Abstract
At present, much attention is paid to the use of antimicrobial peptides (AMPs) of natural and artificial origin to combat pathogens. AMPs have several points that determine their biological activity. We analyzed the structural properties of AMPs, as well as described their mechanism of action and impact on pathogenic bacteria and viruses. Recently published data on the development of new AMP drugs based on a combination of molecular design and genetic engineering approaches are presented. In this article, we have focused on information on the amyloidogenic properties of AMP. This review examines AMP development strategies from the perspective of the current high prevalence of antibiotic-resistant bacteria, and the potential prospects and challenges of using AMPs against infection caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Keywords: amyloidogenic regions, toxicity, antibacterial peptides, proteome, mass spectrometry, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), coronavirus disease 2019 (COVID-19)
1. Introduction
Currently, the relevance of the study and development of antimicrobial molecules is due to the global spread of antibiotic-resistant forms of bacteria, as well as new viral diseases [1,2]. Peptides that exhibit antimicrobial activity have several advantages over conventional drugs, including a broad spectrum of action, slow onset of resistance, and the ability to modulate the host immune response [3]. Several thousand antimicrobial peptides (AMPs) have been discovered, but only a few have been approved by the U.S. Food and Drug Administration (FDA) [4]. Our understanding of the physicochemical properties, the peculiarities of the mechanisms of action of AMPs, modern possibilities of searching and predicting the structure and properties of peptides will define new horizons in the development of new AMPs. Of particular interest is the potential for the use of peptides, which, in addition to pronounced antimicrobial properties, also tend to self-assembly, the formation of supramolecular complexes, and amyloidogenesis [5,6,7]. Traditionally, the amyloidogenic properties of peptides are negatively regarded as contributing to the development of neurodegenerative and progressive metabolic diseases [8,9,10]. However, in recent years, more and more attention has been paid to the study of the functional role of amyloidogenic peptides in the protection of the host and the prospects for their use as antimicrobial agents [11,12]. New data indicate that amyloidogenic peptides can interact with various combinations of pattern recognition receptors (PRRs), which in turn leads to modulation of innate immune cells [13]. Interestingly, such immunomodulatory amyloidogenic peptides can be specific targets for changing entire metabolic pathways. For example, Smole et al. showed that changes in the oligomeric organization of serum amyloid A1 (SAA1) caused by dust mite allergens is recognized by the PRRs and ultimately lead to the development of an immune response [14]. The therapeutic potential of peptide molecules prone to aggregation is truly enormous due to the diversity of their structure and properties, in particular, the ability to form supramolecular complexes with other peptides and proteins, biocompatibility, and biodegradability. It was shown using theoretical and experimental methods that peptide coaggregation is necessary for the destruction of bacterial membranes [15]. Theoretical methods for finding and predicting new AMPs, based on the use of specially designed programs for these purposes, are making an increasing contribution to the development of new AMPs. In turn, we proposed another mechanism of the antimicrobial action of peptides, based on the directed coaggregation of the amyloidogenic peptide with the target protein and the subsequent dysfunction of this protein. Using programs predicting amyloidogenic regions in a protein molecule, sequences of peptides with a tendency to aggregate with the target protein were selected [16]. It is important that the potential for use of peptides combining antimicrobial and amyloidogenic properties is not limited to their use against antibiotic-resistant bacteria. Peptides that tend to coaggregate with viral proteins or prevent the virus from interacting with cell receptors can be used against the spread of coronavirus infection caused by SARS-CoV-2. Currently, significant resources of the scientific community are involved in the search for effective methods of treating coronavirus infection. Many strategies have been proposed, among which peptides play an important role [17]. Interestingly, a similar target effect is a characteristic of AMPs with lectin-like functions (e.g., defensins [18] and hevein-like peptides [19], which can bind to N-glycosylated sites of the viral spike protein or the receptor, which ultimately prevents penetration of the virus into the host cell [20,21]. In general, as will be described below, AMPs have great potential for effective use in the therapy of coronavirus infection [22].
The purpose of this review is to show the possibilities of using the antimicrobial and amyloidogenic properties of peptides to create new therapeutic strategies against antibiotic-resistant bacteria and the spread of viral infections. In the following sections, we describe the physicochemical properties and variety of natural and artificial peptides. We also discuss the known mechanisms of action of antibacterial peptides and our proposed new mechanism associated with directed coaggregation with the target protein of the pathogenic organism. Finally, we present promising results for the search and development of new AMPs, including those against coronavirus infection.
2. Brief History of Research, Physicochemical Properties, Classification and Diversity of Antimicrobial Peptides
Speaking about antimicrobial activity, one cannot fail to note the 1945 Nobel Prize winners in physiology and medicine A. Fleming, E.B. Cheyne, and H. W. Florey, who discovered penicillin and its properties. In addition to the discovery of penicillin, A. Fleming described a protein with bacteriolytic activity—lysozyme.
These and other discoveries preceded the “Golden Age of Medicine”, when millions of lives were saved. Antimicrobial substances play an important role in the functioning of the innate immunity of both humans and living beings [23,24]. A significant number of studies on the mechanisms of immune defense have shown the key role of AMPs in the non-specific defense of the host [25]. AMPs are characteristic of all living things: vertebrates and invertebrates, plants, fungi and bacteria [24,25]. Interest in AMPs is mainly due to the fact that most of the known and available methods of treatment were ineffective due to the rapid emergence and spread of resistance in nosocomial bacterial pathogens, fungi and viruses [26]. At the same time, AMPs or host defense peptides have a potent broad-spectrum antimicrobial action against pathogenic bacteria, fungi, and viruses [27]. Thus, AMPs and their various combinations are considered as promising candidates for drugs against multidrug-resistant bacteria such as P. aeruginosa, S. aureus, K. pneumoniae or A. baumannii [28,29,30].
AMPs are a group of heterogeneous peptides that have been studied for several decades. During this time, researchers concentrated in several areas of research (Table 1). AMP study is based on the discovery of the antimicrobial activity of peptides in fluids and tissues of humans and mammals [31]. Subsequent studies showed that AMPs play an important role in the immune system of all living beings [32]. The development of physicochemical methods for studying molecular processes made it possible to describe the structural features of peptides and to reveal some connections between the properties of AMPs. The next step, which became a consequence of the computer revolution, was the development of algorithms for predicting the antimicrobial properties of peptides. A century and a half after the discovery of AMPs, out of more than 3000 characterized peptides, seven (oritavancin, gramicidin, daptomycin, colistin, vancomycin, dalbavancin, telavancin) are applicable to clinical practice [4]. Recently, the problems of using AMPs in veterinary medicine and medicine have been widely discussed, which stimulates studies of targeted delivery of AMPs (for example, in the form of metal nanoparticles) or the development of species-specific AMPs [33,34,35]. Undoubtedly, further research will continue the existing directions. However, the topic of AMPs is much broader due to the fact that AMPs are capable of exhibiting antitumor, antibiofilm, antiviral, and other types of activity, the number of studies in these areas is increasing [36,37]. The accumulation of scientific experience and a large number of facts in the study of AMP allows one to focus not only on fundamental issues, but on the possible application of AMP in a practical field. Thus, the described areas are an important basis for further main and secondary research in this area.
Table 1.
Research fields for antimicrobial peptides (AMPs).
| Scientific Direction | Description | References |
|---|---|---|
| Search and description of new AMPs | This direction is rather diverse and includes from the characterization of new, as yet unknown AMPs using classic biology methods to the creation of AMP search algorithms based on the structural features of peptides and omix data in silico. | [32,38,39,40,41] |
| Study of physicochemical and structural properties of AMPs | Despite numerous studies of physicochemical properties from 1990 to 2000, much is still unknown about the relationship between physicochemical properties and physiological activity. | [15,42,43,44,45,46] |
| Generalization of structural properties and prediction of peptides with antimicrobial properties | This area includes the creation of databases of known peptides and algorithms for predicting physiological activity. | [47,48,49,50,51,52,53,54] |
| Rational design and structure modification | Bacterial resistance to antibiotics creates a need to develop new or modified old molecules to fight bacterial infections. | [3,4,55,56,57] |
| Study of a narrow biological activity | The specific biological activity of AMP is an important property that requires close attention. | [36,37] |
| Methodical developments | The results of AMP research over the decades have led to the revisions and modifications of experimental procedures. | [58,59] |
| Solving application problems in practice. | Improving the effectiveness of AMP can be realized through targeted delivery using non-materials or a combination of dosage forms. Non-drug forms of AMP are proposed to be used as antiseptics to combat dangerous pathogens. Another possible application of AMPs is in biomarkers of parasitic infections. |
[33,34,35,60,61,62] |
In total, 98% of AMPs are encoded in the genome. These peptides can be constitutively produced or induced to maintain the health of the host [24,33]. AMPs are often modified. A total of 24 types of chemical modifications of AMP are known [63]. Three types of modifications are of greatest interest to researchers of drag design: lipidation, glycosylation, and PEGylation. A feature of these modifications is that they increase the activity and bioavailability, increase the metabolic stability of the molecule, and can promote direct translocation [64,65,66]. Lipidation and glycosylation of molecules can be a modification of both natural and synthetic AMPs. Aliphatic chain in peptides modulates their hydrophobicity, tendency to self-assembly, while the possibility of action through an intracellular receptor is not excluded [67,68,69,70]. Glycated peptides also demonstrate greater biostability, the ability to self-assemble complex frameworks, and increase selectivity [71,72]. AMP PEGylation modulates the rate of biotransformation and also reduces toxicity [73,74]. Modifications serve some important functions. For example, enterotoxin AS-48 undergoes cyclization to maintain the correct molecular structure [75]. The cyclical structure of calata B1 supports the functionality [76]. Halogenation and hydroxylation of the arginine, lysine, and tyrosine residues of styelin D allows one to regulate the range of conditions under which this peptide acts [77].
It should be borne in mind that the modification of AMP affects their effectiveness against specific bacteria in different ways. Thus, an analog of AMP with an alternating cationic/hydrophobic structure was optimized, demonstrating only moderate activity against Gram-positive pathogens in [78]. The attachment of hydrophobic fragments to the N-terminus enhanced the antibacterial effect of the analogs against multidrug-resistant Staphylococcus aureus and Enterococcus faecium. At the same time, modification with hydrophobic fragments led to an increase in activity against Gram-negative Acinetobacter baumannii, while the effectiveness against Escherichia coli (E. coli) strains did not change.
Peptides of different lengths have antimicrobial properties. According to dbAMP data, the polypeptide chain can reach 700 amino acid residues [79]. Interestingly, some peptides with a minimum number of amino acid residues in the chain (5 or less) contain a lipid aliphatic chain [80]. However, 90% of polypeptide chains consist of less than 50 amino acid residues, and most of them contain 20–30 [81]. Considering the hydrophobicity of the molecule, AMPs may consist only of hydrophobic amino acids or not at all. The ratio of hydrophobic amino acids to the total number of residues in the chain may indicate the mechanism of AMP action. Typically, 78% of AMPs have a hydrophobic content in the range of 30–60%. Under physiological conditions, a peptide molecule can be charged depending on its composition. Most AMPs are positively charged—87%, 7% neutral, and 6% negatively charged. Among positively charged peptides, 73% are in the range from +1 to +6, the most pronounced peak corresponds to charge +3. However, the Oncorhyncin II peptide is known to have a total charge of +30. This peptide, isolated from rainbow trout (Oncorhynchus mykiss), is the C-terminal fragment of histone H1, which causes pronounced destabilization of flat lipid bilayers [82]. The combination of positive charge and hydrophobicity leads to the formation of an amphipathic nature (spatial separation of the hydrophobic and charged parts of the molecule) of most AMPs. The positive charge is important for the initial interaction with the negatively charged surface of bacteria membranes. The hydrophobic component of the peptide is required for the subsequent attachment of the peptide to the membrane surface.
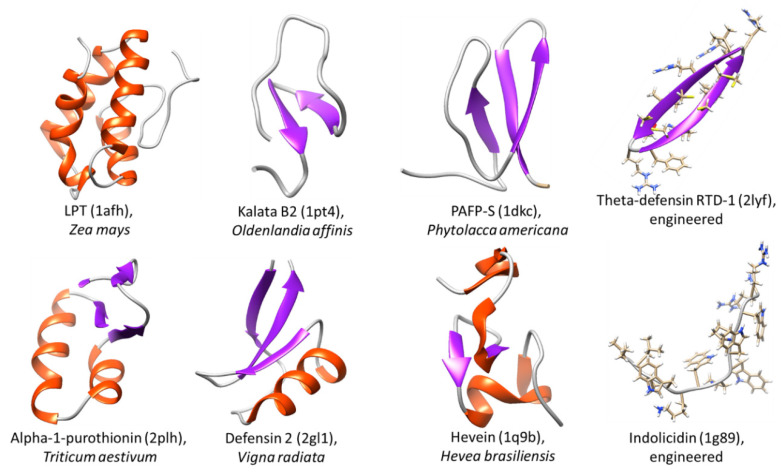
Like proteins, AMPs form various types of secondary structures (Figure 1): α-helical; containing β-strands stabilized by disulfide bonds; including both α-helical and β-strands, cyclic or loop- forming; linear peptides without definite conformation enriched in proline, tryptophan, histidine, arginine, glycine, or their combinations.
Figure 1.
Variety of spatial structures of AMPs.
Interestingly, alpha-helical AMPs with a significant frequency contain a small and flexible glycine residue at positions 7 or 14. The results of the study show that although the hydrophobicity of residue 7 and flexibility of the chain at this position can be modulated to increase selectivity, position 14 is less resistant to substitutions [83]. In the extracellular fluids of insects and frogs, α-helical AMPs are present in excess in the form of an extended or unstructured polypeptide chain conformation [84]. These peptides, interacting with membranes, often form a helical fold. The β-region forming peptides have a wide variety of primary structures. Despite the differences, β-fold AMPs share common features, including an amphipathic composition with well-defined hydrophilic and hydrophobic surfaces. Little is known about the structures of proline, arginine and tryptophan rich peptides. There are several conformations that differ from the above. For example, the proline and arginine-rich AMPs and the tryptophan-rich indolicidines form a type II polyproline helix [85,86].
The size and conformation of peptides affect their interaction with membranes [87,88]. In this regard, much attention is paid to the study of the spatial structure and key motifs of folding of AMPs. Recently, in addition to measuring circular dichroism, a high-resolution NMR method has also been used for these purposes [89]. The secondary structure of peptides can be influenced by addition a fluorescent label, which is usually taken into account when developing AMP analogs [90].
AMPs are characterized by the combination of several types of secondary structures in one molecule. The presence of more than one type of structure in the molecule is observed in 94% of known AMPs and, on the contrary, only 4% of peptides have either only helical or β-structural folds [91].
Summarizing the properties and structural features, AMPs are molecules of a peptide nature with a high content of positively charged and hydrophobic amino acids, with spatial separation of charged and hydrophobic parts of the molecule, synthesized in vivo on ribosomes or enzymatically or in vitro, possessing wide activity against bacteria, fungi, viruses, and protozoa in physiological conditions [23,92].
According to G. Wang [81], the world community recognizes two AMP nomenclatures: the first describes the source of isolation (domain, kingdom, and other systematic categories), the mechanism of biosynthesis and properties; the second is based on biological activity.
For classification by source of isolation, the biological nomenclature proposed by Robert H. Whittaker in 1969 is used. According to this nomenclature, the detected AMPs are distributed as follows: prokaryotes (bacteria and archaea) include 272 and 4 peptides, respectively, protists (protozoa and algae)—8, fungi—13, plants—335, and animals—2043 [81]. As of March 2020, over 70% of the peptides in the CAMPR3 electronic antimicrobial peptide database come from the animal kingdom. It is Interesting that in this kingdom two main sources of AMPs can be distinguished: 50% of all sources are amphibians, 13% are insects [63].
Bacterial peptides are grouped into the bacteriocin class. Among the known AMPs, there are about 6% of them, and the number of bacteriocins is growing rapidly, since new representatives of this class are being developed [93]. The main systematic feature of the classification of bacterial AMPs is the structure of the cell wall of bacteria producing peptides. The subsequent classification is carried out according to the features of the structure of the peptide and its molecular weight. There are three classes of peptides synthesized by Gram-positive bacteria. The first class—lantibiotics—is characterized by the presence of thioether rings in the molecule. There is no such structure in second-class AMP molecules. These peptides are classified into 4 subclasses: pedoicin-like bacteriocins (subclass IIa), two-component bacteriocins (subclass IIb), circular bacteriocins (subclass IIc), and linear unmodified pedicin-like bacteriocins (subclass IId). Gram-negative bacteria are classified in a similar way [81]. Bacteriocin of Gram-negative bacteria is grouped into four classes [94]. Colicins and colicin-like peptides have similar physicochemical properties and differ only in the type of producer bacteria. These AMPs have a high molecular weight and are sensitive to temperature and proteases. The synthesis of colicins is associated with the expression of cell autolysis proteins. The mechanism of action can be directed both to the cell membrane and to intracellular targets. Tailocins are multicomponent peptides with a molecular weight of 20 to 100 kDa, consisting of 8-14 different polypeptide chains homologous to the modules of the bacteriophage tail. The exact mechanism of action of the tailocins is not known, but is directed against a narrow group of related strains. Microcins are low molecular weight peptides (<10 kDa) that are resistant to proteases, extreme pH and temperature, and can have complex post-translational modifications. The mechanism of action can be explained both by the formation of pores in the membrane and by inhibition of DNA gyrase and RNA polymerase [94].
The AMPs of the fungal kingdom are divided into two classes. Peptides isolated from soil fungi of the genera Trichoderma and Emericellopsis form the peptaibole class. These molecules are characterized by the presence in the composition of 15–20 amino acids with a high content of aminoisobutyric acid, and acetic acid residue at the N-terminus and hydroxyamino acid at the C-terminus. The second class is represented by defensin-like peptides, usually containing multiple disulfide bonds.
Antimicrobial plant peptides are classified into 9 groups [81]. The key features of these AMPs are high content of cysteine and glycine in the molecule, as well as the presence of disulfide bridges to increase the stability of the structure in stressful conditions. About 17% of amino acids in plant AMPs are charged mainly due to arginine and lysine.
However, it is interesting to note that residues such as aspartic acid and glutamic acid may also be present. The presence of negatively charged amino acids can play a significant role in the activity against pathogenic bacteria [95].
The classification of animal AMPs is complex, and the number of systematic units is extensive [81]. Amphibians have the following AMP families: magainins, dermaseptins, brevinins, esculentins, japonitsins, nigrocin-2, palustrins, ranatsicins, ranatuerins, and temporins [96,97]. In insects, the cetsropin, defensin, and proline-rich peptide families are well known [98]. With regard to marine invertebrates, Otero-González et al. [99] described AMPs of various types, such as Porifera, Cnidaria, Mollusca, Annelida, Arthropoda, Echinodermata, and Chordata. In mammals, including humans, the main AMP families are defensins and catelicidins [100].
The disadvantage of classification by the source of isolation is the impossibility of assessing the structural similarity of molecules of different systematic categories. Therefore, a kind of artificial system is created; on the bases of genomic and proteomic data, AMPs were divided into 45 families. This classification is based on the similarity of the primary structure of the peptide [91].
Defensins hold a special place among the known families. A superfamily of small (4–6 kDa) cationic peptides secreted by many species, including humans and other mammals, as well as fish, birds, insects, fungi, and plants. The persistence of this superfamily of peptides among evolutionarily young and ancient groups indicates that peptides are an ancient and conservative defense mechanism [101]. Defensins are synthesized in the cell as precursors. Interestingly, the precursor plays a key role in the functioning of the peptide. For example, the α-defensin precursor contains 40 amino acids and is required to maintain a biologically active peptide in an inactive state.
By interacting with a mature peptide, the pro-segment acts as an effective intramolecular inhibitor, avoiding autocytotoxicity [101]. Another widely represented AMP family is thaumatins. Peptides of this family exhibit antifungal activity. Thaumatins have been identified in fungi, plants, and animals (nematodes, ticks and insects). Comparison of thaumatins of animal and plant origin reveals their close similarity, which also indicates the antiquity of both the AMP family and the defense system itself [102]. Typical plant thaumatins have a molecular weight of about 20 kDa, an even number of cysteine residues (10–16), a conserved REDDD sequence in the primary structure, and a conservative three-dimensional organization [103].
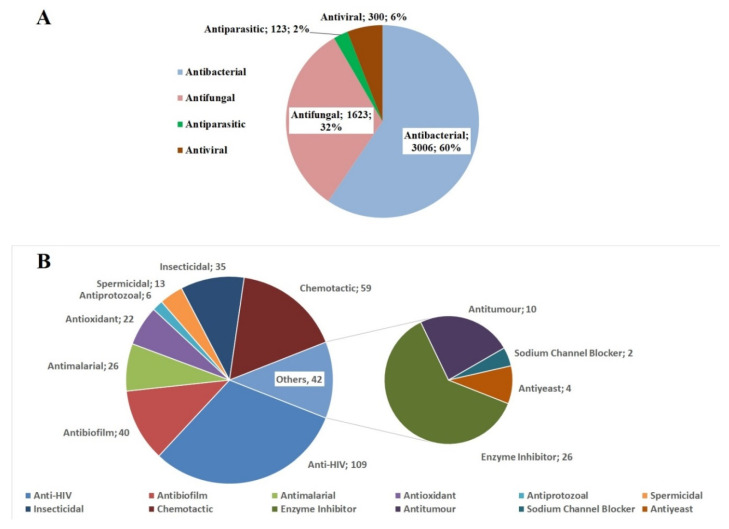
AMPs provide a wide range of biological functions (see Figure 2A). In addition to antimicrobial, antibacterial, antiviral, antifungal, antiparasitic activity, AMPs may have more specific effects (see Figure 2B).
Figure 2.
Distribution of AMPs by their biological activity according to dbAMP data: general (A) and special (B) [79].
Antimicrobial activity is understood as the presence of one or more, and in rare cases all types of activity that suppress the life of viruses, bacteria, fungi or parasites [81]. It is known that the amount of AMP decreases with an increase in the area of their activity. A decrease in the number of activities may be associated with the physicochemical properties of peptides [81]. Indeed, there are very few well known AMPs with all four types of activity. These peptides are amphibian magainin 2, dermseptin C1 and dermseptin C4; insect melittin; human α-defensin HNP-1, cathelicidin LL-37; bovine BMAP-27, BMAP-28 and vegetable calata B2 [96,100,104,105,106,107,108]. The antibacterial activity of peptides is the most common. This activity was found in 31% of the peptides in the dbAMP database. This activity is usually explained by the action of cationic peptides on the membrane [81].
Interestingly, the activity against Gram-negative and Gram-positive bacteria is distributed approximately equally (19% and 22%, respectively). 16% of dbAMP peptides have antifungal activity. This activity often belongs to representatives of plant AMPs [63]. The antiviral activity of AMP is interesting. Cathelicidin LL-37 is known to be active against human immunodeficiency virus 1 (HIV-1), respiratory syncytial virus (RSV) and influenza virus [11,109]. Moreover, viral particles, both coated and uncoated, are exposed. A possible mechanism of action is associated with the ability of the antimicrobial peptide LL37 and similar α-helical AMPs to form nanocrystalline complexes with viral dsRNA. Such complexes are recognized by toll-like receptors, which further induce an immune response [110,111].
It is assumed that the amphipathic nature of AMP allows it to bind to the viral nucleic acid. Antiparasitic peptides make up a relatively small group of AMPs. Although parasitic organisms are usually multicellular eukaryotes, the mechanism of action of antiparasitic peptides involves cell death trough destruction of the cell membrane, as in other AMPs [112]. Peptides in this group are active against various parasites, including Leishmania and Trypanosoma brucei [113].
AMPs may be of particular interest in medicine. The anti-cancer activity of AMP is of special interest. Several studies have shown that some AMPs are cytotoxic to cancer cells [114,115,116]. This discovery is of great importance, since there is a problem of resistance of cancer cells to anticancer drugs [117]. It is not yet clear why some peptides are capable of killing cancer cells while others are not. It is assumed that there is some similarity between cancer and bacterial cells, which allows AMP to specifically bind to them without exhibiting cytotoxicity towards healthy cells [118].
Also, of great interest is the activity of AMPs against microbial biofilms. In addition to drugs, various peptide conjugates can be used to coat catheters to prevent the formation of polymicrobial biofilms [119]. Bacteria in biofilms are coated with a complex mixture of extracellular polymeric substances. In such an environment, bacteria become extremely resistant to common (conventional) antibiotics and the host’s immune response [120]. It has been shown that human cathelicidin LL-37 is able to inhibit the biofilm formation by Francisella novicida [121]. Another artificially modified peptide R17, in addition to inhibition, exhibited dispersing activity against biofilms of Acinetobacter baumannii [122]. The DP7 peptide was shown to inhibit the formation of multidrug-resistant Pseudomonas aeruginosa biofilms [123]. In general, the methods of destruction of biofilms using AMPs are currently being intensively developed [124]. AMPs can be used as a treatment option for patients with recurrent infections who will benefit from high local doses of persistent antimicrobials. An important way to improve AMP is to decrease the value of their minimum inhibitory concentration (MIC) in relation to the pathogen. The approaches to reducing the MIC can be as follows: selection of the cyclization point, modification of amino acid residues taking into account changes in their hydrophobicity, as well as changes in the length of the cationic side chain, and various combinations of these approaches [125].
A contribution to understanding the action of AMP can be made by creating analogues of AMP and studying their action. For example, the study of analogs of cathelicidin allowed researchers to reveal patterns in the manifestation of its antimicrobial action, depending on the characteristics of the amino acid sequence. Thus, Ser9 in cathelicidin LL-23 was replaced by Ala or Val, which made LL-23 hydrophobic and enhanced its antibacterial activity. A decrease in the rate of hydrogen-deuterium exchange from LL-23 to LL-23A9 to LL-23V9 suggests a deeper penetration of LL-23V9 into micelles, which correlates with an increased effect [126]. LL-37, which converts positively charged arginine to neutral citrulline, in contract to the unmodified peptide, does not kill E. coli, indicating that the net positive charge is important for the antibacterial and membrane lysing effects [127]. It has also been shown that citrullination of LL-37 reduces its direct antiviral activity against human rhinovirus. In addition, it is discussed that overexpression of the gene of peptidyl arginine deiminase (PAD) and citrullination of the host defense peptide (HDP) during infection may represent a new evasion mechanism for the virus [128].
Various strategies have been proposed for surface immobilization of a wide variety of materials and local release of AMPs from implant surfaces [129]. For LL-37 a significant loss of antimicrobial function was shown when the peptide was exposed to carbon nanoparticles at low concentrations, and the interaction of nanomaterials with the peptide led to a significant change in the peptide structure [130]. Another study showed that encapsulation of the AMP in dextran nanoparticles increases the residence time in the lungs of the peptide administered via aerosol [131]. In a recent work, the effective localization of natural AMPs on a negatively charged bacterial surface was described using structurally nanoengineered AMP polymers (SNAPPs) [132]. In addition, it was proposed to use cubosomes for local delivery of AMPs [133]. It has been shown that the creation of conjugates based on antibiotics and AMP is effective against Staphylococcus aureus [134]. The green fluorescent protein (GFP) based fusion lantipeptide class I expression system is a robust system with the advantage of direct visualization of expression and purification by GFP fluorescence [135]. Small ubiquitin-related modifier (SUMO) technology is widely used in E. coli expression systems for the production of AMPs. However, E. coli is a pathogenic bacterium that produces endotoxins and often secretes target proteins to form inclusion bodies. E. coli was replaced by the Bacillus subtilis expression system using SUMO technology to synthesize cathelicidin-BF (CBF), an AMP purified from venom of Bungarus fasciatus [136].
3. Mechanisms of Antimicrobial Action of Peptides
3.1. Analysis of Possible Mechanisms
The wide variety of AMPs and their functions indicates the complexity of the defense mechanisms. It is believed that the selectivity of AMPs action is regulated by production inducibility and limited localization at the lesion sites or by the nature of the peptide [84]. The selectivity of AMPs can be determined by the following physicochemical and structural parameters: conformation, charge, hydrophobicity, hydrophobic moment, amphipathicity. It is essential that these molecular parameters are interdependent, therefore, a change in one parameter often leads to compensatory changes in others. Despite the great variety of the primary structure, AMPs form typical elements of the secondary and tertiary structure. These features are important for systematization and classification (see Figure 1).
The total charge of AMPs plays an important role in the initial electrostatic attraction of molecules to the membranes of bacteria and other microorganisms. It is the negative charge that forms those structural features of the bacterial membranes that provide selectivity in relation to host tissues. The presence of phosphatidylglycerol, phosphatidylserine, cardiolipin provides total negative charge, and the presence of lipopolysaccharides, teichoic and teichuronic acids gives an additional negative charge to the membranes of Gram-negative and Gram-positive bacteria. The chemoosmotic potential of a prokaryotic target cell is usually 50% higher than that of mammalian cells. Some authors emphasize the correlation between the total positive charge of the peptide and its antimicrobial activity [137]. However, there is a limit beyond which an increase in the positive charge does not lead to an increase in the activity of the peptide. Indeed, an increase in the charge of the AMP magainin from 3 to 5 leads to an increase in antibacterial activity against Gram-negative and Gram-positive pathogens; however, increasing the charge to 7 significantly increases the hemolytic activity and leads to the loss of antimicrobial activity [138].
Among the physical characteristics reflecting the hemolytic activity of AMPs are amphipathicity and hydrophobic moment. The hydrophobic moment is a quantitative characteristic of amphipathicity, calculated as the vector sum of the hydrophobicity of individual amino acids normalized to an ideal helix [139]. An increase in the hydrophobic moment leads to a significant increase in the permeability and hemolytic activity of model peptides in relation to target membranes. In a study by Dathe et al. [140], a relatively small increase in the hydrophobic moment led to an 8-fold decrease in the amount of peptide required for hemolysis.
Another important characteristic of the interaction of AMPs with membranes is hydrophobicity. This property determines the ability of AMP to be incorporated into the lipid bilayer. In a study by Dathe et al. [140] designed peptides with constant charge, helicity and hydrophobic moment, but differed in hydrophobicity. With a slight change in the hydrophobicity of the molecules, they achieved differences in membrane binding and permeability. This indicates the importance of hydrophobic properties in the interaction of the peptide and the lipid bilayer. Thus, for peptide S3(B), the effects of changing the overall hydrophobicity of the modified peptide were shown due to the introduction of fatty acid fragments of different lengths and in different positions into this cyclic peptide containing a flexible linker [141].
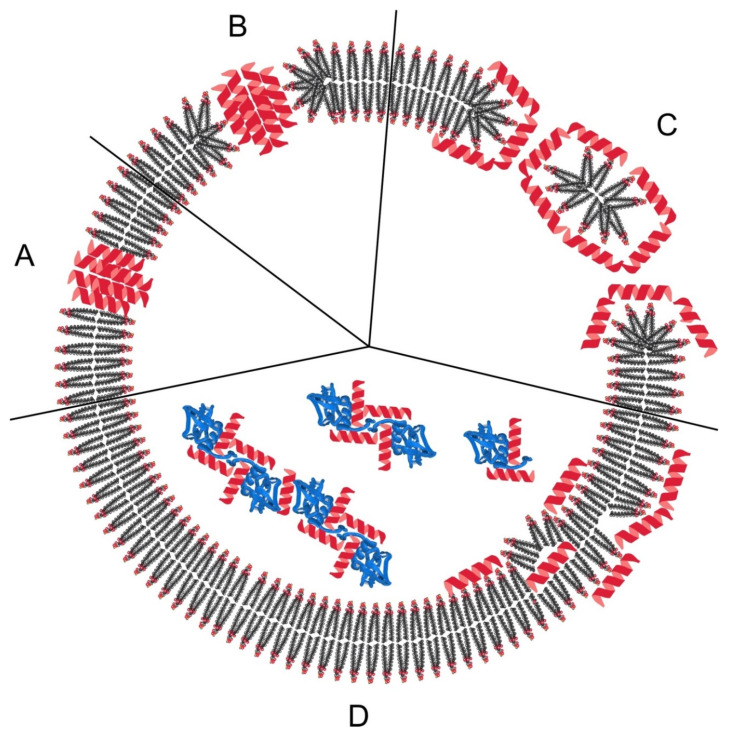
The main activity of AMP is a membranolytic mechanism aimed at disrupting the integrity of the cell membrane and cell wall of bacteria. At the moment, five such mechanisms are known: the threshold concentration mechanism, the conformational phase transition mechanism, the keg mechanism, the toroidal pore mechanism and the “carpet” mechanism. In this review we suggest a new mechanism based on the direct coaggregation of amyloidogenic peptide with its protein or protein silencing mechanism.
3.2. Mechanism of the Threshold Concentration
The mechanism of the threshold concentration includes two stages of the interaction of AMP with the cell. First, the peptide under the action of electrostatic forces binds to phosphate groups in the membrane and accumulates on the surface. After reaching a certain concentration, the second stage begins. Peptides begin to penetrate and pass through the lipid bilayer, binding to intracellular targets. It is assumed that the membrane potential electrophoretically draws cationic peptides into a non-polar membrane environment, thereby reducing the energy barrier for pore formation [142]. In addition, peptides can multimerize and thus form pores. A recent modeling study also showed that the orientation of Mac1 AMP remains transmembrane in bilayers and on micelle surfaces regardless of lipid charge. Thus, as suggested by the authors, the orientation of the peptide apparently, depends more on the curvature of the membrane than on the surface charge [143]. In addition to the concentration of the peptide and the tendency to self-assembly, the phospholipid composition of the membrane and its fluidity can be factors influencing the rate-limiting stage of the process.
3.3. Conformational Phase Transition Mechanism
The conformational phase transition mechanism involves a change in the structure of the peptide molecule after binding to the membrane. Numerous studies using various biophysical methods show that disordered AMPs exhibit extended or random helical conformations in aqueous media under conditions of a bilipid layer [144]. Interestingly, the composition of the membrane can determine the possibility and rate of conformational transition of the peptide. For example, the frog skin PGLa peptide requires a negatively charged phosphatidylglycerol and phosphatidylethanolamine bilayer to make this transition. Compared to helical peptides, β-structured peptides containing disulfide bonds are generally more stable in polar and non-polar solvents. However, some quaternary peptides dissociate upon contact with the membrane surface. The selective toxicity and antimicrobial activity in this case can be explained by monomerization of oligomeric peptides.
3.4. Keg Mechanism
The keg mechanism implies the action of AMP through the formation of pores in the cell membrane. Pore-forming peptides are located in the membrane in a barrel-like form. The keg walls are formed by individual peptides or peptide complexes. The β-regions and α-helices face outward and together with the acyl chains of the bilipid layer form a hydrophobic surface, while the pore channel is formed by a hydrophilic surface [145]. The initial stage of pore formation involves the binding of peptides, probably in the form of monomers, to the membrane surface (see Figure 3A). Upon binding, the peptide can undergo a conformational phase transition, causing a thinning of the membrane due to the redistribution of the polar heads of phospholipid groups. At this stage, the hydrophobic part of the peptide penetrates the membrane. In addition, the peptide accumulates in the bilipid layer until the concentration reaches the threshold required for aggregation and incorporation into the hydrophobic membrane core. The transmembrane configuration of the peptides provides minimal impact on the membrane surface, and the pore expansion is associated with the attachment of peptide monomers. An example of such a mechanism of action was proposed for alamethicin [146,147]. Alamethicin-induced membrane permeability changes stepwise with continuous increase in peptide concentration. This fact indicates the presence of pores of different diameters corresponding to channels consisting of four or more transmembrane peptides.
Figure 3.
Scheme of diversity of mechanisms for antimicrobial action of AMPs. (A) Barrel-stave model; (B) Toroidal pore model; (C) “Carpet” model; (D) Mechanism of direct coaggregation or protein silencing.
3.5. Toroidal Pore Mechanism
One of the well-studied mechanisms of peptide-membrane interactions is the toroidal pore mechanism. The main difference between this mechanism and the «barrel-strand» mechanism is the intercalation of peptides by membrane lipids (Figure 3B). In this model, peptides in the extracellular environment acquire a helical structure due to interaction with a charged and hydrophobic bacterial membrane. Initially, the helices are oriented parallel to the membrane surface, which is confirmed by the data of NMR, fluorescence, and circular dichroism [148]. Hydrophobic residues of bound peptides displace the polar heads of phospholipids, creating a rupture in the hydrophobic region and causing membrane deformation [148]. The resulting pressure and thinning further destabilize the membrane surface, making it more vulnerable to subsequent peptide interactions. When the threshold concentration is reached, peptides are oriented perpendicular to the membrane, which was demonstrated for magainin at a peptide/lipid ratio of 1:30 [84]. At a threshold concentration, peptides tend to self-association, which leads to the formation of a peptide/lipid-supramolecular pore complex. Pore dissociation leads to the penetration (translocation) of peptides into the cytoplasm and (their) binding to intracellular targets. The characteristic features of toroid pores are finite channel lifespan, discrete size, ionic selectivity, and an inverse relationship between peptide stability and peptide charge [149,150,151].
3.6. The “Carpet” Mechanism
The “carpet” mechanism is characterized by membrane damage without penetration of AMP into the cell (Figure 3C). In this model, the target membrane is covered with an extensive layer of peptides. Changes in the mobility of phospholipids due to molecular architecture and membrane lead to and/or a decrease in the barrier properties of the membranes and subsequently to the destruction of membranes [152]. As in other models, peptides initially bind to the membrane mainly through electrostatic interactions, covering the phospholipid bilayer [153]. Upon reaching the threshold concentration of the peptide, the membrane collapses due to energy losses. Thus, membrane disruption occurs without channeling, and peptides do not penetrate into the hydrophobic membrane core. The AMP cecropin P1, which is characterized by such a mechanism of action, is initially oriented parallel to the membrane and does not penetrate into the hydrophobic medium. This orientation destabilizes the packing of phospholipids and causes membrane destruction when a concentrated layer of peptide monomers is formed on the surface [154]. Likewise, the tryptophan-rich peptide indolicidin does not penetrate the membrane according to the data of fluorescence spectroscopy, but it is an effective AMP [155].
In recent years, many authors have discovered intracellular targets for the action of AMPs. For example, the mechanism of action of human β-defensin 3 is not based on the formation of specific transmembrane pores, but on interference with the organization of membrane-bound multienzyme mechanisms, such as the cell wall biosynthesis complex and the electron transport chain [156,157]. Fungal defensin eurocin inhibits cell wall synthesis by binding equimolar to the cell wall precursor lipid II [158]. Comparison of the mechanisms of action of the two lipopeptides showed that the lipopeptide compound INA-Ac-5812 directly destroys the integrity of the membrane, while daptomycin exhibits an antimicrobial effect, binding to methicillin-resistant Staphylococcus aureus cells and interrupting cell wall biosynthesis with followed by delocalization of peptidoglycan components of the biosynthetic apparatus [159,160]. It has been shown that AMPs are capable of altering DNA morphology, with possible effects of nucleic acid disruption and fragmentation [161]. For example, buforin II and takiplesin bind to DNA, pleurocidin and indolicidin inhibit the DNA, RNA and protein synthesis (biosynthetic processes), drosocin and pyrrhocoricin reduce enzymatic activity, mersacidin inhibits the biosynthesis of the cell wall [162]. The study of supramolecular structures formed between DNA and AMPs has demonstrated a correlation between these structures and their ability to activate toll-like receptor 9 (TLR9), an important receptor expressed in cells of the immune system [163]. As mentioned above, AMPs exhibit a wide range of biological activities (Figure 2). Moreover, a number of AMPs are known to perform two or more inhibitory functions, which may indicate the action of one peptide against different targets [164]. Therefore, it is important to consider both membranolytic and intracellular mechanisms of AMP action.
It was noted that the action of some AMPs does not lead to cell lysis. One of these peptides is indolicidine. This peptide inhibits only DNA biosynthesis. The targets of indolicidine are regions of single- or double-stranded DNA with deleted nitrogenous bases. The peptide was found to have a high affinity for double-stranded (ds) [AT], ds [CG] and ds [AG], but less affinity for ds [GT]. The binding of indolicidine to duplex DNA is stabilized by the central Pro-Trp-Trp-Pro motif, thus DNA replication and transcription are inhibited. The ability of indolicidine to inhibit DNA topoisomerase was also noted [165]. Another peptide, microcin 25, synthesized by E. coli, is able to bind to the catalytic site of RNA polymerase, which temporarily stops the elongation of the transcript. It is assumed that microcin 25 can bind to the 3′-end of the newly synthesized template and block the elongation complex of GreA/GreB-dependent transcription [166].
AMPs can also hamper processes of protein synthesis. The translation system can be blocked not only by inhibition of chaperones, translation factors or ribosomes, but also at the transcriptional level. The interaction of the peptide with any of the associated enzymes or effector molecules can interrupt protein biosynthesis. It is reported that Bac7 peptide, isolated from neutrophil granules, in addition to its membranolytic activity, inhibits translation when interacting with ribosomes [167]. Proteome analysis revealed that E. coli treated with Bac7-demonstrated enrichment in a number of metabolic pathways, including nucleic acid metabolism (transport and metabolism of nucleotides) [168].
Another interesting mechanism of the antimicrobial action of peptides is the inhibition of the protein folding system. DnaK is the main bacterial heat shock protein 70 (Hsp70), which plays a key role in the folding process in E. coli cells. It should be also noted that, in addition to inhibiting protein biosynthesis, Bac7 has a high binding constant with DnaK and inhibits the protein folding function in the DnaK-DnaJ-GrpE-ATP molecular chaperone system [169]. Proline-rich AMPs (pyrocorticin isolated from the bug Pyrrhocoris apterus and apidaecin from the beetle Riptortus clavatus) have the DnaK-binding YL/IPRP motif. Both pyrocorticin and apidecin inhibit DnaK and GroEL, and also disrupt the ATPase activity of DnaK [170,171]. Another proline-rich AMP, oncocin, consisting of 19 amino acids and obtained from the antibacterial peptide oncopeltus, exhibits high antibacterial activity against Gram-negative bacteria. The peptide easily penetrates the bacterial membrane without any lytic effect on cell. Oncocin has been shown to form a complex with DnaK, which indicates an inhibitory activity against protein folding [172].
3.7. Mechanism of Direct Coaggregation or Protein Silencing
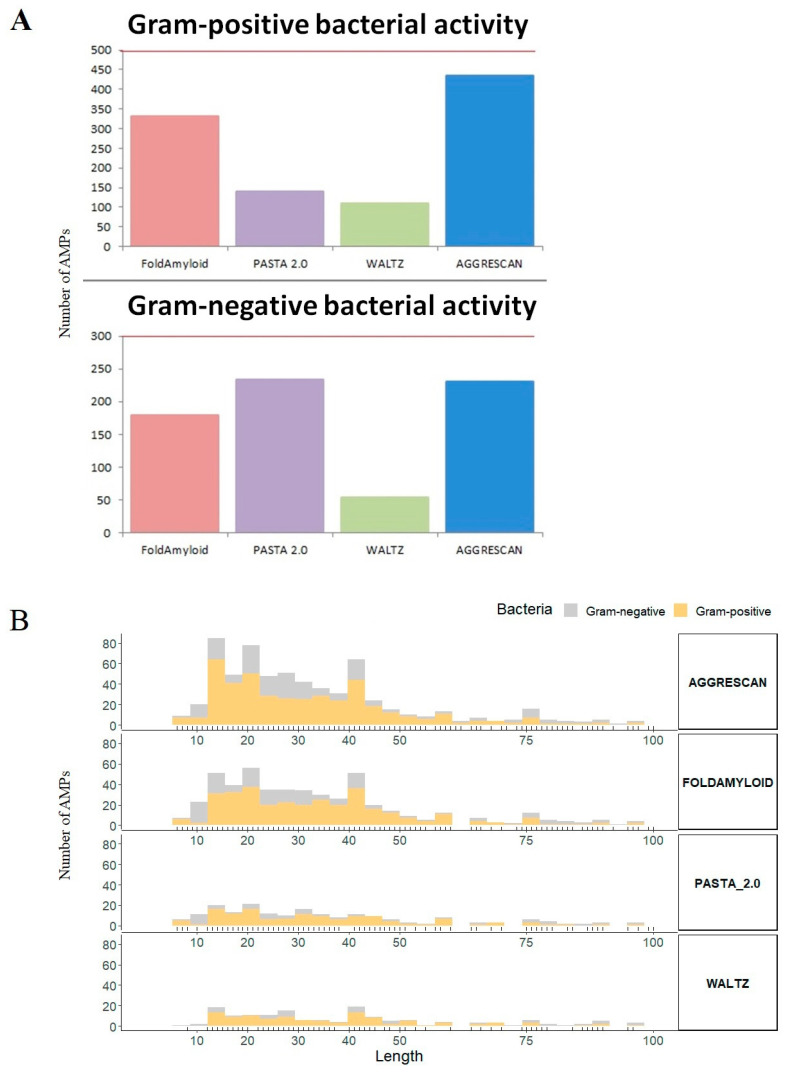
Here we should talk about the mechanisms of the antimicrobial action of amyloidogenic peptides [12]. Most studies of the antimicrobial mechanisms of action of amyloids reveal their destructive effect on cell membranes, for example, through channel/pores formation or membrane thinning [12,173,174,175]. Several studies have demonstrated the formation of antibacterial nanonets by amyloids, which inhibit the spread of microbes [176,177]. AMPs capable of forming hydrogels with mechanical protective properties for the further proliferation of both planktonic bacterial cells and biofilms are of particular interest [178,179]. Using the software AGGRESCAN [180], FoldAmyloid [181], PASTA 2.0 [182] and Waltz [183], we searched for and predicted amyloidogenic regions in a sample of 300 and 499 AMPs showing activity against Gram-negative and Gram-positive bacteria, respectively (Figure 4).
Figure 4.
Number of AMPs with predicted amyloidogenic regions by AGGRESCAN [180], FoldAmyloid [181], PASTA 2.0 [182], and Waltz [183]. (A) General number (the red line showed count peptides analyzed), (B) Length distribution of AMPs.
Figure 4 demonstrates similar patterns of distribution of predicted amyloidogenic regions in AMPs, showing activity against Gram-negative and Gram-positive bacteria, respectively. This may indicate that the ability to aggregate may be combined with antimicrobial action against Gram-negative and Gram-positive bacteria. Interestingly, amyloidogenic regions are predicted in about half of the AMP samples. The median length of the AMP was 30 amino acid residues (a.a.), and the median length of the amyloidogenic region was 17 a.a. residues. At the moment, there is no theory about how antimicrobial activity and aggregation ability are related.
At the same time, we would like to consider a new mechanism of the antimicrobial action of amyloids based on the targeted blocking of the functions of a key protein, the so-called protein silencing. This mechanism is based on the phenomenon of coaggregation of proteins and peptides [184,185,186]. Thus, it is known that human serum albumin is able to form coaggregates with the amyloid β (Aβ) peptide due to binding sites that are evenly distributed over three domains on the protein surface [187,188]. Human serum albumin, like many other multi-domain proteins (immunoglobulin light chains [189,190], titin [191,192], YB1 [193,194], tends to aggregation and is capable of forming fibrils [195].
Moreover, the process of aggregation/fibrillation of human serum albumin involves changes in the tertiary structure of domains II and III [196]. It was shown that human serum albumin itself can prevent the formation of amyloids not only for the Aβ peptide, but also for insulin [197], and the regions of this protein that are responsible for interaction with amyloids were identified [198,199]. Studies of the formation of amyloids and aggregation sites made it possible to hypothesize site-specific coaggregation of the target protein with an amyloidogenic peptide as one of the possible antibacterial mechanisms of AMP action.
The largest multi-domain and functionally important bacterial ribosomal S1 protein was proposed as a target protein, the disruption of which is critical for the life of bacteria [200,201,202]. The full-length S1 protein has a high mobility of individual protein domains and a tendency to aggregation, which creates problems for establishing its three-dimensional structure [203,204]. At the same time, using computational and experimental approaches, we described specific amyloidogenic regions within the domains of the S1 protein, which can be potential sites for modulating the aggregation properties of bacterial ribosomal S1 proteins [9,205,206,207]. Based on the predicted amyloidogenic regions, amyloidogenic peptides were synthesized, some of which exhibited antimicrobial activity in vitro tests [16].
The mechanism proposed by us can be divided into several stages. The first stage is the penetration of the effector molecule into the cytoplasm of the bacterial cell. We assume that the penetration of peptides occurs by analogy with peptides penetrating into cell [208]. Penetration through the bilipid layer can be carried out in several ways. One of the methods involves direct translocation of the peptide across the membrane and its local disturbance without further destruction. [209]. The peptide crosses the bilipid layer and enters the cytoplasm due to the transmembrane potential, pH gradient, and other physicochemical processes [163,210]. Another way of peptide penetration through the membrane can be via by endocytosis and/or macropinocytosis [211,212].
The second stage is the binding of the effector molecule and the target protein. The action of the effector molecule can lead to both bacteriostatic and bactericidal action, depending on the chosen target. The target protein must perform several vital functions in the cell, which are disrupted when this protein coaggregates with an effector peptide. To assess possible binding, various approaches can be used both in silico based on interaction patterns [213,214], and in vitro using spectrophotometry, NMR, or electron microscopy [215,216,217].
The third stage is the aggregation of the effector molecule and the target protein. Self-aggregation of a protein or its coaggregation with an effector peptide should lead to blocking of protein functions and a decrease in the viability of the bacterial cell. In our study, we used methods of thioflavin T fluorescence, electron microscopy, spectrophotometry and mass-spectrometry to assess the amyloidogenic and antibacterial activity of peptides synthesized on the basis of the ribosomal S1 protein from Thermus thermophilus [16]. It has been experimentally demonstrated that amyloidogenic regions of the ribosomal S1 protein can be used as candidates for the development of new antibacterial peptides [16].
Coaggregation of the target and effector can occur by the mechanism of aggregation controlled by nucleation, which has already been demonstrated for the Aβ(1-40) peptide during the formation of amyloid fibrils [218]. Interestingly, Aβ peptides isolated from the brains of Alzheimer’s disease patients have amyloidogenic properties as well as antimicrobial activity, and that incubation of these brain samples with antibodies to Aβ reduced the antimicrobial activity of the peptides. Several studies report that Aβ peptides have demonstrated antibacterial and antifungal activity, as well as antiviral effects, in particular against influenza virus. At the same time, as the authors note, Aβ(1-42) has a greater antimicrobial or antiviral activity than Aβ(1-40), which suggests a possible relationship between the ability of Aβ(1-42) to assemble into oligomers and its antiviral activity, since this peptide has a greater tendency to form oligomers and fibrils than Aβ(1-40) [11].
4. Prediction and Development of New AMPs
4.1. Types of Programm for Prediction and Development of New AMPs
Over the past decades, great efforts have been made to discover AMPs in natural sources using wet biology techniques. Classical chromatographic methods for the detection of natural AMPs have played an important role in shaping the modern view of AMP in terms of biological source, amino acid sequence, three-dimensional structure, and antimicrobial activity. Since the 90s of the twentieth century, there are more and more possibilities for predicting and designing AMP with a specific function, pharmacological target and activity. The first report of such a discovery was associated with human cathelicidin, this peptide was detected using the method of the conservative sequence alignment of the pro-peptide domain [219]. In 2004, Yount and Yeaman discovered a motive for identifying new AMPs [220]. In addition, more efficient genomic and proteomic methods for the identification of peptides with antimicrobial function have been developed for peptide in recent years [221,222]. The application of prediction methods to genomes and proteomes, followed by experimental verification, accelerates the rate of discovery of peptides, which, in turn, allows expanding understanding of the functional nature of these compounds.
The similarity of peptides with and without antimicrobial activity is a bottleneck for bioinformatics methods. To solve this problem, programs for predicting antimicrobial activity based on various mathematic algorithms and information sources are used.
The programs of the first category use only data on the primary structure of mature AMPs. Based on these data, the G-X-C motif was discovered in peptides containing disulfide bonds (defensins) [223]. Using this motif, Yount and Yeaman discovered previously unknown peptides brazzein and charybdotoxin, which are active against bacteria and fungi Candida albicans [220]. A total of 28 new human defensin was identified. Programs in this category determine the length of the peptide, its charge, the content of hydrophobic residues in the molecule, and its amino acid composition. The predictions are made according to the correspondence of these peptide parameters to the range of parameters from the database. Machine learning methods are also used for prediction. Artificial neural networks perform highly accurate predictions based on rules from databases of antimicrobial peptide.
The programs of the second category use highly conserved sequences of amino acid residues of pro-peptides. The amino acid sequences of mature peptides are more variable than the sequences of pro-peptides. This observation lies in the strategy of searching for such peptides [100]. Since the discovery of the first (antimicrobial) peptide, cathelicidin, in 1988, more than 100 such peptides have been identified. Analysis of processing signals allows predicting antimicrobial activity of other classes of AMPs. For example, amphibian antimicrobial peptide precursors also have a common and highly conserved pro-region, usually terminating in a typical Lys-Arg processing signal. This discovery was used as the basis for identifying many amphibian peptides [222,224].
A combination of approaches from previous types of programs is used in the third category. Fjell et al. created a program for prediction based on both pro-peptide sequences and mature peptides [225]. Peptides from Antimicrobial Sequences Database (AMSDb) are classified into several clusters, after which prediction parameters are calculated [226]. The authors identified 146 clusters for mature peptides and 40 clusters for pro-peptides. Using this program, it was possible to achieve 99% prediction accuracy. However, it is unclear how many more clusters can be found based on the most recent collection of naturally occurring AMPs [63], and whether it is possible to make the prediction even better.
The fourth type of programs is based on the search for homologous sequences of enzymes that carry out the transfer or modification of AMPs. LanM is a group of proteins that modify peptides from the lantibiotics family. The search for homologs of this group of proteins led to the discovery of new peptides in this family—haloduracin and lichenicidin [227]. Using this approach, 89 LanM homologues were identified; some of them were found in 61 strains that do not produce lantibiotics. The conserved LanT transporter was used to detect other lantibiotics. Based on the similarity with this protein, Singh and Sareen identified 54 bacterial strains containing LanT homologues [228]. Morton et al. used a protein cluster required for modification, transport, and resistance to bacteriocins for prediction [229]. Using this program, the authors predicted bacteriocin gene blocks for 2773 genomes.
The last type of program uses genetic information about expression, processing, and transport, and also performs comparisons with the already described AMPs. The processing of eukaryotic transcription data using data mining tools has allowed the identification of new AMPs. Lynn et al. identified 9 novel chicken AMPs by searching for homology tags of clustered expressed sequences using BLAST and a hidden Markov model [230]. Amaral et al. identified about a hundred AMPs based on physicochemical characteristics: peptide length, total charge surface, and hydrophobic moment [231]. Proteolytic cleavage of proteins can release AMP. Based on this fact, Torrent et al. developed a theoretical method for identifying potentially active regions with antimicrobial activity [232]. Hellinger et al. achieved significant success by combining transcriptomic and proteomic data [233]. They were able to identify 164 AMPs from single plant: 108 based on transcriptomic data only and 127 based on mass spectrometry data. Perhaps one of the most effective strategies for searching of natural candidates for AMPs is based on in silico methods of analysis of RNAs synthesized after immunization of host cells with bacteria [234].
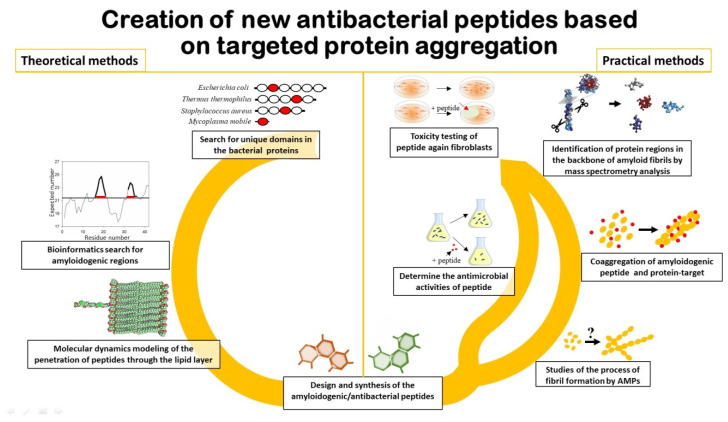
It is important to note that in order to predict AMPs that will act by the mechanism of coaggregation with the target protein (Figure 5), it is first of all necessary to search for amyloidogenic regions in this protein [205,235]. Approaches that include algorithms to predict aggregation/amyloidogenic sites may be useful for this purpose [180,181,182,183]. In addition, information on specific folded/unfolded or rigidity/flexibility regions of the protein may be appropriate for the development of AMP based on the sequence of the target protein [236,237].
Figure 5.
Schematic representation of the main aspects associated with the development of new antibacterial peptides.
It is important to take into account AMP development programs that are used to predict a possible immune response and allergic reactions to a specific peptide structure [238]. This validation is especially important for drug development based on AMPs, in particular, antiviral drugs, which will be discussed in more detail in the final section of this review.
4.2. SARS-CoV-2 Like an Object for Prediction and Development of New AMPs
Coronaviruses (CoVs) are a diverse group of viruses that infect a variety of animals, including live animals, including livestock, poultry, and can cause mild to severe respiratory infections in humans. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a novel highly transmissible, pathogenic coronavirus that emerged at the end of 2019 and triggered a pandemic of new acute respiratory disease, also known as coronavirus disease 2019 (COVID-19). Previously, Severe Acute Respiratory Syndrome Coronavirus 1 (SARS-CoV-1) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) caused an outbreak of unusual viral pneumonia in 2002 and 2012, respectively [239]. 216 countries and regions from all six continents have reported more than 65 million COVID-19 cases, and more than 1.5 million patients have died, according to data from www.worldometer.info on December 4, 2020. The spread of SARS-CoV-2 is currently a serious threat to the health and life of people around the world. The efforts of scientists and specialists are aimed at developing effective methods of treating coronavirus infection. Clinical trials of various treatments for SARS-CoV-2 are currently underway, but none have been approved yet. Thus, SARS-CoV-2 is a relevant object for demonstrating the features of a strategy for the development of AMPs against a specific virus. It is well known that some peptides have antimicrobial activity against viruses. For example, melittin has significant antiviral activity and reduces the infectivity of enterovirus, human immunodeficiency virus (HIV), influenza A viruses, vesicular stomatitis virus (VSV), and some other viruses [240]. Thus, AMPs, derived from cathelicidin, effectively suppress Ebola virus the infection [241].
The genome of SARS-CoV-2 is around 30 kb, and is a positive-sense single-stranded RNA (+ssRNA) that contains six functional open reading frames (ORFs): spike (S), nucleocapsid (N), membrane (M), envelope (E) and replicase (ORF1a/ORF1b) [242]. The SARS-CoV-2 S protein consists of two subunits. The S1 subunit is important for binding to the host cell angiotensin-converting enzyme 2 (ACE2) receptor and the S2 subunit is responsible for the fusion of the virus and the cell membrane [243]. The S1 region is further divided into two functional domains, N-terminal domain (NTD) and three C-terminal domains (CTDs) [244]. CTDs contain a region (amino acids 319–529) of the receptor-binding domain (RBD), which plays a key role in contact with the hACE2 receptor [245]. Fourteen RBD residues bind to the hACE2 receptor. These are Tyr449, Tyr453, Asn487, Tyr489, Gly496, Thr500, Gly502, and Tyr505 are conserved in SARS-CoV-2 RBD, while Leu455, Phe456, Phe486, Gln493, Gln498 and Asn501 are substituted in different forms RBDs of SARS-CoV-1 and SARS-CoV-2 [246]. Biochemical analysis and pseudovirus penetration assay confirmed that the structural features of SARS-CoV-2 RBD have a higher hACE2 binding affinity than SARS-CoV RBDs [247]. The S2 subunit contains regions required for membrane fusion, including two heptad repeats (HR), a transmembrane domain (TM), an internal membrane fusion peptide (FP), and a membrane-proximal external region (MPER) [248].
Infection mechanisms have been investigated and it has been highlighted that the SARS-CoV-2 viral cell-surface spike protein targets hACE2 receptors. Cannalire et al. [249] discussed that the SBP1 peptide has been identified to inhibit the S/ACE2 interaction by targeting the RBD region of the S protein, but the RBD region is more prone to mutation, making it difficult to develop broad-spectrum inhibitors. In this case, the preferred approach is fusion inhibitors targeting the more stable heptad repeats (HRs) involved in the membrane fusion process. Among the peptides described, the EK1C4 lipopeptide exhibits strong S-mediated inhibition of fusion combined with selective broad antiviral activity in cellular assays against SARS-CoV-2 and other relevant CoVs such as MERS-CoV [249]. Another study used a programmatic approach to search and identification based on the hACE2 sequence of ten peptides that have a high potential for interaction with CoV-RBD [250]. Drugs targeting this mechanism are not yet available, but the recent work has demonstrated that peptide hACE2 mimics, creating from the H1 helix and consisting only of natural amino acids, block infection of human lung cells with IC50 in the nM range [251]. It is about eliminating the effect on the renin–angiotensin system and the kinin–kallikrein system in the light of the pathophysiological mechanisms associated with SARS-CoV-2 [252,253].
Similarly, to fusion proteins of other coronaviruses, SARS-CoV-2 S is activated by cellular proteases. It is assumed that the S1 and S2 subunits remain non-covalently linked after cleavage [254]. It is known that furin protease of the host cell cleaves the SARS-CoV-2 at the S1/S2 site. Cleavage at the S1/S2 site (residues 669−688) and subsequent cleavage at the S2’ site (residues 808−820) by the TMPRSS2 protease is important for virus penetration into lung cells [255,256]. Tavassoly et al. presented an interesting view that a peptide that is cut between two sites (S1/S2 and S2′) is detached from the virus and enters the intra- or extracellular environment. Subsequently, this peptide can induce some immunological reactions and act as a functional amyloid. The latter hypothesis was supported by the data of the AGGRESCAN program, which identified sites in the S-CoV peptide responsible for the self-aggregation properties [257]. Because the SARS-CoV-2 spike protein is critical for penetration into the host cell, it could be an interesting target protein option for the development of antibacterial peptides. The identification of regions of a protein molecule prone to aggregation/formation of amyloid fibrils is an important step for the development of AMPs acting on the basis of a coaggregation mechanism. For this reason, we used several bioinformatics tools (FoldAmyloid, PASTA 2.0, Waltz and AGGRESCAN), which were previously used [205,207] to predict amyloidogenic sequences in proteins. The results of predicting amyloidogenic regions for the SARS-CoV-2 S protein by various methods are shown in Table 2.
Table 2.
Amyloidogenic regions predicted by different programs for SARS-CoV-2 spike protein.
| Software | Amyloidogenic Regions for SARS-CoV-2 Spike Protein (https://www.uniprot.org/uniprot/P0DTC2) |
|---|---|
| AGGRESCAN (UAB, Barcelona, Spain http://bioinf.uab.es/aggrescan/) |
1–8, 10–16, 31–38, 40–47, 50–56, 58–70, 86–91, 100–109, 114–133, 140–145, 162–167, 189–208, 231–247, 260–271, 303–311, 331–336, 338–354, 362–383, 390–400, 428–437, 448–457, 484–490, 508–520, 584–588, 590–599, 610–617, 666–671, 688–698, 716–727, 729–745, 751–759, 761–768, 783–787, 797–805, 817–833, 853–861, 864–870, 872–914, 961–982, 1000–1013, 1044–1052, 1058–1068, 1097–1101, 1124–1137, 1171–1179, 1207–1249. |
| PASTA 2.0 (University of Padova, Padova, Italy http://protein.bio.unipd.it/pasta2/) |
1214–1244. |
| Waltz (Switch Laboratory, Leuven, Belgium http://waltz.switchlab.org/index.cgi) |
1–9, 88–95, 115–137, 141–146, 199–204, 261–271, 275–280, 365–378, 447–455, 485–496, 537–544, 608–618, 689–697, 703–727, 798–806, 913–940, 969–974, 1003–1015, 1063–1068, 1117–1122, 1175–1179, 1206–1233. |
| FoldAmyloid (Institute of Protein Research of the Russian Academy of Sciences, Pushchino, Russia http://bioinfo.protres.ru/fold-amyloid/) |
1–10, 34–38, 53–69,100–108,116–120, 126–135, 141–146, 174–178, 190–195, 235–244, 264–269, 274–278, 327–331, 348–353, 366–370, 389–395, 431– 436, 450–457, 487–492, 507–519, 539–543, 560–564, 582–586, 609–613, 691–697, 718–722, 738–743, 751– 755, 780–784, 799–804, 819–825, 875–880, 894–907, 993–999, 1005–1014, 1047–1051, 1060–1067, 1101–1105, 1127–1132, 1210–1239. |
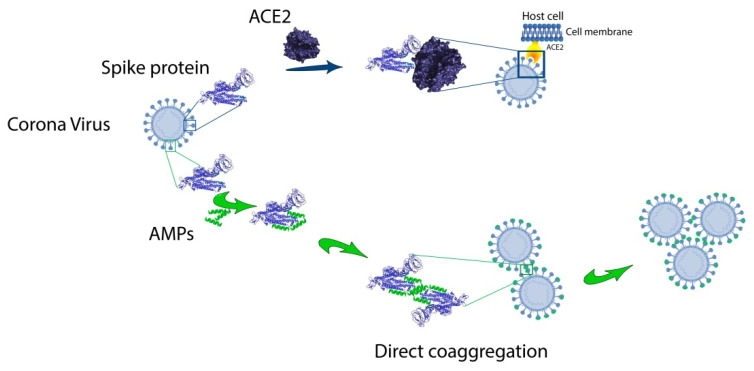
As can be seen from Table 2, amyloidogenic regions identified by any software as amyloidogenic ones are predicted within one amino acid residue, but there are no completely similar predictions for FoldAmyloid, PASTA 2.0, Waltz and AGGRESCAN. However, the prediction of the amyloidogenic region of about 30 amino acid residues at the C-terminus of the SARS-CoV-2 spike protein, common for all four programs, is noteworthy. However, this region corresponds to the transmembrane domain. Interestingly, for the envelope (E) protein in SARS-CoV-1, amyloidogenic sequences at the C-terminus were also previously noted [258]. The same work discusses the possible role of the amyloidogenic sequence in the performance of protein E of its functions of binding to the host cell membranes and formation of ion channels. In our opinion, the amyloidogenic region in the middle of SARS-CoV-2 protein S can be the basis for the development of AMPs acting by the mechanism of directed coaggregation [16]. We hypothesize that anti-CoV peptides can bind to RBD of spike S1 protein (Figure 6). This interaction should reduce the binding affinity of the S1 to the hACE2 receptor of the target cell. A feature of this mechanism is the possibility of viral particles “sticking together”, which can prevent the spread of the virus and reduce the viral load.
Figure 6.
Hypothetical mechanism of direct coaggregation against CoVs.
Based on a similar logic, a number of authors tried to synthesize peptide inhibitors of the Spike–hACE2 interaction against SARS-CoV-1 and SARS-CoV-2 viruses [259,260,261,262,263]. As mentioned above, therapeutic peptides have several disadvantages: low bioavailability, short half-life and toxicity. Therefore, lipidation, PEGylation, glycosylation will be useful for improving the pharmacological properties of peptides. In addition, glycosylation can facilitate the specific recognition of viral particle binding sites. For example, how it happens, C-type lectins recognize pathogen patterns [264,265,266].
Recently, promising technologies for creating a vaccine against COVID-19 have been described. In these works, it is proposed to use antigenic peptides developed on the basis of peptide epitopes of viral proteins to inhibit SARS-CoV-2 infection and based on peptide epitopes of T and B lymphocytes to stimulate the human immune response [267]. High-performance in silico technologies currently allows the development of new approaches to the creation of peptide vaccines. Antigenic epitopes found in viral proteins can simultaneously meet several criteria, including immunodominant regions, non-allergenicity, population coverage, and lack of variability (conservatism) for efficient binding and molecular interaction with HLA (human leukocyte antigen) and TLR (toll-like receptor) alleles [238]. To evaluate the effectiveness of peptide vaccines against SARS-CoV-2, the frequencies of the HLA haplotypes can be used to predict the coverage of the developed vaccine. At the same time, peptides are evaluated based on the frequency of HLA haplotypes or HLA alleles in the target population, peptides with undesirable properties are filtered out, which are expected to be glycosylated, identical to peptides in the human proteome, or rapidly degraded [268] Using in silico methods, epitopes of B cells, T cells, and IFN-gamma present on four structural proteins of the virus were mapped, and multi-epitope peptide-based vaccine was developed [269]. It should be taken into account when developing a peptide vaccine that some peptides can provoke an increased production of interleukin 6. In general, the increased mortality of patients with COVID-19 is associated with the induction of the cytokine storm. The work predicted 222 peptide sequences based on the viral spike protein, which can cause increased production of interleukin 6 (IL-6) [270]. The peptide protein kinase inhibitor CK2, previously proven in cancer, can also be used in the treatment of Covid-19. In a small (20 patient) randomized controlled clinical trial, CIGB-325 peptide reduced the mean number of lung lesions. However, in some patients, CIGB-325 may cause itching, redness, and rashes. In the future, it is planned to test the CIGB-325 peptide on more subjects in various combinations with antiviral drugs [271]. Preliminary encouraging results were obtained with the intravenous administration of the CIGB-258 immunomodulatory peptide to critically or severely ill COVID-19 patients. CIGB-258 significantly reduced the levels of biomarkers associated with hyperinflammation, IL-6 and tumor necrosis factor (TNFα) during treatment [272]. Another work suggests the use of annexin A1 Ac2-26 mimetic peptide, which reduces IL-6 production, pain and exudate, which could be a promising treatment in the fight against COVID-19 [273]. In addition, it is assumed that some short peptides, which that have already been developed and tested in the treatment of other diseases, can be used as inhibitors of the SARS-CoV-2 virus, as well as immunomodulators and bronchoprotectors of the pathological process with COVID-19 [274]. In a recent work, a method for preparing a peptide-mRNA vaccine based on engineered peptides synthesized on the binding sequence of the hACE2 receptor, which targets the RBD of the viral S protein, and simultaneously recruits ubiquitin E3 ligases for subsequent intracellular degradation of SARS-CoV-2 in the proteasome, is presented. In a vaccine, mRNA encodes stable perfused forms of the S protein, so that immune cells recognize them [275].
Drug redirection methods for the treatment of SARS-Cov-2 infection are currently being investigated. 300 peptide-like structures from various databases were selected. Using molecular dynamics modeling and docking analysis, the four peptide-like structures demonstrated strong binding affinity for amino acid residues within the SARS-CoV-2 site, the major protease of Mpro, also called 3CLpro [276,277]. The evolutionary aspect of SARS-CoV-2 should be considered in order to develop peptides that can further target the virus. Thus, in order to select four peptides with strong binding affinity for the main protease SARS-CoV-2, 2765 sequences containing a wide range of mutants from patients with COVID-19 were analyzed by molecular dynamics methods [278]. A computational approach was used to select 15 peptides that showed a higher affinity for RBD of SARS-CoV-2 S protein compared to the α-helix of the hACE2 receptor. It was noted that, in all the detected stable peptide-protein complexes, the Tyr489 and Tyr505 residues in RBD are involved in interaction, which suggests that they are critical for the binding of the discovered antiviral peptides to RBD [279]. After modeling based on the key interacting motifs of the spike protein, four new synthetic SARS-BLOCK™ peptides have been developed and characterized, which can serve as a combined therapeutic, immune and prophylactic agent against SARS-CoV-2. The technology may be interesting if it is possible to show the low cytotoxicity of the developed peptides [280].
Since the COVID-19 pandemic was announced, various options have been proposed to combat the infection. Recently, reviews have appeared that draw attention to the prospects for the development and use of antiviral peptides in the therapy of SARS-CoV-2, SARS-CoV-1, MERS-CoV and other respiratory viruses [281]. In particular, there was a point of view that synthetic or natural AMPs can be used to reduce the viral load on cells by blocking the contacts of the virus with receptors on the cell surface [22]. The use of protein/peptide inhalers, due to their prolonged action, higher efficacy, lower systemic availability and minimal toxicity, may be an effective approach for the treatment of SARS-CoV-2 [282]. Modification of existing natural AMPs is an attractive approach, since it can lead to the creation of new antiviral therapeutic agents [11]. The study shows the promise of using nisin, a dietary AMP produced by lactic acid bacteria, which can bind to hACE2, competitively inhibiting RBD SARS-CoV-2 [283]. Against COVID-19, it is proposed to use an AMP such as lactoferrin (LF), which is an iron-binding glycoprotein. However, it should be noted that the antiviral mechanisms of LF differ from virus to viruses. Lactoferrin can enhance the host’s immunity against viral infection or bind directly to the viral particle or receptors and heparan sulfate proteoglycans on the cell-surface of the host cell [284,285]. Interestingly, a recent review suggests the use of peptide BPP-10c (Glu-Asn-Trp-Pro-His-Pro-Gln-Ile-Pro-Pro) derived from venom of Bothrops jararaca to counteract the effects of COVID-19 [286]. It has been suggested that AMPs that destroy the envelope glycoprotein can be used to stop or disrupt the life cycle of SARS-CoV-2 [287]. AMPs, which are found in the secretion of mesenchymal stem cells (MSCs), have antimicrobial properties, the ability to attenuate cytokine storms, and, therefore are an attractive approach to prevent the COVID-19 pandemic [288].
The influence of the coronavirus on the course of Alzheimer’s disease has been disclosed. It has been shown that Alzheimer’s disease can be triggered by various bacterial diseases, as well as certain viruses, albeit indirectly. The hypothesis is relevant due to the fact that many patients with COVID-19 have central nervous system disorders and cerebrovascular diseases. There is no data on the penetration of the virus into the human central nervous system, but this is important due to the presence of hACE2 expression in human nerve cells, and the virus can also enter the cerebrospinal fluid through the choroid plexus of the ventricles [289]. Interestingly, Aβ(1-42) peptide associated with Alzheimer’s disease may have antiviral and, in general, antimicrobial activity. AMPs have proven to be antiviral therapy that can be effective against infection and be used to reverse the effects of virus-induced inflammation. However, it is known that viral infections are often accompanied by bacterial infections. COVID-19 is no exception, and some patients die from bacterial co-infection. It can be assumed that the SARS-Cov-2 virus has a mechanism to suppress the production of the host’s own AMPs [290].
AMPs have immunomodulatory effects, with some activating and others suppressing inflammation, which should be considered when selecting candidates. To select specific immunogenic epitopes and accelerate vaccine development, the SARS-CoV-2 genes were analyzed for B and T cell epitope candidates. In addition, the authors propose to increase the low immunogenicity of a peptide vaccine by including an adjuvant in its composition and using an effective delivery system based on chitosan or a copolymer of lactic and glycolic acid (PLGA) [291]. It is suggested that peptides derived from the short-palate, lung and nasal epithelial clone-1 (SPLUNC1), alpha-1-antitrypsin (AAT), dornase alfa (DA) and neutralizing human S230 light chain antibodies can be used as anti-adhesion agents, preventing the SARS-CoV-2 virus from interacting with the human host cell. However, the use of the SPLUNC1 peptide may be associated with 33.3% of allergic reactions, based on in silico data [282]. In the review, the authors discuss bioengineering strategies, chemical modifications, and combined approaches to finding solutions that improve the design, development of AMP, and peptide delivery technologies. It has been noted that clinical use of AMP may be hampered by general properties such as sensitivity to environmental conditions, large size, and poor distribution and excretion [292].
5. Conclusions
This review describes various peptides exhibiting antimicrobial properties, their structural and physicochemical features, membranolytic mechanisms of their action and intracellular AMP targets, methods and strategies for the search and creation of new molecules. In this work, we have shown that AMPs have a powerful potential to be used both against the gradually accumulating threat of antibiotic-resistant bacteria and against the suddenly emerging COVID-19 pandemic. Undoubtedly, in the future, the number of antimicrobial peptides used in clinical practice will increase. Improvement of algorithms for prediction and development of new peptides, taking into account the physicochemical properties of already existing AMPs, will contribute here. Additionally, chemical modification and creation of analogues of already well-studied peptides (for example, with membrane binding, amyloidogenic or lectin-like properties) open endless opportunities for the rational design of AMPs with various mechanisms of antimicrobial action. In particular, our proposed new mechanism of antimicrobial action of amyloidogenic peptides, associated with targeted coaggregation with the functional protein of the pathogenic organism, may be of interest for use against a wide range of pathogens including bacteria and viruses. The rapid spread of COVID-19 is the most serious threat to global health this century. It is noteworthy that at present, almost any scientist or just a student can search for the structures of virus inhibitors. So, a cyclic peptide inhibitor of COVID-19 has been described using publicly available software and X-ray crystallographic structures [293]. It is likely that SARS-CoV-2 will occupy a certain niche in the human body and will coexist with us for a long time [294]. Therefore, synthetic or natural AMPs are still an important subject of research [295]. And the search for new antimicrobial drugs remains an urgent problem.
Acknowledgments
We thank A.V. Glyakina and A.P. Kochetov for assistance in the manuscript preparation.
Abbreviations
| Aβ | amyloid β |
| AMP | antimicrobial peptide |
| COVID-19 | coronavirus disease 2019 |
| CoVs | coronaviruses |
| CTD | C-terminal domain |
| E | envelope protein of coronavirus |
| hACE2 | human angiotensin-converting enzyme 2 |
| HLA | human leukocyte antigen |
| HR | heptad repeat |
| IL-6 | interleukin 6 |
| MERS-CoV | middle east respiratory syndrome coronavirus |
| MIC | minimum inhibitory concentration |
| NMR | nuclear magnetic resonance |
| ORF | open reading frames |
| PRR | pattern recognition receptor |
| RBD | receptor-binding domain |
| S | spike protein of coronavirus |
| SARS-CoV-1 | severe acute respiratory syndrome coronavirus 1 |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| TLR | toll-like receptor |
Author Contributions
S.R.K., S.Y.G., A.V.P., M.V.S., S.D.C., A.K.S., and O.V.G.: conceptualization, methodology, writing—original draft preparation, and writing—review and editing. All authors contributed to writing and reviewing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the RUSSIAN SCIENCE FOUNDATION, grant number 18-14-00321.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Christaki E., Marcou M., Tofarides A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020;88:26–40. doi: 10.1007/s00239-019-09914-3. [DOI] [PubMed] [Google Scholar]
- 2.Wu Y., Ho W., Huang Y., Jin D.-Y., Li S., Liu S.-L., Liu X., Qiu J., Sang Y., Wang Q., et al. SARS-CoV-2 is an appropriate name for the new coronavirus. Lancet. 2020;395:949–950. doi: 10.1016/S0140-6736(20)30557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magana M., Pushpanathan M., Santos A.L., Leanse L., Fernandez M., Ioannidis A., Giulianotti M.A., Apidianakis Y., Bradfute S., Ferguson A.L., et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020;20:e216–e230. doi: 10.1016/S1473-3099(20)30327-3. [DOI] [PubMed] [Google Scholar]
- 4.Chen C.H., Lu T.K. Development and Challenges of Antimicrobial Peptides for Therapeutic Applications. Antibiotics. 2020;9:24. doi: 10.3390/antibiotics9010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petkov P., Lilkova E., Ilieva N., Litov L. Self-Association of Antimicrobial Peptides: A Molecular Dynamics Simulation Study on Bombinin. Int. J. Mol. Sci. 2019;20:5450. doi: 10.3390/ijms20215450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aisenbrey C., Amaro M., Pospíšil P., Hof M., Bechinger B. Highly synergistic antimicrobial activity of magainin 2 and PGLa peptides is rooted in the formation of supramolecular complexes with lipids. Sci. Rep. 2020;10:11652. doi: 10.1038/s41598-020-68416-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jang H., Teran Arce F., Ramachandran S., Kagan B.L., Lal R., Nussinov R. Disordered amyloidogenic peptides may insert into the membrane and assemble into common cyclic structural motifs. Chem. Soc. Rev. 2014;43:6750–6764. doi: 10.1039/C3CS60459D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mroczko B., Groblewska M., Litman-Zawadzka A., Kornhuber J., Lewczuk P. Amyloid β oligomers (AβOs) in Alzheimer’s disease. J. Neural Transm. 2018;125:177–191. doi: 10.1007/s00702-017-1820-x. [DOI] [PubMed] [Google Scholar]
- 9.Galzitskaya O.V. Oligomers Are Promising Targets for Drug Development in the Treatment of Proteinopathies. Front. Mol. Neurosci. 2020;12:319. doi: 10.3389/fnmol.2019.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Höppener J.W.M., Ahrén B., Lips C.J.M. Islet Amyloid and Type 2 Diabetes Mellitus. N. Engl. J. Med. 2000;343:411–419. doi: 10.1056/NEJM200008103430607. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh I.-N., Hartshorn K. The Role of Antimicrobial Peptides in Influenza Virus Infection and Their Potential as Antiviral and Immunomodulatory Therapy. Pharmaceuticals. 2016;9:53. doi: 10.3390/ph9030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M., Zhao J., Zheng J. Molecular understanding of a potential functional link between antimicrobial and amyloid peptides. Soft Matter. 2014;10:7425–7451. doi: 10.1039/C4SM00907J. [DOI] [PubMed] [Google Scholar]
- 13.Westwell-Roper C., Verchere C.B. Modulation of Innate Immunity by Amyloidogenic Peptides. Trends Immunol. 2019;40:762–780. doi: 10.1016/j.it.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Smole U., Gour N., Phelan J., Hofer G., Köhler C., Kratzer B., Tauber P.A., Xiao X., Yao N., Dvorak J., et al. Serum amyloid A is a soluble pattern recognition receptor that drives type 2 immunity. Nat. Immunol. 2020;21:756–765. doi: 10.1038/s41590-020-0698-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remington J.M., Liao C., Sharafi M., Ste.Marie E.J., Ferrell J.B., Hondal R.J., Wargo M.J., Schneebeli S.T., Li J. Aggregation State of Synergistic Antimicrobial Peptides. J. Phys. Chem. Lett. 2020;11:9501–9506. doi: 10.1021/acs.jpclett.0c02094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurpe S.R., Grishin S.Y., Surin A.K., Selivanova O.M., Fadeev R.S., Dzhus U.F., Gorbunova E.Y., Mustaeva L.G., Azev V.N., Galzitskaya O.V. Antimicrobial and Amyloidogenic Activity of Peptides Synthesized on the Basis of the Ribosomal S1 Protein from Thermus Thermophilus. Int. J. Mol. Sci. 2020;21:6382. doi: 10.3390/ijms21176382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ansari M.A., Jamal Q.M.S., Rehman S., Almatroudi A., Alzohairy M.A., Alomary M.N., Tripathi T., Alharbi A.H., Adil S.F., Khan M., et al. TAT-peptide conjugated repurposing drug against SARS-CoV-2 main protease (3CLpro): Potential therapeutic intervention to combat COVID-19. Arab. J. Chem. 2020;13:8069–8079. doi: 10.1016/j.arabjc.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehrer R.I. Primate defensins. Nat. Rev. Microbiol. 2004;2:727–738. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- 19.Odintsova T., Shcherbakova L., Slezina M., Pasechnik T., Kartabaeva B., Istomina E., Dzhavakhiya V. Hevein-Like Antimicrobial Peptides Wamps: Structure–Function Relationship in Antifungal Activity and Sensitization of Plant Pathogenic Fungi to Tebuconazole by WAMP-2-Derived Peptides. Int. J. Mol. Sci. 2020;21:7912. doi: 10.3390/ijms21217912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C., Wang S., Li D., Zhao X., Han S., Wang T., Zhao G., Chen Y., Chen F., Zhao J., et al. Lectin-like Intestinal Defensin Inhibits 2019-nCoV Spike binding to ACE2. bioRxiv. 2020 doi: 10.1101/2020.03.29.013490. [DOI] [Google Scholar]
- 21.Jang H., Lee D.-H., Kang H.G., Lee S.J. Concanavalin A targeting N -linked glycans in spike proteins influence viral interactions. Dalton Trans. 2020;49:13538–13543. doi: 10.1039/D0DT02932G. [DOI] [PubMed] [Google Scholar]
- 22.Maiti B.K. Potential Role of Peptide-Based Antiviral Therapy Against SARS-CoV-2 Infection. ACS Pharmacol. Transl. Sci. 2020;3:783–785. doi: 10.1021/acsptsci.0c00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azimova V.T., Potaturkina-Nesterova N.I., Nesterov A.S. Human Endogenous Antimicrobial Peptides. Mod. Probl. Sci. Educ. 2012;5:317–320. [Google Scholar]
- 24.Panteleev P.V. In: Structural-Functional Research of Antimicrobial Peptides of Animal Origin. Shemyakin M.M., Ovchinnikov Y.A., editors. Institute of Bioorganic Chemistry of the Russian Academy of Sciences; Moscow, Russia: 2015. Dissertation for the degree of candidate of chemical sciences. [Google Scholar]
- 25.Phoenix D.A., Dennison S.R., Harris F. Antimicrobial Peptides. Wiley-VCH; Weinheim, Germany: 2013. p. 254. [Google Scholar]
- 26.Mwangi J., Hao X., Lai R., Zhang Z.-Y. Antimicrobial peptides: New hope in the war against multidrug resistance. Zool. Res. 2019;40:488–505. doi: 10.24272/j.issn.2095-8137.2019.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei J., Sun L., Huang S., Zhu C., Li P., He J., Mackey V., Coy D.H., He Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019;11:3919–3931. [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H.R., You D.G., Kim H.K., Sohn J.W., Kim M.J., Park J.K., Lee G.Y., Yoo Y. Romo1-Derived Antimicrobial Peptide Is a New Antimicrobial Agent against Multidrug-Resistant Bacteria in a Murine Model of Sepsis. mBio. 2020;11:e03258-19. doi: 10.1128/mBio.03258-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castiglia F., Zevolini F., Riolo G., Brunetti J., De Lazzari A., Moretto A., Manetto G., Fragai M., Algotsson J., Evenäs J., et al. NMR Study of the Secondary Structure and Biopharmaceutical Formulation of an Active Branched Antimicrobial Peptide. Molecules. 2019;24:4290. doi: 10.3390/molecules24234290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cesa-Luna C., Muñoz-Rojas J., Saab-Rincon G., Baez A., Morales-García Y.E., Juárez-González V.R., Quintero-Hernández V. Structural characterization of scorpion peptides and their bactericidal activity against clinical isolates of multidrug-resistant bacteria. PLoS ONE. 2019;14:e0222438. doi: 10.1371/journal.pone.0222438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones F.E., DeCosse J.J., Condon R.E. Evaluation of “instant” preparation of the colon with povidone-iodine. Ann. Surg. 1976;184:74–79. doi: 10.1097/00000658-197607000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nesa J., Sadat A., Buccini D.F., Kati A., Mandal A.K., Franco O.L. Antimicrobial peptides from Bombyx mori: A splendid immune defense response in silkworms. RSC Adv. 2020;10:512–523. doi: 10.1039/C9RA06864C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Shi X., Zhu Y., Chen Y., Gao M., Gao H., Liu L., Wang L., Mao C., Wang Y. On-demand storage and release of antimicrobial peptides using Pandora’s box-like nanotubes gated with a bacterial infection-responsive polymer. Theranostics. 2020;10:109–122. doi: 10.7150/thno.38388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.León-Buitimea A., Garza-Cárdenas C.R., Garza-Cervantes J.A., Lerma-Escalera J.A., Morones-Ramírez J.R. The Demand for New Antibiotics: Antimicrobial Peptides, Nanoparticles, and Combinatorial Therapies as Future Strategies in Antibacterial Agent Design. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drayton M., Kizhakkedathu J.N., Straus S.K. Towards Robust Delivery of Antimicrobial Peptides to Combat Bacterial Resistance. Molecules. 2020;25:3048. doi: 10.3390/molecules25133048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tornesello A.L., Borrelli A., Buonaguro L., Buonaguro F.M., Tornesello M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules. 2020;25:2850. doi: 10.3390/molecules25122850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umnyakova E.S., Zharkova M.S., Berlov M.N., Shamova O.V., Kokryakov V.N. Human antimicrobial peptides in autoimmunity. Autoimmunity. 2020;53:137–147. doi: 10.1080/08916934.2020.1711517. [DOI] [PubMed] [Google Scholar]
- 38.Moretta A., Salvia R., Scieuzo C., Di Somma A., Vogel H., Pucci P., Sgambato A., Wolff M., Falabella P. A bioinformatic study of antimicrobial peptides identified in the Black Soldier Fly (BSF) Hermetia illucens (Diptera: Stratiomyidae) Sci. Rep. 2020;10:16875. doi: 10.1038/s41598-020-74017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melo-Braga M.N., De Marco Almeida F., dos Santos D.M., de Avelar Júnior J.T., dos Reis P.V.M., de Lima M.E. Antimicrobial Peptides From Lycosidae (Sundevall, 1833) Spiders. Curr. Protein Pept. Sci. 2020;21:527–541. doi: 10.2174/1389203721666200116091911. [DOI] [PubMed] [Google Scholar]
- 40.Smits J.P.H., Ederveen T.H.A., Rikken G., van den Brink N.J.M., van Vlijmen-Willems I.M.J.J., Boekhorst J., Kamsteeg M., Schalkwijk J., van Hijum S.A.F.T., Zeeuwen P.L.J.M., et al. Targeting the Cutaneous Microbiota in Atopic Dermatitis by Coal Tar via AHR-Dependent Induction of Antimicrobial Peptides. J. Investig. Dermatol. 2020;140:415–424.e10. doi: 10.1016/j.jid.2019.06.142. [DOI] [PubMed] [Google Scholar]
- 41.Neshani A., Sedighian H., Mirhosseini S.A., Ghazvini K., Zare H., Jahangiri A. Antimicrobial peptides as a promising treatment option against Acinetobacter baumannii infections. Microb. Pathog. 2020;146:104238. doi: 10.1016/j.micpath.2020.104238. [DOI] [PubMed] [Google Scholar]
- 42.Brunner S.R., Varga J.F.A., Dixon B. Antimicrobial Peptides of Salmonid Fish: From Form to Function. Biology. 2020;9:233. doi: 10.3390/biology9080233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaezi Z., Bortolotti A., Luca V., Perilli G., Mangoni M.L., Khosravi-Far R., Bobone S., Stella L. Aggregation determines the selectivity of membrane-active anticancer and antimicrobial peptides: The case of killerFLIP. Biochim. Biophys. Acta Biomembr. 2020;1862:183107. doi: 10.1016/j.bbamem.2019.183107. [DOI] [PubMed] [Google Scholar]
- 44.Albada B. Tuning Activity of Antimicrobial Peptides by Lipidation. In: Goldfine H., editor. Health Consequences of Microbial Interactions with Hydrocarbons, Oils, and Lipids. Springer International Publishing; Cham, Germany: 2020. pp. 317–334. [Google Scholar]
- 45.dos Santos-Silva C.A., Zupin L., Oliveira-Lima M., Vilela L.M.B., Bezerra-Neto J.P., Ferreira-Neto J.R., Ferreira J.D.C., de Oliveira-Silva R.L., de Pires C.J., Aburjaile F.F., et al. Plant Antimicrobial Peptides: State of the Art, In Silico Prediction and Perspectives in the Omics Era. Bioinform. Biol. Insights. 2020;14:117793222095273. doi: 10.1177/1177932220952739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greco I., Molchanova N., Holmedal E., Jenssen H., Hummel B.D., Watts J.L., Håkansson J., Hansen P.R., Svenson J. Correlation between hemolytic activity, cytotoxicity and systemic in vivo toxicity of synthetic antimicrobial peptides. Sci. Rep. 2020;10:13206. doi: 10.1038/s41598-020-69995-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mercer D.K., Torres M.D.T., Duay S.S., Lovie E., Simpson L., von Köckritz-Blickwede M., de la Fuente-Nunez C., O′Neil D.A., Angeles-Boza A.M. Antimicrobial Susceptibility Testing of Antimicrobial Peptides to Better Predict Efficacy. Front. Cell. Infect. Microbiol. 2020;10:326. doi: 10.3389/fcimb.2020.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huan Y., Kong Q., Mou H., Yi H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020;11:582779. doi: 10.3389/fmicb.2020.582779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan J., Bhadra P., Li A., Sethiya P., Qin L., Tai H.K., Wong K.H., Siu S.W.I. Deep-AmPEP30: Improve Short Antimicrobial Peptides Prediction with Deep Learning. Mol. Ther. Nucleic Acids. 2020;20:882–894. doi: 10.1016/j.omtn.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye G., Wu H., Huang J., Wang W., Ge K., Li G., Zhong J., Huang Q. LAMP2: A major update of the database linking antimicrobial peptides. Database. 2020;2020 doi: 10.1093/database/baaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burdukiewicz M., Sidorczuk K., Rafacz D., Pietluch F., Chilimoniuk J., Rödiger S., Gagat P. Proteomic Screening for Prediction and Design of Antimicrobial Peptides with AmpGram. Int. J. Mol. Sci. 2020;21:4310. doi: 10.3390/ijms21124310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cardoso M.H., Orozco R.Q., Rezende S.B., Rodrigues G., Oshiro K.G.N., Cândido E.S., Franco O.L. Computer-Aided Design of Antimicrobial Peptides: Are We Generating Effective Drug Candidates? Front. Microbiol. 2020;10:3097. doi: 10.3389/fmicb.2019.03097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung C.-R., Jhong J.-H., Wang Z., Chen S., Wan Y., Horng J.-T., Lee T.-Y. Characterization and Identification of Natural Antimicrobial Peptides on Different Organisms. Int. J. Mol. Sci. 2020;21:986. doi: 10.3390/ijms21030986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung C.-R., Kuo T.-R., Wu L.-C., Lee T.-Y., Horng J.-T. Characterization and identification of antimicrobial peptides with different functional activities. Brief. Bioinform. 2020;21:1098–1114. doi: 10.1093/bib/bbz043. [DOI] [PubMed] [Google Scholar]
- 55.Lazzaro B.P., Zasloff M., Rolff J. Antimicrobial peptides: Application informed by evolution. Science. 2020;368:eaau5480. doi: 10.1126/science.aau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhattacharjya S., Straus S.K. Design, Engineering and Discovery of Novel α-Helical and β-Boomerang Antimicrobial Peptides against Drug Resistant Bacteria. Int. J. Mol. Sci. 2020;21:5773. doi: 10.3390/ijms21165773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu L., Shao C., Li G., Shan A., Chou S., Wang J., Ma Q., Dong N. Conversion of Broad-Spectrum Antimicrobial Peptides into Species-Specific Antimicrobials Capable of Precisely Targeting Pathogenic Bacteria. Sci. Rep. 2020;10:944. doi: 10.1038/s41598-020-58014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moghal M.M.R., Hossain F., Yamazaki M. Action of antimicrobial peptides and cell-penetrating peptides on membrane potential revealed by the single GUV method. Biophys. Rev. 2020;12:339–348. doi: 10.1007/s12551-020-00662-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schäfer A.-B., Wenzel M. A How-To Guide for Mode of Action Analysis of Antimicrobial Peptides. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.540898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiao Z., Fu Y., Lei C., Li Y. Advances in antimicrobial peptides-based biosensing methods for detection of foodborne pathogens: A review. Food Control. 2020;112:107116. doi: 10.1016/j.foodcont.2020.107116. [DOI] [Google Scholar]
- 61.Yasir M., Dutta D., Hossain K.R., Chen R., Ho K.K.K., Kuppusamy R., Clarke R.J., Kumar N., Willcox M.D.P. Mechanism of Action of Surface Immobilized Antimicrobial Peptides Against Pseudomonas aeruginosa. Front. Microbiol. 2020;10 doi: 10.3389/fmicb.2019.03053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carratalá J.V., Serna N., Villaverde A., Vázquez E., Ferrer-Miralles N. Nanostructured antimicrobial peptides: The last push towards clinics. Biotechnol. Adv. 2020;44:107603. doi: 10.1016/j.biotechadv.2020.107603. [DOI] [PubMed] [Google Scholar]
- 63.Wang G. Host Defense Peptides and Their Potential as Therapeutic Agents. Springer International Publishing; Berlin/Heidelberg, Germany: 2016. Structural analysis of Amphibian, insect, and plant host defense peptides inspires the design of novel therapeutic molecules; pp. 229–252. [Google Scholar]
- 64.Cochrane S.A., Findlay B., Bakhtiary A., Acedo J.Z., Rodriguez-Lopez E.M., Mercier P., Vederas J.C. Antimicrobial lipopeptide tridecaptin A 1 selectively binds to Gram-negative lipid II. Proc. Natl. Acad. Sci. USA. 2016;113:11561–11566. doi: 10.1073/pnas.1608623113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swiecicki J.-M., Di Pisa M., Lippi F., Chwetzoff S., Mansuy C., Trugnan G., Chassaing G., Lavielle S., Burlina F. Unsaturated acyl chains dramatically enhanced cellular uptake by direct translocation of a minimalist oligo-arginine lipopeptide. Chem. Commun. 2015;51:14656–14659. doi: 10.1039/C5CC06116D. [DOI] [PubMed] [Google Scholar]
- 66.Gottardi D., Hong P.K., Ndagijimana M., Betti M. Conjugation of gluten hydrolysates with glucosamine at mild temperatures enhances antioxidant and antimicrobial properties. LWT Food Sci. Technol. 2014;57:181–187. doi: 10.1016/j.lwt.2014.01.013. [DOI] [Google Scholar]
- 67.Zhang L., Bulaj G. Converting Peptides into Drug Leads by Lipidation. Curr. Med. Chem. 2012;19:1602–1618. doi: 10.2174/092986712799945003. [DOI] [PubMed] [Google Scholar]
- 68.Vasco A.V., Brode M., Méndez Y., Valdés O., Rivera D.G., Wessjohann L.A. Synthesis of Lactam-Bridged and Lipidated Cyclo-Peptides as Promising Anti-Phytopathogenic Agents. Molecules. 2020;25:811. doi: 10.3390/molecules25040811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bellavita R., Falanga A., Buommino E., Merlino F., Casciaro B., Cappiello F., Mangoni M.L., Novellino E., Catania M.R., Paolillo R., et al. Novel temporin L antimicrobial peptides: Promoting self-assembling by lipidic tags to tackle superbugs. J. Enzyme Inhib. Med. Chem. 2020;35:1751–1764. doi: 10.1080/14756366.2020.1819258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoganathan S., Miller S.J. Structure Diversification of Vancomycin through Peptide-Catalyzed, Site-Selective Lipidation: A Catalysis-Based Approach To Combat Glycopeptide-Resistant Pathogens. J. Med. Chem. 2015;58:2367–2377. doi: 10.1021/jm501872s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu J., Sun Z., Yuan Y., Tian X., Liu X., Duan G., Yang Y., Yuan L., Lin H.-C., Li X. Peptide Glycosylation Generates Supramolecular Assemblies from Glycopeptides as Biomimetic Scaffolds for Cell Adhesion and Proliferation. ACS Appl. Mater. Interfaces. 2016;8:6917–6924. doi: 10.1021/acsami.6b00850. [DOI] [PubMed] [Google Scholar]
- 72.McDonald L.C. Vancomycin-Resistant Enterococci Outside the Health-Care Setting: Prevalence, Sources, and Public Health Implications. Emerg. Infect. Dis. 1997;3:311–317. doi: 10.3201/eid0303.970307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gong Y., Andina D., Nahar S., Leroux J.-C., Gauthier M.A. Releasable and traceless PEGylation of arginine-rich antimicrobial peptides. Chem. Sci. 2017;8:4082–4086. doi: 10.1039/C7SC00770A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morris C.J., Beck K., Fox M.A., Ulaeto D., Clark G.C., Gumbleton M. Pegylation of Antimicrobial Peptides Maintains the Active Peptide Conformation, Model Membrane Interactions, and Antimicrobial Activity while Improving Lung Tissue Biocompatibility following Airway Delivery. Antimicrob. Agents Chemother. 2012;56:3298–3308. doi: 10.1128/AAC.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montalbán-López M., Spolaore B., Pinato O., Martínez-Bueno M., Valdivia E., Maqueda M., Fontana A. Characterization of linear forms of the circular enterocin AS-48 obtained by limited proteolysis. FEBS Lett. 2008;582:3237–3242. doi: 10.1016/j.febslet.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 76.Colgrave M.L., Craik D.J. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: The importance of the cyclic cystine knot. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 77.Taylor S.W., Craig A.G., Fischer W.H., Park M., Lehrer R.I. Styelin D, an extensively modified antimicrobial peptide from ascidian hemocytes. J. Biol. Chem. 2000;275:38417–38426. doi: 10.1074/jbc.M006762200. [DOI] [PubMed] [Google Scholar]
- 78.Jahnsen R.O., Sandberg-Schaal A., Frimodt-Møller N., Nielsen H.M., Franzyk H. End group modification: Efficient tool for improving activity of antimicrobial peptide analogues towards Gram-positive bacteria. Eur. J. Pharm. Biopharm. 2015;95:40–46. doi: 10.1016/j.ejpb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 79.Jhong J.H., Chi Y.H., Li W.C., Lin T.H., Huang K.Y., Lee T.Y. DbAMP: An integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. 2019;47:D285–D297. doi: 10.1093/nar/gky1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Greber K.E., Roch M., Rosato M.A., Martinez M.P., Rosato A.E. Efficacy of newly generated short antimicrobial cationic lipopeptides against methicillin-resistant Staphylococcus aureus (MRSA) Int. J. Antimicrob. Agents. 2020;55:105827. doi: 10.1016/j.ijantimicag.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 81.Wang G. Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies. Cabi; Wallingford, UK: 2017. [DOI] [Google Scholar]
- 82.Fernandes J.M.O., Molle G., Kemp G.D., Smith V.J. Isolation and characterisation of oncorhyncin II, a histone H1-derived antimicrobial peptide from skin secretions of rainbow trout, Oncorhynchus mykiss. Dev. Comp. Immunol. 2004;28:127–138. doi: 10.1016/S0145-305X(03)00120-4. [DOI] [PubMed] [Google Scholar]
- 83.Zelezetsky I., Pag U., Sahl H.-G., Tossi A. Tuning the biological properties of amphipathic α-helical antimicrobial peptides: Rational use of minimal amino acid substitutions. Peptides. 2005;26:2368–2376. doi: 10.1016/j.peptides.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 84.Yeaman M.R., Yount N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 85.Boman H.G., Agerberth B., Boman A. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect. Immun. 1993;61:2978–2984. doi: 10.1128/IAI.61.7.2978-2984.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cabiaux V., Agerberths B., Johansson J., Homble F., Goormaghtigh E., Ruysschaert J.-M. Secondary Structure and Membrane Interaction of PR-39, a Pro+Arg-rich Antibacterial Peptide. Eur. J. Biochem. 1994;224:1019–1027. doi: 10.1111/j.1432-1033.1994.01019.x. [DOI] [PubMed] [Google Scholar]
- 87.Malekkhaiat Häffner S., Nyström L., Nordström R., Xu Z.P., Davoudi M., Schmidtchen A., Malmsten M. Membrane interactions and antimicrobial effects of layered double hydroxide nanoparticles. Phys. Chem. Chem. Phys. 2017;19:23832–23842. doi: 10.1039/C7CP02701J. [DOI] [PubMed] [Google Scholar]
- 88.Liu K., Yang L., Peng X., Wang J., Lu J.R., Xu H. Modulation of Antimicrobial Peptide Conformation and Aggregation by Terminal Lipidation and Surfactants. Langmuir. 2020;36:1737–1744. doi: 10.1021/acs.langmuir.9b03774. [DOI] [PubMed] [Google Scholar]
- 89.Biswas K., Ilyas H., Datta A., Bhunia A. NMR Assisted Antimicrobial Peptide Designing: Structure Based Modifications and Functional Correlation of a Designed Peptide VG16KRKP. Curr. Med. Chem. 2020;27:1387–1404. doi: 10.2174/0929867326666190624090817. [DOI] [PubMed] [Google Scholar]
- 90.Zhu J., Pedersen M.D., Ahmed L.S., Abdolalizadeh B., Grell A.-S., Berg J.O., Thulstrup P.W., Franzyk H., Edvinsson L., Sams A., et al. Fluorescent Analogues of Human α-Calcitonin Gene-Related Peptide with Potent Vasodilator Activity. Int. J. Mol. Sci. 2020;21:1343. doi: 10.3390/ijms21041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Waghu F.H., Barai R.S., Gurung P., Idicula-Thomas S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016;44:D1094–D1097. doi: 10.1093/nar/gkv1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zharkova M.S., Orlov D.S., Kokryakov V.N., Shamova O.V. Antimicrobial peptides in mammals: Classification, biological role, prospects for practical application (review article) Biol. Commun. 2014;1:98–114. [Google Scholar]
- 93.Mazumdar A., Haddad Y., Sur V.P., Milosavljevic V., Bhowmick S., Michalkova H., Guran R., Vesely R., Moulick A. Characterization and in vitro Analysis of Probiotic-Derived Peptides Against Multi Drug Resistance Bacterial Infections. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zimina M., Babich O., Prosekov A., Sukhikh S., Ivanova S., Shevchenko M., Noskova S. Overview of Global Trends in Classification, Methods of Preparation and Application of Bacteriocins. Antibiotics. 2020;9:553. doi: 10.3390/antibiotics9090553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nawrot R., Barylski J., Nowicki G., Broniarczyk J., Buchwald W., Goździcka-Józefiak A. Plant antimicrobial peptides. Folia Microbiol. 2014;59:181–196. doi: 10.1007/s12223-013-0280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Amiche M., Ladram A., Nicolas P. A consistent nomenclature of antimicrobial peptides isolated from frogs of the subfamily Phyllomedusinae. Peptides. 2008;29:2074–2082. doi: 10.1016/j.peptides.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 97.Conlon J.M. Reflections on a systematic nomenclature for antimicrobial peptides from the skins of frogs of the family Ranidae. Peptides. 2008;29:1815–1819. doi: 10.1016/j.peptides.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 98.Bulet P., Stocklin R. Insect Antimicrobial Peptides: Structures, Properties and Gene Regulation. Protein Pept. Lett. 2005;12:3–11. doi: 10.2174/0929866053406011. [DOI] [PubMed] [Google Scholar]
- 99.Otero-Gonzáiez A.J., Magalhães B.S., Garcia-Villarino M., López-Abarrategui C., Sousa D.A., Dias S.C., Franco O.L. Antimicrobial peptides from marine invertebrates as a new frontier for microbial infection control. FASEB J. 2010;24:1320–1334. doi: 10.1096/fj.09-143388. [DOI] [PubMed] [Google Scholar]
- 100.Zanetti M. The role of cathelicidins in the innate host defenses of mammals. Curr. Issues Mol. Biol. 2005;7:179–196. [PubMed] [Google Scholar]
- 101.Suarez-Carmona M., Hubert P., Delvenne P., Herfs M. Defensins: “Simple” antimicrobial peptides or broad-spectrum molecules? Cytokine Growth Factor Rev. 2015;26:361–370. doi: 10.1016/j.cytogfr.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 102.Mylonakis E., Podsiadlowski L., Muhammed M., Vilcinskas A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. B Biol. Sci. 2016;371:20150290. doi: 10.1098/rstb.2015.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Petre B., Major I., Rouhier N., Duplessis S. Genome-wide analysis of eukaryote thaumatin-like proteins (TLPs) with an emphasis on poplar. BMC Plant Biol. 2011;11:1–16. doi: 10.1186/1471-2229-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Selsted M.E., Harwig S.S.L., Ganz T., Schilling J.W., Lehrer R.I. Primary structures of three human neutrophil defensins. J. Clin. Investig. 1985;76:1436–1439. doi: 10.1172/JCI112121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 106.Nylén F., Miraglia E., Cederlund A., Ottosson H., Strömberg R., Gudmundsson G.H., Agerberth B. Boosting innate immunity: Development and validation of a cell-based screening assay to identify LL-37 inducers. Innate Immun. 2014;20:364–376. doi: 10.1177/1753425913493338. [DOI] [PubMed] [Google Scholar]
- 107.Fensterseifer I.C.M., Silva O.N., Malik U., Ravipati A.S., Novaes N.R.F., Miranda P.R.R., Rodrigues E.A., Moreno S.E., Craik D.J., Franco O.L. Effects of cyclotides against cutaneous infections caused by Staphylococcus aureus. Peptides. 2015;63:38–42. doi: 10.1016/j.peptides.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 108.Xhindoli D., Pacor S., Benincasa M., Scocchi M., Gennaro R., Tossi A. The human cathelicidin LL-37 - A pore-forming antibacterial peptide and host-cell modulator. Biochim. Biophys. Acta Biomembr. 2016;1858:546–566. doi: 10.1016/j.bbamem.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 109.Barlow P.G., Svoboda P., Mackellar A., Nash A.A., York I.A., Pohl J., Davidson D.J., Donis R.O. Antiviral Activity and Increased Host Defense against Influenza Infection Elicited by the Human Cathelicidin LL-37. PLoS ONE. 2011;6:e25333. doi: 10.1371/journal.pone.0025333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee E.Y., Takahashi T., Curk T., Dobnikar J., Gallo R.L., Wong G.C.L. Crystallinity of Double-Stranded RNA-Antimicrobial Peptide Complexes Modulates Toll-Like Receptor 3-Mediated Inflammation. ACS Nano. 2017;11:12145–12155. doi: 10.1021/acsnano.7b05234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li K., Tao N., Zheng L., Sun T. LL-37 restored glucocorticoid sensitivity impaired by virus dsRNA in lung. Int. Immunopharmacol. 2020;79:106057. doi: 10.1016/j.intimp.2019.106057. [DOI] [PubMed] [Google Scholar]
- 112.Ahmed T.A.E., Hammami R. Recent insights into structure–function relationships of antimicrobial peptides. J. Food Biochem. 2019;43:e12546. doi: 10.1111/jfbc.12546. [DOI] [PubMed] [Google Scholar]
- 113.Jenssen H., Hamill P., Hancock R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mader J.S., Hoskin D.W. Cationic antimicrobial peptides as novel cytotoxic agents for cancer treatment. Expert Opin. Investig. Drugs. 2006;15:933–946. doi: 10.1517/13543784.15.8.933. [DOI] [PubMed] [Google Scholar]
- 115.Chu H.-L., Yip B.-S., Chen K.-H., Yu H.-Y., Chih Y.-H., Cheng H.-T., Chou Y.-T., Cheng J.-W. Novel Antimicrobial Peptides with High Anticancer Activity and Selectivity. PLoS ONE. 2015;10:e0126390. doi: 10.1371/journal.pone.0126390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hanson M.A., Lemaitre B. New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 2020;62:22–30. doi: 10.1016/j.coi.2019.11.008. [DOI] [PubMed] [Google Scholar]
- 117.Li X., Lewis M.T., Huang J., Gutierrez C., Osborne C.K., Wu M.F., Hilsenbeck S.G., Pavlick A., Zhang X., Chamness G.C., et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 2008;100:672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 118.Hoskin D.W., Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gupta S., Thakur J., Pal S., Gupta R., Mishra D., Kumar S., Yadav K., Saini A., Yavvari P.S., Vedantham M., et al. Cholic Acid-Peptide Conjugates as Potent Antimicrobials against Interkingdom Polymicrobial Biofilms. Antimicrob. Agents Chemother. 2019;63:e00520-19. doi: 10.1128/AAC.00520-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Batoni G., Maisetta G., Esin S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta Biomembr. 2016;1858:1044–1060. doi: 10.1016/j.bbamem.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 121.Amer L.S., Bishop B.M., van Hoek M.L. Antimicrobial and antibiofilm activity of cathelicidins and short, synthetic peptides against Francisella. Biochem. Biophys. Res. Commun. 2010;396:246–251. doi: 10.1016/j.bbrc.2010.04.073. [DOI] [PubMed] [Google Scholar]
- 122.Zhang S.K., Song J.W., Gong F., Li S.B., Chang H.Y., Xie H.M., Gao H.W., Tan Y.X., Ji S.P. Design of an α-helical antimicrobial peptide with improved cell-selective and potent anti-biofilm activity. Sci. Rep. 2016;6:27394. doi: 10.1038/srep27394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yin Q., Wu S., Wu L., Wang Z., Mu Y., Zhang R., Dong C., Zhou B., Zhao B., Zheng J., et al. A novel in silico antimicrobial peptide DP7 combats MDR Pseudomonas aeruginosa and related biofilm infections. J. Antimicrob. Chemother. 2020;75:3248–3259. doi: 10.1093/jac/dkaa308. [DOI] [PubMed] [Google Scholar]
- 124.Galdiero E., Lombardi L., Falanga A., Libralato G., Guida M., Carotenuto R. Biofilms: Novel Strategies Based on Antimicrobial Peptides. Pharmaceutics. 2019;11:322. doi: 10.3390/pharmaceutics11070322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Thomsen T., Mendel H., Al-Mansour W., Oddo A., Løbner-Olesen A., Hansen P. Analogues of a Cyclic Antimicrobial Peptide with a Flexible Linker Show Promising Activity against Pseudomonas aeruginosa and Staphylococcus aureus. Antibiotics. 2020;9:366. doi: 10.3390/antibiotics9070366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang G., Elliott M., Cogen A.L., Ezell E.L., Gallo R.L., Hancock R.E.W. Structure, Dynamics, and Antimicrobial and Immune Modulatory Activities of Human LL-23 and Its Single-Residue Variants Mutated on the Basis of Homologous Primate Cathelicidins. Biochemistry. 2012;51:653–664. doi: 10.1021/bi2016266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Al-Adwani S., Wallin C., Balhuizen M.D., Veldhuizen E.J.A., Coorens M., Landreh M., Végvári Á., Smith M.E., Qvarfordt I., Lindén A., et al. Studies on citrullinated LL-37: Detection in human airways, antibacterial effects and biophysical properties. Sci. Rep. 2020;10:2376. doi: 10.1038/s41598-020-59071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Casanova V., Sousa F.H., Shakamuri P., Svoboda P., Buch C., D’Acremont M., Christophorou M.A., Pohl J., Stevens C., Barlow P.G. Citrullination Alters the Antiviral and Immunomodulatory Activities of the Human Cathelicidin LL-37 During Rhinovirus Infection. Front. Immunol. 2020;11:85. doi: 10.3389/fimmu.2020.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kazemzadeh-Narbat M., Cheng H., Chabok R., Alvarez M.M., de la Fuente-Nunez C., Phillips K.S., Khademhosseini A. Strategies for antimicrobial peptide coatings on medical devices: A review and regulatory science perspective. Crit. Rev. Biotechnol. 2020:1–27. doi: 10.1080/07388551.2020.1828810. [DOI] [PubMed] [Google Scholar]
- 130.Findlay F., Pohl J., Svoboda P., Shakamuri P., McLean K., Inglis N.F., Proudfoot L., Barlow P.G. Carbon Nanoparticles Inhibit the Antimicrobial Activities of the Human Cathelicidin LL-37 through Structural Alteration. J. Immunol. 2017;199:2483–2490. doi: 10.4049/jimmunol.1700706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Falciani C., Zevolini F., Brunetti J., Riolo G., Gracia R., Marradi M., Loinaz I., Ziemann C., Cossío U., Llop J., et al. Antimicrobial Peptide-Loaded Nanoparticles as Inhalation Therapy for Pseudomonas aeruginosa Infections. Int. J. Nanomed. 2020;15:1117–1128. doi: 10.2147/IJN.S218966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mukhopadhyay S., Bharath Prasad A.S., Mehta C.H., Nayak U.Y. Antimicrobial peptide polymers: No escape to ESKAPE pathogens-a review. World J. Microbiol. Biotechnol. 2020;36:131. doi: 10.1007/s11274-020-02907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Boge L., Hallstensson K., Ringstad L., Johansson J., Andersson T., Davoudi M., Larsson P.T., Mahlapuu M., Håkansson J., Andersson M. Cubosomes for topical delivery of the antimicrobial peptide LL-37. Eur. J. Pharm. Biopharm. 2019;134:60–67. doi: 10.1016/j.ejpb.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 134.Jelinkova P., Splichal Z., Jimenez Jimenez A.M., Haddad Y., Mazumdar A., Sur V.P., Milosavljevic V., Kopel P., Buchtelova H., Guran R., et al. Novel vancomycin–peptide conjugate as potent antibacterial agent against vancomycin-resistant Staphylococcus aureus. Infect. Drug Resist. 2018;11:1807–1817. doi: 10.2147/IDR.S160975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Van Staden A.D.P., Faure L.M., Vermeulen R.R., Dicks L.M.T., Smith C. Functional Expression of GFP-Fused Class I Lanthipeptides in Escherichia coli. ACS Synth. Biol. 2019;8:2220–2227. doi: 10.1021/acssynbio.9b00167. [DOI] [PubMed] [Google Scholar]
- 136.Luan C., Zhang H.W., Song D.G., Xie Y.G., Feng J., Wang Y.Z. Expressing antimicrobial peptide cathelicidin-BF in Bacillus subtilis using SUMO technology. Appl. Microbiol. Biotechnol. 2014;98:3651–3658. doi: 10.1007/s00253-013-5246-6. [DOI] [PubMed] [Google Scholar]
- 137.Bessalle R., Haas H., Goria A., Shalit I., Fridkin M. Augmentation of the antibacterial activity of magainin by positive-charge chain extension. Antimicrob. Agents Chemother. 1992;36:313–317. doi: 10.1128/AAC.36.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dathe M., Nikolenko H., Meyer J., Beyermann M., Bienert M. Optimization of the antimicrobial activity of magainin peptides by modification of charge. FEBS Lett. 2001;501:146–150. doi: 10.1016/S0014-5793(01)02648-5. [DOI] [PubMed] [Google Scholar]
- 139.Eisenberg D. Three-Dimensional Structure of Membrane and Surface Proteins. Annu. Rev. Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- 140.Dathe M., Wieprecht T., Nikolenko H., Handel L., Maloy W.L., MacDonald D.L., Beyermann M., Bienert M. Hydrophobicity, hydrophobic moment and angle subtended by charged residues modulate antibacterial and haemolytic activity of amphipathic helical peptides. FEBS Lett. 1997;403:208–212. doi: 10.1016/S0014-5793(97)00055-0. [DOI] [PubMed] [Google Scholar]
- 141.Jensen S.K., Thomsen T.T., Oddo A., Franzyk H., Løbner-Olesen A., Hansen P.R. Novel Cyclic Lipopeptide Antibiotics: Effects of Acyl Chain Length and Position. Int. J. Mol. Sci. 2020;21:5829. doi: 10.3390/ijms21165829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tam J.P., Wu C., Yang J.-L. Membranolytic selectivity of cystine-stabilized cyclic protegrins. Eur. J. Biochem. 2000;267:3289–3300. doi: 10.1046/j.1432-1327.2000.01359.x. [DOI] [PubMed] [Google Scholar]
- 143.Le Brun A.P., Zhu S., Sani M.-A., Separovic F. The Location of the Antimicrobial Peptide Maculatin 1.1 in Model Bacterial Membranes. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dathe M., Wieprecht T. Structural features of helical antimicrobial peptides: Their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta Biomembr. 1999;1462:71–87. doi: 10.1016/S0005-2736(99)00201-1. [DOI] [PubMed] [Google Scholar]
- 145.Breukink E., De Kruijff B. The lantibiotic nisin, a special case or not? Biochim. Biophys. Acta Biomembr. 1999;1462:223–234. doi: 10.1016/S0005-2736(99)00208-4. [DOI] [PubMed] [Google Scholar]
- 146.Sansom M.S. The biophysics of peptide models of ion channels. Prog. Biophys. Mol. Biol. 1991;55:139–235. doi: 10.1016/0079-6107(91)90004-C. [DOI] [PubMed] [Google Scholar]
- 147.Yang L., Harroun T.A., Weiss T.M., Ding L., Huang H.W. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 2001;81:1475–1485. doi: 10.1016/S0006-3495(01)75802-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hara T., Kodama H., Kondo M., Wakamatsu K., Takeda A., Tachi T., Matsuzaki K. Effects of peptide dimerization on pore formation: Antiparallel disulfide-dimerized magainin 2 analogue. Biopolymers. 2001;58:437–446. doi: 10.1002/1097-0282(20010405)58:4<437::AID-BIP1019>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 149.Ludtke S.J., He K., Heller W.T., Harroun T.A., Yang L., Huang H.W. Membrane pores induced by magainin. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 150.Matsuzaki K., Mitani Y., Akada K.Y., Murase O., Yoneyama S., Zasloff M., Miyajima K. Mechanism of synergism between antimicrobial peptides magainin 2 and PGLa. Biochemistry. 1998;37:15144–15153. doi: 10.1021/bi9811617. [DOI] [PubMed] [Google Scholar]
- 151.Uematsu N., Matsuzaki K. Polar angle as a determinant of amphipathic α-helix-lipid interactions: A model peptide study. Biophys. J. 2000;79:2075–2083. doi: 10.1016/S0006-3495(00)76455-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lau Q.Y., Li J., Sani M.-A., Sinha S., Li Y., Ng F.M., Kang C., Bhattacharjya S., Separovic F., Verma C., et al. Elucidating the bactericidal mechanism of action of the linear antimicrobial tetrapeptide BRBR-NH2. Biochim. Biophys. Acta Biomembr. 2018;1860:1517–1527. doi: 10.1016/j.bbamem.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 153.Shai Y., Oren Z. From “carpet” mechanism to de-novo designed diastereomeric cell-selective antimicrobial peptides. Peptides. 2001;22:1629–1641. doi: 10.1016/S0196-9781(01)00498-3. [DOI] [PubMed] [Google Scholar]
- 154.Sitaram N., Nagaraj R. Interaction of antimicrobial peptides with biological and model membranes: Structural and charge requirements for activity. Biochim. Biophys. Acta Biomembr. 1999;1462:29–54. doi: 10.1016/S0005-2736(99)00199-6. [DOI] [PubMed] [Google Scholar]
- 155.Rozek A., Friedrich C.L., Hancock R.E.W. Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry. 2000;39:15765–15774. doi: 10.1021/bi000714m. [DOI] [PubMed] [Google Scholar]
- 156.Sass V., Pag U., Tossi A., Bierbaum G., Sahl H.-G. Mode of action of human β-defensin 3 against Staphylococcus aureus and transcriptional analysis of responses to defensin challenge. Int. J. Med. Microbiol. 2008;298:619–633. doi: 10.1016/j.ijmm.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 157.Sass V., Schneider T., Wilmes M., Körner C., Tossi A., Novikova N., Shamova O., Sahl H.-G. Human β-Defensin 3 Inhibits Cell Wall Biosynthesis in Staphylococci. Infect. Immun. 2010;78:2793–2800. doi: 10.1128/IAI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Oeemig J.S., Lynggaard C., Knudsen D.H., Hansen F.T., Nørgaard K.D., Schneider T., Vad B.S., Sandvang D.H., Nielsen L.A., Neve S., et al. Eurocin, a New Fungal Defensin. J. Biol. Chem. 2012;287:42361–42372. doi: 10.1074/jbc.M112.382028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vasilchenko A.S., Julian W.T., Lapchinskaya O.A., Katrukha G.S., Sadykova V.S., Rogozhin E.A. A Novel Peptide Antibiotic Produced by Streptomyces roseoflavus Strain INA-Ac-5812 With Directed Activity Against Gram-Positive Bacteria. Front. Microbiol. 2020;11:556063. doi: 10.3389/fmicb.2020.556063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Grein F., Müller A., Scherer K.M., Liu X., Ludwig K.C., Klöckner A., Strach M., Sahl H.-G., Kubitscheck U., Schneider T. Ca2+-Daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids. Nat. Commun. 2020;11:1455. doi: 10.1038/s41467-020-15257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Overall S.A., Zhu S., Hanssen E., Separovic F., Sani M.-A. In Situ Monitoring of Bacteria under Antimicrobial Stress Using 31P Solid-State NMR. Int. J. Mol. Sci. 2019;20:181. doi: 10.3390/ijms20010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boparai J.K., Sharma P.K. Mini Review on Antimicrobial Peptides, Sources, Mechanism and Recent Applications. Protein Pept. Lett. 2019;27:4–16. doi: 10.2174/0929866526666190822165812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lee E.Y., Zhang C., Di Domizio J., Jin F., Connell W., Hung M., Malkoff N., Veksler V., Gilliet M., Ren P., et al. Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nat. Commun. 2019;10:1012. doi: 10.1038/s41467-019-08868-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hale J.D.F., Hancock R.E.W. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev. Anti-Infect. Ther. 2007;5:951–959. doi: 10.1586/14787210.5.6.951. [DOI] [PubMed] [Google Scholar]
- 165.Le C.F., Fang C.M., Sekaran S.D. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 2017;61:e02340-16. doi: 10.1128/AAC.02340-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Adelman K., Yuzenkova J., La Porta A., Zenkin N., Lee J., Lis J.T., Borukhov S., Wang M.D., Severinov K. Molecular mechanism of transcription inhibition by peptide antibiotic Microcin J25. Mol. Cell. 2004;14:753–762. doi: 10.1016/j.molcel.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 167.Mardirossian M., Grzela R., Giglione C., Meinnel T., Gennaro R., Mergaert P., Scocchi M. The host antimicrobial peptide Bac71-35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem. Biol. 2014;21:1639–1647. doi: 10.1016/j.chembiol.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 168.Ho Y.H., Shah P., Chen Y.W., Chen C.S. Systematic analysis of intracellular-targeting antimicrobial peptides, bactenecin 7, hybrid of pleurocidin and dermaseptin, proline-arginine-rich peptide, and lactoferricin b, by using Escherichia coli proteome microarrays. Mol. Cell. Proteom. 2016;15:1837–1847. doi: 10.1074/mcp.M115.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Scocchi M., Lüthy C., Decarli P., Mignogna G., Christen P., Gennaro R. The Proline-rich Antibacterial Peptide Bac7 Binds to and Inhibits in vitro the Molecular Chaperone DnaK. Int. J. Pept. Res. Ther. 2009;15:147–155. doi: 10.1007/s10989-009-9182-3. [DOI] [Google Scholar]
- 170.Kragol G., Lovas S., Varadi G., Condie B.A., Hoffmann R., Otvos L. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry. 2001;40:3016–3026. doi: 10.1021/bi002656a. [DOI] [PubMed] [Google Scholar]
- 171.Chesnokova L.S., Slepenkov S.V., Witt S.N. The insect antimicrobial peptide, L-pyrrhocoricin, binds to and stimulates the ATPase activity of both wild-type and lidless DnaK. FEBS Lett. 2004;565:65–69. doi: 10.1016/j.febslet.2004.03.075. [DOI] [PubMed] [Google Scholar]
- 172.Knappe D., Zahn M., Sauer U., Schiffer G., Sträter N., Hoffmann R. Rational Design of Oncocin Derivatives with Superior Protease Stabilities and Antibacterial Activities Based on the High-Resolution Structure of the Oncocin-DnaK Complex. ChemBioChem. 2011;12:874–876. doi: 10.1002/cbic.201000792. [DOI] [PubMed] [Google Scholar]
- 173.Kagan B.L., Jang H., Capone R., Teran Arce F., Ramachandran S., Lal R., Nussinov R. Antimicrobial Properties of Amyloid Peptides. Mol. Pharm. 2012;9:708–717. doi: 10.1021/mp200419b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tayeb-Fligelman E., Salinas N., Tabachnikov O., Landau M. Staphylococcus aureus PSMα3 Cross-α Fibril Polymorphism and Determinants of Cytotoxicity. Structure. 2020;28:301–313.e6. doi: 10.1016/j.str.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 175.Schnaider L., Brahmachari S., Schmidt N.W., Mensa B., Shaham-Niv S., Bychenko D., Adler-Abramovich L., Shimon L.J.W., Kolusheva S., DeGrado W.F., et al. Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity. Nat. Commun. 2017;8:1365. doi: 10.1038/s41467-017-01447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Simonson A.W., Aronson M.R., Medina S.H. Supramolecular Peptide Assemblies as Antimicrobial Scaffolds. Molecules. 2020;25:2751. doi: 10.3390/molecules25122751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chairatana P., Nolan E.M. Human α-Defensin 6: A Small Peptide That Self-Assembles and Protects the Host by Entangling Microbes. Acc. Chem. Res. 2017;50:960–967. doi: 10.1021/acs.accounts.6b00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sarkar B., Siddiqui Z., Nguyen P.K., Dube N., Fu W., Park S., Jaisinghani S., Paul R., Kozuch S.D., Deng D., et al. Membrane-Disrupting Nanofibrous Peptide Hydrogels. ACS Biomater. Sci. Eng. 2019;5:4657–4670. doi: 10.1021/acsbiomaterials.9b00967. [DOI] [PubMed] [Google Scholar]
- 179.Yang K., Han Q., Chen B., Zheng Y., Zhang K., Li Q., Wang J. Antimicrobial hydrogels: Promising materials for medical application. Int. J. Nanomed. 2018;13:2217–2263. doi: 10.2147/IJN.S154748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Conchillo-Solé O., de Groot N.S., Avilés F.X., Vendrell J., Daura X., Ventura S. AGGRESCAN: A server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinform. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Garbuzynskiy S.O., Lobanov M.Y., Galzitskaya O.V. FoldAmyloid: A method of prediction of amyloidogenic regions from protein sequence. Bioinformatics. 2010;26:326–332. doi: 10.1093/bioinformatics/btp691. [DOI] [PubMed] [Google Scholar]
- 182.Walsh I., Seno F., Tosatto S.C.E., Trovato A. PASTA 2.0: An improved server for protein aggregation prediction. Nucleic Acids Res. 2014;42:W301–W307. doi: 10.1093/nar/gku399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Oliveberg M. Waltz, an exciting new move in amyloid prediction. Nat. Methods. 2010;7:187. doi: 10.1038/nmeth0310-187. [DOI] [PubMed] [Google Scholar]
- 184.Singh P.K., Maji S.K. Amyloid-Like Fibril Formation by Tachykinin Neuropeptides and Its Relevance to Amyloid β-Protein Aggregation and Toxicity. Cell Biochem. Biophys. 2012;64:29–44. doi: 10.1007/s12013-012-9364-z. [DOI] [PubMed] [Google Scholar]
- 185.Ganesan A., Debulpaep M., Wilkinson H., Van Durme J., De Baets G., Jonckheere W., Ramakers M., Ivarsson Y., Zimmermann P., Van Eldere J., et al. Selectivity of Aggregation-Determining Interactions. J. Mol. Biol. 2015;427:236–247. doi: 10.1016/j.jmb.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 186.Artemova N.V., Stein-Margolina V.A., Bumagina Z.M., Gurvits B.Y. Acceleration of protein aggregation by amphiphilic peptides: Transformation of supramolecular structure of the aggregates. Biotechnol. Prog. 2011;27:846–854. doi: 10.1002/btpr.574. [DOI] [PubMed] [Google Scholar]
- 187.Milojevic J., Melacini G. Stoichiometry and affinity of the human serum albumin-Alzheimer’s Aβ peptide interactions. Biophys. J. 2011;100:183–192. doi: 10.1016/j.bpj.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Litus E.A., Kazakov A.S., Sokolov A.S., Nemashkalova E.L., Galushko E.I., Dzhus U.F., Marchenkov V.V., Galzitskaya O.V., Permyakov E.A., Permyakov S.E. The binding of monomeric amyloid β peptide to serum albumin is affected by major plasma unsaturated fatty acids. Biochem. Biophys. Res. Commun. 2019;510:248–253. doi: 10.1016/j.bbrc.2019.01.081. [DOI] [PubMed] [Google Scholar]
- 189.Raffen R., Dieckman L.J., Szpunar M., Wunschl C., Pokkuluri P.R., Dave P., Wilkins Stevens P., Cai X., Schiffer M., Stevens F.J. Physicochemical consequences of amino acid variations that contribute to fibril formation by immunoglobulin light chains. Protein Sci. 1999;8:509–517. doi: 10.1110/ps.8.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Borgia A., Kemplen K.R., Borgia M.B., Soranno A., Shammas S., Wunderlich B., Nettels D., Best R.B., Clarke J., Schuler B. Transient misfolding dominates multidomain protein folding. Nat. Commun. 2015;6:8861. doi: 10.1038/ncomms9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bobylev A.G., Yakupova E.I., Bobyleva L.G., Galzitskaya O.V., Nikulin A.D., Shumeyko S.A., Yurshenas D.A., Vikhlyantsev I.M. Changes in Titin Structure during Its Aggregation. Mol. Biol. 2020;54:578–585. doi: 10.1134/S0026893320040044. [DOI] [PubMed] [Google Scholar]
- 192.Bobylev A.G., Galzitskaya O.V., Fadeev R.S., Bobyleva L.G., Yurshenas D.A., Molochkov N.V., Dovidchenko N.V., Selivanova O.M., Penkov N.V., Podlubnaya Z.A., et al. Smooth muscle titin forms in vitro amyloid aggregates. Biosci. Rep. 2016;36:e00334. doi: 10.1042/BSR20160066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Guryanov S.G., Selivanova O.M., Nikulin A.D., Enin G.A., Melnik B.S., Kretov D.A., Serdyuk I.N., Ovchinnikov L.P. Formation of Amyloid-Like Fibrils by Y-Box Binding Protein 1 (YB-1) Is Mediated by Its Cold Shock Domain and Modulated by Disordered Terminal Domains. PLoS ONE. 2012;7:e36969. doi: 10.1371/journal.pone.0036969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Selivanova O.M., Guryanov S.G., Enin G.A., Skabkin M.A., Ovchinnikov L.P., Serdyuk I.N. YB-1 is capable of forming extended nanofibrils. Biochemistry. 2010;75:115–120. doi: 10.1134/S0006297910010153. [DOI] [PubMed] [Google Scholar]
- 195.Taboada P., Barbosa S., Castro E., Mosquera V. Amyloid Fibril Formation and Other Aggregate Species Formed by Human Serum Albumin Association. J. Phys. Chem. B. 2006;110:20733–20736. doi: 10.1021/jp064861r. [DOI] [PubMed] [Google Scholar]
- 196.Maciążek-Jurczyk M., Janas K., Pożycka J., Szkudlarek A., Rogóż W., Owczarzy A., Kulig K. Human Serum Albumin Aggregation/Fibrillation and its Abilities to Drugs Binding. Molecules. 2020;25:618. doi: 10.3390/molecules25030618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wasko J., Wolszczak M., Kaminski Z.J., Steblecka M., Kolesinska B. Human Serum Albumin Binds Native Insulin and Aggregable Insulin Fragments and Inhibits Their Aggregation. Biomolecules. 2020;10:1366. doi: 10.3390/biom10101366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ishima Y., Mimono A., Tuan Giam Chuang V., Fukuda T., Kusumoto K., Okuhira K., Suwa Y., Watanabe H., Ishida T., Morioka H., et al. Albumin domain mutants with enhanced Aβ binding capacity identified by phage display analysis for application in various peripheral Aβ elimination approaches of Alzheimer’s disease treatment. IUBMB Life. 2020;72:641–651. doi: 10.1002/iub.2203. [DOI] [PubMed] [Google Scholar]
- 199.Picón-Pagès P., Bonet J., García-García J., Garcia-Buendia J., Gutierrez D., Valle J., Gómez-Casuso C.E.S., Sidelkivska V., Alvarez A., Perálvarez-Marín A., et al. Human Albumin Impairs Amyloid β-peptide Fibrillation Through its C-terminus: From docking Modeling to Protection Against Neurotoxicity in Alzheimer’s disease. Comput. Struct. Biotechnol. J. 2019;17:963–971. doi: 10.1016/j.csbj.2019.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Subramanian A.-R. Structure qnd Functions of Ribosomal Protein S1. Prog. Nucleic Acid Res. Mol. Biol. 1983;28:101–142. doi: 10.1016/S0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- 201.Machulin A., Deryusheva E., Lobanov M., Galzitskaya O. Repeats in S1 Proteins: Flexibility and Tendency for Intrinsic Disorder. Int. J. Mol. Sci. 2019;20:2377. doi: 10.3390/ijms20102377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Deryusheva E.I., Machulin A.V., Matyunin M.A., Galzitskaya O.V. Investigation of the Relationship between the S1 Domain and Its Molecular Functions Derived from Studies of the Tertiary Structure. Molecules. 2019;24:3681. doi: 10.3390/molecules24203681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Loveland A.B., Korostelev A.A. Structural dynamics of protein S1 on the 70S ribosome visualized by ensemble cryo-EM. Methods. 2018;137:55–66. doi: 10.1016/j.ymeth.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Grishin S.Y., Dzhus U.F., Selivanova O.M., Balobanov V.A., Surin A.K., Galzitskaya O.V. Comparative Analysis of Aggregation of Thermus thermophilus Ribosomal Protein bS1 and Its Stable Fragment. Biochemistry. 2020;85:344–354. doi: 10.1134/S0006297920030104. [DOI] [PubMed] [Google Scholar]
- 205.Grishin S.Y., Deryusheva E.I., Machulin A.V., Selivanova O.M., Glyakina A.V., Gorbunova E.Y., Mustaeva L.G., Azev V.N., Rekstina V.V., Kalebina T.S., et al. Amyloidogenic propensities of ribosomal s1 proteins: Bioinformatics screening and experimental checking. Int. J. Mol. Sci. 2020;21:5199. doi: 10.3390/ijms21155199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Galzitskaya O. New mechanism of amyloid fibril formation. Curr. Protein Pept. Sci. 2019;20:630–640. doi: 10.2174/1389203720666190125160937. [DOI] [PubMed] [Google Scholar]
- 207.Surin A.K., Grishin S.Y., Galzitskaya O.V. Identification of Amyloidogenic Regions in the Spine of Insulin Fibrils. Biochemistry. 2019;84:47–55. doi: 10.1134/S0006297919010061. [DOI] [PubMed] [Google Scholar]
- 208.Henriques S.T., Melo M.N., Castanho M.A.R.B. Cell-penetrating peptides and antimicrobial peptides: How different are they? Biochem. J. 2006;399:1–7. doi: 10.1042/BJ20061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Henriques S.T., Castanho M.A.R.B. Consequences of nonlytic membrane perturbation to the translocation of the cell penetrating peptide pep-1 in lipidic vesicles. Biochemistry. 2004;43:9716–9724. doi: 10.1021/bi036325k. [DOI] [PubMed] [Google Scholar]
- 210.Liu B.R., Huang Y.W., Winiarz J.G., Chiang H.J., Lee H.J. Intracellular delivery of quantum dots mediated by a histidine- and arginine-rich HR9 cell-penetrating peptide through the direct membrane translocation mechanism. Biomaterials. 2011;32:3520–3537. doi: 10.1016/j.biomaterials.2011.01.041. [DOI] [PubMed] [Google Scholar]
- 211.Futaki S. Membrane-permeable arginine-rich peptides and the translocation mechanisms. Adv. Drug Deliv. Rev. 2005;57:547–558. doi: 10.1016/j.addr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 212.Fuerst J.A., Sagulenko E. Protein uptake by bacteria. Commun. Integr. Biol. 2010;3:572–575. doi: 10.4161/cib.3.6.13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Verschueren E., Vanhee P., Rousseau F., Schymkowitz J., Serrano L. Protein-peptide complex prediction through fragment interaction patterns. Structure. 2013;21:789–797. doi: 10.1016/j.str.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 214.Trabuco L.G., Lise S., Petsalaki E., Russell R.B. PepSite: Prediction of peptide-binding sites from protein surfaces. Nucleic Acids Res. 2012;40:W423–W427. doi: 10.1093/nar/gks398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Inbar P., Li C.Q., Takayama S.A., Bautista M.R., Yang J. Oligo(ethylene glycol) Derivatives of Thioflavin T as Inhibitors of Protein-Amyloid Interactions. ChemBioChem. 2006;7:1563–1566. doi: 10.1002/cbic.200600119. [DOI] [PubMed] [Google Scholar]
- 216.Tessier P.M., Jinkoji J., Cheng Y.C., Prentice J.L., Lenhoff A.M. Self-interaction nanoparticle spectroscopy: A nanoparticle-based protein interaction assay. J. Am. Chem. Soc. 2008;130:3106–3112. doi: 10.1021/ja077624q. [DOI] [PubMed] [Google Scholar]
- 217.Boassa D., Lemieux S.P., Lev-Ram V., Hu J., Xiong Q., Phan S., Mackey M., Ramachandra R., Peace R.E., Adams S.R., et al. Split-miniSOG for Spatially Detecting Intracellular Protein-Protein Interactions by Correlated Light and Electron Microscopy. Cell Chem. Biol. 2019;26:1407–1416.e5. doi: 10.1016/j.chembiol.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Chi E.Y., Frey S.L., Winans A., Lam K.L.H., Kjaer K., Majewski J., Lee K.Y.C. Amyloid-beta fibrillogenesis seeded by interface-induced peptide misfolding and self-assembly. Biophys. J. 2010;98:2299–2308. doi: 10.1016/j.bpj.2010.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Agerberth B., Gunne H., Odeberg J., Kogner P., Boman H.G., Gudmundsson G.H. FALL-39, a putative human peptide antibiotic, is cysteine-free and expressed in bone marrow and testis. Proc. Natl. Acad. Sci. USA. 1995;92:195–199. doi: 10.1073/pnas.92.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yount N.Y., Yeaman M.R. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. USA. 2004;101:7363–7368. doi: 10.1073/pnas.0401567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Coquet L., Kolodziejek J., Jouenne T., Nowotny N., King J.D., Conlon J.M. Peptidomic analysis of the extensive array of host-defense peptides in skin secretions of the dodecaploid frog Xenopus ruwenzoriensis (Pipidae) Comp. Biochem. Physiol. Part D Genom. Proteom. 2016;19:18–24. doi: 10.1016/j.cbd.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 222.Yang X., Lee W.H., Zhang Y. Extremely abundant antimicrobial peptides existed in the skins of nine kinds of Chinese odorous frogs. J. Proteome Res. 2012 doi: 10.1021/pr200782u. [DOI] [PubMed] [Google Scholar]
- 223.Schutte B.C., Mitros J.P., Bartlett J.A., Walters J.D., Jia H.P., Welsh M.J., Casavant T.L., McCray P.B. Discovery of five conserved β-defensin gene clusters using a computational search strategy. Proc. Natl. Acad. Sci. USA. 2002;99:2129–2133. doi: 10.1073/pnas.042692699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Li Y., Li X., Li H., Lockridge O., Wang G. A novel method for purifying recombinant human host defense cathelicidin LL-37 by utilizing its inherent property of aggregation. Protein Expr. Purif. 2007;54:157–165. doi: 10.1016/j.pep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 225.Fjell C.D., Hancock R.E.W., Cherkasov A. AMPer: A database and an automated discovery tool for antimicrobial peptides. Bioinformatics. 2007;23:1148–1155. doi: 10.1093/bioinformatics/btm068. [DOI] [PubMed] [Google Scholar]
- 226.Tossi A., Sandri L. Molecular Diversity in Gene-Encoded, Cationic Antimicrobial Polypeptides. Curr. Pharm. Des. 2002;8:743–761. doi: 10.2174/1381612023395475. [DOI] [PubMed] [Google Scholar]
- 227.Begley M., Cotter P.D., Hill C., Ross R.P. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl. Environ. Microbiol. 2009;75:5451–5460. doi: 10.1128/AEM.00730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Singh M., Sareen D. Novel LanT Associated Lantibiotic Clusters Identified by Genome Database Mining. PLoS ONE. 2014;9:e91352. doi: 10.1371/journal.pone.0091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Morton J.T., Freed S.D., Lee S.W., Friedberg I. A large scale prediction of bacteriocin gene blocks suggests a wide functional spectrum for bacteriocins. BMC Bioinform. 2015;16:381. doi: 10.1186/s12859-015-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lynn D.J., Higgs R., Gaines S., Tierney J., James T., Lloyd A.T., Fares M.A., Mulcahy G., O′Farrelly C. Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics. 2004;56:170–177. doi: 10.1007/s00251-004-0675-0. [DOI] [PubMed] [Google Scholar]
- 231.Amaral A.C., Silva O.N., Mundim N.C.C.R., De Carvalho M.J.A., Migliolo L., Leite J.R.S.A., Prates M.V., Bocca A.L., Franco O.L., Felipe M.S.S. Predicting antimicrobial peptides from eukaryotic genomes: In silico strategies to develop antibiotics. Peptides. 2012;37:301–308. doi: 10.1016/j.peptides.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 232.Torrent M., Nogués V.M., Boix E. A theoretical approach to spot active regions in antimicrobial proteins. BMC Bioinform. 2009;10:1–9. doi: 10.1186/1471-2105-10-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hellinger R., Koehbach J., Soltis D.E., Carpenter E.J., Wong G.K.S., Gruber C.W. Peptidomics of circular cysteine-rich plant peptides: Analysis of the diversity of cyclotides from viola tricolor by transcriptome and proteome mining. J. Proteome Res. 2015;14:4851–4862. doi: 10.1021/acs.jproteome.5b00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lee J.H., Chung H., Shin Y.P., Kim I.-W., Natarajan S., Veerappan K., Seo M., Park J., Hwang J.S. Transcriptome Analysis of Psacothea hilaris: De Novo Assembly and Antimicrobial Peptide Prediction. Insects. 2020;11:676. doi: 10.3390/insects11100676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Galzitskaya O.V., Garbuzynskiy S.O., Lobanov M.Y. Is it possible to predict amyloidogenic regions from sequence alone? J. Bioinf. Comput. Biol. 2006;4:373–388. doi: 10.1142/S0219720006002004. [DOI] [PubMed] [Google Scholar]
- 236.Galzitskaia O.V., Garbuzinskiy S.O., Lobanov M.I. Prediction of natively unfolded regions in protein chain. Mol. Biol. 2006;40:341–348. doi: 10.1134/S0026893306020166. [DOI] [PubMed] [Google Scholar]
- 237.Mamonova T.B., Glyakina A.V., Galzitskaya O.V., Kurnikova M.G. Stability and rigidity/flexibility—Two sides of the same coin? Biochim. Biophys. Acta Proteins Proteom. 2013;1834:854–866. doi: 10.1016/j.bbapap.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 238.Khairkhah N., Aghasadeghi M.R., Namvar A., Bolhassani A. Design of novel multiepitope constructs-based peptide vaccine against the structural S, N and M proteins of human COVID-19 using immunoinformatics analysis. PLoS ONE. 2020;15:e0240577. doi: 10.1371/journal.pone.0240577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Memariani H., Memariani M., Moravvej H., Shahidi-Dadras M. Melittin: A venom-derived peptide with promising anti-viral properties. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:5–17. doi: 10.1007/s10096-019-03674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yu Y., Cooper C.L., Wang G., Morwitzer M.J., Kota K., Tran J.P., Bradfute S.B., Liu Y., Shao J., Zhang A.K., et al. Engineered Human Cathelicidin Antimicrobial Peptides Inhibit Ebola Virus Infection. iScience. 2020;23:100999. doi: 10.1016/j.isci.2020.100999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Fan Y., Zhao K., Shi Z.-L., Zhou P. Bat Coronaviruses in China. Viruses. 2019;11:210. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Huang Y., Yang C., Xu X.-F., Xu W., Liu S.-W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.He J., Tao H., Yan Y., Huang S.-Y., Xiao Y. Molecular Mechanism of Evolution and Human Infection with SARS-CoV-2. Viruses. 2020;12:428. doi: 10.3390/v12040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ali A., Vijayan R. Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci. Rep. 2020;10:14214. doi: 10.1038/s41598-020-71188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Zhang J., Zeng H., Gu J., Li H., Zheng L., Zou Q. Progress and Prospects on Vaccine Development against SARS-CoV-2. Vaccines. 2020;8:153. doi: 10.3390/vaccines8020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Cannalire R., Stefanelli I., Cerchia C., Beccari A.R., Pelliccia S., Summa V. SARS-CoV-2 Entry Inhibitors: Small Molecules and Peptides Targeting Virus or Host Cells. Int. J. Mol. Sci. 2020;21:5707. doi: 10.3390/ijms21165707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Barh D., Tiwari S., Silva Andrade B., Giovanetti M., Almeida Costa E., Kumavath R., Ghosh P., Góes-Neto A., Carlos Junior Alcantara L., Azevedo V. Potential chimeric peptides to block the SARS-CoV-2 spike receptor-binding domain. F1000Research. 2020;9:576. doi: 10.12688/f1000research.24074.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Karoyan P., Vieillard V., Odile E., Denis A., Guihot A., Luyt C.-E., Gómez-Morales L., Grondin P., Lequin O. Human ACE2 peptide mimics block SARS-CoV-2 Pulmonary Cells Infection. bioRxiv. 2020 doi: 10.1101/2020.08.24.264077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.van de Veerdonk F.L., Netea M.G., van Deuren M., van der Meer J.W., de Mast Q., Brüggemann R.J., van der Hoeven H. Kallikrein-kinin blockade in patients with COVID-19 to prevent acute respiratory distress syndrome. Elife. 2020;9 doi: 10.7554/eLife.57555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hoffmann M., Kleine-Weber H., Pöhlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Tavassoly O., Safavi F., Tavassoly I. Seeding Brain Protein Aggregation by SARS-CoV-2 as a Possible Long-Term Complication of COVID-19 Infection. ACS Chem. Neurosci. 2020 doi: 10.1021/acschemneuro.0c00676. [DOI] [PubMed] [Google Scholar]
- 258.Mukherjee S., Bhattacharyya D., Bhunia A. Host-membrane interacting interface of the SARS coronavirus envelope protein: Immense functional potential of C-terminal domain. Biophys. Chem. 2020;266:106452. doi: 10.1016/j.bpc.2020.106452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Yuan K., Yi L., Chen J., Qu X., Qing T., Rao X., Jiang P., Hu J., Xiong Z., Nie Y., et al. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem. Biophys. Res. Commun. 2004;319:746–752. doi: 10.1016/j.bbrc.2004.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Han D.P., Penn-Nicholson A., Cho M.W. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350:15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zhang G., Pomplun S., Loftis A.R., Tan X., Loas A., Pentelute B.L. Investigation of ACE2 N-terminal fragments binding to SARS-CoV-2 Spike RBD. bioRxiv. 2020 doi: 10.1101/2020.03.19.999318. [DOI] [Google Scholar]
- 264.Lin B., Qing X., Liao J., Zhuo K. Role of Protein Glycosylation in Host-Pathogen Interaction. Cells. 2020;9:1022. doi: 10.3390/cells9041022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Engering A., Geijtenbeek T.B., van Kooyk Y. Immune escape through C-type lectins on dendritic cells. Trends Immunol. 2002;23:480–485. doi: 10.1016/S1471-4906(02)02296-2. [DOI] [PubMed] [Google Scholar]
- 266.Cambi A., Koopman M., Figdor C.G. How C-type lectins detect pathogens. Cell. Microbiol. 2005;7:481–488. doi: 10.1111/j.1462-5822.2005.00506.x. [DOI] [PubMed] [Google Scholar]
- 267.TopuzoĞullari M., Acar T., Pelİt Arayici P., UÇar B., UĞurel E., Abamor E.Ş., ArasoĞlu T., Turgut-Balik D., Derman S. An insight into the epitope-based peptide vaccine design strategy and studies against COVID-19. Turkish J. Biol. Turk Biyol. Derg. 2020;44:215–227. doi: 10.3906/biy-2006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Liu G., Carter B., Bricken T., Jain S., Viard M., Carrington M., Gifford D.K. Computationally Optimized SARS-CoV-2 MHC Class I and II Vaccine Formulations Predicted to Target Human Haplotype Distributions. Cell Syst. 2020;11:131–144.e6. doi: 10.1016/j.cels.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Singh A., Thakur M., Sharma L.K., Chandra K. Designing a multi-epitope peptide based vaccine against SARS-CoV-2. Sci. Rep. 2020;10:16219. doi: 10.1038/s41598-020-73371-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Dhall A., Patiyal S., Sharma N., Usmani S.S., Raghava G.P.S. Computer-aided prediction and design of IL-6 inducing peptides: IL-6 plays a crucial role in COVID-19. Brief. Bioinform. 2020 doi: 10.1093/bib/bbaa259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Cruz L.R., Baladron I., Rittoles A., Diaz P.A., Valenzuela C., Santana R., Vazquez M.M., Garcia A., Chacon D., Thompson D., et al. Treatment with an Anti-CK2 Synthetic Peptide Improves Clinical Response in Covid-19 Patients with Pneumonia. A Randomized and Controlled Clinical Trial. medRxiv. 2020 doi: 10.1101/2020.09.03.20187112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Dominguez Horta M.d.C. CIGB-258 immunomodulatory peptide: A novel promising treatment for critical and severe COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.05.27.20110601. [DOI] [Google Scholar]
- 273.Bonavita A.G. Ac2-26 mimetic peptide of annexin A1 to treat severe COVID-19: A hypothesis. Med. Hypotheses. 2020;145:110352. doi: 10.1016/j.mehy.2020.110352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Khavinson V., Linkova N., Dyatlova A., Kuznik B., Umnov R. Peptides: Prospects for Use in the Treatment of COVID-19. Molecules. 2020;25:4389. doi: 10.3390/molecules25194389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Farshi E. Peptide-mRNA Vaccine for SARS-Cov-2 Journal of Vaccines & Vaccination. J. Vaccines Vaccin. 2020;S4:1–6. doi: 10.35248/2157-7560.20.S4.002. [DOI] [Google Scholar]
- 276.Pant S., Singh M., Ravichandiran V., Murty U.S.N., Srivastava H.K. Peptide-like and small-molecule inhibitors against Covid-19. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1757510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kabra R., Singh S. Evolutionary artificial intelligence based peptide discoveries for effective Covid-19 therapeutics. Biochim. Biophys. Acta Mol. Basis Dis. 2021;1867:165978. doi: 10.1016/j.bbadis.2020.165978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Chowdhury S.M., Talukder S.A., Khan A.M., Afrin N., Ali M.A., Islam R., Parves R., Al Mamun A., Sufian M.A., Hossain M.N., et al. Antiviral Peptides as Promising Therapeutics against SARS-CoV-2. J. Phys. Chem. B. 2020;124:9785–9792. doi: 10.1021/acs.jpcb.0c05621. [DOI] [PubMed] [Google Scholar]
- 280.Watson A., Ferreira L., Hwang P., Xu J., Stroud R. Peptide Antidotes to SARS-CoV-2 (COVID-19) bioRxiv. 2020 doi: 10.1101/2020.08.06.238915. [DOI] [Google Scholar]
- 281.Mahendran A.S.K., Lim Y.S., Fang C.-M., Loh H.-S., Le C.F. The Potential of Antiviral Peptides as COVID-19 Therapeutics. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.575444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Salman S., Shah F.H., Chaudhry M., Tariq M., Akbar M.Y., Adnan M. In silico analysis of protein/peptide-based inhalers against SARS-CoV-2. Future Virol. 2020;15:557–564. doi: 10.2217/fvl-2020-0119. [DOI] [Google Scholar]
- 283.Bhattacharya R., Gupta A.M., Mitra S., Mandal S., Biswas S.R. A natural food preservative peptide nisin can interact with the SARS-CoV-2 spike protein receptor human ACE2. Virology. 2021;552:107–111. doi: 10.1016/j.virol.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Elnagdy S., AlKhazindar M. The Potential of Antimicrobial Peptides as an Antiviral Therapy against COVID-19. ACS Pharmacol. Transl. Sci. 2020;3:780–782. doi: 10.1021/acsptsci.0c00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Lang J., Yang N., Deng J., Liu K., Yang P., Zhang G., Jiang C. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS ONE. 2011;6:e23710. doi: 10.1371/journal.pone.0023710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Gouda A.S., Mégarbane B. Snake venom-derived bradykinin-potentiating peptides: A promising therapy for COVID-19? Drug Dev. Res. 2020 doi: 10.1002/ddr.21732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Shakil M., Hossain A.J., Karal M.A.S., Islam Z., Chowdhury S., Haque A., Alam M. Comparative Time-Kill Assessment of Cationic Antimicrobial Peptideand Fluoroquinolone against Gram-Negative Bacteria. EC Pharmacol. Toxicol. 2020;8:117–124. [Google Scholar]
- 288.Verma Y.K., Verma R., Tyagi N., Behl A., Kumar S., Gangenahalli G.U. COVID-19 and its Therapeutics: Special Emphasis on Mesenchymal Stem Cells Based Therapy. Stem Cell Rev. Rep. 2020 doi: 10.1007/s12015-020-10037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Abate G., Memo M., Uberti D. Impact of COVID-19 on Alzheimer’s Disease Risk: Viewpoint for Research Action. Healthcare. 2020;8:286. doi: 10.3390/healthcare8030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Mirzaei R., Goodarzi P., Asadi M., Soltani A., Aljanabi H.A.A., Jeda A.S., Dashtbin S., Jalalifar S., Mohammadzadeh R., Teimoori A., et al. Bacterial co-infections with SARS-CoV-2. IUBMB Life. 2020;72:2097–2111. doi: 10.1002/iub.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Lim H.X., Lim J., Jazayeri S.D., Poppema S., Poh C.L. Development of multi-epitope peptide-based vaccines against SARS-CoV-2. Biomed. J. 2020 doi: 10.1016/j.bj.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Luong H.X., Thanh T.T., Tran T.H. Antimicrobial peptides–Advances in development of therapeutic applications. Life Sci. 2020;260:118407. doi: 10.1016/j.lfs.2020.118407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Zhang S., Krumberger M., Morris M.A., Parrocha C.M.T., Griffin J.H., Kreutzer A., Nowick J.S. Structure-Based Drug Design of an Inhibitor of the SARS-CoV-2 (COVID-19) Main Protease Using Free Software: A Tutorial for Students and Scientists. ChemRxiv Prepr. Serv. Chem. 2020 doi: 10.26434/chemrxiv.12791954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Zhang R., Eckert T., Lütteke T., Hanstein S., Scheidig A.J., Bonvin A.M., Nifantiev N.E., Kožár T., Schauer R., Enani M.A., et al. Structure-function relationships of antimicrobial peptides and proteins with respect to contact molecules on pathogen surfaces. Curr. Top. Med. Chem. 2016;16:89–98. doi: 10.2174/1568026615666150703120753. [DOI] [PubMed] [Google Scholar]