Abstract
Background and Purpose
Hepatic fibrosis is a public health problem characterized by activation of hepatic stellate cells (HSCs), which triggers excessive production of extracellular matrix (ECM). Inhibition of HSC activation may be an effective treatment. Since various pathways control HSC activation, a combination of drugs with different mechanisms may be more effective than monotherapy.
Methods
Here, we prepared liposomes loaded with curcumin and cyclopamine to inhibit HSC activation. We systematically analyzed the physicochemical characteristics of liposomes loaded with the two drugs, as well as their effects on HSC proliferation, activation and collagen production on gene, protein and cellular levels.
Results
The prepared liposomes helped solubilize both drugs, contributing to their uptake by cells. Liposomes loaded with both drugs inhibited cell proliferation, migration and invasion, as well as induced more apoptosis and perturbed the cell cycle more than the free combination of both drugs in solution or liposomes loaded with either drug alone. Liposomes loaded with both drugs strongly suppressed HSC activation and collagen secretion.
Conclusion
Our results suggest that liposome encapsulation can increase the uptake of curcumin and cyclopamine as well as the synergism between them in anti-fibrosis. This approach shows potential for treating hepatic fibrosis.
Keywords: hepatic stellate cells, HSCs, hepatic fibrosis, cyclopamine, curcumin, synergistic effect
Introduction
Many types of chronic liver injury involve hepatic fibrosis, which often leads to abnormal liver structure and function.1 Hepatic fibrosis can eventually progress to hepatic cirrhosis, which can trigger portal hypertension, liver dysfunction, circulatory disease, and even hepatic carcinoma.2 In 2010, cirrhosis caused 1 million deaths, accounting for 2% of all deaths that year.3 If properly treated early enough, liver fibrosis can be slowed or even reversed before it develops into cirrhosis. However, current treatments are often ineffective.
Hepatic fibrosis involves excessive accumulation of extracellular matrix (ECM), which arises after livery injury activates hepatic stellate cells (HSCs) to transdifferentiate into myofibroblastic HSCs.4 ECM is composed mainly of types I and III collagen, proteoglycans, and fibronectin.5 To prevent the ECM accumulation that results in liver fibrosis, HSC activation can be inhibited.6–9 HSC activation is controlled by multiple pathways, including the hedgehog, matrix metalloproteinase (MMP), platelet-derived growth factor, and inflammatory signaling pathways.4 Therefore, the regulation of only one signaling pathway often cannot inhibit HSCs’ activation efficiently, and the most effective inhibition probably requires a combination of drugs that act through different mechanism.
The hedgehog pathway, for example, plays an important role in the activation, proliferation and apoptosis of HSCs.10−12 Inhibition of this pathway could revert myofibroblasts back into quiescent HSCs.10,13 Therefore, chemical inhibitors of hedgehog signaling are promising treatments for liver fibrosis.14–17 The inhibitor cyclopamine (Cyc) has been shown to restrain HSC activation and viability both in vitro and in vivo.12
Another promising anti-hepatic fibrosis agent is curcumin (Cur), a major curcuminoid from turmeric.8,18,19 Cur effectively mitigates hepatic fibrosis by inhibiting HSC activation through multiple pathways: blocking leptin and succinate signaling, regulating intracellular glucose and its derivatives, modulating lipid metabolism of HSCs, regulating matrix metalloproteinases (MMPs), and activating peroxisome proliferator-activated receptors (PPARγ) signaling.20–22
We reasoned that Cyc and Cur might inhibit HSC activation synergistically, thereby slowing or even reversing hepatic fibrosis. However, both drugs show low solubility in water, posing an obstacle to drug administration and absorption. To get around this problem, we encapsulated both drugs into liposomes, biocompatible systems for delivering hydrophilic and hydrophobic drugs that can easily be scaled up for commercialization. We analyzed the physicochemical characteristics of liposomes loaded with the two drugs and assessed their effects on HSC proliferation, activation and collagen production.
Materials and Methods
Materials
Lipoid S100 was purchased from Lipoid (Ludwigshafen, Germany); N-(Carbonyl-methoxypolyethyleneglycol 2000)-1,2-distearoyl-sn-glycero-3-phosphoethanolamine (mPEG2000-DSPE), from AVT Pharmaceutical (Shanghai, China). Cur was bought from Solarbio Technology (Beijing, China), and Cys was provided by Selleck Chemicals (Houston, TX, USA). Cholesterol, Propidium iodide (PI) and RNA enzyme were obtained from Sigma-Aldrich (St Louis, MO, USA), and 1,1′-dioctadecyl- 3,3,3′,3′-tetramethylindotricarbocyanine iodide (DiD) was purchased from Biotium (Hayward, CA, USA). Acetonitrile (HPLC grade) and methanol (HPLC grade) were purchased from Kemiou (Tianjin, China). Other agents were of analytical grade or better.
Cells and Cell Culture
Rat hepatic stellate cell line HSC-T6 was purchased from the Shanghai Institutes for Biological Sciences (Shanghai, China) and cultured in DMEM medium with high glucose (Hyclone, USA) containing streptomycin (100 µg/mL), penicillin (100 IU/mL) and 10% fetal bovine serum (Fu Meng Gene, Shanghai, China) in a humidified atmosphere containing 5% CO2 at 37°C. HSC-T6 cells in logarithmic phase were used in experiments. Their activation led them to take on morphological features of myofibroblasts (data not shown).
Preparation and Characterization of Drug-Loaded Liposomes
Liposomes containing Cur, Cyc or both were prepared using the thin-film hydration method.23 Lipoid S100, cholesterol, mPEG2000-DSPE and drug (Cur, Cyc or both) in a mass ratio of 3:1: 1:0.18 were dissolved in trichloromethane in a round flask, after which the solvent was removed by rotary evaporation in a water bath at 37°C. The resulting thin film was hydrated in 5% glucose solution, and the suspension was sonicated using a probe (225 W, 1.5 min), resulting in a homogeneous suspension of drug-loaded liposomes. DiD-loaded liposomes were prepared in the same way, except that DiD was used instead of drug(s).
Size distribution and zeta potential of undiluted the prepared liposomes were measured based on dynamic light scattering (DLS) and electro-phoretic light scattering (ELS) technology, respectively (Malvern Nano ZS90, UK). Liposome ultrastructure was analyzed by transmission electron microscopy (H-600, Hitachi, Japan) after staining them with 2% (w/v) phosphotungstic acid for 30 s on copper grids.
Ultrafiltration through a membrane (Merk Millipore, USA) with a molecular weight cut-off of 30 kDa was used to determine entrapment efficiency (EE%) and drug loading (DL%) of Cur and Cyc according to the equations:
 |
(1) |
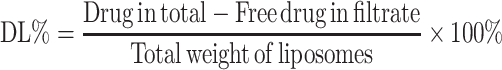 |
(2) |
Drug Release from Liposomes in vitro
Drug release from liposomes was analyzed using dynamic dialysis in 0.01 M phosphate-buffered saline (PBS, pH 7.4) containing 1% (w/w) Tween-80.24,25 Liposomes (1 mL) or drug solution (1 mL) containing 0.6 mg/mL Cur and 0.6 mg/mL Cyc were added inside dialysis bags (Merk Millipore, USA) with a molecular weight cut-off of 8–14 kDa, and the release of free drug into the release medium (4 mL) was measured (n = 3). The dialysis set-ups were shaken on a horizontal shaker at 70 rpm and 37 ± 0.5°C. At pre-set time points from 0.5 h up to 8 d, the entire release medium was collected and replaced with 4 mL of fresh, prewarmed medium. The release medium was diluted up to 5 mL with ethanol, stored at −80°C, then analyzed by high-performance liquid chromatography (Agilent 1260 infinity, USA).
Samples (10 μL for Cur assays, 100 μL for Cyc assays) were injected into the chromatography system equipped with a reverse-phase Kromasil-C18 column (150 mm × 4.6 mm, 5 µm). For Cur analysis, the mobile phase was water (containing 0.1% formic acid) mixed in a 20:80 (v/v) ratio with methanol, and UV absorption of the eluate was measured at 426 nm. For Cyc analysis, the mobile phase was water (containing 0.1% trifluoroacetic acid) mixed in a 55:45 (v/v) ratio with acetonitrile, and UV absorption of the eluate was measured at 209 nm. In both cases, flow rate was 1.0 mL/min at 35°C. Peak areas were recorded for quantification based on validated standard curves.
Uptake of Liposomes by Cells in Culture
As a model of cellular uptake of drugs delivered free in solution or encapsulated in liposomes, the ability of HSC-T6 cells to take up free DiD or DiD-loaded liposomes was analyzed using flow cytometry. Cells were seeded in 12-well plates at a density of 1.7 × 105 cells per well in 1 mL of culture medium. After 24 h, cells were exposed to culture medium containing free DiD or DiD-loaded liposomes; in both cases, the DiD concentration was 100 ng/mL. Then, cells were cultured for another 2 or 4 h, and the medium was removed. Cells were rinsed three times with PBS at 4°C, trypsinized, collected by washing in PBS and centrifuging at 400 g for 3 min. The cell pellet was resuspended in PBS, and mean fluorescence intensity was measured by flow cytometry (Beckman, CA, USA). Each sample contained 1 × 105 cells, and cells cultured with blank medium instead of DiD or DiD-loaded liposomes were processed in parallel as a negative control.
Cytotoxicity
Cytotoxicity of the combination of Cur and Cyc free in solution or loaded into liposomes was assessed in HSC-T6 using the Cell Counting Kit-8 (CCK-8; APExBIO, Shanghai, China). Cells in the logarithmic growth phase were seeded into 96-well plates at a density of 3500 cells per well in 100 μL culture medium, then incubated for 24 h. Cells were exposed for 48 h to 200 μL medium containing liposomes loaded with Cur, Cyc or both, or to the same volume of medium containing both drugs free in solution (Free Cur-Cyc). In all cases, the concentration of each drug was 10 μM. Subsequently, 10 μL of CCK-8 solution was added to each well, and cells were incubated for 4 h at 37°C. Optical density (OD) was measured at 450 nm using a Varioskan Flash multimode plate reader (Thermo, NH, USA). Control wells were seeded with cells and treated with blank medium, while blank wells were not seeded with cells but were treated with blank medium.
Cellular viability was calculated according to the equation:
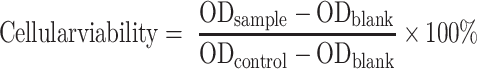 |
(3) |
Synergism between Cur and Cyc in causing cytotoxicity was assessed based on the Q index:26
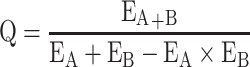 |
(4) |
where EA+B refers to relative viability in the presence of both drugs, and EA or EB to the relative viability of one or the other drug on its own. Q > 1.15 indicates synergism; Q = 0.85–1.15, additivity; Q < 0.85, antagonism.
Cell Cycle Disruption
The distribution of cells in the cell cycle was assessed based on PI staining and flow cytometry.27 HSC-T6 cells were seeded into 12-well plates at the density of 1.7 × 105 cells per well and incubated for 24 h at 37°C. Then the medium was replaced with 1 mL of medium containing liposomes loaded with Cur, Cyc or both, or medium containing the two drugs mixed in free solution. In all cases, the concentration of each drug was 10 μM. Cultures were incubated for another 48 h. Cells exposed to blank medium served as a negative control. Adherent cells were digested with 0.25% (w/w) trypsin, and washed three times with PBS. Then the cells were fixed in cold 75% (v/v) ethanol for 30 min at 4°C, followed by incubation for 15 min in 0.1 mL 0.1% (v/v) Triton X-100 to permeabilize the cytomembrane. Cells were incubated with 20 μg/mL ribonuclease A at 37°C for 30 min, followed by 20 μg/mL PI at 4°C for another 30 min in darkness. Finally, cells (1 × 105 per sample) were analyzed using flow cytometry (Beckman, CA, USA), and DNA histograms were analyzed using ModFit LT (Verify Software House, USA).
Wound Healing
Migration of activated HSCs contributes to liver fibrosis,28 so drug formulations were tested for their ability to inhibit cell migration in a wound-healing assay.29,30 HSC-T6 cells were seeded into 12-well plates at the density of 6 × 104 cells per well, incubated at 37°C for 24 h to obtain confluent monolayers, then scratched across the plate with a sterile 10-μL micropipette tip. The cells were washed three times with PBS, then exposed to serum-free culture medium containing liposomes loaded with Cur, Cyc or both drugs or with the two drugs mixed in free solution. In all cases, the concentration of each drug was 1 μg/mL. Cells treated with serum-free culture medium served as a control. After 24 h of incubation, the medium was replaced with fresh serum-free culture medium and incubated for another 24 h. Wound healing was monitored by microscopy, and the cell-free scratched area after treatment was quantified using Image J software 1.48 (National Institutes of Health, Bethesda, USA) as a percentage of the pre-treatment area.29,30
Apoptosis Induction
Inducing apoptosis in HSCs is one way to inhibit their activation.7 Therefore, we assessed the ability of our drug formulations to induce apoptosis using a double-staining assay based on annexin V-FITC and PI with kit (Wuhan Sanying Biotechnology Co. LTD, Wuhan, China).25 HSC-T6 cells were seeded into 12-well plates at the density of 6 × 104 cells per well and incubated at 37°C for 24 h. Then, culture medium containing liposomes loaded with Cur, Cyc or both drugs or medium containing the two drugs mixed in free solution was added to the wells. In all cases, the concentration of each drug was 1 μg/mL. The cells were incubated for another 48 h, and then cells were collected by trypsinization [0.25% (w/w) trypsin], washed with PBS and centrifuged at 400 g for 3 min for a total of three times, resuspended in binding buffer, and stained for 10 min at room temperature in the darkness with annexin V-FITC and PI. Cells (1 × 105 cells per sample) were immediately analyzed by flow cytometry (Beckman, CA, USA).
Expression of α-SMA and Collagens
HSC activation is associated with high expression of α-SMA and collagens.8,31 Therefore, we examined the ability of our drug formulations to reduce this expression. For experiments ending with Western blotting or quantitative reverse transcription-polymerase chain reaction (RT-PCR), HSC-T6 cells were seeded into 12-well plates at a density of 6 × 104 cells per well and incubated at 37°C for 24 h. Then, culture medium containing liposomes loaded with Cur, Cyc or both drugs or medium containing the two drugs mixed free in solution was added to the well. In all cases, the concentration of each drug was 1 μg/mL. Cultures were incubated for another 48 h, and then cells were trypsinized and pelleted by centrifugation.
In one set of experiments, total RNA was extracted from the cells using Trizol reagent (ApplyGen, Beijing, China), followed by reverse transcription into cDNA using the iScript cDNA synthesis kit (Bio-rad, Hercules, CA, USA) and SYBR Green for quantification. Primers were designed based on mRNA sequences in GenBank and synthesized by Shanghai Shenggong Biotechnology (Shanghai, China). Quantitative RT-PCR was performed on a CFX96 Real-Time system (Bio-Rad, USA) with 18S as an internal control.
In another set of experiments, levels of α-SMA in total cell lysate were quantified using Western blotting as described.31 In brief, total proteins in lysates were resolved via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene nitrocellulose filter membranes. The membranes were then blocked and incubated overnight at 4°C with primary antibodies against α-SMA (Sigma-Aldrich, USA) and β-tubulin (Zen Bio Science, China). Then the membranes were incubated for 2 h at room temperature with horseradish peroxidase-conjugated secondary antibodies. Immunoblots were visualized using the Odyssey system (Li-Cor, Lincoln, NE, USA).
For immunostaining experiments, cells were seeded onto coverslips in the 12-well dishes at a density of 4 × 104 cells per well, then cultured and treated as described above. After the 48-h incubation, cells were rinsed three times with PBS, fixed in 4% paraformaldehyde for 15 min, permeabilized in 0.1% Triton-X 100 for 15 min, blocked with 10% donkey serum, incubated overnight at 4°C with primary antibody against α-SMA (Sigma-Aldrich, USA) and finally incubated for 1 h with FITC-conjugated secondary antibody. Nuclei were counterstained with DAPI. Immunostained cells were examined by confocal laser scanning microscopy (Leica, Wetzlar, Germany).
Collagen Deposition Based on Sirius Red/Fast Green Staining
We also assessed the ability of our drug formulations to influence collagen production by using Sirius Red/Fast Green staining.18,32 HSC-T6 cells were seeded onto coverslips in 12-well plates at a density of 4 × 104 cells per well, then incubated for 24 h. Medium containing liposomes loaded with Cur, Cyc or both drugs or containing the two drugs mixed free in solution was added to the wells. In all cases, the concentration of each drug was 1 μg/mL. Cultures were incubated another 48 h. Cells treated with blank culture medium served as control. Collagen expression was quantified based on Sirius Red/Fast Green staining using a commercial kit (Chondrex, Redmond, Washington state, USA). The wells were photographed with a microscope (Nikon, USA), and their OD at 540 nm and 605 nm was measured using a microplate reader (Biotek Eon, USA). The amounts of collagen and non-collagenous protein were calculated from the following equations:
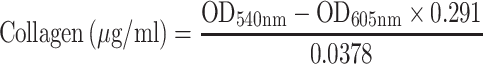 |
(5) |
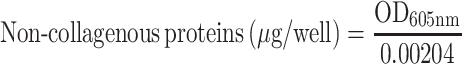 |
(6) |
The results of these calculations were used to determine the ratio of collagen to total protein.
Statistical Analysis
All measurements were made at least in triplicate, and results were presented as mean ± SD. The statistical analysis was performed using Graphpad Prism software. Differences were assessed for significance using the two-tailed Student’s t test, and they were considered significant if the associated P < 0.05.
Results and Discussion
Preparation and Characterization of Liposomes Loaded with Cur and Cyc
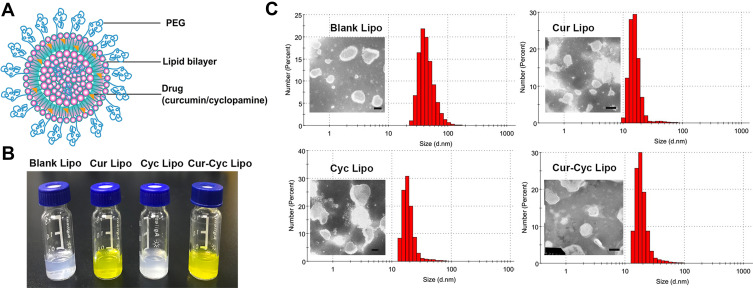
Figure 1A shows a schematic of drug-loaded liposomes, whose opalescent appearance without obvious impurities is shown in Figure 1B. Dynamic light scattering showed that all liposomes were approximately 90 nm in size with a PDI of about 0.20 or less (Figure 1C, Table 1), indicating homogeneous size distribution. Transmission electron microscopy showed liposomes to be spherical and uniform with a size of 80–120 nm (Figure 1C), consistent with dynamic light scattering. Zeta potential was −11.4 mV for empty liposomes and −10.4 mV for Cur-loaded liposomes. In contrast, zeta potential was positive for liposomes containing Cyc alone or together with Cur, which may reflect the alkalinity of Cyc. In liposomes coloaded with Cur and Cyc, average EE% was nearly 100% for Cur and 73.92% for Cyc. The respective drug loading capacities were 3.26% and 2.42%.
Figure 1.
Characterization of liposomes: (A) schematic of liposomes, (B) photographs of prepared liposomes, and (C) transmission electron micrographs and dynamic light scattering data of the liposomes.
Note: Scale bar, 100 µm. Abbreviations: PEG, polyethylene glycol; Cur, curcumin; Cyc, cyclopamine; Lipo, liposomes.
Table 1.
Biophysical Characteristics of Liposomes
| Liposomes | Average Size (d.nm) | PDI | Zeta Potential (mV) | EE (%) | DL (%) |
|---|---|---|---|---|---|
| Blank | 89.17 ± 5.3 | 0.180 ± 0.003 | −11.4 ± 1.2 | NA | NA |
| Loaded with Cur | 88.89 ± 6.2 | 0.192 ± 0.004 | −10.4 ± 2.0 | 99.76 ± 0.19 | 3.89 ± 0.05 |
| Loaded with Cyc | 83.35 ± 4.5 | 0.183 ± 0.003 | 6.67 ± 1.5 | 76.11 ± 1.06 | 2.82 ± 0.16 |
| Loaded with Cur and Cyc | 88.97 ± 3.7 | 0.204 ± 0.002 | 6.93 ± 1.4 | 99.59 ± 0.06; 74.07 ± 0.41 |
3.26 ± 0.00; 2.42 ± 0.01 |
Abbreviations: Cur, curcumin; Cyc, cyclopamine; DL, drug loading; EE, entrapment efficiency; NA, not applicable; PDI, polydispersity index.
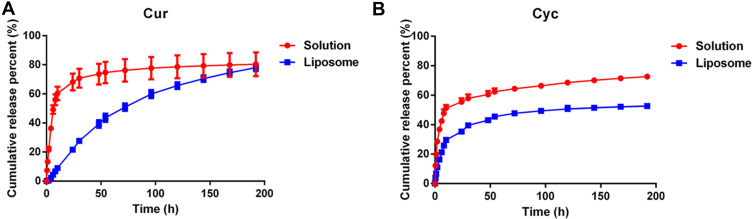
Sustained Cur and Cyc Release from Liposomes in vitro
During dialysis experiments to examine drug release from liposomes, Tween-80 was added at 1% (w/w) to release medium to promote their solubilization. Both free Cur and free Cyc showed rapid drug release profiles (Figure 2). Almost 80% of Cur or Cyc was released in 192 h, including 49.19% of Cur and 42.47% of Cyc during the first 6 h. In contrast, liposome-encapsulated Cur and Cyc showed slower, more sustained release: only 4.38% of Cur and 21.18% of Cyc were released within 6 h, while only 21.62% of Cur and 35.26% of Cyc were released in 24 h. The apparently faster release of Cyc may reflect its lower encapsulation efficiency (76.11% vs 99.76%).
Figure 2.
In vitro release profiles of (A) Cur and (B) Cyc from mixed solution or Cur-Cyc liposomes.
Note: The dialysis buffer was 0.01 M phosphate-buffered saline (pH 7.4) containing 1% (w/w) Tween-80, and the released Cur and Cyc were determined by HPLC method. Abbreviations: Cur, curcumin; Cyc, cyclopamine; HPLC, high-performance liquid chromatography.
The release data were fitted to several mathematical models in an effort to elucidate the release mechanism (Table 2). The Weibull model and Ritger−Peppas model performed comparably well. In the Ritger−Peppas model, a > 1 for Cur, suggesting that its release was driven by Super case-II transport, while that of Cyc was <0.45, suggesting a mechanism of Fickian diffusion.33
Table 2.
Regression Analysis of the Release of Curcumin (Cur) and Cyclopamine (Cyc) from Solution or Liposomes
| Function | Formulation | Drug | Regression Equation | R2 |
|---|---|---|---|---|
| Zero-order: y = a*t+b | Solution | Cur | y = 0.2723t + 43.323 | 0.4923 |
| Cyc | y = 0.2274t + 39.162 | 0.6214 | ||
| liposome | Cur | y = 0.4414t + 7.2066 | 0.919 | |
| Cyc | y = 0.2194t + 21.589 | 0.6724 | ||
| First-order: ln(1-y) = a*t + b | Solution | Cur | ln(1-y) = −0.0069t + 3.9591 | 0.6476 |
| Cyc | ln(1-y) = −0.005t + 4.0872 | 0.7535 | ||
| liposome | Cur | ln(1-y)= −0.0082t + 4.5693 | 0.9879 | |
| Cyc | ln(1-y) = −0.0034t + 4.3517 | 0.7407 | ||
| Higuchi: y = a*t0.5+b | Solution | Cur | y =4.6642t0.5 + 29.854 | 0.6993 |
| Cyc | y =3.7334t0.5 + 28.926 | 0.7992 | ||
| liposome | Cur | y = 6.5835t0.5 −8.5154 | 0.9898 | |
| Cyc | y = 3.5735t0.5 + 11.892 | 0.8636 | ||
| Ritger−Peppas: lny = a*lnt +b | Solution | Cur | lny = 0.3433lnt ± 2.8851 | 0.8205 |
| Cyc | lny = 0.2506lnt± 3.1132 | 0.8838 | ||
| liposome | Cur | lny = 1.0589lnt- 0.7032 | 0.966 | |
| Cyc | lny = 0.4007lnt ± 2.1246 | 0.9214 | ||
| Weibull: lnln(1/(1-y)) = a*lnt +b | Solution | Cur | lnln(1/(1-y)) = 0.03691nt - 2.4055 | 0.9711 |
| Cyc | lnln(1/(1-y)) = 0.33631nt - 1.3787 | 0.9321 | ||
| liposome | Cur | lnln(1/(1-y)) =1.1708lnt - 5.4102 | 0.983 | |
| Cyc | lnln(1/(1-y)) = 0.463lnt - 2.4535 | 0.9443 |
Abbreviations: Cur, curcumin; Cyc, cyclopamine.
Increased Uptake of Cur and Cyc When Delivered in Liposomes
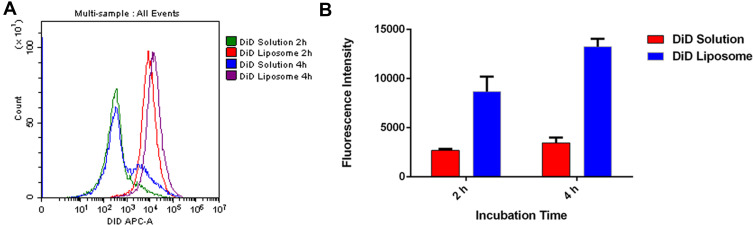
After loading liposomes with the fluorescent dye DiD to allow their tracking,23,34 flow cytometry showed that HSC-T6 cells internalized 3- to 4-fold more DiD after 2 and 4 h when the dye was delivered in liposomes than when it was delivered free in solution (Figure 3). This may reflect the particular uptake pathway for liposomes, which enter the cell mainly through endocytosis and can fuse with intracellular membranes because of their biocompatible phospholipid bilayer.35 This supports the ability of liposomes to solubilize drugs and improve their uptake by cells.
Figure 3.
Uptake of DiD in solution or encapsulated in liposomes by HSC-T6 cells after 2 h and 4 h at 37°C analysed by flow cytometry: (A) representative histogram, (B) quantification of intracellular DiD fluorescence.
Abbreviations: DiD, 1,1ʹ-dioctadecyl- 3,3,3ʹ,3ʹ-tetramethylindotricarbocyanine iodide; HSC-T6, hepatic stellate cells.
Cytotoxicity of Liposomes Loaded with Cur and Cyc
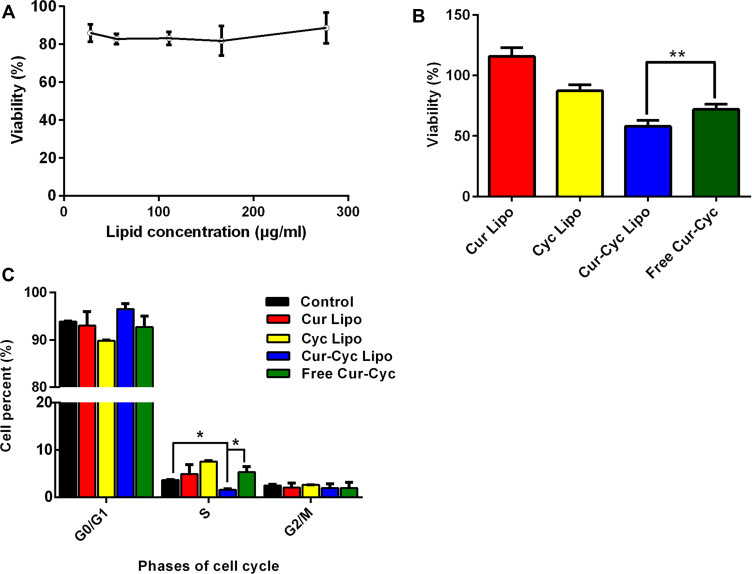
HSC-T6 cells were exposed to blank liposomes at lipid concentrations from 28 to 278 μg/mL. Cell viability remained higher than 80% at all lipid concentrations (Figure 4A), suggesting that the liposomes themselves were well tolerated. Similarly, viability was 87.32% after 48-h incubation with liposomes loaded with 10 μM Cyc and almost 100% after incubation with the corresponding Cur-loaded liposomes (Figure 4B). In contrast, viability was significantly lower with liposomes containing both drugs (57.88% ± 5.12%) than with a mixture of the two free drugs (71.96 ± 4.38%). The Q index was 3.32, suggesting strong synergism between Cur and Cyc in liposomes.
Figure 4.
Ability of liposomes loaded with Cur and Cyc to inhibit HSC-T6 cells proliferation: (A) cytotoxicity of blank liposomes, (B) cytotoxicity of the indicated formulations and (C) flow cytometry analysis of cell cycle distribution after 48-h treatment with the indicated formulations.
Notes: The concentration of each drug in cell treatment was 10 μM. Data are mean ± SD (n=5). *P < 0.05 or **P < 0.01.Abbreviations: Lipo, liposomes; Cur, curcumin; Cyc, cyclopamine; HSC-T6, hepatic stellate cells.
Ability of Liposomes Loaded with Cur and Cyc to Inhibit Proliferation
Cur and Cyc can inhibit HSC proliferation and thereby help reverse their pro-fibrotic effects.12,18 Therefore, we analyzed the effects of drug-loaded liposomes on progression of HSCs through the cell cycle assay (Figure 4C). Liposomes loaded with Cur and Cyc significantly decreased the proportion of cells in S phase in which DNA synthesis is active. However, no significant anti-mitotic effects were observed with liposomes containing only one drug, or with a mixture of the two free drugs. These results are consistent with the results of the MTT assay.
Inhibition of Cell Migration and Invasion by Liposomes Loaded with Cur and Cyc
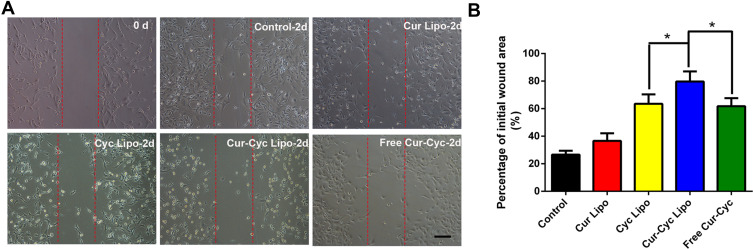
In a wound-healing assay, liposomes loaded with one or both drugs inhibited HSC-T6 cell migration and invasion significantly, while the liposomes loaded with both drugs showed the strongest effects (Figure 5). Since HSCs migration and invasion help drive hepatic fibrosis,27 liposomes loaded with both drugs may have anti-fibrotic potential.
Figure 5.
Ability of liposomes loaded with Cur and Cyc to inhibit HSC-T6 migration in a wound-healing assay: (A) representative photomicrographs of cells before and after treated with indicated formulations for 24 h, (B) quantification of scratch area.
Notes: Scale bar, 100 µm. Data are mean ± SD (n=3). *P < 0.05.Abbreviations: Lipo, liposomes; Cur, curcumin; Cyc, cyclopamine; HSC-T6, hepatic stellate cells.
Induction of Apoptosis by Liposomes Loaded with Cur and Cyc
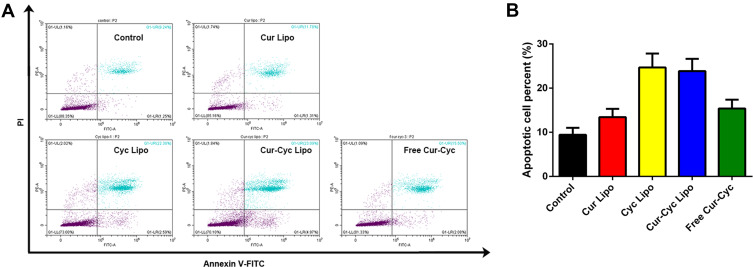
Inducing apoptosis in activated HSCs may exert anti-fibrotic effects,7,36 and such induction has been demonstrated for Cur37 and Cyc.11 In an apoptosis assay based on double staining with annexin V-FITC and PI, the combination of Cur and Cyc in solution was associated with 16.13% of cells in apoptosis, much lower than the corresponding percentages for liposomes loaded only with Cyc (25.8%) or both drugs (25.04%), and higher than the percentage for liposomes loaded only with Cur (10.08%) (Figure 6). These results suggest that delivering the two drugs together in liposomes improves the apoptosis induction effect compared with free drugs in solution. Cyc appeared to contribute more to apoptosis than Cur. We did not detect the synergistic apoptosis induction effect between the two drugs, in contrast to our observations for cytotoxicity.
Figure 6.
Ability of liposomes loaded with Cur and Cyc to induce apoptosis in HSC-T6 cells: (A) representative flow cytometry results, (B) quantification of the percentage of apoptotic cells.
Notes: Cultures were exposed to the indicated formulations for 48 h, then stained with annexin V-FITC and propidium iodide (PI) and analyzed by flow cytometry. The concentration of each drug was 1 μg/mL.Abbreviations: Lipo, liposomes; Cur, curcumin; Cyc, cyclopamine; HSC-T6, hepatic stellate cells.
Suppression of HSCs Activation by Liposomes Loaded with Cur and Cyc
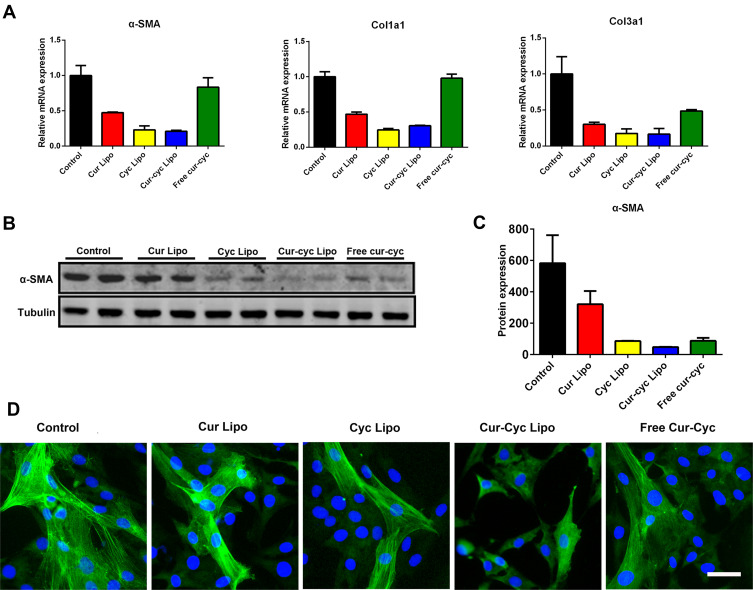
Activation of HSCs is associated with up-expression of α-SMA and collagens.8 Therefore, we tested our drug formulations for their ability to inhibit expression of α-SMA, Col1a1 and Col3a1 based on Western blot, RT-PCR and immunofluorescence staining (Figure 7). After treatment with drug formulations for 48 h, HSC-T6 cells showed decreased expression of fibrotic genes including α-SMA, Col1a1 and Col3a1 (Figure 7A). Liposomes loaded with Cur and Cyc led to 3- to 4-fold lower expression of all three genes than the two drugs mixed in solution. The protein level of α-SMA was significantly lower after treatment with liposomes loaded with both drugs than after treatment with liposomes loaded with either drug alone, and nearly 2-fold lower than the level after treatment with the two drugs mixed in solution (Figure 7B-C). The Q index was 1.00, indicating the addition effect in inhibiting α-SMA expression. These results were corroborated by immunofluorescence staining of α-SMA in HSC-T6 cells (Figure 7D). The green fluorescence stained filamentous α-SMA could be observed apparently in control group cells, while HSC-T6 cells treated with Cur and Cyc coloaded liposomes displayed the weakest signal of α-SMA, further indicated the increased anti-activation effect of HSCs by liposomes loaded with Cur and Cyc.
Figure 7.
Ability of 48-h treatment with liposomes loaded with Cur and Cyc to inhibit expression of α-SMA, Col1a1, and Col3a1 in HSCs-T6 cells: (A) quantitation of mRNA levels, (B-C) Western blotting with quantification of protein levels, and (D) immunofluorescence staining against α-SMA in HSC-T6 cells.
Note: Scale bar, 20 µm.Abbreviations: Lipo, liposomes; Cur, curcumin; Cyc, cyclopamine; HSC-T6, hepatic stellate cells.
Inhibition of Collagen Accumulation by Liposomes Loaded with Cur and Cyc
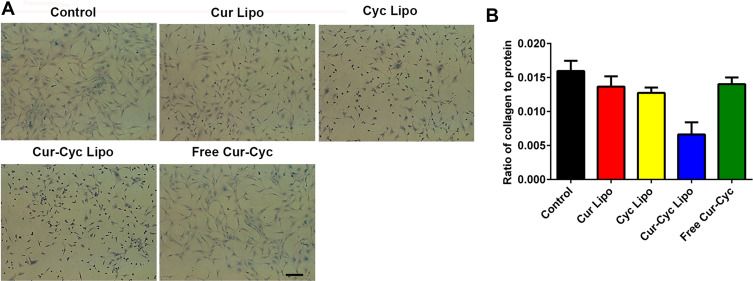
HSC activation is also associated with collagen deposition, which can be detected using Sirius Red/Fast Green staining.18,31,38 Sirius Red specifically binds the [Gly-X-Y]n helical structure of all types of fibrillar collagen, whereas Fast Green binds to non-collagenous proteins.
Figure 8 shows the results of Sirius Red/Fast Green collagen staining on HSC-T6 cells after treated with different formulations for 48 h, and the red color on HSC-T6 cells indicates the presence of collagens. Following 48-h treatment, the extent of collagen deposition in HSC-T6 cells was smallest with liposomes loaded with Cur and Cyc than that with any other treatment (P < 0.01), and the cells in that group became smaller and rounded. Treatment with the two drugs in solution only slightly reduced collagen deposition. Liposomes loaded with Cur and Cyc reduced expression of collagen overall, based on the lower ratio of collagen to total protein (Figure 8B). The other treatments led to slightly lower ratios than in the control cells, which may reflect lower cell numbers because of apoptosis during the 48-h treatment. Therefore, liposomes loaded with Cur and Cyc showed the strongest effect of inhibiting collagen secretion among the tested formulations, which was corresponding to previous results in this study.
Figure 8.
Ability of 48-h treatment with liposomes loaded with Cur and Cyc to inhibit collagen deposition in HSC-T6 cells, based on Sirius Red/Fast Green staining: (A) representative photomicrographs, (B) ratio of collagen to total protein levels.
Note: Scale bar, 100 µm.Abbreviations: Lipo, liposomes; Cur, curcumin; Cyc, cyclopamine; HSC-T6, hepatic stellate cells.
Conclusion
Our results suggest the poorly soluble Cur and Cyc can be efficiently encapsulated into liposomes, which promotes their uptake into HSCs and leads to stronger inhibition of cell proliferation, migration and invasion as well as greater induction of apoptosis than liposomes loaded with either drug alone or than the two drugs mixed in solution. Similar results were observed for HSC activation based on collagen expression and deposition. The co-delivery of the two drugs in liposomes appears to optimize their uptake as well as promote their synergism. This approach shows strong potential for slowing or even reversing hepatic fibrosis, and it merits further studies on safety, efficacy and pharmacokinetics.
Acknowledgments
This work was supported by grant from the National Natural Science Foundation of China (81700538).
Disclosure
The authors declare no conflict of interest.
References
- 1.Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14(7):397–411. [DOI] [PubMed] [Google Scholar]
- 2.Stasi C, Silvestri C, Voller F, Cipriani F. Epidemiology of liver cirrhosis. J Clin Exp Hepatol. 2015;5(3):272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mokdad A, Lopez A, Shahraz S, et al. Liver cirrhosis mortality in 187 countries between 1980 and 2010: a systematic analysis. BMC Med. 2014;12:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134(6):1655–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Z, Ye T, Sun Y, et al. Targeting the vascular and perivascular niches as a regenerative therapy for lung and liver fibrosis. Sci Transl Med. 2017;9(405):eaai8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beljaars L, Meijer DK, Poelstra K. Targeting hepatic stellate cells for cell-specific treatment of liver fibrosis. Front Biosci. 2002;7:e214–222. [DOI] [PubMed] [Google Scholar]
- 7.Elsharkawy AM, Oakley F, Mann DA. The role and regulation of hepatic stellate cell apoptosis in reversal of liver fibrosis. Apoptosis. 2005;10(5):927–939. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Pu S, Liu Q, et al. An integrin-based nanoparticle that targets activated hepatic stellate cells and alleviates liver fibrosis. J Control Release. 2019;303:77–90. [DOI] [PubMed] [Google Scholar]
- 9.Patsenker E, Popov Y, Stickel F, et al. Pharmacological inhibition of integrin alphavbeta3 aggravates experimental liver fibrosis and suppresses hepatic angiogenesis. Hepatology. 2009;50(5):1501–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Choi SS, Michelotti GA, et al. Hedgehog controls hepatic stellate cell fate by regulating metabolism. Gastroenterology. 2012;143(5):1319–1329e1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Omenetti A, Choi S, Michelotti G, Diehl AM. Hedgehog signaling in the liver. J Hepatol. 2011;54(2):366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sicklick JK, Li YX, Choi SS, et al. Role for hedgehog signaling in hepatic stellate cell activation and viability. Lab Invest. 2005;85(11):1368–1380. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, Hao M, Jin HH, et al. Canonical hedgehog signalling regulates hepatic stellate cell-mediated angiogenesis in liver fibrosis. Br J Pharmacol. 2017;174(5):409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Agroudy NN, El-Naga RN, El-Razeq RA, El-Demerdash E. Forskolin, a hedgehog signalling inhibitor, attenuates carbon tetrachloride-induced liver fibrosis in rats. Br J Pharmacol. 2016;173(22):3248–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J, Cao G, Wu X, Cai H, Cai B. Tetramethylpyrazine inhibits activation of hepatic stellate cells through hedgehog signaling pathways in vitro. Biomed Res Int. 2015;2015:603067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyun J, Jung Y. MicroRNAs in liver fibrosis: focusing on the interaction with hedgehog signaling. World J Gastroenterol. 2016;22(29):6652–6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar V, Mondal G, Dutta R, Mahato RI. Co-delivery of small molecule hedgehog inhibitor and miRNA for treating liver fibrosis. Biomaterials. 2016;76:144–156. [DOI] [PubMed] [Google Scholar]
- 18.Rachmawati H, Novel MA, Nisa RM, et al. Co-delivery of curcumin-loaded nanoemulsion and Phaleria macrocarpa extract to NIH 3T3 cell for antifibrosis. J Drug Delivery Sci Technol. 2017;39:123–130. [Google Scholar]
- 19.Chen YN, Hsu SL, Liao MY, et al. Ameliorative effect of curcumin-encapsulated hyaluronic acid-PLA nanoparticles on thioacetamide-induced murine hepatic fibrosis. Int J Environ Res Public Health. 2016;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen G, Wang Y, Li M, et al. Curcumol induces HSC-T6 cell death through suppression of Bcl-2: involvement of PI3K and NF-κB pathways. Eur J Pharm Sci. 2014;65:21–28. [DOI] [PubMed] [Google Scholar]
- 21.She L, Xu D, Wang Z, et al. Curcumin inhibits hepatic stellate cell activation via suppression of succinate-associated HIF-1α induction. Mol Cell Endocrinol. 2018;476:129–138. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y. Curcumin targets multiple pathways to halt hepatic stellate cell activation: updated mechanisms in vitro and in vivo. Dig Dis Sci. 2015;60(6):1554–1564. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Tang J, Fu L, et al. A pH-responsive alpha-helical cell penetrating peptide-mediated liposomal delivery system. Biomaterials. 2013;34(32):7980–7993. [DOI] [PubMed] [Google Scholar]
- 24.Zhang T, Peng Q, San FY, et al. A high-efficiency, low-toxicity, phospholipids-based phase separation gel for long-term delivery of peptides. Biomaterials. 2015;45:1–9. [DOI] [PubMed] [Google Scholar]
- 25.Zhang T, Zheng Y, Peng Q, Cao X, Gong T, Zhang Z. A novel submicron emulsion system loaded with vincristine-oleic acid ion-pair complex with improved anticancer effect: in vitro and in vivo studies. Int J Nanomedicine. 2013;8:1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Z. Addition in drug combination. Acta Pharmacol Sin. 1980;1(1):70–76. [PubMed] [Google Scholar]
- 27.Zhang T, Luo J, Fu Y, et al. Novel oral administrated paclitaxel micelles with enhanced bioavailability and antitumor efficacy for resistant breast cancer. Colloids Surf B. 2017;150:89–97. [DOI] [PubMed] [Google Scholar]
- 28.Yin L, Qi Y, Xu Y, et al. Dioscin inhibits HSC-T6 cell migration via adjusting SDC-4 expression: insights from iTRAQ-based quantitative proteomics. Front Pharmacol. 2017;8:665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jemaa M, Abassi Y, Kifagi C, et al. Reversine inhibits colon carcinoma cell migration by targeting JNK1. Sci Rep. 2018;8(1):11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Z, Feng C, Lu Y, Gao Y, Lin Y, Dong C. Overexpression and biological function of MEF2D in human pancreatic cancer. Am J Transl Res. 2017;9(11):4836–4847. [PMC free article] [PubMed] [Google Scholar]
- 31.Sato Y, Murase K, Kato J, et al. Resolution of liver cirrhosis using vitamin A-coupled liposomes to deliver siRNA against a collagen-specific chaperone. Nat Biotechnol. 2008;26(4):431–442. [DOI] [PubMed] [Google Scholar]
- 32.Mercer KE, Hennings L, Sharma N, et al. Alcohol consumption promotes diethylnitrosamine-induced hepatocarcinogenesis in male mice through activation of the Wnt/-Catenin signaling pathway. Cancer Prev Res. 2014;7(7):675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dash S, Murthy PN, Nath L, Chowdhury P. Kinetic modeling on drug release from controlled drug delivery systems. Acta Pol Pharm. 2010;67(3):217–223. [PubMed] [Google Scholar]
- 34.Li R, Li Y, Zhang J, et al. Targeted delivery of celastrol to renal interstitial myofibroblasts using fibronectin-binding liposomes attenuates renal fibrosis and reduces systemic toxicity. J Control Release. 2020;320:32–44. [DOI] [PubMed] [Google Scholar]
- 35.Khalil I, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58(1):32–45. [DOI] [PubMed] [Google Scholar]
- 36.Li -C-C, Yang C-Z, Li X-M, et al. Hydroxysafflor yellow A induces apoptosis in activated hepatic stellate cells through ERK1/2 pathway in vitro. Eur J Pharm Sci. 2012;46(5):397–404. [DOI] [PubMed] [Google Scholar]
- 37.Shu J, He Y, Lv X, Ye G, Wang L. Curcumin prevents liver fibrosis by inducing apoptosis and suppressing activation of hepatic stellate cells. J Nat Med. 2009;63(4):415–420. [DOI] [PubMed] [Google Scholar]
- 38.Sato M, Mikamo A, Kurazumi H, et al. Ratio of preoperative atrial natriuretic peptide to brain natriuretic peptide predicts the outcome of the maze procedure in mitral valve disease. J Cardiothorac Surg. 2013;8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]