Significance
Understanding species’ evolutionary responses to past climate change is fundamental for improved biodiversity conservation. Species in the same area could exhibit either similar or individualistic evolutionary responses. We tested this hypothesis within the highly biodiverse tropical rain forests of Central Africa. We generated an unparalleled genomic dataset to estimate the evolutionary dynamics of a range of codistributed plant species. We reveal variable, asynchronous, individualistic evolutionary responses driven by different environmental factors and life-history traits. Our results challenge the long-standing view that past climate change led to similar rain forest dynamics within this region. Our study has important implications for the conservation of tropical rain forest biodiversity in general, and in Central Africa in particular.
Keywords: tropical rain forests, climate change, phylogenetics, demography
Abstract
Understanding the evolutionary dynamics of genetic diversity is fundamental for species conservation in the face of climate change, particularly in hyper-diverse biomes. Species in a region may respond similarly to climate change, leading to comparable evolutionary dynamics, or individualistically, resulting in dissimilar patterns. The second-largest expanse of continuous tropical rain forest (TRF) in the world is found in Central Africa. Here, present-day patterns of genetic structure are thought to be dictated by repeated expansion and contraction of TRFs into and out of refugia during Pleistocene climatic fluctuations. This refugia model implies a common response to past climate change. However, given the unrivalled diversity of TRFs, species could respond differently because of distinct environmental requirements or ecological characteristics. To test this, we generated genome-wide sequence data for >700 individuals of seven codistributed plants from Lower Guinea in Central Africa. We inferred species’ evolutionary and demographic histories within a comparative phylogeographic framework. Levels of genetic structure varied among species and emerged primarily during the Pleistocene, but divergence events were rarely concordant. Demographic trends ranged from repeated contraction and expansion to continuous growth. Furthermore, patterns in genetic variation were linked to disparate environmental factors, including climate, soil, and habitat stability. Using a strict refugia model to explain past TRF dynamics is too simplistic. Instead, individualistic evolutionary responses to Pleistocene climatic fluctuations have shaped patterns in genetic diversity. Predicting the future dynamics of TRFs under climate change will be challenging, and more emphasis is needed on species ecology to better conserve TRFs worldwide.
Lower Guinea (Fig. 1) is the most biodiverse floristic region of Central Africa, with about 7,000 vascular plant species, a quarter of which are endemics (1, 2). How Lower Guinean tropical rain forests (TRFs) responded to glacial–interglacial Milankovitch cycles of the Pleistocene (2.58 to 0.01 million years ago [Ma]), in particular during the last glacial maximum (LGM; approximately 24 to 12 Ka), remains highly debated (3–7).
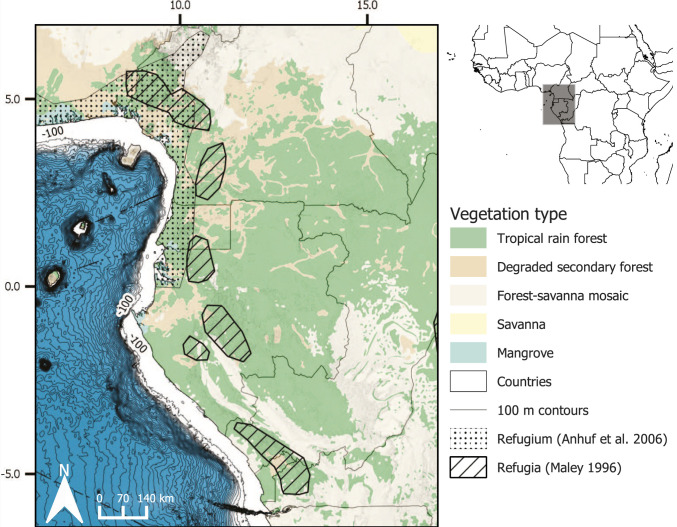
Fig. 1.
Major vegetation types of Lower Guinea and bathymetry of the Gulf of Guinea. Refugial areas proposed by Maley (3) and Anhuf et al. (6) for our study area are shown. Vegetation layers are taken from ref. 19. Inset on the top right shows the area in the context of the African continent. Bathymetry contour values for every 100 m for the Gulf of Guinea (north: 5.5; south: −7.0; west: 6.0; east: 14) were downloaded from ref. 95. The light blue region indicates ocean floor greater than 100 m in depth. Map was drawn with QGIS (v3.123).
Two main hypotheses have been proposed to explain TRF responses to Pleistocene climatic fluctuations in Central Africa, and Lower Guinea in particular (Table 1). TRFs either contracted significantly into discrete forest refugia (3, 7, 8) [hereafter “refugia hypothesis,” or H1 (9)] or did not contract, but persisted in a more diffuse way as open lowland rain forest with a decreased canopy density and height (“no refugia hypothesis,” or H2) (4, 7). In the former hypothesis, refugia are defined as geographic areas that operate on evolutionary time scales, allowing viable populations of a species to persist during climatic fluctuations (10, 11). Species contract into refugia during unfavorable periods and then expand out from them when the climate improves (10, 11). Based on paleovegetation reconstructions (12) and plant or animal endemicity patterns (8, 13), TRFs are suggested to have contracted as much as 90% in Central Africa into a single (6, 14) or several discrete refugial areas (3). In Lower Guinea, most refugia are thought to be located in mountainous regions close to the Atlantic coast (3, 6, 15) (Fig. 1).
Table 1.
Main hypotheses tested with resulting spatial and temporal genetic population expectations under the assumption of climatic fluctuations impacting (or not impacting) species dynamics
| Hypotheses | |||
| Refugia model (H1): During adverse climatic periods (e.g., LGM), TRFs significantly contracted across Lower Guinea into one or several discrete ecologically stable areas surrounded by more arid ecosystems | No refugia model (H2): During adverse climatic periods, TRFs did not contract across Lower Guinea | ||
| Subhypotheses | Strict species-independent | Relaxed species-dependent | Not applicable |
| refugia model (H1a): | refugia model (H1b): | ||
| All species responded in a | Species responded in different | ||
| similar evolutionary way | evolutionary ways dependent | ||
| independent of life-history | on life-history traits | ||
| traits and environmental factors. | and environmental factors | ||
| Genetic structure | High | Low to high | Low |
| within species | |||
| Spatial distribution | Allopatric or | Allopatric or | No prior |
| of genetic clusters | parapatric | parapatric | expectations |
| within species | |||
| Genetic diversity | Higher within | Higher within stable areas | No relationship |
| within species | stable areas | in some species and | |
| no relationship in others | |||
| Demographic history | Evidence of population | Evidence of population | No evidence of |
| within and | contraction (bottleneck) | contraction (bottleneck) and | population contraction |
| among species | and expansion | expansion in some species | (bottlenecks) or |
| and no evidence in others | expansion | ||
| Temporal origin of population | Pleistocene, temporally | Pleistocene, temporally | Not necessarily Pleistocene, |
| structure among species | congruent | incongruent | temporally incongruent. |
| Environmental factors | Factors related to | Different factors across | Different factors across |
| influencing genetic variation | stability explain distribution | species explain distribution | species explain |
| within species | of genetic diversity. | of genetic diversity | distribution of genetic |
| including stability. | diversity. No role | ||
| of stability. | |||
| References | (3, 6, 7) | (7, 16) | (4, 7) |
These expectations are centered around the hypothesis that stable areas or refugia played an important role in the evolutionary dynamics of Lower Guinean rain forest flora. We note that other mechanisms might also influence genetic structure and dynamics. Stability refers to discrete stable areas inferred to harbor populations of a species even during adverse times and are either the traditionally defined refugia or based on inferred habitat or climate stability.
A strict interpretation of the refugia hypothesis (3, 9) implies that all species responded in a similar evolutionary way to shared Pleistocene climatic events, independent of ecological or life-history traits (7). We refer to this as the “strict species-independent refugia” hypothesis (H1a; see Table 1 for details). Alternatively, even under the refugia hypothesis, species could have responded individualistically because of different adaptations, phenotypes, and/or environmental tolerances (7, 16–18). We refer to this as the “relaxed species-dependent refugia” hypothesis (H1b; Table 1). Although population genetic or phylogeographic studies of (usually single) Central African plant species generally favor the refugia (H1) over the no refugia (H2) hypothesis (7, 15), no study has set out to test the concordance of evolutionary histories among multiple plant species under a common comparative framework within Lower Guinea (7).
Here, we inferred the evolutionary histories of a variety of codistributed, lowland rain forest plants, including trees, shrubs, and a liana species, in Lower Guinea (Cameroon, Gabon, and the Republic of Congo) from two ecologically dominant TRF plant families: Arecaceae [palms (20); three species) and Annonaceae [custard-apples (21); four species]. Our species sampling covers a range of habit types, ecological tolerances, and animal-mediated dispersal syndromes (SI Appendix, Tables S1 and S2). We used a comparative phylogeographic approach (22–24) to test how similar evolutionary dynamics are among Lower Guinea TRF plant species.
If species follow the expectations of the refugia hypothesis (H1; Table 1), then, under the strict species-independent refugia hypothesis (H1a), we expect to find high levels of genetic structure, concordance in the timing of the emergence of this structure, and demographic trends showing similar patterns of contraction (i.e., genetic bottlenecks) and expansion of populations across all sampled species, independent of their life-history traits or environmental preferences (7, 25). In contrast, under the relaxed species-dependent refugia hypothesis (H1b; Table 1), we expect to infer responses to Pleistocene climatic events, but no evolutionary or temporal concordance, with each species responding in its own individualistic manner (7, 16, 18). Using family-tailored nuclear-marker baiting kits (26, 27), we sequenced hundreds of nuclear loci in more than 750 individuals. By doing so, we reduced biases linked to the use of different types or few molecular markers, enabling us to undertake evolutionary and temporal analyses with increased accuracy in a common framework.
Results
Generated sequence data were analyzed in four main steps. First, we inferred intraspecific genetic structure for each species using 2,000 (2k) to 6k single-nucleotide polymorphisms (SNPs) (SI Appendix, Table S1). Second, phylogenetic relationships between resulting genetic clusters were reconstructed by using whole-sequence datasets of up to 365 nuclear loci (SI Appendix, Table S1). We used a subset of nuclear loci together with fossil-calibration information to date the origin of intraspecific genetic structure. Third, we inferred the demographic history of each species and their genetic clusters separately. Finally, we investigated associations between intraspecific genetic variation and a range of environmental factors related to geography, climate, stability, and soil characteristics.
Stability Is Linked to Genetic Structure.
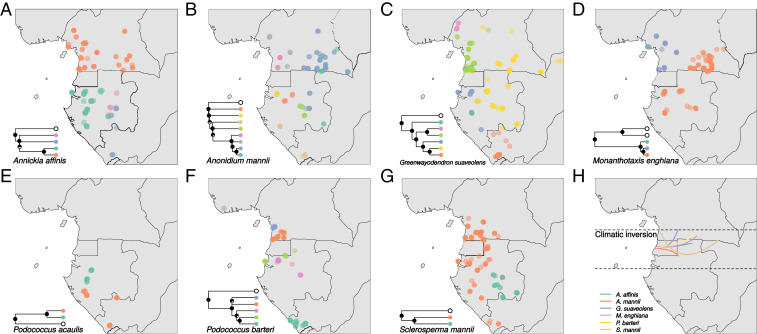
We recovered evidence for significant geographic structure in all species (SI Appendix, Fig. S1). Our genetic clustering analyses using Discriminant Analysis of Principal Components (DAPC) (28) and TESS3 (29) revealed that intraspecific diversity was highly structured, with four species exhibiting four or more distinct genetic clusters (Fig. 2 and SI Appendix, Figs. S2–S4). The topology of individual-level phylogenetic trees built using the entire dataset revealed groupings of individuals that closely matched those from clustering approaches (Fig. 2 and SI Appendix, Figs. S5–S11).
Fig. 2.
Genetic-clustering results of seven Central African rain forest species. For each species, collection points for individuals (where known) are shown, colored by genetic cluster, as inferred with DAPC. To the bottom left of A–G is a phylogenetic tree showing the relationships among genetic clusters (including outgroups in white). Pie charts at nodes on the trees indicate support (posterior probability). H shows the location of major genetic discontinuities associated with the climatic inversion that were inferred for each species by using TESS3, by drawing a line along contact of ancestry coefficients among clusters in interpolation maps (SI Appendix, Fig. S2).
To test the impact of stability on genetic structure across species, we used three main definitions of “stable areas.” The first consisted of the traditional refugia proposed by Maley (3) or Anhuf et al. (6) (Fig. 1) that are common to all species. For the two others, we estimated stable areas based on regional climate and species-specific habitat (30): 1) We identified climatically stable areas at the regional level by calculating differences in temperature and rainfall between the LGM and present (31); and 2) we inferred stable habitats between the LGM and present (e.g., ref. 15) by building ecological niche models (ENMs; see SI Appendix for methodological details) for each species.
Initially, we tested whether traditional refugia (3, 6) were important for determining patterns of present-day genetic diversity (H1). We expected individuals in or close to these traditional refugia to be more genetically diverse (10). We calculated individual-level expected heterozygosity (), observed heterozygosity (), and allelic richness and compared these to distance from traditional refugia (Fig. 1 and SI Appendix, Figs. S12–S18). A significant relationship was found in five of the seven species, depending on the distance and diversity metrics used. Decreasing distance to the coast [representing the single coastal refugium hypothesis of Anhuf et al. (6)] was significantly associated with higher levels of genetic diversity in Annickia affinis and Podococcus barteri. Both concepts of traditional refugia were linked to increased genetic diversity in Anonidium mannii and Sclerosperma mannii. No significant relationship was found for the remaining species. In a second analysis, we used analysis of molecular variance (AMOVA) to test how much genetic variance is explained by the three different definitions of stability defined above. We found that proximity to stable areas generally explained a small amount of genetic variance (10%; SI Appendix, Fig. S19). Even so, stable areas explained a significant amount of variance in all species. Most notably, in A. mannii, all three versions of stability explained large amounts of variance, while for P. barteri, we found that climatic stability explained substantially more variance than the others.
A Common Equatorial Genetic Discontinuity.
We found a conspicuous latitudinal genetic discontinuity at the climatic inversion located 0 to 3° North, in Southern Cameroon/Northern Gabon (Fig. 2H). It was recovered in all species distributed across the equator (6/7) (Fig. 2 and SI Appendix, Figs. S2 and S4). We then examined how important this barrier is to explaining genetic diversity in each species. We performed a hierarchical AMOVA to show that, though ubiquitous, this North–South barrier explains different amounts of variance among species (SI Appendix, Table S3): It is important in A. mannii and P. barteri, but less so in the other species.
Asynchronous Population Divergence.
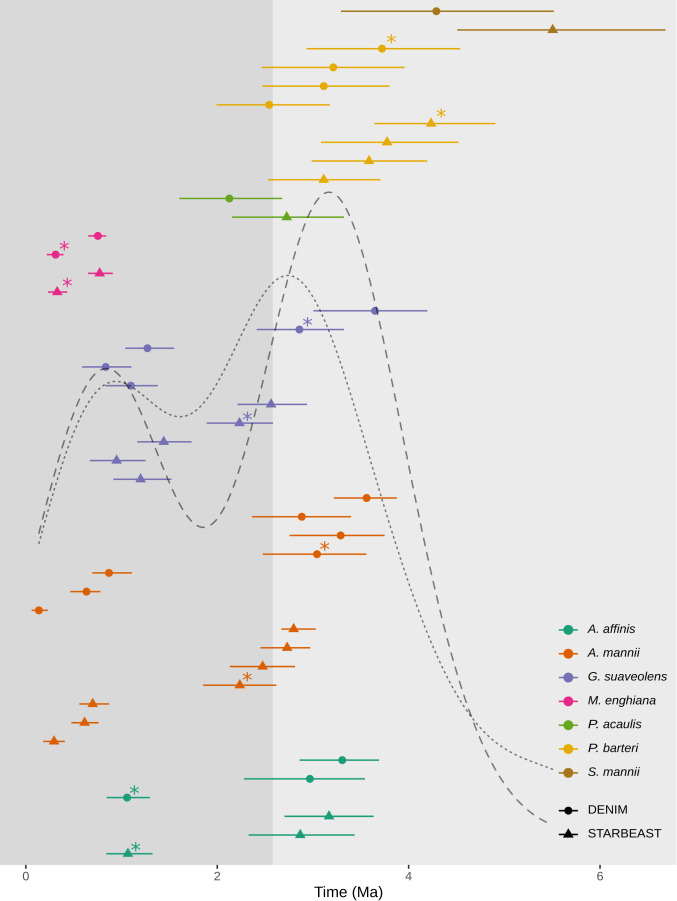
To estimate the timing of population divergence, we took a secondary calibration approach. We first produced time-calibrated backbone trees at a higher taxonomic level, which allowed us to make use of fossils that are not available for the focal groups (SI Appendix, Figs. S20 and S21). We inferred divergence times between genetic clusters using multiple phylogenetic approaches. We built population-level phylogenetic trees using divergence times inferred in backbone trees and the Bayesian coalescent approach StarBEAST (83). We selected the 32 most clock-like loci (32) for each of our species and subsampled five individuals to represent each genetic cluster. Species trees were reasonably well supported (Fig. 2), and their topologies generally reflected individual-level trees (SI Appendix, Figs. S5–S11). For comparison, we also used divergence estimation notwithstanding incomplete lineage sorting (ILS) and migration (DENIM) (33). This approach expands upon our StarBEAST analyses by incorporating low levels of migration when reconstructing species trees. These two approaches produced similar topologies and divergence times in all species (Fig. 3 and SI Appendix, Figs. S24–S29). Population divergence occurred most frequently at two time points—during the Pliocene–Pleistocene boundary (approximately 2.5 Ma), and around 1 Ma. However, the distribution of divergence events through time differed considerably among species, supporting the hypothesis that past climate change affected population divergence in different ways. For example, divergence across the climatic inversion occurred in only one major event per species, most commonly during the Pleistocene (Fig. 3). However, the median ages of these divergence events ranged from 0.3 to 4.2 Ma, indicating that there was likely no single event driving this common pattern.
Fig. 3.
Population divergence times in Lower Guinean rain forest species. Median divergence times estimated by using StarBEAST (triangles) and DENIM (circles) are shown. Lines on either side of these medians represent 95% HPDs for each event. Overlain black lines represent density plots of the median age of population divergence across all species for StarBEAST (dotted) and DENIM (dashed). The change in background color at 2.58 Ma represents the start of the Pleistocene. Stars indicate divergence events between DAPC genetic clusters situated on either side of the North/South climatic inversion.
Two Major Demographic Trends.
Species ranges are thought to have fluctuated repeatedly during the Pleistocene, as species expanded out of and contracted into stable areas. Therefore, under the refugia model, we expected a similar cyclical pattern of increases and decreases (Table 1) in effective population size (). A more constant trend in over this period may indicate that these cycles had less of an impact on rain forest species.
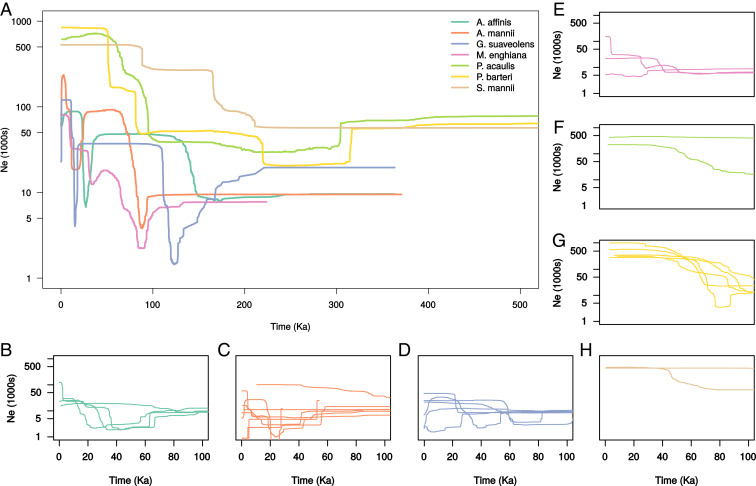
We recovered two major demographic patterns using the Stairway Plot approach (34): First, the three palm species experienced a relatively constant rate of population size increase through time (Fig. 4A); second, the Annonaceae species experienced fluctuating patterns of decline and growth over the last 200,000 years (200 Ka), including evidence for declines close to the LGM in A. affinis, A. mannii, and Greenwayodendron suaveolens. We found evidence of rapid decline in all but one species, which may be representative of population bottlenecks as species’ ranges contracted during glacial periods.
Fig. 4.
Demographic histories of seven Lower Guinean rain forest species. Population size changes were reconstructed by using the Stairway Plot method. Median effective population sizes () over time are shown by colored lines. A shows patterns inferred using all individuals in each species. B–H show demographic trends for each genetic cluster detected within each species, based on the same color code as A. Additional plots, including CIs, can be found in ref. 92.
We also estimated population size change for each genetic cluster within our species (Fig. 4 B–H). In general, these results reflected species-level patterns, but different clusters belonging to the same species did not have similar demographic trends in all cases. For example, in A. affinis, three of four clusters showed evidence of decline and expansion between 20 and 70 Ka, while the final cluster was relatively constant over this time period (Fig. 4B). This indicates that populations from different areas are also responding differently to past climate change. If particular areas acted as cross-species refugia, we might expect to infer similar population size trends in different species (H1a); however, upon examination, we found no common demographic pattern recovered for any particular area.
Distinct Sets of Environmental Factors Explain Patterns in Genetic Diversity Across Species.
We tested how different environmental factors shaped intraspecific genetic variation in each of our species. Again, under the strict species-independent refugia model (H1a), we expected genetic variation across species to be mainly explained by shared stable areas (Table 1). To investigate this, we collated data relating to geographic distribution of individuals, climate, soil type, and stability for each of the seven species (SI Appendix, Table S5). We used a distance-based redundancy analysis (db-RDA) framework (35) to build models and determine the total amount of genetic variance that can be explained by these variables and their relative importance. As expected under H1b, species differed in the total amount of variance explained and by the factors that were most important (Fig. 5). Indeed, for the liana species M. enghiana, stability explained negligible variance, while for the tree species A. affinis stability was shown to be more important (Fig. 5).
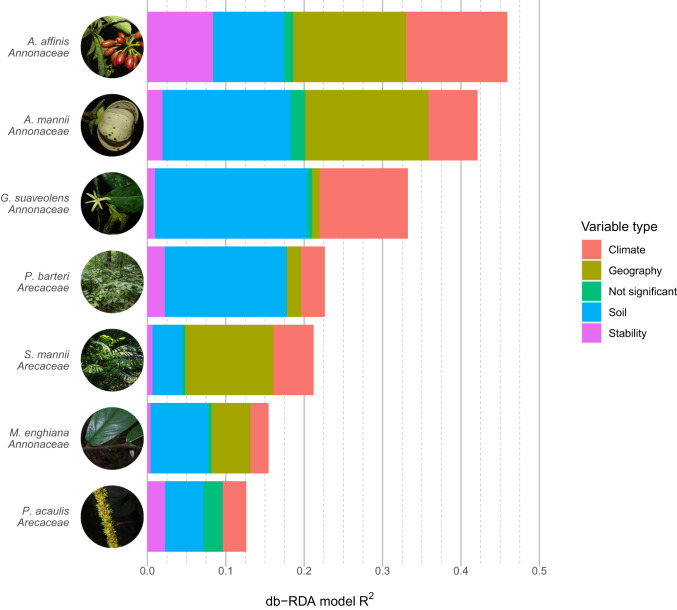
Fig. 5.
A stacked bar plot showing the components of the models from distance-based redundancy analyses. Variables making up the model were assigned to one of four different categories: climate, geography, soil, or stability. There were a small number of variables that were included in the full models, but nonsignificant, which were assigned to their own category. The x axis shows the total of the full model, and each bar is divided to show the relative proportion of variables in the model belonging to each category. See SI Appendix, Table S7 for the amount of variance explained by each variable.
Other major differences among species emerged: For example, soil characteristics explained diversity patterns in G. suaveolens, while the geographic distribution of individuals was key in S. mannii. However, some variables were common across models, shared by up to seven species (SI Appendix, Table S7). A number of these were soil variables, including pH, depth to bedrock, and the presence of particular soil types (inceptisols and ultisols). In terms of present-day environment, precipitation was a common explanatory variable, particularly the amount of precipitation during the driest and warmest periods of the year. Stability in precipitation over time and ENM-based stability were also common to seven and five species respectively, but were generally not important explanatory factors, explaining low amounts of variance.
Discussion
Our study provides a perspective on the impact of the Pleistocene climatic fluctuations on the highly diverse flora of the Lower Guinean TRF. Using a common comparative phylogeographic framework, we tested hypotheses relating to species’ evolutionary responses (Table 1) simultaneously across seven lowland rain forest plant species exhibiting a broad range of life-history traits.
Overall, our results support the refugia hypothesis (H1; Table 1), where species have undergone periods of contraction into stable areas and expansion out of them, mainly during the Pleistocene. Indeed, as for most plant species studied to date in Lower Guinea (7, 15, 24, 36, 37), we recovered generally high levels of population structure: Four species had four or more genetic clusters (Fig. 2 and SI Appendix, Fig. S2). Even when fixing the number of genetic clusters and comparing across our species, we still found geographically disparate patterns of genetic clustering (SI Appendix, Fig. S4). In contrast to the previously mentioned studies, our results are based on the same set of genome-wide markers in each family, rather than small numbers of species-tailored markers, which can be biased toward increased levels of diversity and structuring (38).
Geographically, most genetic clusters within species are parapatric in distribution (Fig. 2). These results are in line with the prediction of past population contraction, leading to genetic differentiation (H1; Table 1), followed by range expansion (7). Secondary contact after expansion out of putative refugia has not led to genetic homogenization. Barriers to gene flow appear to have been maintained since forest contraction, leading to the patterns of genetic structure observed today. However, these clusters do not generally overlap with traditional refugia (Fig. 1), nor are they concordant among species. For all of our sampled species, genetic diversity was linked (SI Appendix, Fig. S19) to at least one or more of our three definitions of stability [traditional, climate, or habitat, (30)], favoring the refugia hypothesis (H1). A previous phylogeographic study of the palm P. barteri based on plastome data also inferred a strong correlation between habitat stability (ENMs) and unique and high genetic diversity (15) in areas along the Atlantic coast. While the same pattern was observed in this study for the same species, climate-based stability actually explained more genetic variance (SI Appendix, Fig. S19) and, therefore, played a more important role than habitat stability in P. barteri.
Using molecular dating, we showed that, in most species, genetic structure originated during the Pleistocene (Fig. 3), when climatic fluctuations increased in magnitude (39). These results are in line with the refugia hypothesis (H1), in which Pleistocene climate fluctuations play a key role in generating intraspecific population structure (7, 10). Pleistocene-dated population structuring was also inferred from other single-species studies in Central African plants (15, 36, 40, 41) and animals (17, 42, 43). Estimated divergence times within G. suaveolens, a species sampled here, based on complete plastome data, resulted in older estimates (3.5 to 4.5 Ma) of the emergence of population structure (44) than those inferred here (Fig. 3). These results were used to suggest a pre-Pleistocene origin of population structure in the region. These age discrepancies are probably the result of differences in the evolutionary dynamics of chloroplast and nuclear DNA. Indeed, age estimates of population divergences based of nuclear microsatellites in G. suaveolens (41) were similar to those recovered in the present study.
Our results, however, do not fit with a strict species-independent refugia hypothesis (H1a; Table 1). As suggested for frogs in Lower Guinea (17) and in other TRF regions, such as the Neotropics (45–48), this hypothesis appears too simplistic to explain current patterns in genetic diversity across the Lower Guinean TRF flora. Rather, our data support the relaxed species-dependent refugia hypothesis (H1b; Table 1), with our sampled plant species showing individualistic responses to Pleistocene climate fluctuations (16). In the Lower Guinean flora, this was also suggested based on single-species population-structure analyses in tree, herb, and liana species (7, 37). We note that sea-level drops during the LGM (of up to 100 m) in the Gulf of Guinea did not lead to important increases of continental land (less than 70 km; Fig. 1) and, thus, had limited impact on species evolutionary dynamics, in contrast to other TRF regions (47, 49).
A case in point of individualistic responses pertains to the temporal origin of the North–South genetic discontinuity (7) (Fig. 2H). This barrier has often been recovered in plants (7, 15, 24, 50), but seems to be less common in animals (17, 42, 43). We show that these northern/southern splits are dated to four significantly different time periods (nonoverlapping highest posterior densities [HPDs]; Fig. 3), mainly during the Pleistocene (3/4 time periods). This dominant genetic pattern in Lower Guinean plants is, thus, not the result of a single shared event (7). Instead, this discontinuity may result from similar, but asynchronous, processes linked to species-dependent traits, such as differences in phenology associated with the inverted seasons on either side of the barrier (7) or limited dispersal linked to differences in ecological niches of populations [e.g., immigrant inviability (50, 51)]. In addition, we recovered just a single, major divergence event across the inversion in each species (Fig. 3). This further supports the hypothesis that colonization across this climatic barrier is a rare event in the Lower Guinean flora (50). Nevertheless, this genetic discontinuity appears to break down further inland. Genetic clusters recovered for three species sampled in Eastern Cameroon/Gabon did not show a genetic structuring along this divide (Fig. 2 B–D and SI Appendix, Fig. S2 B–D). The causal mechanism behind this pattern remains unclear, but it could be related to reduced environmental differences between northern and southern regions further inland.
We inferred the demographic histories of all seven species, revealing two major patterns (Fig. 4). We found that Annonaceae species experienced a somewhat cyclical pattern of increases and decreases, as might be expected in a response to climatic fluctuations (Table 1). Pleistocene demographic changes, including recent demographic expansions (41), gradual declines (36), and population bottlenecks (52), have been found in several other Central African plants. Palms exhibited a general lack of population decline over the last 300 Ka (Fig. 4), which is also commonly inferred in other flora and fauna from TRFs worldwide (36, 53–56). This suggests a muted effect of Pleistocene climatic fluctuations on the long-term demographic trajectory of these species. Our results point to a disparity between plant families where glacial periods had a greater effect on Annonaceae species than palms. However, the palm dataset contained less sequence data than Annonaceae (270 kb versus 740 kb; SI Appendix, Table S1), and it may be that this reduction in data limits our ability to detect rapidly fluctuating patterns of . Genetic clusters with conspicuously stable population sizes were found in some species, but not others (Fig. 4 B–H), and the locations of those stable populations differed—lending further support to the relaxed species-specific hypothesis (H1b).
In addition to inferring the origin and dynamics of genetic structure, we tested what environmental factors have influenced the distribution of genetic diversity across species. Under a strict refugia hypothesis (H1a), we expected stability to be the main explanatory factor across species (Table 1). However, our distance-based redundancy analyses (35) did not support that prediction, as factors explaining genetic variance differed considerably across species (Fig. 5). Stability was recovered as a significant explanatory variable for each species, supporting our previous results. However, it was not the most important factor (largest sum of R2) for any species (Fig. 5). For some species, soil variables were the most important (M. enghiana, G. suaveolens, P. acaulis, and P. barteri), whereas for others, it was climate and/or geography (A. affinis, A. mannii, and S. mannii). Overall, our analyses showed that each species possesses a unique set of environmental variables that explains patterns in genetic variation (Fig. 5), favoring our hypothesis H1b.
Finally, given our variety of sampled species (SI Appendix, Table S2), we show that differences in life-history traits and phenotypes resulted in alternative evolutionary histories, supporting previous studies (18, 48, 57). However, it was also suggested that species sharing life-history traits or phenotypes, such as habit or dispersal syndromes, could respond in a similar evolutionary manner to past climate change (7). We show that this isn’t necessarily true. For example, the three tree species sampled here show differences in terms of spatial genetic structure and factors explaining the distribution of diversity (Figs. 2 A–C and 5), though the temporal origin of this structure did exhibit some similarities (Fig. 3). Differences were also evident for species with potentially similar dispersal syndromes. Given fruit morphology (58, 59) and field observation (ref. 60 and personal observation by T.L.P.C.), A. mannii and S. mannii are often dispersed by elephants (SI Appendix, Table S2), which can travel long distances and have a strong effect on population structure and recruitment (61, 62). However, population structure and evolutionary dynamics of these species fall at the two extremes of our study (Figs. 2 B and G, 3, and 4 A, C, and H), although geography is an important factor in both species (Fig. 5).
TRFs contain an estimated 50% of the world’s plant biodiversity (63). As much as one-third of the tropical African flora is potentially threatened with extinction (64), so it is critical that we understand the origins of diversity in this region (65). Our study gives a temporal context to the evolutionary dynamics for multiple Lower Guinean Central African TRF plant species. The idiosyncratic nature of genetic structure, population divergence, demographic histories, and the environmental traits underlying patterns of diversity across our seven species demonstrates that each has responded to past climate change differently, favoring a relaxed species-dependent refugia hypothesis (7, 16–18). We show that similar patterns of genetic structure across species, such as the North–South discontinuity, are likely the result of different underlying causal factors (66) (i.e., asynchronous origins [Fig. 3] and varied demographic histories [Fig. 4]). Therefore, rather than aiming for general patterns, it may be more pertinent to understand individual species’ ecology and evolutionary history, before combining these to understand how TRFs have responded to past climate change. Our framework would allow similar studies in Southeast Asian or Amazonian rain forests to investigate whether similar, species-dependent patterns are found. Our understanding of individualistic responses will be critical if we are to predict general responses to ongoing climate change and has important implications if we want to conserve those species most at risk.
Materials and Methods
Sample Collection, Library Preparation, and DNA Sequencing.
We sampled and sequenced a total of 725 individuals from four Annonaceae and three palm (Arecaceae) species across most of their ranges in Cameroon, Gabon, and Republic of the Congo (one specimen of P. barterii from Nigeria). At each collection site, we surveyed for the presence of the seven species, sampling them if found. This typically resulted in three to seven species per sampling location. These species are lowland rain forest restricted, but cover a variety of life forms, such as trees (A. affinis, A. mannii, and G. suaveolens), understory shrubs (P. barterii, P. acaulis, and S. mannii), and one liana species (M. enghiana). They also cover a range of different ecologies (inundated rain forest, terra firme, lowland, and premontane [ranges under 1,300 m]) and dispersal syndromes (elephant, rodent, ruminant, monkey, or bird dispersed). These syndromes were inferred from fruit morphology because few studies are available with concrete data; e.g., ref. 60. See SI Appendix, Table S2 for details. Methodology for library preparation, DNA sequencing, and read cleaning followed that of ref. 27 for both palms and Annonaceae species.
SNP Calling.
To call SNPs, we first used SECAPR [version (v) 1.1.4 (67)] to build a psuedoreference for each species to map reads against. After filtering out low coverage and paralogous loci, consensus contigs were built and combined to form a reference file. We mapped our cleaned, paired reads to these new, species-specific references using BWA [v0.7.12 (68)]. Duplicates were removed, and we called SNPs using the program HaplotypeCaller in GATK [v4.0 (69)]. We filtered SNPs by mapping quality (40%) depth (25), quality by depth (2), average depth across individuals (10), and minor allele frequency (0.01) to filter SNPs using bcftools [v1.8 (70)]. We kept only biallelic SNPs and excluded monomorphic sites.
Population Structure and Diversity.
We initially conducted Mantel tests between pairs of con-specific individuals using SNP data to identify whether broad-scale patterns of isolation-by-distance existed. We then performed DAPC (28) to assign individuals to genetic clusters. We used the function find.clusters in the R package “adegenet” (71) to infer the number of clusters using successive K-means with 100,000 iterations per value of k up to k = 20. We used the Bayesian information criterion (BIC) to identify the best-fitting number of clusters by examining when BIC values began increasing or change in BIC flattened off. We also conducted DAPC analyses for all values from k = 2 to k = 8. For P. acaulis, k = 1 and k = 2 had similar BIC values, and we selected k = 2 to keep at least one intraspecific divergence event per species for downstream phylogenetic analyses based on clustering results. We then used the function dapc (28) to define the diversity among the clusters identified, choosing the optimum number of axes for DAPC, initially selecting a number of axes that maximized cross-validation score and then using the function optim.a.score to select a final number of axes to use.
A second clustering approach, TESS3, differed from DAPC by incorporating geographic location data when inferring population clusters (29). TESS3 was implemented by using the R package “tess3r” with the projected least-squares algorithm and a maximum k of 10. In some species, a small number of SNPs were removed from the dataset, as they caused errors during file conversion to Latent Factor Mixed Models filetype. We identified the appropriate number of clusters by examining the cross-validation score for each value of k. Interpolations of ancestry coefficients were displayed on geographic maps by using the function plot.tess3Q.
We also calculated per-individual values of expected heterozygosity (), observed heterozygosity (), and allelic richness and compared these to distance from Maley’s refugia (calculated as distance from an approximately central point) and distance from the coastline. We performed a linear regression for each summary statistic and distance measure.
AMOVA.
This approach can be used to test how much genetic variation can be explained by a given stratification of the data. We performed two sets of AMOVAs. First, we used a hierarchical approach where we grouped individuals by genetic cluster (DAPC), then assigned these clusters to North or South. This classification was based on whether the cluster was mostly North or South of the species’ genetic discontinuity at the climatic inversion. As a result, this comparison was not possible with P. acaulis and S. mannii. Second, we grouped individuals by their proximity to stable areas. We performed separate analyses based on three different criteria: 1) distance from Maley’s refugia; 2) climatic stability (31) calculated from change in annual mean temperature and annual rainfall; and 3) stable areas, as calculated from ENMs (see below) for present and past climate. See SI Appendix for further methodological details.
Contig Assembly and Multisequence Alignment.
We used the program HybPiper (v1.2) (72) to process our cleaned data, as described in ref. 27, for phylogenetic inference. This produced “supercontigs” of the target exons plus associated off-target sequence data for each target locus. We aligned each set of supercontigs using MAFFT (v7.305) (73) with the –auto option and cleaned these alignments with GBLOCKS (v0.91b) (74) using the default parameters and all allowed gap positions. If putative paralogous sequences were flagged by HybPiper, we inferred gene trees of the corresponding loci using RAxML (v8.2.9) (75). When sequences concerning more than three individuals were flagged for a locus, we examined whether the “main” and alternative sequences formed separate clades. If so, this locus was classified as a true paralog and discarded from the dataset. To identify a suitable set of loci for reliable phylogenetic inference, we selected only those supercontigs that had 75% of their exon length reconstructed in 75% of individuals. We did this for each of our phylogenetic analyses, based on the target species (27).
Maximum-Likelihood Trees.
After suitable loci were identified, we filled any missing individuals in each alignment with an empty sequence. We then concatenated all aligned loci using the pxcat function in the program phyx (76). Separate GTR + GAMMA models were assigned to each locus using a partition file. We performed a rapid bootstrap analysis (100 replicates) followed by a thorough maximum-likelihood search in RAxML (v8.2.9) (75). This was done separately for each species, using all sequenced individuals, in addition to one to three closely related outgroup taxa.
Backbone Trees.
Dating intraspecific divergence events is difficult, as there are rarely fossils available to calibrate the tree. To find a solution to this issue and date divergence events in each of our seven focal species, we built two backbone trees, one for palms and one for Annonaceae, to be used in secondary calibration. For our Annonaceae backbone tree, we sampled 20 taxa from across all subfamilies and included a single individual from each Annonaceae focal species, producing alignments for phylogenetic inference using HybPiper, as detailed above.
We built gene trees for each locus using RAxML (v8.2.9) (75) as above and rooted our trees on Anaxagorea (27). We then estimated how clock-like each locus was by calculating root-to-tip variance for each gene tree using the program SortaDate (32). We selected the 32 loci with the lowest root-to-tip variance and used this subset for our molecular dating analysis.
We performed molecular dating using BEAST2 (v2.4.4) (77). Trees were linked across partitions, and the appropriate substitution models for each partition were identified by using modeltest-ng (v0.1.3) (78). Separate relaxed lognormal clock models were assigned to each partition. We used a uniform prior distribution from 89 to 112 Ma on the root, as recommended (79, 80), based on fossil evidence from Futabanthus and Endressinia.
The analysis was run for 100 million generations, sampling every 10,000 generations to generate a posterior distribution of 10,000 trees. Effective sample-size values were examined by using Tracer (v1.7) (81), and the appropriate amount of burn-in was removed. Multiple runs were performed and combined by using logcombiner. Treeannotator was used to generate a maximum-clade credibility tree, keeping target node heights.
We repeated this process for 23 palm taxa, spread across subfamilies Arecoideae, Ceroxyloideae, and Coryphoideae and including two or three samples for each focal species. We rooted trees on Coryphoideae taxa (21). We used three fossils for calibration, following priors in Couvreur et al. (21), but modifying the prior for crown Hyphaeninae due to a recently published fossil, Hyphaeneocarpon indicum (82) (SI Appendix, Table S4).
Dated StarBEAST and DENIM Trees.
To date divergence among genetic clusters in our focal species, we used a Bayesian approach involving the multispecies coalescent, implemented in StarBEAST (83). To improve mixing, we used the StarBEASTWithSTACEYOps template in BEAST2. When building a tree for each species, we included the closest outgroup taxa from the backbone trees. We randomly chose five individuals per genetic cluster in the focal species and defined each set of five as a different population. Subsetting in this way was previously shown to represent general patterns well (50). Outgroup individuals were assigned to a separate species (multiple species if necessary).
Like for the backbone trees, 32 loci were selected for each species based on root-to-tip variance. We used a constant population function and a coalescent exponential population tree model. Given that our analyses were among very close relatives and that we limited root-to-tip variance, we chose to use strict clock models for each locus. These models are appropriate for recently diverged lineages, given the expected low substitution-rate variation and improved computational feasibility (84, 85) when using relatively large sequence datasets. We also tested using lognormal uncorrelated relaxed clock models for each locus for A. affinis and found similar ages to those obtained with the strict clock (SI Appendix, Fig. S30).
We added uniform priors on the root where the upper and lower bounds were set to the 95% HPD intervals from the relevant node in the appropriate backbone tree. The analysis was then run as specified previously for the backbone trees. Given that the Podococcus are sister species, we analyzed them together, while all other species were analyzed separately.
We also used DENIM (33) to account for migration among different genetic clusters. We used the same input dataset as for StarBEAST, modifying priors in accordance with the DENIM manual. This yielded topologies and divergence times contingent on migration events between taxa for individual loci, which can be compared to StarBEAST results. Initial runs revealed that uniform divergence-time priors were incompatible with palm species, so normal priors were used instead. In this case, the 2.5% and 97.5% values of the normal distribution encompassed the 95% HPD intervals of the corresponding node in the palm backbone tree. See SI Appendix for more information on our dating approaches.
ENMs.
We constructed ENMs for each of the seven species using the R package “biomod2” (86) and climatic, altitude, and soil variables (SI Appendix, Table S5). We collated known occurrence points for each species from our field data, including additional individuals beyond those that were sequenced for genetic data (1,691 individuals total; SI Appendix). The background area for each species consisted of a 5° buffer around occurrence points. We inferred ENMs using five different model algorithms: generalized linear model, boosted regression trees, artificial neural networks, random forest, and MAXENT.Phillips (maxent). We evaluated model fit by using Cohen’s kappa (KAPPA) and true skill statistic (TSS). Following this, we performed ensemble modeling using all models. We then projected models (of all algorithms) into the past, performing a further ensemble modeling step. Finally we converted ensemble projections to binary presencevabsence maps using KAPPA and TSS, allowing us to identify areas suitable for each species under a given statistic and present or past climate model. Further information on our ENM approach can be found in SI Appendix, and details of model adequacy can be found in SI Appendix, Table S6.
Demographic Inference.
We used Stairway Plot (34) (v2), a model-flexible approach that uses site-frequency spectra (SFS), to infer changes in effective population size () over time. We generated VCF files for each cluster, but did not apply a minor-allele frequency filter to avoid skewing the SFS. Stairway Plot uses SNP counts to infer changes in , so the removal of SNPs with missing data may skew counts. We generated SFS for each cluster by calculating the minor-allele frequency at each SNP and multiplying this by the mean number of haploid samples, following Burgarella et al. (87). This resulted in a new SFS that used all observed site frequencies and minimized the number of SNPs removed. The total number of samples was slightly reduced based on the amount of missing data. Breakpoints were calculated as recommended in the manual. We used two-thirds of sites for training and performed 200 bootstraps. The number of observed sites was calculated as the total length of the SECAPR pseudoreference (see above) for each species. We used an angiosperm wide mutation rate of 5.35 sites per year (88) and a generation time of 15 y, based on the generation time of the Annonaceae species Annona crassiflora (89). For palms, we used a mutation rate of 2.5 and a generation time of 10 y (90). A longer generation time of up to 50 y has been proposed for some of our study species (91). Previous work in A. affinis (50) has shown that increasing generation time does not change demographic trends and has only minor effects on the timing of events, though was markedly scaled down as a result, so values of should be interpreted with caution.
db-RDA.
We used db-RDA (35) to uncover how genetic variation could be attributed to different geographic, climatic, soil, and stability variables. Genetic distances were calculated by using principal coordinates analysis and used as a response variable. We used Moran Eigenvector’s Maps (MEMs) to summarize spatial structure in the data (referred to as geography throughout). Explanatory variables included those variables used for ENMs, MEMs, several variables related to stable areas, and US Department of Agriculture soil taxonomy (SI Appendix, Table S5). We pruned correlated variables from each dataset and then built db-RDA models for each species by adding and removing variables in order to maximize the explained variance. Further details about this approach can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Prof. Moutsambote, Raoul Niangadouma, and Théophile Ayole for help in the field. We also thank Olivier Hardy and his team for sharing samples of G. suaveolens. We thank the Institut de Recherche pour le Développement (IRD) itrop High-Performance Computing (HPC) (South Green Platform) at IRD Montpellier for providing HPC resources that have contributed to the research results reported within this paper. We thank the Centre National de la Recherche Scientifique et Technologique (CENAREST), the Agence National des Parques Nationaux (ANPN), and Prof. Bourobou Bourobou for research permits (AR0020/16 and AR0036/15 [CENAREST] and AE16014 [ANPN]). Fieldwork in Cameroon was undertaken under the “accord cadre de cooperation” between the IRD and Ministère de la Recherche Scientifique et de l’Innovation. We thank Prof. Bouka Biona of the Institut National de Recherche en Sciences Excates et Naturelles of the Republic of Congo for research permits.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001018117/-/DCSupplemental.
Data Availability.
Data associated with this manuscript are stored on Dryad (92). Scripts used for analyses can be found on GitHub (93). Sequence data have been deposited in the Sequence Read Archive (94).
References
- 1.Linder H. P., et al. , The partitioning of Africa: Statistically defined biogeographical regions in sub-Saharan Africa. J. Biogeogr. 39, 1189–1205 (2012). [Google Scholar]
- 2.Droissart V., et al. , Beyond trees: Biogeographical regionalization of tropical Africa. J. Biogeogr. 45, 1153–1167 (2018). [Google Scholar]
- 3.Maley J., The African rain forest—main characteristics of changes in vegetation and climate from the Upper Cretaceous to the quaternary. Proc. Roy. Soc. Edinb. B Biol. Sci. 104, 31–73 (1996). [Google Scholar]
- 4.Cowling S. A., et al. , Simulated glacial and interglacial vegetation across Africa: Implications for species phylogenies and trans-African migration of plants and animals. Global Change Biol. 14, 827–840 (2008). [Google Scholar]
- 5.Lézine A. M. M., Izumi K., Kageyama M., Achoundong G., A 90,000-year record of Afromontane forest responses to climate change. Science 363, 177–181 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Anhuf D., et al. , Paleo-environmental change in Amazonian and African rainforest during the LGM. Palaeogeogr. Palaeoclimatol. Palaeoecol. 239, 510–527 (2006). [Google Scholar]
- 7.Hardy O. J., et al. , Comparative phylogeography of African rain forest trees: A review of genetic signatures of vegetation history in the Guineo-Congolian region. Compt. Rendus Geosci. 345, 284–296 (2013). [Google Scholar]
- 8.Diamond A. W., Hamilton A. C., The distribution of forest passerine birds and quaternary climatic change in tropical Africa. J. Zool. 191, 379–402 (1980). [Google Scholar]
- 9.Haffer J., Speciation in Amazonian forest birds. Science 165, 131–137 (1969). [DOI] [PubMed] [Google Scholar]
- 10.Hewitt G., The genetic legacy of the Quaternary ice ages. Nature 405, 907–913 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Bennett K. D., Provan J., What do we mean by ‘refugia’? Quat. Sci. Rev. 27, 2449–2455 (2008). [Google Scholar]
- 12.Bonnefille R., “Rainforest responses to past climatic tropical Africa” in Tropical Rainforest Responses to Climatic Change, Flenley J., Bush M., eds. (Springer, Berlin, Germany, 2007), pp. 117–170. [Google Scholar]
- 13.Sosef M. S. M., “Studies in Begoniaceae V. Refuge begonias: Taxonomy, phylogeny and historical biogeography of Begonia sect. Loasibegonia and sect. Scutobegonia in relation to glacial rain forest refuges in Africa” (Wageningen Agricultural University Papers, Wageningen University, Wageningen, Netherlands, 1994), vol. 94.
- 14.Dupont L. M., Donner B., Schneider R., Wefer G., Mid-Pleistocene environmental change in tropical Africa began as early as 1.05 Ma. Geology 29, 195–198 (2001). [Google Scholar]
- 15.Faye A., et al. , Phylogeography of the genus Podococcus (Palmae/Arecaceae) in Central African rain forests: Climate stability predicts unique genetic diversity. Mol. Phylogenet. Evol. 105, 126–138 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Stewart J. R., Lister A. M., Barnes I., Dalén L., Refugia revisited: Individualistic responses of species in space and time. Proc. Roy. Soc. B. Biol. Sci. 277, 661–671 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell R. C., et al. , Idiosyncratic responses to climate-driven forest fragmentation and marine incursions in reed frogs from Central Africa and the Gulf of Guinea Islands. Mol. Ecol. 26, 5223–5244 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Zamudio K. R., Bell R. C., Mason N. A., Phenotypes in phylogeography: Species’ traits, environmental variation, and vertebrate diversification. Proc. Natl. Acad. Sci. U.S.A. 113, 8041–8048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillaumet J. L., Chevillotte H., Valton C., Les forêts tropicales humides Africaines, 1: 6 000 000; Format 115 × 75 cm (Institut de Recherche pour le Développement, Bondy, France, 2009).
- 20.Couvreur T. L. P., Baker W. J., Tropical rain forest evolution: Palms as a model group. BMC Biol. 11, 48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couvreur T. L. P., Forest F., Baker W. J., Origin and global diversification patterns of tropical rain forests: Inferences from a complete genus-level phylogeny of palms. BMC Biol. 9, 44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moritz C., Faith D. P., Comparative phylogeography and the identification of genetically divergent areas for conservation. Mol. Ecol. 7, 419–429 (1998). [Google Scholar]
- 23.Hickerson M. J., Meyer C. P., Testing comparative phylogeographic models of marine vicariance and dispersal using a hierarchical Bayesian approach. BMC Evol. Biol. 8, 322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heuertz M., Duminil J., Dauby G., Savolainen V., Hardy O. J., Comparative phylogeography in rainforest trees from lower Guinea, Africa. PLoS One 9, e84307 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carnaval A. C., Hickerson M. J., Haddad C. F., Rodrigues M. T., Moritz C., Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science 323, 785–789 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Heyduk K., Trapnell D. W., Barrett C. F., Leebens-Mack J., Phylogenomic analyses of species relationships in the genus Sabal (Arecaceae) using targeted sequence capture. Biol. J. Linn. Soc. 117, 106–120 (2016). [Google Scholar]
- 27.Couvreur T. L. P., et al. , Phylogenomics of the major tropical plant family annonaceae using targeted enrichment of nuclear genes. Front. Plant Sci. 9, 1941 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jombart T., Devillard S., Balloux F., Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 11, 94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caye K., Deist T. M., Martins H., Michel O., François O., TESS3: Fast inference of spatial population structure and genome scans for selection. Mol. Ecol. Resour. 16, 540–548 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Ashcroft M. B., Identifying refugia from climate change. J. Biogeogr. 37, 1407–1413 (2010). [Google Scholar]
- 31.Blach-Overgaard A., Kissling W. D., Dransfield J., Balslev H., Svenning J. C., Multimillion-year climatic effects on palm species diversity in Africa. Ecology 94, 2426–2435 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Smith S. A., Brown J. W., Walker J. F., So many genes, so little time: A practical approach to divergence-time estimation in the genomic era. PLoS One 13, e0197433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones G. R., Divergence estimation in the presence of incomplete lineage sorting and migration. Syst. Biol. 68, 19–31 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Liu X., Fu Y. X., Exploring population size changes using SNP frequency spectra. Nat. Genet. 47, 555–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Q., Edwards D. L., Knowles L. L., Integrative testing of how environments from the past to the present shape genetic structure across landscapes. Evolution 67, 3386–3402 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Duminil J., et al. , Late Pleistocene molecular dating of past population fragmentation and demographic changes in African rain forest tree species supports the forest refuge hypothesis. J. Biogeogr. 42, 1443–1454 (2015). [Google Scholar]
- 37.Ley A. C., et al. , Comparative phylogeography of eight herbs and lianas (Marantaceae) in central African rainforests. Front. Genet. 5, 403 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Väli U., Einarsson A., Waits L., Ellegren H., To what extent do microsatellite markers reflect genome-wide genetic diversity in natural populations? Mol. Ecol. 17, 3808–3817 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Dupont L., Orbital scale vegetation change in Africa. Quat. Sci. Rev. 30, 3589–3602 (2011). [Google Scholar]
- 40.Debout G. D., Doucet J. L., Hardy O. J., Population history and gene dispersal inferred from spatial genetic structure of a Central African timber tree, Distemonanthus benthamianus (Caesalpinioideae). Heredity 106, 88–99 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piñeiro R., Dauby G., Kaymak E., Hardy O. J., Pleistocene population expansions of shade-tolerant trees indicate fragmentation of the African rainforest during the Ice Ages. Proc. Biol. Sci. 284, 20171800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Portik D. M., et al. , Evaluating mechanisms of diversification in a Guineo-Congolian tropical forest frog using demographic model selection. Mol. Ecol. 26, 5245–5263 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Charles K. L., et al. , Sky, sea, and forest islands: Diversification in the African leaf-folding frog Afrixalus paradorsalis (Anura: Hyperoliidae) of the lower Guineo-Congolian rain forest. J. Biogeogr. 45, 1781–1794 (2018). [Google Scholar]
- 44.Migliore J., et al. , Pre-Pleistocene origin of phylogeographical breaks in African rain forest trees: New insights from Greenwayodendron (Annonaceae) phylogenomics. J. Biogeogr. 46, 212–223 (2019). [Google Scholar]
- 45.Colinvaux P. A., De Oliveira P. E., Bush M. B., Amazonian and neotropical plant communities on glacial time-scales: The failure of the aridity and refuge hypotheses in Quat. Sci. Rev. 19, 141–169 (2000). [Google Scholar]
- 46.da Rocha D. G., Kaefer I. L., What has become of the refugia hypothesis to explain biological diversity in Amazonia? Ecol. Evol. 9, 4302–4309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leite Y. L. R., et al. , Neotropical forest expansion during the last glacial period challenges refuge hypothesis. Proc. Natl. Acad. Sci. U.S.A. 113, 1008–1013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paz A., Ibáñez R., Lips K. R., Crawford A. J., Testing the role of ecology and life history in structuring genetic variation across a landscape: A trait-based phylogeographic approach. Mol. Ecol. 24, 3723–3737 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Raes N., et al. , Historical distribution of Sundaland’s Dipterocarp rainforests at Quaternary glacial maxima. Proc. Natl. Acad. Sci. U.S.A. 111, 16790–16795 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helmstetter A. J., et al. , Phylogenomic data reveal how a climatic inversion and glacial refugia shape patterns of diversity in an African rain forest tree species. bioRxiv. 10.1101/807727 (24 October 2019). [DOI] [Google Scholar]
- 51.Nosil P., Vines T. H., Funk D. J., Reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution 59, 705–719 (2005). [PubMed] [Google Scholar]
- 52.Lowe A. J., Harris D., Dormontt E., Dawson I. K., Testing putative African tropical forest refugia using chloroplast and nuclear DNA phylogeography. Trop. Plant Biol. 3, 50–58 (2010). [Google Scholar]
- 53.Morgan K., et al. , Comparative phylogeography reveals a shared impact of Pleistocene environmental change in shaping genetic diversity within nine Anopheles mosquito species across the Indo-Burma biodiversity hotspot. Mol. Ecol. 20, 4533–4549 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Allen J. M., et al. , Primate DNA suggests long-term stability of an African rainforest. Ecol. Evol. 2, 2829–2842 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Batalha-Filho H., Cabanne G. S., Miyaki C. Y., Phylogeography of an Atlantic forest passerine reveals demographic stability through the last glacial maximum. Mol. Phylogenet. Evol. 65, 892–902 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Paz A., et al. , Phylogeography of Atlantic Forest glassfrogs (Vitreorana): When geography, climate dynamics and rivers matter. Heredity 122, 545–557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duminil J., et al. , Can population genetic structure be predicted from life-history traits? Am. Nat. 169, 662–672 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Lissambou B. J., et al. , Taxonomic revision of the African genus Greenwayodendron (Annonaceae). PhytoKeys 114, 55–93 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le Thomas A., “Annonacées” in Flore du Gabon, Aubréville A., Ed. (Muséum National d’Histoire Naturelle, Paris, France, 1969), vol. 16, pp. 1–371. [Google Scholar]
- 60.Gautier-Hion A., et al. , Fruit characters as a basis of fruit choice and seed dispersal in a tropical forest vertebrate community. Oecologia 65, 324–337 (1985). [DOI] [PubMed] [Google Scholar]
- 61.Blake S., Deem S. L., Mossimbo E., Maisels F., Walsh P., Forest elephants: Tree planters of the Congo. Biotropica 41, 459–468 (2009). [Google Scholar]
- 62.Terborgh J., et al. , Megafaunal influences on tree recruitment in African equatorial forests. Ecography 39, 180–186 (2016). [Google Scholar]
- 63.Eiserhardt W. L., Couvreur T. L. P., Baker W. J., Plant phylogeny as a window on the evolution of hyperdiversity in the tropical rainforest biome. New Phytol. 214, 1408–1422 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Stévart T., et al. , A third of the tropical African flora is potentially threatened with extinction. Sci. Adv. 5, eaax9444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pauls S. U., Nowak C., Bálint M., Pfenninger M., The impact of global climate change on genetic diversity within populations and species. Mol. Ecol. 22, 925–946 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Soltis D. E., Morris A. B., McLachlan J. S., Manos P. S., Soltis P. S., Comparative phylogeography of unglaciated eastern North America. Mol. Ecol. 15, 4261–4293 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Andermann T., Cano Á., Zizka A., Bacon C., Antonelli A., SECAPR—A bioinformatics pipeline for the rapid and user-friendly processing of targeted enriched Illumina sequences, from raw reads to alignments. PeerJ Preprints 10.7287/peerj.preprints.26477v3 (13 February 2018). [DOI] [PMC free article] [PubMed]
- 68.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McKenna A., et al. , The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H., Barrett J., A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jombart T., Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Johnson M. G., et al. , HybPiper: Extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl. Plant Sci. 4, 1600016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Castresana J., Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- 75.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown J. W., Walker J. F., Smith S. A., Phyx: Phylogenetic tools for Unix. Bioinformatics 33, 1886–1888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bouckaert R., et al. , BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Darriba D., Posada D., Kozlov A. M., Stamatakis A., Flouri T., ModelTest-NG : A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37, 291–294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pirie M. D., Doyle J. A., Dating clades with fossils and molecules: The case of Annonaceae. Bot. J. Linn. Soc. 169, 84–116 (2012). [Google Scholar]
- 80.Thomas D. C., et al. , The historical origins of palaeotropical intercontinental disjunctions in the pantropical flowering plant family Annonaceae. Perspect. Plant Ecol. Evol. Systemat. 17, 1–16 (2015). [Google Scholar]
- 81.Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A., Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matsunaga K. K. S., Manchester S. R., Srivastava R., Kapgate D. K., Smith S. Y., Fossil palm fruits from India indicate a Cretaceous origin of Arecaceae tribe Borasseae. Bot. J. Linn. Soc. 190, 260–280 (2019). [Google Scholar]
- 83.Heled J., Drummond A. J., Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27, 570–580 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weir J. T., Schluter D., Calibrating the avian molecular clock. Mol. Ecol. 17, 2321–2328 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Brown R. P., Yang Z., Rate variation and estimation of divergence times using strict and relaxed clocks. BMC Evol. Biol. 11, 271 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thuiller W., Lafourcade B., Engler R., Araujo M. B., BIOMOD-A platform for ensemble forecasting of species distributions. Ecography 32, 369–373 (2009). [Google Scholar]
- 87.Burgarella C., et al. , A Western Sahara centre of domestication inferred from pearl millet genomes. Nat. Ecol. Evol. 2, 1377–1380 (2018). [DOI] [PubMed] [Google Scholar]
- 88.De La Torre A. R., Li Z., Van De Peer Y., Ingvarsson P. K., Contrasting rates of molecular evolution and patterns of selection among gymnosperms and flowering plants. Mol. Biol. Evol. 34, 1363–1377 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Collevatti R. G., Telles M. P., Lima J. S., Gouveia F. O., Soares T. N., Contrasting spatial genetic structure in Annona crassiflora populations from fragmented and pristine savannas. Plant Systemat. Evol. 300, 1719–1727 (2014). [Google Scholar]
- 90.Gros-Balthazard M., et al. , The discovery of wild date palms in Oman reveals a complex domestication history involving centers in the Middle East and Africa. Curr. Biol. 27, 2211–2218.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Baker T. R., et al. , Fast demographic traits promote high diversification rates of Amazonian trees. Ecol. Lett. 17, 527–536 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Helmstetter A. J., Béthune K., Kamdem N. G., Sonké B., Couvreur T. L. P., Data from: Individualistic evolutionary responses of central African rain forest plants to Pleistocene climatic fluctuations. Dryad. 10.5061/dryad.1rn8pk0rk. Deposited 16 November 2020. [DOI] [PMC free article] [PubMed]
- 93.Helmstetter A. J., Béthune K., Kamdem N. G., Sonké B., Couvreur T. L. P., AFRODYN. Github. https://github.com/ajhelmstetter/afrodyn. Deposited 13 November 2020.
- 94.Helmstetter A. J., Béthune K., Kamdem N. G., Sonké B., Couvreur T. L. P., Annickia affinis: AFRODYN data. Sequence Read Archive. http://www.ncbi.nlm.nih.gov/bioproject/PRJNA541395. Deposited 15 November 2020.
- 95.General Bathymetric Chart of the Oceans (GEBCO) Compilation Group, GEBCO 2020 grid. 10.5285/a29c5465-b138-234d-e053-6c86abc040b9. Accessed 24 November 2020. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this manuscript are stored on Dryad (92). Scripts used for analyses can be found on GitHub (93). Sequence data have been deposited in the Sequence Read Archive (94).