Abstract
In October 2020, a highly pathogenic avian influenza (HPAI) subtype H5N8 virus was identified from a fecal sample of a wild mandarin duck (Aix galericulata) in South Korea. We sequenced all eight genome segments of the virus, designated as A/Mandarin duck/Korea/K20-551-4/2020(H5N8), and conducted genetic characterization and comparative phylogenetic analysis to track its origin. Genome sequencing and phylogenetic analysis show that the hemagglutinin gene belongs to H5 clade 2.3.4.4 subgroup B. All genes share high levels of nucleotide identity with H5N8 HPAI viruses identified from Europe during early 2020. Enhanced active surveillance in wild and domestic birds is needed to monitor the introduction and spread of HPAI via wild birds and to inform the design of improved prevention and control strategies.
Keywords: highly pathogenic avian influenza virus, wild bird, surveillance, H5N8
1. Introduction
Highly pathogenic avian influenza (HPAI) subtype H5Nx viruses have been causing substantial losses to the poultry industry and public health concerns since the detection of A/Goose/Guangdong/1/1996(H5N1) (Gs/GD) from domestic poultry in southern China. The descendent Gs/GD-lineage H5 viruses have evolved into 10 genetically independent hemagglutinin (HA) clades (0–9) and subclades [1]. Wild birds, especially wild waterfowl of Anatidae family, are known as a natural reservoir of avian influenza virus (AIV) [2]. Migratory wild birds have been widely suggested to contribute to long-distance transmission and reassortment of HPAI along their migration flyways while they move between wintering and breeding sites seasonally [3,4,5,6,7].
Since the first identification of HPAI H5N5 virus in China in 2008, Gs/Gd-lineage clade 2.3.4 HPAI H5Nx viruses have evolved into subclades 2.3.4.1–2.3.4.4 [8]. Particularly, the novel clade 2.3.4.4 HPAI H5Nx viruses containing multiple neuraminidase (NA) subtypes including H5N2, H5N5, H5N6, and H5N8 have been causing numerous outbreaks in wild birds and poultry in broad geographical regions including Asia, Europe, Africa, and North America since 2014 [9]. The H5 genes of clade 2.3.4.4 HPAI H5Nx viruses were phylogenetically classified into 4 subgroups (A–D) [10]. Subgroup A and B viruses have spread over a broad geographic region due to migratory wild birds [9], and subgroup C and D viruses have been circulating mainly in China and neighboring countries in Asia [9,10,11].
During May–June 2016, novel reassortant clade 2.3.4.4 subgroup B H5N8 viruses containing internal genes of Eurasian low-pathogenic avian influenza (LPAI) viruses were identified in wild birds in Uvs Lake, Russia and Qinghai Lake, China [12,13]. These viruses had undergone further reassortments with prevailing LPAI viruses in wild birds and spread to Europe, Africa, and Asia in winter 2016–17 following the migration of wild birds [14,15,16,17,18]. In addition, these viruses further spread over large geographical areas in Asia, Africa, and Europe through wild birds and poultry trade in 2017–2018 winter [19,20,21,22].
The clade 2.3.4.4 subgroup B H5Nx viruses caused multiple outbreaks in South Korea in winter 2016–17 and 2017–18, which were most likely introduced by wild waterfowl [18,19,20,21,22,23]. The clade 2.3.4.4 subgroup B strains responsible for the outbreaks in South Korea had not been detected in the country since April 2018, despite large-scale active surveillance targeting both wild birds and poultry. In the present study, we report isolation of a clade 2.3.4.4 subgroup B HPAI subtype H5N8 virus from a fecal sample of a mandarin duck (Aix galericulata) collected in October 2020 during active wild bird surveillance for AIV in South Korea. We sequenced all eight genome segments of the virus, designated as A/Mandarin duck/Korea/K20-551-4/2020(H5N8), and conducted genetic characterization and comparative phylogenetic analysis to track its origin.
2. Materials and Methods
2.1. Sample Collection
We collected a total of 150 fresh fecal samples in wild bird habitat near Cheongmi Stream (GPS 37°07′09.0″ N 127°24′01.0″ E) in Anseong, South Korea, for active surveillance purposes. The fecal samples were stored at 4–8 ℃ and analyzed within 12 h.
2.2. Virus Isolation
Fecal samples were placed in phosphate-buffered saline (PBS) containing 400 mg/mL gentamicin and thoroughly homogenized by vortexing for 1 min. The supernatant of samples were filtered using a 0.45 µm Minisart Syringe Filter (Sartorius, Göttingen, Germany) and inoculated into 10-day-old specific-pathogen-free (SPF) embryonated chicken eggs. After 72 h of incubation at 37 ℃, the allantoic fluids were harvested and tested for hemagglutinin activity using 10% chicken red blood cells. RNA was extracted from the hemagglutinin-activity-positive allantoic fluid using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction and screened for the matrix (M) gene of avian influenza virus using real-time reverse transcription-PCR (rRT-PCR) as previously described [24].
2.3. Sequencing
Reverse transcription was conducted using SuperScript IV First-Strand Synthesis System (Invitrogen, Carslbad, CA, USA) and the eight gene segments were amplified as previously described using TaKaRa Ex Taq (Takara Bio Inc., Kusatsu, Japan) according to the manufacturer’s instruction [25]. The PCR products were purified using GeneJET Gel Extraction Kit (Thermo Scientific, Waltham, MA, USA) or QIAquick PCR Purification kit (Qiagen, Hilden, Germany) and sequenced using the ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The genome sequences have been deposited in GISAID with isolate ID EPI_ISL_666687.
2.4. Phylogenetic Analysis
The sequences of the highly pathogenic and low-pathogenic avian influenza viruses were retrieved from the GISAID Epiflu (https://platform.gisaid.org/) database and the NCBI’s Influenza Virus Resource at GenBank (https://www.ncbi.nlm.nih.gov/genomes/FLU) (acknowledgement of the research laboratories that contributed these data is provided in Supplementary Table S1) for comparative phylogenetic analysis. Complete coding regions of the retrieved sequences were aligned using MAFFT (https://mafft.cbrc.jp/alignment/software/). The maximum-likelihood (ML) phylogenetic tree for each genome segment was constructed using RAxML v8 [26] using the general time-reversible (GTR) nucleotide substitution model and discrete gamma distribution with 1000 rapid bootstrap replicates.
2.5. Host Species Identification
The host species of the AIV-positive fecal sample was identified using DNA barcoding technique as previously described [27]. Briefly, host DNA was extracted using QIAamp Fast DNA Stool Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. The mitochondrial cytochrome oxidase (COI) gene was amplified using Aves-F (5′-GCATGAGCAGGAATAGTTGG-3′) and Aves-R (5′-AAGATGTAGACTTCTGGGTG-3′) primers and sequenced using the ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA).
3. Results and Discussion
On 24 October in 2020, AIV virus was isolated from the fecal sample of a mandarin duck which was confirmed based on the rRT-PCR and DNA barcoding results. The isolate A/Mandarin duck/Korea/K20-551-4/2020(H5N8), hereafter referred to as 551-4/2020 virus, was identified as an HPAI virus on the basis of multiple basic amino acids at the HA proteolytic cleavage site (PLREKRRKR/G). The GISAID database BLAST searches showed that all eight gene segments of the 551-4/2020 virus shared high nucleotide identity (>99.05%) with HPAI H5N8 viruses identified from poultry farms and wild birds in Europe in early 2020 (Table 1).
Table 1.
Nucleotide sequence identities between the A/Mandarin duck/Korea/K20-551-4/2020(H5N8) virus and nearest virus homologs in the GISAID Epiflu database (as of 2 November 2020).
| Gene 1 | Accession No. | Virus | % Identity |
|---|---|---|---|
| PB2 | EPI1691234 | A/chicken/Germany-BW/AI00049/2020 (H5N8) | 99.56 |
| EPI1669674 | A/hawk/Poland/003/2020 (H5N8) | ||
| PB1 | EPI1721422 | A/duck/Hungary/1565_20VIR749-2/2020 (H5N8) | 99.34 |
| PA | EPI1718616 | A/buzzard/Germany-SN/AI00285/2020 (H5N8) | 99.58 |
| HA | EPI1691237 | A/chicken/Germany-BW/AI00049/2020 (H5N8) | 99.65 |
| NP | EPI1721665 | A/turkey/Hungary/1020_20VIR749-1/2020 (H5N8) | 99.33 |
| EPI1691238 | A/chicken/Germany-BW/AI00049/2020 (H5N8) | ||
| EPI1665425 | A/white-fronted goose/Germany-BB/AI00018/2020 (H5N8) | ||
| NA | EPI1691239 | A/chicken/Germany-BW/AI00049/2020 (H5N8) | 99.08 |
| EPI1669671 | A/turkey/Poland/23/2019 (H5N8) | ||
| MP | EPI1721683 | A/turkey/Germany-ST/AI00352/2020 (H5N8) | 99.80 |
| EPI1718620 | A/turkey/Germany-NI/AI00334/2020 | ||
| EPI1691240 | A/chicken/Germany-BW/AI00049/2020 | ||
| EPI1669680 | A/hawk/Poland/003/2020 | ||
| EPI1669672 | A/turkey/Poland/23/2019 | ||
| EPI1665427 | A/white-fronted goose/Germany-BB/AI00018/2020 | ||
| NS | EPI1719046 | A/turkey/Czech Republic/3071/2020 (H5N8) | 99.05 |
1 PB2, basic polymerase 2; PB1, basic polymerase 1; PA, acidic polymerase; HA, hemagglutinin; NP, nucleoprotein; NA, neuraminidase; MP, matrix protein; NS, nonstructural protein.
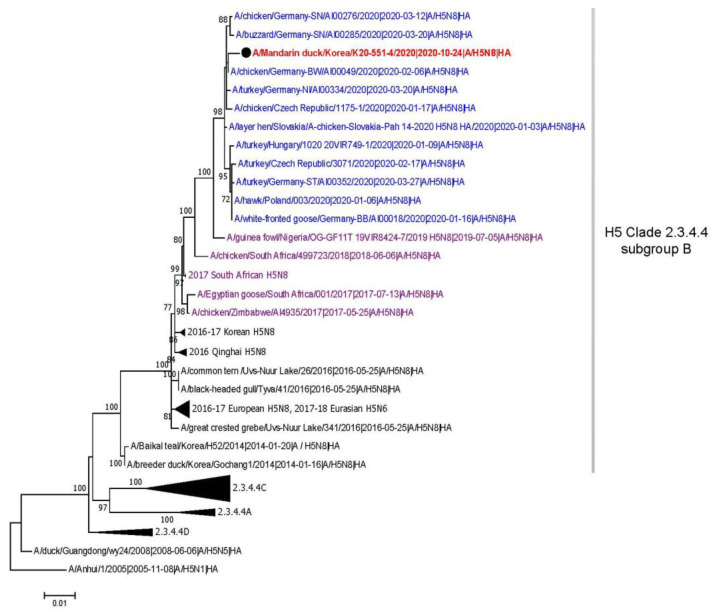
In the (ML) phylogenies, all the eight gene segments of 551-4/2020 virus were closely clustered together with the clade 2.3.4.4 subgroup B subtype H5N8 viruses detected from poultry and wild birds in Europe during early 2020 (Figure 1, Supplementary Figure S1). All genes of the 551-4/2020 virus were phylogenetically distinct from those of clade 2.3.4.4b H5 HPAI viruses isolated in South Korea during 2016–2018.
Figure 1.
Maximum-likelihood phylogenetic tree of hemagglutinin gene. The A/Mandarin duck/Korea/K20-551-4/2020(H5N8) is indicated with black circle and colored with red. Taxa colored with blue and purple indicate H5N8 highly pathogenic avian influenza (HPAI) viruses identified in Europe during early 2020 and Africa during 2017–2019, respectively. Bootstrap values over 70% are shown next to the branches. Scale bar indicates nucleotide substitutions per site. Collapsed branches are shown in black triangles.
South Korea is located in the East Asian–Australasian flyway and serves as a wintering and stopover site for wild migratory birds [7,28,29]. Wetland near the Cheongmi Stream from where the 551-4/2020 virus was isolated is a wintering site of wild migratory Anatidae birds including mallard (Anas platyrhynchos), spot-billed duck (Anas poecilorhyncha), and common teal (Anas crecca) and also the previous outbreak site of clade 2.3.4.4 subgroup B H5N6 in winter 2017–18 [23]. During the sampling near Cheongmi Stream, we observed flocks of spot-billed ducks and mandarin ducks.
Mandarin ducks are native to the Far and Southeast of Asia including China, North and South Korea, Japan, and Eastern Russia along the East Asian–Australasian flyway. Some mandarin ducks are resident in South Korea, and others are passage migrants that occasionally overwinter in Korea and move to and from southern Japan [30]. The identifications of HPAI H5 viruses from mandarin ducks have been reported multiple times in South Korea; clade 2.3.2.1 H5N1 in 2010 [31], clade 2.3.4.4 subgroup A H5N8 in December 2014 [32], clade 2.3.4.4 subgroup C H5N6 in October 2016 [33], and clade 2.3.4.4 subgroup B H5N6 in November 2017 [23]. Previous pathogenicity studies of clade 2.3.4.4 H5Nx viruses in mandarin ducks demonstrated that mandarin ducks can excrete virus without showing clinical signs or mortality and also transmit virus to other ducks by contact, indicating they can serve as healthy carriers of clade 2.3.4.4 H5Nx viruses [34,35,36]. Because the Mandarin duck is not a long-distance migrating species, the other species of long-distance migrants were suspected as an intermediate vector for this long-distance transmission.
The timing and location of the 551-4/2020 virus isolation, lack of identification of related clade 2.3.4.4 subgroup B H5Nx virus in South Korea in recent two and half years, and phylogenetic analysis and BLAST search results collectively suggest that migratory wild birds might have introduced the 551-4/2020 virus into South Korea during the fall migration of wild birds. However, the detailed transmission route of this virus could not be determined because of the lack of wild bird surveillance data during spring and summer 2020. It is most likely that the subgroup B H5Nx viruses belonging to clade 2.3.4.4 affected Europe during early 2020 and were then maintained in wild birds before being introduced into South Korea in fall 2020. Previous studies showed that numerous wild waterfowl species can be infected by clade 2.3.4.4 viruses without detectable clinical signs [37,38], and this biological characteristic makes difficult to detect viruses from wild waterfowl, supporting the hypothesis that wild birds in Eurasia maintained the virus and long-distance migrants introduced the virus into South Korea.
Considering the continued emergence of novel reassortant viruses and global dissemination of the clade 2.3.4.4 HPAI H5Nx viruses, enhanced active surveillance in wild and domestic birds is required to monitor the introduction, spread, and reassortment of HPAI and to inform the design of improved prevention and control strategies.
Acknowledgments
We thank the technical support of Hee-Su Lee, Do-Hyeok Choi, Yun-Jeong Choi, and Kyu-Cheol Choi at Konkuk University.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/12/12/1389/s1, Figure S1: Maximum-likelihood phylogenetic trees of eight gene segments. The A/Mandarin duck/Korea/K20-551-4/2020(H5N8) is indicated with black circle. Bootstrap values over 70% are shown next to the branches. Scale bar indicates nucleotide substitutions. (A) Polymerase basic 2 (PB2); (B) polymerase basic 1 (PB1); (C) polymerase acidic (PA); (D) hemagglutinin (HA); (E) nucleoprotein (NP); (F) neuraminidase (NA); (G) matrix (M); (H) nonstructural (NS). Table S1: The sequences from the GISAID Epiflu Database and NCBI’s Influenza Virus Resources at GenBank, on which this research is based.
Author Contributions
Conceptualization, D.-H.L. and C.-S.S.; methodology, S.J.; validation, D.-H.L., J.-H.K. and C.-S.S.; formal analysis, S.J. and S.-H.L.; investigation, S.J., Y.-J.K., S.-H.L., A.Y.C., T.-H.K., J.-E.P. and S.-I.L.; data curation, S.J.; writing—original draft preparation, S.J.; writing—review and editing, D.-H.L., J.-H.K. and C.-S.S.; visualization, S.J.; supervision, C.-S.S.; project administration, C.-S.S.; funding acquisition, C.-S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Bio and Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (no. NRF-2018M3A9H4056535). This paper was written as part of Konkuk University’s research support program for its faculty on sabbatical leave in 2017.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. OIE. FAO. H5N1 Evolution Working Group Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1) Emerg. Infect. Dis. 2008;14:e1. doi: 10.3201/eid1407.071681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webster R.G., Yakhno M., Hinshaw V.S., Bean W.J., Murti K.G. Intestinal influenza: Replication and characterization of influenza viruses in ducks. Virology. 1978;84:268–278. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/MMBR.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen B., Munster V.J., Wallensten A., Waldenstrom J., Osterhaus A.D., Fouchier R.A. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 5.Kilpatrick A.M., Chmura A.A., Gibbons D.W., Fleischer R.C., Marra P.P., Daszak P. Predicting the global spread of H5N1 avian influenza. Proc. Natl. Acad. Sci. USA. 2006;103:19368–19373. doi: 10.1073/pnas.0609227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keawcharoen J., van Riel D., van Amerongen G., Bestebroer T., Beyer W.E., van Lavieren R., Osterhaus A.D., Fouchier R.A., Kuiken T. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1) Emerg. Infect. Dis. 2008;14:600–607. doi: 10.3201/eid1404.071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian H., Zhou S., Dong L., Van Boeckel T.P., Cui Y., Newman S.H., Takekawa J.Y., Prosser D.J., Xiao X., Wu Y., et al. Avian influenza H5N1 viral and bird migration networks in Asia. Proc. Natl. Acad. Sci. USA. 2015;112:172–177. doi: 10.1073/pnas.1405216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu M., Liu W., Cao Y., Peng D., Wang X., Wan H., Zhao G., Xu Q., Zhang W., Song Q., et al. Novel reassortant highly pathogenic avian influenza (H5N5) viruses in domestic ducks, China. Emerg. Infect. Dis. 2011;17:1060–1063. doi: 10.3201/eid/1706.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee D.H., Bertran K., Kwon J.H., Swayne D.E. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J. Vet. Sci. 2017;18(Suppl. 1):269–280. doi: 10.4142/jvs.2017.18.S1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee D.H., Bahl J., Torchetti M.K., Killian M.L., Ip H.S., DeLiberto T.J., Swayne D.E. Highly Pathogenic Avian Influenza Viruses and Generation of Novel Reassortants, United States, 2014–2015. Emerg. Infect. Dis. 2016;22:1283–1285. doi: 10.3201/eid2207.160048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bi Y., Chen Q., Wang Q., Chen J., Jin T., Wong G., Quan C., Liu J., Wu J., Yin R., et al. Genesis, Evolution and Prevalence of H5N6 Avian Influenza Viruses in China. Cell Host Microbe. 2016;20:810–821. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 12.Li M., Liu H., Bi Y., Sun J., Wong G., Liu D., Li L., Liu J., Chen Q., Wang H., et al. Highly Pathogenic Avian Influenza A(H5N8) Virus in Wild Migratory Birds, Qinghai Lake, China. Emerg. Infect. Dis. 2017;23:637–641. doi: 10.3201/eid2304.161866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee D.H., Sharshov K., Swayne D.E., Kurskaya O., Sobolev I., Kabilov M., Alekseev A., Irza V., Shestopalov A. Novel Reassortant Clade 2.3.4.4 Avian Influenza A(H5N8) Virus in Wild Aquatic Birds, Russia, 2016. Emerg. Infect. Dis. 2017;23:359–360. doi: 10.3201/eid2302.161252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fusaro A., Monne I., Mulatti P., Zecchin B., Bonfanti L., Ormelli S., Milani A., Cecchettin K., Lemey P., Moreno A., et al. Genetic Diversity of Highly Pathogenic Avian Influenza A(H5N8/H5N5) Viruses in Italy, 2016–2017. Emerg. Infect. Dis. 2017;23:1543–1547. doi: 10.3201/eid2309.170539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beerens N., Heutink R., Bergervoet S.A., Harders F., Bossers A., Koch G. Multiple Reassorted Viruses as Cause of Highly Pathogenic Avian Influenza A(H5N8) Virus Epidemic, the Netherlands, 2016. Emerg. Infect. Dis. 2017;23:1974–1981. doi: 10.3201/eid2312.171062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pohlmann A., Starick E., Harder T., Grund C., Hoper D., Globig A., Staubach C., Dietze K., Strebelow G., Ulrich R.G., et al. Outbreaks among Wild Birds and Domestic Poultry Caused by Reassorted Influenza A(H5N8) Clade 2.3.4.4 Viruses, Germany, 2016. Emerg. Infect. Dis. 2017;23:633–636. doi: 10.3201/eid2304.161949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yehia N., Naguib M.M., Li R., Hagag N., El-Husseiny M., Mosaad Z., Nour A., Rabea N., Hasan W.M., Hassan M.K., et al. Multiple introductions of reassorted highly pathogenic avian influenza viruses (H5N8) clade 2.3.4.4b causing outbreaks in wild birds and poultry in Egypt. Infect. Genet. Evol. 2018;58:56–65. doi: 10.1016/j.meegid.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y.I., Park S.J., Kwon H.I., Kim E.H., Si Y.J., Jeong J.H., Lee I.W., Nguyen H.D., Kwon J.J., Choi W.S., et al. Genetic and phylogenetic characterizations of a novel genotype of highly pathogenic avian influenza (HPAI) H5N8 viruses in 2016/2017 in South Korea. Infect. Genet. Evol. 2017;53:56–67. doi: 10.1016/j.meegid.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Baek Y.G., Lee Y.N., Lee D.H., Cheon S.H., Kye S.J., Park Y.R., Si Y.J., Lee M.H., Lee Y.J. A novel reassortant clade 2.3.4.4 highly pathogenic avian influenza H5N6 virus identified in South Korea in 2018. Infect. Genet. Evol. 2020;78:104056. doi: 10.1016/j.meegid.2019.104056. [DOI] [PubMed] [Google Scholar]
- 20.Fusaro A., Zecchin B., Vrancken B., Abolnik C., Ademun R., Alassane A., Arafa A., Awuni J.A., Couacy-Hymann E., Coulibaly M.B., et al. Disentangling the role of Africa in the global spread of H5 highly pathogenic avian influenza. Nat. Commun. 2019;10:5310. doi: 10.1038/s41467-019-13287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatesh D., Brouwer A., Goujgoulova G., Ellis R., Seekings J., Brown I.H., Lewis N.S. Regional Transmission and Reassortment of 2.3.4.4b Highly Pathogenic Avian Influenza (HPAI) Viruses in Bulgarian Poultry 2017/18. Viruses. 2020;12:605. doi: 10.3390/v12060605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pohlmann A., Hoffmann D., Grund C., Koethe S., Hussy D., Meier S.M., King J., Schinkothe J., Ulrich R., Harder T., et al. Genetic Characterization and Zoonotic Potential of Highly Pathogenic Avian Influenza Virus A(H5N6/H5N5), Germany, 2017–2018. Emerg. Infect. Dis. 2019;25:1973–1976. doi: 10.3201/eid2510.181931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon J.H., Jeong S., Lee D.H., Swayne D.E., Kim Y.J., Lee S.H., Noh J.Y., Erdene-Ochir T.O., Jeong J.H., Song C.S. New Reassortant Clade 2.3.4.4b Avian Influenza A(H5N6) Virus in Wild Birds, South Korea, 2017–2018. Emerg. Infect. Dis. 2018;24:1953–1955. doi: 10.3201/eid2410.180461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spackman E., Senne D.A., Bulaga L.L., Myers T.J., Perdue M.L., Garber L.P., Lohman K., Daum L.T., Suarez D.L. Development of real-time RT-PCR for the detection of avian influenza virus. Avian Dis. 2003;47(Suppl. 3):1079–1082. doi: 10.1637/0005-2086-47.s3.1079. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann E., Stech J., Guan Y., Webster R.G., Perez D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 26.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee D.H., Lee H.J., Lee Y.J., Kang H.M., Jeong O.M., Kim M.C., Kwon J.S., Kwon J.H., Kim C.B., Lee J.B., et al. DNA barcoding techniques for avian influenza virus surveillance in migratory bird habitats. J. Wildl. Dis. 2010;46:649–654. doi: 10.7589/0090-3558-46.2.649. [DOI] [PubMed] [Google Scholar]
- 28.Palm E.C., Newman S.H., Prosser D.J., Xiao X., Ze L., Batbayar N., Balachandran S., Takekawa J.Y. Mapping migratory flyways in Asia using dynamic Brownian bridge movement models. Mov. Ecol. 2015;3:3. doi: 10.1186/s40462-015-0029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman S.H., Iverson S.A., Takekawa J.Y., Gilbert M., Prosser D.J., Batbayar N., Natsagdorj T., Douglas D.C. Migration of whooper swans and outbreaks of highly pathogenic avian influenza H5N1 virus in eastern Asia. PLoS ONE. 2009;4:e5729. doi: 10.1371/journal.pone.0005729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lever C. The Mandarin Duck. Poyser; London, UK: 2013. [Google Scholar]
- 31.Lee D.H., Park J.K., Youn H.N., Lee Y.N., Lim T.H., Kim M.S., Lee J.B., Park S.Y., Choi I.S., Song C.S. Surveillance and isolation of HPAI H5N1 from wild Mandarin Ducks (Aix galericulata) J. Wildl. Dis. 2011;47:994–998. doi: 10.7589/0090-3558-47.4.994. [DOI] [PubMed] [Google Scholar]
- 32.Kwon J.H., Lee D.H., Swayne D.E., Noh J.Y., Yuk S.S., Erdene-Ochir T.O., Hong W.T., Jeong J.H., Jeong S., Gwon G.B., et al. Highly Pathogenic Avian Influenza A(H5N8) Viruses Reintroduced into South Korea by Migratory Waterfowl, 2014–2015. Emerg. Infect. Dis. 2016;22:507–510. doi: 10.3201/eid2203.151006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon J.H., Lee D.H., Swayne D.E., Noh J.Y., Yuk S.S., Erdene-Ochir T.O., Hong W.T., Jeong J.H., Jeong S., Gwon G.B., et al. Reassortant Clade 2.3.4.4 Avian Influenza A(H5N6) Virus in a Wild Mandarin Duck, South Korea, 2016. Emerg. Infect. Dis. 2017;23:822–826. doi: 10.3201/eid2305.161905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang H.M., Lee E.K., Song B.M., Heo G.B., Jung J., Jang I., Bae Y.C., Jung S.C., Lee Y.J. Experimental infection of mandarin duck with highly pathogenic avian influenza A (H5N8 and H5N1) viruses. Vet. Microbiol. 2017;198:59–63. doi: 10.1016/j.vetmic.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Kwon J.H., Noh Y.K., Lee D.H., Yuk S.S., Erdene-Ochir T.O., Noh J.Y., Hong W.T., Jeong J.H., Jeong S., Gwon G.B., et al. Experimental infection with highly pathogenic H5N8 avian influenza viruses in the Mandarin duck (Aix galericulata) and domestic pigeon (Columba livia domestica) Vet. Microbiol. 2017;203:95–102. doi: 10.1016/j.vetmic.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Son K., Kim Y.K., Oem J.K., Jheong W.H., Sleeman J.M., Jeong J. Experimental infection of highly pathogenic avian influenza viruses, Clade 2.3.4.4 H5N6 and H5N8, in Mandarin ducks from South Korea. Transbound. Emerg. Dis. 2018;65:899–903. doi: 10.1111/tbed.12790. [DOI] [PubMed] [Google Scholar]
- 37.Kwon J.H., Lee D.H., Swayne D.E., Noh J.Y., Yuk S.S., Jeong S., Lee S.H., Woo C., Shin J.H., Song C.S. Experimental infection of H5N1 and H5N8 highly pathogenic avian influenza viruses in Northern Pintail (Anas acuta) Transbound. Emerg. Dis. 2018;65:1367–1371. doi: 10.1111/tbed.12872. [DOI] [PubMed] [Google Scholar]
- 38.van den Brand J.M.A., Verhagen J.H., Veldhuis Kroeze E.J.B., van de Bildt M.W.G., Bodewes R., Herfst S., Richard M., Lexmond P., Bestebroer T.M., Fouchier R.A.M., et al. Wild ducks excrete highly pathogenic avian influenza virus H5N8 (2014–2015) without clinical or pathological evidence of disease. Emerg. Microbes Infect. 2018;7:67. doi: 10.1038/s41426-018-0070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.