Abstract
Acyclovir is the drug of choice for the treatment of herpes simplex virus (HSV) infections. Acyclovir-resistant HSV strains may emerge, especially during long-term drug use, and subsequently cause difficult-to-treat exacerbations. Previously, we set up a novel treatment approach, based on enzymatically synthesized pools of siRNAs, or siRNA swarms. These swarms can cover kilobases-long target sequences, reducing the likelihood of resistance to treatment. Swarms targeting the UL29 essential gene of HSV-1 have demonstrated high efficacy against HSV-1 in vitro and in vivo. Here, we assessed the antiviral potential of a UL29 siRNA swarm against circulating strains of HSV-1, in comparison with acyclovir. All circulating strains were sensitive to both antivirals, with the half-maximal inhibitory concentrations (IC50) in the range of 350–1911 nM for acyclovir and 0.5–3 nM for the UL29 siRNA swarm. Additionally, we showed that an acyclovir-resistant HSV-1, devoid of thymidine kinase, is highly sensitive to UL29 siRNA treatment (IC50 1.0 nM; Imax 97%). Moreover, the detected minor variations in the RNAi target of the HSV strains had no effect on the potency or efficacy of UL29 siRNA swarm treatment. Our findings support the development of siRNA swarms for the treatment of HSV-1 infections, in order to circumvent any potential acyclovir resistance.
Keywords: herpes simplex virus, antiviral, RNAi, siRNA, acyclovir, acyclovir resistance
1. Introduction
Acyclovir (ACV) and other structurally related analogues of deoxyguanosine are well-established antivirals for treatment of herpes simplex virus (HSV) infection, with high specificity and low toxicity [1,2]. In certain recurrent herpes infections, prophylactic ACV treatment is needed to prevent potential exacerbations caused by the reactivation of HSV from its latent state, which the virus establishes after primary, lytic infection. However, if patients require such long-term prophylactic treatment, as is needed by those suffering from herpes keratitis, ACV-resistant HSV strains can emerge not only in immunocompromised, but also in immunocompetent individuals [3,4,5,6]. The ACV resistance of HSV-1 is mainly caused by mutations in the UL23 gene encoding the HSV thymidine kinase [7,8]. The prevalent inter- and intrastrain genetic variability in UL23 among circulating strains of HSV-1 [7] causes differences in ACV sensitivity between and among circulating clinical HSV-1 strains [6,9].
Small interfering RNA (siRNA) drugs are recognized as potential antivirals for treatment of virus infections [10]. Antiviral siRNAs are, however, challenged by their limited target sequence, usually covering only 20–25 base pairs (bp), making them susceptible to variations in viral genomes and to emergence of escape mutants. To enable better tolerance for genetic diversity and better treatment for medication-resistant viral infections, we have developed siRNA swarms, which are pools of enzymatically synthesized Dicer substrate siRNAs that can cover even several kilobases of the viral targets [11]. The individual Dicer substrate siRNA molecules in the swarm are 25 bp long, and upon introduction to the cells, are processed by the endogenous human Dicer, resulting in enhanced efficacy and potency [12,13]. Moreover, siRNA swarms may be directed against multiple viral genes simultaneously, when chimeric constructs are used as sources for the siRNA swarm synthesis [14]. Antiviral siRNA swarms have previously proven efficient against various viruses and viral strains [11,13,14]. Recently, siRNA swarms were developed for increased antiviral efficacy and plasma stability with incorporated modified nucleotides [15].
We have previously developed siRNA swarms against HSV-1 [13,15,16,17,18], which potentially provide novel options for therapy as topical treatments of herpes keratitis and other superficial infections. Of the studied HSV target genes, the essential UL29 gene has proven most suitable [18]. The gene codes for the single stranded DNA-binding protein ICP8, which is required throughout the biphasic DNA replication of HSV-1 [19]. As a target for RNAi, the UL29 gene has proven efficient for various recombinant, reference and clinical HSV-1 strains, both in vitro [18] and in vivo [16,20], with no emerging resistance over multiple passages in vitro [18].
Here, our aim is to assess the antiviral potential of the UL29 siRNA swarm against circulating HSV-1 strains and a thymidine kinase (tk) deficient HSV-1 [21], in comparison with ACV. We show that treatment with the UL29 siRNA swarm inhibits 98% or more of viral production of every studied circulating strain of HSV-1, and that all studied circulating strains were susceptible to the treatment with only minimal deviation in sensitivity. Furthermore, we show that the tk-deficient HSV-1 was susceptible to treatment with the UL29 siRNA swarm, suggesting the potential of UL29-targeted RNAi in treatment of ACV-resistant herpes infections.
2. Materials and Methods
2.1. Cells
Vero cells (CCL-81, ATCC, Manassas, VA, USA), used for all experiments, were maintained in M199 medium (Gibco, Waltham, MA, USA) with 5% fetal bovine serum (FBS) and gentamycin.
2.2. HSV-1 Strains
Seventeen anonymous archival HSV isolates were derived from clinical specimens sent to a local clinical virology diagnostic service (Virus Diagnostic Unit of the Turku University Hospital, research permit #J10/17). The isolates were confirmed as HSV-1 by immunoperoxidase rapid culture assay [22]. In addition to these clinical isolates, HSV-1 reference strains, 17+ and F, as well as an ACV-resistant, tk-deficient strain (HSV-1 (Δ305), [21]) were studied. For a summary of viruses used, please see Table 1. All viruses were propagated into supernatant stocks in Vero cells, as described previously [18]. The stocks of the circulating strains were of low passage (passage #1–4) and had viral titers varying from 3 × 105 to 3 × 108 plaque forming units (pfu) per mL.
Table 1.
HSV-1 strains used in this study.
| Strain Code | Sample No. | Sample | Gender | Genbank Accession 1 |
|---|---|---|---|---|
| HSV-1 (17+) | - | - | - | JN555585.1 |
| HSV-1 (F) | - | - | - | GU734771.1 |
| HSV-1 (Δ305) | - | - | - | - |
| F-14g | H12114 | genital | F | MH999844.1 |
| F-15 | H12115 | - | F | - |
| F-17 | H12117 | blister | F | MH99845.1 |
| F-18g | H12118 | genital | F | MH99847.1 |
| F-19 | H12119 | lip | F | - |
| M-15 | H1215 | blister | M | MH99846.1 |
| M-10 | H15110 | - | M | - |
| F-12 | H15112 | blister | F | - |
| F-15g | H15115 | genital | F | - |
| F-17/ | H15117 | facial | F | - |
| M-19 | H15119 | blister | M | MH999850.1 |
| M-17 | H1517 | genital | M | - |
| M-19/ | H1519 | - | F | - |
| M-01 | H151V1 | blister | - | - |
| M-02 | H151V2 | blister | - | - |
| M-03 | H151V3 | blister | - | - |
| M-04 | H151V4 | blister | - | - |
1 Indicated when the whole genome sequence is available.
2.3. Acyclovir Sensitivity
ACV sensitivity was studied by a plaque reduction assay on Vero cells [23]. For five of the virus isolates (F-14g, F-17, F-18g, M-15, M-19), as well as for HSV-1 (Δ305) and HSV-1 (17+), previously published data of acyclovir sensitivity [9] were used and were thus not repeated here in vitro. The assay was conducted on a 96-well plate in duplicates with infections of 100 pfu/well at four hours post treatment (hpt), and quantified three days post infection (dpi) by counting the plaques. The acyclovir concentrations ranged from 0.03 to 128 µg/mL, corresponding approximately to 0.1 and 570 µM, respectively. The upper limit of susceptibility was considered to be the half maximal inhibitory concentration (IC50) value of 1.90 μg/mL (8.43 μM). The reduction of viral growth (%) by ACV was determined against untreated, infected samples.
2.4. Sensitivity to RNAi with the UL29 siRNA Swarm
The antiviral UL29-specific siRNA swarm, targeting a 653 bp sequence of the essential UL29 gene, was synthesized as previously described [13,17,18]. The transfections of the siRNA swarm, at different concentrations of the siRNA, were performed in 40–60% confluent Vero cell cultures on 96-well plates using Lipofectamine RNAiMAX (#13778-150; Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The concentration of the siRNA swarm ranged from 0.4 to 100 nM, as specified in the Results section. Four hours post treatment (hpt), the cells were infected with 100 pfu of each of the viral strains, as previously described [18]. Our data represent at least five biological replicates per RNA concentration for each virus. At 3 dpi, the virus was quantified by titration of the culture supernatants on Vero cells using six parallel wells for each dilution. The inhibition of virus production by the anti-UL29 siRNA swarm was determined as a reduction of viral growth (%) in comparison with mock samples transfected with water.
2.5. Sequencing of the UL29 Swarm Target of the Strains
The UL29 siRNA swarm target area (nucleotides 59,954–59,302 as according to HSV-1 (17+) sequence, Genbank accession number JN555585.1) was sequenced from the strains of which there was yet no sequence data available. The sequencing of the strains was conducted by amplifying the target sequence from viral stocks using Phusion polymerase (Thermo Scientific, Waltham, MA, USA) and the primers CCTGCACGCTGGGGG and CAGTGCCACGGGGTGTTC. The products were analyzed and purified using agarose gel electrophoresis and submitted for sequencing (LightRun, Eurofins Genomics, Denmark). The siRNA swarm target sequences of the other strains were obtained from Genbank (Table 1). The Genbank accession numbers of the novel target sequences are MW287999-MW288011.
2.6. Data Analysis
The IC50 values and maximum inhibition rates (Imax) were calculated using a nonlinear, sigmoidal fit in Origin version b9.3.2.303 (OriginLab Corporation, Northhampton, MA, USA). The correlation analyses were done using Spearman’s nonparametric correlation analysis in GraphPad Prism version 8.4.3. (GraphPad Software Inc., San Diego, CA, USA).
3. Results
Seventeen HSV-1 strains circulating in Finland (Table 1), two common reference strains, HSV-1 (17+) and HSV-1 (F), as well as a tk-deficient virus HSV-1 (Δ305) [21] were tested for sensitivity to treatment with ACV or a UL29 siRNA swarm. The antiviral assays determining the sensitivities were conducted on Vero cells with 100 pfu per well on a 96-well plate format. UL29 siRNA swarm-related toxicity was evaluated in Vero cells, and was minimal, remaining similar throughout the used siRNA concentration range (Figure S1).
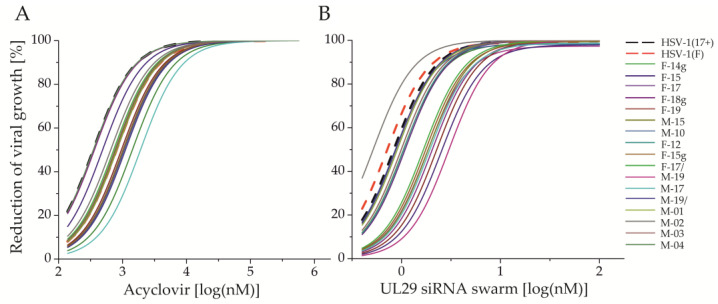
None of the studied circulating HSV-1 strains was resistant to ACV (Table 2) and all were dose-responsive to ACV treatment (Figure 1A). Their ACV sensitivity (half maximal inhibitory concentration, IC50) ranged from 349.7 to 1911.2 nM (0.08 to 0.43 µg/mL), with an average of 844.0 nM (standard deviation, SD 379.1 nM) (Table 2). All studied circulating and reference strains were susceptible to treatment with UL29 siRNA swarm (Table 2). The sensitivity (IC50) of circulating strains to UL29 siRNA swarm treatment ranged from 0.5 nM to 3.0 nM, with an average of 1.3 nM (SD 0.68 nM), whereas the reference strains HSV-1 (17+) and HSV-1 (F) had sensitivities of 0.8 and 0.7 nM, respectively (Table 2). All of the strains demonstrated dose-responsiveness to UL29 siRNA swarm treatment (Figure 1B).
Table 2.
Finnish clinical HSV-1 isolates and their sensitivities (IC50) to ACV or to UL29 siRNA treatment.
| HSV-1 Strain | Sample No. | IC50 ACV (nM) 1 | IC50, UL29 Swarm (nM) 1 |
|---|---|---|---|
| HSV-1 (17+) | - | 349.7 2 | 0.8 |
| HSV-1 (F) | - | 803.3 | 0.7 |
| HSV-1 (Δ305) | - | 25468.8 2 | 1.0 |
| F-14g | H12114 | 1425.8 2 | 1.4 |
| F-15 | H12115 | 488.2 | 1.0 |
| F-17 | H12117 | 995.0 2 | 2.5 |
| F-18g | H12118 | 526.1 2 | 1.1 |
| F-19 | H12119 | 921.1 | 1.1 |
| M-15 | H1215 | 784.9 2 | 2.3 |
| M-10 | H15110 | 1023.2 | 1.9 |
| F-12 | H15112 | 1016.1 | 0.9 |
| F-15g | H15115 | 895.1 | 1.1 |
| F-17/ | H15117 | 527.9 | 0.9 |
| M-19 | H15119 | 644.9 2 | 1.7 |
| M-17 | H1517 | 1911.2 | 3.0 |
| M-19/ | H1519 | 1079.8 | 0.5 |
| M-01 | H151V1 | 624.0 | 0.9 |
| M-02 | H151V2 | 559.3 | 0.9 |
| M-03 | H151V3 | 575.1 | 0.5 |
| M-04 | H151V4 | 349.7 | 1.0 |
| Average of circulating strains | 844.0 | 1.3 | |
| Standard deviation of circulating strains | 379.1 | 0.7 | |
1 The IC50 values were determined in Vero cells and are expressed as nM values. The infection was done 4 h post treatment, using 100 plaque forming units/well of each virus. 2 ACV sensitivity of the strain according to Bowen et al., 2019 [9].
Figure 1.
Inhibition of viral shedding in response to (A) acyclovir (ACV) or (B) UL29 siRNA swarm. The fitted curves represent circulating clinical (solid lines, N = 17) or reference strains (dashed lines, N = 2) of HSV-1. In all antiviral assays, Vero cells were treated with the antiviral agent on 96-well plates, infected 4 h post treatment (hpt) with 100 plaque forming units (pfu) per well, and finally quantified for reduction of viral production into the culture medium at 72 hpt. The dose is shown as log (nM) for both antivirals. The response is shown as inhibition percentage (%) in comparison to untreated cells for ACV and to water-treated cells for the UL29 siRNA swarm. The ACV plaque reduction assay was conducted in duplicates, and the antiviral siRNA swarm assay with five or more biological replicates for each concentration.
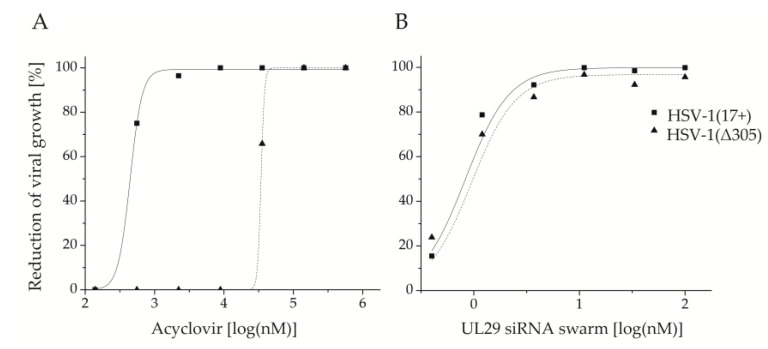
HSV-1 (Δ305) is ACV-resistant (IC50: 25,468.8 nM) due to the lack of a functional tk gene (Table 2, Figure 2A). However, HSV-1 (Δ305) was clearly susceptible to treatment with UL29 siRNA swarm (IC50: 1.0 nM) (Table 2), with similar dose-responsiveness to the reference strain HSV-1 (17+) (Figure 2B). The UL29 siRNA swarm treatment specificity in Vero cells was tested for both HSV-1 (Δ305) and HSV-1 (17+) using a nonspecific siRNA swarm (Figure S2). Even at the highest siRNA swarm concentration used for the antiviral assays (100 nM), treatment with the nonspecific siRNA swarm did not lead to any reduction of viral growth (in comparison with water treatment) with either of the viruses.
Figure 2.
The thymidine kinase (UL23) defective HSV-1 strain (Δ305), and reduction of its growth in response to elevating concentrations of (A) acyclovir (ACV) or (B) UL29 siRNA swarm. HSV-1 (Δ305) is shown as a dotted line and triangles. The corresponding curve of the reference strain HSV-1 (17+) is shown with a solid line and squares. Both the UL29 siRNA swarm and ACV were studied for prophylactic efficacy, with treatment given four hours before infection. The assays were conducted on 96-well plates with a virus dose of 100 pfu per well. The dose is shown as log(nM) for both antivirals and the response is shown as inhibition percentage (%) versus untreated cells for ACV and versus water-treated for the UL29 siRNA swarm. The ACV plaque reduction assay was conducted in duplicates, and the antiviral siRNA swarm assay with five or more biological replicates for each concentration.
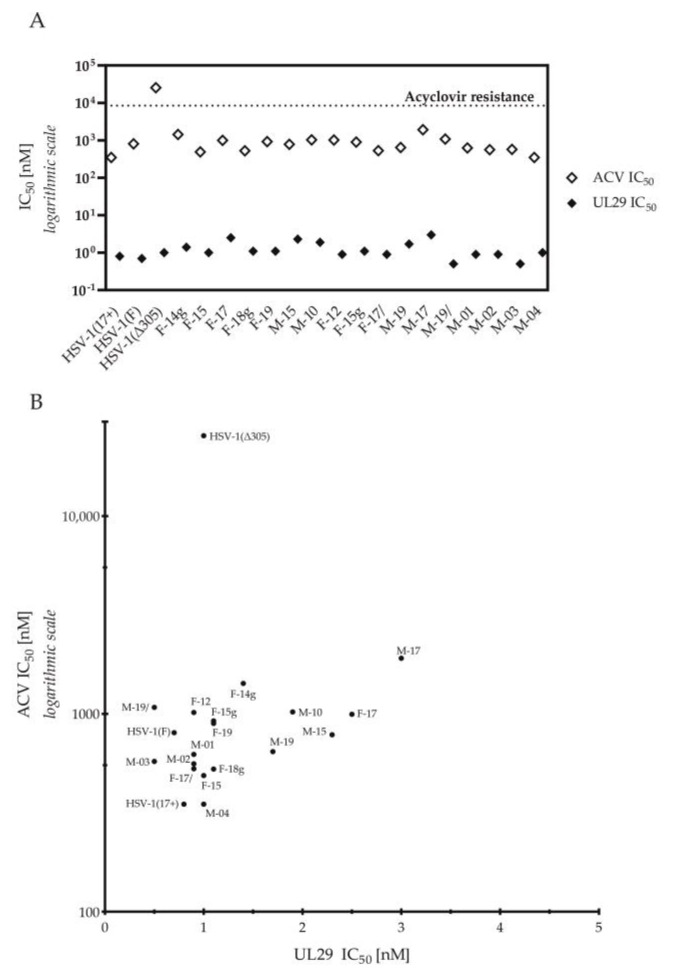
Out of all tested viruses, only HSV-1 (Δ305) exceeded our limit of acyclovir resistance (Figure 3A), clearly deviating from the other viruses with a more than fifteen-fold higher ACV IC50 value (Figure 3B). Still, the sensitivity of HSV-1 (Δ305) to treatment with UL29 siRNA swarm was similar to other strains (Table 2, Figure 3). Altogether, the efficacies of ACV or the UL29 siRNA swarm varied only modestly among the circulating strains (Figure 1 and Figure 3A). Furthermore, the genital sample-derived HSV-1 strains, F14g, F18g, and F15g, were similar to the other circulating strains in both ACV and UL29 siRNA swarm sensitivity (Table 2, Figure 3) and in dose-response profiles (Figure 1). In comparison with other circulating strains, M-17 was less sensitive to both treatments (Figure 3B). Nevertheless, no correlation between the ACV sensitivities and UL29 siRNA swarm sensitivities of the circulating strains was detected (Figure 3B) (Spearman’s r = 0.19, p = 0.49, nonsignificant).
Figure 3.
Comparison of the antiviral susceptibility of the strains. (A) Acyclovir (ACV) and UL29 RNAi sensitivities (IC50) of HSV-1 strains. The dotted line shows the limit of ACV resistance (1.9 µg/mL, 8.43 µM). (B) A scatter plot analysis for correlation of ACV and UL29 sensitivities. The sensitivity (IC50) of the HSV-1 strains for ACV is shown on the y-axis and for UL29 siRNA swarm shown on the x-axis. The values are shown as nM. ACV sensitivity is plotted in logarithmic scale. There was no significant correlation between the ACV sensitivities and UL29 siRNA swarm sensitivities of the circulating strains (p = 0.49) (Spearman’s r = 0.19).
The UL29 siRNA swarm target sequence, which is based on the HSV-1 (17+) UL29 gene sequence (JN555585.1; nucleotides 59,954–59,302), was determined for all strains of which there is not a Genbank accession available. A majority of the strains had over 99.7% similarity with the siRNA swarm target sequence, yet only M-19/had complete similarity with the HSV-1 (17+) UL29 target. F-18g, with 1.08 single nucleotide polymorphisms per 100 nucleotides in the UL29 siRNA swarm target area, differed the most (Table 3). The detected differences in the RNAi target sequence did not affect the UL29 swarm RNAi efficacy, as no correlation between the sequence similarity and sensitivity to the UL29 siRNA swarm or the maximal inhibitory efficacy of the UL29 siRNA swarm could be detected (Figure S3).
Table 3.
Maximal inhibition of viral shedding (Imax) in response to UL29 siRNA treatment and sequence similarity with the UL29 siRNA swarm target.
| Strain | Imax, UL29 Swarm (%) 1 | Sequence Similarity (%) 2 | Differences per 100 Nucleotides 2 |
|---|---|---|---|
| HSV-1 (17+) | 99.85 | 100.0 | 0.00 |
| HSV-1 (F) | 99.79 | 99.85 | 0.15 |
| HSV-1 (Δ305) | 96.75 | 99.21 | 0.79 |
| F-14g | 97.34 | 99.85 | 0.15 |
| F-15 | 98.68 | 99.84 | 0.16 |
| F-17 | 97.95 | 99.70 | 0.31 |
| F-18g | 99.81 | 98.93 | 1.08 |
| F-19 | 99.22 | 99.85 | 0.15 |
| M-15 | 100.00 | 99.85 | 0.15 |
| M-10 | 100.00 | 99.85 | 0.15 |
| F-12 | 98.23 | 99.85 | 0.15 |
| F-15g | 100.00 | 99.85 | 0.15 |
| F-17/ | 99.78 | 99.85 | 0.15 |
| M-19 | 99.89 | 99.85 | 0.15 |
| M-17 | 100.00 | 99.85 | 0.15 |
| M-19/ | 98.69 | 100.0 | 0 |
| M-01 | 100.00 | 99.84 | 0.16 |
| M-02 | 99.81 | 99.69 | 0.31 |
| M-03 | 100.00 | 99.84 | 0.16 |
| M-04 | 100.00 | 99.85 | 0.15 |
1 Determined in Vero cells and expressed as reduction of viral production (%). The infection was done 4 h post treatment, using 100 plaque forming units/well of each virus. 2 In comparison to the UL29 siRNA swarm target: Nucleotides 59,954–59,302 according to the Genbank accession JN555585.1 [13]. An existing Genbank sequence was used for alignments when available.
4. Discussion
In the current study, we evaluated the sensitivity of seventeen Finnish circulating strains of HSV-1 to both acyclovir and to RNAi with a UL29 siRNA swarm [13,15,16,17,18]. The target for our RNAi was the essential UL29 gene, whereas the sensitivity of a given HSV strain to ACV is dependent on its thymidine kinase or DNA polymerase genes [2,6,7]. All the studied viruses, including the ACV-resistant tk-deficient HSV-1 (Δ305), proved to be susceptible to the UL29 siRNA swarm.
Previously, we have shown the efficacy of the UL29 siRNA swarm in epithelial, retinal, and neuronal cell types [13,15,17,18]. Here, in order to compare the antiviral efficacies of the UL29 siRNA swarm and ACV, we utilized Vero cells for all antiviral assays, as they are commonly used for determining HSV-1 susceptibility to antivirals [9,23]. Furthermore, due to the minimal interferon response of Vero cells, they serve well in predicting the sequence-specific silencing of the viral replication in antiviral RNAi. We show that Vero cells are compatible with UL29-specific antiviral siRNA swarm treatment (Figure 1, Figure 2 and Figure 3), without any cytotoxicity (Figure S1) or nonspecific antiviral efficacy (Figure S2). Altogether, the detected antiviral efficacy of the UL29 siRNA swarm in Vero cells is similar to that previously detected in human epithelial cells [13,17,18], favoring the translationality of the results derived from Vero cells.
For our RNAi target, we decided to exploit the UL29 gene, instead of targeting the nonessential thymidine kinase (UL23 gene). SiRNAs targeting genes, such as UL23, which are not essential for viral replication in many settings [21], would not serve as novel antivirals against HSV-1, and might even select for mutant viral strains. Moreover, as simultaneous substantial mutations in UL29 and UL23 have not been reported to our knowledge, the UL29-targeted antiviral RNAi therapy is a promising alternative for the treatment of HSV-1 infections with decreased response to ACV due to mutations in the UL23 gene.
All seventeen circulating HSV-1 strains were susceptible and dose-responsive to ACV (Table 2, Figure 1A). The tk-deficient HSV-1(Δ305) was the only one of the studied viruses resistant to ACV (Table 2, Figure 2A and Figure 3A), as none of the circulating strains exceeded the limit of resistance (Figure 3A). Altogether, the ACV sensitivity of the strains was consistent with previous literature, and the ACV IC50 value of HSV-1(Δ305) was well in line with other published ACV-resistant strains [4,6,9].
The UL29 siRNA swarm led to the reduction of viral growth in a dose-responsive manner (Figure 1B), similarly to ACV. All reference and circulating strains were shown to be susceptible, with little to no deviation in the IC50 value (Table 2, Figure 1B and Figure 3). Moreover, the UL29 siRNA swarm treatment reduced the growth and shedding of all circulating strains by more than 98% (Figure 1B, Table 3) at well-tolerated concentrations (Figure S1).
Supporting our intent to develop siRNA swarms for treatment of ACV-resistant herpes, we observed the susceptibility of an ACV-resistant virus to UL29 siRNA swarm treatment (Figure 2B). The responsiveness to the treatment was similar to that of the reference strain HSV-1 (17+) (Figure 2B), which has served as the sequence template for the UL29 siRNA swarm [13]. As mutations in the UL23 sequence are the most prevalent cause of ACV resistance in circulating strains of HSV-1 [7,8], the capability to treat UL23-deficient variants of HSV-1 with an siRNA swarm enhances the potential of the treatment approach.
Notably, M-17, the least sensitive circulating strain to both ACV and to UL29 siRNA swarm treatment (Figure 3B), was eliminated almost completely (99.8%) by UL29 siRNA swarm treatment (Table 3). Moreover, no significant correlation between ACV and UL29-targeted RNAi sensitivities was detected (Figure 3B), indicating that the efficacy of the UL29 siRNA swarm is independent of ACV sensitivity of circulating strains, as was confirmed by its potency and efficacy against the ACV-resistant HSV-1(Δ305) (Figure 2B, Figure S2).
Additionally, we showed that the chosen target area of the UL29 siRNA swarm has minimal but existing diversity among the studied strains (Table 3). The differences in sequence, which ranged from 0.15 to 1.08 nucleotide differences per 100 nucleotides (Table 3), did not have an effect on the UL29 siRNA swarm efficacy or potency (Figure S3), consistent with the potential of the siRNA swarm approach to overcome viral escape mutants in therapeutic use.
In this proof-of-concept study, we provided evidence supporting the treatment of ACV-resistant HSV-1 infections with an siRNA swarm. Furthermore, we confirmed the compatibility of Vero cells with siRNA therapy, as well as the dose-responsivity of HSV-1 to the UL29 siRNA swarm. Collectively, our results support further development of siRNA swarm therapies for difficult-to-treat topical HSV infections, as all the tested circulating clinical HSV-1 strains were susceptible to UL29 siRNA swarm treatment with high efficacy and sensitivity, regardless of slight variations in target sequence. Our results indicate that UL29-targeted siRNA swarms can be used to treat ACV-resistant herpes infections with a low probability for emergence of escape mutants. Taken together, the UL29 siRNA swarm is highly promising for future treatment of HSV-1 infections, independent of the ACV susceptibility of the causative viral strain.
Acknowledgments
Ritva Kajander, Marja-Leena Mattila and Tanja Westerholm are acknowledged for technical assistance. Bernard Roizman, the University of Chicago, is acknowledged for the HSV-1 (Δ305) virus. The facilities and expertise of the HiLIFE Biocomplex unit at the University of Helsinki, a member of Instruct-ERIC Centre Finland, FINStruct, and Biocenter Finland are gratefully acknowledged.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/12/12/1434/s1, Figure S1: Viability of Vero cells after UL29 siRNA swarm treatment. Figure S2: Comparison of inhibition of viral shedding with specific and nonspecific siRNA swarm treatment in Vero cells using a reference strain (HSV-1 (17+)) and an ACV-resistant virus (HSV-1 (Δ305)). Figure S3: UL29 siRNA swarm potency or efficacy do not correlate with RNAi target sequence similarities.
Author Contributions
Conceptualization, K.K., H.P. and V.H.; Methodology, K.K., J.L., J.G., L.M.L., M.C.N., J.O., T.L., F.F., A.A.L., M.M.P., V.H.; Formal Analysis, K.K., L.M.L., J.G.; Investigation, K.K., J.L., J.G., L.M.L., M.C.N., J.O., T.L., F.F., A.A.L.; Resources, V.H., T.V., M.M.P.; Data Curation, K.K., L.M.L., J.G.; Writing—Original Draft Preparation, K.K., V.H.; Writing—Review and Editing, K.K., J.L., J.G., L.M.L., M.C.N., H.P., J.O., T.L., F.F., A.A.L., M.M.P., T.V., V.H.; Visualization, K.K., J.G., L.M.L.; Supervision, V.H., H.P., M.M.P.; Project Administration, V.H., M.M.P.; Funding Acquisition, V.H., J.L., M.M.P. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by a grant #170046 from the Jane and Aatos Erkko Foundation. The Drug Research Doctoral Programme (University of Turku), Sigrid Juselius Foundation, Academy of Finland (grant #331627) and Maud Kuistila Memorial Foundation are acknowledged for funding.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Elion G.B., Furman P.A., Fyfe J.A., de Miranda P., Beauchamp L., Schaeffer H.J. Selectivity of action of an antiherpetic agent, 9-(2-hydroxyethoxymethyl) guanine. Proc. Natl. Acad. Sci. USA. 1977;74:5716–5720. doi: 10.1073/pnas.74.12.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elion G.B. Mechanism of action and selectivity of acyclovir. Am. J. Med. 1982;73:7–13. doi: 10.1016/0002-9343(82)90055-9. [DOI] [PubMed] [Google Scholar]
- 3.Duan R., de Vries R.D., Osterhaus A.D., Remeijer L., Verjans G.M. Acyclovir-resistant corneal HSV-1 isolates from patients with herpetic keratitis. J. Infect. Dis. 2008;198:659–663. doi: 10.1086/590668. [DOI] [PubMed] [Google Scholar]
- 4.Duan R., de Vries R.D., van Dun J.M., van Loenen F.B., Osterhaus A.D., Remeijer L., Verjans G.M. Acyclovir susceptibility and genetic characteristics of sequential herpes simplex virus type 1 corneal isolates from patients with recurrent herpetic keratitis. J. Infect. Dis. 2009;200:1402–1414. doi: 10.1086/606028. [DOI] [PubMed] [Google Scholar]
- 5.van Velzen M., van de Vijver D.A., van Loenen F.B., Osterhaus A.D., Remeijer L., Verjans G.M. Acyclovir prophylaxis predisposes to antiviral-resistant recurrent herpetic keratitis. J. Infect. Dis. 2013;208:1359–1365. doi: 10.1093/infdis/jit350. [DOI] [PubMed] [Google Scholar]
- 6.Pan D., Kaye S.B., Hopkins M., Kirwan R., Hart I.J., Coen D.M. Common and new acyclovir resistant herpes simplex virus-1 mutants causing bilateral recurrent herpetic keratitis in an immunocompetent patient. J. Infect. Dis. 2014;209:345–349. doi: 10.1093/infdis/jit437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt S., Bohn-Wippert K., Schlattmann P., Zell R., Sauerbrei A. Sequence analysis of herpes simplex virus 1 thymidine kinase and DNA polymerase genes from over 300 clinical isolates from 1973 to 2014 finds novel mutations that may be relevant for development of antiviral resistance. Antimicrob. Agents Chemother. 2015;59:4938–4945. doi: 10.1128/AAC.00977-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimberlin D.W., Coen D.M., Biron K.K., Cohen J.I., Lamb R.A., McKinlay M., Emini E.A., Whitley R.J. Molecular mechanisms of antiviral resistance. Antiviral. Res. 1995;26:369–401. doi: 10.1016/0166-3542(95)00027-J. [DOI] [PubMed] [Google Scholar]
- 9.Bowen C.D., Paavilainen H., Renner D.W., Palomaki J., Lehtinen J., Vuorinen T., Norberg P., Hukkanen V., Szpara M.L. Comparison of herpes simplex virus 1 strains circulating in Finland demonstrates the uncoupling of whole-genome relatedness and phenotypic outcomes of viral infection. J. Virol. 2019;93:e01824-18. doi: 10.1128/JVI.01824-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levanova A., Poranen M.M. RNA Interference as a prospective tool for the control of human viral Infections. Front. Microbiol. 2018;9:2151. doi: 10.3389/fmicb.2018.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nygårdas M., Vuorinen T., Aalto A.P., Bamford D.H., Hukkanen V. Inhibition of coxsackievirus B3 and related enteroviruses by antiviral short interfering RNA pools produced using phi6 RNA-dependent RNA polymerase. J. Gen. Virol. 2009;90:2468–2473. doi: 10.1099/vir.0.011338-0. [DOI] [PubMed] [Google Scholar]
- 12.Kim D.H., Behlke M.A., Rose S.D., Chang M.S., Choi S., Rossi J.J. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 13.Romanovskaya A., Paavilainen H., Nygardas M., Bamford D.H., Hukkanen V., Poranen M.M. Enzymatically produced pools of canonical and Dicer-substrate siRNA molecules display comparable gene silencing and antiviral activities against herpes simplex virus. PLoS ONE. 2012;7:e51019. doi: 10.1371/journal.pone.0051019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang M., Österlund P., Westenius V., Guo D., Poranen M.M., Bamford D.H., Julkunen I. Efficient inhibition of avian and seasonal influenza A viruses by a virus-specific Dicer-substrate small interfering RNA swarm in human monocyte-derived macrophages and dendritic cells. J. Virol. 2019;93:e01916-18. doi: 10.1128/JVI.01916-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levanova A.A., Kalke K.M., Lund L.M., Sipari N., Sadeghi M., Nyman M.C., Paavilainen H., Hukkanen V., Poranen M.M. Enzymatically synthesized 2’-fluoro-modified Dicer-substrate siRNA swarms against herpes simplex virus demonstrate enhanced antiviral efficacy and low cytotoxicity. Antiviral. Res. 2020;182:104916. doi: 10.1016/j.antiviral.2020.104916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paavilainen H., Lehtinen J., Romanovskaya A., Nygårdas M., Bamford D.H., Poranen M.M., Hukkanen V. Topical treatment of herpes simplex virus infection with enzymatically created siRNA swarm. Antivir. Ther. 2017;22:631–637. doi: 10.3851/IMP3153. [DOI] [PubMed] [Google Scholar]
- 17.Paavilainen H., Romanovskaya A., Nygårdas M., Bamford D.H., Poranen M.M., Hukkanen V. Innate responses to small interfering RNA pools inhibiting herpes simplex virus infection in astrocytoid and epithelial cells. Innate. Immun. 2015;21:349–357. doi: 10.1177/1753425914537921. [DOI] [PubMed] [Google Scholar]
- 18.Paavilainen H., Lehtinen J., Romanovskaya A., Nygårdas M., Bamford D.H., Poranen M.M., Hukkanen V. Inhibition of clinical pathogenic herpes simplex virus 1 strains with enzymatically created siRNA pools. J. Med. Virol. 2016;88:2196–2205. doi: 10.1002/jmv.24578. [DOI] [PubMed] [Google Scholar]
- 19.Roizman B., Knipe D.M., Whitley R.J. Herpes Simplex Viruses. In: Knipe D.M., Howley P.M., Cohen J.I., Griffin D.E., Lamb R.A., Martin M.A., Racaniello V.R., et al., editors. Fields Virology. 6th ed. Volume 2. Lippincott, Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 1823–1897. [Google Scholar]
- 20.Palliser D., Chowdhury D., Wang Q.Y., Lee S.J., Bronson R.T., Knipe D.M., Lieberman J. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439:89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 21.Post L., Mackem S., Roizman B. Regulation of a genes of herpes simplex virus: Expression of chimeric genes produced by fusion of thymidine kinase with a gene promoters. Cell. 1981;24:555–565. doi: 10.1016/0092-8674(81)90346-9. [DOI] [PubMed] [Google Scholar]
- 22.Ziegler T., Waris M., Rautiainen M., Arstila P. Herpes simplex virus detection by macroscopic reading after overnight incubation and immunoperoxidase staining. J. Clin. Microbiol. 1988;26:2013–2017. doi: 10.1128/JCM.26.10.2013-2017.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tardif K.D., Jorgensen S., Langer J., Prichard M., Schlaberg R. Simultaneous titration and phenotypic antiviral drug susceptibility testing for herpes simplex virus 1 and 2. J. Clin. Virol. 2014;61:382–386. doi: 10.1016/j.jcv.2014.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.