Significance
Much of Earth’s volcanism occurs in the deep sea, yet little is known about the microbial communities inhabiting such extreme and dynamic systems. Using a multidisciplinary approach to study distinct hydrothermal systems at Brothers submarine arc volcano, we provide insights into how microbial community composition and function reflect subtly different fluid chemistries resulting from subsurface fluid interactions with distinct alteration mineral assemblages. These variations can be traced to the subsurface hydrogeologic history beneath Brothers volcano. Further, we show that these systems represent oases of phylogenetically diverse Archaea and Bacteria. Our findings highlight the importance of geologic legacy in understanding drivers of microbial diversity, assembly, and evolution and may have insights into processes that drove early diversification of life on Earth.
Keywords: metagenomics, deep-sea hydrothermal, thermophiles, Archaea, volcanics
Abstract
Hydrothermally active submarine volcanoes are mineral-rich biological oases contributing significantly to chemical fluxes in the deep sea, yet little is known about the microbial communities inhabiting these systems. Here we investigate the diversity of microbial life in hydrothermal deposits and their metagenomics-inferred physiology in light of the geological history and resulting hydrothermal fluid paths in the subsurface of Brothers submarine volcano north of New Zealand on the southern Kermadec arc. From metagenome-assembled genomes we identified over 90 putative bacterial and archaeal genomic families and nearly 300 previously unknown genera, many potentially endemic to this submarine volcanic environment. While magmatically influenced hydrothermal systems on the volcanic resurgent cones of Brothers volcano harbor communities of thermoacidophiles and diverse members of the superphylum “DPANN,” two distinct communities are associated with the caldera wall, likely shaped by two different types of hydrothermal circulation. The communities whose phylogenetic diversity primarily aligns with that of the cone sites and magmatically influenced hydrothermal systems elsewhere are characterized predominately by anaerobic metabolisms. These populations are probably maintained by fluids with greater magmatic inputs that have interacted with different (deeper) previously altered mineral assemblages. However, proximal (a few meters distant) communities with gene-inferred aerobic, microaerophilic, and anaerobic metabolisms are likely supported by shallower seawater-dominated circulation. Furthermore, mixing of fluids from these two distinct hydrothermal circulation systems may have an underlying imprint on the high microbial phylogenomic diversity. Collectively our results highlight the importance of considering geologic evolution and history of subsurface processes in studying microbial colonization and community dynamics in volcanic environments.
Submarine volcanoes account for ∼75% of global volcanic activity (1), contributing significantly to ocean biological productivity (2) and mineral resources (3). The Kermadec portion of the Kermadec–Tonga intraoceanic volcanic arc has more than 30 major volcanoes, of which 80% are hydrothermally active, making it the most active intraoceanic arc in the world (4). Hydrothermal activity associated with these arc volcanoes is commonly dominated by the discharge of magmatic volatiles, in contrast to midocean ridge vent systems, which are dominated by seawater circulation through oceanic crust. Brothers volcano on the Kermadec arc is unusual in that it hosts both types of hydrothermal systems. At the Upper Cone (UC) and Lower Cone (LC) sites inside the caldera (Fig. 1), relatively low-temperature (<120 °C), highly acidic (pH to 1.9) acid-sulfate fluids derived from disproportionation of magmatic sulfur gases (SO2 and H2S) discharge from native sulfur mounds, and extensive Fe oxyhydroxide crusts are common. By contrast, only ∼3 km away, high-temperature (≤320 °C), less acidic (pH >2.8), but metal-rich fluids are expelled from ≤20-m-tall chimneys composed of significant Cu-Zn-Ba ± Au mineralization at the Northwest Caldera Wall (NWC) and Upper Caldera Wall (UCW) sites (5–8) (Fig. 1).
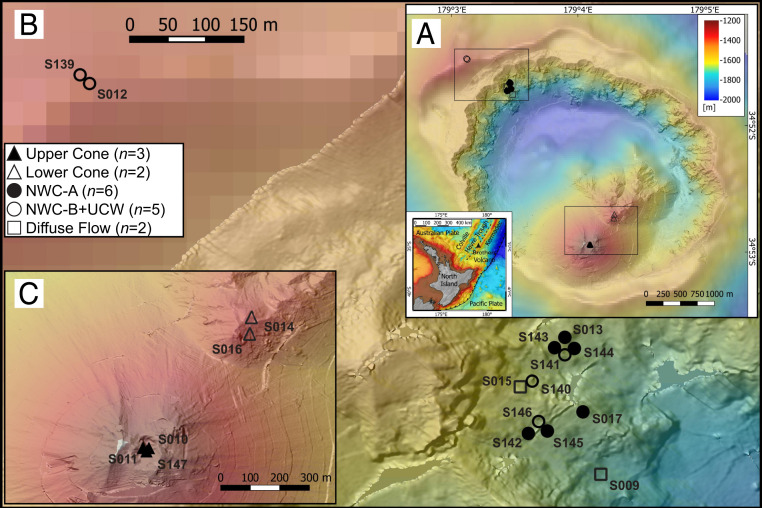
Fig. 1.
Location of hydrothermal vent sampling sites from Brothers volcano. (A) Overall bathymetric map of the volcano. (Inset) Location north of New Zealand. (B) Detail of NWC and UCW sites. (C) Detail of UC and LC sites. Total samples per area are indicated with n values.
Changes in the hydrothermal fluid regime at Brothers volcano have been linked to the evolution of the caldera (9). In the precaldera stage, the volcano hosted a hydrothermal system dominated by magmatic volatiles and metal-rich brines. The magmatic volatiles mixed with seawater as they ascended toward the seafloor to produce low-pH, vapor-rich, metal-poor fluids, very much like those discharging at the Cone sites today. The metal-rich brines were segregated and temporarily sequestered in the subsurface. As the volcano grew through the eruption of thick layers of volcaniclastics, caldera collapse eventually occurred, allowing seawater to infiltrate the volcano through fault-controlled permeability. This seawater interacted with wallrock and the segregated brines and transported the associated metals to the seafloor to discharge high-temperature, moderately acidic, metal-rich fluids, forming Cu-Zn-Au–rich chimneys on the NWC and UCW. The recent International Ocean Discovery Program Expedition 376 on the NWC (9, 10) showed that, while the shallower parts of the volcanic sequence are characterized by an alteration mineral assemblage indicative of wallrock reactions with seawater, deeper (underlying) parts of the stratigraphy record evidence of earlier alteration reactions with the magmatically influenced hydrothermal fluids (9). After caldera collapse, a larger resurgent cone (UC) developed followed by a more recent but smaller subsidiary cone (LC) that today both host modern magmatically influenced hydrothermal systems (6, 7).
While several studies have analyzed the microbial diversity of Brothers volcano (11, 12), the impact of both its complex geological evolution and the existence of two proximal but distinct hydrothermal systems on the microbial community diversity is unknown. Studies of terrestrial hydrothermal environments have shown that the geological history of a system impacts microbial community assembly (13) and can be the underlying cause for biogeographically distinct microbial populations (14). Likewise, at deep-sea hydrothermal vent systems, differences in subsurface hydrothermal processes produce fluids of contrasting chemistry that result in different microbial communities associated with the diffuse venting fluids (15, 16) and mineral deposits (17, 18) that develop as the hydrothermal fluids mix with cold seawater. Here, we use amplicon and metagenome sequencing to show that Brothers volcano supports at least three distinct and phylogenetically diverse microbial communities that reflect the complex subsurface hydrogeology of the system.
Results and Discussion
Microbial Community Diversity of Brothers Volcano Hydrothermal Deposits Reveals Two Distinct Communities on the Caldera Wall.
To explore the patterns of microbial diversity associated with hydrothermal deposits from the geologically and geochemically different areas at Brothers volcano, we collected hydrothermal deposits from 16 actively discharging hydrothermal vents and two diffuse flow samples from the UCW, NWC, LC, and UC sites (Fig. 1, Dataset S1A, and SI Appendix, Fig. S1). Wherever possible, hydrothermal fluids from vent orifices were collected to examine possible geochemical drivers of differences in microbial community assemblies (Dataset S1B).
Similar to previous analyses of Brothers vent fluids (6, 7), fluids from the NWC and UCW feature chemistry indicative of seawater and rock interactions at high temperatures (∼300 °C). These vent fluids reveal subcritical phase separation with chloride values above and below background seawater concentrations (540 mmol/kg), with most being greater than seawater. Magnesium and sulfate have been removed from the end-member fluids, and potassium, calcium, trace metals, and sulfide are all elevated (Dataset S1B). In contrast, samples from the cone sites do not show phase separation and indicate a greater degree of magmatic degassing (as compared to water–rock interactions) with large releases of CO2, H2, and shallow mixing of gases and seawater. While both cones have vent field systems primarily showing magmatic degassing, fluids sampled in our study at the UC were hotter at 200 °C than previously recorded (7), compared to the 45 to 60 °C fluids from the LC. The pH in the UC samples was pH 2.1 ± 0.1 and higher in the LC samples (pH 5.4 ± 0.3).
Initially we assessed the microbial community diversity and composition associated with the deposits (and one fluid sample) from the different Brothers volcano sites using 16S ribosomal RNA (rRNA) gene amplicon sequencing (Dataset S1C). The taxonomic composition associated with the magmatically influenced cone sites differed from that of the NWC and UCW sites. Furthermore, there also appeared to be two distinct communities associated with the different caldera wall sites, NWC-A, and NWC-B+UCW (Fig. 2A; NWC-A versus NWC-B+UCW, analysis of similarities [ANOSIM] R = 0.896, P = 0.002). Several of these different communities were almost adjacent sites, for example S145 and S146 (Fig. 1). Despite their close proximity, the microbial community diversity of the NWC-A samples clustered somewhat with samples from the cone sites, while those from NWC-B+UCW formed a unique cluster (Fig. 2A).
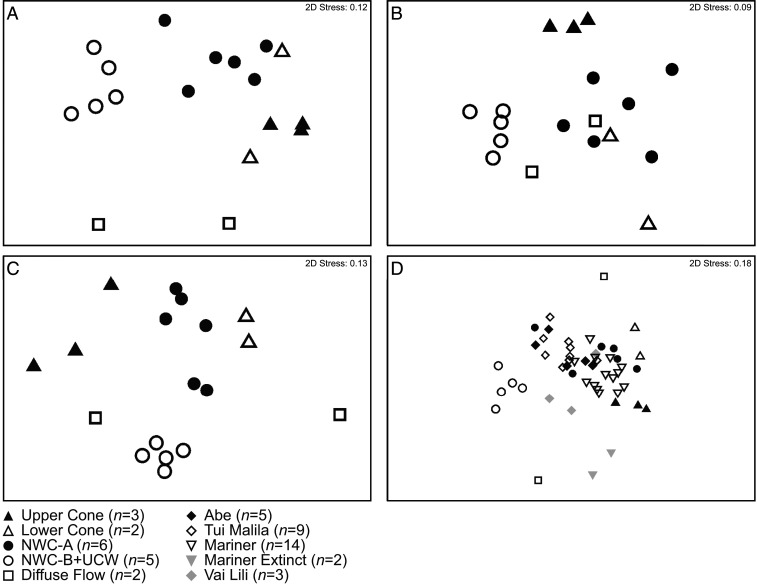
Fig. 2.
Nonmetric multidimensional scaling (NDMS) plots of (A) Brothers volcano community composition, (B) functional gene diversity, (C) MAG phylogenetic diversity, and (D) Brothers volcano and ELSC-VFR deep-sea hydrothermal vent community diversity. Plots are based on Bray–Curtis matrices of (A) community composition of Brothers volcano samples constructed from amplicon sequence variants (ASVs) of 16S rRNA gene sequences at a depth of 3,000 sequences per sample; (B) relative abundance of select functional genes and gene categories described in the text (n = 123 genes/gene categories) and shown in Dataset S2B; (C) relative abundance of medium- to high-quality MAGs based on normalized read coverage of taxa assigned by GTDB-Tk (n = 251 taxonomic assignations). Relative abundance data are available in Dataset S3D; and (D) community composition of Brothers volcano and ELSC-VFR hydrothermal vent samples based on ASVs of 16S rRNA gene sequences at a depth of 3,000 sequences per sample. Points closer together on the ordination plot are more similar to each other. The number of samples at each area is indicated as an n value.
Based on similarity percentages breakdown (SIMPER) analysis (Dataset S1D), the Epsilonbacteraeota and Deltaproteobacteria* (in NWC-A) and the Gammaproteobacteria, Aquificae, Aigarchaeota/Geothermarchaeota group (SILVA nomenclature Aigarchaeales) and Hydrothermarchaeota (in NWC-B+UCW) are some of the taxa that contribute to the compositional differences seen between these distinct Caldera Wall communities. The shared Epsilonbacteraeota between the cone sites and NWC-A drive some of the similarity in composition between these communities. Thermoacidophiles, like members of Thermoplasmata, are also shared between the cone sites and NWC-A. In addition, members of the Candidate Phyla Radiation (CPR) and the Diapherotrites, Aenigmarchaeota, Nanoarchaeota, Nanohaloarchaeota (DPANN) Archaea (19) contribute to differences in community structure between the UC and LC and the other sites.
Geochemical Energy Calculations Confirm Importance of Sulfide Oxidation at All Sites.
Fluid chemistry could be one potential factor driving the above observed community structure differences. The notable differences between the distribution of the Epsilonbacteraeota and the Gammaproteobacteria at the Brothers sites suggest that these taxa may be examples of ecological indicators of the environmental conditions driving the microbial composition shifts. In general, Epsilonbacteraeota occupy niches of lower oxygen and higher sulfide concentrations, while Gammaproteobacteria thrive in higher oxygen and lower sulfide levels (20). Unfortunately, due to insufficient geochemical data for the fluids associated with the samples, especially from NWC-A (only one dilute sample for S142) and NWC-B+UCW (four samples; Dataset S1B), we could not determine a statistically relevant correlation between end-member fluid chemistry and the observed microbial community structure. Further, it is possible that the observed differences in microbial community composition on adjacent sites on the NWC are due to adaptation to subtlety different chemistries (not detectable through bulk chemistry measurements) as a result of fluids following different pathways and intersecting different subsurface rock assemblages. Nonetheless, catabolic energy availability calculations for autotrophs of measured vent fluids upon mixing with seawater confirmed that sulfide oxidation was likely the most favorable energy-yielding metabolism at all sites examined, with iron oxidation likely representing an additional important potential process at the caldera wall sites (21). Furthermore, modeling the fluid geochemistry from NWC-B samples suggested that hydrogen oxidation may provide an additional source of energy for these communities.
Functional Metagenomic Diversity Tracks Microbial Community Diversity.
These thermodynamic calculations provided initial information about the potential energy availability for autotrophy in the Brothers volcano communities. However, natural microbial communities are functionally more complex and usually represent a continuum of other processes in addition to autotrophy, such as syntrophy, symbiosis, and heterotrophy, all contributing to the overall microbial community composition. To determine to what extent differences in community composition (NWC-A, NWC-B+UCW, UC, and LC) resemble differences in functional complexity, we sequenced the metagenomes of all 18 samples (Dataset S2A) and identified genes coding for proteins involved in energy conservation, mediating key steps of biogeochemical cycles, and indicative of oxygen sensitivity (Fig. 3 A and B, Dataset S2B, and SI Appendix, Fig. S2 A–F), including the functional processes explored through thermodynamic modeling.
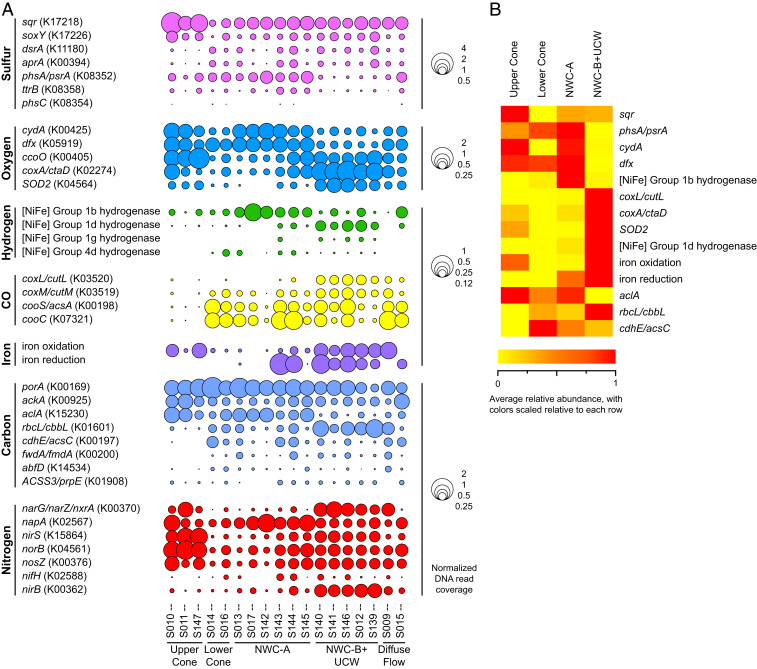
Fig. 3.
Relative normalized abundance of metabolic and functional genes involved in energy generation, oxygen tolerance, and biogeochemical cycling from Brothers volcano metagenome assemblies, including those discussed in the text. (A) Bubbles represent single genes or iron metabolism gene categories listed in Dataset S2B. Bubble size indicates normalized abundance, calculated by dividing the summed coverage for each gene/gene category in an assembly by the average summed coverage for 14 single-copy marker genes. Full gene names are shown in Dataset S2B. (B) Heat map shows the average normalized abundance of genes/gene categories discussed in the text. Normalized abundance was determined as described above and subsequently averaged across all assemblies at each particular area (UC, n = 3 assemblies; LC, n = 2 assemblies; NWC-A, n = 6 assemblies; NWC-B+UCW, n = 5 assemblies). Coloring is scaled relative to each row, with red indicating the highest average relative abundance of the four areas and yellow indicating the lowest average relative abundance. Normalized abundance of genes and gene categories and full gene names are shown in Dataset S2B.
The level of similarity based on the normalized abundance patterns of 123 functional genes/gene categories in the assemblies (Fig. 2B) followed a pattern similar to that of the amplicon diversity (Fig. 2A), with NWC-A affiliating more with the cone sites and NWC-B+UCW forming an independent cluster (NWC-A versus NWC-B+UCW, ANOSIM R = 0.872, P = 0.002). While some of the functional genes confirm the thermodynamic model predictions, namely the importance of sulfide oxidation (sulfide-quinone oxidoreductase, sqr; SI Appendix, Fig. S2A) at all sites, other genes provide insights into potential functional differences that may be driving the differences in community diversity between sites (Fig. 3 A and B and SI Appendix, Fig. S2 A–F). For example, genes associated with metabolic processes in low-oxygen or anoxic environments (22, 23) are all in higher relative abundance in the NWC-A samples, including the bd-type terminal oxidase (cydA), superoxide reductase (dfx), and the [NiFe] Group 1b hydrogenase, which couples hydrogen oxidation to sulfate, fumarate, or nitrate reduction in certain anaerobes (24) (SI Appendix, Fig. S2 B and C). Additionally, NWC-A communities have a lower relative abundance of the microaerophile-associated [NiFe] Group 1d hydrogenase gene (24) and the putative aerobic-type xanthine/carbon monoxide dehydrogenase family (coxL) and superoxide dismutase (SOD2) (Fig. 2 B–D). By contrast, the NWC-B+UCW communities appear to be functionally more diverse, possessing a range of genes related to growth under anaerobic, microaerophilic, and aerobic conditions. This suggests that these communities occupy more complex geochemical gradients, or that geothermal fluid inputs are mixed or temporally variable. Furthermore, iron oxidation and iron reduction potential appear to be variable across all sites (SI Appendix, Fig. S2D). While thermodynamic models suggest that sulfide oxidation is likely the dominant energy-conserving process in the UC and LC communities, genes encoding proteins involved in iron oxidation (based on FeGenie, candidate iron oxidase cyc2) were also detected, suggesting that iron oxidation (of iron oxides) may also support some populations in these communities.
While the NWC-A community members have a greater potential for anaerobic growth, perhaps as a result of more magmatic inputs in the fluids that support these communities, and the mixing with oxygenated seawater enriches for communities with both anaerobic and aerobic metabolisms in NWC-B+UCW communities, we did not identify significant shifts in carbon metabolism pathways between the sites (SI Appendix, Fig. S2E), although there were some subtle differences. Common to most samples were genes for pathways such as the anaerobic reductive acetyl-CoA/Wood–Ljungdahl pathway (cdhABCDE, and fmd/fwdABCDEF, fwdG in Archaea) and fermentation (e.g., acs and porA). However, the marker gene for the typically anaerobic/microaerophilic reductive tricarboxylic acid (rTCA) cycle (aclA) was further enriched at the UC and NWC-A sites and the gene for the aerotolerant Calvin–Benson–Bassham (CBB) cycle (rbcL/cbbL) was relatively higher in abundance at the NWC-B+UCW. The shifts in aclA and rbcL/cbbL may mirror the differences in relative abundance of Epsilonbacteraeota and Gammaproteobacteria at these sites, respectively. The Epsilonbacteraeota are typically prevalent in lower-oxygen/higher-sulfide environments where the rTCA cycle for carbon fixation is advantageous (25), while the Gammaproteobacteria are typically found in higher-oxygen/lower-sulfide environments (20) and use the CBB cycle for carbon fixation (25). Genes for the dicarboxylate/4-hydroxybutyrate and 3-hydroxypropionate/4-hydroxybutyrate cycles (abfD) and the 3-hydroxypropionate bicycle (prpE) were also detected in low relative abundance.
Which Taxa Contribute to the Observed Functional Differences between Sites?
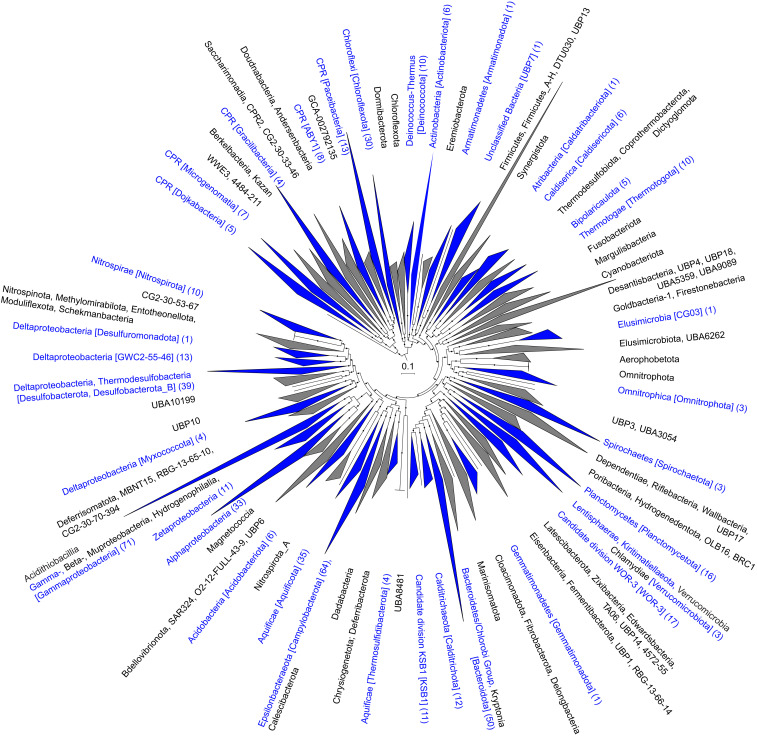
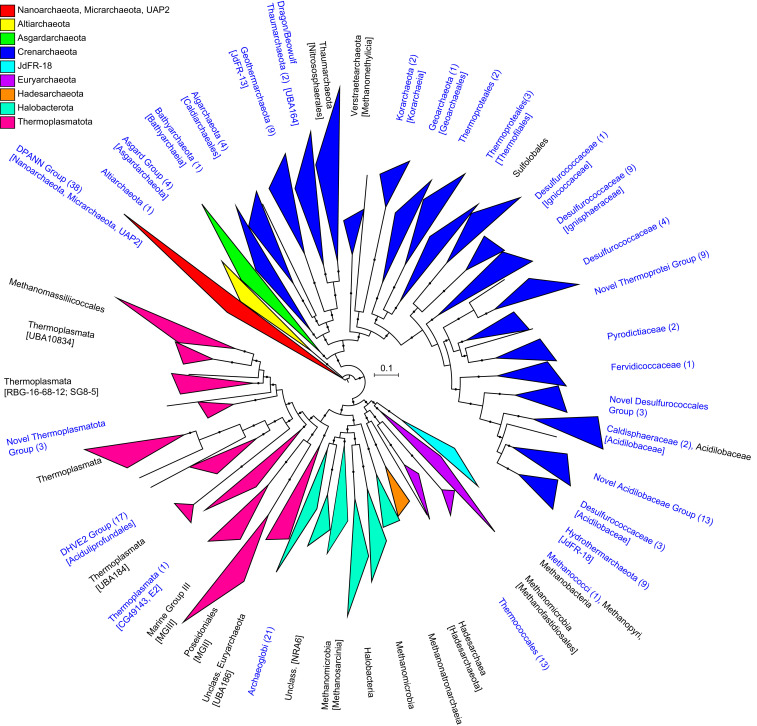
In order to explore further which taxa were most likely contributing to the functional differences between the different Brothers volcano sites, we used the metagenome assemblies to reconstruct 701 medium- to high-quality draft genomes (MAGs) according to recently established standards (26) (Dataset S3A and SI Appendix, Fig. S3). Of these, 517 MAGs belonged to the Bacteria and represented about 65 new families and 202 new genera without cultured representatives based on GTDB-Tk (27) and average amino acid identity (AAI) analyses (Fig. 4, Dataset S3 B and C, and see Materials and Methods and SI Appendix, SI Text and SI Methods), and the 184 archaeal MAGs represented about 27 new families and 83 new potential genera (Fig. 5, Dataset S3 B and C, and SI Appendix, Fig. S4).
Fig. 4.
Maximum-likelihood phylogenetic reconstruction of bacterial metagenome-assembled genomes from Brothers volcano using GTDB-Tk. The tree was constructed using 120 bacterial marker genes. Branch support was determined with the SH test, and support values from 0.8 to 1.0 are indicated with filled circles. Collapsed taxonomic clades are shown as triangles, with blue triangles indicating clades containing Brothers volcano MAGs with the number of MAGs shown in parentheses. Clades with Brothers MAG are labeled using NCBI taxonomy followed by GTDB-Tk taxonomy in brackets, while gray clades without Brothers MAGs are shown with GTDB-Tk taxonomy only. Due to the extensive reordering of the Tree of Life within GTDB-Tk taxonomy, the NCBI taxonomy is approximate and represents large-scale taxa equivalencies. The scale bar indicates expected amino acid substitutions per site. The uncollapsed phylogenetic tree used to create this figure is available online (28) at https://itol.embl.de/shared/alrlab and on FigShare in phyloXML format (21) at https://doi.org/10.6084/m9.figshare.c.5099348.
Fig. 5.
Maximum-likelihood phylogenetic tree of archaeal MAGs from Brother volcano, constructed in GTDB-Tk using 122 archaeal marker genes. SH branch support (0.8 to 1.0) is shown with dark circles. Major clades are collapsed into triangles, and triangle coloring is based on GTDB-Tk phylum-level taxonomic classifications. Individual clades are labeled using NCBI taxonomy with GTDB-Tk nomenclature shown in brackets, and novel clades are named using GTDB-Tk taxonomy. Taxonomic groups containing Brothers volcano MAGs are indicated by blue text, with the number of MAGs shown in parentheses. The scale bar shows expected substitutions per amino acid. The uncollapsed tree used to create this figure is available as SI Appendix, Fig. S4 and at iTOL (29) (https://itol.embl.de/shared/alrlab).
While MAG diversity (National Center for Biotechnology Information [NCBI] and Genome Taxonomy Database [GTDB] nomenclature in Dataset S3B) might not necessarily be a proxy for whole-community composition (30), in our study similar diversity patterns were observed for MAG and 16S rRNA gene amplicon data (SI Appendix, Fig. S5 A and B); the functional data also tracked these patterns (Fig. 2 A–C). When MAG diversity differences were explored statistically, the analysis further confirmed that the MAGs obtained from NWC-A samples were significantly different from those found at NWC-B+UCW (NWC-A versus NWC-B+UCW, ANOSIM R = 0.931, P = 0.002; Fig. 2C). Putative anaerobes related to the genera Archaeoglobus, Thermococcus, Hippea, Nitratifractor, and Mesoaciditoga and the family Caldisericaceae were prevalent MAGs from the NWC-A samples, while putative microaerophilic Aquificae and Thermoprotei (Aeropyrum-related) and putative carboxydotrophic Hydrothermarchaeota MAGs characterized the NWC-B+UCW samples (Dataset S3D). Furthermore, from the LC site samples in particular a diversity of previously undescribed members of the DPANN superphylum were assembled (Datasets S3E and S4 and SI Appendix, Figs. S5C and S6 and SI Text). One DPANN lineage common to several sites was the Woesearchaeaota. We successfully enriched for a novel woesesarchaeote with only Bacteria (Thermales and Aquificales) in the coculture (Dataset S5A and SI Appendix, SI Text). The reduced metabolic repertoire of the assembled metagenome of this woesearchaeote suggests that it forms a symbiotic association with bacterial hosts (Dataset S5B).
Several of the MAGs most likely contributed to the functional differences described above (data available at FigShare (21). For example, 143 (67%) of NWC-A MAGs, including members of the Epsilonbacteraeota, Thermodesulfobacteria, Archaeoglobi, Thermoprotei, Thermococci, Thermoplasmata, and several DPANN, all encoded for superoxide reductase (dfx), which is often prevalent in anaerobes (22), while very few (n = 40, or 19%) MAGs encoded the superoxide dismutase (SOD2) typically found in aerobes. By contrast, over 200 (59%) of the MAGs from NWC-B+UCW encoded for SOD2. Likewise, the genes for terminal oxidases also pointed to the enriched patterns seen in the assemblies, where the high-oxygen-adapted, low-affinity aa3-type heme-copper oxidase (coxA) (23) was detected in 56% of the NWC-B+UCW MAGs but only in 24% of the NWC-A MAGs. Conversely, the low-oxygen-adapted, high-affinity bd-type terminal oxidase (cydA) (23) was identified in 43% of the NWC-A MAGs but only in 26% of the NWC-B+UCW MAGs. The coxA distribution in the NWC-B+UCW assembled genomes was associated with the Gammaproteobacteria, Thermoprotei, and Aquificae and to a lesser extent with Heimdallarchaeota, Aigarchaeota, Geothermarchaeota, and Hydrothermarchaeota. The cydA genes from NWC-A were mostly associated with members of the Epsilonbacteraeota (such as Desulfurellales), Deltaproteobacteria/Thermodesulfobacteria and, to a lesser degree, with the Thermococci. Furthermore, the gene encoding for the putative large subunit of the aerobic-type xanthine/carbon monoxide dehydrogenase family (coxL) was only present in 4% of NWC-A MAGs, while it was detected in 18% of NWC-B+UCW MAGs (including MAGs affiliated with members of the Alphaproteobacteria, Chloroflexi, Calditrichaeota, Thermoprotei, and members of the candidate division KSB1). The differences between the [NiFe] Group 1b (oxygen-sensitive) and [NiFe] Group 1d hydrogenases (oxygen-tolerant) from the NWC-A and NWC-B+UCW tracked with anaerobic Epsilonbacteraeota in NWC-A ([NiFe] Group 1b hydrogenases, 7% of MAGs) and microaerophilic Aquificae and the candidate division KSB1 in NWC-B+UCW ([NiFe] Group 1d hydrogenases, 13% of MAGs). Further, the enrichment in CO2 fixation pathways, namely the rTCA cycle and the CBB pathway, could be linked to Epsilonbacteraeota MAGs in NWC-A and the prevalence of Gammaproteobacteria MAGs in NWC-B+UCW, respectively.
Hydrothermal Magmatic Inputs Drive Similarities in Microbial Composition at Brothers Volcano and the Backarc Mariner Vent Field.
Collectively, the amplicon, MAG, and functional data point to the NWC-A sites supporting mostly anaerobic communities, while those associated with the NWC-B+UCW sites are more mixed with respect to oxygen usage potential (anaerobic, microaerophilic, and aerobic). Given that the microbial communities at the cone sites track more closely with the NWC-A communities, we hypothesized that the NWC-A community and functional diversity may be influenced by hydrothermal fluids with mostly magmatic inputs, while those of NWC-B+UCW have a stronger oxygenated seawater (due to mixing) signature. Recent drilling on the wall of the caldera has documented the influence of both types of hydrothermal fluids through the presence of mineral assemblages indicative of rock reactions with acid-sulfate-type fluids intercalated and overprinted by assemblages resulting from reactions with seawater-dominated fluids (9).
In order to explore this further, we tested whether NWC-A microbial communities would be more similar in structure to microbial communities from other magmatically influenced deep-sea vents, such as those associated with the Mariner vent field on the Eastern Lau Spreading Center–Valu Fa Ridge (ELSC-VFR) (17). Here, the deep-sea vent fields (ABE, Tui Malila, Mariner, and Vai Lili) run north to south along a basalt to andesite gradient (31). Chemical species such as H2S, Fe, and Mn decrease southward whereas pH and alkalinity increase, with the exception of the southern Mariner vent field, where fluids are significantly hotter, more acidic and metal-rich, and richer in dissolved gases such as H2S and CO2 relative to the other vent fields, consistent with the greater input of magmatic gases (32). Indeed, when comparing our Brothers volcano amplicon diversity data with that from ELSC-VFR, the NWC-A communities are more similar to those from Mariner (Mariner versus NWC-A, ANOSIM R = 0.414, P = 0.003), while the NWC-B+UCW communities are unique to Brothers volcano (NWC-A versus NWC-B+UCW, ANOSIM R = 0.896, P = 0.002; Fig. 2D). Thus, it is likely that the differences in the microbial communities on the caldera wall reflect subtle differences (sometimes undetectable by bulk geochemical measurements) in temporal fluxes of fluid mixing and fluid paths in the subsurface (Fig. 6), much like differences observed in the Yellowstone caldera where acid and alkaline hot springs can exit adjacent to each other (33, 34).
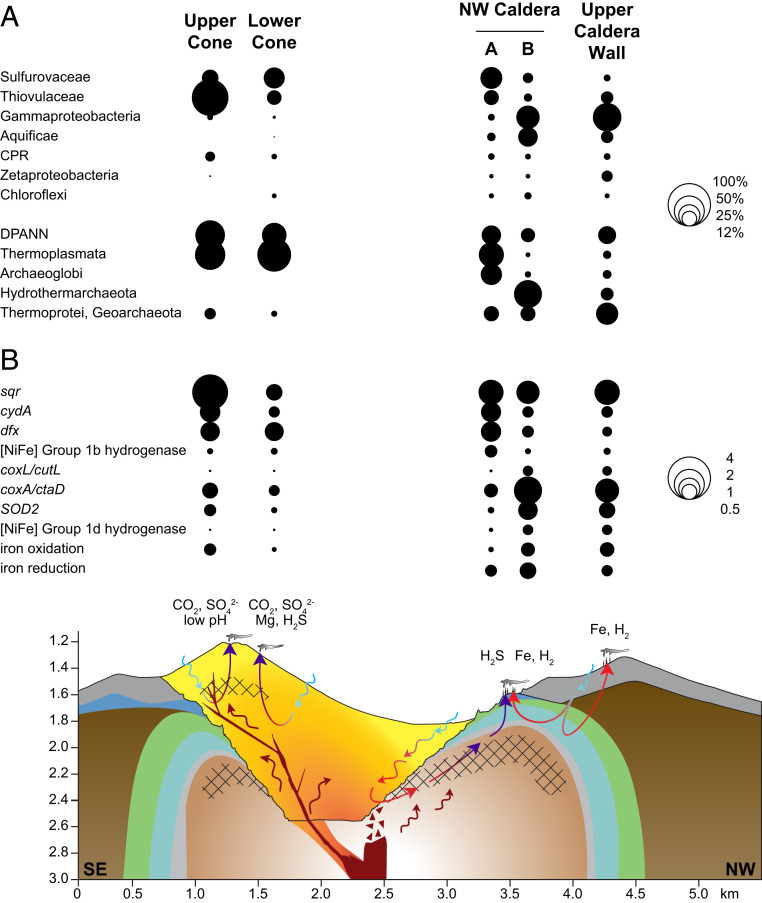
Fig. 6.
The proposed influence of subsurface hydrogeological processes on the microbial phylogenetic and functional diversity associated with Brothers volcano and its hydrothermal systems. Schematic cross-section adapted from de Ronde et al. (9) from southeast to northwest. Overall, the shallow subsurface is influenced by seawater-dominated hydrothermal circulation (blue arrows = recharge; red arrows = discharge). Recharge of seawater occurs both at a volcano scale through the caldera floor and faults along the caldera wall and more locally at the 100- to 200-m scale at the vent sites [see Caratori Tontini et al. (35)]. Red arrows denote heated (modified) seawater after reaction with the rocks; purple arrows represent hot, magmatically influenced fluids; red-brown arrows represent magmatic volatiles. Cross-hatching represents zones of inferred magmatic salt [see de Ronde et al. (9)]. (A) Bubble plots of the average relative abundance of key taxa (NCBI taxonomy) amplicons discussed in the text from each area are shown, with bubble size corresponding to percent average relative abundance (Dataset S1C; UC, n = 3 samples; LC, n = 2 samples; NWC-A, n = 6 samples; NWC-B, n = 3 samples; UCW, n = 2 samples). The average relative abundance of Archaea was calculated separately using only archaeal taxa, and bubble size represents the percent average relative abundance of Archaea (UC, n = 2 samples; LC, n = 2 samples; NWC-A, n = 6 samples; NWC-B, n = 3 samples; UCW, n = 2 samples). (B) Associated enrichment of key functional genes where bubble size corresponds to relative abundances as described for Fig. 3A (Dataset S2B), averaged across samples at each area. Enrichment of carbon dioxide (CO2), sulfate (SO42−), magnesium (Mg), hydrogen sulfide (H2S), iron (Fe) and hydrogen (H2) and pH shifts are shown for each site (Dataset S1B). The combination of different geochemical environments intersected by the circulating hydrothermal fluids, and different scales of recharge, may explain some of the shifts in diversity of the microbial communities observed at Brothers volcano.
Subsurface Heat Flow Patterns Help Explain the Observed Microbial Differences between Sites.
The collective phylogenetic, functional, and metagenomic differences of the microbial communities associated with hydrothermal deposits at Brothers volcano (Fig. 6) correspond to the complexity in the subseafloor hydrogeology at Brothers volcano. This is supported by recent heat flow studies that show both caldera-scale (2 to 3 km) upflow and recharge and more local-scale (100 to 200 m) heat flow patterns (35). The geological evolution of Brothers volcano and the accompanying changes in the subsurface fluid regime (e.g., phase separation, water–rock reactions, mixing of differently sourced fluids, etc.) produce geochemical gradients that provide a range of geochemical disequilibria that support a diversity of Archaea and Bacteria. In particular, we posit that the NWC-A communities are supported by a mix of magmatically influenced and seawater-influenced hydrothermal fluids, while the NWC-B+UCW communities are supported by shallower seawater-derived hydrothermal fluids. Differences in these microbial communities may be due to adaptation to subtly different chemistries, generated as hydrothermal fluids follow different paths and interact with both the earlier (deeper) acid-sulfate conditions and the more recent (shallow overprinting) modified seawater-dominated conditions. The high phylogenetic diversity of members of the DPANN superphylum (SI Appendix, Figs. S4C and S5), especially in the vigorously venting resurgent LC, may suggest that in these environments symbiotic interactions play an important role in shaping the microbial community diversity.
Together, our findings from Brothers volcano point to the importance of considering magmatic activity, the geological legacy, and the subsurface hydrologic fluid regime when assessing drivers of microbial community composition, assembly, and evolution. Likewise, a recent study from Yellowstone hot springs attributed the presence of a unique diverse archaeal-dominated anaerobic community to past seismic events that released acidic magmatic volatiles into hot-spring fluids (36). Microbial communities can also respond rapidly to fluctuating environments (37), adapting uniquely to temperature and geochemical fluctuations (16), which could also explain the functional diversity seen in the NWC-B+UCW microbial communities at Brothers volcano. Collectively, these findings show that microbial community diversity can reflect complex subsurface hydrothermal processes, both past and present. As we continue to study these systems and their microbial communities more carefully, we may be able to use the microbial community diversity as indicators of potential subsurface magmatic and hydrothermal processes.
Materials and Methods
Sample Collection, DNA Extraction, and Hydrothermal Fluid Chemistry Analysis.
Hydrothermal deposits were collected from the Brothers volcano (−34.8708° S, 179.0667° E) NW Caldera, UCW, and LC and UC sites during the research vessel (R/V) Thomas Thompson March 2018 TN350 expedition using the remotely operated vehicle (ROV) Jason. Water samples were collected using isobaric gas-tight fluid samplers (38) and major water samplers (39). Additional deposits were collected from the ELSC-VFR during the R/V Roger Revelle (RR1507) expedition in April/May 2015. Once shipboard, deposits were processed as described previously (17). DNA was extracted from homogenized deposits using the DNeasy PowerSoil kit (Qiagen). For DNA from water, 1 L of water was filtered using a 0.2-μm Sterivex filter (Merck) and DNA was extracted as described by Anderson et al. (40).
Geochemical Measurements.
pH was measured onboard at thermal equilibrium with a Metrohm 780 pH meter calibrated daily with standard buffers. A Hach digital titrator and prepared titrant solutions of HCl and NaOH (Fisherbrand 0.1 M appropriately diluted and calibrated) were used for alkalinity/acidity titrations. Sulfide was measured using a modified Cline (methylene blue) method and a HACH DR2800 spectrophotometer. Appropriate dilutions were made with efforts to minimize air exposure and loss of analyte. Onshore, major cations and metal concentrations were analyzed by inductively coupled plasma optical emission spectroscopy using acidified and filtered splits with appropriate dilutions and matrix-matched standards. Check standards were run every 10 samples and verified to be within 5% of expected values, and samples were run in triplicate. Major anion samples were filtered and refrigerated prior to analysis with a Dionex ICS3000 equipped with an AG/AS-18 column. Concentrations of dissolved H2 and CH4 in vent fluid samples were analyzed using on-board gas chromatography within 12 h after recovery of fluid samples. Volatiles were extracted by a syringe headspace extraction. A Thermo Scientific Trace GC Ultra gas chromatograph, equipped with a packed Molsieve 60/80 column (Sigma-Aldrich), was operated with N2 as carrier gas at 50 °C. The concentration of H2 was quantified with a thermal conductivity detector and the concentration of CH4 was quantified using a flame ionization detector. The device was calibrated on a daily basis with reference gases of 1.02 mol % H2 or 0.987 mol % CH4 in a N2 matrix. Samples for determination of dissolved CO2 were stored upside down in preweighted He-filled and subsequently evacuated glass serum vials to avoid atmospheric CO2 contamination. CO2 concentrations were determined onshore using a Thermo Scientific Trace GC Ultra equipped with a packed HaySep 80/100 column (Sigma-Aldrich) and operated with helium as carrier gas at 50 °C. The system was calibrated with reference gas of pure CO2 with 99.995 mol %. Each sample was measured in triplicate and a control standard was measured prior to each sample block. The accuracy and precision of the measurements are within 10%. The method is described in Reeves et al. (41) and was adopted with an improved calculation procedure for headspace liquid–vapor partitioning. End-member fluid compositions were calculated for the NW Caldera and Upper Caldera based on Mg = 0 extrapolations from background seawater concentrations. The measured vent fluid compositions were used to calculate the Gibbs energies for catabolic reactions in mixtures of vent fluid and seawater following the procedure outlined in Amend et al. (42) and shown in Dataset S1B.
Amplicon Amplification and Sequencing of Environmental Samples.
Microbial communities were characterized by sequencing the V4 region of the 16S rRNA gene following the Earth Microbiome Protocol (43) with primers 515F-Y (5′-GTGYCAGCMGCCGCGGTAA-3′) (44) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Twelve-base-pair barcodes were added to primer 806R. Triplicate 25-μL PCR reactions were prepared with 12.5 μL of GoTaq Master Mix (Promega Corp.), 0.5 μL of forward and reverse primers (0.2 μM final concentration), 10.5 μL of nuclease-free PCR water (Promega Corp.), and 1 μL of template DNA. Thermal cycling conditions began with an initial denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 45 s, 50 °C for 60 s, and 72 °C for 90 s. The final extension was at 72 °C for 10 min. After amplification, PCR products were pooled, visualized using gel electrophoresis, and then quantified using the Quant-iT Picogreen dsDNA kit (Invitrogen). Three hundred nanograms of each product were pooled, cleaned using the Invitrogen PureLink PCR purification kit (Carlsbad), and quantified using the Qubit dsDNA High Sensitivity (HS) kit (Invitrogen). Sequencing was performed on an Illumina MiSeq using the v2 chemistry (2 × 150 base pairs [bp]) at California State University, Northridge, CA.
Amplicon Sequence Analysis.
Demultiplexed amplicon sequence data were imported into QIIME2 v.2019.748 (45). The reads were joined using vsearch, quality-filtered, and denoised using deblur (46). Diversity analyses (alpha and beta diversity) were performed at a depth of 3,000 sequences per sample. Taxonomic assignments were performed using the SILVA database classification (v.132) (47) using the QIIME2 Naive Bayes classifier trained for the V4 region (Silva-132-99-515-806-nb-classifier.qza). All QIIME2 commands are available on FigShare.
Metagenome Sequencing of Environmental Samples.
Metagenomic libraries were constructed using Nextera DNA Library Prep kits (Illumina) according to the manufacturer’s protocols. Paired-end sequencing (2 × 150 bp) of the libraries was done at the Oregon State University Center for Genome Research and Computing on the Illumina HiSeq 3000 platform.
Metagenome Assembly.
Metagenome reads were trimmed and cleaved of Illumina sequencing adapters and barcodes using Trimmomatic v.0.36 (48). Reads were interleaved, and both interleaved and unpaired reads were assembled into contigs using MEGAHIT v.1.1.1-2-g02102e1 (49) with the parameters –min-contig-len 2000 –k-min 31 –k-max 151 and –k-step 20. In order to evaluate different assembly approaches, the reads of three MEGAHIT assemblies with the lowest ratio of assembly length to clean reads (S009, S017, and S142) were also assembled using metaSPAdes v.3.10.1 (50) with the following options: -k 33, 55, 77, 99, 127 –only-assembler. Only contigs ≥2,000 bp were retained. The quality of the MEGAHIT assemblies was in general significantly better than those obtained using metaSPAdes (best assembly length in all three using MEGAHIT, and higher maximum and mean contig lengths in two) and thus MEGAHIT assemblies were chosen for further metagenomic analyses. To generate differential read coverage profiles, reads were mapped to the contigs with Bowtie2 v.2.2.9 (51) and parsed using SAMtools v.1.3.1 (52).
Annotation of Metagenome Assemblies.
To assess functional differences between samples, proteins on assembled contigs from each metagenome were predicted using Prodigal v.2.6.3 (53) and annotated with the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (54) using GhostKOALA (55). KEGG orthology (KO) hits corresponding to catalytic hydrogenase subunits (K00436, K00440, K05922, K06281, K12142, K14090, K14106, K14123, K14126, K18016, K18332, and K23549) were further characterized using HydDB (24), and only hydrogenases passing HydDB quality standards were retained. Gene categories involved in iron transport and metabolism were independently annotated using FeGenie (56). HydDB-validated hydrogenase hits (n = 21 genes), FeGenie-annotated iron gene categories (n = 7 categories), and a subset of KO hits (n = 95 genes) involved in key energy and carbon acquisition processes and oxygen sensitivity were extracted for further analysis. Each functional gene in the analysis was assigned a coverage value using contig coverage profiles (discussed above). The summed coverage for each functional gene was normalized by the average total coverage for 14 single-copy marker genes (K02950, K02992, K02906, K02926, K02890, K02874, K02931, K02994, K02952, K02948, K02871, K02961, K02881, and K02986), as previously described (57). Normalized values were also averaged across samples from each area (UC, LC, NWC-A, and NWC-B+UCW), and a heat map was constructed using Morpheus (https://software.broadinstitute.org/morpheus).
Metagenome Binning.
Contigs (≥2,000 bp) were binned into draft MAGs with MetaBAT v.0.32.4 (58) based on tetranucleotide frequency and differential read coverage. CheckM (59) was used to estimate MAG completion and contamination, and Phylosift v.1.0.1 (60) was used to screen for 16 ribosomal protein genes (61) (rpL2-6, rpL14-16, rpL18, rpL22, rpL24, rpS3, rpS8, rpS10, rpS17, and rpS19). Downstream analysis was restricted to MAGs containing at least six of the above-mentioned ribosomal proteins and with ≥50% completion and ≤10% contamination. Relative abundance was estimated for each MAG using the differential read profiles generated for each individual contig. Read coverage for medium- to high-quality MAGs was summed and normalized across each sample. Combined with taxonomy assignments, normalized read coverage was used as a proxy for normalized relative abundance of Bacteria and Archaea represented by medium- to high-quality MAGs.
Curation of Representative Genomes.
Representative MAGs belonging to taxonomic clades of interest were selected for additional curation using Emergent Self-Organizing Maps (62) with the tetramerFreq script package (https://github.com/tetramerFreqs/Binning) (63). After calculating the tetranucleotide frequency of individual MAGs and multiple reference genomes, ESOM maps were trained using the K-batch option with the recommended number of rows and columns. MAGs were manually rebinned, and outlier contigs with <90% confidence were removed. These curated MAGs are denoted on the phylogenetic trees (discussed below) by an asterisk.
Phylogenetic Analyses.
As a framework for this study, genome bins were used to construct maximum-likelihood phylogenetic trees for the Bacteria (n = 21,780 taxa and 5,035 amino acid positions) and Archaea (n = 1,371 taxa and 5,024 amino acid positions) using the identify, align, and infer steps of the GTDB-Tk v.0.2.2 de novo workflow (data release 86 v.3, WAG+GAMMA model) (27). Branch support values were computed using the Shimodaira–Hasegawa (SH) test. Additional phylogenetic trees were constructed of the Chloroflexi (n = 67 taxa and 2,375 amino acid positions) and the Aquificae (n = 48 taxa and 2,373 amino acid positions) using 16 ribosomal proteins (61), which were aligned individually using Muscle v.3.8.425 (64) and concatenated, and positions containing ≥90% gaps were also removed. Alignments were then subjected to maximum-likelihood phylogenetic analysis in RAxML v.8.2.8 (65) (PROTGAMMA+LG model) with 1,000 replicate bootstraps. GTDB-Tk trees and ribosomal protein trees were viewed and rooted with Interactive Tree of Life v.5 (66). Furthermore, we performed phylogenetic analyses of DPANN superphylum MAGs using 48 marker proteins identified in 400 archaeal genomes (67) from the Archaeal Clusters of Orthologous Genes (arCOG) database (68). All markers were individually aligned with MAFFT v.7.407 (69) (mafft-linsi), trimmed with BMGE v.1.12 (70) (settings: -m BLOSUM30 -h 0.55), and concatenated using catfasta2phyml.pl (https://github.com/nylander/catfasta2phyml). IQ-TREE v.1.6.7 (71) was used to perform maximum-likelihood analysis (C60+LG+F+R mixture model, n = 400 taxa and 9,461 amino acid positions) with an ultrafast bootstrap approximation and SH-like approximate likelihood tests, each run with 1,000 bootstrap replicates.
Taxonomic Classification and Novel Taxonomic Rank Designations.
MAGs were classified using the GTDB-Tk v.0.2.2 (data release 86 v.3) classify workflow (27) and with the NCBI taxonomy browser. In order to explore novel family and genus level ranks for MAGs, we used both the GTDB-Tk workflow and average amino acid identity (AAI) matrices (using the Enveomics AAI Matrix Tool, enve-omics.ce.gatech.edu/g-matrix/) to inform our decision. In general, we used the GTDB-Tk assignment first and confirmed the ranking with AAI. In a few cases, compelling evidence based on AAI similarity superseded the GTDB-Tk ranking. For comparison, archaeal taxonomic ranks were also determined using AAI comparisons between MAGs and reference genomes. AAI matrices for the DPANN superphylum were generated separately using CompareM v.0.0.23 (https://github.com/dparks1134/CompareM).
Nomenclature.
We recognize the archaeal and bacterial nomenclature is in flux. Here we primarily use the NCBI-assigned taxonomies or the GTDB-recommended taxonomy in parentheses. Dataset S3B provides both nomenclatures for reference. We refer to the Epsilonbacteraeota (formerly Epsilonproteobacteria; GTDB Campylobacterota) throughout the text using detailed nomenclature proposed by Waite et al. (72), which is largely congruent with GTDB data release 86 v.3 taxonomy at the class level and lower taxonomic ranks. Detailed taxonomy of the DPANN superphylum MAGs is also included in the phylogenetic tree depicted in SI Appendix, Fig. S6.
Annotation and Comparison of MAGs.
Open reading frames (ORFs) within MAGs were predicted using Prodigal v.2.6.3 (53) as implemented in Prokka v.1.13 (73). For comparison with assembly annotation data, the MAGs were annotated as described above using GhostKoala (55), HydDB (24), and FeGenie (56).
Statistical Analyses.
To assess site differences at Brothers volcano, Bray–Curtis similarity matrices were generated in PRIMER v.6.1.13 (74) using the relative abundance of key functional genes/gene categories identified in the assemblies (n = 123 genes/gene categories) and the relative abundance of medium- to high-quality MAGs based on normalized read coverage and GTDB-Tk taxonomy (n = 251 taxonomic assignations). Bray–Curtis dissimilarity matrices comparing communities from Brothers volcano samples and from both Brothers volcano and ELSC-VFR samples were also generated in QIIME2 as part of the “core-metrics-phylogenetic” workflow in the “diversity” plugin, at a sampling depth of 3,000 sequences per sample. Similarity and distance matrices were used to create nonmetric multidimensional scaling (NMDS) plots in PRIMER. One-way ANOSIM was performed on each matrix to identify statistically significant differences between sites, and one-way SIMPER analysis was used to identify specific contributors to the similarities and differences between sites where applicable.
Supplementary Material
Acknowledgments
We thank the captain and crew of the R/V Thompson and the engineers from Woods Hole Oceanographic Institution for the successful operation of ROV Jason. The project was funded by NSF grants OCE‐1558356 (Principal Investigator S.E.H.) and OCE-1558795 (Principal Investigator A.-L.R.). S.L. received a grant from the University of Brest to work in the A.-L.R. laboratory. A travel fund from Interridge enabled A.D. to participate on the R/V Thompson cruise. Funding for this work for C.E.J.d.R., F.C.T., V.K.S., and L.C.S. was provided by the New Zealand government. A.S. was supported by the Swedish Research Council (Vetenskapsrådet starting grant 2016-03559 to A.S.) and the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Dutch Research Council) Foundation of the Netherlands Organization for Scientific Research (Women In Science Excel [WISE] fellowship to A.S.). A.-L.R. and E.S.J. thank Rika Anderson for helpful methodological discussions and Sean Sylva for assistance in shipboard geochemical analysis.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
*These have been recently reclassified, as Desulfobacterota, Waite et al., Int. J. Syst. Evol. Microbiol. DOI: 10.1099/ijsem.0.004213.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019021117/-/DCSupplemental.
Data Availability.
Amplicon reads from Brothers volcano and ELSC-VFR are available from the NCBI Sequence Read Archive (see Dataset S1 A and E for accession nos.). Metagenome reads are also deposited in the Sequence Read Archive (see Dataset S2A for accesson nos.), and metagenome-assembled genomes are available on GenBank (see Dataset S3B for accession nos.). Additional data are available on FigShare at https://doi.org/10.6084/m9.figshare.c.5099348 (21).
References
- 1.Crisp J. A., Rates of magma emplacement and volcanic output. J. Volcanol. Geotherm. Res. 20, 177–211 (1984). [Google Scholar]
- 2.Wilson S. T., et al. , Kīlauea lava fuels phytoplankton bloom in the North Pacific Ocean. Science 365, 1040–1044 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Hannington M., Jamieson J., Monecke T., Petersen S., Beaulieu S., The abundance of seafloor massive sulfide deposits. Geology 39, 1155–1158 (2011). [Google Scholar]
- 4.de Ronde C. E. J., Massoth G. J., Baker E. T., Lupton J. E., “Submarine hydrothermal venting related to volcanic arcs” in Volcanic, Geothermal, and Ore-Forming Fluids: Rulers and Witnesses of Processes within the Earth, Simmons S. F., Graham I., Eds. (Society of Economic Geologists, Inc., 2005), pp. 91–110. [Google Scholar]
- 5.Berkenbosch H. A., de Ronde C. E. J., Gemmell J. B., McNeill A. W., Goemann K., Mineralogy and formation of black smoker chimneys from Brothers submarine volcano, Kermadec arc. Econ. Geol. 107, 1613–1633 (2012). [Google Scholar]
- 6.Kleint C., et al. , Geochemical characterization of highly diverse hydrothermal fluids from volcanic vent systems of the Kermadec intraoceanic arc. Chem. Geol. 528, 119289 (2019). [Google Scholar]
- 7.de Ronde C. E. J., et al. , Submarine hydrothermal activity and gold-rich mineralization at Brothers volcano, Kermadec arc, New Zealand. Miner. Depos. 46, 541–584 (2011). [Google Scholar]
- 8.de Ronde C. E. J., et al. , Evolution of a submarine magmatic-hydrothermal system: Brothers volcano, southern Kermadec Arc, New Zealand. Econ. Geol. 100, 1097–1133 (2005). [Google Scholar]
- 9.de Ronde C. E. J., Humphris S. E., Höfig T. W., Reyes A. G., Critical role of caldera collapse in the formation of seafloor mineralization: The case of Brothers volcano. Geology 47, 762–766 (2019). [Google Scholar]
- 10.de Ronde C. E. J., Humphris S. E., Hofig T. W., The Expedition 376 Scientists, “Brothers Arc Flux” in Proceedings of the International Ocean Discovery Program, 376 (International Ocean Discovery Program, College Station, TX, 2019). [Google Scholar]
- 11.Stott M. B., et al. , Culture-independent characterization of a novel microbial community at a hydrothermal vent at Brothers volcano, Kermadec arc, New Zealand. J. Geophys. Res. Solid Earth 113, B08S06 (2008). [Google Scholar]
- 12.Takai K., et al. , Variability in microbial communities in black smoker chimneys at the NW Caldera vent field, Brothers volcano, Kermadec arc. Geomicrobiol. J. 26, 552–569 (2009). [Google Scholar]
- 13.Payne D., et al. , Geologic legacy spanning >90 years explains unique Yellowstone hot spring geochemistry and biodiversity. Environ. Microbiol. 21, 4180–4195 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Takacs-Vesbach C., Mitchell K., Jackson-Weaver O., Reysenbach A.-L., Volcanic calderas delineate biogeographic provinces among Yellowstone thermophiles. Environ. Microbiol. 10, 1681–1689 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Galambos D., Anderson R. E., Reveillaud J., Huber J. A., Genome-resolved metagenomics and metatranscriptomics reveal niche differentiation in functionally redundant microbial communities at deep-sea hydrothermal vents. Environ. Microbiol. 21, 4395–4410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opatkiewicz A. D., Butterfield D. A., Baross J. A., Individual hydrothermal vents at Axial Seamount harbor distinct subseafloor microbial communities. FEMS Microbiol. Ecol. 70, 413–424 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Flores G. E., et al. , Inter-field variability in the microbial communities of hydrothermal vent deposits from a back-arc basin. Geobiology 10, 333–346 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Nakagawa S., et al. , Variability in microbial community and venting chemistry in a sediment-hosted backarc hydrothermal system: Impacts of subseafloor phase-separation. FEMS Microbiol. Ecol. 54, 141–155 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Hug L. A., et al. , A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto M., Takai K., Sulfur metabolisms in epsilon- and gamma-proteobacteria in deep-sea hydrothermal fields. Front. Microbiol. 2, 192 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reysenbach A.-L., et al. , Additional files for ‘Complex subsurface hydrothermal fluid mixing at a submarine arc volcano supports distinct and highly diverse microbial communities’. Figshare. 10.6084/m9.figshare.c.5099348. Deposited 20 November 2020. [DOI] [PMC free article] [PubMed]
- 22.Sheng Y., et al. , Superoxide dismutases and superoxide reductases. Chem. Rev. 114, 3854–3918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris R. L., Schmidt T. M., Shallow breathing: Bacterial life at low O2. Nat. Rev. Microbiol. 11, 205–212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Søndergaard D., Pedersen C. N. S., Greening C., HydDB: A web tool for hydrogenase classification and analysis. Sci. Rep. 6, 34212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg I. A., Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl. Environ. Microbiol. 77, 1925–1936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowers R. M. et al.; Genome Standards Consortium , Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaumeil P.-A., Mussig A. J., Hugenholtz P., Parks D. H., GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reysenbach A.-L., et al. , Brothers volcano GTDB-Tk Bacteria tree. Interactive Tree of Life. https://itol.embl.de/shared/alrlab. Deposited 6 November 2019.
- 29.Reysenbach A.-L., et al. , Brothers volcano GTDB-Tk Archaea tree. Interactive Tree of Life. https://itol.embl.de/shared/alrlab. Deposited 19 August 2020.
- 30.Tessler M., et al. , Large-scale differences in microbial biodiversity discovery between 16S amplicon and shotgun sequencing. Sci. Rep. 7, 6589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrini V. L., Tivey M. K., Carbotte S. M., Martinez F., Roman C., Variable morphologic expression of volcanic, tectonic, and hydrothermal processes at six hydrothermal vent fields in the Lau back-arc basin. Geochem. Geophys. Geosyst. 9, Q07022 (2008). [Google Scholar]
- 32.Tivey M. K., et al. , Links from mantle to microbe at the Lau integrated study site: Insights from a back-arc spreading center. Oceanography 25, 62–77 (2012). [Google Scholar]
- 33.Colman D. R., Lindsay M. R., Amenabar M. J., Boyd E. S., The intersection of geology, geochemistry, and microbiology in continental hydrothermal systems. Astrobiology 19, 1505–1522 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Fournier R. O., Geochemistry and dynamics of the Yellowstone National park hydrothermal system. Annu. Rev. Earth Planet. Sci. 17, 13–53 (1989). [Google Scholar]
- 35.Caratori Tontini F., Tivey M. A., de Ronde C. E. J., Humphris S. E., Heat flow and near-seafloor magnetic anomalies highlight hydrothermal circulation at Brothers volcano caldera, southern Kermadec arc, New Zealand. Geophys. Res. Lett. 46, 8252–8260 (2019). [Google Scholar]
- 36.Colman D. R., Lindsay M. R., Boyd E. S., Mixing of meteoric and geothermal fluids supports hyperdiverse chemosynthetic hydrothermal communities. Nat. Commun. 10, 681 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward L., et al. , Microbial community dynamics in Inferno Crater Lake, a thermally fluctuating geothermal spring. ISME J. 11, 1158–1167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seewald J. S., Doherty K. W., Hammar T. R., Liberatore S. P., A new gas-tight isobaric sampler for hydrothermal fluids. Deep. Res. Part I Oceanogr. Res. Pap. 49, 189–196 (2002). [Google Scholar]
- 39.Von Damm K. L., et al. , Chemistry of submarine hydrothermal solutions at 21 °N, East Pacific rise. Geochim. Cosmochim. Acta 49, 2197–2220 (1985). [Google Scholar]
- 40.Anderson R. E., et al. , Genomic variation in microbial populations inhabiting the marine subseafloor at deep-sea hydrothermal vents. Nat. Commun. 8, 1114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeves E. P., et al. , Geochemistry of hydrothermal fluids from the PACMANUS, Northeast Pual and Vienna Woods hydrothermal fields, Manus Basin, Papua New Guinea. Geochim. Cosmochim. Acta 75, 1088–1123 (2011). [Google Scholar]
- 42.Amend J. P., McCollom T. M., Hentscher M., Bach W., Catabolic and anabolic energy for chemolithoautotrophs in deep-sea hydrothermal systems hosted in different rock types. Geochim. Cosmochim. Acta 75, 5736–5748 (2011). [Google Scholar]
- 43.Caporaso J. G., et al. , Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parada A. E., Needham D. M., Fuhrman J. A., Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Bolyen E., et al. , Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amir A., et al. , Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quast C., et al. , The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li D., et al. , MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102, 3–11 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Nurk S., Meleshko D., Korobeynikov A., Pevzner P. A., metaSPAdes: A new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H., A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyatt D., et al. , Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M., Sato Y., Morishima K., BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Garber A. I., et al. , FeGenie: A comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front. Microbiol. 11, 37 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan L., et al. , Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. U.S.A. 109, E1878–E1887 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kang D. D., Froula J., Egan R., Wang Z., MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3, e1165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darling A. E., et al. , PhyloSift: Phylogenetic analysis of genomes and metagenomes. PeerJ 2, e243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anantharaman K., et al. , Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7, 13219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ultsch A., Mörchen F., “ESOM-Maps: Tools for clustering, visualization, and classification with Emergent SOM” (Technical Report, Dept. of Mathematics and Computer Science, University of Marburg, Marburg, Germany, 2005).
- 63.Dick G. J., et al. , Community-wide analysis of microbial genome sequence signatures. Genome Biol. 10, R85 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Letunic I., Bork P., Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaremba-Niedzwiedzka K., et al. , Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Makarova K. S., Wolf Y. I., Koonin E. V., Archaeal clusters of orthologous genes (arCOGs): An update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life 5, 818–840 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katoh K., Misawa K., Kuma K., Miyata T., MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Criscuolo A., Gribaldo S., BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waite D. W., et al. , Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front. Microbiol. 8, 682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Clarke K. R., Gorley R. N., Primer v6: User Manual/Tutorial (PRIMER-E Ltd, 2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Amplicon reads from Brothers volcano and ELSC-VFR are available from the NCBI Sequence Read Archive (see Dataset S1 A and E for accession nos.). Metagenome reads are also deposited in the Sequence Read Archive (see Dataset S2A for accesson nos.), and metagenome-assembled genomes are available on GenBank (see Dataset S3B for accession nos.). Additional data are available on FigShare at https://doi.org/10.6084/m9.figshare.c.5099348 (21).