Abstract
The evolutionary epidemiology, resistome, virulome and mobilome of thirty-one multidrug resistant Klebsiella pneumoniae clinical isolates from the northern Vila Real region of Portugal were characterized using whole-genome sequencing and bioinformatic analysis. The genomic population structure was dominated by two main sequence types (STs): ST147 (n = 17; 54.8%) and ST15 (n = 6; 19.4%) comprising four distinct genomic clusters. Two main carbapenemase coding genes were detected (blaKPC-3 and blaOXA-48) along with additional extended-spectrum β-lactamase coding loci (blaCTX-M-15, blaSHV-12, blaSHV-27, and blaSHV-187). Moreover, whole genome sequencing enabled the identification of one Klebsiella variicola KPC-3 producer isolate previously misidentified as K. pneumoniae, which in addition to the blaKPC-3 carbapenemase gene, bore the chromosomal broad spectrum β-lactamase blaLEN-2 coding gene, oqxAB and fosA resistance loci. The blaKPC-3 genes were located in a Tn4401b transposon (K. variicola n = 1; K. pneumoniae n = 2) and Tn4401d isoform (K. pneumoniae n = 28). Overall, our work describes the first report of a blaKPC-3 producing K. variicola, as well as the detection of this species during infection control measures in surveillance cultures from infected patients. It also highlights the importance of additional control measures to overcome the clonal dissemination of carbapenemase producing clones.
Keywords: Klebsiella pneumoniae, Klebsiella variicola, KPC-3, OXA-48, Gram-negative, molecular epidemiology, carbapenemase, whole-genome sequencing, Enterobacteriaceae, Portugal
1. Introduction
Klebsiella pneumoniae (Kp) has emerged as an increasingly resistant pathogen and has shown an ability to express several intrinsic and acquired mechanisms making the species multi-resistant to clinical antimicrobial classes. Using sequencing and phylogenetic analysis, isolates in the Kp complex have been classified into seven phylo-groups (Kp1 to Kp7) [1,2]. The phenotypic and biochemical characteristics of the Kp complex are extremely similar under standard microbiological conditions, which makes it difficult to distinguish phylo-groups by biochemical tests and routine microbiological methods. The inaccurate identification of Kp complex phylo-groups is leaving gaps in knowledge and has clinical implications within health care systems. K. variicola (Kv) isolates are included in phylogenetic group Kp3 and are becoming increasingly significant from a clinical standpoint and associated with increased mortality [3,4]. Of great concern is that Kp and Kv can co-exist in the same host. Garza-Ramos et al. identified two isolates from an infected Mexican patient: one susceptible Kv isolate and a second carbapenemase-producing Kp isolate [5]. Usually, Kv isolates display low antibiotic resistance rates; however, this pattern has changed over time due to an increase in reports of multidrug-resistant isolates, including extended-spectrum β-lactamase (ESBL) (blaCTX-M-15) or carbapenemase (blaKPC-2, blaNDM-5, blaIMI-2) producing isolates [6,7]. Moreover, reports of multidrug-resistant and hypervirulent Kv isolates have been increasing from other different sources, such as plants and environmental isolates, which highlights a key role of the environment as reservoirs of antimicrobial resistance genes and hypervirulent isolates [3,8].
Recently, it has been reported that Kv display a large spectrum of virulence factors, similar to Kp, encoded by genes located in chromosomal gene clusters or in genetic accessory elements. Additionally, it has been shown using genomic and phylogenetic analyses that these gene clusters and elements confer infective potential for community-acquired isolates [9,10,11]. The mechanisms underpinning the virulence of pathogenic bacteria as well as those determining antibiotic resistance are important and widely studied topics in clinical microbiology. However, due to the lack of a suitable identification approach for routine microbiological use and the potential impact of species of the Klebsiella complex on clinical outcomes and settings, the aim of this study was to analyze the whole-genome sequences of Kp isolates from a hospital in the northern region of Portugal to determine their relatedness and to detect possible high-risk clones, antimicrobial resistance markers and putative virulence factors.
2. Materials and Methods
2.1. Bacterial Isolates
Thirty-one isolates of Kp-producing carbapenemases, from 28 patients identified by Centro Hospitalar de Trás os Montes e Alto Douro (CHTMAD) in Vila Real, Portugal, were characterized using whole-genome sequencing and bioinformatic analysis. The biological samples collected were inoculated on specific culture medium CHROMagar mSuperCARBA media (bioMérieux, La Balme-les-Grotte, France) for detection of carbapenemases for rectal and nasal swabs, and additional Columbia agar with 5% sheep blood plates for clinical specimens (bioMérieux, La Balme-les-Grotte, France). Identification and antibiotic susceptibility testing were performed using the VITEK Microbial Detection System (bioMérieux, La Balme-les-Grotte, France). The European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines were used for interpretation of antimicrobial susceptibility testing (v.8) [12]. Phenotypic KPC carbapenemase detection was performed with the double-disc synergy test with boronic acid inhibition [13].
2.2. Whole-Genome Sequencing
DNA for whole genome sequencing were extracted from cultures grown overnight in Mueller–Hinton agar, using the NZY Tissue gDNA Isolation kit (NZYTech, Lisboa, Portugal), as per the manufacturer’s recommendations. Sequencing libraries were prepared using Covaris and the New England Biolabs (NEB) NEBNext Ultra DNA Library Prep Kit (NEB, Ipswich, MA, USA E7370) following the manufacturer’s recommended protocol and sequenced using an Illumina HiSeq 4K platform with paired-end 2 × 151 bp reads. De novo assembly was performed using the Unicycler assembly pipeline (https://github.com/rrwick/Unicycler) with SPAdes (v.3.8) software [14]. All assemblies were carried out with a k-mer length of 127 bp and contig N50 ranged between 147.5 and 504.6 Kbp, contig count between 54 and 297 Kbp, and the longest contig between 156.4 and 1046.0 Kbp. Genomic sequences were deposited on the European Nucleotide Archive under Study PRJEB41591.
2.3. Phylogenetic Analysis
Phylogenetic reconstruction from high-quality SNPs obtained by mapping raw sequencing reads to the reference genome of Kp IA65 (GenBank Accession NZ_CP030070.1). The reference genome was selected using refRank (https://gitlab.com/s.fuchs/refRank) from a total of 289 Kp genomes available at GenBank with an assembly level of “complete”. Trimmomatic (v0.36) software was used for read trimming and quality control, where reads <36 bp length and with an average PHRED score below 20 in 4 bp sliding windows were excluded. Surviving reads were mapped to the reference genome using the Burrows–Wheeler Aligner tool (BWA-MEM algorihm) [15,16]. BAM deduplication and read realignment around indels were done using Picard Tools and GATK v.3.6 [17]. Variant calling was performed using both SAMtools/BCFtools and GATK (UnifiedGenotyper) software and only concordant variants were retained for further downstream analysis. The following filtering steps were performed to obtain a set of high-quality SNPs: (i) coverage-validation, a missing call assigned if coverage depth did not reach a minimum of 20 reads or if none of the nucleotides accounted for 75% of the total coverage; (ii) only reference genome positions yielding 49/50 bp unique k-mers were retained in order to remove SNP positions associated with low mappability; (iii) sites displaying an excess of 10% missed calls were removed; (iv) SNP sites within 10 bp were removed (pruning) [18]. A total of 64,106 high quality SNPs were identified across the 31 isolates included in the study and concatenated into pseudo DNA molecules for phylogenetic inference. The Generalized Time Reversible model allowing for a proportion of invariable sites was selected upon fitting different nucleotide substitution models to this dataset (R/Phangorn package;—log likelihood: 426,121.7703; AIC weight: 0.49881). A maximum-likelihood phylogenetic tree was constructed using PhyML as implemented in Seaview and tree topology statistically assessed using the approximate Likelihood Ratio Test (aLRT) [19,20]. The Interactive Tree of Life tool (iTOL, https://itol.embl.de/) [21] was used to annotate the tree and a minimum spanning tree was also constructed using Phyloviz (http://online2.phyloviz.net/users/login) and the goeBURST algorithm therein implemented using the pairwise comparison analysis method [22]. Genomic clusters were defined as a group of two or more isolates within 23 SNPs [23].
2.4. In Silico MLST, Plasmid Replicons, Drug Resistance Associated Genes and Capsular Types
In silico Multilocus Sequence Typing MLST was carried out using MLST 1.8 (https://cge.cbs.dtu.dk/). Abricate software was used for plasmid replicon detection using the PlasmidFinder database (https://cge.cbs.dtu.dk/), with the following cut-off values: minimum of 60% coverage and 95% identity (80% for Col-like plasmids) [24]. Antimicrobial resistance genes were screened using the AMRFinder software (https://ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/AMRFinder/) along with the NCBI Bacterial Antimicrobial Resistance Reference Gene Database (Accession PRJNA313047) using a 60% coverage and 95% identity thresholds. The genetic context of blaCTX-M-15, blaKPC-3 and blaOXA-48 was investigated and compared across different isolates by aligning the respective contigs using the MAFFT 7 online server [25]. Reconstruction of the genetic context was performed using BLAST (blastn) across full length contigs, or on the region comprising up to 10 Kbp upstream and downstream of the resistance gene of interest, against the GenBank nucleotide collection. Tn4401 isoform identification was performed using BLAST and sequence alignment against Tn4401b and Tn4401a (GenBank Accessions KT378598 and EU16011, respectively). Capsular type (K-locus) and O-locus types were predicted using Kaptive Web (https://github.com/kelwyres/Kaptive-Web). SpeciesFinder 2.0 was used to confirm/validate initial phenotypic-based identification [26].
3. Results
3.1. Whole Genome Sequencing Enables Identification of K. variicola Misidentified as K. pneumoniae
The study comprises a total of 31 contemporary Klebsiella spp. isolates recovered in a Northern Central Hospital. All isolates were initially identified as Kp based on phenotypical biochemical testing (VITEK) and were subjected to WGS with the objective of gaining further insight on the evolutionary epidemiology, resistome, mobilome and virulome of these Klebsiella complex species. Upon whole genome sequencing and in silico species confirmation, all but one isolate was identified as Kp (n = 30). The remaining isolate was identified as Kv (n = 1) with high confidence by SpeciesFinder (v2.0). Eighteen of the 31 isolates were obtained from six clinical specimens: urine (n = 10), blood (n = 2), respiratory tract secretions by double-lumen catheter (RTS/DLC) (n = 2); catheter (n = 2); biopsy (n = 1) and intra-operative swab (IOSwab; n = 1) (Table 1). According to control measures established by Infection Prevention and Control Commission of the Hospital, 13 out of the 31 Kp isolates were recovered from rectal swabs from four colonized patients (P3, P8, P20, P27) upon admittance to the hospital and from eight colonized patients staying on the wards: Medicine (P1, P12, P28); intensive care unit (ICU) (P2, P26); Surgical (P6, P16) and Nephrology (P7). In addition, patient 28 was also colonized with Kv5163. Five patients attending the emergency were infected with Kp isolates obtained from urine (P4, P14, P17), blood (P15) and sputum (P20), which suggests colonization/infection outside the hospital.
Table 1.
Characteristics of the 31 isolates recovered from 28 patients colonized/infected by K. pneumoniae and K. variicola isolates.
| ID | Isolates | Patient | Sample | Hospital Ward | Date | ST |
|---|---|---|---|---|---|---|
| 5141 | K. pneumoniae | P1 | Rectal swab | Medicine | 2 May 2018 | ST147 |
| 5142 | K. pneumoniae | P2 | Rectal swab | ICU | 3 May 2018 | ST147 |
| 5144 | K. pneumoniae | P3 | Rectal swab | Emergency | 4 May 2018 | ST147 |
| 5147 | K. pneumoniae | P4 | Urine | Emergency | 6 May 2018 | ST147 |
| 5151 | K. pneumoniae | P5 | Dialysis liquid | Dialysis Clinic | 15 May 2018 | ST147 |
| 5152 | K. pneumoniae | Purulent exudate | Dialysis Clinic | 15 May 2018 | ST147 | |
| 5143 | K. pneumoniae | P6 | Rectal swab | Surgery | 16 May 2018 | ST147 |
| 5160 | K. pneumoniae | P7 | Rectal swab | Neurology | 25 May 2018 | ST147 |
| 5161 | K. pneumoniae | P8 | Rectal swab | Emergency | 26 May 2018 | ST147 |
| 5155 | K. pneumoniae | P9 | Urine | Oncology Clinic | 29 May 2018 | ST147 |
| 5158 | K. pneumoniae | P10 | Urine | Surgery | 4 June 2018 | ST147 |
| 5164 | K. pneumoniae | P11 | Urine | Medicine | 4 June 2018 | ST147 |
| 5167 | K. pneumoniae | P12 | Rectal swab | Medicine | 4 June 2018 | ST147 |
| 5168 | K. pneumoniae | P13 | Urine | Outpatient Clinic | 11 June 2018 | ST147 |
| 5173 | K. pneumoniae | P14 | Urine | Emergency | 17 June 2018 | ST147 |
| 5174 | K. pneumoniae | P15 | Blood | Emergency | 19 June 2018 | ST147 |
| 5175 | K. pneumoniae | P16 | Rectal swab | Surgery | 19 June 2018 | ST147 |
| 5146 | K. pneumoniae | P17 | Urine | Emergency | 3 May 2018 | ST15 |
| 5149 | K. pneumoniae | P18 | Hepatic liquid | Surgery | 9 May 2018 | ST15 |
| 5154 | K. pneumoniae | P19 | Blood | Hematology | 28 May 2018 | ST15 |
| 5159 | K. pneumoniae | P20 | Rectal swab | Emergency | 25 May 2018 | ST15 |
| 5156 | K. pneumoniae | Sputum | Emergency | 30 May 2018 | ST15 | |
| 5157 | K. pneumoniae | P21 | Biopsy | Plastic Surgery | 4 June 2018 | ST15 |
| 5166 | K. pneumoniae | P22 | Urina | Nephrology clinic | 6 June 2018 | ST15 |
| 5148 | K. pneumoniae | P23 | Urine | ICU | 7 May 2018 | ST34 |
| 5150 | K. pneumoniae | P24 | Urine | Orthopaedic | 13 May 2018 | ST1079 |
| 5153 | K. pneumoniae | P25 | RTS/DLC a | Nephrology | 21 May 2018 | ST29 |
| 5162 | K. pneumoniae | P26 | Rectal swab | ICU | 29 May 2018 | ST461 |
| 5165 | K. pneumoniae | P27 | Rectal swab | Emergency | 5 June 2018 | ST307 |
| 5163 | K. variicola | P28 | Rectal swab | Medicine | 29 May 2018 | ST4197 |
| 5172 | K. pneumoniae | Rectal swab | Medicine | 18 June 2018 | ST280 |
a Respiratory tract secretions by double-lumen catheter.
3.2. Genomic Population Structure and High-Resolution Phylogenomic Analysis Unveils Multiple Healthcare-Associated Transmission Clusters
To ascertain the genomic population structure, in silico MLST identification was carried out for each isolate in order to assign them to different sequence types (ST). Among the 30 Kp, eight STs were detected: ST147 (n = 17 isolates, 16 patients), ST15 (n = 7, 6 patients), ST34 (n = 1), ST1029 (n = 1), ST29 (n = 1), ST46 (n = 1), ST307 (n = 1), and ST280 (n = 1). Each of the P5 and P20 patients had two isolates assigned to the ST147 and ST15, respectively. Moreover, patient P28, admitted to the Medicine ward, was colonized with Kv ST4197 and after two weeks a Kp ST280 strain was identified.
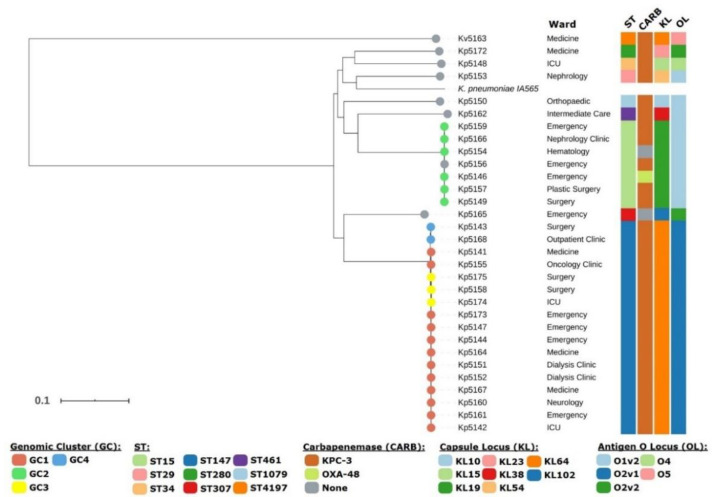
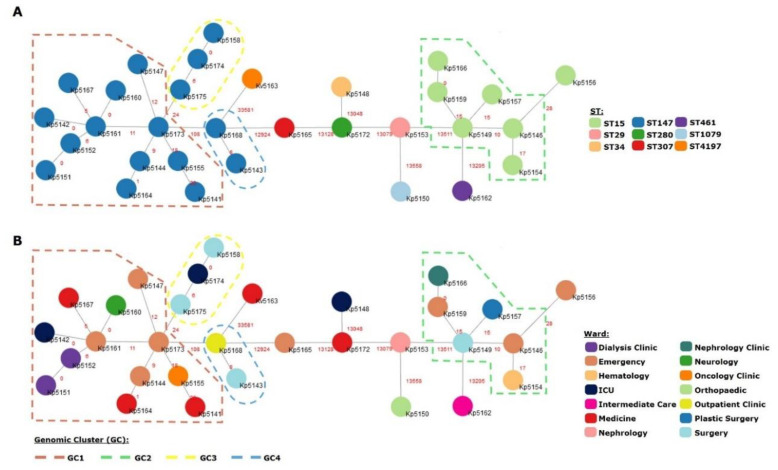
High-resolution phylogenomic analysis based on 64,106 high quality SNPs enabled the construction of a phylogenetic tree whose topological structure was congruent with the MLST-based classification with Kv5163 phylogenetically placed farther apart from all Kp isolates included in the study (Figure 1). Both ST147 and ST15 strains formed monophyletic clades and no association with a specific ward was identified. Interestingly, Kp5156 and Kp5159, both isolated from P20, did not form a monophyletic clade and were separated by 35 SNPs, suggesting different infection/colonization events with a ST15 strain (Figure 2). Contrarily, Kp5151 and Kp5152, did form a monophyletic clade and were indistinguishable through the present phylogenetic scenario (distance: 0 SNPs). Using a 23 SNP cutoff to delineate clusters enabled the identification of four genomic clusters (GC1-4) with the two largest clusters, GC1 (n = 12) and GC2 (n = 6), being comprised by ST147 and ST15 isolates, respectively (Figure 2). Both GC1 and GC2 show a high dispersal through different wards of the studied hospital but were more prevalent from patients attending the Emergency as 4/12 and 3/6 isolates from GC1 and GC2 were in fact isolated from patients in this ward (Figure 1).
Figure 1.
Genome-wide phylogenetic tree of 30 K. pneumoniae (Kp) isolates and one K. variicola (Kv) isolate analyzed in the present study. The phylogeny depicted is based on 64,106 high quality SNPs obtained from mapping raw sequence reads against the reference strain Kp IA65 also present in the tree. The tree is topologically structured by sequence type (ST) and is annotated with the ward, ST, capsular locus type (KL), and antigen O locus type (OL) according to the legend. Colored tree tips highlight the different genomic clusters (GCs) to which the isolate has been assigned or if it is non-clustered (light grey).
Figure 2.
Minimum spanning trees (MST) constructed using the goeBURST algorithm with the pairwise comparison method for all isolates included in the study. The MSTs are shown annotated with the genomic clusters and node coloring reflect the ST (A) or ward from which the isolate was obtained (B). Four different GCs (GC1-4) were detected and highlighted on the tree reflecting ongoing transmission across the wards from the studied hospital.
The two other GCs are shown to involve ST147 strains (GC3 and GC4). Noteworthily, GC3 isolates are in fact separated from GC1 isolates by 24 SNPs, one SNP above the threshold for cluster assignment and likely highlights a group of ST147 strains derived from GC1, unsampled cases or missed links in the transmission chains that may take the form of undetected colonized patients. The fact that GC4 isolates form a monophyletic clade nested within the GC1 branch of the tree lends further support to this hypothesis (Figure 1).
3.3. KPC-3 as the Main Driver of Carbapenem Resistance in Northern Portugal and First Description of KPC-3 Producing K. variicola
According to antimicrobial susceptibility (Supplementary Table S1) and regarding the resistance genes found in Kp, we identified two carbapenemase-coding genes (blaKPC-3 and blaOXA-48), four genes coding for extended-spectrum β-lactamases (ESBLs), namely blaCTX-M-15, blaSHV-12, blaSHV-27, blaSHV-187), and eight genes coding for narrow/broad spectrum beta-lactamases (blaTEM-1, blaTEM-33, blaSHV-1, blaSHV-11, blaSHV-26, blaSHV-28, blaOXA-1, blaOXA-9). The aac, aad and aph aminoglycoside-modifying enzyme genes were detected in most (27/31; 87.1%) of the isolates, except in three Kp and one Kv. Trimethoprim–sulfamethoxazole resistance was encoded by sul and dfr genes. The sul1 and sul2 genes were detected in 13 isolates each, and one isolate had both genes. Moreover, the four isolates without aminoglycoside-modifying enzyme genes did not have sul and dfr genes. The qnr, oqxAB and aac(6′)-lb-cr5 plasmid-mediated quinolone resistance (PMQR) genes and mutations in chromosomal gyrA or parC loci were detected in these isolates. The fosA gene, which codes for fosfomycin resistance, was identified in all isolates; chloramphenicol acetyltransferase genes were also identified: catA1 (n = 4), catA2 (n = 1), and catB3 (n = 4). The rifampicin adenosine diphosphate (ADP) ribosylating transferase arr−3 gene was detected in 10 isolates (Table 2). The Kv5163 had the chromosomal broad spectrum β-lactamase blaLEN-2, the blaKPC-3, oqxAB and fosA genes.
Table 2.
Resistance determinants of thirty-one multi-resistant K. pneumoniae and K. variicola isolates.
| Isolates ID | Sequence Type | Genomic Cluster | bla_Carb | bla_ESBL | bla_narrow Spectrum | Aminoglycoside | aac(6′)-Ib-cr5 | qnr | Phe | Rif | Fos | oqxAB | tmp | qacEdelta1 | sul | tet | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kp5141 | ST147 | GC1 | KPC-3 | CTX-M-15 | SHV-11 | TEM-1 | OXA-1 | aph(6)-Id; aph(3″)-Ib | aac(3)-IIa | aac(6′)-Ib-cr5 | catB3 | fosA | oqxAB | dfrA14 | sul2 | ||||
| Kp5142 | ST147 | GC1 | KPC-3 | SHV-11 | aadA1 | fosA | oqxAB | dfrA1 | qacEdelta1 | sul1 | |||||||||
| Kp5144 | ST147 | GC1 | KPC-3 | SHV-11 | aadA16 | aac(6′)-Ib-cr5 | qnrB6 | - | arr-3 | fosA | oqxAB | dfrA27 | sul1 | ||||||
| Kp5147 | ST147 | GC1 | KPC-3 | - | SHV-11 | aadA16 | aac(6′)-Ib-cr5 | - | arr-3 | fosA | oqxAB | dfrA27 | qacEdelta1 | sul1 | |||||
| Kp 5151 | ST147 | GC1 | KPC-3 | - | SHV-11 | aadA16 | aac(6′)-Ib-cr5 | qnrB6 | - | arr-3 | fosA | oqxAB | dfrA27 | qacEdelta1 | sul1 | ||||
| Kp5152 | ST147 | GC1 | KPC-3 | - | SHV-11 | aadA16 | aac(6′)-Ib-cr5 | qnrB6 | - | arr-3 | fosA | oqxAB | dfrA27 | qacEdelta1 | sul1 | ||||
| Kp5143 | ST147 | GC4 | KPC-3 | SHV-12 | TEM-1 | aph(6)-Id; aph(3″)-Ib | qnrB1 | fosA | oqxAB | dfrA14 | qacEdelta1 | sul2 | tet(A) | ||||||
| Kp5160 | ST147 | GC1 | KPC-3 | - | SHV-11 | aadA1 | - | - | fosA | oqxAB | dfrA1 | qacEdelta1 | sul1 | ||||||
| Kp5161 | ST147 | GC1 | KPC-3 | - | SHV-11 | aadA1 | - | - | fosA | oqxAB | dfrA1 | qacEdelta1 | sul1 | ||||||
| Kp5155 | ST147 | GC1 | KPC-3 | SHV-11 | TEM-33 | - | - | - | fosA | oqxAB | - | ||||||||
| Kp5158 | ST147 | GC1 | KPC-3 | - | SHV-1 | aadA16 | aac(6′)-Ib-cr5 | qnrB6 | - | arr-3 | fosA | oqxAB | dfrA27 | qacEdelta1 | sul1 | ||||
| Kp5164 | ST147 | GC1 | KPC-3 | - | SHV-11 | aadA16 | aac(6′)-Ib-cr5 | qnrB6 | - | arr-3 | fosA | oqxAB | dfrA27 | qacEdelta1 | sul1 | ||||
| Kp5167 | ST147 | GC1 | KPC-3 | - | SHV-11 | aadA1 | - | - | fosA | oqxAB | dfrA1 | qacEdelta1 | sul1 | ||||||
| Kp5168 | ST147 | GC1 | KPC-3 | SHV-12 | TEM-1 | OXA-9 | aph(6)-Id; aph(3″)-Ib | aac(6′)-Ib; aadA1 | qnrB1 | - | - | fosA | oqxAB | dfrA14 | sul2 | tet(A) | |||
| Kp5173 | ST147 | GC1 | KPC-3 | - | SHV-11 | aadA16 | aac(6′)-Ib-cr5 | qnrB6 | - | arr-3 | fosA | oqxAB | dfrA27 | qacEdelta1 | sul1 | ||||
| Kp5174 | ST147 | GC3 | KPC-3 | - | SHV-1 | aadA16 | aac(6′)-Ib-cr5 | qnrB6 | - | arr-3 | fosA | oqxAB | dfrA27 | qacEdelta1 | sul1 | ||||
| Kp5175 | ST147 | GC3 | KPC-3 | - | SHV-11 | aadA16 | aac(6′)-Ib-cr5 | qnrB6 | - | arr-3 | fosA | oqxAB | dfrA27 | qacEdelta1 | sul1 | ||||
| Kp5146 | ST15 | GC2 | OXA-48 | CTX-M-15 | SHV-28 | TEM-1 | OXA-1 | aph(6)-Id; aph(3″)-Ib | aac(3)-IIa | aac(6′)-Ib-cr5 | qnrB1 | catA1; catB3 | - | fosA | oqxAB | dfrA14 | sul2 | ||
| Kp5149 | ST15 | GC2 | KPC-3 | - | SHV-28 | TEM-1 | OXA-9 | aph(6)-Id; aph(3″)-Ib | aac(6′)-Ib; aadA1 | - | - | fosA | oqxAB | dfrA14 | sul2 | ||||
| Kp5154 | ST15 | GC2 | CTX-M-15 | SHV-28 | TEM-1 | aph(6)-Id; aph(3″)-Ib | aac(3)-IIa | qnrB1 | catA1; catB3 | - | fosA | oqxAB | dfrA14 | sul2 | |||||
| Kp5159 | ST15 | GC2 | KPC-3 | - | SHV-28 | TEM-1 | OXA-9 | aph(6)-Id; aph(3″)-Ib | aadA1 | qnrB1 | catA1 | - | fosA | oqxAB | dfrA14 | sul2 | tet(A) | ||
| Kp5156 | ST15 | KPC-3 | CTX-M-15 | TEM-1 | OXA-9 | aph(6)-Id; aph(3″)-Ib | aac(6′)-Ib; aadA1 | qnrB1 | - | - | fosA | oqxAB | dfrA14 | sul2 | |||||
| Kp5157 | ST15 | GC2 | KPC-3 | - | SHV-28 | TEM-1 | OXA-9 | aph(6)-Id; aph(3″)-Ib | aac(6′)-Ib; aadA1 | - | - | fosA | oqxAB | dfrA14 | sul2 | ||||
| Kp5166 | ST15 | GC2 | KPC-3 | - | SHV-28 | TEM-1 | OXA-9 | aph(6)-Id; aph(3″)-Ib | aadA1 | qnrB1 | catA1 | - | fosA | oqxAB | dfrA14 | sul2 | tet(A) | ||
| Kp5148 | ST34 | KPC-3 | - | SHV-26 | - | - | - | fosA | oqxA10; oqxB19 | - | |||||||||
| Kp5150 | ST1079 | KPC-3 | - | SHV-1 | TEM-1 | OXA-9 | aph(6)-Id; aph(3″)-Ib | aac(6′)-Ib; aadA1 | - | - | fosA | oqxA; oqxB19 | dfrA14 | sul2 | |||||
| Kp5153 | ST29 | KPC-3 | - | SHV-187 | TEM-1 | OXA-9 | aph(6)-Id; aph(3″)-Ib | aac(6′)-Ib; aadA1 | qnrS1 | - | - | fosA | oqxA; oqxB25 | dfrA14 | sul2 | ||||
| Kp5162 | ST451 | KPC-3 | - | SHV-1 | - | - | - | fosA | oqxA10; oqxB25 | - | |||||||||
| Kp5165 | ST307 | CTX-M-15 | SHV-28 | TEM-1 | aph(6)-Id; aph(3″)-Ib | aac(3)-IIa; aadA16 | aac(6′)-Ib-cr5 | qnrB6 | catA2 | arr-3 | fosA | oqxA; oqxB19 | dfrA27 | qacEdelta1 | sul1;sul2 | tet(D) | |||
| Kv5163 | ST4197 | KPC-3 | - | LEN-2 | - | - | - | fosA | oqxA; oqxB15 | - | |||||||||
| Kp5172 | ST280 | KPC-3 | CTX-M-15 | SHV-27 | TEM-1 | OXA-1; OXA-9 | aph(6)-Id; aph(3″)-Ib | aac(3)-IIa; aac(6′)-Ib; aadA1 | aac(6′)-Ib-cr5 | qnrB1 | catB3 | - | fosA | oqxA; oqxB19 | dfrA14 | sul2 | tet(A) | ||
Inspection of the genetic environments of blaKPC-3, blaCTX-M-15, and blaOXA-48 revealed twenty-five out of the 28 KPC-3 producing isolates had the same genetic environment as part of the Tn4401d isoform, which is characterized by a 68 bp deletion between istB and the blaKPC gene when compared with the Tn4401b isoform [27]. This Tn4401b isoform was identified in the remaining three isolates Kp5148 (ST34), Kp5162 (ST451) and Kv5163. The co-existence of Kv5163 with Kp5172 in same patient is supportive of distinct dissemination events regarding the origin of the blaKPC-3 in these strains.
The blaCTX-M-15 gene was carried on an ISEcp1 insertion sequence (IS) element and was detected in six isolates belonging to ST15 (n = 3 isolates) and in each one of ST147, ST307, ST280. The blaOXA-48 gene, detected in Kp5146 (ST15) isolate, was inserted into an IncL/M-type plasmid, a replicon exclusively found in this strain, with 100% coverage and identity to reference replicon type under accession no. JN626286. Part of the plasmid (IncL/M p5146) was found in a contig where the region of blaOXA-48 gene was identical to pOXA48a [28], described as having part of Tn1999 upstream and a lysR gene downstream encoding a regulatory protein. In addition, in this Kp5146 isolate, four replicons were found: IncFIB(K)Kpn3; IncFIB(Mar); IncFII(pMET) and ColRNAI_1. Using the criterion of >95% nucleotide identity for large plasmids and >80% for Col-like plasmids and >96% coverage with the reference replicon sequences [24], among the 31 isolates five major replication loci were detected, namely IncFIB(K) (n = 24, 77.4%), IncFIA(HI1) and IncFII-1-pKP91 (n = 18, 58.1%), IncN-1 (n = 17, 54.8%). Sixteen replicons were found among 31 isolates (Table 3). Eight (n = 8) out 16 Kp ST147 isolates had as part of IncN-1 plasmid a complex sul1-type that contain a partial duplication of the 3′-CS. Between both copies of the 3′-CS there is a variable region harbouring a qnrB6 gene and, in the region between the 5′CS and the first 3′CS we detected the presence of the aac(6′)-Ib-cr5; arr-3; dfrA27 and aadA16 gene cassettes. The IncFIB group was the most abundantly distributed by all ST types (including Kv5163) represented by four subvariants (IncFIB (pQil)_1; IncFIB (K)_1_Kpn3; IncFIB (pKPHS1); IncFIB (Mar)_1_pNDM).
Table 3.
Distribution of plasmid replicons identified among 31 whole genome sequenced K.pneumoniae and K. variicola isolates by PlasmidFinder database.
| K. pneumoniae | K. variicola | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Replicon | ST147 ** | ST15 | ST461 | ST1079 | ST29 | ST307 | ST280 | ST34 | ST4197 | Identity/% | N° Isolates/% |
| IncFIA (HI1) | 6 | 7 | 1 | 1 | 1 | 1 | 1 | 94.59–100 | 18/58.1 | ||
| IncFIB (pQil)_1_pQil | 1 * | 1 * | 100 | 02/06.4 | |||||||
| IncFIB (K)_1_Kpn3 | 12 (9 *) | 7 * | 1 | 1 | 1 * | 1 | 1 * | 98.93–100 | 24/77.4 | ||
| IncFIB (pKPHS1) | 14 | 97.32–100 | 14/45.5 | ||||||||
| IncFIB (Mar)_1_pNDM | 1 | 1 | 1 * | 99.77–100 | 03/09.7 | ||||||
| IncFII_1_pKP91 | 10 | 5 | 1 | 1 | 1 | 85.47–88.55 | 18/58.1 | ||||
| IncFII (Yp) | 6 | 5 | 1 | 1 | 1 | 85.71 | 14/45.5 | ||||
| IncFII (pMET)_1_pMET1 | 1 | 98.09 | 01/03,2 | ||||||||
| IncR_1 | 4 (2 *) | 2 * | 1 | 99.2–100 | 07/22.5 | ||||||
| IncN_1 | 16 | 1 | 99.42 | 17/54.8 | |||||||
| IncL/M (pOXA-48) | 1 * | 100 | 01/03.2 | ||||||||
| IncX4_1 | 1 | 99.73 | 01/03.2 | ||||||||
| repA-pKPC-2 | 1 | 99.71 | 01/03.2 | ||||||||
| Col (MG828)1 | 1 | 93.51 | 01/03.2 | ||||||||
| ColRNAI_1 | 4 (2 *) | 6 | 1 | 1 | 1 | 1 | 85.5–100 | 14/45.5 | |||
| ColpVC_1 | 1 | 98.45 | 01/03.2 | ||||||||
* The number followed by * represents the isolates with 100% nucleotide coverage and the identity to reference replicon type. ** number of isolates for STs: ST147-17 isolates; ST15-7 isolates; the remaining 1 isolate each has ST type.
3.4. Virulome of K. pneumoniae Highlights the Occurrence of Multiple Virulence Factors and ST-Specific Patterns
Regarding the virulence genes found in Kp and Kv isolates, the distribution of capsular locus (KL) types denoted specific associations with ST, where each KL type occurred only in one ST type and was common to all isolates belonging to same ST type (Table 4). The exception was Kv5163 with KL64, similar to Kp ST147 isolates but with diverging antigen O locus (OL) types O5 and O2v1, respectively. The O1v2 types were dispersed throughout the Kp ST15, Kp ST1079, Kp ST29 and Kp ST461.
Table 4.
Distribution of virulent determinants against thirty-one multiresistant K. pneumoniae and K. variicola isolates.
| Isolates ID | Sequence Type | wzi | K_locus | O_locus | Enterobactin * | Ferric Iron ** | Ybt *** | Type I Fimbriae | Type 3 Fimbriae | Type VI SSE |
|---|---|---|---|---|---|---|---|---|---|---|
| Kp5141 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5142 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5144 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5147 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5151 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5152 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5143 | ST147 | wzi64 | KL64 | O2v1 | entB | |||||
| Kp5160 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5161 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5155 | ST147 | wzi64 | KL64 | O2v1 | entB | |||||
| Kp5158 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5164 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5167 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5168 | ST147 | wzi64 | KL64 | O2v1 | entB | fimH | mrkD_12 | tle1; tli1 | ||
| Kp5173 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5174 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5175 | ST147 | wzi64 | KL64 | O2v1 | entB | ybtQ | fimH | mrkD_12 | tle1; tli1 | |
| Kp5146 | ST15 | wzi19 | KL19 | O1v2 | entB | kfuA_3 | ybtQ | fimH | ||
| Kp5149 | ST15 | wzi19 | KL19 | O1v2 | entB | kfuA_3 | ybtQ | fimH | ||
| Kp5154 | ST15 | wzi19 | KL19 | O1v2 | entB | kfuA_3 | ybtQ | fimH | ||
| Kp5159 | ST15 | wzi19 | KL19 | O1v2 | entB | kfuA_3 | ybtQ | fimH | ||
| Kp5156 | ST15 | wzi19 | KL19 | O1v2 | entB | kfuA_3 | fimH | |||
| Kp5157 | ST15 | wzi19 | KL19 | O1v2 | entB | kfuA_3 | fimH | |||
| Kp5166 | ST15 | wzi19 | KL19 | O1v2 | entB | kfuA_3 | ybtQ | fimH | ||
| Kp5148 | ST34 | wzi50 | KL15 | O4 | entB | fimH | tle1; tli1 | |||
| Kp5150 | ST1079 | wzi236 | KL10 | O1v2 | entB | fimH | ||||
| Kp5153 | ST29 | wzi115 | KL54 | O1v2 | entB | fimH | ||||
| Kp5162 | ST451 | wzi38 | KL38 | O1v2 | entB | kfuA_26 | fimH | |||
| Kp5165 | ST307 | wzi173 | KL102 | O2v2 | entB | fimH | tle1; tli1 | |||
| Kv5163 | ST4197 | - | KL64 | O5 | entB | kfuA_8 | fimH | tle1; tli1 | ||
| Kp5172 | ST280 | wzi82 | KL23 | O2v2 | entB | fimH | tle1; tli1 |
* entB gene represents the Enterobactin gene cluster (entA; entB; entC; entD; entE; entF; fepA; fepB; fepC; fepD; fepG; fes; ybdA); ** kfuA gene represents the kfuABC operon; *** ybtQ gene represents the Yersiniabactin gene cluster (fyuA; irp1; irp2; ybtA; ybtE; ybtP; ybtQ; ybtS; ybtT; ybtU; ybtX).
Both bacterial species Kp and Kv showed a profile of virulence-associated determinants, specifically fimbrial adhesins (fimH-1), and siderophore entB (enterobactin). The iroE and iutA-18 genes were found in all strains, however the other genes integrating the iucABCD and iroBCDN siderophore gene clusters were not detected. Further analysis on the genetic context of these two genes and its distribution across other Enterobactereaceae species revealed that these genes are likely co-localized to the chromosome and show a wide distribution: these iroE homologues were detected across the chromosomes of K. pneumoniae, K. quasipneumoniae, K. variicola and K. quasivariicola (phylogroups Kp1-4) [2]; the iutA homologues showed an even wider distribution that included the former species but also Raoultella spp, Citrobacter spp., Kluyvera spp., etc. with varying homology identities. The yersiniabactin siderophore cluster represented by ybtQ gene was identified in Kp ST147 and ST15 genomes (Table 4). However, the iron-uptake system siderophore (kfu) was present in Kp ST15, Kp ST461 and Kv ST4197 strains but not Kp ST147. The Type VI secretion system (T6SS) mediated type-1 fimbriae expression, cell adherence, invasion, and in vivo colonization, is represented by type VI membrane-targeting phospholipase effector Tle1 and Tli1 immunity proteins [29,30] and was detected in 19 isolates, the majority of which were ST147 strains (Table 4).
4. Discussion
Genomic sequences of thirty-one isolates initially identified as Kp were used for phylogenetic analyses and to investigate their genomic content for antimicrobial resistance genes, plasmid replicon types and virulence genes. Through whole genome sequencing we were able to correctly identify a KPC-3-producing Kv isolate previously misidentified as Kp during routine laboratory diagnosis. The present study thus highlights the presence of Kp and Kv isolates from the same patient at different time-points. The correct identification of the Klebsiella species can be technically challenging for most clinical microbiology laboratories since several species share a similar colony morphology and biochemical profile. A second potential case of colonization by Kv was detected in a patient in this same hospital setting, and also identified as Kp, but was excluded from the study due to mixed infection with another Kp strain (data not shown). This latter isolate yielded a larger genome assembly with duplicate resistance gene sequences mapping to distinct contigs that despite several attempts were not possible to isolate. The misidentified Kv isolate and the mixed colonization described stress the emerging importance of Kv as a host for drug resistance determinants and the existing technical challenges that may underestimate its epidemiological importance. The presence of blaLEN-2, a broad-spectrum b-lactamase that confers resistance to ampicillin is characteristic for Kv strains [31]. The oqxAB, fosA, blaSHV or blaLEN genes are ubiquitous and considered the ancestral resistome from the Klebsiella complex [32,33]. Carbapenemase-producing Kv isolates have been described in different countries, namely KPC-2 in USA, Europe and in Asia [3]. In Portugal the widespread distribution of KPC-3 among Kp isolates in different hospitals was associated to successful high-risk clones ST147 [34]; ST307; ST15 [27] and ST14 [35,36] located in a Tn4401d isoform. The genetic context of blaKPC is known to have contributed greatly to the spread of KPC producers in many countries, and the emergence of non-CG258 KPC-Kp isolates in France was linked to dissemination of these clones from Portugal [37]. To our knowledge, we report the first KPC-3 producing Kv isolate with the blaKPC-3 gene mapped to the Tn4401b isoform, a less common genetic environment also found in two additional Kp isolates from this study (Kp5148 and Kp5162) which also suggests a potential scenario for cross-species lateralization of KPC-3.
Herein, the population structure found across these isolates was dominated by two of the high-risk clones mentioned above: ST147 (n = 17) and ST15 (n = 7). Both strains formed monophyletic clades comprising all transmission clusters found in the present study. The identification of these genomic clusters reflecting ongoing transmission was performed on the basis of a SNP distance threshold of 23 SNPs as previously reported [23]. In fact, 23/30 (76.7%) Kp isolates were within one of the four genomic clusters identified and therefore highlight the highly clonal nature of this population structure and its dissemination across multiple wards in the studied setting. This level of uncontrolled dissemination stresses the importance of additional infection control measures such as the rapid detection of colonization and hygiene/decontamination of possible contaminated environmental sources. Additionally, the ability to discriminate at the genome-wide level allowed for the discrimination within ST147 strains of two isolates that are here shown to be epidemiologically unrelated to the rest of the ST147 strains (distance: 108 SNPs) further stressing the low discriminatory power of MLST.
A diverse set of plasmids has been characterized in Kp and although these can vary in terms of length and incompatibility types, IncFIIK and IncFIBK are considered the most prevalent across this species [38]. In this study, these and other Inc. groups co-existed across different STs. Across ST147 (n = 17 isolates) strains a marked variation was observed for the number of replicons found per isolate (n = 2–6; 11.8% and 35.3%, respectively), whereas for ST15 (n = 7) strains the number of replicons per isolate was lower with three different replicon types per isolate as the predominant value (57%). In contrast, the Kv5163 isolate had five detectable replicons of which FIB(K)Kpn3 and ColRNAI _1 were the predominant replicons found for Kp, while from the three remaining replicons only IncFII(pMet) was exclusively detected in the Kv strain. Furthermore, for ST147 strains, the blaKPC−3 was colocalized to an IncN-1 plasmid whose repN showed 100% identity with plasmid R46 identified in Salmonella serovar Typhimurium (GenBank accession number AY046276). The same genetic background for blaKPC-3 was described in isolates identified on non-hospitalized patients (three different community laboratories) in Portugal [34]. The results obtained in this study also highlight its putative importance as a genetic platform for cross species mobilization of carbapenem resistance determinants. Moreover, also colocalized to this IncN-1 plasmid, a complex sul1-type Class1 integron was found, harboring in region 5′ CS five gene cassettes the aac(6′)-Ib-cr5; arr-3; dfrA27 and aadA16 and following the 2nd copy of 3′ CS of the sixth qnrB6 gene cassette. This latter structure appears to be absent from the KPC-3-bearing IncN plasmid detected in Salmonella serovar Typhimurium (accession number AY046276).
A different set of ST147 (n = 4) strains was found to include the sul2 gene on a class 1 integon associated to aac(6′)-Ib, aadA1, blaOXA-9, blaTEM-1, aph(6)-Id, aph(3”)-Ib gene cassettes. Interestingly, this genetic profile was found for all ST15 isolates, although with three variants regarding aminoglycoside resistance gene cassettes aac(6′)-Ib, aadA1 (3 isolates), aac(3)-IIa (2 isolates) and aadA1 (2 isolates). The sul2 has been previously described on small non-conjugative plasmids in association with the aph(6)-Id, aph(3″)-Ib gene although its presence has also been reported on large plasmids, some of which are transmissible and carrying several resistance determinants [39]. Although we could not phase the location of this gene with the replicon type, in this study sul2 was detected in contigs as large as ≥30 Kbp.
Kp and Kv infections have limited therapeutic options due to extensive drug resistance and successful immune escape mechanisms of these pathogens, which exhibit a high number of virulence factors. The leading virulence factors are mostly associated with capsular serotype [40] and the LPS (O antigen) O1 and O2 serotypes are the most common underpinning 50–68% of all Klebsiella spp. infections [40]. Twenty-nine out of the 30 isolates included in the study had the O1 and O2 serotypes associated with variants v1 or v2 while Kp5148 (ST34) and Kv5163 belonged to the O4 and O5 serotypes, respectively. The O1 strains are generally more resistant to the bactericidal effect of human serum, which has been attributed to the d-galactan II component in O1 antigen [41] which is not present in the O4 and O5 strains and there are clinical data suggesting that these antigens play a role in virulence or pathogenicity. The high prevalence observed for O1/O2 serotypes across the studied isolates is paralleled by a recent study in a Lisbon hospital and is congruent with a selective advantage by these serotypes [27]. However, while the Lisbon study mentioned demonstrated that the ST147 strains from this study and the ones from Lisbon (both KPC-3 producers) exhibit the same KL and OL types, the ST15 strains herein characterized differ in respect to these serotypes. This observation may be associated with the fact that ST15 strains detected recently in Lisbon were ESBL producers but not carbapenemase producing strains and highlights the serotype switching event that is herein associated with the phylogenetic branch of carbapenemase producing ST15 strains in the northern region of Portugal.
Additionally, microbial adherence and invasion of host cells are critical steps in the infection process and comprise additional virulence determinants with most clinical Kp isolates expressing two types of fimbrial adhesins, type 1 and type 3 fimbriae. Twenty-eight Kp and the Kv isolates had the fim gene cluster coding for the type 1 fimbriae which mediates adhesion to mannose-containing structures. Moreover, all but two ST147 isolates (Kp5143 and Kp5155) were concomitantly positive for the mrk gene cluster (type 3 fimbriae). The type 3 fimbriae encoded by the mrk gene cluster is characterized by their ability to mediate adhesion to different structures, through MrkD adhesin, as well as role in biofilm formation, by MrkA adhesin [42].
The presence of multiple iron transport systems in Kp suggests that iron acquisition systems are needed during infection. Four iron transport systems, entB, ybtS, iutA, and kfu are usually present, although a single siderophore-dependent iron acquisition system may be sufficient for bacterial colonization. The siderophore enterobactin (Ent) are common to all Kp and conserved in the chromosome as core genes. Nevertheless, the enhanced virulence in the majority of Kp lineages associated with liver abscess carry virulence plasmid encoding iuc and iro genes inserted in specific siderophore systems, aerobactin (Iuc) and salmochelin (Iro) [43,44]. The iucABCDiutA siderophore gene cluster was first reported on pCoIV-K30 in E. coli and on pLVPK of K. pneumoniae CG43 [43]. The iroBCDEN gene cluster, first described in Salmonella enterica, and was also found either on the chromosome or a transmissible plasmid in the uropathogenic Escherichia coli [45] and in Kp four structures of the salmochelin (iro) locus were described, one of them without the iroE gene [44]. The allele iutA-18 showed 100% coverage with 93.73–95% identity with the core chromosomal paralog of iutA encoding a TonB-dependent siderophore receptor. In addition, the iroE gene found herein is located in the chromosome and showed 100% coverage with 98.83–99.25% identity with an alpha/beta hydrolase. We believe that these are homologous genes acquired throughout the evolutionary history of the Klebsiella genus but unrelated to the iucABCDiutA and iroBCDEN gene clusters.
The iron uptake system (kfu) contributes to invasive disease and could play a part in the pathogenicity of bloodstream infections caused by Kv [46]. In this study, all Kp ST15 strains and the Kv5163 isolate carried the kfu gene, which was not present in Kp ST147 isolates. However, the ST147 strains and Kv5163 had the type VI secretion system (T6SS), which is widely implicated in microbial antagonism and mediates interactions with host eukaryotic cells and in the environment in competition with other species. This could explain of presence of this type VI secretion system in Kv5163 isolate.
5. Conclusions
The present study contributes with novel data on the genomic epidemiology, its resistome, mobilome and virulome of carbapenemase producing K. pneumoniae in a hospital in northern Portugal. To our knowledge this is the largest study involving carbapenemase producing isolates from Portugal carried out at the genome-wide level while simultaneously comprising the first report of a KPC-3 producing K. variicola worldwide with the blaKPC-3 gene being located in the rather uncommon horizontal gene transfer element Tn4401b isoform. This work further highlights the technical challenges associated with K. variicola identification at the species level during routine laboratory diagnosis with WGS providing a correct identification and the importance for more strict control measures to gain further control on the highly clonal dissemination.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/12/1986/s1, Table S1: Antimicrobial susceptibility of 30 K. pneumoniae isolates and one K. variicola isolate identified from 28 patients.
Author Contributions
Conceptualization, A.D.; data curation, J.P. (João Perdigão); formal analysis, J.P. (João Perdigão), C.C. and A.D.; investigation, J.P. (João Perdigão), C.C., R.E., A.M., A.S., J.P. (Jody Phelan), S.C., T.G.C., E.C., M.J.S. and A.D.; methodology, J.P. (João Perdigão) and C.C.; resources, S.C., T.G.C., E.C. and M.J.S.; software, A.S., J.P. (Jody Phelan), S.C. and T.G.C.; supervision, A.D.; validation, João Perdigão, A.S., J.P. (Jody Phelan), S.C. and T.G.C.; writing—original draft, C.C. and A.D.; writing—review and editing, J.P. (João Perdigão), C.C., T.G.C. and A.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by UID/DTP/04138/2019 and UIDB/04033/2020 from Fundação para a Ciência e Tecnologia (FCT), Portugal. J.P. (João Perdigão) is supported by FCT through Estímulo Individual ao Emprego Científico (CEECIND/00394/2017). T.G.C. received funding from the MRC UK (Grant no. MR/K000551/1, MR/M01360X/1, MR/N010469/1, MR/R020973/1, MR/R025576/1, MR/S01988X/1, MR/S03563X/1) and BBSRC UK (BB/R013063/1). S.C. received funding from the BloomsburySet, Medical Research Council UK grants (MR/R020973/1, MR/R025576/1, MR/S01988X/1, MR/S03563X/1) and the BBSRC UK (BB/R013063/1). J.P. (Jody Phelan) is funded by the Medical Research Council UK grants (MR/S03563X/1).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brisse S., Verhoef J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int. J. Syst. Evol. Microbiol. 2001;51:915–924. doi: 10.1099/00207713-51-3-915. [DOI] [PubMed] [Google Scholar]
- 2.Rodrigues C., Passet V., Rakotondrasoa A., Diallo T.A., Criscuolo A., Brisse S. Description of Klebsiella africanensis sp. nov., Klebsiella variicola subsp. tropicalensis subsp. nov. and Klebsiella variicola subsp. variicola subsp. nov. Res. Microbiol. 2019;170:165–170. doi: 10.1016/j.resmic.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Medina N., Barrios-Camacho H., Duran-Bedolla J., Garza-Ramos U. Klebsiella variicola: An emerging pathogen in humans. Emerg. Microbes Infect. 2019;8:973–988. doi: 10.1080/22221751.2019.1634981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maatallah M., Vading M., Kabir M.H., Bakhrouf A., Kalin M., Nauclér P., Brisse S., Giske C.G. Klebsiella variicola Is a Frequent Cause of Bloodstream Infection in the Stockholm Area, and Associated with Higher Mortality Compared to K. pneumoniae. PLoS ONE. 2014;9:e113539. doi: 10.1371/journal.pone.0113539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garza-Ramos U., Moreno-Dominguez S., Hernandez-Castro R., Silva-Sanchez J., Barrios H., Reyna-Flores F., Sanchez-Perez A., Carrillo-Casas E.M., Sanchez-Leon M.C., Moncada-Barron D. Identification and Characterization of Imipenem-Resistant Klebsiella pneumoniae and Susceptible Klebsiella variicola Isolates Obtained from the Same Patient. Microb. Drug Resist. 2016;22:179–184. doi: 10.1089/mdr.2015.0181. [DOI] [PubMed] [Google Scholar]
- 6.Liu L., Feng Y., Tang G., Lin J., Huang W., Qiao F., Zong Z. Carbapenem-resistant Isolates of the Klebsiella pneumoniae Complex in Western China: The Common ST11 and the Surprising Hospital-specific Types. Clin. Infect. Dis. 2018;67:S263–S265. doi: 10.1093/cid/ciy662. [DOI] [PubMed] [Google Scholar]
- 7.Hopkins K.L., Findlay J., Doumith M., Mather B., Meunier D., D’Arcy S., Pike R., Mustafa N., Howe R., Wootton M., et al. IMI-2 carbapenemase in a clinical Klebsiella variicola isolated in the UK. J. Antimicrob. Chemother. 2017;72:2129–2131. doi: 10.1093/jac/dkx103. [DOI] [PubMed] [Google Scholar]
- 8.Rosenblueth M., Martinez L., Silva J., Martinez-Romero E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst. Appl. Microbiol. 2004;27:27–35. doi: 10.1078/0723-2020-00261. [DOI] [PubMed] [Google Scholar]
- 9.Potter R.F., Lainhart W., Twentyman J., Wallace M.A., Wang B., Burnham C.A., Rosen D.A., Dantas G. Population Structure, Antibiotic Resistance, and Uropathogenicity of Klebsiella variicola. mBio. 2018;9 doi: 10.1128/mBio.02481-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrade B.G., de Veiga Ramos N., Marin M.F., Fonseca E.L., Vicente A.C. The genome of a clinical Klebsiella variicola strain reveals virulence-associated traits and a pl9-like plasmid. FEMS Microbiol. Lett. 2014;360:13–16. doi: 10.1111/1574-6968.12583. [DOI] [PubMed] [Google Scholar]
- 11.Guo Y., Zhai Y., Zhang Z., Li D., Wang Z., Li J., He Z., Hu S., Kang Y., Gao Z. Complete Genomic Analysis of a Kingdom-Crossing Klebsiella variicola Isolate. Front. Microbiol. 2018;9:2428. doi: 10.3389/fmicb.2018.02428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The European Committee on Antimicrobial Susceptibility Testing . Breakpoint Tables for Interpretation of MICs and Zone Diameters—Version 8.0: European Society of Clinical Microbiology and Infectious Diseases. EUCAST; Växjö, Sweden: 2018. [Google Scholar]
- 13.Tsakris A., Kristo I., Poulou A., Themeli-Digalaki K., Ikonomidis A., Petropoulou D., Pournaras S., Sofianou D. Evaluation of boronic acid disk tests for differentiating KPC-possessing Klebsiella pneumoniae isolates in the clinical laboratory. J. Clin. Microbiol. 2009;47:362–367. doi: 10.1128/JCM.01922-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coll F., Phelan J., Hill-Cawthorne G.A., Nair M.B., Mallard K., Ali S., Abdallah A.M., Alghamdi S., Alsomali M., Ahmed A.O., et al. Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis. Nat. Genet. 2018;50:307–316. doi: 10.1038/s41588-017-0029-0. [DOI] [PubMed] [Google Scholar]
- 19.Anisimova M., Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 20.Schliep K.P. Phangorn: Phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nascimento M., Sousa A., Ramirez M., Francisco A.P., Carrico J.A., Vaz C. PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017;33:128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- 23.Sherry N.L., Lane C.R., Kwong J.C., Schultz M., Sait M., Stevens K., Ballard S., Goncalves da Silva A., Seemann T., Gorrie C.L., et al. Genomics for Molecular Epidemiology and Detecting Transmission of Carbapenemase-Producing Enterobacterales in Victoria, Australia, 2012 to 2016. J. Clin. Microbiol. 2019;57 doi: 10.1128/JCM.00573-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carattoli A., Zankari E., Garcia-Fernandez A., Voldby Larsen M., Lund O., Villa L., Moller Aarestrup F., Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen M.V., Cosentino S., Lukjancenko O., Saputra D., Rasmussen S., Hasman H., Sicheritz-Ponten T., Aarestrup F.M., Ussery D.W., Lund O. Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 2014;52:1529–1539. doi: 10.1128/JCM.02981-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perdigao J., Modesto A., Pereira A.L., Neto O., Matos V., Godinho A., Phelan J., Charleston J., Spadar A., de Sessions P.F., et al. Whole-genome sequencing resolves a polyclonal outbreak by extended-spectrum beta-lactam and carbapenem-resistant Klebsiella pneumoniae in a Portuguese tertiary-care hospital. Microb. Genom. :2020. doi: 10.1099/mgen.0.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel L., Bonnin R.A., Nordmann P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 2012;56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L., Ye M., Li X., Li J., Deng Z., Yao Y.F., Ou H.Y. Identification and Characterization of an Antibacterial Type VI Secretion System in the Carbapenem-Resistant Strain Klebsiella pneumoniae HS11286. Front. Cell Infect. Microbiol. 2017;7:442. doi: 10.3389/fcimb.2017.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh P.F., Lu Y.R., Lin T.L., Lai L.Y., Wang J.T. Klebsiella pneumoniae Type VI Secretion System Contributes to Bacterial Competition, Cell Invasion, Type-1 Fimbriae Expression, and In Vivo Colonization. J. Infect. Dis. 2019;219:637–647. doi: 10.1093/infdis/jiy534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fonseca E.L., Ramos N.D., Andrade B.G., Morais L.L., Marin M.F., Vicente A.C. A one-step multiplex PCR to identify Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae in the clinical routine. Diagn. Microbiol. Infect. Dis. 2017;87:315–317. doi: 10.1016/j.diagmicrobio.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Huynh B.T., Passet V., Rakotondrasoa A., Diallo T., Kerleguer A., Hennart M., Lauzanne A., Herindrainy P., Seck A., Bercion R., et al. Klebsiella pneumoniae carriage in low-income countries: Antimicrobial resistance, genomic diversity and risk factors. Gut. Microbes. 2020;11:1287–1299. doi: 10.1080/19490976.2020.1748257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez F., Rudin S.D., Marshall S.H., Coakley P., Chen L., Kreiswirth B.N., Rather P.N., Hujer A.M., Toltzis P., van Duin D., et al. OqxAB, a quinolone and olaquindox efflux pump, is widely distributed among multidrug-resistant Klebsiella pneumoniae isolates of human origin. Antimicrob. Agents Chemother. 2013;57:4602–4603. doi: 10.1128/AAC.00725-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodrigues C., Bavlovic J., Machado E., Amorim J., Peixe L., Novais A. KPC-3-Producing Klebsiella pneumoniae in Portugal Linked to Previously Circulating Non-CG258 Lineages and Uncommon Genetic Platforms (Tn4401d-IncFIA and Tn4401d-IncN) Front. Microbiol. 2016;7:1000. doi: 10.3389/fmicb.2016.01000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caneiras C., Calisto F., Jorge da Silva G., Lito L., Melo-Cristino J., Duarte A. First Description of Colistin and Tigecycline-Resistant Acinetobacter baumannii Producing KPC-3 Carbapenemase in Portugal. Antibiotics. 2018;7:96. doi: 10.3390/antibiotics7040096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caneiras C., Lito L., Mayoralas-Alises S., Diaz-Lobato S., Melo-Cristino J., Duarte A. Virulence and resistance determinants of Klebsiella pneumoniae isolated from a Portuguese tertiary university hospital centre over a 31-year period. Enferm. Infecc. Microbiol. Clin. 2019;37:387–393. doi: 10.1016/j.eimc.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Bonnin R.A., Jousset A.B., Chiarelli A., Emeraud C., Glaser P., Naas T., Dortet L. Emergence of New Non-Clonal Group 258 High-Risk Clones among Klebsiella pneumoniae Carbapenemase-Producing K. pneumoniae Isolates, France. Emerg. Infect. Dis. 2020;26:1212–1220. doi: 10.3201/eid2606.191517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyres K.L., Lam M.M.C., Holt K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020;18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 39.Enne V.I., Livermore D.M., Stephens P., Hall L.M. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet. 2001;357:1325–1328. doi: 10.1016/S0140-6736(00)04519-0. [DOI] [PubMed] [Google Scholar]
- 40.Paczosa M.K., Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsieh P.F., Wu M.C., Yang F.L., Chen C.T., Lou T.C., Chen Y.Y., Wu S.H., Sheu J.C., Wang J.T. D-galactan II is an immunodominant antigen in O1 lipopolysaccharide and affects virulence in Klebsiella pneumoniae: Implication in vaccine design. Front. Microbiol. 2014;5:608. doi: 10.3389/fmicb.2014.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Martino P., Cafferini N., Joly B., Darfeuille-Michaud A. Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 2003;154:9–16. doi: 10.1016/S0923-2508(02)00004-9. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y.T., Chang H.Y., Lai Y.C., Pan C.C., Tsai S.F., Peng H.L. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Lam M.M., Wyres K.L., Judd L.M., Wick R.R., Jenney A., Brisse S., Holt K.E. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018;10:77. doi: 10.1186/s13073-018-0587-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorsa L.J., Dufke S., Heesemann J., Schubert S. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: Evidence for horizontal transfer of a chromosomal virulence factor. Infect. Immun. 2003;71:3285–3293. doi: 10.1128/IAI.71.6.3285-3293.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma L.C., Fang C.T., Lee C.Z., Shun C.T., Wang J.T. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J. Infect. Dis. 2005;192:117–128. doi: 10.1086/430619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.