Abstract
Neutrophils are primary host innate immune cells defending against pathogens. One proposed mechanism by which neutrophils prevent the spread of pathogens is NETosis, the extrusion of cellular DNA resulting in neutrophil extracellular traps (NETs). The protease neutrophil elastase (NE) has been implicated in the formation of NETs through proteolysis of nuclear proteins leading to chromatin decondensation. In addition to NE, neutrophils contain three other serine proteases that could compensate if the activity of NE was neutralized. However, whether they do play such a role is unknown. Thus, we deployed recently described specific inhibitors against all four of the neutrophil serine proteases (NSPs). Using specific antibodies to the NSPs along with our labeled inhibitors, we show that catalytic activity of these enzymes is not required for the formation of NETs. Moreover, the NSPs that decorate NETs are in an inactive conformation and thus cannot participate in further catalytic events. These results indicate that NSPs play no role in either NETosis or arming NETs with proteolytic activity.
Keywords: activity-based probes, neutrophil extracellular traps, NETosis, pyroptosis, neutrophil, cell death, protease, serine protease, protease inhibitor
Neutrophils are short-lived cells that act as frontline defenders of the innate immune response. Neutrophils neutralize microbial infections or other endogenous or exogenous stimuli using a combination of responses including phagocytosis, an oxidative burst and release of antimicrobial peptides and proteins (1). The same stimuli can also lead to the extrusion of decondensed chromatin from the cell nucleus, and even mitochondria (2), forming fibrous weblike structures called neutrophil extracellular traps (NETs) that are decorated with histones and antimicrobial agents (3). The process of NET formation (NETosis) has been defined as a type of regulated cell death (4). With the extrusion of DNA from the cell, NETosis stands in marked contrast to two other well-studied types of lytic cell death: pyroptosis and necroptosis (5). Mechanistically, NET release requires an oxidative burst and peptidyl arginine deiminase 4 (PAD4)–mediated histone citrullination (6). The neutrophil serine protease (NSP) elastase (NE) has been implicated in NET formation through translocation to the nucleus, where it may hydrolyze histones, leading to chromatin decondensation (7–9). NE is one of four NSPs stored in an active form in neutrophil azurophil granules (10). Pyroptosis is a lytic form of cell death executed by proinflammatory caspases that results in release of cytokines and other damage-associated molecular patterns. Although pyroptosis is generally described in monocytes and macrophages, it is a cell fate that also awaits neutrophils (11). Pyroptosis results from the limited cleavage of gasdermin D (GSDMD) to release the lytic N-terminal domain (12–14) that is thought to form pores in the plasma membrane, leading to lysis and release of cellular components (15, 16). In monocytic cells inflammatory caspases are the triggers of pyroptosis (17), but in neutrophils the NSPs NE and cathepsin G (CatG) also produce the signature lytic fragment of GSDMD (11, 18). The NSP PR3 has a similar substrate specificity to NE, whereas NSP4 has a distinct specificity for cleaving after arginine (19, 20). Both have been implicated in the modulation of inflammatory mediators, but neither has been implicated in NETosis or pyroptosis (19, 21).
We hypothesized that other NSPs may be involved in NETosis, and to test this hypothesis we employed a recently described set of highly selective inhibitors of each NSP (22) to determine whether they have a role in NET formation.
Results
Selective inhibition of NSPs minimally influences NETosis
NETosis was originally defined as DNA released from neutrophils following treatment with phorbol 12-myristate 13-acetate (PMA) and IL-1β (3, 23), and to a lesser extent with bacteria (Streptococcus aureus) or lipopolysaccharide (LPS) (3). Accordingly, we analyzed the extent of DNA extrusion from neutrophils induced with different stimulants: Candida albicans (24), Escherichia coli strain JM109, PMA, LPS, TNF-α, IL-1β, IFN-γ. We developed a microplate-based assay incorporating cell impermeable SYTOXTM Green as an indicator of released DNA. To eliminate a potential effect of SYTOXTM Green in NET formation, readings were taken at indicated time points in a separate plate and the increase in fluorescence was monitored up to 370 min. We observed that PMA leads to extensive DNA extrusion, whereas C. albicans, E. coli, LPS, TNF-α and IL-1β, and IFN-γ less potently activated NETosis (Fig. 1A), in agreement with previous findings (25).
Figure 1.
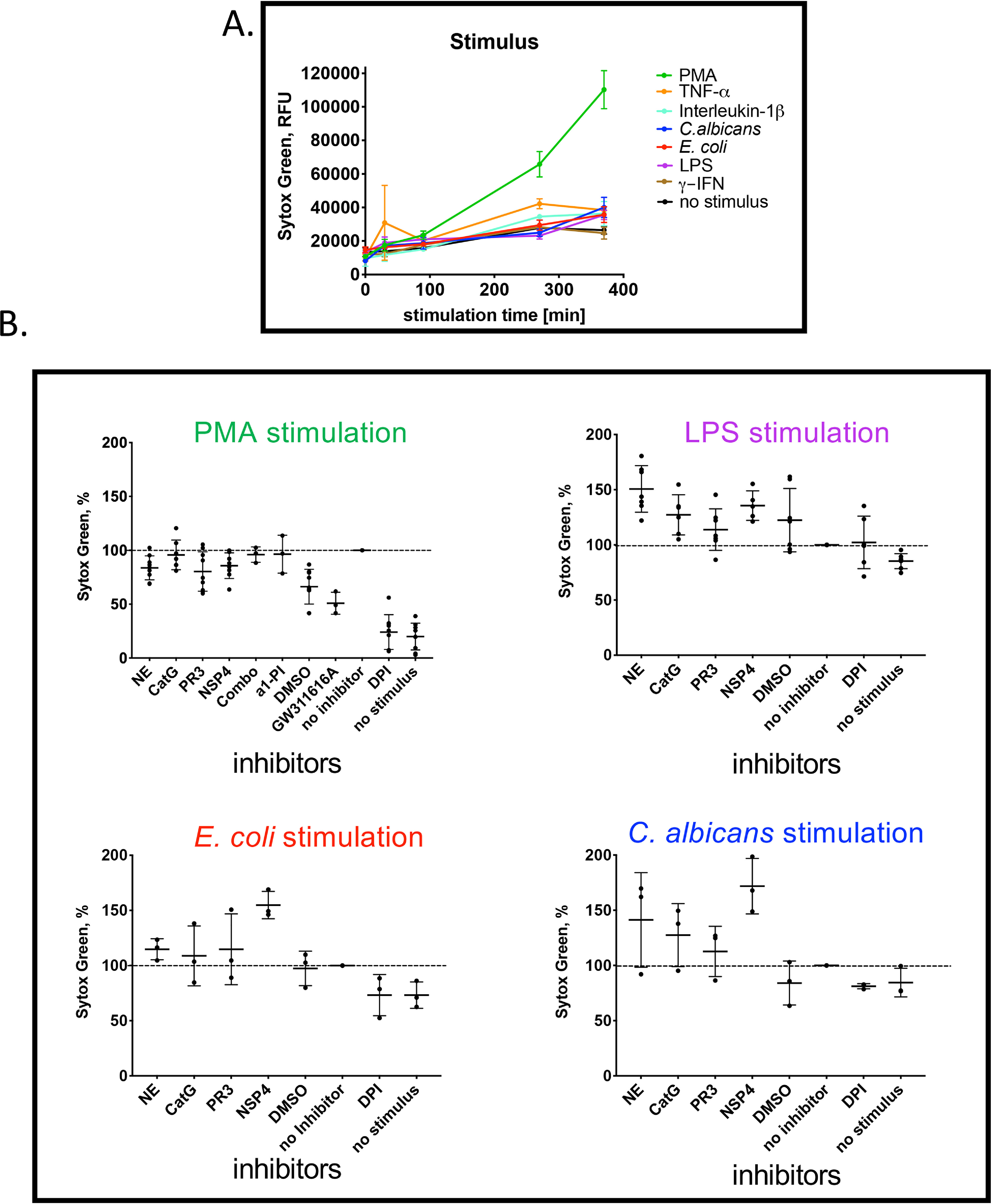
NETosis induction and inhibition. A, kinetics of NET formation in freshly isolated neutrophils induced with the indicated stimuli. At the indicated time points SYTOXTM Green was added to quantitate DNA release by reading the fluorescence at 532 nm. B, influence of NSP inhibitors on NET formation. Neutrophils were induced with the indicated stimuli for 4 h in the presence of 2.5 μm of the inhibitors of the indicated enzymes, individually or in combination (Combo). Released DNA was measured using SYTOXTM Green by reading at 532 nm. Each stimulus, color coded as in panel A, resulted in DNA release. A no-inhibitor control was set at 100% and other samples were normalized to this control. Values are mean ± S.D. of at least three biological replicates.
NE is considered to be a key mediator in the mechanism of NETosis (7, 8, 26), but the role of the three other NSPs had not been addressed previously because of the lack of specific inhibitors. Having recently developed highly specific peptidyl inhibitors of NSPs that gain access to the active forms of these enzymes (22), we asked the question of which NSPs were dominant in driving NETosis. We tested the engagement of individual NSPs using neutrophils isolated from human peripheral blood with our specific NSP inhibitors, which we have previously demonstrated to be cell permeable (22). Each inhibitor is composed of a tetrapeptide recognition element that endows high specificity, an N-terminal biotin or fluorescence tag for detection, and a C-terminal electrophilic warhead (diphenyl α-aminophosphonate, PO3Ph2) that covalently attaches to the catalytic nucleophile; the structure and validation of these probes are described in previous work (22). We also employed DPI as a positive control and the elastase inhibitor GW311616A previously described to attenuate NETosis (7). We previously calculated the concentration of each active NSP (measured in 107 cells/ml) ranged from 230 to 340 nm for NE, 150 to 530 nm for PR3, 75 to 188 nm for CatG, and 17 to 34 nm for NSP4 (20). To ensure complete saturation of each of the NSPs by their specific inhibitors we used a large excess (2.5 μm). Neutrophils were added to a plate containing inhibitors and treated with PMA, LPS, E. coli (J109), or C. albicans for 4 h. As a positive NETosis-blocking control, neutrophils were treated with diphenyleneiodonium (DPI), a NADPH oxidase inhibitor (23). The amount of released DNA was measured after 4 h (Fig. 1B).
NET formation is known to be dependent on NADPH oxidative burst, and accordingly was blocked by DPI. Compound GW311616A, a protease inhibitor previously reported to attenuate NETosis (7), had a small effect inhibiting DNA extrusion by about 50% (Fig. 1B). However, our selective NSP inhibitors had minimal impact on the release of DNA. We used α1-antitrypsin (α1-PI), the endogenous cognate inhibitor of NE (28), as a non–cell permeable control, and likewise we observed no decrease in DNA release (Fig. 1B). To test for redundancy in NSPs we combined all four inhibitors and again failed to see any inhibition of DNA release (Fig. 1B, panel 1). Similarly, NSP inhibitors failed to block DNA extrusion following stimulation of neutrophils with LPS, E. coli (JM109), and C. albicans for 4 h. Thus, in contrast to previous reports (8, 26), we were not able to observe an influence of NSPs in NET formation, at a 4-h time point.
These data allow us to conclude that NSPs play a minimal role, if any, in the stimulus-dependent expulsion of DNA from neutrophils, addressed with highly specific inhibitors of NSPs.
NSPs are in NETs in an inactive conformation
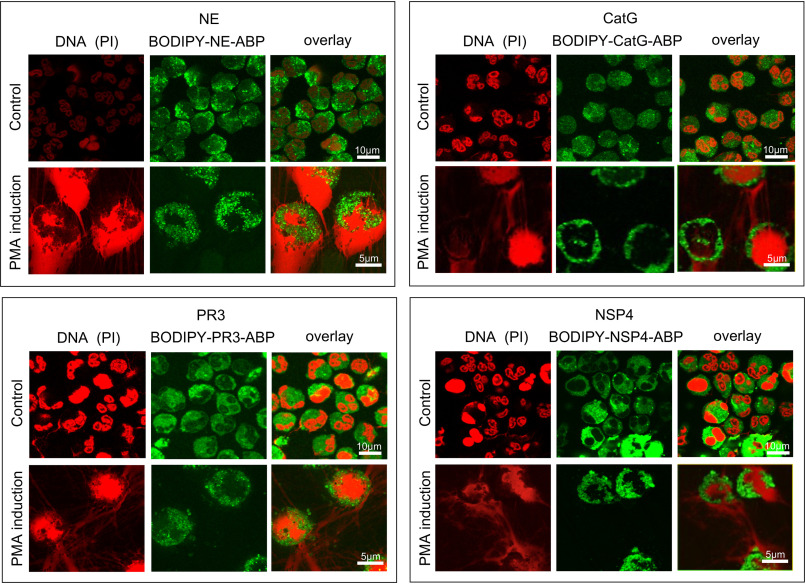
NSPs are known to be present in NET structures (3, 8, 29), but not necessarily in an active form (30). To determine the activity status of NET-associated NSPs following induction of NETosis, we stimulated fresh neutrophils for 3 h with PMA to induce NET formation, followed by 30-min incubation with BODIPY-tagged NSP probes to label the proteolytically active form. After washing, fixing, and DNA staining we imaged the active NSPs and DNA by confocal microscopy. The probes appeared to be completely absent from NETs, and bright punctate fluorescence originating from probes was directed to intracellular granular compartments (Fig. 2). In some neutrophils the nucleus appears somewhat intact but devoid of DNA; the punctate pattern of probe staining is retained.
Figure 2.
Active NSPs remain intracellular and do not decorate NETs. Fresh neutrophils (1 × 106 cells/ml) were stimulated for 3 h with PMA to induce NETosis and afterward treated with 100 nm of the indicated NSP-selective BODIPY-labeled probes (green) for 30 min followed by fixation and addition of propidium iodide (red) to visualize DNA. Neutrophils were mounted and imaged by confocal microscopy. See also Movies S3a–S3c for Z-stack views of the data.
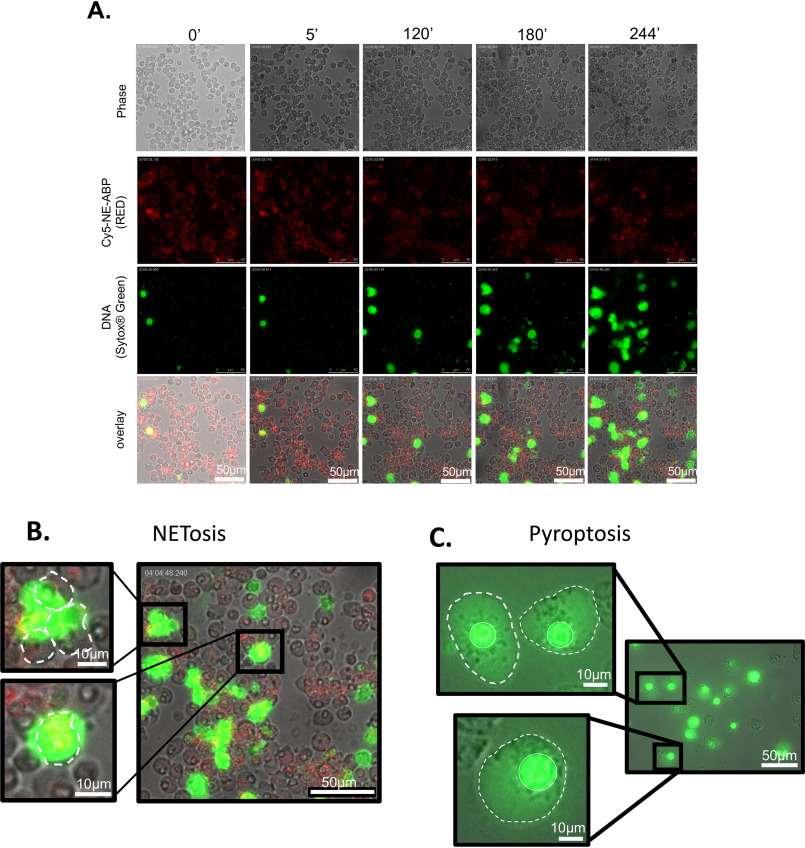
We considered that we may have missed transient association of active NSPs with NETs, and to this end we observed changes in enzyme localization over time by live cell imaging. Freshly isolated human neutrophils were simultaneously treated with PMA, SYTOXTM Green, and a Cy5-labeled NE probe, and the morphology changes and DNA release were monitored for 4 h. Images were captured at 5-min intervals (Movie S1) and representative frames are shown in Fig. 3A. The probe is internalized immediately (fluorescent signal observed at time 0). We observed NET formation beginning at 120 min. However, at no time did we observe the NE probe associated with NETs. We conclude that active NE is not associated with NETs, even transiently. As in the previous 3-h time point (Fig. 2), active NE remained within the cells.
Figure 3.
NETosis versus pyroptosis. A, time course of NET formation in PMA-treated neutrophils co-incubated with a Cy5-labeled NE probe (red) and SYTOX TM Green to mark DNA. B and C, nuclear DNA is extruded in NETosis (B) and contained within the nucleus during pyroptosis induced by treatment of macrophages by LPS and ATP (C). In the images, the nucleus is marked by a continuous white line and the cell membrane with a dashed line. Panel B intentionally duplicates the 244 min time point image from panel A to provide an enlarged display of the field captured in the image.
Importantly, during NETosis, DNA is extruded from the nucleus outside the cell and the amount of nuclear DNA within the nucleus dramatically decreased (Fig. 2). Another form of cell death common to phagocytic cells is pyroptosis (31). In contrast to NETosis, pyroptosis results in cell lysis where the nucleus is retained in the cell (Fig. 3, B and C). NE (18) and CatG (11) are both implicated in pyroptotic death of neutrophils. Consequently, we considered that it would be possible to confuse pyroptosis with NETosis. A key distinction between these two forms of cell death is whether DNA is retained within the lysing cell (pyroptosis) or expelled from the lysing cell (NETosis) (Fig. 3, B and C and Movie S2).
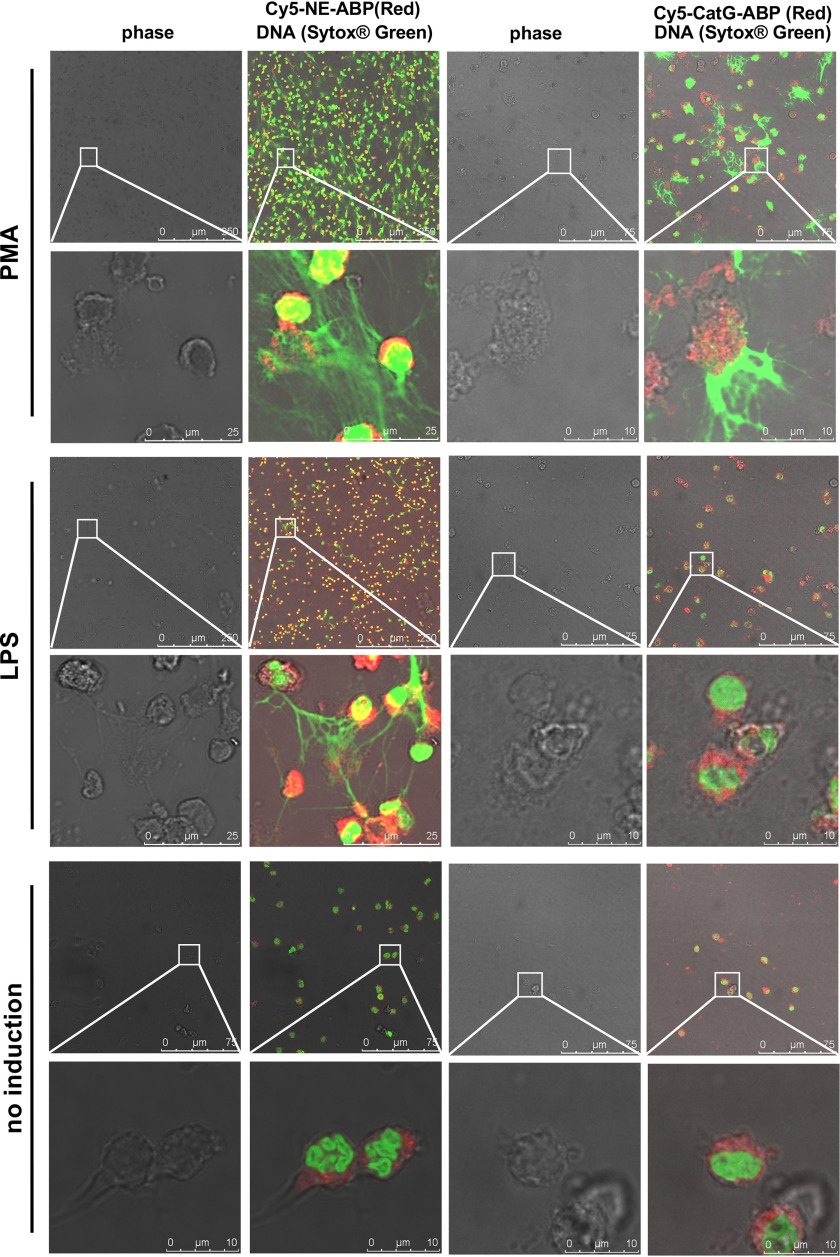
It is noteworthy that the location of NSPs following NETosis was stimulus-independent because both PMA and LPS treatment revealed cell-associated active NE and CatG, but no activity in NETs, as defined by probe binding (Fig. 4 and Movies S3a–S3c).
Figure 4.
Localization of active NSPs in neutrophils after induction with different stimuli. Neutrophils were treated as indicated, allowed to settle on coverslips, and incubated with Cy5-labeled specific probes (red) as indicated. Slides were fixed and DNA stained with SYTOXTM Green. Images are representative of three separate donors.
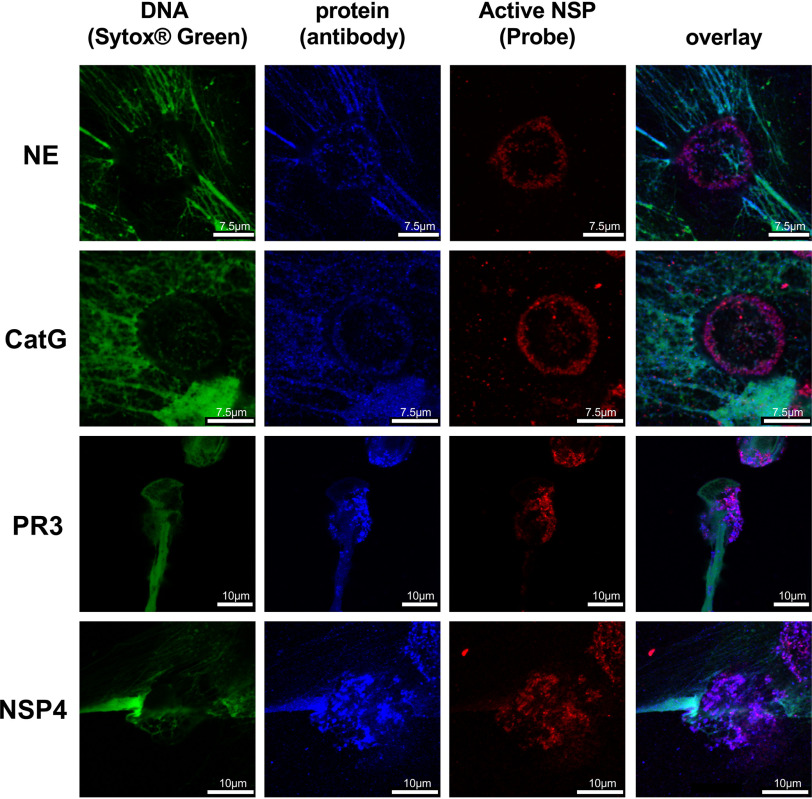
When PMA-stimulated neutrophils were imaged with specific antibodies, we observed co-staining of antibodies and the active NSPs, but also association of the antibodies with NETs (Fig. 5). Our data are consistent with previous studies in which antibodies were applied to visualize NSPs on NETs (3, 23), but we now demonstrate that, at least in vitro, NET-associated NSPs are captured in an inactive form.
Figure 5.
Specific antibodies reveal that NSPs are bound to NETs, but in an inactive form. Images represent neutrophils induced with PMA and treated with the Cy5-labeled specific NSP probes (red), NSP-specific antibodies imaged with Alexa FluorTM 568 coupled secondaries (blue) followed by fixation and addition of SYTOXTM Green to stain DNA (green). In all cases probe staining is punctate and not associated with DNA, extracellular or intracellular. However, each NSP antigen substantially decorates DNA, revealing that NET (DNA)-associated NSPs are inactive.
Discussion
NETs have been proposed to trap, immobilize, and kill bacteria and fungi (3). The mechanism of NETosis involves reactive oxygen species formation through NADPH oxidase activation, nuclear envelope rupture, mixing of nuclear and cytosolic components, and release of nuclear DNA (3, 23). Based on the use of a small molecule inhibitor, NE has been implicated in NETosis via hydrolysis of histone H4 leading to chromosome decondensation (8). However, NE released from neutrophils undergoing NETosis is bound in an inactive form in NETs (30), raising the question of whether additional evidence would support a role for NE in NETosis. To address this question we utilized highly efficient, selective inhibitors of NSPs in paradigms of NETosis induction. None of the inhibitors were able to block NETosis induced by any stimuli (LPS, PMA, E. coli, C. albicans), despite evidence that the inhibitors are able to penetrate living neutrophils. This conclusion is consistent with genetic evidence that NE-deficient mice can form NETs, but at odds with a previous study (8) implicating NE in the generation of NETs. The conclusions of the forgoing study were based largely on the attenuation of NETosis by GW311616A, which we could replicate (7). However, we do not think GW311616A is targeting NE because our highly selective cell-permeable NSP inhibitors failed to inhibit NETosis. Accordingly, we propose that GW311616A might have nonNE off target activity.
In terms of neutrophil lytic events, genetic evidence implicates both NE and CatG in neutrophil pyroptosis (11, 18), but we show, by using highly selective inhibitors, that neither are involved in NETosis. It is possible to conflate NETosis with pyroptosis (Fig. 3), which may explain the discrepancy between our conclusion and that of the previous study (8).
NETosis is characterized by nuclear delobulization and subsequent nuclear swelling, which is in contrast to pyroptosis and necroptosis, where minimal nuclear expansion is observed even though cellular swelling is evident. Necroptosis and pyroptosis counteract microbial invasion by removal of the replicative niche of pathogens through cell lysis. However, the lytic nature of these death mechanisms also means that intracellular damage-associated molecular patterns are released and can act in an immunostimulatory manner (32) leading to acute organ failure and septic shock (33). If left unchecked, both pyroptosis and necroptosis can lead to autoimmune and autoinflammatory disorders. NETs on the other hand are speculated to trap pathogens on the extruded DNA fibers. However, they are also regarded as a source of self-antigens and are increasingly implicated in cancer progression (34, 35).
Previous studies have demonstrated that NSPs are released from neutrophils and decorate NETs (3, 23, 29). Our studies confirm this with the important caveat that the enzymes are displayed in inactive forms. Because DNA is negatively charged and NSPs are basic, pI of over 9 for NE (36), over 12 for CatG (37), and predicted pI of 7.79 for PR3 and 9.15 for NSP4 (calculated for active enzyme structures, ExPASy Database), we predict that ionic interactions with DNA in NETs could account for enzyme inhibition. This is supported by evidence showing that DNA can inhibit the activity of CatG (38) and NE (39) in vitro. Alternatively, we cannot rule out the presence of endogenous inhibitors that may prevent probe binding. Whatever the mechanism that restrains activity of NSPs when associated with NETs will be important to consider in studies where NSPs are implicated in specific biological events, for example in the processing of proinflammatory cytokines (40).
Experimental procedures
Neutrophils were isolated as described elsewhere (22) from fresh blood from healthy donors obtained from Scripps Clinic, La Jolla, CA. Procedures were reviewed and approved by both the Sanford Burnham Prebys Medical Discovery Institute and The Scripps Research Institute internal review boards (IRB-16-6789) and abided by the Declaration of Helsinki principles. All researchers working with isolated neutrophils completed a required blood-borne pathogens plan which was reviewed by the Sanford Burnham Prebys Medical Discovery Institute's internal review board.
Induction of NETosis with different stimuli
A volume of 80 μl of 1 × 106 neutrophils/ml in RPMI/FBS were added to wells of a 96-well black plate (Corning®), followed by stimulation with PMA (100 nm), LPS (5 mg/ml), fMLP (100 nm), C. albicans (m.o.i. 100), E. coli (m.o.i. 100), TNF-α (10 nm), IL-1β (10 nm), or INF-γ (50 nm) for up to 370 min. SYTOXTM Green was added to a concentration of 6.26 μm followed by measurement of fluorescence at 532 nm, using a ClarioStar® (BMG Labtech). All incubations were at 37°C in 5% CO2 in RPMI1640 (without phenol red) supplemented with 10% dye-free serum.
NETosis inhibition assay
NSP inhibitors and other compounds, at the concentrations indicated in the figures, were added to neutrophils (1 × 106 cells/ml) followed by stimulation (100 nm PMA, 5 mg/ml LPS, C. albicans (m.o.i. 100), or E.coli J109 (m.o.i. 100)) and DNA fluorescence measurements with SYTOXTM Green (532 nm, ClarioStar®, BMG Labtech) after 4 h. Data were collected from 3–10 donors and normalized for each donor separately.
Microscopy
Fresh neutrophils in RPMI/FBS were added to sterile coverslips coated with poly-d-lysine, stimulated with PMA (100 nm) or LPS (5 mg/ml) for the indicated time, followed by the indicated NSP probe at 100 nm final concentration for 30 min. Supernatant was removed by aspiration and the coverslips were washed with 0.5% Triton X-100 in Dulbecco's PBS (DPBS) and fixed for 40 min with 4% paraformaldehyde (Sigma Aldrich). Coverslips were blocked with 10% BSA in Hanks' Balanced Salt Solution for 60 min and washed with DPBS. Where indicated, coverslips were treated with primary antibodies (1:200 dilution) in a solution of 3% BSA, 0.5% Triton X-100 in DPBS (rabbit anti-PR3 (Abcam, ab21592), rabbit anti-CatG (Abcam, ab131407), rabbit anti-NE (Abcam, ab21595), and rabbit anti-NSP4 (Abcam, ab156095) overnight at 4°C. Secondary antibody (AF568-conjugated anti-rabbit 1:1000 dilution in 3% BSA, 0.5% Triton X-100 in DPBS,) was added and incubated for 1 h, followed by washing 3 times in a solution of 3% BSA, 0.5% Triton X-100 in DPBS. Propidium iodide or SYTOXTM Green were added for 5 min and washed three times with DPBS. Coverslips were mounted in Fluoromount G and dried for 2 h. Neutrophils were imaged at 63× using Leica confocal microscope with 488 nm (for BODIPY FL, SYTOXTM Green), 552 nm (for Alexa FluorTM 568) and 638 nm (for Cy5) lasers.
Time course of NET formation in Cy5-NE-ABP–labeled neutrophils
A volume of 200 μl of 5 × 106 neutrophils/ml in 2% FBS in RPMI medium were placed in the well and incubated for 30 min in 5% CO2 at 37°C, followed by 50 nm PMA stimulation and simultaneously 200 nm probe and 25 μm SYTOXTM Green addition. Then the neutrophils were analyzed up to 244 min at 63× using Leica confocal microscope with 488 nm and 638 nm lasers (accordingly for SYTOXTM Green and Cy5). Data were analyzed in a LasX software.
Pyroptosis
The RAW 264.7 murine macrophage cell line was obtained from ATCC. Additional copies of the pyroptosis modulator ASC were introduced by lentiviral transfection and selected against puromycin resistance. Single cell clones were grown and tested for amount of ASC expression (27). For live cell microscopy, cells were seeded at the concentration of 2 × 105 cells/ml the previous night. Macrophages were primed with 100 ng/ml of LPS (Sigma Aldrich) for 4 h in DMEM containing 10% FBS. Cells were then washed with PBS and 500 nm SYTOXTM Green (Thermo Fisher) in OptiMEM (Gibco) was added. Pyroptosis was induced by addition of 5 mm ATP (Sigma Aldrich). Cells were imaged on an Olympus IX71 and images collected with PictureFrame software. Movies were generated with Fiji.
Data availability
All raw data are available at request by contacting Dr. Paulina Kasperkiewicz (paulina.kasperkiewicz@pwr.edu.pl) from Wroclaw University of Science and Technology. All remaining data are contained within the main text or in the supporting information.
Supplementary Material
Acknowledgments
We thank Daniel Kirchhofer, Ph.D., for the kind gift of the NSP4 protease.
This article contains supporting information.
Author contributions—P. K., A. H., and G. S. S. conceptualization, P. K., A. H., S. J. S., and G. S. S. formal analysis; P. K., A. H., T. J., S. K., S. J. S., and G. S. S. investigation; P. K., A. H., and S. J. S. visualization; P. K. and A. H. methodology; P. K., A. H., and G. S. S. writing-original draft; A. H., T. J., S. K., S. J. S., and G. S. S. data curation; T. J., S. K., S. J. S., and G. S. S. writing-review and editing; M. D. and G. S. S. resources; M. D. and G. S. S. supervision; M. D. and G. S. S. funding acquisition; G. S. S. validation; G. S. S. project administration.
Funding and additional information—This work was supported by National Institutes of Health Grants R01 GM99040 and P30 CA30199 (to G. S. S.) and by National Science Centre Grant 2014/14/M/ST5/00619 in Poland (to M. D.). This work was also supported by a START scholarship from the Foundation for Polish Science (to P. K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- NET
- neutrophil extracellular trap
- NE
- neutrophil elastase
- NSP
- neutrophil serine protease
- GSDMD
- gasdermin D
- CatG
- cathepsin G
- PMA
- phorbol 12-myristate 13-acetate
- LPS
- lipopolysaccharide
- DPI
- diphenyleneiodonium
- m.o.i.
- multiplicity of infection.
References
- 1. Nauseef W. M., and Borregaard N. (2014) Neutrophils at work. Nat. Immunol. 15, 602–611 10.1038/ni.2921 [DOI] [PubMed] [Google Scholar]
- 2. Yousefi S., Mihalache C., Kozlowski E., Schmid I., and Simon H. U. (2009) Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ. 16, 1438–1444 10.1038/cdd.2009.96 [DOI] [PubMed] [Google Scholar]
- 3. Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y., and Zychlinsky A. (2004) Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- 4. Urban C. F., Reichard U., Brinkmann V., and Zychlinsky A. (2006) Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol. 8, 668–676 10.1111/j.1462-5822.2005.00659.x [DOI] [PubMed] [Google Scholar]
- 5. Salvesen G. S., Hempel A., and Coll N. S. (2016) Protease signaling in animal and plant-regulated cell death. FEBS J. 283, 2577–2598 10.1111/febs.13616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leshner M., Wang S., Lewis C., Zheng H., Chen X. A., Santy L., and Wang Y. (2012) PAD4 mediated histone hypercitrullination induces heterochromatin decondensation and chromatin unfolding to form neutrophil extracellular trap-like structures. Front. Immunol. 3, 307 10.3389/fimmu.2012.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Metzler K. D., Goosmann C., Lubojemska A., Zychlinsky A., and Papayannopoulos V. (2014) A myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Rep. 8, 883–896 10.1016/j.celrep.2014.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Papayannopoulos V., Metzler K. D., Hakkim A., and Zychlinsky A. (2010) Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 191, 677–691 10.1083/jcb.201006052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farley K., Stolley J. M., Zhao P., Cooley J., and Remold-O'Donnell E. (2012) A serpinB1 regulatory mechanism is essential for restricting neutrophil extracellular trap generation. J. Immunol. 189, 4574–4581 10.4049/jimmunol.1201167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dewald B., Rindler-Ludwig R., Bretz U., and Baggiolini M. (1975) Subcellular localization and heterogeneity of neutral proteases in neutrophilic polymorphonuclear leukocytes. J. Exp. Med. 141, 709–723 10.1084/jem.141.4.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burgener S. S., Leborgne N. G. F., Snipas S. J., Salvesen G. S., Bird P. I., and Benarafa C. (2019) Cathepsin G inhibition by serpinb1 and serpinb6 prevents programmed necrosis in neutrophils and monocytes and reduces GSDMD-driven inflammation. Cell Rep. 27, 3646–3656.e3645 10.1016/j.celrep.2019.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F. (2015) Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- 13. Kayagaki N., Stowe I. B., Lee B. L., O'Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q. T., Liu P. S., Lill J. R., Li H., Wu J., Kummerfeld S., et al. (2015) Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- 14. He W. T., Wan H., Hu L., Chen P., Wang X., Huang Z., Yang Z. H., Zhong C. Q., and Han J. (2015) Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 25, 1285–1298 10.1038/cr.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aglietti R. A., Estevez A., Gupta A., Ramirez M. G., Liu P. S., Kayagaki N., Ciferri C., Dixit V. M., and Dueber E. C. (2016) GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. U. S. A. 113, 7858–7863 10.1073/pnas.1607769113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ding J., Wang K., Liu W., She Y., Sun Q., Shi J., Sun H., Wang D. C., and Shao F. (2016) Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 10.1038/nature18590 [DOI] [PubMed] [Google Scholar]
- 17. Miao E. A., Leaf I. A., Treuting P. M., Mao D. P., Dors M., Sarkar A., Warren S. E., Wewers M. D., and Aderem A. (2010) Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11, 1136–1142 10.1038/ni.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kambara H., Liu F., Zhang X., Liu P., Bajrami B., Teng Y., Zhao L., Zhou S., Yu H., Zhou W., Silberstein L. E., Cheng T., Han M., Xu Y., and Luo H. R. (2018) Gasdermin D exerts anti-inflammatory effects by promoting neutrophil death. Cell Rep. 22, 2924–2936 10.1016/j.celrep.2018.02.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perera N. C., Schilling O., Kittel H., Back W., Kremmer E., and Jenne D. E. (2012) NSP4, an elastase-related protease in human neutrophils with arginine specificity. Proc. Natl. Acad. Sci. U. S. A. 109, 6229–6234 10.1073/pnas.1200470109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Korkmaz B., Moreau T., and Gauthier F. (2008) Neutrophil elastase, proteinase 3 and cathepsin G: Physicochemical properties, activity and physiopathological functions. Biochimie (Paris) 90, 227–242 10.1016/j.biochi.2007.10.009 [DOI] [PubMed] [Google Scholar]
- 21. Wiedow O., and Meyer-Hoffert U. (2005) Neutrophil serine proteases: Potential key regulators of cell signalling during inflammation. J. Intern. Med. 257, 319–328 10.1111/j.1365-2796.2005.01476.x [DOI] [PubMed] [Google Scholar]
- 22. Kasperkiewicz P., Altman Y., D'Angelo M., Salvesen G. S., and Drag M. (2017) Toolbox of fluorescent probes for parallel imaging reveals uneven location of serine proteases in neutrophils. J. Am. Chem. Soc. 139, 10115–10125 10.1021/jacs.7b04394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fuchs T. A., Abed U., Goosmann C., Hurwitz R., Schulze I., Wahn V., Weinrauch Y., Brinkmann V., and Zychlinsky A. (2007) Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 176, 231–241 10.1083/jcb.200606027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Urban C. F., Ermert D., Schmid M., Abu-Abed U., Goosmann C., Nacken W., Brinkmann V., Jungblut P. R., and Zychlinsky A. (2009) Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 5, e1000639 10.1371/journal.ppat.1000639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoppenbrouwers T., Autar A. S. A., Sultan A. R., Abraham T. E., van Cappellen W. A., Houtsmuller A. B., van Wamel W. J. B., van Beusekom H. M. M., van Neck J. W., and de Maat M. P. M. (2017) In vitro induction of NETosis: Comprehensive live imaging comparison and systematic review. PLoS One 12, e0176472 10.1371/journal.pone.0176472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zabieglo K., Majewski P., Majchrzak-Gorecka M., Wlodarczyk A., Grygier B., Zegar A., Kapinska-Mrowiecka M., Naskalska A., Pyrc K., Dubin A., Wahl S. M., and Cichy J. (2015) The inhibitory effect of secretory leukocyte protease inhibitor (SLPI) on formation of neutrophil extracellular traps. J. Leukoc. Biol. 98, 99–106 10.1189/jlb.4AB1114-543R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Proell M., Gerlic M., Mace P. D., Reed J. C., and Riedl S. J. (2013) The CARD plays a critical role in ASC foci formation and inflammasome signalling. Biochem. J. 449, 613–621 10.1042/BJ20121198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Travis J., and Salvesen G. S. (1983) Human plasma proteinase inhibitors. Ann. Rev. Biochem. 52, 655–709 10.1146/annurev.bi.52.070183.003255 [DOI] [PubMed] [Google Scholar]
- 29. O'Donoghue A. J., Jin Y., Knudsen G. M., Perera N. C., Jenne D. E., Murphy J. E., Craik C. S., and Hermiston T. W. (2013) Global substrate profiling of proteases in human neutrophil extracellular traps reveals consensus motif predominantly contributed by elastase. PLoS One 8, e75141 10.1371/journal.pone.0075141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kasperkiewicz P., Poreba M., Snipas S. J., Parker H., Winterbourn C. C., Salvesen G. S., and Drag M. (2014) Design of ultrasensitive probes for human neutrophil elastase through hybrid combinatorial substrate library profiling. Proc. Natl. Acad. Sci. U. S. A. 111, 2518–2523 10.1073/pnas.1318548111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vande Walle L., and Lamkanfi M. (2016) Pyroptosis. Curr. Biol. 26, R568–R572 10.1016/j.cub.2016.02.019 [DOI] [PubMed] [Google Scholar]
- 32. Frank D., and Vince J. E. (2019) Pyroptosis versus necroptosis: Similarities, differences, and crosstalk. Cell Death Differ. 26, 99–114 10.1038/s41418-018-0212-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aglietti R. A., and Dueber E. C. (2017) Recent insights into the molecular mechanisms underlying pyroptosis and Gasdermin family functions. Trends Immunol. 38, 261–271 10.1016/j.it.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 34. Albrengues J., Shields M. A., Ng D., Park C. G., Ambrico A., Poindexter M. E., Upadhyay P., Uyeminami D. L., Pommier A., Küttner V., Bružas E., Maiorino L., Bautista C., Carmona E. M., Gimotty P. A., et al. (2018) Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science 361, eaao4227 10.1126/science.aao4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cools-Lartigue J., Spicer J., McDonald B., Gowing S., Chow S., Giannias B., Bourdeau F., Kubes P., and Ferri L. (2013) Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J. Clin. Invest. 123, 3446–3458 10.1172/JCI67484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Korkmaz B., and Gauthier F. (2013) Elastase-2/leukocyte elastase. in Handbook of Proteolytic Enzymes (Rawlings N. D., and Salvesen G., eds.), pp. 2653–2661, Academic Press, Cambridge, MA [Google Scholar]
- 37. Salvesen G. S. (2013) Cathepsin G. in Handbook of Proteolytic Enzymes (Rawlings N. D., and Salvesen G., eds.), pp. 2661–2666, Academic Press, Cambridge, MA [Google Scholar]
- 38. Duranton J., Boudier C., Belorgey D., Mellet P., and Bieth J. G. (2000) DNA strongly impairs the inhibition of cathepsin G by α1-antichymotrypsin and α1-proteinase inhibitor. J. Biol. Chem. 275, 3787–3792 10.1074/jbc.275.6.3787 [DOI] [PubMed] [Google Scholar]
- 39. Belorgey D., and Bieth J. G. (1995) DNA binds neutrophil elastase and mucus proteinase inhibitor and impairs their functional activity. FEBS Lett. 361, 265–268 10.1016/0014-5793(95)00173-7 [DOI] [PubMed] [Google Scholar]
- 40. Clancy D. M., Henry C. M., Sullivan G. P., and Martin S. J. (2017) Neutrophil extracellular traps can serve as platforms for processing and activation of IL-1 family cytokines. FEBS J. 284, 1712–1725 10.1111/febs.14075 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data are available at request by contacting Dr. Paulina Kasperkiewicz (paulina.kasperkiewicz@pwr.edu.pl) from Wroclaw University of Science and Technology. All remaining data are contained within the main text or in the supporting information.