathways to deal with extreme ecological environments, especially stress. Many deep-sea microbes are hypertrophic or pressure-sensitive. Existing research methods limit the cultivation of these steps [2].
Over the past fifty years, more than 30,000 marine natural products have been discovered, of which about 2% are derived from deep-sea microorganisms [3]. Based on our review of the literature, the number of marine natural products from deep-sea have increased since then, but they are still a small percentage of the total amount found. Also, we found that most recent researches on bioactive secondary metabolites are derived from bacteria and fungi in deep-sea environment. So, this review mainly covers natural products from deep-sea derived fungi and bacteria who were almost isolated from sediment or sea water. Among the natural products, people pay the most attention to compounds with antibacterial activity, especially for their application in the field of biotechnology and pharmaceuticals. The discovery of antibiotics with new structures is very important for dealing with the spread of resistant bacteria [4]. To a large extent, it is related to the isolation and cultivation of unknown deep-sea microorganisms and the discovery of related secondary metabolites. Although it is supposed that microorganisms are huge in number and rich in diversity in these environments [5], few have been characterized so far [6]. In fact, the discovery of deep-sea microbial diversity can lead to the discovery of compounds with new biological activities, further promoting the drug development process [7]. The first antibacterial natural product isolated from deep-sea sediments was a glial toxin produced by the metabolism of a fungus species, Penicillium sp., isolated from the Seto Inland Sea, Japan, which inhibits the growth of Gram-positive bacteria Staphylococcus aureus and Bacillus subtilis [8].
In the past three years, the development of deep-sea exploration and molecular biology provided technical support for the exploitation of deep-sea microbial natural products. To enable researchers to better understand the research work in these fields, this article summarized the characteristics of secondary metabolites isolated from deep-sea microorganisms and their biological activities in 2018–2020, as well as research methods for diversity of secondary metabolites from deep-sea microorganisms.
2. Secondary Metabolites from Deep-Sea Derived Fungi
Recent studies have shown that fungi from the extreme environments are potential producers for clinically important natural products [9] and may be the next frontier of drug discovery [10].
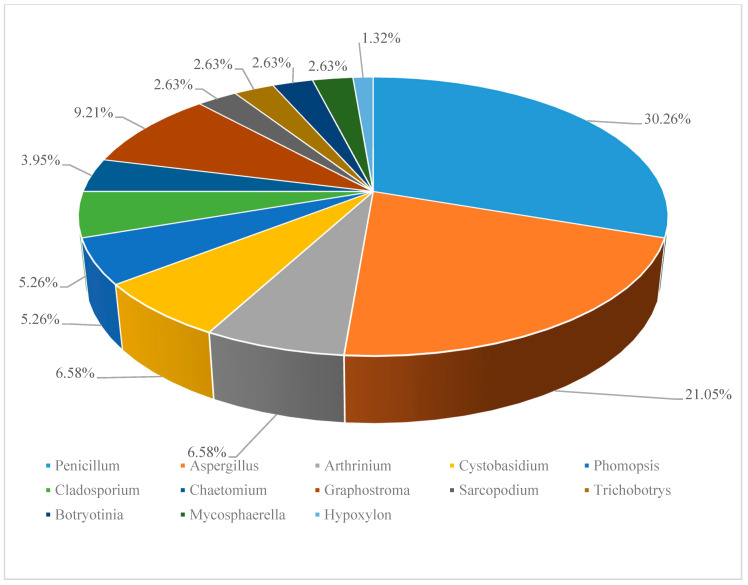
According to the references, 829 (194 novel) natural products were discovered from deep-sea derived fungi in the past three years, 79 among which showed biological activities (Table 1). Most of these compounds were isolated from species of two genera of fungi, Penicillium sp. (23, accounting for 30.26% of the total compounds) and Aspergillus sp. (16, accounting for 21.05% of the total compounds) (Figure 1).
Table 1.
Bioactive Natural products from deep-sea derived fungi in 2018–2020.
| Bioactivity | Fungal Species | Structural Class | Number | Depth (m) | Region | Source | Reference |
|---|---|---|---|---|---|---|---|
| antiallergic | Botryotinia fuckeliana | diterpenoid | 79 | 5572 | Western Pacific Ocean | sea water | [38] |
| antibacterial | Aspergillus penicillioides | steroid | 32, 33 | 2038 | South China Sea | sediment | [23] |
| antibacterial | Mycosphaerella sp. SCSIO z059 | iron (III) chelator | 34 | 1330 | Okinawa Trough | sediment | [24] |
| antibacterial | Mycosphaerella sp. SCSIO z059 | dimerum acid | 35 | 1330 | Okinawa Trough | sediment | [24] |
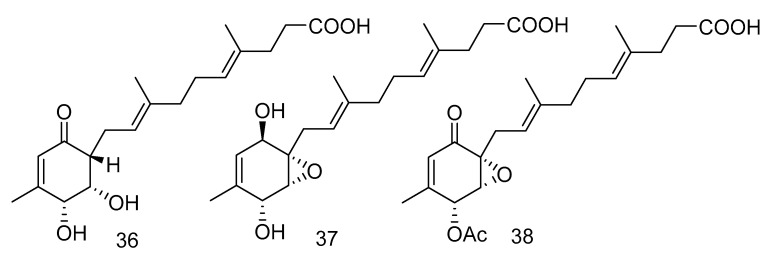
| antibacterial | Penicillium sp. YPGA11 | polyketide | 36–38 | 4500 | West Pacific | sea water | [25] |
| antibacterial | Penicillium canescens SCSIO z053. | polyketide | 39, 40 | 1387 | Okinawa Trough | sediment | [26] |
| antibacterial | Aspergillus versicolor | sesquiterpene | 41–44 | 1487 | South China Sea | sediment | [27] |
| antibacterial | Aspergillus fumigatus | alkaloid | 45, 46 | 3614 | India Ocean | sediment | [28] |
| antibacterial | Penicillium biourgeianum | alkaloid | 47 | 2226 | South China Sea | sediment | [29] |
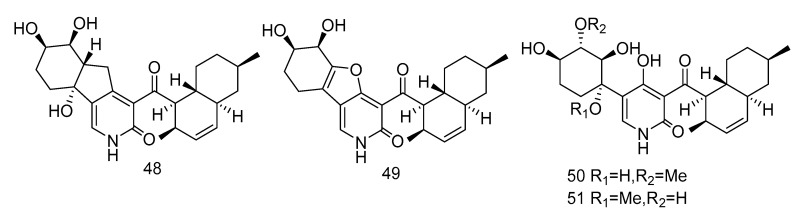
| antibacterial | Arthrinium sp. UJNMF0008 | alkaloid | 25, 48–51 | 3858 | South China Sea | sediment | [19] |
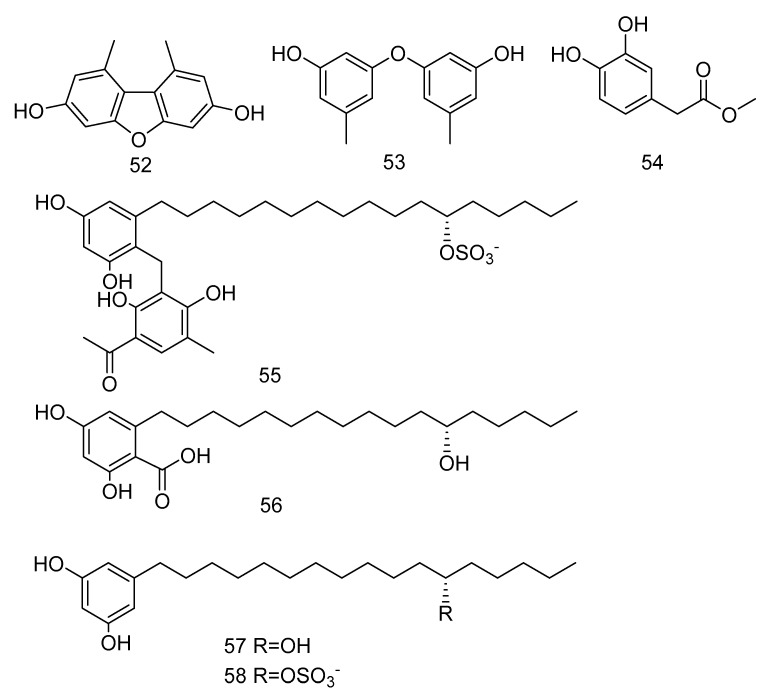
| antibacterial | Aspergillus sp. SCSIO06786 | phenol | 52–56 | 4762 | India Ocean | sediment | [30] |
| antibacterial | Penicillium crustosum * | phenol | 55–58 | 526 | Prydz Bay | sediment | [31] |
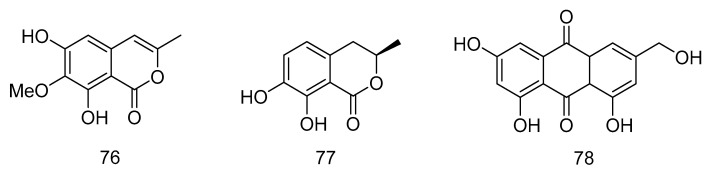
| antifood allergic | Graphostroma sp. MCCC 3A00421 | polyketide | 76–78 | 2721 | Atlantic | hydrothermal sulfide | [37] |
| antifungal | Aspergillus fumigatus | alkaloid | 63 | 3614 | India Ocean | sediment | [28] |
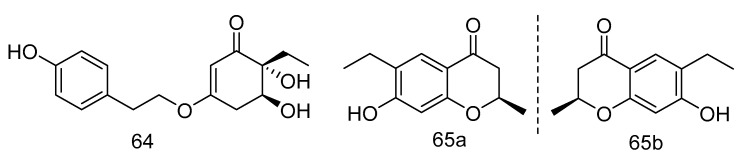
| anti-inflammatory | Trichobotrys effuse | polyketide | 64, 65 | 1428 | South China Sea | sediment | [34] |
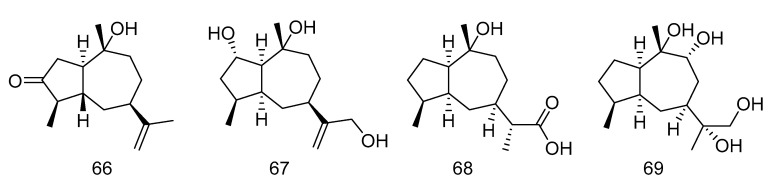
| anti-inflammatory | Graphostroma sp. MCCC 3A00421 | sesquiterpenoid | 66–69 | 2721 | Atlantic | hydrothermal sulfide | [35] |
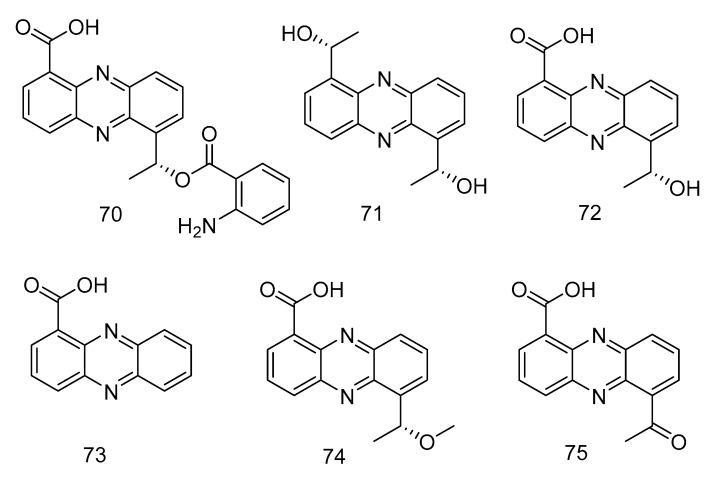
| anti-inflammatory | Cystobasidium laryngis | alkaloid | 70–75 | 4317 | India Ocean | sediment | [36] |
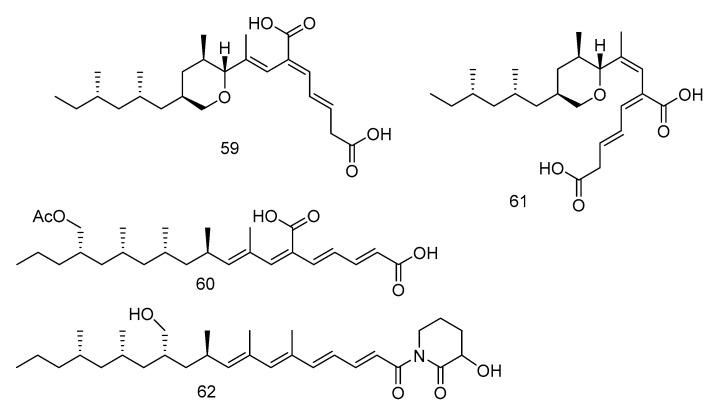
| antituberculosis | Aspergillus fischeri | polyketide | 60–63 | 3000 | India Ocean | sediment | [33] |
| cytotoxic | Penicillum citreonigrum | polyketide | 1, 2 | 2910 | Southeast India Ocean | sediment | [11] |
| cytotoxic | Phomopsis lithocarpus | polyketide | 3, 4 | 3606 | India Ocean | sediment | [12] |
| cytotoxic | Chaetomium globosum | polyketide | 5, 6 | 2476 | South China Sea | sediment | [13] |
| cytotoxic | Penicillium chrysogenum | polyketide | 7–15 | 2076 | South Atlantic Ocean | sediment | [14] |
| cytotoxic | Hypoxylon rubiginosum | polyketide | 16–19 | 4188 | South China Sea | sediment | [15] |
| cytotoxic | Penicillium griseofulvum | sesquiterpene | 20 | 1420 | India Ocean | sediment | [16] |
| cytotoxic | Botryotinia fuckeliana | diterpene | 21 | 5572 | West Pacific | sea water | [17] |
| cytotoxic | Phomopsis tersa | meroterpenoid | 22, 23 | 3000 | India Ocean | sediment | [18] |
| cytotoxic | Chaetomium globosum | alkaloid | 24 | 2476 | South China Sea | sediment | [13] |
| cytotoxic | Arthrinium sp. UJNMF0008 | alkaloid | 25 | 3858 | South China Sea | sediment | [19] |
| cytotoxic | Cladosporium sphaerospermum | alkaloid | 26 | 4571 | East India Ocean | sediment | [20] |
| cytotoxic | Cladosporium sphaerospermum | alkaloid | 27–29 | 6562 | Mariana Trench | sediment | [21] |
| cytotoxic | Sarcopodium sp. FKJ-0025 | phenol | 30, 31 | 200 | Kagoshima coast | sediment | [22] |
* refers to a mixed culture of the Antarctic deep-sea-derived fungus Penicillium crustosum PRB-2 and the mangrove-derived fungus Xylaria sp. HDN13-249.
Figure 1.
Distribution of deep-sea derived fungi in secondary metabolites discovery. (*Data based on the statistics of references [11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38]).
2.1. Antitumoral Secondary Metabolites
Compounds 1–32 were isolated from deep-sea fungi from different sea areas and showed varying degrees of antitumor activity.
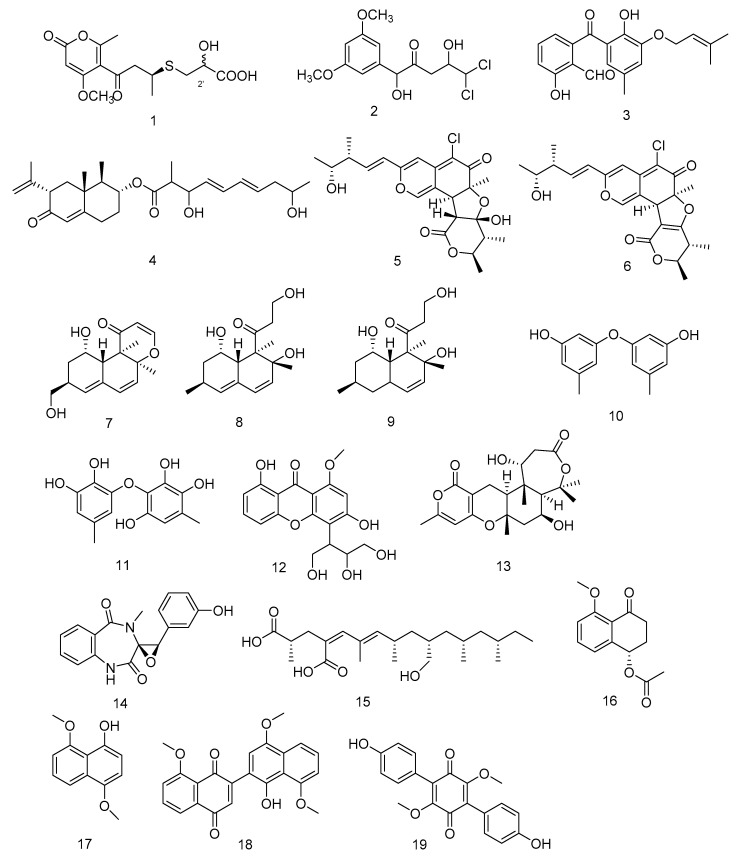
Compounds 1–4 (Figure 2) are all polyketides. New compounds 2-hydroxyl-3pyrenocine-thio propanoic acid (1) and 5, 5-dichloro-1-(3, 5dimethoxyphenyl)-1, 4-dihydroxypentan-2-one (2), containing sulfur or chlorine atoms, were isolated from the ethyl acetate extract of Penicillum citreonigrum. Compound 1 existed in the form of C-2′ epimers with a ratio of 1:2 (2′R:2′S). It showed potent cytotoxicity against human hepatocellular carcinoma cell (HCC) line Bel7402 and human fibrosarcoma cell line HT1080. The IC50 (50% inhibiting concentration) values were 7.63 ± 1.46 and 10.22 ± 1.32 µM, respectively. Compound 2 exhibited an IC50 value of 16.53 ± 1.67 µM against human fibrosarcoma tumor cell HT1080 [11]. Benzophenone derivatives tenellone H (3) and compound AA03390 (4) were isolated from the extract of a fungus Phomopsis lithocarpus derived from Indian Ocean sediments, compound 3 exhibited moderate cytotoxic activity against human HCC line and human non-small cell lung cancer cell (NSCLC) line A549, with IC50 values of 16.0 ± 0.1 and 17.6 ± 0.3 µM, respectively. Compound 4 showed weak cytotoxic activity against human HCC cell line HepG-2, human breast cancer cell line MCF-7, human neuronal cancer cell line SF-268 and human NSCLC line A549 [12].
Figure 2.
Structures of polyketides with antitumor activity.
Chaetomium globosum HDN151398 was isolated from deep-sea sediments in the South China Sea, and its metabolites chaetomugilin A (5) and chaetomugilin C (6) (Figure 2) showed broad-spectrum cytotoxic activities. Compound 5 exhibited significant cytotoxic activity against human promyelocytic leukemia cell line HL-60 and human colorectal cancer cells HCT-116, with IC50 values of 6.4 and 6.1 µM, respectively. While compound 6 exhibited IC50 values of 6.6 and 5.7 µM against HL-60 and HCT-116, respectively [13].
Peniciversiols A (7), decumbenone A (8), decumbenone B (9), 3, 3′-dihydroxy-5, 5′-dimethyldiphenyl ether (10), violaceol-II (11), 3, 8-dihydroxy-4-(2, 3-dihydroxy-1-hydroxymethylpropyl)-1-methoxyxanthone (12), asperdemin (13), cyclopenol (14) and radiclonic acid (15)were isolated from the ethyl acetate extract of Penicillium chrysogenum MCCC 3A00292.Their structures were shown in Figure 2. Compound 7 was a versiol-type analogue featuring a 2, 3-dihydropyran-4-one ring and showed significant cytotoxic activity against human Bladder cancer cell line BIU-87 with the IC50 value of 10.21 μM. Meanwhile compounds 10, 14 and 15 also had selective inhibition against BIU-87 with IC50 values of 16.41, 8.34 and 12.47 μM, respectively. Compounds 8, 9, 11 and 16 exhibited selective inhibitory effect against human esophageal cell line ECA109 with IC50 values of 12.41, 15.60, 8.95 and 7.70 μM, respectively. Compounds 12–15 had selective inhibition against human Hepatocellular carcinoma cell line BEL-7402. The IC50 values were 15.94, 12.75, 7.81 and 13.75, respectively [14].
Hypoxylon rubiginosum FS521, a higher fungi species, was isolated from deep-sea sediments in the South China Sea. From its ethyl acetate extract, hypoxone A (16), 4, 8-dimethoxy-1-naphthol (17), 1′-hydroxy-4′, 8, 8′-trimethoxy[2, 2′]binaphthalenyl-1, 4-dione (18) and 3, 6-dimethylatromentin (19) were isolated. Compounds 17 and 18 were new natural products. Compound 18 exhibited significant selective inhibitory effects against human glioblastoma carcinoma cell line SF-268, human breast cancer cell line MCF-7, human liver cancer cell line HepG-2 and human lung cancer cell line A549 with IC50 values of 1.9, 3.2, 2.5, and 5.0 μM, respectively. Compounds 16, 17 and 19 showed weak inhibition against the four tumor cell lines with IC50 values ranging from 18.89 to 69.62 μM [15].
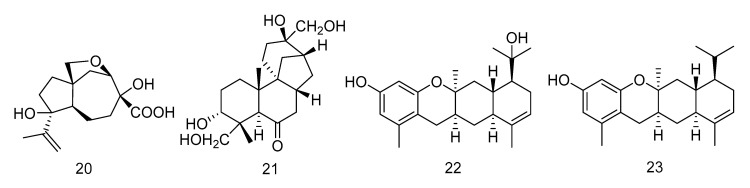
Penigrisacid D (20) is a sesquiterpene isolated from the extract of Penicillium griseofulvum, which showed a weak inhibitory activity against esophageal cancer cell line ECA-109, with an IC50 value of 28.7 µM [16]. Aphidicolin A8 (21) is a diterpene isolated from the extract of Botryotinia fuckeliana derived from Western Pacific seawater samples, which induced human bladder cancer cell T24 and human promyelocytic leukemia cell HL-60 apoptosis by DNA damage, with the IC50 values of 2.5 μM and 6.1 μM, respectively [17]. Photeroids A (22) and B (23) were isolated from the extract of the fungus Phomopsis tersa. They are both heteroterpenes containing a 6/6/6/6 tetracyclic system which forms Ortho-Quinone methides (o-QMs) intermediates through a rare Diels-Alder reaction. Compounds 22 and 23 (Figure 3) showed moderate cytotoxicity against four human cancer cell lines: SF-268, MCF-7, HepG-2 and A549 [18].
Figure 3.
Structures of terpenoids with antitumor activity.
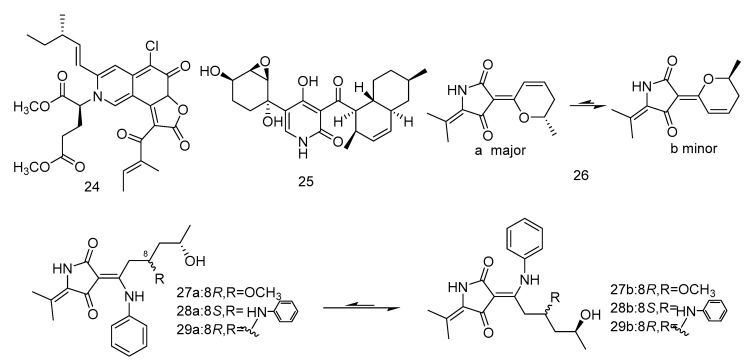
N-glutarylchaetoviridins (24) is an azaphilone alkaloid containing glutamine residues isolated from Chaetomium globosum HDN151398. Compound 24 exhibited significant cytotoxic activity against human gastric cancer cell line MGC-803 and human ovarian cancer cell line HO-8910, with IC50 values of 6.6 and 9.7 µM, respectively [13]. Apiosporamide (25) is also an alkaloid, isolated from the extract of Arthrinium sp. UJNMF0008 derived from sediments of the South China Sea. It exihibited cytotoxity against two human osteosarcoma cell lines (U2OS and MG63) with IC50 values of 19.3 and 11.7 µM [19].
Cladodionen (26) was isolated from the extract of Cladosporium sphaerospermum derived from the Indian Ocean deep sea sediments, and cladosins I–K (27–30) (Figure 4) were isolated from the extract of Cladosporium sphaerospermum derived from Mariana Trench. Compound 26 showed cytotoxic activity against human promyelocytic leukemia cell line HL-60, with the IC50 value of 28.6 µM [20]. Compounds 27–29 showed different levels of cytotoxic activity against human chronic myelogenous leukemia cell line K562 and human promyelocytic leukemia cell line HL-60, with IC50 values ranging from 2.8 to 7.8 µM [21].
Figure 4.
Structures of alkaloids with antitumor activity.
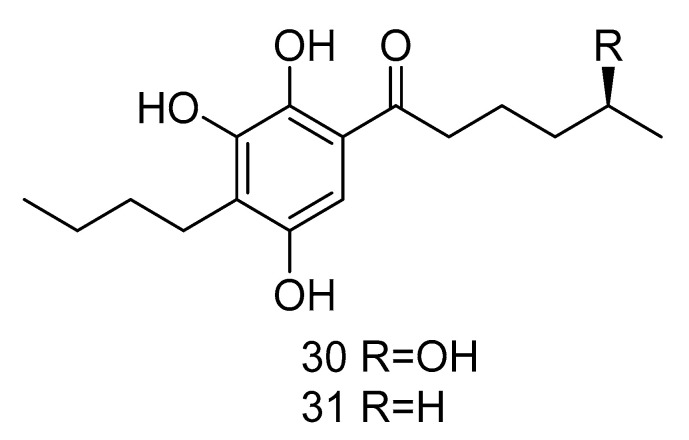
Sarcopodinols A (30) and B (31) (Figure 5) were isolated from the fungus Sarcopodium sp. FKJ-0025 isolated from coastal sediments of Kagoshima. Compound 30 had weak cytotoxity towards human T-lymphocytic leukemia Jurkat cell line, with the IC50 value of 47 μg/mL. Compound 31 exhibited an IC50 value of 37 μg/mL against HL-60 cell line, 47 μg/mL against Jurkat cell line, and 66 μg/mL against human pancreatic cancer Panc1 cell [22].
Figure 5.
Structures of other compounds with antitumor activity.
2.2. Antmicrobial Secondary Metabolites
2.2.1. Antibacterial Secondary Metabolites
Compounds 32–62 are secondary metabolites with antibacterial activity isolated from fungi extracts from different deep-sea environments.
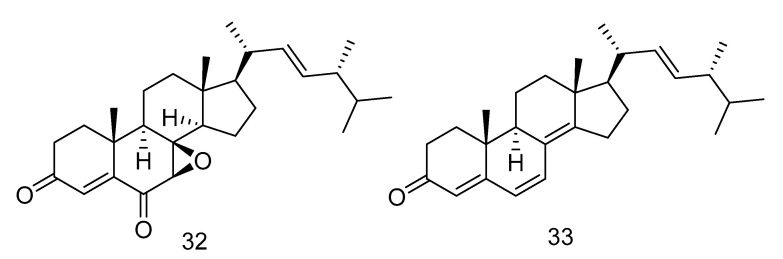
7β, 8β-epoxy-(22E, 24R)-24-methylcholesta-4, 22-diene-3, 6-dione (32) and ergosta-4, 6, 8(14), 22-tetraene-3-one (33) (Figure 6) were steroids isolated from the extract of Aspergillus penicillioides SD-311 from deep sea-sediment collected of the South China Sea. Compound 32 could inhibit Vibrio anguillarum with the MIC (minimum inhibitory concentration) value of 32.0 mg/mL. And compound 33 showed antibacterial activity against Edwardsiella tarda and Micrococcus luteus with MIC values both of 16 mg/mL [23].
Figure 6.
Structures of steroids with antibacterial activity.
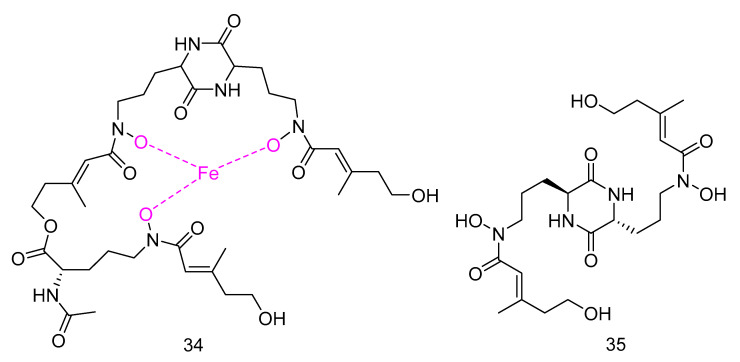
Mycosphazine A (34) was isolated from the extract of Mycosphaerella sp. SCSIO z059. It is a new a new iron(III) chelator of coprogen-type siderophore which could greatly promote the biofilm formation of Bacillus amyloliquefaciens with the rate of about 249% at concentration of 100 μg·mL−1. Its alkaline hydrolysate was a new epimer of dimerum acid, mycosphazine B (35) (Figure 7) which showed the same activity with the rate of about 524% at concentration of 100 μg·mL−1 [24].
Figure 7.
Structures of Mycosphazine A and B.
Peniginsengins C–E (36–38) (Figure 8) were new farnesylcyclohexenones isolated from the extract of Penicillium sp. YPGA11 from the sea water in the Yapu Trench. They showed activity against methicillin-resistant Staphylococcus aureus (Methicillin-resistant Staphylococcus aureus (MRSA), and anti-methicillin-sensitive Staphylococcus aureus (Methicillin-Sensitive Staphylococcus aureus, MSSA), with the MICvalues ranging from 8 μg/mL to 64 μg/mL [25].
Figure 8.
Structures of farnesylcyclohexenones with antibacterial activity.
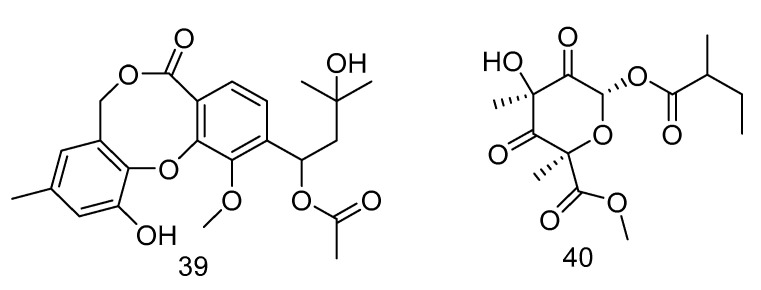
Canescenin A–B (39–40) (Figure 9) were isolated from the extract of Penicillium canescens SCSIO z053 derived from the deep-sea sediment of Okinawa Trough. Both of the compounds showed weak antibacterial activities toward B. amyloliquefaciens and P. aeruginosa at 100 μM [24].
Figure 9.
Structures of Canescenin A–B.
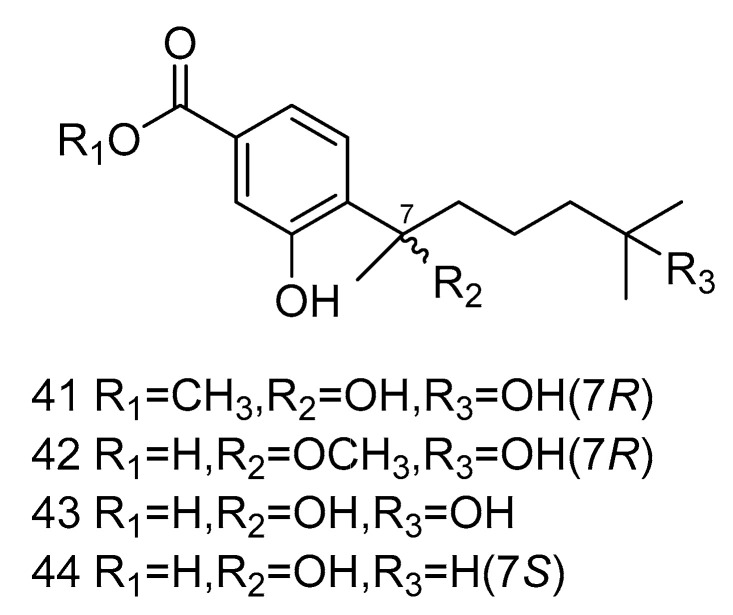
Four bisabolane-type sesquiterpenoid derivatives ent-aspergoterpenin C (41), 7-O-methylhydroxysydonic acid (42), hydroxysydonic acid (43) and sydonic acid (44) (Figure 10) were isolated from Aspergillus versicolor derived from the deep-sea sediment of South China Sea. Compounds 41–42 had strong antibacterial activity, whose MIC values against Escherichia coli, Edwardsiella tarda, Vibrio harveyi and Vibrio parahaemolyticus were all below or equal to 8.0 µg/mL. Moreover, compound 43 exihibited antibacterial activities against Aeromonas hydrophilia, Escherichia coli, Vibrio anguillarum and Vibrio harveyi, with the MIC value of 4.0 µg/mL, equal to the positive control Chloramphenicol [25].
Figure 10.
Structures of bisabolane-type sesquiterpenoid with antibacterial activity.
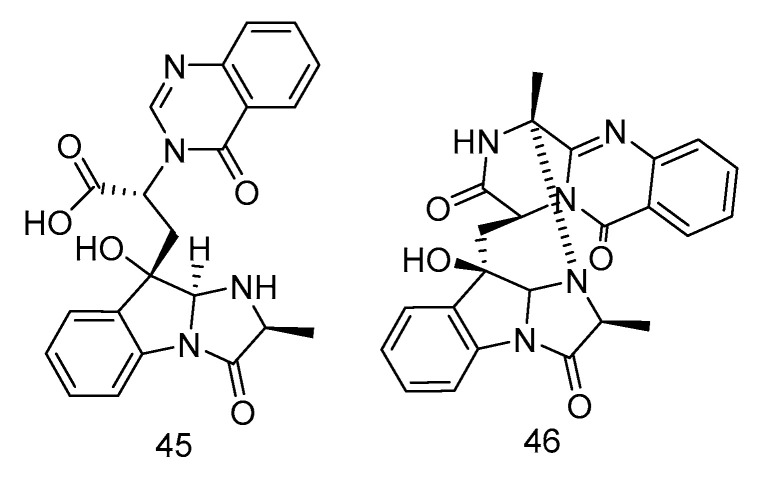
Fumigatosides E (45) and F (46) (Figure 11) were quinazoline-containing indole alkaloids, isolated from the extract of Aspergillus fumigatus from the deep-sea sediments of the Indian Ocean. Both of them showed potent antibacterial activities. The MIC values of compound 45 against Acinetobacter baumannii ATCC 19606, Acinetobacter baumannii ATCC 15122, Staphylococcus aureus ATCC 16339 and Klebsiella pneumoniae ATCC 14578 were: 12.5 ± 0.042, 6.25 ± 0.035, 6.25 ± 0.13, 12.5 ± 0.098 µg/mL, and the MIC value of compound 46 against Acinetobacter baumannii was 6.25 ± 0.033 µg/mL [26].
Figure 11.
Structures of quinazoline-containing indole alkaloids with antibacterial activity.
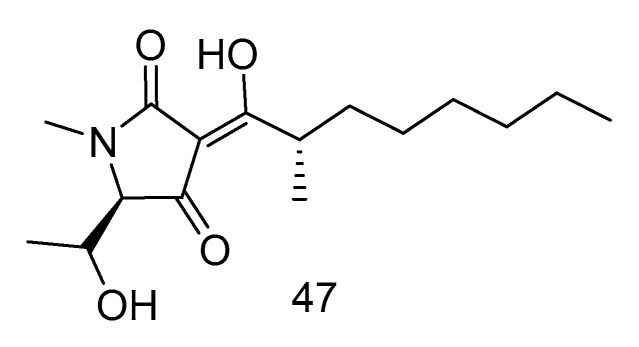
Penicillenol A2 (47) (Figure 12) was isolated from the extract of Penicillium biourgeianum isolated from the sediments of the South China Sea with an inhibitory effect on MSSA. The diameter of inhibitory zone (ZD) is 6.75 ± 0.25 mm. Besides, the synergy of compound 47 with penicillin G sodium (Pen), cefotaxime sodium (Ctx) and oxacillin sodium (Oxa) was studied by plate count and Kirby-Bordisk diffusion method. It was found that, in comparison with the control group, the reduction of bacteria in the experimental group using Pen (10 U mL−1), Ctx (15 U mL−1) and Oxa (1 U mL−1) was less than 1 log CFU/mL. Compared with using compound 30 alone, the reduction of viable bacteria in the experimental group using both the above drugs and compound 47 was greater than or equal to 2 log CFU/mL. Therefore, the combination of compound 30 and β-lactam antibiotics had a synergistic effect, which can increase the sensitivity of MRSA to β-lactam antibiotics [27].
Figure 12.
Structure of Penicillenol A2.
Pyridone alkaloids, apiosporamide (25) and arthpyrones F–K (48–51) (Figure 13), were isolated from the extract of Arthrinium sp. UJNMF0008 from the sediments of South China Sea, which showed moderate to strong antibacterial activity against Mycobacterium smegmatis and Staphylococcus aureus, with the IC50 values ranging from 1.66–42.8 µM [19].
Figure 13.
Structures of pyridone alkaloids with antibacterial activity.
3, 5-dimethoxytoluene (52), 3, 3′-dihydroxy-5, 5′-dimethyldiphenyl ether (53), 3, 4-dihydroxyphenylacetic acid methyl ester (54) (Figure 14) were isolated from the extract of Aspergillus sp. SCSIO06786 from deep-sea sediments in the Indian Ocean. Compounds 52 and 53 with 50 μg/disc showed inhibition zones against S. aureus, MRSA and E. faecalis. Compound 54 with 50 μg/disc inhibited the growth of MRSA. In addition, their MIC was tested and the results showed that it was between 3.13–12.5 μg/mL [30]. Penixylarins B–C (55–56), 1, 3-dihydroxy-5-(12-hydroxyheptadecyl)benzene (57), and 1, 3dihydroxy-5-(12-sulfoxyheptadecyl)benzene (58) (Figure 14) were isolated from a mixed culture of the Antarctic deep-sea-derived fungus Penicillium crustosum PRB-2 with a fungus Xylaria sp. HDN13-249. Compounds 55–58 showed activities against Bacillus tuberculosis, B. subtilis or Vibrio parahaemolyticus, MIC values ranging from 6.25 to 100 μM. Among them, the MIC value of compound 56 against B. tuberculosis was 6.25 μM, showing the anti-tuberculosis potential [31].
Figure 14.
Structures of phenols with antibacterial activity.
The tyrosine phosphatase (Mptp) secreted by Mycobacterium tuberculosis is an important virulence factor of Mycobacterium tuberculosis and recognized to be an important target to treat tuberculosis. Tyrosine phosphatase is secreted by Mycobacterium tuberculosis, which has two functional phosphatases, PTP A and B (MptpA and MptpB) and enters the cytoplasm of macrophages, preventing the activation of the host’s immune system and regulating the survival of the bacilli in the host [32]. Compounds 59–62 (Figure 15) were polyacrylate derivatives with long hydrophobic chains, isolated from the extract of Aspergillus fischeri derived from deep sea sediments in Indian Ocean. Compounds 59–62 inhibited M. tuberculosis protein tyrosine phosphatase B (MptpB) through non-competitive inhibition, with IC50 values of 5.1, 12, 4.0 and 11 μM, respectively [33].
Figure 15.
Structures of polypropionate derivatives with antituberculosis activity.
2.2.2. Antifungal Secondary Metabolites
Quinazoline-containing indole alkaloid, fumigatoside F (63) (Figure 16), was isolated from the extract of Aspergillus fumigatus derived from deep-sea sediments of the Indian Ocean. The MIC values against Fusarium oxysporum f. sp. cucumerinu and Fusarium oxysporum f. sp. momordicae were 25 ± 0.04 and 1.565 ± 0.098 µg/mL, respectively [28].
Figure 16.
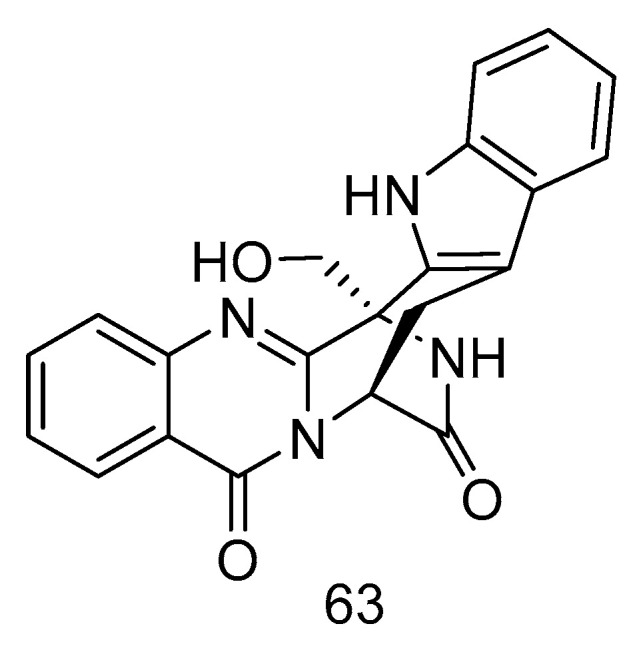
Structure of fumigatoside F.
2.3. Secondary Metabolites with Other Bioactivities
In addition to antitumoral and antimicrobial activity, secondary metabolites of deep-sea fungi reported in recent years also have anti-inflammatory and anti-food allergic activities. Compounds 64–69 were secondary metabolites from deep-sea fungi with anti-inflammatory activity.
Trieffusols C (64) and D (65) (Figure 17) were isolated from the extract of Trichobotrys effuse from deep-sea sediments of the South China Sea with inhibition of nitric oxide (NO) production in murine macrophages. Their IC50 values were 51.9 and 55.9 μM, which is equivalent to the positive control aminoguanidine (IC50: 24.8 μM) [34].
Figure 17.
Molecular structures of Trieffusols C and D.
Graphostromane D (66), graphostromane F (67), graphostromane I (68) and (1R, 4S, 5S, 7S, 9R, 10S, 11R)-guaiane-9, 10, 11, 12-tetraol (69) (Figure 18) were sesquiterpenoids isolated from the extract of Graphostroma sp. MCCC 3A00421 derived from hydrothermal sulfide deposit. Compound 50 showed anti-infammatory activity against LPS-induced NO production in RAW264.7 macrophages, with an IC50 value of 14.2 μM, even stronger than that of positive contrast. Meanwhile compound 66, 68 and 69 exhibited weak anti-infammatory activities, with the IC50 values of 72.9, 79.1, and 88.2 μM [35].
Figure 18.
Molecular structures of sesquiterpenoids.
Phenazine derivatives, 6-[1-(2-aminobenzoyloxy)ethyl]-1-phenazinecarboxylic acid (70), saphenol (71), (R)-saphenic acid (72), phenazine-1-carboxylic acid (73), 6-(1-hydroxyehtyl)phenazine-1-carboxylic acid (74) and 6-acetyl-phenazine-1-carboxylic acid (75) (Figure 19), were isolated from the extract of Cystobasidium laryngis, which inhibits the NO in mouse macrophage RAW 264.7 cells induced by Lipopolysaccharide (LPS), and does not affect the viability of RAW 264.7 cells at a concentration of up to 30 µg/mL. Compounds 72, 74, 75 showed similar inhibitory effects, of which compound 74 had the most obvious inhibitory effect, with the concentration for 50% of maximal effect (EC50) value of 19.6 µM. Methylated compound 74 and oxidized compound 75 showed no significant differences in activity mentioned above, but their strength was twice as strong as compound 70 (EC50 = 46.8 µM) substituted with 2-aminobenzoic acid. In addition, when there is no functional group substitution at C-6 of phenazine (73, EC50 = 76.1 μM), the activity is the lowest [36].
Figure 19.
Structures of phenazine derivatives with anti-inflammatory activity.
The occurrence of food allergic diseases may be related to excessive immune response. Allergens are usually harmless foods such as milk, eggs, fish, peanuts and grains [39]. Acute hypersensitivity is triggered by factors released by mast cells when allergens interact with membrane-bound immune proteins (IgE) [40].
Polyketides 76–78(Figure 20) were isolated from Graphostroma sp. MCCC 3A00421 derived from hydrothermal sulfide, which showed antifood allergic activity. Reticulol (76) showed effective inhibition of immunoglobulin E-mediated rat basophilic leukemia-2H3 cells (RBL-2H3) degranulation, with an IC50 value of 13.5 µM, which was about seven times stronger than the commercially available anti-food allergy drug loratadine (IC50 = 91.6 µM), while 7, 8-dihydroxy -3-methyl-3, 4-dihydroisocoumarin (77) and hydroxyemodin (78) showed weaker effects, with IC50 values of 154.1 and 139.3 µM [37].
Figure 20.
Structures of polyketides with antifood allergic activity.
Botryotin A (79) (Figure 21) was isolated from Botryotinia fuckeliana derived from deep-sea water of the Western Pacific Ocean, which showed moderate antiallergic effect with the IC50 value of 0.2 mM [38].
Figure 21.
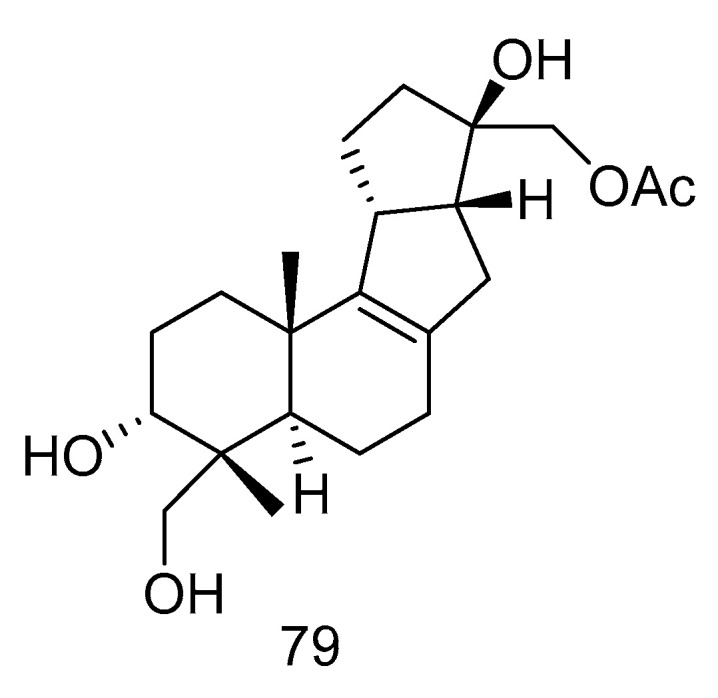
Structure of Botryotin A.
3. Secondary Metabolites from Deep-Sea Derived Bacteria
Bacteria from deep-sea sediments are a good source of marine natural products, and their secondary metabolites are usually novel in structure with significant biological activities [41,42,43,44,45]. In particular, actinomycetes are currently proven to be the most important sources of biologically active natural products with clinical or pharmaceutical applications [46]. According to the references, 40 (16 novel) natural products were discovered from deep-sea derived bacteria in the past three years, 19 among which showed biological activities (Table 2).
Table 2.
Bioactive Natural products from deep-sea derived bacteria in 2018–2020.
| Bioactivity | Bacterial Species | STRUCTRUAL CLASS | Number | Depth (m) | Region | Source | Reference |
|---|---|---|---|---|---|---|---|
| anti-allergic effect | Saccharopolyspora cebuensis | polyketide | 98 | 2875 | Atlantic | sediment | [47] |
| cytotoxic | Saccharopolyspora cebuensis | polyketide | 80 | 2875 | Atlantic | sediment | [47] |
| cytotoxic | Nonomuraea sp. AKA32 | polyketide | 81–83 | 800 | Sagami Bay | sea water | [48] |
| cytotoxic | Ochrobactrum sp. OUCMDZ-2164 | polyketide | 84 | 2000 | South China Sea | sea water | [49] |
| cytotoxic | Ochrobactrum sp. OUCMDZ-2164 | polyketide | 85 | 2000 | South China Sea | sea water | [50] |
| cytotoxic | Streptomyces cyaneofuscatus | alkaloid | 86, 87 | 2000 | Biscay Bay | solitary coral | [51] |
| antibacterial | Streptomyces sp. SCSIO ZS0098 | peptide | 88 | 3000 | South China Sea | sediment | [52] |
| antibacterial | Streptomyces atratus | peptide | 89 | 3536 | South China Sea | sediment | [53] |
| antibacterial | Streptomyces cyaneofuscatus | polyketide | 90, 91 | 1500 | Avilés submarine Canyon | gorgonian coral | [54] |
| antibacterial | Nocardiopsis sp. HB-J378 | polyketide | 92–94 | —— | —— | Theonella sp. | [55] |
| antibacterial | Streptomycetes sp. strain SMS636. | alkaloid | 95, 96 | 3000 | South China Sea | sediment | [56] |
| anti-BCG | Streptomycetes sp. strain SMS636. | alkaloid | 96 | 3000 | South China Sea | sediment | [56] |
| inhibit the cell damage | Alcanivorax sp. SHA4 | alkaloid | 97 | 5180 | West Atlantic | sediment | [57] |
3.1. Antitumoral Secondary Metabolites
Compounds 80–87 all showed potent cytotoxic activity.
Cebulactam A2 (80) (Figure 22) was a polyketide isolated from the extract of Saccharopolyspora cebuensis derived from Atlantic deep-sea sediments, which had a weak antiproliferative effect on human cervical cancer cell Hela and human lung cancer cell H1299, the inhibition rates (20.00 μg/mL) were 35.0 and 31.0%, respectively [47].
Figure 22.
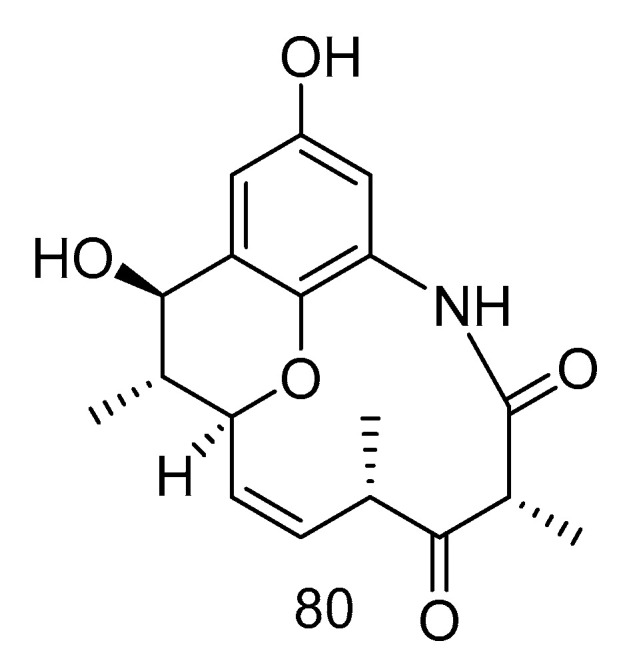
Structure of Cebulactam A2.
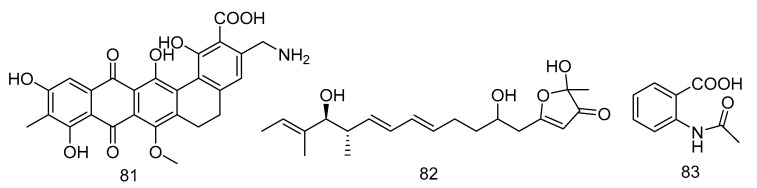
Akazamicin (81), actinofuranone C (82) and N-formylanthranilic acid (83) (Figure 23) were isolated from the extract of Nonomuraea sp. AKA32 derived from seawater of Sagami Bay in Japan. Compounds 81 and 82 showed the same level of cytotoxic activity against B16 melanoma cells, with IC50 values of 1.7 and 1.2 µM, respectively. Compound 83 showed about 10 times lower cytotoxic activity than that of compounds 81 and 82. The cytotoxic activity of these three compounds against human liver cancer cells Hep G2 and human colorectal adenocarcinoma cells Caco-2 was not obvious, with IC50 values ranging from 10 to 200 μM [48].
Figure 23.
Structures of aromatic polyketides with antitumor activity.
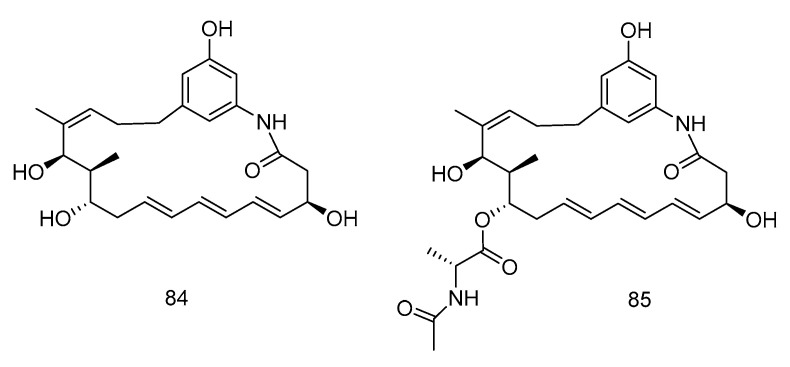
Trienomycins J–H (84–85) (Figure 24) were isolated from the extract of Ochrobactrum sp. OUCMDZ-2164 derived from deep-sea water of the South China Sea. Compound 84 exhibited antitumor activity against human breast cancer cells (MCF-7) with 61.5% inhibition rate at 10 μmol/L [49]. Compound 85 showed cytotoxic activity against human lung carcinoma cell line (A549) and human leukemia cell line (K562) with IC50 values of 15 and 23 µM, respectively [50].
Figure 24.
Structures of Ansamycins with antitumor activity.
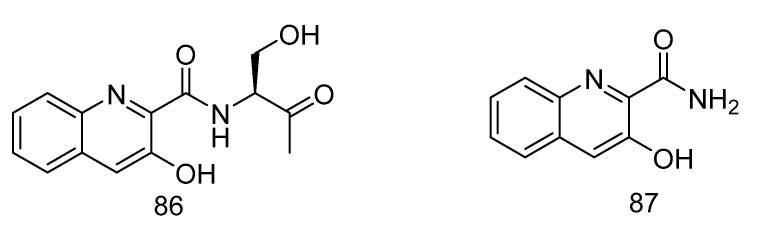
(S)-3-hydroxy-N-(1-hydroxy-3-oxobutan-2-yl) quinoline-2-carboxamide (86) and 3-hydroxyquinoline-2-carboxamide (87) (Figure 25), were isolated from a solitary coral derived Streptomyces cyaneofuscatus from Biscay Bay of north Atlantic. The IC50 values towards human liver cancer cell HepG2 were 15.6 and 51.5 µM, respectively [51].
Figure 25.
Structures of (S)-3-hydroxy-N-(1-hydroxy-3-oxobutan-2-yl) quinoline-2-carboxamide and 3-hydroxyquinoline-2-carboxamide.
3.2. Antimicrobial Secondary Metabolites
Compounds 88–96 all showed potent antibacterial activity.
Aborycin (88) was a lasso peptide isolated and identified from the deep-sea-derived microbe Streptomyces sp. SCSIO ZS0098 which was isolated from the deep-sea sediments of the South China Sea. Shao et al. [52] identified the aborycin biosynthetic gene cluster (abo) on the basis of genomic sequence analysis, and then heterologously expressed in Streptomyces coelicolor to obtain compound 86. The compound had moderate bacteriostatic activity against 13 Staphylococcus aureus strains from various sources, with MIC values between 8.0–128 µg/mL. The MIC values of compound 88 against Enterococcus faecalis and Bacillus thuringiensis were 8.0 µg/mL and, 2.0 µg/mL, respectively. In addition, compound 88 had significant antibacterial activity against the poultry pathogen Enterococcus enterococci (MIC = 0.5 µg/mL) [44]. Atratumycin (89) was also a peptide isolated from the extract of Streptomyces atratus from deep-sea sediments of the South China Sea, which is a cyclic dipeptide that has activity against Mycobacterium tuberculosis, whose MIC values were 3.8 and 14.6 µM against M. tuberculosis H37Ra and H37Rv [53].
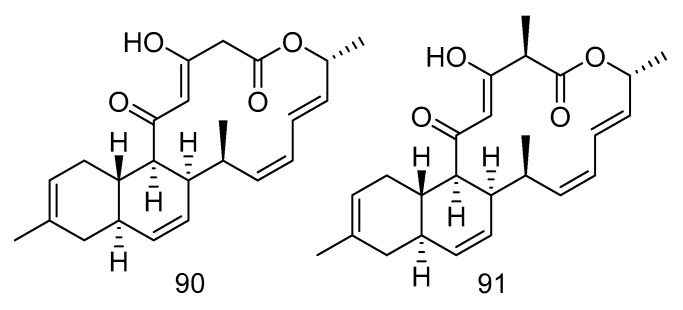
Compounds 90–94 are all polyketides. Anthracimycin B (90) and anthracimycin (91) (Figure 26) were isolated from the extract of Streptomyces cyaneofuscatus isolated from a gorgonian coral collected in the 1500 m Avilis submarine canyon. They were sensitive to Gram-positive pathogens MRSA, MSSA, vancomycin-sensitive Enterococcus faecium and vancomycin-sensitive Enterococcus faecalis and all showed strong antibacterial effects. The MIC value of compound 90 was less than 0.03 µg/mL, and the MIC value of compound 91 was between 0.125–8 µg/mL. Compound 90 also had anti-tuberculosis activity, with the MIC value of 1–2 µg/mL [54].
Figure 26.
Structures of anthracimycin B and anthracimycin.
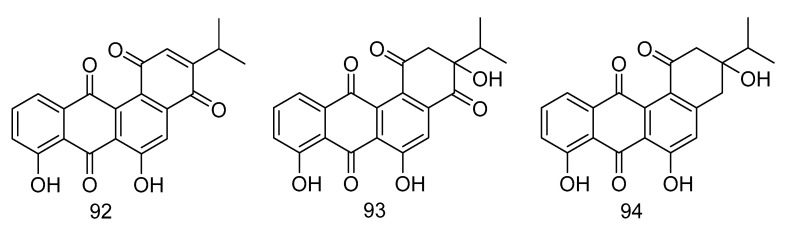
Nocardiopsistins A–C (92–94) (Figure 27) were isolated from the extract of Nocardiopsis sp. HB-J378 isolated from a deep-sea sponge Theonella sp. Compound 93 had the same MIC (3.12 µg/mL) as the positive control chloramphenicol, while compounds 92 and 94 had moderate anti-MRSA activity (MIC = 12.5 µg/mL) [55].
Figure 27.
Structures of nocardiopsistins A–C.
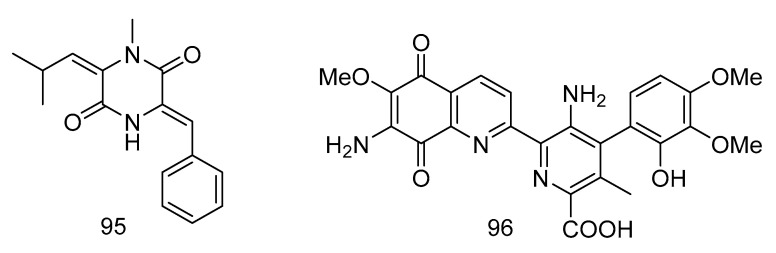
1-N-methyl-(E, Z)-albonoursin (95) and streptonigrin (96) (Figure 28) were alkaloids isolated from the extract of Streptomycetes sp. strain SMS636 from deep-sea sediments in the South China Sea. Compound 95 showed moderate antibacterial activity towards Staphylococcus aureus and MRSA, with MIC values of 12.5 and 25 µg/mL, respectively. The MIC value of compound 96 was 0.78 µg/mL for Staphylococcus aureus and MRSA, and had anti-BCG (Bacillus Calmette-Guérin, BCG) activity with a MIC value of 1.25 µg/mL [56].
Figure 28.
Structures of 1-N-methyl- (E, Z)-albonoursin and streptonigrin.
3.3. Other Bioactive Secondary Metabolites
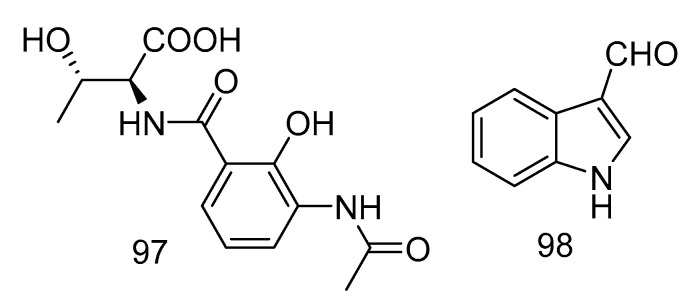
Acantimycic acid (97) was an alkaloid with good neuroprotection. It was isolated from the extract of Alcanivorax sp. SHA4 from deep-sea sediments of the Western Pacific and could inhibit the cell damage caused by glutamic acid to PC12 cells. The protective effect was more obvious at low concentration [57]. Indol-3-carbaldehyde (98) was isolated from the extract of Saccharopolyspora cebuensis derived from Atlantic deep-sea sediments. It showed weak anti-allergic effect with the IC50 value of 55.75 μg/mL [47]. The structures of compounds 97 and 98 are shown in Figure 29.
Figure 29.
Structures of acantimycic acid and Indol-3-carbaldehyde.
4. Research Methods for Diversity of Secondary Metabolites from Deep-Sea Microorganisms
4.1. Isolation and Cultivation of Deep-Sea Microorganisms
Marine microorganisms have the following characteristics: (1) can grow and/or form spores in the marine environment; (2) form a symbiotic relationship with other marine organisms; or (3) adapt and evolve at the genetic level or have metabolic activity in the marine environment [58]. It is estimated that the diversity of marine fungi exceeds 10,000 species [59,60], but so far only about 1250 species have been described [61,62]. However, deep-sea microbial research starts late for its difficulties in collection and cultivation, so people face more challenges in the exploration of its secondary metabolites.
4.1.1. Sample Pretreatment
In natural samples without pretreatment, the isolation frequency of bacteria is higher than that of fungi [63]. Different pretreatment methods should be adopted for different target strains to improve the isolation efficiency.
Because the actinomycete spores have a certain heat resistance, dry and wet heat treatment can effectively reduce other bacterial contamination [64,65,66,67,68]. Dry heat treatment can inactivate bacteria, and at the same time induce the germination of actinomycetes spores to a certain extent; the principle of wet heat treatment is to denature and inactivate non-target strain proteins in the sample by heating in a water bath. In addition to heat treatment, the commonly used pretreatment methods include chemical reagent treatment [69], differential centrifugation [70] and so on.
Microwave treatment can not only significantly increase the number of isolated alkaliphilic and halophilic marine actinomycetes, but also significantly increase the isolation of rare marine actinomycetes. Ding et al. [71] used 120 W, 2450 MHz microwave and ice-water mixture to process one part of the suspension in the treatment of sea mud samples. After gradient dilution, they were applied to three separate media. In the seven samples after microwave treatment, the number of rare alkaliphilic marine actinomycetes in four samples and the halophilic marine actinomycetes in three samples increased significantly. Therefore, microwave processing also has certain application value.
4.1.2. Medium Selection and Improvement
When the strains are separated, the medium as a nutrient source plays an important role in the growth and metabolism of the strains. Different culture media provide different carbon and nitrogen sources for different microorganisms to grow, so it is necessary to select the appropriate culture media for microorganism screening.
For fungi, we usually use common media such as: Potato Dextrose Agar Medium (PDA), Czapek Dox Agar Medium (CDA), Sabouraud Dextrose Agar Medium (SDA), Corn Meal Agar Medium (CMA), Malt Extract Agar Medium (MEA), yeast malt agar medium (YM), etc. He et al. [72] used the above-mentioned six media (all added chloramphenicol and streptomycin sulfate to inhibit growth of bacteria) to separate samples from the deep-sea sediment samples of Yapu Trench. In their study, YM media was the best from the perspective of the isolation ability of six different media, which obtained nine kinds of fungi; followed by PDA which allowed the retrieval of eight different fungal species. The worst were CMA (three kinds) and CDA (two kinds).
For bacteria, according to the main components of the medium, it can be divided into marine agar medium (MA), synthetic medium for selective isolation of actinomycetes (Actinomycete Isolation Agar, AIA), starch medium, natural ingredient medium, high salt medium and other media (Table 3). Chen et al. [73] used the 23 media in the table to isolate bacteria in the 4000 m deep-sea sediments of the South China Sea, and most of natural products from the strains obtained from the deep sea sediment environment, are antibiotic, cytotoxins, with high efficiency enzyme activity, and tolerant for unfavorable environment, degradation of refractory pollutants and other characteristics suitable for the unique marine extreme environment.
Table 3.
Different kinds of medium and their components.
| Type of Medium | Name of Medium | Composition of Medium |
|---|---|---|
| marine agar medium (MA) | MA; BD Difco™ | ① marine bacteria medium2216E |
| MAB | ② 50%MA medium | |
| MAE | ③ 20%MA medium | |
| MAJ | ④ 10%MA medium | |
| Actinomycete Isolation Agar (AIA) | AIA; BD Difco™ | ① selective medium for actinomycetes |
| AIAB | ② 50%AIA medium | |
| AIAE | ③ 20%AIA medium | |
| natural ingredient medium | acid microbial medium (AM) | ① MgSO4 7 H2O 0.50 g, (NH4)2SO4 0.40 g, K2HPO4 0.20 g, KCl 0.10 g, FeSO4·7H2O 0.01 g, yeast extract 0.25 g |
| Maltose-Yeast-Peptone Medium (MYP) | ② maltose extract 5.0 g, yeast extract 5.0 g, peptone 5.0 g, NaCl 3.0 g | |
| nutrient medium® | ③ peptone 10.0 g, yeast extract 5.0 g, maltose extract 5.0 g, casein amino acid 5.0 g, beef extract 2.0 g, glycerin 2.0 g, Tween 80 50.0 mg, MgSO4·7H2O 1.0 g | |
| R2AB; BD Difco™ | ④ 50%R2A medium | |
| R2AJ | ⑤ 5%R2A medium | |
| starch medium | MA starch medium (MAS) | ① MA, 1% (m/V) soluble starch |
| 50%MA starch medium (MABS) | ② MAB, 1% (m/V) soluble starch | |
| 20%MA starch medium (MAES) | ③ MAE, 1% (m/V) soluble starch | |
| 10%MA starch medium (MAJS) | ④ MAJ, 1% (m/V) soluble starch | |
| 5%MA starch medium (MATS) | ⑤ 5% MA, 1% (m/V) soluble starch | |
| high-salinity medium | high-salinity AIA medium (AIAS) | ① AIA 10.0 g, NaCl 100.0 g, crude salt extract5.0 g, SrCl2 2.0 g |
| high-salinity beef extract medium (BFSM) | ② beef extract 2.0 g, CaCO3 1.0 g, crude salt extract 5.0 g, Na2MoO4 5.0 g, soluble starch 2.0 g, NaCl 100.0 g | |
| high-salinity casein medium (CAAM) | ③ casein hydrolysate 1.0 g, KCl 2.0 g, MgSO4·7H2O 2.0 g, NaCl 100.0 g, crude salt extract 10.0 g, gluconate 1.0 g; trisodium citrate1.0 g, yeast extract 1.0 g, KMnO4 2.0 g (sterilize alone) | |
| high-salinity iron-containing medium (YJSF) | ④ MA 15.0 g, CaCO3 5.0 g, NaCl 100.0 g, FeCl2 0.5 g (filter sterilization), FeSO4 0.5 g (filter sterilization) | |
| other medium | SN | ① NaNO3 0.75 g, K2HPO4 0.0159 g, EDTA-2Na 0.0056 g, Na2CO3 0.0104 g, 50% sea water, Vitamin B12 0.001 g(filter sterilization), cyano trace metal solution 1 × 10−6 sterilize alone (acetic acid 6.25 g, ammonium ferric citrate 6.0 g, MnCl2·4H2O 1.4 g, Na2MoO4·2H2O 0.39 g, Co(NO3) 2·6H2O 0.025 g, ZnSO3·7H2O 0.222 g) |
| ZANT | ② NaHCO3 2.0 g, NaH2PO4·2H2O 0.05 g, NaNO3 0.5 g, CaCl2 0.02 g, MgSO4·7H2O 0.05 g, KCl 0.1 g, A5 solution 1 × 10−6(H3BO3 2.86 g, MnCl·4H2O 1.80 g, ZnSO4·7H2O 0.22 g, Na2MoO4·2H2O 0.3 g, CuSO4·5H2O 0.08 g) |
4.2. Screening Methods of Deep-Sea Natural Products
One of the keys to develop and utilize biological resources is how to obtain bioactive natural products from cultivable deep-sea microorganisms. Traditional natural product activity screening method is also suitable for the activity screening of deep-sea microbial metabolites, which mainly tracks the active substances in the cultivation broth. In addition, commonly used methods include model screening for specific target modeling and evaluation, and gene screening based on microbial natural product synthesis gene clusters.
4.2.1. In Vivo Screening Methods
In vivo screening models mainly refer to animal models and Serum pharmacology models. Animal models can mimic clinical features such as physiology and pathology similar to those of patients. Serum pharmacology models can help prove the true positive compounds, whether they are original drugs or metabolites [74]. Therefore, in vivo experiments have an irreplaceable role in activity screening. However, due to its time-consuming, low throughput, and large sample consumption, it is less used in preliminary screening.
4.2.2. Cell and Receptor/Enzyme Model Screening Methods
Cell and receptor/enzyme model screening is used for target screening, and is usually established as a specific and effective model on pharmacology at the cellular or molecular level.
Compared with simple chemical methods, evaluation of biological activity of natural products based on cell models can not only simulate the human physiological environment, but it can also explore and evaluate the biological activity and mechanism of natural products at multiple targets; compared with animal experiments in vivo, it not only shorten the experimental time, but greatly reduces the experimental cost [75]. The most commonly used cell models are human cancer cell lines, such as: A54, MCF7, HepG2, Caov-3, PANC-1 and so on. In addition, there are other models at the cellular level to test other bioactivities. Xu et al. used hemolysis assay on sheep red blood cell to test the anti-complement activity of 42 strains of marine actinomycetes isolated from Dalian Xinghai Bay mud samples, and further isolated three small molecular compounds with weak anti-complement activity from extract of strain DUT11 [76].
Receptors or enzymes related to various physiological and pathological processes in the body are considered to be one of the main targets of drug action [74]. Liu et al. tested IC50 values of the polypropionate derivatives against MptpB to show their antituberculosis activities [33]. ACE2 has been shown to be the main receptor for SARS-CoV S protein to infect cells [77]. Deng et al. showed that baicalin had an inhibitory activity against ACE with the IC50 value of 2.24 mM [78].
4.2.3. Virtual and Gene Screening Methods
Virtual screening based on compound structural diversity, that is, using computer programs to screen bioactive compounds from existing virtual libraries. And compounds with higher chemical structure spatial diversity are more suitable for virtual library establishment [79].
Gene screening breaks through the traditional active screening model. Because the secondary metabolite synthetic gene clusters with similar structures have a certain degree of similarity, the strains that produce the target compound can be obtained from nature by screening specific gene clusters.
Polyketide compounds are catalyzed by a type of polyketide synthetase (PKS) which is widely present in nature. Polyketide synthetase can generally be divided into three types according to its protein structure and catalytic mechanism, namely type I, type II and type III [80]. Type I PKS includes type I modular PKS (bacteria) and type I repeat PKS (fungi). A typical type I module PKS is a multifunctional complex enzyme composed of modules. Each module contains a unique and non-repetitive structural domain, which mainly contains acyltransferase (acyltransferase, AT), β-keto synthase (ketosynthase, KS) responsible for catalyzing the formation of carbon-carbon bonds and extending the main chain, and acyltransferase cylcar-rier protein (acyltransferase cylcar-rier protein, ACP) responsible for receiving and transporting acyl units provided by the AT domain), these three domains constitute the smallest catalytic module and are also the three essential domains of PKS.
Non-ribosomal peptide synthetase (NRPS) is also widely present in bacteria, fungi and plants, and uses different amino acids as substrates to catalyze the production of condensed peptides. NRPS is mainly composed of different independent modules. Each module contains an adenylation structural functional domain (andeylation, A) that selects and activates special amino acids, and loads aminoacyl residues into the sulfhydryl structural functional domain (thiolation, T), and the condensation domain (C) of peptide compounds that polymerize activated amino acids to produce amides.
In fungi, the genes encoding PKS and NRPS can be aggregated to produce type I repeat PKS units (KS, AT, DH, CMeT, KR and ACP domains) and NRPS units (A, T and C domains) PKS-NRPS hybrid enzyme. PKS-NRPS has the function of catalyzing the combination of PKS products and NRPS products, thereby producing more abundant natural products-PKS-NRPS hybrid compounds [81]. Such PKS-NRPS hybrid compounds are often a class of natural products with complex and diverse structures and a wide range of biological activities. They not only play an important role in the survival and prosperity of the host in the natural environment, but also is an important source for the discovery of active lead compounds with potential applications.
Jiang et al. [82] applied the type I polyketide synthase (PKS-I) gene screening system and DNA sequence similarity comparison to select positive Actinoplanes sp. from 32 strains of marine actinomycetes. Actinoplanes sp. FIM060065 was one of them. And from its fermentation broth researchers obtained a macrolide compound homogenous to tiacumicin B by High Performance Liquid Chromatography (HPLC) preparation, which showed strong antibacterial activity towards Gram-positive bacteria, such as Clostridium difficile, Streptococcus pneumoniae, Bifidobacterium, etc. Vanessa Rédou et al. [83] isolated 124 filamentous fungi and 59 yeasts from sediments in the Canterbury basin of New Zealand. The PKS-NRPS analysis results showed that there was no PKS-NRPS hybrid gene in the yeast genome; compared with yeast, filamentous fungal isolates from deep seabed sediments have greater bioactive compound synthesis potential, but they have fewer bioactive compound genes than those isolated from shallower depths.
4.3. Secondary Metabolite Discovery Based on Synthetic Biology
Synthetic biology is a multidisciplinary disruptive study leading a new generation of biotechnology revolution, in which gene editing takes an important part. Gene editing has unique advantages in establishing an artificially regulated biosynthetic system, further mining new natural product resources of actinomycetes, solving the bottleneck of existing natural products and developing derivatives. Recently, CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 system, known as “magic scissor”, was found to improve the efficiency of gene editing.
Elizabeth J. Culp et al. [84] applied CRISPR/Cas9 system to 11 actinomycete strains, knocked out the common streptomycin and streptomycin genes, produced a variety of hidden rare antibiotics, and constructed a platform that can be widely used to stimulate the potential of microbial secondary metabolism. Indra Roux et al. [85] established the first CRISPRa system for filamentous fungi and discovered the mic cluster product, dehydromicroperfuranone. Meanwhile, factors affecting the efficiency of the system was also studied.
5. Conclusions and Perspective
Although the research on secondary metabolites of deep-sea microbes started later than that in other environments [9], it has drawn much more attention, and natural products with novel structures and good biological activities have been discovered in the past three years. From our literature review, fungi seem to be the focus of most isolations from the deep-sea for bioprospection of metabolites with biological activities; also, producers of higher diversity and amount of compounds. Among bacteria, actinomycetes seem to be studied more deeply in natural product research, and they have shown the potential to become biological resources with novel structures and good biological activities. When it comes to structural classes, polyketides showed a broad spectrum of bioactivities, such as antitumor, antibacterial, anti-inflammatory and antifood allergic.
The rapid development of deep sea exploration and bioinformatics has provided solid technical support for the chemical diversity and bioactivity diversity of secondary metabolites of deep sea microorganisms, but there are still many challenges, such as activation of specific biosynthetic gene clusters and heterologous expression, directed transformation of synthetic gene clusters, design of virtual screening libraries for natural products, and how to solve the problem of yield of active compounds.
Deep-sea microbial natural product resources are still a virgin land that needs to be developed urgently. Reasonable and green applied research will contribute more power to drug discovery.
Author Contributions
Y.-N.W. prepared the manuscript; L.-H.M. and B.-G.W. supervised the research work and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA22050401), and the Natural Science Foundation of China (Grant No. 41976090). L.-H.M. thanks the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2017250). B.-G.W. acknowledges the support of Taishan Scholar Project from Shandong Province.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tortorella E., Tedesco P., Esposito F.P., January G.G., Fani R., Jaspars M., Pascale D. Antibiotics from Deep-Sea Microorganisms: Current Discoveries and Perspectives. Mar. Drugs. 2018;16:355. doi: 10.3390/md16100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jebbar M., Franzetti B., Girard E., Oger P. Microbial diversity and adaptation to high hydrostatic pressure in deep-sea hydrothermal vents prokaryotes. Extremophiles. 2015;19:721–740. doi: 10.1007/s00792-015-0760-3. [DOI] [PubMed] [Google Scholar]
- 3.Skropeta D., Wei L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014;31:999–1025. doi: 10.1039/C3NP70118B. [DOI] [PubMed] [Google Scholar]
- 4.Ventola C.L. The Antibiotic Resistance Crisis: Part 1: Causes and threats. Pharm. Ther. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 5.Mahé S., Rédou V., Le Calvez T., Vandenkoornhuyse P., Burgaud G. The Ecological Genomics of Fungi. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2013. Fungi in Deep-Sea Environments and Metagenomics; pp. 325–354. [Google Scholar]
- 6.Sharma S., Fulke A.B., Chaubey A. Bioprospection of marine actinomycetes: Recent advances, challenges and future perspectives. Acta Oceanol. Sin. 2019;38:1–17. doi: 10.1007/s13131-018-1340-z. [DOI] [Google Scholar]
- 7.Bhatnagar I., Kim S.-K. Immense Essence of Excellence: Marine Microbial Bioactive Compounds. Mar. Drugs. 2010;8:2673–2701. doi: 10.3390/md8102673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okutani K. Gliotoxin produced by a strain of Aspergillus isolated from marine mud. Nippon. Suisan Gakkaishi. 1977;43:995–1000. doi: 10.2331/suisan.43.995. [DOI] [Google Scholar]
- 9.Muhammad Z.A., Ma Y.N., Xue Y.R., Liu C.H. Deep-Sea Fungi Could Be the New Arsenal for Bioactive Molecules. Mar. Drugs. 2020;18:9. doi: 10.3390/md18010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrar M., Ullah M.W., Manan S., Farooq U., Rafiq M., Hasan F. Fungi from the extremes of life: An untapped treasure for bioactive compounds. Appl. Microbiol. Biotechnol. 2020;104:2777–2801. doi: 10.1007/s00253-020-10399-0. [DOI] [PubMed] [Google Scholar]
- 11.Tang X., Liu S.-Z., Yan X., Tang B.-W., Fang M., Wang X., Wu Z., Qiu Y.-K. Two New Cytotoxic Compounds from a Deep-Sea Penicillum citreonigrum XT20-134. Mar. Drugs. 2019;17:509. doi: 10.3390/md17090509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J.-L., Liu H., Chen Y., Tan H.-B., Guo H., Xu L.-Q., Li S.-N., Huang Z.-L., Li H., Gao X.-X., et al. Highly Substituted Benzophenone Aldehydes and Eremophilane Derivatives from the Deep-Sea Derived Fungus Phomopsis lithocarpus FS508. Mar. Drugs. 2018;16:329. doi: 10.3390/md16090329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun C., Ge X., Shah M., Zhou L., Yu G., Che Q., Zhang G., Peng J., Gu Q.-Q., Zhu T.-J., et al. New Glutamine-Containing Azaphilone Alkaloids from Deep-Sea-Derived Fungus Chaetomium globosum HDN151398. Mar. Drugs. 2019;17:253. doi: 10.3390/md17050253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu S., Xia M., Chen M., Liu X., Li Z., Xie Y., Shao Z., Zhang G. Cytotoxic Polyketides Isolated from the Deep-Sea-Derived Fungus Penicillium chrysogenum MCCC 3A00292. Mar. Drugs. 2019;17:686. doi: 10.3390/md17120686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., Chen Y., Liu Z., Guo B., Gao X., Liu H., Zhang W. Cytotoxic Secondary Metabolites from a Sea-Derived Fungal Strain of Hypoxylon rubiginosum FS521. Chin. J. Org. Chem. 2020;40:1367. doi: 10.6023/cjoc201912012. [DOI] [Google Scholar]
- 16.Xing C., Xie C.-L., Xia J.-M., Liu Q., Lin W.-X., Ye D.-Z., Liu G., Yang X. Penigrisacids A–D, Four New Sesquiterpenes from the Deep-Sea-Derived Penicillium griseofulvum. Mar. Drugs. 2019;17:507. doi: 10.3390/md17090507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niu S., Xia J.-M., Li Z., Yang L.-H., Yi Z.-W., Xie C.-L., Peng G., Luo Z.-H., Shao Z., Yang X. Aphidicolin Chemistry of the Deep-Sea-Derived Fungus Botryotinia fuckeliana MCCC 3A00494. J. Nat. Prod. 2019;82:2307–2331. doi: 10.1021/acs.jnatprod.9b00705. [DOI] [PubMed] [Google Scholar]
- 18.Chen S.C., Liu Z.M., Tan H.B., Chen Y.C., Zhu S., Liu H.X., Zhang W.M. Photeroids A and B, unique phenol–sesquiterpene meroterpenoids from the deep-sea-derived fungus Phomopsis tersa. Org. Biomol. Chem. 2020;18:642–645. doi: 10.1039/C9OB02625H. [DOI] [PubMed] [Google Scholar]
- 19.Bao J., Zhai H., Zhu K., Yu J.-H., Zhang Y., Wang Y.-Y., Jiang C.-S., Zhang X.-Y., Zhang Y., Zhang H. Bioactive Pyridone Alkaloids from a Deep-Sea-Derived Fungus Arthrinium sp. UJNMF0008. Mar. Drugs. 2018;16:174. doi: 10.3390/md16050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang X., Huang Z.-H., Ma X., Qi S. Unstable Tetramic Acid Derivatives from the Deep-Sea-Derived Fungus Cladosporium sphaerospermum EIODSF 008. Mar. Drugs. 2018;16:448. doi: 10.3390/md16110448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z., He X., Wu G., Liu C., Lu C., Gu Q., Che Q., Zhu T., Zhang G., Li D. Aniline-Tetramic Acids from the Deep-Sea-Derived Fungus Cladosporium sphaerospermum L3P3 Cultured with the HDAC Inhibitor SAHA. J. Nat. Prod. 2018;81:1651–1657. doi: 10.1021/acs.jnatprod.8b00289. [DOI] [PubMed] [Google Scholar]
- 22.Matsuo H., Nonaka K., Nagano Y., Yabuki A., Fujikura K., Takahashi Y., Ōmura S., Nakashima T. New metabolites, sarcopodinols A and B, isolated from deep-sea derived fungal strain Sarcopodium sp. FKJ-0025. Biosci. Biotechnol. Biochem. 2018;82:1323–1326. doi: 10.1080/09168451.2018.1467264. [DOI] [PubMed] [Google Scholar]
- 23.Chi L., Yang S.-Q., Li X.-M., Li X.-D., Wang B.-G., Li X. A new steroid with 7β,8β-epoxidation from the deep sea-derived fungus Aspergillus penicillioides SD-311. J. Asian Nat. Prod. Res. 2020:1–8. doi: 10.1080/10286020.2020.1791096. [DOI] [PubMed] [Google Scholar]
- 24.Huang Z.-H., Liang X., Qi S. A new iron(III) chelator of coprogen-type siderophore from the deep-sea-derived fungus Mycosphaerella sp. SCSIO z059. Chin. J. Nat. Med. 2020;18:243–249. doi: 10.1016/s1875-5364(20)30029-7. [DOI] [PubMed] [Google Scholar]
- 25.Cheng Z., Xu W., Liu L., Li S., Yuan W., Luo Z., Zhang J., Cheng Y., Li Q. Peniginsengins, B–E, New Farnesylcyclohexenones from the Deep Sea-Derived Fungus Penicillium sp. YPGA11. Mar. Drugs. 2018;16:358. doi: 10.3390/md16100358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dasanayaka S.A.H.K., Nong X.-H., Liang X., Liang J.-Q., Amin M., Qi S. New dibenzodioxocinone and pyran-3,5-dione derivatives from the deep-sea-derived fungus Penicillium canescens SCSIO z053. J. Asian Nat. Prod. Res. 2020;22:338–345. doi: 10.1080/10286020.2019.1575819. [DOI] [PubMed] [Google Scholar]
- 27.Li X.-D., Yin X., Li X., Wang B., Li X. Antimicrobial Sesquiterpenoid Derivatives and Monoterpenoids from the Deep-Sea Sediment-Derived Fungus Aspergillus versicolor SD-330. Mar. Drugs. 2019;17:563. doi: 10.3390/md17100563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salendra L., Luo X., Lin X., Liao S., Wang J., Zhou X., Yang B., Liu Y. Bioactive Novel Indole Alkaloids and Steroids from Deep Sea-Derived Fungus Aspergillus fumigatus SCSIO 41012. Molecules. 2018;23:2379. doi: 10.3390/molecules23092379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S., Mou Q., Xu X., Qianqian M., Leung P.H.M. Synergistic antibacterial activity between penicillenols and antibiotics against methicillin-resistant Staphylococcus aureus. R. Soc. Open Sci. 2018;5:172466. doi: 10.1098/rsos.172466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang X., Lin X., Zhou X., Yang B., Tian X., Wang J., Xu S., Liu Y. New quinoline alkaloid and bisabolane-type sesquiterpenoid derivatives from the deep-sea-derived fungus Aspergillus sp. SCSIO06786. Fitoterapia. 2020;140:104406. doi: 10.1016/j.fitote.2019.104406. [DOI] [PubMed] [Google Scholar]
- 31.Yu G., Sun Z., Peng J., Zhu M., Che Q., Zhang G., Zhu T., Gu Q., Li D. Secondary Metabolites Produced by Combined Culture of Penicillium crustosum and a Xylaria sp. J. Nat. Prod. 2019;82:2013–2017. doi: 10.1021/acs.jnatprod.9b00345. [DOI] [PubMed] [Google Scholar]
- 32.Xiao Z.E., Lin S.E., Huang X.S., Xia G.P., Li H.X., Lu Y.J., She Z.G. The mPtpB Enzyme Inhibitors Derived from the Mangrove Fungi of the South China Sea. Abstracts of Papers; Proceedings of the Ninth National Conference on Marine Biotechnology and Innovative Drugs; ChiFeng, China. 6 August 2014. [Google Scholar]
- 33.Liu Z., Wang Q., Li S., Cui H., Sun Z., Chen D., Lu Y.-J., Liu H., Zhang W. Polypropionate Derivatives with Mycobacterium tuberculosis Protein Tyrosine Phosphatase B Inhibitory Activities from the Deep-Sea-Derived Fungus Aspergillus fischeri FS452. J. Nat. Prod. 2019;82:3440–3449. doi: 10.1021/acs.jnatprod.9b00834. [DOI] [PubMed] [Google Scholar]
- 34.Chen S.-C., Liu Z., Chen Y., Tan H.-B., Li S.-N., Liu H., Zhang W.-M., Zhu S. Highly Substituted Phenol Derivatives with Nitric Oxide Inhibitory Activities from the Deep-Sea-Derived Fungus Trichobotrys effuse FS524. Mar. Drugs. 2020;18:134. doi: 10.3390/md18030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H.-S., Kang J.S., Choi B.-K., Shin H.J., Lee H.-S., Lee Y.-J., Lee J., Shin H.J. Phenazine Derivatives with Anti-Inflammatory Activity from the Deep-Sea Sediment-Derived Yeast-Like Fungus Cystobasidium laryngis IV17-028. Mar. Drugs. 2019;17:482. doi: 10.3390/md17080482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niu S., Xie C.-L., Xia J.-M., Luo Z.-H., Shao Z., Yang X. New anti-inflammatory guaianes from the Atlantic hydrotherm-derived fungus Graphostroma sp. MCCC 3A00421. Sci. Rep. 2018;8:530. doi: 10.1038/s41598-017-18841-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niu S., Liu Q., Xia J.-M., Xie C.-L., Luo Z.-H., Shao Z., Liu G., Yang X. Polyketides from the Deep-Sea-Derived Fungus Graphostroma sp. MCCC 3A00421 Showed Potent Antifood Allergic Activities. J. Agric. Food Chem. 2018;66:1369–1376. doi: 10.1021/acs.jafc.7b04383. [DOI] [PubMed] [Google Scholar]
- 38.Niu S., Xie C.-L., Xia J.-M., Liu Q.-M., Peng G., Liu G., Yang X. Botryotins A−H, tetracyclic diterpenoids representing three carbon skeletons from a Deep-Sea-Derived. Org. Lett. 2019;22:580–583. doi: 10.1021/acs.orglett.9b04332. [DOI] [PubMed] [Google Scholar]
- 39.Bochner B.S., Rothenberg M.E., Boyce J.A., Finkelman F. Advances in mechanisms of allergy and clinical immunology in 2012. J. Allergy Clin. Immunol. 2013;131:661–667. doi: 10.1016/j.jaci.2012.12.676. [DOI] [PubMed] [Google Scholar]
- 40.Kay A.B. Overview of ‘Allergy and allergic diseases: With a view to the future’. Br. Med Bull. 2000;56:843–864. doi: 10.1258/0007142001903481. [DOI] [PubMed] [Google Scholar]
- 41.Abdel-Mageed W.M., Milne B.F., Wagner M., Schumacher M., Sandor P., Pathom-Aree W., Goodfellow M., Bull A.T., Horikoshi K., Ebel R., et al. Dermacozines, a new phenazine family from deep-sea dermacocci isolated from a Mariana Trench sediment. Org. Biomol. Chem. 2010;8:2352–2362. doi: 10.1039/c001445a. [DOI] [PubMed] [Google Scholar]
- 42.Huang H., Yang T., Ren X., Liu J., Song Y., Sun A., Ma J., Wang B., Zhang Y., Huang C., et al. Cytotoxic Angucycline Class Glycosides from the Deep Sea Actinomycete Streptomyces lusitanus SCSIO LR32. J. Nat. Prod. 2012;75:202–208. doi: 10.1021/np2008335. [DOI] [PubMed] [Google Scholar]
- 43.Li S., Tian X., Niu S., Zhang W., Chen Y.-C., Zhang H., Yang X., Zhang W., Li W., Zhang S., et al. Pseudonocardians A–C, New Diazaanthraquinone Derivatives from a Deap-Sea Actinomycete Pseudonocardia sp. SCSIO 01299. Mar. Drugs. 2011;9:1428–1439. doi: 10.3390/md9081428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato S., Iwata F., Yamada S., Kawahara H., Katayama M. Usabamycins A-C: New anthramycin-typeanalogues from a marine-derived actinomycete. Bioorganic Med. Chem. Lett. 2011;21:7099–7101. doi: 10.1016/j.bmcl.2011.09.086. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W., Liu Z., Li S., Yang T., Zhang Q., Ma L., Tian X., Zhang H., Huang C., Zhang S., et al. Spiroindimicins A–D: New Bisindole Alkaloids from a Deep-Sea-Derived Actinomycete. Org. Lett. 2012;14:3364–3367. doi: 10.1021/ol301343n. [DOI] [PubMed] [Google Scholar]
- 46.Bérdy J. Bioactive Microbial Metabolites. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 47.Xie C.-L., Niu S., Xia J.-M., Peng K., Zhang G.-Y., Yang X.-W. Saccharopolytide A, a new cyclic tetrapeptide with rare 4-hydroxy-proline moieties from the deep-sea derived actinomycete Saccharopolyspora cebuensis MCCC 1A09850. Nat. Prod. Res. 2017;32:1627–1631. doi: 10.1080/14786419.2017.1392956. [DOI] [PubMed] [Google Scholar]
- 48.Yang T., Yamada K., Zhou T., Harunari E., Igarashi Y., Terahara T., Kobayashi T., Imada C. Akazamicin, a cytotoxic aromatic polyketide from marine-derived Nonomuraea sp. J. Antibiot. 2019;72:202–209. doi: 10.1038/s41429-018-0139-7. [DOI] [PubMed] [Google Scholar]
- 49.Wang C., Cui T.X., Wang D.Y., Zhu W.M. Trienomycin J, a new ansamycin from deep-sea derived bacterium Ochrobactrum sp. Chin. Tradit. Herb. Drugs. 2019;23:5661–5665. [Google Scholar]
- 50.Fan Y., Wang C., Wang L., Chairoungdua A., Piyachaturawat P., Fu P., Zhu W. New Ansamycins from the Deep-Sea-Derived Bacterium Ochrobactrum sp. OUCMDZ-2164. Mar. Drugs. 2018;16:282. doi: 10.3390/md16080282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ortiz-López F.J., Alcalde E., Sarmiento-Vizcaíno A., Díaz C., Cautain B., García L.A., Blanco G., Reyes F. New 3-Hydroxyquinaldic Acid Derivatives from Cultures of the Marine Derived Actinomycete Streptomyces cyaneofuscatus M-157. Mar. Drugs. 2018;16:371. doi: 10.3390/md16100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shao M., Ma J., Li Q., Ju J. Identification of the Anti-Infective Aborycin Biosynthetic Gene Cluster from Deep-Sea-Derived Streptomyces sp. SCSIO ZS0098 Enables Production in a Heterologous Host. Mar. Drugs. 2019;17:127. doi: 10.3390/md17020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Z.-J., Wei X., He J., Sun C., Ju J., Ma J. Characterization of the Noncanonical Regulatory and Transporter Genes in Atratumycin Biosynthesis and Production in a Heterologous Host. Mar. Drugs. 2019;17:560. doi: 10.3390/md17100560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martín J., Martín J., Sarmiento-Vizcaíno A., De La Cruz M., García L.A., Blanco G., Reyes F. Anthracimycin B, a Potent Antibiotic against Gram-Positive Bacteria Isolated from Cultures of the Deep-Sea Actinomycete Streptomyces cyaneofuscatus M-169. Mar. Drugs. 2018;16:406. doi: 10.3390/md16110406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu D., Nepal K.K., Chen J., Harmody D., Zhu H., McCarthy P.J., Wright A.E., Wang G. Nocardiopsistins A-C: New angucyclines with anti-MRSA activity isolated from a marine sponge-derived Nocardiopsis sp. HB-J378. Synth. Syst. Biotechnol. 2018;3:246–251. doi: 10.1016/j.synbio.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu X., Han J., Lin R., Polyak S.W., Song F. Two New Piperazine-Triones from a Marine-Derived Streptomycetes sp. Strain SMS636. Mar. Drugs. 2019;17:186. doi: 10.3390/md17030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang D.S. Master’s Thesis. Zhejiang University; Hangzhou, China: 2019. Research on the Active Secondary Metabolites of Three Marine Microorganisms Based on Different Fermentation Methods. [Google Scholar]
- 58.Pang K.L., Overy P.D., Jones E.B.G., da Luz Calado M., Burgaud G., Walker K.A., Johnson A.J., Kerr G.R., Cha H.J., Gerald F.B. ‘Marine fungi’ and ‘marine-derived fungi’ in natural product chemistry research: Toward a new consensual definition. Fungal Biol. Rev. 2016;30:163–175. doi: 10.1016/j.fbr.2016.08.001. [DOI] [Google Scholar]
- 59.Jones E.G. Are there more marine fungi to be described? Bot. Mar. 2011;54:343–354. doi: 10.1515/bot.2011.043. [DOI] [Google Scholar]
- 60.Jones E.B.G., Pang K.L. Marine Fungi and Fungal-Like Organisms. De Gruyter; Berlin, Germany: 2012. pp. 1–13. [Google Scholar]
- 61.Jones E.G., Suetrong S., Sakayaroj J., Bahkali A.H., Abdel-Wahab M.A., Boekhout T., Pang K.-L. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Divers. 2015;73:1–72. doi: 10.1007/s13225-015-0339-4. [DOI] [Google Scholar]
- 62.Raja H.A., Miller A.N., Pearce C.J., Oberlies N.H. Fungal identification using molecular tools: A primer for the natural products research community. J. Nat. Prod. 2017;80:756–770. doi: 10.1021/acs.jnatprod.6b01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seong C.N., Choi J.H., Baik K.S. An improved selective isolation of rare Actinomycetes from forest soil. J. Microbiol. 2001;39:17–23. [Google Scholar]
- 64.Wang H.Y., Liu J., Zhao S.J. Study on the isolation of marine actinomycetes in the sediments of Nanji Island. Mar. Sci. 2010;34:48–51. [Google Scholar]
- 65.Wang F., Xu X.X., Qu Z., Wang C., Lin H.P., Xie Q.Y., Ruan J.S., Sun M., Hong K. Nonomuraea wenchangensis sp.nov. isolated from mangrove rthizosphere soil. Int. J. Syst. Evol. Microbiol. 2011;61:1304–1308. doi: 10.1099/ijs.0.025742-0. [DOI] [PubMed] [Google Scholar]
- 66.Xi L.J., Zhang L.M., Ruan J.S., Huang Y. Micromonospora rhizosphaerae sp. nov. isolated from mangrove rhizosphere soil. Int. J. Syst. Evol. Microbiol. 2011;61:320–324. doi: 10.1099/ijs.0.020461-0. [DOI] [PubMed] [Google Scholar]
- 67.Lin L., Tan Y., Chen F.F., Zhou H.X., Wang Y.G., He W.Q., Wang Y. Diversity of cultivable actinomycetes in sediments of Laohutan, Bohai Sea, Dalian. Acta Microbiol. Sin. 2011;51:262–269. [PubMed] [Google Scholar]
- 68.Xi L.J., Zhang L.M., Ruan J.S., Huang Y. Nonomuraea maritima sp. nov., isolated from coastal sediment. Int. J. Syst. Evol. Microbiol. 2011;61:2740–2744. doi: 10.1099/ijs.0.028266-0. [DOI] [PubMed] [Google Scholar]
- 69.Li J., Yang J., Zhu W.Y., He J., Tian X.P., Xie Q., Zhang S., Li W.J. Nocardiopsis coralliicola sp. nov., isolated from the gorgonian coral, Menella praelonga. Int. J. Syst. Evol. Microbiol. 2012;62:1653–1658. doi: 10.1099/ijs.0.035402-0. [DOI] [PubMed] [Google Scholar]
- 70.Maldonado L.A., Stach J.E.M., Pathom-Aree W., Ward A.C., Bull A.T., Goodfellow M. Diversity of cultivable actinobacteria in geographically widespread marine sediments. Antonie van Leeuwenhoek. 2005;87:11–18. doi: 10.1007/s10482-004-6525-0. [DOI] [PubMed] [Google Scholar]
- 71.Ding Y.B., Cai C.J., Mu Y.L., Shan Y.Q., Lu X.H., Jiang Q. The effect of microwave treatment on the separation of alkali and halophilic marine actinomycetes. Bull. Microbiol. 2012;39:407–414. [Google Scholar]
- 72.He G.Y., Xu W., Guo S.S., Liu W.H., Luo Z.H. Study on the cultivable fungal diversity and denitrification ability of deep-sea sediments in Yapu Trench. J. Appl. Oceanogr. 2018;37:230–238. [Google Scholar]
- 73.Chen R.W., Wang K.X., He Y.Q., Tian X.P., Long L.J. Diversity of culturable bacteria in a deep-sea sediment sample from the South China Sea. Biol. Resour. 2018;40:321–333. [Google Scholar]
- 74.Wang B., Deng J., Gao Y., Zhu L., He R., Xu Y. The screening toolbox of bioactive substances from natural products: A review. Fitoterapia. 2011;82:1141–1151. doi: 10.1016/j.fitote.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 75.Bu L.N. Master’s Thesis. Lanzhou University; Lanzhou, China: 2013. Screening and Evaluation of Active Ingredients of Natural Products Based On in vitro Cell Model Screening. [Google Scholar]
- 76.Xu X.N., Cui H.T., Chen L.Y., Su C., Bai F.W., Zhao X.Q. Screening of marine actinomycetes with anti-complement activity and isolation of their active substances. J. Appl. Environ. Biol. 2018;24:1295–1300. [Google Scholar]
- 77.Ou H.L., Li L.J. Research progress of renin angiotensin system in newly emerging respiratory infectious diseases. Mod. Pract. Med. 2019;1:1–3. [Google Scholar]
- 78.Deng Y.F., Aluko R.E., Jin Q., Zhang Y., Yuan L.J. Inhibitory activities of baicalin against renin and angiotensin-converting enzyme. Pharm. Biol. 2011;50:401–406. doi: 10.3109/13880209.2011.608076. [DOI] [PubMed] [Google Scholar]
- 79.Yibo L. Designing natural product-like virtual libraries using deep molecule generative models. J. Chin. Pharm. Sci. 2018;27:451–459. doi: 10.5246/jcps.2018.07.046. [DOI] [Google Scholar]
- 80.Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 2003;7:285–295. doi: 10.1016/S1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 81.Evans B.S., Robinson S.J., Kelleher N.L. Surveys of non-ribosomal peptide and polyketide assembly lines in fungi and prospects for their analysisin vitro and in vivo. Fungal Genet. Biol. 2011;48:49–61. doi: 10.1016/j.fgb.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang H.L., Fang Z.K., Chen M.H., Peng F., Jiang H., Lian Y.Y. Gene screening of macrolide-producing bacteria and research on their metabolites. Nat. Prod. Res. Dev. 2017;29:1895–1899. [Google Scholar]
- 83.Rédou V., Navarri M., Meslet-Cladière L., Barbier G., Burgaud G. Species Richness and Adaptation of Marine Fungi from Deep-Subseafloor Sediments. Appl. Environ. Microbiol. 2015;81:3571–3583. doi: 10.1128/AEM.04064-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Culp E.J., Yim G., Waglechner N., Wang W., Pawlowski A.C., Wright G.D. Hidden antibiotics in actinomycetes can be identified by inactivation of gene clusters for common antibiotics. Nat. Biotechnol. 2019;37:1149–1154. doi: 10.1038/s41587-019-0241-9. [DOI] [PubMed] [Google Scholar]
- 85.Roux I., Woodcraft C., Hu J., Wolters R., Gilchrist C.L.M., Chooi Y.-H. CRISPR-Mediated Activation of Biosynthetic Gene Clusters for Bioactive Molecule Discovery in Filamentous Fungi. ACS Synth. Biol. 2020;9:1843–1854. doi: 10.1021/acssynbio.0c00197. [DOI] [PubMed] [Google Scholar]