Abstract
Spinal muscular atrophy (SMA), a leading genetic cause of infant death, is a neurodegenerative disease characterized by the selective loss of particular groups of motor neurons (MNs) in the anterior horn of the spinal cord with progressive muscle wasting. SMA is caused by a deficiency of the survival motor neuron (SMN) protein due to a homozygous deletion or mutation of the SMN1 gene. However, the molecular mechanisms whereby the SMN complex regulates MN functions are not fully elucidated. Emerging studies on SMA pathogenesis have turned the attention of researchers to RNA metabolism, given that increasingly identified SMN-associated modifiers are involved in both coding and non-coding RNA (ncRNA) processing. Among various ncRNAs, microRNAs (miRNAs) are the most studied in terms of regulation of posttranscriptional gene expression. Recently, the discovery that miRNAs are critical to MN function and survival led to the study of dysregulated miRNAs in SMA pathogenesis. Circulating miRNAs have drawn attention as a readily available biomarker due to their property of being clinically detectable in numerous human biofluids through non-invasive approaches. As there are recent promising findings from novel miRNA-based medicines, this article presents an extensive review of the most up-to-date studies connecting specific miRNAs to SMA pathogenesis and the potential applications of miRNAs as biomarkers and therapeutic targets for SMA.
Keywords: biomarker, microRNA, RNA-binding protein, spinal muscular atrophy, survival motor neuron protein, therapeutic target
Introduction
Pathogenesis of spinal muscular atrophy
Spinal muscular atrophy (SMA) is an autosomal recessive neuromuscular disorder caused by the degeneration of alpha motor neurons (MNs) in the spinal cord leading to muscle atrophy and weakness. Despite being recognized as a rare disease with an estimated worldwide incidence of ~1/10,000 live births, SMA accounts for the second most common autosomal recessive disease and the most common monogenic disorder causing early infant death.1,2 The carrier frequency varies from 1 in 38 to 1 in 72 among different ethnic groups, with a pan-ethnic average of 1 in 54.3,4
From a pathological viewpoint, SMA results from an insufficient level of a 38 kDa protein, called survival motor neuron (SMN), due to a homozygous deletion or mutation of the Survival of Motor Neuron 1 (SMN1) gene. Studies indicate that two genes encode SMN protein in humans: SMN1 and a 99% identical copy in sequence, SMN2, which is fundamentally differentiated by a single nucleotide change (C to T) in exon 7.1,4 Such a variant causes exon 7 exclusion in most transcripts (90%) of SMN2, called SMN△7. Consequently, SMN2 can produce only 10% of full-length (FL) SMN mRNA and its product: functional SMN protein. Given that residual FL-SMN2 transcripts can compensate for the defect in SMN1 to a limited extent, SMA severity is inversely related to SMN2 copy number; that is, the higher the copy number, the less severe the SMA phenotype. However, such SMN2 copy number-relevant phenotype–genotype correlation can be affected by other factors. Recent studies have shown that other cellular mechanisms, like positive or negative disease modifiers, may also involve modulation of SMA clinical severity. For example, rare SMN2 variants (c.859G > C), and several independent modifiers such as plastin 3 or neurocalcin delta, can modify phenotypic severity.4 Conclusively, the deficiency in SMN1 causes SMA, whose phenotype is partially compensated by retained SMN2 copy number.
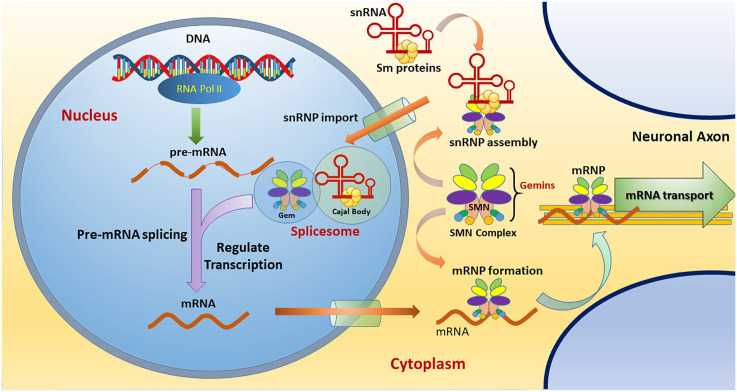
SMN is a multifactorial protein, expressed ubiquitously in both the cytoplasm and nucleus of cells. Notably, SMN is known to serve as a core component in assembling ribonucleoprotein (RNP), which involves several essential cellular pathways, including DNA repair and pre-mRNA splicing.5,6 The SMN complex, composed of SMN, Gemins 2–8, and UNR-interacting protein (UNRIP), binds to the Sm proteins and uridine-rich small nuclear RNAs (snRNAs), thereby constructing small nuclear RNPs (snRNPs) that assemble into a spliceosome in the nucleus.7,8 Besides functioning in snRNP assembly, the SMN complex is known to play a role in regulating the pre-mRNA splicing machinery by facilitating arginine methylation of specific splicing-related proteins in the nucleus (Figure 1).6,9 SMN also functions in restoring damaged DNA and transporting mRNA along the axon of MNs.10,11 In animal models, a positive correlation between snRNP production and SMA phenotypes has been demonstrated, and SMA severity can be even ameliorated by delivering a separate snRNP complex without an SMN component.12–14 This finding suggests that an autoregulatory feedback loop exists, where SMN levels might influence the pre-mRNA splicing of SMN, probably through manipulating a common pathway of snRNP biogenesis.15 Collectively, current evidence points out that the defective assembly of the SMN–RNP complex leads to the majority of pathognomonic features of SMA.
Figure 1.
The cellular functions of SMN complex. The SMN complex is composed of SMN, Gemins 2–8, and UNRIP. In the cytoplasm, the SMN complex functions to assemble Sm proteins onto the snRNAs to create an active snRNP. Then, the SMN–snRNP complex is imported into the nucleus to form a spliceosome that functions in transcriptional regulation, especially pre-mRNA splicing. In the cytoplasm, the SMN complex can assemble an mRNP that acts in mRNA transport along the neuronal axon.
mRNP, messenger ribonucleoprotein; SMN, survival motor neuron; snRNA, small nuclear RNA; snRNP, small nuclear ribonucleoprotein; UNRIP, UNR-interacting protein.
However, it is still unclear whether the pathogenesis of SMA is caused by a specific pattern or a combination of dysregulated effects. The cell-autonomous effects due to SMN deficiency are the main causes of MNs degeneration; however, this does not explain the full SMA phenotype, implying the involvement of not only dysregulated neural networks but other non-neuronal cell types in SMA pathology.16–18 Interestingly, recent studies of SMA animal and cell models suggest that MN survival and functionality relies highly on glial cells, which might play an essential role in neuronal communication and neuroinflammation.19 Moreover, three independent pieces of research also identified immune organ defects in SMA models.20 This evidence suggests that SMA could also be a disease of the neuroinflammatory pathway.
Biogenesis of non-coding RNAs and microRNAs
The interpretation of genetic data and genome-wide analysis of gene regulatory networks have been traditionally protein-centric. However, new sequencing technology of mammalian transcriptomes revealed that more than 50% of RNA transcripts do not possess protein-coding elements, and are thus termed non-coding RNAs (ncRNAs).21,22 These pervasive ncRNAs are diverse in biogenesis, action mode, and functional features.23 While well-studied “housekeeping” ncRNAs are known to be involved in the spliceosomal and translational machinery, increasing efforts are now being focused on the sophisticated class of “regulatory” ncRNAs that affect predominantly the expression or regulation of protein-coding genes.24 Increasing evidence of biochemical and genetic pieces have suggested an essential role for regulatory ncRNAs in developmental and pathological contexts.25 Furthermore, a growing body of study also suggests that ncRNAs function as epigenetic modifiers, mediating fine-tuning of the process of neuronal development, differentiation, biochemical pathways, and synaptogenesis.26–28
The ncRNAs can be classified broadly based on their size. Regulatory ncRNAs can be further divided into two groups based on their lengths, as small or long ncRNAs. Small ncRNAs are shorter than 200 nucleotides (nt) in size, including microRNA (miRNA, 22–25 nt), Piwi-interacting RNA (piRNA, 21–35 nt), small nucleolar RNA (snoRNA, 60–170 nt), and transfer RNA (tRNA, 70–100 nt).29 Among various small ncRNAs, miRNAs are the most studied in regulating developments and pathology in humans. One of the main factors for this is that miRNA expression profiles usually demonstrate marked changes with regards to a pathological status, implying that miRNAs act as important disease modifiers.30,31
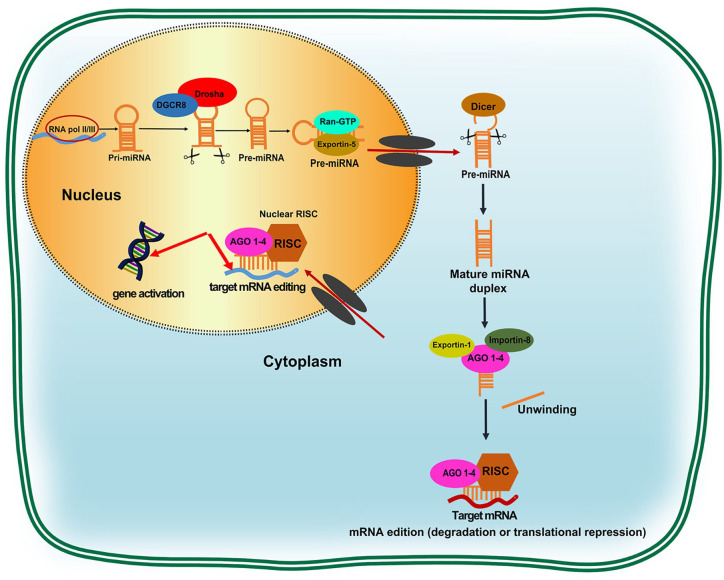
Figure 2 illustrates the canonical pathway of miRNA biogenesis in detail. MiRNAs are endogenous single-chain RNA molecules transcribed initially by the intranuclear enzymes, RNA polymerase II or III, into hairpin-form primary transcripts (pri-miRNA) of over 1 kb in length. These pri-miRNA transcripts are specially recognized and then cleaved by the “microprocessor complex” comprising RNAse IIII enzyme-Drosha ribonuclease and its cofactor, the DiGeorge syndrome critical region gene (DGCR8). The result of this cleavage is a 60–70 nt precursor molecule (pre-miRNA) generated in the nucleus. Exportin5 then transports the pre-miRNA into the cytoplasm, where a second cleavage by the Dicer ribonuclease occurs, which, acting in conjunction with other factors, gives rise to a miRNA/miRNA duplex of ~22–25 nt nucleotides. One of the chains of this duplex, the so-called guide chain, is incorporated into the Argonauta (AGO) protein. Human-specific AGO protein 1–4, loaded with the guidewire, is then incorporated into the RNA-induced silencing complex (RISC), where, through compatibility through the 3′-untranslated region (3′UTR) region, it binds to the target mRNA, causing further translational repressions and/or modifications of target mRNAs.31,32 However, the stability of the mature miRNA molecule is controlled by many cis- and trans-acting factors, the formation of protein complexes, and exposure to nucleases.33
Figure 2.
Canonical pathway of microRNA biogenesis. pri-miRNA is transcribed by RNA pol II/III and then cleaved by Drosha and its cofactor DGCR8 to form hairpin-form pre-miRNA. Pre-miRNA is then transported by Exportin5 into the cytoplasm and then cleaved by Dicer, producing an miRNA/miRNA duplex. One chain of miRNA duplex, called the guide chain, is incorporated into the AGO protein, while the transient chain will be degraded. The guiding chain of miRNA loaded with AGO 1–4 is then incorporated into the RISC, which induces translational repression and degradation of the mRNA targets. Furthermore, by integrating with exportin-1 and importin-8, the miRNA-AGO complex can also be translocated into the nucleus, where miRNAs can function in gene activation or an unconventional manner, regulating intranuclear mRNA biogenesis.
AGO, Argonauta; DGCR8, DiGeorge syndrome chromosomal region 8; miRNA, microRNA; pre-miRNA, precursor miRNA; pri-miRNA, primary miRNA; RISC, RNA-induced silencing complex; RNA pol II/III, RNA polymerase II or III.
Intriguingly, mature miRNAs in the nucleus suggest that miRNAs can travel between the cytoplasm and nucleus. MiRNAs and AGO proteins are known to be translocated into the nucleus through some nuclear export receptors, as exportin-1 and importin-8. In the nucleus, miRNAs can engage in gene activation or in an atypical approach to modify the metabolism and activities of miRNAs (Figure 2).34
The role of miRNAs in motor neuron development, survival, and degeneration
The developmental patterns of the central nervous system (CNS) depend largely on a sophisticated direction through the spatial and temporal expression of transcriptional modulators that gradually restrain cell potential to determine final neuronal identity and connectivity. It has recently been suggested that miRNAs may play a pivotal role in defining spatial boundaries and temporal transitions of developing neurons through their ability to epigenetic regulation.35,36 Currently, more than 2000 miRNAs have been identified in humans, and it is believed that they collectively regulate more than one-third of the genes in the genome37; however, only a few known miRNA regulating embryonic patterning and development of neurons have been reported.38 This circumstance could have two main explanations: (1) a large number of miRNAs belong to diverse families, with solitary members working irrelevantly; and (2) as miRNAs might work primarily to fine-tune developmental transitions, their incapacity may not cause predominant deficits or phenotypic changes but instead more insidious defects affecting subgroups of cells observed at developmental boundaries.39
Several mouse models with miRNA-knockouts have demonstrated significant neuropathological phenotypes,35 and miRNAs have shown to be able to promote reprogramming of human fibroblasts into neuronal cells as well as spinal MNs.40 Emerging evidence also indicates specific miRNAs involved in a diverse of neurological disorders; for example, miR-34b and miR-9 in Huntington’s disease, miR-128a/miR-24/let-7b in mood disorder, miR-206 and miR- 153 in Alzheimer’s disease, and miR-189 in Tourette’s syndrome.41–44 In the spinal cord, it has been suggested that miRNAs might fine-tune neuronal progenitor patterning, cell fate determination, and vitality during MN development.39,40 The proposed link between MNs and miRNA first came from the finding that miR-196 is involved in spinal MN patterning during embryogenesis by targeting Hoxb8.45 Subsequently, increasing numbers of miRNAs have been identified in diverse associations with MN developments, from neural progenitor patterning to cell fate differentiation and regulation of postmitotic MNs development and survival.27,46–48 Given that miRNAs are a pivotal component of the genetic program regulating MN development and acquirement of subtype-specific characteristics, we can speculate how a subtle alteration in miRNA expression might affect MN development and survival.
Concerning the pathogenic role of miRNA in MN degeneration, previous research reported that deletion of Dicer – an enzyme responsible for miRNA biogenesis – in mice spinal MNs by applying Olig2-Cre or ChAT-Cre produced hallmark phenotypes mimicking motor neuron diseases (MND).49–51 These fundamental findings set up the critical role of miRNA participating in MN degeneration. Indeed, an increasing number of miRNAs, termed motomiRs, are proposed to be involved in the maintenance, regeneration, development, and survival of MNs.34 With advances in molecular biology, researchers have extended the role of miRNAs to a diverse range of MN processes, including subtype specification, cytoskeletal integrity, synapse plasticity, neurotransmitter release, and neurite (axon) growth.27,28,52
According to their tissue-specific properties, miRNAs can function with either neuroprotective or disease-promoting effects.53 Nevertheless, aberrant miRNA expression is increasingly identified as a culprit of degeneration in spinal MN. There are several commonalities of SMA with other MND, in particular amyotrophic lateral sclerosis (ALS). Both these two most common MND share a similar pathognomonic pathway, exhibiting aberrant RNA-mediated gene expression, including the processing of both pre-mRNA and miRNA.54 Given the aforementioned multifaceted roles of miRNAs in controlling MN development, it is reasoned that the role of miRNA has been increasingly associated with the pathogenesis of ALS and SMA. Accordingly, several neuronal developmentally associated (i.e. miR-9, miR-124, and miR-133) and MN- enriched (i.e. miR-183, miR-218, and miR-17~92) miRNAs have been suggested as being pivotal to the pathomechanisms of SMA and ALS.46,48,51,52,55,56 Moreover, several neuronal-specific miRNAs have been proposed to be causal for other spinal diseases, including miR-196a in spinal and bulbar muscular atrophy (SBMA) and miR-21/miR-431/miR-138 in axonal degeneration of sensory neurons.57–60
Associations between SMA and miRNAs
MiRNAs associated with the pathogenesis of ALS have been studied extensively and reviewed systemically elsewhere.61–63 On the other hand, miRNAs have been linked to SMA pathogenesis increasingly in studies that suggest miRNAs might act as (1) an essential modulator of SMN-mediated molecular pathways; (2) applicable biomarkers representing SMA progress or treatment response; (3) potential therapeutic targets for SMA.39,55,64 The hypothesis for the link between miRNA and SMA might elucidate the diverse pathways affected by SMN deficiency. A single miRNA can modulate multiple genes concurrently, and might regulate whole genetic networks by modifying the expression of a specific protein. Nevertheless, whether aberrant expression of miRNAs is responsible for the direct pathognomonic cause, or just a consequence reflecting disease progression of SMA, is still unclear, and further investigations are continuing.
Interactions between SMN and RNA-binding proteins in modulating miRNA metabolism and activity
No direct evidence ties an SMN-miRNA signature because SMN protein per se does not possess any known binding domains for RNA or miRNA. Therefore, the SMN complex might instead interact with a specific RNA-binding protein (RBP) to affect miRNA biogenesis.65,66 Indeed, mounting research has reported the intersection of SMN with various RBPs involved in different modifications for mRNA transcription and translation, among other aspects of RNA metabolism.39,67 Similar to the SMN protein complex, miRNAs can also participate in the formation of various RBP or RNP complexes affecting mRNAs functions.68 For example, two components of SMN complex, Gemin3 and Gemin4, are known to integrate with several miRNAs to create a novel miRNA-binding protein called miRNP.69,70 This miRNP complex can further integrate with AGO2, serving as a core component in RISC, which mediates miRNA biogenesis and mRNA post-transcriptional regulation.71 However, it seems that SMN does not bind directly to the miRNP complex; therefore, whether the miRNP is functionally crucial in SMA pathogenesis requires further investigation.
Notably, besides the regulatory function of SMN in RNA processing, updated studies have deciphered potential associations of SMN and RBP in regulating miRNA biogenesis (Table 1), in particular TAR DNA-binding protein-43 (TDP-43), fused in sarcoma/translocated in liposarcoma (FUS/TLS), fragile X mental retardation protein (FMRP), and KH-type splicing regulatory protein (KSRP).54,72–74 SMN might participate in miRNA–RBP or miRNP complex formation, which further regulates miRNA biogenesis and metabolism. The defective SMN protein may dysregulate the MN-specific miRNA or miRNP complex, leading to MN death.71,75 Collectively, the proposed mechanisms by which SMN-associated RBPs might participate in miRNA biogenesis include (1) promoting Drosha recruitment onto specific miRNA loci; (2) engaging in components of the Drosha and Dicer complexes; (3) acting as regulators of the RISC complex.66,76,77 As a consequence, a dysfunctional SMN–RBP complex related to defective SMN protein under the SMA pathological background might cause dysregulated processing of miRNAs and pre-mRNA splicing. Identifying the interaction between SMN and its associated RBPs involved in miRNAs biogenesis can help to elucidate unknown pathogenesis underlying SMA.
Table 1.
Proposed SMN-associated RNA-binding proteins affecting miRNA biogenesis and function.
| SMN-associated RBPs | Roles in post-transcriptional modification of mRNA | Roles in miRNA biogenesis and functions | Neuronal modeling mechanism | Association with miRNAs reported in MND78 | Stress granule formation | References |
|---|---|---|---|---|---|---|
| TDP-43a,b | Regulating transportation, transcription and translation of mRNA, and modulation of pre-mRNA splicing | • Binding to the Dicer and Drosha complexes • Modulating pri-miRNA generation |
Differentiation and development of neuron, synaptic plasticity/formation, neurite outgrowth | miR-9, miR-125b, miR-132 | Yes; SG assembly |
Wang et al.79, Kawahara and Mieda-Sato80, Ederle and Dormann81 |
| FUS/TLSa | Regulating transportation, transcription and translation of mRNA, and modification of pre-mRNA splicing | Facilitating recruitment of Drosha complex | Differentiation and survival of neuron, synaptic plasticity/formation, neurite outgrowth | miR-9, miR-125b, miR-132 | Yes; SG assembly |
Yamazaki et al.54, Ederle and Dormann81, Morlando et al.82 |
| FMRP | mRNA metabolism, translational modification | Binding with Dicer and AGO onto RISC | Differentiation and maintenance of neuron, synaptic plasticity/formation, neurite outgrowth | miR-125 a/b, miR-132 | Yes; SG assembly |
Piazzon et al.72, Jin et al.83, Edbauer et al.84 |
| KSRP | mRNA metabolism, translational modification | Binding with Drosha and Dicer complexes to facilitate miRNA maturation and regulate mRNA metabolism | Functioning in the regulation of mRNA stability in neurons and glial cells and affecting axonal outgrowth | let-7a (NMJ), miR-206 | Yes; SG assembly |
Tadesse et al.73, Trabucchi et al.85, Amirouche et al.86 |
| HuD (ELAVL4, ELAV like RNA binding protein 4) | Regulate stability, transport, translation of mRNA; participating in mTOR pathway | Binding onto RISC to regulate miRNA biogenesis | Regulating axon outgrowth, and synaptic plasticity | miR-129 | Yes; SG assembly |
Fukao et al.87, Loffreda et al.88, Sosanya et al.89, Hao le et al.90 |
| IMP1 (IGF2BP1; ZBP1), | Maintaining mRNA stability by halting miRNA-mediated silencing | Binding to AGO2 to modulate miRNA biogenesis | Axon outgrowth | miR-183 let-7 | Unknown | Fallini et al.76, Gardiner et al.91, Degrauwe et al.92 |
| TIAL1 (also referred to as TIAR) | Pre-mRNA splicing, translation | Reducing Dicer catalytic activity in cells | Regulation of SMN exon 7 splicing, maintaining MN health | To be determined | Yes; SG assembly |
Emde et al.62, Singh et al.93 |
TDP-43 and FUS/TLS proteins interact with pre-mRNA molecules and define their fate by modulating splicing, transportation, and translation.
TDP43 can co-localize with Drosha, a miRNA-processing enzyme, suggesting its potential to participate in miRNA processing.
FMRP, fragile X mental retardation protein; FUS/TLS, fused sarcoma/translocated in liposarcoma; IMP1, insulin-like growth factor 2 mRNA-binding protein 1; KSRP, KH-type splicing regulatory protein; miRNA, microRNA; MN, motor neuron; MND, motor neuron disease; NMJ, neuromuscular junction; RBP, RNA-binding protein; RISC, RNA-induced silencing complex; SG, stress granule; SMA, spinal muscular atrophy; SMN, survival motor neuron; TIAL1, T-cell-restricted intracellular antigen-like 1; TDP-43, TAR DNA-binding protein-43.
The SMN–MiRNA interaction in SMA pathogenesis
The potential ways that miRNAs are involved in the pathogenesis of SMA are just beginning to be identified. Recent studies have demonstrated aberrant miRNAs expression in SMA, and several of these experiments have proposed the SMA-miRNA signature in different SMA models,39,64 which we summarize in Table 2. Recent transcriptome profiling has discovered a growing number of specific miRNAs linked to survival, synaptic plasticity/formation, endoplasmic reticulum (ER) stress, and ribosomal RNA binding in different SMA animal and cell models.55,94,95 Several potential molecular mechanisms underlying an SMA-miRNA signature have been hypothesized: (1) as the SMN complex functions in mRNA pre-splicing and editing, deficiency of SMN might also cause defects in proteins responsible for miRNA biogenesis, leading to aberrant miRNA expression34; (2) the SMN complex is known to participate in miRNP complex formation, which further incorporates with RISC to regulate miRNA biogenesis69–71; (3) the SMN complex has been found in the stress granules (SGs), and a low level of SMN is known to prevent SG formation.8,96 Furthermore, several SMN-associated RBPs, including TDP-43, FUS/TLS, FMRP, and KSRP, are known to be involved in the SG formation (Table 1).66 Because these SG-related RBPs are also involved in miRNA biogenesis, such an indirect SMN-miRNA signature may affect cellular stress responses, which could regulate MN survival11,62,94,97; (4) degradation of the SMN complex might occur via the ubiquitin-proteasome pathway.98,99 Interestingly, some studies have demonstrated that miRNAs might modulate SMN expression by affecting the expression of ubiquitin enzymes.52,100,101
Table 2.
SMA-related miRNAs discussed in this review.
| miRNA | Role in MN development and degeneration | Targets with interaction | Research models/expression Profiles | Involvements in SMA pathomechanisms | Dysregulation in other MND and potential links | References |
|---|---|---|---|---|---|---|
| miR-1 (muscle-specific) | Myogenesis by regulating muscle transcription factors, such as MyoD and myogenin | HDAC4, Pax7 | • Patient serum • Increased |
Possible similar mechanism found in ALS, as to promote myogenesis | • Yes, in ALS animal model and patient • May be involved in myogenesis or myelination process in the spinal cord |
Bonanno et al.102 |
| miR-2 | Neuronal development and function; correct NMJ functioning | CHRM2, m2R | • Caenorhabditis elegans model, SMA mouse model • Decreased |
Defected NMJ function | None reported | O’Hern et al.103 |
| miR-9 | MN subtype determination; neuron dendritic outgrowth; regulate synaptic function | OC1, NEFH, REST, Map1b, MCPIP1 | • Mouse, patient fibroblast, patient serum • Decreased in the spinal cord, but increased in skeletal muscle |
• Dysregulated expression in MNs differentiated from embryonic stem cells • Regulation of motor subtype determination via FOXP1 patterning • miR-9 can delay neurite outgrowth in vitro and impair the radial neuronal migration in embryonic mouse neocortex in vivo |
• Yes, in ALS animal model and patients • May be involved in neural stem cells proliferation, distribution, and differentiation |
Haramati et al.50, Catapano et al.104, Wang et al.105 |
| miR-23a | Neuroprotective properties; regulate axonal development; suppress skeletal muscle atrophy | Atrogin1, MuRF1 (maybe, no direct target experiment was verified by luciferase assay) | • SMA patient iPSCs, SMA mouse model • Decreased |
• Prevent the astrocyte-conditioned media-induced MN loss in vitro
• In mice model, enhanced miR-23a expression via virus vector increased MN soma size and muscle fiber area, and reduced NMJ defects |
• Yes, dysregulated in ALS animal model and patient, • May be involved in neuronal apoptotic pathway and mitochondrial function |
Kaifer et al.106 |
| miR-100-5p | Abnormal proliferation of neural progenitors aberrant cell cycle | IGF1R, possible | • SMNΔ7 mouse NSCs • Decreased |
Decreased miR-100-5p in SMAΔ7 mice neural stem cells induces high IGF1R, excessive proliferation of neural progenitors, and prevents appropriate exit of the cell cycle. | None reported | Luchetti et al.107 |
| miR-132 (possible) | Neuron dendritic outgrowth and synaptic plasticity; neovascularization, may cause ischemic pathology in both skeletal muscle and spinal cord of SMA model | Dysregulated expression due to TDP43 interaction with Dicer (data from ALS), p250GAP | • SMA mice, patient serum • Decreased in the spinal cord, but increased in skeletal muscle |
• Expression is dysregulated in TDP-43-deficient ALS • Neuronal morphology and cognition in ALS • Delay neurite outgrowth in vitro and impair radial neuronal migration in embryonic mouse neocortex in vivo • Involved in synaptic plasticity • Process of neovascularization: may induce vascular defects in both skeletal muscle and spinal cord of SMA patients |
• Yes, in ALS animal model and patients • May be involved in FUS or TDP-43 dysregulated pathway |
Catapano et al.104 |
| miR-133 a/b (muscle-specific) | Proliferation of myoblasts by repressing Serum Response Factor (SRF) development and maturation of NMJ | SRF, UCP2 | • SMA Patient serum • Increased |
Possible similar myogenesis mechanism found in ALS, including: • Sustains myoblast proliferation • Participates in regulating the myogenic fate determination • Regulate NMJ synaptic function |
• Yes, in ALS animal model and patients • May be involved in myogenesis, NMJ function, or myelination process (neurofilaments) in the ALS spinal cord |
Bonanno et al.102 |
| miR-146 | MN loss caused by astrocyte-mediated pathology through NFκB signaling | GDNF, NOTCH2, GATA transcription factors, | • Patient iPSCs • Decreased |
• miR-146 levels are influenced both directly and indirectly by SMN1 levels. SMN re-expression decreases miR-146a levels nearly to control levels • The NFκB pathway is an inducer of miR-146a |
• Yes, in ALS animal model and patients • May be involved in the regulation of Neurofilament mRNA expression in ALS |
Sison et al.108 |
| miR-183 | Protein synthesis; axonal outgrowth | mTOR pathway | • Mouse, cortical neurons, patient fibroblast • Increased |
Increased miR-183 and reduced local axonal translation of mTOR in SMN-deficient neurons | None reported | Kye et al.109 |
| miR-206 (muscle-specific) | Myofiber formation; satellite cell differentiation; neuroprotective role in re-innervation of muscle endplates after acute nerve injury | Axis of HDAC4-FGFBP1 Pola1, BDNF, Cx43, Pax3/7, Fstl1, and Utrn; Notch3 and Igfbp5 | • SMA mouse, patient serum • Increased in both spinal cord and skeletal muscle of mouse; increased in patient serum |
Endogenously increased miR-206, with HDAC4 protein reduction and increased FGFBP1 mRNA, activates neuroprotective mechanism in muscle cells to increase re-innervation of muscle endplates | • Yes, in ALS animal model and patients • May be involved in myogenesis or neuroprotection in NMJ regeneration of ALS |
Bonanno et al.102, Catapano et al.104, Valsecchi et al.110 |
| miR-335-5p | Control of differentiation or self-renewal of mouse embryonic stem cells | MEST, OCT4, RB1 | • SMN∆7 mouse NSCs, human iPSCs • Decreased |
Possible epigenetic regulation through methylation to affect cell differentiation | None reported | Luchetti et al.107, Murdocca et al.111 |
| miR-375 | Neurogenesis and protects neurons from apoptosis in response to DNA damage | p53, PAX6, CCND2 | • Human neural progenitor cell cultures • Decreased |
MNs from an SMA patient have shown low miR-375, high p53 protein, and higher susceptibility to DNA damage-induced apoptosis | None reported but may share the same pathomechanism of p53-mediated apoptosis with ALS | Bhinge et al.112 |
| miR-431 | Regulation of MN axon neurite outgrowth | Chodl: a type 1 transmembrane protein and member of the c-type lectin domain-containing family | • Mouse MN culture, patient fibroblasts and iPSCs • Increased |
Increased miR-431 regulates MN neurite length by targeting chondrolectin involved in MN axon outgrowth | None reported | Wertz et al.113 |
ALS, amyotrophic lateral sclerosis; Chodl, chondrolectin; IGF1R, insulin-like growth factor 1 receptor; iPSCs, induced pluripotent stem cells; miRNA, microRNA; MN, motor neuron; MND, motor neuron diseases; NEFH, neurofilament heavy subunit; NMJ, neuromuscular junction; NSCs, neural stem cells; SMA, spinal muscular atrophy.
Roles of miRNAs in non-motor neuron tissues in SMA pathogenesis
Increasing evidence extends the pathogenic effect of SMN deficiency beyond MNs to include additional cells both within and outside the CNS, whereby numerous peripheral organs and tissues demonstrate pathological changes both in preclinical models and patients.17,18,114–116 Theoretically, these non-MN affections may be reflected by an altered level of circulating miRNAs. It remains unclear whether dysregulated miRNAs and its target are cell-context dependent, given that compromised ubiquitously expressing SMN may just affect miRNA homeostasis selectively in different tissues.64,113,117 However, emerging studies show that alteration of specific miRNAs in non-MN tissues of muscle, glial cells, and the neuromuscular junction (NMJ) might correlate with SMA severity, which implies a non-autonomous effect of miRNAs on SMA pathogenesis.103,110 For example, muscle-specific miRNAs (myomiRs) such as miR-1, miR-206, and miR-133 a/b, and astrocyte-produced miR-146a have been reported to be possibly implicated in SMA pathogenesis among different SMA models and human samples (Table 2).102,104,108,110 Notably, several therapies targeting the SMN-independent pathway (non-SMN enhancing) are undergoing clinical or preclinical trials, including neuroprotective agents, skeletal muscle enhancers, and NMJ facilitators. In the future, enthusiastic identification of both MN-intrinsic motomiRs and none-MN-intrinsic miRNAs, for example, myomiRs and their targets, might provide authentic biomarkers as novel therapeutic approaches for SMA.
MiRNA as circulating biomarkers for SMA
While advances in developing therapeutic approaches for SMA are cautiously optimistic, they also raise new challenges, particularly about identifying reliable outcome measures for preclinical and clinical trials.2,118 Reliable molecular biomarkers would not only help stratify SMA patients with heterogeneous phenotypes into homogeneous prognostic groups, but would also improve the statistical power of clinical trials, reduce trial durations and costs, and reveal therapeutic effects in specific types of SMA patients.53,95 Accordingly, researchers have been prompted to identify more authentic biomarkers of SMA to facilitate patient classification, follow disease progression, and better monitor responses to therapeutic approaches using minimally invasive procedures.
Two approved SMA therapies either use antisense oligonucleotides (ASO) or virus-mediated gene transfer exhibited promising outcomes.119,120 However, not all treated SMA patients responded equally well to these novel therapies. The only identified factor influencing treatment response is the time, called the therapeutic window, between the first recorded symptoms of SMA patients and the initial administration of the therapeutic agent.121 However, this sole factor would not seem to satisfactorily explain the wide variation in patient outcomes, even those receiving early treatment.122,123 Any potential outcome measures to reflect the treatment response of these two novel therapies remain to be tested.124 Answering this question is imperative, as early identification of good patient responders through multiple sensitive outcome measures can decrease the cost of innovative therapeutics.125,126
Until recently, the evaluation of therapeutic response for SMA relied primarily on clinical outcomes, including motor function, electrophysiological tests, and respiratory functions.119,120,128,129 However, fluctuating inter-rater or intra-rater variabilities of motor function measurement is problematic, and usually confounded by diverse care methods and age groups. As disease progression in SMA is typically slow, clinical outcome measures may lack sensitivity to detect significant motor function improvements in 1–2 years of follow up during a randomized clinical trial.130 It is conceivable that molecular biomarkers may represent more objective measures of treatment efficacy. Identification of a set of reliable circulating biomarkers from biofluids such as serum or cerebrospinal fluid (CSF) that can be correlated with the parameters of motor function, for example, the Hammersmith Functional Motor Scale – a commonly used clinical parameter in SMA trials – in SMA trials might help stratify the variability among patients.104,127,131
In the beginning, quantification of SMN mRNA or protein levels was the molecular biomarker applied most commonly in monitoring SMA patient therapeutic response. However, these do not necessarily correlate with disease severity and may not reliably reflect disease progression.132,133 Recently, plasma phosphorylated neurofilament heavy chain (pNF-H) has been proposed as a promising molecular biomarker to reflect disease activity or treatment response in children with type 1 SMA.134,135 pNF-H is expressed exclusively in neurons and released into extracellular fluids upon axon degeneration. However, although pNF-H might efficiently monitor neurodegeneration, there are still some limitations of its application: (1) as pNF-H is highly neuron-specific,136 it may not reflect the health of extra-MN tissues like muscles and NMJ, which could also play a role in therapeutic response in SMA patients; (2) finding increased released pNF-H relies on the slower disease progression in SMA patients other than type 1, which might interfere with the detection of changes in CSF composition in a short evaluation period131; (3) pNF-H levels, unfortunately, have not proven to be a reliable biomarker to correlate with motor function improvement in an elder group of adolescent and adult SMA type 2 and type 3 patients.137,138
Another potential candidate molecular biomarker for SMA is serum creatinine, which correlates with SMA type, SMN2 copy number, motor function, and denervation severity.139 However, creatinine production not only varies between individuals and over time in association with changes in muscle mass, liver function, and diet, but it has also been shown to undergo renal tubular secretion in addition to glomerular filtration.140 Moreover, two recent studies found liver damage in a mouse model of severe SMA,116 and kidney damage with renal tubular dysfunction in infants with type 1 SMA.141 In addition to decreased muscle mass, serum creatinine levels in SMA patients may also be affected by liver or renal tubular dysfunction, therefore reducing the reliability of using serum creatinine solely as an authentic SMA biomarker.
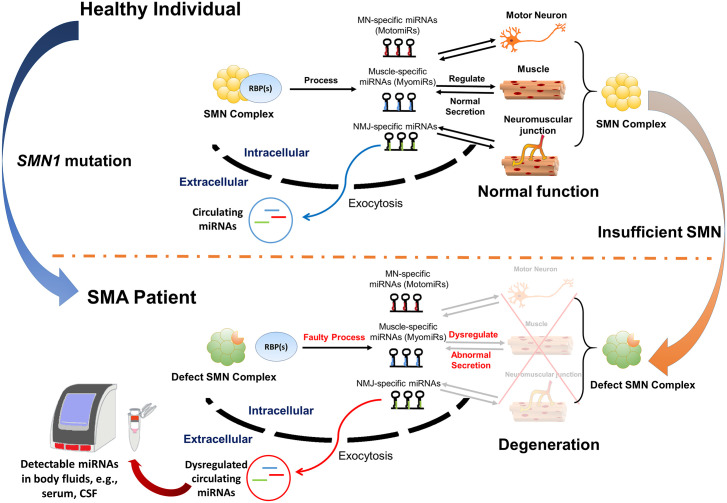
MiRNAs have gained attention as an easily accessible biomarker due to their features of being clinically detectable in many biofluids – such as CSF, serum/plasma, saliva, and urine – using non-invasive methods.95 MiRNAs can be secreted actively from a cell or by leaking through the membrane in response to various stimuli and insults, resulting in varying circulating miRNA levels in biofluids that are relatively stable, making the application of miRNAs biomarkers and therapeutic targets more attractive. Indeed, miRNAs are now being used as novel clinical biomarkers for the prognosis of several diseases, including various solid or liquid malignancies, cardiovascular diseases, and neurodegenerative disorders.30,142,143 In ALS patients, altered miRNA expression has been reported in human specimens of spinal cord, brain, induced pluripotent stem cells (iPSC), muscle, serum, and CSF.63,144 Recently, miRNAs have also been proposed as potential biomarkers in several clinical trials for neuromuscular disorders.145–148 As SMN is known to be involved in miRNA expression, and it entails circulating miRNAs as diagnostic or prognostic biomarkers to reflect the SMA pathology per se or therapeutic effects (Figure 3). Of importance, due to the versatile roles of miRNA in regulating generation/degeneration not only in MNs but in muscles and NMJ, researchers can detect level changes in different tissue-specific miRNAs simultaneously to monitor disease progression or the therapeutic response. Various tissue-specific miRNAs, such as miR-1, miR-9, miR-132, miR-206, miR-183, miR-375, and miR-1331a/b, have been investigated further as potentially reliable SMA biomarkers (Table 2).102,104,109,112 An early discernment of good patient responders through multiple sensitive outcome measures, including circulating miRNAs, may decrease the cost burden of innovative therapeutics.125,126 However, several critical challenges remain, such as unraveling inconsistencies due to human subject variability and technical issues related to the relative fragility of miRNAs.30,95
Figure 3.
Hypothetical model of miRNA-mediated biomarkers detection in SMA pathological background. In a healthy individual, the various tissue-specific miRNAs, including motomiRs, myomiRs, and NMJ-specific miRNAs (indicated in red, blue, and green, respectively) are normally produced by the functional SMN complex integrating with various RBP that involve miRNA biogenesis. These miRNAs may maintain a normal development/function of MN, muscles, NMJ, or reciprocally; these tissues may secrete relative miRNAs with normal expression profiles. However, in an SMA patient with SMN deficiency, dysregulated expression profiles of miRNAs may be caused by the failure of SMN–RBP interaction or degenerations of SMN-dependent tissues as MN, muscles, and NMJ. Theoretically, changes of miRNA expression profiles in the SMA pathological background might be detected through extracellular circulating miRNAs.
miRNA, microRNA; MN, motor neurons; NMJ, neuromuscular junctions; RBP, RNA-binding protein; SMA, spinal muscular atrophy; SMN, survival motor neuron.
Therapeutic potentials of miRNA for SMA
The ncRNAs may represent a unique therapeutic entry point for disease as it can regulate multiple signaling pathways rather than the classical one gene, one target approach. A recent study showed that treatment with ASO targeting SMN2 splicing is sufficient to restore miR-9, miR-132 and miR-206 levels in SMA mice.104 Besides, another study showed that knockdown of a neuron-enriched long ncRNA, SMN-AS1, results in both transcriptional activations of SMN promoter as well as modulation of SMN splicing by suppressing the epigenetic Polycomb repressive complex-2.149 Interestingly, combining therapeutic ASO targeting SMN-AS1 with SMN2 splice-switching oligonucleotides can synergically enhance SMN expression and ameliorate the phenotype of the SMA mice model. This evidence is of considerable interest since specific miRNAs can modulate the expression of several target genes. It also implied that miRNA intervention might supplement the effect of ASO therapy in SMA patients. Manipulating expression of target miRNA via ASO or decoy molecules, such as miRNA sponges, or overexpression of gain-of-function miRNA via viral vector delivery, may provide an alternative treatment strategy for SMA.
Delivering miR-196a via an adeno-associated virus (AAV) vector has been found to ameliorate phenotypes of the SBMA mice model, suggesting a promising strategy to apply a disease-specific miRNA for MND treatment.57 Notably, the miR-206–HDAC4–FGFBP1 signaling pathway has been proposed to maintain NMJ integrity and plasticity, which is also postulated to be the pathogenic mechanism occurring in SMA.110,150 In this context, it is intriguing that histone deacetylase inhibitors (HDACIs) have been proposed as a therapeutic candidate for SMA.151 The reinnervation property of miR-206 and its ability to repair NMJ following nerve injury need to be exploited in depth to develop potential therapeutic strategies. Excitingly, a very recent study showed that injection of a self-complementary AAV9 viral vector to reintroduce miR-23a into the Smn2B/– SMA mouse model could increase MN size, reduce NMJ pathology, and extend survival.106 The detailed mechanisms underlying how miR-23a-mediated target pathways lead to SMA pathology have yet to be characterized. However, these findings suggest that a specific cohort of miRNAs might cause MN vulnerability in SMA, and identification of those miRNA culprits and their targets could provide a new treatment strategy for SMA.
Last but not least, not a few challenges must be overcome, making it complex to use miRNA for therapeutic purposes. One of these issues is the administration route: these molecules can be delivered naked to target tissues. Considering the capability of molecules to cross the blood–brain barrier, intrathecal injection represents the most effective administration route to date, but is more invasive; therefore, systemic administration should also be considered. Furthermore, it must be considered that miRNAs often act through multiple pathways, which may pose a risk of off-target effects – a frequently considerable side effect of molecular therapy. According to their cellular developmental status, the timing of miRNA regulation must also be reckoned with, as miRNA levels should be manipulated at the correct development stage.
Conclusion
miRNAs play a crucial role in MN development, SMA pathogenesis and in determining selective MN vulnerability. The application of high-throughput RNA sequencing technologies, single-cell qRT-PCR, and proteomic approaches have allowed a comprehensive comparison of MNs generated via different strategies, providing additional insights into SMA pathogenesis. Furthermore, identification of potential SMA-associated miRNA and their corresponding chaperone RBPs may not only provide biomarkers monitoring disease progression but also represent potential therapeutic targets. However, significant challenges remain, for example, possible inconsistencies in results due to human subject variability and technical issues related to the relative fragility of miRNAs. Although recent progress in producing and understanding MNs has been remarkable, substantial challenges remain. The complex transcriptome networks regulated by miRNAs can be found in both health and disease. Recruiting a large cohort of SMA patients compared with healthy controls can increase the power of biomarker studies on miRNA and further promote advancement of this field.
Footnotes
Author contributions: T.-H. C contributed conceptualization, data curation, original draft preparation, review and editing to this work.
Funding: The author disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: this research was funded by the Ministry of Science and Technology (Taiwan), grant number MOST108-2314-B-037-073; Kaohsiung Medical University Hospital in Kaohsiung, Taiwan, grant number KMUH-108-8R47.
Conflict of interest statement: The author declare that there is no conflict of interest.
ORCID iD: Tai-Heng Chen  https://orcid.org/0000-0001-7713-3627
https://orcid.org/0000-0001-7713-3627
References
- 1. Darras BT. Spinal muscular atrophies. Pediatr Clin N Am 2015; 62: 743–766. [DOI] [PubMed] [Google Scholar]
- 2. Messina S. New directions for SMA therapy. J Clin Med 2018; 7: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farrar MA, Park SB, Vucic S, et al. Emerging therapies and challenges in spinal muscular atrophy. Ann Neurol 2017; 81: 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wirth B, Karakaya M, Kye MJ, et al. Twenty-five years of spinal muscular atrophy research: from phenotype to genotype to therapy, and what comes next. Annu Rev Genomics Hum Genet 2020; 21: 231–261. [DOI] [PubMed] [Google Scholar]
- 5. Donlin-Asp PG, Bassell GJ, Rossoll W. A role for the survival of motor neuron protein in mRNP assembly and transport. Curr Opin Neurobiol 2016; 39: 53–61. [DOI] [PubMed] [Google Scholar]
- 6. Chaytow H, Huang YT, Gillingwater TH, et al. The role of survival motor neuron protein (SMN) in protein homeostasis. Cell Mol Life Sci 2018; 75: 3877–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raimer AC, Gray KM, Matera AG. SMN - a chaperone for nuclear RNP social occasions? RNA Biol 2017; 14: 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singh RN, Howell MD, Ottesen EW, et al. Diverse role of survival motor neuron protein. Biochim Biophys Acta Gene Regul Mech 2017; 1860: 299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gruss OJ, Meduri R, Schilling M, et al. UsnRNP biogenesis: mechanisms and regulation. Chromosoma 2017; 126: 577–593. [DOI] [PubMed] [Google Scholar]
- 10. Murray LM, Beauvais A, Gibeault S, et al. Transcriptional profiling of differentially vulnerable motor neurons at pre-symptomatic stage in the Smn (2b/–) mouse model of spinal muscular atrophy. Acta Neuropathol Commun 2015; 3: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fallini C, Bassell GJ, Rossoll W. Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res 2012; 1462: 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gabanella F, Butchbach ME, Saieva L, et al. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One 2007; 2: e921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Workman E, Saieva L, Carrel TL, et al. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum Mol Genet 2009; 18: 2215–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Winkler C, Eggert C, Gradl D, et al. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev 2005; 19: 2320–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jodelka FM, Ebert AD, Duelli DM, et al. A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum Mol Genet 2010; 19: 4906–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tu WY, Simpson JE, Highley JR, et al. Spinal muscular atrophy: factors that modulate motor neurone vulnerability. Neurobiol Dis 2017; 102: 11–20. [DOI] [PubMed] [Google Scholar]
- 17. Nash LA, Burns JK, Chardon JW, et al. Spinal muscular atrophy: more than a disease of motor neurons? Curr Mol Med 2016; 16: 779–792. [DOI] [PubMed] [Google Scholar]
- 18. Simone C, Ramirez A, Bucchia M, et al. Is spinal muscular atrophy a disease of the motor neurons only: pathogenesis and therapeutic implications? Cell Mol Life Sci 2016; 73: 1003–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abati E, Citterio G, Bresolin N, et al. Glial cells involvement in spinal muscular atrophy: could SMA be a neuroinflammatory disease? Neurobiol Dis 2020; 140: 104870. [DOI] [PubMed] [Google Scholar]
- 20. Deguise MO, Kothary R. New insights into SMA pathogenesis: immune dysfunction and neuroinflammation. Ann Clin Transl Neurol 2017; 4: 522–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011; 25: 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu G, Mattick JS, Taft RJ. A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle 2013; 12: 2061–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 2014; 9: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 2016; 96: 1297–1325. [DOI] [PubMed] [Google Scholar]
- 26. Chen JA, Huang YP, Mazzoni EO, et al. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron 2011; 69: 721–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thiebes KP, Nam H, Cambronne XA, et al. miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1-Lhx3. Nat Commun 2015; 6: 7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Briggs JA, Wolvetang EJ, Mattick JS, et al. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron 2015; 88: 861–877. [DOI] [PubMed] [Google Scholar]
- 29. Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet 2009; 5: e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Basak I, Patil KS, Alves G, et al. microRNAs as neuroregulators, biomarkers and therapeutic agents in neurodegenerative diseases. Cell Mol Life Sci 2016; 73: 811–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 2019; 20: 5–20. [DOI] [PubMed] [Google Scholar]
- 32. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014; 15: 509–524. [DOI] [PubMed] [Google Scholar]
- 33. Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol 2010; 17: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hawley ZCE, Campos-Melo D, Droppelmann CA, et al. MotomiRs: miRNAs in motor neuron function and disease. Front Mol Neurosci 2017; 10: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bartel DP. Metazoan microRNAs. Cell 2018; 173: 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wolter JM, Le HH, Linse A, et al. Evolutionary patterns of metazoan microRNAs reveal targeting principles in the let-7 and miR-10 families. Genome Res 2017; 27: 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 38. Rajman M, Schratt G. MicroRNAs in neural development: from master regulators to fine-tuners. Development 2017; 144: 2310–2322. [DOI] [PubMed] [Google Scholar]
- 39. Chen T-H, Chen J-A. Multifaceted roles of microRNAs: from motor neuron generation in embryos to degeneration in spinal muscular atrophy. Elife 2019; 8: e50848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abernathy DG, Kim WK, McCoy MJ, et al. MicroRNAs induce a permissive chromatin environment that enables neuronal subtype-specific reprogramming of adult human fibroblasts. Cell Stem Cell 2017; 21: 332–348.e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tan L, Yu JT, Tan L. Causes and consequences of microRNA dysregulation in neurodegenerative diseases. Mol Neurobiol 2015; 51: 1249–1262. [DOI] [PubMed] [Google Scholar]
- 42. Lau P, de Strooper B. Dysregulated microRNAs in neurodegenerative disorders. Semin Cell Dev Biol 2010; 21: 768–773. [DOI] [PubMed] [Google Scholar]
- 43. Abelson JF, Kwan KY, O’Roak BJ, et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science 2005; 310: 317–320. [DOI] [PubMed] [Google Scholar]
- 44. Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues Clin Neurosci 2014; 16: 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Asli NS, Kessel M. Spatiotemporally restricted regulation of generic motor neuron programs by miR-196-mediated repression of Hoxb8. Dev Biol 2010; 344: 857–868. [DOI] [PubMed] [Google Scholar]
- 46. Amin ND, Bai G, Klug JR, et al. Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science 2015; 350: 1525–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li CJ, Hong T, Tung YT, et al. MicroRNA filters hox temporal transcription noise to confer boundary formation in the spinal cord. Nat Commun 2017; 8: 14685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoye ML, Koval ED, Wegener AJ, et al. MicroRNA profiling reveals marker of motor neuron disease in ALS models. J Neurosci 2017; 37: 5574–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen JA, Wichterle H. Apoptosis of limb innervating motor neurons and erosion of motor pool identity upon lineage specific dicer inactivation. Front Neurosci 2012; 6: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haramati S, Chapnik E, Sztainberg Y, et al. miRNA malfunction causes spinal motor neuron disease. Proc Natl Acad Sci U S A 2010; 107: 13111–13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tung YT, Peng KC, Chen YC, et al. Mir-17 approximately 92 confers motor neuron subtype differential resistance to ALS-associated degeneration. Cell Stem Cell 2019; 25: 193–209.e197. [DOI] [PubMed] [Google Scholar]
- 52. Tung YT, Lu YL, Peng KC, et al. Mir-17 approximately 92 governs motor neuron subtype survival by mediating nuclear PTEN. Cell Rep 2015; 11: 1305–1318. [DOI] [PubMed] [Google Scholar]
- 53. De Paola E, Verdile V, Paronetto MP. Dysregulation of microRNA metabolism in motor neuron diseases: novel biomarkers and potential therapeutics. Noncoding RNA Res 2019; 4: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamazaki T, Chen S, Yu Y, et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep 2012; 2: 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kye MJ, Goncalves ID. The role of miRNA in motor neuron disease. Front Cell Neurosci 2014; 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pegoraro V, Merico A, Angelini C. MyomiRNAs dysregulation in ALS rehabilitation. Brain Sci 2019; 9: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miyazaki Y, Adachi H, Katsuno M, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med 2012; 18: 1136–1141. [DOI] [PubMed] [Google Scholar]
- 58. Liu C-M, Wang R-Y, Saijilafu, et al. MicroRNA-138 and SIRT1 form a mutual negative feedback loop to regulate mammalian axon regeneration. Genes Dev 2013; 27: 1473–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu D, Murashov AK. MicroRNA-431 regulates axon regeneration in mature sensory neurons by targeting the Wnt antagonist Kremen1. Front Mol Neurosci 2013; 6: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Strickland IT, Richards L, Holmes FE, et al. Axotomy-induced miR-21 promotes axon growth in adult dorsal root ganglion neurons. PLoS One 2011; 6: e23423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eitan C, Hornstein E. Vulnerability of microRNA biogenesis in FTD-ALS. Brain Res 2016; 1647: 105–111. [DOI] [PubMed] [Google Scholar]
- 62. Emde A, Eitan C, Liou L-L, et al. Dysregulated miRNA biogenesis downstream of cellular stress and ALS-causing mutations: a new mechanism for ALS. EMBO J 2015; 34: 2633–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ravnik-Glavac M, Glavac D. Circulating RNAs as potential biomarkers in amyotrophic lateral sclerosis. Int J Mol Sci 2020; 21: 1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Magri F, Vanoli F, Corti S. miRNA in spinal muscular atrophy pathogenesis and therapy. J Cell Mol Med 2018; 22: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bertrandy S, Burlet P, Clermont O, et al. The RNA-binding properties of SMN: deletion analysis of the zebrafish orthologue defines domains conserved in evolution. Hum Mol Genet 1999; 8: 775–782. [DOI] [PubMed] [Google Scholar]
- 66. Ravanidis S, Kattan FG, Doxakis E. Unraveling the pathways to neuronal homeostasis and disease: mechanistic insights into the role of RNA-binding proteins and associated factors. Int J Mol Sci 2018; 19: 2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li DK, Tisdale S, Lotti F, et al. SMN control of RNP assembly: from posttranscriptional gene regulation to motor neuron disease. Semin Cell Dev Biol 2014; 32: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Barbee SA, Estes PS, Cziko AM, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 2006; 52: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dostie J, Mourelatos Z, Yang M, et al. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 2003; 9: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mourelatos Z, Dostie J, Paushkin S, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev 2002; 16: 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cauchi RJ. SMN and Gemins: ‘we are family’ … or are we?: insights into the partnership between Gemins and the spinal muscular atrophy disease protein SMN. Bioessays 2010; 32: 1077–1089. [DOI] [PubMed] [Google Scholar]
- 72. Piazzon N, Rage F, Schlotter F, et al. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J Biol Chem 2008; 283: 5598–5610. [DOI] [PubMed] [Google Scholar]
- 73. Tadesse H, Deschenes-Furry J, Boisvenue S, et al. KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum Mol Genet 2008; 17: 506–524. [DOI] [PubMed] [Google Scholar]
- 74. Sun S, Ling SC, Qiu J, et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat Commun 2015; 6: 6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA 2004; 10: 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fallini C, Rouanet JP, Donlin-Asp PG, et al. Dynamics of survival of motor neuron (SMN) protein interaction with the mRNA-binding protein IMP1 facilitates its trafficking into motor neuron axons. Dev Neurobiol 2014; 74: 319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Goncalves I, Brecht J, Thelen MP, et al. Neuronal activity regulates DROSHA via autophagy in spinal muscular atrophy. Sci Rep 2018; 8: 7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang L, Zhang L. Circulating microRNAs as diagnostic biomarkers for motor neuron disease. Front Neurosci 2020; 14: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang IF, Reddy NM, Shen CK. Higher order arrangement of the eukaryotic nuclear bodies. Proc Natl Acad Sci U S A 2002; 99: 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci U S A 2012; 109: 3347–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ederle H, Dormann D. TDP-43 and FUS en route from the nucleus to the cytoplasm. FEBS Lett 2017; 591: 1489–1507. [DOI] [PubMed] [Google Scholar]
- 82. Morlando M, Dini Modigliani S, Torrelli G, et al. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J 2012; 31: 4502–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jin P, Zarnescu DC, Ceman S, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci 2004; 7: 113–117. [DOI] [PubMed] [Google Scholar]
- 84. Edbauer D, Neilson JR, Foster KA, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 2010; 65: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Trabucchi M, Briata P, Garcia-Mayoral M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 2009; 459: 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Amirouche A, Tadesse H, Miura P, et al. Converging pathways involving microRNA-206 and the RNA-binding protein KSRP control post-transcriptionally utrophin A expression in skeletal muscle. Nucleic Acids Res 2014; 42: 3982–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fukao A, Mishima Y, Takizawa N, et al. MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol Cell 2014; 56: 79–89. [DOI] [PubMed] [Google Scholar]
- 88. Loffreda A, Rigamonti A, Barabino SM, et al. RNA-binding proteins in the regulation of miRNA activity: a focus on neuronal functions. Biomolecules 2015; 5: 2363–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sosanya NM, Huang PP, Cacheaux LP, et al. Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J Cell Biol 2013; 202: 53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hao le T, Duy PQ, An M, et al. HuD and the survival motor neuron protein interact in motoneurons and are essential for motoneuron development, function, and mRNA regulation. J Neurosci 2017; 37: 11559–11571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gardiner AS, Twiss JL, Perrone-Bizzozero NI. Competing interactions of RNA-binding proteins, microRNAs, and their targets control neuronal development and function. Biomolecules 2015; 5: 2903–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Degrauwe N, Suva ML, Janiszewska M, et al. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev 2016; 30: 2459–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Singh NN, Seo J, Ottesen EW, et al. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol Cell Biol 2011; 31: 935–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Vanderweyde T, Youmans K, Liu-Yesucevitz L, et al. Role of stress granules and RNA-binding proteins in neurodegeneration: a mini-review. Gerontology 2013; 59: 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Viswambharan V, Thanseem I, Vasu MM, et al. miRNAs as biomarkers of neurodegenerative disorders. Biomark Med 2017; 11: 151–167. [DOI] [PubMed] [Google Scholar]
- 96. Zou T, Yang X, Pan D, et al. SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress. Cell Mol Neurobiol 2011; 31: 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Volk N, Shomron N. Versatility of microRNA biogenesis. PLoS One 2011; 6: e19391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chang HC, Hung WC, Chuang YJ, et al. Degradation of survival motor neuron (SMN) protein is mediated via the ubiquitin/proteasome pathway. Neurochem Int 2004; 45: 1107–1112. [DOI] [PubMed] [Google Scholar]
- 99. Wishart TM, Mutsaers CA, Riessland M, et al. Dysregulation of ubiquitin homeostasis and β-catenin signaling promote spinal muscular atrophy. J Clin Invest 2014; 124: 1821–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Smrt RD, Szulwach KE, Pfeiffer RL, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 2010; 28: 1060–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kwon DY, Dimitriadi M, Terzic B, et al. The E3 ubiquitin ligase mind bomb 1 ubiquitinates and promotes the degradation of survival of motor neuron protein. Mol Biol Cell 2013; 24: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bonanno S, Marcuzzo S, Malacarne C, et al. Circulating myomiRs as potential biomarkers to monitor response to nusinersen in pediatric SMA patients. Biomedicines 2020; 8: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. O’Hern PJ, do Carmo G, Gonçalves I, Brecht J, et al. Decreased microRNA levels lead to deleterious increases in neuronal M2 muscarinic receptors in spinal muscular atrophy models. Elife 2017; 6: e20752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Catapano F, Zaharieva I, Scoto M, et al. Altered levels of microRNA-9, -206, and -132 in spinal muscular atrophy and their response to antisense oligonucleotide therapy. Mol Ther Nucleic Acids 2016; 5: e331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang LT, Chiou SS, Liao YM, et al. Survival of motor neuron protein downregulates miR-9 expression in patients with spinal muscular atrophy. Kaohsiung J Med Sci 2014; 30: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kaifer KA, Villalón E, O’Brien BS, et al. AAV9-mediated delivery of miR-23a reduces disease severity in Smn2B/-SMA model mice. Hum Mol Genet 2019; 28: 3199–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Luchetti A, Ciafre SA, Murdocca M, et al. A perturbed microRNA expression pattern characterizes embryonic neural stem cells derived from a severe mouse model of Spinal Muscular Atrophy (SMA). Int J Mol Sci 2015; 16: 18312–18327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sison SL, Patitucci TN, Seminary ER, et al. Astrocyte-produced miR-146a as a mediator of motor neuron loss in spinal muscular atrophy. Hum Mol Genet 2017; 26: 3409–3420. [DOI] [PubMed] [Google Scholar]
- 109. Kye MJ, Niederst ED, Wertz MH, et al. SMN regulates axonal local translation via miR-183/mTOR pathway. Hum Mol Genet 2014; 23: 6318–6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Valsecchi V, Boido M, De Amicis E, et al. Expression of muscle-specific miRNA 206 in the progression of disease in a murine SMA model. PLoS One 2015; 10: e0128560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Murdocca M, Ciafre SA, Spitalieri P, et al. SMA human iPSC-derived motor neurons show perturbed differentiation and reduced miR-335-5p expression. Int J Mol Sci 2016; 17: 1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bhinge A, Namboori SC, Bithell A, et al. MiR-375 is essential for human spinal motor neuron development and may be involved in motor neuron degeneration. Stem Cells 2016; 34: 124–134. [DOI] [PubMed] [Google Scholar]
- 113. Wertz MH, Winden K, Neveu P, et al. Cell type-specific miR-431 dysregulation in a motor neuron model of spinal muscular atrophy. Hum Mol Genet 2016; 25: 2168–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hua Y, Liu YH, Sahashi K, et al. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev 2015; 29: 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Somers E, Lees RD, Hoban K, et al. Vascular defects and spinal cord hypoxia in spinal muscular atrophy. Ann Neurol 2016; 79: 217–230. [DOI] [PubMed] [Google Scholar]
- 116. Szunyogova E, Zhou H, Maxwell GK, et al. Survival Motor Neuron (SMN) protein is required for normal mouse liver development. Sci Rep 2016; 6: 34635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Laird AS, Mackovski N, Rinkwitz S, et al. Tissue-specific models of spinal muscular atrophy confirm a critical role of SMN in motor neurons from embryonic to adult stages. Hum Mol Genet 2016; 25: 1728–1738. [DOI] [PubMed] [Google Scholar]
- 118. Chen T-H. New and developing therapies in spinal muscular atrophy: from genotype to phenotype to treatment and where do we stand? Int J Mol Sci 2020; 21: 3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mendell JR, Al-Zaidy S, Shell R, et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med 2017; 377: 1713–1722. [DOI] [PubMed] [Google Scholar]
- 120. Finkel RS, Mercuri E, Darras BT, et al. Nusinersen versus Sham control in infantile-onset spinal muscular atrophy. N Engl J Med 2017; 377: 1723–1732. [DOI] [PubMed] [Google Scholar]
- 121. Kariya S, Obis T, Garone C, et al. Requirement of enhanced survival motoneuron protein imposed during neuromuscular junction maturation. J Clin Invest 2014; 124: 785–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lowes LP, Alfano LN, Arnold WD, et al. Impact of age and motor function in a phase 1/2A study of infants with SMA type 1 receiving single-dose gene replacement therapy. Pediatr Neurol 2019; 98: 39–45. [DOI] [PubMed] [Google Scholar]
- 123. De Vivo DC, Bertini E, Swoboda KJ, et al. Nusinersen initiated in infants during the presymptomatic stage of spinal muscular atrophy: interim efficacy and safety results from the phase 2 NURTURE study. Neuromuscul Disord 2019; 29: 842–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Sumner CJ, Crawford TO. Two breakthrough gene-targeted treatments for spinal muscular atrophy: challenges remain. J Clin Invest 2018; 128: 3219–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Taga A, Maragakis NJ. Current and emerging ALS biomarkers: utility and potential in clinical trials. Expert Rev Neurother 2018; 18: 871–886. [DOI] [PubMed] [Google Scholar]
- 126. Kolb SJ, Coffey CS, Yankey JW, et al. Baseline results of the NeuroNEXT spinal muscular atrophy infant biomarker study. Ann Clin Transl Neurol 2016; 3: 132–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Gidaro T, Servais L. Nusinersen treatment of spinal muscular atrophy: current knowledge and existing gaps. Dev Med Child Neurol 2019; 61: 19–24. [DOI] [PubMed] [Google Scholar]
- 128. Mercuri E, Darras BT, Chiriboga CA, et al. Nusinersen versus Sham control in later-onset spinal muscular atrophy. N Engl J Med 2018; 378: 625–635. [DOI] [PubMed] [Google Scholar]
- 129. Finkel R, Bertini E, Muntoni F, et al. 209th ENMC international workshop: outcome measures and clinical trial readiness in spinal muscular atrophy 7-9 November 2014, Heemskerk, The Netherlands. Neuromuscul Disord 2015; 25: 593–602. [DOI] [PubMed] [Google Scholar]
- 130. Saffari A, Kolker S, Hoffmann GF, et al. Novel challenges in spinal muscular atrophy - how to screen and whom to treat? Ann Clin Transl Neurol 2019; 6: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kessler T, Latzer P, Schmid D, et al. Cerebrospinal fluid proteomic profiling in nusinersen-treated patients with spinal muscular atrophy. J Neurochem 2020; 153: 650–661. [DOI] [PubMed] [Google Scholar]
- 132. Sumner CJ, Kolb SJ, Harmison GG, et al. SMN mRNA and protein levels in peripheral blood: biomarkers for SMA clinical trials. Neurology 2006; 66: 1067–1073. [DOI] [PubMed] [Google Scholar]
- 133. Wadman RI, Stam M, Jansen MD, et al. A comparative study of SMN protein and mRNA in blood and fibroblasts in patients with spinal muscular atrophy and healthy controls. PLoS One 2016; 11: e0167087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Darras BT, Crawford TO, Finkel RS, et al. Neurofilament as a potential biomarker for spinal muscular atrophy. Ann Clin Transl Neurol 2019; 6: 932–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Olsson B, Alberg L, Cullen NC, et al. NFL is a marker of treatment response in children with SMA treated with nusinersen. J Neurol 2019; 266: 2129–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Yuan A, Rao MV, Veeranna, et al. Neurofilaments and neurofilament proteins in health and disease. Cold Spring Harb Perspect Biol 2017; 9: a018309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Totzeck A, Stolte B, Kizina K, et al. Neurofilament heavy chain and tau protein are not elevated in cerebrospinal fluid of adult patients with spinal muscular atrophy during loading with nusinersen. Int J Mol Sci 2019; 20: 5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Faravelli I, Meneri M, Saccomanno D, et al. Nusinersen treatment and cerebrospinal fluid neurofilaments: an explorative study on spinal muscular atrophy type 3 patients. J Cell Mol Med 2020; 24: 3034–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Alves CRR, Zhang R, Johnstone AJ, et al. Serum creatinine is a biomarker of progressive denervation in spinal muscular atrophy. Neurology 2020; 94: e921–e931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Sandilands EA, Dhaun N, Dear JW, et al. Measurement of renal function in patients with chronic kidney disease. Br J Clin Pharmacol 2013; 76: 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Nery FC, Siranosian JJ, Rosales I, et al. Impaired kidney structure and function in spinal muscular atrophy. Neurol Genet 2019; 5: e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Carter JV, Galbraith NJ, Yang D, et al. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer 2017; 116: 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Eyileten C, Wicik Z, De Rosa S, et al. MicroRNAs as diagnostic and prognostic biomarkers in ischemic stroke-a comprehensive review and bioinformatic analysis. Cells 2018; 7: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Joilin G, Leigh PN, Newbury SF, et al. An overview of microRNAs as biomarkers of ALS. Front Neurol 2019; 10: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Toivonen JM, Manzano R, Olivan S, et al. MicroRNA-206: a potential circulating biomarker candidate for amyotrophic lateral sclerosis. PLoS One 2014; 9: e89065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Alexander MS, Kunkel LM. Skeletal muscle microRNAs: their diagnostic and therapeutic potential in human muscle diseases. J Neuromuscul Dis 2015; 2: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Perry MM, Muntoni F. Noncoding RNAs and duchenne muscular dystrophy. Epigenomics 2016; 8: 1527–1537. [DOI] [PubMed] [Google Scholar]
- 148. Israeli D, Poupiot J, Amor F, et al. Circulating miRNAs are generic and versatile therapeutic monitoring biomarkers in muscular dystrophies. Sci Rep 2016; 6: 28097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. d’Ydewalle C, Ramos DM, Pyles NJ, et al. The antisense transcript SMN-AS1 regulates SMN expression and is a novel therapeutic target for spinal muscular atrophy. Neuron 2017; 93: 66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Williams AH, Valdez G, Moresi V, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 2009; 326: 1549–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Swoboda KJ, Scott CB, Reyna SP, et al. Phase II open label study of valproic acid in spinal muscular atrophy. PLoS One 2009; 4: e5268. [DOI] [PMC free article] [PubMed] [Google Scholar]