Significance
The synthesis of a eukaryotic messenger RNA molecule involves the association of RNA polymerase and dozens of accessory proteins on DNA. We used differently colored fluorescent dyes to tag DNA, RNA polymerase II, and the elongation factor Spt4/5 in yeast nuclear extract and then observed the assembly and dynamics of individual molecules of the proteins with single DNA molecules by microscopy. The observations quantitatively define an overall pathway by which transcription complexes form and evolve during activator-dependent transcription. They also suggest how Spt4/5 dynamics might promote efficient RNA production.
Keywords: CoSMoS, Saccharomyces cerevisiae, Gal4-VP16
Abstract
In eukaryotes, RNA polymerase II (RNApII) transcribes messenger RNA from template DNA. Decades of experiments have identified the proteins needed for transcription activation, initiation complex assembly, and productive elongation. However, the dynamics of recruitment of these proteins to transcription complexes, and of the transitions between these steps, are poorly understood. We used multiwavelength single-molecule fluorescence microscopy to directly image and quantitate these dynamics in a budding yeast nuclear extract that reconstitutes activator-dependent transcription in vitro. A strong activator (Gal4-VP16) greatly stimulated reversible binding of individual RNApII molecules to template DNA. Binding of labeled elongation factor Spt4/5 to DNA typically followed RNApII binding, was NTP dependent, and was correlated with association of mRNA binding protein Hek2, demonstrating specificity of Spt4/5 binding to elongation complexes. Quantitative kinetic modeling shows that only a fraction of RNApII binding events are productive and implies a rate-limiting step, probably associated with recruitment of general transcription factors, needed to assemble a transcription-competent preinitiation complex at the promoter. Spt4/5 association with transcription complexes was slowly reversible, with DNA-bound RNApII molecules sometimes binding and releasing Spt4/5 multiple times. The average Spt4/5 residence time was of similar magnitude to the time required to transcribe an average length yeast gene. These dynamics suggest that a single Spt4/5 molecule remains associated during a typical transcription event, yet can dissociate from RNApII to allow disassembly of abnormally long-lived (i.e., stalled) elongation complexes.
As the initiating event in eukaryotic gene expression, mRNA transcription by RNA polymerase II (RNApII) is regulated at several steps (1–3). First, gene-specific transcription activators recognize specific DNA binding sites upstream of the transcription start site. Once bound, these activators recruit histone acetyltransferases and chromatin remodelers to move promoter-proximal nucleosomes, making the DNA accessible for RNApII preinitiation complex (PIC) formation. Activators further accelerate preinitiation complex (PIC) formation by recruiting the basal initiation factors via contacts with the Mediator/RNApII complex and TFIID. Once assembled, the PIC undergoes several ATP-dependent rearrangements, including promoter melting and phosphorylation of the RNApII largest subunit Rpb1. Phosphorylation of the Rpb1 C-terminal domain (CTD) releases Mediator, facilitating conversion to a transcription elongation complex (EC). During elongation, a series of CTD kinases and phosphatases creates dynamic phosphorylation patterns that differentially recruit mRNA processing and chromatin modifying enzymes to early and late stages of transcription. Transcription can be regulated at the level of PIC assembly, by controlling the transition to elongation, or by early termination events that attenuate gene expression.
The PIC-to-EC transition requires the exchange of initiation and elongation factors that contact various surfaces of RNApII. For example, structural studies show that the elongation factor Spt4/5 (also known as DRB sensitivity-inducing factor [DSIF] in metazoans) and the initiation factor TFIIE occupy overlapping space on RNApII. The archaeal Spt5 and TFE (TFIIE) homologs compete for binding to the RNAP clamp, a mobile domain that regulates template DNA movement into or out of the polymerase active site cleft (4). Similarly, the TFIIF initiation factor and Paf1 complex elongation factor may also bind RNApII mutually exclusively (5).
Eukaryotic Spt5 consists of a NusG N-terminal (NGN) domain, three to five C-terminal Kyprides-Ouzounis-Woese (KOW) domains, and a C-terminal region (CTR) that is phosphorylated during transcription elongation (6). In a manner analogous to its bacterial homolog NusG, the Spt5 NGN and KOW1 domains bridge the gap across the RNApII cleft in the EC (5). Spt4, although nonessential for viability in yeast and not conserved in bacteria, dimerizes with the NGN domain to stabilize this interaction. A large body of data shows that Spt4/5 acts to positively regulate elongation (6). Both bacterial and yeast Spt5 homologs increase the RNApII elongation rate by reducing transcriptional pausing (6). Human Spt4/5 overcomes promoter-proximal pausing induced by the Negative Elongation Factor, NELF (7). Chromatin immunoprecipitation experiments in Saccharomyces cerevisiae show decreased RNApII density at the 3′ end of genes in strains mutated in Spt4/5 (8–11). Stimulation of elongation by Spt4/5 may involve multiple mechanisms, including stabilization of a more processive RNApII conformation, steric blockage of template release, interaction between KOW domains and the RNA transcript (12, 13), and recruitment of additional elongation factors to the KOW and CTR domains (6, 14–16).
To study the factors that drive transcription activation, PIC formation, and transition to elongation, we have used quantitative mass spectrometry to analyze transcription complexes in vitro. Complexes were assembled on an immobilized DNA transcription template in yeast nuclear extract (17). This system uses a model promoter consisting of the CYC1 core promoter driven by five upstream Gal4 binding sites. We used the synthetic protein Gal4-VP16, which fuses the Gal4 DNA binding domain to the herpes simplex virus VP16 activation domain, resulting in a powerful transcription activator in both mammalian (18) and yeast systems (19). We previously showed that the Gal4 DNA binding domain, when fused to either VP16 or the native yeast Gcn4 activation domain, strongly recruits the NuA4 and SAGA histone acetyltransferases, the Swi/Snf remodeling complex, and Mediator and other PIC components, to both naked DNA and chromatinized templates (17). Furthermore, this system faithfully recapitulates the transition to elongation and the exchange of factors mediated by the Rpb1 CTD phosphorylation cycle (20). While this system has proven useful for studying dynamics, the time resolution of the mass spectrometry experiments was on the order of minutes rather than seconds. Furthermore, since the transcription complexes were analyzed in bulk, results represent the averaged behaviors of the molecular ensemble, yielding only incomplete information about the pathways of complex assembly.
To provide increased time resolution and the ability to monitor reaction pathways of activated transcription events on individual DNA molecules, here we adapt the immobilized template assay for single-molecule light microscopy. Colocalization single-molecule spectroscopy (CoSMoS) is a multiwavelength single-molecule fluorescence method well suited for characterizing molecular mechanisms (21, 22). We have previously used this approach to analyze the pathways of molecular complex formation and function in bacterial transcription and pre-mRNA splicing (22–29). By fluorescently labeling the endogenous RNApII and Spt4/5 in nuclear extract, as well as the DNA template, with three differently colored dyes, we can monitor the arrival and departure of the individual protein molecules from the template DNA. Because these experiments are done in nuclear extracts, they complement other single-molecule studies of RNApII transcription that used a limited set of purified factors (30). As the full complement of nuclear proteins is present, the experimental system can recapitulate a more complete set of transcription processes and thus provide a better approximation of events in vivo. Conversely, the approach overcomes some of the technical challenges of single-molecule studies of transcription in live cells (31), allowing more complete kinetic analysis of multiple components associating simultaneously with an isolated DNA molecule leading up to and during the synthesis of a single RNA molecule.
Our experiments, together with numerical modeling of the dynamics seen in the experiments, quantitatively define a minimal kinetic mechanism for activator-dependent transcription in this system. The analysis distinguishes productive and nonproductive RNApII binding and defines key rate-limiting steps in initiation. Furthermore, the experiments reveal reversible association of Spt4/5 with transcription elongation complexes and suggest that these dynamics are tuned to the timescale of yeast gene transcription.
Results
A System for Imaging RNApII and Spt4/5 Binding to Promoter DNA.
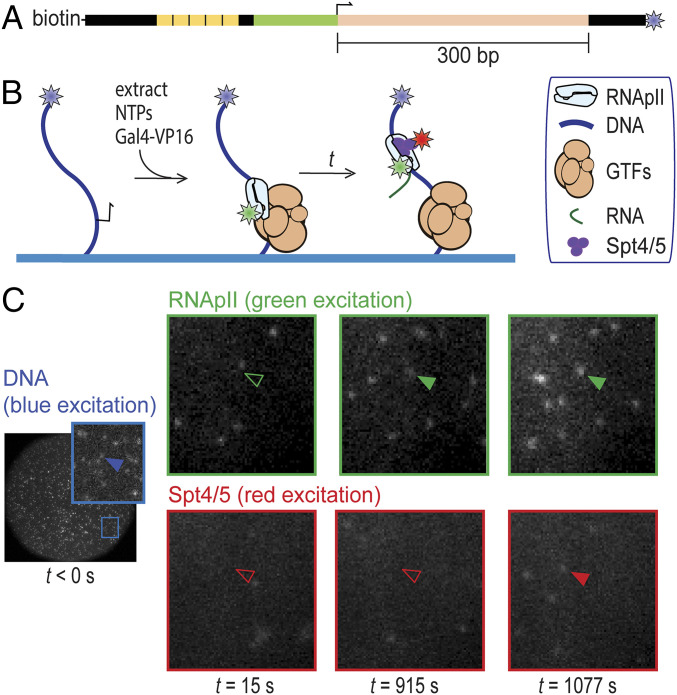
To detect transcription complex formation by single-molecule fluorescence, we constructed a S. cerevisiae strain in which the Rpb1 subunit of RNApII is fused to a C-terminal SNAP tag (32) and the Spt5 subunit of Spt4/5 is fused to a C-terminal dihydrofolate reductase (DHFR) tag (33). The tag-containing strains displayed robust growth and the tagged proteins were expressed at levels similar to the corresponding untagged proteins in the parental strain (SI Appendix, Fig. S1). Rpb1-SNAP in the extract was labeled with SNAP-Surface 549, a SNAP substrate conjugated to the green-excited dye DY549. Imaged transcription reactions also included Cy5-trimethoprim (Cy5-TMP) to noncovalently label the DHFR-tagged Spt5 subunit of Spt4/5 with the red-excited dye Cy5. To observe activated transcription, we used a DNA template containing five Gal4 binding sites upstream of the CYC1 core promoter, which drives a downstream 300-bp cassette encoding a G-less RNA (Fig. 1A). The multiple Gal4 sites were included to increase the extent of activation (34) so that activated transcription predominates over any basal transcription on this template. On supercoiled (SI Appendix, Fig. S1 C and D) or linear templates (20), transcripts were detected upon addition of the transcription activator Gal4-VP16 (18) and the nucleoside triphosphates (NTPs) ATP, UTP, CTP, and 3′-O-methyl-GTP. The omission of GTP suppresses production of nonspecific transcripts initiating from cryptic promoters and DNA ends.
Fig. 1.
Detection of RNApII and Spt4/5 binding to individual surface-tethered DNA488 molecules in Rpb1SNAP549/Spt5DHFR-Cy5 S. cerevisiae nuclear extract. (A) Schematic of DNA488 transcription template. This DNA contains five upstream Gal4 binding sites (yellow) and the CYC1 core promoter (green) with its transcription start site (bent arrow), followed by a 300-bp cassette (pink) encoding a G-less RNA. The template has attached biotin and AF488 dye (blue star) moieties. (B) Experimental scheme. DNA488 molecules immobilized on the surface of a flow chamber (blue) were at time t = 0 incubated with yeast nuclear extract containing dye (stars)-labeled proteins Rpb1SNAP549 and Spt5DHFR-Cy5 along with unlabeled general transcription factors (GTFs) and other nuclear proteins. Reactions were supplemented with recombinant Gal4-VP16 activator. RNApII and Spt4/5 binding to DNA were detected as colocalization of spots of green- and red-excited fluorescence at locations of blue-excited DNA spots. (C) Images of the same microscope field of view (65 × 65 µm) in the red-, green-, and blue-excited fluorescence channels taken at various times before (blue) and after (red and green) extract addition at time t = 0. Insets show magnified views of the marked region. Absence or presence of a spot of fluorescence colocalized with a particular DNA molecule are shown by open and filled arrowheads, respectively.
To visualize RNApII and Spt4/5 association with individual DNA molecules, transcription templates were PCR amplified using an upstream oligonucleotide labeled with biotin and downstream oligo carrying a blue-excited dye (Fig. 1A). The templates were tethered to the surface of a glass flow chamber and then observed by micromirror multiwavelength total internal reflection fluorescence microscopy (21). DNA locations were mapped in the blue channel (DNA488), as were a randomly selected control set of “no-DNA” spots used to monitor background binding. Labeled extract, Gal4-VP16, and NTPs (ATP, UTP, CTP, and 3′-O-methyl-GTP) were then added to begin the reaction (Fig. 1B). Candidate binding events for RNApII and/or Spt4/5 were detected in the green (Rpb1SNAP549) and red (Spt5DHFR-Cy5) fluorescence channels, respectively (Fig. 1C).
Activator Dependence of RNApII Recruitment.
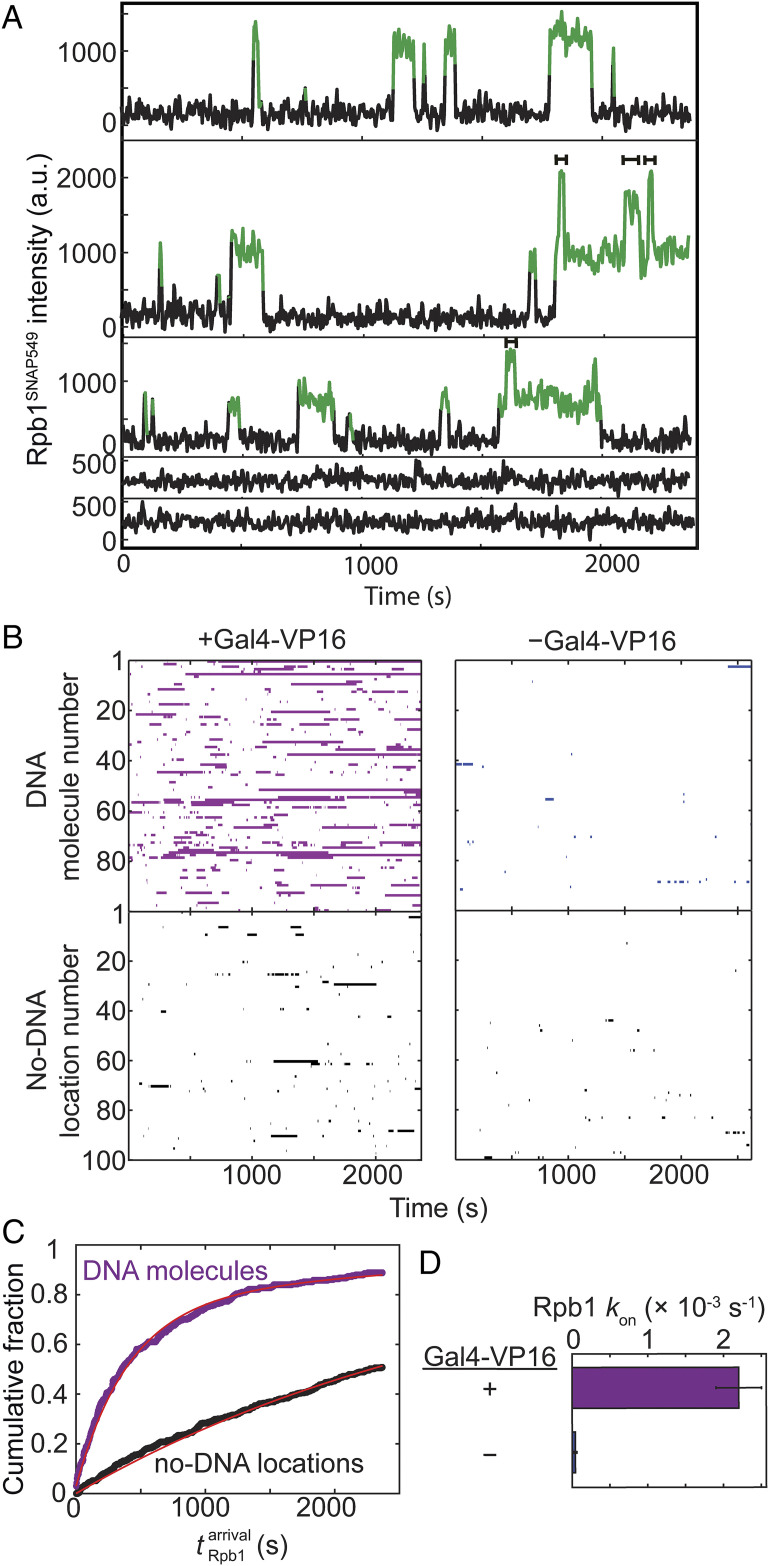
Time records of Rpb1SNAP549 fluorescence at individual DNA locations showed association of single or sometimes two or more RNApII molecules with the template (Fig. 2A). Of n = 331 DNA488 molecules observed in a representative experiment, 294 (89%) exhibited binding of at least one Rpb1SNAP549 molecule over the course of the 40-min recording. Control no-DNA locations showed far fewer Rpb1SNAP549 binding events (Fig. 2 A and B), confirming that most binding is DNA specific.
Fig. 2.
Kinetics of RNApII association with promoter DNA in the experiment shown in Fig. 1. (A) Example time records showing Rpb1SNAP549 fluorescence colocalized at the positions of three different single DNA488 molecules (Top three records) and two control locations without visible DNA488 molecules (Bottom two). Green points designate times at which there was a colocalized spot of Rpb1SNAP549 fluorescence as detected by an objective spot-recognition algorithm (60). Brackets mark time intervals during which more than one Rpb1SNAP549 molecule was present. (B) Rastergrams of Rpb1SNAP549 colocalization at the locations of 100 randomly selected DNA488 molecules (Top) and 100 randomly selected control no-DNA locations (Bottom) from the same recording. Each horizontal line of the plot is data from a single location showing times with (colored bars) or without (white space) a colocalized Rpb1SNAP549 fluorescent spot. Data are from the experiment of Fig. 1 (purple) plus a negative control experiment in which no Gal4-VP16 activator was added (blue). (C) Cumulative distribution of the time intervals prior to the first Rpb1SNAP549 association seen on each DNA488 molecule (purple) and at no-DNA488 control locations (black) in the presence of Gal4-VP16, along with fits (red) to an exponential binding model (a single rate-limiting step). Parameters of this and analogous fits to the negative control and the numbers of observations for all fits are given in SI Appendix, Table S1. (D) Apparent first-order rate constants (± SE) for Rpb1 association with template DNA derived from the model fits.
Measurement of the delay time from the addition of extract to first observed binding Rpb1SNAP549 on each DNA488 molecule revealed an exponential distribution with an apparent first-order association rate constant of (2.2 ± 0.3) × 10−3 s−1 (Fig. 2 C and D). This number usually was within a factor of ∼2.5 in experiments using different extracts, including those where Rpb1 was tagged with DHFR instead of SNAP, suggesting that extract batch and tag identity did not greatly alter binding kinetics (SI Appendix, Table S1). These results are consistent with initial RNApII association being dominated by a single rate-limiting step under these experimental conditions. The binding rate constant was reduced by more than an order of magnitude in the absence of Gal4-VP16 (Fig. 2 B and D, blue), demonstrating that the vast majority of RNApII DNA binding detected in this system is activator dependent.
The mean lifetime for RNApII occupancy of DNA in the presence of NTPs and activator was 79 ± 2 s (SE; n = 1,259), although some individual Rpb1 molecules remained associated with DNA for hundreds of seconds (Fig. 2 A and B). However, multiple molecules of Rpb1SNAP549 were sometimes observed simultaneously on an individual DNA (Fig. 2A), so the average lifetime of individual molecules may be shorter. Multiple RNApII molecules might reflect simultaneous presence of initiation and elongation complexes, binding at sequences other than the promoter, or DNA-end binding. When NTPs were absent, we still observed significant RNApII association with DNA (including some multiple binding); a similar result was obtained in the presence of NTPs when the inhibitor α-amanitin was added (SI Appendix, Fig. S2 and Table S1). These results are consistent with the ability of RNApII to participate in preinitiation complexes under these conditions, even though NTP depletion or α-amanitin both block productive transcription.
Spt4/5 Binding Is Specific to Elongation Complexes.
In the same experiment monitoring Rpb1SNAP549, the arrival of Spt4/5 was visualized by colocalization of Spt5DHFR-Cy5 with the DNA488 template molecules (Figs. 1 and 3A and SI Appendix, Fig. S3). Neither RNApII nor Spt4/5 kinetics were affected by tagging of the other protein (SI Appendix, Tables S1 and S2), suggesting that the tagged proteins behave similarly to wild type.
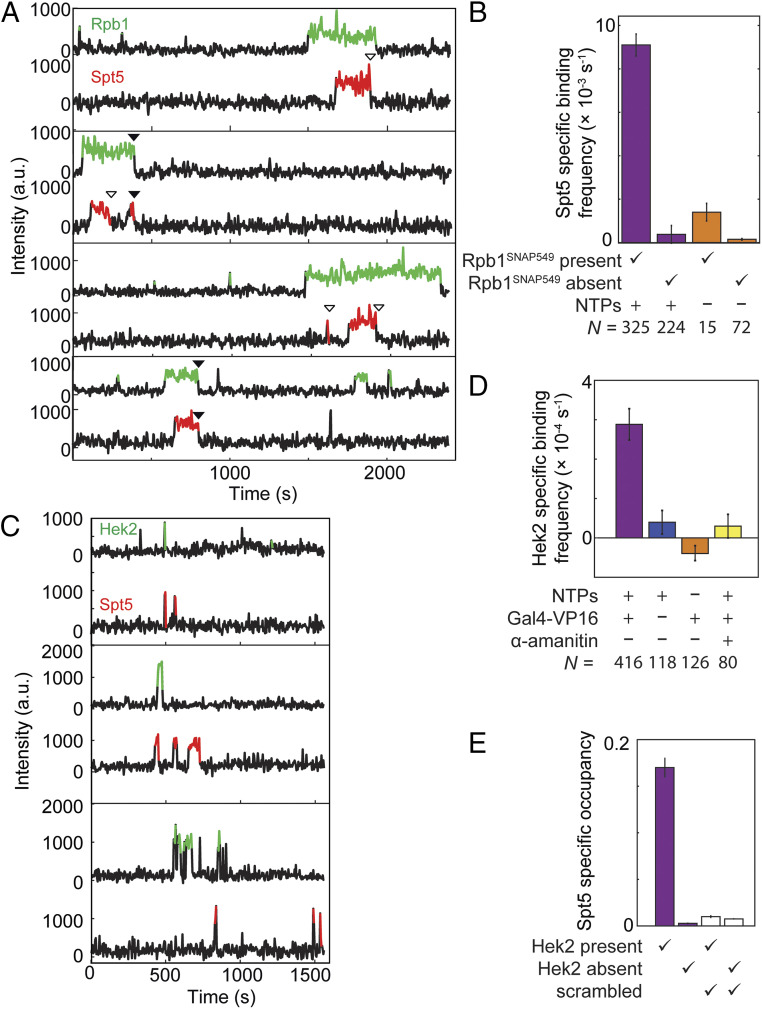
Fig. 3.
Correlation of Spt4/5 binding at individual DNA molecules with binding of RNApII (A and B) or Hek2 (C–E). (A) Example time records of Rpb1SNAP549 and Spt5DHFR-Cy5 fluorescence at four individual DNA488 locations, taken from the Rpb1SNAP549/Spt5DHFR-Cy5 experiment shown in Fig. 1. Colored intervals indicate times at which a fluorescent Rpb1SNAP549 (green) and/or Spt5DHFR-Cy5 (red) spot colocalized to the DNA488 molecule being monitored. Each Spt5DHFR-Cy5 departure is marked according to whether it occurred before (open triangles) or simultaneously with (closed triangles) Rpb1SNAP549 departure. Additional example records are shown in SI Appendix, Fig. S3. (B) DNA-specific binding frequencies (± SE) of Spt5DHFR-Cy5, calculated separately for time intervals when Rpb1SNAP549 was present or absent at the DNA. DNA-specific binding frequency is calculated as the frequency seen at DNA locations that is in excess of the background nonspecific binding seen at no-DNA locations. (C) Examples of time records of Hek2SNAP549 and Spt5DHFR-Cy5 fluorescence over time at three individual DNA488 locations taken from an experiment using a dual-tagged Hek2SNAP549/Spt5DHFR-Cy5 yeast nuclear extract in the presence of NTPs and Gal4-VP16. (D) DNA-specific binding frequencies (± SE) of Hek2SNAP549 under transcription conditions (purple) and in negative controls (blue, orange, and yellow); see SI Appendix, Table S3. (E) Mean fraction (± SEM) of time that a DNA488 molecule had a colocalized Spt5DHFR-Cy5, after correction for nonspecific surface binding. Separate analyses were conducted for time points at which Hek2SNAP549 was present or absent (SI Appendix, Table S4). Data may underestimate the true occupancy of DNA locations by Spt4/5 due to possible incomplete labeling of Spt5DHFR-Cy5, but this factor is constant across all four bars. Open bars show results of a scrambled negative control analysis of the same data (SI Appendix, Materials and Methods) showing that the correlation of Spt5 binding with Hek2 binding is not a statistical artifact. Data in D (purple bar) and E are aggregated from the experiment in C and one additional replicate.
Spt4/5 binding at the location of a particular DNA molecule almost always occurred while RNApII was detected on that DNA (Fig. 3A and SI Appendix, Fig. S3). In particular, Spt5DHFR-Cy5 DNA-specific binding (i.e., binding in excess of the background seen at no-DNA locations) occurred >10-fold more frequently when an Rpb1SNAP549 spot was present than when absent (Fig. 3B, purple bars). The rare occurrences of Spt5DHFR-Cy5 binding without Rpb1SNAP549 on the DNA (33 of 549 total Spt5DHFR-Cy5 binding events) may reflect nonspecific binding to the slide surface or specific binding to unlabeled or photobleached Rpb1SNAP549. Based on these observations, at least 94 ± 1% of the RNApII molecules in the extract were fluorescent, consistent with observations that dye labeling of Rpb1SNAP was saturated under the labeling conditions used (SI Appendix, Fig. S4). Even though Spt4/5 contacts both RNApII protein and template DNA in ECs (13, 35), the colocalization data suggest that Spt4/5 recruitment to DNA requires the presence of RNApII. In agreement with this idea, we almost invariably (288 of 312 instances) saw that RNApII arrived on DNA before Spt4/5 when complexes that contained both proteins were formed.
Consistent with a direct interaction between Spt4/5 and RNApII, 42 out of 312 colocalized Rpb1SNAP549/Spt5DHFR-Cy5 spots departed from the DNA simultaneously, within experimental time resolution (i.e., within ±1 video frame; Fig. 3A, filled arrows), as expected for an RNApII•Spt4/5 complex dissociating from the DNA as a unit. In most other events (241 of the 270 colocalized Rpb1SNAP549 and Spt5DHFR-Cy5 spots without simultaneous departure), Spt4/5 departed before RNApII dissociation. Based on all these results, we conclude that most (and possibly all, if the exceptions reflect Rpb1SNAP549 photobleaching) Spt4/5 binding is dependent on the presence of DNA-bound RNApII.
Some key Spt4/5 contacts with RNApII seen in EC structural models appear prohibited in the PIC due to obstruction by TFIIE or other PIC components (13, 36, 37), and multiple observations suggest that Spt4/5 associates stably with ECs but not PICs (20, 38). Consistent with this suggestion, we saw that the frequency with which Spt5DHFR-Cy5 associated with a DNA containing Rpb1SNAP549 was more than sixfold higher in the presence of added NTPs than in NTP depletion conditions in which ECs cannot form [(9.4 ± 0.5) ×10−3 s−1 vs. (1.4 ± 0.1) ×10−3 s−1, respectively; Fig. 3B]. Simultaneous occupancy of a DNA by multiple Spt5DHFR-Cy5 molecules was almost never seen (2 out of 331 DNA molecules examined). Taken together, these data suggest that Spt4/5 is interacting with ECs and that at most one EC is typically present on each DNA in these experiments.
As an independent test for Spt5 binding to ECs, we used an Spt5DHFR-Cy5/Hek2SNAP549 nuclear extract to monitor colocalization of Spt4/5 with Hek2, an RNA binding protein known to associate with elongation complexes generated in the nuclear extract system used here (20, 39). In contrast to Spt5DHFR-Cy5, Hek2SNAP549 association with surface-tethered DNA locations was much shorter-lived (Fig. 3C), possibly due to low affinity or competition by other RNA binding proteins for nascent transcript binding. Nevertheless, Hek2SNAP549 colocalization with DNA was transcription dependent, as no significant binding over background was detected in controls lacking activator or NTPs or containing α-amanitin (Fig. 3D). Thus, we propose that DNA-specific presence of Hek2SNAP549 can serve as a proxy for detection of an EC-associated transcript. Consistent with Spt4/5 binding being restricted to ECs, the probability of Spt5DHFR-Cy5 being present simultaneously with Hek2SNAP549 was more than 10-fold higher than that for Spt5DHFR-Cy5 presence during intervals when Hek2SNAP549 was absent (Fig. 3E). To exclude the possibility that the apparent association between Spt4/5 and Hek2 presence resulted from coincidental independent binding events, the Spt5DHFR-Cy5 and Hek2SNAP549 occupancy records from different individual DNAs molecules were randomly paired and the analysis repeated (“scrambled” in Fig. 3E and SI Appendix, Table S4). The correlation was completely lost, confirming that the observed Hek2 and Spt4/5 colocalization represents binding to the same molecular complex. Thus, the Hek2 data provide additional support that the observed Spt4/5 binding is specific for ECs.
Dynamics of Initiation and Elongation Complex Formation.
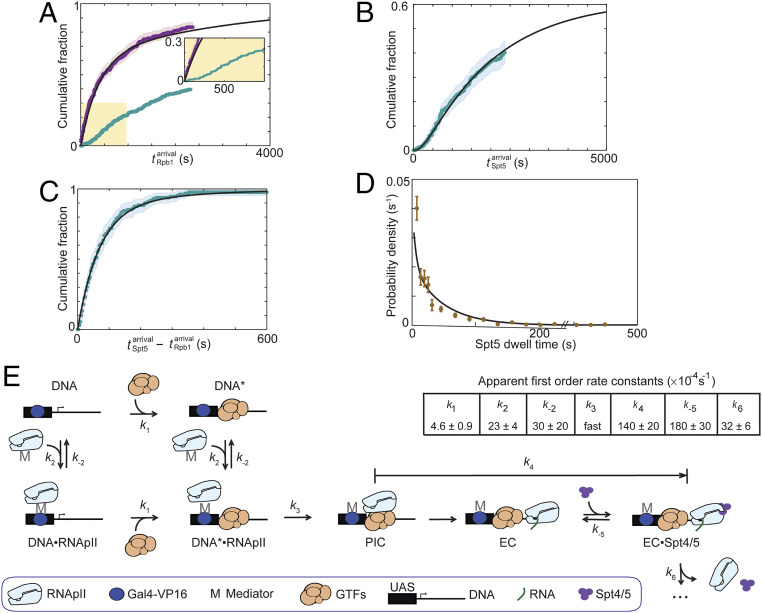
Despite most DNAs having one or more Rpb1SNAP549 binding events (Figs. 2C and 4A, purple), only a subpopulation of these events showed colocalization with Spt5DHFR-Cy5 (examples are marked “p” in SI Appendix, Fig. S3). As discussed above, the exponential distribution of initial Rpb1SNAP549 binding times (Fig. 2C) indicates a single rate-limiting step for association. Strikingly, this was not the case for times to the first productive RNApII associations (defined as the first Rpb1SNAP549 binding event during which Spt5DHFR-Cy5 was also colocalized; Fig. 4A, turquoise). These productive events exhibited a nonexponential distribution that increases steeply only after a distinct lag time of ∼100 s. Such a distribution is inconsistent with a simple model in which all RNApII associations have the same chance of forming an Spt4/5-containing EC. Instead, they suggest an additional slow event must occur during incubation of DNA template with extract to allow elongation complex production. In such a situation, later binding RNApII molecules are more likely to produce elongation complexes, resulting in the observed lag kinetics.
Fig. 4.
RNApII and Spt4/5 binding dynamics during activated transcription initiation in Rpb1SNAP549/Spt5DHFR-Cy5 extract. (A) Cumulative distribution of n = 268 time intervals measured from the addition of extract (t = 0) until the first Rpb1SNAP549 binding was observed at each DNA molecule. Separate curves show the distributions for all Rpb1SNAP549 first binding events (purple; same data as in Fig. 2C); and for the n = 129 first productive events (i.e., the first Rpb1SNAP549 binding event during which Spt5DHFR-Cy5 was also seen to bind) (turquoise). Inset: Magnified view. (B) Cumulative distribution of n = 129 time intervals from addition of extract until Spt5DHFR-Cy5 binding in the first productive events. (C) Cumulative distribution of n = 129 time differences between the first Spt5DHFR-Cy5 binding and Rpb1SNAP549 binding in the first productive events. Note expanded time scale compared to A and B. Shading in A–C indicates 95% confidence interval. (D) Distribution (probability density ± SE) of Spt5DHFR-Cy5 dwell times during productive binding events (brown; n = 312). (E) Minimal kinetic scheme consistent with the data in A–D and the rate constants determined by globally fitting those data to the scheme. Complexes represented in the schematics are plausible suggestions but are not intended to convey conclusive details of molecular structures or interactions. In the scheme shown, RNApII can bind either at the upstream Gal4 binding sites (UAS), presumed to be via an indirect interaction through Mediator and Gal4-VP16, or at the promoter, presumed to require a minimal set of RNApII general transcription factors (GTFs). Arrows represent the rate-limiting step in each transformation; additional non-rate-limiting processes are not shown and are ignored in the quantitative modeling. Rate constants k1, k2, k−2, and k4 were determined by global fitting of the model to data underlying A–C, and k−5 and k6 by fitting to the data underlying D and the measured partition ratio (SI Appendix, Materials and Methods). k3 was fixed to a value (1 s−1) faster than the experimental time resolution because the data show no evidence for a rate limitation by this step. For simplicity, this illustration does not show steps in which Rpb1SNAP549 or Spt5DHFR-Cy5 molecules bound nonspecifically to the slide surface, but these kinetic processes (SI Appendix, Fig. S7) were included in the determination of the rate constants shown. Black lines in A–D show the distributions calculated from the model using the rate constant values in E.
The distribution of times to first binding of Spt5DHFR-Cy5 while Rpb1SNAP549 is present, which we interpret as indicative of a full elongation complex, shows an even longer (∼300 s) lag than productive Rpb1SNAP549 association (Fig. 4B and SI Appendix, Fig. S5). In contrast, the time intervals between Rpb1SNAP549 and Spt5DHFR-Cy5 binding in productive events were short, with the majority less than 100 s (Fig. 4C). These intervals were exponentially distributed, consistent with a single rate-limiting process occurring between RNApII binding and elongation complex formation.
Spt4/5 Exchange on Elongation Complexes.
To complete the analysis of Spt4/5 dynamics, we examined the fates of the DNA-bound RNApII•Spt4/5 complexes. The lifetimes of Spt5DHFR-Cy5 fluorescence spots in these productive complexes displayed an exponential distribution (Fig. 4D); that is, they behaved kinetically like a single-molecular species (SI Appendix, Materials and Methods). As described earlier, Rpb1SNAP549 and Spt5DHFR-Cy5 fluorescence spots sometimes disappeared from DNA simultaneously (within ±1 video frame, e.g., closed arrows in Fig. 3A and SI Appendix, Fig. S3), consistent with dissociation of a unitary RNApII•Spt4/5 complex. However, more often Spt5DHFR-Cy5 fluorescence disappeared while Rpb1SNAP549 fluorescence remained (e.g., open arrows in Fig. 3A and SI Appendix, Fig. S3; 77 ± 2% of 312 RNApII•Spt4/5 complexes observed). Control experiments showed that Spt5DHFR-Cy5 photobleaching and dissociation of the Cy5-TMP dye are not significant factors in limiting Spt5DHFR-Cy5 bound lifetimes (SI Appendix, Fig. S6). Thus, loss of Spt5DHFR-Cy5 fluorescence while Rpb1SNAP549 remains signifies dissociation of Spt4/5 from the RNApII elongation complex. Indeed, some DNA-bound RNApII molecules that lost Spt5DHFR-Cy5 were subsequently seen to bind another Spt5DHFR-Cy5 molecule (Fig. 3A and SI Appendix, Fig. S3). Of 49 DNA molecules exhibiting only a single Rpb1SNAP549 binding event, 15 showed multiple sequential Spt5DHFR-Cy5 binding and release events. Together, these data indicate that Spt4/5 binding to elongation complexes is reversible, with individual elongation complexes able to undergo multiple cycles of Spt4/5 binding and release in vitro.
Quantitative Kinetic Model for Initiation Complex Formation and Spt4/5 Recruitment and Release.
The relative kinetics of RNApII and Spt4/5 association with the DNA template (Fig. 4 A–C) suggest a reaction mechanism in which 1) reversible RNApII associations with DNA early in the experiment are dominated by the formation of complexes that cannot go on to elongate until 2) a slower step occurs that is needed to make the promoter competent for PIC and EC formation, after which 3) subsequent initiation of RNA synthesis and binding of Spt4/5 to the elongation complex are comparatively quick. To aid interpretation, we used the quantitative kinetic data to formulate a minimal scheme consistent with these results (Fig. 4E). The rate constants given in the figure were determined by fitting the data summarized in Fig. 4 A–D with the model, and the fit curves predicted by the model and rate constants are shown in those same panels. The data do not speak directly to the structures of the molecular species in the model, but we have proposed plausible candidates for the identities of the different DNA-associated RNApII complexes based on literature data. As illustrated (Fig. 4E), the scheme embodies the following assumptions:
1) Because Rpb1SNAP549 does not bind DNA in the control reaction lacking Gal4-VP16, we show all states of the model with this activator bound. A step encompassing Gal4-VP16 binding to the upstream Gal4 binding sites was not explicitly included since Gal4 binding is likely to be fast and occupancy high, given the concentration of Gal4-VP16 used and the five Gal4 binding sites in the template (34, 40).
2) The model includes an additional step (k1) needed for productive RNApII binding (i.e., the ability to eventually bind Spt4/5). This step renders the DNA competent (DNA*) for PIC and EC formation and probably represents binding to the core promoter of a subset of the RNApII general transcription factors. In particular, it is known that at least TBP and TFIIB must be present before RNApII can proceed on the pathway to PIC formation (2, 41). Alternative simpler mechanisms lacking the nonproductive RNApII binding intermediate (DNA•RNAPII) did not produce satisfactory agreement with the data.
3) We use the same rate constant for RNApII binding (k2) whether or not the minimal general transcription factors are present. The distribution of times until the first Rpb1SNAP549 binding to a given DNA molecule fits a single-exponential model (Fig. 2B), consistent with a single rate-limiting step. As RNApII recruitment is completely dependent on Gal4-VP16, we speculate that the initial binding involves association of a Mediator-RNApII complex with the DNA-bound activator. We model this step as reversible, with the dissociation rate constant (k−2) also being independent of general transcription factors.
4) While our earlier immobilized template assays suggest Gal4-VP16 promotes basal factor binding (20), the current model presumes that activator is saturating on templates. It is therefore agnostic as to whether the activator increases the rate of promoter competence (k1) in addition to increasing the rate of RNApII recruitment.
5) The k1 and k2 processes are assumed to be independent. This is equivalent to saying that the basal factor binding (or other event) that triggers promoter competence is unaffected by the presence of RNApII linked via Gal4-VP16 activator to the upstream activating sequence (UAS).
6) A number of possible additional steps unnecessary for fitting the data were omitted for simplicity. These include the reverse of the k1 and k3 steps (discussed below). Activator-independent binding of RNApII to general transcription factors at the promoter (DNA* → PIC in Fig. 4E) was also omitted, given the observed strong dependence of RNApII binding on Gal4-VP16.
7) The model does not include possible binding of a second RNApII molecule at the UAS when a first RNApII molecule is already engaged as a PIC or EC. This pathway would provide a simple explanation for the occasional observation of multiple Rpb1 molecules simultaneously bound to the same DNA (Fig. 2A, brackets). However, when there are simultaneous multiple Rpb1SNAP549 bindings on a DNA template, it is impossible to conclusively link the individual binding and dissociation events. Therefore, these multiple binding events were not easily usable for kinetic modeling.
Several mechanistic insights emerge from the rate constants determined for the model. The slowest step in the mechanism is k1, the step leading to the DNA* state competent for transcription. We propose this step involves binding of one or more of the general transcription factors needed for subsequent PIC formation. Rate constant k2 is larger than k1, implying that most (but not all) RNApII binding that occurs early in the reaction forms state DNA•RNApII. Because k−2 is greater than k1 by approximately sixfold, most DNA•RNApII complexes dissociate back to the DNA state, with only a fraction continuing on to the DNA*•RNApII intermediate. Thus, the model explains why RNApII binding early in the reaction is mostly nonproductive. The two classes of RNApII binding events in the model, to DNA or to DNA*, explain the difference between the exponential kinetics seen for total first Rpb1SNAP549 binding events (Fig. 4A, purple), versus the lag in the cumulative distribution of times for the first productive Rpb1SNAP549 binding events (Fig. 4A, turquoise).
Once RNApII is bound to a competent promoter (DNA*•RNApII), RNApII can in principle either dissociate to restore DNA* (k−2) or convert into a PIC (k3). The latter step may involve transfer of Mediator-RNApII from the activator to the TBP-TFIIB-TATA box complex, separation of RNApII from Mediator, or incorporation of later basal factors (TFIIF, TFIIE, and/or TFIIH). Because k3 is much greater than k−2, most complexes that get to the DNA*•RNAPII state go on to produce PICs, consistent with the idea that once all components are present, PIC formation is fast. In the model, reversibility of this step was not required to fit the data, so k−3 was set to zero for simplicity.
In our system, Spt4/5 serves to indicate the presence of an EC, but the transition from PIC to EC involves multiple molecular events, including DNA unwinding (42), release of basal factors, and binding of multiple elongation factors, all of which might precede Spt4/5 binding. Time intervals between Rpb1SNAP549 and Spt5DHFR-Cy5 bindings appear as a single exponential distribution (Fig. 4C), consistent with a single predominant rate-limiting step between productive RNApII binding and Spt4/5 binding. This interval is therefore modeled as a single aggregate step with rate constant k4. Future experiments may be able to better define which molecular event is rate limiting.
Discussion
While there has been remarkable recent progress in our understanding of the structures of RNApII transcription complexes, far less is known about the dynamics of transitions between the different structural states. Here we report the use of CoSMoS to directly observe the dynamics with which single fluorescently labeled molecules of RNApII, the elongation factor Spt4/5, and the mRNA binding protein Hek2 interact on a transcription template DNA. In contrast to most other single-molecule studies, these experiments were carried out in nuclear extracts, which should more closely approximate conditions in the nucleus than do systems reconstituted from RNApII and a subset of purified transcription factors. Using a template carrying multiple Gal4 binding sites upstream of the well-characterized CYC1 core promoter, we observe that RNApII binding and subsequent transcription are highly dependent on the exogenous activator Gal4-VP16. Analysis of RNApII binding dynamics in this strongly activated transcription reaction reveals distinct classes of productive and nonproductive binding events and suggests there is a rate-limiting process that makes the promoter competent for PIC formation. Kinetic modeling suggests that initial binding of RNApII to the activator, possibly as an RNApII-Mediator complex, is relatively fast. The slower competence step is likely to reflect association of one or more general transcription factors. Future CoSMoS experiments can rigorously test these modeling-derived hypotheses by labeling additional components of the transcription machinery.
The reported experiments capture the transition from initiation to elongation, which we previously analyzed by quantitative mass spectrometry (20). To detect EC formation in the single-molecule experiments, we initially tried hybridization of fluorescently labeled probe oligonucleotides to the nascent RNA, as has been done in single-molecule transcription experiments using purified factors (27, 43, 44). Probe detection of nascent transcript was ineffective in extract, perhaps because RNA binding proteins interfere with hybridization. As an alternative, we fluorescently labeled the endogenous mRNA binding protein Hek2, which is highly enriched in ECs on this template (20). We observed that Spt4/5 occupancy is strongly correlated with Hek2 presence (Fig. 3E) and is nucleotide dependent (Fig. 3B), and therefore conclude that Spt4/5 binding is confined to ECs, consistent with our mass spectrometry observations.
The kinetics of RNApII basal transcription initiation (i.e., initiation that occurs in the absence of activators) has been previously examined in vitro, including by single-molecule methods (41–43, 45, 46). Such studies typically observe transcription from only a small fraction of template molecules, although this can be greatly improved by preformation of PICs before initiating RNA synthesis (47–49). Here we focused on the dynamics of activated transcription, which is thought to be the dominant mode of gene expression in vivo. As observed for Gal4-dependent promoters in vivo (50), transcription in our system is highly activator dependent. High efficiency is suggested by the observation that in most experiments which included Gal4-VP16 the majority of template molecules bound RNApII.
Chromatin immunoprecipitation experiments show Spt4/5 cross-linking throughout transcribed regions (4, 51). This result is consistent with either stable incorporation of Spt4/5 into the EC or a dynamic interaction in which Spt4/5 dissociates and reassociates during synthesis of a single transcript. In the CoSMoS experiments, when we saw Spt4/5 dissociate before RNApII it was not unusual to observe rebinding of another Spt4/5 molecule to RNApII, likely indicating dynamic interaction. The average length of a yeast pre-mRNA is 1.5 kb (52), so 45 to 90 s are needed for RNApII to transcribe an average gene if the mean elongation rate is roughly 1 to 2 kb min−1 (53, 54). This time range aligns with the average time measured for Spt5DHFR-Cy5 to dissociate from elongation complexes, 1/k−5 = 56 ± 9 s (Fig. 4E). We speculate that Spt4/5 binding times may be tuned to match the average EC lifetime. Thus, a single Spt4/5 molecule would stay attached to the EC throughout a normal, unimpeded transcription cycle. On the other hand, an excessively long interval of pausing or backtracking may cause the EC to persist beyond the average lifetime of Spt4/5 binding, allowing dissociation of Spt4/5 and favoring termination of these RNApII molecules.
RNA polymerases have been shown to participate in phase-separated clusters or condensates in living cells (55, 56). In the experiments reported here, we see no evidence for groups or clusters of RNApII molecules simultaneously arriving at or simultaneously departing from DNA. This behavior is consistent with single-molecule observations of transcription in living yeast cells (50) and greatly facilitates quantitative analysis of the kinetic mechanisms of transcription.
In recent years, researchers have reconstituted numerous molecular complexes representing intermediates in RNApII transcription initiation and transcript elongation, processing, and termination. High-resolution three-dimensional structures of the complexes show how transcription factors, DNA template, and RNApII interact, and the structures suggest mechanisms by which the interactions are remodeled to coordinate and regulate mRNA production. In contrast to this wealth of structural information, we know less about the arrangement of these intermediates in the reaction pathway and even less about the dynamics of the reactions that drive the transcription machinery through successive intermediates. The approach used here represents a generally applicable tool to elucidate the dynamic mechanisms of transcript production by RNApII and its regulation.
Materials and Methods
Yeast Strains, Plasmids, and Oligonucleotides.
S. cerevisiae strains used in this study are listed in SI Appendix, Table S5. SNAP- or DHFR-tagged fusion strains were constructed by PCR amplification of a cassette encoding the SNAP/DHFR protein and a selectable marker. The DHFR-containing plasmid was generously provided by Aaron Hoskins, University of Wisconsin, Madison, WI (22). Oligonucleotides (IDT Ultramers) for PCR had 50 nt of homology to the target gene C-terminal coding region and 20 nt of homology to the fusion cassette. The amplified fragment was transformed into yeast with selection for the marker. In-frame fusion protein expression and stability was confirmed by immunoblotting for the target protein (e.g., SI Appendix, Fig. S1 A and B), and in the case of SNAP fusions, by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) after labeling with SNAP-Surface 549 (New England Biolabs). All tagged strains grew in YPD media; tagged strain growth rates and those of their parental strain YF702 or YF4 are reported in SI Appendix, Fig. S1E.
Yeast Nuclear Extracts.
Yeast nuclear extracts were prepared as previously described (17). For SNAP fusion strains, the protocol was modified in that nuclear protein pellets were resuspended in 1 to 2 mL of buffer C′ (20 mM Hepes, pH 7.6, 10 mM MgSO4, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 10% glycerol, 3 mM dithiothreitol (DTT), and 1 µg mL−1 each of aprotinin, leupeptin, pepstatin A, and antipain). (In pilot experiments, the 20% glycerol concentration and phenylmethylsulfonyl fluoride used in ref. 17 were found to reduce SNAP labeling.) To fluorescently label extracts with SNAP fusion proteins, SNAP-Surface 549 was added with gentle vortexing to the resuspended pellet at a final concentration of 0.5 μM (unless otherwise specified), and the mixture was incubated at 4 °C for 1 h on a rotator. All extracts were dialyzed against 3 × 0.5 L buffer C′ supplemented with 75 mM (NH4)2SO4. For SNAP-labeled extracts, unreacted SNAP-Surface 549 was further depleted as described (57) with some modifications. Specifically, 0.25 volumes of SNAP-agarose beads (SI Appendix, Materials and Methods) was added to the dialyzed extract and incubated at 4 °C for 1 h on a rotator, after which the beads were removed by centrifugation at 1,000 × g for 2 min at 4 °C. All extracts were aliquoted, frozen in liquid N2, and stored at –80 °C. Labeling of SNAP fusion proteins and depletion of free dye was confirmed by SDS-PAGE followed by fluorescence imaging on a Typhoon imager (GE Healthcare). Extracts were checked to verify transcription activity (e.g., SI Appendix, Fig. S1 C and D). If DNA-specific Spt5DHFR-Cy5 binding occurred at a frequency of <0.5 × 10−4 s−1, the extract was considered to have inactive or proteolytically cleaved Spt5 and was not used.
DNA Template.
Transcription template (Fig. 1A) was prepared by PCR from plasmid SB649 (20) using primers 5′-biotin-GTTGGGTAACGCCAGGG-3′ and 5′-Alexa488-GGAAACAGCTATGACATG-3′ (IDT) and Platinum Taq DNA Polymerase (Invitrogen). The PCR product was purified using DNA SizeSelector-I SPRI magnetic beads (Aline Biosciences) according to the manufacturer’s instructions. The template contained five tandem repeats of 5′-CGGAGGACAGTACTCCG-3′, a consensus Gal4 binding sequence (58).
CoSMoS Transcription Experiments.
Single-molecule fluorescence microscopy experiments were conducted at 20 to 23 °C, 1 frame s−1 (except as otherwise noted) on a micromirror total internal fluorescence microscope at excitation wavelengths of 488, 532, and 633 nm (21) using laser powers incident to the micromirror of 700, 400, and 200 μW, respectively. Focus was automatically maintained as described (59). Transcription reactions were observed in glass flow chambers passivated with a mPEG-SG2000:biotin-PEG-SVA5000 (Laysan Bio) 200:1 wt/wt mixture as described in ref. 27. Streptavidin-coated fluorescent beads (T-10711, Molecular Probes) to serve as fiducial markers for stage drift correction (60) were added to the flow chamber at a dilution of ∼1:400,000 in transcription buffer (TB; 20 mM potassium acetate, 20 mM Hepes, pH 7.6, 1 mM ethylenediaminetetraacetic acid [EDTA], 5 mM magnesium acetate) supplemented with 1 mg mL−1 bovine serum albumin (TB+BSA). The chamber was subsequently incubated with 0.013 mg/mL NeutrAvidin (31000, Thermo Fisher) in TB+BSA for 45 s and then flushed with three chamber volumes (3 × 20 µL) of TB+BSA. We then introduced ∼10 pM DNA488 in TB+BSA and incubated until 200 to 400 fluorescent spots were visible under 488-nm excitation. After moving the stage to a new field of view, the transcription reaction was started by introducing a mixture of yeast nuclear extract, TB, and other reagents with the following final concentrations: extract (6.6 mg mL−1 final protein concentration), O2-scavenging system (59) (0.9 units mL−1 protocatechuate dioxygenase, 5 mM protocatechuate), 2 mM DTT, triplet state quenchers (61) (0.5 mM propyl gallate, 1 mM Trolox, 1 mM 4-nitrobenzyl alcohol, 0.5% dimethylsulfoxide), ATP regeneration system (10 mM phosphocreatine. 0.1 mg mL−1 [35 units ml−1] creatine kinase), 20 nM Cy5-TMP (22) (only if Spt5DHFR was present), 0 or 4 µg mL−1 Gal4-VP16 activator, nucleotides (400 µM each ATP, CTP, and UTP plus 100 µM 3′-O-methyl-GTP to aid in stalling the RNApII at the end of the G-less cassette), and buffer components (100.5 mM potassium acetate, 24.1 mM Hepes, pH 7.6, 0.4 mM Tris-acetate, 1 mM EDTA, 5 mM magnesium acetate, 2 mM MgSO4, 0.2 mM EGTA, 15 mM (NH4)2SO4, 4.25% glycerol). We monitored labeled extract component association to and dissociation from the recorded surface-tethered DNA488 locations by exciting individually (one-color experiments) or simultaneously (two-color experiments) with the 532- and 633-nm lasers.
For the minus-NTPs (–NTPs) control experiments, nucleotides and ATP regeneration components were omitted and residual ATP was depleted by the addition of 20 mM glucose and 40 units mL−1 hexokinase (Sigma H4502). This treatment is presumed to also deplete other NTPs as a result of NTP pool equilibration by nucleoside diphosphate kinase activity in the extract. In inhibitor controls, α-amanitin was added at 10 μg mL−1. For the photobleaching control experiment (SI Appendix, Fig. S6E), we conducted three separate transcription experiments with different duty ratios of the 633-nm laser at 200 µW.
Data Analysis and Kinetic Modeling.
See SI Appendix, Materials and Methods for data analysis and kinetic modeling.
Supplementary Material
Acknowledgments
We thank Judy Ciyue Shen and Yujin Chun for constructing yeast strains used in this work and members of the S.B. and J.G. laboratories for advice and comments on the manuscript. Research reported in this publication was supported by the National Institute of General Medical Sciences and the National Cancer Institute of the NIH under awards R01 GM046498 and R01GM056663 (to S.B.), R01GM081648 (to J.G.), and R01CA246500 (to S.B. and J.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011224117/-/DCSupplemental.
Data Availability.
Source data for the single-molecule experiments are provided as “interval” files that can be read and manipulated by the Matlab program imscroll, which is publicly available in GitHub: https://github.com/gelles-brandeis/CoSMoS_Analysis. The source data are archived in Zenodo (DOI: 10.5281/zenodo.4065399).
References
- 1.Thomas M. C., Chiang C. M., The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Sikorski T. W., Buratowski S., The basal initiation machinery: Beyond the general trancription factors. Curr. Opin. Chem. Biol. 21, 344–351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luse D. S., The RNA polymerase II preinitiation complex. Through what pathway is the complex assembled? Transcription 5, e27050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer A., et al. , Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17, 1272–1278 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Vos S. M., et al. , Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 560, 607–612 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Werner F., A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J. Mol. Biol. 417, 13–27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wada T., et al. , DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12, 343–356 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morillon A., et al. , Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115, 425–435 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Quan T. K., Hartzog G. A., Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4-Spt5 in transcription. Genetics 184, 321–334 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Booth G. T., Wang I. X., Cheung V. G., Lis J. T., Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast. Genome Res. 26, 799–811 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viladevall L., et al. , TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol. Cell 33, 738–751 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirtreiter A., et al. , Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 38, 4040–4051 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehara H., et al. , Structure of the complete elongation complex of RNA polymerase II with basal factors. Science 357, 921–924 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Lindstrom D. L., et al. , Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23, 1368–1378 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mooney R. A., Schweimer K., Rösch P., Gottesman M., Landick R., Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 391, 341–358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lidschreiber M., Leike K., Cramer P., Cap completion and C-terminal repeat domain kinase recruitment underlie the initiation-elongation transition of RNA polymerase II. Mol. Cell. Biol. 33, 3805–3816 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sikorski T. W., et al. , Proteomic analysis demonstrates activator- and chromatin-specific recruitment to promoters. J. Biol. Chem. 287, 35397–35408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadowski I., Ma J., Triezenberg S., Ptashne M., GAL4-VP16 is an unusually potent transcriptional activator. Nature 335, 563–564 (1988). [DOI] [PubMed] [Google Scholar]
- 19.Ranish J. A., Yudkovsky N., Hahn S., Intermediates in formation and activity of the RNA polymerase II preinitiation complex: Holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13, 49–63 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joo Y. J., Ficarro S. B., Chun Y., Marto J. A., Buratowski S., In vitro analysis of RNA polymerase II elongation complex dynamics. Genes Dev. 33, 578–589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman L. J., Chung J., Gelles J., Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys. J. 91, 1023–1031 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoskins A. A., et al. , Ordered and dynamic assembly of single spliceosomes. Science 331, 1289–1295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun J. E., Friedman L. J., Gelles J., Moore M. J., Synergistic assembly of human pre-spliceosomes across introns and exons. eLife 7, e37751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stumper S. K., et al. , Delayed inhibition mechanism for secondary channel factor regulation of ribosomal RNA transcription. eLife 8, e40576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tetone L. E., et al. , Dynamics of GreB-RNA polymerase interaction allow a proofreading accessory protein to patrol for transcription complexes needing rescue. Proc. Natl. Acad. Sci. U.S.A. 114, E1081–E1090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harden T. T., et al. , Bacterial RNA polymerase can retain σ70 throughout transcription. Proc. Natl. Acad. Sci. U.S.A. 113, 602–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman L. J., Gelles J., Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation. Cell 148, 679–689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harden T. T., et al. , Alternative transcription cycle for bacterial RNA polymerase. Nat. Commun. 11, 448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shcherbakova I., et al. , Alternative spliceosome assembly pathways revealed by single-molecule fluorescence microscopy. Cell Rep. 5, 151–165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H., Larson D. R., What have single-molecule studies taught us about gene expression? Genes Dev. 30, 1796–1810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z., Tjian R., Measuring dynamics of eukaryotic transcription initiation: Challenges, insights and opportunities. Transcription 9, 159–165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keppler A., et al. , A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Gallagher S. S., Sable J. E., Sheetz M. P., Cornish V. W., An in vivo covalent TMP-tag based on proximity-induced reactivity. ACS Chem. Biol. 4, 547–556 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carey M., Lin Y. S., Green M. R., Ptashne M., A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature 345, 361–364 (1990). [DOI] [PubMed] [Google Scholar]
- 35.Meyer P. A., et al. , Structures and functions of the multiple KOW domains of transcription elongation factor Spt5. Mol. Cell. Biol. 35, 3354–3369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plaschka C., et al. , Transcription initiation complex structures elucidate DNA opening. Nature 533, 353–358 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Murakami K., et al. , Structure of an RNA polymerase II preinitiation complex. Proc. Natl. Acad. Sci. U.S.A. 112, 13543–13548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grohmann D., et al. , The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol. Cell 43, 263–274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasegawa Y., Irie K., Gerber A. P., Distinct roles for Khd1p in the localization and expression of bud-localized mRNAs in yeast. RNA 14, 2333–2347 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y., North J. A., Rose S. D., Poirier M. G., Nucleosomes accelerate transcription factor dissociation. Nucleic Acids Res. 42, 3017–3027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horn A. E., Kugel J. F., Goodrich J. A., Single molecule microscopy reveals mechanistic insight into RNA polymerase II preinitiation complex assembly and transcriptional activity. Nucleic Acids Res. 44, 7132–7143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomko E. J., Fishburn J., Hahn S., Galburt E. A., TFIIH generates a six-base-pair open complex during RNAP II transcription initiation and start-site scanning. Nat. Struct. Mol. Biol. 24, 1139–1145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Revyakin A., et al. , Transcription initiation by human RNA polymerase II visualized at single-molecule resolution. Genes Dev. 26, 1691–1702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z., Revyakin A., Grimm J. B., Lavis L. D., Tjian R., Single-molecule tracking of the transcription cycle by sub-second RNA detection. eLife 3, e01775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z., et al. , Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes Dev. 30, 2106–2118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fazal F. M., Meng C. A., Murakami K., Kornberg R. D., Block S. M., Real-time observation of the initiation of RNA polymerase II transcription. Nature 525, 274–277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murakami K., et al. , Formation and fate of a complete 31-protein RNA polymerase II transcription preinitiation complex. J. Biol. Chem. 288, 6325–6332 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Damodaren N., et al. , Def1 interacts with TFIIH and modulates RNA polymerase II transcription. Proc. Natl. Acad. Sci. U.S.A. 114, 13230–13235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujiwara R., Damodaren N., Wilusz J. E., Murakami K., The capping enzyme facilitates promoter escape and assembly of a follow-on preinitiation complex for reinitiation. Proc. Natl. Acad. Sci. U.S.A. 116, 22573–22582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donovan B. T., et al. , Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J. 38, e100809 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim M., Ahn S.-H., Krogan N. J., Greenblatt J. F., Buratowski S., Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23, 354–364 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miura F., et al. , Absolute quantification of the budding yeast transcriptome by means of competitive PCR between genomic and complementary DNAs. BMC Genomics 9, 574 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zenklusen D., Larson D. R., Singer R. H., Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15, 1263–1271 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mason P. B., Struhl K., Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17, 831–840 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Cramer P., Organization and regulation of gene transcription. Nature 573, 45–54 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Ladouceur A.-M., et al. , Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid-liquid phase separation. Proc. Natl. Acad. Sci. U.S.A. 117, 18540–18549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haraszti R. A., Braun J. E., “Preparation of SNAPf-beads for colocalization single-molecule spectroscopy (CoSMoS) of RNA-protein complexes” in RNA Spectroscopy: Methods and Protocols, Arluison V., Wien F., Eds. (Methods in Molecular Biology, Springer, 2020), pp. 17–22. [DOI] [PubMed] [Google Scholar]
- 58.Giniger E., Varnum S. M., Ptashne M., Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell 40, 767–774 (1985). [DOI] [PubMed] [Google Scholar]
- 59.Crawford D. J., Hoskins A. A., Friedman L. J., Gelles J., Moore M. J., Visualizing the splicing of single pre-mRNA molecules in whole cell extract. RNA 14, 170–179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friedman L. J., Gelles J., Multi-wavelength single-molecule fluorescence analysis of transcription mechanisms. Methods 86, 27–36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dave R., Terry D. S., Munro J. B., Blanchard S. C., Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 96, 2371–2381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for the single-molecule experiments are provided as “interval” files that can be read and manipulated by the Matlab program imscroll, which is publicly available in GitHub: https://github.com/gelles-brandeis/CoSMoS_Analysis. The source data are archived in Zenodo (DOI: 10.5281/zenodo.4065399).