Abstract
Background
The sinus node (SN) is the primary pacemaker of the heart. SN myocytes possess distinctive action potential morphology with spontaneous diastolic depolarization because of a unique expression of ion channels and Ca2+‐handling proteins. MicroRNAs (miRs) inhibit gene expression. The role of miRs in controlling the expression of genes responsible for human SN pacemaking and conduction has not been explored. The aim of this study was to determine miR expression profile of the human SN as compared with that of non‐pacemaker atrial muscle.
Methods and Results
SN and atrial muscle biopsies were obtained from donor or post‐mortem hearts (n=10), histology/immunolabeling were used to characterize the tissues, TaqMan Human MicroRNA Arrays were used to measure 754 miRs, Ingenuity Pathway Analysis was used to identify miRs controlling SN pacemaker gene expression. Eighteen miRs were significantly more and 48 significantly less abundant in the SN than atrial muscle. The most interesting miR was miR‐486‐3p predicted to inhibit expression of pacemaking channels: HCN1 (hyperpolarization‐activated cyclic nucleotide‐gated 1), HCN4, voltage‐gated calcium channel (Cav)1.3, and Cav3.1. A luciferase reporter gene assay confirmed that miR‐486‐3p can control HCN4 expression via its 3′ untranslated region. In ex vivo SN preparations, transfection with miR‐486‐3p reduced the beating rate by ≈35±5% (P<0.05) and HCN4 expression (P<0.05).
Conclusions
The human SN possesses a unique pattern of expression of miRs predicted to target functionally important genes. miR‐486‐3p has an important role in SN pacemaker activity by targeting HCN4, making it a potential target for therapeutic treatment of SN disease such as sinus tachycardia.
Keywords: ion channels, microRNAs, pacemaker of the heart, sinus node disease
Subject Categories: Basic Science Research, Translational Studies, Mechanisms, Ion Channels/Membrane Transport, Gene Therapy
Nonstandard Abbreviations and Acronyms
- AP
action potential
- HCN
hyperpolarization‐activated cyclic nucleotide‐gated
- IPA
Ingenuity Pathway Analysis
- miR
microRNA
- qPCR
quantitative polymerase chain reaction
- RA
right atrium
- SN
sinus node
- Tbx
T‐box
- 3′‐UTR
3 prime untranslated region
Clinical Perspective
What Is New?
This is the first study to investigate the expression of key microRNAs and their predicated targets important for pacemaking in the human heart.
MicroRNA‐486‐3p directly inhibits hyperpolarization‐activated cyclic nucleotide‐gated 4 and thereby reduces action potential generation by the sinus mode.
What Are the Clinical Implications?
MicroRNA expression has an important role in establishing tissue‐specific gene expression profiles in the sinus node and enhances our understanding of the molecular makeup of the human sinus node and its function in health and disease.
The effect of microRNA‐486‐3p on sinus node beating rate makes it a potential target for manipulating pacemaking and could have therapeutic implications for the treatment of inappropriate sinus node tachycardia and for biological pacemaker development.
The sinus node (SN) is the primary pacemaker of the heart and is located at the junction of the superior vena cava with the right atrium (RA). It is an extensive crescent shaped structure and its 3‐dimensional anatomy has recently been shown within the whole ex vivo human heart. 1 , 2 , 3 , 4 The SN myocytes possess distinctive action potential morphology, with a phase (phase 4) of slow, spontaneous, diastolic depolarization ultimately responsible for pacemaking. A unique expression of ion channels and Ca2+‐handling proteins in the SN (described in the human by Chandler et al 5 ) is responsible for 2 main mechanisms that synergistically generate this pacemaker potential during phase 4—the membrane voltage and Ca2+ clocks. 6
Recent extensive work on non‐coding molecules has begun to recognise their robust gene regulatory functions. One family of non‐coding molecules are the microRNA (miR) family. miRs are ≈18 to 24 nucleotide single stranded RNAs, which regulate mRNA translation into functional protein through post‐transcriptional repression 7 by complementary nucleotide binding to the 3 prime untranslated region (3′UTR) recruiting the target gene into RNA‐induced silencing complexes to be rendered translationally incompetent. miRs negatively regulate gene expression at the post‐transcriptional level by degradation of mRNA or translational repression, 8 and have been implicated in cardiac development and pathophysiological processes such as cardiac hypertrophy, fibrosis, arrhythmias, and heart failure. 9 , 10 , 11 , 12 , 13 There is some evidence that miRs are involved in the SN function: in the mouse, the key pacemaker gene, HCN4 (hyperpolarization‐activated cyclic nucleotide‐gated 4), is under the control of miRs following athletic training. 14 The aim of this work was to determine if differences in miR expression between the SN and neighboring atrial muscle in the human can explain the differences in the expression of pacemaker genes in the 2 tissues and why the SN shows pacemaking and the atrial muscle does not.
Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Human Tissue
The human specimens used in this study are described in detail in Table S1. They were obtained, dissected and frozen by co‐authors in Australia (PM) and Romania (FP) under their local ethical approval procedures. No informed consent was required. Specimens were stored under the Human Tissue Act 2004.
Upon arrival of donor hearts to the Prince Charles Hospital (in a cardiologic solution on ice) and cadavers (10–35 hours from death) to National Institute of Legal Medicine right atrial/venocaval blocks (an example is shown in Figure 1C) were removed and trimmed, immediately frozen in liquid nitrogen or −60°C liquid isopentane and stored in freezers (−60° to −80°C). Specimens were transported on dry ice to the University of Manchester where the frozen samples were stored at −80°C.
Figure 1. MicroRNAs significantly more or less expressed in the human SN in comparison with right atrial muscle.
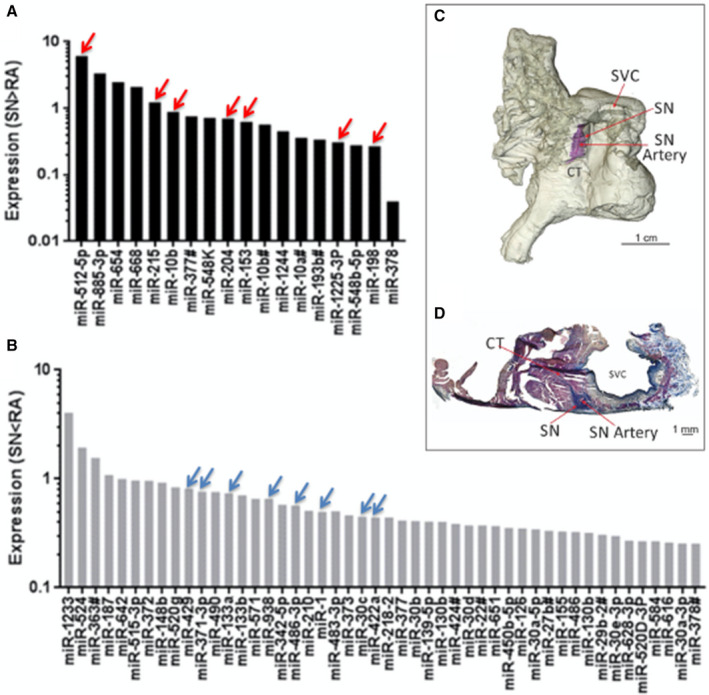
A, Expression of 18 microRNAs that are significantly more abundant in the sinus node (SN) compared with atrial muscle. B, Expression of 48 microRNAs that are significantly less abundant in the SN compared with atrial muscle (n=7; P<0.05). C, Three‐dimensional model of the human right atrium showing the location of the SN around the SN artery (similar to Stephenson et al, 2017 study). 1 The SN is stained blue by the Masson trichrome stain, because of its high content of fibrous tissue. D, Masson trichrome stained section through the SN cut perpendicular to the crista terminalis showing the location of the SN around the SN artery. See Table for key microRNAs indicated by red and blue arrows. CT indicates crista terminalis; miR, microRNA; SN, sinus node; and SVC, superior vena cava.
Animal Tissue
Male Wistar‐Hanover rats (Charles River UK Ltd.; 230–250 g) were euthanized in accordance with the guidelines of the Animal (Scientific Procedures) Act 1986 and the local ethical committee of the University of Manchester.
Histology
To identify the location of the SN, histology was performed on 10‐ to 30‐μm thick cryosections, which were cut perpendicular to the crista terminalis from each specimen listed in Table S1. The cryosections were stained with Masson trichrome to identify the location of the SN as previously described 15 , 16 (Figure 1). Sections were fixed overnight at room temperature in Bouin solution (Sigma) and then rinsed 3 times in 70% alcohol for 10 minutes. Sections were: stained with Celestine blue for 5 minutes and washed in tap water for 10 minutes; stained with Cole alum hematoxylin for 10 minutes and washed for 15 minutes in tap water; stained with acid fuchsin for 5 minutes and washed for 30 minutes in tap water; and placed in phosphomolybdic acid for 5 minutes, drained and then stained with methyl green for 1 minute and washed for 20 minutes in tap water. Sections were then dehydrated using a graded series of alcohol as follows: 70% alcohol (1 minute), 90% alcohol (1 minute), 100% alcohol (twice for 2 minutes). Finally, sections were placed in histoclear solution for 5 minutes and mounted with distyrene, plasticizer, and xylene (Thermo Fisher). Histological sections were visualized with a light microscope (Zeiss LSM5) and an Axiocam camera (Zeiss) and collected with Axiovision software (Zeiss).
Immunohistochemistry
To further confirm the location of the SN, immunohistochemistry experiments were performed on sections neighbouring those used for histological verification. Tissue sections were fixed in 10% neutral buffer formalin (Sigma) for 30 minutes and washed 3 times for 10 minutes in 0.01 mol/L PBS (Sigma). Tissue sections were then treated with 0.1% Triton X‐100 for 30 minutes, washed 3 times for 10 minutes in PBS and then blocked with 1% bovine serum albumin (Sigma) in PBS for 1 hour at room temperature. Sections were then incubated overnight at room temperature with primary antibodies diluted in 1% bovine serum albumin in PBS. The antibodies used in immunohistochemical experiments are listed in Table S2. Sections were washed 3 times in PBS after incubation with the primary antibody and then incubated with a secondary antibody conjugated to fluorescence markers diluted 1:100 in 1% bovine serum albumin in PBS for 2 hours at room temperature. The sections were washed 3 times in PBS and mounted using Vectashield mounting medium (Vector Laboratories) and coverslips sealed. Immunofluorescence was detected by a confocal microscope (Zeiss LSM5, Zeiss Microscopy) and images were taken with Pascal software (Zeiss Microscopy).
Tissue Sampling and RNA Extraction
The SN area was identified by the presence of the SN artery, a large amount of connective tissue, positive staining for HCN4. After identification of the location of the SN by histology and immunohistochemistry, total RNA was isolated from small SN biopsies taken around the SN artery and an area of right atrial pectinate muscle remote from the SN and flash frozen in liquid N2. 2 The tissue biopsies from the SN and atrial muscle were homogenized with an Ika T10 homogenizer (Ika Werke) for 1 minute, and the mirVana miRNA Isolation Kit (Applied Biosystems) with phenol (Life Technologies) was used for RNA isolation according to the manufacturer's instructions. RNA was treated with deoxyribonuclease (Ambion), and RNA purity (260/280 ratio), concentration, and RNA integrity number were measured using Agilent 2100 Bioanalyzer (Agilent). Quality and quantity of RNA extracted from each specimen can be found in Table S1.
Reverse Transcription, Preamplification, and Quantitative Polymerase Chain Reaction for miRs
One‐hundred and eighty‐five nanograms RNA from each sample were reverse‐transcribed using the TaqMan microRNA Reverse Transcription Kit (ThermoFisher). The product of this reaction (2.5 μL) was preamplified with Megaplex PreAmp Primers (ThermoFisher). The primers were divided into pool A and B, each pool containing 380 stem‐looped reverse transcription primers and TaqMan PreAmp Master Mix (Applied Biosystems) in a 25‐μL polymerase chain reaction. The preamplification cycles were run as follows: 95°C (10 minutes), 55°C (2 minutes), and 75°C (2 minutes) followed by 12 cycles of 95°C (15 seconds) and 60° (4 minutes). The cDNA products for each sample were diluted to 100 μL with 0.1x Tris buffer and EDTA, ethylenediaminetetraacetic acid, molecule (pH=8.0). Ten microliters of the diluted cDNA were used for quantitative polymerase chain reaction (qPCR) using TaqMan Array Human MicroRNA A+B Cards Set v3.0 (ThermoFisher) for 754 human microRNAs. 7900HT Fast Real‐Time PCR System (Applied Biosystems) was used for qPCR. The reaction conditions were as follows: 92°C for 10 minutes, 40 cycles of 97°C for 1 second, 60°C for 20 seconds. RQ Manager (Applied Biosystems) was used to obtain the average threshold cycle values. RealTime StatMiner (Integromics) was used for differential expression analysis. GeNorm stability assessment of the suitability of the housekeepers for the analysis of cards A and B was used. Housekeepers RNU44 and RNU48 were selected to analyze card A; RNU44 and U6 small nuclear RNA were used for card B. Statistical analysis of the expression levels was performed using Benjamini‐Hochberg test and P<0.05 values were assumed as significant.
Reverse Transcription and qPCR for mRNAs
Total RNA was extracted using RNeasy Micro Kit (Qiagen) according to manufacturer's instructions. Two hundred and fifty‐four ng RNA from each sample was reverse transcribed to cDNA with SuperScript VILO cDNA Synthesis Kit (ThermoFisher) in 20 μL reactions. The cDNA samples were run on a qPCR Veriti 96‐well thermal cycler (Life Technologies) in accordance with the manufacturer's recommended protocol. HCN4 and voltage‐gated calcium channel (Cav)1.3 Quantitect primers were used (Qiagen) for qPCR using a 7900HT Fast Real‐Time PCR System (Applied Biosystems). The reaction conditions were as follows: 50°C for 2 minutes, 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, 60°C for 1 minute. mRNA expression was analyzed using the delta threshold cycle method. RQ Manager (Applied Biosystems) was used to obtain the average threshold cycle values and GAPDH was used for normalization.
Ingenuity Pathway Analysis Bioinformatics
Sixty‐six miRs listed in Table S3 were joined with the mRNA data for the SN and atrial muscle from the study by Chandler et al, 5 and analyzed using Ingenuity Pathway Analysis (IPA, Qiagen) to identify potential interactions and relationships between the miRs and mRNAs involved in the membrane and Ca2+ clock pacemaker mechanisms. IPA uses data from TarBase database, miRecords (mirecords.biolead.org), TargetScan (www.targetscan.org/) and rna22 (cm.jefferson.edu/rna22) to predict if any of the miRs potentially target the mRNAs based on conserved 8mer (≥0.8 conserved branch length) and 7mer sites that match the seed region of each miR.
Next Generation Sequencing
Next generation sequencing was performed at the Genomic Technologies Core Facility at the University of Manchester on RNA samples collected from frozen human SN preparations. SN samples were collected from the area around the SN artery and right atrial muscle samples from the pectinate muscles remote from the SN region. Samples were collected from 3 human specimens (Specimens 8, 9, 10 in Table S1). Quantity and integrity of the RNA samples were measured using a 2200 TapeStation (Agilent Technologies) to ensure their suitability. Subsequently, TruSeq Stranded mRNA assays (Illumina) were used to produce libraries of more stable, single‐stranded cDNA as follows. Total RNA was purified to polyadenylated mRNA via magnetic separation technology, which works through hybridization of covalent interactions of oligo d(T)25 to poly (A) regions present in most eukaryotic mRNA. The mRNA sequences were fragmented into parts via divalent cations at higher temperature, and random primers were used to reverse transcribe the mRNA fragments into single‐stranded cDNA. DNA polymerase and ribonuclease H‐mediated the synthesis of the second cDNA strand produced from RNA oligonucleotides, originating from the 5´ end of the mRNA. The final cDNA library was generated by an addition of a single “A” base, binding of adapters to the fragments and purification and enrichment via a polymerase chain reaction. The cDNA libraries were incorporated into a multiplex system using the adapters; they were then pooled and clustered using a cBlot instrument (Illumina). Optical flow‐cells containing the mRNA samples were then paired‐end sequenced and mRNA was quantified through repeating 76 cycles twice, using a HiSeq4000 instrument (Illumina). Bcl2fastq software (2.17.1.14) was used to generate an mRNA expression database for each individual SN versus atrial muscle and calculate fold difference in expression between the 2 regions.
Luciferase Reporter Gene Assay
Rat cardiac H9C2 cells (ATCC, UK) were maintained in DMEM (Invitrogen), containing 10% fetal bovine serum, and 1% penicillin‐streptomycin. H9C2 cells were seeded at a density of 5×105 cells per well in 24‐well plates 24 hour before the transfection experiment. Cells were transfected with 500 ng HCN4 or CaV1.3 3′UTR‐containing plasmid (see below for description of the plasmid) and 1 μg miR‐486‐3p, scrambled miR, or culture media only. Lipofectamine 2000 (Invitrogen, UK) transfection agent was used in accordance with the manufacturer's instructions. The DNA‐lipofectamine transfection complex was incubated for 20 minutes at room temperature and then 37°C and 5% CO2 for 24 hours, followed by washing with PBS and lysis via passive lysis buffer (Promega) on a rocker for 20 minutes. Luciferase reporter gene activity, which is directly proportional to mRNA expression of the target gene, was assessed using the luciferase assay system (Promega). A Lumat LB9507 luminometer (Berthold Technologies) was used to measure the bioluminescent activity in 10 μL cell lysate. Each assay was performed in quadruplicate and repeated 3 independent times. The luciferase assay activity was normalised to Renilla (Promega) activity and expressed as a ratio.
Sinus Node Preparations Used for Extracellular Potential Recording
Three‐month‐old male Wistar–Hannover rats weighing 230 to 250 g were used. Animals had free access to food and water and were maintained under standard laboratory conditions in a temperature‐controlled room (22°C) with a 12:12 hour light:dark lighting regime. Animals were humanely euthanized by a Schedule 1 procedure (concussion and cervical dislocation) in accordance with UK Home Office regulations under the Animals (Scientific Procedures) Act 1986 under an institutional licence held at the University of Manchester.
Rats were humanely euthanised via CO2 inhalation and cervical dislocation. The heart was dissected out and placed in 37°C Tyrode's solution (containing NaCl 120 mmol/L, CaCl2 1.2 mmol/L, KCl 4 mmol/L, MgSO4·7H2O 1.3 mmol/L, NaH2PO4·2H2O 1.2 mmol/L, NaHCO3 25.2 mmol/L, glucose 5.8 mmol/L). The entire right atrium was then dissected and opened along the anterior atrial wall and anterior superior vena cava so that the posterior intercaval region remained intact (Figure 2A).
Figure 2. Functional effects of microRNA‐486‐3p transfection of ex vivo rat sinus node preparations.
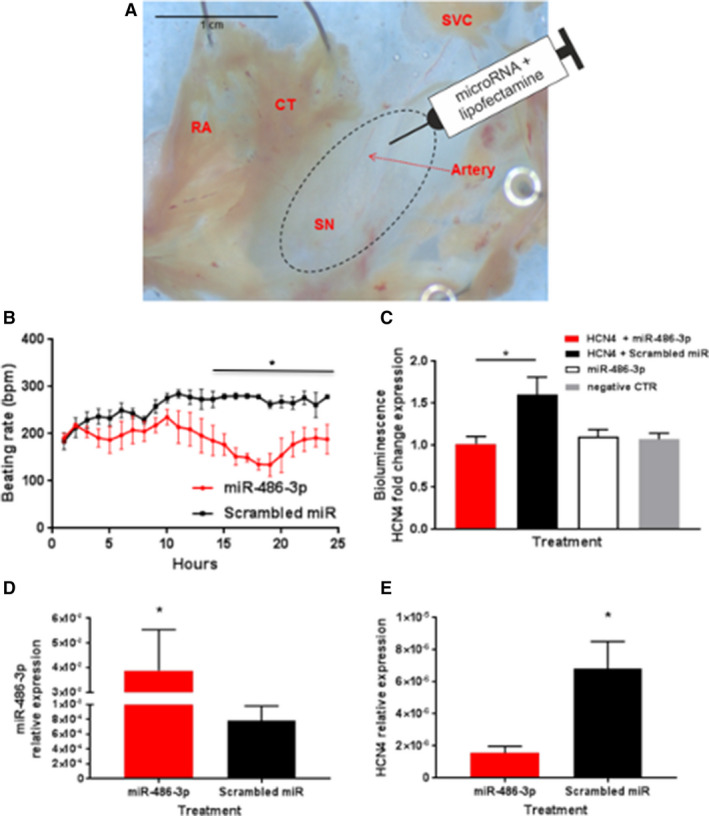
A, Typical sinus node (SN) preparation. Site of injection around the SN artery shown. B, Beating rate of SN preparations in the 24 hours after injection with microRNA‐486‐3p or scrambled microRNA (means±SEM, n=9). C, Luciferase bioluminescence recorded 24 hours after transfection of H9C2 cells with 500 ng with HCN4 3 prime untranslated region ‐containing plasmid and 1 μg microRNA‐486‐3p or scrambled microRNA. As 2 control groups, cells were transfected with microRNA‐486‐3p alone or remained untransfected (ie, an equivalent volume of culture medium was added in place of transfected oligos or 3 prime untranslated region plasmid). These 2 control groups did not include the luciferase reporter. Means±SEM shown (n=3 batches of cells with 4 replicates; *P≤0.05). D and E, Quantitated polymerase chain reaction experiments showing expression of microRNA‐486‐3p, (D) and HCN4 (hyperpolarization‐activated cyclic nucleotide‐gated). (E) mRNA in SN preparations 24 hours after microRNA‐486‐3p transfection (means±SEM; n=4; *P≤0.05). CT indicates crista terminalis; HCN4, hyperpolarization‐activated cyclic nucleotide‐gated; miR, microRNA; SN, sinus node; and SVC, superior vena cava.
The dissection medium was changed to Modified Eagle's Minimum Essential Medium, containing 5% fetal bovine serum (Life Technologies), and preparations incubated at 37°C/5% CO2. The culture medium was changed 8 hours after injection (see below) to Advanced DMEM/F‐12 medium (Life Technologies), containing 10% fetal bovine serum and 1% penicillin‐streptomycin (Sigma). Preparations were kept for 24 hours.
Sinus Node Preparations Used for Sharp Microelectrode Recordings
Ten‐week‐old male Wistar rats weighing 280 to 300 g were used. Animals were kept in an animal house and had free access to food and water and were maintained under standard laboratory conditions with a 12:12 hour light:dark lighting regime. Animals were humanely culled. They were heparinized (100 IU/100 g, intraperitoneal injection), anesthetized with isoflurane (3.5%) and decapitated in accordance with European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes (Council of Europe No 123, Strasbourg 1985) and approved by the Ethics Committee of the National Medical Research Center of Cardiology Institute of Experimental Cardiology.
The chest was opened RA with intercaval SN region was rapidly excised and pinned with endocardial side up to the bottom of a 5 mL perfusion chamber filled with physiological (Tyrode) solution of the following composition (in mmol/L): NaCl 118.0, KCl 2.7, NaH2PO4 2.2, MgCl2 1.2, CaCl2 1.2, NaHCO3 25.0, glucose 11.0, pH 7.4±0.2 bubbled by 95% O2 and 5% CO2 gas mixture. The constant perfusion with flow rate of 10 mL/min at 37°C was started immediately after the preparation.
Spontaneously evoked SN pacemaker derived action potentials (APs) were recorded with glass microelectrodes (10–20 MΩ) filled with 3 mol/L KCl, connected to Warner intracellular electrometer (IE‐210, Warner Instruments, USA) from the endocardial side of the preparations. Signal was digitized at 10 kHz sampling rate with analog‐digital converter (E‐154, ADC L‐card, Russia). The rate of spontaneous AP was calculated using PowerGraph (PowerGraph 3.3 Professional, version 3.3.8, DISoft) and MiniAnalysis software (Synaptosoft, USA, version 6.0.7). The preparations were equilibrated for 30 minutes before recording of control APs in 3 to 4 mm2 region surrounding SN artery bifurcation. Only APs with a diastolic depolarization (35–55 mV/s) and a slow rate of the AP upstroke (<15 V/s) were considered as pacemaker.
After the control recording, the SN region was injected with Tyrode solution (control), lipofectamine or miR‐486‐3p transfection mixture. The 5 µL of transfection mixture was delivered via glass microelectrode (tip diameter <50 μm) connected to Narishige micromanipulator and microinjection syringe pump (Harward Apparatus, PHD ultra) with a constant rate 1 μL/min, the injection was repeated for 5 times to cover the 2 to 4 mm2 of SN region and to deliver 25 μL of the transfection mixture in total. Pacemaker APs from the same sites in SN preparations were recorded at least for 5 minutes immediately, 2, 4, and 6 hours after the injections. The rate of spontaneous APs as beats per minute was calculated.
Transfection Injection
≈2 μL transfection mixture, containing 6 μL 1x Modified Eagle's Minimum Essential Medium, reduced serum medium (Life Technologies), 1.5 μL Lipofectamine RNAmix (Life Technologies), and 2.5 μL miR‐486‐3p (MC12986; Life Technologies), was injected at the bifurcation of the SN artery with a 10 μL NanoFil syringe (World Precision Instruments). Control preparations were transfected with Cy3‐labeled pre‐miR negative control (ThermoFisher). All transfection was performed immediately after dissection of the preparations at the time of the initial culture. The beating rate was measured 24 hours later, ie, post injection.
Plasmids
Human HCN4 (NCBI Reference Sequence: NM_005477.2; HmiT088528‐MT06) and Cav1.3 (NCBI Reference Sequence: NM_000720.2; HmiT054373‐MT06) 3′UTR‐containing plasmids were purchased from GeneCopoeia (Rockville, MD, USA). The pEZX‐MT06 luciferase miR expression vector contained reporter genes for luciferase and Renilla luciferase. The amplification of the plasmids was performed as follows: DH5α Escherichia coli cells (Sigma) were transformed with the plasmids. A single bacterial colony transfected with the plasmid was incubated in 2 mL LB medium, containing 100 μg/mL ampicillin overnight at 37°C, shaking at 150 rpm. Plasmid DNA was purified from the transduced E coli using PureLink Plasmid Kit (Thermo Fisher) according to the manufacturer's protocol. Restriction digest, to confirm the presence of the correct ligation of the miR 3′UTR inserts in the pEZX‐MT06 vector, was then performed. One microgram of each plasmid was incubated with restriction endonuclease enzymes EcoRV and HindIII (New England Biolabs) overnight at 37°C. DNA gel electrophoresis was then performed to confirm the presence of the expected DNA fragments.
Extracellular Potential Recording
To record and monitor automaticity of the ex vivo SN preparations, extracellular potentials were recorded from the right atrial appendage using 2 0.15‐mm diameter stainless steel electrodes (ADInstruments) as previously described by Morris et al. 17 In addition, the culture medium surrounding the preparation was grounded with a 0.15 mm wire earth electrode. Extracellular potentials were continuously recorded using PowerLab 4/35, 4‐channel recorder, and LabChart v7 software (ADInstruments). The average beating rate of the SN preparation was calculated via the detection of a deflection >2 SD of a paced beat. Recordings were collected for 24 hours and the SN preparation retained for further experiments.
Statistical Analysis
Mean±SEM values are shown. Significant differences were identified with 1‐way ANOVA and/or paired t tests. A difference was assumed to be significant at P≤0.05 or P≤0.001.
The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the article as written.
The presented data can be available from the corresponding author upon request.
Results
Differential MicroRNA Expression in Human Sinus Node and Atrial Muscle
The expression profile of miRs in the SN and neighboring right atrial muscle was mapped using qPCR and a TaqMan assay for human miRs. Out of the 754 human miRs examined, 18 were significantly more abundant (Figure 1A) and 48 significantly less abundant (Figure 1B) in the SN than the atrial muscle. The location of the SN tissue in which the miR expression was measured is shown in Figure 1C and 1D and the miRs that are significantly differentially expressed in the SN and atrial muscle are summarized in Table S3.
Ingenuity Pathway Analysis
IPA software was used to predict which of the miRs that are differentially expressed between the SN and atrial muscle may be involved in the differential expression of key ion channels, Ca2+‐handling molecules and connexins involved in the regulation of the membrane, and Ca2+ clock pacemaker mechanisms. The predictions were based on miR‐to‐mRNA sequence binding probability, in combination with miR expression data for the human SN and atrial muscle from this study (Table S3) and expression of mRNA for key ion channels, Ca2+‐handling proteins and connexins for the human SN, and atrial muscle from Chandler et al, 2009; upregulation of an miR is expected to lead to downregulation of its target mRNA and vice versa. TargetScan Human and/or TarBase software was used to predict target mRNA; rna22 software was used to predict the number of binding sites for an miR on an mRNA. The predicted miR‐mRNA relationships are summarized in Table and are shown graphically in Figure 1 (arrows). Of the 18 miRs that were more abundant in the SN than the atrial muscle, 7 were predicted to target physiologically relevant pacemaker mRNAs that were expressed at lower levels in the SN compared with the atrial muscle (Figure 1 arrows; Table). The 7 miRs were: miR‐10b‐5p, predicted to target connexin 40; miR‐153, predicted to target Cav1.2, ryanodine receptor 2, and Nav1.5; miR‐198, predicted to target Cav1.2, ryanodine receptor 2, and ether‐a‐go‐go‐related gene (ERG); miR‐204, predicted to target Cav1.2; miR‐215, predicted to target Kv1.4; miR‐512‐5p, predicted to target connexin 43; and miR‐1225‐3p, predicted to target Nav1.5 (Figure 1, arrows). Of the 48 miRs that were more abundant in the atrial muscle than the SN, 8 were predicted to target physiologically relevant pacemaker mRNAs that were expressed at lower levels in the atrial muscle compared with the SN (Figure 1, arrows; Table). The 8 miRs were: miR‐1‐3p, predicted to target HCN1, HCN4 and T‐box (Tbx)3; miR‐30c, predicted to target HCN1 and Cav1.3; miR‐133a, predicted to target HCN4; miR‐371‐3p, predicted to target Cav1.3 and connexin 45; miR422a, predicted to target Tbx3; miR‐429, predicted to target Tbx18; miR‐486‐3p, predicted to target Cav1.3, HCN1, and HCN4; and miR‐938, predicted to target Cav1.3 (Figure 1, arrows).
Table 1.
Summary of Predicted MicroRNA‐mRNA Interactions in the Human Sinus Node
| miR | Expression of miR in Sinus Node (vs Right Atrial Muscle) | Predicted Target | Expression of Target mRNA in Sinus Node (vs Right Atrial Muscle) | No. of Binding Sites for miR on Predicted Target mRNA |
|---|---|---|---|---|
| miR 1‐3p | ↓ | Tbx3 | ↑ | 1 |
| HCN1 | ↑ | 1 | ||
| HCN4 | ↑ | 1 | ||
| miR 10b‐5p | ↑ |
Gja5 (Connexin 40) |
↓ | 1 |
| miR 30c‐5p | ↓ | HCN1 | ↑ | 1 |
| HCN4 | ↑ | 1 | ||
|
Cacna1g Cav1.3 |
↑ | 1 | ||
| miR 133a‐3p | ↓ | HCN4 | ↑ | 5 |
| miR 153‐3p | ↑ | Scn5a (Nav1.5) | ↓ | 1 |
|
Cacna1c (Cav1.2) |
↓ | 1 | ||
| RyR2 | ↓ | 2 | ||
| miR 198 | ↑ | Cacna1c (Cav1.2) | ↓ | 2 |
| Kcnh2 (ERG) | ↓ | 7 | ||
| RyR2 | ↓ | 1 | ||
| miR 204‐5p | ↑ | Cacna1c (Cav1.2) | ↓ | 1 |
| miR 215‐5p | ↑ | Kcna4 (Kv1.4) | ↓ | 1 |
| miR 371‐3p | ↓ | Cacna1d (Cav1.3) | ↑ | 1 |
| Gjc1 (Connexin 45) | ↑ | 5 | ||
| miR 422a | ↓ | Tbx3 | ↑ | 1 |
| miR 429 | ↓ | Tbx18 | ↑ | 1 |
| miR 486‐3p | ↓ | HCN1 | ↑ | 6 |
| HCN4 | ↑ | 7 | ||
| Cacna1d (Cav1.3) | ↑ | 6 | ||
| Cacna1g (Cav3.1) | ↑ | 1 | ||
| miR 512‐5p | ↑ | Gja1 (Connexin 43) | ↓ | 1 |
| miR 938 | ↓ | Cacna1d (Cav1.3) | ↑ | 9 |
| miR 1225‐3p | ↑ | Scn5a (Nav1.5) | ↓ | 2 |
The data are based on the expression of miRs (microRNAs) from this study and the expression of selected mRNAs important for pacemaking from Chandler et al. 5 The columns from left to right show the miRs, expression of the miRs in the sinus node vs the right atrial muscle, the gene names (ion channel names shown in parenthesis) of targets predicted by TargetScan Human and/or TarBase software, expression of the predicted targets in the sinus node vs the atrial muscle, and the number of potential binding sites on the target for each miR (predicted by rna22 software). Note that upregulation of an miR is expected to result in a downregulation of the target and vice versa. ERG indicates ether‐a‐go‐go‐related gene; Gja, gap junction protein ahpha 5; HCN1, hyperpolarization‐activated cyclic nucleotide‐gated, miR, microRNA; RyR, ryanodine receptor 2; and Tbx, t‐box.
miR‐486‐3p Effect on SN Beating Rate
Four miRs were predicted to target the pacemaker channel, HCN4, and the L‐type Ca2+ channel, Cav1.3, (Table) and this may reflect the importance of these pacemaker channels. Three miRs were predicted to target the alternative pacemaker channel, HCN1, and the alternative L‐type Ca2+ channel, Cav1.2 (Table). Two miRs were predicted to target the Na+ channel, Nav1.5, the ryanodine receptor, ryanodine receptor 2, and the SN transcription factor, Tbx3 (Table). Finally, 1 miR was predicted to target a T‐type Ca2+ channel, Cav3.1, 2 K+ channels (Kv1.4 and ERG), 3 connexins (40, 43, and 45) and the SN transcription factor, Tbx18 (Table). Arguably the most important pacemaker channel in the SN is HCN4 and consequently it was of special interest. Of the miRs potentially targeting HCN4, miR‐486‐3p was of special interest, because it was predicted by IPA to have 7 binding sites on HCN4 mRNA in the human (Table). To confirm that miR‐486‐3p can target HCN4, experiments were performed on the ex vivo rat SN preparation. McGahon et al showed that the human miR‐486‐3p sequence is conserved in other species, including the rat. 18 Also analysis showed that rat HCN4 mRNA has predicted binding sites for miR‐486‐3p. This suggests that the ex vivo rat SN preparation is suitable to validate HCN4 mRNA as a target of miR‐486‐3p. miR 486‐3p (in a transfection mixture) was injected into the SN (Figure 2A) and the preparation was maintained in culture for 24 hours. Preparations injected with a non‐functional scrambled miR were used as controls. miR‐486‐3p significantly reduced the beating rate of the ex vivo rat SN preparations by ≈35% 15 hours after transfection and this change was maintained over the rest of the 24‐hour period of culture (Figure 2B). The effect of miR‐486‐3p on SN beating rate appeared to develop a few hours after transfection (Figure 2B).
In another set of experiments, using sharp microelectrode recordings of the electrical activity in the RA/SN preparations, we observed that after injections of the SN region (n=5) with miR‐486‐3p, the rate of the SN‐derived spontaneous APs in the RA decreased gradually from 250±30 down to 32±21 beats per minute±SEM (Figure 3). This gradual reduction was observed from 2 hours after injection and was maximal at 6 hours post‐injection. The rate of APs was only 13±9% (P<0.0001, n=5) of the initial rate before injections calculated both on the basis of the SN or RA recordings (Figure 3). After injections with Tyrode (n=5) and lipofectamine (n=5) there was no change to the rate of the SN‐derived spontaneous APs (Figure 3).
Figure 3. The intracellular recordings of action potentials from rat sinus node preparations after microRNA‐486‐3p transfection ex vivo.
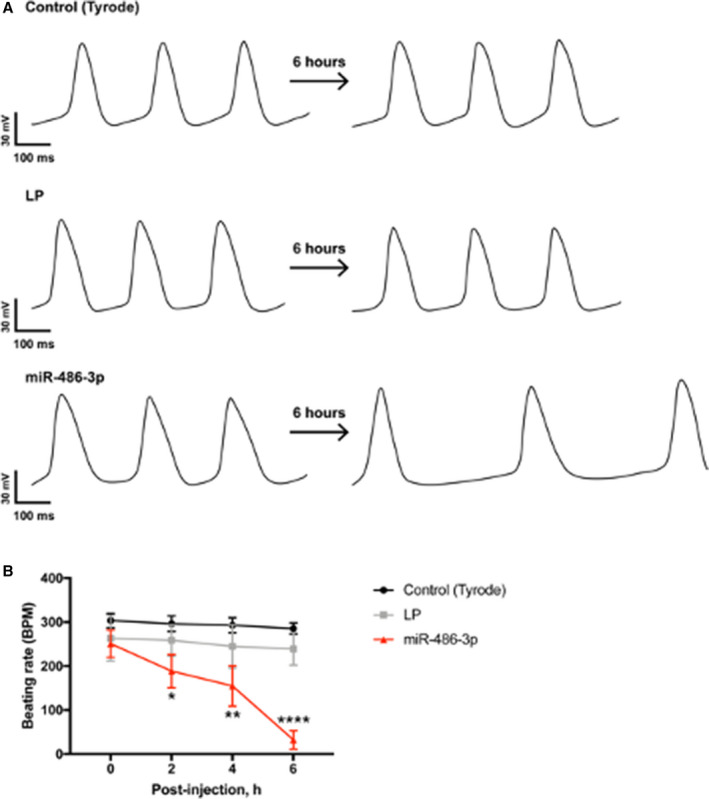
A, Representative example of action potentials recorded intracellularly in the sinus node before and 6 hours after injection with Tyrode (control), lipofectamine, and microRNA‐486‐3p transfection mixture. B, Beating rate of sinus node preparations in 6 hours after the injection (means±SEM, n=5 for each group). AP indicates action potential; LP, lipofectamine; and miR, microRNA. *P<0.05, **P<0.005, ****P<0.0001 (2‐way ANOVA with Tukey multiple comparison test).
miR‐486‐3p Effect on HCN4 Expression
To investigate whether miR‐486‐3p can affect HCN4 expression as predicted, a luciferase reporter gene assay was performed. The predicted binding sites for miR‐486‐3p are in the 3′‐UTR of human HCN4. Therefore, the 3′‐UTR of human HCN4 was introduced as the 3′‐UTR of the luciferase gene. The resulting plasmid was transfected into rat cardiac H9C2 cells. Following transcription and translation, the expression of the luciferase protein (surrogate of HCN4 expression) was measured by the resulting luciferase bioluminescence. Bioluminescence was significantly less from cells transfected with the plasmid and miR‐486‐3p than from cells transfected with the plasmid and scrambled (non‐functional) miR (Figure 2C). As expected, bioluminescence was also low if the cells were not transfected with the HCN4 plasmid but only transfected with miR‐486‐3p or neither transfected with the plasmid nor miR‐486‐3p (Figure 2C). This suggests that miR‐486‐3p can control HCN4 expression.
To confirm that miR‐486‐3p can affect HCN4 expression, experiments were conducted on the ex vivo rat SN preparation. Preparations were injected (Figure 2A) with miR‐486‐3p or scrambled miR. As expected, miR‐486‐3p expression was greater in preparations in which the SN was transfected with miR‐486‐3p rather than scrambled miR (Figure 2D). HCN4 mRNA expression was significantly reduced in preparations injected with miR‐486‐3p (as compared with preparations injected with the scrambled miR). This shows that miR‐486‐3p controls HCN4 mRNA expression in the sinus node (miR‐486‐3p must promote HCN4 mRNA degradation). In the same ex vivo rat SN preparations, HCN4 protein expression was assessed by immunolabeling of HCN4 protein in thin cryosections through the SN (Figure 4A, 4B, and 4E). Labeling was significantly reduced in preparations injected with miR‐486‐3p (as compared with preparations injected with the scrambled miR) (Figure 4A, 4B, and 4E). Cell membrane preservation and integrity over the 24‐hour incubation period was confirmed by Caveolin‐3 immunolabeling (Figure 4C, 4D, and 4F).
Figure 4. Effect of exogenous expression of microRNA‐486‐3p on HCN4 protein expression in the rat sinus node.
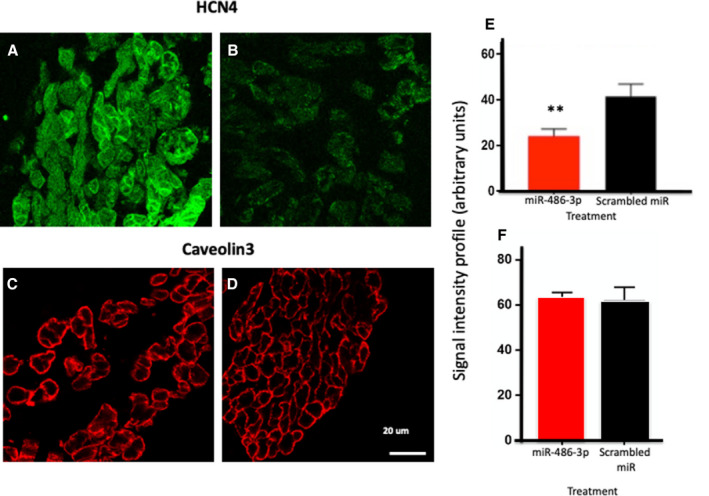
A and B, Immunolabeling of HCN4 (hyperpolarization‐activated cyclic nucleotide‐gated) protein in the rat sinus node before (A) and after (B) 24 hours after transfection with microRNA‐486‐3p. C and D, Immunolabeling of Caveolin‐3 (plasma membrane marker) in the rat sinus node before (A) and after (B) 24 hours after transfection with scrambled microRNA. E, HCN4 protein signal intensity in the rat sinus node 24 hours after transfection with microRNA‐486‐3p or scrambled microRNA (means±SEM; n=5; **P≤0.005). F, Caveolin‐3 protein signal intensity in the rat sinus node 24 hours after transfection with microRNA‐486‐3p or scrambled microRNA (means±SEM; n=4; no significance difference was observed). HCN4 indicates hyperpolarization‐activated cyclic nucleotide‐gated; and miR, microRNA.
Transcription Factors are Also Involved in Differential Gene Expression Between the Human Sinus Node and Atrial Muscle
Differential gene expression between the human sinus node and atrial muscle will not exclusively be the result of a differential expression of miRs—a differential expression of transcription factors will also be involved. Expression of transcription factors was investigated using next generation sequencing. The transcriptomes of 3 human SN and corresponding right atrial muscle samples were sequenced. The expression of the 3060 most abundant human mRNAs was investigated and principal component analysis confirmed distinct SN and atrial muscle mRNA profiles (Figure 5A); 1238 mRNAs had significantly higher expression in the SN (log2 fold change >1, P<0.05), 1357 had significantly higher expression in the atrial muscle (log2 fold change <1, P<0.05), and 465 were not significantly different between the 2 tissues (−1 <log2 fold change >1) (Figure 5B, red dots). IPA software was used to identify transcription factors and potential relationships with either miRs or mRNAs in this study; 68 transcription factors were significantly more expressed in the SN, and 60 were more significantly expressed in the atrial muscle. Six of the differentially expressed transcription factors have potential relationships with either the miRs or mRNAs in this study. These are Islet1, short stature homeobox 2, Tbx3, and Tbx18, which were significantly more highly expressed in the SN, and Tbx5 and NK2 homeobox 5, which were significantly more highly expressed in the atrial muscle (Figures 5B and 6). The predicted relationships between these transcription factors and either miRs or mRNAs are shown in Figure 7. This network shows 24 potential links (direct or indirect) between miRs and ion channels, but only 3 between transcription factors and ion channels. This raises the question of whether much of the ion channel regulation occurs at the post‐transcriptional level via miRs. However, miRs themselves are under the control of transcription factors as shown in Figure 7.
Figure 5. Next generation sequencing for RNAseq data of 3 human sinus node and 3 right atria specimens from the same hearts.
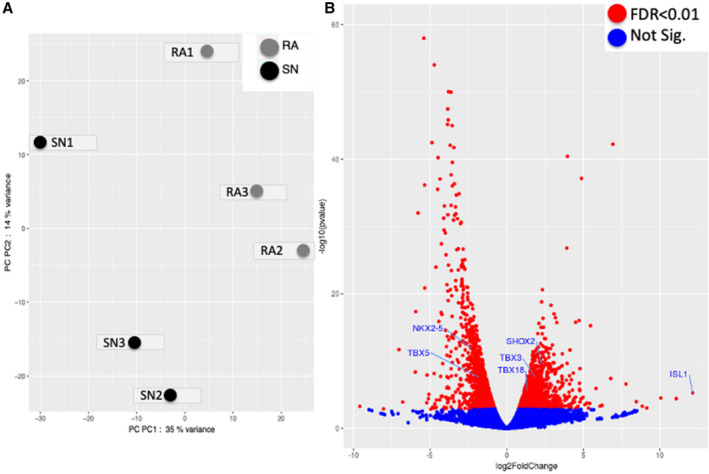
A, Principal component (PC) analysis scatter‐plot of RNAseq data from 3 human sinus node (SN) (black dots) and 3 right atrium (RA) (grey dots) specimens. The graph shows that there are 2 distinct groups of tissue, namely the SN (SN1 to SN3 cluster close on the left of x‐axis) and RA (RA1 to RA3 cluster closer close on the right of x‐axis), that have significant differential gene expression. B, Volcano plot of all RNAseq data with 6 “embryonic” transcription factors annotated on the plot. Log2FoldChange P≤0 indicates genes less expressed in the SN vs RA and vice versa. RA indicates right atrium; and SN, sinus node. Significantly more/less expressed genes (illustrated in red) based on false discover rate <0.01. FDR indicates false discovery rate; RA, right atrium; and sinus node.
Figure 6. “Embryonic” transcription factors significantly more or less expressed in the human adult sinus node in comparison with right atrial muscle.
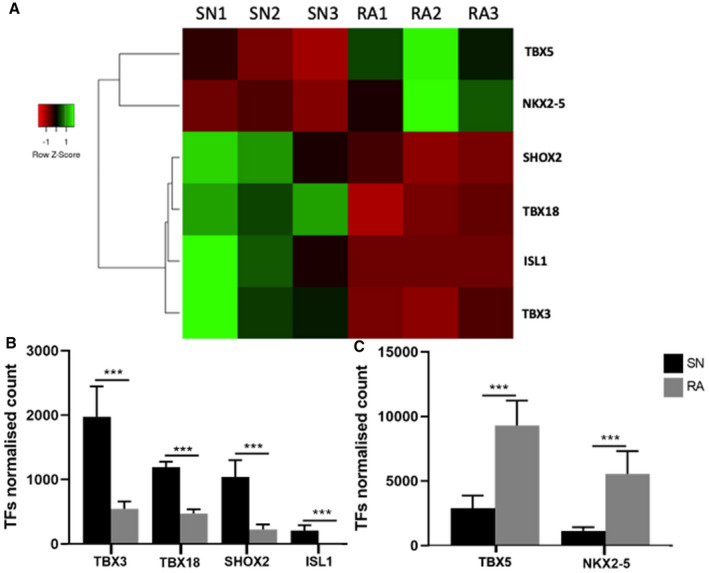
Data were analyzed via Bcl2fastq software (2.17.1.14). Transcription factors (TFs) normalized counts are the mean of the DESeq2 normalized read counts for each TF in the sinus node (SN) and right atrium (RA). A, Heat maps showing relationship across 6 SN and RA samples (SN1‒SN3 and RA1‒RA3) for the expression of 6 TFs. Three SNs cluster together and 3 RAs cluster together for the expression of these 6 TFs. B and C, Means±SEM; n=3; ***P≤0.001 of 6 TFs in SN and RA. Graphs were created via GraphPad Prism 7.0. Nkx2‐5 indicates NK2 homeobox 5; RA, right atrium; Shox; short stature homeobox; SN, sinus node; Tbx, T‐box; and TF, transcription factor.
Figure 7. Ingenuity Pathway Analysis predictions of interactions between “embryonic” transcription factors, microRNAs, ion channels, and gap junction subunits.
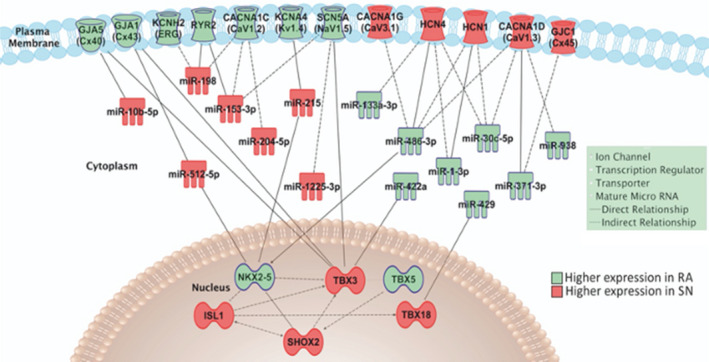
Transcription factors are shown in the nucleus, microRNAs in the cytoplasm, and ion channels in the surface membrane. For simplicity, RYR2 (ryanodine receptor 2) is shown in the surface membrane rather than the sarcoplasmic reticulum. HCN4 indicates hyperpolarization‐activated cyclic nucleotide‐gated; miR, microRNA; NKX2‐5, NK2 homeobox 5; RA, right atrium; SHOX; short stature homeobox; SN, sinus node; and TBX, T‐box.
Discussion
This study shows, for the first time, a distinct expression pattern of miRs in the human SN compared with that of the right atrial muscle that is predicted to affect the expression of target molecules responsible for the pacemaker mechanisms in the heart. This study shows 66 differentially expressed miRs (Figure 1). The differentially expressed miRs are predicted to target mRNAs that have been reported to be differentially expressed in these tissues. 5
In this study, we found 7 miRs (miR‐10b‐5p, miR‐153‐3p, miR‐198, miR‐204‐5p, miR‐215‐5p, miR‐512‐5p, and miR‐1225‐3p) expressed at higher levels in the SN compared with atrial muscle that are predicted to bind to and thus downregulate molecules that are known to be expressed at lower levels in the SN 5 : the Na+ channel, Nav1.5, the L‐type Ca2+ channel, Cav1.2, 2 K+ channels (Kv1.4 and ERG), the RYR2 (ryanodine receptor 2), and 2 connexins (40 and 43). The absence of Nav1.5 (and the corresponding absence of the Na+ current, I Na) in the SN explains why the upstroke of the SN action potential is slow and the absence of the 2 high conductance connexins, 40 and 43, in the SN explains the poor electrical coupling in the SN (essential to protect the SN from the hyperpolarizing influence of the neighboring atrial muscle). 19
Eight miRs (miR‐1‐3p, miR‐30c‐5p, miR‐133a‐3p, miR‐371‐3p, miR‐422a, miR‐429, miR‐486‐3p, and miR‐938) that were expressed at higher levels in the atrial muscle compared with the SN are predicted to inhibit molecules that are important for pacemaking: 2 pacemaker HCN channels (HCN1 and HCN4), 2 pacemaker Ca2+ channels (Cav1.3 and Cav3.1), and the low conductance connexin isoform, connexin 45. If the predicted actions of the miRs are correct, this helps to explain why the atrial muscle does not normally show pacemaker activity; conversely, the low expression of these miRs in the SN helps to explain why the SN does show pacemaking. Of these miRs and their predicted effects, miR‐486‐3p and its predicted effect on HCN4 were the focus, because of the importance of HCN4 and the number of predicted binding sites on HCN4 for miR‐486‐3p. A luciferase reporter gene assay confirmed that miR‐486‐3p can potentially control expression of HCN4 (Figure 2), and miR‐486‐3p, when ectopically expressed in the SN, reduced HCN4 mRNA and protein levels (Figures 2 and 4), and reduced the SN beating rate (Figure 2) In a recent study, we showed that miR‐486‐3p is upregulated in the SN in a mouse model of athletic training. 14 Following athletic training, the downregulation of HCN4 and the corresponding ionic current (I f) in the SN and the consequent sinus bradycardia was attributed to an upregulation of miR‐486‐3p and miR‐423‐5p 14 ; in this study, a luciferase reporter gene assay showed that miR‐486‐3p was able to regulate mouse HCN4 expression—therefore, miR‐486‐3p is able to regulate both human (this study) and mouse 14 HCN4 expression.
In this study, we also identified key “embryonic” transcription factors that are differentially expressed in the adult human SN versus atrial muscle and with predicted links to either the miRs or ion channels of interest in the SN (Figures 5, 6, and 7). This suggests that both transcription factors and miRs are responsible for the unique gene expression pattern of the SN. Furthermore, transcription factors may frequently act via miRs.
Conclusions
The human SN possesses a unique pattern of expression of miRs. Some of the differentially expressed miRs are predicted to target genes that are important for pacemaking, such as HCN1, HCN4, Cav1.3, and Cav3.1. The action of miRs is complex with interactions between multiple miRs, transcription factors, and target genes. It has been confirmed that miR‐486‐3p has an important role in regulating SN pacemaker activity. miR‐486‐3p directly inhibits HCN4 and thereby reduces action potential generation by the SN, making it a potential target for manipulating pacemaking in therapeutic treatment of sinus node disease. For example, inappropriate sinus node tachycardia is currently treated using ivabradine to block HCN4 20 and use of miR‐486‐3p could be alternative strategy. This study provides novel insights into the mechanisms controlling SN gene expression vital for its role as the primary pacemaker in the heart.
Sources of Funding
This work was supported by Leducq Foundation (THE FANTACY 19CVD03), the British Heart Foundation program grant RG/18/2/33392; National Institutes of Health (grants HL115580 and HL135109); Russian Science Foundation grant 19‐15‐00163.
Disclosures
None.
Supporting information
Tables S1–S3
(J. Am. Heart Assoc. 2020;9:e016590 DOI: 10.1161/JAHA.120.016590.)
For Sources of Funding and Disclosures, see page 14.
References
- 1. Stephenson RS, Atkinson A, Kottas P, Perde F, Jafarzadeh F, Bateman M, Iaizzo PA, Zhao JC, Zhang HG, Anderson RH, et al. High resolution 3‐dimensional imaging of the human cardiac conduction system from microanatomy to mathematical modeling. Sci Rep. 2017;7:7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li N, Csepe TA, Hansen BJ, Dobrzynski H, Higgins RS, Kilic A, Mohler PJ, Janssen PM, Rosen MR, Biesiadecki BJ, et al. Molecular mapping of sinoatrial node HCN channel expression in the human heart. Circ Arrhythm Electrophysiol. 2015;8:1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Csepe TA, Zhao J, Hansen BJ, Li N, Sul LV, Lim P, Wang Y, Simonetti OP, Kilic A, Mohler PJ, et al. Human sinoatrial node structure: 3d microanatomy of sinoatrial conduction pathways. Prog Biophys Mol Biol. 2016;120:164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li N, Hansen BJ, Csepe TA, Zhao J, Ignozzi AJ, Sul LV, Zakharkin SO, Kalyanasundaram A, Davis JP, Biesiadecki BJ, et al. Redundant and diverse intranodal pacemakers and conduction pathways protect the human sinoatrial node from failure. Sci Transl Med. 2017;9:eaam5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chandler NJ, Greener ID, Tellez JO, Inada S, Musa H, Molenaar P, DiFrancesco D, Baruscotti M, Longhi R, Anderson RH, et al. Molecular architecture of the human sinus node insights into the function of the cardiac pacemaker. Circulation. 2009;119:1562–1575. [DOI] [PubMed] [Google Scholar]
- 6. Lakatta EG, DiFrancesco D. What keeps us ticking: a funny current, a calcium clock, or both? J Mol Cell Cardiol. 2009;47:157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Griffiths‐Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post‐transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. [DOI] [PubMed] [Google Scholar]
- 9. Boettger T, Braun T. A new level of complexity the role of micrornas in cardiovascular development. Circ Res. 2012;110:1000–1013. [DOI] [PubMed] [Google Scholar]
- 10. Romaine SPR, Tomaszewski M, Condorelli G, Samani NJ. MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart. 2015;101:921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Torrente AG, Mesirca P, Neco P, Rizzetto R, Dubel S, Barrere C, Sinegger‐Brauns M, Striessnig J, Richard S, Nargeot J, et al. L‐type Cav1.3 channels regulate ryanodine receptor‐dependent Ca2+ release during sino‐atrial node pacemaker activity. Cardiovasc Res. 2016;109:451–461. [DOI] [PubMed] [Google Scholar]
- 12. Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79:562–570. [DOI] [PubMed] [Google Scholar]
- 13. Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. [DOI] [PubMed] [Google Scholar]
- 14. D'Souza A, Pearman CM, Wang YW, Nakao S, Logantha S, Cox C, Bennett H, Zhang Y, Johnsen AB, Linscheid N, et al. Targeting miR‐423‐5p reverses exercise training‐induced HCN4 channel remodeling and sinus bradycardia. Circ Res. 2017;121:1058–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chandler N, Aslanidi OV, Buckley D, Inada S, Birchall S, Atkinson A, Kirk D, Monfredi O, Molenaar P, Anderson R, et al. Computer three‐dimensional anatomical reconstruction of the human sinus node and a novel paranodal area. Anat Rec (Hoboken). 2011;294:970–979. [DOI] [PubMed] [Google Scholar]
- 16. Sanchez‐Quintana D, Cabrera JA, Farre J, Climent V, Anderson RH, Ho SY. Sinus node revisited in the era of electroanatomical mapping and catheter ablation. Heart. 2005;91:189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morris GM, D'Souza A, Dobrzynski H, Lei M, Choudhury M, Billeter R, Kryukova Y, Robinson RB, Kingston PA, Boyett MR. Characterization of a right atrial subsidiary pacemaker and acceleration of the pacing rate by HCN over‐expression. Cardiovasc Res. 2013;100:160–169. [DOI] [PubMed] [Google Scholar]
- 18. McGahon MK, Yarham JM, Daly A, Guduric‐Fuchs J, Ferguson LJ, Simpson DA, Collins A. Distinctive profile of isomir expression and novel microRNAs in rat heart left ventricle. PLoS One. 2013;8:e65809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dobrzynski H, Anderson RH, Atkinson A, Borbas Z, D'Souza A, Fraser JF, Inada S, Logantha SJ, Monfredi O, Morris GM, et al. Structure, function and clinical relevance of the cardiac conduction system, including the atrioventricular ring and outflow tract tissues. Pharmacol Ther. 2013;139:260–288. [DOI] [PubMed] [Google Scholar]
- 20. Baruscotti M, Bucchi A, Milanesi R, Paina M, Barbuti A, Gnecchi‐Ruscone T, Bianco E, Vitali‐Serdoz L, Cappato R, DiFrancesco D. A gain‐of‐function mutation in the cardiac pacemaker HCN4 channel increasing cAMP sensitivity is associated with familial inappropriate sinus tachycardia. Eur Heart J. 2017;38:280–288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S3


